- 1College of Biotechnology and Pharmaceutical Engineering, Nanjing Tech University, Nanjing, China
- 2Bioresource Institute for Healthy Utilization, Zunyi Medical University, Zunyi, China
- 3Key Laboratory of Basic Pharmacology of Ministry of Education and Joint International Research Laboratory of Ethnomedicine of Ministry of Education, Zunyi Medical University, Zunyi, China
Dendrobine is a representative component of Dendrobium nobile, and its pharmacological effects have been extensively studied. Trichoderma longibrachiatum MD33 was isolated from the stem of Dendrobium nobile which can produce dendrobine. In order to understand the effect of Methyl Jasmonate (MeJA) on the production of dendrobine, transcriptome analysis was performed after MeJA treatment in the MD33 and control groups. The dendrobine production of MeJA (20 μmol/L) treatment group was 44.6% higher than that of control. In this study, the RNA sequencing technology was applied, a total of 444 differentially expressed genes (DEGs) in the control and MeJA treatment groups, including 226 up-regulated genes and 218 down-regulated genes. The Kyoto Encyclopedia of Genes and Genomes annotation showed that numbers of DEGs were associated with the putative alkaloid biosynthetic pathway in T Trichoderma longibrachiatum MD33. Several MVA pathway enzyme-coding genes (isopentenyl-diphosphate Delta-isomerase, iphosphomevalonate decarboxylase and farnesyl diphosphate synthase) were found to be differentially expressed, suggesting an active precursor supply for alkaloid biosynthesis after MeJA treatment, in other wise, dendrobine may synthesis through the MVA pathway in MD33. Numerous MeJA-induced P450 family genes, aminotransferase genes and methyltransferase genes were identified, providing several important candidates to further elucidate the dendrobine biosynthetic pathway of T. longibrachiatum MD33. Furthermore, several MeJA-induced transcription factors (TFs) encoding genes were identified, suggesting a complex genetic network affecting the dendrobine in T. longibrachiatum MD33. These findings reveal the regulation mechanism underlying the MeJA-induced accumulation of dendrobine in T. longibrachiatum MD33.
Introduction
Dendrobine is a sesquiterpene alkaloid mainly found in the medicinal orchid Dendrobium nobile, which is considered to possess therapeutic properties. It is a representative component of the D. nobile species (Zhang et al., 2021; Li et al., 2022). Dendrobine possesses numerous of pharmacological properties, including analgesic and antipyretic properties, the ability to regulate blood glucose and blood pressure (Bai et al., 2020), antitumor activity (Song et al., 2018), and inhibition of cardiovascular system and gastrointestinal tract (Mou et al., 2021). It has also demonstrated promising therapeutic effects in Alzheimer’s disease (Nie et al., 2018; Xiong et al., 2021). Dendrobine significantly reduces the ischemic damage caused by oxygen-glucose deprivation/reperfusion (OGD/RP) to brain slices and has a major protective effect on primary neurons of OGD/RP (Liu et al., 2021). However, given the slow growth rate of the D. nobile species, dendrobine is seldom produced in large amounts. The quantity of D. nobile dendrobine available is insufficient to meet the present industrial and research needs (Jiang et al., 2018; Zhang Y. et al., 2018).
In 1980s, plant endophytic fungi were found and recognized. These fungi live in the tissues and organs of plants without causing evident illness. Paclitaxel producing short leaf Taxus was the first endophytic fungi found in medical plants, after which different kinds of endophytic fungi were found (Stierle et al., 1993). Among these, the taxol content of endophytic fungus ne-32 (Pestalotipsis microspora) obtained from Taxus is more than 1,000 times higher than that of the host (Strobel et al., 1996), Thereafter, endophytic fungi producing camptothecin, podophyllotoxin, vinblastine, huperzine A, ginsenoside, and Dendrobium polysaccharide were consecutively isolated. There are at least one million species of plant endophytic fungi (Fagan, 2002) and some medicinal plant endophytic fungi produce a certain amount of “plant-derived” bioactive substances. Hence, we researched and isolated several dendrobine-producing endophytic fungi from D. nobile, of which T. longibrachiatum MD33 produced the highest amount of dendrobine (Sarsaiya et al., 2019b).
A dendrobine metabolic pathway in the Dendrobium plant has been defined based on transcriptome sequencing and data analysis. The mechanism by which the mycorrhizal fungus MF23 increased dendrobine content in D. nobile seedlings was analyzed using the transcriptome, the dendrobine synthetic pathway was proposed for the first time, and key modifying enzymes such as cytochrome P450, aminotransferase, and methyltransferase were identified (Li et al., 2017). Chen et al. (2019) determined the expression levels of P450 family, aminotransferase, and methyltransferase genes activated by MeJA. MeJA induces a significant number of transcription factor-coding genes. The effector stress culture is a very useful research model for alkaloid active compounds. Examining the link between products and gene expression under regulated settings enables direct identification of functional genes or gene clusters involved in alkaloid production pathway.
The phytohormone (MeJA) has been extensively used to stimulate secondary metabolite production in medicinal plants (Xiaori et al., 2018). MeJA acts as a signaling molecule various secondary metabolites, notably alkaloids (Zhang X. N. et al., 2018). However, the mechanism by which MeJA induces accumulation of sesquiterpene alkaloids in T. longibrachiatum MD33 remains unclear. Numerous differentially expressed genes (DEGs) were identified in this study to clarify the regulatory mechanism driving the accumulation of sesquiterpene alkaloids in T. longibrachiatum MD33 triggered by MeJA, which may provide a source of dendrobine.
Materials and Methods
Fungal Strain, Culture Conditions, and Methyl Jasmonate Treatment
The Trichoderma longibrachiatum MD33 was isolated from stem segments of a single wild D. nobile (Sarsaiya et al., 2019a,2020b) and stored at the laboratory of Nanjing Tech University’s Institute of Bioresources Engineering. After activating the stored fungus for 48 h, it was transferred to PD medium and cultivated at 28°C for 36 h. Thereafter 0.1180 g MeJA (95%) was completely dissolved in pure ethanol, transferred to a 10 ml volumetric flask and dilute with deionized water to 50 mmol/L MeJA. Methyl Jasmonate (MeJA) was added to PD medium at 0, 5, 20, and 35 μmol/L (250 ml Erlenmeyer flask, 100 ml PD medium), eight bottles per group. The pH value at the beginning was neutral. The block at the colony’s edge where the new hypha forms was removed and put into each flask using a 6 mm punch. The culture conditions used with the strain were neutral pH, 28°C, 120 rpm rotation speed, and 15 days (Sarsaiya et al., 2020a). The growth of MD33 were shown in Figure 1. There were three biological replicates for each group.

Figure 1. The growth of Trichoderma longibrachiatum MD33 MeJA treatment group and control group (18 days). (A) MD33 growth in MeJA treatment group. From the third day, the color of liquid gradually deepened, from light yellow to brown, the biomass of MD33 increased significantly from the second day, in the seventh day, the biomass reached the maximum and remained basically unchanged. (B) MD33 growth in Control group. In the first 4 days, the biomass remained at a low level and the color of liquid was limpid, from the fifth day to the eighteenth day, the color gradually changed from light yellow to brown. From the eighth day, the biomass reached the maximum.
Determination of Dendrobine in MD33
Five bottles of Trichoderma longibrachiatum MD33 were used for dendrobine extraction and detection. The following steps were used: The fungi cultured in shake flasks were filtered, dried to constant weight at 45°C, and ground to a powder. Thereafter 50 ml chloroform was added to the powder, transferred to a separation funnel for static stratification after 30 min of ultrasonic extraction, distilled and the lower solvent layer was dried under reduced pressure (rotary evaporator SHZ-III model). The dried powder was dissolved in 5 ml methanol centrifuged at 1,000 rpm for 15 min. Thereafter 1 ml solution was filtered via a 0.22 μm filter membrane, and a sample was injected for detection.
Analysis of Dendrobine in MD33 by LC/MS
The LC-MS technique was used to characterize non-volatile and thermally fragile compounds. The detection of dendrobine was performed using a UHPLC system (Thermo Fisher Scientific Dionex Ultimate 3000, Golden Valley, Minnesota, United States) with a column (150 × 2.1 mm,1.9 μm) and a mobile phase consisting of 0.1% formic acid: acetonitrile at 95:5 (v/v) with a flow rate of 0.3 ml/min, column temperature of 40°C, feed volume 2 μl, a sheath gas flow rate of 35 arbitrary units, an auxiliary gas flow rate of 15 arbitrary units, a spray voltage of 3.5 kV, a capillary temperature of 350°C, an aux gas heater temperature of 300°C, the detection range was 100–1,500 m/Z, the scanning mode was full scan/positive ion scan, and the resolution was MS full scan 70,000 full width at half maxima (FWHM).
To provide a scientific rational for MeJA-induced dendrobine accumulation in MD33, a standard curve was established and the contents of dendrobine were evaluated under control and MeJA conditions. For the standard curve, 1.00 mg of dendrobine standard was accurately weighed, dissolved in methanol to prepare 1 g/L mother liquor, and then diluted into standard solutions at concentrations of 1, 10, 100, 500, 1,000, and 1,500 ng/L. The linear regression equation and standard curve were constructed using the LC-MS detection results. The regression curve equation of dendrobine is y = 0.6726x + 23.205, R2 = 0.9948 (Figure 2A). Each sample detected for three times.
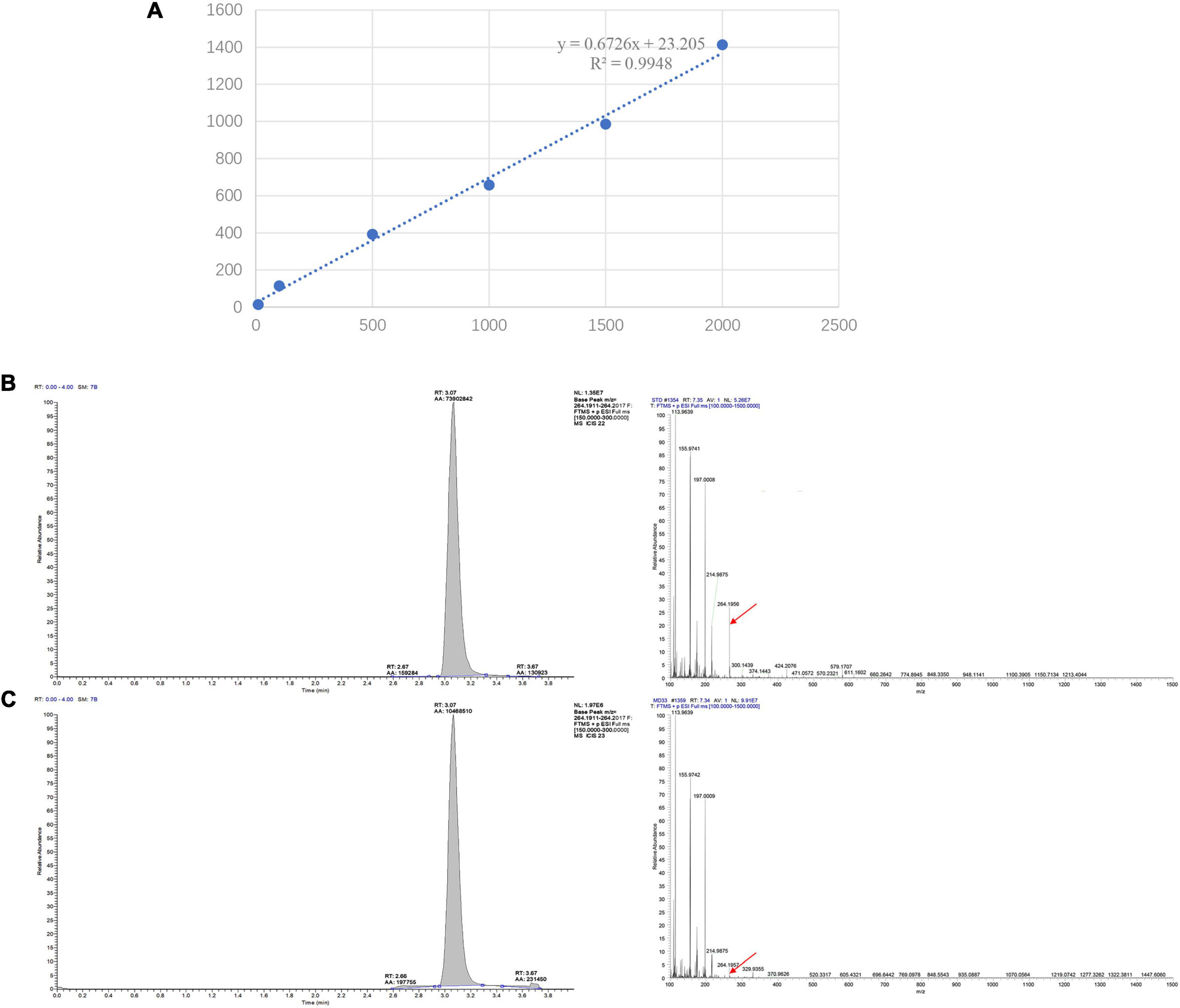
Figure 2. LC-MS/MS chromatographs for the detection of dendrobine. (A) LC-MS detection standard curve of Dendrobine. The regression curve equation of dendrobine is y = 0.6726x + 23.205, R2 = 0.9948. (B) T. longibrachiatum MD33 intracellular dendrobine. (C) Dendrobium nobile stem dendrobine (molecular weight: 264.195).
RNA Isolation and Construction of cDNA Libraries
The cultivated Trichoderma longibrachiatum MD33 was treated according to the LC/MS findings (Step 2.1 and Step 2.2). Three flasks from each group were randomly selected and filtered. Prior to RNA extraction, the hypha samples were snap-frozen in liquid nitrogen and stored at –80°C.
Total RNA was extracted from hypha using the RNeasy mini kit (QIAGEN, Cat. No. 74104, Germany) and genomic DNA was removed using DNase. Then RNA quality was determined by Bioanalyzer 2100 and the RNA 6000 Nano LabChip kit (Agilent, CA, United States). Only high-quality RNA sample (OD260/280 = 1.8–2.2, OD260/230 ≥ 2.0, RIN ≥ 6.5) was used A SureSelect strand-specific RNA library preparation kit from Agilent Technologies was used to generate an RNA-seq library according to the manufacturer’s procedure using 2 μg of total RNA. The Agilent 2100 Bioanalyzer and the ABI Step One Plus Real-Time PCR system was used to assess the quality of the RNA-seq library. Thereafter, sequencing was performed on the Illumina HiSeq 4000 platform. The transcriptome library was prepared by KEGENE Company (Shandong, China). There were three biological replicates for each group.
De novo Assembly and Annotation
The raw FASTQ data was analyzed for quality using FastQC and filtered to remove reads with sequencing adaptors, unknown nucleotides (Nradio > 5%), and poor quality (quality scores 30). Due to the unavailability of a reference genomic sequence, de novo assembly was used to create the transcripts. The redundant sequences were eliminated using the CORSET software suite, and the longest unigenes were further spliced. All assembled unigenes were aligned against the non-redundant (Nr), nucleotide (Nt), protein family (Pfam), Gene Ontol-ogy (GO), SwissProt, and KOG/COG, Kyoto Encyclopedia of Genes and Genomes (KEGG) databases with a threshold of E-value < 10–10.
Differentially Expressed Genes Analysis
In this study, the expression level of all transcripts was standardized using RSEM and Bowtie2 with default parameters. Ballgown was used for Differentially Expressed Gene (DEG) investigation. An FDR value of (0.05) and a log2FC value of 1 were used as the criteria for DEG screening. The DEGs were then analyzed for GO and KEGG enrichment.
RNA-Seq Validation by q-RT PCR
qRT-PCR was performed to validate the accuracy of RNA-seq data. GAPDH gene was used as an internal reference. The foldchange in gene expression was calculated using the comparative Ct method (1 + E–△△Ct) (Pfaffl, 2001). Total RNA samples were extracted from the hyphae using RNeasy mini kit (QIAGEN, Cat: 74104, Germany). The cDNA synthesis kit (TSINGKE, TSK302M) was used to synthesize cDNA from 1 μg of RNA. For gene expression analysis, the Applied Biosystems StepOnePlus™ Real-Time PCR system was used. qRT-PCR amplification was performed in 20 μl reactions containing 10 μl 2 × TSINKE® Master qPCR Mix (SYBR GREEN), 20 ng cDNA, and 0.4 μM of each primer (Supplementary Table 5). The PCR reaction conditions were: 95°C for 15 min, followed by 40 cycles of 95°C for 15 s, 60°C for 30 s. After final annealing (72°C, 5 min) and redenaturation (95°C, 30 s), a melt curve analysis was done by increasing from 60 to 95°C at 0.5°C intervals.
Results
Determination of Dendrobine Contents
The LC-MS results revealed that standard chemical reference dendrobine was found at a retention time of 3.05 and a molecular weight of 264.195. From the MD33 dendrobine the dendrobine peak was recorded at 3.07 with a molecular weight of 264.195 (Figures 2B,C). LC-MS detection results showed that, the content of dendrobine in MeJA treatment groups were higher than control group. When the concentration of MeJA was 20 μmol/L, the content of dendrobine was the highest (0.68 μg/L), which increased 44.7% compared with the control group (0.47 μg/L) (Figure 3A). indicating that MeJA plays an important role in dendrobine accumulation in MD33.
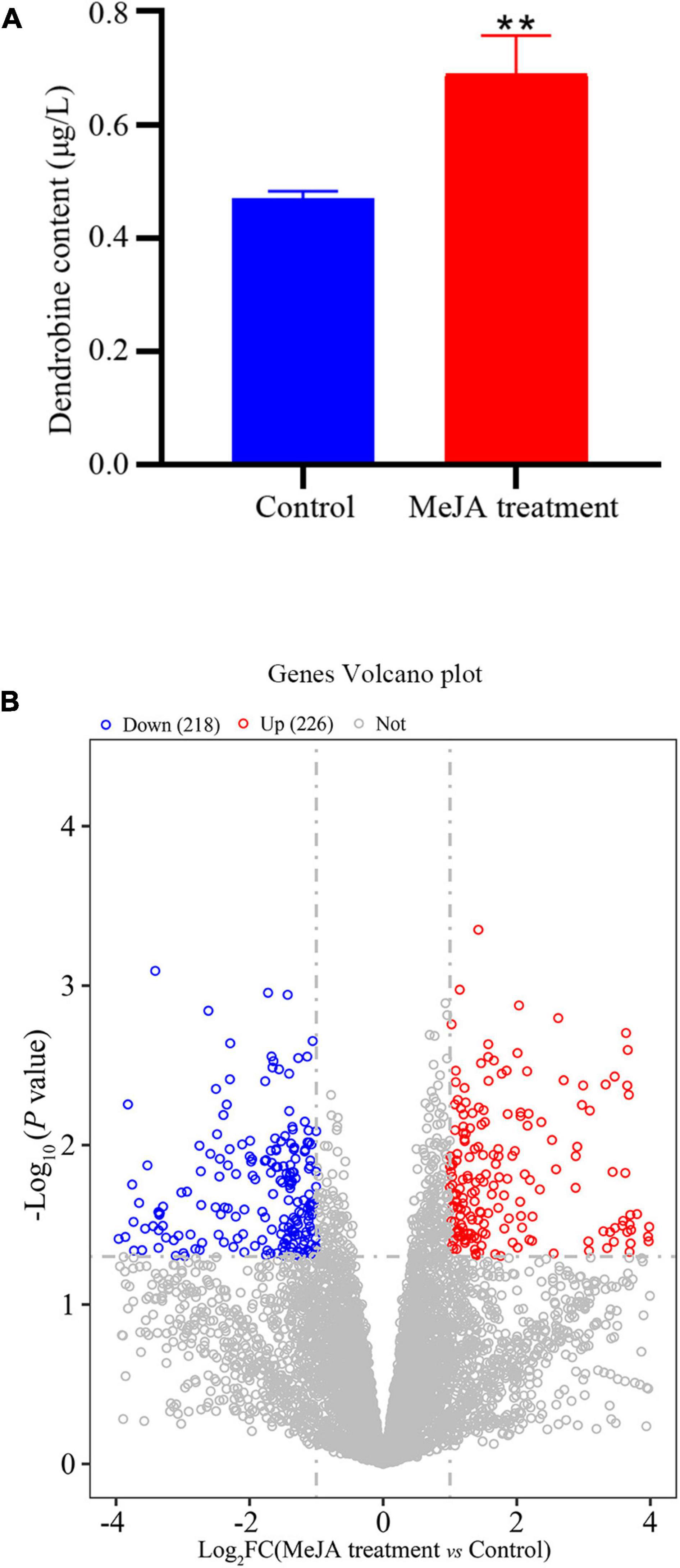
Figure 3. Transcriptional variation between Control and MeJA treatment groups. (A) In the MeJA treatment group dendrobine content increased 44.6%, (B) significance analysis of all DEGs between the strain MeJA treatment MD33 and Control by a volcano plot. **p < 0.01.
De novo Assembly and Unigene Annotation
Three distinct RNA-seq libraries were generated utilizing RNA from the control and MeJA treated groups. A total of 274,332,098 pure reads were isolated from 288,958,158 raw reads in this study, equal to 41.16 Gb of clean bases (Supplementary Table 1). The Q20/Q30 fraction, the N proportion, and the GC percentages were 98.73/95.64, 0.00, and 58.05%, respectively. The clean sequences were then de novo assembled and clustered into 21,331 (median length = 3,850 bp) and 4,477 (median length = 3,326 bp) genes, respectively, using Trinity (Supplementary Tables 2, 3). We analyzed the size distributions of transcripts and genes, respectively. For transcripts, the majority (76.22%) were > 2,000 bp in length, 23.00% were 500–2,000 bp in length, and 0.77% were 500 bp in length. For genes, 42.93% were > 2,000 bp in length, 54.75% were 500–2,000 bp on length, and 2.32% < 500 bp in length (Supplementary Table 2). All identified genes were annotated using several databases. The NR database contained 6,037 genes, the KO database contained 1,765 genes, the KEGG database contained 1,761 genes, the KO database contained 2,920 genes, the SwissProt database contained 4,885 genes, the Pfam database contained 5,666 genes, the GO database contained 4,677 genes, and the KOG database contained 5,777 genes (Supplementary Table 4).
Gene Ontology and Kyoto Encyclopedia of Genes and Genomes Classification of Differentially Expressed Genes
To ascertain the likely function of these DEGs in Trichoderma longibrachiatum MD33, we performed a Gene Ontol-ogy (GO) enrichment analysis, which revealed that these DEGs were classified into three primary categories: biological process (BP), cellular component (CC), and molecular function (MF). The terms “oxidation-reduction process,” “protein transport,” and “cell cycle” were the most frequently used in the biological process category; “nucleus,” “membrane,” and “cytoplasm” were frequently used in the cellular component category; and a large percentage of genes were associated with “metal ion binding,” “nucleotide binding,” and “hydrolase activity” in the molecular function category (Figure 4A). Moreover 1,761 genes were classified into 18 Kyoto Encyclopedia of Genes and Genome (KEGG) secondary pathways. The majority of pathways were classified as “Metabolism” (866 genes) or “Genetic Information Processing” (695 genes). Moreover, 190 genes belonged to the “amino acid metabolism” pathway, 139 genes belonged to the “carbohydrate metabolism” pathway, 126 genes belonged to the “metabolism of cofactors and vitamins” pathway, 106 genes belonged to the “lipid metabolism” pathway, and 28 genes belonged to the “metabolism of terpenoids and polyketides” pathway in metabolism, 302 genes belonged to the “translation” route in “Genetic Information Processing,” whereas 205 genes belonged to the “folding, sorting, and degradation” pathway (Figure 4B).
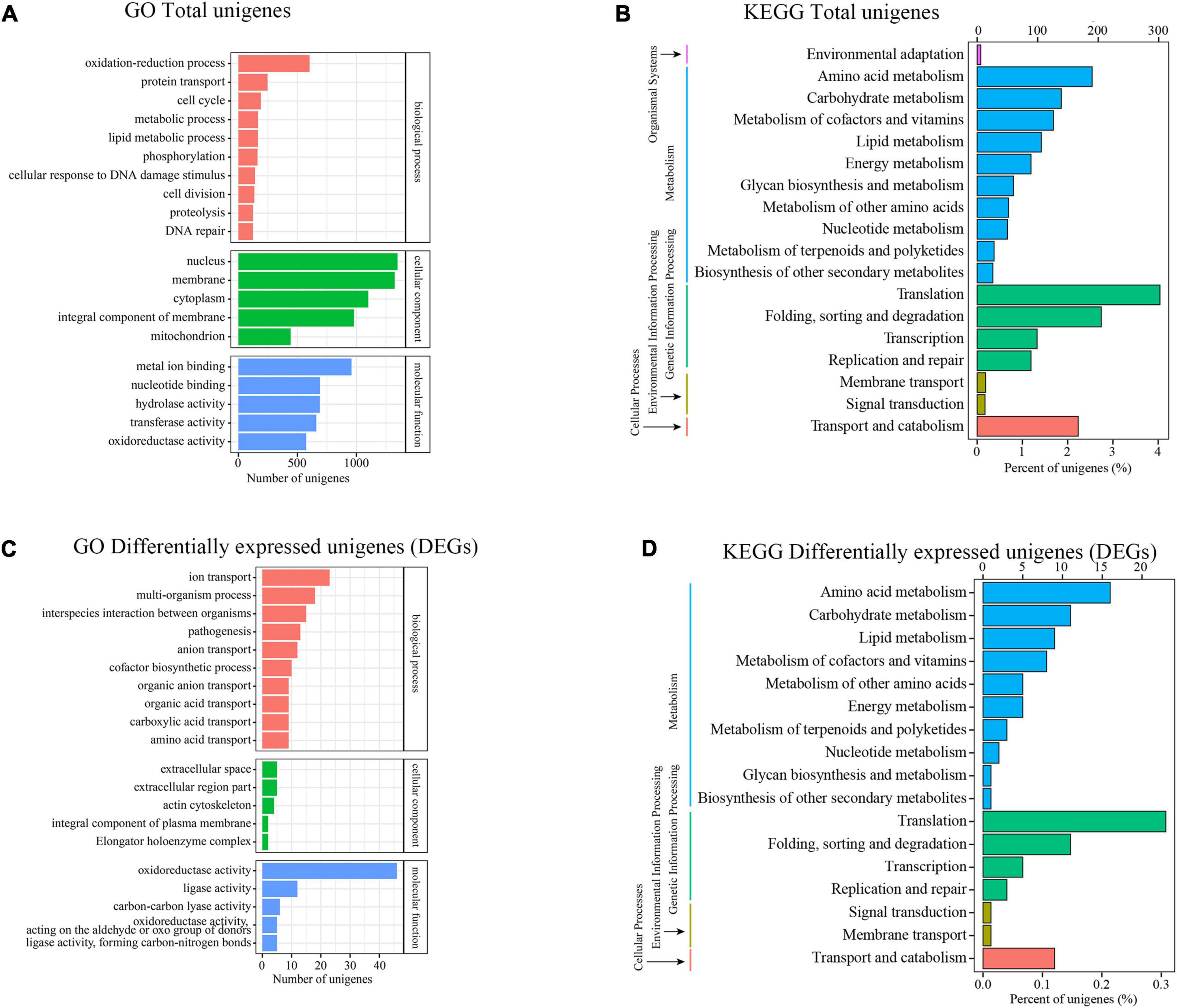
Figure 4. Transcriptional analysis and DEGs differential expressions between Control and MeJA treatment groups. (A) GO enrichment analysis (Total unigenes). (B) KEGG enrichment analysis (Total unigenes). (C) GO enrichment analysis (Differentially expressesunigenes). (D) KEGG enrichment analysis (Differentially expressesunigenes).
Differentially Expressed Genes After Methyl Jasmonate Treatment
The DEGs that increased considerably after MeJA treatment were regarded as MeJA-regulated genes (Supplementary Table 6). We performed a significance analysis on the DEGs and displayed the results using a volcano diagram. Notably, 444 DEGs were identified between the control and MeJA treatment groups. Among these significant DEGs, 226 (50.9%) were up-regulated, while the remaining 218 (49.1%) were down-regulated (Figure 3B). The examination of GO enrichment revealed that the most enriched GO keywords were “oxidoreductase activity,” “ion transport,” “multi-organism,” “interspecies interaction between organisms,” “pathogenesis,” “anion transport,” and “ligase activity” (Figure 4C). Additionally, genes were allocated to several KEGG metabolic pathways (Supplementary Table 4). Enrichment analysis showed that the significantly enriched KEGG pathways were “Translation,” “Amino acid metabolism,” “Folding,” “sorting and degradation,” “Carbohydrate metabolism,” “Transport and catabolism,” “Lipid metabolism,” “Metabolism of cofactors and vitamins” (Figure 4D).
Differentially Expressed Genes Involved in Secondary Metabolite Biosynthesis
The DEGs analysis of secondary metabolites revealed that, 10 clusters were up-regulated compares to the control group. These clusters are involved in secondary metabolite production and degradation, caffeine metabolism, glucosinolate biosynthesis, and novobiocin biosynthesis. There was a significant difference in Phenylpropanoid production (p < 0.05), and the function of the divergent gene may be beta-glucosidase. The unchanged clusters were involved in the synthesis of tropane, piperidine, and pyridine alkaloid (Supplementary Table 5).
Expression Changes of Genes in MVA Pathway
The upstream biosynthesis routes for sesquiterpene intermediate products have been extensively researched, and are preserved in plants (Li et al., 2017). Dendrobium alkaloids are mainly sesquiterpene alkaloids, which are derived from MVA, and the MEP route would act as a source of isoprene units in the production of dendrobine (Chen et al., 2019). Three DEGs, isopentenyl-diphosphate Delta isomerase (IDI), dishomevalonate decarboxylase (MVD), and farnesyl diphosphate synthase (FDPS), were mapped to the MVA pathway in this study (Figure 5).
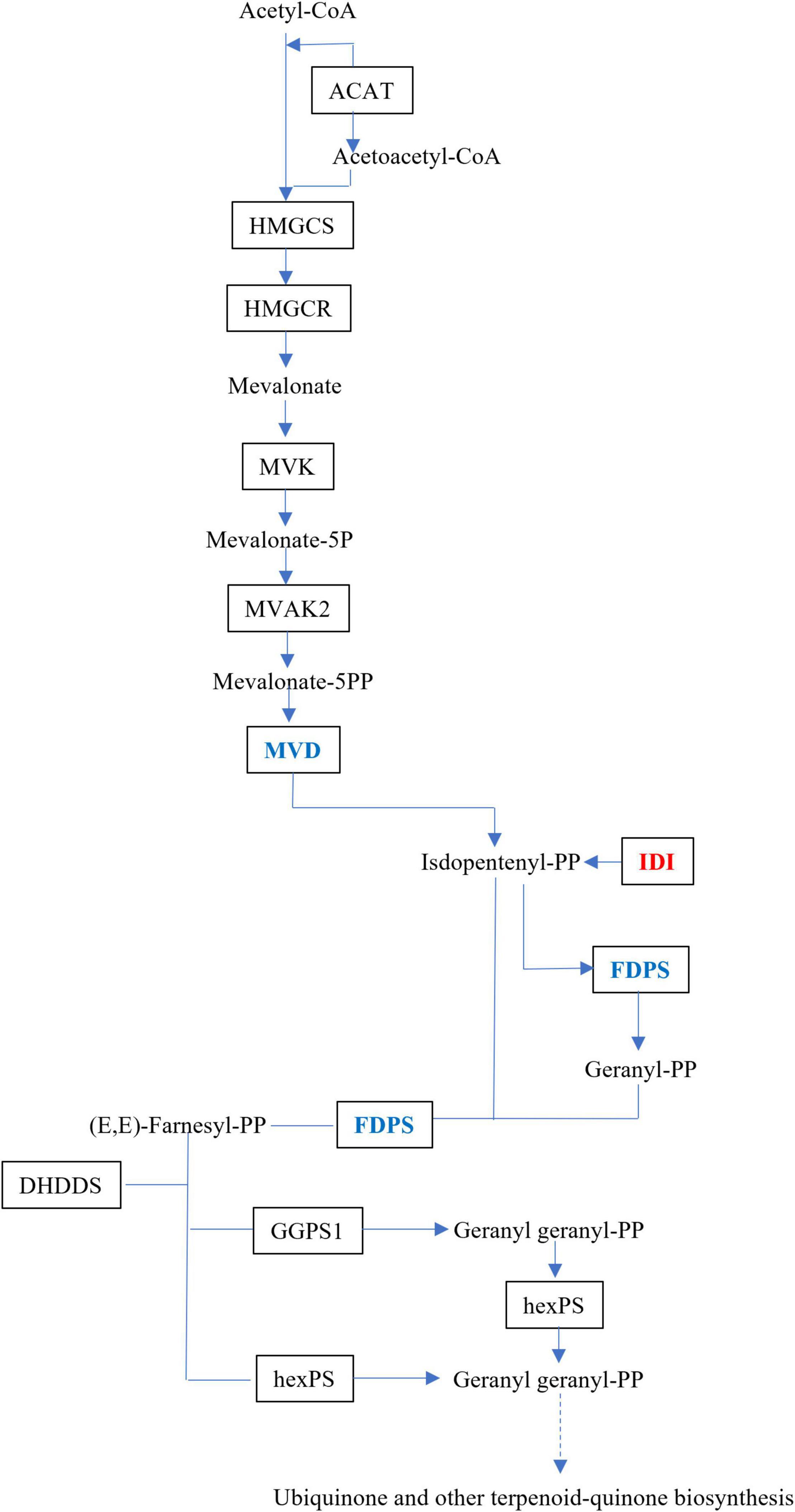
Figure 5. Schematic illustration of MVA biosynthetic pathways in MD33. Abbreviations, as quoted in the figure, are as follows. MVD, diphosphomevalonate decarboxylase; IDI, isopentenyl-diphosphate Delta-isomerase. Expression changes of the genes associated with MVA pathway in the MeJA treatment group and Control. Red indicates up-regulated genes and green indicates down-regulated genes.
Comparison of the Expression Levels of P450 Family Methyltransferase and Aminotransferase Genes
By examining the DEGs, we identified 51 potential P450 unigenes, nine of which were novel. MD33 with and without MeJA treatment is shown in Table 1. Cluster-4787.0, Cluster-514.0, Cluster-2347, Cluster-5538.0, and Cluster-5724.0 were found to be up-regulated, while Cluster-1305.0, Cluster-3418.0, Cluster-1102.0, and Cluster-3538.0 were found to be down-regulated (Figure 6 and Table 1).
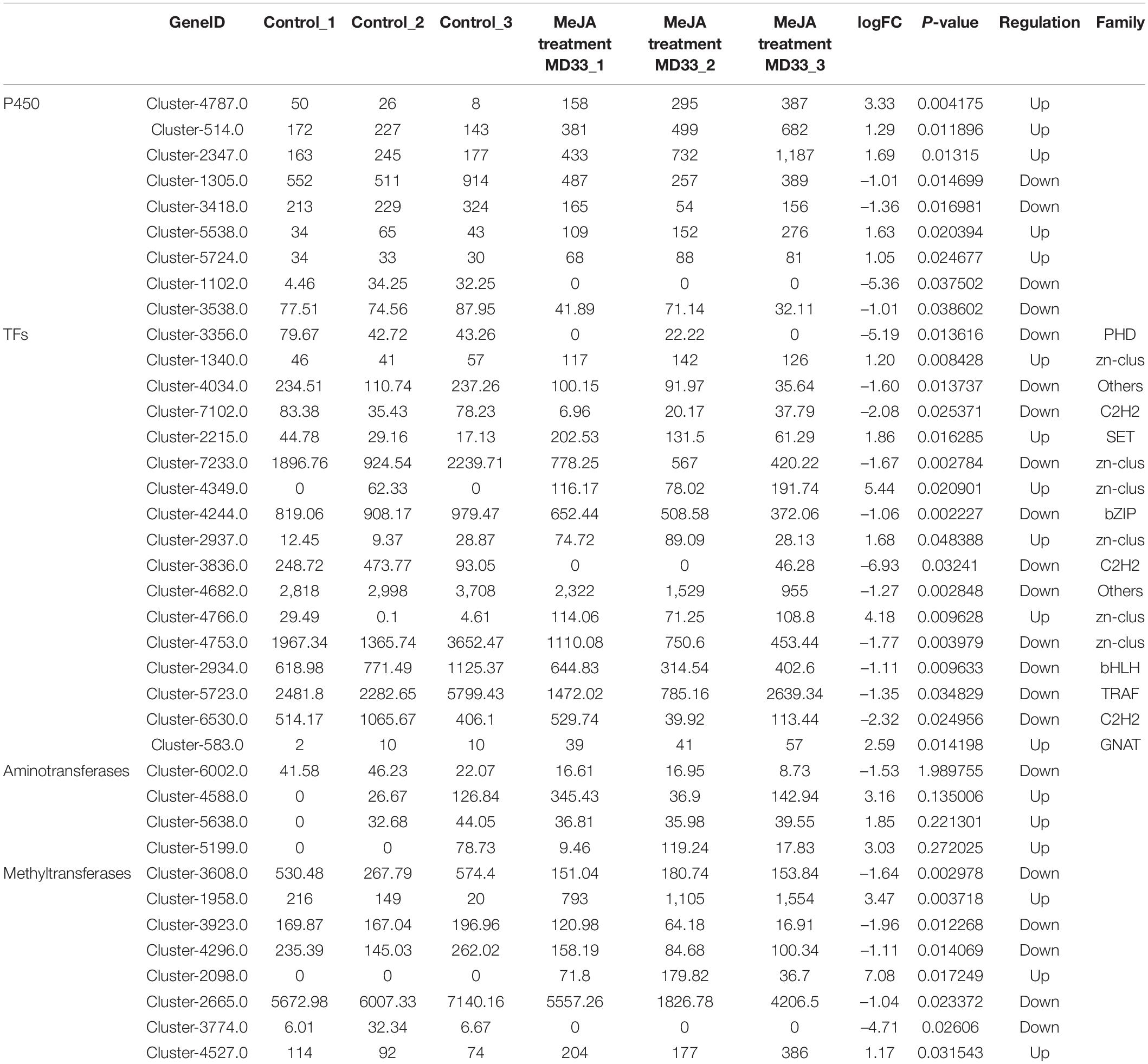
Table 1. The comparation of P450, TFs, aminotransferases and methyltransferases between control and MeJA treatment MD33 groups.
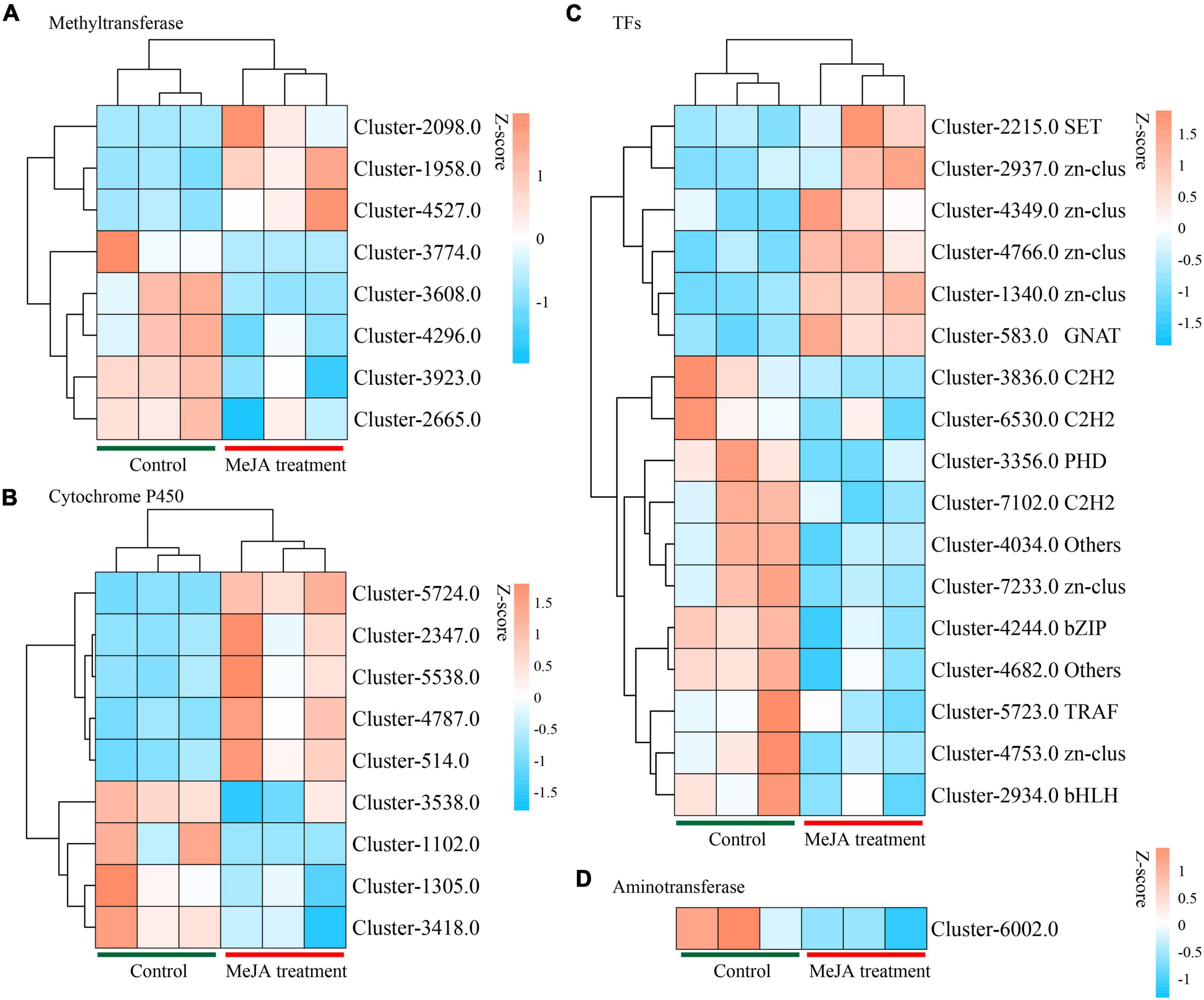
Figure 6. Heat map analysis of genes in backbone post-modification. (A) Heat map analysis of methyltransferases. (B) Heat map analysis of Cytochrome P450 family. (C) Heat map analysis of TFs. (D) Heat map analysis of Aminotransferase.
Aminotransferase and methyltransferase were investigated as two additional key enzymes involved in post-modification. Clustering analysis indicated that only one aminotransferase (Cluster-6002.0) was down-regulated (p < 0.05), but Cluster-4588.0 (log2 fold change = 3.16-fold), Cluster-5638.0 (log2 fold change = 1.85-fold), and Cluster-5199.0 (log2 fold change = 3.02-fold) were up-regulated after MD33 MeJA treatment (Table 1). According to the reference genome (T. longibrachiatum ATCC 18648) (Shi et al., 2020), Cluster-6002.0 encodes a glutamate-1-semialdehyde aminotransferase. The metabolic pathways implicated may include heme (k01845) production and porphyrin and chlorophyll metabolism (ko00860). Heme and porphyrin are pyrrole chemicals with a similar structure to dendrobine. Hence this gene may be implicated in the transaminase function during dendrobine production (Table 1).
By analyzing the DEGs, we obtained 140 putative methyltransferase unigenes, of which eight unigenes (Cluster-1958.0, Cluster-2098.0, and Cluster-4527.0 increased expression, Cluster-3608.0, Cluster-3923.0, Cluster-4296.0, Cluster-2665.0, and Cluster-3774.0 decreased expression) expressed differently (p < 0.05) between MeJA treatment MD33 and control MD33 (Figure 6 and Supplementary Table 1). The primary molecular functions were hydrolase and methyltransferase activities.
Differentially Expressed Genes Related to Transcription Factors
Various Transcription Factors (TFs) were reported to participate in the biosynthesis of secondary metabolites in plants. In this study, 484 putative TF genes belonging to 31 major TF families were identified, and 17 genes belonging to 11 major TF families were analyzed (Figure 6). The Zn-clus (six genes) and the zinc finger protein contained the maximum members. Among these TF genes, three Zn-Clus genes, one SET gene, one C3H gene, and 1 GNAT gene were found to be up-regulated in the MeJA treatment MD33 group (Table 1).
q-RT PCR Validation of Altered Genes
A qRT-PCR assay with independent samples from the control and MeJA treatment groups was used to verify the expression changes of several key genes involved in the dendrobine biosynthesis pathway. In total 10 genes, including one MVA pathway genes, two P450 family genes, two methyltransferase genes, three aminotransferase genes, and two TF genes, were selected to confirm the RNA-seq data. The expression levels of these selected genes were consistent with the RNA-seq results (Figure 7 and Supplementary Table 5).
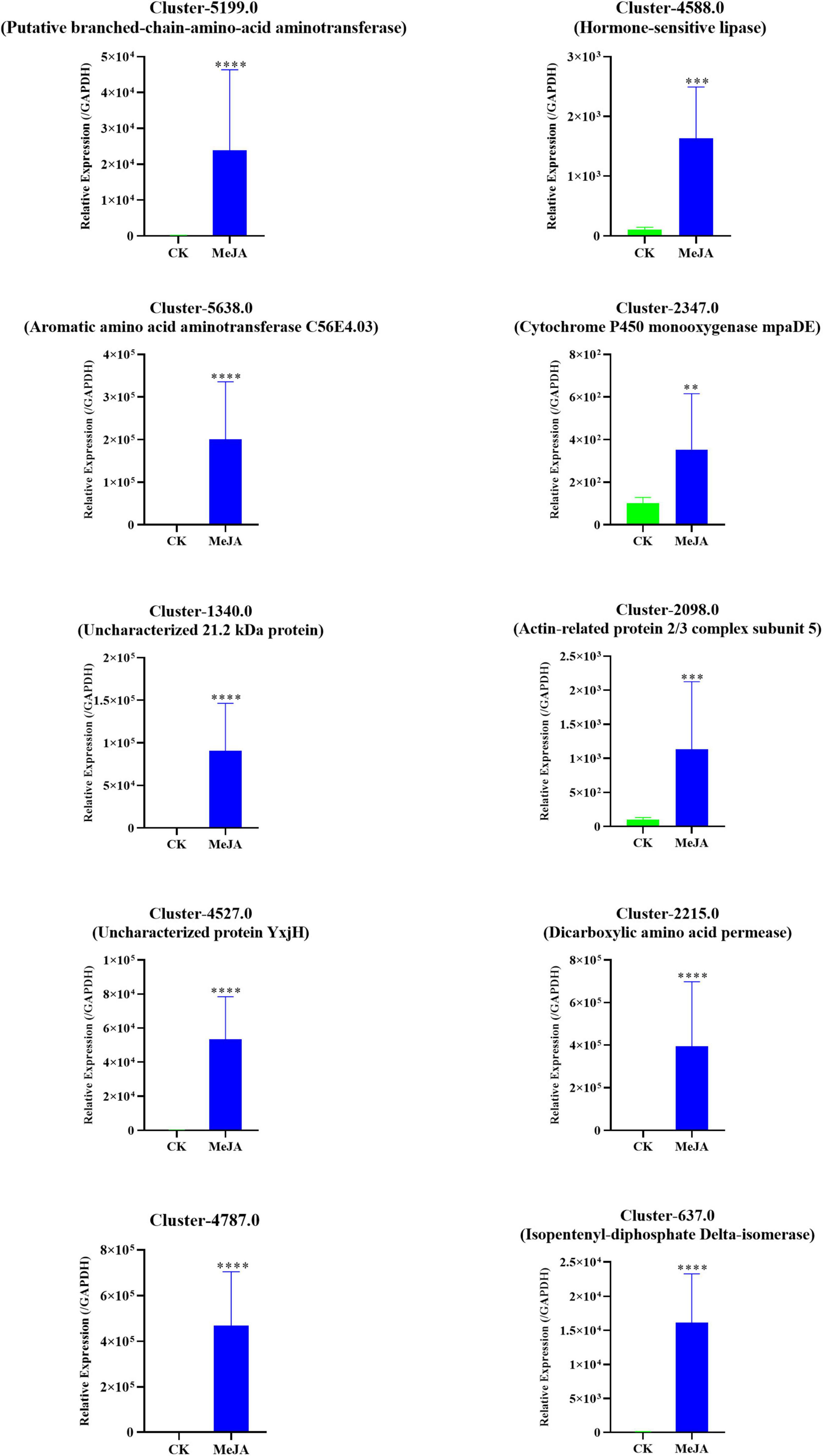
Figure 7. qRT-PCR verification of the RNA-seq data of 10 selected DEGs in the MeJA treatment group and Control. The related expression levels of the key genes involved in the MeJA-induced accumulation of alkaloids in MD33. The expression level of MeJA changed significantly between MeJA and control group. **p< 0.01, ***p < 0.001, ****p < 0.0001.
Discussion
The primary active element in Dendrobium nobile is dendrobine, which is classified as a sesquiterpenoid alkaloid or terpenoid indole alkaloid (TIA) (Wang et al., 2020). Alkaloids were identified and reported for the first time in Dendrobium in 1932 (Suzuki et al., 1932; Chen and Chen, 1935). Recent studies have shown that the alkaloids found in Dendrobium nobile and Dendrobium officinale have significant antioxidant and pharmacological activities (Xu et al., 2017; Huang et al., 2019). As a common elicitor, MeJA was widely used in D. nobile and D. officinal to induce the accumulation of alkaloids (Chen et al., 2019; Zhang et al., 2022). Furthermore, MeJA can also promote the production of secondary metabolites in parasites (Chen et al., 2022) and fungi (Liang et al., 2015; Xu et al., 2021). However, the mechanism by which MeJA induces alkaloid accumulation in dendrobine-producing fungi has not been reported.
Given the same sesquiterpene backbone of these alkaloids, it was previously assumed that dendrobine-type alkaloids had comparable production routes. Dendrobine was produced using aminotransferases and methyltransferases after the sesquiterpene skeleton was generated (Yuan et al., 2018). Cytochromes P450s (CYP450s) play a critical role in the biosynthesis of sesquiterpene alkaloids (dendrobine) (Coon, 2005; Mou et al., 2021). Numerous CYP450s have been identified in D. officinale (Guo et al., 2013; Chen et al., 2019), D. nobile (Jin-Ling et al., 2017; Li et al., 2017), and D. huoshanense (Yuan et al., 2018). These were shown to be associated with many dendrobine synthesis pathways. For instance, CYP4 and CYP71 may catalyze monooxygenase and hydroxylation processes, respectively (Jin-Ling et al., 2017). Four putative genes (Cluster-4787.0, Cluster-514.0, Cluster-5538.0, and Cluster-5724.0) were identified in our RNA-seq data from three clans: CYP526, CYP65, and CYP531. Clan CYP65 was found to be the most abundant with 56 protein entries in Trichoderma, CYP526 was found to participate in secondary metabolic processes (Sonia et al., 2018) and CY (Moktali et al., 2012). This data suggests a novel function for the CYP526 and CYP65 clans in dendrobine accumulation. In this study, we found three MeJA-induced aminotransferase up-regulated genes and 15 MeJA-induced methyltransferase up-regulated genes (Chen et al., 2019). These genes have been annotated to play a role in the dendrobine biosynthesis pathway in T. longibrachiatum MD33.
There is increasing evidence that TFs are involved in the biosynthesis of various alkaloids, including bHLH, ERF, Orca, Zct, Gbf, and WRKY (Goklany et al., 2013). Some of these alkaloids exhibit Jasmonate reactivity, suggesting that the JA signaling cascade is involved in alkaloid production (Goklany et al., 2013; Yamada and Sato, 2013). The Fungal Transcription Factors Database FTFD1 contains information on 61 TF families, including bHLH, bZIP, C2H2 zinc finger, Zn2Cys6, and the zinc finger type GATA. These TFs have been identified in T. atroviride, T. virens, and T. reesei (Qian et al., 2021), and are involved in amino acid and vitamin synthesis, carbon and nitrogen metabolism, meiosis, and morphogenesis (Macpherson et al., 2006). Following MeJA treatment, many TFs were up-regulated, including zn-clus, SET, C3H, and GNAT. These differentially expressed TFs showed that they may play a role in dendrobine production.
Conclusion
A total of 7,067 DEGs were identified in this study, od which some were linked to potential alkaloid biosynthesis pathway in Trichoderma longibrachiatum MD33. Unlike plants, the MVA pathway only regulates genes of the MD33 gene family. This indicates that MeJA treatment provides precursors for production of alkaloids. Additionally, multiple MeJA-induced P450 family genes, aminotransferase genes, and methyltransferase genes were identified, offering several prospective candidates for elucidating the probable alkaloid biosynthesis pathway of T. longibrachiatum MD33. These findings contribute to the understanding of the regulatory mechanism underlying MeJA-induced alkaloid accumulation in T. longibrachiatum MD33.
Data Availability Statement
The RNA-seq used in this study have been deposited in the Sequence Read Achieve (SRA) of the NCBI database under the BioProject accession number: PRJNA809435.
Author Contributions
QJ, LW, HJ, and SS performed the experiments. JC designed and supervised the project. XQ helped for the data curation. FS and LJ arranged the materials for experiments. QJ, LW, and SS wrote and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Jiangsu Synergetic Innovation Center for Advanced Bio-Manufacture (No. XTD1825), the Science and Technology Projects of Guizhou Province, China [No. (2017)5733-028], the Talent Team of Guizhou Education Department [No. (2014)39], Guizhou Science and Technology Corporation Platform Talents Fund [Nos. (2017)5733-001 and CK-1130-002], and the Project of Honghuagang District project of Guizhou Province, China [No. (2018)10].
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.890733/full#supplementary-material
Footnotes
References
Bai, J., Guo, Q. X., Zhang, J., Huang, J., and Huang, Z. H. (2020). Metabolic Profile of Dendrobine in Rats Determined by Ultra-high-performance Liquid Chromatography/Quadrupole Time-of-flight Mass Spectrometry. Comb. Chem. High. Throughput Screen 23, 1364–1376. doi: 10.2174/1386207323666201023115744
Chen, C., Liu, F., Zhang, K., Niu, X., Zhao, H., Liu, Q., et al. (2022). MeJA-responsive bHLH transcription factor LjbHLH7 regulates cyanogenic glucoside biosynthesis in Lotus japonicus. J. Exp. Bot. 73, 2650–2665. doi: 10.1093/jxb/erac026
Chen, K. K., and Chen, A. L. (1935). The alkaloid of chin-shih-hu. J. Biol. Chem. 111, 653–665. doi: 10.1016/S0021-9258(18)75010-2
Chen, Y., Wang, Y., Lyu, P., Chen, L., Shen, C., and Sun, C. (2019). Comparative transcriptomic analysis reveal the regulation mechanism underlying MeJA-induced accumulation of alkaloids in Dendrobium officinale. J. Plant Res. 132, 419–429. 01099-6 doi: 10.1007/s10265-019-01099-6
Coon, M. J. (2005). CYTOCHROME P450: nature’s Most Versatile Biological Catalyst. Ann. Rev. Pharmacol. Toxicol. 45, 1–25. doi: 10.1146/annurev.pharmtox.45.120403.100030
Fagan, F. W. F. (2002). Fungal Endophytes: common Host Plant Symbionts but Uncommon Mutualists. Integr. Compar. Biol. 42, 360–368. doi: 10.2307/3884691
Goklany, S., Rizvi, N. F., Loring, R. H., Cram, E. J., and Lee-Parsons, C. W. T. (2013). Jasmonate-dependent alkaloid biosynthesis in Catharanthus Roseus hairy root cultures is correlated with the relative expression of Orca and Zct transcription factors. Biotechnol. Progr. 29, 1367–1376. doi: 10.1002/btpr.1801
Guo, X., Li, Y., Li, C., Luo, H., Wang, L., Qian, J., et al. (2013). Analysis of the Dendrobium officinale transcriptome reveals putative alkaloid biosynthetic genes and genetic markers. Gene 527, 131–138. doi: 10.1016/j.gene.2013.05.073
Huang, J., Huang, N., Zhang, M., Nie, J., and Shi, J. (2019). Dendrobium alkaloids decrease Aβ by regulating α- and β-secretases in hippocampal neurons of SD rats. PeerJ. 7(Suppl. 1), e7627. doi: 10.7717/peerj.7627
Jiang, C., Luo, Y., Yuan, Y., Dong, X., Zhao, Y., and Huang, L. (2018). Conventional octaplex PCR for the simultaneous identification of eight mainstream closely related Dendrobium species. Industr. Crops Produc. 112, 569–576. doi: 10.1016/j.indcrop.2017.12.048
Jin-Ling, L., Zhao, Z., Hong-Chang, L, Luo, C-L., Wang, H-L., et al. (2017). Influence of light intensity and water content of medium on total dendrobine of Dendrobium nobile Lindl. Asian Pac. J. Trop. Med. 11 81–86. doi: 10.1016/j.apjtm.2017.10.015
Li, D. D., Zheng, C. Q., Zhang, F., and Shi, J. S. (2022). Potential neuroprotection by Dendrobium nobile Lindl alkaloid in Alzheimer’s disease models. Neural Regener. Res. 17, 972–977. doi: 10.4103/1673-5374.324824
Li, Q., Ding, G., Li, B., and Guo, S. X. (2017). Transcriptome Analysis of Genes Involved in Dendrobine Biosynthesis in Dendrobium nobile Lindl. Infected with Mycorrhizal Fungus MF23 (Mycena sp.). Sci. Rep. 7:316. doi: 10.1038/s41598-017-00445-9
Liang, S., Li, G., Zhang, X., Ren, A., and Zhao, M. (2015). The regulation of methyl jasmonate on hyphal branching and GA biosynthesis in Ganoderma lucidum partly via ROS generated by NADPH oxidase. Fungal Genet. Biol. 81, 201–211. doi: 10.1016/j.fgb.2014.12.002
Liu, J., Han, Y., Zhu, T., Yang, Q., Wang, H. M., and Zhang, H. (2021). Dendrobium nobile Lindl. polysaccharides reduce cerebral ischemia/reperfusion injury in mice by increasing myeloid cell leukemia 1 via the downregulation of miR-134. Neuroreport 32, 177–187. doi: 10.1097/wnr.0000000000001562
Macpherson, S., Larochelle, M., and Turcotte, B. (2006). A Fungal Family of Transcriptional Regulators: the Zinc Cluster Proteins. Microbiol. Mol. Biol. Rev. 70, 583–604. doi: 10.1128/mmbr.00015-06
Moktali, V., Park, J., Fedorova-Abrams, N. D., Park, B., Choi, J., Lee, Y. H., et al. (2012). Systematic and searchable classification of cytochrome P450 proteins encoded by fungal and oomycete genomes. BMC Genom. 13:525. doi: 10.1186/1471-2164-13-525
Mou, Z. M., Zhao, Y., Ye, F., Shi, Y. N., Kennelly, E. J., Chen, S. Y., et al. (2021). Identification, Biological Activities and Biosynthetic Pathway of Dendrobium Alkaloids. Front. Pharmacol. 12:605994. doi: 10.3389/fphar.2021.605994
Nie, J., Jiang, L. S., Zhang, Y., Tian, Y., Li, L. S., Lu, Y. L., et al. (2018). Dendrobium nobile Lindl. Alkaloids Decreases the Level of Intracellular beta-Amyloid by Improving Impaired Autolysosomal Proteolysis in APP/PS1 Mice. Front. Pharmacol. 9:1479. doi: 10.3389/fphar.2018.01479
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Qian, X., Jin, H., Chen, Z. J., Dai, Q. Q., Sarsaiya, S., Qin, Y. T., et al. (2021). Comparative Transcriptome Analysis of Genes Involved in Sesquiterpene Alkaloid Biosynthesis in Trichoderma longibrachiatum MD33 and UN32. Front. Microbiol. 12:800125. doi: 10.3389/fmicb.2021.800125
Sarsaiya, S., Jain, A., Fan, X., Jia, Q., and Chen, J. (2020a). New Insights into Detection of a Dendrobine Compound From a Novel Endophytic Trichoderma longibrachiatum Strain and Its Toxicity Against Phytopathogenic Bacteria. Front. Microbiol. 11:337. doi: 10.3389/fmicb.2020.00337
Sarsaiya, S., Jain, A., Jia, Q., Fan, X. K., Shu, F. X., Chen, Z. W., et al. (2020b). Molecular Identification of Endophytic Fungi and Their Pathogenicity Evaluation Against Dendrobium nobile and Dendrobium officinale. Int. J. Mol. Sci. 21:316. doi: 10.3390/ijms21010316
Sarsaiya, S., Jia, Q., Fan, X., Jain, A., and Chen, J. (2019a). First Report of Leaf Black Circular Spots on Dendrobium nobile Caused by Trichoderma longibrachiatum in Guizhou Province, China. Plant Dis. 103. doi: 10.1094/PDIS-03-19-0672-PDN
Sarsaiya, S., Shi, J. S., and Chen, J. S. (2019b). A comprehensive review on fungal endophytes and its dynamics on Orchidaceae plants: current research, challenges, and future possibilities. Bioengineered 10, 316–334. doi: 10.1080/21655979.2019.1644854
Shi, J. C., Shi, W. L., Zhou, Y. R., Chen, X. L., and Song, X. Y. (2020). The Putative Methyltransferase TlLAE1 Is Involved in the Regulation of Peptaibols Production in the Biocontrol Fungus Trichoderma longibrachiatum SMF2. Front. Microbiol. 11:1267. doi: 10.3389/fmicb.2020.01267
Song, T. H., Chen, X. X., Kai-Fai, L. C., Cho-Wing, S. S., Feng, Y. B., Yang, Z. J., et al. (2018). Dendrobine targeting JNK stress signaling to sensitize chemotoxicity of cisplatin against non-small cell lung cancer cells in vitro and in vivo. Phytomedicine 53, S0944711318302046. doi: 10.1016/j.phymed.2018.06.018
Sonia, C., Mehetre, S. T., Ravindra, B., Alan, K., Andrea, A., Grigoriev, I. V., et al. (2018). Genome-wide analysis of cytochrome P450s of Trichoderma spp.: annotation and evolutionary relationships. Fungal Biol. Biotechnol. 5:12. 018-0056-3 doi: 10.1186/s40694-
Stierle, A., Strobel, G., and Stierle, D. (1993). Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of Pacific yew. Science 260:214. doi: 10.1126/science.8097061
Strobel, G. A., Hess, W. M., Ford, E., Sidhu, R. S., and Yang, X. (1996). Taxol from fungal endophytes and the issue of biodiversity. J. Industr. Microbiol. 17, 417–423. doi: 10.1007/BF01574772
Suzuki, H., Keimatsu, I., and Ito, M. (1932). Über die Alkaloide der chinesischen Droge “Chin-Shih-Hu&rdquo. I. Mitteil. YAKUGAKU ZASSHI 52, 996–1009. doi: 10.1248/yakushi1881.52.11_996
Wang, Z., Zhao, M., Cui, H., Li, J., and Wang, M. (2020). Transcriptomic Landscape of Medicinal Dendrobium Reveals Genes Associated With the Biosynthesis of Bioactive Components. Front. Plant Sci. 11:391. doi: 10.3389/fpls.2020.00391
Xiaori, Z., Xinyue, L., Xiujun, L., Yujia, Z., Shangguo, F., Chunna, Y., et al. (2018). Comparative Metabolomic and Proteomic Analyses Reveal the Regulation Mechanism Underlying MeJA-Induced Bioactive Compound Accumulation in Cutleaf Groundcherry (Physalis angulata L.) Hairy Roots. J. Agricult. Food Chem. 66, 6336–6347. doi: 10.1021/acs.jafc.8b02502
Xiong, T. W., Liu, B., Wu, Q., Xu, Y. Y., Liu, P., Wang, Y., et al. (2021). Beneficial effects of Dendrobium nobile Lindl. Alkaloids (DNLA) on anxiety and depression induced by chronic unpredictable stress in rats. Brain Res. 1771:147647. doi: 10.1016/j.brainres.2021.147647
Xu, L. A., Yr, B., Jin, J. A., Yj, A., Qy, B., Sl, A., et al. (2021). The inhibitory mechanism of methyl jasmonate on Aspergillus flavus growth and aflatoxin biosynthesis and two novel transcription factors are involved in this action. Food Res. Int. 140:110051. doi: 10.1016/j.foodres.2020.110051
Xu, Y. Y., Xu, Y. S., Wang, Y., Wu, Q., and Shi, J. S. (2017). Dendrobium nobile Lindl. alkaloids regulate metabolism gene expression in livers of mice. J. Pharm. Pharmacol. 69, 1409–1417. doi: 10.1111/jphp.12778
Yamada, Y., and Sato, F. (2013). Transcription Factors in Alkaloid Biosynthesis - ScienceDirect. Int. Rev. Cell Mol. Biol. 305, 339–382. doi: 10.1016/B978-0-12-407695-2.00008-1
Yuan, Y., Yu, M., Jia, Z., Song, X.e, Liang, Y., and Zhang, J. (2018). Analysis of Dendrobium huoshanense transcriptome unveils putative genes associated with active ingredients synthesis. BMC Genom. 19:978. doi: 10.1186/s12864-018-5305-6
Zhang, B., Niu, Z., Li, C., Hou, Z., Xue, Q., Liu, W., et al. (2022). Improving large-scale biomass and total alkaloid production ofDendrobium nobileLindl. using a temporary immersion bioreactor system and MeJA elicitation. Plant Methods 18:10. doi: 10.1186/s13007-022-00843-9
Zhang, X. N., Liu, J., Liu, Y., Wang, Y., Abozeid, A., Yu, Z. G., et al. (2018). Metabolomics Analysis Reveals that Ethylene and Methyl Jasmonate Regulate Different Branch Pathways to Promote the Accumulation of Terpenoid Indole Alkaloids in Catharanthus roseus. J. Nat. Produc. 81, 335–342. doi: 10.1021/acs.jnatprod.7b00782
Zhang, Y., Wang, H., Mei, N., Ma, C., Lou, Z., Lv, W., et al. (2018). Protective effects of polysaccharide from Dendrobium nobile against ethanol-induced gastric damage in rats. Int. J. f Biol. Macromol. 107, 230–235. doi: 10.1016/j.ijbiomac.2017.08.175
Keywords: dendrobine, Trichoderma longibrachiatum MD33, differentially expressed genes, MVA pathways, P450 family
Citation: Jia Q, Wang L, Qian X, Jin H, Shu F, Sarsaiya S, Jin L and Chen J (2022) Transcriptome Analysis of Dendrobine Biosynthesis in Trichoderma longibrachiatum MD33. Front. Microbiol. 13:890733. doi: 10.3389/fmicb.2022.890733
Received: 06 March 2022; Accepted: 08 June 2022;
Published: 01 August 2022.
Edited by:
Dipesh Dhakal, University of Florida, United StatesReviewed by:
Monika Bielecka, Wrocław Medical University, PolandPing Su, China Academy of Chinese Medical Sciences, China
Feng-Juan Mou, Southwest Forestry University, China
Copyright © 2022 Jia, Wang, Qian, Jin, Shu, Sarsaiya, Jin and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jishuang Chen, biochenjs@njtech.edu.cn
 Qi Jia1,2
Qi Jia1,2