- 1Genomics and Genetic Engineering Laboratory of Ornamental Plants, College of Agriculture and Biotechnology, Zhejiang University, Hangzhou, Zhejiang, China
- 2Laboratory of Flower Bulbs, Department of Landscape Architecture, Zhejiang Sci-Tech University, Hangzhou, Zhejiang, China
Bulblet initiation, including adventitious bud initiation and bulblet formation, is a crucial process for lily and other bulbous flowers that are commercially propagated by vegetative means. Here, by a hybrid strategy combining Pacific Biosciences (PacBio) full-length sequencing and Illumina RNA sequencing (RNA-seq), high-quality transcripts of L. brownii (Lb) and its variety, L. brownii var. giganteum (Lbg), during in vitro bulblet initiation were obtained. A total of 53,576 and 65,050 high-quality non-redundant full-length transcripts of Lbg and Lb were generated, respectively. Morphological observation showed that Lbg possessed a stronger capacity to generate bulblets in vitro than Lb, and 1 mg L−1 2,4-dichlorophenoxyacetic acid (2,4-D) significantly increased bulblet regeneration rate in two lilies. Screening of differentially expressed transcripts (DETs) between different stages and Mfuzz analysis showed 0 DAT to 1 DAT was the crucial stage with the most complex transcriptional change, with carbohydrate metabolism pathway was significantly enriched. In addition, 6,218 and 8,965 DETs were screened between the 2,4-D-treated group and the control group in Lbg and Lb, respectively. 2,4-D application had evident effects on the expression of genes involved in auxin signaling pathway, such as TIRs, ARFs, Aux/IAAs, GH3s and SAURs. Then, we compared the expression profiles of crucial genes of carbohydrate metabolism between different stages and different treatments. SUSs, SUTs, TPSs, AGPLs, GBSSs and SSs showed significant responses during bulblet initiation. The expression of CWINs, SUTs and SWEETs were significantly upregulated by 2,4-D in two lilies. In addition, 2,4-D increased the expression of starch degradation genes (AMYs and BAMs) and inhibited starch synthesis genes (AGPLs, GBSSs and SSs). SBEs were significantly upregulated in Lbg but not in Lb. Significant co-expression was showed between genes involved in carbohydrate metabolism and auxin signaling, together with transcription factors such as bHLHs, MYBs, ERFs and C3Hs. This study indicates the coordinate regulation of bulblet initiation by carbohydrate metabolism and auxin signaling, serving as a basis for further studies on the molecular mechanism of bulblet initiation in lily and other bulbous flowers.
1 Introduction
Bulbous flowers, which are highly popular in the world floriculture market, are usually commercially propagated by vegetative means, especially by bulbs, to maintain phenotypic uniformity and genetic purity. Bulblet initiation, including the process of adventitious/axillary bud initiation and bulblet formation, has been reported in multiple bulbous plants from plant tissues in vitro (Van Aartrijk and Blom-Barnhoorn, 1984; Li et al., 2014; Ren et al., 2017; Lv et al., 2020). Lily (Lilium spp.), a perennial monocotyledon of the family Liliaceae, is one of the major bulbous crops in the floriculture industry with high ornamental, medical and edible value (Lee et al., 2013; Xu et al., 2017; Wu et al., 2019). Due to its strong bulblet formation capacity in vitro, lily is considered as an appropriate experimental material for the study of bulblet initiation and its underlying mechanisms. Studies on bulblet initiation in lily have focused mainly on the influence of wounds, temperature treatment and exogenous phytohormones (Van Aartrijk and Blom-Barnhoorn, 1984), changes in endogenous carbohydrate and hormone contents, and the expression level of genes involved in carbohydrate and phytohormone metabolism (Li et al., 2014; Wu et al., 2020).
Carbohydrate metabolism plays a vital role in bulblet initiation of multiple bulbous flowers. Therein, starch metabolism has been reported to be ubiquitously involved. Storage starch in mother scales could act as a carbon source for bulblet initiation. At the bulblet appearance and enlargement stage, the enzymes involved in the starch synthetic direction, such as ADP-glucose Pyrophosphorylase (AGPase, EC 2.7.7.27), Starch Synthase (SS, EC 2.4.1.21), Starch Branching Enzyme (SBE, EC 2.4.1.18) and Granule-bound Starch Synthase (GBSS, EC 2.4.1.242), showed a decreasing trend in mother scales but higher gene expression levels in newly formed bulblets, while the enzyme in the starch cleavage direction, Starch Debranching Enzyme (DBE, EC 3.2.1.10), showed higher expression levels in scales than in bulblets in lily (Li et al., 2014). Similarly, in Lycoris, soluble sugars derived from starch degradation in the outer scales were transported into the inner scales and promote bulblet initiation and development through starch synthesis, especially through AGPases (Xu et al., 2020a). Sucrose, the main form of transported sugar in higher plants, were also considered to regulate bulblet initiation. Sucrose Synthase (SUS, EC 2.4.1.13) and Invertase (INV, EC 3.2.1.26, including Cell Wall Invertase (CWIN), Vacuolar Invertase (VIN) and Cytoplasmic Invertase (CIN)), mainly hydrolyzing sucrose, presented higher expression levels in mother scales and bulblets at stages of bulblet appearance and enlargement in lily (Li et al., 2014). A clear shift was observed from CWIN-catalyzed to SUS-catalyzed sucrose cleavage patterns, meanwhile, sucrose unloading pathway changed from apoplasmically to symplasimically at the key shoot-to-bulblet transition stage in Lilium Oriental Hybrids ‘Sorbonne’ (Wu et al., 2021). Similarly, CWIN and SUS exhibited exactly opposite expression patterns during the competence stage of bulblet regeneration in Lycoris (Ren et al., 2021). The above results indicated that the transition from bud initiation to bulblet enlargement was usually accompanied by the change of dominant sucrose unloading pathway.
Auxin, a key phytohormone, regulates diverse aspects of plant growth and developmental processes through its dynamic differential distribution (Vanneste and Friml, 2009). The biosynthesis of indole-3-acetic acid (IAA), the main naturally occurring auxin, in higher plants requires two steps: first, tryptophan is converted to indole-3-pyruvate (IPA) by Tryptophan Aminotransferase (TAA) or Tryptophan Aminotransferase Related (TAR); second, IAA is produced from IPA by YUC family (YUC) (Zhao, 2012). A previous study revealed that adventitious bulblets of lily were formed at the basal edge of the explant under tissue culture conditions, which caused by basipetal auxin transport (Van Aartrijk and Blom-Barnhoorn, 1984). Different auxin concentrations showed different effects on the process of bulblet formation. Auxin likely promoted the initiation of bulbils and then inhibited further bulbil formation in lily (Yang et al., 2017). In Lycoris, endogenous IAA content showed an increase and then a decrease during bulblet initiation and development, which were consistent with the expression patterns of genes involved in IAA synthesis and signal transduction (Xu et al., 2020a).
To obtain more genetic information, Illumina next-generation sequencing (NGS) technology has been widely used in the study of the process of bulblet formation in multiple bulbous plants, including Lilium (Li et al., 2014; Yang et al., 2017), Lycoris (Ren et al., 2022) and sweet potato (Ipomoea batatas) (Firon et al., 2013). In Lilium, transcriptome analysis was used to elucidate the molecular mechanism of bulblet/bulbil formation and development. (Li et al., 2014; Yang et al., 2017). However, the early stage of bulblet formation in Lilium and even in bulbous flowers remains unclear. Recently, the single-molecule real-time (SMRT) sequencing technology of the PacBio system has offered a new third-generation sequencing platform, which possesses advantages such as long read lengths (length > 10 kb), high consensus accuracy and a low degree of bias (Hu et al., 2022), which is an available and reliable strategy to generate more accurate and comprehensive genetic information. A recent study obtained Lilium Oriental Hybrids ‘Sorbonne’ transcriptome during induction of aerial bulbil using the combination of SMRT and NGS technology (Li et al., 2022).
Here, we explore the in vitro bulblet initiation process of Lilium brownii (Lb) and Lilium brownii var. giganteum (Lbg), a variant of Lb (Li et al., 2007), through careful morphological observation, and then divided the process into four stages. Then, a hybrid strategy combining Pacific Biosciences (PacBio) full-length sequencing and Illumina sequencing was conducted. Through differentially expression transcript (DET) screening and Mfuzz analysis, we identified key metabolic pathways and candidate genes during bulblet initiation. In addition, DET screening was also performed between the 2,4-dichlorophenoxyacetic acid (2,4-D)-treated group and the control group, to explore how 2,4-D, a synthetic auxin analog, affect the process of bulblet initiation. Furthermore, we hypothesized that carbohydrate metabolism and auxin signaling coordinately regulate bulblet initiation, with several transcriptional factors (TFs) involved in the regulation of this process. Our findings provide a comprehensive understanding of the molecular mechanism underlying the process of bulblet initiation in lily.
2 Materials and methods
2.1 Plant materials and growth conditions
Bulblet induction experiments were conducted at the Physiology & Molecular Biology Laboratory of Ornamental Plants and Tissue Culture Laboratory of Ornamental Plants at Zhejiang University, Hangzhou (118°21’-120°30’E, 29°11′-30°33’N), China. In vitro seedlings of Lb and Lbg were cultured at 25 ± 2°C under a 12:12 h light:dark photoperiod with 60 μmol photons m-2 s-1. Healthy outer scales without damage were removed carefully from fresh in vitro bulbs (4~6 cm in circumference) and then cultured for 14 days on basal Murashige and Skoog (MS) medium (Murashige and Skoog, 1962) containing 6% sucrose and 0.3% Phytagel (P8169, Sigma-Aldrich, St. Louis, MO, USA) (pH 5.8), to which was added 0 mg L−1 or 1 mg L−1 2,4-D, with the adaxial side facing upward. Each treatment contained three biological replicates, and each replicate included 120 scales.
2.2 Morphological and histological observation
Morphological changes of bulblet initiation in Lb and Lbg were observed under a stereomicroscope (SZM745T, OPLENIC, China). The regeneration rate and propagation efficiency of bulblets were calculated as follows: Regeneration rate = number of scales that produced adventitious buds/total number of scales; Propagation efficiency = total number of produced buds/total number of scales. Representative data were supported by three biological replicates, each containing 120 repeats. Transverse sections of scales at the proximal end where adventitious buds initiated were stained with periodic acid-Schiff (PAS) and Naphthol Yellow S as previously described (Ren et al., 2017), and then observed using an upright light microscope (Eclipse E100, Nikon, Japan).
2.3 Sample collection
The bulblet initiation process of Lb and Lbg was divided into four crucial stages according to the results of morphological and histological observations: stage of scale detachment (0 DAT; DAT, days after the treatment of detaching the scale from the basal plate), stage of wounding response and early regeneration competence (1 DAT), stage of adventitious bud initiation (8 DAT) and stage of adventitious bud swelling and bulblet formation (14 DAT). The entire scales used for bulblet induction at 0 DAT, 1 DAT, 8 DAT and 14 DAT were sampled, frozen in liquid nitrogen and stored at -80°C for total RNA extraction. Sampling was performed with three biological replicates for each stage.
2.4 Generation of the full-length reference transcripts for Lb and Lbg
Total RNA of scale samples at four stages of Lb and Lbg cultured on 2,4-D-free (0 mg L-1 2,4-D) and 2,4-D-containing (1 mg L-1 2,4-D) medium was extracted using an EASYspin Plus Complex RNA Kit (RN53, Aidlab Bio, China) according to the manufacturer’s instructions. Total RNA from each sample was equally mixed to generate a pool and then synthesized to first-strand cDNA using Clontech SMARTer PCR cDNA Synthesis Kit (Clontech, Mountain View, CA, USA). Large-scale PCR was performed using the BluePippin™ Size Selection System (Sage Science, Beverly, MA, USA). The SMRTbell template libraries were constructed and then sequenced on the PacBio Sequel platform. The following methods for generating full-length reference transcripts referenced Li et al. (2022) with some modifications. The high-quality full-length transcripts were removed rebundancy using CD-HIT v4.6.142 (Li and Godzik, 2006). BUSCO 5.2.0 was used to assess the quality and completeness of the reference transcripts using the official BUSCO datasets (liliopsida_odb10) (Manni et al., 2021).
Total RNA of scale samples was sequenced with the Illumina Xten 4000 platform (Illumina, San Diego, CA, USA). Quality control (QC) for each Illumina transcriptome was performed by fastp (v0.19.7) with default parameters (Chen et al., 2018). Clean reads were separately mapped to their corresponding reference transcripts by Bowtie2 (v2.3.4), and the expression levels of each transcript, including reads count and fragments per kilobase million (FPKM), were calculated by RSEM (v1.3.1) (Li and Dewey, 2011; Langmead and Salzberg, 2012).
2.5 Transcript annotation
Each transcript was annotated by the eggNOG-mapper webserver (http://eggnog-mapper.embl.de/) with an e-value of ≤ 1e-5 and identity of ≥ 60% with Viridiplantae (green plants) selected as the taxonomic scope (Huerta-Cepas et al., 2019; Cantalapiedra et al., 2021). As a result, transcripts were functionally annotated based on the following databases: Pfam, NCBI nonabundant (NR), Kyoto Encyclopedia of Genes and Genomes (KEGG) and Gene Ontology (GO). The PlantTFDB v5.0 database (Jin et al., 2017) was used to predict TFs and their homologous genes in Arabidopsis thaliana of the full-length reference transcripts in Lb and Lbg. Venn diagrams were generated using Evenn (http://www.ehbio.com/test/venn/#/, Chen et al., 2021).
2.6 DET screening and enrichment analysis
Differential expression analysis was performed using R package DESeq2 (v1.32.0) (Love et al., 2014). Transcripts with a false discovery rate (FDR) ≤ 0.05 and a threshold of |log2-fold changes| ≥ 2 were recognized as DETs. Each group of DETs was calculated for KEGG pathways by clusterProfiler (v4.0.5) using an p-value of 0.05 to find significant enrichments (Wu et al., 2021). Time-series cluster analysis was performed by the Mfuzz package (v2.50.0) in R software, with the low expression level transcripts with FPKM ≤ 5 removed.
2.7 Quantitative real-time PCR validation
To confirm the results of the transcript expression levels from RNA-Seq, six DETs of Lbg and six DETs of Lb were selected for expression analysis using quantitative real-time PCR (qRT-PCR). Total RNA (1 μg) of each sample was reverse transcribed by the PrimeScript™ RT reagent Kit with gDNA Eraser (RR047A, TaKaRa, Dalian, China). The diluted (1:30) cDNA was used as the template for qRT-PCR analysis. Gene-specific primers were designed by the NCBI Primer-BLAST tool (Ye et al., 2012) and are listed in Supplementary Table S2. Then, qRT-PCR was performed with TB Green™ Premix Ex Taq™ Kit (RR420A, TaKaRa, Dalian, China) in a Bio-Rad Connect™ Optics Module (Bio-Rad, CA, USA). All reactions were conducted in triplicate, and the 2–ΔΔCT method was applied to calculate the relative expression level using GAPDH as the reference gene.
2.8 Nonstructural carbohydrate content assay
Total starch contents of scales at different stages were measured using a Starch Content Kit (A148-1-1, Nanjing Jiancheng Bioengineering Institute, Nanjing, China) following the manufacturer’s protocol. Three replicates were included in each assay. Sucrose, fructose, and glucose contents were measured by high-performance liquid chromatography (HPLC) (e2695, Waters, MA, USA) equipped with Refractive Index (RI) Detector 2414 (Waters) according to the previous method in Liu et al. (2020).
2.9 Determination of IAA concentration
IAA extraction and quantification were performed using a previously described method with slight modifications (Guo et al., 2016). Briefly, frozen scale sample (100 mg) of each treatment at each stage was weighed in a 10-mL centrifuge tube, and homogenized in 1 mL of ethyl acetate that had been spiked with D5-IAA (C/D/N Isotopes) as an internal standard at a final concentration of 100 ng mL−1. The tubes were centrifuged at 12 000 rpm for 10 min at 4°C. The resulting supernatant was dried by blowing under N2. The residue was resuspended in 0.5 mL of 70% (v/v) methanol and centrifuged, and the supernatants were then analyzed in a triple quadrupole mass spectrometer (6470, Agilent Technologies, CA, USA).
2.10 Statistical analysis
One-way analysis of variance (ANOVA) was used to compare differences among different indices or treatments via SPSS 26.0 (IBM Corp., Armonk, NY, USA). Correlation analyses between gene expression data in Lb and Lbg were performed using Pearson’s two-tailed tests and visualized by Cytoscape v3.7.1 (Shannon et al., 2003).
3 Results
3.1 Stage division according to morphological and histological observation
Throughout the bulblet initiation and development process (0 DAT to 49 DAT), the regeneration rate and propagation efficiency were calculated. It is showed that visible adventitious buds occurred at 8 DAT, and the number of newly formed bulblets was significantly increased until 14 DAT and then tended to plateau until 25 DAT or later in both Lbg and Lb (Figure 1A). In addition, 1 DAT was considered as the crutial wounding response stage according to our previous studies. Based on morphological observations, browning substances began to accumulate in the transverse section at 1 DAT (Figures 1B3, B8). At 8 DAT, adventitious buds formed at the adaxial side rather than the abaxial side of the scale, mainly around lateral vascular bundles (Figures 1B4, B9; Supplementary Figures S1B, E). Then, adventitious buds swelled, and visible bulblets occurred at 14 DAT (Figures 1B5, B10; Supplementary Figures S1C, F). Taken together, we divided the early bulblet initiation process into four stages: 0 DAT (scale detachment), 1 DAT (wounding response and early regeneration competence), 8 DAT (adventitious bud initiation) and 14 DAT (bud swelling and bulblet formation). Moreover, the bulblet regeneration rate and propagation efficiency of Lbg were both significantly higher than those of Lb from 8 DAT to 49 DAT (Figure 1A). At 14 DAT, the regeneration rate and propagation efficiency of Lbg were 0.619 and 1.677, respectively, significantly higher (p < 0.001 and p < 0.01, respectively) than those in Lb (0.236 and 1.114, respectively) (Figure 1A). The above results indicated that Lbg possesses a stronger capacity to generate bulblets in vitro than Lb.
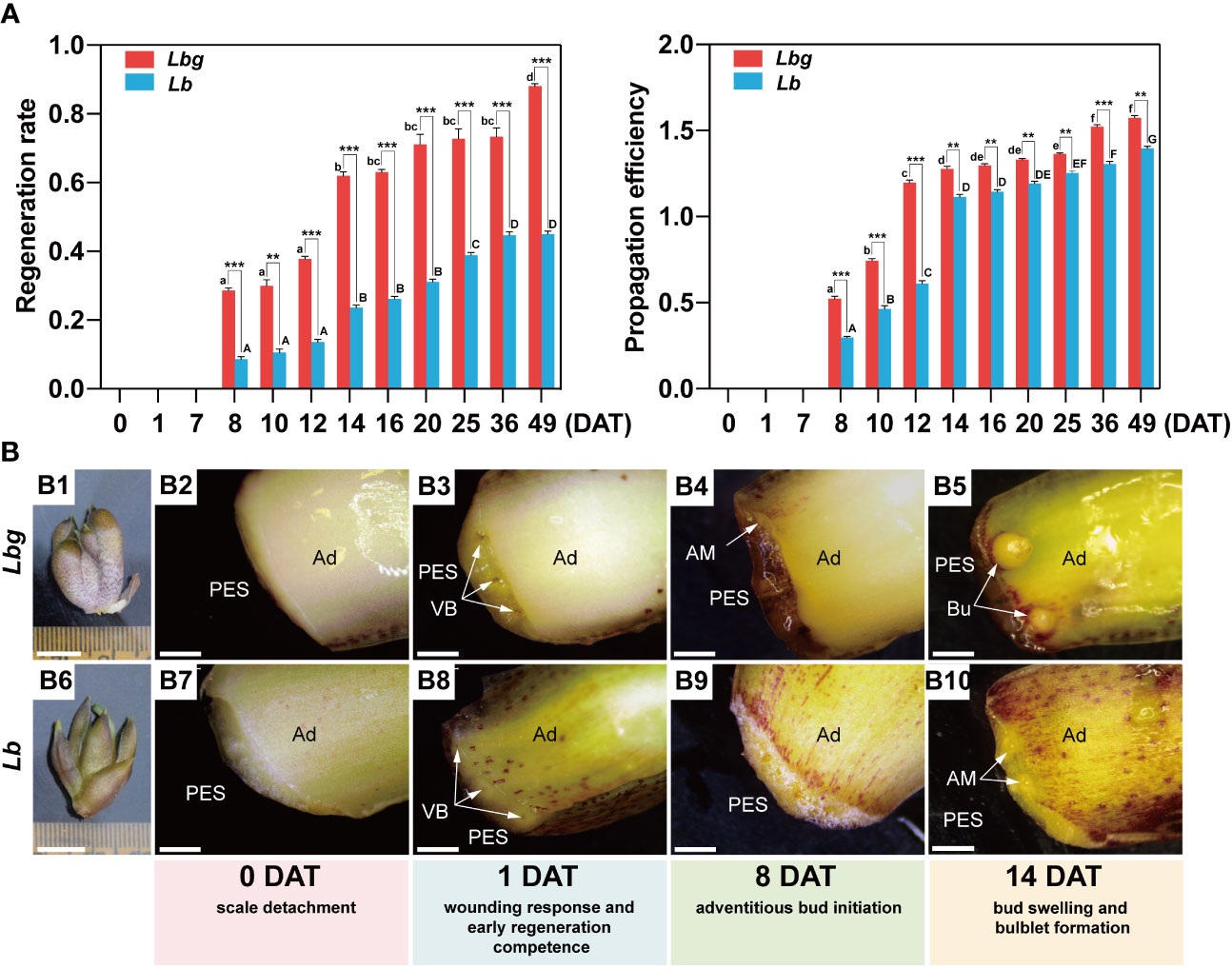
Figure 1 Morphological observation during in vitro bulblet initiation in Lbg and Lb. (A) Regeneration rate and propagation efficiency during in vitro bulblet initiation. Regeneration rate, number of scales that produced adventitious buds/total number of scales. Propagation efficiency, total number of produced buds/total number of scales. Lowercase and uppercase letters represent significant differences (p < 0.001) for relevant parameters within Lbg and Lb, respectively. Asterisks indicate significant differences for relevant parameters between Lbg and Lb (**Differences significant at p < 0.01; ***Differences significant at p < 0.001). Representative data were supported by three biological replicates containing 120 repeats each. (B) In vitro bulb of which scales were used for bulblet induction (B1, B6) and key stages during bulblet initiation in Lbg (B2-B5) and Lb (B7-B10). PES, proximal end of scale; Ad, adaxial side of scale; VB, vascular bundle; AM, adventitious meristem; Bu, bulblet. The white arrows represent vascular bundles (B3 and B4), adventitious meristems (B4 and B10) or bulblets (B9). Bars, 1 cm (B1, B6) and 1 mm (B2-B5, B7-B10).
3.2 Quality assessment of obtained reference transcripts
Full-length transcriptome sequencing was conducted to generate complete and accurate gene information during bulblet initiation (Figure 2A). The reference transcripts for Lbg were first obtained with 53,576 nonredundant transcripts of an average length of 3,108 bp, and 80.79% of the Lbg clean reads obtained by Illumina RNA-Seq mapped to the Lbg reference transcripts (Supplementary Table S1). Similarly, the Lb full-length reference transcripts were obtained, consisting of 65,050 nonredundant transcripts of an average length of 2,965 bp, with 81.85% Lb clean reads mapped to them (Supplementary Table S1). Moreover, the Lbg and Lb reference transcripts had 73.1% and 75.9% of the conserved plant genes by BUSCO 5.2.0, respectively (Supplementary Table S1). These results confirmed the reliability of these two transcriptomes for downstream analysis. The length of transcripts ranged from 294 bp to 13,738 bp in Lbg and from 292 bp to 14,486 bp in Lb, with a median length of 2,139 and 2,021 bp and a mean length of 3,108 and 2,965 bp in Lbg and Lb, respectively (Figure 2B). Moreover, a total of 38,725 (72.3%) and 45,967 (70.7%) transcripts were annotated to the NR, GO, KEGG and Pfam databases in Lbg and Lb, respectively (Figure 2C). Among them, 13,174 and 15,472 transcripts were annotated in all the four databases.
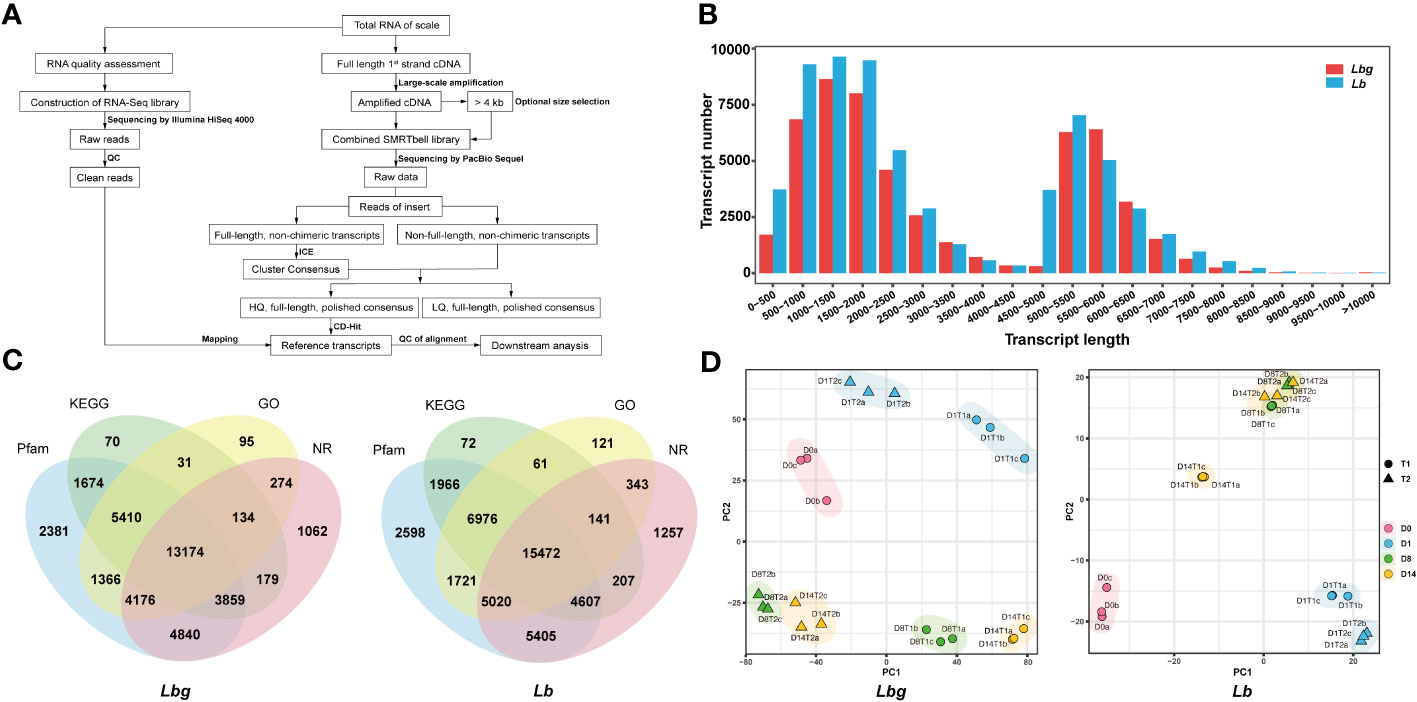
Figure 2 Reference transcripts generation of lily samples. (A) Workflow of lily sample sequencing. (B) Distribution of the length of reference transcripts. (C) Functional annotation of reference transcripts by Pfam, KEGG, GO and Nr databases. (D) Principal component analysis (PCA) plot of the samples. T1 and T2 represent samples of the control group and 2,4-D-treated group, respectively. D0, D1, D8 and D14 represent samples of 0 DAT, 1 DAT, 8 DAT and 14 DAT, respectively. The letters (a-c) represent three biological replicates.
Principal component analysis (PCA) showed that there were obvious differences of expression patterns among different stages under the same treatment conditions, except for 8 DAT and 14 DAT in Lb in the medium supplemented with 1 mg L-1 2,4-D (Figure 2D). In addition, a large deviation was also observed between the 2,4-D treatment group and the control group at the same stage. The results of sample clustering were consistent with the PCA results (Supplementary Figure S2). The above results further validated that obvious differences existed among stages during bulblet initiation, and 2,4-D influenced this process in both Lbg and Lb.
Six transcripts of Lbg (Isoform 22016, 21863, 28761, 46833, 29525 and 20315) and six transcripts of Lb (Isoform 31916, 25438, 39080, 36307, 34089 and 27602) were randomly selected for qRT-PCR to validate the differential expression by RNA-Seq. The results showed that the differential expression levels of these selected transcripts by qRT-PCR were highly consistent with those obtained by RNA-Seq (Supplementary Figure S3), confirming the reliability and accuracy of the RNA-Seq data.
3.3 Stage-specific DET screening and Mfuzz analysis revealed possible events of different stages
To identify transcriptional changes between distinct bulblet initiation stages, we compared the transcript expression profiles of adjacent stages in each lily, including 1 DAT versus (vs) 0 DAT (Group 1), 8 DAT vs 1 DAT (Group 2) and 14 DAT vs 8 DAT (Group 3). In total, 11,964 and 18,834 stage-specific DETs were screened in Lbg and Lb, respectively (Figures 3A, B). Clearly, Group 1 had the largest number of DETs among the three groups in both Lbg and Lb (Figures 3A, B), indicating that there were more complex transcriptional changes from 0 DAT to 1 DAT than in the following stages.
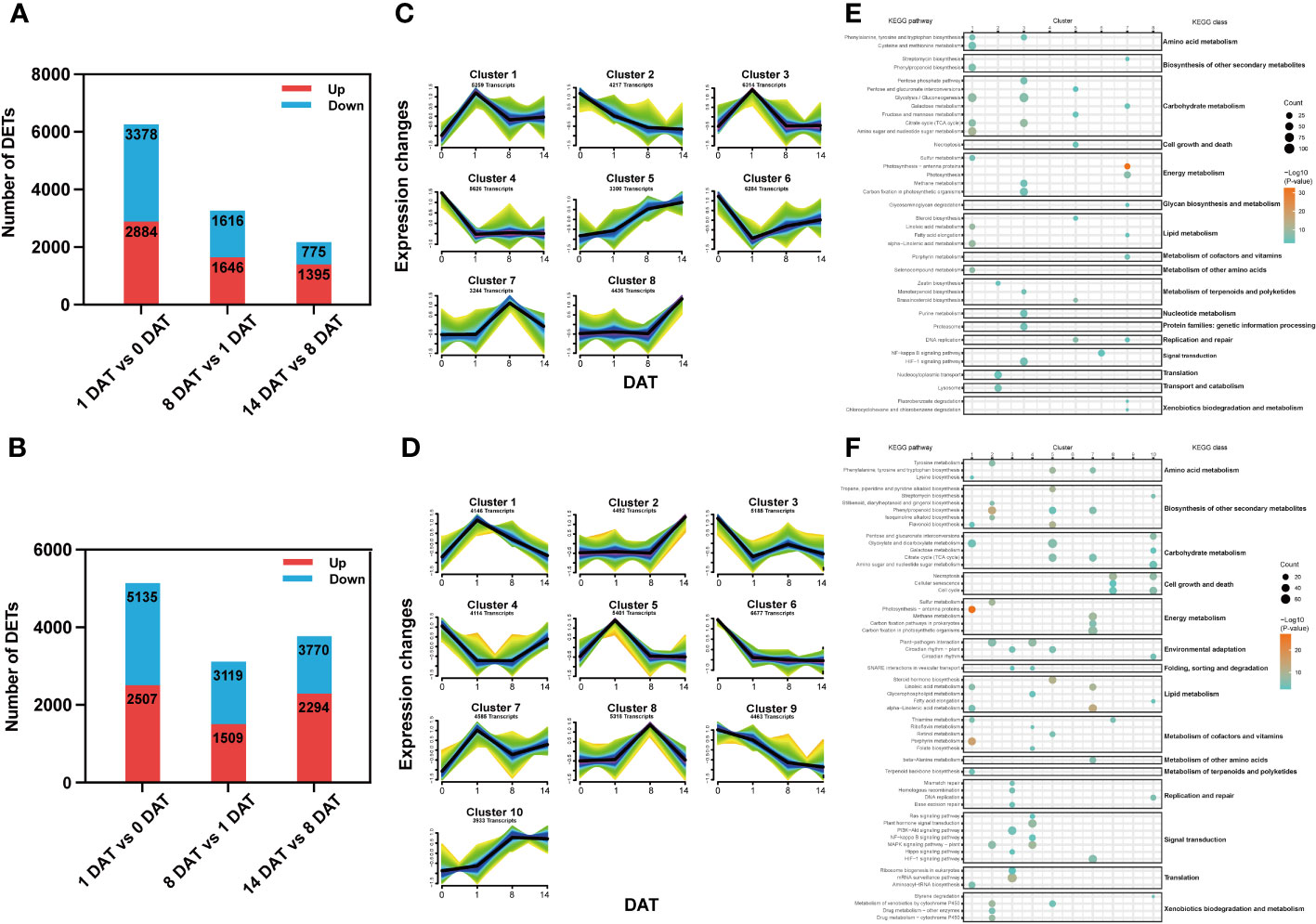
Figure 3 Stage-specific DETs screening and Mfuzz analysis of reference transcripts. (A, B) Number of DETs (|log2-fold changes| ≥ 2) identified between different stages in Lbg (A) and Lb (B). (C, D) Stage-specific DETs were classified into eight clusters of Lbg (C) and ten clusters of Lb (D) through Mfuzz analysis. (E, F) Bubble charts of KEGG pathway enrichment analysis of each cluster in Lbg (E) and Lb (F).
To define the temporal characteristics of the transcript dataset, we performed clustering analysis of 41,680 and 48,314 transcripts by Mfuzz in Lbg and Lb, respectively. The transcripts were divided into eight and ten clusters in Lbg and Lb, respectively (Figures 3C, D), and KEGG pathway enrichment analysis of these clusters was conducted (Figures 3E, F). The transcripts of clusters 1 and 3 in Lbg and clusters 5 and 7 in Lb could be candidates for the early response to scale detachment, since the highest expression of these clusters was exhibited at 1 DAT. The pathway “citrate cycle (TCA cycle)” (map00020), which is involved in carbohydrate metabolism, was upregulated in all the four candidate clusters (Figures 3E, F). The transcripts of cluster 7 in Lbg and cluster 8 in Lb were highly expressed at 8 DAT, with “DNA replication” (map03030) and “glycosaminoglycan degradation” (map00531) enriched in cluster 7 (Lbg), and “cell cycle” (map04110), “cellular senescence” (map04218) and “necroptosis” (map04217) enriched in cluster 8 (Lb), indicating that this stage could be associated with cell growth and death (Figures 3E, F). Moreover, the expression levels of transcripts in cluster 8 (Lbg) and cluster 2 (Lb) were increased at 14 DAT, and transcripts in cluster 2 (Lb) were mainly enriched in “biosynthesis of other secondary metabolites” and “xenobiotics biodegradation and metabolism” classes, which might play important functional roles in the bulblet swelling stage (Figures 3E, F).
3.4 2,4-D treatment promoted the process of in vitro bulblet initiation
To explore the effect of the exogenous application of 2,4-D on in vitro bulblet initiation, 1 mg L-1 2,4-D was added to the medium for bulblet induction. Results showed that the regeneration rate was significantly higher (p < 0.001) in 2,4-D-treated group than in the control group in both Lbg and in Lb (Figure 4A). More specifically, the 2,4-D treatment increased the regeneration rate by 1.84-fold in Lbg and 3.55-fold in Lb at 8 DAT, and 1.36-fold in Lbg and 1.51-fold in Lb at 14 DAT, indicating that the promotion effect of 2,4-D was stronger in Lb. Next, we screened 6,218 (3,175 upregulated, 3,043 downregulated) and 8,965 (5,382 upregulated, 3,583 downregulated) DETs between 2,4-D-treated group and the control group at 1 DAT, 8 DAT and 14 DAT in Lbg and Lb, respectively (Figure 4B). KEGG pathway enrichment analysis showed that, with 2,4-D treatment, “plant hormone signal transduction” (map04075) was upregulated in all the three stages in Lb and was also upregulated at 1 DAT and 8 DAT in Lbg (Figure 4C), indicating that 2,4-D could promote phytohormone responsiveness throughout the bulblet initiation process. Thus, 2,4-D had a promoting effect on in vitro bulblet initiation, and this effect might be closely related to phytohormone signal transduction.
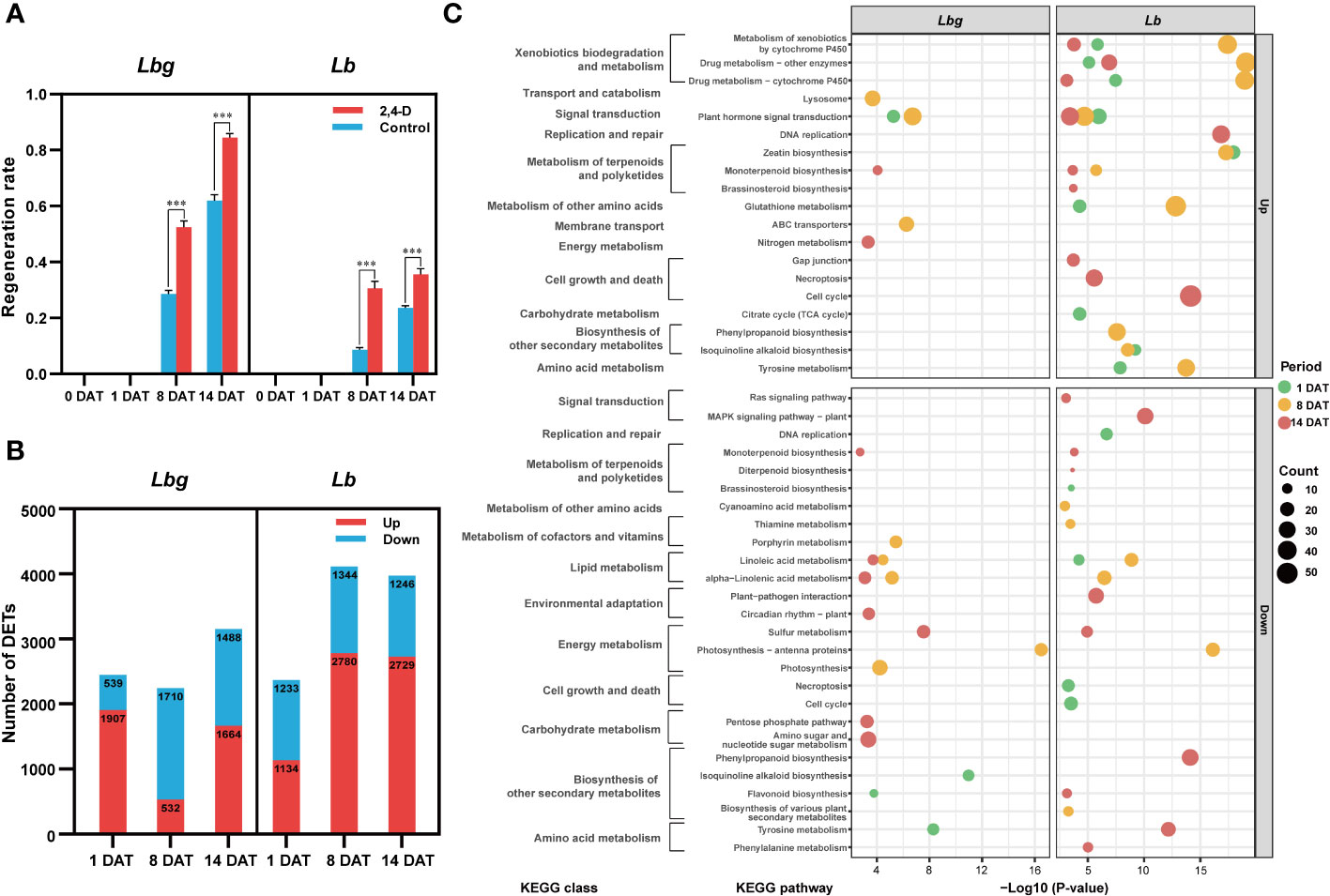
Figure 4 2,4-D-related DETs screening between the control group and 1 mg L-1 2,4-D-treated group of reference transcripts. (A) Regeneration rate of the control group and 2,4-D-treated group during in vitro bulblet initiation. Regeneration rate = number of scales that produced adventitious buds/total number of scales. ***Differences significant at p < 0.001. (B) Number of DETs (|log2-fold changes| ≥ 2) identified between the control group and 2,4-D-treated group. (C) Bubble charts of KEGG pathway enrichment analysis of 2,4-D-related DETs.
3.5 Changes in auxin-related genes during bulblet initiation
Considering the effect of auxin on bulblet initiation in various bulbous flowers, and the “plant hormone signal transduction” pathway was in response to exogenous 2,4-D application, we focused on the expression patterns of all screened DETs involved in auxin biosynthesis and signaling (Figure 5A). It is obvious that all screed DETs encoding Transporter Inhibitor Response 1 (TIR1) (six in Lbg and four in Lb) were significantly downregulated (p < 0.001) at 1 DAT in both Lbg and Lb (Figures 5B, C). Particularly, in Lb, the downregulation of four TIR1s (Isoform 33167, 34316, 32513 and 25870) were significantly enhanced (p < 0.001) by 2,4-D treatment (Figure 5C). Besides, 2,4-D significantly promoted (p < 0.001) the expression of two Tryptophan Aminotransferase (TAA) and Tryptophan Aminotransferase Related (TAR) (Isoform 49388 and 40427) in Lb, which could promote the endogenous synthesis of auxin (Figure 5C). Correspondingly, the content of endogenous IAA in the scales was significantly higher (p < 0.05) under 2,4-D treatment than that of the control group at 8 DAT and 14 DAT in Lb (Supplementary Figure S4). These changes above may lead to a stronger increase of regeneration rate in Lb than in Lbg.
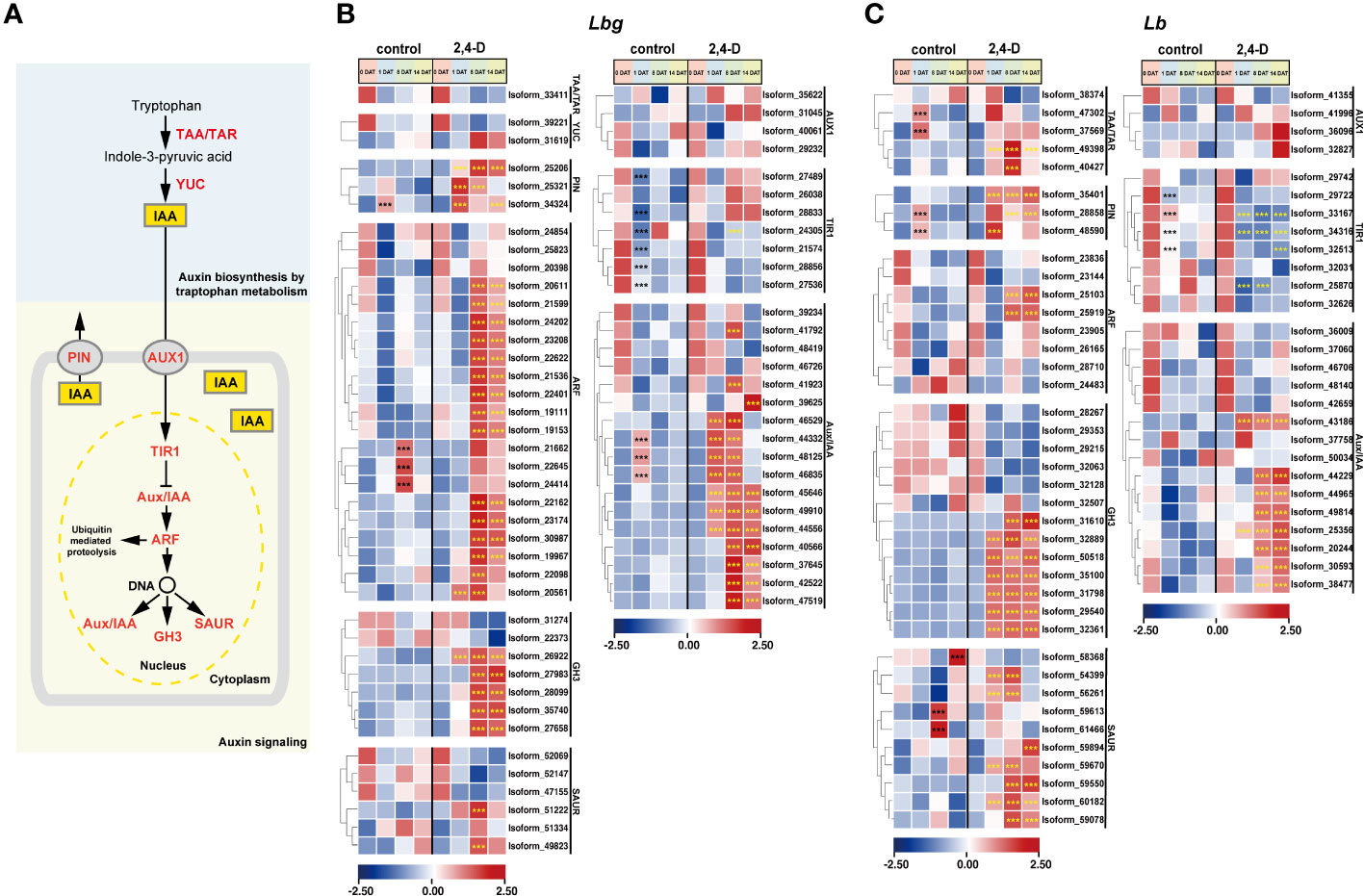
Figure 5 Expression patterns of stage-specific and 2,4-D related DETs involved in pathways of auxin biosynthesis and signaling. (A) Pathway of indole-3-acetic acid (IAA) biosynthesis and signaling. TAA, tryptophan aminotransferase; TAR, tryptophan aminotransferase related; YUC, YUC family; PIN, PIN-formed protein family; AUX1, AUX1/LAX symporters; TIR1, transporter inhibitor response 1, Aux/IAA, indole-3-acetic acid inducible; ARF, auxin response factor; GH3, GH3 family; SAUR, small auxin upregulated RNA. (B, C) Expression patterns of DETs involved in auxin biosynthesis and signaling in Lbg (B) and Lb (C). ***Differences significant at p < 0.001. The black asterisk represents a significant difference compared to 0 DAT in the control group. The yellow asterisk represents a significant difference in the 2,4-D-treated group compared to the control group at the same stage.
Three Auxin Response Factors (ARFs) of Lbg (Isoform 21662, 22645 and 24414) were significantly (p < 0.001) upregulated at 8 DAT in the control group, and 15 ARFs in Lbg and two ARFs in Lb were significantly (p < 0.001) upregulated at 8 DAT with 2,4-D treatment, following the downregulation of TIR1s (Figures 5B, C), indicating that 2,4-D could promote auxin signaling pathway during bulblet initiation. The significantly higher expression levels of DETs (p < 0.001) encoding some GH3 (five in Lbg and seven in Lb), Small Auxin Upregulated RNA (SAUR) (two in Lbg and seven in Lb) and Indole-3-acetic Acid Inducible (Aux/IAA) (14 in Lbg and eight in Lb) in the 2,4-D-treated group than in the control group at 1 DAT or 8 DAT further support the above idea (Figures 5B, C). Moreover, 2,4-D also promoted all screed DETs encoding PIN-formed protein family (PIN) (three in Lbg and three in Lb) after 1 DAT (Figures 5B, C). Overall, the promotion of auxin signaling pathway by 2,4-D application might contribute to the enhancement of bulblet regeneration ability.
3.6 Changes in key genes involved in sucrose and starch metabolism during bulblet initiation
Sucrose and starch metabolism was repeatedly reported to play an essential role in bulblet initiation in various bulbous flowers. Here, we focused on the expression patterns of key enzymes, transporters, and regulators involved in sucrose and starch metabolism pathway during bulblet initiation (Figure 6A). Without 2,4-D treatment, all screened DETs encoding Sucrose Synthase (SUS) (16 in Lbg and 17 in Lb) were significantly upregulated (p < 0.001) at 1 DAT or 8 DAT (Figures 6B, C). One Cell Wall Invertase (CWIN) in Lbg (Isoform 29531) were upregulated at 1 DAT and then downregulated, and the expression level of four CWINs (Isoform 29531, 28761 in Lbg and Isoform 39080, 34537 in Lb) were significantly increased (p < 0.001) in the 2,4-D-treated group (Figures 6B, C). Correspondingly, the content of glucose, one of the hexose hydrolytic products of sucrose, in the scales was significantly higher (p < 0.05) in the 2,4-D treated group than that in the control group in both Lbg and Lb (Supplementary Figure S4). Three Trehalose 6-Phosphate Synthases (TPSs) in Lbg (Isoform 36005, 19880 and 20796) and five TPSs in Lb (Isoform 24997, 32954, 34599, 24963 and 43346) were significantly upregulated (p < 0.001) at 1 DAT, 8 DAT or 14 DAT (Figures 6B, C). Notably, the expression patterns of some Sucrose Transporters (SUTs) and SWEET Sucrose-Efflux Transporters (SWEETs) were strongly affected by 2,4-D application. All screened differentially expressed SUTs (two in Lbg and two in Lb) were significantly upregulated (p < 0.001) at 1 DAT, among them, the upregulation of one SUT in Lbg (Isoform 31091) and two SUTs (Isoform 36404 and 36445) in Lb was significantly enhanced (p < 0.001) by 2,4-D (Figures 6B, C). In addition, there was no significant change in the expression level of three SWEETs in Lbg (Isoform 42147, 46206 and 42002) and one in Lb (Isoform 48829) without 2,4-D during bulblet initiation, but these SWEETs significantly upregulated (p < 0.001) at 8 DAT with 2,4-D application (Figures 6B, C).
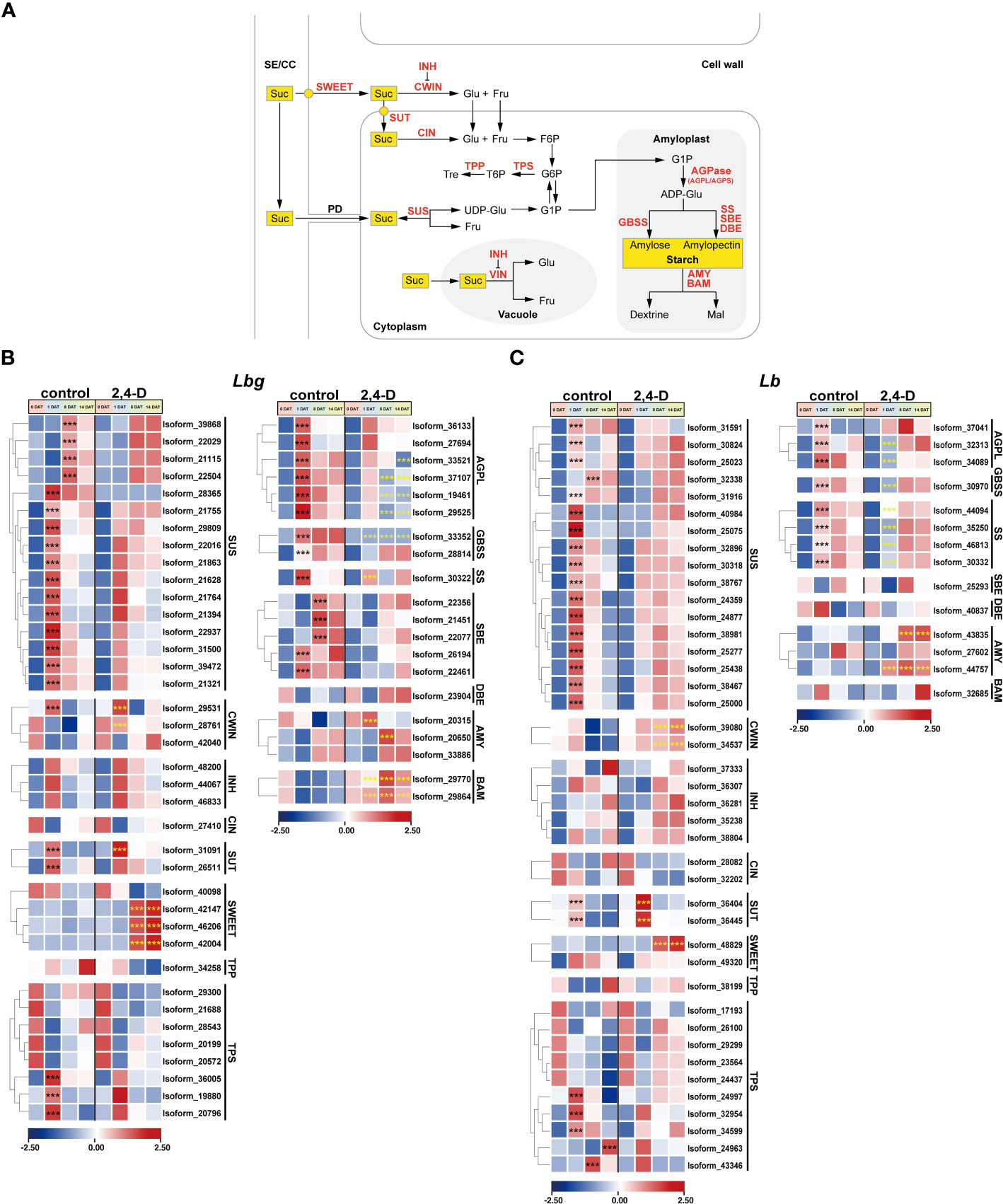
Figure 6 Expression patterns of stage-specific and 2,4-D related DETs involved in sucrose and starch metabolism pathway. (A) Sucrose and starch metabolism pathway. SE/CC, sieve element/companion cell complex; PD, plasmodesma; Suc, sucrose; Glu, glucose; Fru, fructose; Tre, trehalose; Mal, maltose; UDP-Glu, UDP-glucose; F6P, fructose-6-phosphate; G6P, glucose-6-phosphate; G1P, glucose-1-phosphate; ADP-Glu, ADP-Glucose; SWEET, SWEET sucrose-efflux transporter family; SUT, sucrose transporter; SUS, sucrose synthase; CWIN, cell wall invertase; VIN, vacuolar invertase; INH, invertase inhibitor; CIN, cytoplasmic invertase; TPP, trehalose 6-phosphate phosphatase; TPS, trehalose 6-phosphate synthase; AGPL/AGPS, large/small subunit of ADP-glucose pyrophosphorylase (AGPase); GBSS, granule-bound starch synthase; SS, starch synthase; SBE; starch branching enzyme; DBE, starch debranching enzyme; AMY, amylase, BAM, β-amylase. (B, C) Expression patterns of DETs involved in sucrose and starch metabolism in Lbg (B) and Lb (C). ***Differences significant at p < 0.001. The black asterisk represents a significant difference compared to 0 DAT in the control group. The yellow asterisk represents a significant difference in the 2,4-D-treated group compared to the control group at the same stage.
Several genes encoding key enzymes involved in starch synthesis were upregulated during bulblet initiation. All screened DETs encoding large subunit of AGPase (AGPL), Granule-Bound Starch Synthase (GBSS), and Starch Synthase (SS) were significantly upregulated (p < 0.001) at 1 DAT without 2,4-D, but some of these upregulations were inhibited (Isoform 33352 in Lbg), attenuated (Isoform 33521, 37101, 19461, 29525, 30322 in Lbg and Isoform 44094 in Lb) or delayed (Isoform 28814 in Lbg and Isoform 32313, 32089, 30970, 44094, 35250, 46813, 30332 in Lb) in the 2,4-D-treated group (Figures 6B, C), which indicated that 2,4-D suppressed starch synthesis during bulblet initiation. On the contrary, the expression levels of some Amylases (AMYs) (Isoform 20315, 20650 in Lbg and Isoform 43835, 44457 in Lb) and β-Amylases (BAMs) (Isoform 29770, 29864 in Lbg), which were involved in starch degradation, were significantly upregulated (p < 0.001) by 2,4-D (Figures 6B, C), indicating that 2,4-D promoted starch degradation. In Lbg, the starch content of scales was significantly decreased (p < 0.05) by 2,4-D at 1 DAT, 8 DAT and 14 DAT (Supplementary Figure S4). Similar decreases were also observed in Lb, but there were no significant differences (p < 0.05) (Supplementary Figure S4). In addition, we identified five significantly upregulated Starch Branching Enzymes (SBEs) (p < 0.001) in Lbg (Isoform 22356, 21451, 22077, 26194 and 22461) but not in Lb (Figures 6B, C).
3.7 Identification of potential regulators of in vitro bulblet initiation
In total, we identified 1209 and 1363 TFs from stage-specific DETs and 2,4-D-related DETs in Lbg and Lb, respectively. In stage-specific TFs, the number of MYB, bHLH and ERF ranked in the top three in Lbg, and MYB, bHLH and WRKY ranked in the top three in Lb (Figure 7A). In 2,4-D-related TFs, the three most were bHLH, GRAS and C3H in Lbg, and bHLH, C3H and ERF in Lb (Figure 7B). Given these, the expression patterns of TFs belonging to these TF families were analyzed, including 25 bHLHs, 24 GRASs, 22 C3Hs, 21 MYBs and 25 ERFs in Lbg, and 28 bHLHs, 25 C3Hs, 23 ERFs, 31 MYBs and 23 WRKYs in Lb (Figures 7C, D).
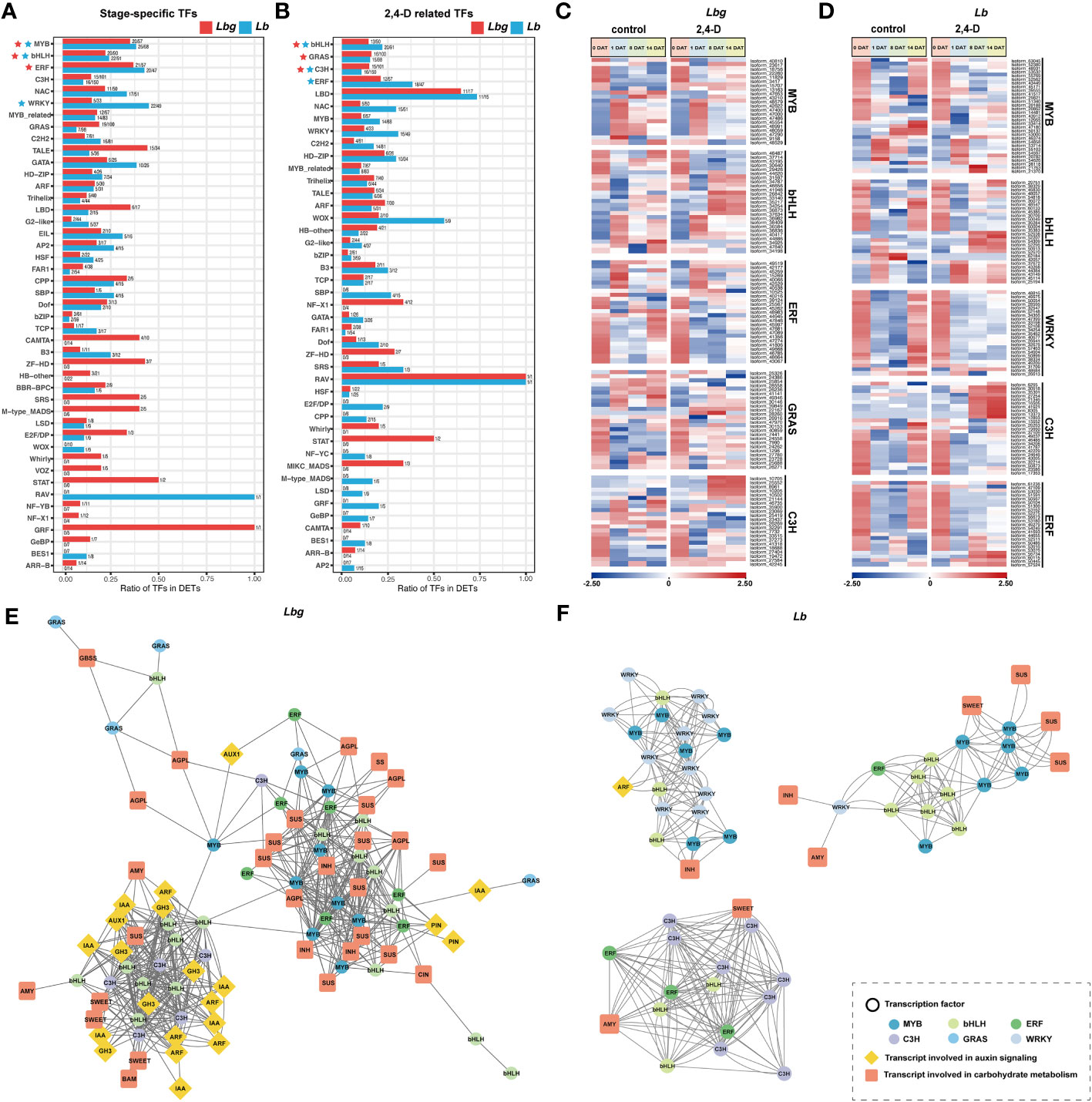
Figure 7 (A) TF families identified from the stage-specific DETs. (B) TF families identified from the 2,4-D-related DETs. The TF families in (A) and (B) rank according to the number of differentially expressed TFs they contained. (C) Express patterns of differentially expressed TFs belonging to MYB, bHLH, ERF, GRAS and C3H families in Lbg. (D) Express patterns of differentially expressed TFs belonging to MYB, bHLH, WRKY, C3H and ERF families in Lb. (E, F) Correlation analysis of DETs involved in auxin signaling and carbohydrate metabolism, and differentially expressed TFs in Lbg (E) and Lb (F). The correlation analysis was conducted with Pearson’s two-tailed test. DETs with significant correlations (|r| ≥ 0.8. r, Pearson correlation coefficient) were linked. stering analysis of reference transcripts of Lbg (A) and Lb (B).
Further, the correlation analysis (|r| ≥ 0.8. r, Pearson correlation coefficient) between these candidate TFs and DETs involved in auxin signaling and carbohydrate metabolism was conducted. In Lbg, we found six bHLHs, seven ERFs, eight MYBs and one GRAS were co-expressed with ten SUSs, two INHs, one CIN, three AGPLs and two PINs (Figure 7E). Eight bHLHs and five C3Hs were co-expressed with three SWEETs, two AMYs, one BAM, one SUS and one AUX1, six Aux/IAAs, five ARFs and five GH3s (Figure 7E). In Lb, the expression level of six MYBs and six bHLHs were co-expressed with three SUSs and one SWEET (Figure 7F). Eight C3Hs three ERFs and three bHLHs were co-expressed with an AMY and a SWEET (Figure 7F). Ten WRKYs, six MYBs, three bHLHs and one ARF were co-expressed (Figure 7F). Above all, the above-mentioned TFs are possibly involved in the regulation of auxin signaling and carbohydrate metabolism, thus regulate the process of in vitro bulblet initiation.
4 Discussion
4.1 High-quality full-length transcripts of lily during in vitro bulblet initiation were constructed by a hybrid sequencing strategy
Tissue culture is a main asexual reproduction method for many bulbous crops and has a significant advantage in promoting regeneration efficiency and shortening the breeding and propagation cycle (Bakhshaie et al., 2016). Adventitious bud initiation and bulblet formation are critical steps during micropropagation of bulbous flowers, especially for direct organogenesis via shoot induction, which depends heavily on efficient nutritional allocation and hormone regulation (Xu et al., 2020a; Ren et al., 2022). Although many reports have been published on the process of bulblet formation and development in the lily (De Klerk, 2012; Li et al., 2014; Wu et al., 2021), there are relatively few reports on the detailed mechanism of early bulblet initiation.
In the present study, comprehensive full-length transcriptomes of Lbg and Lb were obtained through PacBio Iso-seq together with Illumina short-read sequencing during in vitro bulblet initiation. To date, Illumina sequencing technology has been widely applied to explore the process of bulblet formation and development in lily (Li et al., 2014; Du et al., 2017; Yang et al., 2017; Lazare et al., 2019). Recently, a full-length transcriptome of Lilium Oriental Hybrids ‘Sorbonne’ was generated (Li et al., 2022). Here, we conducted construct two high-quality full-length reference transcriptomes (Lbg and Lb). The N50 of transcripts of Lbg and Lb was 5,422 bp and 5,199 bp, respectively, and the mean length of transcripts of Lbg and Lb were 3,108 bp and 2,965 bp, respectively (Supplementary Table S1). These data contribute to further studies on molecular mechanisms of bulblet formation and other biological process of lily.
4.2 Exogenous 2,4-D application promote in vitro bulblet initiation by enhancing auxin signaling
Recent studies have indicated that auxin contributes to bulblet initiation in several bulbous flowers (Yang et al., 2017; Xu et al., 2020a). The content of IAA, the main naturally occurring auxin, increased consistently during the process of bulblet initiation and development in Lycoris, and exogenous IAA improved bulblet growth (Zhao, 2012; Xu et al., 2020a). In the present study, 1 mg L-1 2,4-D significantly increase the regeneration rate of in vitro bulblet in Lbg and Lb (Figure 5A). Generally, auxin binds to TIR1 nuclear receptors, and then the auxin signal is modulated by the quantitative and qualitative responses of the Aux/IAAs and ARFs (Chapman and Estelle, 2009; Hayashi, 2012). ARFs are crucial regulators involved in the auxin signaling pathway, and then induce three major families: SAUR, GH3 and Aux/IAA genes (Guilfoyle and Hagen, 2007; Hayashi, 2012). In a previous study, differentially expressed TIRs, ARFs, and SAURs were identified during bulblet in Lycoris through transcriptome analysis (Xu et al., 2020a). Here, we found that 2,4-D affected the expression of genes involved in auxin signaling, thus promoted in vitro bulblet initiation. The obvious downregulations were found in many differentially expressed TIR1s at 1 DAT in both Lbg and Lb, which could be enhanced by 2,4-D application (Figures 5B, C). In addition, many ARFs, SAUR, GH3 and Aux/IAAs, which were not apparently responsive in the control group during bulblet initiation, were significantly upregulated in the 2,4-D-treated group (Figures 5B, C). Particularly, three ARFs in Lbg were significantly upregulated at 8 DAT in the control group (Figure 5B). Taken together, we proposed ARFs as key response factors during in vitro bulblet initiation in lily.
The distribution of auxin in cells depends largely on auxin transport, especially auxin efflux, which is directed by the polar subcellular localization of the PIN1 auxin efflux transporter in the plasma membrane (Vanneste and Friml, 2009; Hayashi, 2012). The expression and gradual polarization of PINs induced by auxin promote the formation of new vascular strands originating from the position of auxin application (Sauer et al., 2006). Three PIN genes were significantly upregulated during bulblet initiation in Lycoris (Xu et al., 2020a). In our study, all differentially expressed PINs were upregulated by 2,4-D in both Lbg and Lb (Figures 5B, C), indicating that 2,4-D might promote auxin transportation to promote bulblet initiation.
4.3 Starch and sucrose metabolism are crucial processes during in vitro bulblet initiation
The process of bulblet initiation involves carbohydrate transport from the source to the sink. The starch storage in the mother scales and exogenous carbon supply can be considered as the carbon source, and the basal of the mother scale, where the bulblets initiate, act as the sink tissue. Sucrose is unloaded from the phloem into sink cells either apoplasmically or symplasmically, then utilized to produce energy for cellular process (Ruan, 2014; Figure 6A).
Starch accounts for approximately 70% of the dry weight of lily bulbs (Wu et al., 2021). Starch synthesis is considered to be a crucial pathway for bulblet initiation. Several studies have indicated that whether a meristem can produce scale primordia depends on its capacity to accumulate starch (Bourque et al., 1987; Wu et al., 2021). In Lycoris, abscisic acid (ABA) upregulated the expression level of LrSS1, LrSS2, and LrGBSS1 genes, which could enhance carbohydrate accumulation in the bulblets, thus promoted their development (Xu et al., 2020b). Similarly, starch synthesis was positively correlated with bulbil formation in Lilium lancifolium with upregulation of AGPL, SS, GBSS and SBE (Yang et al., 2017). In the present study, all screened DETs encoding AGPL, GBSS, and SS were upregulated at 1 DAT or 8 DAT without 2,4-D in both Lbg and Lb (Figures 6B, C), indicating the enhancement of starch synthesis process. Especially, five significantly upregulated SBEs were identified in Lbg (Figures 6B, C), indicating that Lbg might have a stronger ability of starch accumulation for bulblet initiation than Lb. During bulblet formation, starch is degraded in the mother scales and synthesized at the bulblet regeneration site and in the newly formed bulblets. In Lilium, the enzymes involved in starch synthetic direction, such as AGPase, GBSS, SS, and SBE, showed a decreasing trend in mother scales but an increasing trend in bulblets during bulblet formation (Li et al., 2014; Wu et al., 2021). Moreover, starch content in basal scales and basal plates of Lycoris (the major sites of bulblet regeneration) showed a rapid decline during bulblet initiation in the efficient bulblet regeneration system (Ren et al., 2021). Here, we found in the 2,4-D-treated group, the more efficient group for in vitro bulblet initiation, key enzymes involved in starch synthesis (AGPL, SS and GBSS) were downregulated, while key enzymes involved in starch degradation (AMY and BAM) were upregulated compared to the control group (Figures 6B, C). In addition, 2,4-D reduced the starch content in the scales during bulblet initiation (Supplementary Figure S4). Taken together, we suggested that 2,4-D accelerate the starch degradation process to increase carbon supply for newly bulblet initiation.
Soluble sugars in mother scales were transported into the region where bulblets were initiated to supply the follow-up bulblet development (Xu et al., 2020a). Sucrose is the dominant transport form of sugars in higher plants (Lalonde et al., 1999; Rolland et al., 2006). SUS and CWIN are considered the most important sucrose hydrolases involved in the sucrose unloading pathway (Ruan, 2014; Figure 6A). SUS contribute to starch synthesis and accumulation, functioning during the later bulblet initiation and development (Yang et al., 2017; Wu et al., 2021). In Lilium, SUSs and INVs were both highly expressed in the mother scales and bulblets during bulblet emergence and swelling (Li et al., 2014). Similarly, SUSs and INVs were greatly upregulated accompanied by a decrease in sucrose content in mother scales during bulblet initiation in Lycoris (Xu et al., 2020a). In this study, 16 SUSs in Lbg and 17 SUSs in Lb were upregulated at 1 DAT or 8 DAT, and one CWIN in Lbg and two CWINs in Lb were upregulated at 1 DAT and then downregulated (Figures 6B, C). For example, SUS and CWIN often presented an opposite expression pattern during bulblet initiation, and this change was considered to be a possible sign of the transition from bulblet initiation to development (Ren et al., 2021; Wu et al., 2021). In particular, CWINs were highly expressed during the early bulblet initiation stage and produced glucose, which might act as sugar signaling rather than carbon resources (Wu et al., 2021). In Lycoris, the more highly LsCWIN2 was expressed, the more bulblets were produced (Ren et al., 2021). Here, the expression levels of CWINs in Lbg and Lb were significantly increased by 2,4-D, showing a possible role of CWINs in increasing bulblet regeneration rate. Recent study showed that LbgCWIN1 significantly upregulated endogenous starch was degraded during in vitro bulblet initiation in lily (Gao et al., 2023), indicating that CWIN can be selected as a candidate gene subsequent function verification.
Interestingly, the application of 2,4-D in the medium for bulblet induction had significant effects on the expression of genes involved in carbohydrate metabolism, especially SUTs and SWEETs (Figure 6A). SWEETs probably mediate sucrose efflux from SE/CC to apoplasm, and then sucrose can be taken up by SUTs, which are key steps proceeding phloem unloading (Ruan, 2014). The upregulation of one SUT in Lbg and two SUTs in Lb was significantly enhanced by 2,4-D at 1 DAT (Figures 6B, C). Three SWEETs in Lbg and one SWEET had no significant expression change in Lb without 2,4-D during bulblet initiation, but significantly upregulated at 8 DAT and 14 DAT with 2,4-D application (Figures 6B, C). Thus, 2,4-D might facilitate in vitro bulblet initiation mainly through promoting sucrose unloading from the SE/CC to the sink cells, and SWEETs and SUTs can be considered as good candidates for future functional studies.
4.4 Candidate TFs might be involved in the regulation of in vitro bulblet initiation
Although carbohydrate metabolism and the auxin signaling pathway have been respectively demonstrated to participate in bulblet initiation, their cooperative function during this process has not yet been reported. Here, we found that in Lbg, three SWEETs, two AMYs, one BAM and one SUS were co-expressed with one AUX1, six Aux/IAAs, five ARFs and five GH3s, and ten SUSs, two INHs, one CIN and three AGPLs were co-expressed with two PINs (Figure 7E), indicating the possible coregulation by carbohydrate metabolism and auxin signaling of in vitro bulblet initiation in lily. A recent study showed that bHLH, bZIP, WRKY, TCP, MYB, YABBY, NAC, C2H2 were identified to be involved in bulbil induction of ‘Sorbonne’ lily by coexpression analysis (Li et al., 2022). In Lycoris, coexpression analysis revealed that transcripts encoding WOX14, MYB117, and ULT1 coexpressed with LsCWIN2, and transcripts encoding ERFs were coexpressed with LsSUS4 during bulb vegetative production (Ren et al., 2022). In the present study, MYB, bHLH, ERF, C3H, GRAS, WRKY families were identified to be candidate regulators of in vitro bulblet initiation. Among them, several TFs might be involved in the regulation the expression of key enzymes in carbohydrate metabolism during in vitro bulblet initiation in both Lbg and Lb, for example, C3Hs and bHLHs were co-expressed with AMYs and SWEETs, and some MYBs and bHLHs were co-expressed with SUSs in both Lbg and in Lb (Figures 7E, F). The above results may assist in the understanding of molecular mechanism of lily bulblet initiation.
Data availability statement
The data presented in the study are deposited in the GenBank repository, accession number PRJNA933000.
Author contributions
LZ, YW and ZMR designed the experiment. LZ, CG, YCX, YL and XX carried out the experiment. CG and LC analyzed the data. CG drafted the manuscript. ZR, YW and YPX revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the Zhejiang Science and Technology Major Program on Agricultural New Variety Breeding (2021C02071-6) and the National Natural Science Foundation of China (Grant Nos. 32002071, 32101571).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1236315/full#supplementary-material
Supplementary Table 1 | Statistical summary of the reference transcriptome.
Supplementary Table 2 | Primers of selected transcripts for quantitative real-time PCR validation.
References
Bakhshaie, M., Khosravi, S., Azadi, P., Bagheri, H., van Tuyl, J. M. (2016). Biotechnological advances in lilium. Plant Cell Rep. 35 (9), 1799–1826. doi: 10.1007/s00299-016-2017-8
Bourque, J. E., Miller, J. C., Park, W. D. (1987). Use of an in vitro tuberization system to study tuber protein gene expression. In Vitro Cell. Dev. Biol. 23 (5), 381–386. doi: 10.1007/BF02620996
Cantalapiedra, C. P., Hernández-Plaza, A., Letunic, I., Bork, P., Huerta-Cepas, J. (2021). EggNOG-mapper v2: Functional Annotation, Orthology Assignments, and Domain Prediction at the Metagenomic Scale. Mol. Biol. Evol. 38 (12), 5825–5829. doi: 10.1093/molbev/msab293
Chapman, E. J., Estelle, M. (2009). Mechanism of auxin-regulated gene expression in plants. Annu. Rev. Genet. 43 (1), 265–285. doi: 10.1146/annurev-genet-102108-134148
Chen, T., Zhang, H., Liu, Y., Liu, Y. X. (2021). EVenn: Easy to create repeatable and editable Venn diagrams and Venn networks online. J. Genet. Genomics 48 (9), 863–866. doi: 10.1016/j.jgg.2021.07.007
Chen, S., Zhou, Y., Chen, Y., Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34 (17), i884–i890. doi: 10.1093/bioinformatics/bty560
De Klerk, G.-J. (2012). Micropropagation of bulbous crops: technology and present state. Floricult. Ornamental Biotechnol. 6, 1–8.
Du, F., Fan, J., Wang, T., Wu, Y., Grierson, D., Gao, Z., et al. (2017). Identification of differentially expressed genes in flower, leaf and bulb scale of lilium oriental hybrid ‘Sorbonne’ and putative control network for scent genes. BMC Genomics 18 (1), 899. doi: 10.1186/s12864-017-4303-4
Firon, N., LaBonte, D., Villordon, A., Kfir, Y., Solis, J., Lapis, E., et al. (2013). Transcriptional Profiling of Sweetpotato (Ipomoea batatas) Roots Indicates Down-regulation of Lignin Biosynthesis and Up-regulation of Starch Biosynthesis at an Early Stage of Storage Root Formation. BMC Genomics 14, 460. doi: 10.1186/1471-2164-14-460
Gao, C., Li, S., Xu, Y., Liu, Y., Xia, Y., Ren, Z., et al. (2023). Molecular cloning, characterization and promoter analysis of LbgCWIN1 and its expression profiles in response to exogenous sucrose during in vitro bulblet initiation in lily. Hortic. Plant J. doi: 10.1016/j.hpj.2022.09.009
Guilfoyle, T. J., Hagen, G. (2007). Auxin response factors. Curr. Opin. Plant Biol. 10 (5), 453–460. doi: 10.1016/j.pbi.2007.08.014
Guo, Z., Wang, F., Xiang, X., Ahammed, G. J., Wang, M., Onac, E., et al. (2016). Systemic induction of photosynthesis via illumination of the shoot apex is mediated sequentially by phytochrome B, auxin and hydrogen peroxide in tomato. Plant Physiol. 172 (2), 1259–1272. doi: 10.1104/pp.16.01202
Hayashi, K. (2012). The interaction and integration of auxin signaling components. Plant Cell Physiol. 53 (6), 965–975. doi: 10.1093/pcp/pcs035
Hu, X. G., Zhuang, H., Lin, E., Borah, P., Du, M., Gao, S., et al. (2022). Full-Length Transcriptome Sequencing and Comparative Transcriptomic Analyses Provide Comprehensive Insight into Molecular Mechanisms of Cellulose and Lignin Biosynthesis in Cunninghamia lanceolata. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.883720
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernández-Plaza, A., Forslund, S. K., Cook, H., et al. (2019). EggNOG 5.0: A hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47 (D1), D309–D314. doi: 10.1093/nar/gky1085
Jin, J., Tian, F., Yang, D. C., Meng, Y. Q., Kong, L., Luo, J., et al. (2017). PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 45 (D1), D1040–D1045. doi: 10.1093/nar/gkw982
Lalonde, S., Boles, E., Hellmann, H., Barker, L., Patrick, J. W., Frommer, W. B., et al. (1999). The dual function of sugar carriers: transport and sugar sensing. Plant Cell 11 (4), 707–726. doi: 10.1105/tpc.11.4.707
Langmead, B., Salzberg, S. L. (2012). Fast gapped-read alignment with bowtie 2. Nat. Methods 9 (4), 357–359. doi: 10.1038/nmeth.1923
Lazare, S., Bechar, D., Fernie, A. R., Brotman, Y., Zaccai, M. (2019). The proof is in the bulb: glycerol influences key stages of lily development. Plant J. 97 (2), 321–340. doi: 10.1111/tpj.14122
Lee, E., Yun, N., Jang, Y. P., Kim, J. (2013). Lilium lancifolium thunb. Extract attenuates pulmonary inflammation and air space enlargement in a cigarette smoke-exposed mouse model. J. Ethnopharmacol. 149 (1), 148–156. doi: 10.1016/j.jep.2013.06.014
Li, G., Chen, Z., Yan, F. (2007). Lilium brownii var. giganteum: A New Variety of Lilium from Wenling, Zhejiang. J. Zhejiang Forest. Coll. 06, 767–768.
Li, B., Dewey, C. N. (2011). RSEM: accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinf. 12, 323. doi: 10.1186/1471-2105-12-323
Li, W., Godzik, A. (2006). Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22 (13), 1658–1659. doi: 10.1093/bioinformatics/btl158
Li, J., Sun, M., Li, H., Ling, Z., Wang, D., Zhang, J., et al. (2022). Full-length transcriptome-referenced analysis reveals crucial roles of hormone and wounding during induction of aerial bulbils in lily. BMC Plant Biol. 22 (1), 415. doi: 10.1186/s12870-022-03801-8
Li, X., Wang, C., Cheng, J., Zhang, J., da Silva, J. A. T., Liu, X., et al. (2014). Transcriptome Analysis of Sarbohydrate Metabolism during Bulblet Formation and Development in Lilium davidii var. unicolor. BMC Plant Biol. 14 (1), 358. doi: 10.1186/s12870-014-0358-4
Liu, B., Wang, X., Cao, Y., Arora, R., Zhou, H., Xia, Y. (2020). Factors affecting freezing tolerance: a comparative transcriptomics study between field and artificial cold acclimations in overwintering evergreens. Plant J. 103, 2279–2300. doi: 10.1111/tpj.14899
Love, M. I., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15 (12), 550. doi: 10.1186/s13059-014-0550-8
Lv, X., Zhang, D., Min, R., Li, S., Li, Z., Ren, Z., et al. (2020). Effects of Exogenous Sucrose on Bulblet Formation of Lycoris sprengeri in vitro. Acta Hortic. 47 (8), 1475–1489. doi: 10.16420/j.issn.0513-353x.2019-0415
Manni, M., Berkeley, M. R., Seppey, M., Simão, F. A., Zdobnov, E. M. (2021). BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol. Biol. Evol. 38 (10), 4647–4654. doi: 10.1093/molbev/msab199
Murashige, T., Skoog, F. (1962). A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol. Plant 15 (3), 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Ren, Z., Xia, Y., Zhang, D., Li, Y., Wu, Y. (2017). Cytological analysis of the bulblet initiation and development in lycoris species. Sci. Hortic. 218, 72–79. doi: 10.1016/j.scienta.2017.02.027
Ren, Z., Xu, Y., Lvy, X., Zhang, D., Gao, C., Lin, Y., et al. (2021). Early sucrose degradation and the dominant sucrose cleavage pattern influence lycoris sprengeri bulblet regeneration in vitro. Int. J. Mol. Sci. 22 (21), 11890. doi: 10.3390/ijms222111890
Ren, Z., Zhang, D., Jiao, C., Li, D., Wu, Y., Wang, X., et al. (2022). Comparative transcriptome and metabolome analyses identified the mode of sucrose degradation as a metabolic marker for early vegetative propagation in bulbs of lycoris. Plant J. 112 (1), 115–134. doi: 10.1111/tpj.15935
Rolland, F., Baena-Gonzalez, E., Sheen, J. (2006). Sugar sensing and signaling in plants: conserved and novel mechanisms. Annu. Rev. Plant Biol. 57, 675–709. doi: 10.1146/annurev.arplant.57.032905.105441
Ruan, Y. L. (2014). Sucrose metabolism: gateway to diverse carbon use and sugar signaling. Annu. Rev. Plant Biol. 65 (1), 33–67. doi: 10.1146/annurev-arplant-050213-040251
Sauer, M., Balla, J., Luschnig, C., Wisniewska, J., Reinöhl, V., Friml, J., et al. (2006). Canalization of auxin flow by aux/IAA-ARF-dependent feedback regulation of PIN polarity. Genes Dev. 20 (20), 2902–2911. doi: 10.1101/gad.390806
Shannon, P., Markiel, A., Ozier, O., Baliga, N. S., Wang, J. T., Ramage, D., et al. (2003). Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13 (11), 2498–2504. doi: 10.1101/gr.1239303
Van Aartrijk, J., Blom-Barnhoorn, G. J. (1984). Adventitious bud formation from bulb-scale explants of lilium speciosum thunb. In vitro interacting effects of NAA, TIBA, wounding, and temperature. J. Plant Physiol. 116 (5), 409–416. doi: 10.1016/S0176-1617(84)80132-7
Vanneste, S., Friml, J. (2009). Auxin: A trigger for change in plant development. Cell 136 (6), 1005–1016. doi: 10.1016/j.cell.2009.03.001
Wu, T., Hu, E., Xu, S., Chen, M., Guo, P., Dai, Z., et al. (2021). ClusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation 2 (3), 100141. doi: 10.1016/j.xinn.2021.100141
Wu, Y., Ma, Y., Li, Y., Zhang, L., Xia, Y. (2019). Plantlet Regeneration from Primary Callus Cultures of Lilium brownii F.E.Br. Ex Miellez var. giganteum G. Y. Li and Z. H. Chen, a Rare Bulbous Germplasm. In Vitro Cell. Dev. Biol.: Plant 55 (1), 44–59. doi: 10.1007/s11627-018-09955-1
Wu, Y., Ren, Z., Gao, C., Sun, M., Li, S., Min, R., et al. (2020). Change in sucrose cleavage pattern and rapid starch accumulation govern lily shoot-to-bulblet transition in vitro. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.564713
Xu, J., Li, Q., Li, Y., Yang, L., Zhang, Y., Cai, Y. (2020b). Effect of exogenous gibberellin, paclobutrazol, abscisic acid, and ethrel application on bulblet development in lycoris radiata. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.615547
Xu, J., Li, Q., Yang, L., Li, X., Wang, Z., Zhang, Y. (2020a). Changes in Carbohydrate Metabolism and Endogenous Hormone Regulation during Bulblet Initiation and Development in Lycoris radiata. BMC Plant Biol. 20, 180. doi: 10.1186/s12870-020-02394-4
Xu, L., Yang, P., Feng, Y., Xu, H., Cao, Y., Tang, Y., et al. (2017). Spatiotemporal transcriptome analysis provides insights into bicolor tepal development in lilium “Tiny padhye”. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.00398
Yang, P., Xu, L., Xu, H., Tang, Y., He, G., Cao, Y., et al. (2017). Histological and Transcriptomic Analysis during Bulbil Formation in Lilium lancifolium. Front. Plant Sci. 8. doi: 10.3389/fpls.2017.01508
Ye, J., Coulouris, G., Zaretskaya, I., Cutcutache, I., Rozen, S., Madden, T. L. (2012). Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinf. 13, 134. doi: 10.1186/1471-2105-13-134
Keywords: starch synthesis and degradation, sucrose unloading, auxin signaling, full-length sequencing, Lilium
Citation: Gao C, Zhang L, Xu Y, Liu Y, Xiao X, Cui L, Xia Y, Wu Y and Ren Z (2023) Full-length transcriptome analysis revealed that 2,4-dichlorophenoxyacetic acid promoted in vitro bulblet initiation in lily by affecting carbohydrate metabolism and auxin signaling. Front. Plant Sci. 14:1236315. doi: 10.3389/fpls.2023.1236315
Received: 07 June 2023; Accepted: 01 September 2023;
Published: 20 September 2023.
Edited by:
Yan Zhao, Shandong Agricultural University, ChinaReviewed by:
XingWen Zhou, Fujian University of Technology, ChinaYun-peng Du, Beijing Academy of Agricultural and Forestry Sciences, China
Copyright © 2023 Gao, Zhang, Xu, Liu, Xiao, Cui, Xia, Wu and Ren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ziming Ren, zimingren@zju.edu.cn; Yun Wu, yunwu@zju.edu.cn
†Present address: Lin Zhang, Department of Ophthalmology and Neurobiology, University of California, Los Angeles, Los Angeles, CA, United States
‡These authors have contributed equally to this work
 Cong Gao
Cong Gao