
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Nutr., 21 December 2022
Sec. Nutrition and Food Science Technology
Volume 9 - 2022 | https://doi.org/10.3389/fnut.2022.1077893
This article is part of the Research TopicApplication of Nanosensors For the Detection and Analysis of Food ContaminantsView all 4 articles
Herein, we developed a dual fluorescent aptasensor based on mesoporous silica to simultaneously detect sulfadimethoxine (SDM) and oxytetracycline (OTC) in animal-derived foods. We immobilized two types of aptamers modified with FAM and CY5 on the silica surface by base complementary pairing reaction with the cDNA modified with a carboxyl group and finally formed the aptasensor detection platform. Under optimal conditions, the detection range of the aptasensor for SDM and OTC was 3–150 ng/mL (R2 = 0.9831) and 5–220 ng/mL (R2 = 0.9884), respectively. The limits of detection for SDM and OTC were 2.2 and 1.23 ng/mL, respectively. The limits of quantification for SDM and OTC were 7.3 and 4.1 ng/mL, respectively. Additionally, the aptasensor was used to analyze spiked samples. The average recovery rates ranged from 91.75 to 114.65% for SDM and 89.66 to 108.94% for OTC, and all coefficients of variation were below 15%. Finally, the performance and practicability of our aptasensor were confirmed by HPLC, demonstrating good consistency. In summary, this study was the first to use the mesoporous silica-mediated fluorescence aptasensor for simultaneous detection of SDM and OTC, offering a new possibility to analyze other antibiotics, biotoxins, and biomolecules.
Farmers often use sulfadimethoxine (SDM) and oxytetracycline (OTC) as antimicrobial treatments for animals due to their low cost and high efficacy (1, 2). However, some studies have shown that SDM and OTC residues in animals enter the human body through the food chain, causing allergies, renal failure, liver function impairment, and other hazards to the body (3, 4). Therefore, the European Commission and China set the maximum residue level of SDM and OTC in food to 100 ng/mL (5, 6). At present, the methods of detecting antibiotic residues usually include high-performance liquid chromatography (HPLC) (7), HPLC coupled with mass spectrometry (HPLC-MS) (8), enzyme-linked immunoassay (ELISA) (9), and capillary electrophoresis (10). These methods have the advantages of high efficiency, sensitivity, and specificity. However, their disadvantages are evident. HPLC and HPLC-MS can only detect one sample at a time and are not suitable for mass detection of samples. Moreover, the antibodies required by ELISA are highly affected by the environment, and the capillary electrophoresis equipment is costly. Therefore, developing a novel high-throughput, low-cost, and stable detection method for antibiotic residues in animal-derived foods is imperative.
Sensors based on aptamers as signal probes are of particular interest due to their good selectivity for target molecules and stability in complex physicochemical environments (11). Recently, various aptasensors have been used to detect SDM and OTC residues, such as chemiluminescence (12, 13), electrochemical (14, 15), colorimetric (16, 17), fluorescence (18, 19). These methods have the advantages of high sensitivity and specificity but can only be used to detect a single target. Therefore, Díaz-García et al. created a gold-based colorimetric assay to simultaneously detect SDM and OTC residues in milk (20). However, due to the physical and chemical properties of gold nanoparticles, which are easily affected by the surrounding detection environment, this method has shortcomings such as poor reproducibility and inconsistent detection results. Therefore, constructing a new multiplexed aptasensor to detect SDM and OTC residues in animal-derived foods is urgent.
Fluorescent aptasensors based on porous materials (carbon nanotubes, mesoporous silica, graphene oxide, etc.) are widely used for the monitoring of contaminants in food because of their high specificity and high accuracy (21–23). Among them, silicon nanoparticles have the advantages of simple preparation, easy surface modification, non-toxicity and good thermal stability, and are widely used to construct aptasensors to detect antibiotic residues (24–26). However, most studies have relied on mesoporous silica interacting with aptamers to create molecular gates, isolating fluorescein molecules in the pores and finally releasing the fluorescein molecules by target-specific binding to the corresponding aptamers. These methods depend on the pore size (27, 28). If the pore size is too large, the sealing of the molecular gate will be poor. If it is too small, it will be difficult for the fluorescein molecules to enter the molecular gate. This effect increases the difficulty of preparing mesoporous silica (29, 30). Hence, using the easy modification of the surface of mesoporous silica to immobilize fluorescein molecules on the silica surface can effectively avoid the loss of fluorescein molecules and improve the sensor’s accuracy.
Based on these ideas, we proposed for the first time a dual fluorescent signal aptasensor based on aminoaminated mesoporous silica (MSN-NH2) to simultaneously detect SDM and OTC residues in animal-derived foods. First, we used MSN-NH2 as a loading platform for aptamers. Then, the fluorophore-modified aptamer was attached to silica using the carboxyl-modified cDNA as a linker. Finally, the aptamers were released from the silica surface when the test sample contained SDM and OTC. The quantitative detection of SDM and OTC was realized according to the fluorescence intensity in the supernatant. The whole detection process was efficient and sensitive, providing a broad prospect for quantitative detection of SDM and OTC.
The SDM, OTC, sulfadiazine, sulfamethoxypyridazine, kanamycin, doxycycline, 3-amino-propyltriethoxysilane, hexadecyl trimethyl ammonium bromide (CTAB), Tetraethyl orthosilicate (TEOs), and other chemical reagents were purchased from Aladdin Co., Ltd., (Shanghai, China).
The characterization images of MSN and MSN-NH2 particle size were obtained from JEM-2100F (Japan). The amino modification success of mesoporous silica was verified by Spectrum 100 (PerkinElmer, USA) and UV-2450 (Shimadzu, Kyoto, Japan). The accuracy of the aptasensor was confirmed by HPLC (Ultimate 3000).
All the nucleotide sequences below were synthesized by Sangong Biotechnology Co., Ltd., (Shanghai, China).
Sulfadimethoxine (SDM) aptamer 5′FAM-GAGGGCAACG AGTGTTTATAGA-3′, (31).
Oxytetracycline (OTC) aptamer 5′-GAGCCGGGCGCGGT ACGGGTACTGGTA-CY5-3′ (32).
Linker: 5′COOH-TCTATAAACACACTCGTTGCCCTCT TTTTTTCTCGGCCCGCGCCATGCCCATGACCAT-3′
The MSN composite was synthesized based on previous reports with minor modifications (24, 27, 33). Briefly, 1.0 g of CTAB was dissolved in 480 mL of deionized water and stirred slowly at 80°C until a clear solution was obtained. Then, we added 3.5 mL NaOH (2M) and 5.0 mL tetraethyl orthosilicate to the above mixture dropwise, stirred vigorously for 2 h, and cooled to room temperature. Finally, the white precipitate was centrifuged at 15,000 rpm for 10 min and successively washed three times with ethanol and water to remove any solvent remaining in the pores. The washed products were dried under a vacuum overnight at 60°C to obtain MSNs containing CTAB templates. The prepared MSNs were cauterized at a high temperature for a long time to remove unreacted CTAB.
First, 0.5 g of MSNs were ultrasonically dispersed in 100 mL of ethanol, then 2.5 mL of 3-amino-propyltriethoxysilane was added. The mixture was stirred slowly and uniformly at room temperature for 24 h. Next, the suspension was centrifuged at 8,000 rpm for 15 min, and the MSN-NH2 was washed three times with ethanol and ultrapure water to remove excess 3-amino-propyltriethoxysilane. The product was vacuum-dried overnight at 60°C. Finally, the dry white MSN-NH2 powder was stored at room temperature for later use.
First, 40 mg of MSN-NH2 was sonicated in 500 μL phosphate buffer (PBS, 0.1 M, PH = 7.4) for 5 min to disperse uniformly. To form the MSN-NH2/cDNA complex, 70 μL of cDNA was added to the above mixture and incubated for 3 h at 25°C with shaking. The reacted solution was centrifuged at 15,000 rpm for 5 min, and the MSN-NH2/cDNA was washed with PBS to remove excess cDNA. Subsequently, 500 μL of PBS was added again to form an MSN-NH2/cDNA complex suspension. Next, 50 μL of SDM (15 μM) FAM-aptamer and 75 μL of OTC (10 μM) CY5-aptamer were added, and the cells were incubated with shaking in the dark for 1 h. It was centrifuged again, the pellet was washed with PBS, and the MSN-NH2/cDNA/aptamer complex was formed. Finally, the obtained pellet was suspended in PBS and stored at 4°C for further use.
The linear relationship between the target concentration and the fluorescence intensity of the aptasensor was established as follows: 5 μL of the above particle suspension was added to 200 μL of PBS with different concentrations of SDM (0, 4.6875, 9.375, 18.75, 37.5, 75, and 150 ng/mL) and OTC (0, 5.625, 11.25 22.5, 55, 110, and 220 ng/mL). It was then reacted in the dark at 25°C for 1 h. Finally, the reaction mixture was centrifuged at 15,000 rpm for 5 min, and the supernatant was retained. Correlations between the fluorescence intensity in the supernatant and SDM and OTC concentrations were calculated using ORIGIN (FAM: λex = 492 nm and λem = 518 nm; CY5: λex = 649 nm and λem = 670 nm). The limit of detection (LOD) of the dual fluorescent aptasensor was calculated according to the equation: LOD = 3SD/slope. The SD represented the standard deviation of the fluorescence intensity in the control group. The slope comprised the linear relationship between the fluorescence intensity of the target and the dual fluorescence aptasensor at different concentrations (11).
The practical application capability of the aptasensor was verified by adding known concentrations (50, 100, and 150 ng/mL) of SDM and OTC to milk and honey. Milk and honey were purchased from local supermarkets and verified to be free of SDM and OTC by HPLC. Sample pretreatment was performed as previously described (34–37). Recovery and coefficient of variation were chosen as performance parameters for the aptasensor and were validated by HPLC. The recovery formula was: (measured concentration/added concentration) x 100%. Each concentration was measured five times (12).
The schematic diagram of the silica-based dual fluorescent aptasensor for simultaneous detection of SDM and OTC is shown in Figure 1. The carboxyl-modified cDNA was immobilized on the silica surface by forming an amide bond with MSN-NH2. The SDM and OTC aptamers, labeled with FAM and CY5, respectively, were used as signaling probes, captured by cDNA via the Watson-Crick base pairing principle, and immobilized on the silica surface. Finally, a dual-fluorescent aptasensor with MSN-NH2/cDNA/aptamer as a composite material was formed. In the presence of SDM and OTC, the aptamer was released into the supernatant, resulting in decreased fluorescence of the aptasensor detection platform and increased fluorescence in the supernatant. The final fluorescence value in the supernatant was linearly correlated with the target concentration after centrifugation, resulting in the detection of SDM and OTC.
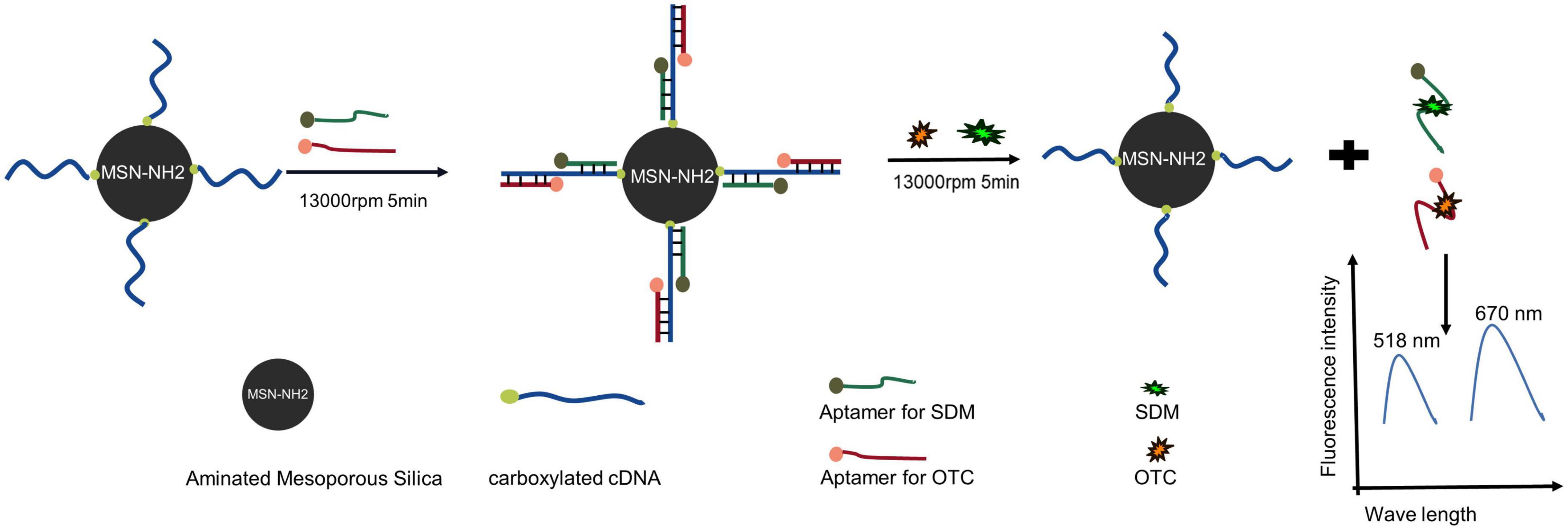
Figure 1. Schematic diagram of a dual fluorescent aptasensor based on MSN-NH2 to detect sulfadimethoxine (SDM) and oxytetracycline (OTC).
The transmission electron microscope (TEM) images of MSN and MSN-NH2 are presented in Figures 2A, B. The average size of the synthesized MSN and MSN-NH2 was 50 nm. The FTIR spectra of MSN, MSN-CTAB, and MSN-NH2 are shown in Figure 3A. The blue line shows distinct absorption peaks at 2,921 and 2,851 cm–1 due to the residual CTAB on the surface of MSN (27). After high-temperature burning, the black line has no clear peak in this band, indicating that CTAB was removed. After adding APTES, the red line appeared at 1,636 and 1,554 cm–1, representing the bending stretching vibration of amide and the characteristic peaks of NH2, respectively, which indicated that the amino group was modified on the surface of MSN. Additionally, the carboxyl group-modified cDNA has a carboxyl group characteristic peak at 210 nm, and the MSN-NH2/cDNA combination produces an amide bond, resulting in a significant increase in the absorbance at 210 nm, which also indicates that the amino group of MSN was successfully modified (Figure 3B).
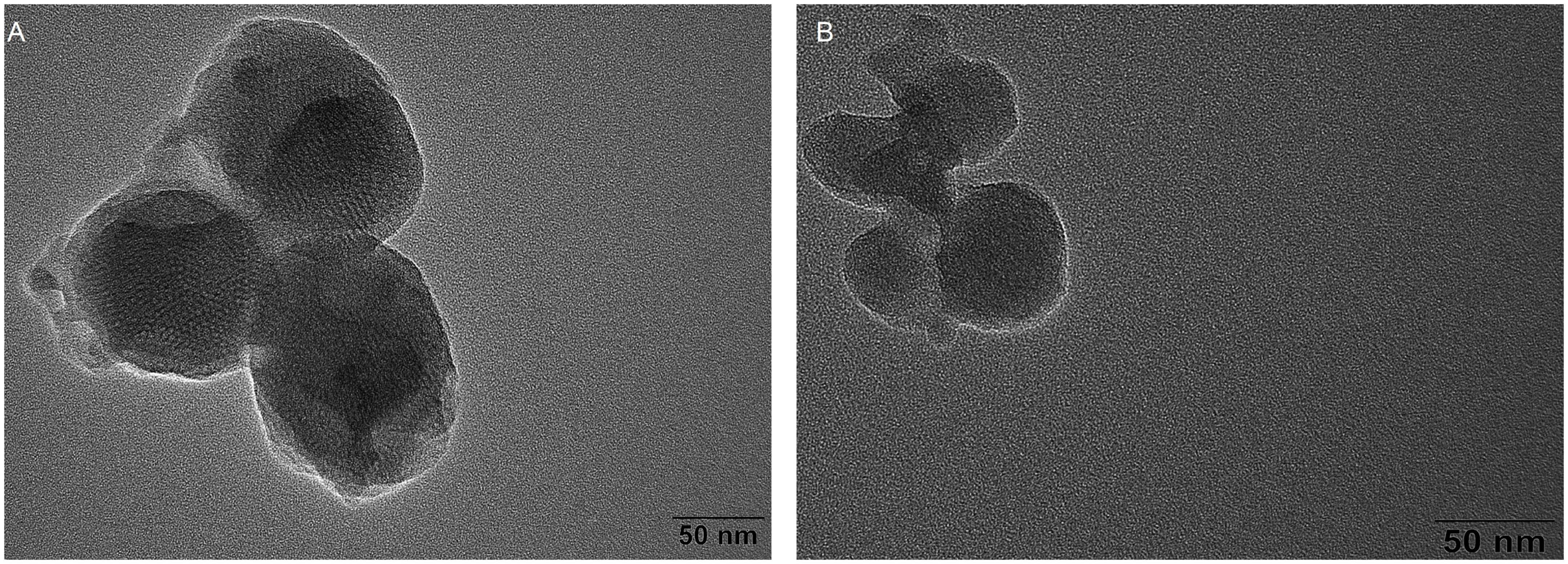
Figure 2. (A) Transmission electron microscope (TEM) micrograph of MSN; (B) TEM micrograph of MSN-NH2.
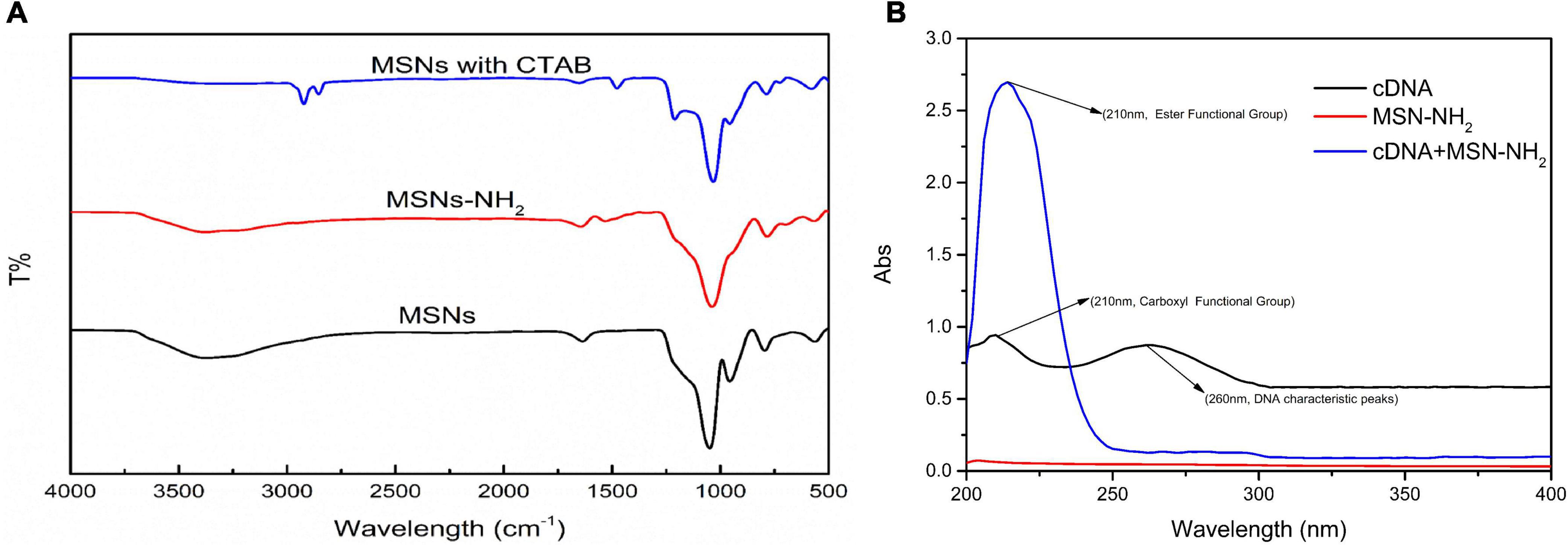
Figure 3. (A) FTIR spectra of MSN-NH2, cDNA, and cDNA/MSN-NH2; (B) UV-Vis spectra of supernatant of pure aptamer, aptamer/MSN, and aptamer/MSN-NH2 complexes.
To improve the performance of the dual fluorescent aptasensor, we optimized the aptamer concentration, aptamer/target incubation time, and cDNA/aptamer reaction time based on the fluorescence intensity. Besides, to verify the influence of SDM and OTC aptamers on the sensor performance, all optimization conditions were divided into incubating aptamers simultaneously or alone with MSN-NH2/cDNA.
Aptamer concentrations varied from 100 to 700 nM. When the two tapes of aptamers reacted with the MSN-NH2/cDNA complex alone, the fluorescence intensity reached the maximum at 450 and 550 nM (Figure 4A). When the two tapes of aptamers were simultaneously complexed with MSN-NH2/cDNA, the aptamer concentrations corresponding to the maximum fluorescence intensity increased by 50 and 100 nM, respectively (Figure 4B). These results indicated that, with increasing aptamer concentrations, the competitiveness of the two aptamers with MSN-NH2/cDNA complexes gradually increased.
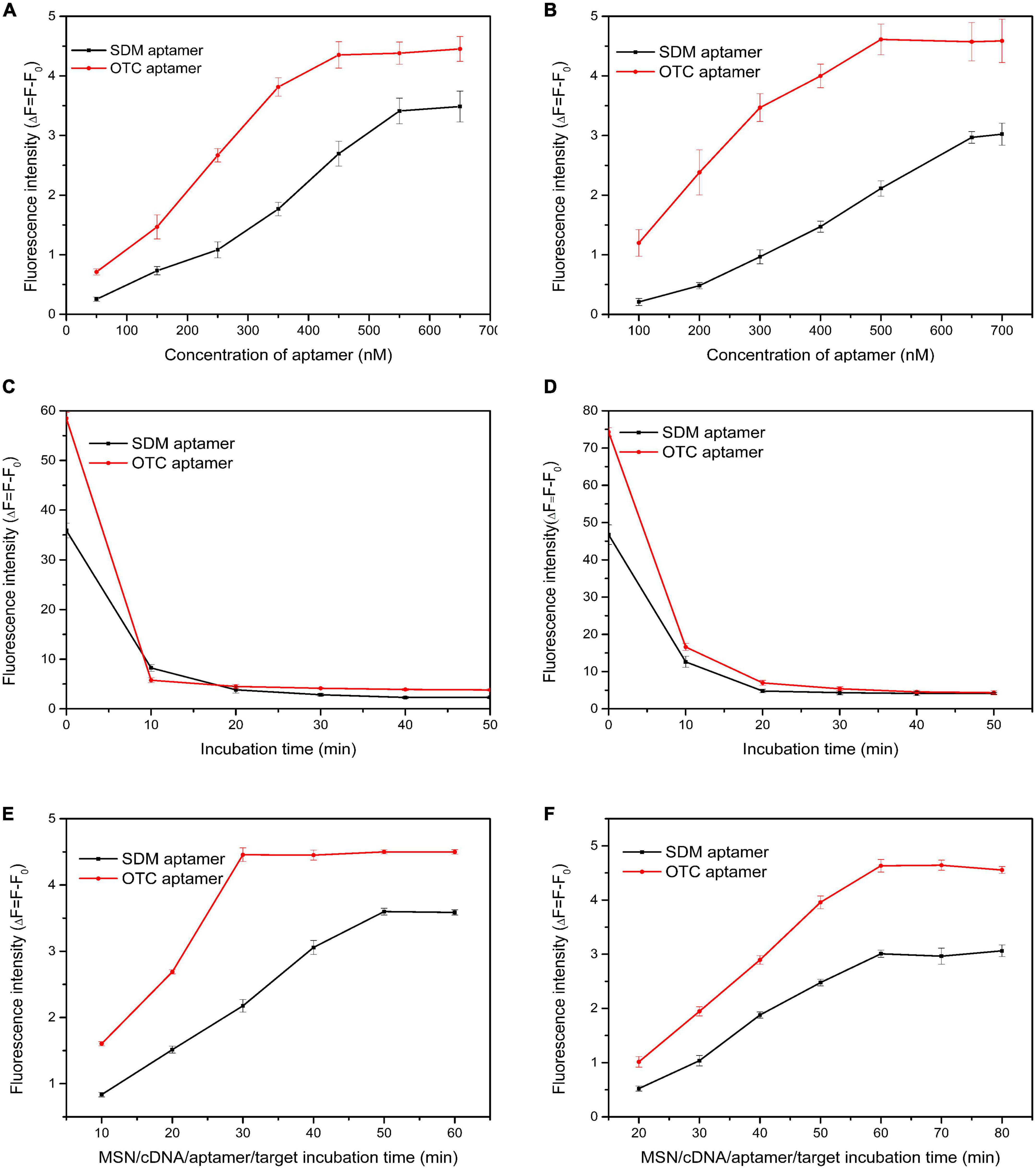
Figure 4. (A,B) Optimization of aptamer concentration; (C,D) optimization of incubation time of aptamer/cDNA; (E,F) optimization of incubation time for aptamer/target. The concentrations of sulfadimethoxine (SDM) and oxytetracycline (OTC) were set at 100 ng/mL. (Panels A,C,E: SDM and OTC aptamers were added separately; panels B,D,F: SDM and OTC aptamers were added simultaneously).
The aptamer/cDNA incubation time varied from 0 to 50 min. The aptamer reacted most rapidly with cDNA within 10 min, and the fluorescence value changed the fastest (Figures 4C, D). From 10 to 20 min, the fluorescence value did not change much (Figure 4C), indicating that the reaction of the two aptamers alone with the MSN-NH2/cDNA complex was saturated at 10 min. When the aptamers reacted simultaneously with the MSN-NH2/cDNA complex, they tended to be stable after 20 min (Figure 4D).
The aptamer/target incubation time varied from 10 to 80 min. As shown in Figure 4E, when the two types of aptamers were incubated with MSN-NH2/cDNA alone, the OTC and SDM aptamers fully reacted with the target at 30 and 50 min, respectively. As shown in Figure 4F, when the two types of aptamers were incubated with MSN-NH2/cDNA at the same time, the fluorescence intensity reached the maximum at 60 min, and the maximum fluorescence value also decreased accordingly. These results indicated that when aptamers are present simultaneously, they will affect each other to a certain extent.
Under optimized experimental conditions, SDM and OTC were simultaneously detected with the proposed method. The limits of detection and linear ranges of the two aptamers were also verified when added individually in order to exclude the effect of competition on experimental precision when both aptamers were added simultaneously. When the two types of aptamers were added alone, the linear relationship between different OTC and SDM concentrations and the fluorescence value are shown in Figures 5A, B: y = 0.0436x + 0.248 (OTC 2.5–320 ng/mL), y = 0.0321x + 0.342 (SDM 2.5–180 ng/mL). The LOD was 3.81 ng/mL for OTC and 0.64 ng/mL for SDM. When the two types of aptamers were added simultaneously, the increase in target concentration and the linear change of its fluorescence value are shown in Figures 5C, D: y = 0.045x + 0.273 (OTC: 5–220 ng/mL), y = 0.0269x + 0.136 (SDM: 3–150 ng/mL). The LOD was 1.23 ng/mL for OTC and 2.2 ng/mL for SDM. The limits of quantification for SDM and OTC were 7.3 and 4.1 ng/mL, respectively. These results showed that the different addition methods of aptamers did not affect the accuracy of system.
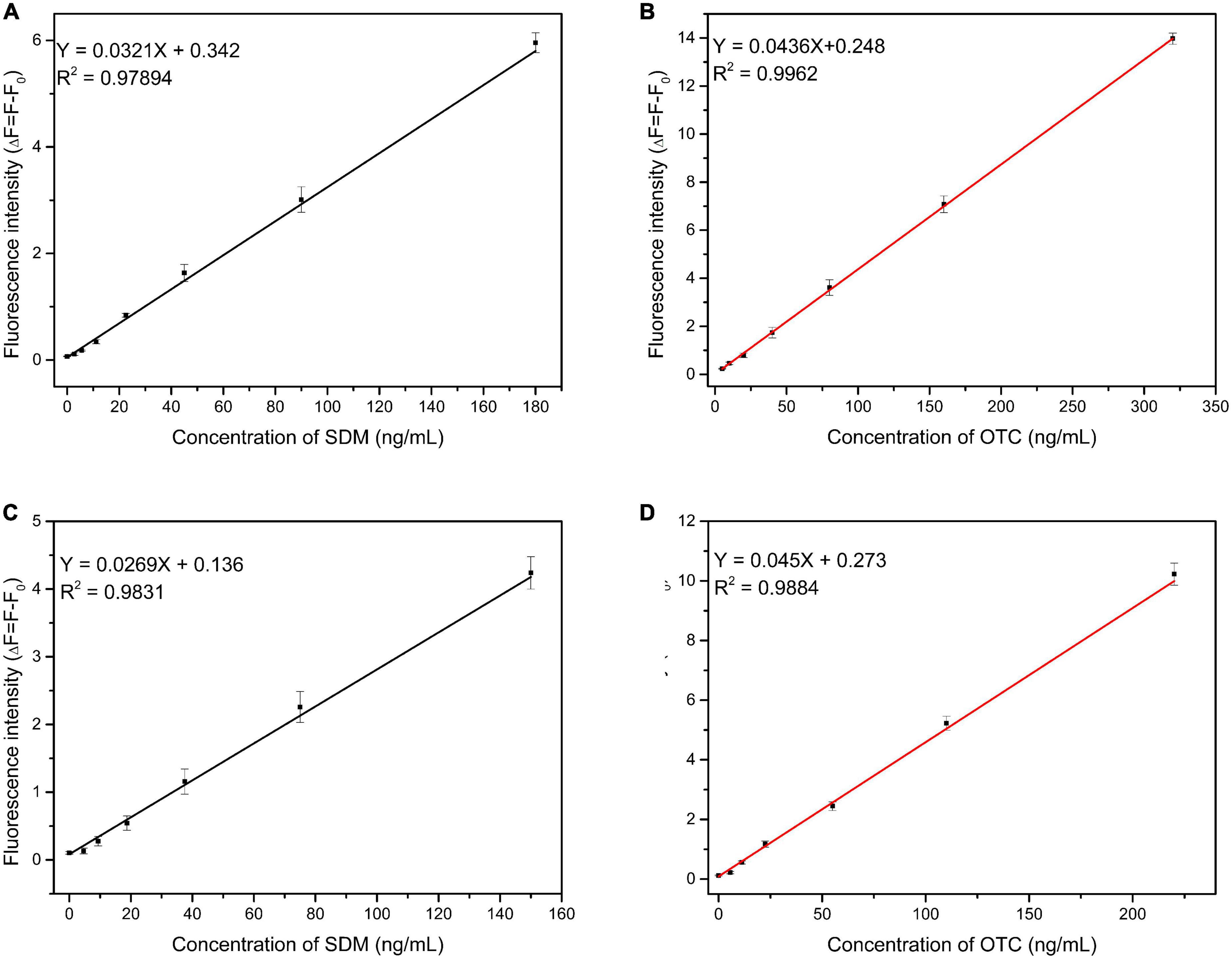
Figure 5. Linearity between fluorescence intensity and target concentration. (A,B): Two types of aptamers were added separately; (C,D): two types of aptamers were added together. (The concentrations of sulfadimethoxine (SDM) and oxytetracycline (OTC) were set at 100 ng/mL).
In additation, several antibiotics at 100 ng/mL were used to evaluate the specificity of the proposed method to detect SDM and OTC. As shown in Figure 6, although the fluorescence values change significantly in the separate and mixed systems, the aptasensor remains significantly specific for SDM and OTC compared to several of the remaining antibiotics. Furthermore, the fluorescence values of the mixture of SDM and OTC did not change much from that of the individual systems, indicating that SDM and OTC do not interact. Therefore, this method has great application prospects for the simultaneous detection of SDM and OTC in foods.
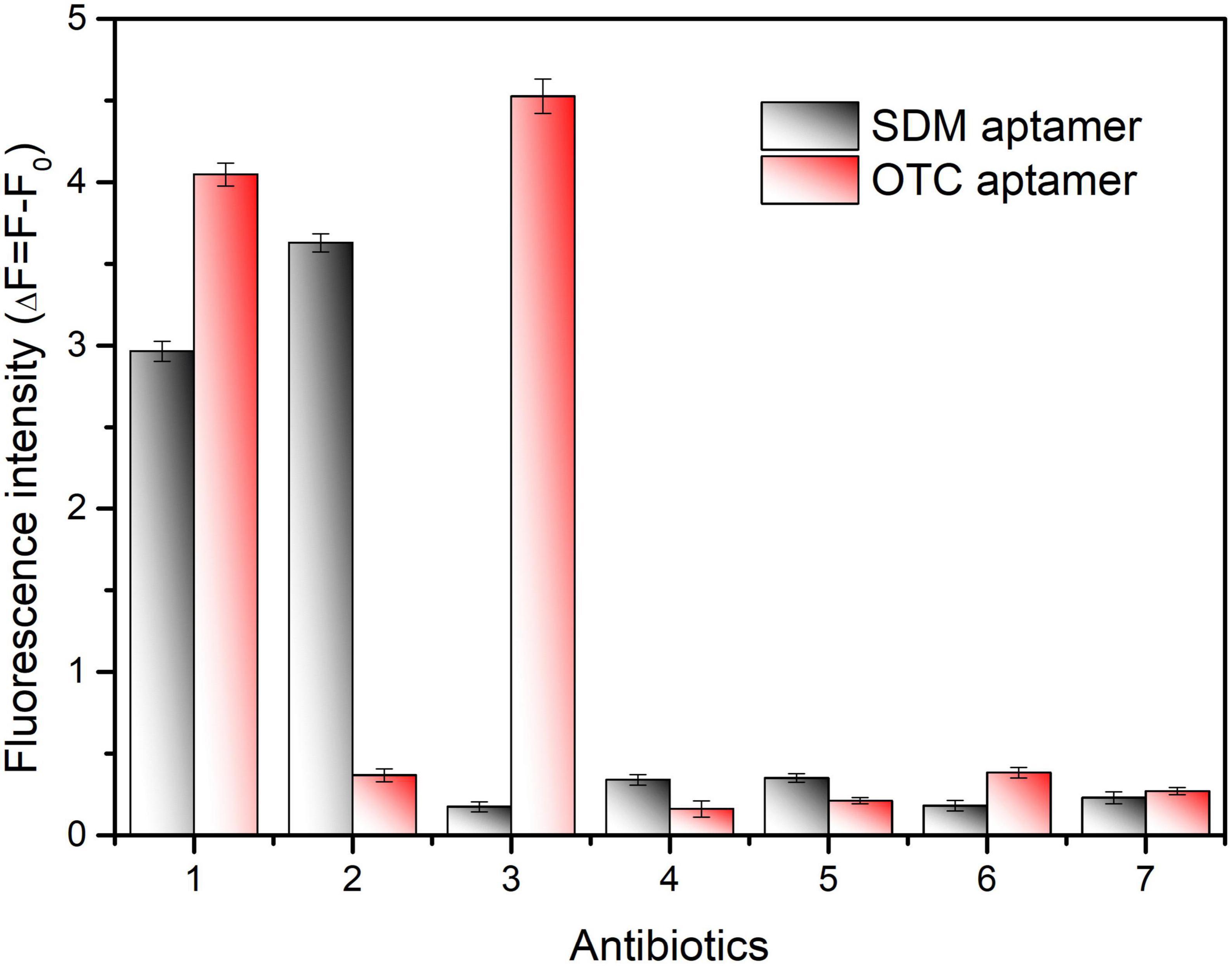
Figure 6. Specificity validation of the dual fluorescent aptasensor for sulfadimethoxine (SDM) and oxytetracycline (OTC). The concentration of all antibiotics was set to 100 ng/mL. (1) SDM + OTC, (2) SDM, (3) OTC, (4) Sulfamethoxypyridazine, (5) Sulfadiazine, (6) Doxycycline, (7) Kanamycin.
In order to verify the detection effect of real samples, the proposed method was applied to the determination in milk and honey samples. As shown in Table 1, the recoveries of SDM and OTC in milk were 92.32–106.53%, 98.81–107.82%, respectively; the recoveries of SDM and OTC in honey were 91.75–114.65%, 89.66–108.94%, respectively. The relative standard deviation for detecting SDM and OTC ranged from 4.89 to 14.65% and 3.78 to 11.12%, respectively. The accuracy and precision of the method were verified to be within acceptable limits. As shown in Table 2, the proposed method exhibits higher sensitivity, and a wider linear detection range than previous methods for the detection of SDM and OTC. Additionally, the detection results of this method were positively correlated with the HPLC results, indicating that the proposed method is reliable for the simultaneous detection of SDM and OTC.
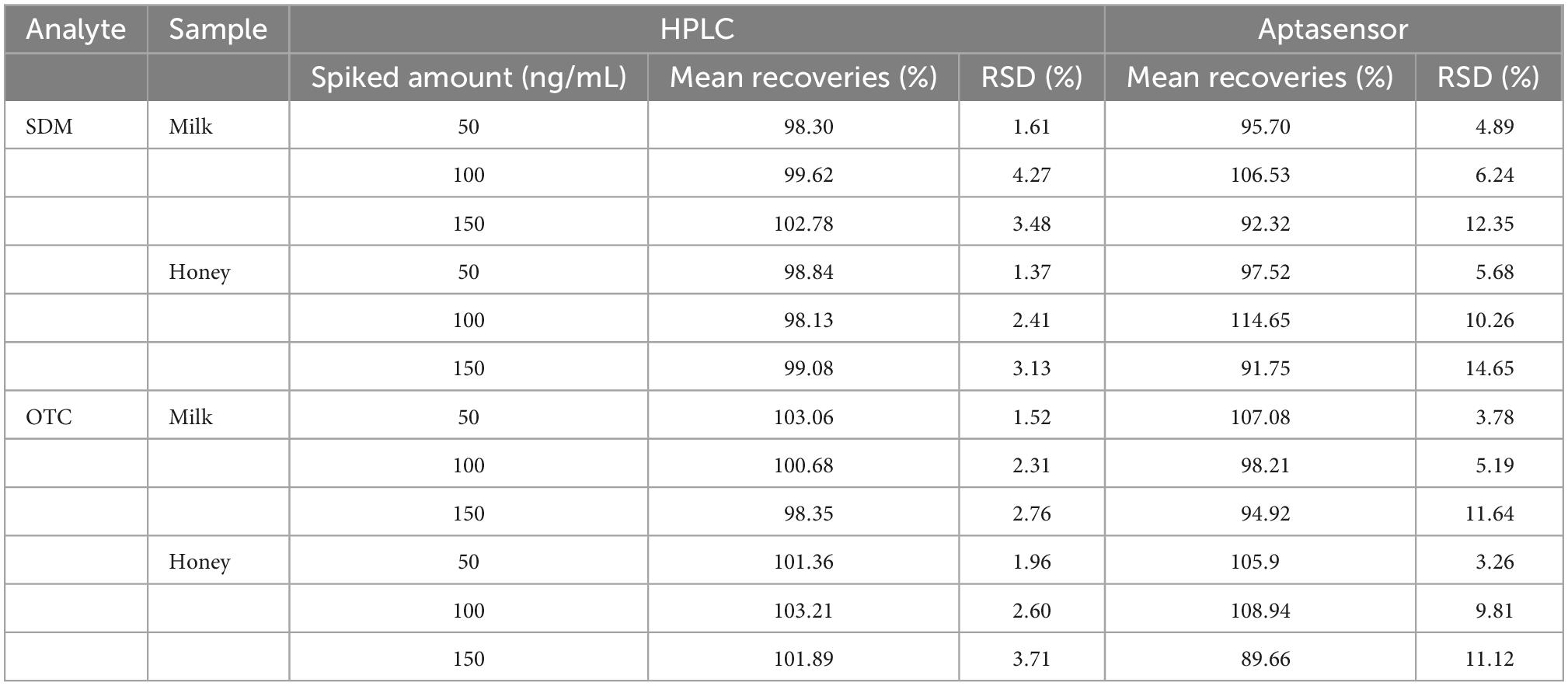
Table 1. A brief overview of recently reported methods for the determination of sulfadimethoxine (SDM) and oxytetracycline (OTC).
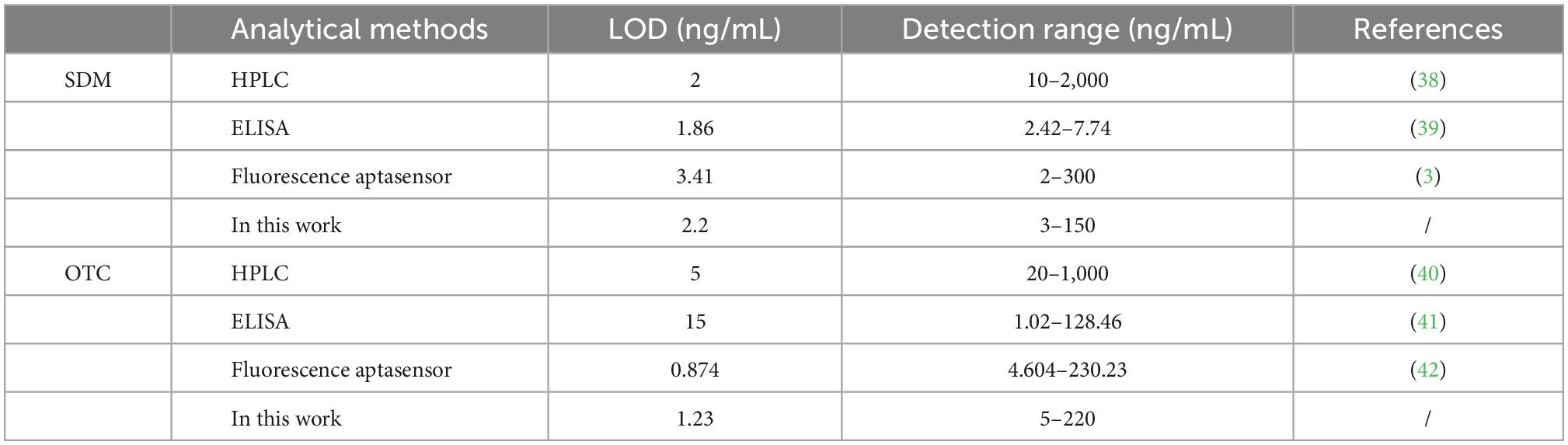
Table 2. Recoveries and coefficients of variation of spiked samples and comparison between aptasensor and high performance liquid chromat ography (HPLC) results (n = 5).
Herein, we established a novel dual fluorescent aptasensor based on MSN-NH2 for simultaneous detection of SDM and OTC in edible tissues. The two types of aptamers were labeled with FAM and CY5 fluorophores, respectively. SDM and OTC competed with the cDNA for the corresponding aptamer to make the fluorophore off the silica surface to achieve the quantitative target detection according to the fluorescence intensity in the supernatant. In addition, the accuracy of the proposed method is ensured whether the aptamers are added alone or simultaneously. Finally, the proposed method demonstrated excellent performance in real sample assays with high recoveries and good coefficients of variation. To the best of our knowledge, a strategy based on the immobilization of two aptamers on a silica surface by cDNA has not been reported. The rational design of this method comprises a strategy to simultaneously detect multiple antibiotic residues in food, laying the foundation for future aptasensor detection of multiple targets.
The original contributions presented in this study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
JT: conceptualization, methodology, and writing. XZ: methodology, validation, investigation, and writing. SJ: methodology, investigation, and reviewed the manuscript. MC, SW, ZZ, XN, and YF: methodology, validation, and investigation. TL: supervision and project administration. All authors have contributed to the article, read, and agreed to the published version of the manuscript.
This work was financially supported by the National Natural Science Foundation of China (Grant No. 31671939), the Postgraduate Scientific Research and Innovation Project of Chongqing Municipal Education Commission (CYS21273), the Chongqing of University Innovation Research Group Project (No. CXQT20031), and the Chongqing Natural Science Foundation project (cstc2021jcyj-msmX0314).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
1. Kim HJ, Jeong MH, Park HJ, Kim WC, Kim JE. Development of an immunoaffinity chromatography and HPLC-UV method for determination of 16 sulfonamides in feed. Food Chem. (2016) 196:1144–9. doi: 10.1016/j.foodchem.2015.10.014
2. Alnassrallah MN, Alzoman NZ, Almomen A. Qualitative immunoassay for the determination of tetracycline antibiotic residues in milk samples followed by a quantitative improved HPLC-DAD method. Sci Rep. (2022) 12:114502. doi: 10.1038/s41598-022-18886-2
3. Chen XX, Lin ZZ, Hong CY, Yao QH, Huang ZY. A dichromatic label-free aptasensor for sulfadimethoxine detection in fish and water based on AuNPs color and fluorescent dyeing of double-stranded DNA with SYBR Green I. Food Chem. (2020) 309:125712. doi: 10.1016/j.foodchem.2019.125712
4. Al-Shaalan NH, Nasr JJ, Shalan S, El-Mahdy AM. Inspection of antimicrobial remains in bovine milk in egypt and saudi arabia employing a bacteriological test kit and HPLC-MS/MS with estimation of risk to human health. PLoS One. (2022) 17:e0267717. doi: 10.1371/journal.pone.0267717
5. Bahreyni A, Luo H, Ramezani M, Alibolandi M, Soheili V, Danesh N, et al. A fluorescent sensing strategy for ultrasensitive detection of oxytetracycline in milk based on aptamer-magnetic bead conjugate, complementary strand of aptamer and PicoGreen. Spectrochim Acta A. (2021) 246:119009. doi: 10.1016/j.saa.2020.119009
6. Chen Y, Guo L, Liu L, Song S, Kuang H, Xu C. Ultrasensitive immunochromatographic strip for fast screening of 27 sulfonamides in honey and pork liver samples based on a monoclonal antibody. J Agric Food Chem. (2017) 65:8248–55. doi: 10.1021/acs.jafc.7b03190
7. Alipanahpour DE, Ghaedi M, Mehrabi F, Tayebi L. Highly selective magnetic dual template molecularly imprinted polymer for simultaneous enrichment of sulfadiazine and sulfathiazole from milk samples based on syringe-to-syringe magnetic solid-phase microextraction. Talanta. (2021) 232:122449. doi: 10.1016/j.talanta.2021.122449
8. Chiesa LM, Nobile M, Panseri S, Biolatti B, Cannizzo FT, Pavlovic R, et al. A liquid chromatography-tandem mass spectrometry method for the detection of antimicrobial agents from seven classes in calf milk replacers: validation and application. J Agric Food Chem. (2016) 64:2635–40. doi: 10.1021/acs.jafc.6b00155
9. He J, Liu G, Jiang M, Xu L, Kong F, Xu Z. Development of novel biomimetic enzyme-linked immunosorbent assay method based on Au@SiO2 nanozyme labelling for the detection of sulfadiazine. Food Agr Immunol. (2020) 31:341–51. doi: 10.1080/09540105.2020.1728234
10. Li X, Yang Y, Miao J, Yin Z, Zhai Y, Shi H, et al. Determination of sulfa antibiotic residues in river and particulate matter by field-amplified sample injection-capillary zone electrophoresis. Electrophoresis. (2020) 41:1584–91. doi: 10.1002/elps.202000122
11. Yan XL, Yang LL, Tang JM, Wen X, Chen XY, Zheng XL, et al. High-sensitive FAM labeled aptasensor based on Fe3O4/Au/g-C3N4 for the detection of sulfamethazine in food matrix. Biosensors. (2022) 12:759. doi: 10.3390/bios12090759
12. Wang Y, Rink S, Baeumner A, Seidel MJ. Microfluidic flow-injection aptamer-based chemiluminescence platform for sulfadimethoxine detection. Microchim Acta. (2022) 189:117. doi: 10.1007/s00604-022-05216-6
13. Zhang H, Fang C, Wu S, Duan N, Wang Z. Upconversion luminescence resonance energy transfer-based aptasensor for the sensitive detection of oxytetracycline. Anal Biochem. (2015) 489:44–9. doi: 10.1016/j.ab.2015.08.011
14. Bai Z, Chen Y, Li F, Zhou Y, Yin H, Ai S. Electrochemical aptasensor for sulfadimethoxine detection based on the triggered cleavage activity of nuclease P1 by aptamer-target complex. Talanta. (2019) 204:409–14. doi: 10.1016/j.talanta.2019.06.035
15. Peng B, Zhang Z, Tang L, Ouyang X, Zhu X, Chen L, et al. Self-powered photoelectrochemical aptasensor for oxytetracycline cathodic detection based on a dual z-scheme WO3/g-C3N4/MnO2 Photoanode. Anal Chem. (2021) 93:9129–38. doi: 10.1021/acs.analchem.1c00929
16. Wang A, Zhao H, Chen X, Tan B, Zhang Y, Quan X. A colorimetric aptasensor for sulfadimethoxine detection based on peroxidase-like activity of graphene/nickel@palladium hybrids. Anal Biochem. (2017) 525:92–9. doi: 10.1016/j.ab.2017.03.006
17. Xu Y, Lu C, Sun Y, Shao Y, Cai Y, Zhang Y, et al. A colorimetric aptasensor for the antibiotics oxytetracycline and kanamycin based on the use of magnetic beads and gold nanoparticles. Microchim Acta. (2018) 185:548. doi: 10.1007/s00604-018-3077-y
18. Zhao Y, Gao B, Sun P, Liu J, Liu J. Metal and pH-dependent aptamer binding of tetracyclines enabling highly sensitive fluorescence sensing. Biosensors. (2022) 12:717. doi: 10.3390/bios12090717
19. Chen XX, Lin ZZ, Hong CY, Zhong HP, Yao QH, Huang ZY. Label-free fluorescence-based aptasensor for the detection of sulfadimethoxine in water and fish. Appl Spectros. (2019) 73:294–303. doi: 10.1177/0003702818799100
20. Diaz-Garcia V, Contreras-Trigo B, Rodriguez C, Coelho P, Oyarzun P. A simple yet effective preanalytical strategy enabling the application of aptamer-conjugated gold nanoparticles for the colorimetric detection of antibiotic residues in raw milk. Sensors Basel. (2022) 22:1281. doi: 10.3390/s22031281
21. Xing G, Li N, Lin H, Shang Y, Pu Q, Lin JM. Microfluidic biosensor for one-step detection of multiplex foodborne bacteria ssDNA simultaneously by smartphone. Talanta. (2023) 253:123980. doi: 10.1016/j.talanta.2022.123980
22. Zhang HJ, Zou X, Chen WY, Sun Q, Gao EQ. A Cu-functionalized MOF and multi-walled carbon nanotube composite modified electrode for the simultaneous determination of hydroquinone and catechol. Anal Methods-UK. (2022) 14:3961–9. doi: 10.1039/d2ay01230h
23. Dong S, Shi QY, He KL, Wu JW, Zhu ZX, Feng JG. A simple aptamer SERS sensor based on mesoporous silica for the detection of chlorpyrifos. Foods. (2022) 11:1331. doi: 10.3390/foods11213331
24. Tan H, Guo T, Zhou H, Dai H, Yu Y, Zhu H, et al. A simple mesoporous silica nanoparticle-based fluorescence aptasensor for the detection of zearalenone in grain and cereal products. Anal Bioanal Chem. (2020) 412:5627–35. doi: 10.1007/s00216-020-02778-3
25. Ma N, Ren X, Wang H, Kuang X, Fan D, Wu D, et al. Ultrasensitive controlled release aptasensor using thymine-Hg2+-Thymine mismatch as a molecular switch for Hg2+ detection. Anal Chem. (2020) 92:14069–75. doi: 10.1021/acs.analchem.0c03110
26. Song Q, Peng M, Wang L, He D, Ouyang J. A fluorescent aptasensor for amplified label-free detection of adenosine triphosphate based on core-shell Ag@SiO2 nanoparticles. Biosens Bioelectron. (2016) 77:237–41. doi: 10.1016/j.bios.2015.09.008
27. Tan H, Ma L, Guo T, Zhou H, Chen L, Zhang Y, et al. A novel fluorescence aptasensor based on mesoporous silica nanoparticles for selective and sensitive detection of aflatoxin B1. Anal Chim Acta. (2019) 1068:87–95. doi: 10.1016/j.aca.2019.04.014
28. Emrani AS, Danesh NM, Ramezani M, Taghdisi SM, Abnous K. A novel fluorescent aptasensor based on hairpin structure of complementary strand of aptamer and nanoparticles as a signal amplification approach for ultrasensitive detection of cocaine. Biosens Bioelectron. (2016) 79:288–93. doi: 10.1016/j.bios.2015.12.025
29. Chen W, Glackin CA, Horwitz MA, Zink JI. Nanomachines and other caps on mesoporous silica nanoparticles for drug delivery. Acc Chem Res. (2019) 52:1531–42. doi: 10.1021/acs.accounts.9b00116
30. Kankala RK, Han YH, Na J, Lee CH, Sun Z, Wang SB, et al. Nanoarchitectured structure and surface biofunctionality of mesoporous silica nanoparticles. Adv Mater. (2020) 32:e1907035. doi: 10.1002/adma.201907035
31. Song K, Jeong E, Jeon W, Jo H, Ban C. A coordination polymer nanobelt (CPNB)-based aptasensor for sulfadimethoxine. Biosens Bioelectron. (2012) 33:113–9. doi: 10.1016/j.bios.2011.12.034
32. Birader K, Kumar P, Tammineni Y, Barla JA, Reddy S, Suman P. Colorimetric aptasensor for on-site detection of oxytetracycline antibiotic in milk. Food Chem. (2021) 356:129659. doi: 10.1016/j.foodchem.2021.129659
33. Wang X, Zhang L, Hao A, Shi Z, Dai C, Yang Y, et al. Silica-coated silver nanoparticles decorated with fluorescent CdTe quantum dots and DNA aptamers for detection of tetracycline. ACS Appl Nano Mater. (2020) 3:9796–803. doi: 10.1021/acsanm.0c01890
34. Mudili V, Makam SS, Sundararaj N, Siddaiah C, Gupta VK, Rao PV. A novel IgY-aptamer hybrid system for cost-effective detection of SEB and its evaluation on food and clinical samples. Sci Rep. (2015) 5:15151. doi: 10.1038/srep15151
35. Kumar H, Bhardwaj K, Kaur T, Nepovimova E, Kuca K, Kumar V, et al. Detection of bacterial pathogens and antibiotic residues in chicken meat: a review. Foods. (2020) 9:1504. doi: 10.3390/foods9101504
36. You X, Zhang G, Chen Y, Liu D, Ma D, Zhou J, et al. A novel electrochemical immunosensor for the sensitive detection of tiamulin based on staphylococcal protein A and silver nanoparticle-graphene oxide nanocomposites. Bioelectrochemistry. (2021) 141:107877. doi: 10.1016/j.bioelechem.2021.107877
37. Zhang J, Xu L, Jiang H, Xu C, Liu W, Wu T, et al. Development of an immunoassay for the detection of copper residues in pork tissues. Biosensors. (2021) 11:235. doi: 10.3390/bios11070235
38. Chen Z, Yu C, Xi J, Tang S, Bao T, Zhang J. A hybrid material prepared by controlled growth of a covalent organic framework on amino-modified MIL-68 for pipette tip solid-phase extraction of sulfonamides prior to their determination by HPLC. Microchim Acta. (2019) 186:393. doi: 10.1007/s00604-019-3513-7
39. Zeng Y, Liang D, Zheng P, Zhang Y, Wang Z, Mari G, et al. A simple and rapid immunochromatography test based on readily available filter paper modified with chitosan to screen for 13 sulfonamides in milk. J. Dairy Sci. (2020) 104:126–33. doi: 10.3168/jds.2020-18987
40. Saleh H, Elhenawee M, Hussien E, Ahmed N, Ibrahim A. Validation of HPLC-UV multi-residue method for the simultaneous determination of tetracycline, oxytetracycline, spiramycin and neospiramycin in raw milk. Food Anal Method. (2021) 14:36–43. doi: 10.1007/s12161-020-01838-9
41. Chen Y, Kong D, Liu L, Song S, Kuang H, Xu C. Development of an elisa and immunochromatographic assay for tetracycline, oxytetracycline, and chlortetracycline residues in milk and honey based on the class-specific monoclonal antibody. Food Anal Method. (2016) 9:905–14. doi: 10.1007/s12161-015-0262-z
Keywords: fluorescent aptasensor, sulfadimethoxine, oxytetracycline, silica nanoparticles, quantitative detection
Citation: Tang J, Zheng X, Jiang S, Cao M, Wang S, Zhou Z, Nie X, Fang Y and Le T (2022) Dual fluorescent aptasensor for simultanous and quantitative detection of sulfadimethoxine and oxytetracycin residues in animal-derived foods tissues based on mesoporous silica. Front. Nutr. 9:1077893. doi: 10.3389/fnut.2022.1077893
Received: 23 October 2022; Accepted: 06 December 2022;
Published: 21 December 2022.
Edited by:
Mahmood Alizadeh Sani, Tehran University of Medical Sciences, IranReviewed by:
Ali Falahati, Yazd University, IranCopyright © 2022 Tang, Zheng, Jiang, Cao, Wang, Zhou, Nie, Fang and Le. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tao Le, ✉ bGV0YW9AY3FudS5lZHUuY24=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.