- State Key Laboratory of Rice Biology and Breeding, China National Rice Research Institute, Hangzhou, China
3-Ketoacyl-CoA synthase (KCS) is the key rate-limiting enzyme for the synthesis of very long-chain fatty acids (VLCFAs) in plants, which determines the carbon chain length of VLCFAs. However, a comprehensive study of KCSs in Oryza sativa has not been reported yet. In this study, we identified 22 OsKCS genes in rice, which are unevenly distributed on nine chromosomes. The OsKCS gene family is divided into six subclasses. Many cis-acting elements related to plant growth, light, hormone, and stress response were enriched in the promoters of OsKCS genes. Gene duplication played a crucial role in the expansion of the OsKCS gene family and underwent a strong purifying selection. Quantitative Real-time polymerase chain reaction (qRT-PCR) results revealed that most KCS genes are constitutively expressed. We also revealed that KCS genes responded differently to exogenous cadmium stress in japonica and indica background, and the KCS genes with higher expression in leaves and seeds may have functions under cadmium stress. This study provides a basis for further understanding the functions of KCS genes and the biosynthesis of VLCFA in rice.
Introduction
Very long-chain fatty acids (VLCFAs) are molecules with a hydrocarbon chain containing more than 18 carbon atoms, which are important components of plant cell membrane lipids, cutin wax, and seed storage lipids (Scott et al., 2022). VLCFAs and their derivatives play an important role in plant growth and development, signal transduction, and adverse stress. Disorders in the expression of genes involved in the synthesis of VLCFAs lead to phenotypic consequences, ranging from cell dedifferentiation to embryo lethality (De Bigault Du Granrut and Cacas, 2016; Zhukov and Popov, 2022). VLCFA exists in cell membrane phospholipids and sphingolipids, especially in phosphatidylserine (PS) and phosphatidylethanolamine (PE), participating in intercellular signal transduction to regulate plant growth and development (Boutte and Jaillais, 2020). VLCFAs are precursors of plant cuticle and cutin waxes in epidermal cells, which attach to plant surfaces and are the first line of defense against external stresses (Kunst and Samuels, 2003; Li et al., 2018). VLCFAs can be contained in developing seeds, accounting for up to two-thirds of the total amount of FA. There, they can be in the composition of triacylglycerols (TAGs), playing a key role in seed germination (Cahoon and Li-Beisson, 2020).
The biosynthesis of VLCFA is the elongation of fatty acids from C18 chains to C26–C34 chains via fatty acid elongase (FAE) complex in the endoplasmic reticulum (ER); FAE is composed of four major enzymes such as 3-ketoacyl-CoA synthetase (called as β-ketoacyl-CoA synthetase, KCS), trans-2,3-enoyl CoA reductase (ECR), 3-hydroxacyl-CoA dehydratase (HCD), and 3-ketoacyl-CoA reductase (KCR) (Haslam and Kunst, 2013). In this process, C16:0 or C18:0 or C18:1, the product of de novo fatty acid synthesis, is used as a substrate, which is catalyzed through four consecutive reactions in ER, namely, condensation, reduction, dehydration, and secondary reduction; two carbons atoms are added in each cycle (Leonard et al., 2004).
There are two types of non-homologous condensing enzymes involved in fatty acid elongation in organisms: one of these is FAE1-like 3-ketoacyl-CoA synthases (KCS-type enzymes), and the other is ELONGATION DEFECTIVE-LIKE proteins (ELO-LIKEs). ELO-LIKEs are found in many organisms such as humans, plants, and yeasts, whereas KCSs are only found in plants and protists (Stenback et al., 2022). Several studies have been performed on the KCS gene family in plants. There are 21 KCS genes in Arabidopsis thaliana (Joubès et al., 2008), 26 KCS genes in Zea mays (Campbell et al., 2019), 30 KCS genes in Arachis hypogaea (Huai et al., 2020), and 58 KCS genes in Gossypium hirsutum (Xiao et al., 2016). In Arabidopsis thaliana, 21 members are divided into four subfamilies according to the amino acid sequence homology: KCS1-like, FDH-like, FAE1-like, and CER6 (Costaglioli et al., 2005), and 21 members are classified into eight subclasses according to their duplication history, genomic organization, protein topology, and 3D modeling (Joubès et al., 2008).
KCS is not only the rate-limiting enzyme in the elongation process of VLCFAs but also has substrate specificity and tissue specificity, which determines the rate of product formation and the carbon chain length of VLCFA. The function of KCS gene in Arabidopsis thaliana has been thoroughly studied. For example, KCS18/FAE1, which is specifically expressed in seeds, catalyzes the elongation of C18 to C20 and C22 VLCFAs. KCS4 is involved in the differential accumulation of polyunsaturated TAGs under stress (Luzarowska et al., 2023). The mutants do not contain C20 and C22 VLCFAs and lead to C18 accumulation (James et al., 1995); KCS2 and KCS20 are highly expressed in root endothelium, mainly producing C22 and C24 VLCFAs (Lee et al., 2009); KCS5 and KCS6 (CER6) play important roles in C24–C28 VLCFAs (Millar et al., 1999), and KCS3–KCS6 module affects wax synthesis (Huang et al., 2023); KCS9 was the highest expressed in Arabidopsis stem epidermal cells, and the C24 VLCFA of the mutant was significantly reduced (Kim et al., 2013). At present, there are few studies on the function of KCS genes in rice. SD38 is involved in the elongation of C24:0 VLCFA, and the mutant plants are semi-dwarf (Zhang et al., 2022). Two KCS genes, ONI1 and ONI2, were specifically expressed in the shoot apical meristem, and mutants with abnormal VLCFA composition resulted in death of plant seedlings (Tsuda et al., 2013). WSL1 is involved in wax biosynthesis in leaves and leaf sheaths (Yu et al., 2008; Zhou et al., 2021). WSL4/HMS1 is involved in C22:0 VLCFA elongation; its functional deficiency leads to less wax in leaves, shorter plants and fewer tillers, affecting the formation of pollen walls (Gan et al., 2017; Chen et al., 2020).
Cadmium (Cd) is considered as one of the most toxic metals for plant and can cause severe damage both to environment and human. The contamination of Cd in Chinese agricultural soils is quite prevalent, and about a quarter of the soil samples exceed China’s national standard, which are mainly located in the Yangtze River Delta (Cheng et al., 2023). The average of Cd content in brown rice in a survey was slightly higher than milled rice samples and rice Cd content in 35.1% of total 208 cultivars exceed the rice Cd limit (0.2 mg/kg), which were collected in South China (Liu et al., 2023). In many plants, one of the metabolic adaptions to Cd tolerance is related to modifications of fatty acid; the elongation of fatty acid is one way of modifications (Zhukov and Shumskaya, 2020). Similar effect of accumulation of VLCFAs was observed in Noccaea caerulescens and tomato plants (Ben Ammar et al., 2008; Zemanova et al., 2015).
Genome-wide identification of gene family provides the basis for further functional analysis. Due to the extensive application of large-scale plant genome sequencing and bioinformatics technology, KCS gene families of many species have been identified, but the KCS gene family members of rice have not yet been identified. More than 3.5 billion people in the world rely on rice as a staple food and livelihood (Huang et al., 2017). Therefore, in this study, we systematically identified and analyzed the characteristics of rice KCS gene family by bioinformatics methods, and the expression levels of OsKCS in different tissues were also investigated. Under Cd stress, the expression profiles of OsKCS are different in japonica and indica. This study provided useful information for further investigating the molecular functions of KCS genes in rice.
Materials and methods
Plant materials and treatment
In this study, the rice variety is Nipponbare and 9311. Seeds were rinsed with distilled water, and then germinated at 28°C under dark conditions. After 48h, seedlings with a root length of approximately 0.5 cm were moved to hydroponic culture boxes (day/night temperatures of 28°C/22°C, light/dark photoperiod of 12h/12h, and light intensity of 18,000 Lx). At the one-leaf stage, the seedlings were treated with nutrient solution. At the two-leaf stage, Cd stress experiments were performed. The CdCl2 solutions (20 μmol/L) prepared with nutrient solution were used to simulate Cd stress, and nutrient solution without CdCl2 was used as the control. After 3h and 6h of treatment, seedlings were selected for each sample, and quickly stored at -80°C until analysis. The experiment was performed in triplicate.
Identification of KCS genes in rice
In order to identify and characterize KCS gene family in rice genome, gff3, proteins, CDS, and genome files were downloaded from Ensembl Plants (http://plants.ensembl.org/Oryza_sativa/Info/Index). Twenty-one identified KCS protein sequences of Arabidopsis thaliana were downloaded from Arabidopsis genome database TAIR (https://www.arabidopsis.org/). We performed two methods, which are Basic Local Alignment Search Tool for proteins (BLASTp) and Hidden Markov Models (HMMER) search tool, to identify KCS genes in rice genome. The BLASTp (BLAST 2.7.1+) was performed based on protein homologous alignment with default mode using the Arabidopsis KCS protein sequences to obtain the candidate KCS genes in rice genome (E-value < 10−10). The Hidden Markov Model files corresponding to the conserved domain (Accession No.: PF08392 and PF08541) were downloaded from database Pfam (http://pfam.xfam.org/). HMMSEARCH was used to retrieve rice candidate sequences (E-value < 10−20) containing KCS conserved domains (FAE1_CUT1_RppA and ACP_syn_III_C). Results of these two methods were checked and merged, and the redundancies were manually removed to obtain the candidate KCS genes in rice. The candidate sequences were submitted into the NCBI Conserved Domains (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi) and SMART (http://smart.embl-heidelberg.de/) to confirm the domains in rice KCS proteins (Letunic et al., 2012).
Physiochemical properties, alignment and phylogenetic analysis
The OsKCS genes’ physical and chemical properties, namely, molecular weight (M.W.), amino acid (aa) length, isoelectric point (pl), instability index, aliphatic index (Ai), and grand average of hydropathicity (GRAVY) were evaluated by using the ExPASY-Prot (http://web.expasy.org/protparam/). The phylogenetic analysis was performed by aligning the KCS protein sequences of rice by MEGA software. The aligned sequences were subjected to neighbor-joining (NJ) tree construction using the MEGA software with 1,000 bootstrap replications and all other parameters were set to default (Hall, 2013).
Chromosome locations, gene structures, and motif analysis
The chromosome locations and structures of OsKCS genes were retrieved from rice genome annotation files, and the conserved motifs of OsKCS protein sequences were predicted by using MEME (MEME 5.1.0) (Bailey et al., 2009). The OsKCS gene structures, chromosome locations, and conserved motifs were visualized by TBtools software (Bailey et al., 2009).
Cis-regulatory element analysis of promoters
The 2,000 bp genomic sequences upstream of the transcription start site of OsKCS genes were extracted from the genomic DNA sequences. Since the upstream regions of some genes overlap with other genes, the upstream regions of these genes were shortened. The promoter sequences were submitted to the PlantCARE database (https://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to predict cis-regulatory elements (Lescot et al., 2002).
Synteny analysis and Ka/Ks values calculation
The tandem and segmental duplication or whole-genome duplication (WGD) provides new insights into genes development and genome progression. The duplication events of OsKCS genes and the syntenic relationships of KCS genes between rice and maize were analyzed using MCScanX toolkit (Wang et al., 2012), and KCS relationships between the target species were visualized by using Circos (Krzywinski et al., 2009). Nonsynonymous (Ka) and synonymous (Ks) values and the Ka/Ks ratios of gene pairs were calculated by ParaAT 2.0 (Zhang et al., 2012) and KaKs_Calculator (Zhang et al., 2006), Ks value could be used as molecular clock to reckon the time after gene replication event, Ka/Ks ratio has been used to determine the type of gene selection during evolution. Ka/Ks = 1, Ka/Ks > 1, and Ka/Ks < 1 represent natural, positive, and purifying selections, respectively (Hurst, 2002). The divergence time was calculated with the formula: T = Ks/r; r = 6.5 × 10−9) (Quraishi et al., 2011).
Gene expression analysis based on RNA-seq data
To examine the expression pattern of KCS genes under Cd stress, three RNA samples of Nipponbare and 9311 were sequenced on the HiSeq 4000 platform (Illumina) for transcriptome analysis by Novogene Technology (Beijing, China) to obtain clean reads. Differentially Expressed Genes (DEGs) were identified by a false discovery rate ≤ 0.05 and an absolute value of the log2 ratio ≥ 1. Using the RNA-seq data, the absolute FPKM of the OsKCS were obtained, and the R software was used for statistics and visualization. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were performed by R packages: clusterProfiler.
RNA extraction and qRT–PCR analysis
Total RNA was extracted from different plant tissues using Trizol reagents (Invitrogen, Carlsbad, CA, USA), RNA was reverse transcribed to cDNA using the ReverTra Ace qPCR-RT kit (Toyobo, Osaka, Japan), and qPCR was performed using SYBR Green Real-Time PCR Master Mix (Toyobo).
Results
Identification of KCS genes in rice
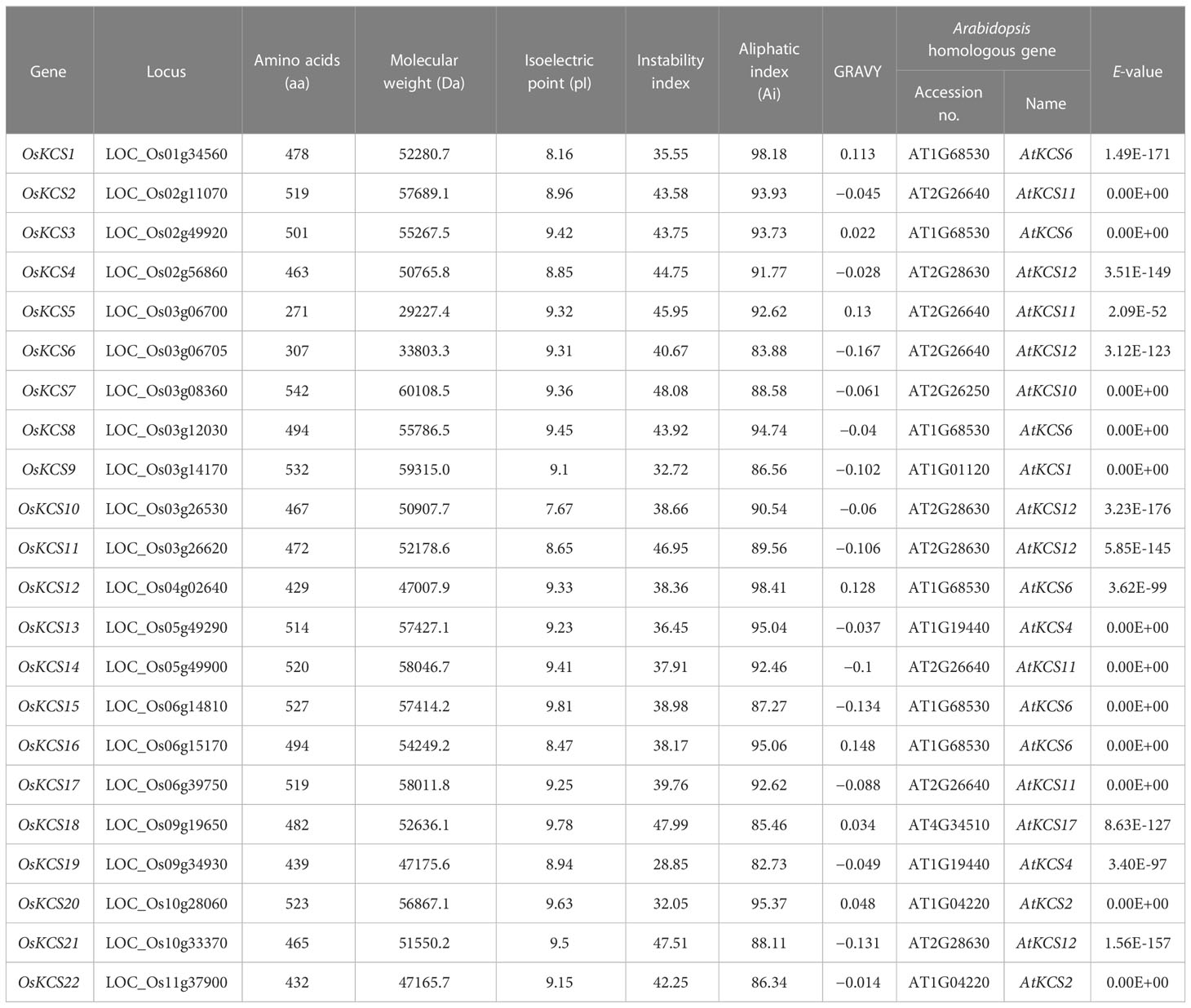
Twenty-two KCS genes in rice genome were identified after removing redundant and repetitive sequences from BLASTp and HMMER results. To explore the phylogenetic relationships of the KCS family, an NJ phylogenetic tree was constructed using the full-length protein sequences of 22 OsKCSs and 21 AtKCSs. Results demonstrated that KCS proteins were classified into eight clades based on phylogenetic relationship, whereas, KCS proteins in rice were divided into six subclasses: α, γ, δ, ϵ, ζ and η (Figure 1). KCS genes in rice genome were named OsKCS1 to OsKCS22 according to their chromosomal locations. GO and KEGG annotation analysis of the OsKCS genes was performed to further understand the possible roles of OsKCS genes in molecular function, cellular component, and biological process at the molecular levels. GO and KEGG enrichment analysis showed that 22 OsKCS genes were all enriched in fatty acid biosynthetic process and involved in fatty acid elongation (Supplementary Table S1). Furthermore, the results of physiochemical properties showed that OsKCS genes varied in their properties such as the protein length varied from 271 (OsKCS5) to 542 aa (OsKCS7), as well as the molecular weights ranged from 29.23 to 60.11 kD; the isoelectric point (pI) of OsKCS proteins also varied ranged from 7.67 (OsKCS10) to 9.81 (OsKCS15), and the protein instability indexes of 11 OsKCS proteins were smaller than 40, indicating that these proteins are stable. The remaining OsKCS proteins’ instability indexes were greater than 40; most KCS proteins are hydrophilic. The KCS genes with the highest homology to the OsKCS genes and E-values were obtained in Arabidopsis thaliana (Table 1).
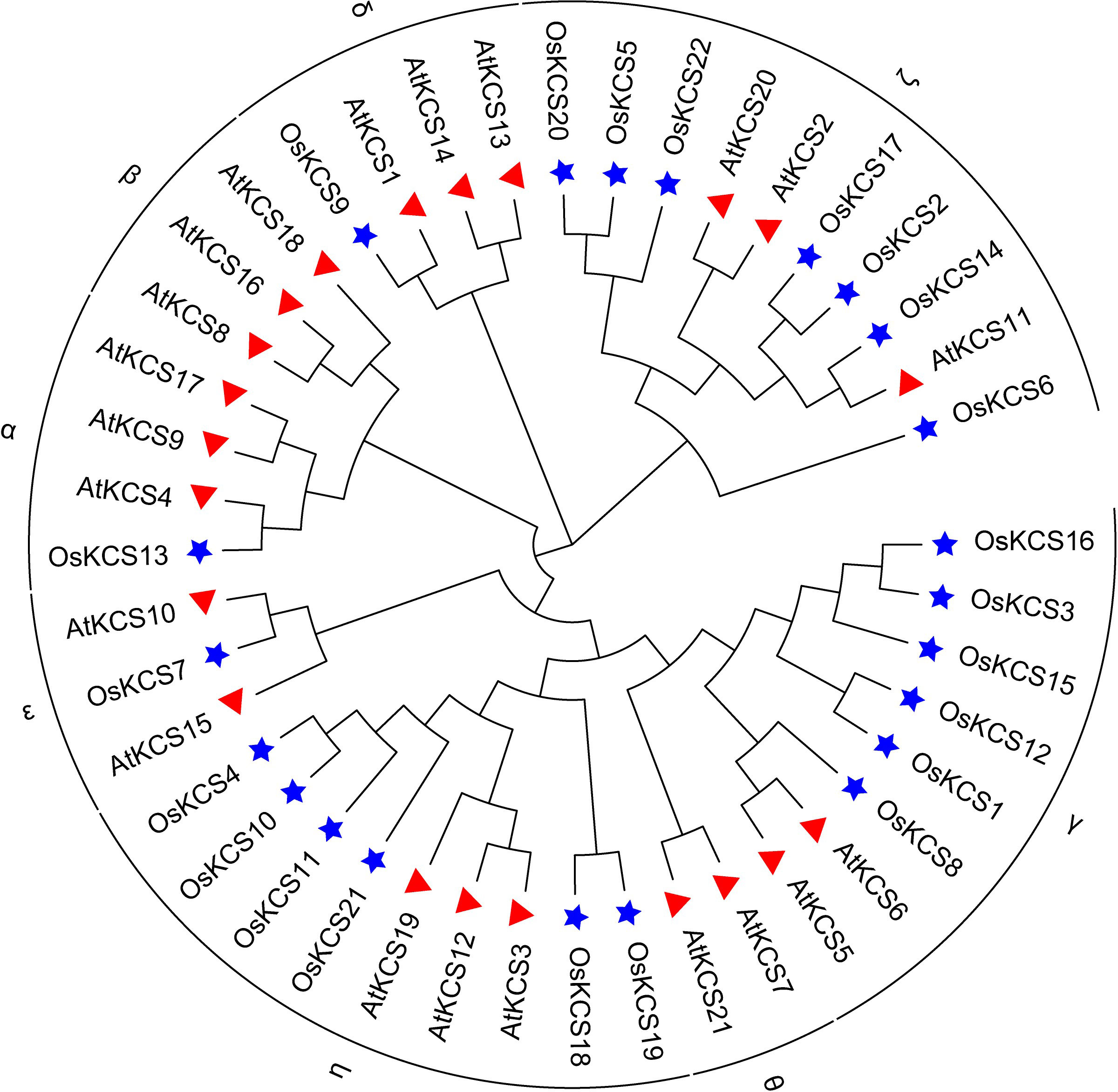
Figure 1 Phylogenetic analysis of the KCS genes in Oryza sativa (Os) and Arabidopsis thaliana (At) by the neighbor-joining method. The KCSs were clustered into eight clades; each member of the KCSs was annotated by (★ for Os) and (▲ for At), respectively.
Gene structures and conserved motif analysis of OsKCS
OsKCS proteins were classified into four main groups (KCS1-like, FAE1-like, CER6-like, and FDH-like) (Figure 2A). Most of the OsKCS gene family members contained 1–2 exons, of which 11 OsKCS genes did not contain introns (Figure 2B). With the exception of OsKCS5, which has only four motifs, most of the conserved motifs of the OsKCS gene family members have the same types, numbers, and orders (Figure 2C). CDD and SMART search tools for domains verification was used and found that OsKCS proteins contained two domains such as FAE1_CUT1_RppA [(PF08392) FAE1/Type III polyketide synthase-like protein domain] and ACP_syn_III_C [(PF08541) 3-Oxoacyl-acyl-carrier protein (ACP) synthase III C terminal domain], which were main conserved domains in KCS proteins (Figure 2D), almost all genes contain these two domains except for OsKCS12.
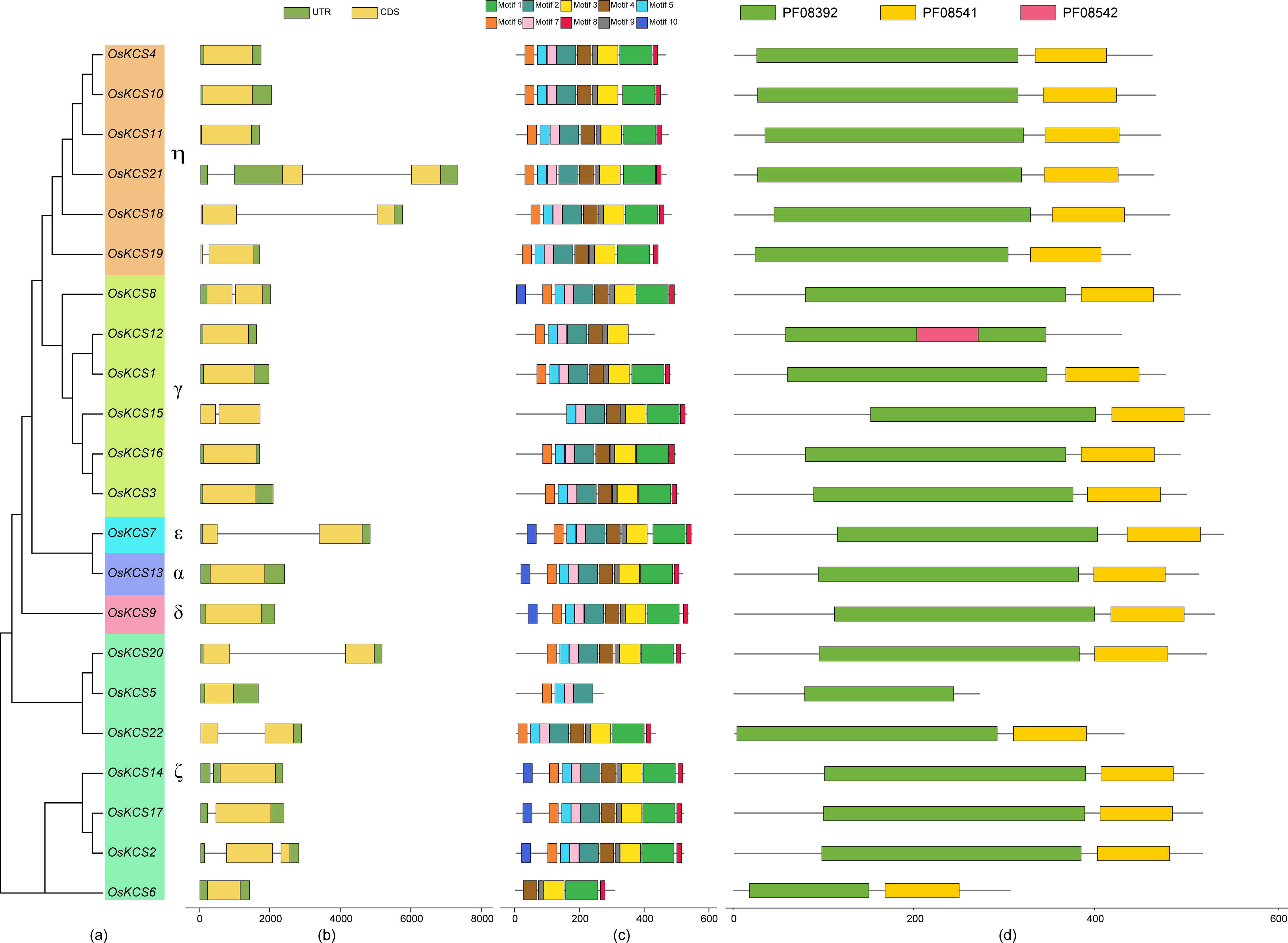
Figure 2 The unrooted phylogenetic tree, conserved motifs, and gene structure of OsKCS. (A) The neighbor-joining tree on the left composed of 22 KCS proteins from rice. (B) OsKCS genes structures: yellow color indicates the exons, the green color shows the untranslated 5′ and 3′ regions. (C) Conserved motifs were represented via boxes and different colors represents different motifs. (D) The function conserved domains of OsKCS genes. PF08392: FAE1_CUT1_RppA; PF08541: ACP_syn_III_C; PF08542: ACP_syn_III.
Cis-regulatory element analysis of OsKCS
A total of 18 cis-regulatory elements were predicted in the upstream 2,000 bp from the transcription start sites of OsKCSs (Figure 3), which were widely involved in the growth biological process, hormonal responsiveness, light responsiveness, and stress response. A list of genomic location and names/annotations of 22 OsKCSs and the respective upstream genes will be provided to know whether any of the KCS-upstream gene blocks are paralogous (Supplementary Table S2). Among them, MYB transcription factors involved in plant biological process were identified in all OsKCSs. Four types of hormone-responsive elements were also found, namely, auxin-responsive elements (TGA-elements) in nine OsKCSs, MeJA-responsive elements (CGTCA-motif and TGACG-motif) in 20 OsKCSs, salicylic acid–responsive elements (TCA-element) in seven OsKCSs, and ABA-responsive elements (ABRE) in 20 OsKCSs. Furthermore, there were meristem expression-related elements (CAT-box) in seven OsKCSs and gliadin metabolism-related elements in seven OsKCSs. The results of cis-regulatory element analysis indicate that OsKCS may be expressed in different growth environments, hormones, and stress treatments. However, many motifs have not been functionally verified, and whether these motifs confer unique functions on OsKCS remains to be further studied.
Figure 3 Cis-elements found in the promoter region of OsKCS genes. (Left): The number and function classification of cis-acting element in each OsKCS genes. (Right): Distribution of 18 identified cis-acting elements in each OsKCS; elements are represented by the boxes in different colors.
Chromosome distribution and synteny relationship of OsKCS
Except for chromosomes 7, 8, and 12, all of the OsKCS genes were unevenly distributed on nine of 12 chromosomes (Figure 4A). The largest number of OsKCS genes (OsKCS5–OsKCS11) appeared on chromosome 3, followed by chromosomes 2 and 6 (three genes each), chromosomes 5, 9, and 10 (two genes each). Synteny analysis was used to understand the evolution and expansion mechanism of OsKCS gene family in the rice genome and the genomes of other species. The results of gene duplication analysis indicated that there were only two OsKCS gene pairs (OsKCS2/OsKCS17 and OsKCS3/OsKCS15), both were segmentally duplicated on chromosomes 2 and 6 (Figure 4B). There were no tandem duplications in tight regions on chromosomes 3 and 6. Ka/Ks value is used to evaluate the evolution of coding sequences and determine the type of selection pressure after duplication. The Ka/Ks values of the two gene pairs were smaller than 0.05, indicating that these genes had gone through purifying selection (Supplementary Table S3). The results of the divergence time indicated that the duplication process between the segmental OsKCS genes was estimated to be 6 million years ago, and the evolutionary mechanism of OsKCS was conserved during evolution. In order to further understand the evolutionary origins and orthologous relationship of KCS gene family, the synteny analysis was performed among four representative plant species (two dicots: Arabidopsis and Glycine max; two monocots: Zea mays and Triticum aestivum) (Figure 5). Sixteen and 40 orthologous pairs were found between Zea mays and Triticum aestivum, respectively. Two and 0 orthologous pairs were found between Glycine max and Arabidopsis, respectively. The huge differences in homologous gene pairs between dicots and monocots suggested that KCS genes may be formed after the differentiation of monocots and dicots.
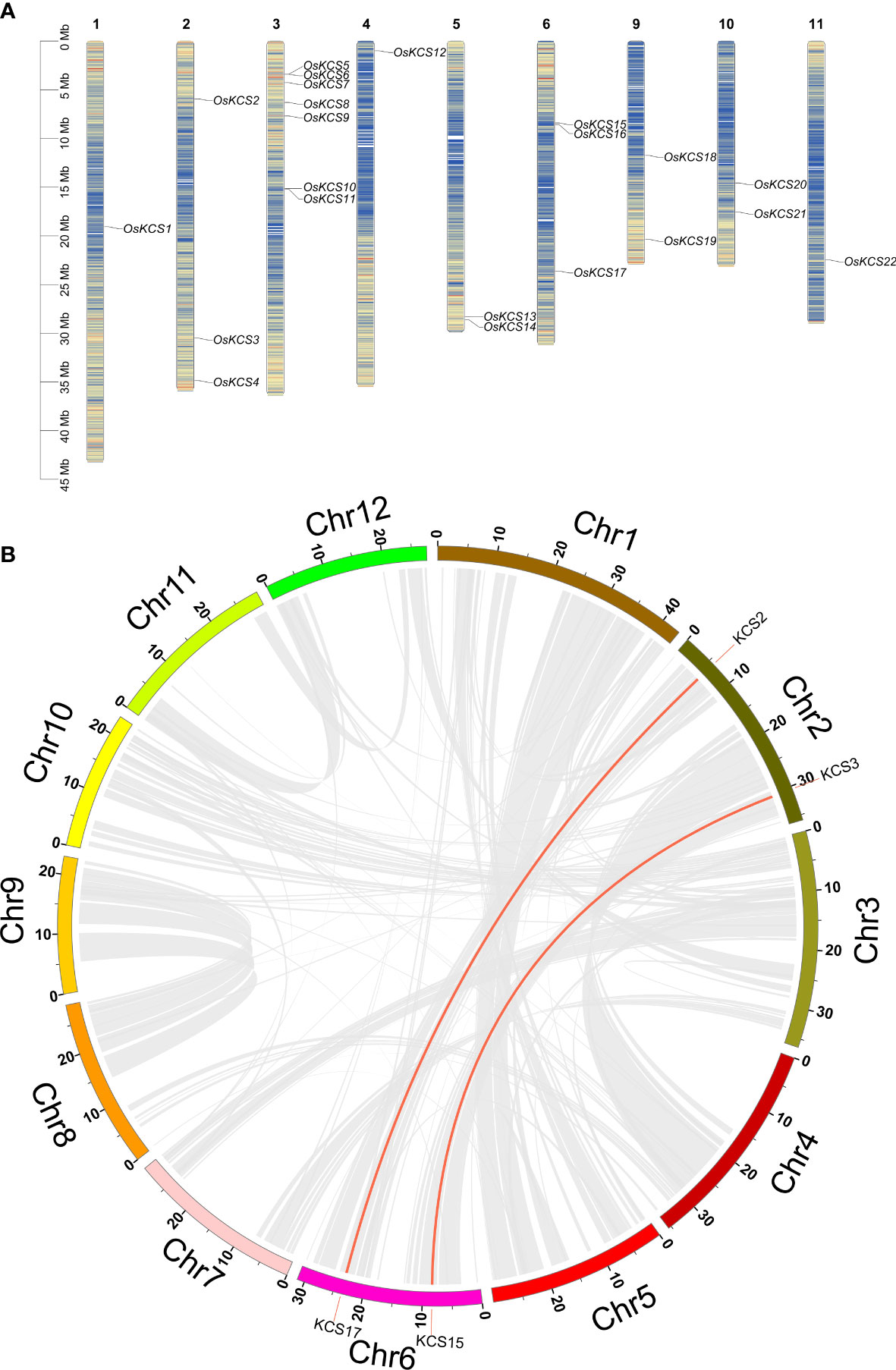
Figure 4 Chromosome distribution and synteny relationship of OsKCS gene family. (A) Chromosome location of 22 OsKCS genes in rice. (B) Circle map of the duplication gene pairs of the OsKCS genes. The background gray lines show all the syntenic blocks in the rice genome, and the red lines show the segmental duplication link regions among OsKCS genes.
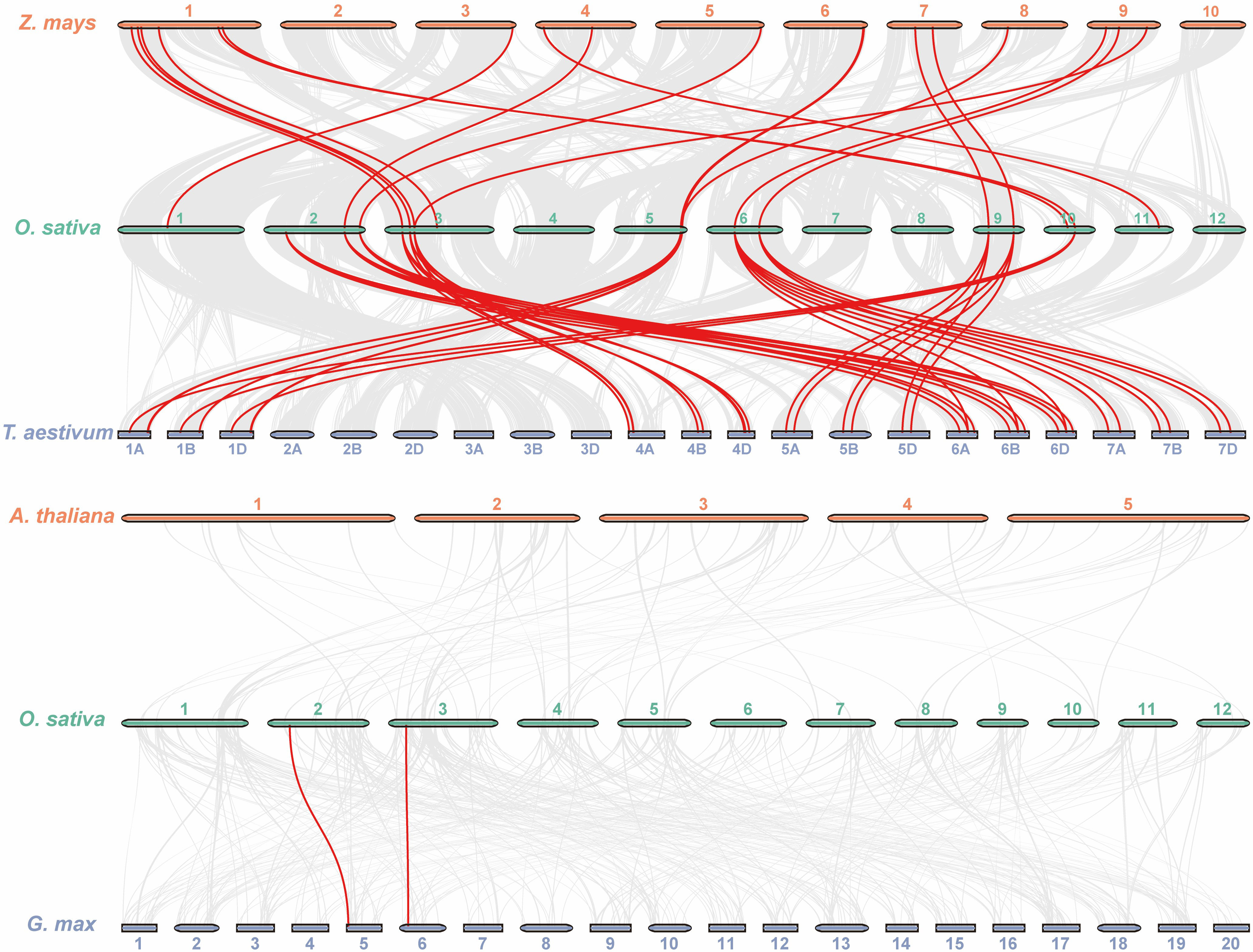
Figure 5 Synteny analysis of KCS genes between rice and four representative plant species. The red lines highlight the syntenic KCS gene pairs; the gray lines in the background represent the collinear blocks in rice that are orthologous to the other plant genomes.
Expression characteristics of OsKCS
To investigate the rice KCS genes expression patterns in different tissues of rice plants, we analyzed rice transcript expression (RNA-seq data) in four different tissues; this included the expression in the root, leaf, panicle, and mature seed (Supplementary Figure S1). In order to further investigate the functions of OsKCS genes involved in different developing stages, qRT-PCR was performed on all KCS gene members (Figure 6). These results indicated that the predictions of expression profiles of most KCS genes were consistent with the qPCR data. KCS genes were specifically expressed in leaves (two genes), stem (six genes), panicle (three genes), and seeds (11 genes), respectively, and most of the KCS genes (OsKCS4, OsKCS6, OsKCS9, OsKCS13, OsKCS16, OsKCS17, and OsKCS22) expressed in seeds were highly expressed in the early stage of growth and development, suggesting that these OsKCS genes played distinct roles during the development of grain.
Expression patterns of OsKCS under cadmium stress based on RNA-seq
Based on the analysis of cis-elements in the promoter of OsKCS genes, all of these genes were hypothesized to respond to stress. Cadmium (Cd) is considered as one of the most toxic metals for plant. In order to analyze OsKCS involved in the response to Cd stress, we analyzed the RNA-seq data to evaluate the response of 22 OsKCS to Cd treatment in indica and japonica. All OsKCSs were differently expressed under Cd stress. In japonica Nipponbare background, after 3h treatment, the expression of 12 genes was significantly down-regulated, and the expression of these genes was different between 3h and 6h treatment (Figure 7). For example, OsKCS8, OsKCS13, OsKCS14, and OsKCS17 were up-regulated after 3h treatment, the expression of these genes decreased gradually in 6h but was still higher than 0h (control). In indica 9311 background, 15 genes were up-regulated and seven genes were down-regulated continuously with the extension of treatment, seven genes were down-regulated and two genes were up-regulated under 6h of Cd treatment, 13 genes were up-regulated under 3h of Cd treatment. Whether in indica or japonica, OsKCS2, OsKCS5, OsKCS6, OsKCS8, OsKCS13, OsKCS17, OsKCS18, and OsKCS21 were up-regulated gradually with the extension of treatment time. The expression of OsKCS19 and OsKCS20 was opposite in indica and japonica after Cd treatment. The results showed that most OsKCSs were sensitive to the Cd.
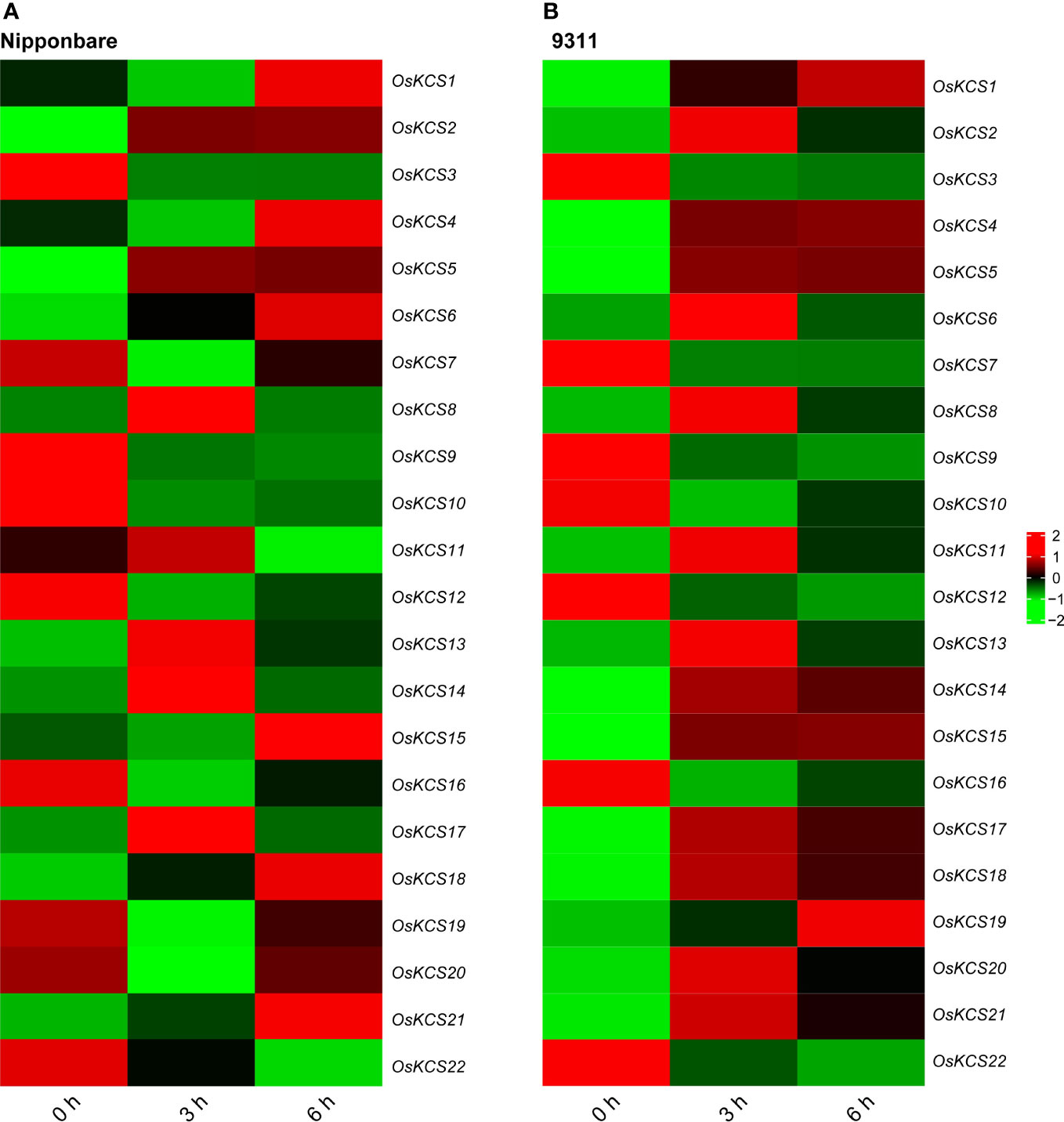
Figure 7 Expression patterns of OsKCS genes in shoots under exogenous cadmium treatment. (A, B) Heatmaps of gene expression levels treated with 20 μM Cd in japonica Nipponbare and indica 9311.
Discussion
The β-ketoacyl-CoA synthase (KCS) family plays an important role in regulating plant growth and development and resisting abiotic stress (De Bigault Du Granrut and Cacas, 2016; Batsale et al., 2021). The identification and analysis of the OsKCS gene family at the genome level could provide a theoretical basis for functional characterization. In the current study, a total of 22 OsKCS genes were identified in the rice genome; the number of identified KCS genes in the rice genome was slightly higher compared with Arabidopsis thaliana (21), but smaller than that of Zea mays (26) and Brassica campestris (46), which may be due to the differences in genome size and the time when the duplication event occurred. The specific domain (FAE1_CUT1_RppA) was conserved in OsKCS proteins and all of KCS genes have this domain (Huai et al., 2020; Tong et al., 2021; Rizwan et al., 2022), indicating that 22 KCS proteins of rice containing the active motif involved in the elongation of VLCFAs.
Using sequence alignment and phylogenetic tree construction, the grouping and evolutionary relationships of the rice KCS gene family were determined. The phylogenetic tree categorized the rice KCS gene family into six subclasses. The grouping and clustering of KCS proteins were caused by differences in protein sequences. Twenty-one KCSs in Arabidopsis thaliana are classified into eight subclasses: α, β, γ, δ, ϵ, ζ, η and θ (Joubès et al., 2008). The KCSs in subclasses α, β, γ, δ and ϵ are known to possess catalytic activity, because they could display activity in various heterologous yeast expression systems (Blacklock and Jaworski, 2006; Paul et al., 2006). However, the KCS3 in Arabidopsis thaliana belongs to subclass θ, and it plays a negative regulator of wax metabolism by reducing the enzymatic activity of KCS6, a key KCS involved in wax production (Huang et al., 2023). KCSs in Oryza sativa were not classified into β and θ; it is possible that most of the KCS in rice have catalytic activity, which needs further experimental verification. There were some differences in the conserved motifs of OsKCS proteins among different subclasses. The differences in the distribution of these conserved motifs indicated the different functions of OsKCS genes. The differences in gene structure might play a role in gene evolution. The intron–exon structure of the 22 OsKCS genes is different, and the differences in structure will lead to different functions (Xu et al., 2012).
Cis-regulatory elements play an essential role in gene’s spatiotemporal expression, and further in regulating plant growth and development, as well as in coordination and adaptation to the environment (Priest et al., 2009). The cis-regulatory element analysis in OsKCS genes was performed and found that MYB transcription factor binding sites existed in the promoter regions of its members. Previous studies have also shown that MYB transcription factors may regulate KCS and further regulate VLCFA and plant function in growth and development (Raffaele et al., 2008; Xu et al., 2022; Yang et al., 2023).
The KCS gene family in plants has formed a huge gene family through duplication and has accumulated many mutations after a long evolution, leading to the differentiation of gene functions. Some studies believed that the KCS genes in plants are the results of large-scale duplication events such as WGDs or segmental chromosomal duplications (Guo et al., 2016). The rice genome did not contain any tandem KCS genes but contained four segmental duplication genes. The large-scale duplication events of KCS genes in rice are smaller than that in Arabidopsis and Passiflora (Rizwan et al., 2022). Therefore, the dominating duplication mode of KCS genes in rice appeared to be WGDs or segmental duplications. The number of orthologous pairs of KCS genes between Grapevine and Arabidopsis is more than that between Oryza and Arabidopsis(Zheng et al., 2023).
Measurements of the adverse effects of heavy metal on plants generally are related with seed germination, root length, and morphologic growth. A study showed that the fatty acid composition (C18) of leaves was also correlated with heavy metals accumulation in soils, and the fatty acid composition of leaves could be used as an indicator of the adverse effects of heavy metals on plants. (Verdoni et al., 2001). Under heavy metal stress, HMA3 overexpressing transgenic plants displayed a higher quantity of fatty acids (C18-20)s in seed (Park et al., 2015). These studies indicated that the fatty acid composition was altered after metal stress. VLCFAs are synthesized in the ER through C18 via FAE complex. The expression levels of eight genes (OsKCS2, OsKCS5, OsKCS6, OsKCS8, OsKCS13, OsKCS17, OsKCS18, and OsKCS21) were gradually up-regulated with the extension of treatment time and were higher in leaves and seeds compared with other tissues. The OsKCS with high expression in leaves and seeds might play a role in cadmium stress.
Conclusions
In this study, a total of 22 OsKCS genes were identified in the rice genome. OsKCS is divided into six subclasses, and the physiochemical features of KCS protein were different. OsKCS gene family has undergone purification selection. The qRT-PCR based expression suggested that OsKCS genes are specifically expressed in different tissues. The KCS genes in indica and japonica with high expression in leaves and seeds might play roles under Cd stress. These findings provide a basis for further studies on the functions of KCS genes in rice. In future studies, we will further explore the role of OsKCS genes in VLCFA.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Author contributions
Conceptualization, LY and SKH; methodology, JF, JW, LY, and SZH; validation, LY, JF, LZ, BX, and YZ; formal analysis, JW, LY and YJC; resources, ZS, XW, YC, FZ, and GS; data curation, GS, LW, and LX; writing—original draft preparation, LY and JF; writing—review and editing, LY, JW, and SKH; supervision, SKH, PH, and ST. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Zhejiang Provincial Science and Technology Project (LDQ23C130001, 2020R51007), the Key Research and Development Program of Zhejiang Province (2022C02011, 2021C02056-1), the National Natural Science Foundation of China (32188102, 32071991, and 31972961).
Acknowledgments
The authors would like to express thanks to everyone who contributed to this work.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2023.1222288/full#supplementary-material
Supplementary Figure 1 | Expression pattern of OsKCS genes in panicle, seed, root, leaf in RNA-seq.
Supplementary Table 1 | Enrichment analysis of OsKCS genes.
References
Bailey, T. L., Boden, M., Buske, F. A., Frith, M., Grant, C. E., Clementi, L., et al. (2009). MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 37 (Web Server issue), W202–W208. doi: 10.1093/nar/gkp335
Batsale, M., Bahammou, D., Fouillen, L., Mongrand, S., Joubès, J., Domergue, F. (2021). Biosynthesis and functions of very-long-chain fatty acids in the responses of plants to abiotic and biotic stresses. Cells 10 (6), 1284. doi: 10.3390/cells10061284
Ben Ammar, W., Nouairi, I., Zarrouk, M., Jemal, F. (2008). The effect of cadmium on lipid and fatty acid biosynthesis in tomato leaves. Biologia 63 (1), 86–93. doi: 10.2478/s11756-008-0002-6
Blacklock, B. J., Jaworski, J. G. (2006). Substrate specificity of Arabidopsis 3-ketoacyl-CoA synthases. Biochem. Biophys. Res. Commun. 346 (2), 583–590. doi: 10.1016/j.bbrc.2006.05.162
Boutte, Y., Jaillais, Y. (2020). Metabolic cellular communications: feedback mechanisms between membrane lipid homeostasis and plant development. Dev. Cell 54 (2), 171–182. doi: 10.1016/j.devcel.2020.05.005
Cahoon, E. B., Li-Beisson, Y. (2020). Plant unusual fatty acids: learning from the less common. Curr. Opin. Plant Biol. 55, 66–73. doi: 10.1016/j.pbi.2020.03.007
Campbell, A. A., Stenback, K. E., Flyckt, K., Hoang, T., Perera, M. A. D., Nikolau, B. J. (2019). A single-cell platform for reconstituting and characterizing fatty acid elongase component enzymes. PloS One 14 (3), e0213620. doi: 10.1371/journal.pone.0213620
Chen, H., Zhang, Z., Ni, E., Lin, J., Peng, G., Huang, J., et al. (2020). HMS1 interacts with HMS1I to regulate very-long-chain fatty acid biosynthesis and the humidity-sensitive genic male sterility in rice (Oryza sativa). New Phytol. 225 (5), 2077–2093. doi: 10.1111/nph.16288
Cheng, Y., Ma, J., Li, S., Tang, Q., Shi, W., Liang, Y., et al. (2023). Dietary cadmium health risk assessment for the Chinese population. Environ. Sci. Pollut. Res. Int. doi: 10.1007/s11356-023-28199-0
Costaglioli, P., Joubes, J., Garcia, C., Stef, M., Arveiler, B., Lessire, R., et al. (2005). Profiling candidate genes involved in wax biosynthesis in Arabidopsis thaliana by microarray analysis. Biochim. Biophys. Acta 1734 (3), 247–258. doi: 10.1016/j.bbalip.2005.04.002
De Bigault Du Granrut, A., Cacas, J. L. (2016). How very-long-chain fatty acids could signal stressful conditions in plants? Front. Plant Sci. 7, 1490. doi: 10.3389/fpls.2016.01490
Gan, L., Zhu, S., Zhao, Z., Liu, L., Wang, X., Zhang, Z., et al. (2017). Wax Crystal-Sparse Leaf 4, encoding a beta-ketoacyl-coenzyme A synthase 6, is involved in rice cuticular wax accumulation. Plant Cell Rep. 36 (10), 1655–1666. doi: 10.1007/s00299-017-2181-5
Guo, H. S., Zhang, Y. M., Sun, X. Q., Li, M. M., Hang, Y. Y., Xue, J. Y. (2016). Evolution of the KCS gene family in plants: the history of gene duplication, sub/neofunctionalization and redundancy. Mol. Genet. Genomics 291 (2), 739–752. doi: 10.1007/s00438-015-1142-3
Hall, B. G. (2013). Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 30 (5), 1229–1235. doi: 10.1093/molbev/mst012
Haslam, T. M., Kunst, L. (2013). Extending the story of very-long-chain fatty acid elongation. Plant Sci. 210, 93–107. doi: 10.1016/j.plantsci.2013.05.008
Huai, D., Xue, X., Li, Y., Wang, P., Li, J., Yan, L., et al. (2020). Genome-wide identification of peanut KCS genes reveals that AhKCS1 and AhKCS28 are involved in regulating VLCFA contents in seeds. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00406
Huang, M., Tang, Q.-Y., Ao, H.-J., Zou, Y.-B. (2017). Yield potential and stability in super hybrid rice and its production strategies. J. Integr. Agric. 16 (5), 1009–1017. doi: 10.1016/s2095-3119(16)61535-6
Huang, H., Yang, X., Zheng, M., Chen, Z., Yang, Z., Wu, P., et al. (2023). An ancestral role for 3-KETOACYL-COA SYNTHASE3 as a negative regulator of plant cuticular wax synthesis. Plant Cell 35 (6), 2251–2270. doi: 10.1093/plcell/koad051
Hurst, L. D. (2002). The Ka/Ks ratio: diagnosing the form of sequence evolution. Trends Genet. 18 (9), 486. doi: 10.1016/s0168-9525(02)02722-1
James, D. W., Jr., Lim, E., Keller, J., Plooy, I., Ralston, E., Dooner, H. K. (1995). Directed tagging of the Arabidopsis FATTY ACID ELONGATION1 (FAE1) gene with the maize transposon activator. Plant Cell 7 (3), 309–319. doi: 10.1105/tpc.7.3.309
Joubès, J., Raffaele, S., Bourdenx, B., Garcia, C., Laroche-Traineau, J., Moreau, P., et al. (2008). The VLCFA elongase gene family in Arabidopsis thaliana: phylogenetic analysis, 3D modelling and expression profiling. Plant Mol. Biol. 67 (5), 547–566. doi: 10.1007/s11103-008-9339-z
Kim, J., Jung, J. H., Lee, S. B., Go, Y. S., Kim, H. J., Cahoon, R., et al. (2013). Arabidopsis 3-ketoacyl-coenzyme a synthase9 is involved in the synthesis of tetracosanoic acids as precursors of cuticular waxes, suberins, sphingolipids, and phospholipids. Plant Physiol. 162 (2), 567–580. doi: 10.1104/pp.112.210450
Krzywinski, M., Schein, J., Birol, I., Connors, J., Gascoyne, R., Horsman, D., et al. (2009). Circos: an information aesthetic for comparative genomics. Genome Res. 19 (9), 1639–1645. doi: 10.1101/gr.092759.109
Kunst, L., Samuels, A. L. (2003). Biosynthesis and secretion of plant cuticular wax. Prog. Lipid Res. 42 (1), 51–80. doi: 10.1016/s0163-7827(02)00045-0
Lee, S. B., Jung, S. J., Go, Y. S., Kim, H. U., Kim, J. K., Cho, H. J., et al. (2009). Two Arabidopsis 3-ketoacyl CoA synthase genes, KCS20 and KCS2/DAISY, are functionally redundant in cuticular wax and root suberin biosynthesis, but differentially controlled by osmotic stress. Plant J. 60 (3), 462–475. doi: 10.1111/j.1365-313X.2009.03973.x
Leonard, A. E., Pereira, S. L., Sprecher, H., Huang, Y. S. (2004). Elongation of long-chain fatty acids. Prog. Lipid Res. 43 (1), 36–54. doi: 10.1016/s0163-7827(03)00040-7
Lescot, M., Déhais, P., Thijs, G., Marchal, K., Moreau, Y., Van de Peer, Y., et al. (2002). PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 30 (1), 325–327. doi: 10.1093/nar/30.1.325
Letunic, I., Doerks, T., Bork, P. (2012). SMART 7: recent updates to the protein domain annotation resource. Nucleic Acids Res. 40 (Database issue), D302–D305. doi: 10.1093/nar/gkr931
Li, C., Haslam, T. M., Kruger, A., Schneider, L. M., Mishina, K., Samuels, L., et al. (2018). The beta-Ketoacyl-CoA synthase HvKCS1, encoded by Cer-zh, plays a key role in synthesis of barley leaf wax and germination of barley powdery mildew. Plant Cell Physiol. 59 (4), 806–822. doi: 10.1093/pcp/pcy020
Liu, Q., Lu, W., Bai, C., Xu, C., Ye, M., Zhu, Y., et al. (2023). Cadmium, arsenic, and mineral nutrients in rice and potential risks for human health in South China. Environ. Sci. Pollut. Res. Int. 30 (31), 76842–76852. doi: 10.1007/s11356-023-27857-7
Luzarowska, U., Russ, A. K., Joubes, J., Batsale, M., Szymanski, J., Thirumalaikumar, V. P., et al. (2023). Hello darkness, my old friend: 3-KETOACYL-COENZYME A SYNTHASE4 is a branch point in the regulation of triacylglycerol synthesis in Arabidopsis thaliana. Plant Cell 35 (6), 1984–2005. doi: 10.1093/plcell/koad059
Millar, A. A., Clemens, S., Zachgo, S., Giblin, E. M., Taylor, D. C., Kunst, L. (1999). CUT1, an Arabidopsis gene required for cuticular wax biosynthesis and pollen fertility, encodes a very-long-chain fatty acid condensing enzyme. Plant Cell 11 (5), 825–838. doi: 10.1105/tpc.11.5.825
Park, W., Feng, Y., Kim, H., Suh, M. C., Ahn, S. J. (2015). Changes in fatty acid content and composition between wild type and CsHMA3 overexpressing Camelina sativa under heavy-metal stress. Plant Cell Rep. 34 (9), 1489–1498. doi: 10.1007/s00299-015-1801-1
Paul, S., Gable, K., Beaudoin, F., Cahoon, E., Jaworski, J., Napier, J. A., et al. (2006). Members of the Arabidopsis FAE1-like 3-ketoacyl-CoA synthase gene family substitute for the Elop proteins of Saccharomyces cerevisiae. J. Biol. Chem. 281 (14), 9018–9029. doi: 10.1074/jbc.M507723200
Priest, H. D., Filichkin, S. A., Mockler, T. C. (2009). Cis-regulatory elements in plant cell signaling. Curr. Opin. Plant Biol. 12 (5), 643–649. doi: 10.1016/j.pbi.2009.07.016
Quraishi, U. M., Abrouk, M., Murat, F., Pont, C., Foucrier, S., Desmaizieres, G., et al. (2011). Cross-genome map based dissection of a nitrogen use efficiency ortho-metaQTL in bread wheat unravels concerted cereal genome evolution. Plant J. 65 (5), 745–756. doi: 10.1111/j.1365-313X.2010.04461.x
Raffaele, S., Vailleau, F., Leger, A., Joubes, J., Miersch, O., Huard, C., et al. (2008). A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20 (3), 752–767. doi: 10.1105/tpc.107.054858
Rizwan, H. M., Shaozhong, F., Li, X., Bilal Arshad, M., Yousef, A. F., Chenglong, Y., et al. (2022). Genome-Wide Identification and Expression Profiling of KCS Gene Family in Passion Fruit (Passiflora edulis) Under Fusarium kyushuense and Drought Stress Conditions. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.872263
Scott, S., Cahoon, E. B., Busta, L. (2022). Variation on a theme: the structures and biosynthesis of specialized fatty acid natural products in plants. Plant J. 111 (4), 954–965. doi: 10.1111/tpj.15878
Stenback, K. E., Flyckt, K. S., Hoang, T., Campbell, A. A., Nikolau, B. J. (2022). Modifying the yeast very long chain fatty acid biosynthetic machinery by the expression of plant 3-ketoacyl CoA synthase isozymes. Sci. Rep. 12 (1), 13235. doi: 10.1038/s41598-022-17080-8
Tong, T., Fang, Y.-X., Zhang, Z., Zheng, J., Zhang, X., Li, J., et al. (2021). Genome-wide identification and expression pattern analysis of the KCS gene family in barley. Plant Growth Regul. 93 (1), 89–103. doi: 10.1007/s10725-020-00668-3
Tsuda, K., Akiba, T., Kimura, F., Ishibashi, M., Moriya, C., Nakagawa, K., et al. (2013). ONION2 fatty acid elongase is required for shoot development in rice. Plant Cell Physiol. 54 (2), 209–217. doi: 10.1093/pcp/pcs169
Verdoni, N., Mench, M., Cassagne, C., Bessoule, J. J. (2001). Fatty acid composition of tomato leaves as biomarkers of metal-contaminated soils. Environ. Toxicol. Chem. 20 (2), 382–388. doi: 10.1002/etc.5620200220
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40 (7), e49. doi: 10.1093/nar/gkr1293
Xiao, G. H., Wang, K., Huang, G., Zhu, Y. X. (2016). Genome-scale analysis of the cotton KCS gene family revealed a binary mode of action for gibberellin A regulated fiber growth. J. Integr. Plant Biol. 58 (6), 577–589. doi: 10.1111/jipb.12429
Xu, G., Guo, C., Shan, H., Kong, H. (2012). Divergence of duplicate genes in exon-intron structure. Proc. Natl. Acad. Sci. USA 109 (4), 1187–1192. doi: 10.1073/pnas.1109047109
Xu, X., Li, M., Zou, J. X., Zheng, Y. S., Li, D. D. (2022). EgMYB108 regulates very long-chain fatty acid (VLCFA) anabolism in the mesocarp of oil palm. Plant Cell Rep. 41 (6), 1449–1460. doi: 10.1007/s00299-022-02868-9
Yang, H., Zhang, M., Li, X., Zhu, Z., Xu, R., Zhu, F., et al. (2023). CsERF003, CsMYB7 and CsMYB102 promote cuticular wax accumulation by upregulating CsKCS2 at fruit ripening in Citrus sinensis. Scientia Hortic. 310, 111744. doi: 10.1016/j.scienta.2022.111744
Yu, D., Ranathunge, K., Huang, H., Pei, Z., Franke, R., Schreiber, L., et al. (2008). Wax Crystal-Sparse Leaf1 encodes a beta-ketoacyl CoA synthase involved in biosynthesis of cuticular waxes on rice leaf. Planta 228 (4), 675–685. doi: 10.1007/s00425-008-0770-9
Zemanova, V., Pavlik, M., Kyjakova, P., Pavlikova, D. (2015). Fatty acid profiles of ecotypes of hyperaccumulator Noccaea caerulescens growing under cadmium stress. J. Plant Physiol. 180, 27–34. doi: 10.1016/j.jplph.2015.02.012
Zhang, Z., Li, J., Zhao, X. Q., Wang, J., Wong, G. K., Yu, J. (2006). KaKs_Calculator: calculating Ka and Ks through model selection and model averaging. Genomics Proteomics Bioinf. 4 (4), 259–263. doi: 10.1016/S1672-0229(07)60007-2
Zhang, X., Wang, Y., Wang, X., Zhu, Z., Zhang, X., Jia, L., et al. (2022). A very-long-chain fatty acids synthesis gene, SD38, influences plant height by activating ethylene biosynthesis in rice. Plant J. 112 (4), 1084–1097. doi: 10.1111/tpj.15998
Zhang, Z., Xiao, J., Wu, J., Zhang, H., Liu, G., Wang, X., et al. (2012). ParaAT: a parallel tool for constructing multiple protein-coding DNA alignments. Biochem. Biophys. Res. Commun. 419 (4), 779–781. doi: 10.1016/j.bbrc.2012.02.101
Zheng, H., Liang, Y., Hong, B., Xu, Y., Ren, M., Wang, Y., et al. (2023). Genome-scale analysis of the grapevine KCS genes reveals its potential role in male sterility. Int. J. Mol. Sci. 24 (7), 6510. doi: 10.3390/ijms24076510
Zhou, H., Xia, D., Li, P., Ao, Y., Xu, X., Wan, S., et al. (2021). Genetic architecture and key genes controlling the diversity of oil composition in rice grains. Mol. Plant 14 (3), 456–469. doi: 10.1016/j.molp.2020.12.001
Zhukov, A., Popov, V. (2022). Synthesis of C20-38 fatty acids in plant tissues. Int. J. Mol. Sci. 23 (9), 4731. doi: 10.3390/ijms23094731
Keywords: rice, very long-chain fatty acids, β-ketoacyl-CoA synthase, gene family, cadmium stress
Citation: Yang L, Fang J, Wang J, Hui S, Zhou L, Xu B, Chen Y, Zhang Y, Lai C, Jiao G, Sheng Z, Wei X, Shao G, Xie L, Wang L, Chen Y, Zhao F, Hu S, Hu P and Tang S (2023) Genome-wide identification and expression analysis of 3-ketoacyl-CoA synthase gene family in rice (Oryza sativa L.) under cadmium stress. Front. Plant Sci. 14:1222288. doi: 10.3389/fpls.2023.1222288
Received: 14 May 2023; Accepted: 03 July 2023;
Published: 24 July 2023.
Edited by:
Wensheng Wang, Chinese Academy of Agricultural Sciences, ChinaReviewed by:
Wenguang Wang, Shandong Agricultural University, ChinaWenbang Tang, Hunan Agricultural University, China
Sandip Das, University of Delhi, India
Copyright © 2023 Yang, Fang, Wang, Hui, Zhou, Xu, Chen, Zhang, Lai, Jiao, Sheng, Wei, Shao, Xie, Wang, Chen, Zhao, Hu, Hu and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shikai Hu, aHVzaGlrYWlAY2Fhcy5jbg==; Peisong Hu, aHVwZWlzb25nMThAMTYzLmNvbQ==; Shaoqing Tang, c3F0YW5nQDEyNi5jb20=
†These authors share first authorship
 Lingwei Yang
Lingwei Yang Junchao Fang†
Junchao Fang† Guiai Jiao
Guiai Jiao Zhonghua Sheng
Zhonghua Sheng Xiangjin Wei
Xiangjin Wei Gaoneng Shao
Gaoneng Shao Ling Wang
Ling Wang Fengli Zhao
Fengli Zhao Shikai Hu
Shikai Hu