- 1NORCE Norwegian Research Centre AS, Department of Climate & Environment, Tromsø, Norway
- 2Department of Crop Science, Federal University of Santa Catarina, Florianópolis, Brazil
CRISPR/Cas9-based ribonucleoprotein (RNP)-mediated system has the property of minimizing the effects related to the unwanted introduction of vector DNA and random integration of recombinant DNA. Here, we describe a platform based on the direct delivery of Cas9 RNPs to soybean protoplasts for genetic screens in knockout gene-edited soybean lines without the transfection of DNA vectors. The platform is based on the isolation of soybean protoplasts and delivery of Cas RNP complex. To empirically test our platform, we have chosen a model gene from the soybean genetic toolbox. We have used five different guide RNA (gRNA) sequences that targeted the constitutive pathogen response 5 (CPR5) gene associated with the growth of trichomes in soybean. In addition, efficient protoplast transformation, concentration, and ratio of Cas9 and gRNAs were optimized for soybean for the first time. Targeted mutagenesis insertion and deletion frequency and sequences were analyzed using both Sanger and targeted deep sequencing strategies. We were able to identify different mutation patterns within insertions and deletions (InDels) between + 5 nt and –30 bp and mutation frequency ranging from 4.2 to 18.1% in the GmCPR5 locus. Our results showed that DNA-free delivery of Cas9 complexes to protoplasts is a useful approach to perform early-stage genetic screens and anticipated analysis of Cas9 activity in soybeans.
Introduction
The use of the Cas protein and evolutionary components of type II bacteria were the first CRISPR-associated effectors to be widely used in genetic engineering. For this type of bacteria, defense and survival are conditions that generate cellular responses and activate protein domains (HNH, RuvC, and PI) responsible for recognizing PAM sequences in the DNA of organisms invading the bacteria (in Streptococcus pyogenes: NGG). Cas9 cuts the DNA strand, and the invasion is stopped (Nishimasu et al., 2014).
For gene editing via the CRISPR/Cas9 system, essential components, such as Cas9 endonuclease and crRNA from a sequenced genome, can be easily synthesized. The opportunity for manipulation in laboratory routines is vast, for example, in agricultural improvement where CRISPR/Cas9 makes it possible to more quickly achieve the selection, alteration, and mutagenesis of genetic traits of interest (Makarova et al., 2011; Yu et al., 2016). The system as a biotech tool works by fusing crRNA and tracrRNA (trans-activating crRNA) to create a single “guide RNA” with 20 base pairs that correspond to the target DNA. After the complementary sequence, Cas9 can operate by cutting double-stranded DNA (Jinek et al., 2013; Jiang and Doudna, 2017; Murovec et al., 2018; Manghwar et al., 2020).
After the double-strand break, it is natural for the ends to ligate, at this point occurs the signalization to repair by non-homologous end joining (NHEJ). This pathway can cause error-prone repair which can occur just by adding or deleting DNA nucleotides or interrupting the reading frame of a gene that could affect a protein (Hsu et al., 2013; Osakabe and Osakabe, 2015; Yu et al., 2021; Zhang et al., 2021). On the other side, using a homologous template, it is possible to direct homologous end joining – via homologous-directed repair pathways (HDR); in this case, the model can be used with an endogenous or exogenous template (La Russa and Qi, 2015; Wang et al., 2016).
The CRISPR/Cas9 system has often been delivered with Agrobacterium transformation vectors where the DNA fragment encoding the guide RNA targeting the gene of interest and the endonuclease-coding sequence are usually cloned in a transfer DNA (T-DNA) (Zhang et al., 2019; Liu et al., 2020). For targeted mutagenesis in plant cells, the strategy integrates the T-DNA in the plant genome where it constitutively expresses the CRISPR machinery (Zhang et al., 2019; Dalla Costa et al., 2020; Liu et al., 2020; Rodrigues et al., 2021). Although vectors have been widely used as a delivery tool, their application is often associated with side effects including the off-target cleavage and random insertion of foreign DNA into the genome (Amirkhanov and Stepanov, 2019). Besides that, these delivery strategies can be more exigent in techniques and equipment (Nicolia et al., 2021). Alternatively, the DNA-free RNP (preassembled Cas9-gRNA complex) delivery approach is less complex and can be delivered directly into living cells. In plants, the removal of the cell wall (cellulose, polysaccharides, hemicellulases, and pectins) with enzymatic treatment possibilities the CRISPR delivery by polyethylene glycol (PEG) (Metje-Sprink et al., 2019). This approach was demonstrated as efficient in protoplasts of several plant species including Arabidopsis thaliana, rice (Oryza sativa), lettuce (Lactuca sativa) (Woo et al., 2015), tobacco (Nicotiana tabacum and N. attenuata), petunia (Petunia × hybrida) (Subburaj et al., 2016; Yu et al., 2021), corn (Zea mays) (Sant’Ana et al., 2020), grapevine (Vitis vinifera), apple (Malus × domestica) (Malnoy et al., 2016), wheat (Triticum aestivum) (Liang et al., 2017), tomato (lat. Solanum lycopersicum) (Nicolia et al., 2021), cabbage (Brassica oleracea), Chinese cabbage (Brassica rapa; Murovec et al., 2018), banana (Musa spp.) (Wu et al., 2020), and pepper (Capsicum annum) (Kim et al., 2020).
In soybean, the most recent studies were related, which described an optimized PEG-calcium-mediated transformation method to do transient gene expression into soybean protoplasts (Wu and Hanzawa, 2018). The genome edition uses preassembled binary vectors with CRISPR/Cas9 delivered by PEG into protoplasts (Sun et al., 2015), and the use of type V of CRISPR with Cpf1 endonucleases proteins (LbCpf1 and AsCpf1) and crRNAs preassembly delivered into the protoplast by PEG (Kim et al., 2017, 2020). The CPR5 gene (Glyma_06g14800) that regulates the growth of trichomes was also used for knockout via the CRISPR/Cas9 system successfully, eliminating the gene using somatic embryos transformed by a plasmid with gRNA and preassembled Staphylococcus pyogenes (Spy) Cas9 incorporated into a vector for biolistic transformation (Campbell et al., 2019).
To contribute to genome editing approaches in soybean, we have developed a DNA-free friendly plant genome editing platform where the CRISPR/Cas9 system and PEG are delivered in protoplasts. We use the knowledge since available to target Glyma_06g145800 a single copy gene, without repetitive genome sequence, involved in cell division and endoreduplication in trichomes. The results of targeted deep and Sanger sequencing analysis showed an InDel mutagenesis efficiency of 4.2 to 18.1% for the targeted distinct sites of endogenous Glycine max CPR5 locus (GmCPR5). This approach is useful for generating mutant cell lines in protoplasts without the possibility of backbone integration and for investigations into the complexity and interactions between cellular physiological responses to gene editing by CRISPR/Cas9.
Materials and Methods
Platform Development
This paper describes a platform for genetic screens in gene-edited soybean lines. The focus of the platform is the development of knockout gene-edited cell lines without the transfection of DNA-based vectors. To enable genomic screens and investigations, the platform consists of delivering CRISPR as RNP reagents to protoplasts with the aid of PEG solutions (Figure 1). In addition to establishing a pipeline for editing genes in soybean, the platform also works to improve the effectiveness of CRISPR techniques, enhance delivery methods, and create infrastructure and resources to enable their use on large- and small-scale assays. Importantly, the platform can also be expanded to include other species, other types of molecular profiling, and other CRISPR-delivery methodologies. The platform was developed by two different laboratories which served as a validation for the protocols applied.
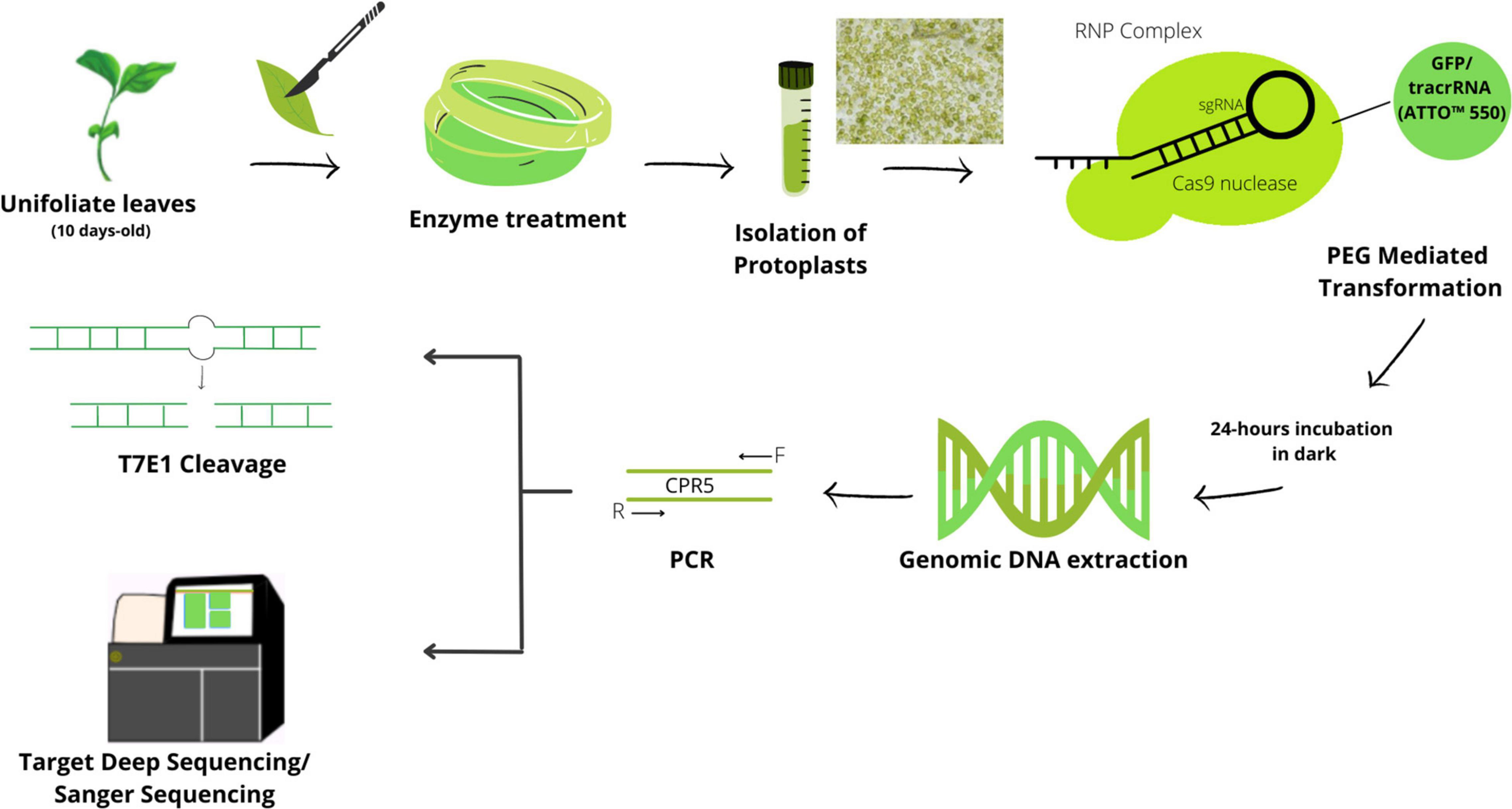
Figure 1. Schematic protocol for CRISPR/Cas9-mediated DNA-free/transient genome editing in the soybean. The protoplasts were isolated from young seedlings of unifoliate leaf strips with enzyme solution. Isolated protoplasts were transformed with preassembled CRISPR-RNP complexes via the PEG-mediated method. The transformed protoplasts were subjected to either genomic DNA extraction or fluorescence-activated cell sorting (FACS) of Cas9-GFP expressing protoplasts for an enrichment followed by DNA extraction or to cultivation. Target sites are amplified by PCR followed by T7E1 validation of mutation and high-throughput sequencing for estimation of mutation efficiency.
Plant Material and Growth Conditions
Seeds of non-transgenic soybean varieties (Glycine max) were sown and grown in soil (composed of peat, vermiculite, organic waste, and limestone) for 10 days in a growth chamber (Enviro Plant®). Growing conditions were standardized to 60% humidity, 25°C temperature, and photoperiod (10/14-h light/dark cycle). Plants were subjected to regular sunlight at a photosynthetic flux of 625 Umol/m2/s–1 and photosynthetic active radiation (PAR) of 1,200 lumens/m2. Unifoliolate green leaves were used for protoplast isolation.
Selection of Target Region and Guide RNA Design
CPR5 gene (Glyma_06g15080; gene ID: 100791857) was selected as a model gene in this study. The CPR5 gene has only a single copy in the soybean genome (NCBI GenBank Glycine_max_v4.0) and a phenotype associated with the growth of trichomes (Campbell et al., 2019). According to available sequence information of CPR5 in the NCBI and SoyBase databases1, a partial CPR5 locus flanking the targeted site was amplified and sequenced by the Sanger platform (ABI3500xl Applied Biosystems) with identical matching results from the Glycine max CPR5 NCBI sequence. In order to design the gRNAs from the gene, four online prediction software, namely CRISPOR (Concordet and Haeussler, 2018), CRISPR RGEN Tools (Bae et al., 2014; Park et al., 2015), CRISPRdirect (Naito et al., 2015), and CHOPCHOP v3 (Labun et al., 2019), were used. Three different target sites corresponding to the exons 1, 2, and 4 of CPR5 locus were chosen which ranked as top among the potential five gRNA targets in most software (Figure 2). For the selection of efficient gRNAs, predicted cleavage efficiency score, high GC% (content between 40 and 70%), high out-of-frame scores or frameshift rate (complete knockout efficiency on-target), and minimum number of mismatches at off-target sites were taken into consideration. The list of designed gRNAs is shown in Supplementary Table 1. In vitro transcription of sgRNAs and synthesis were carried out using the HiScribe™ Quick T7 High Yield RNA Synthesis Kit (New England Biolabs, Ipswich, MA, United States) according to the manufacturer’s instructions.
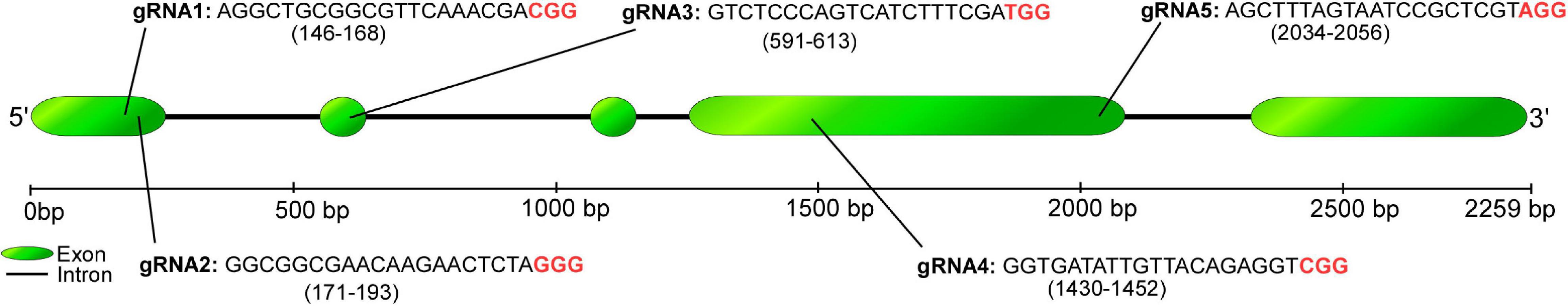
Figure 2. Schematic representation of the GmCPR5 locus and design of gRNAs. The location of target sites was shown by engineered gRNAs 1, 2, 3, 4, and 5 along with their sequences. The PAM motifs are indicated in red.
In vitro Cleavage Assay
Cas9 RNP activity was determined in vitro using 20 μl reactions containing tracrRNA [1 μg/μl], each crRNA [1 μg/μl], and 1.35 μl of duplex buffer for preassembly at 95°C for 5 min. Cas9 [10 μg/μg] and NEB 3 buffer (1×) (Integrated DNA Technologies, Coralville, IA, United States) were then added and incubated at 25°C for 15 min. The reaction was stopped with 0.5 μl of proteinase K (8,000 μ/μl). The target site containing 300 ng of PCR product was amplified (Platinum PCR SuperMix High Fidelity, Invitrogen) using the following cycling conditions: 2 min at 94°C, followed by 30 cycles of 30 s at 94°C, and 1 min at 68°C, and then visualized by running a 2% agarose gel and staining with GelRed® for UV imaging. Primers to amplify the target DNA region were designed by the PrimerQuest tool (Integrated DNA Technologies Inc., Skokie, IL, United States), and their annealing temperature and amplicon sizes are available in Supplementary files (Supplementary Table 2).
Protoplast Isolation
Protoplasts were isolated from 10-day-old grown seedlings. After removing their midribs, 10–12 young “unexpanded” unifoliate leaves were sliced transversely into 0.2–0.4 mm thick slices and placed into 20 ml of enzyme solution containing Viscozyme® L (0.8%) + Celluclast® 1.5L (0.4%) + Pectinex® Ultra SPL (0.4%) mixtures (Sigma-Aldrich, Germany), MES (5 mM), and mannitol (9%) in cell and protoplast washing (CPW) salts (Frearson et al., 1973) at pH 5.8. The enzyme digestion was carried out in a gyratory shaker at 60 rpm for 2, 4, 6, and 8 h in dark conditions. After enzymatic digestion of cell walls, the solution was filtered through a 0.45-μm nylon mesh and the filtrate was further diluted with 10 ml of CPW salts with 9% mannitol (9M CPW) to stop the enzyme reaction. Then, the protoplast cells were pelleted by centrifugation at 100 × g for 5 min, and harvested cells were further washed three times by resuspending them with a 10 ml of 9M CPW, followed by a centrifugation at 100 g for 5 min. Washed cells were resuspended with 1–2 ml of 9M CPW, and 10 μl of this suspension was loaded into the Neubauer cell chamber for calculating the number of protoplasts under a light microscope. The resuspended cells (1–2 ml) were further dispersed in 9–8 ml of 9M CPW solution and rested at 4°C for 1 h before the viability counting and the PEG transfection were conducted. The viability of protoplasts was determined according to a previous study (Adedeji et al., 2020); briefly, a 100 μl of cells was incubated with 2 μl of 0.5% of fluorescein diacetate (FDA) (Sigma-Aldrich, Germany) at 25°C for 5 min and observed under a fluorescence microscope.
Protoplast Transfection With Cas9 Ribonucleoprotein (RNP)
Each transfection experiment consisted of 1 × 106 protoplasts in MMG solution (4 mM MES, 0.4 M Mannitol, and 15 mM MgCl). RNP complexes were composed of 30 μ g gRNA [1 μg/μl] and 10 μ g Cas9 [10 μg/μl] molecules at a 3:1 molar ratio. The reagents were purchased from Integrated DNA Technologies (IDT) (California, United States) and Sigma-Aldrich (Darmstadt, Germany), and the complexes were mixed, following the manufacturer’s instructions. The complex was then mixed with protoplast suspensions followed by an equal volume of freshly prepared PEG solution (40% PEG-4000, 0.4 M mannitol, and 0.1 M CaNO3) which was added and mixed by gentle shaking. Cells were incubated at room temperature in the dark for 20 min. The transfection was induced at 25°C for 23 min in darkness and stopped by a gradual dilution and gentle mixing of the reaction content with ascending series of 9M CPW (0.6, 1.2, 2.4, and 4.8 ml) over a 30-s period at 2-min intervals. Transfected protoplasts were centrifuged at 100 × g for 7 min, and the pellet was retained. Then, the protoplast pellets were resuspended in 1 ml of KP8 liquid medium (Kao, 1977) containing 3% sucrose and 9% mannitol at pH of 5.8 and incubated at 25°C in darkness for 16–24 h prior to DNA extraction. For microscopic fluorescent analysis, protoplasts were transfected with either GPF-tagged Cas9 (Sigma-Aldrich, Germany) or fluorescently labeled tracrRNA–ATTO™ 550 (IDT, United States).
Mutation Detection and Sequencing
Genomic DNA was extracted from protoplast transfectants after 24 h using Plant DNAzol™ Reagent (Invitrogen Co., Carlsbad, CA, United States) according to the manufacturer’s instructions. CRISPR-targeted sites in GmCPR5 loci were amplified from genomic DNA using designed primers as described in vitro cleavage assay. Amplified PCR products were subjected to T7 endonuclease I (T7E1) assay after denaturation and re-anneal process. The T7EI assay was performed as per the manufacturer’s instructions for the Alt-R® Genome Editing Detection Kit (Integrated DNA Technologies, Coralville, IA, United States). T7E1-digested PCR products were resolved on a 2% agarose gel. PCR products were further analyzed by targeted deep sequencing using the Illumina NovaSeq™ 6000 platform at Novogene Europe (Cambridge Science Park, United Kingdom). Mutation patterns at cleavage sites were analyzed by the Cas-Analyzer program in CRISPR RGEN Tools2 and calculated according to the previous study (Subburaj et al., 2016). Sanger sequencing was performed with purified PCR product (1 ng/μl) using ExoSAP-IT Express (Thermo Fisher Scientific) following the manufacturer’s instructions: forward primer (3.2 μM), 1 μl of BigDye Terminator v3, 4 μl sequencing buffer, and 13 μl water. Sanger was performed (3500×, Genetic Analyzer, Applied Biosystems, São Paulo, Brazil). To determine and characterize the types of insertions and deletions (InDels) at the target location, DECODR (Bloh et al., 2021) was used. Protoplasts edited with CRISPR/Cas9 were compared to their negative controls and also sequenced.
Microscopic Analyses
Bright-field images of isolated protoplasts were captured using phase contrast on a Zeiss Primovert compact inverted microscope. Fluorescence images were obtained with the confocal laser scanning microscope Zeiss LSM800 using a 488-nm diode laser for green fluorescent protein (GFP)-labeled and FDA-stained cells. For transmitted light detection images, the electronically switchable illumination and detection module (ESID) was used.
Results
Protoplast Isolation and Transfection
In this study, we used 10-day-old unifoliate leaves for protoplast isolation and further cell editing (Figures 3A–F). By using the 1 × VCP enzyme solution, we obtained a maximum yield of 1.5 × 106 cells after 4 h of incubation, in which 77% of cells were alive during the viability test using FDA. The results obtained suggest optimized conditions for fast isolation of protoplast cells that can be used for the study of knocked out gene regulation. The visualization of RNP complex internalization into cells was verified using ATTO-labeled TracrRNA and GFP-labeled Cas9 (Figures 4A–F). Internalization efficiency was calculated with an unsupervised eye to approximately 38% after 18 h of incubation.
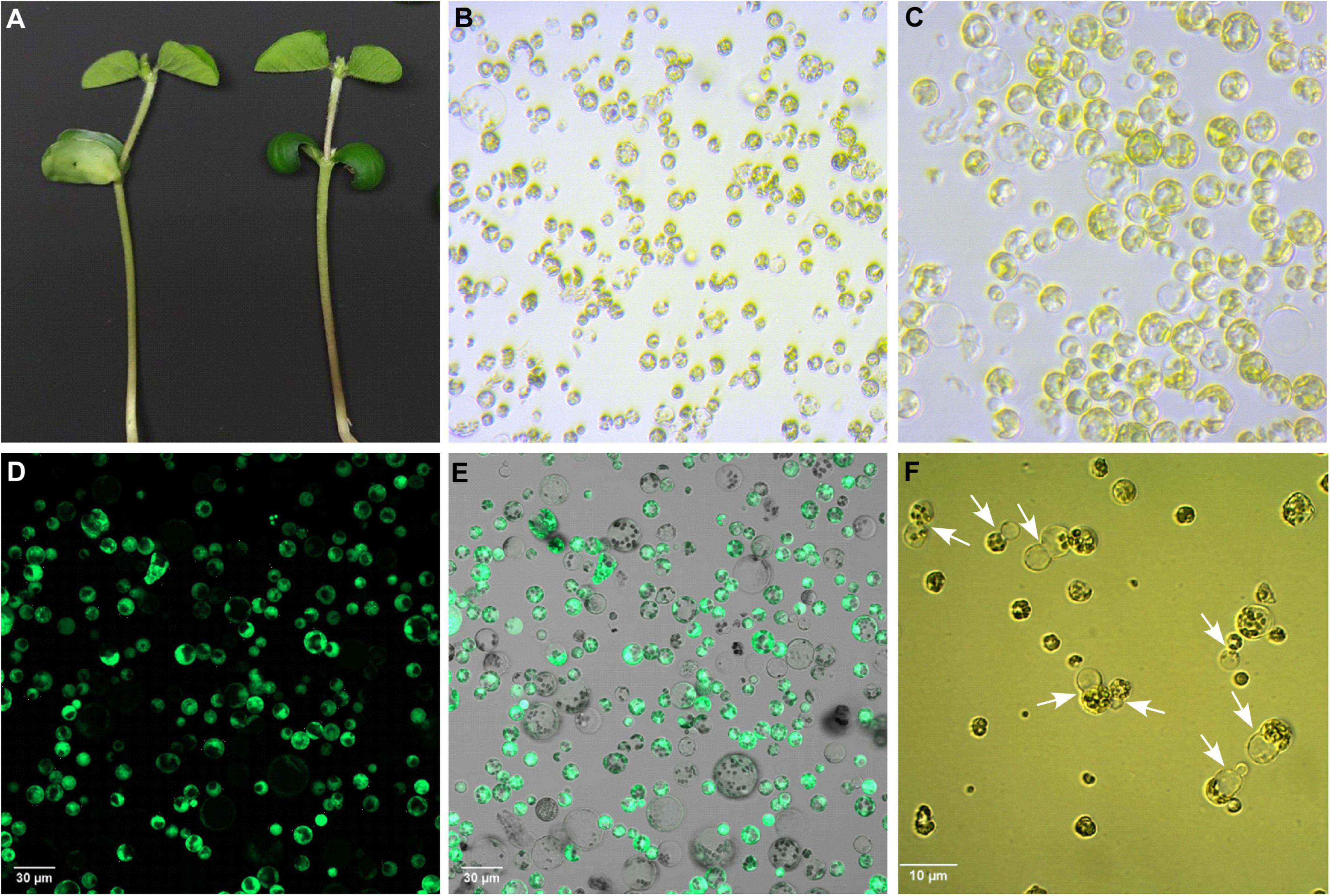
Figure 3. Isolation, purification, and cultivation of protoplasts from Glycine max cv. OAC Bayfield. (A) Unifoliate leaves of 10-day-old soybean seedlings. (B) Freshly isolated protoplasts. (C) Washed and purified protoplasts. (D) FDA-stained protoplasts subjected to confocal fluorescence microscopy to visualize viable cells (GFP-positive). (E) Merged image of green channel (GFP) and ESID channel (transmitted light detection) showing all protoplasts. (F) Protoplasts undergoing cell division (indicated by white arrows) in culture medium after 3 days of isolation.
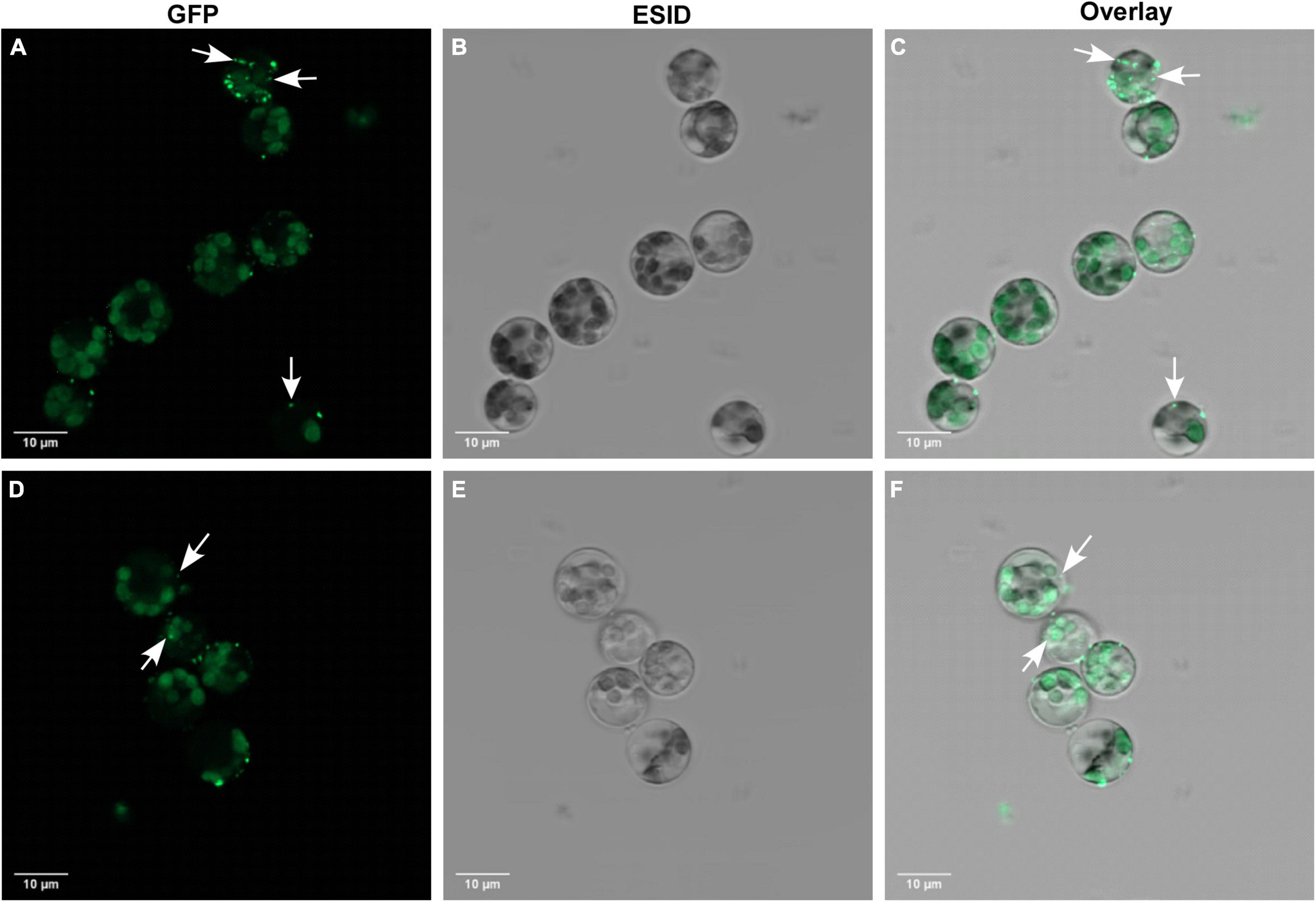
Figure 4. Cellular localization of GFP-Cas9 RNP complexes in transfected protoplasts from Glycine max cv Bayfield. (A,D) Confocal fluorescence microscopy showing GFP-Cas9 located inside transfected protoplasts using the green channel. White arrows indicate internalized localization of GFP-Cas9. (B,E) The same protoplasts as depicted in (A,D) showing bright-field images using the ESID channel. (C,F) Are overlay images of green and ESID channels. White arrows indicate internalized localization of GFP-Cas9.
In vitro Cleavage of Soybean Genomic Target Sites
Three partial genomic regions in the CPR5 loci with flanking exons 1, 2, and 4 were analyzed and confirmed by Sanger sequencing (NCBI accessions: OK631878, OK631879, and OK631880), and subsequently, five sgRNAs were designed. Each of the sgRNAs is 20 nucleotides in length, and they pair with their corresponding 20 nucleotides at target sites in GmCPR5 locus to aid CRISPR/Cas9 system to make site-specific DSBs. To determine the specificity of the CRISPR-RNP complexes (recombinant Cas9 + in vitro transcribed gRNAs), in vitro cleavage assay was performed. The 657 bp PCR amplicon of GmCPR5 for gRNA1 was cleaved into ∼ 397and ∼ 260 bp as expected. For gRNA2, digestion of 931 bp PCR product generated two fragments of ∼ 600 and ∼ 331 bp. Likewise, cleaved fragments of 522 and 138 bp in gRNA3 (660 bp), 763 and 190 bp in gRNA4 (953 bp), and 439 and 47 bp in gRNA5 (486 bp) were noted in the cleavage assay (Figure 5 and Supplementary Table 2). Our results show that the designed sgRNAs were able to efficiently cleave at their corresponding target regions of CPR5 (Figure 5).
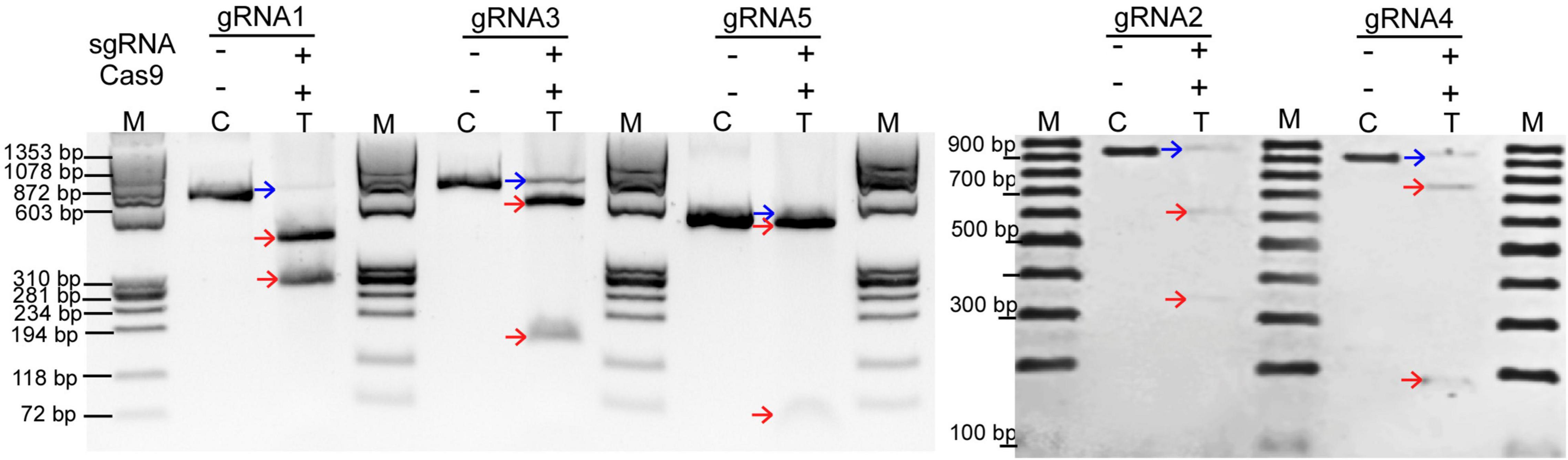
Figure 5. In vitro cleavage assay. The in vitro transcribed or purchased sgRNAs at GmCPR5 loci were mixed with SpCas9 and PCR templates of target sites (gRNA1–5) for in vitro digestion and resolved on 2% agarose gel. Lanes M, DNA ladders; C, PCR wild type (control untreated); T, treated with sgRNAs and SpCas9. The parental and cleaved fragments are indicated with blue and red arrows, respectively.
Targeted Mutagenesis of Soybean Using CRISPR/Cas9 Ribonucleoproteins
Ribonucleoprotein complexes (Cas9 protein + gRNA) were transfected into the soybean protoplasts using the above-mentioned protoplast transformation system to make site-directed mutations in GmCPR5. After 24 h of transfection, the genomic DNA was extracted from control and transfectant protoplasts for mutation T7E1 assay. T7E1 digestion assay showed the appearance of cleaved DNA products for all the sgRNA-transfected samples (Figure 6). This confirmed that there were induced InDel mutations at the corresponding targeted sites within the GmCPR5 locus, whereas in the negative controls, WT and Cas9, no cleavages were detected.
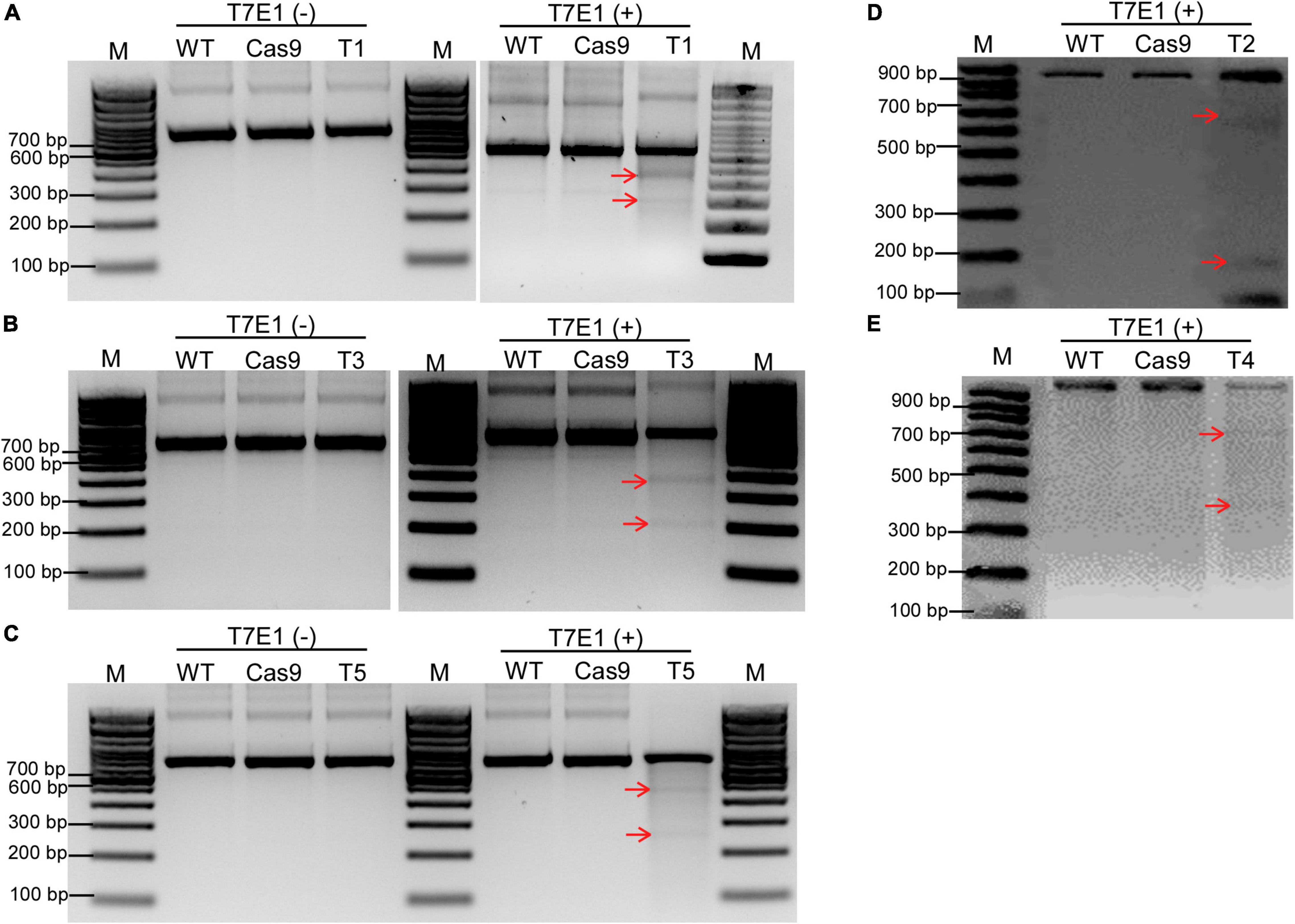
Figure 6. Detection of site-directed mutagenesis at target sites on GmCPR5 loci using direct delivery of RGEN RNPs. (A–E) T7E1 digestion resulting gel images for the transformants of T1–T5. Lanes M, DNA ladder; WT, untransformed wild type (control) to each target site; Cas9, transformed with SpCas9 only; T1–T5, transformed with RNPs; T3T7E1(–), negative control (undigested). Cleaved fragments are indicated with red arrows. Images 6E and 6F have been processed for better visualization of bands. This process is in accordance with Frontiers Policies and Publication Ethics Guidelines.
Mutation Range Characterization in Soybean Edited Cell Lines
In order to calculate the mutation frequency and characterize the site-targeted mutation patterns in the GmCPR5 gene locus, we performed the analysis of sequencing results obtained from targeted deep sequencing (for gRNAs 1, 3, and 5) and also Sanger sequencing (for gRNAs 2 and 4) for the genomic DNA from targeted protoplast transformants. The designed nested PCR primers and their corresponding amplicons from control and protoplast transformants are provided in supplementary files (Supplementary Table 2 and Supplementary Figure 1). The obtained raw sequencing data for RNP transformants of T1, T3, and T5 are available under accession number PRJNA785774 at the National Center for Biotechnology Information (NCBI) BioProject. The targeted deep sequencing results showed that the T1, T3, and T5 RNP transformants were found to possess a range of mutations, such as InDels, in their corresponding target site, whereas no significant mutations were observed in wild-type, negative control samples (Table 1). Some of the RNPs produced an equal proportion of insertions and deletions (T1) or either only insertions (T3) or deletions (T5) with higher frequencies. Based on the number of insertions and deletions for each RNPs, the calculated ratio of deletion to insertion was found as about 49.2:50.8 in the three target sites. Furthermore, by analyzing the total number of mutated sequences in contrast to the total number of obtained reads, the results showed that the three different gRNAs (gRNA1, 3, and 5) generated mutation frequencies ranging from 5.5 to 18.1% with an average mutation frequency of 12.9 ± 3.1% in the GmCPR5 locus.
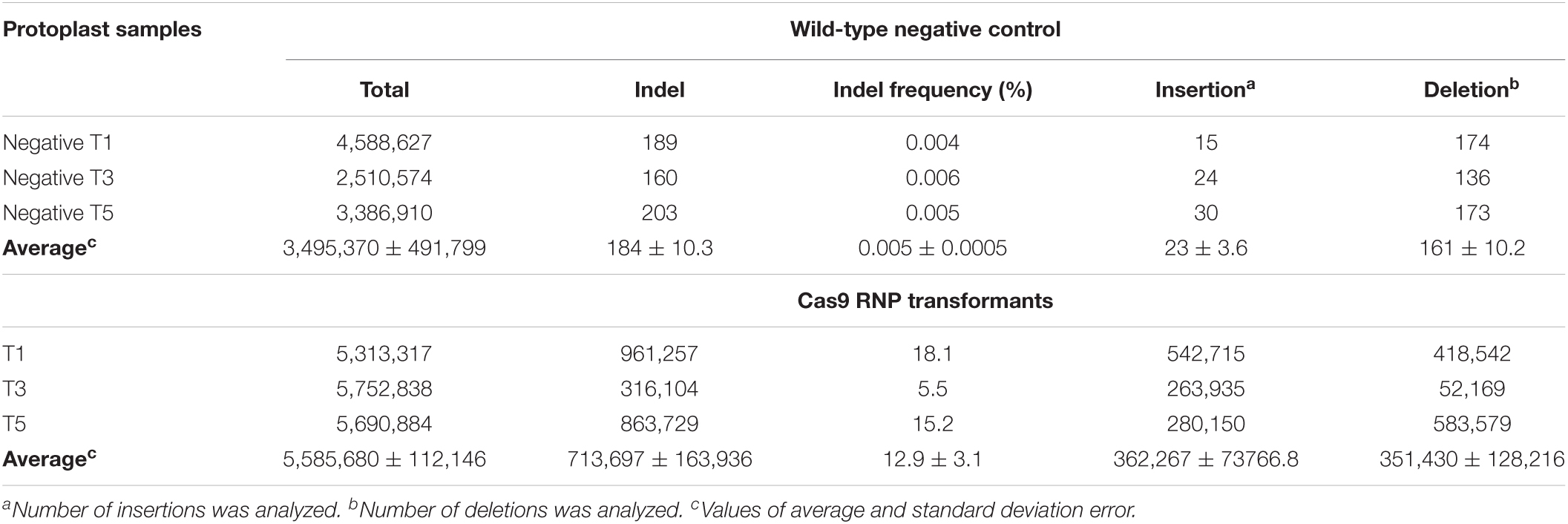
Table 1. Estimation of mutation rate in GmCPR5 gene sequences in wild-type non-transformed and transformed protoplasts by targeted deep sequencing in soybean protoplasts using direct delivery of RNP’s.
The targeted deep sequencing results from T1, T3, and T5 transformants showed a range of mutational profiles of which we selected five. The five most contributing mutation sequencing patterns (highest frequencies) are presented in Figure 7A. These five most frequent alleles themselves contributed 11, 4.2, and 11.1% of the total mutation rates for T1, T3, and T5, respectively (Figure 7A and Table 1). It was observed that the targeted sites were mutated with InDels ranging from + 1 to –6 nt in length, in which T5 produced a maximum of –6 bp deletion. Interestingly, we have the insertion of an adenine as one of the most frequent outcomes in gRNAs 1 (7.1% with 382020 reads) and 5 (4.2% with 236764 reads) analyzed by target deep sequencing (Figure 7A). In addition, all mutant GmCPR5 alleles derived from T1, T3, and T5 were compared with wild-type GmCPR5 for further characterization. The results indicated that these mutations were frameshift types, which would produce in-frame premature stop codons at mRNA and cause loss of function in GmCPR5 alleles.
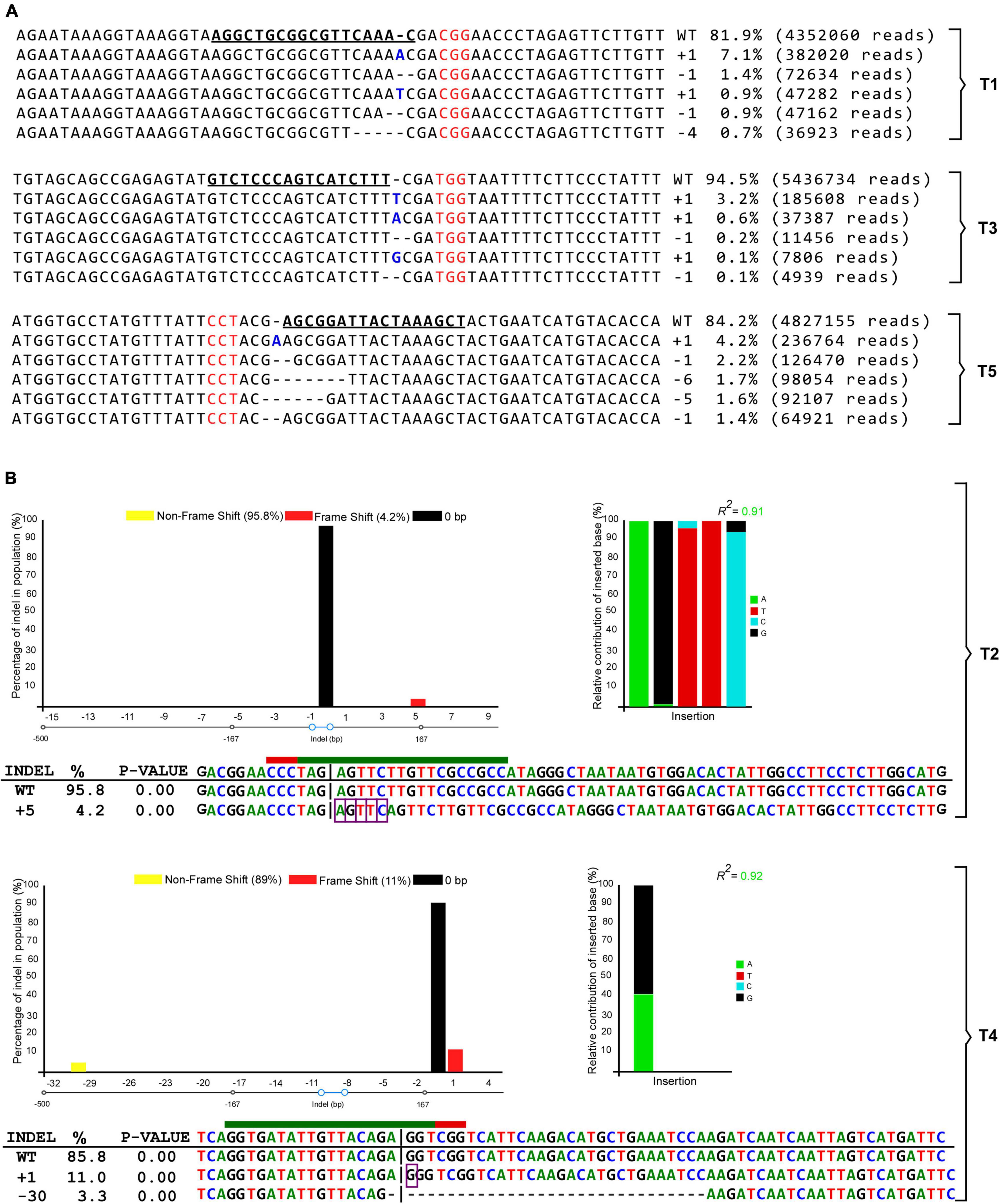
Figure 7. Mutation patterns observed by targeted deep and Sanger sequencing for the corresponding target sites at GmCPR5 loci are shown. (A) Distribution of the five most frequent alleles along with their mutation pattern, contribution percentage, and read count observed with the Cas-Analyzer around targeted sites in GmCPR5 for T1, T3, and T5. Wild-type (WT) nuclease target sequences were in bold and underlined. PAM sites are denoted by the red font. Insertions and deletions are shown in bold font (blue) and horizontal dashed lines, respectively. (B) The result of editing efficiency and mutation patterns analyzed with DECODR software for T2 and T4. The top panels display the graphs for the InDel distribution rate. The bottom panel shows the list of deconvoluted InDel-containing sequences as alignments along with InDel types and percentages (%). Insertion (highlighted with purple rectangles) and deletion (marked with horizontal dashed lines) of mutations are shown in alignments. A 20 bp target and 3 bp PAM site are depicted with green and red lines, respectively.
To assess the CRISPR-induced mutations at targeted sites of T2 and T4 transformants, we have PCR-amplified and Sanger-sequenced each of the targeted sites. The obtained chromatogram files from Sanger sequencing are provided in Supplementary File 1. The corresponding sequencing chromatograms were analyzed by DECODR online CRISPR analysis software. Our Sanger sequencing results showed that RNP transformants of T2 and T4 samples had an InDel rate of 4.2 and 14.3%, respectively (Figure 7B). Interestingly, the sequencing analysis of T2 showed the highest contributing mutational profile of + 5 bp (AGTTC) insertion (4.2%) at the targeted site (Figure 6B), whereas the T4 had the highest contributing sequence variant of + 1 bp (G) insertion (11%) followed by a −30 bp (GGTCGGTCATTCAAGACATGCTGAAATCC) deletion (3.3%) sequence contribution. Upon characterizing the mutant alleles, + 5 bp (T2) and + 1 bp (T4) were identified as frameshift types that would affect the reading frame and cause complete loss of functions in GmCPR5, whereas −30 bp (T4) were found to be a mutant with in-frame deletion in the target site which could alter or partially eliminate protein function of GmCPR5 (Figure 7B).
Overall, the results of both targeted deep and Sanger sequencing analysis demonstrate that the five different targeted distinct sites (T1–5) had mutation frequencies ranging from 4.2 to 18.1% in the GmCPR5 locus (Table 1 and Figure 7). Among all the RNP-induced mutations, + 1 insertions at the fourth nucleotide upstream of the PAM sites were prevalent and most frequently observed in all the targeted sites except for T2 (Figure 7).
Discussion
DNA-Free Platform Performance in Soybean
Soybean methods, already known, have been used for the edition by CRISPR including the construction of binary vectors with the insertion of T-DNA (Chen et al., 2021), biolistic bombardment (Sun et al., 2015), and electroporation treatments (Liu et al., 2019a). Here, we report for the first time that CRISPR/Cas9 was used in the DNA-free target mutagenesis of soybean material by RNP delivery into protoplasts without the application of any DNA vectors (Figures 1–6). The results obtained from targeted deep and Sanger sequencing analysis revealed that at five target sites (T1-5), we successfully mutated the GmCPR5 locus at frequencies ranging from 4.2 to 18.1% (Table 1 and Figure 7). The mutation frequency observed in our study is similar to frequencies observed by other DNA-containing gene-edited platforms for soybean. The vector Agrobacterium tumefaciens carrying CRISPR/Cas9 and gRNAs was utilized to knockout the male-sterile gene (GmAMS1) in soybean cotyledons; in this case, the mutation frequency was 25% (Chen et al., 2021). In another study, the same vector, with gRNA/Cas9, was applied to get knockout the soybean flowering gene (GmFT2a), and the mutations obtained were between 12.1 and 15.8% (Cai et al., 2018). Knockout obtained in genes related to fatty acid (FAD2-2 omega-6) by CRISPR/Cas9-mediated in the binary vector Agrobacterium tumefaciens produced 21% maximum mutation efficiency (Amin et al., 2019). Using the CRISPR-Cas9 system in the Agrobacterium rhizogenes mediated into soybean protoplasts using PEG, the mutation frequencies obtained in the three gene targets (Glyma06g14180, Glyma08g02290, and Glyma12g37050) ranged from 14.7 to 20.2% (Sun et al., 2015). The Agrobacterium transformation targeting the glucosyltransferase soybean gene (Glyma07g14530) achieved a maximum mutation efficiency of 21%, while in events in which transformation was performed by biolistic, the frequency was only 12.5% (Jacobs et al., 2015). Our review of the literature showed that gene editing in soybean, regardless of the techniques applied, did not reach efficiency greater than 25% and these were quite variable depending on the target gene sequence, the sgRNA sequence, and material/tissue type (Tsai et al., 2015; Zischewski et al., 2017).
Despite the common understanding that NHEJ mechanisms will trigger insertions and deletions 3 bp after PAM NGG sites (Shrivastav et al., 2008), we showed different patterns which do not seem to follow that rule. The NHEJ repair after DNA cleavage produced different mutation patterns, and it was observed that the target sites were mutated with InDels ranging from + 1 to –7 nt in length. We also observed big deletions outside the NHEJ repair. The screening in the sequence made by Sanger sequencing revealed that the gRNA 5 produced a deletion with –30 bp (3.3%) (Figure 7B). Other studies also observed the deletion of larger sequences when samples were analyzed using Sanger sequencing approaches. A range of deletion sizes from –7 to –77 bp in two different cultivars (DT26 and Maverick) was detected after the knockout of three GmGOLS genes (GmLox1, GmLox2, and GmLox3). The seeds obtained after knockout showed significant shifts in stachyose, raffinose, verbascose, and sucrose (Do and Chu, 2020). Using Agrobacterium strains to edit and get knockout of lipoxygenase-free soybean genes, different mutation patterns ranging from + 1 to –8 nt were found in PCR products sequenced by Sanger (Wang et al., 2020).
A knockout using the CRISPR system in Agrobacterium-mediated gene to flowering-related genes (E1) in soybean produced two different deletion patterns. The mutations identified by sequence peaks showed that homozygous mutations at the target sites were 11 bp deletion and 40 bp deletion. The authors related that this mutation frameshift resulted in premature translation termination codons of 79 amino acids (aa) and 88 aa, respectively (Han et al., 2019). The CRISPR/Cas9 system was used to gene edit the target gene (GmJAG1) predicted to modulate yield in the low-latitude was applied in soybean. Although the authors do not mention the delivery method, the sequencing results obtained by Sanger showed that in the T1 segregation population, there was deletion (−4 bp) and lost start codons (Cai et al., 2021). Interestingly, our gRNA2 results diverge from a previous study that first designed this gRNA sequence. Campbell et al. (2019), using an expression vector and biolistic transformation, obtained four different allelic combinations that ranged from –2 to –21 bp of deletion using gRNA2 (Campbell et al., 2019). In our study, using the same gRNA sequence, we did not obtain any series with deletion; on the contrary, we have obtained an insertion of + 5 bp (AGTTC) at the target site. These results can demonstrate that the differences found may be related to the genotype used, tissue issue, delivery method, or even the analytical methods such as the sequencing platform and the software used for the deconvolution of Sanger sequencing data. Overall, in this study, we demonstrated that our editing platform is (1) effective for in-frame mutations in the trichome gene; (2) satisfactory mutation efficiency which is suitable for gene expression studies; and (3) advantageous toward the absence of external DNA backbone integration into the genome.
DNA-Free Gene Editing in Other Species
RNP-mediated genome engineering has been demonstrated in protoplasts for various model plants and crop species including Arabidopsis, tobacco, rice (Woo et al., 2015), Petunia (Subburaj et al., 2016; Yu et al., 2021), apple, grape (Malnoy et al., 2016), maize (Sant’Ana et al., 2020), potato (González et al., 2020), cabbage (Murovec et al., 2018), chickpea (Badhan et al., 2021), and soybean (Kim et al., 2017; Kim and Choi, 2021).
Direct delivery of DNA-free recombinant Cas9 proteins was found to be as efficient or even more efficient in some cases compared to other techniques, and it also limits the unwanted off-target mutations. Synthesis of unique sgRNAs, prediction of unique target sites, and the molar ratio of Cas9:sgRNA would also facilitate the on-target specificity of CRISPR/Cas9. In order to achieve a higher mutation rate in endogenous CPR5 in the soybean protoplast system, we have tried a fixed Cas9 ratio along with several different sgRNA ratios (1:1, 1:2, and 1:3) in a similar way to previous studies (Woo et al., 2015; Subburaj et al., 2016). We found that the only ratio of 1: 3 (10 μg Cas9 + 30 μg sgRNA) was very suitable, in which we confirmed the induced mutations during T7E1 analysis (Figure 6), while other ratios showed only negative results. This suggests that adjustment of the molar ratio of Cas9 to sgRNA would be a crucial factor to achieve a higher mutation rate of interest genes as noted in recent studies (Malnoy et al., 2016; Murovec et al., 2018).
In this study, with a fixed Cas9 to sgRNA ratio (1:3), the mutation rates of 4.2–18.1% were noted from sequencing results including target deep and Sanger assays in soybean protoplasts within 24 h of transfection (Table 1 and Figure 7). The editing efficiency in this study corresponded well with many previous reports. The editing efficiency has been observed to greatly vary in plant protoplasts using the RNP-mediated CRISPR/Cas9 system. Editing efficiencies could likely be attributed to explant, species, sgRNA efficiency, Cas protein activity, transformation, and detection methods. Using PEG-mediated protoplast assays, editing frequencies of 16, 44, and 19% have been reported in Arabidopsis, tobacco, and rice, respectively (Woo et al., 2015). In garden petunia, 5.30–17.83% (Nitrate reductase) and 9.99–26.72% (flavone 3’ hydroxylase) of editing frequencies were obtained (Subburaj et al., 2016; Yu et al., 2021). In cabbage species, a minimum of 0.09% (Brassica oleracea var. capitata f. alba) and a maximum of 24.51% (Brassica rapa subsp. pekinensis) have been observed (Murovec et al., 2018). Likewise, mutation frequencies of 0.5–6.9% in apple, 0.1% in grapes (Malnoy et al., 2016), 0.19–0.92% in cavendish banana (Wu et al., 2020), 0.5–11.3% in sweet pepper (Kim et al., 2020), and 0.85–5.85% in maize (Sant’Ana et al., 2020) have been reported in protoplasts.
LbCas12a-RNP (previously named Cpf1) has been considered an alternative approach to the SpCas9-RNP system (Swarts and Jinek, 2018) because of its smaller protein size and induction of large deletions. In this study, using the SpCas9-RNP system, we obtained the highest mutation frequency of 18.1% in soybean leaf mesophyll protoplast for T1 RNP (Table 1). It is significantly higher than the previously reported mutation frequency of 11.7% for FAD2-1A in soybean leaf mesophyll protoplast by the cpf1-RNP system (Kim et al., 2017). Furthermore, characterizing the mutation patterns, the designed five sgRNAs (T1–T5) were successfully induced the InDels at target sites, which are 1–7 (target deep) and 1–30 bp long (Sanger sequencing) that would change the open reading frame of GmCPR5 and cause loss of their function. These observed mutation patterns and their sizes corresponded well with the previous genome editing studies using CRISPR/Cas9 (Woo et al., 2015; Subburaj et al., 2016; Sant’Ana et al., 2020). These results demonstrate that the direct DNA-free delivery of CRISPR RNPs to soybean protoplast is improvised in this study which could produce mutations on targeted distinct sites of endogenous target genes through DSBs. Currently, we developed and optimized an efficient genome editing platform in soybean; in addition, future studies on the regeneration of whole plantlets from CRISPR/Cas9-edited protoplast cells will facilitate the development of DNA-free genome editing of soybean and its related legume crops.
DNA-Free Editing as a Tool for Genetic Screening in Plants
In this study, we demonstrated a DNA-free genome editing approach to edit the endogenous GmCPR5 locus using CRISPR/Cas9-based technology. We used RNP-mediated CRISPR/Cas9 system as a safe and effective tool to make site-directed mutations. The Agrobacterium-mediated and particle bombardment-mediated transformation methods are also commonly used to deliver the plasmid DNA carrying CRISPR (Cas9 and sgRNA) reagents into plant tissues and cells. However, these methods are limited by their unwanted off-target mutations, caused by random integration of CRISPR expression cassettes into genomes and followed by genome damage (Banakar et al., 2019; Liu et al., 2019b). Furthermore, the continuous expression of integrated transgene cassettes could have resulted in continuous damaging of genomic DNA which leads to off-target mutations (Hashimoto et al., 2016). The random integration of genome editing components into the recipient genome would also be considered as genetically modified organism (GMO) and raise concerns among enforcement institutions (Zhang et al., 2015). The plasmid-mediated transformation of CRISPR/Cas9 into living cells often needs optimized compatibility of promoters and terminators in the expression system. In some cases, the DNA-based expression of Cas9 protein has been found toxic to living cells (Morgens et al., 2017; Foster et al., 2018). To overcome the above-mentioned drawbacks, transiently expressed plasmid DNA carrying Cas9 nucleases along with sgRNA(s) has been successfully delivered into plant cells (Zhang et al., 2016). Alternatively, the direct delivery of DNA-free proteins, such as the RNP complex (preassembled Cas9 protein and sgRNA), could also be delivered into living cells. Using RNPs has been found to decrease off-target effects as it could be easily degraded by cell endogenous proteases and nucleases (Kim et al., 2014). Organisms edited by RNPs also are not restricted by GMO rules as it involves using recombinant DNA (Araki and Ishii, 2015; Wolt et al., 2016). Recent CRISPR/Cas9 studies on soybean have successfully produced trait-specific knockout lines using Agrobacterium-mediated transformation methods which could be considered GMO (Han et al., 2019; Wang et al., 2020).
CRISPR/Cas9-based RNP-mediated system has been more effective in terms of preparation, delivery, screening of CRISPR components, and generating target-specific mutations at the targeted locus to produce transgene-free engineered plants (Subburaj et al., 2016; Yu et al., 2021). By using RNPs, site-directed mutations in plants could easily be genotyped, suggesting that they are sensitive and easy to approach (Liang et al., 2018). Most studies have exploited the NHEJ-mediated genome editing using RNP-mediated CRISPR/Cas9 system, which usually creates imprecise small InDels. However, in maize, RNPs were used to make site-directed mutations through the HDR pathway by introducing donor DNA templates (Svitashev et al., 2016). A study in Arabidopsis reported a 223 bp deletion using RNPs (Woo et al., 2015), indicating that metabolic engineering of plants is feasible in the future by inducing large deletions in the genome. RNP-mediated editing relies on the perfect delivery method, compared with various RNP delivery methods such as electroporation (Lee et al., 2020), lipofection (Liu et al., 2020), and particle bombardment (Svitashev et al., 2016; Liang et al., 2017; Banakar et al., 2020). PEG-mediated transfection was noted as a predominantly used method as it has also been successfully demonstrated for various model plants and crop species (Woo et al., 2015; Subburaj et al., 2016; Kim et al., 2017; Sant’Ana et al., 2020; Yu et al., 2021). The PEG-mediated method could be more efficient, cost-effective, and simple in terms of delivering RNPs into plant cells than other methods like particle bombardment which requires specific instruments and optimized parameters (Banakar et al., 2020). The process of regeneration of protoplasts is highly necessary for the recovery of genome-edited plants through RNP-mediated genome editing. However, it could still be possible to evaluate the efficacy of CRISPR systems at cell level for a new plant species, having unoptimized protocol for the regeneration process. Alternative RNP transformation methods like de novo meristem induction could help bypass the regeneration process if any plant lacks robust methods of protoplast regeneration (Maher et al., 2020). For soybean, the protocol for regeneration of protoplast cells is available (Wei and Xu, 1988; Dhir et al., 1991). With the well-established method of direct delivery of engineered RNPs in this study, it would be feasible to breed novel traits of soybean and other related bean species without the use of any stable transformation methods.
Conclusion
In summary, we describe here CRISPR/Cas9-based gene editing in soybean leaf protoplasts from young seedlings transformed with preassembled CRISPR-RNP mediated by PEG as a fast and low-cost approach to developing mutant lines for plant biology and biotechnology studies. Although the mutation efficiency was found to vary according to each sgRNA utilized at the gene (Glyma06g15080), the targeted deep and Sanger sequencing showed a range of mutational profiles (ranging from 4.2 to 18.1%) that resulted in frameshift types predicted to cause a premature stop codon at mRNA and cause loss of function in GmCPR5 alleles. Combining confocal fluorescence microscopy to visualize viable cells and stained CRISPR cells is an important checkpoint to improve the targeted mutagenesis. Transformed protoplasts stained can also be subjected to fluorescence-activated cell sorting (FACS) providing enrichment mutants cells that could be used for cell embryogenic cultivation. This, despite a great challenge, presents potential in the face of increasingly future studies on regeneration. Finally, this platform used here as a proof of concept can also be used as a strategy to apply transient genes and study the function and regulation of the genes.
Data Availability Statement
The original contributions presented in this study are publicly available. This data can be found here: NCBI, PRJNA785774.
Author Contributions
SS initiated, designed, and conducted the analyses. CZ helped to initiate the work and co-designed the experiments. JN and AH contributed to the data preparation. RN and SA-T supervised the research. All authors contributed to the article and approved the submitted version.
Funding
This project was funded by the Research Council of Norway (Project Number: 301911—FOODPRINT: Traceability and labeling of gene-edited products in the food chain). CZ, AH, and RN received scholarships from Conselho Nacional de Pesquisa Científica in Brazil.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We would like to thank Kenneth Bowitz Larsen at The Advanced Microscopy Core Facility, Department of Medical Biology at The Arctic University of Tromsø, for his technical support with the confocal microscopy. We would also like to thank Idun Merete Grønsberg for helping to set up the safety certificates for the laboratory.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2022.939997/full#supplementary-material
Supplementary Figure 1 | Agarose gel photographs of PCR amplicons during targeted deep and Sanger sequencing. (A,B) Gel images of first (A) and second (B) rounds of multiplex nested PCR products for targeted deep sequencing using primers listed in Supplementary Table 2. (C) Gel images of PCR amplicons for Sanger sequencing. Lanes M, DNA ladders; T1–T5, Protoplast transformants; C, untransformed wild type (control); T, transformed with RNPs. The expected size of PCR fragments is indicated with red arrows.
Supplementary File 1 | Chromatogram files in ABI format from Sanger sequencing assay for the control and protoplast transformants of T2 and T4.
Abbreviations
9M CPW, 9% mannitol with CPW salts; Aa, amino acids; CPR5, constitutive pathogen response 5; CPW, cell and protoplast washing; crRNA, CRISPR RNA; ESID, electronically switchable illumination and detection module; FACS, fluorescence-activated cell sorting; FDA, fluorescein diacetate; GFP, green fluorescent protein; GMO, genetically modified organism; gRNA, guide RNAs, InDels, insertions and deletions; NCBI, National Center for Biotechnology Information; NHEJ, non-homologs end joining; PEG, polyethylene glycol; RNP, ribonucleoproteins; T7E1, T7 endonuclease I; T-DNA, transfer DNA.
Footnotes
References
Adedeji, O. S., Naing, A. H., and Kim, C. K. (2020). Protoplast isolation and shoot regeneration from protoplast-derived calli of Chrysanthemum cv. White ND Plant Cell Tiss. Organ. Cult. 141, 571–581. doi: 10.1007/s11240-020-01816-3
Amin, N., Ahmad, N., Wu, N., Pu, X., Ma, T., Du, Y., et al. (2019). CRISPR-Cas9 mediated targeted disruption of FAD2 – 2 microsomal omega-6 desaturase in soybean (Glycine max. L). BMC Biotechnol. 19:9. doi: 10.1186/s12896-019-0501-2
Amirkhanov, R. N., and Stepanov, G. A. (2019). Systems of Delivery of CRISPR/Cas9 Ribonucleoprotein Complexes for Genome Editing. Russ. J. Bioorganic. Chem. 45, 431–437. doi: 10.1134/S1068162019060025
Araki, M., and Ishii, T. (2015). Towards social acceptance of plant breeding by genome editing. Trends Plant Sci. 20, 145–149. doi: 10.1016/j.tplants.2015.01.010
Badhan, S., Ball, A. S., and Mantri, N. (2021). First Report of CRISPR/Cas9 Mediated DNA-Free Editing of 4CL and RVE7 Genes in Chickpea Protoplasts. Int. J. Mol. Sci. 22:396. doi: 10.3390/ijms22010396
Bae, S., Kweon, J., Kim, H. S., and Kim, J. S. (2014). Microhomology-based choice of Cas9 nuclease target sites. Nat. Methods 11, 705–706. doi: 10.1038/nmeth.3015
Banakar, R., Eggenberger, A. L., Lee, K., Wright, D. A., Murugan, K., Zarecor, S., et al. (2019). High-frequency random DNA insertions upon co-delivery of CRISPR-Cas9 ribonucleoprotein and selectable marker plasmid in rice. Sci. Rep. 9:19902. doi: 10.1038/s41598-019-55681-y
Banakar, R., Schubert, M., Collingwood, M., Vakulskas, C., Eggenberger, A. L., and Wang, K. (2020). Comparison of CRISPR-Cas9/Cas12a Ribonucleoprotein Complexes for Genome Editing Efficiency in the Rice Phytoene Desaturase (OsPDS) Gene. Rice 13:4. doi: 10.1186/s12284-019-0365-z
Bloh, K., Kanchana, R., Bialk, P., Banas, K., Zhang, Z., Yoo, B. C., et al. (2021). Deconvolution of Complex DNA Repair (DECODR): establishing a Novel Deconvolution Algorithm for Comprehensive Analysis of CRISPR-Edited Sanger Sequencing Data. CRISPR J. 4, 120–131. doi: 10.1089/crispr.2020.0022
Cai, Y., Chen, L., Liu, X., Guo, C., Sun, S., Wu, C., et al. (2018). CRISPR/Cas9-mediated targeted mutagenesis of GmFT2a delays flowering time in soya bean. Plant Biotechnol. J. 16, 176–185. doi: 10.1111/pbi.12758
Cai, Z., Xian, P., Cheng, Y., Ma, Q., Lian, T., Nian, H., et al. (2021). CRISPR/Cas9-mediated gene editing of GmJAGGED1 increased yield in the low-latitude soybean variety Huachun 6. Plant Biotechnol. J. 19, 1898–1900. doi: 10.1111/pbi.13673
Campbell, B. W., Hoyle, J. W., Bucciarelli, B., Stec, A. O., Samac, D. A., Parrott, W. A., et al. (2019). Functional analysis and development of a CRISPR/Cas9 allelic series for a CPR5 ortholog necessary for proper growth of soybean trichomes. Sci. Rep. 9:14757. doi: 10.1038/s41598-019-51240-7
Chen, X., Yang, S., Zhang, Y., Zhu, X., Yang, X., Zhang, C., et al. (2021). Generation of male-sterile soybean lines with the CRISPR / Cas9 system. Crop J. 9, 1270–1277. doi: 10.1016/j.cj.2021.05.003
Concordet, J. P., and Haeussler, M. (2018). CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res. 46, W242–W245. doi: 10.1093/nar/gky354
Dalla Costa, L., Piazza, S., Pompili, V., Salvagnin, U., Cestaro, A., Moffa, L., et al. (2020). Strategies to produce T-DNA free CRISPRed fruit trees via Agrobacterium tumefaciens stable gene transfer. Sci. Rep. 10:20155. doi: 10.1038/s41598-020-77110-1
Dhir, S. K., Dhir, S., and Widholm, J. M. (1991). Plantlet regeneration from immature cotyledon protoplasts of soybean (Glycine max L.). Plant Cell Rep. 10, 39–43. doi: 10.1007/BF00233030
Do, P. T., and Chu, H. H. (2020). CRISPR / Cas9-Mediated Knockout of Galactinol Synthase-Encoding Genes Reduces Raffinose Family Oligosaccharide Levels in Soybean Seeds. Front. Plant Sci. 11:612942. doi: 10.3389/fpls.2020.612942
Foster, A. J., Martin-Urdiroz, M., Yan, X., Wright, H. S., Soanes, D. M., and Talbot, N. J. (2018). CRISPR-Cas9 ribonucleoprotein-mediated co-editing and counterselection in the rice blast fungus. Sci. Rep. 8:14355. doi: 10.1038/s41598-018-32702-w
Frearson, E. M., Power, J. B., and Cocking, E. C. (1973). The isolation, culture and regeneration of Petunia leaf protoplasts. Dev. Biol. 33, 130–137. doi: 10.1016/0012-1606(73)90169-3
González, M. N., Massa, G. A., Andersson, M., Turesson, H., Olsson, N., Fält, A. S., et al. (2020). Reduced Enzymatic Browning in Potato Tubers by Specific Editing of a Polyphenol Oxidase Gene via Ribonucleoprotein Complexes Delivery of the CRISPR/Cas9 System. Front. Plant Sci. 10:1649. doi: 10.3389/fpls.2019.01649
Han, J., Guo, B., Guo, Y., Zhang, B., Wang, X., and Qiu, L.-J. (2019). Creation of Early Flowering Germplasm of Soybean by CRISPR/Cas9 Technology. Front. Plant Sci. 10:1446. doi: 10.3389/fpls.2019.01446
Hashimoto, M., Yamashita, Y., and Takemoto, T. (2016). Electroporation of Cas9 protein/sgRNA into early pronuclear zygotes generates non-mosaic mutants in the mouse. Dev. Biol. 418, 1–9. doi: 10.1016/j.ydbio.2016.07.017
Hsu, P. D., Scott, D. A., Weinstein, J. A., Ran, F. A., Konermann, S., Agarwala, V., et al. (2013). DNA targeting specificity of RNA-guided Cas9 nucleases. Nat. Biotechnol. 31, 827–832. doi: 10.1038/nbt.2647
Jacobs, T. B., Lafayette, P. R., Schmitz, R. J., and Parrott, W. A. (2015). Targeted genome modifications in soybean with CRISPR/Cas9. BMC Biotechnol. 15:16. doi: 10.1186/s12896-015-0131-2
Jiang, F., and Doudna, J. A. (2017). CRISPR – Cas9 Structures and Mechanisms. Annu. Rev. Biophys. 46, 505–529. doi: 10.1146/annurev-biophys-062215-010822
Jinek, M., East, A., Cheng, A., Lin, S., Ma, E., and Doudna, J. (2013). RNA-programmed genome editing in human cells. eLife 2:e00471. doi: 10.7554/eLife.00471
Kao, K. N. (1977). Chromosomal behaviour in somatic hybrids of soybean-Nicotiana glauca. Molec. Gen. Genet. 150, 225–230. doi: 10.1007/BF00268120
Kim, H., and Choi, J. (2021). A robust and practical CRISPR/crRNA screening system for soybean cultivar editing using LbCpf1 ribonucleoproteins. Plant Cell Rep. 40, 1059–1070. doi: 10.1007/s00299-020-02597-x
Kim, H., Choi, J., and Won, K. H. (2020). A stable DNA-free screening system for CRISPR/RNPs-mediated gene editing in hot and sweet cultivars of Capsicum annuum. BMC Plant Biol. 20:449. doi: 10.1186/s12870-020-02665-0
Kim, H., Kim, S. T., Ryu, J., Kang, B. C., Kim, J. S., and Kim, S. G. (2017). CRISPR/Cpf1-mediated DNA-free plant genome editing. Nat. Commun. 8:14406. doi: 10.1038/ncomms14406
Kim, S., Kim, D., Cho, S. W., Kim, J., and Kim, J. S. (2014). Highly efficient RNA-guided genome editing in human cells via delivery of purified Cas9 ribonucleoproteins. Genome Res. 24, 1012–1019. doi: 10.1101/gr.171322.113
La Russa, M. F., and Qi, L. S. (2015). The New State of the Art: cas9 for Gene Activation and Repression. Mol. Cell. Biol. 35, 3800–3809. doi: 10.1128/mcb.00512-15
Labun, K., Montague, T. G., Krause, M., Torres Cleuren, Y. N., Tjeldnes, H., and Valen, E. (2019). CHOPCHOP v3: expanding the CRISPR web toolbox beyond genome editing. Nucleic Acids Res. 47, W171–W174. doi: 10.1093/nar/gkz365
Lee, M. H., Lee, J., Choi, S. A., Kim, Y. S., Koo, O., Choi, S. H., et al. (2020). Efficient genome editing using CRISPR-Cas9 RNP delivery into cabbage protoplasts via electro-transfection. Plant Biotechnol. Rep. 14, 695–702. doi: 10.1007/s11816-020-00645-2
Liang, Z., Chen, K., Li, T., Zhang, Y., Wang, Y., Zhao, Q., et al. (2017). Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat. Commun. 8:14261. doi: 10.1038/ncomms14261
Liang, Z., Chen, K., Yan, Y., Zhang, Y., and Gao, C. (2018). Genotyping genome-edited mutations in plants using CRISPR ribonucleoprotein complexes. Plant Biotechnol. J. 16, 2053–2062. doi: 10.1111/pbi.12938
Liu, J., Gunapati, S., Mihelich, N. T., Stec, A. O., Michno, J. M., and Stupar, R. M. (2019a). Genome Editing in Soybean with CRISPR/Cas9. Methods Mol. Biol. 1917, 217–234. doi: 10.1007/978-1-4939-8991-1_16
Liu, J., Nannas, N. J., Fu, F. F., Shi, J., Aspinwall, B., Parrott, R., et al. (2019b). Genome-Scale Sequence Disruption Following Biolistic Transformation in Rice and Maize. Plant Cell 31, 368–383. doi: 10.1105/tpc.18.00613
Liu, W., Rudis, M. R., Cheplick, M. H., Millwood, R. J., Yang, J. P., Ondzighi-Assoume, A. C., et al. (2020). Lipofection-mediated genome editing using DNA-free delivery of the Cas9/gRNA ribonucleoprotein into plant cells. Plant Cell Rep. 39, 245–257. doi: 10.1007/s00299-019-02488-w
Maher, M. F., Nasti, R. A., Vollbrecht, M., Starker, C. G., Clark, M. D., and Voytas, D. F. (2020). Plant gene editing through de novo induction of meristems. Nat. Biotechnol. 38, 84–89. doi: 10.1038/s41587-019-0337-2
Makarova, K. S., Haft, D. H., Barrangou, R., Brouns, S. J. J., Mojica, F. J. M., Wolf, Y. I., et al. (2011). Evolution and classification of the CRISPR–Cas systems. Nat. Publ. Gr. 9, 467–477. doi: 10.1038/nrmicro2577
Malnoy, M., Viola, R., Jung, M. H., Koo, O. J., Kim, S., Kim, J. S., et al. (2016). DNA-Free Genetically Edited Grapevine and Apple Protoplast Using CRISPR/Cas9 Ribonucleoproteins. Front. Plant Sci. 7:1904. doi: 10.3389/fpls.2016.01904
Manghwar, H., Li, B., Ding, X., Hussain, A., Lindsey, K., Zhang, X., et al. (2020). CRISPR/Cas Systems in Genome Editing: methodologies and Tools for sgRNA Design, Off-Target Evaluation, and Strategies to Mitigate Off-Target Effects. Adv. Sci. 7:1902312. doi: 10.1002/advs.201902312
Metje-Sprink, J., Menz, J., Modrzejewski, D., and Sprink, T. (2019). DNA-Free genome editing: past, present and future. Front. Plant Sci. 9:1957. doi: 10.3389/fpls.2018.01957
Morgens, D. W., Wainberg, M., Boyle, E. A., Ursu, O., Araya, C. L., Tsui, C. K., et al. (2017). Genome-scale measurement of off-target activity using Cas9 toxicity in high-throughput screens. Nat. Commun. 8:15178. doi: 10.1038/ncomms15178
Murovec, J., Guček, K., Bohanec, B., Avbelj, M., and Jerala, R. (2018). DNA-Free Genome Editing of Brassica oleracea and B. rapa Protoplasts Using CRISPR-Cas9 Ribonucleoprotein Complexes. Front. Plant Sci. 9:1594. doi: 10.3389/fpls.2018.01594
Naito, Y., Hino, K., Bono, H., and Ui-Tei, K. (2015). CRISPRdirect: software for designing CRISPR/Cas guide RNA with reduced off-target sites. Bioinformatics 31, 1120–1123. doi: 10.1093/bioinformatics/btu743
Nicolia, A., Andersson, M., Hofvander, P., Festa, G., and Cardi, T. (2021). Tomato protoplasts as cell target for ribonucleoprotein (RNP)- mediated multiplexed genome editing. Plant Cell Tiss. Organ. Cult. 144, 463–467. doi: 10.1007/s11240-020-01954-8
Nishimasu, H., Ran, F. A., Hsu, P. D., Konermann, S., Shehata, S. I., Dohmae, N., et al. (2014). Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell 156, 935–949. doi: 10.1016/j.cell.2014.02.001
Osakabe, Y., and Osakabe, K. (2015). Genome editing with engineered nucleases in plants. Plant Cell Physiol. 56, 389–400. doi: 10.1093/pcp/pcu170
Park, J., Bae, S., and Kim, J. S. (2015). Cas-Designer: a web-based tool for choice of CRISPR-Cas9 target sites. Bioinformatics 31, 4014–4016. doi: 10.1093/bioinformatics/btv537
Rodrigues, S. D., Karimi, M., Impens, L., Van Lerberge, E., Coussens, G., Aesaert, S., et al. (2021). Efficient CRISPR-mediated base editing in Agrobacterium spp. Proc. Natl. Acad. Sci. U.S.A. 118:e2013338118. doi: 10.1073/pnas.2013338118
Sant’Ana, R., Caprestano, C. A., Nodari, R. O., and Agapito-Tenfen, S. Z. (2020). PEG-Delivered CRISPR-Cas9 Ribonucleoproteins System for Gene-Editing Screening of Maize Protoplasts. Genes 11:1029. doi: 10.3390/genes11091029
Shrivastav, M., De Haro, L. P., and Nickoloff, J. A. (2008). Regulation of DNA double-strand break repair pathway choice. Cell Res. 18, 134–147. doi: 10.1038/cr.2007.111
Subburaj, S., Chung, S. J., Lee, C., Ryu, S. M., Kim, D. H., Kim, J. S., et al. (2016). Site-directed mutagenesis in Petunia × hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep. 35, 1535–1544. doi: 10.1007/s00299-016-1937-7
Sun, X., Hu, Z., Chen, R., Jiang, Q., Song, G., Zhang, H., et al. (2015). Targeted mutagenesis in soybean using the CRISPR-Cas9 system. Sci. Rep. 5:10342. doi: 10.1038/srep10342
Svitashev, S., Schwartz, C., Lenderts, B., Young, J. K., and Mark Cigan, A. (2016). Genome editing in maize directed by CRISPR-Cas9 ribonucleoprotein complexes. Nat. Commun. 7:13274. doi: 10.1038/ncomms13274
Swarts, D. C., and Jinek, M. (2018). Cas9 versus Cas12a/Cpf1: structure-function comparisons and implications for genome editing. Wiley Interdiscip. Rev. RNA. 9:e1481. doi: 10.1002/wrna.1481
Tsai, S. Q., Zheng, Z., Nguyen, N. T., Liebers, M., Topkar, V. V., Thapar, V., et al. (2015). GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat. Biotechnol. 33, 187–198. doi: 10.1038/nbt.3117
Wang, H., La Russa, M., and Qi, L. S. (2016). CRISPR/Cas9 in Genome Editing and Beyond. Annu. Rev. Biochem. 85, 227–264. doi: 10.1146/annurev-biochem-060815-014607
Wang, J., Kuang, H., Zhang, Z., Yang, Y., Yan, L., Zhang, M., et al. (2020). Generation of seed lipoxygenase-free soybean using CRISPR-Cas9. Crop J. 8, 432–439. doi: 10.1016/j.cj.2019.08.008
Wei, Z. M., and Xu, Z. H. (1988). Plant regeneration from protoplasts of soybean (Glycine max L.). Plant Cell Rep. 7, 348–351. doi: 10.1007/BF002699
Wolt, J. D., Wang, K., and Yang, B. (2016). The Regulatory Status of Genome-edited Crops. Plant Biotechnol. J. 14, 510–518. doi: 10.1111/pbi.12444
Woo, J. W., Kim, J., Kwon, S. I., Corvalán, C., Cho, S. W., Kim, H., et al. (2015). DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 33, 1162–1164. doi: 10.1038/nbt.3389
Wu, F., and Hanzawa, Y. (2018). A Simple Method for Isolation of Soybean Protoplasts and Application to Transient Gene Expression Analyses. J. Vis. Exp. 131:57258. doi: 10.3791/57258
Wu, S., Zhu, H., Liu, J., Yang, Q., Shao, X., Bi, F., et al. (2020). Establishment of a PEG-mediated protoplast transformation system based on DNA and CRISPR/Cas9 ribonucleoprotein complexes for banana. BMC Plant Biol. 20:425. doi: 10.1186/s12870-020-02609-8
Yu, J., Tu, L., Subburaj, S., Bae, S., and Lee, G. J. (2021). Simultaneous targeting of duplicated genes in Petunia protoplasts for flower color modification via CRISPR-Cas9 ribonucleoproteins. Plant Cell Rep. 40, 1037–1045. doi: 10.1007/s00299-020-02593-1
Yu, L., Batara, J., and Lu, B. (2016). “Application of Genome Editing Technology to MicroRNA Research in Mammalians,” in Modern Tools for Genetic Engineering, ed. M. S. D. Kormann (London: Intech Open), doi: 10.5772/6433013
Zhang, X. H., Tee, L. Y., Wang, X. G., Huang, Q. S., and Yang, S. H. (2015). Off- target Effects in CRISPR/Cas9-mediated Genome Engineering. Mol. Ther. Nucleic Acids 4:e264. doi: 10.1038/mtna.2015.37
Zhang, Y., Iaffaldano, B., and Qi, Y. (2021). CRISPR ribonucleoprotein-mediated genetic engineering in plants. Plant Commun. 2:100168. doi: 10.1016/j.xplc.2021.100168
Zhang, Y., Liang, Z., Zong, Y., Wang, Y., Liu, J., Chen, K., et al. (2016). Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 7:12617. doi: 10.1038/ncomms12617
Zhang, Z., Hua, L., Gupta, A., Tricoli, D., Edwards, K. J., Yang, B., et al. (2019). Development of an Agrobacterium-delivered CRISPR/Cas9 system for wheat genome editing. Plant Biotechnol. J. 17, 1623–1635. doi: 10.1111/pbi.13088
Keywords: genetically modified organism, gene editing, mutagenesis, transgenesis, target deep sequencing, breeding, genetic screening, genetically modified plants
Citation: Subburaj S, Zanatta CB, Nunn JAL, Hoepers AM, Nodari RO and Agapito-Tenfen SZ (2022) A DNA-Free Editing Platform for Genetic Screens in Soybean via CRISPR/Cas9 Ribonucleoprotein Delivery. Front. Plant Sci. 13:939997. doi: 10.3389/fpls.2022.939997
Received: 09 May 2022; Accepted: 20 June 2022;
Published: 12 July 2022.
Edited by:
Ljudmilla Timofejeva, Department of Gene Technology, Tallinn University of Technology, EstoniaReviewed by:
Rukmini Mishra, Centurion University of Technology and Management, IndiaChannakeshavaiah Chikkaputtaiah, North East Institute of Science and Technology (CSIR), India
Copyright © 2022 Subburaj, Zanatta, Nunn, Hoepers, Nodari and Agapito-Tenfen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sarah Zanon Agapito-Tenfen, saag@norceresearch.no
†These authors have contributed equally to this work
 Saminathan Subburaj
Saminathan Subburaj