
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Physiol. , 29 September 2023
Sec. Invertebrate Physiology
Volume 14 - 2023 | https://doi.org/10.3389/fphys.2023.1270751
Predatory stink bugs derive from phytophagous stink bugs and evolved enhanced predation skills. Neuropeptides are a diverse class of ancient signaling molecules that regulate physiological processes and behavior in animals, including stink bugs. Neuropeptide evolution might be important for the development of predation because neuropeptides can be converted to venoms that impact prey. However, information on neuropeptide signaling genes in predatory stink bugs is lacking. In the present study, neuropeptide signaling genes of Picromerus lewisi, an important predatory stink bug and an effective biological agent, were comprehensively identified by transcriptome analysis, with a total of 59 neuropeptide precursor genes and 58 potential neuropeptide receptor genes found. In addition, several neuropeptides and their receptors enriched in salivary glands of P. lewisi were identified. The present study and subsequent functional research contribute to an in-depth understanding of the biology and behavior of the predatory bugs and can provide basic information for the development of better pest management strategies, possibly including neuropeptide receptors as insecticide targets and salivary gland derived venom toxins as novel killing moleculars.
Neuropeptides are a large diverse class of signaling molecules with key roles in insect physiology and behavior (Schoofs et al., 2017). Neuropeptides mediate their effects mainly through G protein-coupled receptors (GPCRs), receptor guanylyl cyclases (RGCs), and receptor tyrosine kinases (RTKs) (Caers et al., 2012). Given their essential regulatory functions and high specificity, neuropeptide signaling systems have been considered promising targets for “green” pest control (Verlinden et al., 2014; Audsley and Down, 2015). In order to develop a “green” insecticide based off this system, scientists should seek out unique neuropeptide signaling pathways that are absent in beneficial predatory insects. To this end, recent improvements in genome, transcriptome and proteome analysis have led to the discovery of neuropeptides and their receptors in a number of insects, including several beneficial species, which will provide valuable information for the development of novel insecticides with high selectivity.
Heteropteran insects (true bugs) are an ideal group to explore the evolution of trophic strategies (Walker et al., 2016). Heteropterans have diverse feeding strategies. The group includes phytophages (such as Pentatomomorpha and Miridae), entomophages (such as Nepomorpha, Enicocephalomorpha, Leptopodomorpha, Gerromorpha, and Dipsocoromorpha), and hematophages (such as Triatominae and Cimicidae) (Walker et al., 2016; Walker et al., 2018). Entomophagy is also present in some Pentatomomorpha and Miridae groups, with a reversal to predation from phytophagy (Walker et al., 2016). To date, sets of neuropeptides have been analyzed in several heteropterans, including three phytophagous bugs, Lygus hesperus (Miridae) (Christie et al., 2016; Hull et al., 2021), Nezara viridula (Pentatomidae) and Halyomorpha halys (Pentatomidae) (Lavore et al., 2018), and four hematophagous Reduviidae bugs, Rhodnius prolixus, Triatoma dimidiata, T. infestans, and T. pallidipennis (Ons et al., 2011; Ons et al., 2016). Comprehensive identification of neuropeptide receptors has only been reported for five hematophagous bugs, R. prolixus, T. dimidiata, T. pallidipennis, T. infestans (Ons et al., 2016), and Cimex lectularius (Benoit et al., 2016), and two phytophagous bugs, N. viridula (Lavore et al., 2018) and Apolygus lucorum (Gao et al., 2021). To date, no comprehensive study of neuropeptide signaling genes in predatory heteropterans has been reported. Identifying and comparing neuropeptide sets of diverse bugs is important for finding pest specific targets that do not negatively impact beneficial natural enemies like predatory stink bugs.
The predatory stink bug Picromerus lewisi Scott (Hemiptera: Pentatomidae) is widely distributed in China and other Asian regions (Lin et al., 2000). It has been selected as an excellent biological control agent for a wide range of agricultural and forest insect pests, such as Lepidoptera larvae (Mu et al., 2022). Currently, P. lewisi has been successfully mass produced by natural enemy factories in China and has been shown to have good control of many important pests, such as Spodoptera frugiperda (Wang et al., 2019b; Tang et al., 2019). Predatory stink bugs use their salivary venom to paralyze prey and initiate extra-oral digestion. The salivary glands with venom components (also called venom glands) of predatory stink bugs are likely to be derived from the salivary glands of related non-venomous stink bug species (Cohen, 1990; Cohen, 1995; Walker et al., 2016). Insect salivary systems are influenced by neuropeptides and neurohormones secreted from endocrine organs and various neuronal cells (Spit et al., 2012). For example, in the cockroach, Periplanea americana, the SMYamide neuropeptide gene is specifically expressed in the neurons innervating the salivary glands and functions as a hormone to generate action potentials during feeding (Veenstra, 2020). In addition, insect salivary glands also have endocrine functions, producing neuropeptides and neurohormones to regulate other tissues and organs (Li et al., 2022b; Titos et al., 2023). A salivary gland-derived peptide, Sgsf, has been identified in Drosophila as an endocrine factor secreted into the hemolymph that systemically regulates larval growth (Li et al., 2022b). What’s more, there are a few interesting cases where invertebrate neuropeptides have been recruited into salivary glands as venom toxins to affect prey, such as TKs from the cephalopods Eledone moschata and Octopus vulgaris (Champagne and Ribeiro, 1994; Kanda et al., 2003).
Given the state of the field described above, our fundamental hypotheses are that: 1) Insect neuropeptides might be repurposed into novel insecticides; 2) finding a neuropeptide pathway that is different or absent in beneficial predators, could lead to insecticides that do not hurt beneficial predators; 3) natural enemy evolution might repurpose neuropeptides as venom toxins, causing them to be expressed in salivary glands. To begin testing these hypotheses, we identified neuropeptides and their receptors in P. lewisi using transcriptome data analysis. In addition, the expression patterns of these genes in heads, guts and salivary glands were analyzed. The fundamental genetic information of the neuropeptide signaling system will be helpful to better understand the biology and ecology of the predatory bugs and to develop better pest management strategies utilizing bothe natural enemies and novel synthetic chemistries.
In our previous study, full-length transcriptome and RNA-seq transcriptome analysis of P. lewisi were jointly performed (Li et al., 2022a). However, no more than 20 neuropeptide precursor transcripts were found in the full-length transcriptome, which might be because a number of short precursor transcripts were polished during sequence clustering of raw reads obtained from Iso-seq sequencing. Herein, for the identification of neuropeptide signaling genes of P. lewisi, we performed de novo re-assembly of the RNA-seq transcriptome data that were generated from the previous study (Li et al., 2022a). Briefly, 100 fifth-instar nymphs starved for about 6 h were sampled for total RNA extraction from salivary glands (SG), guts (G), antennae (A), legs (L), and heads without antennae and salivary glands (H), with three independent biological replicates for each tissue/part sample. RNA samples were extracted using TRIzol reagent (Invitrogen, USA) and about 3 μg RNA per sample was used as the template for cDNA synthesis and Illumina sequencing using Ilumina NovaSeq 6000 (Illumina, USA). A total of fifteen libraries with approximately 20 million sequence clean reads each were generated and deposited in the National Center for Biotechnology Information (NCBI) Short Read Archive (SRA) under accession numbers of SRR20681617∼SRR20681631 (Wang et al., 2019a). All libraries were pooled and then assembled into unigenes using the Trinity software (v2.6.6), with min_kmer_cov set to 2 by default and all other parameters set default. A Benchmarking Universal Single-Copy Orthologs (BUSCO) analysis was performed to assess the completeness of the assembly (Simão et al., 2015). All unigenes were de novo annotated against Nr/Nt (NCBI non-redundant protein/nucleotide sequences), Pfam (Protein family), KOG/COG (Clusters of Orthologous Groups of proteins), Swiss-Prot (A manually annotated and reviewed protein sequence database), KEGG (Kyoto Encyclopedia of Genes and Genomes), and GO (Gene Ontology) databases.
Differential expression analysis of transcripts from SG, G and H with three biological replicates each was performed using the DESeq2 R package (1.20.0). The Fragments per kilobase of transcript per million fragments mapped (FPKM) value was calculated to estimate the expression level of each transcript from each library. The Benjamini-Hochberg approach was used to adjust p values (P_adj) to control the false discovery rate. Genes with P_adj values <0.05 and | log2 (fold change) | > 1 were designated as differentially expressed genes (DEGs). Heat maps showing gene expression profiles were constructed based on Log10 (FPKM +1) values.
To validate the transcriptome data, qRT-PCR analysis of eighteen genes had been performed (Li et al., 2022a). Briefly, eight cytochrome P450 monooxygenase (CYP) genes, six carboxyl/choline esterase (CCE) genes, and four glutathione S-transferase (GST) genes were selected, and the EF1A gene was used as the candidate reference gene. The RNA samples for qRT-PCR were the same as those for RNA-seq sequencing. Significant consistency was found between the expression profiles obtained by qRT-PCR and RNA-Seq (Li et al., 2022a).
Local tBLASTn searches were performed to predict the genes of neuropeptide precursors and their receptors from the reassembled P. lewisi transcriptome. Amino acid sequences of known neuropeptide precursors from H. halys (Lavore et al., 2018), L. hesperus (Hull et al., 2021), Nilaparvata lugens (Tanaka et al., 2014), and R. prolixus (Ons et al., 2011; Ons et al., 2016) were used as reference queries. Sequences of neuropeptide GPCR receptors from N. lugens (Tanaka et al., 2014), N. viridula (Lavore et al., 2018), A. lucorum (Gao et al., 2021), and R. prolixus (Ons et al., 2016) were collected. Neuropeptide RGC and RTK receptor sequences from Drosophila melanogaster and Aedes aegypti were obtained from Kong et al. (2021). What’s more, sequences of some potential novel neuropeptide precursors, agatoxin-like peptide (ALP), Carausius neuropeptide-like precursor (CNP), parathyroid hormone (PTH), PaOGS36577, and RFLamide (RFLa), were identified based on their homologous genes from Periplaneta americana (Zeng et al., 2021) or Tribolium castaneum (Xie et al., 2020). In addition, sequences of few novel identified insect neuropeptide receptors, for PTH (Xie et al., 2020), CNMamide (CNMa) (Jung et al., 2014) and elevenin (Ele) (Uchiyama et al., 2017), were also used as reference queries. The BLAST E-value threshold for neuropeptides was 1.0, and the hits were manually checked based on the characteristics of putative mature active peptide sequences. The E-value threshold for receptors was 10−5. Besides, the remaining neuropeptide sequences not included in our custom P. lewisi transcriptome database, were also searched using a public Sequence Read Archive (SRA) database (SRR10134979) via the NCBI tBLASTn program.
A well-established workflow was used to predict the potential active peptides of P. lewisi. Briefly, the presence of signal peptides was predicted using the online program SignalP 5.0 (https://services.healthtech.dtu.dk/services/SignalP-5.0/). Prohormone cleavage sites and post-translational modifications were identified based on the information presented in Veenstra (Veenstra, 2000) and/or by homology to known arthropod peptides.
Multiple alignments of amino acid sequences were performed using the online program MAFFT version 7 (https://mafft.cbrc.jp/alignment/server/, “G-INS-1” progressive method setting). For the alignments of neuropeptides, the putative active peptides or the amino acid sequences removing the putative signal peptides were adopted. For the alignments of neuropeptide receptors, full-length sequences were used when available, or partial sequences were used. Amino acid identity was subsequently determined and the alignment result was visualized using GeneDoc version 2 (Nicholas et al., 1997). Sequence logos of the aligned peptide sequences were generated using the online program WebLogo (http://weblogo.berkeley.edu/logo.cgi) (Crooks et al., 2004).
Maximum likelihood (ML) phylogenetic analysis was conducted using IQ-TREE (Minh et al., 2020). In a first run, the ModelFinder function was employed to determine the best-fit model using Bayesian Information Criterion (Darriba et al., 2019). In a second run, ML phylogenetic tree was constructed using a ultrafast (UF) bootstrap test with 3000 replicates and default settings to reduce overestimation of bootstrap support (-bnni) (Hoang et al., 2017). Phylogenetic trees were visualized using the Interactive Tree Of Life (iTOL) web server (https://itol.embl.de/) (Letunić and Bork, 2021).
Two species-specific neuropeptide transcripts, Crustacean cardioactive peptide (CCAP) and Orcokinin B (OKB), were amplified by PCR and sequenced to verify their reliability. Primers were designed to amplify the coding sequences (Supplementary Table S1). Five fifth-instar nymphs of P. lewisi used for cloning were supplied by the Fenggang County Natural Enemy Breeding Center of the Guizhou Tobacco Company Zunyi Branch, Zunyi, Guizhou Province, China. Total RNA was isolated using TRIzol reagent (Invitrogen, United States). RNA quality and quantity were determined using the RNA Nano 6000 Assay Kit in the Bioanalyzer 2,100 system (Agilent Technologies, United States). Total RNA was treated with DNase I (Invitrogen, United States) to remove any residual genomic DNA. cDNA were synthesized from 1 μg of the total RNA using a SMARTer PCR cDNA Synthesis Kit (Takara Bio United States, Inc, United States). Neuropeptide precursor transcripts were amplified in a 50-uL reaction system using 2 × Phanta Flash Master Mix (Vazyme, China). The PCR procedure was set as follows: pre-denaturation at 98°C for 30 s, 35 cycles of 98°C for 10 s, 55°C for 5 s and 72°C for 5 s, and termination at 72°C for 1 min. The PCR products were cut from the gel, purified and sequenced by Beijing Tsingke Biotech Co., Ltd.
The raw sequences of 15 biosamples from five tissues/parts with three biological repeats each were re-assembled using the de novo assembly procedure, resulting in a total of 62,183 unigenes with a mean length of 1,203 bp, and an N50 length of 2,040 bp (Supplementary Table S2). BUSCO analysis showed a high degree of completeness (87.0%) in the assembly (Supplementary Figure S1). 41.9% of the assembled unigenes were annotated in at least one database and 64.1% of the annotated unigenes were best matched to their H. halys homologs (Supplementary Table S2).
A total of 59 neuropeptide precursors were identified in the P. lewisi transcriptome, including several novel neuropeptides, CNP, PTH, PaOGS36577, and RFLa (Table 1; Supplementary Data S1). Glycoprotein hormone beta 5 (GPB5), allatostatin C (AST-C), and trissin were not found in the P. lewisi transcriptome. Although AST-C was not identified, potential homologs of AST-CC and AST-CCC were found in P. lewisi and other hemipterans (Table 2). GPB5 was identified from H. halys and R. prolixus but still not found in N. viridula and L. hesperus (Table 2). Adipokinetic hormone (AKH) and sulfakinin (SK) were not found in our custom transcriptome but were identified in NCBI SRA data (SRR10134979). 50 neuropeptide transcripts have full-length sequences and the remaining nine non-full-length sequences include AKH, ALP1, ALP2, arginine-vasopressin-like peptide (AVLP), natalisin (NTL), OKA, OKB, PTH and SK (Supplementary Data S1). Most of the predicted P. lewisi active neuropeptides are identical or highly similar to their homologous peptides from H. halys or other bugs (Table 1), whereas a few peptides have low similarity to their homologs, such as OKB, CCAP, prothoracicotropic hormone (PTTH) and neuroparsin A5 (NPA5) (with no more than 70% identity) (Figure 1). To increase the confidence in the putative P. lewisi-specific neuropeptide precursors, the coding sequences of the OKB and CCAP transcripts were targeted for RT-PCR amplification. Amplimers of the expected sizes were obtained for these two transcripts (730 bp and 195 bp for OKB and CCAP, respectively; Supplementary Figure S1). All of the cloned products were found to have 100% nucleotide identity with the in silico sequences from the transcriptomic data.
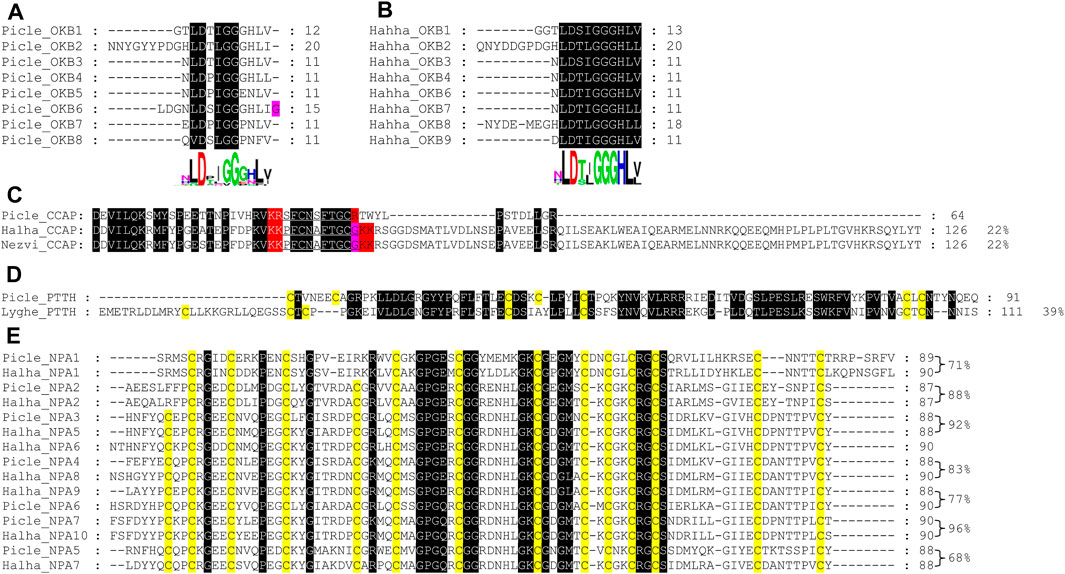
FIGURE 1. Multiple sequence alignment of four neuropeptides from P. lewisi and other bugs. (A,B) OKB active peptides from P. lewisi and H. halys. (C) CCAP precursors cutting signal peptides from P. lewisi, H. halys and N. viridula. (D) PTTH precursors cutting signal peptides from P. lewisi and L. hesperus. (E) NPA precursors cutting signal peptides from P. lewisi and H. halys. Full conservative residues are shaded in black background. Predicted convertase cleavage sites are shaded in red. Glycine residues are shaded in pink. Conserved cysteine residues are shaded in yellow. Identity values between the P. lewisi precursor and its ortholog are shown at the end.
The P. lewisi OKB precursor can encode eight potential active peptides that share the DXI/LGGG consensus sequence (Figure 1A). The H. halys OKB precursor can also encode eight distinct active peptides. Only one of the P. lewisi OKB active peptides shares the identical sequence (NLDTIGGGHLV) with an H. halys OKB peptide, whereas other P. lewisi OKB peptides had 45%∼90% identity with their H. halys orthologues (Figures 1A∼1B).
CCAP is known as the most conserved arthropod neuropeptide, with an identical amino acid sequence, PFCNAFTGCamide, found in all insects examined. Unexpectedly, the mature CCAP peptide predicted from P. lewisi (SFCNSFTGC) has two variant residues and lacks a C-terminal glycine residue (Figure 1C). The same CCAP peptide sequence was also found in P. lewisi RNA-seq SRA reads released in NCBI (e.g., SRR10134979.5654305.2).
The P. lewisi PTTH transcript has features typical of other insect PTTHs, encoding a putative active peptide containing seven cystine residues that form inter- and intra-chain disulfide bonds. PTTH has also been found in another heteropteran species, L. hesperus, but it is absent in a number of heteropteran species such as H. halys, N. viridula, and R. prolixus (Table 2). The P. lewisi PTTH peptide shares 39% amino acid identity with that of L. hesperus (Figure 1D).
A total of seven P. lewisi NPA transcripts were found (Supplementary Data S1). The P. lewisi NPA precursors can encode potential active peptides with a length of approximately 80 amino acid residues and 12∼14 cystine residues forming 6∼7 intrachain disulfide bridges (Figure 1E). Sequence alignment of the NPA peptides showed that Picle_NPA5 shares a relatively low identity (68%) with its H. halys orthologue, whereas other P. lewisi NPA peptides had 71%∼96% identity with their H. halys orthologs (Figure 1E).
A heatmap based on FPKM values of 57 neuropeptide genes (excluding AKH and SK, which were identified from the NCBI SRA database) in heads, salivary glands, and guts of P. lewisi, is shown in Table 1. A total of 22 neuropeptide genes were found to be expressed in salivary glands using a criterion of more than 1 FPKM in at least one repeat (Supplementary Table S3). Among them, six neuropeptides, OKB, CCAP, OKA, diuretic hormone 31 (DH31), ALP2, and ecdysis triggering hormone (ETH), showed FPKM values higher than ten in at least one repeat (Supplementary Table S3).
Compared to heads, only two neuropeptide genes (OKB and CCAP) were significantly more highly expressed in salivary glands, with log2 (fold) >1 and P_adj <0.05 (Table 1). OKB had the highest FPKM value (3228.1) in the salivary glands among all the identified neuropeptide genes and its relative expression level in the salivary glands compared to heads was 190.6-fold. CCAP was the second most highly expressed neuropeptide gene in the salivary glands (FPKM value = 109.3) and it was also more highly expressed in the salivary glands than heads, with an expression fold of 134.9. OKB, CNMa, AST-CC and CCHamide 2 (CCHa2) were significantly upregulated in guts than heads of P. lewisi (Table 1).
Using homology to research against our transcriptome data producted a total of 58 potential neuropeptide receptor genes (Table 3; Supplementary Data S2), including 41 family A GPCRs (rhodopsin-like receptors), seven family B GPCRs (secretin-like receptors), six RGCs and four RTKs. Eighteen neuropeptide receptor transcripts were partial sequences and the remaining 40 were full-length (Table 3; Supplementary Data S2). The family A neuropeptide GPCRs of P. lewisi can be classified into 27 groups based on their putative ligands: Receptors for AKH, AKH/corazonin-related peptide (ACP), AST-C, allatotropin (AT), CCAP, CNMa, corazonin (Crz), Ele, myosuppressin (MS), NTL, proctolin (Pro), SIFamide (SIFa), Short neuropeptide F (sNPF), SK, capability/cardioacceleratory peptide 2b (CAPA), ETH, pyrokinin (PK), AST-A, FMRFamide (FMRFa), tachykinins (TKs), AST-B, leucokinin (LK), bursicon (Bur), GPA2/GPB5, neuropeptide F (NPF), insulin-like peptide (ILP), and orphan (Table 3; Figure 2). No orthologous gene encoding the receptors for AVLP, RYamide (RYa) or trissin was identified in the P. lewisi transcriptome. The family B neuropeptide GPCRs of P. lewisi can be subdivided into four groups: receptors for DH31, DH44, pigment dispersing factor (PDF) and PTH (Table 3; Figure 2). Among RGCs, eclosion hormone receptor (EHR), Neuropeptide-like precursor receptor (NPLPR), and four orphan RGCs were found in P. lewisi (Table 3; Figure 3). In addition to RTKs, one PTTH receptor (PTTHR), two insulin-like peptide receptors (InR1 and InR2) and one orphan RTK receptor were identified (Table 3; Figure 3).
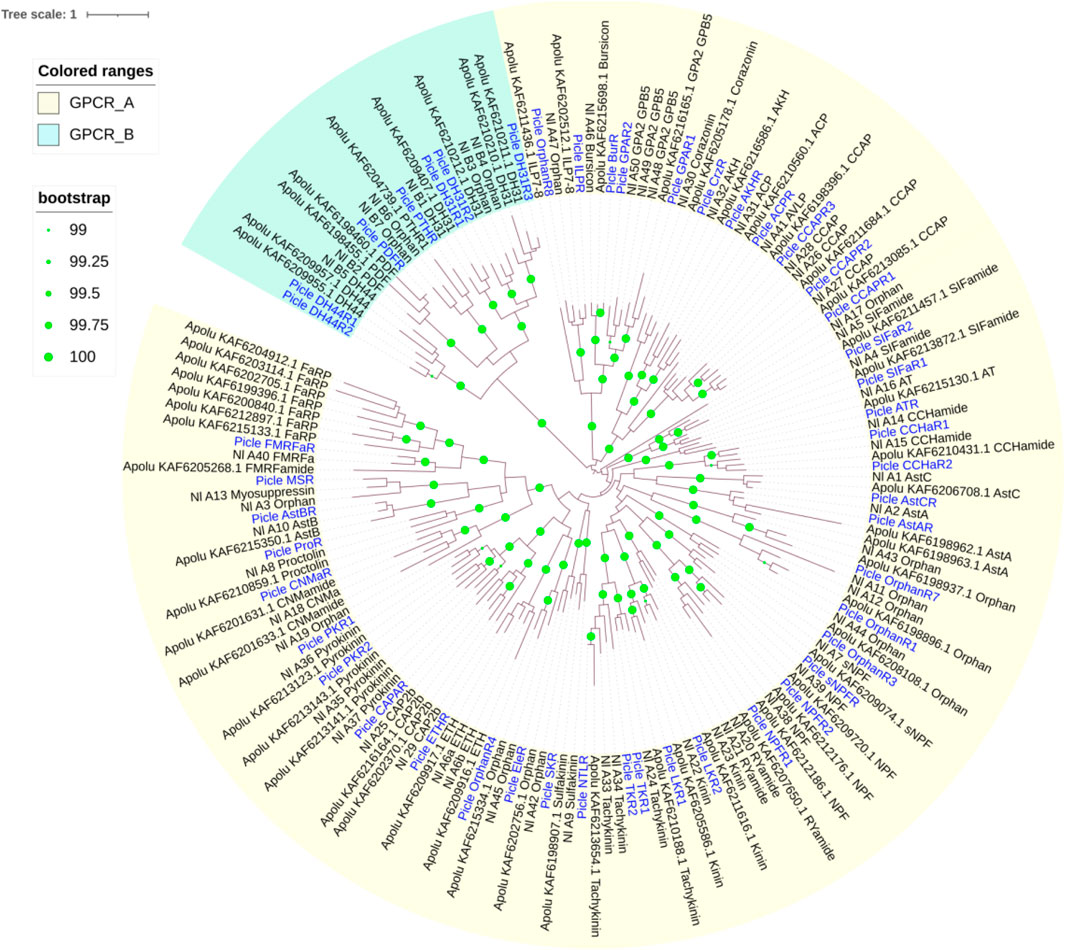
FIGURE 2. Phylogenetic tree of neuropeptide GPCR receptors from P. lewisi (gene IDs beginning “Picle”), A. lucorum (Apolu) and N. lugens (Nl). The ultrafast (UF) bootstrap value with more than 99% was marked in the tree. The P. lewisi gene names are marked with a blur color.
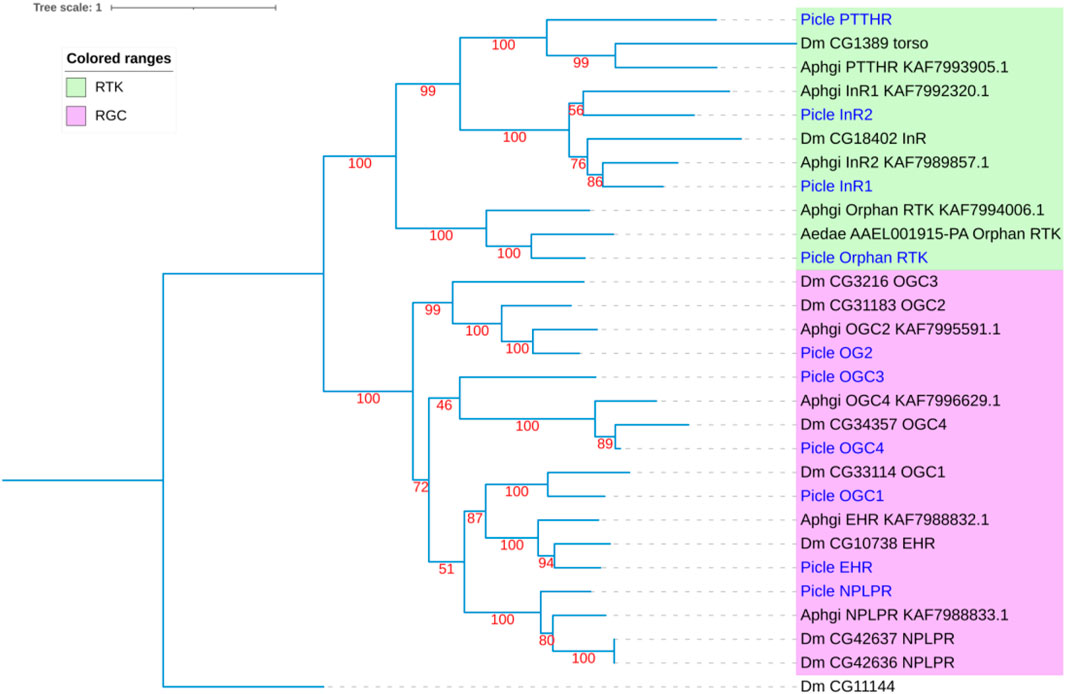
FIGURE 3. Phylogenetic tree of neuropeptide receptor guanylyl cyclases (RGCs) and receptor tyrosine kinases (RTKs) from P. lewisi (gene IDs beginning with “Picle”), D. melanogaster (Dm), Aphidius gifuensi (Aphgi), and Aedes aegypti (Aedae). The UF bootstrap value is marked in the tree. The P. lewisi gene names are marked with a blue color.
A heatmap based on FPKM values of 57 neuropeptide receptor genes (excluding AKHR, which was identified from the trinity sequences rather than the unigene sequences of our custom transcriptome) in heads, guts and salivary glands is shown in Table 3. A total of 15 out of 57 neuropeptide receptors were expressed in P. lewisi salivary glands with the FPKM values higher than one in at least one repeat, among which two receptor genes (EHR and NPLPR) presented the FPKM value higher than 10 in one repeat (Supplementary Table S4). Compared to heads, receptors for EH, NPLP, CAPA, GPA2/GPB5, Ele, DH44, and ILP, and one orphan RTK receptor were more highly expressed in the salivary glands than heads, with log 2 (fold) >1 and P_adj <0.05 (Table 3). Receptors for CAPA, NPLP, and GPA2/GPB5, and one orphan RTK receptor were significantly more highly expressed in guts than heads (Table 3).
In the present study, neuropeptide signaling genes were systematically identified in the predatory stink bug P. lewisi, with a total of 59 neuropeptide precursor genes and 58 potential neuropeptide receptor genes found. The number of neuropeptide precursors and their receptors identified in P. lewisi is similar to that found in H. halys and higher than those in other heteropterans such as N. viridula, L. hesperus, and R. prolixus (Christie et al., 2016; Lavore et al., 2018; Hull et al., 2021). In P. lewisi, nearly all of the neuropeptide signaling genes identified in other heteropterans were found, with the exception of GPB5 and RYaR. GPB5 and RYaR were not detected in the P. lewisi transcriptome, likely due to sample bias inherent to transcriptomes as opposed to genomes.
Tissue-specific expression profiles of neuropeptides and their receptors in P. lewisi provide basic information for in-depth studies of the biology and ecology of this important biological control species. The majority of neuropeptide precursor genes and their receptor genes in P. lewisi were found to be predominantly expressed in heads, indicating that most of neuropeptide signaling systems may act mainly as neuroregulators in the predatory bugs and/or derive from tissues in the head. A few neuropeptide signaling genes were also expressed in salivary glands and guts. These may play critical roles in the regulation of feeding and digestion. Neuropeptides in insect guts are involved in the regulation of feeding processes, including food choice, appetite, digestion, metabolism and excretion (Abou El Asrar et al., 2020). DH31, CCHa-1, CCHa-2, NPF, and CNMa are well-known insect neuropeptides associated with the gut-brain axis, regulating appetite, sleep and even courtship (Sano et al., 2015; Kim et al., 2021; Lin et al., 2022; Malita et al., 2022; Titos et al., 2023). In P. lewisi, three neuropeptides (CNMa, ASTCC and CCHa2) and five receptor genes were significantly upregulated in the guts when compared to heads, suggesting that they may have functions in feeding regulation.
Multiple neuropeptides and their receptors were identified as genes expressed in insect salivary glands. In the flyAtlas database (https://motif.mvls.gla.ac.uk/FlyAtlas2/), ion transport peptide (ITP), ILP6, NPLP2, NPLP4, and receptors for Crz, GPA2/GPB5, ILPs and NPLP1 were expressed in Drosophila salivary glands (Nässel and Zandawala, 2019). DH31, CCAP, CCAPR, AST-B, Pro and NPLP1 were expressed in R. prolixus salivary glands (Ons, 2017). FMRFa, AstA, SIFa and TK have also been detected in the salivary glands of other insects (Audsley and Weaver, 2009; Veenstra, 2020; Yu et al., 2020). Receptors for PDF, AST-A, DH44, TK, sNPF and DH31 were expressed at higher levels in the salivary glands of the ectoparasitoid, Habrobracon hebetor (Yu et al., 2020). In P. lewisi, three neuropeptide genes (CCAP and OKB) and eight receptors were enriched in salivary glands. In all these cases, the physiological functions of only a few neuropeptides and their receptors in insect salivary glands have been determined. More research is needed on this topic to describe specific functions.
Several insect neuropeptides have been implicated in the neural control of salivary production or secretion, such as FMRFa from the blow fly Calliphora vomitoria (Duve et al., 1992) and the kissing bug R. prolixus (Ons, 2017). In R. prolixus, AST-B, Pro, CCAP, and CCAPR have also been detected in processes innervating salivary glands, indicating their involvement in the hormonal control of salivary production or secretion (Ons, 2017). In ticks, several neuropeptides have been identified to be expressed in innervations of salivary glands, such as AST-B, DH31, Ele, ELeR, NPLP1, OKA, PDF, SIFa, and SIFaR (Sterkel et al., 2011; Simo et al., 2012; Ladislav et al., 2015; Kim et al., 2018; Vancová et al., 2019; Guerrib et al., 2023). CCAP, OKA, DH31, and NPLPR, which were highly expressed in P. lewisi, indicated their possible functions in neural control of the salivary system, as consistent with previous studies, however, expression of OKB and EHR in venom gland or salivary glands has not been reported in other insects so far.
Limited studies provide evidence for the functions of neuropeptides derived from invertebrate salivary glands as endocrine factors that regulate other tissues and organs. In addition, some of these salivary peptides might be venom toxins which alter the physiology of another species. A attractive study demonstrated that a peptide Sgsf expressed in Drosophila salivary glands can be secreted into the hemolymph and regulate Dilp2 secretion in the brain (Li et al., 2022b). Nässel et al. (2019) reviewed that TKs produced by salivary glands of mosquitos and cephalopods have been identified as exogenous vasodilators or paralyzing agents that can be delivered to prey. Determining whether neuropeptides expressed in the salivary glands of P. lewisi could be endogenous endocrine factors or exogenous venom peptides is important because venom peptides might be utilized as novel insecticidal peptides and/or signaling neuropeptides that their receptors might become useful targets for insecticides.
Our study presented species- and tissue-specific expression patterns of the neuropeptide signaling system in the predatory bugs, which will be used to generate testable functional genetic hypotheses in future studies. The most noteworthy neuropeptide is P. lewisi OKB, which was the most highly expressed neuropeptide in P. lewisi salivary glands. The expression pattern of OKB in the nervous system and intestine has also been reported in D. melanogaster, Bombyx mori, T. castaneum, R. prolixus, and Blattella germanica (Chen et al., 2015; Jiang et al., 2015; Ons et al., 2015; Wulff et al., 2017; Wang et al., 2019a). However, expression of OKB in salivary glands has not been reported in other insect species to date. OKs were first discovered with the myotropic activity in the crayfish Orconectes limosus (Stangier et al., 1992). One study suggested a role of OKB awakening behavior in T. castaneum (Ladislav et al., 2015). There is plenty of evidence for the recruitment of neuropeptides into animal venoms (Nässel et al., 2019; Sachkova et al., 2020; Goudarzi et al., 2023). Therefore, we would like to determine whether OKBs could be recruited as venom peptides in the salivary glands of P. lewisi and injected into prey to regulate myotropic activity or behavior. From an evolutionary perspective, the high genetic diversity of OKB peptide sequences possibly supports the repurposing of neuropeptides into venom peptides. A well-known example is TKs, another highly diverse and pleiotropic neuropeptide class, which have been convergently recruited into the venom or salivary glands of venomous invertebrates to affect prey (Nässel et al., 2019). This interesting evolutionary path of neuropeptide recruitment for novel toxins has also been recently revealed in the sea anemone Nematostella vectensis (ShK-like peptides) and the caterpillar Acharia stimulea (RF-amide peptides) (Sachkova et al., 2020; Goudarzi et al., 2023). Nevertheless, we cannot exclude the possibility that OKB peptides are endogenous endocrine factors. The OK receptor has not yet been identified in any species, therefore efforts to identify the OK receptor in insects will be crucial to understanding the role of OKBs in the salivary glands of P. lewisi.
The most unexpected result in the present study was finding a very atypical CCAP in P. lewisi. To our knowledge, CCAP is the identical neuropeptide (PFCNAFTGC-NH2) in all examined insect species. CCAP from P. lewisi (SFCNSFTGC) identified in the present study has two variant amino acid residues and no amidation at the C-terminus. Thus our findings indicate a highly novel primary structure for this peptide which we will seek to confirm through further studies utilizing liquid chromatography tandem mass spectrometry analysis. CCAP is mainly expressed in the central nervous system and/or in the intestine in most of the determined insects, such as D. melanogaster and R. prolixus (Lee et al., 2011; Shi et al., 2019). In R. prolixus, CCAP and its receptor have also been detected to be present in salivary glands (Lee et al., 2011; Lee and Lange, 2011; Lee et al., 2013). However, salivary gland-specific expression of CCAP in P. lewisi has not been reported in other insects to date. CCAP plays a crucial role in numerous biological and physiological processes in insects, mainly including the regulation of heart contraction, ecdysis, and feeding (Sakai et al., 2006; Estevez-Lao et al., 2013; Lee et al., 2013; Marco et al., 2018; Shi et al., 2019; Shen et al., 2021; Verbakel et al., 2021; Shi et al., 2022a), but its potential physiological function in the salivary glands of R. prolixus is still unknown. In P. lewisi, three putative CCAP receptors were identified, which were highly identical to their homologs (>90% identities) and highly expressed in heads, indicating that salivary gland-derived CCAP of P. lewisi could be considered as an endogenous endocrine factor. It is worth mentioning that the presence of the atypical CCAP in P. lewisi venom as a toxin cannot be ruled out given an intriguing example of a CCAP-related peptide discovered in the venom of Conus villepinii and having the activity of decreasing the heart frequency in Drosophila larvae (Möller et al., 2010).
The insect neuropeptidergic system has been considered as an ideal target for the development of greener pest control strategies. Comparative genomics and transcriptomics provide useful information for appropriate design strategies to develop target-specific insecticidal molecules that could successfully control pests while protecting beneficial species. To date, insecticidal activity and biosafety have been demonstrated for a few insect neuropeptides and their analogues, such as kinins, proctolin, CAPA, and TKs (Shi et al., 2022b). For example, insect kinins and their analogues were determined to exhibit high efficacy against aphids, however, they showed safety to an aphid predator, the common green lacewing Chrysoperla carnea, based on the transcriptome analysis information that insect kinins were not found in C. carnea (Shi et al., 2022b). Based on the comparative analysis of gene sets of known neuropeptides and their receptors between P. lewisi and other heteropterans, P. lewisi harbors almost all kinds of neuropeptide signaling system identified in other heteropteran species. Because of the conservation of this signaling system, it seems difficult to develop green pest control strategies based on neuropeptide systems putatively lost in benefical predatory bugs. Although sequence alignment of mature peptides between P. lewisi and its close herbivorous bug species showed several neuropeptides like OKB, CCAP, and PTTH, which are highly diverse in bugs, these neuropeptide signaling classes could be potential targets for the development of highly selective insecticidal agents needs to be further determined.
In the present study, a total of 59 neuropeptide precursors and 58 potential neuropeptide receptor genes were identified through transcriptomic analysis. The present study also revealed a set of neuropeptides and their receptors that were enriched in the salivary glands of P. lewisi, providing basic information for in-depth study on repurposing neuropeptides and their receptors into insecticides and targets.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SRR20681617∼SRR20681631.
The manuscript presents research on animals that do not require ethical approval for their study.
WL: Conceptualization, Methodology, Project administration, Writing–original draft. ZL: Methodology, Validation, Writing–review and editing. XY: Software, Writing–review and editing. XW: Investigation, Writing–review and editing. MY: Investigation, Writing–review and editing. CH: Funding acquisition, Supervision, Writing–review and editing. YH: Conceptualization, Data curation, Project administration, Software, Supervision, Writing–original draft.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the grants from Key Project of Guizhou Provincial Science and Technology Foundation (ZK[2023]023), Guizhou Provincial Tobacco Company Zunyi Branch (Science and Technology Major Project No. 2021XM01), and the grant from Guizhou Tobacco Company Qianxinan Company (Science and Technology Project No. 2022-08).
We gratefully acknowledge Dr. John F. Beckmann (Auburn University) for his careful revisions for our manuscript. We would like to thank the Fenggang County Natural Enemy Breeding Center at Zunyi city for providing the tested insects.
Authors ZL, XY, and CH were employed by Guizhou Provincial Tobacco Company Zunyi Branch.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fphys.2023.1270751/full#supplementary-material
Abou El Asrar R., Cools D., Vanden Broeck J. (2020). Role of peptide hormones in insect gut physiology. Curr. Opin. Insect Sci. 41, 71–78. doi:10.1016/j.cois.2020.07.004
Audsley N., Down R. E. (2015). G protein coupled receptors as targets for next generation pesticides. Insect Biochem. Mol. Biol. 67, 27–37. doi:10.1016/j.ibmb.2015.07.014
Audsley N., Weaver R. J. (2009). Neuropeptides associated with the regulation of feeding in insects. Gen. Comp. Endocrinol. 162, 93–104. doi:10.1016/j.ygcen.2008.08.003
Benoit J. B., Adelman Z. N., Reinhardt K., Dolan A., Poelchau M., Jennings E. C., et al. (2016). Unique features of a global human ectoparasite identified through sequencing of the bed bug genome. Nat. Commun. 7, 10165. doi:10.1038/ncomms10165
Caers J., Verlinden H., Zels S., Vandersmissen H. P., Vuerinckx K., Schoofs L. (2012). More than two decades of research on insect neuropeptide GPCRs: an overview. Front. Endocrinol. (Lausanne) 3, 151. doi:10.3389/fendo.2012.00151
Champagne D. E., Ribeiro J. M. (1994). Sialokinin I and II: vasodilatory tachykinins from the yellow fever mosquito Aedes aegypti. Proc. Natl. Acad. Sci. U. S. A. 91, 138–142. doi:10.1073/pnas.91.1.138
Chen J., Choi M. S., Mizoguchi A., Veenstra J. A., Kang K., Kim Y. J., et al. (2015). Isoform-specific expression of the neuropeptide orcokinin in Drosophila melanogaster. Peptides 68, 50–57. doi:10.1016/j.peptides.2015.01.002
Christie A. E., Hull J. J., Richer J. A., Geib S. M., Tassone E. E. (2016). Prediction of a peptidome for the Western tarnished plant bug Lygus hesperus. Gen. Comp. Endocrinol. 243, 22–38. doi:10.1016/j.ygcen.2016.10.008
Cohen A. C. (1995). Extra-oral digestion in predaceous terrestrial Arthropoda. Annu. Rev. Entomol. 40, 85–103. doi:10.1146/annurev.en.40.010195.000505
Cohen A. C. (1990). Feeding adaptations of some predaceous Hemiptera. Ann. Entomol. Soc. Am. 83, 1215–1223. doi:10.1093/aesa/83.6.1215
Crooks G. E., Hon G. C., Chandonia J., Brenner S. E. (2004). WebLogo: A sequence logo generator. Genome Res. 14, 1188–1190. doi:10.1101/gr.849004
Darriba D., Posada D., Kozlov A. M., Stamatakis A., Morel B., Flouri T. (2019). ModelTest-NG: A new and scalable tool for the selection of DNA and protein evolutionary models. Mol. Biol. Evol. 37, 291–294. doi:10.1093/molbev/msz189
Duve H., Johnsen A. H., Sewell J. C., Scott A. G., Orchard I., Rehfeld J. F., et al. (1992). Isolation, structure, and activity of -Phe-Met-Arg-Phe-NH2 neuropeptides (designated calliFMRFamides) from the blowfly Calliphora vomitoria. Proc. Natl. Acad. Sci. U. S. A. 89, 2326–2330. doi:10.1073/pnas.89.6.2326
Estevez-Lao T. Y., Boyce D. S., Honegger H. W., Hillyer J. F. (2013). Cardioacceleratory function of the neurohormone CCAP in the mosquito Anopheles gambiae. J. Exp. Biol. 216, 601–613. doi:10.1242/jeb.077164
Gao H., Li Y., Wang M., Song X., Tang J., Feng F., et al. (2021). Identification and expression Aanalysis of G protein-coupled receptors in the Miridae insect Apolygus lucorum. Front. Endocrinol. 12, 773669. doi:10.3389/fendo.2021.773669
Goudarzi M. H., Eagles D. A., Lim J., Biggs K. A., Kotze A. C., Ruffell A. P., et al. (2023). Venom composition and bioactive RF-amide peptide toxins of the saddleback caterpillar, Acharia stimulea (Lepidoptera: limacodidae). Biochem. Pharmacol. 213, 115598. doi:10.1016/j.bcp.2023.115598
Guerrib F., Ning C., Mateos-Hernandéz L., Rakotobe S., Park Y., Hajdusek O., et al. (2023). Dual SIFamide receptors in Ixodes salivary glands. Insect Biochem. Mol. Biol. 158, 103963. doi:10.1016/j.ibmb.2023.103963
Hoang D. T., Chernomor O., von Haeseler A., Minh B. Q., Vinh L. S. (2017). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi:10.1093/molbev/msx281
Hull J. J., Gross R. J., Brent C. S., Christie A. E. (2021). Filling in the gaps: A reevaluation of the Lygus hesperus peptidome using an expanded de novo assembled transcriptome and molecular cloning. Gen. Comp. Endocrinol. 303, 113708. doi:10.1016/j.ygcen.2020.113708
Jiang H., Kim H. G., Park Y. (2015). Alternatively spliced orcokinin isoforms and their functions in Tribolium castaneum. Insect Biochem. Mol. Biol. 65, 1–9. doi:10.1016/j.ibmb.2015.07.009
Jung S. H., Lee J. H., Chae H. S., Seong J. Y., Park Y., Park Z. Y., et al. (2014). Identification of a novel insect neuropeptide, CNMa and its receptor. FEBS Lett. 588, 2037–2041. doi:10.1016/j.febslet.2014.04.028
Kanda A., Iwakoshi-Ukena E., Takuwa-Kuroda K., Minakata H. (2003). Isolation and characterization of novel tachykinins from the posterior salivary gland of the common octopus Octopus vulgaris. Peptides 24, 35–43. doi:10.1016/s0196-9781(02)00274-7
Kim B., Kanai M. I., Oh Y., Kyung M., Kim E. K., Jang I. H., et al. (2021). Response of the microbiome-gut-brain axis in Drosophila to amino acid deficit. Nature 593, 570–574. doi:10.1038/s41586-021-03522-2
Kim D., Šimo L., Park Y. (2018). Molecular characterization of neuropeptide elevenin and two elevenin receptors, IsElevR1 and IsElevR2, from the blacklegged tick, Ixodes scapularis. Insect Biochem. Mol. Biol. 101, 66–75. doi:10.1016/j.ibmb.2018.07.005
Kong X., Li Z. X., Gao Y. Q., Liu F. H., Chen Z. Z., Tian H. G., et al. (2021). Genome-wide identification of neuropeptides and their receptors in an aphid endoparasitoid wasp, Aphidius gifuensi. Insects 12, 745. doi:10.3390/insects12080745
Ladislav R., Ladislav S., Akira M., Mirko S., Yoonseong P., Dusan Z. (2015). Orcokinin-like immunoreactivity in central neurons innervating the salivary glands and hindgut of ixodid ticks. Cell Tissue Res. 360, 209–222. doi:10.1007/s00441-015-2121-z
Lavore A., Perez-Gianmarco L., Esponda-Behrens N., Palacio V., Catalano M. I., Rivera-Pomar R., et al. (2018). Nezara viridula (Hemiptera: pentatomidae) transcriptomic analysis and neuropeptidomics. Sci. Rep. 8, 17244. doi:10.1038/s41598-018-35386-4
Lee D., Orchard I., Lange A. B. (2013). Evidence for a conserved CCAP-signaling pathway controlling ecdysis in a hemimetabolous insect, Rhodnius prolixus. Front. Neurosci. 7, 207. doi:10.3389/fnins.2013.00207
Lee D. H., Lange A. B. (2011). Crustacean cardioactive peptide in the chagas' disease vector, Rhodnius prolixus: presence, distribution and physiological effects. Gen. Comp. Endocrinol. 174, 36–43. doi:10.1016/j.ygcen.2011.08.007
Lee D. H., Paluzzi J. P., Orchard I., Lange A. B. (2011). Isolation, cloning and expression of the Crustacean Cardioactive Peptide gene in the Chagas' disease vector, Rhodnius prolixus. Peptides 32, 475–482. doi:10.1016/j.peptides.2010.06.035
Letunić I., Bork P. (2021). Interactive tree of Life (iTOL) v5: an online tool for phylogenetic tree display and annotation. Nucleic Acids Res. 49, W293–W296. doi:10.1093/nar/gkab301
Li W., Wang X., Jiang P., Yang M., Li Z., Huang C., et al. (2022a). A full-length transcriptome and gene expression analysis of three detoxification gene families in a predatory stink bug, Picromerus lewisi. Front. Physiol. 13, 1016582. doi:10.3389/fphys.2022.1016582
Li Z., Qian W., Song W., Zhao T., Yang Y., Wang W., et al. (2022b). A salivary gland-secreted peptide regulates insect systemic growth. Cell Rep. 38, 110397. doi:10.1016/j.celrep.2022.110397
Lin H. H., Kuang M. C., Hossain I., Xuan Y., Beebe L., Shepherd A. K., et al. (2022). A nutrient-specific gut hormone arbitrates between courtship and feeding. Nature 602, 632–638. doi:10.1038/s41586-022-04408-7
Lin Y., Long J., Zhang S., Lin Z. (2000). A checklist of asopinae from China (Hemiptera: pentatomidae). Jiangxi Plant Prot. 2, 36–39.
Malita A., Kubrak O., Koyama T., Ahrentlov N., Texada M. J., Nagy S., et al. (2022). A gut-derived hormone suppresses sugar appetite and regulates food choice in Drosophila. Nat. Metab. 4, 1532–1550. doi:10.1038/s42255-022-00672-z
Marco H. G., Katali O. K. H., Gade G. (2018). Influence of aminergic and peptidergic substances on heart beat frequency in the stick insect Carausius morosus (Insecta, Phasmatodea). Arch. Insect Biochem. Physiol. 98, e21469. doi:10.1002/arch.21469
Minh B. Q., Schmidt H. A., Chernomor O., Schrempf D., Woodhams M. D., von Haeseler A., et al. (2020). IQ-TREE 2: new models and efficient methods for phylogenetic inference in the genomic era. Mol. Biol. Evol. 37, 1530–1534. doi:10.1093/molbev/msaa015
Möller C., Melaun C., Castillo C., Díaz M. E., Renzelman C. M., Estrada O., et al. (2010). Functional hypervariability and gene diversity of cardioactive neuropeptides. J. Biol. Chem. 285, 40673–40680. doi:10.1074/jbc.M110.171397
Mu Y. L., Zhang C. H., Zhang Y. J., Yang L., Chen X. S. (2022). Characterizing the complete mitochondrial genome of Arma custos and Picromerus lewisi (Hemiptera: pentatomidae: asopinae) and conducting phylogenetic analysis. J. Insect Sci. 22, 6. doi:10.1093/jisesa/ieab105
Nässel D. R., Zandawala M., Kawada T., Satake H. (2019). Tachykinins: neuropeptides that are ancient, diverse, widespread and functionally pleiotropic. Front. Neurosci. 13, 1262. doi:10.3389/fnins.2019.01262
Nässel D. R., Zandawala M. (2019). Recent advances in neuropeptide signaling in Drosophila, from genes to physiology and behavior. Prog. Neurobiol. 179, 101607. doi:10.1016/j.pneurobio.2019.02.003
Nicholas K. B., Nicholas H. B., Deerfield D. W. (1997). GeneDoc: analysis and visualization of genetic variation. EMBNEW News 4, 14.
Ons S., Belles X., Maestro J. L. (2015). Orcokinins contribute to the regulation of vitellogenin transcription in the cockroach Blattella germanica. J. Insect Physiol. 82, 129–133. doi:10.1016/j.jinsphys.2015.10.002
Ons S., Lavore A., Sterkel M., Wulff J. P., Sierra I., Martinez-Barnetche J., et al. (2016). Identification of G protein coupled receptors for opsines and neurohormones in Rhodnius prolixus. Genomic and transcriptomic analysis. Insect Biochem. Mol. Biol. 69, 34–50. doi:10.1016/j.ibmb.2015.05.003
Ons S. (2017). Neuropeptides in the regulation of Rhodnius prolixus physiology. J. Insect Physiol. 97, 77–92. doi:10.1016/j.jinsphys.2016.05.003
Ons S., Sterkel M., Diambra L., Urlaub H., Rivera-Pomar R. (2011). Neuropeptide precursor gene discovery in the Chagas disease vector Rhodnius prolixus. Insect Mol. Biol. 20, 29–44. doi:10.1111/j.1365-2583.2010.01050.x
Sachkova M. Y., Landau M., Surm J. M., Macrander J., Singer S. A., Reitzel A. M., et al. (2020). Toxin-like neuropeptides in the sea anemone Nematostella unravel recruitment from the nervous system to venom. Proc. Natl. Acad. Sci. U. S. A. 117, 27481–27492. doi:10.1073/pnas.2011120117
Sakai T., Satake H., Takeda M. (2006). Nutrient-induced alpha-amylase and protease activity is regulated by crustacean cardioactive peptide (CCAP) in the cockroach midgut. Peptides 27, 2157–2164. doi:10.1016/j.peptides.2006.04.009
Sano H., Nakamura A., Texada M. J., Truman J. W., Ishimoto H., Kamikouchi A., et al. (2015). The nutrient-responsive hormone CCHamide-2 controls growth by regulating insulin-like peptides in the brain of Drosophila melanogaster. PLoS Genet. 11, e1005209. doi:10.1371/journal.pgen.1005209
Schoofs L., De Loof A., Van Hiel M. B. (2017). Neuropeptides as regulators of behavior in insects. Annu. Rev. Entomol. 62, 35–52. doi:10.1146/annurev-ento-031616-035500
Shen C. H., Jin L., Fu K. Y., Guo W. C., Li G. Q. (2021). Crustacean cardioactive peptide as a stimulator of feeding and a regulator of ecdysis in Leptinotarsa decemlineata. Pestic. Biochem. Physiol. 175, 104838. doi:10.1016/j.pestbp.2021.104838
Shi Y., Liu T. Y., Ding B. Y., Niu J., Jiang H. B., Liu T. X., et al. (2022a). Crustacean cardioactive peptide and its receptor modulate the ecdysis behavior in the pea aphid, Acyrthosiphon pisum. J. Insect Physiol. 137, 104364. doi:10.1016/j.jinsphys.2022.104364
Shi Y., Liu T. Y., Pei Y. X., Jiang H. B., Dou W., Smagghe G., et al. (2019). Crustacean cardioactive peptide (CCAP) of the oriental fruit fly, Bactrocera dorsalis (Diptera: tephritidae): molecular characterization, distribution and its potential roles in larva-pupa ecdysis. Peptides 122, 169929. doi:10.1016/j.peptides.2018.02.007
Shi Y., Pandit A., Nachman R. J., Christiaens O., Davies S. A., Dow J. A. T., et al. (2022b). Transcriptome analysis of neuropeptides in the beneficial insect lacewing (Chrysoperla carnea) identifies kinins as a selective pesticide target: A biostable kinin analogue with activity against the peach potato aphid Myzus persicae. J. Pest Sci. 96, 253–264. doi:10.1007/s10340-022-01511-6
Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M. (2015). BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212. doi:10.1093/bioinformatics/btv351
Simo L., Zitnan D., Park Y. (2012). Neural control of salivary glands in ixodid ticks. J. Insect Physiol. 58, 459–466. doi:10.1016/j.jinsphys.2011.11.006
Spit J., Badisco L., Verlinden H., Van Wielendaele P., Zels S., Dillen S., et al. (2012). Peptidergic control of food intake and digestion in insects1This review is part of a virtual symposium on recent advances in understanding a variety of complex regulatory processes in insect physiology and endocrinology, including development, metabolism, cold hardiness, food intake and digestion, and diuresis, through the use of omics technologies in the postgenomic era. Can. J. Zool. 90, 489–506. doi:10.1139/z2012-014
Stangier J., Hilbich C., Burdzik S., Keller R. (1992). Orcokinin: A novel myotropic peptide from the nervous system of the crayfish, Orconectes limosus. Peptides 13, 859–864. doi:10.1016/0196-9781(92)90041-z
Sterkel M., Urlaub H., Rivera-Pomar R., Ons S. (2011). Functional proteomics of neuropeptidome dynamics during the feeding process of Rhodnius prolixus. J. Proteome Res. 10, 3363–3371. doi:10.1021/pr2001012
Tanaka Y., Suetsugu Y., Yamamoto K., Noda H., Shinoda T. (2014). Transcriptome analysis of neuropeptides and G-protein coupled receptors (GPCRs) for neuropeptides in the brown planthopper Nilaparvata lugens. Peptides 53, 125–133. doi:10.1016/j.peptides.2013.07.027
Tang Y., Wang M., Chen H., Wang Y., Zhang H., Chen F., et al. (2019). Predatory capacity and behavior of Picromerus lewisi Scott against Spodoptera frugiperda higher instar larve. Chin. J. Biol. Control 35, 698–703. doi:10.16409/j.cnki.2095-039x.2019.04.005
Titos I., Juginovic A., Vaccaro A., Nambara K., Gorelik P., Mazor O., et al. (2023). A gut-secreted peptide suppresses arousability from sleep. Cell 186, 1382–1397 e21. doi:10.1016/j.cell.2023.02.022
Uchiyama H., Maehara S., Ohta H., Seki T., Tanaka Y. (2017). Elevenin regulates the body color through a G protein-coupled receptor NlA42 in the brown planthopper Nilaparvata lugens. Gen. Comp. Endocrinol. 258, 33–38. doi:10.1016/j.ygcen.2017.07.017
Vancová M., Bílý T., Nebesářová J., Grubhoffer L., Bonnet S., Park Y., et al. (2019). Ultrastructural mapping of salivary gland innervation in the tick Ixodes ricinus. Sci. Rep. 9, 6860. doi:10.1038/s41598-019-43284-6
Veenstra J. A. (2000). Mono- and dibasic proteolytic cleavage sites in insect neuroendocrine peptide precursors. Arch. Insect Biochem. Physiol. 43, 49–63. doi:10.1002/(SICI)1520-6327(200002)43:2<49:AID-ARCH1>3.0.CO;2-M
Veenstra J. A. (2020). The neuropeptide SMYamide, a SIFamide paralog, is expressed by salivary gland innervating neurons in the American cockroach and likely functions as a hormone. Peptides 136, 170466. doi:10.1016/j.peptides.2020.170466
Verbakel L., Lenaerts C., Abou El Asrar R., Zandecki C., Bruyninckx E., Monjon E., et al. (2021). Prothoracicostatic activity of the ecdysis-regulating neuropeptide Crustacean Cardioactive Peptide (CCAP) in the desert locust. Int. J. Mol. Sci. 22, 13465. doi:10.3390/ijms222413465
Verlinden H., Vleugels R., Zels S., Dillen S., Lenaerts C., Crabbé K., et al. (2014). Receptors for neuronal or endocrine signalling molecules as potential targets for the control of insect pests. Adv Insect Phys 46, 167–303. doi:10.1016/B978-0-12-417010-0.00003-3
Walker A. A., Hernandez-Vargas M. J., Corzo G., Fry B. G., King G. F. (2018). Giant fish-killing water bug reveals ancient and dynamic venom evolution in Heteroptera. Cell Mol. Life Sci. 75, 3215–3229. doi:10.1007/s00018-018-2768-1
Walker A. A., Weirauch C., Fry B. G., King G. F. (2016). Venoms of heteropteran insects: A treasure trove of diverse pharmacological toolkits. Toxins 8, 43. doi:10.3390/toxins8020043
Wang P., Zhao Q., Qiu Z., Bi S., Wang W., Wu M., et al. (2019a). The silkworm (Bombyx mori) neuropeptide orcokinin is involved in the regulation of pigmentation. Insect Biochem. Mol. Biol. 114, 103229. doi:10.1016/j.ibmb.2019.103229
Wang Y., Wang M., Zhang H., Zhao X., Yin Y., Li X., et al. (2019b). Predation potential of adult of Picromerus lewisi (fallou) on larvae of Spodoptera frugiperda. Chin. J. Biol. Control 35, 691–697. doi:10.16409/j.cnki.2095-039x.2019.05.006
Wulff J. P., Sierra I., Sterkel M., Holtof M., Van Wielendaele P., Francini F., et al. (2017). Orcokinin neuropeptides regulate ecdysis in the hemimetabolous insect Rhodnius prolixus. Insect Biochem. Mol. Biol. 81, 91–102. doi:10.1016/j.ibmb.2017.01.003
Xie J., Sang M., Song X., Zhang S., Kim D., Veenstra J. A., et al. (2020). A new neuropeptide insect parathyroid hormone iPTH in the red flour beetle Tribolium castaneum. PLoS Genet. 16, e1008772. doi:10.1371/journal.pgen.1008772
Yu K., Xiong S., Xu G., Ye X., Yao H., Wang F., et al. (2020). Identification of neuropeptides and their receptors in the ectoparasitoid, Habrobracon hebetor. Front. Physiol. 11, 575655. doi:10.3389/fphys.2020.575655
Keywords: Picromerus lewisi, predatory stink bug, neuropeptide, neuropeptide receptor, transcriptome, salivary gland
Citation: Li W, Li Z, Yang X, Wang X, Yang M, Huang C and He Y (2023) Transcriptome analysis reveals salivary gland-specific neuropeptide signaling genes in the predatory stink bug, Picromerus lewisi. Front. Physiol. 14:1270751. doi: 10.3389/fphys.2023.1270751
Received: 01 August 2023; Accepted: 19 September 2023;
Published: 29 September 2023.
Edited by:
Hongbo Jiang, Southwest University, ChinaReviewed by:
Pawel Marciniak, Adam Mickiewicz University, PolandCopyright © 2023 Li, Li, Yang, Wang, Yang, Huang and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunyang Huang, hcy_0701@126.com; Yueping He, heyp@mail.hzau.edu.cn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.