- 1Department of Biotechnology, Al-Farabi Kazakh National University, Almaty, Kazakhstan
- 2Institute for Bioengineering, FH Aachen University of Applied Sciences, Jülich, Germany
- 3Department of Internal Diseases, Kazakh National Medical University Named After S.D. Asfendiyarov, Almaty, Kazakhstan
- 4School of Food and Biological Engineering, Shaanxi University of Science and Technology, Xi’an, China
The oral cavity is a key biocenosis for many distinct microbial communities that interact with both the external environment and internal body systems. The oral microbiota is a vital part of the human microbiome. It has been developed through mutual interactions among the environment, host physiological state, and microbial community composition. Indigenious microbiota of the oral cavity is one of the factors that prevent adhesion and invasion of pathogens on the mucous membrane, i.e., the development of the infectious process and thereby participating in the implementation of one of the mechanisms of local immunity–colonization resistance. The balance between bacterial symbiosis, microbial virulence, and host resistance ensures the integrity of the oral cavity. In this review we have tried to address how nutritional factors influence integrity of the oral indigenous microbiota and its involvement in colonization resistance.
Introduction
The oral cavity is considered a unique ecological system, creating favorable conditions for the vital activity of manifold commensal microorganisms that may reside either as planktonic cells or inhabit the biofilms (1). These microbial communities contribute to oral and systemic health by maintaining homeostasis and modulating the immune system (2). The oral cavity becomes colonized with a microbiota, the composition and characteristics of which reflect the local aspects, including potential nutrients, receptors for adhesion, oxygen levels, microbial competitors/collaborators, and local innate and adaptive immune factors. The presence of certain nutrients can lead to a defined spatial architecture within the oral biofilms and contribute to colonization resistance.
Colonization resistance of the oral cavity is a complex, multifaceted phenomenon and characterizes the ability of the resident microbial community to oppose the invasion by exogenous microorganisms. The colonization level, in general, depends on how well the host oral cavity is suitable for growth and on the physiological requirements of the microorganisms. Several principal factors known as “colonization barriers” control the microbial background in direct or indirect pathways (3, 4). Qualitative and quantitative changes in microbiome accompanying the oral cavity’s diseases have been characterized and studied in sufficient detail over the past decade (5, 6).
Especially prominent is the “mucous barrier,” which consists of mechanical, humoral, and other factors protecting the mucous membrane from the colonization of harmful microorganisms (7). In general, the mucous membranes of the lips, palate, cheeks, tongue, gums, teeth, and saliva provide a favorable environment for the growth and reproduction of a wide range of microorganisms. These surfaces are typically densely colonized by complex microbial communities interacting through sophisticated biochemical and biophysical mechanisms (8). Many published studies mention antagonistic activity and adhesiveness of the resident microbiome as the main factors in maintaining intestinal colonization resistance (9). However, the colonization resistance of the oral cavity mediated by nutritional factors remains poorly understood in many aspects. Therefore, the present review furnishes a brief overview the main mechanisms and factors responsible for nutrient-related colonization resistance of the oral microbiota.
Materials and methods
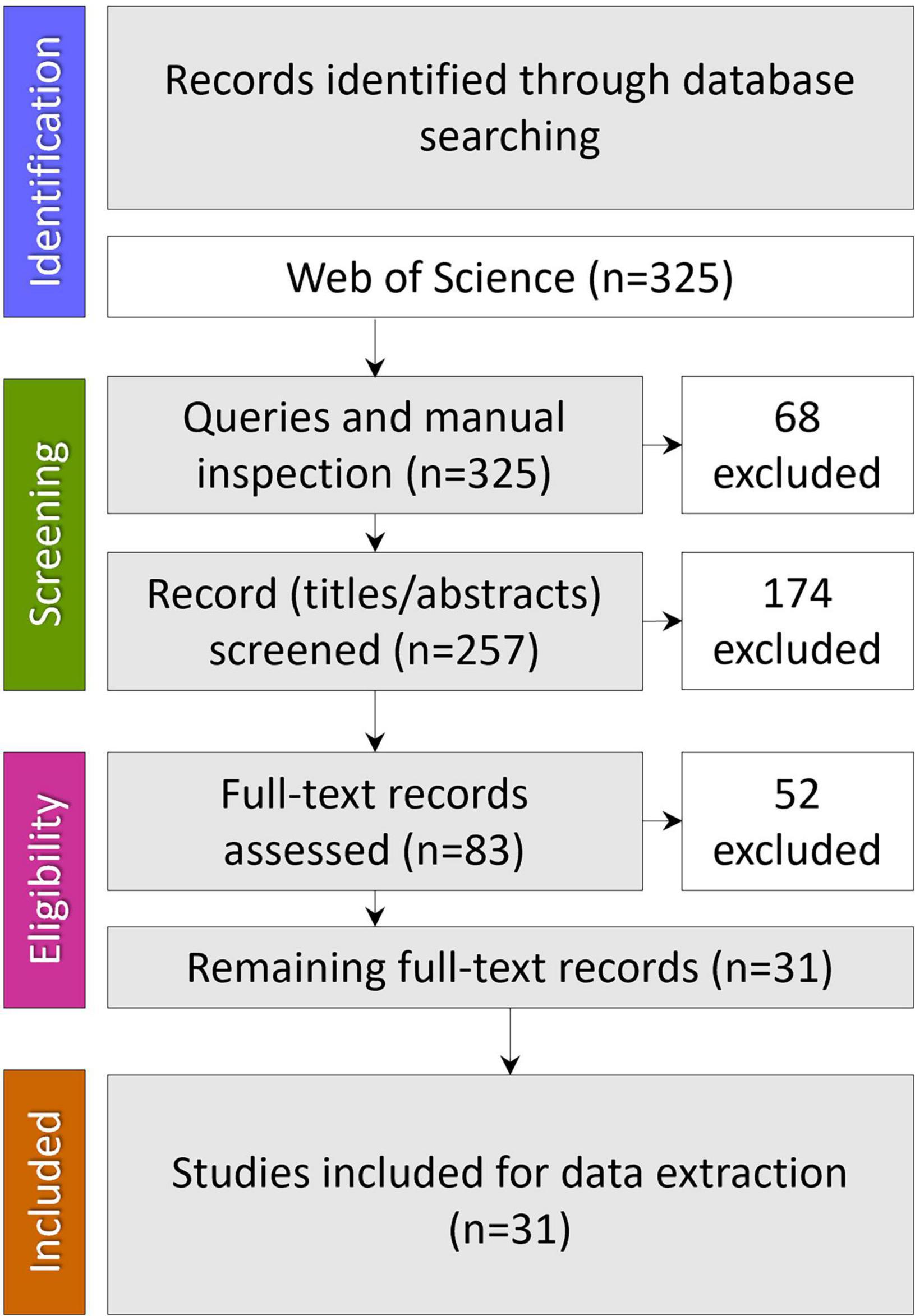
A comprehensive literature search was carried out in the online version of the Science Citation Index Expanded (SCI-EXPANDED) from the Web of Science (WoS) database. WoS was chosen as it covers multidisciplinary areas being the oldest citation database. No time restrictions were placed on these searches, and only articles published in English were retrieved. The date when all searches were last performed was September 23, 2022. The search strategy combined three search strings: #1 “oral microbiome” OR “oral microbiota” OR “oral microbiocenosis” OR “oral microbial communities”; #2 “colonization resistance” OR “bacterial interference”; #3 “nutrition” OR “diet” and combining these by “AND” to obtain only the intersection. Results were imported into a bibliographic referencing tool (EndNote 20) and assessed for relevance and quality, removing articles that have no relation to the review topic. Finally, the query results were manually checked before excluding duplicates (Figure 1).
Results
Our set of queries identified three hundred twenty-five records; after manual inspection and excluding the duplicates, 257 remained. Of these, 174 articles were excluded at the title/abstract level and 52 at the full-text assessment level. Thirty-one were found to meet inclusion criteria to describe the nutritional factors influencing microbiota-mediated colonization resistance of the oral cavity and were used in the analysis.
The evaluation of the keywords in the included studies is valuable to provide a detailed picture of the review topic, reflecting the research hotspots in the current discipline. Here, the publication keyword analysis to word cloud visualization (Biblioshiny app from the Bibliometrix-R package) revealed that the most common keywords of the thirty-one studies were oral, microbiota, biofilm, formation, saliva, colonization, and resistance (Figure 2). This emphasizes that most studies have focused on biofilm formation and colonization resistance in the oral cavity, as well as saliva’s role in determining the oral microbiota composition.
Colonization and principal composition of the oral microbiome
The mean total surface area of the mouth is 214.7 ± 12.9 cm2 (10), and the mean surface area of the oral mucosa is 196.96 ± 24.20 cm2 (11). Each of the anatomical surfaces of the oral cavity, including tooth, gingival sulcus, tongue, hard and soft palates, tonsils, and saliva, is covered with a conglomerate of microorganisms (12, 13). In addition, a large number of microbes are located on the back of the tongue, in the cracks, crevices, and fissures of the tonsils, and in gingival pockets (14).
Intestinal colonization gets its origin from the oral cavity. As the oral cavity is in constant contact with the external environment, it is populated by microorganisms representing a complex biocenosis. In other words, microorganisms making up the microbiocenosis of the oral cavity are intrinsically diverse in their abundance and properties (15). Various microbial taxonomic groups colonize the oral cavity as a kind of ecological niche involving biochemical, immunological, and other interactions with the host. The evolutionary complex and symbiotic communities of microorganisms are therefore specific for a particular area of the mucosal surfaces.
Approximately 99% of all bacteria live together as a biofilm, forming spatially and functionally complex communities (16). Biofilms act as protective shells, making inhabitants more resistant to physical, chemical, and biological factors in comparison with planktonic (free-floating) bacteria (17, 18). In addition to that the biofilm polymers provide adhesion, stabilization, and nutrient flows within the biofilm (19).
Microbial populations in the oral cavity can be divided into resident and transient groups. The resident (indigenous) microbiome includes relatively constant species characteristic of a certain oral biotope and the age of the host. An indigenous microbiome can be divided into core (shared by all host organisms) and variable (due to physiological and biochemical differences between individuals) categories. The transient (exogenous) microbiome consists of non-pathogenic or opportunistic microorganisms that populate the oral cavity for a limited period without causing disease (20). In case of violations or loss of the indigenous microflora, members of the transient can replace the “vacant” niche of a specific biotope that subsequently can contribute to the development of pathology.
The Human Oral Microbiome Database (HOMD1) has been created to systematize the bacteria in the human oral cavity, which includes both members of normal microflora and pathogens. HOMD collected 16S rRNA gene sequences from oral prokaryote species into a curated phylogeny-based database. The HOMD contains approximately 772 microbial species, where 70% are culturable, and 57% of which are officially named. Most of the HOMD-listed bacterial species belong to transient microflora since they are not capable of long-term survival under special conditions of the oral cavity. The 16S rDNA profiling of the healthy cavity categorized the inhabitant bacteria into six broad phyla, namely, Firmicutes, Actinobacteria, Proteobacteria, Fusobacteria, Bacteroidetes, and Spirochaetes constituting 96% of total oral bacteria (21).
Streptococcus is the most abundant genus in the oral cavity (8). In the HOMD, the Streptococcus genus is represented by 37 species, of which 29 are named, four are not named, and four are lost. The species of Streptococcus occupy a specific niche in the oral cavity and thus play a key role in establishing and shaping the oral microbiota (22). S. gordonii and S. oralis are among the first microorganisms that colonize the oral cavity, followed by cryogenic S. sanguinis, S. mutans, and S. sobrinus, initiating biofilm formation (21). Other pioneer organisms include Actinomyces spp. Granulicatella adiacens, Abiotrophia defectiva, Gemella spp., and Rothia (23). Diverse molecular forces, including hydrogen bonds, hydrophobic interactions, calcium bridges, van der Waals forces, acid-base interactions, and electrostatic interactions, contribute to the attachment of pioneer bacteria to the salivary acquired pellicle (a layer of proteins and glycoproteins of salivary origin that tightly coat the tooth surface) (24). The early colonizers are predominantly members of the normal microbiota, and just a few are known to be directly responsible for disease development (20). Species of Streptococcus initiate numerous cooperative and antagonistic bacterial interactions within the dental microbial community. Thus, mainly streptococci determine and shape the composition of later colonizers in the oral biofilm and greatly impact the health or disease status of the host (22, 25). Polymicrobial colonization and biofilm development have been well-described and depicted by D. Verma et al. (21).
The role of the normal oral microbiota
Normal microbiota performs protective functions due to indirect antagonism toward pathogenic and opportunistic microorganisms, particularly by preventing colonization of mucous membranes and diminishing the penetration of microbes, microbial toxins, and xenobiotics into the host organism (26, 27). Additionally, the functions of normal oral human microbiota include:
• the regulation of the gas composition of the intestine and other cavities of the host;
• morphokinetic effect;
• the production of enzymes involved in the metabolism of proteins, carbohydrates, lipids, and nucleic acids;
• production of biologically active compounds (vitamins, antibiotics, toxins, hormones, etc.); immunogenic role;
• participation in the recirculation of bile acids, cholesterol, and other macromolecules; mutagenic/antimutagenic role;
• detoxification of exogenous and endogenous substrates and metabolites;
• source of endogenous infection and,
storage of microbial plasmid and chromosomal genes (28–30).
Nutritional factors influencing the oral microbiota
Saliva is the medium by which the host “supplies” its resident microorganisms with nutrients, including amino acids, proteins, glycoproteins, peptides, and vitamins. In addition, a host-derived nutrient, gingival crevicular fluid (GCF), favors the growth and activity of microorganisms in the oral cavity. In a smaller proportion, the gingival crevice, through GCF secretion, contributes with additional nutrients such as albumin and heme-containing molecules as a source of vital iron (31). Host hormones, such as sex steroid hormones, cholesterol, and catecholamines, delivered through saliva can also be utilized by resident bacteria (32). Many studies suggest that these hormones have the potential to modulate the composition of the oral microbiome (33, 34).
Despite the obvious impact of diet on the oral microbiome, relatively scant information is available regarding this. This can partly be explained by the fact that the primary substrates for oral microbial growth are endogenous nutrients provided by saliva, tissue excludes, GCF, degenerating host cells, or other bacterial metabolites (35).
Studies by Hatakka et al. and Jiang et al. have shown no difference in the growth rates of oral bacteria in the presence or absence of food, indicating no relationship between diet and the composition of oral bacterial communities (36, 37). In contrast, another very recent study by W. G. Wade revealed differences in salivary metabolomic profiles in relation to diet type (omnivorous, ovo-lacto-vegetarian, or vegetarians) (38).
Reduced food intake and fasting periods may affect microbiome-based colonization resistance. The salivary flow and secretion stasis due to dehydration or decreased oral water intake retrograde bacterial migration and colonization (39, 40). Fasting has also been found to be associated with oral cytokine levels caused by resident and transient microbiome (41).
Oral colonization resistance
The composition of microbial communities in different biotopes of the oral cavity is determined by environmental and biological factors, giving rise to synergistic or antagonistic relationships (Figure 3). Especially antagonistic relationships between different groups of microbes can be induced by various factors [such as lack of saliva, its bactericidal substances, stimulants (e.g., smoking), increased sugar content, acidic microenvironment] that alter the microbial community structure subsequently impacting colonization resistance (8).
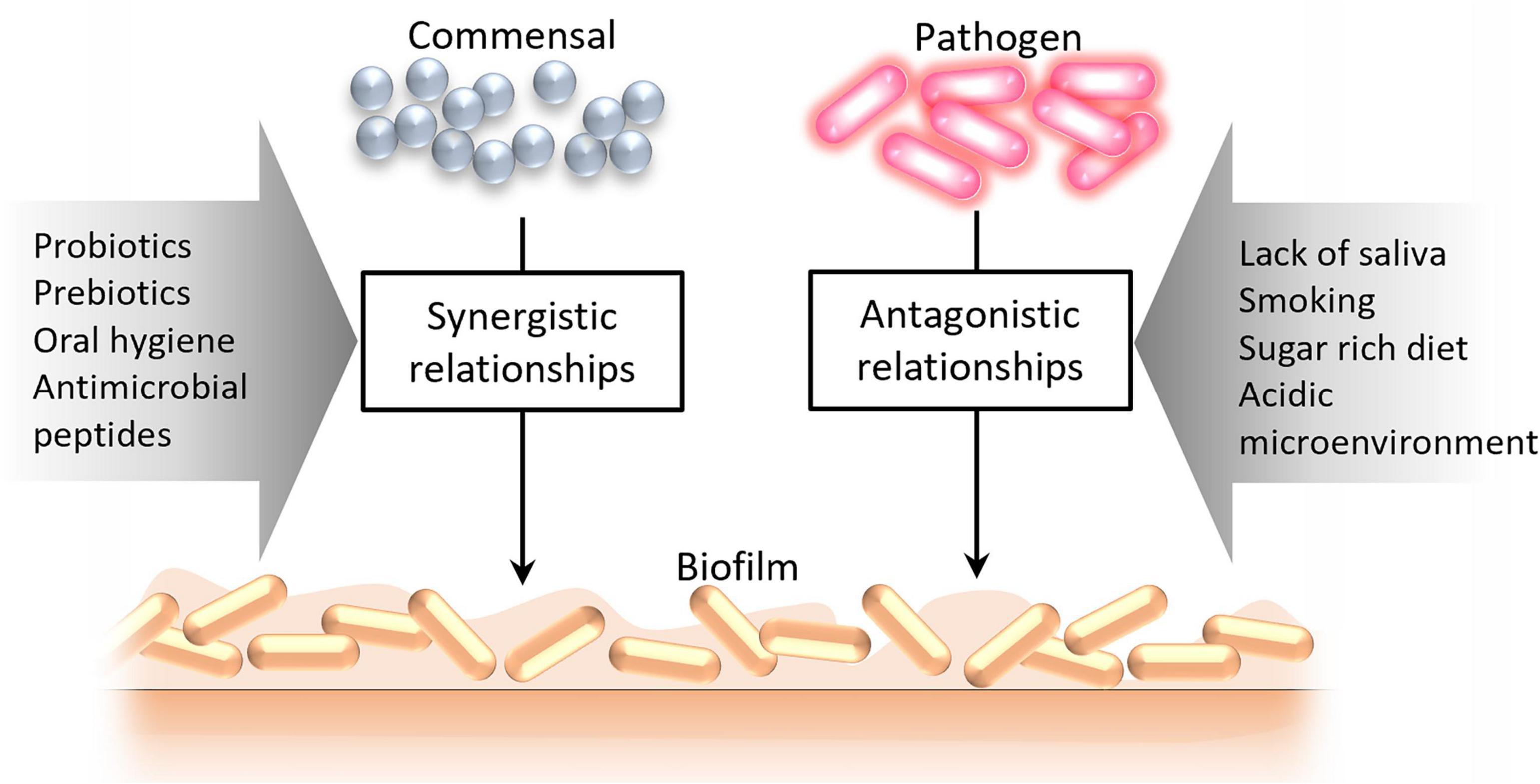
Figure 3. Factors leading to the formation of synergistic and antagonistic relationships between oral microorganisms.
Colonization resistance (also known as bacterial interference) refers to a set of mechanisms providing individual specificity and stability to the microbial community and preventing the host surfaces colonization by pathogens. The term “colonization resistance” was coined by D. van der Waaij (42) who also pointed out that the normal microbiota being a combination of many microbiocenoses characterized by a certain composition and occupying a particular biotope in the human body, plays deciding role in such resistance (43).
In the case of weakened colonization resistance, the fraction of “core” bacteria resident for the surfaces of the human body reduces, while the number and spectrum of potentially pathogenic microorganisms increase. This can lead to their translocation to internal organs and even to the development of purulent-inflammatory processes (44, 45).
Germ-free animals are the primary models showing the pivotal role of resident microflora in colonization resistance and overall health. For example, P. D. Marsh has shown that the absence of resident microbiome has a negative impact on its host, leading to thinning of the intestinal walls, ill-developed villi, poor nutrient absorption, vitamin deficiencies, caecum enlargement, etc., (46). Later, further experimental validations of the role of normal microflora in preventing infections for different microbial models have reported (47–49).
According to Marsh and Percival (50) the mechanisms mediating colonization resistance can be divided into (a) competition for nutrients, (b) competition for attachment sites, (c)production of antagonistic compounds, and (d) creation of adverse environmental conditions for exogenous microorganisms (Figure 4). Here, we would like to update and address the abovementioned factors in more detail.
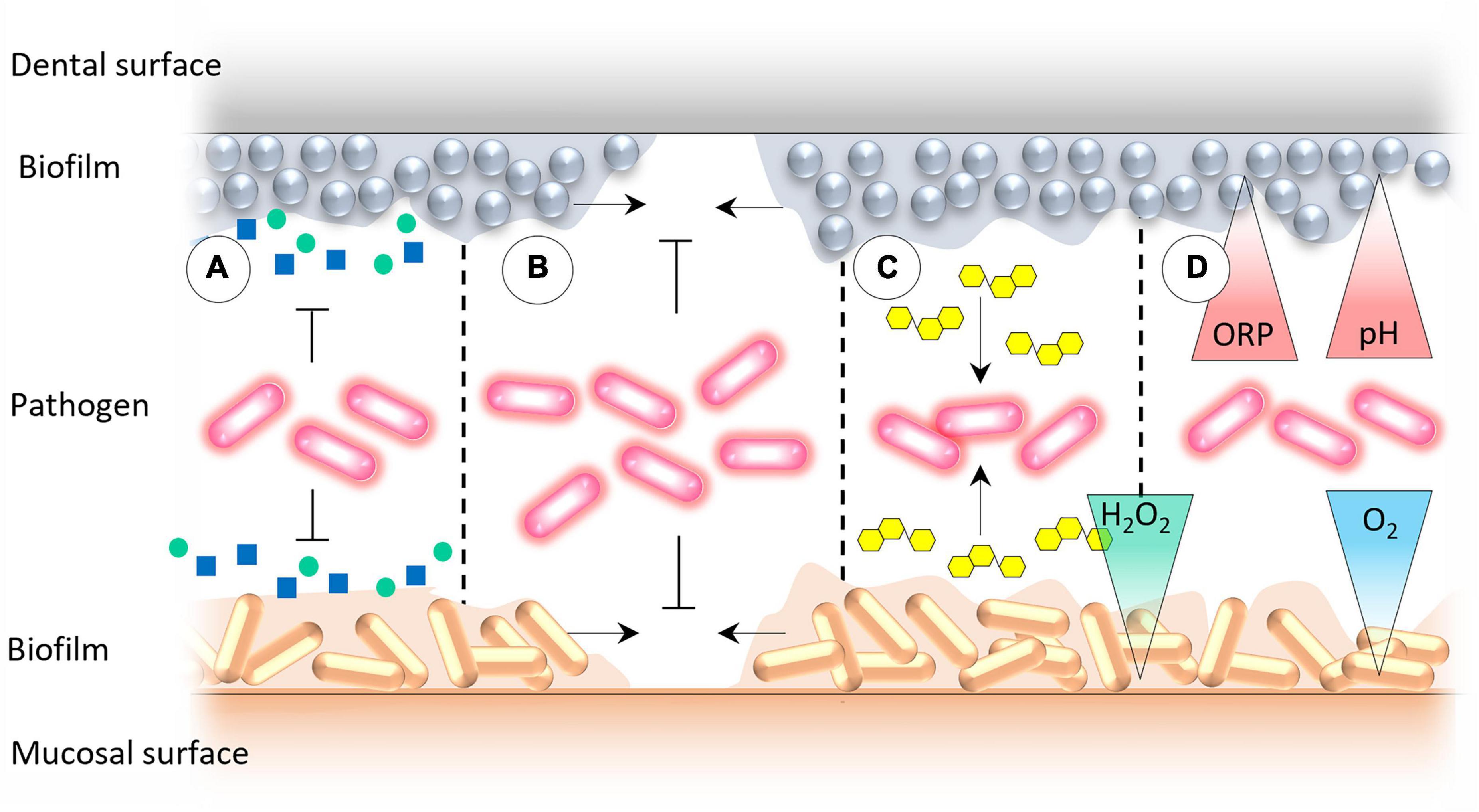
Figure 4. The mechanisms that underpin the oral colonization resistance: (A) Competition for essential nutrients and co-factors for microbial growth, (B) competition for binding sites for microbial attachment on mucosal and dental surfaces, (C) production of antagonistic compounds by the resident oral microbiota, and (D) creation of adverse environmental conditions that prevent the growth of exogenous microorganisms.
Nutrient-related factors of colonization resistance
Temperature, pH, atmospheric conditions, salinity, redox potential, sheer and mechanical forces, chemical exposure from hygiene practices, and water of saliva affect the formation of biofilms in the oral cavity (51). However, overall microbial biomass and its composition become considerably controlled by competition for nutritional substances that can be utilized at a low redox potential conditions and in the presence of metabolic inhibitors synthesized by oral microorganisms (52). Salivary amino acids, peptides, GCF, and glycoproteins (such as mucin) are the main limiting nutrient sources for bacteria inhabiting dental plaques. Saliva is the primary nutrient source for bacteria that reside in supragingival biofilm, while GCF provides nutrients for bacteria of the subgingival biofilms (53).
The exact composition of saliva, and therefore the availability of particular nutrients, displays significant interindividual differences as well as high temporary variability (54). In general, resident bacteria outcompete periodontopathogens in the uptake of these nutrients (55–57). However, periodontitis-associated microorganisms destruct tissue through degradation of the extracellular matrix, which leads to additional release of specific nutrients (heme-containing compounds, sources of amino acids, and iron). These nutrients are carried into the gingival crevice through GCF, which favors the atypical growth of asaccharolytic and proteolytic microorganisms with iron-acquisition capacity in the subgingival region (15).
“Food sharing” through bacterial metabolic products also strongly shapes the microbial composition, by encouraging the growth of some species while averting others. For instance, lactic acid, produced by Streptococcus and Actinomyces in the mouth as a result of carbohydrate fermentation can be utilized by Veillonella, allowing for menadione production that is, in turn, important for the growth of Porphyromonas and Prevotella. Fusobacterium produces fatty acids that are used by Treponema. Porphyromonas cal also cooperate with Treponema to generate end metabolites that are utilized by Mogibacterium timidum (58).
As shown by Van Hoogmoed et al., some conventional oral commensals, such as S. sanguinis, S. cristatus, S. salivarius, S mitis, and A. naeslundii, decrease the ability of a pathogen Porphyromonas gingivalis to adhere to the substrate and retrieve essential nutrients (59). Under optimal conditions, L. lactis, a member of the normal oral microbiota, produces nisin, a bacteriocin that mitigates pathogen-mediated oral tumorigenesis (36, 60). Numerous mutualistic nutritional behaviors have also been observed for bacteria growing in saliva as their sole nutrient source (37, 61).
One of the emerging therapeutic approaches could be the introduction of probiotic bacteria which may prevent pathogen colonization in the oral cavity by limiting their adhesion and producing antimicrobials that selectively target disease-associated bacteria (62, 63). For instance, Streptococcus salivarius displayed properties compatible with their potential use as probiotics antagonizing Streptococcus pyogenes (64–66). However, the main disadvantage here is that the presence of probiotic bacteria can be only transient since they are not indigenous to the oral cavity. Therefore, one more attractive therapeutic strategy could be the nutritional stimulation of indigenous bacteria, promoting oral health.
Indeed, prebiotic substances often induce desirable microbial composition and activity changes, thus delivering local health benefits (67–69). For example, Slomka et al. (69) observed that potential oral prebiotic compounds, such as beta-methyl-D-galactoside and N-acetyl-D-mannosamine selectively stimulate beneficial commensal bacteria of the resident oral microbiome while suppressing the growth of pathogenic bacteria.
Colonization resistance affects the oral ecology in health and disease by preventing or modulating the prevalence of specific microbial groups. Many clinical studies revealed that nutritional compounds are the key factors that alter the oral microbial composition by colonizing microbial biofilms, co-aggregating, and competing with pathogenic bacteria, subsequently reducing/replacing their numbers (Table 1).
Other types of colonization resistance
Competition for microbial attachment sites on mucosal and dental surfaces
A large number of bacterial species appear to exhibit specific tropism in relation to various anatomical surfaces of the oral cavity. By examining 40 bacterial species, Mager et al. (80) have shown that bacteria that inhabit numerous oral cavity surfaces use very many different receptors and adhesion molecules that define the formation of biofilms (29). Resident oral bacteria form a robust and tight biofilm on the surface of mucous membranes, hindering the adhesion of foreign microorganisms. The most prominent mechanism inhibiting biofilm formation and inducing detachment of extrinsic bacteria from the native biofilm is known as biosurfactant action. Several in vitro studies indicated that many bacterial species, especially Streptococcus, rely predominantly on this method to prevent foreign colonization of the oral cavity (59, 81, 82). This and other attachment-related strategies are briefly summarized in Table 2.
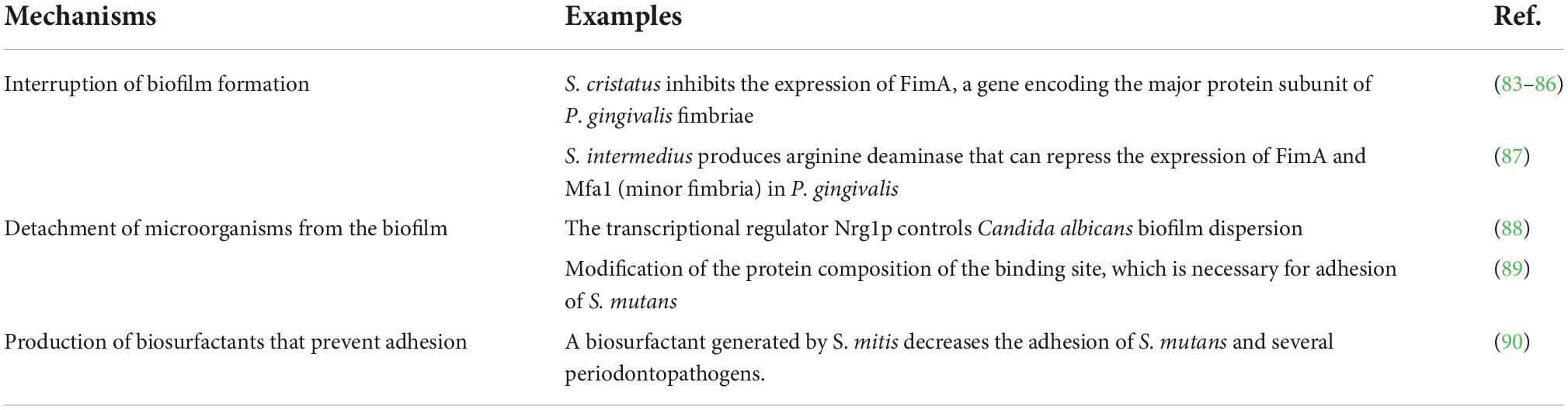
Table 2. The mechanisms involved in the competition for attachment sites of colonization resistance.
Production of antagonistic compounds (inhibitory metabolites)
End products of metabolism of resident microflora are also used for effective protection against extraneous colonization. The antagonism of microorganisms that make up the normal microbiota concerning potentially pathogenic bacteria is due to the production of bacteriocins, lysozyme, and other substances (Table 3).
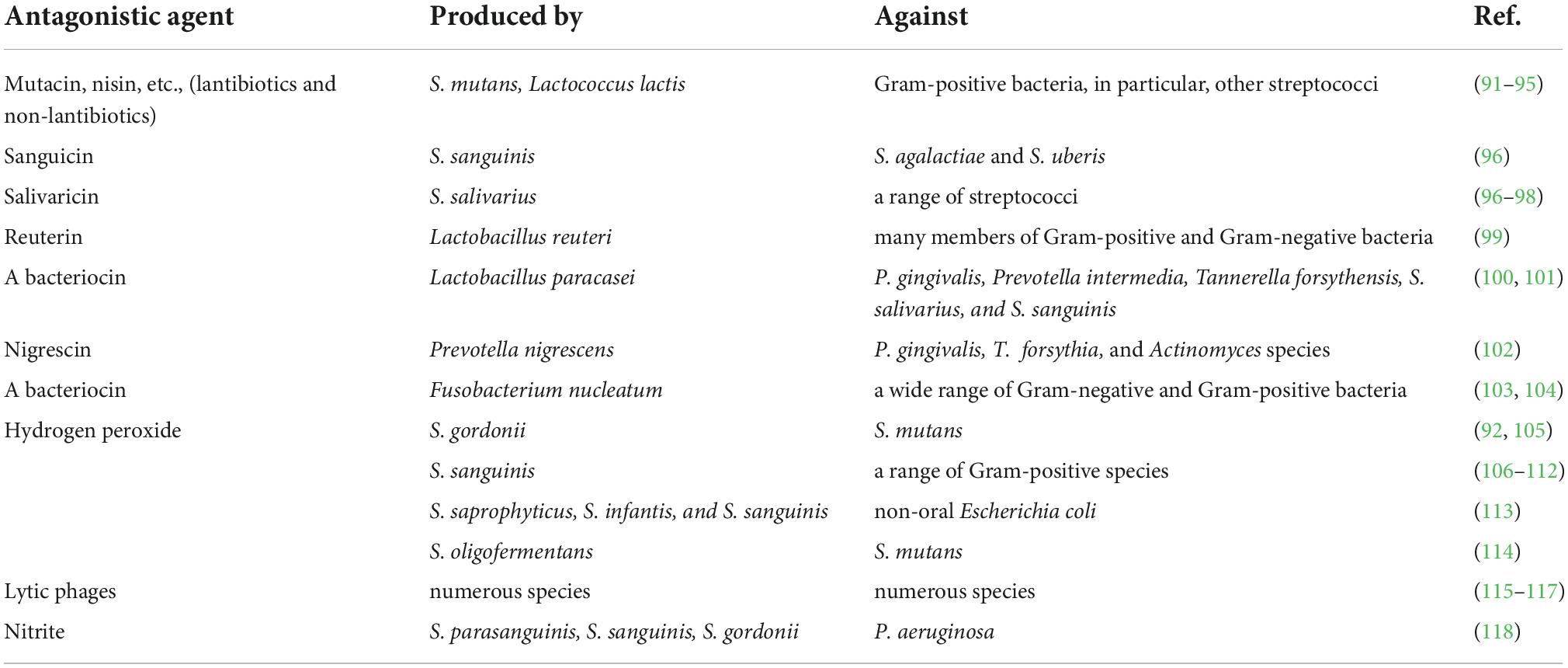
Table 3. Production of inhibitory factors by the resident oral microflora that contribute to “colonization resistance”.
Various bacteriophages represent very abundant and interesting group of oral antimicrobial agents. Oral phages are able to invade many other bacteria besides their putative bacterial hosts. Therefore, phages strongly shape the ecology of oral bacterial communities, accelerate their molecular diversity and help to acquire new gene functions (115–117).
In addition to the metabolic antimicrobials of the microbiota listed in the Table 2, various organic acids should be mentioned, such as short-chain fatty acids (SCFAs), that may act as inhibitory factors (119, 120). Though SCFAs are mainly produced in the intestines, they also contribute to preventing colonization by pathogenic microorganisms in the oral cavity (55).
Creation of microenvironments that inhibit the growth of exogenous bacterial species
Here the competition is enabled due to altered environmental conditions, such as pH, oxygen pressure, redox potential, etc., in oral biofilms. The members of Lactobacillus and Streptococcus are the powerful acid producers, making the local pH drop as low as 4.5, thus dramatically suppressing the growth of all acid-sensitive bacteria (121). Suppression of S. sanguinis by mixture of organic acids produced by S. mutans has been mentioned in many studies as well (112, 122, 123).
The factors affecting/influencing colonization patterns
In healthy people, the microbial composition of the oral cavity depends on the physiological and ecological aspects of the host, such as age, nutrition preferences, oral hygiene, anatomical features of the oral cavity, hormonal status, general somatic state, etc., (124). The richness and composition of the oral microbiome are relatively stable due to moisture availability, the constant presence of antimicrobial substances (nisin, diplococcin, acidophilus, lactocidin, lactolin lysozyme, amylases, immunoglobulins A, G, M), organic acids (lactic, acetic, ketoglutaric and succinic) and the state of general cellular and humoral immunity (12).
As described in the previous parts, the colonization resistance is determined by factors of microbial, exogenous, and host origin (125, 126).
Microbial factors
Each human individual is characterized by a specific genetically determined spectrum of microorganisms. As we already saw, the normal microflora plays a vital role in the antimicrobial defense system of the oral cavity. The term “normal” indicates a microbial population that colonizes various ecological niches of the healthy oral cavity and takes part in the metabolism of nutrition, protects against highly virulent bacteria by blocking receptors of epithelial cells from adhesion of pathogens, stimulates the immune response, and produces biologically active substances which regulate metabolic processes (127).
Exogenous factors
A person’s normal microflora should also be always considered in the context of the whole organism and its environment. The oral cavity is the very beginning of the digestive tract and serves as the “primary portal” for chemical substances and foreign microorganisms. Therefore, numerous external factors affecting the body also affect the microflora. The most pronounced and well-characterized phenomena depend on colonization resistance from smoking status, alcohol consumption, diet (quality and quantity), socioeconomic status, and antibiotic use (125). Smoking is a major environmental factor associated with the pathophysiology of oral diseases. Toxic components in cigarette impact oral microbiota directly or indirectly through oxygen deprivation, immunosuppression, biofilm formation, or other potential mechanisms, leading to loss of colonization resistance (128). Despite different sampling sites, numerous studies have shown the predominance of Fusobacterium nucleatum and F. naviforme in oral from smokers compared with non-smokers (129). Alcohol consumption may also affect oral microbiota composition affecting functional microbial pathways. Thomas et al. observed the reduced bacterial richness in the oral biofilm of alcohol drinkers (130). Liao et al. found that the genus Prevotella and Moryella were significantly enriched in drinkers; meanwhile, the genus Lautropia, Haemophilus, and Porphyromonas were depleted significantly (131). Studies indicate that socioeconomic status may alter oral microbiota community structure and higher diversity (132, 133). The oral microbiota is a major reservoir of antibiotic-resistant bacteria; many studies have demonstrated that using amoxicillin, erythromycin, and tetracycline changes oral microbiota composition and enriches bacteria resistant to antibiotics (134, 135).
Host factors
Host mechanisms involved in the colonization resistance phenomenon include mucosal desquamation, the antimicrobial effect of secrets, the composition and quantity of mucin, oxygen tension along with the thickness of the biofilm, the pH of the medium, the rate of renewal, maturation and metabolism of mucosal epithelium, innate, and adaptive immune mechanisms, etc., (136). The immune factors, in turn, can involve macrophage activity, lysozyme, lactoferrin, other bactericidal substances of leukocytes, as well as a variety of immunoglobulins, primarily IgA, which prevent microbial adhesion and thus promote the removal of extraneous microorganisms to the external environment (137). The antibacterial potential of saliva on the one side and the number of microorganisms in the oral cavity on the other side exist in dynamic balance. Any infringement of the former leads to disturbances of the normal microflora and the emergence of pathogenic microorganisms by developing various types of pathology in the oral cavity. However, the main functional properties of the host antimicrobial system of saliva not only include suppression of microflora but also effectively control its qualitative and quantitative composition at a level sufficient to maintain microbiocenosis (31, 138).
Global health relevance
Dental infection and antibiotic resistance remain important global health concerns with significant morbidities. There are convincing scientific pieces of evidence that impaired oral health potentiates the severity of numerous systemic diseases, such as endocarditis, diabetes mellitus, osteoporosis, and tumors (139–142). Severe microbial oral infection and subsequent inflammation, along with meningitis and endocarditis, are reported to be associated with cerebral infractions among male patients (143).
As mentioned above, microbial populations in the oral cavity are of two major types: the resident and transient microbiome; their delicate balance is essential for normal oral functions. Poor oral hygiene, smoking or chewing tobacco, inadequate nutrition, and overuse of antibiotics, not only can disrupt such homeostatic balance between oral resident and transient microbiome, but can also induce antimicrobial resistance (144, 145). Since normal oral microbiota exerts defensive functions against opportunistic harmful microorganisms, developing an approach to restore normal oral microbiota in infectious and inflammatory diseases would likely reduce the oral burden of diseases. The administration of healthy fecal microbiota to restore colonization resistance and displace multi-drug resistant (MDR) bacteria is already a commonly used therapeutic practice (146–148). Whether a similar approach could be employed to restore normal oral microbiota in oral diseases is an area that requires further experimental, theoretical and ethical validation.
A better understanding of microbiota-mediated colonization resistance of the oral cavity would promote rational dental care, and minimize oral-infection related chronic debilitating pathologies, which is also a global health concern. As frequently mentioned, the mouth is the gateway to total body wellness; consequently, the oral microbiome is likely to influence the overall health of an individual. Therapeutic manipulation of the oral microbiome in a patient, by targeting harmful species, to maintain healthier oral status in a community will further assist in the maintenance of good health and well-being, in general.
Conclusion
The abundance and composition of the oral microbial communities are characterized by the constancy and integrity of the relationships along with the antagonistic and stimulatory effects between microorganisms and their hosts. Colonization resistance is one of the phenomena of local immunity, which depends on a combination of factors that prevent the adhesion and reproduction of exogenous bacteria on dental and mucous surfaces. A certain role in this belongs to resident microflora, which is a potent inhibitor of pathogens and synergist for commensals of the same ecological niche. The antagonistic effect of normal oral microflora is due to the significant adhesive and colonizing ability of resident microbial species, as well as the production of specific substances that inhibit the growth of transient pathogens. Nutritional factors may also modulate microbiota-mediated colonization resistance. So far, the available evidence to assess the real impact of different nutrients on the colonization resistance of the oral microbiome is still insufficient, and more studies are needed.
Author contributions
All authors contributed to the planning, writing, scientific content, reviewing, editing of this document, and approved the submitted version.
Acknowledgments
The authors thank Eva-Maria Geenen from the Aachen University of Applied Sciences (Germany) for the valuable comments and helpful input.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
1. Cornick S, Tawiah A, Chadee K. Roles and regulation of the mucus barrier in the gut. Tissue Barriers. (2015) 3:e982426. doi: 10.4161/21688370.2014.982426
2. Zhang Y, Wang X, Li H, Ni C, Du Z, Yan F. Human oral microbiota and its modulation for oral health. Biomed Pharmacother. (2018) 99:883–93. doi: 10.1016/j.biopha.2018.01.146
3. Lawley TD, Walker AW. Intestinal colonization resistance. Immunology. (2013) 138:1–11. doi: 10.1111/j.1365-2567.2012.03616.x
4. Buffie CG, Pamer EG. Microbiota-mediated colonization resistance against intestinal pathogens. Nat Rev Immunol. (2013) 13:790–801. doi: 10.1038/nri3535
5. Jia G, Zhi A, Lai PFH, Wang G, Xia Y, Xiong Z, et al. The oral microbiota–a mechanistic role for systemic diseases. Br Dent J. (2018) 224:447–55. doi: 10.1038/sj.bdj.2018.217
6. Sampaio-Maia B, Caldas IM, Pereira ML, Pérez-Mongiovi D, Araujo R. The oral microbiome in health and its implication in oral and systemic diseases. Adv Appl Microbiol. (2016) 97:171–210. doi: 10.1016/bs.aambs.2016.08.002
7. Sharma N, Bhatia S, Sodhi AS, Batra N. Oral microbiome and health. AIMS Microbiol. (2018) 4:42. doi: 10.3934/microbiol.2018.1.42
8. Gao L, Xu T, Huang G, Jiang S, Gu Y, Chen F. Oral microbiomes: more and more importance in oral cavity and whole body. Protein Cell. (2018) 9:488–500. doi: 10.1007/s13238-018-0548-1
9. Dewhirst FE, Chen T, Izard J, Paster BJ, Tanner ACR, Yu W-H, et al. The human oral microbiome. J Bacteriol. (2010) 192:5002–17. doi: 10.1128/JB.00542-10
10. Collins LMC, Dawes C. The surface area of the adult human mouth and thickness of the salivary film covering the teeth and oral mucosa. J Dent Res. (1987) 66:1300–2. doi: 10.1177/00220345870660080201
11. Naumova EA, Dierkes T, Sprang J, Arnold WH. The oral mucosal surface and blood vessels. Head Face Med. (2013) 9:1–5. doi: 10.1186/1746-160X-9-8
12. Lu M, Xuan S, Wang Z. Oral microbiota: a new view of body health. Food Sci Hum Wellness. (2019) 8:8–15. doi: 10.1016/j.fshw.2018.12.001
13. Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. J Clin Microbiol. (2005) 43:5721–32. doi: 10.1128/JCM.43.11.5721-5732.2005
14. Deo PN, Deshmukh R. Oral microbiome: unveiling the fundamentals. J Oral Maxillofac Pathol. (2019) 23:122. doi: 10.4103/jomfp.JOMFP_152_19
15. Chattopadhyay I, Verma M, Panda M. Role of oral microbiome signatures in diagnosis and prognosis of oral cancer. Technol Cancer Res Treat. (2019) 18:1533033819867354. doi: 10.1177/1533033819867354
16. Lasserre JF, Brecx MC, Toma S. Oral microbes, biofilms and their role in periodontal and peri-implant diseases. Materials. (2018) 11:1802. doi: 10.3390/ma11101802
17. Nadell CD, Xavier JB, Foster KR. The sociobiology of biofilms. FEMS Microbiol Rev. (2008) 33:206–24. doi: 10.1111/j.1574-6976.2008.00150.x
18. Digel I, Akimbekov NS, Kistaubayeva A, Zhubanova AA. Microbial sampling from dry surfaces: current challenges and solutions. In: G Artmann, A Artmann, A Zhubanova, I Digel editors. Biological, Physical and Technical Basics of Cell Engineering. Singapore: Springer (2018). p. 421–56. doi: 10.1007/978-981-10-7904-7_19
19. Berger D, Rakhamimova A, Pollack A, Loewy Z. Oral biofilms: development, control, and analysis. High Throughput. (2018) 7:24. doi: 10.20944/preprints201808.0174.v1
20. Wright CJ, Burns LH, Jack AA, Back CR, Dutton LC, Nobbs AH, et al. Microbial interactions in building of communities. Mol Oral Microbiol. (2013) 28:83–101. doi: 10.1111/omi.12012
21. Verma D, Garg PK, Dubey AK. Insights into the human oral microbiome. Arch Microbiol. (2018) 200:525–40. doi: 10.1007/s00203-018-1505-3
22. Abranches J, Zeng L, Kajfasz JK, Palmer SR, Chakraborty B, Wen ZT, et al. Biology of oral streptococci. Microbiol Spectr. (2018) 6:GPP3-0042-2018. doi: 10.1128/microbiolspec.GPP3-0042-2018
23. Jenkinson HF. Beyond the oral microbiome. Environ Microbiol. (2011) 13:3077–87. doi: 10.1111/j.1462-2920.2011.02573.x
24. Hannig C, Hannig M. The oral cavity—a key system to understand substratum-dependent bioadhesion on solid surfaces in man. Clin Oral Invest. (2009) 13:123–39. doi: 10.1007/s00784-008-0243-3
25. Kreth J, Merritt J, Qi F. Bacterial and host interactions of oral streptococci. DNA Cell Biol. (2009) 28:397–403. doi: 10.1089/dna.2009.0868
26. Sicard J-F, Le Bihan G, Vogeleer P, Jacques M, Harel J. Interactions of intestinal bacteria with components of the intestinal mucus. Front Cell Infect Microbiol. (2017) 7:387. doi: 10.3389/fcimb.2017.00387
27. Yin W, Wang Y, Liu L, He J. Biofilms: the microbial “protective clothing” in extreme environments. Int J Mol Sci. (2019) 20:3423. doi: 10.3390/ijms20143423
28. Rowland I, Gibson G, Heinken A, Scott K, Swann J, Thiele I, et al. Gut microbiota functions: metabolism of nutrients and other food components. Eur J Nutr. (2018) 57:1–24. doi: 10.1007/s00394-017-1445-8
29. Mohajeri MH, Brummer RJM, Rastall RA, Weersma RK, Harmsen HJM, Faas M, et al. The role of the microbiome for human health: from basic science to clinical applications. Eur J Nutr. (2018) 57:1–14. doi: 10.1007/s00394-018-1703-4
30. Wang B, Yao M, Lv L, Ling Z, Li L. The human microbiota in health and disease. Engineering. (2017) 3:71–82. doi: 10.1016/J.ENG.2017.01.008
31. Marsh PD, Do T, Beighton D, Devine DA. Influence of saliva on the oral microbiota. Periodontology 2000. (2016) 70:80–92. doi: 10.1111/prd.12098
32. Kumar PS. Sex and the subgingival microbiome: do female sex steroids affect periodontal bacteria? Periodontology 2000. (2013) 61:103–24. doi: 10.1111/j.1600-0757.2011.00398.x
33. Jentsch HFR, März D, Krüger M. The effects of stress hormones on growth of selected periodontitis related bacteria. Anaerobe. (2013) 24:49–54. doi: 10.1016/j.anaerobe.2013.09.001
34. Roberts A, Matthews JB, Socransky SS, Freestone PP, Williams PH, Chapple IL. Stress and the periodontal diseases: effects of catecholamines on the growth of periodontal bacteria in vitro. Oral Microbiol Immunol. (2002) 17:296–303. doi: 10.1034/j.1399-302X.2002.170506.x
35. Kato I, Vasquez A, Moyerbrailean G, Land S, Djuric Z, Sun J, et al. Nutritional correlates of human oral microbiome. J Am Coll Nutr. (2017) 36:88–98. doi: 10.1080/07315724.2016.1185386
36. Wu J, Xie H. Role of Arginine deiminase of Streptococcus cristatus in Porphyromonas gingivalis colonization. Antimicrob Agents Chemother. (2010) 54:4694–8. doi: 10.1128/AAC.00284-10
37. Foster JS, Kolenbrander PE. Development of a multispecies oral bacterial community in a saliva-conditioned flow cell. Appl Environ Microbiol. (2004) 70:4340–8. doi: 10.1128/AEM.70.7.4340-4348.2004
38. Wade WG. Resilience of the oral microbiome. Periodontol 2000. (2021) 86:113–22. doi: 10.1111/prd.12365
39. Joachim MV, Ghantous Y, Zaaroura S, Alkeesh K, Zoabi T, Abu El-Na’aj I. Does fasting during Ramadan increase the risk of the development of sialadenitis? BMC Oral Health. (2020) 20:156. doi: 10.1186/s12903-020-01139-x
40. Graef FA, Celiberto LS, Allaire JM, Kuan MTY, Bosman ES, Crowley SM, et al. Fasting increases microbiome-based colonization resistance and reduces host inflammatory responses during an enteric bacterial infection. PLoS Pathog. (2021) 17:e1009719. doi: 10.1371/journal.ppat.1009719
41. Sarkar A, Kuehl MN, Alman AC, Burkhardt BR. Linking the oral microbiome and salivary cytokine abundance to circadian oscillations. Sci Rep. (2021) 11:2658. doi: 10.1038/s41598-021-81420-3
42. Van der Waaij D, Berghuis-de Vries JM, Lekkerkerk-Van der Wees JEC. Colonization resistance of the digestive tract in conventional and antibiotic-treated mice. Epidemiol Infect. (1971) 69:405–11. doi: 10.1017/S0022172400021653
43. Jandhyala SM, Talukdar R, Subramanyam C, Vuyyuru H, Sasikala M, Reddy DN. Role of the normal gut microbiota. World J Gastroenterol. (2015) 21:8787. doi: 10.3748/wjg.v21.i29.8787
44. Hegde S, Lin Y-M, Golovko G, Khanipov K, Cong Y, Savidge T, et al. Microbiota dysbiosis and its pathophysiological significance in bowel obstruction. Sci Rep. (2018) 8:1–12. doi: 10.1038/s41598-018-31033-0
45. Yu LC-H. Microbiota dysbiosis and barrier dysfunction in inflammatory bowel disease and colorectal cancers: exploring a common ground hypothesis. J Biomed Sci. (2018) 25:1–14. doi: 10.1186/s12929-018-0483-8
46. Marsh PD. Role of the oral microflora in health. Microb Ecol Health Dis. (2000) 12:130–7. doi: 10.1080/089106000750051800
47. Uzbay T. Germ-free animal experiments in the gut microbiota studies. Curr Opin Pharmacol. (2019) 49:6–10. doi: 10.1016/j.coph.2019.03.016
48. Neumann E, Vieira LQ, Nicoli JR. Chapter 1 – Germ-free animals as a tool to study indigenous microbiota. In: J Faintuch, S Faintuch editors. Microbiome and Metabolome in Diagnosis, Therapy, and other Strategic Applications. Cambridge, MA: Academic Press (2019). p. 3–11. doi: 10.1016/B978-0-12-815249-2.00001-4
49. Kennedy EA, King KY, Baldridge MT. Mouse microbiota models: comparing germ-free mice and antibiotics treatment as tools for modifying gut bacteria. Front Physiol. (2018) 9:1534. doi: 10.3389/fphys.2018.01534
50. Marsh PD, Percival RS. The oral microflora–friend or foe? Can we decide? Int Dent J. (2006) 56(4 Suppl. 1):233–9. doi: 10.1111/j.1875-595X.2006.tb00107.x
51. Avila M, Ojcius DM, Yilmaz O. The oral microbiota: living with a permanent guest. DNA Cell Biol. (2009) 28:405–11. doi: 10.1089/dna.2009.0874
52. Elli M, Zink R, Rytz A, Reniero R, Morelli L. Iron requirement of Lactobacillus spp. in completely chemically defined growth media. J Appl Microbiol. (2000) 88:695–703. doi: 10.1046/j.1365-2672.2000.01013.x
53. Jakubovics NS, Kolenbrander PE. The road to ruin: the formation of disease-associated oral biofilms. Oral Dis. (2010) 16:729–39. doi: 10.1111/j.1601-0825.2010.01701.x
54. Jakubovics NS. Saliva as the sole nutritional source in the development of multispecies communities in dental plaque. Microbiol Spectr. (2015) 3:MBP-0013-2014. doi: 10.1128/microbiolspec.MBP-0013-2014
55. Teughels W, Loozen G, Quirynen M. Do probiotics offer opportunities to manipulate the periodontal oral microbiota? J Clin Periodontol. (2011) 38(Suppl. 11):159–77. doi: 10.1111/j.1600-051X.2010.01665.x
56. Wang HL, Greenwell H, Bissada NF. Crevicular fluid iron changes in treated and untreated periodontally diseased sites. Oral Surg Oral Med Oral Pathol. (1990) 69:450–6. doi: 10.1016/0030-4220(90)90378-6
57. Smith VH, Pippin DJ. Implications of resource-ratio theory for oral microbial ecology. Eur J Oral Sci. (1998) 106(2 Pt 1):605–15. doi: 10.1046/j.0909-8836..t01-4-.x
58. Miyakawa H, Nakazawa F. Role of asaccharolytic anaerobic gram-positive rods on periodontitis. J Oral Biosci. (2010) 52:240–4. doi: 10.1016/S1349-0079(10)80027-1
59. Van Hoogmoed CG, Geertsema-Doornbusch GI, Teughels W, Quirynen M, Busscher HJ, Van der Mei HC. Reduction of periodontal pathogens adhesion by antagonistic strains. Oral Microbiol Immunol. (2008) 23:43–8. doi: 10.1111/j.1399-302X.2007.00388.x
60. Kamarajan P, Ateia I, Shin JM, Fenno JC, Le C, Zhan L, et al. Periodontal pathogens promote cancer aggressivity via TLR/MyD88 triggered activation of Integrin/FAK signaling that is therapeutically reversible by a probiotic bacteriocin. PLoS Pathog. (2020) 16:e1008881. doi: 10.1371/journal.ppat.1008881
61. Kolenbrander PE. Multispecies communities: interspecies interactions influence growth on saliva as sole nutritional source. Int J Oral Sci. (2011) 3:49–54. doi: 10.4248/IJOS11025
62. Hatakka K, Ahola AJ, Yli-Knuuttila H, Richardson M, Poussa T, Meurman JH, et al. Probiotics reduce the prevalence of oral candida in the elderly–a randomized controlled trial. J Dent Res. (2007) 86:125–30. doi: 10.1177/154405910708600204
63. Jiang Q, Stamatova I, Kari K, Meurman JH. Inhibitory activity in vitro of probiotic lactobacilli against oral Candida under different fermentation conditions. Benef Microbes. (2015) 6:361–8. doi: 10.3920/BM2014.0054
64. Guglielmetti S, Taverniti V, Minuzzo M, Arioli S, Stuknyte M, Karp M, et al. Oral bacteria as potential probiotics for the pharyngeal mucosa. Appl Environ Microbiol. (2010) 76:3948–58. doi: 10.1128/AEM.00109-10
65. Horz HP, Meinelt A, Houben B, Conrads G. Distribution and persistence of probiotic Streptococcus salivarius K12 in the human oral cavity as determined by real-time quantitative polymerase chain reaction. Oral Microbiol Immunol. (2007) 22:126–30. doi: 10.1111/j.1399-302X.2007.00334.x
66. Tagg JR. Prevention of streptococcal pharyngitis by anti-Streptococcus pyogenes bacteriocin-like inhibitory substances (BLIS) produced by Streptococcus salivarius. Indian J Med Res. (2004) 119(Suppl.):13–6.
67. Laleman I, Yilmaz E, Ozcelik O, Haytac C, Pauwels M, Herrero ER, et al. The effect of a streptococci containing probiotic in periodontal therapy: a randomized controlled trial. J Clin Periodontol. (2015) 42:1032–41. doi: 10.1111/jcpe.12464
68. Teughels W, Durukan A, Ozcelik O, Pauwels M, Quirynen M, Haytac MC. Clinical and microbiological effects of Lactobacillus reuteri probiotics in the treatment of chronic periodontitis: a randomized placebo-controlled study. J Clin Periodontol. (2013) 40:1025–35. doi: 10.1111/jcpe.12155
69. Slomka V, Hernandez-Sanabria E, Herrero ER, Zaidel L, Bernaerts K, Boon N, et al. Nutritional stimulation of commensal oral bacteria suppresses pathogens: the prebiotic concept. J Clin Periodontol. (2017) 44:344–52. doi: 10.1111/jcpe.12700
70. Jobbins J, Bagg J, Parsons K, Finlay I, Addy M, Newcombe RG. Oral carriage of yeasts, coliforms and staphylococci in patients with advanced malignant disease. J Oral Pathol Med. (1992) 21:305–8. doi: 10.1111/j.1600-0714.1992.tb01016.x
71. Generalova E, Vahitova L, Gayfullina R, Rizvanova F, Vahitov H. Minimally invasive methods of diagnosis of bronchitis severity in children. Res J Pharm Biol Chem Sci. (2016) 7:2879–83.
72. Petrushanko TA, Chereda VV, Loban GA. The relationship between colonization resistance of the oral cavity and individual -typological characteristics of personality: dental aspects. Wiad Lek. (2017) 70:754–7.
73. Skrypnyk M, Petrushanko T, Neporada K, Vynnyk N, Petrushanko V, Skrypnyk R. Colonization resistance of oral mucosa in individuals with diverse body mass index. J Stomatol. (2022) 75:171–5. doi: 10.5114/jos.2022.119168
74. Petrushanko TA, Tchereda VV, Loban GA. [The screening diagnostic of micro ecological disorders of oral cavity]. Klin Lab Diagn. (2014) 6:48–50.
75. Grudianov AI, Dmitrieva NA, Fomenko EV. [Use of probiotics Bifidumbacterin and Acilact in tablets in therapy of periodontal inflammations]. Stomatologiia. (2002) 81:39–43.
76. Tsubura S, Mizunuma H, Ishikawa S, Oyake I, Okabayashi M, Katoh K, et al. The effect of Bacillus subtilis mouth rinsing in patients with periodontitis. Eur J Clin Microbiol Infect Dis. (2009) 28:1353. doi: 10.1007/s10096-009-0790-9
77. Krasse P, Carlsson B, Dahl C, Paulsson A, Nilsson A, Sinkiewicz G. Decreased gum bleeding and reduced gingivitis by the probiotic Lactobacillus reuteri. Swed Dent J. (2006) 30:55–60.
78. Çaglar E, Sandalli N, Twetman S, Kavaloglu S, Ergeneli S, Selvi S. Effect of yogurt with Bifidobacterium DN-173 010 on salivary mutans streptococci and lactobacilli in young adults. Acta Odontol Scand. (2005) 63:317–20. doi: 10.1080/00016350510020070
79. Näse L, Hatakka K, Savilahti E, Saxelin M, Pönkä A, Poussa T, et al. Effect of long-term consumption of a probiotic bacterium, Lactobacillus rhamnosus GG, in milk on dental caries and caries risk in children. Caries Res. (2001) 35:412–20. doi: 10.1159/000047484
80. Mager DL, Ximenez-Fyvie LA, Haffajee AD, Socransky SS. Distribution of selected bacterial species on intraoral surfaces. J Clin Periodontol. (2003) 30:644–54. doi: 10.1034/j.1600-051X.2003.00376.x
81. Sliepen I, Hofkens J, Van Essche M, Quirynen M, Teughels W. Aggregatibacter actinomycetemcomitans adhesion inhibited in a flow cell. Oral Microbiol Immunol. (2008) 23:520–4. doi: 10.1111/j.1399-302X.2008.00456.x
82. Sliepen I, Van Essche M, Loozen G, Van Eldere J, Quirynen M, Teughels W. Interference with Aggregatibacter actinomycetemcomitans: colonization of epithelial cells under hydrodynamic conditions. Oral Microbiol Immunol. (2009) 24:390–5. doi: 10.1111/j.1399-302X.2009.00531.x
83. Wang BY, Wu J, Lamont RJ, Lin X, Xie H. Negative correlation of distributions of Streptococcus cristatus and Porphyromonas gingivalis in subgingival plaque. J Clin Microbiol. (2009) 47:3902–6. doi: 10.1128/JCM.00072-09
84. Xie H, Lin X, Wang BY, Wu J, Lamont RJ. Identification of a signalling molecule involved in bacterial intergeneric communication. Microbiology. (2007) 153(Pt 10):3228–34. doi: 10.1099/mic.0.2007/009050-0
85. Lin X, Lamont RJ, Wu J, Xie H. Role of differential expression of streptococcal arginine deiminase in inhibition of fimA expression in Porphyromonas gingivalis. J Bacteriol. (2008) 190:4367–71. doi: 10.1128/JB.01898-07
86. Xie H, Hong J, Sharma A, Wang BY. Streptococcus cristatus ArcA interferes with Porphyromonas gingivalis pathogenicity in mice. J Periodontal Res. (2012) 47:578–83. doi: 10.1111/j.1600-0765.2012.01469.x
87. Christopher AB, Arndt A, Cugini C, Davey ME. A streptococcal effector protein that inhibits Porphyromonas gingivalis biofilm development. Microbiology. (2010) 156(Pt 11):3469–77. doi: 10.1099/mic.0.042671-0
88. Uppuluri P, Pierce CG, Thomas DP, Bubeck SS, Saville SP, Lopez-Ribot JL. The transcriptional regulator Nrg1p controls Candida albicans biofilm formation and dispersion. Eukaryot Cell. (2010) 9:1531–7. doi: 10.1128/EC.00111-10
89. Haukioja A, Loimaranta V, Tenovuo J. Probiotic bacteria affect the composition of salivary pellicle and streptococcal adhesion in vitro. Oral Microbiol Immunol. (2008) 23:336–43. doi: 10.1111/j.1399-302X.2008.00435.x
90. van Hoogmoed CG, van Der Kuijl-Booij M, van Der Mei HC, Busscher HJ. Inhibition of Streptococcus mutans NS adhesion to glass with and without a salivary conditioning film by biosurfactant- releasing Streptococcus mitis strains. Appl Environ Microbiol. (2000) 66:659–63. doi: 10.1128/AEM.66.2.659-663.2000
91. Merritt J, Qi F. The mutacins of Streptococcus mutans: regulation and ecology. Mol Oral Microbiol. (2012) 27:57–69. doi: 10.1111/j.2041-1014.2011.00634.x
92. Ashby MT, Kreth J, Soundarajan M, Sivuilu LS. Influence of a model human defensive peroxidase system on oral streptococcal antagonism. Microbiology. (2009) 155(Pt 11):3691–700. doi: 10.1099/mic.0.031310-0
93. Hossain MS, Biswas I. Mutacins from Streptococcus mutans UA159 are active against multiple streptococcal species. Appl Environ Microbiol. (2011) 77:2428–34. doi: 10.1128/AEM.02320-10
94. Qi F, Chen P, Caufield PW. Purification of mutacin III from group III Streptococcus mutans UA787 and genetic analyses of mutacin III biosynthesis genes. Appl Environ Microbiol. (1999) 65:3880–7. doi: 10.1128/AEM.65.9.3880-3887.1999
95. Qi F, Chen P, Caufield PW. The group I strain of Streptococcus mutans, UA140, produces both the lantibiotic mutacin I and a nonlantibiotic bacteriocin, mutacin IV. Appl Environ Microbiol. (2001) 67:15–21. doi: 10.1128/AEM.67.1.15-21.2001
96. Jakubovics NS, Yassin SA, Rickard AH. Community interactions of oral streptococci. Adv Appl Microbiol. (2014) 87:43–110. doi: 10.1016/B978-0-12-800261-2.00002-5
97. Wescombe PA, Upton M, Renault P, Wirawan RE, Power D, Burton JP, et al. Salivaricin 9, a new lantibiotic produced by Streptococcus salivarius. Microbiology. (2011) 157(Pt 5):1290–9. doi: 10.1099/mic.0.044719-0
98. Upton M, Tagg JR, Wescombe P, Jenkinson HF. Intra- and interspecies signaling between Streptococcus salivarius and Streptococcus pyogenes mediated by SalA and SalA1 lantibiotic peptides. J Bacteriol. (2001) 183:3931–8. doi: 10.1128/JB.183.13.3931-3938.2001
99. Kang MS, Oh JS, Lee HC, Lim HS, Lee SW, Yang KH, et al. Inhibitory effect of Lactobacillus reuteri on periodontopathic and cariogenic bacteria. J Microbiol. (2011) 49:193–9. doi: 10.1007/s12275-011-0252-9
100. Pangsomboon K, Kaewnopparat S, Pitakpornpreecha T, Srichana T. Antibacterial activity of a bacteriocin from Lactobacillus paracasei HL32 against Porphyromonas gingivalis. Arch Oral Biol. (2006) 51:784–93. doi: 10.1016/j.archoralbio.2006.03.008
101. Pangsomboon K, Bansal S, Martin GP, Suntinanalert P, Kaewnopparat S, Srichana T. Further characterization of a bacteriocin produced by Lactobacillus paracasei HL32. J Appl Microbiol. (2009) 106:1928–40. doi: 10.1111/j.1365-2672.2009.04146.x
102. Kaewsrichan J, Douglas CW, Nissen-Meyer J, Fimland G, Teanpaisan R. Characterization of a bacteriocin produced by Prevotella nigrescens ATCC 25261. Lett Appl Microbiol. (2004) 39:451–8. doi: 10.1111/j.1472-765X.2004.01608.x
103. Ribeiro-Ribas RN, de Carvalho MA, Vieira CA, Apolônio AC, Magalhães PP, Mendes EN, et al. Purification and partial characterization of a bacteriocin produced by an oral Fusobacterium nucleatum isolate. J Appl Microbiol. (2009) 107:699–705. doi: 10.1111/j.1365-2672.2009.04250.x
104. Portrait V, Cottenceau G, Pons AM. A Fusobacterium mortiferum strain produces a bacteriocin-like substance(s) inhibiting Salmonella enteritidis. Lett Appl Microbiol. (2000) 31:115–7. doi: 10.1046/j.1365-2672.2000.00774.x
105. Zeng L, Farivar T, Burne RA. Amino sugars enhance the competitiveness of beneficial commensals with Streptococcus mutans through multiple mechanisms. Appl Environ Microbiol. (2016) 82:3671–82. doi: 10.1128/AEM.00637-16
106. Hillman JD, Shivers M. Interaction between wild-type, mutant and revertant forms of the bacterium Streptococcus sanguis and the bacterium Actinobacillus actinomycetemcomitans in vitro and in the gnotobiotic rat. Arch Oral Biol. (1988) 33:395–401. doi: 10.1016/0003-9969(88)90196-3
107. Hillman JD, Socransky SS, Shivers M. The relationships between streptococcal species and periodontopathic bacteria in human dental plaque. Arch Oral Biol. (1985) 30:791–5. doi: 10.1016/0003-9969(85)90133-5
108. Ryan CS, Kleinberg I. Bacteria in human mouths involved in the production and utilization of hydrogen peroxide. Arch Oral Biol. (1995) 40:753–63. doi: 10.1016/0003-9969(95)00029-O
109. Holmberg K, Hallander HO. Interference between gram-positive microorganisms in dental plaque. J Dent Res. (1972) 51:588–95. doi: 10.1177/00220345720510025801
110. Holmberg K, Hallander HO. Production of bactericidal concentrations of hydrogen peroxide by Streptococcus sanguis. Arch Oral Biol. (1973) 18:423–34. doi: 10.1016/0003-9969(73)90167-2
111. Kreth J, Giacaman RA, Raghavan R, Merritt J. The road less traveled - defining molecular commensalism with Streptococcus sanguinis. Mol Oral Microbiol. (2017) 32:181–96. doi: 10.1111/omi.12170
112. Kreth J, Merritt J, Shi W, Qi F. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. J Bacteriol. (2005) 187:7193–203. doi: 10.1128/JB.187.21.7193-7203.2005
113. He X, McLean JS, Guo L, Lux R, Shi W. The social structure of microbial community involved in colonization resistance. ISME J. (2014) 8:564–74. doi: 10.1038/ismej.2013.172
114. Tong H, Chen W, Merritt J, Qi F, Shi W, Dong X. Streptococcus oligofermentans inhibits Streptococcus mutans through conversion of lactic acid into inhibitory H2O2: a possible counteroffensive strategy for interspecies competition. Mol Microbiol. (2007) 63:872–80. doi: 10.1111/j.1365-2958.2006.05546.x
115. Pride DT, Salzman J, Haynes M, Rohwer F, Davis-Long C, White RA III, et al. Evidence of a robust resident bacteriophage population revealed through analysis of the human salivary virome. ISME J. (2012) 6:915–26. doi: 10.1038/ismej.2011.169
116. Dalmasso M, de Haas E, Neve H, Strain R, Cousin FJ, Stockdale SR, et al. Isolation of a novel phage with activity against Streptococcus mutans biofilms. PLoS One. (2015) 10:e0138651. doi: 10.1371/journal.pone.0138651
117. Edlund A, Santiago-Rodriguez TM, Boehm TK, Pride DT. Bacteriophage and their potential roles in the human oral cavity. J Oral Microbiol. (2015) 7:27423. doi: 10.3402/jom.v7.27423
118. Scoffield JA, Wu H. Oral streptococci and nitrite-mediated interference of Pseudomonas aeruginosa. Infect Immun. (2015) 83:101–7. doi: 10.1128/IAI.02396-14
119. Morrison DJ, Preston T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. (2016) 7:189–200. doi: 10.1080/19490976.2015.1134082
120. Flint HJ, Duncan SH, Scott KP, Louis P. Links between diet, gut microbiota composition and gut metabolism. Proc Nutr Soc. (2015) 74:13–22. doi: 10.1017/S0029665114001463
121. Xue J, Xiao X. Ecology of oral infectious diseases. In: L Li, editor. Infectious Microecology: Theory and Applications. Berlin: Springer (2014). p. 227–92. doi: 10.1007/978-3-662-43883-1_10
122. Dashper SG, Reynolds EC. Effects of organic acid anions on growth, glycolysis, and intracellular pH of oral streptococci. J Dent Res. (2000) 79:90–6. doi: 10.1177/00220345000790011601
123. Loesche WJ. Role of Streptococcus mutans in human dental decay. Microbiol Rev. (1986) 50:353–80. doi: 10.1128/mr.50.4.353-380.1986
124. Lamont RJ, Koo H, Hajishengallis G. The oral microbiota: dynamic communities and host interactions. Nat Rev Microbiol. (2018) 16:745–59. doi: 10.1038/s41579-018-0089-x
125. Bagg J. Can the colonisation resistance of the oral microflora be reduced? Microb Ecol Health Dis. (1990) 3:v–viii. doi: 10.3109/08910609009140117
126. Mashima I, Nakazawa F. The interaction between Streptococcus spp. and Veillonella tobetsuensis in the early stages of oral biofilm formation. J Bacteriol. (2015) 197:2104–11. doi: 10.1128/JB.02512-14
127. Ilyin VK, Kiryukhina NV. Disruption of the colonization resistance syndrome in humans in altered habitats and its prevention. Acta Naturae. (2014) 6:10–8. doi: 10.32607/20758251-2014-6-2-10-18
128. Macgregor ID. Effects of smoking on oral ecology. A review of the literature. Clin Prev Dent. (1989) 11:3–7.
129. Lee Y-H, Chung SW, Auh QS, Hong S-J, Lee Y-A, Jung J, et al. Progress in oral microbiome related to oral and systemic diseases: an update. Diagnostics. (2021) 11:1283. doi: 10.3390/diagnostics11071283
130. Thomas AM, Gleber-Netto FO, Fernandes GR, Amorim M, Barbosa LF, Francisco AL, et al. Alcohol and tobacco consumption affects bacterial richness in oral cavity mucosa biofilms. BMC Microbiol. (2014) 14:250. doi: 10.1186/s12866-014-0250-2
131. Liao Y, Tong XT, Jia YJ, Liu QY, Wu YX, Xue WQ, et al. The effects of alcohol drinking on oral microbiota in the Chinese population. Int J Environ Res Public Health. (2022) 19:5729. doi: 10.3390/ijerph19095729
132. Renson A, Jones HE, Beghini F, Segata N, Zolnik CP, Usyk M, et al. Sociodemographic variation in the oral microbiome. Ann Epidemiol. (2019) 35:73–80.e2. doi: 10.1016/j.annepidem.2019.03.006
133. Belstrøm D, Holmstrup P, Nielsen CH, Kirkby N, Twetman S, Heitmann BL, et al. Bacterial profiles of saliva in relation to diet, lifestyle factors, and socioeconomic status. J Oral Microbiol. (2014) 6:23609. doi: 10.3402/jom.v6.23609
134. Ready D, Roberts AP, Pratten J, Spratt DA, Wilson M, Mullany P. Composition and antibiotic resistance profile of microcosm dental plaques before and after exposure to tetracycline. J Antimicrob Chemother. (2002) 49:769–75. doi: 10.1093/jac/dkf005
135. Patangia DV, Anthony Ryan C, Dempsey E, Paul Ross R, Stanton C. Impact of antibiotics on the human microbiome and consequences for host health. Microbiol Open. (2022) 11:e1260. doi: 10.1002/mbo3.1260
136. Malys MK, Campbell L, Malys N. Symbiotic and antibiotic interactions between gut commensal microbiota and host immune system. Medicina. (2015) 51:69–75. doi: 10.1016/j.medici.2015.03.001
137. Moser C, Pedersen HT, Lerche CJ, Kolpen M, Line L, Thomsen K, et al. Biofilms and host response - helpful or harmful. APMIS. (2017) 125:320–38. doi: 10.1111/apm.12674
138. Olsen I, Yamazaki K. Can oral bacteria affect the microbiome of the gut? J Oral Microbiol. (2019) 11:1586422. doi: 10.1080/20002297.2019.1586422
139. Liccardo D, Cannavo A, Spagnuolo G, Ferrara N, Cittadini A, Rengo C, et al. Periodontal disease: a risk factor for diabetes and cardiovascular disease. Int J Mol Sci. (2019) 20:1414. doi: 10.3390/ijms20061414
140. Sanz M, Ceriello A, Buysschaert M, Chapple I, Demmer RT, Graziani F, et al. Scientific evidence on the links between periodontal diseases and diabetes: consensus report and guidelines of the joint workshop on periodontal diseases and diabetes by the International diabetes Federation and the European Federation of Periodontology. Diabetes Res Clin Pract. (2018) 137:231–41. doi: 10.1016/j.diabres.2017.12.001
141. Maisonneuve P, Amar S, Lowenfels AB. Periodontal disease, edentulism, and pancreatic cancer: a meta-analysis. Ann Oncol. (2017) 28:985–95. doi: 10.1093/annonc/mdx019
142. Michaud DS, Lu J, Peacock-Villada AY, Barber JR, Joshu CE, Prizment AE, et al. Periodontal disease assessed using clinical dental measurements and cancer risk in the ARIC study. J Natl Cancer Inst. (2018) 110:843–54. doi: 10.1093/jnci/djx278
143. Syrjänen J, Peltola J, Valtonen V, Iivanainen M, Kaste M, Huttunen JK. Dental infections in association with cerebral infarction in young and middle-aged men. J Intern Med. (1989) 225:179–84. doi: 10.1111/j.1365-2796.1989.tb00060.x
144. Baker JL, Edlund A. Exploiting the oral microbiome to prevent tooth decay: has evolution already provided the best tools? Front Microbiol. (2018) 9:3323. doi: 10.3389/fmicb.2018.03323
145. Mira A. Oral microbiome studies: potential diagnostic and therapeutic implications. Adv Dent Res. (2018) 29:71–7. doi: 10.1177/0022034517737024
146. Weingarden AR, Vaughn BP. Intestinal microbiota, fecal microbiota transplantation, and inflammatory bowel disease. Gut Microbes. (2017) 8:238–52. doi: 10.1080/19490976.2017.1290757
147. Narula N, Kassam Z, Yuan Y, Colombel JF, Ponsioen C, Reinisch W, et al. Systematic review and meta-analysis: fecal microbiota transplantation for treatment of active ulcerative colitis. Inflamm Bowel Dis. (2017) 23:1702–9. doi: 10.1097/MIB.0000000000001228
Keywords: oral microbiota, biofilm, colonization resistance, nutrition, microbial composition, competition, synergism
Citation: Akimbekov NS, Digel I, Yerezhepov AY, Shardarbek RS, Wu X and Zha J (2022) Nutritional factors influencing microbiota-mediated colonization resistance of the oral cavity: A literature review. Front. Nutr. 9:1029324. doi: 10.3389/fnut.2022.1029324
Received: 27 August 2022; Accepted: 06 October 2022;
Published: 20 October 2022.
Edited by:
Md. Anwarul Azim Majumder, The University of the West Indies, Cave Hill, BarbadosReviewed by:
Fatme Al Anouti, Zayed University, United Arab EmiratesAhmad Faisal Ismail, International Islamic University Malaysia, Malaysia
Copyright © 2022 Akimbekov, Digel, Yerezhepov, Shardarbek, Wu and Zha. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nuraly S. Akimbekov, YWtpbWJla292Lm51cmFseUBrYXpudS5lZHUua3o=
 Nuraly S. Akimbekov
Nuraly S. Akimbekov Ilya Digel
Ilya Digel Adil Y. Yerezhepov1
Adil Y. Yerezhepov1 Raiymbek S. Shardarbek
Raiymbek S. Shardarbek