
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci., 19 September 2023
Sec. Plant Abiotic Stress
Volume 14 - 2023 | https://doi.org/10.3389/fpls.2023.1261238
 Hengchun Cao1,2,3
Hengchun Cao1,2,3 Qiuzhen Tian1,2,3
Qiuzhen Tian1,2,3 Ming Ju1,2,3
Ming Ju1,2,3 Yinghui Duan1,2,3
Yinghui Duan1,2,3 Guiting Li1,2,3
Guiting Li1,2,3 Qin Ma1,2,3
Qin Ma1,2,3 Haiyang Zhang1,2,3*
Haiyang Zhang1,2,3* Xianmei Zhang3,4*
Xianmei Zhang3,4* Hongmei Miao1,2,3*
Hongmei Miao1,2,3*Plant U-box (PUB) proteins belong to a class of ubiquitin ligases essential in various biological processes. Sesame (Sesamum indicum L.) is an important and worldwide cultivated oilseed crop. However few studies have been conducted to explore the role of PUBs in drought tolerance in sesame. This study identified a total of 56 members of the sesame PUB family (SiPUB) genes distributed unevenly across all 13 chromosomes. Based on phylogenetic analysis, all 56 SiPUB genes were classified into six groups with various structures and motifs. Cis-acting element analysis suggested that the SiPUB genes are involved in response to various stresses including drought. Based on RNA-seq analysis and quantitative real-time PCR, we identified nine SiPUB genes with significantly different expression profiles under drought stress. The expression patterns of six SiPUB genes in root, leaf and stem tissues corroborated the reliability of the RNA-seq datasets. These findings underscore the importance of SiPUB genes in enhancing drought tolerance in sesame plants. Our study provides novel insights into the evolutionary patterns and variations of PUB genes in sesame and lays the foundation for comprehending the functional characteristics of SiPUB genes under drought-induced stress conditions.
With the escalation of global climate change, the impact of the environment on crop production is becoming increasingly severe (Vicente-Serrano et al., 2020). Drought is recognized as a significant environmental stressor, and its impact on crop production surpasses the cumulative yield losses caused by all pathogens combined (Gupta et al., 2020; Hoover et al., 2021). Drought stresses can occur at any stage of plant growth, leading to water deficit and harsh reactions. Many signaling pathways in plants are activated under drought stress, regulating the expression of different genes and generating multiple defense proteins and protective molecules. For example, abscisic acid (ABA), an essential component of plant response to abiotic stresses, including drought, activates various genes through SnPK2, triggering stomatal closure and improving water balance (Cheng et al., 2017; Lin Z. et al., 2021). In Arabidopsis thaliana, BRL3 allows plant growth and survival under drought stress by promoting the accumulation of osmoprotectant metabolites in root tissues (Fàbregas et al., 2018). Therefore, it is necessary to understand the molecular mechanism of plant responses to drought stress and develop drought-resistant crops.
The Ubiquitin-Proteasome System (UPS) is a major pathway that regulates the post-translational level of proteins and is essential in plant development and plant-environment interactions (Bing et al., 2016). The classic UPS contains several members, including ubiquitin (Ub), Ub-activating enzyme (E1), Ub-binding enzyme (E2), Ub ligase (E3), deubiquitinating enzyme (DUB), and 26S proteasome (Bhattacharyya et al., 2014). E3 can specifically recognize the ubiquitin target proteins, which are crucial in the ubiquitin pathway (Hu and Hochstrasser, 2016). E3 can be classified into four categories according to their functional domains: RING, HECT, U-box, and cullin. The U-box domain is conserved in nearly all eukaryotes with a length of ~70 amino acids. Plant U-box (PUB) proteins are central to many biological processes, such as seed germination, hormonal response, and biotic and abiotic stresses (Hu and Hochstrasser, 2016). Numerous crops involve the diverse functions of PUBs in response to various abiotic stresses, including cold, salinity, high temperature, and drought (Trujillo, 2018; Cui et al., 2023). Under normal conditions, E3 can inhibit the drought stress signaling pathways, thus eliminating negative regulators that respond to stimulation or reduction of the stress signaling pathway (Al-Saharin et al., 2022). In addition, E3 may act as positive feedback factors to enhance signaling under drought stresses and eliminate stress-induced signaling pathways when conditions return to normal, allowing the plant grow further (Liu et al., 2021).
Many studies have demonstrated the significant impact of PUBs on numerous crops subjected to drought stress. For example, by degrading leucine-rich repeat protein 1 (LRR1) and kinase 7 (KIN7), AtPUB11 negatively regulated drought tolerance (Chen et al., 2021). AtPUB22 and AtPUB23 are negative regulators of drought tolerance in A. thaliana. The functional loss of these two genes could significantly improve drought tolerance, while their overexpression would decrease it. Further experiments indicated that AtPUB22 and AtPUB23 could synergistically control drought signaling pathways, mainly because their double mutants displayed greater tolerance (Cho et al., 2008). In poplar, PalPUB79 might positively regulate ABA-dependent drought tolerance through the ubiquitination of PalWRKY77. The overexpression of PalPUB79 enhances drought tolerance, while the PalPUB79 RNAi lines display more sensitivity (Tong et al., 2021). The hot pepper CaPUB1 exerts a negative regulatory effect on drought responses in both A. thalina and Oryza sativa. The overexpression of CaPUB1 leads to the development of drought-sensitive phenotypes in plants (Cho et al., 2006; Hye Jo et al., 2016). In addition, OsPUB41 acts as a crucial negative regulator in drought tolerance in rice (Seo et al., 2021).
Sesame (Sesamum indicum L.) is one of the oldest oilseed crops within the genus Sesamum of the Pedaliaceae family (Ashri, 1998; Zhang et al., 2013). Due to its high oil (~58%) and protein content (~25%) and its excellent antioxidant and acidic properties, the sesame seed is commonly referred to as the “oilseed queen” (Bedigian and Harlan, 1986; Zhang et al., 2019). Compared to other oilseed crops, such as soybean (Glycine max), sunflower (Helianthus annuus L.), and oilseed rape (Brassica napus), sesame has higher oil content and tolerance to drought and high temperatures and can display high yields in tropical and subtropical climates (Zhang et al., 2013; Zhang et al., 2019). Even though sesame can survive in semi-arid conditions, its nutrition quality and seed yield can be affected by drought stress at the vegetative and reproductive growth stages (You et al., 2019). To our knowledge, the key role of the PUB gene family in plantlet growth and stress response has not been fully identified in sesame. In this study, we conducted a systematic characterization of SiPUB genes and a detailed analysis of their evolution and structures. Based on an RNA-seq dataset, we investigated the expression profiles of SiPUBs under drought stress and confirmed their differential expression by quantitative real-time PCR (qRT-PCR). Our study provides valuable information for further studying the evolution and function of PUB genes in sesame.
To identify potential members of the PUB gene family, this study utilized our latest sequenced sesame genome data (GenBank under the accession MBSK00000000 in BioProject no. PRJNA315784) (Miao et al., 2021). The seed file of the U-box domain (PF04564) was obtained from the Protein Families Database (Pfam) v35.0 (Finn et al., 2014). HMMER (v3.3.2) was used to search the U-box domain in the sesame protein sequences (Potter et al., 2018). Additionally, we performed homologous searching using BLAST+ (v2.8.1) based on the PUB genes in Arabidopsis (Camacho et al., 2009). Finally, we used PfamScan (v1.6) to confirm the presence of the U-box domain in the candidate genes (Li et al., 2015). Only SiPUB genes with a complete the U-box conserved domain could be considered for further analysis.
MAFFT (v7.505) was employed to perform multiple sequence alignment in this study (Katoh and Standley, 2013). The phylogenetic tree was constructed by the FastTree v2.1.11 with a bootstrap of 1000 replications using the recommended models (Price et al., 2010). The software FigTree (v1.4.4) was used to display the phylogenetic tree of SiPUB proteins (http://tree.bio.ed.ac.uk/software/Figtree/). Sequence alignments were visualized and displayed using the online LaTeX editor Overleaf (https://www.overleaf.com/). To explore the diversity of SiPUBs, we obtained the amino acid sequences of PUBs in rice and A. thaliana from previous studies (Wiborg et al., 2008; Zeng et al., 2008).
To identify the conserved motifs of SiPUBs, we utilized the Multiple Expectation Maximization for Motif Elicitation (MEME) Suite web server with parameters set to maximum motifs of 10 (Bailey et al., 2009). TBtools was applied to perform the intron-exon gene structures with the annotation files of sesame (Chen et al., 2020). The 2,000bp upstream sequence of SiPUB genes was extracted as the putative promoter sequence, and then utilized in the PlantCare database to identify cis-acting elements with default parameters (Lescot et al., 2002). The sequence length, molecular weight (MW), and isoelectric point (pI) were obtained on the ExPasy website (Duvaud et al., 2021).
The software of TBtools was used for pairwise comparisons of the genomes of sesame, rice, and A. thaliana (Chen et al., 2020). The online tool WoLF PSORT was employed to predict the subcellular localization of SiPUB proteins (Horton et al., 2007).The synteny regions and chromosome distribution were performed by MCScanX with default parameters and then used Circos to visualize these results (Krzywinski et al., 2009; Wang et al., 2012). The chromosome location map was generated using MG2C v2.1 (http://mg2c.iask.in/mg2c_v2.1/), based on the positions of SiPUBs (Chao et al., 2021).
To analyze the expression levels under drought stress, we obtained the Fragments Per Kilobase of exon model per Million mapped fragments (FPKM) matrix data of drought-tolerant genotype TEX-1 and drought-sensitive genotype VEN-1 from the NCBI database (accession number GSE148340) (Song et al., 2021). The log2 fold change (log2FC) of each treatment was calculated and applied to the R package “pheatmap” to generate the expressed heatmap.
To investigate the 3D structures of SiPUBs, we employed the RCSB Protein Data Bank (RCSB PDB) through comparative modeling (Rose et al., 2017). PyMOL (v2.5.4) was used to visualize and analyze 3D structures of the proteins (Yuan et al., 2017). Furthermore, the online resource Search Tool for the Retrieval of Interacting Genes (STRING) was used to predict protein-protein interactions (PPIs) with an interaction score of 0.4 or higher (Szklarczyk et al., 2021). This threshold ensures a high level of confidence in the connections between nodes and lines. Cytoscape (v3.9.1) software was utilized for visualization and analysis of the PPI networks (Saito et al., 2012).
The sesame seeds of drought-sensitive YM1 (Henan No. 1) and YM 5 (Yuzhi No. 3), and drought-tolerant YM19 (Zhengheizhi No.1) and YM20 (Zhengheizhi No.4) were collected from our experimental field (the sesame germplasm orchard of Henan Sesame Research Center at Yuanyang in Henan province). Then, the sesame seeds were surface-sterilized in 3% sodium hypochlorite solution for 7 min, followed by four times washing with sterile water. Twelve seeds were sown in each plot with an equal weight of sterilized vermiculite, and ten seedlings were selected per pot during the stage when the first pair of true leaves emerged. The seedlings were cultivated in an HP1500GS-B type artificial climate incubator. The cultivation conditions were set as follows: a photoperiod of 15h light and 9h darkness, temperature at 28°C, relative humidity of 70%, and a light intensity of 20,000 lux. Both the control group and the treatment group were watered with a quantitative nutrient solution in each pot. The seedlings were cultivated until the stage of two pairs of true leaves. The treatment group was subjected to drought stress treatment without watering, while the control group was maintained regular watering conditions. Samples from both the control and treatment groups were collected at 8, 9, 10, and 12 days.
The RNA extraction from three tissues (root, stem, and leaves) of control and treatment was performed using the Plant Total RNA Isolation Kit Plus (FOREGENE, China). Subsequently, the PrimeScript™ RT reagent kit (Takara Bio, China) was utilized to reverse transcribe the extracted RNA into cDNA. For qRT-PCR analysis, the Roche LightCycler® 480 II instrument (Roche, Mannheim, Germany) was employed with LightCycler® SYBR GREEN I Master Mix kit (Roche, China). We designed 7 pairs of specific primers (Supplementary Table S1) using Primer6.0 software. The relative expression levels of SiPUBs were estimated using the 2−ΔΔCT method, with the sesame β-tubulin gene (Sindi_2728600) serving as the internal reference. These analyses were carried out in three independent biological replicates and three technical replicates of each biological replicate.
In this study, 56 PUB genes with their complete U-box domain were obtained (Supplementary Table S2). These PUB genes were renamed SiPUB1 to SiPUB56 based on their location orders on the 13 sesame chromosomes. To characterize these SiPUB genes, we analyzed the length of their open reading frame (ORF) and the expected protein sequence, molecular weight (MW), isoelectric points (pI), and subcellular and chromosomal location (Supplementary Table S2). The CDS length of SiPUB genes varied from 843bp (SiPUB12) to 4,500bp (SiPUB52), with an average of 1,817bp. SiPUB52 is the longest U-box protein in sesame and comprises 1,499 amino acids, whereas SiPUB12 is the shortest, with 280 amino acids (Supplementary Table S2). MW of the 56 proteins ranged from 32.20 to 167.04 kDa, with an average of 67.06 kDa. The pI value ranged from 5.21 (SiPUB28) to 8.91 (SiPUB18). Based on their pI, 30 PUBs were classified as acidic proteins, and 26 as basic proteins. Additionally, 42 SiPUBs were grouped as hydrophilic (GRAVY <0) and 14 as hydrophobic (GRAVY>0). The most hydrophilic protein was SiPUB43, and the most hydrophobic was SiPUB6. The subcellular location of SiPUBs was predicted by WoLF PSORT, indicating that most PUB proteins were located on the nuclear (20), cytoplasm (16), chloroplast (12), and plasma membrane (4), except three located in the mitochondrion and one located in the endoplasmic reticulum (Supplementary Table S2).
To verify the various evolution characters of the PUB gene family, 192 PUB proteins (56 from sesame, 76 from rice, and 60 from A. thaliana) (Supplementary Table S3) were used to construct a phylogenetic tree (Figure 1). The phylogenetic analysis indicated that these PUB proteins were divided into five subgroups. Ten SiPUBs belong to the same subgroup as Arabidopsis and rice U-box proteins (Group I), with possible similar functions. Group II was the largest, with 55 PUB genes, whereas Group III had the smallest number, with 11 PUB genes. We found that sesame and Arabidopsis PUB genes exhibited a relatively closer relationship compared to rice. For example, Group IV comprised 19 PUB genes, among which four sesame (SiPUB1, SiPUB20, SiPUB5, and SiPUB10) and four Arabidopsis (AtPUB40, AtPUB38, AtPUB39, and AtPUB41) genes were clustered together, whereas one sesame PUB gene (SiPUB52) was clustered with ten rice PUB genes (OsPUB76, OsPUB73, OsPUB26, OsPUB60, OsPUB71, OsPUB77, OsPUB17, OsPUB18, OsPUB19, and OsPUB20).
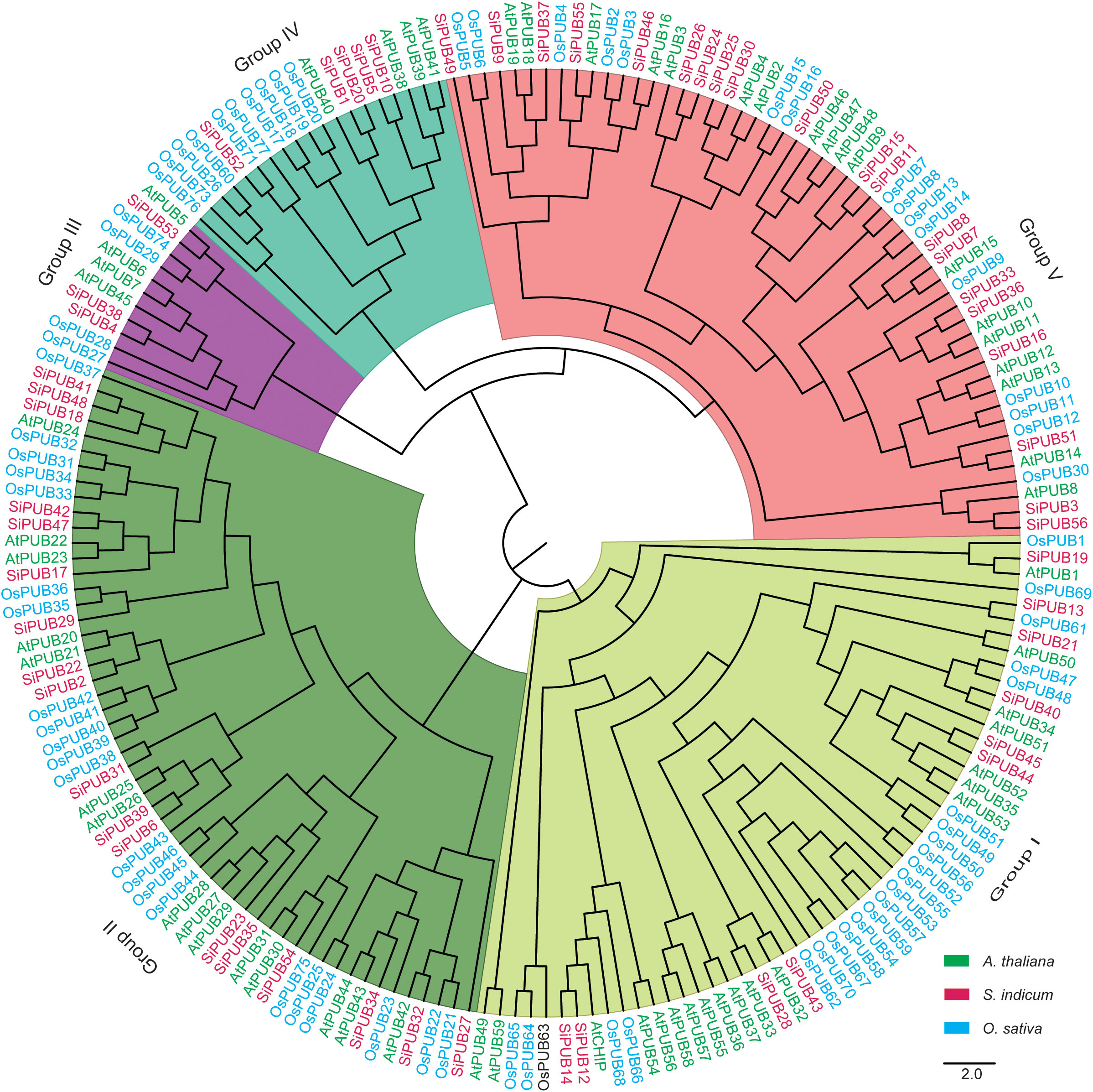
Figure 1 Phylogenetic tree of PUB gene family. The phylogenetic tree was constructed using the maximum likelihood (ML) method with 1,000 bootstraps. All of 192 PUB proteins are clustered into five distinct subgroups, namely Group I-V clades in different colors. U-box proteins in A. thaliana, S. indicum, and O. sativa are labeled in green, pink, and blue, respectively.
To better understand the phylogenetic relationships of PUB genes in sesame, we inferred a phylogenetic tree using FastTree with the Maximum Likelihood (ML) method (Minh et al., 2020). The topology of the phylogenetic tree was divided into six groups (Figure 2A), among which Group 6 was the largest with 15 genes, followed by Groups 1 and 3 with 10 and 11 genes respectively. Group 4 was the smallest, with four genes. Among these SiPUB proteins, SiPUB52 displayed an apparent difference from other SiPUBs, and was not clustered into any groups in the phylogenetic tree (Supplementary Table S2). The findings are consistent with the phylogenetic analysis of PUB genes in the three abovementioned species, as depicted in Figure 1, further validating the reliability of the SiPUB phylogenetic analysis performed in this study.
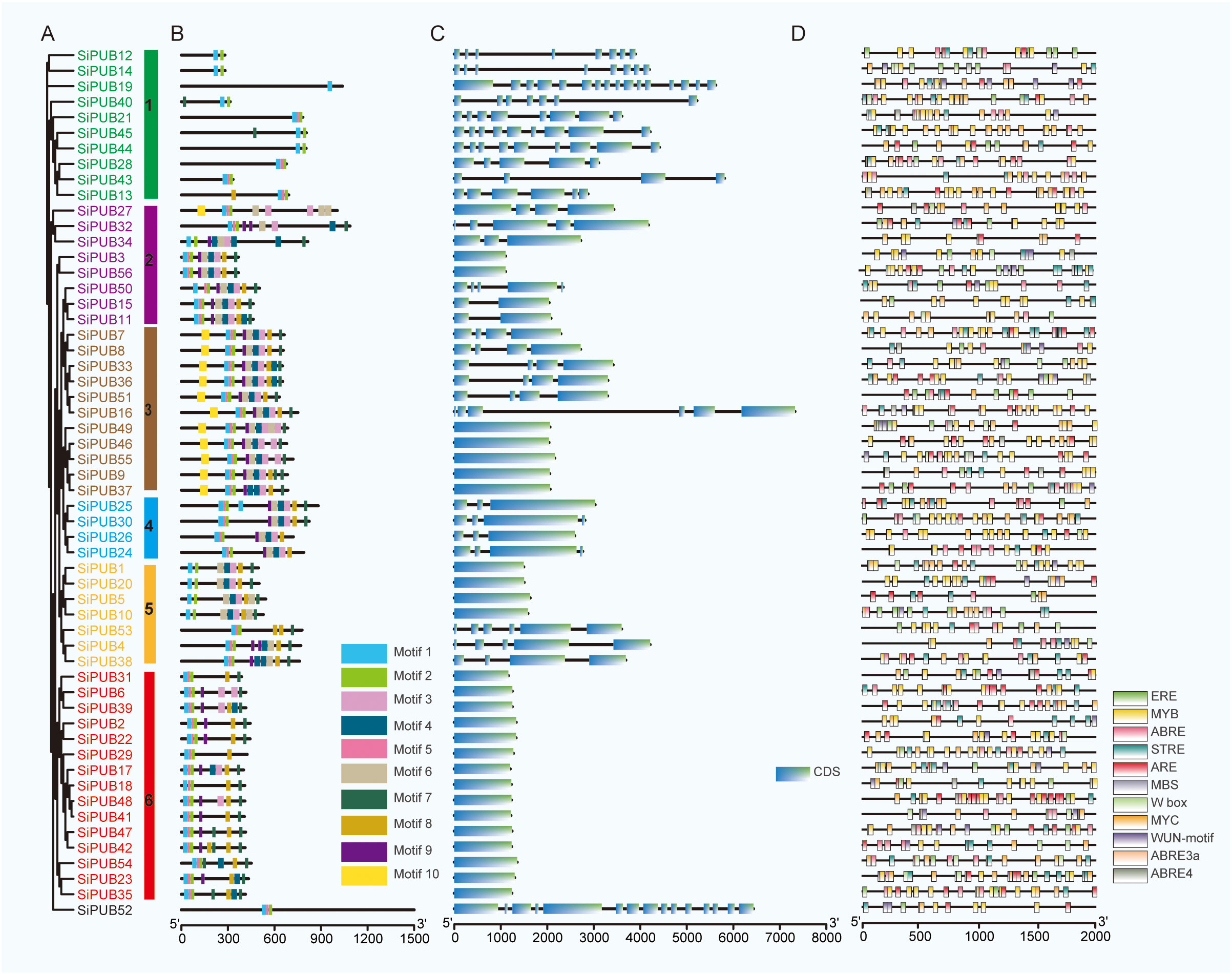
Figure 2 Structural comparison and phylogenetic tree of SiPUBs. (A) Phylogenetic tree of the 56 SiPUBs generated using the maximum likelihood (ML) method. Six distinct groups are shown in different colored squares with gene names. (B) Distribution of conserved motifs in SiPUB proteins. Ten different motifs are predicted using MEME software. The scale bar on the right indicates the distance. (C) Structure comparison of SiPUB genes. Intron and exon are shown in black line and blue rectangle, respectively. (D) Distribution of cis-acting elements associated with drought stress in the putative promoter of SiPUB genes. Eleven elements in the putative promoters of the SiPUB gene family are shown. The color scale on the right represents the distance between cis-acting elements.
To further investigate the relationships between the 56 SiPUB proteins containing a U-box domain (Supplementary Table S4) in the phylogenetic tree, we evaluated the conserved motifs using the program MEME. Ten motifs were identified in 56 SiPUB genes, named Motifs 1 to 10 (Figure 2B), among which Motifs 1, 2, 5, and 7 were encoded by most SiPUB genes and reflected the high motif conservation in SiPUB proteins. Motifs 1 and 2 were determined as conserved U-box sequences necessary to maintain the U-box structure and support the ubiquitin linkage activity. Other domains, such as ARM, PK_Tyr_Ser-Thr, Pkinase, ANAPC3, Ufd2P_core, and WD40, were also identified in sesame U-box proteins. Motif 6 could be a part of the ARM conserved domain, with the U-box-ARM subfamily being the most common type in the U-box family (Supplementary Table S4). Furthermore, all the genes in the same phylogenetic group encoded similar conserved motifs, indicating possible similar functions. For example, within Group 3, 11 genes encoded ten distinct motifs (Motifs 1 to 10). These motifs were conserved in quantity and sequential arrangements across the encoded proteins. Many highly conserved amino acids were also detected in the sesame U-box domain (Supplementary Figure S1), such as the hydrophobic amino acids of proline (P), phenylalanine (F), and isoleucine (I), the acidic amino acid aspartic acid (D), and the basic amino acid basic amino acid arginine (R), cysteine (C), and serine (S). These amino acids likely have a crucial function in forming ionic or hydrogen bonds and stabilizing the U-box domain.
To understand the composition and function of U-box domains, we analyzed the gene structure of the 56 SiPUB genes using conserved sequences and exon/intron positions (Figure 2C). The number of exons varied from 1 to 16 in SiPUBs, suggesting a complex RNA splicing process. Genes from the same group had similar exon/intron structures. Group 6 PUB genes had the fewest exons (1), while Group 1 genes had the most (average 8). The gene structure and conserved motif analysis results were consistent with the SiPUB phylogenetic tree, confirming the accuracy of the phylogenetic tree classification.
According to the location in the sesame genome, 56 PUB genes were unevenly mapped across 13 chromosomes (Figure 3A). SiChr.3 had 13 PUB genes, SiChrs.1 and 5 had seven SiPUBs each, and only one PUB gene was located on SiChrs.7 and 12. The transcription direction of 28 PUBs was the same as the sequence provided by the genome, and the other 28 were reversely transcribed. Next, we performed a homolog analysis of SiPUB using MCScanX software. Thirty genes were assigned to whole genome duplication (WGD) and segmental duplication, while 23 were derived from dispersed duplication blocks (Figure 3B). In addition, three genes were assigned to tandem duplication blocks (Supplementary Table S5). No genes were derived from proximal and singleton duplication blocks. Therefore, WGD and segmental duplication events might expand the PUB gene family in sesame. Most duplicated genes might be silenced over time, but a few purified selected genes still retain. In the sesame PUB gene family, 251 homologous gene pairs were identified, including 56 homologous genes (Figure 3B). Furthermore, we detected a collinear relationship between sesame, Arabidopsis, and rice (Figure 3C). As a result, 45 orthologous gene pairs were identified between SiPUBs and AtPUBs, of which 8 AtPUBs had two or three orthologous copies in the sesame genome. Meanwhile, 25 orthologous gene pairs were identified between SiPUBs and OsPUBs, fewer than those between sesame and Arabidopsis, suggesting that SiPUB genes have a closer relationship with Arabidopsis PUBs than rice ones. Identifying these orthologous genes with potential similar functions can promote the study of SiPUBs in the future.
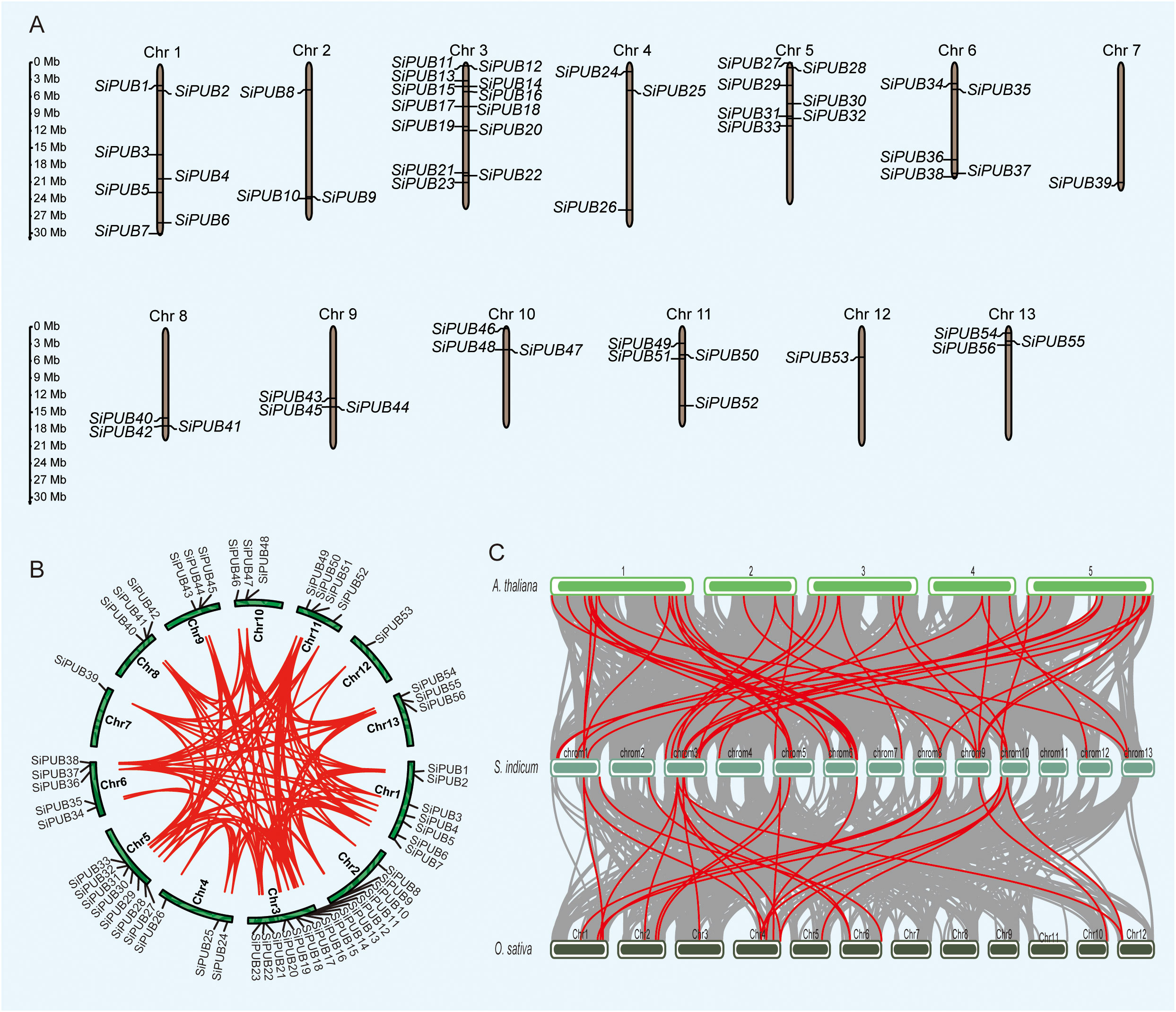
Figure 3 Distribution and synteny results of SiPUB genes in sesame genome. (A) Distribution and chromosomal localization of SiPUB genes. Green columns represent sesame chromosomes. (B) The synteny analysis and distribution pattern of SiPUB genes. Red lines indicate synteny gene pairs within the PUB gene family in sesame. (C) Collinearity analysis of the genomes of A. thaliana, S. indicum, and O. sativa. Gray lines represent the overall co-collinearity of all genes between two species. Red lines represent the collinearity specific within the PUB gene family.
Cis-acting elements play a crucial role in gene regulation. To explore the transcriptional regulation of SiPUBs, we predicted the cis-acting elements present within the 2,000bp region upstream of the coding sequence using the Plant-CARE database (Supplementary Table S6) (Figure 2D). Our analysis revealed a total of 8,819 occurrences spanning 95 distinct types of cis-acting elements within the promoter regions of the 56 SiPUBs. The count of cis-acting elements exhibited variability, ranging from 95 (SiPUB24) to 235 (SiPUB37), with an average of 157. These elements can be categorized into seven distinct functional groups: hormone-responsive elements, elements associated with plant growth and development, stress-responsive elements, light-responsive elements, site-binding elements, promoter-related elements, and other functional elements. The results revealed that most SiPUB promoters contained a CAAT or TATA box, which were commonly found cis-acting elements in the promoter and enhancer regions. The specific elements associated with light response, hormone response, promoter-related functions, and developmental processes widely spread among SiPUBs, suggesting a potential role for SiPUBs in various biological activities. Moreover, we identified a diverse set of 16 cis-acting elements associated with environmental stress responses. For instance, 53 SiPUBs contained the MYB element, which is involved in environmental adaptation and secondary metabolic regulation. Forty-six SiPUBs harbored the AAGAA-motif of the ABA elements, while 54 contained the ABA-responsive element (ARE) and as-1, which are associated with various stresses, including hypoxia, salicylic acid, cold, drought, and pathogen attacks. Notably, we detected that all 56 SiPUB genes contained 15 intriguing cis-acting elements predominantly associated with drought and other environmental stresses (Supplementary Table S6; Figure 2D). These results indicate that SiPUB genes are specifically involved in functions related to environmental stresses.
To further explore the function of SiPUB genes in response to drought stress, we analyzed the expression profiles of all 56 SiPUBs based on the four groups of RNA-Seq transcriptome data on sesame available on the NCBI database. Thirty-four (60.7%) SiPUBs displayed differential expression under drought stress. Next, we analyzed the expression heatmaps of the 34 SiPUB genes with the log2FC values (Figure 4). The differential multiple matrices of these SiPUBs are displayed in Supplementary Table S7. Nine genes were differentially expressed with a |log2FC| > 1 under four drought treatments (Supplementary Figure S2). The results suggested that the SiPUB genes are widely involved in drought tolerance in sesame.
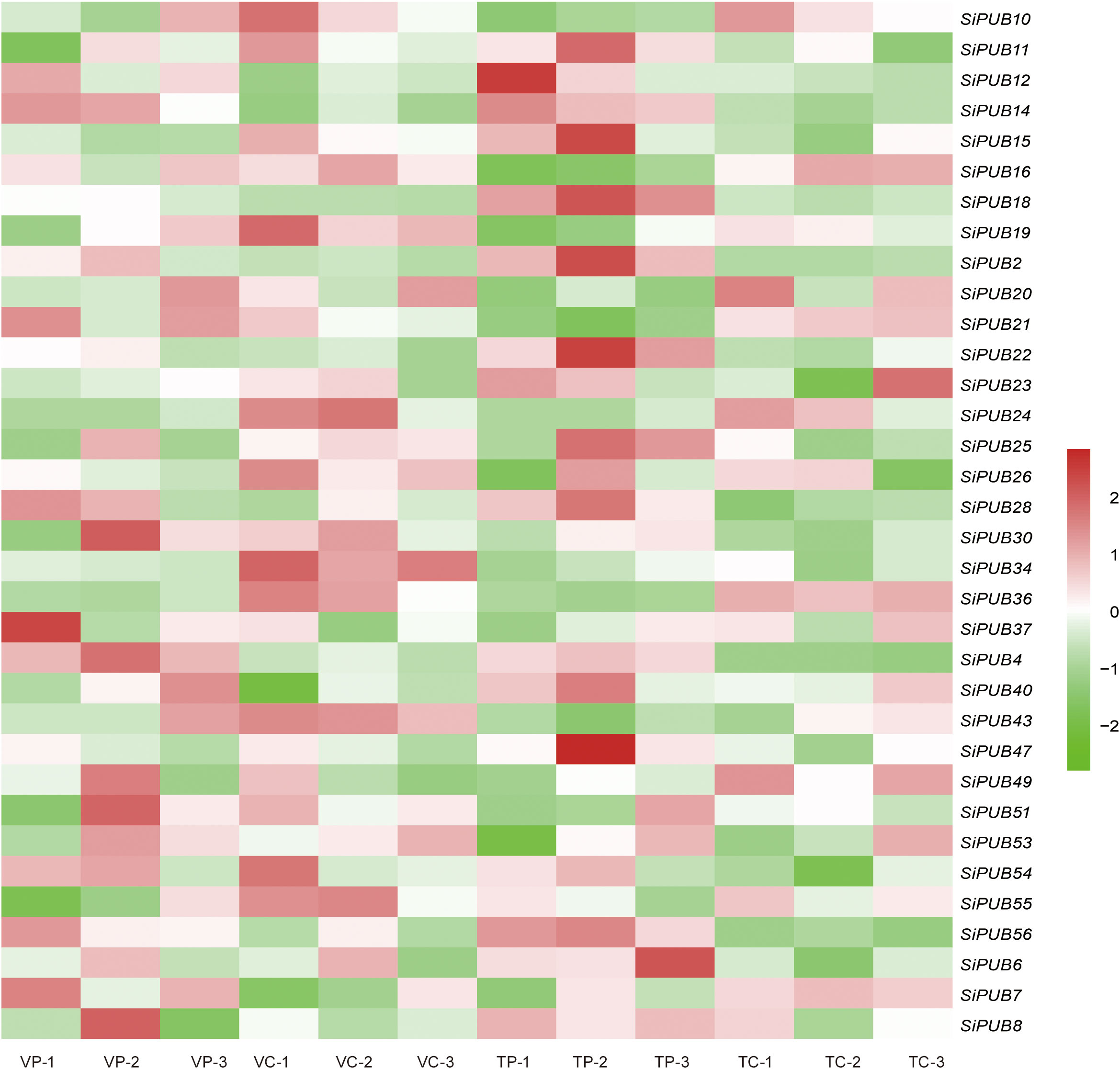
Figure 4 Expression analysis of the SiPUB genes under drought stress. Heatmaps are generated using 4 groups of public RNA-seq data. VP and TP indicate the treatments of drought-tolerant genotype TEX-1 (T) and the drought-sensitive genotype VEN-1 (V) under drought stress. VC and TC indicate the controls of drought-tolerant genotype TEX-1 (T) and the drought-sensitive genotype VEN-1 (V) under normal environment. The color code represents the log2-transformed FPKM value. High and low expression levels are shown in dark red and bright green, respectively.
To validate the reliability of the transcriptome sequence analysis, we selected 6 SiPUB genes (SiPUB2, SiPUB14, SiPUB17, SiPUB18, SiPUB22, and SiPUB47) based on their distinct expression patterns observed in the transcriptome data and performed qRT-PCR validation.The expression levels of these six genes were assayed across three different tissues in two drought- tolerant (YM19 and YM20) and two drought-sensitive sesame accessions (YM1 and YM5) (Figure 5). All six SiPUB genes displayed diverse expression patterns across the various sesame tissues. In the root tissue (Figure 5A), the expression level of all 6 SiPUB genes increased during the 8-day drought exposure in drought-tolerant and drought-sensitive varieties. Among those, SiPUB47 exhibited a notably higher expression level and surpassed the other five SiPUB genes by a significant margin (four to eight times). In the stem tissue (Figure 5B), the expression level of SiPUB2 and SiPUB22 was lower in the drought-sensitive varieties than in the drought-tolerant plants. The two genes showed a general down-regulation trend. Conversely, SiPUB14 exhibited a consistent upregulation in response to drought stress across all samples. SiPUB18 displayed the significantly lowest expression level compared to the other five SiPUB genes. In leaf tissues (Figure 5C), SiPUB47 and SiPUB14 displayed the significantly highest expression level compared to the other four SiPUBs. On the other hand, SiPUB17 exhibited the lowest expression, with levels more than 10 times lower than those of the other SiPUB genes. The qRT-PCR results of the 6 SiPUB genes indicated different expression profiles and matched the expression patterns observed in the abovementioned transcriptomic analyses. The results suggest that the SiPUB gene family potentially functions in different tissues and contributes to drought tolerance capabilities in different cultivated sesame varieties.
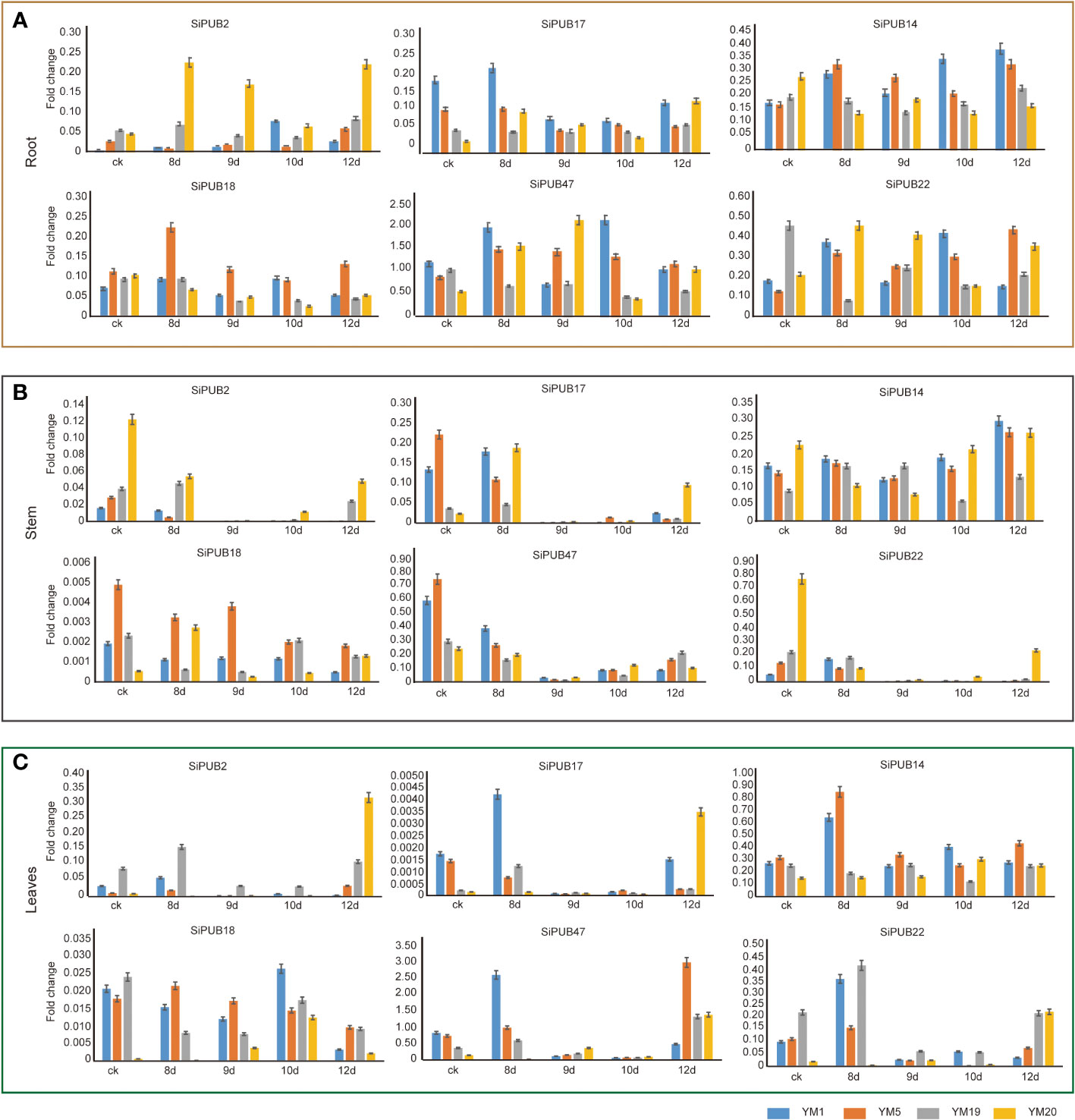
Figure 5 Validation of 6 SiPUB genes by qRT-PCR in different tissues of four sesame accessions under drought treatment. The root (A), stem (B), and leaves (C) of two drought- tolerant sesame accessions (YM19 and YM20) and two drought-sensitive accessions (YM1 and YM5) are treated under 8d, 9d, 10d, and 12d for qRT-PCR validation. X-axis represents different treat time. Y-axis represents the relative expression level of each PUB gene. YM1, YM5, YM19, and YM20 are shown in blue, orange, grey, and yellow column, respectively.
To further differentiate the higher structural characteristics of SiPUB proteins, we performed an RCSB PDB analysis and successfully determined a set of four distinct structural types for the 56 SiPUB proteins (Figure 6A). A significant majority of SiPUB proteins (95%, 53 out of 56) were categorized under the PDB IDs of 1T1H and 7C96. SiPUB12 and SiPUB14 showed a 2C2L structure, while SiPUB19 was classified under the 1WGM structure (Supplementary Table S8). All of these four structures exhibited common structures with a central α-helix (α1), a C-terminal helix (α2), a small antiparallel β-sheet (β1 and β2), and two distinct loops (loop1 and loop2) (Figure 6A-a).
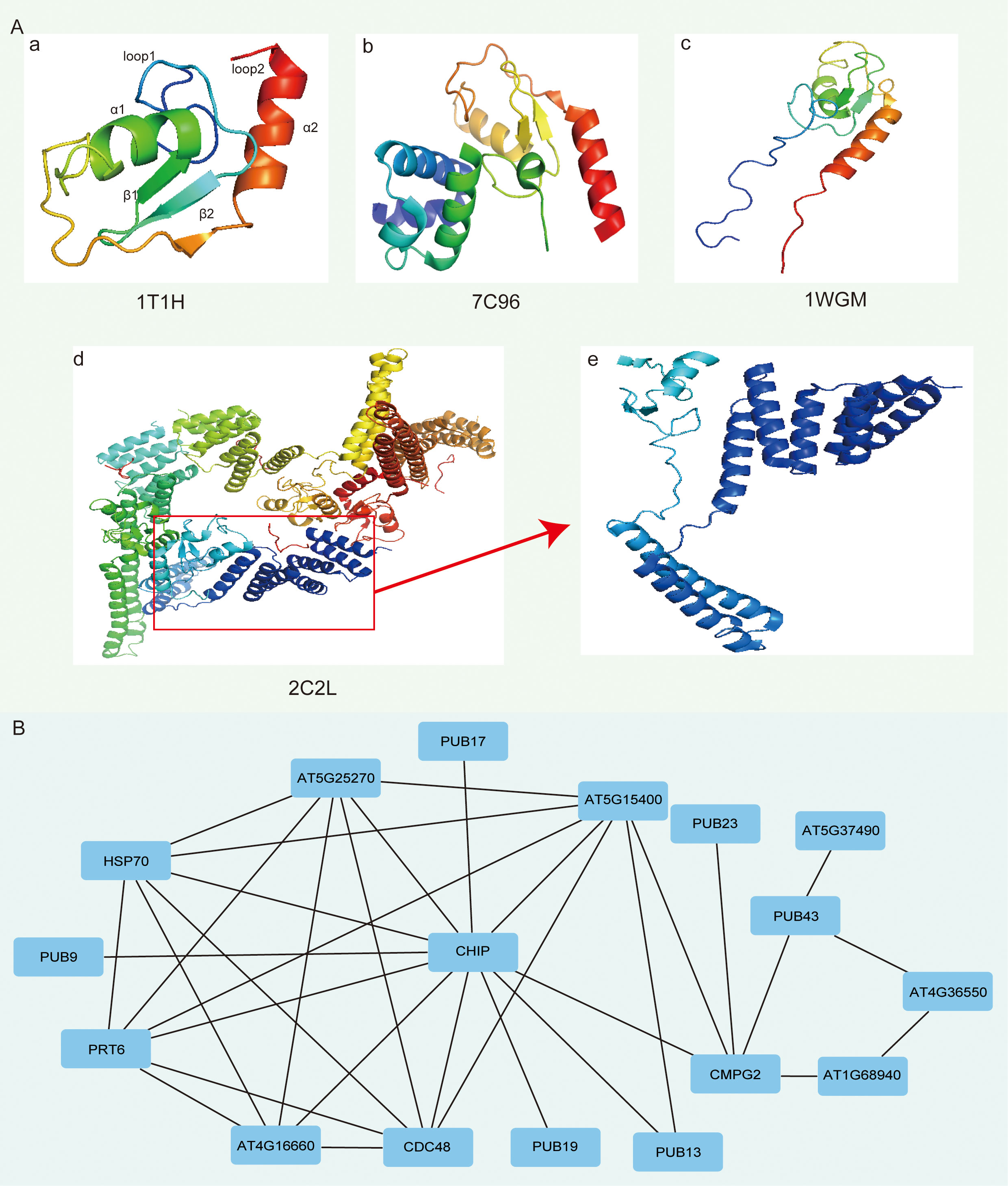
Figure 6 Three-dimensional (3D) structure and protein-protein interaction (PPI) networks of SiPUB proteins. (A) Four structure models of SiPUBs. 1T1H (a), 7C96 (b), 1WGM (c), and 2C2L (d) are determined using RCSC PDB. (B) PPI analysis of SiPUBs with homologous proteins in Arabidopsis (AtPUBs). The homologous annotation of each protein between SiPUB and AtPUB is conducted using the STRING database.
To explore a potential interaction between the proteins, we conducted an interaction network analysis of the SiPUB proteins by comparing them with their homologs in A. thaliana (AtPUBs). Of the 60 AtPUBs examined, 34 (57%) exhibited homology with the 56 SiPUBs (Supplementary Table S9). Based on the STRING database, the PUB proteins displayed extensive connections and interactions with 11 specific proteins, such as HSP70, CMPG2, and other stressed-related proteins (Figure 6B). The intricate protein networks of protein-protein interactions enhance our understanding of molecular mechanisms and biological processes involving SiPUBs.
The PUB gene family plays a significant role in response to various abiotic stresses and has been identified in numerous plant species. Extensive research suggests that PUB genes participate in drought tolerance in many plants, such rice, Arabidopsis, and wheat (Qin et al., 2020; Seo et al., 2021). Sesame is an ancient oilseed crop with high tolerance to drought and high temperature. Identifying the function and expression patterns of PUB genes in response to drought will supply a key aid for genetics and breeding research in sesame and other crops. In this study, we screened SiPUB genes from the sesame reference genome (var. Yuzhi 11) and validated the potential function of SiPUB genes in drought stress responses in drought-tolerant and drought-susceptible accessions for the first time.
We screened all sesame PUB genes based on combined results of HMM and BLAST searches. Fifty-eight candidate SiPUB genes were preliminarily identified in the whole genome. We used the PfamScan tool (v1.6) to verify the accuracy of the candidate PUB genes. After the filtering process, two candidate genes were excluded from consideration due to the absence of a U-box domain or repeat sequences. As a result, a total of 56 non-redundant SiPUB genes were ultimately obtained. All the SiPUB genes displayed a complete U-box domains. However, the number of PUB genes in sesame (56) is relatively lower than in rice (76) (Zeng et al., 2008), soybean (125) (Wang et al., 2015), barley (67) (Ryu et al., 2019), cotton (93~208) (Lu et al., 2020), and banana (91) (Hu et al., 2018). The variation in the genome size with evolution might attribute to the variation in the number of PUB genes in different species.
In sesame, 56 SiPUBs exhibited an uneven distribution across the 13 chromosomes and could be categorized into six groups based on multi-alignments and phylogenetic analysis (Figures 2A, 3A). Phylogenetic analysis including other plant species indicated that 192 PUB proteins from three species (56 from sesame, 76 from rice, and 60 from Arabidopsis) were classified into five groups (I-V) (Figure 1). All five groups displayed substantial similarities due to the common presence of the core U-box domain. The grouping pattern in the phylogenetic tree exhibited a high resemblance to other species, such as Chinese cabbage, cotton, and banana (Wang et al., 2015; Hu et al., 2018; Lu et al., 2020). Notably, sesame and Arabidopsis PUB genes were clustered together in a subclade. Additionally, we observed 45 orthologous gene pairs between SiPUBs and AtPUBs, surpassing the number between SiPUBs and OsPUBs (23) (Figure 3C). Our findings suggest a closer evolutionary relationship between sesame and Arabidopsis than with rice. SiPUBs might possess similar functions to their orthologous counterparts in Arabidopsis (Cho et al., 2008; Chen et al., 2021).
Notably, SiPUB genes within the same group displayed similar gene structures, whereas structural variations were observed among different groups (Figures 2B, C). SiPUBs possess a conserved U-box domain, which is typically associated with ARM, anaphase-promoting complex subunit 3 (ANAPC3), and WD40 repeats (Figure 2B). Notably, many biologically functional PUB proteins have been characterized as U-box proteins with ARM repeats (Wang et al., 2020; Wang et al., 2021). Among the identified SiPUB genes, 24 genes were found to exclusively contain the U-box domain, while 22 genes possessed the U-box domain and ARM repeats (Supplementary Table S4). ARM repeats primarily mediate interaction with substrates, indicating that ubiquitination occurs in response to interactions (Shu and Yang, 2017; Al-Saharin et al., 2022). Based on the diverse gene structures identified, we inferred that SiPUB genes in sesame likely have various biological functions.
In addition, diverse domains were present among the SiPUB genes within the same group (Figure 2B). Group 1 comprises ten genes, most of which exhibit the presence of the U-box domain and other domains, such as ANAPC3 repeats, protein tyrosine and serine/threonine kinase (PK_Tyr_Ser-Thr), protein kinase (Pkinase), and ubiquitin elongating factor core (Ufd2P_core) family. ANAPC3 belongs to the anaphase-promoting complex subunits, a multiprotein subunit E3 ubiquitin ligase complex that controls chromosome segregation and mitotic exit in eukaryotes (Peters, 2002; Vodermaier et al., 2003; Peters, 2006). The PK_Tyr_Ser-Thr and Pkinase domains are part of the receptor-like kinase (RLK) gene families, which have essential roles in resistance signaling pathways against various abiotic stresses. Previous studies have suggested that these domains could have stress-responsive functions under drought or Fusarium graminearum treatments in wheat (Yan et al., 2023). SiPUB52, which possesses a WD40 repeat in its structure, was found to be distinct from other SiPUBs and did not cluster into any of the identified groups in the species tree (Figure 2A). A LIN gene containing U-box and WD40 repeat domains is central in the nodulation process of rhizobia and controls the early infection of rhizobia in leguminous plants (Kiss et al., 2009). Similar observations have been reported in other crops, suggesting that the diverse functions of PUBs may be associated with different motifs (Lu et al., 2020; Wang et al., 2021). Therefore, SiPUB52 might also conduct a similar function in sesame and potentially contribute to disease resistance.
Regulatory elements for a gene are typically found in its upstream region, with their functional impact generally extending within a range of 2,000 base pairs (Doane and Elemento, 2017). Cis-acting elements within gene promoter regions play a vital role in the regulation of gene expression across various crops, particularly in response to multiple abiotic stresses and plant growth (Zhang et al., 2020; Riaz et al., 2021; Rehman et al., 2022; Cui et al., 2023). The cis-acting elements of the putative promoters of PUB genes are involved in many key biological processes, including stress responses, hormonal regulation, and crop growth and development (Wang et al., 2021; Chen et al., 2022). In our study, all 56 SiPUB genes contained stress-related elements in their putative promoter regions (Figure 2D). The number of stress-related element types in these SiPUB genes varied from 3 (SiPUB15) to 11 (SiPUB16, SiPUB18, and SiPUB25), suggesting a role for PUB genes in stress tolerance in sesame. Among these cis-acting elements, 18 involved in environmental stress responses, such as ARE, dehydration-responsive element (DRE), and MYB, were included (Figure 2D). ARE is a crucial regulator of plant drought stress response and could enhance crops’ drought tolerance by regulating the ARE-dependent ABA signaling (Fujita et al., 2005; Roca Paixão et al., 2019). DRE is an important transcription factor in regulating responses to abiotic stress in many plants, such as Brassica napus and rice (Zhao et al., 2006; Khan et al., 2017; Huang et al., 2018). The transgenic expression of LcDREB3a from Leymus chinensis has been reported to enhance drought and salt tolerance in Arabidopsis (Xianjun et al., 2011). DREB2 has also been proven to improve the drought tolerance of Broussonetia papyrifera (Sun et al., 2014). The MYB family of transcription factors are widely involved in many plants, such as Pyrus betulaefolia, barley, and wheat, in their adaptive responses to drought stress (Bi et al., 2016; Li et al., 2017; Alexander et al., 2019). Further evidence indicated that MYBs act as mediators of ABA action in response to drought stress and could be a valuable target for stress tolerance engineering in crop improvement (Baldoni et al., 2015; Xu et al., 2019).
Our study detected that most SiPUB genes (53 out of 56) contained MYB elements. Moreover, several other stress-responsive cis-regulatory elements, including W-box, MBS, TC-rich repeats, as-1 elements, and WUN-motif, were also present in the SiPUB genes. These findings imply that SiPUB genes could play a crucial role in various mechanisms related to abiotic stress tolerance. Therefore, they can offer promising prospects for future breeding endeavors on improving abiotic stress tolerance in sesame.
Based on public RNA-seq datasets, we identified nine SiPUB genes differentially regulated under drought stress conditions and exhibiting a potential function in response to drought (Figures 4, S2). In rice, OsPUB67 is positively correlated with drought tolerance (Qin et al., 2020). We found that SiPUB28 and SiPUB43 were grouped with 11 OsPUB and 2 AtPUB genes (Group I in Figure 1) and presented a similar potential to OsPUB67 in drought tolerance. Similarly, in barley, several HvPUBs have been identified as negative or positive regulators of drought response (Ryu et al., 2019). These findings reflect the similar functions of PUB genes in drought stress responses across various plant species.
In Phyllostachys edulis, PePUB60 and PePUB120 are upregulated under drought stress conditions (Zhou et al., 2021). In Arabidopsis, AtPUB46 is a positive regulator of drought response, as overexpression of this gene resulted in increased tolerance, while knockout mutants exhibited hypersensitivity to drought (Adler et al., 2018). In this article, we observed that three SiPUB genes (SiPUB11, SiPUB15, and SiPUB50) clustered into one branch with four AtPUBs (AtPUB9, AtPUB46, AtPUB47, and AtPUB49) and displayed roles possibly similar to AtPUB46 in drought tolerance. These studies suggest a central role for SiPUB genes in drought stress adaptation in sesame (Wang et al., 2021; Chen et al., 2022).
To explore the functions of PUB genes in sesame, we investigated the expression levels of six SiPUB genes in three different tissues under different drought treatments using qRT-PCR (Figure 5). The results indicated that five SiPUB genes (SiPUB2, SiPUB17, SiPUB18, SiPUB47, and SiPUB22) exhibited higher expression levels in sesame roots (Figure 5A) than in the stem (Figure 5B) and leaf tissues (Figure 5C). In Arabidopsis, mutations in AtPUB22 and AtPUB23 enhanced drought tolerance compared to wild-type plants (Cho et al., 2008). In our analysis, SiPUB17, SiPUB42, and SiPUB47, which were clustered together with AtPUB22 and AtPUB23 in Group II (Figure 1), exhibited more than 50% homology at the amino acid level to AtPUB22 and AtPUB23. AtPUB17 is a positive regulator of plant defense and stress signaling responses, whereas the closely related rice orthologues OsPUB2 and OsPUB3 act as positive regulators, specifically in temperature stress responses (Yang et al., 2006; Byun et al., 2017).
In addition, based on our phylogenetic analysis, two SiPUB genes (SiPUB46 and SiPUB55) were clustered together with the previously mentioned three genes in group V (Figure 1). Interestingly, we observed that SiPUB55 was downregulated in the drought-sensitive variety VEN-1 under drought stress, but was upregulated in the drought-tolerant variety TEX-1 (Figure 4). We infer that SiPUB55 might potentially serve as a negative regulator in response to drought stress in sesame. Moreover, SiPUB14 displayed the highest expression in leaves, more than twice times as high as in the root and stem tissues (Figure 5). This strong concordance between the qRT-PCR results and transcriptome analyses provides compelling evidence for the crucial role of SiPUB genes in drought stress regulation in sesame.
In this article, we analyzed the 3D structure of PUBs to comprehend the sequence patterns, functions, binding sites, and interactions of candidate proteins with other targets. These models provide valuable insights into the structural characteristics and potential functions of the SiPUB proteins in sesame. According to the RCSC PDB database, four comparative homology models for 56 SiPUB proteins were available. The significant sequence identity ranged from 31% to 93% (Figure 6A; Supplementary Table S8). All four models shared common structural features of PUBs, including two helices (α1 and α2), two small antiparallel β-sheets (β1 and β2), and two distinct loops (loop1 and loop2) (Figure 6A). Most SiPUB proteins (95%) were associated with PDB IDs 1T1H and 7C96. These structures are also present in AtPUB14 and GmPUB13 (Andersen et al., 2004; Lin Y. et al., 2021). Moreover, two PUB proteins (SiPUB12 and SiPUB14) corresponded to the part residues of 2C2L, while only SiPUB19 matched the PDB ID 1WGM (Figure 6A(c)). The structural similarity among these proteins suggests the conserved overall fold and potential function. The screened network of SiPUB putative interacting proteins in sesame and Arabidopsis on the STRING database indicated a high conservation of PUB proteins (Figure 6B). As the confidence score of these functional proteins was set at 0.4, the connections between the nodes and lines had a high degree of credibility. For the extensively connected network between PUBs and specific proteins, AtCHIP (homologs of SiPUB12/14) interacted with a key component and connected with 11 other proteins. AtCHIP positively regulates the homeostasis of caseinolytic peptidase (Clp) proteolytic subunits, thereby maximizing the production of functional chloroplasts (Wei et al., 2015). In addition, AT5G25270, PRT6, and CDC48 are all ubiquitin-related proteins, indicating that PUB genes have broad connections with other ubiquitin genes and are central in the ubiquitination process. In the future, we shall perform extensive analysis of the above key SiPUB genes involved in the interacting protein network and decipher the regulation patterns of PUB genes in response to drought stress in sesame and other crops.
We identified 56 SiPUB members in the sesame genome and comprehensively analyzed their distribution, phylogenetic evolution, structure, and expression profiles. All 56 SiPUB genes were classified into six distinct groups. U-box motifs and 18 key cis-acting elements were detected in putative promoters of SiPUB genes. Expression profile comparison analysis of SiPUB genes using transcriptome data and qRT-PCR in drought-tolerant and drought-susceptible accessions validated the potential function of those genes in response to drought. These findings contribute to a comprehensive understanding of the regulatory mechanism of PUB genes in drought stress in sesame.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
The sesame varieties Henan No. 1, Yuzhi No. 3, Zhengheizhi No.1, and Zhengheizhi No.4 used in this study were planted and grown in Yuanyang, Henan Province, China. All the studies are not involved in ethics problems.
HC: Data curation, Software, Writing – original draft, Funding acquisition, Investigation, Methodology, Visualization. QT: Data curation, Validation, Writing – review & editing. MJ: Data curation, Investigation, Writing – review & editing. YD: Investigation, Writing – review & editing. GL: Data curation, Writing – review & editing. QM: Investigation, Writing – review & editing. HZ: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing – review & editing. XZ: Conceptualization, Funding acquisition, Project administration, Supervision, Writing – review & editing. HM: Conceptualization, Funding acquisition, Project administration, Resources, Writing – original draft, Writing – review & editing.
The authors declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Key Laboratory of Specific Oilseed Crops Genomics of Henan Province. This work was financially supported by the earmarked fund of China Agriculture Research System of MOF and MARA (CARS-14), the Key Project of Science and Technology in Henan Province (201300110600), Zhongyuan Science and Technology Innovation Leading Talent Plan (224200510021), the Shennong Laboratory First Class Program (SN01-2022-04), the Henan Province Specific Professor Position Program (SPPP2022), the Innovation Scientists and Technicians Troop Construction Projects of Henan Province (2023TD04), Key Research and Development Project of Henan Province (221111520400) and Science and Technology Foundation for The Excellent Youth Scholars of Henan Academy of Agricultural Sciences (2022YQ14).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1261238/full#supplementary-material
Supplementary Figure S1 | Multiple alignments of U-box domains of SiPUB proteins. The protein sequences were aligned using MAFFT, and the resulting alignments were visualized and displayed using Overleaf. The U-box domains were highlighted with a red frame to facilitate their identification.
Supplementary Figure S2 | A venn diagram of the relationship between differentially expressed gene (DEG) groups. The numbers in each section of the Venn diagram indicated the number of DEGs in each respective DEG group. The Venn diagram provided a visual representation of the overlapping and unique DEGs among the different DEG groups.
Supplementary Table S1 | Primers for qRT-PCR analysis of selected PUB genes in sesame.
Supplementary Table S2 | Summary of the detailed characterization of 56 PUB genes in sesame.
Supplementary Table S3 | A list of AtPUBs and OsPUBs used in phylogenetic analysis with SiPUBs.
Supplementary Table S4 | The Pfam analysis of SiPUB proteins in this study.
Supplementary Table S5 | The tandem duplication block types of 56 SiPUB genes.
Supplementary Table S6 | Cis-Acting elements analysis in the 2 kb promoters of SiPUB genes.
Supplementary Table S7 | Transcriptomic data for the expression analysis of drought-tolerant genotype TEX-1(T) and drought-sensitive genotype VEN-1(V).
Supplementary Table S8 | The three-dimensional (3D) structure analysis of 56 SiPUB proteins.
Supplementary Table S9 | The homologous relationships between AtPUB and SiPUB genes.
Adler, G., Mishra, A. K., Maymon, T., Raveh, D., Bar-Zvi, D. (2018). Overexpression of Arabidopsis ubiquitin ligase AtPUB46 enhances tolerance to drought and oxidative stress. Plant Sci. 276, 220–228. doi: 10.1016/j.plantsci.2018.08.018
Alexander, R. D., Wendelboe-Nelson, C., Morris, P. C. (2019). The barley transcription factor HvMYB1 is a positive regulator of drought tolerance. Plant Physiol. Biochem. 142, 246–253. doi: 10.1016/j.plaphy.2019.07.014
Al-Saharin, R., Hellmann, H., Mooney, S. (2022). Plant E3 ligases and their role in abiotic stress response. Cells 11 (5), 890. doi: 10.3390/cells11050890
Andersen, P., Kragelund, B. B., Olsen, A. N., Larsen, F. H., Chua, N.-H., Poulsen, F. M., et al. (2004). Structure and biochemical function of a prototypical Arabidopsis U-box domain *. J. Biol. Chem. 279 (38), 40053–40061. doi: 10.1074/jbc.M405057200
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME Suite: tools for motif discovery and searching. Nucleic Acids Res. 37 (suppl_2), W202–W208. doi: 10.1093/nar/gkp335
Baldoni, E., Genga, A., Cominelli, E. (2015). Plant MYB transcription factors: their role in drought response mechanisms. Int. J. Mol. Sci. 16 (7), 15811–15851. doi: 10.3390/ijms160715811
Bedigian, D., Harlan, J. R. (1986). Evidence for cultivation of sesame in the ancient world. Economic Bot. 40 (2), 137–154. doi: 10.1007/BF02859136
Bhattacharyya, S., Yu, H., Mim, C., Matouschek, A. (2014). Regulated protein turnover: snapshots of the proteasome in action. Nat. Rev. Mol. Cell Biol. 15 (2), 122–133. doi: 10.1038/nrm3741
Bi, H., Luang, S., Li, Y., Bazanova, N., Morran, S., Song, Z., et al. (2016). Identification and characterization of wheat drought-responsive MYB transcription factors involved in the regulation of cuticle biosynthesis. J. Exp. Bot. 67 (18), 5363–5380. doi: 10.1093/jxb/erw298
Bing, G., Miroslav, R., Figueiredo-Pereira, M. E., Christopher, C. (2016). The ubiquitin-proteasome system: potential therapeutic targets for Alzheimer's disease and spinal cord injury. Front. Mol. Neurosci. 9, 4. doi: 10.3389/fnmol.2016.00004
Byun, M. Y., Cui, L. H., Oh, T. K., Jung, Y.-J., Lee, A., Park, K. Y., et al. (2017). Homologous U-box E3 ubiquitin ligases OsPUB2 and OsPUB3 are involved in the positive regulation of low temperature stress response in rice (Oryza sativa L.). Front. Plant Sci. 8, 16. doi: 10.3389/fpls.2017.00016
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinf. 10 (1), 421. doi: 10.1186/1471-2105-10-421
Chao, J., Li, Z., Sun, Y., Aluko, O. O., Wu, X., Wang, Q., et al. (2021). MG2C: A user-friendly online tool for drawing genetic maps. Mol. Horticult 1 (1), 1–4. doi: 10.1186/s43897-021-00020-x
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13 (8), 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, C., Wang, C., Li, J., Gao, X., Huang, Q., Gong, Y., et al. (2022). Genome-wide analysis of U-box E3 ubiquitin ligase family in response to ABA treatment in Salvia miltiorrhiza. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.829447
Chen, X., Wang, T., Rehman, A. U., Wang, Y., Qi, J., Li, Z., et al. (2021). Arabidopsis U-box E3 ubiquitin ligase PUB11 negatively regulates drought tolerance by degrading the receptor-like protein kinases LRR1 and KIN7. J. Integr. Plant Biol. 63 (3), 494–509. doi: 10.1111/jipb.13058
Cheng, C., Wang, Z., Ren, Z., Zhi, L., Yao, B., Su, C., et al. (2017). SCFAtPP2-B11 modulates ABA signaling by facilitating SnRK2.3 degradation in Arabidopsis thaliana. PloS Genet. 13 (8), e1006947–e1006947. doi: 10.1371/journal.pgen.1006947
Cho, S. K., Chung, H. S., Ryu, M. Y., Park, M. J., Lee, M. M., Bahk, Y.-Y., et al. (2006). Heterologous expression and molecular and cellular characterization of caPUB1 encoding a hot pepper U-box E3 ubiquitin ligase homolog. Plant Physiol. 142 (4), 1664–1682. doi: 10.1104/pp.106.087965
Cho, S. K., Ryu, M. Y., Song, C., Kwak, J. M., Kim, W. T. (2008). Arabidopsis PUB22 and PUB23 are homologous U-box E3 ubiquitin ligases that play combinatory roles in response to drought stress. Plant Cell 20 (7), 1899–1914. doi: 10.1105/tpc.108.060699
Cui, J., Ren, G., Bai, Y., Gao, Y., Yang, P., Chang, J. (2023). Genome-wide identification and expression analysis of the U-box E3 ubiquitin ligase gene family related to salt tolerance in sorghum (Sorghum bicolor L.). Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1141617
Doane, A. S., Elemento, O. (2017). Regulatory elements in molecular networks. Wiley Interdiscip. Reviews: Syst. Biol. Med. 9 (3), e1374. doi: 10.1002/wsbm.1374
Duvaud, S., Gabella, C., Lisacek, F., Stockinger, H., Ioannidis, V., Durinx, C. (2021). Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 49 (W1), W216–W227. doi: 10.1093/nar/gkab225
Fàbregas, N., Lozano-Elena, F., Blasco-Escámez, D., Tohge, T., Martínez-Andújar, C., Albacete, A., et al. (2018). Overexpression of the vascular brassinosteroid receptor BRL3 confers drought resistance without penalizing plant growth. Nat. Commun. 9 (1), 4680–4680. doi: 10.1038/s41467-018-06861-3
Finn, R. D., Bateman, A., Clements, J., Coggill, P., Eberhardt, R. Y., Eddy, S. R., et al. (2014). Pfam: the protein families database. Nucleic Acids Res. 42 (D1), D222–D230. doi: 10.1093/nar/gkt1223
Fujita, Y., Fujita, M., Satoh, R., Maruyama, K., Parvez, M. M., Seki, M., et al. (2005). AREB1 is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell 17 (12), 3470–3488. doi: 10.1105/tpc.105.035659
Gupta, A., Rico-Medina, A., Caño-Delgado, A. I. (2020). The physiology of plant responses to drought. Science 368 (6488), 266–269. doi: 10.1126/science.aaz7614
Hoover, D. L., Pfennigwerth, A. A., Duniway, M. C. (2021). Drought resistance and resilience: The role of soil moisture–plant interactions and legacies in a dryland ecosystem. J. Ecol. 109 (9), 3280–3294. doi: 10.1111/1365-2745.13681
Horton, P., Park, K.-J., Obayashi, T., Fujita, N., Harada, H., Adams-Collier, C., et al. (2007). WoLF PSORT: protein localization predictor. Nucleic Acids Res. 35 (suppl_2), W585–W587. doi: 10.1093/nar/gkm259
Hu, H., Dong, C., Sun, D., Hu, Y., Xie, J. (2018). Genome-wide identification and analysis of U-box E3 ubiquitin-protein ligase gene family in banana. Int. J. Mol. Sci. 19 (12), 3874. doi: 10.3390/ijms19123874
Hu, R., Hochstrasser, M. (2016). Recent progress in ubiquitin and ubiquitin-like protein (Ubl) signaling. Cell Res. 26 (4), 389–390. doi: 10.1038/cr.2016.43
Huang, L., Wang, Y., Wang, W., Zhao, X., Qin, Q., Sun, F., et al. (2018). Characterization of transcription factor gene OsDRAP1 conferring drought tolerance in rice. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00094
Hye Jo, M., Ye Jin, J., Bin Goo, K., Woo Taek, K. (2016). CaPUB1, a hot pepper U-box E3 ubiquitin ligase, confers enhanced cold stress tolerance and decreased drought stress tolerance in transgenic rice (Oryza sativa L.). Mol Cells 39 (3), 250–257. doi: 10.14348/molcells.2016.2290
Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30 (4), 772–780. doi: 10.1093/molbev/mst010
Khan, M. R., Khan, I., Ibrar, Z., Léon, J., Naz, A. A. (2017). Drought-responsive genes expressed predominantly in root tissues are enriched with homotypic cis-regulatory clusters in promoters of major cereal crops. Crop J. 5 (3), 195–206. doi: 10.1016/j.cj.2016.10.001
Kiss, E., Oláh, B. R., Kaló, P. T., Morales, M., Heckmann, A. B., Borbola, A., et al. (2009). LIN, a novel type of U-box/WD40 protein, controls early infection by Rhizobia in legumes. Plant Physiol. 151 (3), 1239–1249. doi: 10.1104/pp.109.143933
Krzywinski, M., Schein, J., Birol, İ., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: An information aesthetic for comparative genomics. Genome Res 19 (9), 1639–1645. doi: 10.1101/gr.092759.109
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 (1), 325–327. doi: 10.1093/nar/30.1.325
Li, W., Cowley, A., Uludag, M., Gur, T., McWilliam, H., Squizzato, S., et al. (2015). The EMBL-EBI bioinformatics web and programmatic tools framework. Nucleic Acids Res. 43 (W1), W580–W584. doi: 10.1093/nar/gkv279
Li, K., Xing, C., Yao, Z., Huang, X. (2017). PbrMYB21, a novel MYB protein of Pyrus betulaefolia, functions in drought tolerance and modulates polyamine levels by regulating arginine decarboxylase gene. Plant Biotechnol. J. 15 (9), 1186–1203. doi: 10.1111/pbi.12708
Lin, Y., Hu, Q., Zhou, J., Yin, W., Yao, D., Shao, Y., et al. (2021). Phytophthora sojae effector Avr1d functions as an E2 competitor and inhibits ubiquitination activity of GmPUB13 to facilitate infection. Proc. Natl. Acad. Sci. 118 (10), e2018312118. doi: 10.1073/pnas.2018312118
Lin, Z., Li, Y., Wang, Y., Liu, X., Ma, L., Zhang, Z., et al. (2021). Initiation and amplification of SnRK2 activation in abscisic acid signaling. Nat. Commun. 12 (1), 2456. doi: 10.1038/s41467-021-22812-x
Liu, R., Xia, R., Xie, Q., Wu, Y. (2021). Endoplasmic reticulum-related E3 ubiquitin ligases: Key regulators of plant growth and stress responses. Plant Commun. 2 (3), 100186. doi: 10.1016/j.xplc.2021.100186
Lu, X., Shu, N., Wang, D., Wang, J., Chen, X., Zhang, B., et al. (2020). Genome-wide identification and expression analysis of PUB genes in cotton. BMC Genomics 21 (1), 213. doi: 10.1186/s12864-020-6638-5
Miao, H., Zhang, H., Kole, C. (2021). The Sesame Genome (Cham Switzerland: Springer International Publishing). doi: 10.1007/978-3-319-98098-0
Minh, B. Q., Schmidt, H. A., Chernomor, O., Schrempf, D., Woodhams, M. D., von Haeseler, A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37 (5), 1530–1534. doi: 10.1093/molbev/msaa015
Peters, J.-M. (2002). The anaphase-promoting complex: proteolysis in mitosis and beyond. Mol. Cell 9 (5), 931–943. doi: 10.1016/S1097-2765(02)00540-3
Peters, J.-M. (2006). The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 7 (9), 644–656. doi: 10.1038/nrm1988
Potter, S. C., Luciani, A., Eddy, S. R., Park, Y., Lopez, R., Finn, R. D. (2018). HMMER web server: 2018 update. Nucleic Acids Res. 46 (W1), W200–W204. doi: 10.1093/nar/gky448
Price, M. N., Dehal, P. S., Arkin, A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments. PloS One 5 (3), e9490. doi: 10.1371/journal.pone.0009490
Qin, Q., Wang, Y., Huang, L., Du, F., Zhao, X., Li, Z., et al. (2020). A U-box E3 ubiquitin ligase OsPUB67 is positively involved in drought tolerance in rice. Plant Mol. Biol. 102 (1), 89–107. doi: 10.1007/s11103-019-00933-8
Rehman, S., Rashid, A., Manzoor, M. A., Li, L., Sun, W., Riaz, M. W., et al. (2022). Genome-wide evolution and comparative analysis of superoxide dismutase gene family in Cucurbitaceae and expression analysis of Lagenaria siceraria under multiple abiotic stresses. Front. Genet. 12. doi: 10.3389/fgene.2021.784878
Riaz, M. W., Lu, J., Shah, L., Yang, L., Chen, C., Mei, X. D., et al. (2021). Expansion and molecular characterization of AP2/ERF gene family in wheat (Triticum aestivum L.). Front. Genet. 12. doi: 10.3389/fgene.2021.632155
Roca Paixão, J. F., Gillet, F.-X., Ribeiro, T. P., Bournaud, C., Lourenço-Tessutti, I. T., Noriega, D. D., et al. (2019). Improved drought stress tolerance in Arabidopsis by CRISPR/dCas9 fusion with a Histone AcetylTransferase. Sci. Rep. 9 (1), 8080. doi: 10.1038/s41598-019-44571-y
Rose, P. W., Prlić, A., Altunkaya, A., Bi, C., Bradley, A. R., Christie, C. H., et al. (2017). The RCSB protein data bank: integrative view of protein, gene and 3D structural information. Nucleic Acids Res. 45 (D1), D271–D281. doi: 10.1093/nar/gkw1000
Ryu, M. Y., Cho, S. K., Hong, Y., Kim, J., Kim, J. H., Kim, G. M., et al. (2019). Classification of barley U-box E3 ligases and their expression patterns in response to drought and pathogen stresses. BMC Genomics 20 (1), 326. doi: 10.1186/s12864-019-5696-z
Saito, R., Smoot, M. E., Ono, K., Ruscheinski, J., Wang, P.-L., Lotia, S., et al. (2012). A travel guide to Cytoscape plugins. Nat. Methods 9 (11), 1069–1076. doi: 10.1038/nmeth.2212
Seo, D. H., Lee, A., Yu, S. G., Cui, L. H., Min, H. J., Lee, S. E., et al. (2021). OsPUB41, a U-box E3 ubiquitin ligase, acts as a negative regulator of drought stress response in rice (Oryza Sativa L.). Plant Mol. Biol. 106 (4), 463–477. doi: 10.1007/s11103-021-01158-4
Shu, K., Yang, W. (2017). E3 ubiquitin ligases: ubiquitous actors in plant development and abiotic stress responses. Plant Cell Physiol. 58 (9), 1461–1476. doi: 10.1093/pcp/pcx071
Song, Q., Joshi, M., Wang, S., Johnson, C. D., Joshi, V. (2021). Comparative analysis of root transcriptome profiles of sesame (Sesamum indicum L.) in response to osmotic stress. J. Plant Growth Regul. 40 (4), 1787–1801. doi: 10.1007/s00344-020-10230-0
Sun, J., Peng, X., Fan, W., Tang, M., Liu, J., Shen, S. (2014). Functional analysis of BpDREB2 gene involved in salt and drought response from a woody plant Broussonetia papyrifera. Gene 535 (2), 140–149. doi: 10.1016/j.gene.2013.11.047
Szklarczyk, D., Gable, A. L., Nastou, K. C., Lyon, D., Kirsch, R., Pyysalo, S., et al. (2021). The STRING database in 2021: customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 49 (D1), D605–D612. doi: 10.1093/nar/gkab835
Tong, S., Chen, N., Wang, D., Ai, F., Liu, B., Ren, L., et al. (2021). The U-box E3 ubiquitin ligase PalPUB79 positively regulates ABA-dependent drought tolerance via ubiquitination of PalWRKY77 in Populus. Plant Biotechnol. J. 19 (12), 2561–2575. doi: 10.1111/pbi.13681
Trujillo, M. (2018). News from the PUB: plant U-box type E3 ubiquitin ligases. J. Exp. Bot. 69 (3), 371–384. doi: 10.1093/jxb/erx411
Vicente-Serrano, S. M., Quiring, S. M., Peña-Gallardo, M., Yuan, S., Domínguez-Castro, F. (2020). A review of environmental droughts: Increased risk under global warming? Earth Sci Rev. 201, 102953. doi: 10.1016/j.earscirev.2019.102953
Vodermaier, H. C., Gieffers, C., Maurer-Stroh, S., Eisenhaber, F., Peters, J.-M. (2003). TPR subunits of the anaphase-promoting complex mediate binding to the activator protein CDH1. Curr. Biol. 13 (17), 1459–1468. doi: 10.1016/S0960-9822(03)00581-5
Wang, C., Duan, W., Riquicho, A. R., Jing, Z., Liu, T., Hou, X., et al. (2015). Genome-wide survey and expression analysis of the PUB family in Chinese cabbage (Brassica rapa ssp. pekinesis). Mol. Genet. Genomics 290 (6), 2241–2260. doi: 10.1007/s00438-015-1075-x
Wang, C., Song, B., Dai, Y., Zhang, S., Huang, X. (2021). Genome-wide identification and functional analysis of U-box E3 ubiquitin ligases gene family related to drought stress response in Chinese white pear (Pyrus bretschneideri). BMC Plant Biol. 21 (1), 235. doi: 10.1186/s12870-021-03024-3
Wang, Y., Tang, H., DeBarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40 (7), e49–e49. doi: 10.1093/nar/gkr1293
Wang, K., Yang, Q., Lanhuang, B., Lin, H., Shi, Y., Dhanasekaran, S., et al. (2020). Genome-wide investigation and analysis of U-box Ubiquitin–Protein ligase gene family in apple: Expression profiles during Penicillium expansum infection process. Physiol. Mol. Plant Pathol. 111, 101487. doi: 10.1016/j.pmpp.2020.101487
Wei, J., Qiu, X., Chen, L., Hu, W., Hu, R., Chen, J., et al. (2015). The E3 ligase AtCHIP positively regulates Clp proteolytic subunit homeostasis. J. Exp. Bot. 66 (19), 5809–5820. doi: 10.1093/jxb/erv286
Wiborg, J., O'Shea, C., Skriver, K. (2008). Biochemical function of typical and variant Arabidopsis thaliana U-box E3 ubiquitin-protein ligases. Biochem. J. 413 (3), 447–457. doi: 10.1042/BJ20071568
Xianjun, P., Xingyong, M., Weihong, F., Man, S., Liqin, C., Alam, I., et al. (2011). Improved drought and salt tolerance of Arabidopsisthaliana by transgenic expression of a novel DREB gene from Leymuschinensis. Plant Cell Rep. 30 (8), 1493–1502. doi: 10.1007/s00299-011-1058-2
Xu, Z., Wang, M., Guo, Z., Zhu, X., Xia, Z. (2019). Identification of a 119-bp promoter of the maize sulfite oxidase gene (ZmSO) that confers high-level gene expression and ABA or drought inducibility in transgenic plants. Int. J. Mol. Sci. 20 (13), 3326. doi: 10.3390/ijms20133326
Yan, J., Su, P., Meng, X., Liu, P. (2023). Phylogeny of the plant receptor-like kinase (RLK) gene family and expression analysis of wheat RLK genes in response to biotic and abiotic stresses. BMC Genomics 24 (1), 224. doi: 10.1186/s12864-023-09303-7
Yang, C.-W., González-Lamothe, R., Ewan, R. A., Rowland, O., Yoshioka, H., Shenton, M., et al. (2006). The E3 ubiquitin ligase activity of Arabidopsis PLANT U-BOX17 and its functional tobacco homolog ACRE276 are required for cell death and defense. Plant Cell 18 (4), 1084–1098. doi: 10.1105/tpc.105.039198
You, J., Zhang, Y., Liu, A., Li, D., Wang, X., Dossa, K., et al. (2019). Transcriptomic and metabolomic profiling of drought-tolerant and susceptible sesame genotypes in response to drought stress. BMC Plant Biol. 19 (1), 267. doi: 10.1186/s12870-019-1880-1
Yuan, S., Chan, H. S., Hu, Z. (2017). Using PyMOL as a platform for computational drug design. Wiley Interdiscip. Reviews: Comput. Mol. Sci. 7 (2), e1298. doi: 10.1002/wcms.1298
Zeng, L.-R., Park, C. H., Venu, R. C., Gough, J., Wang, G.-L. (2008). Classification, expression pattern, and E3 ligase activity assay of rice U-box-containing proteins. Mol. Plant 1 (5), 800–815. doi: 10.1093/mp/ssn044
Zhang, H., Miao, H., Ju, M. (2019). “Potential for adaptation to climate change through genomic breeding in sesame,” in Genomic Designing of Climate-Smart Oilseed Crops. Ed. Kole, C. (Cham: Springer International Publishing), 371–440.
Zhang, H., Miao, H., Wang, L., Qu, L., Liu, H., Wang, Q., et al. (2013). Genome sequencing of the important oilseed crop Sesamum indicumL. Genome Biol. 14 (1), 401. doi: 10.1186/gb-2013-14-1-401
Zhang, T., Wu, A., Yue, Y., Zhao, Y. (2020). uORFs: important cis-regulatory elements in plants. Int. J. Mol. Sci. 21 (17), 6238. doi: 10.3390/ijms21176238
Zhao, T.-J., Sun, S., Liu, Y., Liu, J.-M., Liu, Q., Yan, Y.-B., et al. (2006). Regulating the drought-responsive element (DRE)-mediated signaling pathway by synergic functions of trans-active and trans-inactive DRE binding factors in Brassica napus. J. Biol. Chem. 281 (16), 10752–10759. doi: 10.1074/jbc.M510535200
Keywords: ubiquitin, Sesamum indicum, PUB, drought tolerance, gene family
Citation: Cao H, Tian Q, Ju M, Duan Y, Li G, Ma Q, Zhang H, Zhang X and Miao H (2023) Genome-wide analysis of the U-box E3 ubiquitin ligase family role in drought tolerance in sesame (Sesamum indicum L.). Front. Plant Sci. 14:1261238. doi: 10.3389/fpls.2023.1261238
Received: 19 July 2023; Accepted: 01 September 2023;
Published: 19 September 2023.
Edited by:
Neelakantan Arumugam, Pondicherry University, IndiaReviewed by:
Muhammad Waheed Riaz, Zhejiang Agriculture and Forestry University, ChinaCopyright © 2023 Cao, Tian, Ju, Duan, Li, Ma, Zhang, Zhang and Miao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Haiyang Zhang, emhhbmdoYWl5YW5nQHp6dS5lZHUuY24=; Xianmei Zhang, bGhzZXNhbWVAMTI2LmNvbQ==; Hongmei Miao, bWlhb2hvbmdtZWljaGluYUAxNjMuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.