
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Nutr. , 23 February 2022
Sec. Food Chemistry
Volume 9 - 2022 | https://doi.org/10.3389/fnut.2022.851402
This article is part of the Research Topic Microbial Bioactive Lipids: Their Identification, Biosynthesis and Biochemistry View all 10 articles
A correction has been applied to this article in:
Corrigendum: Key Enzymes in Fatty Acid Synthesis Pathway for Bioactive Lipids Biosynthesis
Dietary bioactive lipids, one of the three primary nutrients, is not only essential for growth and provides nutrients and energy for life's activities but can also help to guard against disease, such as Alzheimer's and cardiovascular diseases, which further strengthen the immune system and maintain many body functions. Many microorganisms, such as yeast, algae, and marine fungi, have been widely developed for dietary bioactive lipids production. These biosynthetic processes were not limited by the climate and ground, which are also responsible for superiority of shorter periods and high conversion rate. However, the production process was also exposed to the challenges of low stability, concentration, and productivity, which was derived from the limited knowledge about the critical enzyme in the metabolic pathway. Fortunately, the development of enzymatic research methods provides powerful tools to understand the catalytic process, including site-specific mutagenesis, protein dynamic simulation, and metabolic engineering technology. Thus, we review the characteristics of critical desaturase and elongase involved in the fatty acids' synthesis metabolic pathway, which aims to not only provide extensive data for enzyme rational design and modification but also provides a more profound and comprehensive understanding of the dietary bioactive lipids' synthetic process.
Lipid plays a critical role in maintaining the normal function of growth and metabolism, which is not only an essential factor for the fluidity of the plasma membrane but is also a carrier to store material and energy (1). Long chain polyunsaturated fatty acids (LCPUFAs) are the main active and functional components in lipid. With the continuous improvement of people's life quality, more and more attention is paid to the intake and proportion of various kinds of LCPUFAs in the daily diet (2). The fatty acids in the daily diet are mainly consisted of saturated ones and unsaturated ones. According to the different positions of unsaturated double bonds, LCPUFAs can be mainly divided into ω3 and ω6 series, such as docosahexaenoic acid (DHA, C22:6Δ4,7,10,13,16,19 ω3), eicosapentaenoic acid (EPA, C20:4Δ5,8,11,14,17, ω3), and arachidonic acid (AA, C20:4Δ5,8,11,14, ω6) (3). These LCPUFAs also play a crucial role in not only growth and brain development but also in preventing cardiovascular diseases, hypertension, and diabetes (4). Therefore, the Food and Agriculture Organization of the United Nations (FAO) and the World Health Organization (WHO) issued a joint statement that the intake of LCPUFAs fatty acids daily should not be <1.3 g (5). However, mammals can't de novo synthesize these LCPUFAs, which make getting supplements from diet to be particularly important (6).
In daily diet, the intake of LCPUFAs is the primary driver of deep-sea fish and vegetable oil. Adapt to a low-temperature marine environment, deep-sea fish oil is rich in LCPUFAs, enriched from the marine microalgae (7, 8). Some plants such as soybeans, flax, and peanut, can store lipid in their seed, from which humans could extract and harvest linoleic acid (LA, 18:2Δ9,12, ω6) and α-linolenic acid (ALA, C18:3Δ9,12,15, ω3) rich oil (9, 10). However, harvesting the LCPUFAs from plants and deep-sea fish is a time-consuming process, which was also limited by climate, land and ecological environment (11, 12). Furthermore, the plant could not provide LCPUFAs of DHA and ARA, while overfishing caused more deadly consequences, including destructive fishing practices, killing off the fish, and breaking the ecological balance (13). Thus, with the explosion of population, the LCPUFAs harvest speed from these two approaches can no longer meet market demand needs. Fortunately, the development of LCPUFAs from oleaginous microorganisms and microalgae provides an alternative source for increasing market demand. At present, many LCPUFAs' production processes have been developed, including Schizochytrium sp., Mortierella alpina, Thraustochytrids and Yarrowia lipolytica (14–17). Compared with LCPUFAs from plant and fish oil, the LCPUFAs driver from microorganisms and microalgae own many advantages. On the one hand, it broke the loose of climate and ground; on the other hand, higher proliferation, growth and LCPUFAs rich oil accumulation rate enhance the productivity and low the cost. Thus, the development LCPUFAs rich oil strategies attract more attention from scientists worldwide, promoting the development and research rate significantly.
Based on the previous studies, the synthesis approaches of LCPUFAs mainly consisted of the fatty acid synthesis pathway (FAS) and polyketosynthase (PKS) pathway (18, 19). Compared with PKS, the FAS pathway is more extensive and typical in all oleogenic microorganisms. As shown in Figure 1, many kinds of fatty acid desaturases and elongases play an essential role in synthesizing LCPUFAs, which perform the functions of introducing double bonds and extending the carbon chain. Thus, this paper reviews the features of and advances of these critical enzymes in LCPUFAs synthetic pathway. We also discuss the challenge and the most promising breakthrough direction of enzyme in LCPUFAs synthetic pathway, which aims to provide detailed information and novel ideas for the follow-up research in this field.
Δ9-desaturase is a central enzyme to synthesize the long chain monounsaturated fatty acids (LCMUFAs) from long chain saturated fatty acids (LCSFAs), which catalyze stearic acid (SA, 18:0) and palmitic acid (PA; C16:0) to oleic acid (OA, 18:1Δ9) and palmitoleic acid (POA, C16:1Δ9) (Figure 1). Scientists also have identified and characterized the various Δ9-fatty acid desaturase gene from Rhodotorula toruloides (20) and Pseudomonas sp. (21) (Table 1). These Δ9-desaturases shared three histidine-conserved boxes that would be the catalytic position of Δ9 fatty acid desaturase. The RtΔ9FAD protein was also predicted to have four possible transmembrane domains (54).
As the obstacles in harvest crystal structures of fatty acid desaturases located in the membrane, homology modeling could predict the three-dimensional structures. The docking studies could be employed to predict the active center of the desaturases. Predicted by docking results, His34, His71, and, His206 were the possible residues to contact with the docked palmitic acid, located in the first, second, and third conserved histidine boxes (21). Another study of homology modeling and docking also showed that four α-helices constitute the catalytic site and transmembrane domain in Δ9-desaturase from Arthrospira platensis (55). Beyond that, 3D structure modeling studies revealed that three variant amino acids (F160, A223, and L156) in domain Δ9-desaturase encoded by gene PtAAD narrow the space of substrate binding center, explaining the reason of substrate preference of this especial Δ9-desaturase for palmitic acid (22, 23) (Table 2).
By overexpressing the RtΔ9FAD gene into R. toruloides, the transformant produces 5-fold more oleic acid content in total amount (54). At the same time, Liu and his colleague (23) reported a Δ9-desaturase from Phaeodactylum tricornutum to prefer palmitic acid-ACP as a substrate to promote the palmitoleic acid under average and stress culture conditions. Rau and his colleague also find that the Δ9-desaturase from Aurantiochytrium sp. T66 could accept palmitic acid and stearic acid as substrates (59). The reasonable explanation for substrate preference was also given out through homology modeling and docking in the last paragraph.
Δ12-desaturase is a vital enzyme in the first step of LCPUFAs synthesized from LCMUFAs, which catalyze oleic acid (OA, 18:1Δ9) to linoleic acid (LA, 18:2Δ9,12) (Figure 1). Scientists also have identified and characterized the various Δ12-fatty acid desaturase gene from Isochrysis galbana (24), Acanthamoeba castellanii (25), Chlamydomonas sp. (26), Calendula officinalis (27), Helianthus annuus (28), Chlorella vulgaris (29), P. tricornutum (30), and Haematococcus pluvialis (31), whose detailed information is listed in Table 1. These Δ12-desaturases also share three conserved histidine boxes, which are considered the catalytic center and are critical for desaturase activity.
Different environment stressors will influence the FAD2 (coding Δ12-desaturase) transcription of the oleaginous microorganism. In Y. lipolytica, the low temperature or substrate (n-alkanes or oleic acid) induce the upregulation of Δ12-desaturase (60). Similarly, low temperature (15°C), high salinity (salinity of 62 and 93%), and nitrogen starvation (220 μmol/L) upregulate the abundance of IgFAD2 transcript in I. galbana as well (24).
Overexpress Δ12-desaturase in oleogenic microorganisms could enhance the product accumulation significantly. After heterologous overexpress Δ12-desaturase from M. alpina or Fusarium verticillioides in the oleaginous yeast Rhodosporidium toruloides, linoleic acid concentration increased up to 1.3 g/L, which was 5-fold higher than that in the parent strain (61). Overexpression of endogenous RtFAD2 in R. toruloides also improved lipid and linoleic acid (32). Conversely, the growth rate was slower at 12°C under the deletion of FAD2 gene (coding Δ12-desaturase) in Y. lipolytica, which was recovered by the addition of 18:2, not 18:1.
Δ6-desaturase catalyze linoleic acid (LA, C18:2Δ9,12) and α-linolenic acid (ALA, C18:3Δ9,12,15) to γ-linolenic acid (GLA, C18:3Δ6,9,12) and produce stearidonic acid (SDA, C18:4Δ6,9,12,15). Scientists also have identified and characterized the various Δ6-desaturase from Glossomastix chrysoplasta (33), Pythium sp. (39), Isochrysis sp. (42), and Mucor sp. (43). Araki and his colleague identified a novel gene encoding Δ6- desaturase from Rhodococcus sp. Different from others, this desaturase preferred saturated fatty acids as substrates and catalyzed hexadecanoic acid to cis-6-hexadecenoic acid (38) (Table 1).
By analyzing the sequence of the Δ6-desaturase from Mucor sp., Jiang et al. found that −919 to −784 bp in the promoter region plays a vital role in the high activity of Δ6-desaturase (62). Compared with native Δ6-desaturase in Pythium sp., the codon-optimized strategy could markedly enhance Δ-6 desaturated products, in which the substrate conversion rates of LA and ALA increased from 5.4 and 4.2% to 62.7 and 60.9%, respectively (39). Zhu et al. (63) found that overexpression of endogenous Δ6-desaturase significantly enhances the eicosapentaenoic acid accumulation in P. tricornutum, compared with overexpression of heterologous one (64). However, the other scientist reported another result that differed massively in M. alpina. First, the Δ-6 desaturase from M. alpina (57) and M. pusilla (65) has the characteristic of significant substrate preference, in which the MpFADS6 (from M. pusilla) and MaFADS6-I (from M. alpina) prefer to ALA and LA, respectively. Zhang et al. (34) also isolated and identified a Δ6-desaturase from Pythium splendens with a preference to LA. Second, they further introduced the exogenous gene encoding ALA-preferring Δ6-desaturase from M. pusilla into the M. alpina, EPA yield was increased from 22.99 ± 2.7 mg/L in WT M. alpina up to 588.5 ± 29.6 mg/L in engineered one in 5-L fermentation, in which peony seed oil (0.1%) and peony seed meal (50 g/L) were exogenously added as a substitution to ALA (66). Last, they introduced the exogenous gene from Thalassiosira pseudonana (41), encoding a higher Δ6-desaturase activity (TpFADS6) for ALA, to the high ALA producer of Dunaliella salina (40). After performing culture conditions optimization, the EPA concentration increased from 1.6 ± 0.2 to 554.3 ± 95.6 mg/L, 343.8-fold higher than that in the wild-type strain (40). Beyond that, the expression of IsFAD6 (encoding Δ6-desaturase) was upregulated in high salinity, low temperature, and high nitrogen deficiency culture condition, indicating IsFAD6 respond to the various abiotic stresses (61). At the same time, heterologous expression Δ6-desaturase in Nannochloropsis oceanica enhanced both growth and photosynthetic efficiency (67).
Scientists also focused on studying detailed characteristics to harvest a deeper insight into the mechanism. Song et al. (68) demonstrated that amino acid residues 114–174, 206–257, and 258–276 play a vital role in substrate recognition for Δ6-desaturase. Shi et al. (57) also found that MpFADS6 (from M. pusilla) and MaFADS6-I (from M. alpina) showed a difference in substrate preference. Further studies based on the domain swapping approach reveal that sequences between the histidine boxes I and II played a pivotal role in which mutation of G194, E222, M227, and V399/I400 cause a significant decrease in the ALA conversion rate of MpFADS6 (57) (Table 2). By employing site-directed mutation, the scientist found that mutants Q409R and M242P lost the desaturation function, while mutants F419V and A374Q weakened the catalytic activities. Combined with molecular modeling, they reveal that electronic transfer in the catalytic process correlated with histidine-conserved region III, while desaturation is highly correlated with histidine-conserved regions I and II (37) (Table 2). These results from site-specific mutagenesis and molecular modeling bridge the gap between structure and the catalytic mechanism of these desaturases.
Δ5-desaturase catalyze dihomo-γ-linolenic acid (DGLA, C20:3Δ8,11,14) and eicosatetraenoic acid (ETA, C20:4Δ8,11,14,17) to arachidonic acid (AA, C20:4Δ5,8,11,14) and eicosapentaenoic acid (EPA, C20:4Δ5,8,11,14,17). Scientists have also identified and characterized the various Δ5-desaturase genes from Paramecium tetraurelia (44), Ostreococcus tauri (44), Ostreococcus lucimarinus (44), Thraustochytrium sp. (46), and Oblongichytrium sp. (45). The amino acid sequence from this Δ5-desaturase was significantly homologous, containing three conserved histidine boxes and a cytochrome b5 domain (Table 1).
Tavares et al. (44) carried out a research on substrate preferences and desaturation efficiencies of Δ5-desaturase from P. tetraurelia, O. tauri, and O. lucimarinus. Their results also demonstrated that Δ5-desaturase from O. tauri, O. lucimarinus, M. alpina, and P. tetraurelia prefer the substrates bound in phospholipid to a promiscuous acyl carrier substrate, while Δ5-desaturase from L. major was an acyl coenzyme A-dependent.
Heterologous expression Δ5-desaturase from Thraustochytrium aureum in Aurantiochytrium limacinum triggers increase of AA and EPA by 4.6- and 13.2-fold, which is driven by the thraustochytrid ubiquitin promoter (47). After disrupting the Δ5-desaturase in M. alpina, scientists achieve a higher percentage of DGLA (40.1%) accumulation in total lipid (69). At the same time, overexpressing the Δ5-desaturase in P. tricornutum exhibited a significant increment of unsaturated fatty acids, EPA (increase by 58%), and neutral lipid content (increase up to 65%) (70). Thus, these results demonstrated the critical role of Δ5-desaturase in catalyzing the DGLA and ETA to AA and EPA, respectively.
Δ4-desaturase catalyze docosatetraenoic acid (DTA, C22:4Δ7,10,13,16) and docosapentaenoic acid (DPA, C22:5Δ7,10,13,16,19) to docosapentaenoic acid (DPA, C22:5Δ4,7,10,13,16) and docosahexaenoic acid (DHA, C22:6Δ4,7,10,13,16,19). As very important LCPUFAs, more researchers focus on another more efficient PKS pathway to synthesize DHA from Schizochytrium sp. However, the scientist also identified this desaturase encoding gene from I. galbana (49), Isochrysis sphaerica (50), Pavlova lutheri (52), and Pavlova viridis (53). Among them, the one from P. lutheri desaturated DTA (C22:4Δ7,10,13,16) and DPA, (C22:5Δ7,10,13,16,19) (52), while the others from I. galbana and I. sphaerica prefer DPA (C22:5Δ7,10,13,16,19) as substrate (49, 50) (Table 1).
Some desaturase can convert ω-6 fatty acids to ω-3 fatty acids, which is named ω-3 desaturase. Δ15-desaturase is a kind of ω3-desaturase with C18 fatty acid as substrate, which catalyzes linoleic acid (LA, 18:2Δ9,12) and γ-linolenic acid (GLA, C18:3Δ6,9,12) to α-linolenic acid (ALA, C18:3Δ9,12,15) and stearidonic acid (SDA, C18:4Δ6,9,12,15), respectively. Scientists found that low temperature and high salinity could motivate the upregulation of CiFAD3 (coding Δ15-desaturase) expression in Chlamydomonas reinhardtii (71).
Substrate preference was an exciting topic determined by the structure and amino acid in the enzyme's binding site. Scientists found that Δ15-desaturase from Riftia pachyptila (72) and M. alpina (73) show a preference for C18 fatty acids, while Δ17-desaturase from Pythium aphanidermatum (74) display a higher catalytic activity for C20 fatty acids (Table 3). On combining site directed mutagenesis, homology modeling, and molecular docking, scientists revealed that the W106 and V137 related to substrate recognition (mutations in these amino acids significantly decreased the enzyme activity), and the A44, M156, and W291 residues related to the higher desaturation activity for C20 substrates (mutations in these amino acids markedly increase the conversion rate of AA). Beyond that, the amino acids residues that bind to CoA groups govern substrate preference (58) (Table 2). Scientists also found that the threonine residue located in the fourth transmembrane was essential for the typical structure and function of Δ15-desaturase in C. reinhardtii, and the mutations in this site resulted in varying degrees of activity weaken (1) (Table 2).
Δ17-desaturase is another kind of ω3-desaturase with C20 fatty acid as a substrate which catalyzes dihomo-γ-linolenic acid (DGLA, C20:3Δ8,11,14) and arachidonic acid (AA, C20:4Δ5,8,11,14) to eicosatetraenoic acid (ETA, C20:4Δ8,11,14,17) and eicosapentaenoic acid (EPA, C20:4Δ5,8,11,14,17), respectively. Considerable efforts have been focused on identified and characteristic Δ17-desaturase, which could convert 20°C ω-6 fatty acids to ω-3 fatty acids. Scientists have identified the Δ17-desaturase from P. aphanidermatum (74), Phytophthora sojae (74), Phytophthora ramorum (74), Saprolegnia diclina (76), and Phytophthora infestans (77) (Table 3). Among them, Δ17-desaturase from S. diclina, P. aphanidermatum, P. sojae, and Phytophthora ramorum exhibited a great preference to convert AA to EPA. Thus, Ge and his colleague (78) transformed the Δ17-desaturase encoding gene from P. aphanidermatum into M. alpina to achieve EPA production with a 49.7% conversion rate of AA. Tang and his colleague identified a new Δ17-desaturase from Phytophthora parasitica, exhibiting high activity for C20 substrate (conversion rate was 70% for AA) and week activity for C18 substrate. They further introduce the gene PPD17 encoding this Δ17-desaturase into the M. alpina, resulting in the conversion of AA to EPA (1.9 g/L) (79). In our previous work, a Δ17-desaturase encoding gene from S. diclina was also introduced into the genome of the Schizochytrium sp. through homologous recombination. Compared with the wild-type strains, the ω-3/ω-6 ratio in fatty acid in genetically modified strains increased from 2.1 to 2.58, and 3% of DPA was converted to DHA (80).
Δ6-elongase is a kind of fatty acid elongase with C20 fatty acid as a substrate which catalyze γ-linolenic acid (GLA, C18:3Δ6,9,12) and stearidonic acid (SDA, C18:4Δ6,9,12,15) to dihomo-γ-linolenic acid (DGLA, C20:3Δ8,11,14) and eicosatetraenoic acid (ETA, C20:4Δ8,11,14,17), respectively. Yu et al. (81) identified a Δ6-elongase localized to the endoplasmic reticulum, whose expression level was enhanced by nitrogen starvation. Jeennor et al. (82) identified a Δ6-elongase gene from Pythium sp., exhibiting a high specificity for C18 PUFAs with a double bond at Δ6-position. Co-overexpression of Δ9-desaturase, Δ12-desaturase, and Δ6-elongase in Aspergillus oryzae, scientists achieve success in enhancing free dihomo-γ-linolenic acid production with a yield of 284 mg/L (83). Shi et al. (84) identified a Δ6-elongase N. oceanica, which applied its elongated function on C18 PUFAs with a double bond at Δ6-position. This elongase encoding gene not only attenuated DGLA, ARA, and EPA content, but also enhanced GLA content, supporting the vital role of this enzyme in the exclusive ω-6 pathway of EPA biosynthesis (84) (Table 4).
Δ5-elongase is a kind of fatty acid elongase with C20 fatty acid as substrate which catalyze arachidonic acid (AA, C20:4Δ5,8,11,14) and eicosapentaenoic acid (EPA, C20:4Δ5,8,11,14,17) to Docosatetraenoic acid (DTA, C22:4Δ7,10,13,16) and Docosapentaenoic acid (DPA, C22:5Δ7,10,13,16,19), respectively. Employing fluorescent protein as an indicator, Niu et al. (87) found the Δ5-elongase from Pavlova viridis located in the endoplasmic reticulum. Robert and his colleague identified a Δ-5 elongase from Pavlova salina and characterized the exclusive elongase function for EPA. Furthermore, the scientist heterologous expressed the gene encoding Δ5-elongase in the moss Physcomitrella patens from the algae Pavlova sp. and harvested 4.51 mg/l Docosapentaenoic acid (DPA, C22:5Δ7,10,13,16,19) from endogenous arachidonic acid (89). Jiang et al. identified a Δ5-elongase from P. tricornutum, exhibiting a substrate preference for EPA. Co-expressed the Δ5-elongase and Δ4-desaturase in Pichia pastoris, they successfully construct a pathway for docosahexaenoic acid biosynthesis (88) (Table 4). There also exists another alternative pathway for Δ6-elongase/Δ6-desaturase, which is called Δ9-elongase/Δ8-desaturase pathway originated from euglenoid species (74). Δ9-elongase is a kind of fatty acid elongase with C18 fatty acid as a substrate which catalyze linoleic acid (LA, C18:2Δ9,12) and α-linolenic acid (ALA, C18:3Δ9,12,15) to eicosadienoic acid (EDA, 20:2 C18:3Δ11,14) and eicosatrienoic acid (ERA, C20:4Δ11,14,17), respectively. Then, Δ8-desaturase further catalyze EDA and ERA to dihomo-γ-linolenic acid (DGLA, C20:3Δ8,11,14) and eicosatetraenoic acid (ETA, C20:4Δ8,11,14,17). However, compared with other elongase and desaturase, there is relatively little research about these enzymes.
The desaturases, introduce a double-bound between the carboxyl (end) and the original double bonds (front) in the substrate, were defined as front-end desaturase, including Δ4, Δ5, Δ6, Δ9, and Δ12 desaturase. Thus, these enzymes share similar characteristics in a structure. First, these enzymes consisted of hydrophobic transmembrane regions and hydrophilic nontransmembrane regions. However, they still exhibit differences in evolution. For example, the number of the transmembrane regions was different in each enzyme, in which the number is 2, 3, and 4 for Δ4, Δ6, and Δ8 desaturase, respectively. Second, these enzymes are fellowed by cytochrome b5-like binding domain with “HPGG” motif in the N-terminus, which is reserved for cytochrome b5 and NAD(P)H as electron donors (31, 51). Third, nontransmembrane regions contain three histidine-rich motifs, which were considered for the binding position of di-iron and are critical for desaturase activity (21, 31). The three histidine-rich motifs that were highly conservative consisted of HXXXH, HXXHH, and (H/Q)XXHH in this enzyme, which demonstrated the similarity and homology of them in origin. Thus, the mutation of the amino acid close to the substrate-binding region (three histidine-rich motifs) will cause a significant impact on desaturase activity and substrate preference. As shown in Table 2, the mutations of these functional domains (F160L, A223T, and L156M), located at the bottom of substrate binding position, cause the substrate preference changes from C16:0 to C18:0 (23).
The structure of ω3-desaturase was similar to that of front-end desaturase, which consisted of hydrophobic transmembrane regions and hydrophilic nontransmembrane regions (79). In detail, ω3-desaturase included three highly conservative histidine-rich motifs (HXXXH, HXXHH, and (H/Q)XXHH) and more than four transmembrane domains (1, 26). The three highly conservative histidine-rich motifs were also predicted as the critical position of di-iron for substrate binding. However, cytb5 “HPGG” motif was not located in the N terminal of the enzyme. Beyond that, many amino acids were conservative in this ω3-desaturase. As shown in Table 2, T286 in Δ15-desaturase from Chlamydomonas reinhardtii was responsible for catalytic activity (1) while A44S, W106, E111, M156, V137, W291M, T322S, and F353H in Δ15-desaturase in Δ15-desaturase from Mortierella alpina was responsible for substrate preference (58).
The characteristic of amino acid sequence exhibit a significant difference with that of desaturase, which may also lead to the difference in structure. In detail, elongase contains seven transmembrane regions and three/five different kinds of conservative motifs. The motifs in Δ5-elongase consisted of a histidine-rich box (SFLHVYHHV), a tyrosine-rich box (YLTQAQLVQF), and a conserved motif (MYXYY) in (87). However, Δ6-elongase contains four motifs (KxxExxDT, QxxFLHxYHH, NxxxHxxMYxYY, and TxxQxxQ) and a histidine-rich catalytic motif (and HVYHH) (82). These conservative motifs were essential for di-iron binding and responsible for enzyme activity.
Enzyme structure model and information are collected from protein crystals, which is hindered by a lack of time and labor resources. These difficulties are especially marked in transmembrane protein, including fatty acid desaturase and elongase, widely distributed in cytomembrane, endoplasmic reticulum, and chloroplast membrane. Currently, only two kinds of three-dimensional structures of the membrane fatty acid desaturases are available, one is the mouse stearoyl-CoA desaturase (Figure 2A, PDB ID: 4YMK), and the other is human integral membrane stearoyl-CoA desaturase (Figure 2B, PDB ID: 4ZYO). Thus, homology modeling and docking are the essential methods to analyze the structure of bacterial membrane fatty acid desaturases and elongase in the current state (21). Herein, we summarized the research approach of homology modeling and docking based on the previous work (21–23, 55). First, after homology analyzes the amino acid sequence, the scientist can determine templates with higher homology and resolution in the protein database (e.g., Protein Data Bank, PDB). Second, scientists began to predict the structure based on the crystal structure of templates by employing some structure prediction software (e.g., Swiss Model and Modeler). Third, a scientist will assess the structure from many predictions. After score and energy minimization, some software will select the optimal structural model from the candidate. Fourth, the active site of the structural model was predicted using the software. Last, the docking simulations process was performed by the software of Autodock. The scientist could run the docking result through Gromacs and harvest stable conformation information with a substrate binding in the active site, which was then evaluated using RMSD (Root Mean Squared Deviation). Combined with site-specific mutagenesis, a scientist could match the performance of the mutant with structure variation, which could provide a reasonable explanation for the mutation (22, 58).
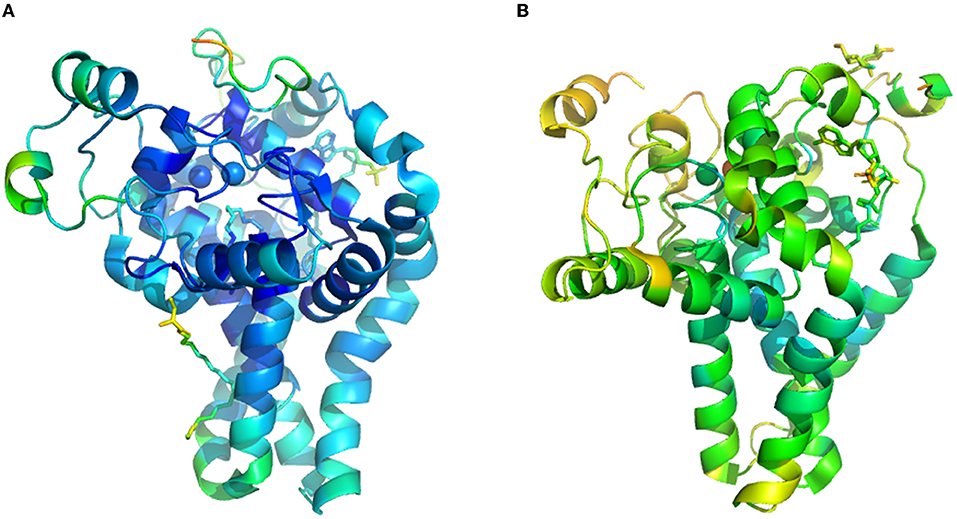
Figure 2. The crystal structure of transmembrane fatty acid desaturases. (A), crystal structure of mouse stearoyl-CoA desaturase (PDB ID: 4YMK). (B), human integral membrane stearoyl-CoA desaturase (PDB ID: 4ZYO).
Dietary bioactive lipids are important nutriments to maintain the typical metabolic status of the organism. As the practical component in lipid, the crucial role is self-evident of LCPUFAs in boosting the development of the brain and immune system and preventing cardiovascular disease. More importantly, what chip is to the electronic equipment, an enzyme is to the cell factory. These fatty acid synthesis related enzymes play a vital role in the desaturation and elongation of the carbon chain. These enzymes exist widely in plants, fungus, microalgae, and bacteria located in the cell and/or organelle membrane, including endoplasmic reticulum and chloroplast. Thus, the Open Reading Frames and amino acid sequence of these enzymes share a high degree of homology among the genera, which also provide evidence for the origins and evolutionary processes. Employing site-specific mutagenesis, homology modeling, and docking studies, scientists reveal that the structure made by the amino acids at a specific site contributes to substrate preference and catalytic activity. Many enzyme engineering strategies used for high content LCPUFAs rich oil synthesis were also summarized. Thus, the review of these important advances on desaturase and elongase could not only provide extensive data for enzyme rational design and modification but also light up the way for the efficiency LCPUFAs rich oil production.
As shown in Table 2, many researchers have created the desaturase mutants with single-site and multisite. Combining the catalytic performance of each mutant with the results from homologous modeling, they can deduce the position of crucial amino acids related to the catalytic activity and substrate preference. However, it is a nonrational and time-consuming process that much work was an indispensable part of harvesting positive mutants. Moreover, most of the time, we do not obtain positive mutants, fortunately. So, it is essential to utilize these hard-earned data, whether positive or negative. Thus, the database could be constructed from positive and negative catalytic performance as listed in Table 2, which could serve as a sample of artificial intelligence (AI) learning. After training and learning are repeated, it will obtain the undisclosed objective laws and provide us a better experimental scheme potential. At last, the AI model will guide us in moving in the right direction and approach the enzyme with higher catalytic activity quickly (90, 91).
Scientists have identified many kinds of desaturase and elongase from different organisms and analyzed their amino acid composition and substrate preference. However, the lacke of three-dimensional crystal structure data still restricts the comprehensive and deep analysis of these enzymes, which remain an excellent challenge for researcher. On the one hand, the cryo-electron microscopy technique has been widely applied in analyzing transmembrane protein, which could provide a reliable experimental basis for modeling (92). On the other hand, the rapid-developed AI techniques (such as AlphaFold and RoseTTAFold) are also a powerful tool for protein structure prediction, which could guide enzyme design and modifications (93, 94). Combined crystal structure analysis with molecular dynamics simulation, we can explore more information from catalytic efficiency, structure change, electron and proton transfer, which could provide a rational strategy to enhance catalytic performance, change substrate preference, improve catalytic stability. Novel technology, including nanopore (95), scanning tunneling microscope-break junction (96), atomic force microscope (97), and optical tweezers (98) will enable scientists to go a step further in enzyme research. Beyond that, metabolic engineering equipped with machine learning techniques (neural network and Bayesian optimization etc.) (99, 100), and omics analysis (101, 102) could also be employed to design, regulate, and optimize the metabolic pathway for high-efficiency LCPUFAs rich oil production. In the future, comprehensive interdisciplinary research will become the theme and contribute to enzymatic research.
X-YZ, Y-HZ, A-FX, and B-SF planned the manuscript, wrote and revised it. They were helped by A-HZ in revision and writing. All authors contributed to the article and approved the submitted version.
The National Postdoctoral Program for Innovative Talents (No. BX20200197) and National Natural Science Foundation of China (No. 21978245).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors would like to thank the National Postdoctoral Program for Innovative Talents (No. BX20200197) and National Natural Science Foundation of China (No. 21978245) for their support.
1. Lim JM, Vikramathithan J, Hwangbo K, Ahn JW, Park YI, Choi DW, et al. Threonine 286 of fatty acid desaturase 7 is essential for omega-3 fatty acid desaturation in the green microalga Chlamydomonas reinhardtii. Front Microbiol. (2015) 6:1–8. doi: 10.3389/fmicb.2015.00066
2. Zhang Y, Zhang TY, Liang Y, Jiang LZ, Sui XN. Dietary bioactive lipids: a review on absorption, metabolism, and health properties. J Agr Food Chem. (2021) 69:8929–43. doi: 10.1021/acs.jafc.1c01369
3. Duan JJ, Song YY, Zhang X, Wang CJ. Effect of omega-3 polyunsaturated fatty acids-derived bioactive lipids on metabolic disorders. Front Physiol. (2021) 12:646491. doi: 10.3389/fphys.2021.646491
4. Ji XJ, Ren LJ, Huang H. Omega-3 biotechnology: a green and sustainable process for omega-3 fatty acids production. Front Bioeng Biotech. (2015) 3:158. doi: 10.3389/fbioe.2015.00158
5. FAO/WHO (Food and Agriculture Organization of the United Nations/ World Health Organization). Fats and oils in human nutrition. Report of a joint expert consultation. Food and Agriculture Organization of the United Nations, Rome, Italy (1994)s.
6. Leonard AE, Pereira SL, Sprecher H, Huang YS. Elongation of long chain fatty acids. Prog Lipid Res. (2004) 43:36–54. doi: 10.1016/S0163-7827(03)00040-7
7. Palmerini CA, Mazzoni M, Giovinazzo G, Arient G. Blood lipids in Antarctic and in temperate-water fish species. J Membr Biol. (2009) 230:125–31. doi: 10.1007/s00232-009-9192-2
8. Sahena F, Zaidul ISM, Jinap S, Saari N, Jahurul HA, Abbas KA, et al. PUFAs in fish: extraction, fractionation, importance in health. Compr Rev Food Sci F. (2009) 8:59–74. doi: 10.1111/j.1541-4337.2009.00069.x
9. Liu C, Chen FS, Xia YM. Composition and structural characterization of peanut crude oil bodies extracted by aqueous enzymatic method. J Food Compos Anal. (2022) 105:104238. doi: 10.1016/j.jfca.2021.104238
10. Belury MA, Raatz SS, Conrad Z. Modeled substitution of traditional oils with high oleic acid oils decreases essential fatty acid intake in children. Am J Clin Nutr. (in press). doi: 10.1093/ajcn/nqab407
11. Sun XM, Ren LJ, Bi ZQ, Ji XJ, Zhao QY, Jiang L, et al. Development of a cooperative two-factor adaptive-evolution method to enhance lipid production and prevent lipid peroxidation in Schizochytrium sp. Biotechnol Biofuels. (2018) 11:65. doi: 10.1186/s13068-018-1065-4
12. Carlson KM, Heilmayr R, Gibbs HK, Noojipady P, Burns DN, Morton DC, et al. Effect of oil palm sustainability certification on deforestation and fire in Indonesia. P Natl Acad Sci USA. (2018) 115:121–6. doi: 10.1073/pnas.1704728114
13. Barclay WR, Meager KM, Abril JR. Heterotrophic production of long chain omega-3 fatty acids utilizing algae and algaelike microorganisms. J Appl Phycol. (1994) 6:123–9. doi: 10.1007/BF02186066
14. Jiang JY, Zhu SY, Zhang YT, Sun XM, Hu XC, Huang H, et al. Integration of lipidomic and transcriptomic profiles reveals novel genes and regulatory mechanisms of Schizochytrium sp. in response to salt stress.s Bioresource Technol. (2019) 294:122231. doi: 10.1016/j.biortech.2019.122231
15. Rayaroth A, Tomar RS, Mishra RK. One step selection strategy for optimization of media to enhance arachidonic acid production under solid state fermentation. LWT-Food Sci Technol. (2021) 152:112366. doi: 10.1016/j.lwt.2021.112366
16. Ma W, Wang YZ, Nong FT, Du F, Xu YS, Huang PW. An emerging simple and effective approach to increase the productivity of thraustochytrids microbial lipids by regulating glycolysis process and triacylglycerols' decomposition. Biotechnol Biofuels. (2021) 14:247. doi: 10.1186/s13068-021-02097-4
17. Lu R, Cao LZ, Wang KF, Ledesma-Amaro R, Ji XJ. Engineering Yarrowia lipolytica to produce advanced biofuels: Current status and perspectives. Bioresource Technol. (2021) 341:125877. doi: 10.1016/j.biortech.2021.125877
18. Wang QT, Feng YB, Lu YD, Xin Y, Shen C, Wei L, et al. Manipulating fatty-acid profile at unit chain-length resolution in the model industrial oleaginous microalgae nannochloropsis. Metab Eng. (2021) 66:157–66. doi: 10.1016/j.ymben.2021.03.015
19. Ji XJ, Zhang AH, Nie ZK, Wu WJ, Ren LJ, Huang H. Efficient arachidonic acid-rich oil production by Mortierella alpina through a repeated fed-batch fermentation strategy. Bioresource Technol. (2014) 170:356–60. doi: 10.1016/j.biortech.2014.07.098
20. Zhu Z, Zhang S, Liu H, Shen H, Lin X, Yang F, et al. A multi-omic map of the lipid-producing yeast Rhodosporidium toruloides. Nat Commun. (2012) 3:1–11. doi: 10.1038/ncomms2112
21. Garba L, Yussoff MAM, Abd Halimi KB, Ishak SNH, Ali MSM, Oslan SN, et al. Homology modeling and docking studies of a delta 9-fatty acid desaturase from a cold-tolerant Pseudomonas sp AMS8. Peer J. (2018) 6:e4347. doi: 10.7717/peerj.4347
22. Smith R, Jouhet J, Gandini C, Nekrasov V, Marechal E, Napier JA. Plastidial acyl carrier protein Delta 9-desaturase modulates eicosapentaenoic acid biosynthesis and triacylglycerol accumulation in Phaeodactylum tricornutum. Plant J. 106:1247–59. doi: 10.1111/tpj.15231
23. Liu BL, Sun Y, Hang W, Wang XD, Xue JN, Ma RY, et al. Characterization of a novel acyl-ACP Δ9 desaturase gene responsible for palmitoleic acid accumulation in a diatom Phaeodactylum tricornutum. Front Microbiol. (2020) 11:584589. doi: 10.3389/fmicb.2020.584589
24. Han XT, Wang S, Zheng L, Liu WS. Identification and characterization of a delta-12 fatty acid desaturase gene from marine microalgae Isochrysis galbana. Acta Oceanol Sin. (2019) 38:107–13. doi: 10.1007/s13131-019-1354-1
25. Sayanova O, Haslam R, Guschina I, Lloyd D, Christie WW, Harwood JL, et al. A bifunctional delta 12, delta 15-desaturase from Acanthamoeba castellanii directs the synthesis of highly unusual n-1 series unsaturated fatty acids. J Bio Chem. (2006) 281:36533–41. doi: 10.1074/jbc.M605158200
26. Zhang PY, Liu SH, Cong BL, Wu GT, Liu CL, Lin XZ, et al. A novel omega-3 fatty acid desaturase involved in acclimation processes of polar condition from Antarctic ice algae Chlamydomonas sp. ICE-L. Mar Biotechnol. (2011) 13:393–401. doi: 10.1007/s10126-010-9309-8
27. Qiu X, Reed DW, Hong H, MacKenzie SL, Covello PS. Identification and analysis of a gene from calendula officinalis encoding a fatty acid conjugase. Plant Physiol. (2001) 125:847–55. doi: 10.1104/pp.125.2.847
28. Martinez-Rivas JM, Sperling P, Luehs W, Heinz E. Spatial and temporal regulation of three different microsomal oleate desaturase genes (FAD2) from normal-type and high-oleic varieties of sunflower (Helianthus annuus L). Mol Breed. (2001) 8:159–68. doi: 10.1023/A:1013324329322
29. Lu YD, Chi XY, Yang QL, Li ZX, Liu SF, Gan QH, et al. Molecular cloning and stress-dependent expression of a gene encoding Δ12-fatty acid desaturase in the Antarctic microalga Chlorella vulgaris NJ-7. Extremophiles. (2009) 13:875–84. doi: 10.1007/s00792-009-0275-x
30. Domergue F, Spiekermann P, Lerchl J, Beckmann C, Kilian O, Kroth PG, et al. New insight into Phaeodactylum tricornutum fatty acid metabolism cloning and functional characterization of plastidial and microsomal delta12-fatty acid desaturases. Plant Physiol. (2003) 131:1648–60. doi: 10.1104/pp.102.018317
31. Zhang L, Chen WB, Yang SP, Zhang YB, Xu JL, Yang DJ, et al. Identification and functional characterization of a novel delta 12 fatty acid desaturase gene from Haematococcus pluvialis. J Ocean U China. (2020) 19:1362–70. doi: 10.1007/s11802-020-4418-0
32. Wu CC, Ohashi T, Kajiura H, Sato Y, Misaki R, Honda K, et al. Functional characterization and overexpression of delta 12-desaturase in the oleaginous yeast Rhodotorula toruloides for production of linoleic acid-rich lipids. J Biosci Bioeng. (2021) 131:631–9. doi: 10.1016/j.jbiosc.2021.02.002
33. Hsiao TY, Holmes B, Blanch HW. Identification and functional analysis of a delta-6 desaturase from the marine microalga Glossomastix chrysoplasta. Mar Biotechnol. (2007) 9:154–65. doi: 10.1007/s10126-006-6075-8
34. Zhang R, Zhu Y, Ren L, Zhou P, Hu J, Yu L. Identification of a fatty acid Δ6-desaturase gene from the eicosapentaenoic acid-producing fungus Pythium splendens RBB-5. Biotechnol Lett. (2013) 35:431–8. doi: 10.1007/s10529-012-1101-6
35. Zhang L, Cao HS, Ning P, Zhou ZG. Functional characterization of a Δ6 fatty acid desaturase gene and its 5′-upstream region cloned from the arachidonic acidrich microalga Myrmecia incisa Reisigl (Chlorophyta). J Oceanol Limnol. (2018) 36:2308–21. doi: 10.1007/s00343-019-7305-z
36. Laoteng K, Ruenwai R, Tanticharoen M, Cheevadhanarak S. Genetic modification of essential fatty acids biosynthesis in Hansenula polymorpha. FEMS Microbiol Lett. (2005) 245:169–78. doi: 10.1016/j.femsle.2005.03.006
37. Cui J, Chen H, Tang X, Zhang H, Chen YQ, Chen W. Consensus mutagenesis and computational simulation provide insight into the desaturation catalytic mechanism for delta 6 fatty acid desaturase. Biochem Bioph Res Co. (2022) 586:74–80. doi: 10.1016/j.bbrc.2021.11.050
38. Araki H, Hagihara H, Takigawa H, Tsujino Y, Ozaki K. Novel genes encoding hexadecanoic acid delta 6-desaturase activity in a Rhodococcus sp. Curr Microbiol. (2016) 73:646–51. doi: 10.1007/s00284-016-1106-9
39. Jeennor S, Cheawchanlertfa P, Suttiwattanakul S, Panchanawaporn S, Chutrakul C, Laoteng K. The codon-optimized Δ6-desaturase gene of Pythium sp. as an empowering tool for engineering n3/n6 polyunsaturated fatty acid biosynthesis. BMC Biotechnol. (2015) 15:82. doi: 10.1186/s12896-015-0200-6
40. Shi HS, Luo X, Wu RN, Yue XQ. Production of eicosapentaenoic acid by application of a delta-6 desaturase with the highest ALA catalytic activity in algae. Microb Cell Fact. (2018) 17:2–15. doi: 10.1186/s12934-018-0857-3
41. Tonon T, Sayanova O, Michaelson LV, Qing R, Harvey D, Larson TR, et al. Fatty acid desaturases from the microalga Thalassiosira pseudonana. FEBS J. (2005) 272:3401–12. doi: 10.1111/j.1742-4658.2005.04755.x
42. Thiyagarajan S, Arumugam M, Senthil N, Vellaikumar S, Kathiresan S. Functional characterization and substrate specificity analysis of delta6-desaturase from marine microalga Isochrysis sp. Biotechnol Lett. (2018) 40:577–84. doi: 10.1007/s10529-017-2501-4
43. Michinaka Y, Aki T, Shimauchi T, Nakajima T, Kawamoto S, Shigeta S, et al. Differential response to low temperature of two delta 6 fatty acid desaturases from Mucor circinelloides. Appl Microbiol Biot. (2003) 62:362–8. doi: 10.1007/s00253-003-1326-3
44. Tavares S, Grotkjaer T, Obsen T, Haslam RP, Napier JA, Gunnarsson N. Metabolic engineering of Saccharomyces cerevisiae for production of eicosapentaenoic acid, using a novel detal 5-desaturase from Paramecium tetraurelia. Appl Environ Microb. (2011) 77:1854–61. doi: 10.1128/AEM.01935-10
45. Kumon Y, Kamisaka Y, Tomita N, Kimura K, Uemura H, Yokochi T, et al. Isolation and characterization of a delta 5-desaturase from Oblongichytrium sp. Biosci Biotech Bioch. (2008) 72:2224–7. doi: 10.1271/bbb.80235
46. Huang JZ, Jiang XZ, Xia XF, Yu AQ, Mao RY, Chen XF, et al. Cloning and functional identification of delta5 fatty acid desaturase gene and its 5 '-upstream region from marine fungus Thraustochytrium sp. FJN-10. Mar Biotechnol. 13:12–21. doi: 10.1007/s10126-010-9262-6
47. Kobayashi T, Sakaguchi K, Matsuda T, Abe E, Hama Y, Hayashi M, et al. Increase of eicosapentaenoic acid in Thraustochytrids through Thraustochytrid Ubiquitin promoter-driven expression of a fatty acid delta 5 desaturase gene. Appl Environ Microb. (2011) 77:3870–6. doi: 10.1128/AEM.02664-10
48. Thiyagarajan S, Khandelwal P, Senthil N, Vellaikumar S, Arumugam M, Dubey AA, et al. Heterologous production of polyunsaturated fatty acids in E. coli using delta 5-desaturase gene from microalga Isochrysis sp. Appl Biochem Biotech. (2021) 193:869–83. doi: 10.1007/s12010-020-03460-1
49. Shi TL, Yu AQ, Li M, Ou XY, Xing LJ, Li MC. Identification of a novel C22-D4-producing docosahexaenoic acid (DHA) specific polyunsaturated fatty acid desaturase gene from Isochrysis galbana and its expression in Saccharomyces cerevisiae. Biotechnol Lett. (2012) 34:2265–74. doi: 10.1007/s10529-012-1028-y
50. Guo B, Jiang ML, Wan X, Gong YM, Liang Z, Hu CJ. Identification and heterologous expression of a δ4-fatty acid desaturase gene from Isochrysis sphaerica. J Microbiol Biotechnol. (2013) 23:1413–21. doi: 10.4014/jmb.1301.01065
51. Ahmann K, Heilmann M, Feussner I. Identification of a delta 4-desaturase from the microalga Ostreococcus lucimarinus. Eur J Lipid Sci Tech. (2011) 113:832–40. doi: 10.1002/ejlt.201100069
52. Tonon T, Harvey D, Larson TR, Graham IA. Identification of a very long chain polyunsaturated fatty acid delta4-desaturase from the microalga Pavlova lutheri. FEBS Lett. (2003) 553:440–4. doi: 10.1016/S0014-5793(03)01078-0
53. Xu Y, Niu Y, Kong J. Heterologous overexpression of a novel delta-4 desaturase gene from the marine microalga Pavlova viridis in Escherichia coli as a Mistic fusion. World J Microbiol Biotechnol. (2011) 27:2931–7. doi: 10.1007/s11274-011-0776-5
54. Tsai YY, Ohashi T, Wu CC, Bataa D, Misaki R, Limtong S, et al. Delta-9 fatty acid desaturase overexpression enhanced lipid production and oleic acid content in Rhodosporidium toruloides for preferable yeast lipid production. J Biosci Bioeng. (2019) 127:430–40. doi: 10.1016/j.jbiosc.2018.09.005
55. Ben Amor F, Ben Hlima H, Abdelkafi S, Fendri I. Insight into Arthrospira platensis delta 9 desaturase: a key enzyme in poly-unsaturated fatty acid synthesis. Mol Biol Rep. (2018) 45:1873–9. doi: 10.1007/s11033-018-4333-2
56. Sakuradani E, Abe T, Matsumura K, Tomi A, Shimizu S. Identification of mutation sites on delta 12 desaturase genes from Mortierella alpina 1S-4 mutants. J Biosci Bioeng. (2009) 107:99–101. doi: 10.1016/j.jbiosc.2008.10.011
57. Shi HS, Chen HQ, Gu ZN, Song Y, Zhang H, Chen W, et al. Molecular mechanism of substrate specificity for delta 6 desaturase from Mortierella alpina and Micromonas pusilla. J Lipid Res. (2015) 56:2309–21. doi: 10.1194/jlr.M062158
58. Rong CC, Chen HQ, Wang MX, Gu ZN, Zhao JX, Zhang H, et al. Molecular mechanism of substrate preference for ω-3 fatty acid desaturase from Mortierella alpina by mutational analysis and molecular docking. Appl Microbiol Biotechnol. (2018) 102:9679–89. doi: 10.1007/s00253-018-9321-x
59. Rau EM, Aasen IM, Ertesvåg H. A non-canonical delta 9-desaturase synthesizing palmitoleic acid identified in the thraustochytrid Aurantiochytrium sp. T66.s Appl Microbiol Biot. (2021) 105:75. doi: 10.1007/s00253-021-11562-x
60. Tezaki S, Iwama R, Kobayashi S, Shiwa Y, Yoshikawa H, Ohta A, et al. Detal 12-fatty acid desaturase is involved in growth at low temperature in yeast Yarrowia lipolytica. Biochem Biophy Res Co. (2017) 488:165–70. doi: 10.1016/j.bbrc.2017.05.028
61. Wang YN, Zhang SF, Pötter M, Sun WY, Li L, Yang XB, et al. Overexpression of Δ12-fatty acid desaturase in the oleaginous yeast Rhodosporidium toruloides for production of linoleic acid-rich lipids. Appl Biochem Biotechnol. (2016) 180:1497–507. doi: 10.1007/s12010-016-2182-9
62. Jiang XZ, Liu HJ, Niu YC, Qi F, Zhang ML, Huang JZ. Functional identification and regulatory analysis of detal 6-fatty acid desaturase from the oleaginous fungus Mucor sp. EIM-10. Biotechnol Lett. (2017) 39:453–61. doi: 10.1007/s10529-016-2268-z
63. Zhu BH, Tu CC, Shi HP, Yang GP, Pan KH. Overexpression of endogenous delta-6 fatty acid desaturase gene enhances eicosapentaenoic acid accumulation in Phaeodactylum tricornutum. Process Biochem. (2017) 57:43–9. doi: 10.1016/j.procbio.2017.03.013
64. Chen G, Qu SJ, Wang Q, Bian F, Peng ZY, Zhang Y, et al. Transgenic expression of delta-6 and delta-15 fatty acid desaturases enhances omega-3 polyunsaturated fatty acid accumulation in Synechocystis sp. PCC6803. Biotechnol Biofuels. (2014) 7:2–10. doi: 10.1186/1754-6834-7-32
65. Petrie JR, Shrestha P, Mansour MP, Nichols PD, Liu Q, Singh SP. Metabolic engineering of omega-3 long-chain polyunsaturated fatty acids in plants using an acyl-CoA Δ6-desaturase with ω3-preference from the marine microalga Micromonas pusilla. Metab Eng. (2010) 12:233–40. doi: 10.1016/j.ymben.2009.12.001
66. Shi HS, Chen HQ, Gu ZN, Zhang H, Chen W, Chen YQ. Application of a delta-6 desaturase with α-linolenic acid preference on eicosapentaenoic acid production in Mortierella alpina. Microb Cell Fact. (2016) 15:2–14. doi: 10.1186/s12934-016-0516-5
67. Yang F, Yuan W, Ma Y, Balamurugan S, Li HY, Fu S, et al. Harnessing the lipogenic potential of detal 16-desaturase for simultaneous hyperaccumulation of lipids and polyunsaturated fatty acids in Nannochloropsis oceanica. Front Mar Sci. (2019) 6:682. doi: 10.3389/fmars.2019.00682
68. Song LY, Zhang Y, Li SF, Hu J, Yin WB, Chen YH, et al. Identification of the substrate recognition region in the Δ6-fatty acid and Δ8-sphingolipid desaturase by fusion mutagenesis. Planta. (2014) 239:753–63. doi: 10.1007/s00425-013-2006-x
69. Kikukawa H, Sakuradani E, Ando A, Okuda T, Shimizu S, Ogawa J. Microbial production of dihomo-gamma-linolenic acid by delta 5-desaturase gene-disruptants of Mortierella alpina 1S-4. J Biosci Bioeng. (2016) 122:22–6. doi: 10.1016/j.jbiosc.2015.12.007
70. Peng KT, Zheng CN, Xue J, Chen XY, Yang WD, Liu JS, et al. Delta 5 fatty acid desaturase upregulates the synthesis of polyunsaturated fatty acids in the marine diatom Phaeodactylum tricornutum. J Arg Food Chem. (2014) 62:8773–6. doi: 10.1021/jf5031086
71. Nguyen HM, Cuine S, Beyly-Adriano A, Legeret B, Billon E, Auroy P, et al. The green microalga Chlamydomonas reinhardtii has a single omega-3 fatty acid desaturase that localizes to the chloroplast and impacts both plastidic and extraplastidic membrane lipids. Plant Physiol. (2013) 163:914–28. doi: 10.1104/pp.113.223941
72. Liu HL, Wang H, Cai SY, Zhang HB. A Novel ω3-desaturase in the deep sea giant tubeworm Riftia pachyptila. Mar Biotechnol. (2017) 19:345–50. doi: 10.1007/s10126-017-9753-9
73. Wang M, Chen H, Ailati A, Chen W, Chilton FH, Lowther WT, et al. Substrate specificity and membrane topologies of the iron-containing ω3 and ω6 desaturases from Mortierella alpina. Appl Microbiol Biotechnol. (2017) 102:211–23. doi: 10.1007/s00253-017-8585-x
74. Xue Z, He H, Hollerbach D, Macool DJ, Yadav NS, Zhang H, et al. Identification and characterization of new detal-17 fatty acid desaturases. Appl Microbiol Biotechnol. (2013) 97:1973–85. doi: 10.1007/s00253-012-4068-2
75. Wang L, Chen W, Feng Y, Ren Y, Gu Z, Chen H, et al. Genome characterization of the oleaginous fungus Mortierella alpina. PLoS ONE. (2011) 6:e28319. doi: 10.1371/journal.pone.0028319
76. Okuda T, Ando A, Negoro H, Muratsubaki T, Kikukawa H, Sakamoto T, et al. Eicosapentaenoic acid (EPA) production by an oleaginous fungus Mortierella alpina expressing heterologous the detal 7-desaturase gene under ordinary temperature. Eur J Lipid Sci Technol. (2015) 117:1919–27. doi: 10.1002/ejlt.201400657
77. Fu Y, Fan XZ, Li XZ, Wang H, Chen HJ. The desaturase opin17 from phytophthora infestans converts arachidonic acid to eicosapentaenoic acid in CHO cells. Appl Biochem Biotech. (2013) 171:975–88. doi: 10.1007/s12010-013-0332-x
78. Ge CF, Chen HQ, Mei TT, Tang X, Chang LL, Gu ZN, et al. Application of a ω-3 desaturase with an arachidonic acid preference to eicosapentaenoic acid production in Mortierella alpina. Front Bioeng Biotechnol. (2018) 5:1–10. doi: 10.3389/fbioe.2017.00089
79. Tang X, Chen HQ, Mei TT, Ge CF, Gu ZN, Zhang H, et al. Characterization of an omega-3 desaturase from phytophthora parasitica and application for eicosapentaenoic acid production in Mortierella alpina. Front Microbiol. (2018) 9:1878. doi: 10.3389/fmicb.2018.01878
80. Ren LJ, Zhuang XY, Chen SL, Ji XJ, Huang H. Introduction of ω-3 desaturase obviously changed the fatty acid profile and sterol content of Schizochytrium sp. J Agric Food Chem. (2015) 63:9770–6. doi: 10.1021/acs.jafc.5b04238
81. Yu SY, Li H, Tong M, Ouyang LL, Zhou ZG. Identification of a Δ6 fatty acid elongase gene for arachidonic acid biosynthesis localized to the endoplasmic reticulum in the green microalga Myrmecia incisa Reisigl. Gene. (2012) 493:219–27. doi: 10.1016/j.gene.2011.11.053
82. Jeennor S, Cheawchanlertfa P, Suttiwattanakul S, Panchanawaporn S, Chutrakul C, Laoteng K. Novel elongase of Pythium sp with high specificity on delta (6)-18C desaturated fatty acids. Biochem Bioph Res Co. (2014) 450:507–12. doi: 10.1016/j.bbrc.2014.06.004
83. Tamano K, Yasunaka Y, Kamishima M, Itoh A, Miura A, Kan E, et al. Enhancement of the productivity of free dihomo-gamma-linolenic acid via co-overexpression of elongase and two desaturase genes in Aspergillus oryzae. J Biosci Bioeng. (2020) 130:480–8. doi: 10.1016/j.jbiosc.2020.07.004
84. Shi Y, Liu MJ, Pan YF, Hu HH, Liu J. Δ6 Fatty acid elongase is involved in eicosapentaenoic acid biosynthesis via the ω6 pathway in the marine alga Nannochloropsis oceanica. J Agric Food Chem. (2021) 69:9837–48. doi: 10.1021/acs.jafc.1c04192
85. Poliner E, Pulman JA, Zienkiewicz K, Childs K, Benning C, Farre EM, et al. A toolkit for Nannochloropsis oceanica CCMP1779 enables gene stacking and genetic engineering of the eicosapentaenoic acid pathway for enhanced long-chain polyunsaturated fatty acid production. Plant Biotechnol J. (2018) 16:298–309. doi: 10.1111/pbi.12772
86. Robert SS, Petrie JR, Zhou XR, Mansour MP, Blackburn SI, Green AG, et al. Isolation and characterisation of a delta 5-fatty acid elongase from the marine microalga Pavlova salina. Mar Biotechnol. (2009) 11:410–8. doi: 10.1007/s10126-008-9157-y
87. Niu Y, Kong J, Fu LY, Yang J, Xu Y. Identification of a novel C20-elongase gene from the marine microalgae Pavlova viridis and its expression in Escherichia coli. Mar Biotechnol. (2009) 11:17–23. doi: 10.1007/s10126-008-9116-7
88. Jiang ML, Guo B, Wan X, Gong YM, Zhang YB, Hu CJ. Isolation and characterization of the Diatom Phaeodactylum Δ5-Elongase gene for transgenic LC-PUFA production in Pichia pastoris. Mar Drugs. (2014) 12:1317–34. doi: 10.3390/md12031317
89. Kaewsuwan S, Bunyapraphatsara N, Cove DJ, Quatrano RS, Chodok P. High level production of adrenic acid in Physcomitrella patens using the algae Pavlova sp delta (5)-elongase gene. Bioresource Technol. (2010) 101:4081–8. doi: 10.1016/j.biortech.2009.12.138
90. Hie Brian L, Yang Kevin K. Adaptive machine learning for protein engineering. Curr Opin Struc Biol. (2021) 72:145–52. doi: 10.1016/j.sbi.2021.11.002
91. Pertusi DA, Moura ME, Jeffryes JG, Prabhu S, Biggs BW, Tyo KEJ. Predicting novel substrates for enzymes with minimal experimental effort with active learning. Metab Eng. (2017) 44:171–81. doi: 10.1016/j.ymben.2017.09.016
92. Gauto DF, Estrozi LF, Schwieters CD, Effantin G, Macek P, Sounier R, et al. Integrated NMR and cryo-EM atomic-resolution structure determination of a half-megadalton enzyme complex. Nature Commun. (2019) 10:2697. doi: 10.1038/s41467-019-10490-9
93. Tunyasuvunakool K, Adler J, Wu Z, Green T, Zielinski M, Zidek A, et al. Highly accurate protein structure prediction for the human proteome. Nature. (2021) 596:590–6. doi: 10.1038/s41586-021-03828-1S
94. Baek M, DiMaio F, Anishchenko I, Dauparas J, Ovchinnikov S, Lee GR, et al. Accurate prediction of protein structures and interactions using a three-track neural network. Science. (2021) 373:871–6. doi: 10.1126/science.abj8754
95. Cao MY, Wang H, Tang HR, Zhao DD, Li YX. Enzyme-encapsulated zeolitic imidazolate frameworks formed inside the single glass nanopore: catalytic performance and sensing application. Anal Chem. (2021) 93:12257–64. doi: 10.1021/acs.analchem.1c01790
96. Zhuang XY, Zhang AH, Qiu SY, Tang C, Zhao SQ, Li HC, et al. Coenzyme coupling boosts charge transport through single bioactive enzyme junctions. IScience. (2020) 23:101001. doi: 10.1016/j.isci.2020.101001
97. Liu JY, Ghanizadeh H, Li XM, Han ZY, Qiu YW, Zhang Y, et al. study of the interaction, morphology, and structure in trypsin-epigallocatechin-3-gallate complexes. Molecule. (2021) 26:4567. doi: 10.3390/molecules26154567
98. Zhuang XY, Wu Q, Zhang AH, Liao LX, Fan BS. Single-molecule biotechnology for protein researches. Chinese J Chem Eng. (2021) 30:212–24. doi: 10.1016/j.cjche.2020.10.031
99. Kumar P, Adamczyk PA, Zhang XL, Andrade RB, Romero PA, Ramanathan P, et al. Active and machine learning-based approaches to rapidly enhance microbial chemical production. Metab Eng. (2021) 67:216–26. doi: 10.1016/j.ymben.2021.06.009
100. Czajka JJ, Oyetunde T, Tang YJ. Integrated knowledge mining, genome-scale modeling, and machine learning for predicting Yarrowia lipolytica bioproduction. Metab Eng. (2021) 67:227–36. doi: 10.1016/j.ymben.2021.07.003
101. Kennes-Veiga DM, Gonzalez-Gil L, Carballa M, Lema JM. Enzymatic cometabolic biotransformation of organic micropollutants in wastewater treatment plants: a review. Bioresource Technol. (2022) 344:126291. doi: 10.1016/j.biortech.2021.126291
Keywords: bioactive lipids, desaturase, elongase, fatty acid synthesis pathway, oleogenic microorganisms
Citation: Zhuang X-Y, Zhang Y-H, Xiao A-F, Zhang A-H and Fang B-S (2022) Key Enzymes in Fatty Acid Synthesis Pathway for Bioactive Lipids Biosynthesis. Front. Nutr. 9:851402. doi: 10.3389/fnut.2022.851402
Received: 09 January 2022; Accepted: 25 January 2022;
Published: 23 February 2022.
Edited by:
Xiao-Jun Ji, Nanjing Tech University, ChinaReviewed by:
Hu-Hu Liu, Hunan Agricultural University, ChinaCopyright © 2022 Zhuang, Zhang, Xiao, Zhang and Fang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ai-Hui Zhang, emhhbmdhaWh1aUB4bXUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.