
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Microbiol., 09 May 2022
Sec. Microbial Physiology and Metabolism
Volume 13 - 2022 | https://doi.org/10.3389/fmicb.2022.878110
Triterpenoids are crucial active ingredients of Ganoderma lucidum (G. lucidum) with various health benefits. Yet the low biosynthesis greatly restricts their industrial applications, novel metabolic engineering strategies are needed to further enhance Ganoderma triterpenoids production. Transcription factors play vital roles in the metabolic regulation of terpenoids, which are still scarce to study in G. lucidum. Herein, a transcription factor GlbHLH5 (GenBank No. MZ436906.1) potential for metabolic regulation of Ganoderma triterpenes was identified for the first time. MeJA could increase Ganoderma triterpenoids biosynthesis, and GlbHLH5 significantly responded to MeJA induction, suggesting GlbHLH5 is a new target for Ganoderma triterpenoids overproduction. The regulatory effect of the newly identified target was further validated by homologous gene overexpression and silence in G. lucidum. It’s demonstrated that overexpression of GlbHLH5 significantly increased triterpenoids accumulation and the key enzyme genes transcription in the biosynthetic pathway, while silencing it displayed the opposite effect, indicating GlbHLH5 could positively regulate the triterpenoids biosynthesis by activating the synergistic expression of key enzyme genes in the biosynthetic pathway. Consequently, GlbHLH5 was identified as a positive regulator and novel metabolic target for Ganoderma triterpenoids biosynthesis, it sheds new lights on the regulatory effect regulation and synthetic biology of Ganoderma triterpenoids.
Ganoderma lucidum (G. lucidum). Karst is a precious and famous large fungus as a food and medicine homologous. It is rich in active polysaccharides (Hu et al., 2019), triterpenoids (He et al., 2019), nucleosides (Chen Y. et al., 2012), sterols (Xu et al., 2021), and other substances with extremely high nutritional and medicinal values (Li et al., 2019a; Zhao et al., 2019). Ganoderma triterpenoids are the main active ingredients of G. lucidum. They are also known as ganoderic acids, which refer to a class of triterpenoids containing carboxyl and polyhydroxyl structures (Hu et al., 2020). As the crucial bioactive components, they have been recognized by the public for their functions of regulating the homeostasis of the body (Grienke et al., 2011), improving endocrine disorders (Zhao et al., 2012), and improving sleep quality (Yao et al., 2021). However, as a kind of secondary metabolites, the content of Ganoderma triterpenoids is relatively low in the raw material, which greatly restricts its development and application. It’s momentous to enhance active triterpenoids production by metabolically regulating their biosynthesis. Although improvement of Ganoderma triterpenoids production had been achieved by optimizing fermentation substrates and process parameters, adding exogenous elicitors, and overexpressing the key enzyme genes in the triterpenoid’s biosynthetic pathway, novel metabolic engineering strategies are needed to further enhance Ganoderma triterpenoids production. Transcription factors play vital roles in the metabolic regulation of terpenoids, which are still scarce to study in G. Lucidum (Dhandapani et al., 2020; Wang et al., 2021). How to increase the biosynthesis of Ganoderma triterpenoids is a problem worthy of attention (Niego et al., 2021).
Jasmonic acids (JAs) are important signal molecules within plant cells, which regulate the expression of defense proteins and the synthesis of secondary substances through interactions with transcription factors (Li et al., 2021). The bHLH transcription factor is regarded as a core member of the JA signaling pathway. The genes involved in a variety of terpenoids biosynthetic pathways such as SmTAT1 (Peng et al., 2020), SmCYP98A14 (Di et al., 2013), AaCYP71AV1 (Li et al., 2019b), and AaDBR2 (Fu et al., 2021) are positively regulated by bHLH transcription factors. Therefore, bHLH is a positive regulator of terpenoids biosynthesis mediated by the JAs signal. It was found that exogenous MeJA could increase the biosynthesis of terpenoids in G. Lucidum by the signaling crosstalk between nitric oxide(NO) and reactive oxygen species (ROS) (Ren et al., 2010; Shi et al., 2020), but the mechanism of its transcriptional regulation remains unclear. Herein, we firstly discovered an endogenous bHLH member GlbHLH5 responded to JAs in G. lucidum. Through bioinformatics analysis, and an excessive and silent expression of this gene in G. lucidum itself, we verified its role in the regulation of Ganoderma triterpenoids biosynthesis, and finally clarified the regulation mechanism of Ganoderma triterpenoid metabolism. It will provide new insights into the metabolic regulatory network associated with triterpenoids biosynthesis in G. lucidum, and define a new tool for improving Ganoderma triterpenoids production by burgeoning synthetic biology as well.
The Ganoderma lucidum strain (G. lucidum CGMCC5.26) used in this experiment was purchased from the Institute of Microbiology, Chinese Academy of Sciences, the DH5a competent cells were purchased from Vazyme, and AH109 competent cells were purchased from Shanghai Weidi Biotechnology Co., Ltd. The pGBKT7 vector was presented by Dr. Bingcong Xing from Zhejiang A&F University.
Fungal complete medium (CYM) and fungal protoplast cell wall regeneration medium (MYG) were used to cultivate G. lucidum. CYM medium was composed of 1% maltose, 2% glucose, 0.2% yeast extract, 0.2% peptone, 0.05% MgSO4⋅7H2O, 0.05% K2HPO4 (Zhou et al., 2014). MYG medium was composed of 1% glucose, 0.5% maltose, 0.5% yeast powder, 2% agar powder, dissolved in mannitol. LB medium: 1% tryptone, 0.5% yeast powder, 1% NaCl, 2% agar powder (solid), pH = 7.0.
The primers were designed according to the CDS region of the GlbHLH5 gene (Supplementary Table 1) and synthesized by Sangon Biotech. The total RNA of Ganoderma lucidum hyphae was extracted according to the instructions of the column-type fungal total RNA extraction and purification kit (Sangon Biotech, Shanghai, China). Agarose gel electrophoresis was used to detect the integrity of the total RNA, and a micro-ultraviolet spectrophotometer was used to detect the concentration and purity. Following the instructions of the PrimeScript™ RT Master Mix kit (Sangon Biotech, Shanghai, China), the total RNA was reversed into cDNA which was further used as a template for PCR amplification of the coding region of the GlbHLH5 gene, and the PCR amplification system was as follows: PrimeSTAR Max Premix (2X) 25 μL, GlbHLH5-F 2.5 μL, GlbHLH5-R 2.5 μL, cDNA template 1.0 μL, ddH2O 19 μL. PCR was performed according to the manufacture’s instruction of PrimeSTAR® Max DNA Polymerase (Takara, Japan) using the following protocol: 98°C, 5 min, 1 cycle; 98°C, 10 s, 55°C, 30 s, 72°C, 1 min, 30 cycles; 72°C, 5 min. After the PCR products were detected by 1.5% agarose gel electrophoresis, they were sent to Zhejiang Youkang Biotechnology Co., Ltd. for sequencing.
A bHLH gene GlbHLH5 (GenBank No. MZ436906.1) was obtained by comparing the G. lucidum genome database1 and the Pfam database (number: PF00010) (Chen S. et al., 2012; Qian et al., 2013; Finn et al., 2016; Chen X. et al., 2021). Online analysis software ORF Finder and Conserved Domain-search of the NCBI database were used to analyze the open reading frame (ORF) and conserved domains of the gene. ProtParam was performed to analyze the physicochemical properties of the GlbHLH5 encoded protein. At last, highly similar protein sequences of other species were downloaded from the NCBI protein database, and a phylogenetic tree was constructed to perform the phylogenetic analysis by MEGA 7.0. PlantCARE was used to perform online promoter cis-acting element analysis on the nucleic acid sequences of 2 000 bp upstream of three key genes HMGR (GenBank No. EU263989) (Shang et al., 2008), SQS (GenBank No. DQ494674) (Zhao et al., 2007), and LS (GenBank No. GQ169528) (Shang et al., 2010) in the triterpenoid synthesis pathway of Ganoderma lucidum, and TBtools software was used for visual display.
The sequenced gene was translated, and the properties of the protein encoded by the GlbHLH5 gene were analyzed. Using the method of homologous recombination, the GlbHLH5 gene was integrated into the pET-32a vector, and the recombinant plasmid was transferred into Escherichia coli DH5a competent cells, cultured at 37°C overnight, and a single colony was picked for PCR verification and sent to the company for sequencing. Plasmid extraction was performed on the correctly sequenced recombinant-positive strains, and the plasmid DNA was transferred into E. coli BL21 (DE3) competent cells, cultured overnight at 37°C, and single colonies were picked and inoculated in Amp-resistant LB liquid medium, and then cultured at 37°C and 200 r⋅min–1 until the bacterial density (OD600) reached 0.8. Subsequently, 0.5 mmol⋅L–1. IPTG was added to induce 12 h at 16, 25, and 37°C, and the bacterial liquid was collected and detected by SDS-PAGE.
To investigate the transcriptional activation of GlbHLH5, pGBKT7 (pBD) was used to construct yeast one-hybrid system pBD-GlbHLH5 vector by EcoRI and BamHI. After the sequencing results were correct, the plasmid pBD-GlbHLH5 and the positive control pGBKT7 were transformed into yeast AH109, respectively. The constructed plasmid was coated on the defect medium SD/-Trp, cultured at 30°C for 3 days, and then transferred to SD/-Trp/-His/-Ade + x-a-Gal for color reaction.
The 50 μmol L–1 MeJA was added to the fermentation broth of G. lucidum cultured for 5 days, and no MeJA treatment was used as a control. The triterpenoid content of G. lucidum was determined at 0, 2, 4, 6, 12, 24, and 48 h after MeJA induction. The relative expression of GlbHLH5 was determined by qRT-PCR, and the response of the key enzyme genes in the triterpenoid’s biosynthetic pathways to MeJA induction was also investigated. Each experiment was repeated in three replicates, and the data are all expressed as mean ± SD (Gharari et al., 2020).
The GlbHLH5 sequence and overexpression vector pBARGPEI were analyzed, and primers containing BamHI and EcoRI sites were designed (Supplementary Table 1). The pBARGPEI was digested with BamHI and EcoRI, and recovered and purified for homologous recombination with GlbHLH5 containing homologous arms (Song et al., 2021). The recombinant product was transferred into E. coli DH5a competent cells and cultured overnight at 37°C. Single colonies were picked for PCR verification of bacterial solution and sent to the company for sequencing. The plasmid was extracted from the bacterial solution with correct sequencing. The strains WT, pBARGPEI-GFP and pBARGPEI-GlbHLH5-GFP were inoculated in the fungal medium containing CaCl2, and the sterilized coverslip was inserted into the medium at 1 cm around the inoculation block. After the mycelia grew on the coverslip, the coverslip was stained with DAPI, and rinsed with PBS for several times. The positions of GFP protein, GlbHLH5-GFP fusion protein and nucleus in cells were observed by inverted fluorescence microscope.
According to the GlbHLH5 sequence and the overexpression vector pBARGPEI, primers containing restriction sites BamHI and EcoRI (Supplementary Table 1) were designed. PBARGPEI was digested with BamHI and EcoRI with double restriction enzymes, recovered and purified, and then homologously recombined with GlbHLH5 containing homology arms. The recombined product was transferred to E. coli DH5a competent cells, cultured overnight at 37°C, and the single colonies were picked on Amp resistant plates. Then, bacterial liquid PCR and sequencing were performed to verify whether the recombinant vector was successfully constructed. At last the recombinant plasmids were transformed into protoplast of G. lucidum.
According to pSilent-1 plasmid and GlbHLH5 silencing sequences, suitable restriction sites were selected to design the primers (Supplementary Table 1). The pSilent-1 and GlbHLH5 silencing sequences were double digested with Xhol and HindIII, and the digested products were ligated and transformed and sequenced to form the intermediate vector pS-1. Then, the intermediate vector and GlbHLH5 reverse silencing sequence were doubly digested with SphI and KpnI, and the GlbHLH5 reverse silencing sequence was insert into the downstream of the IT sequence of the vector to finally construct the pSilent-GlbHLH5 recombinant plasmid with an inverted repeat “hairpin structure.”
The overexpression and silencing vectors constructed above were transferred into G. lucidum protoplasts by a PEG-mediated method (Liu and Friesen, 2012), and the transformants were screened by hygromycin (G-Clone, Beijing, China) resistance, and the well-growing strains were further identified by colony PCR and the detection of the GlbHLH5 gene expression.
The expression levels of GlbHLH5, HMGR, SQS, and LS genes were determined by qRT-PCR with 18S rRNA as the internal reference gene, and the primers used were shown in Supplementary Table 1. qRT-PCR was performed according to the manufacture’s instruction of ChamQ Universal SYBR qPCR Master Mix (Vazyme, Nanjing, China) using the following protocol: 95°C, 30 s, 1 cycle; 95°C, 10 s, 60°C, 30 s, 40 cycles. Salkowski reaction was firstly used for qualitative detection of triterpenoid content in G. lucidum, and further quantitative determination was conducted by the vanillin-perchloric acid chromogenic method (Chen Z. et al., 2021).
GraphPad Prism 8.0 software was used for statistical analysis, in which the qRT-PCR results used 18S rRNA as an internal reference, and the relative expression level of genes was calculated by the 2–△△ Ct method, and then the t-test was used to analyze the significance of the difference. “*” means the difference is significant (P < 0.05) and “**” means the difference is extremely significant (P < 0.01).
The cDNA of G. lucidum was used as a template for PCR amplification. After sequencing, the full length of the Ganoderma bHLH transcription factor cDNA was 1008 bp, and the sequence had been uploaded to the NCBI database. The obtained gene sequence number was MZ436906.1 and named GlbHLH5. The relative molecular weight of GlbHLH5 is 85113.08, the theoretical PI value is 4.98, and it could encode a protein with a molecular weight of about 36.58 kDa. The SMART analysis and multiple comparisons of amino acids showed that it contained an HLH domain composed of 51 amino acids. The phylogenetic analysis indicated that GlbHLH5 was closely related to Ganoderma sinensis transcription factor GsPIL31529.1 (Figure 1).
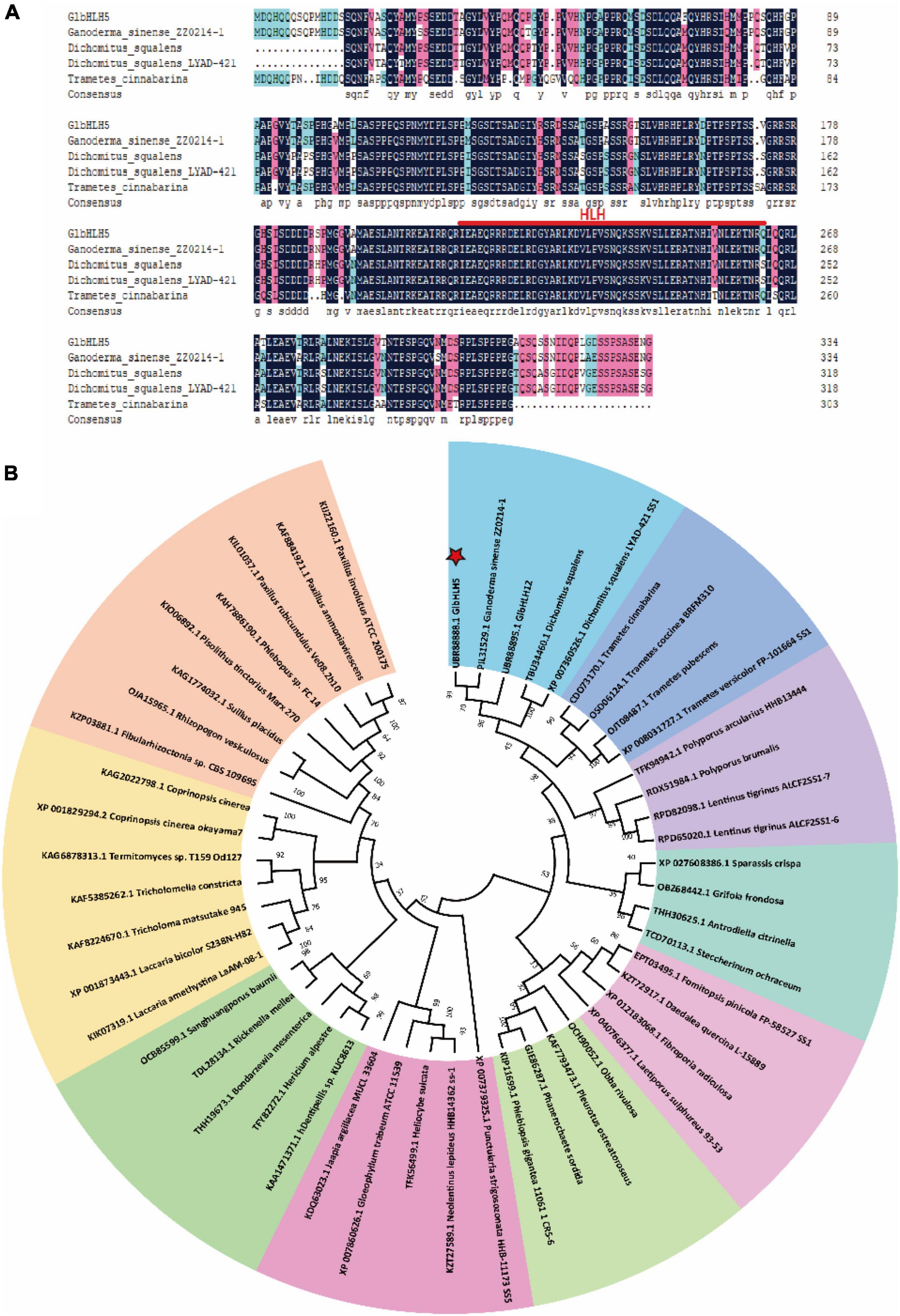
Figure 1. Bioinformatics analysis of GlbHLH5. (A) The comparison analysis of the amino acid sequence of GlbHLH5 and other homologous proteins. (B) The adjacent tree of protein homologs, the percentage value of the leader program for 1,000 repetitions is displayed at the branch point.
On-line protein analysis showed that there were three secondary structures of GlbHLH5 gene. The proportion of α-helix (blue), extended strand (red) and random coil (purple) were 26.57, 5.97, and 67.46%, respectively (Figure 2A-a). SWISS-MODEL was used to predict the three-dimensional structure of GlbHLH5 protein online. The three-dimensional structure was mainly α-helix and random coil, which was consistent with the predicted secondary structure of GlbHLH5 protein (Figure 2A-b). Subsequently, the complete coding sequence of GlbHLH5 was cloned into the pET-32a vector and transformed into DH5a competent cells. The recombinant plasmid pET-32a-GlbHLH5 was sequenced and extracted. Then, the recombinant plasmid pET-32a-GlbHLH5 was transformed into E. coli BL21 (DE3) competent cells. The empty vector pET-32a was used as the control. The recombinant cells were collected after IPTG induction for 12 h. After centrifugation, the cells were suspended in Tris-HCl for ultrasonic fragmentation and lysis. The supernatant was taken for SDS-PAGE detection after denaturation. The results showed that the pET-32a-GlbHLH5 strain inducted by IPTG for 12 h could all express the protein with a size of about 36.58 KDa (Figure 2B). With the increase of induction temperature, the protein expression levels firstly increased and then decreased, and the expression level of the target protein was the highest under the induction condition of 25°C.

Figure 2. Structure prediction and prokaryotic expression of GlbHLH5 protein. (A) Secondary (a) and three-dimensional (b) structure prediction of GlbHLH5 Protein. (B) Prokaryotic expression of GlbHLH5 Protein.
The complete coding sequence of GlbHLH5 gene was cloned into the pGBKT7 (pBD) vector using the yeast single hybridization system, and the constructed pBD-GlbHLH5 vector was identified by colonies PCR and sequencing. The sequenced recombinant plasmid was transformed into the yeast AH109 strain to observe its growth on the Kan-resistant plate. The results showed that pBD empty vector and pBD-GlbHLH5 transformants could grow well on SD/-Trp plates, while only pBD-GlbHLH5 could grow well on SD/-Trp/-His/-Ade solid medium supplemented with 3-AT (aminotriazole) (Figure 3). Because AT is His synthase inhibitor, it can reduce the leakage expression of His synthase in yeast AH109 strain and improve the selection conditions. The above results show that GlbHLH5 has strong transcriptional activation activity.
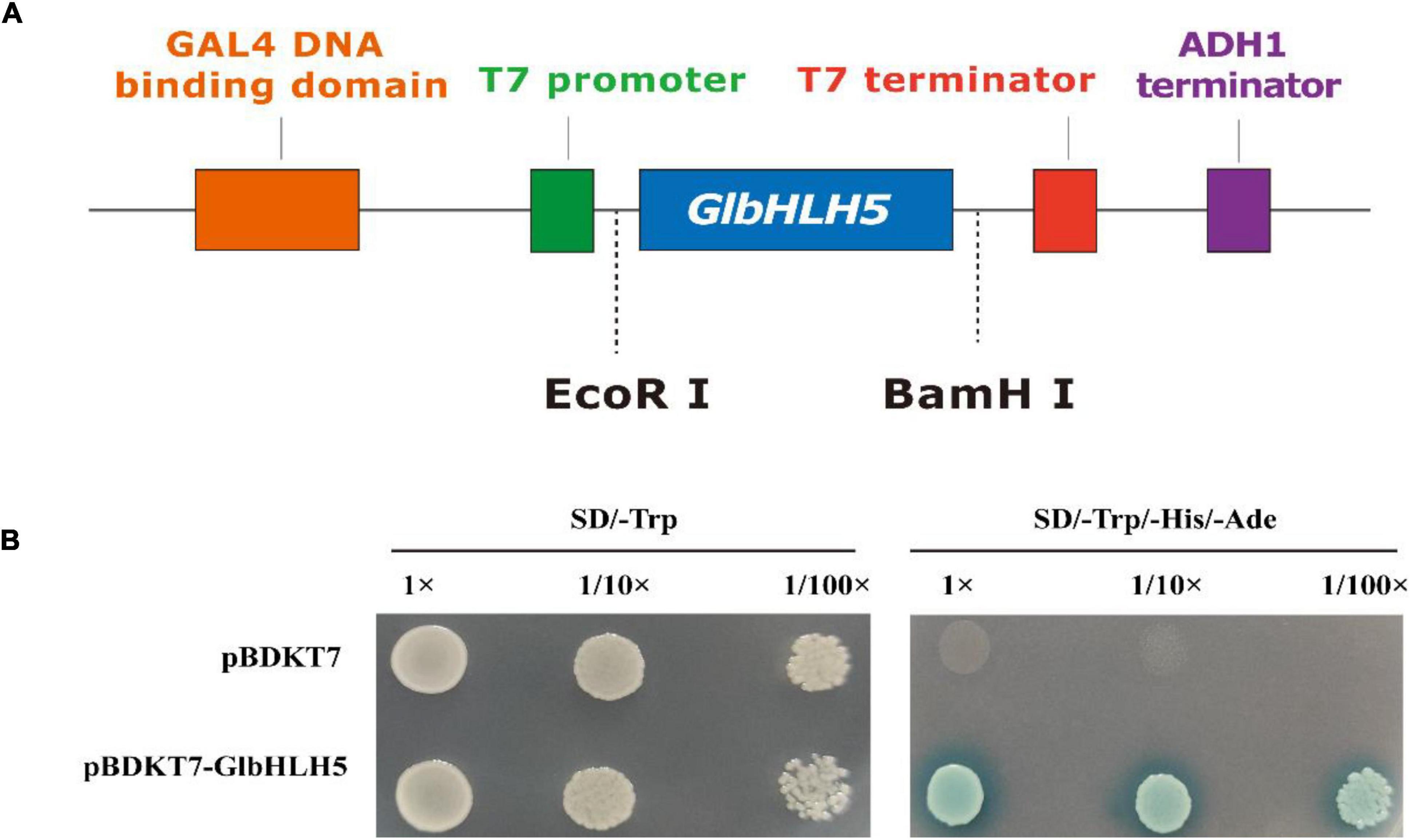
Figure 3. Transcriptional activation analysis of GlbHLH5. (A) The construction of the vector pGBKT7-GlbHLH5. (B) Transcription activation activity of GlbHLH5. 3-AT was added at a final concentration of 0.5 mmol L–1.
Compared with the control group, adding exogenous MeJA significantly increased the content of Ganoderma triterpenoids, and the difference was extremely significant after 12 h of induction (P < 0.01) (Figure 4A). To further clarify whether GlbHLH5 participates in the regulation of Ganoderma triterpenoids biosynthesis by responding to jasmonic acid signals, qRT-PCR was used to determine the response of GlbHLH5 and key genes HMGR, SQS, LS in the biosynthetic pathway of Ganoderma triterpenoids to MeJA induction. After 4 h of induction with 50 μmol L–1 MeJA, the expression of GlbHLH5 increased significantly (P < 0.01), which was twice that of the control group (Figure 4B); compared to the control group, the expression of HMGR increased significantly (P < 0.05) after 4 h of induction, and reached its peak at 12 h of induction (Figure 4C); after 6 h of induction, the expression of SQS was extremely significantly (P < 0.01) higher than the control group, the expression level reached its peak at 12 h after induction (Figure 4D); the expression of LS was extremely significantly (P < 0.01) higher than the control group after 4 h of induction (Figure 4E). To sum up, the relative expression levels of the three key genes in the triterpenoids biosynthetic pathway all reached their peak values at 12 h after MeJA induction, which was consistent with the induced expression pattern of GlbHLH5. At the same time, the correlation analysis showed that the expression of GlbHLH5 has a very significant (P < 0.01) positive correlation with the three key genes HMGR, SQS, and LS (Figure 4F). Promoter cis-acting elements analysis of the three key genes shown that they all contained multiple G-box, which is the binding site of bHLH transcription factors. It was suggested that GlbHLH5 transcription factor might be involved in triterpenoid biosynthesis in G. lucidum by regulating HMGR, SQS, and LS gene expressions. The regulatory effect of GlbHLH5 in Ganoderma triterpenoids biosynthesis will be further analyzed and verified by transgenic technology.
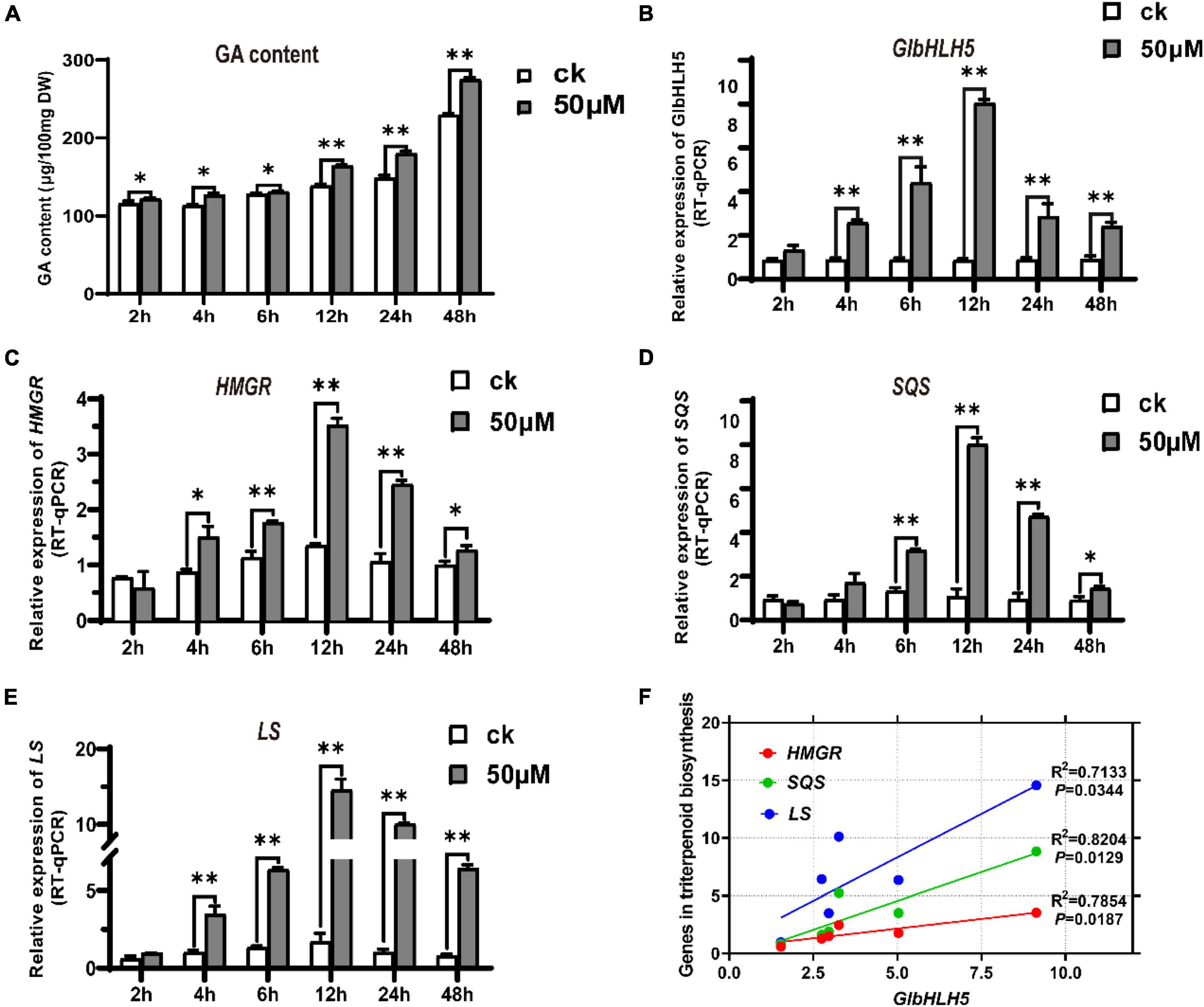
Figure 4. MeJA induced triterpenoids accumulate and expressions of GlbHLH5 along with key genes in the triterpenoids biosynthetic pathway in Ganoderma lucidum. (A) Changes of triterpenoids content induced by MeJA in G. lucidum. (B–E) Relative expression levels of GlbHLH5 and key genes in the triterpenoids biosynthetic pathway. (F) Correlation analysis between GlbHLH5 and key genes in triterpenoids biosynthesis pathway. “*” means the difference is significant (P < 0.05) and “**” means the difference is extremely significant (P < 0.01).
The online software WoLF PSORT was used to analyze GlbHLH5 transcription factor, and the prediction showed that it was mainly located in the nucleus. The fluorescent protein was observed by laser confocal microscope, and the pBARGPEI-GFP was used as the control group, and the pBARGPEI-GlbHLH5-GFP was used as the experimental group. The results showed that in the presence of Ca2+, the nucleus of pBARGPEI-GlbHLH5-GFP strain had obvious fluorescence, while that of WT strain had no fluorescence (Figure 5). It was indicated that GlbHLH5 transcription factor mainly located in the nucleus of G. lucidum, which was consistent with the predicted results, indicating that GlbHLH5 plays a role in the nucleus.
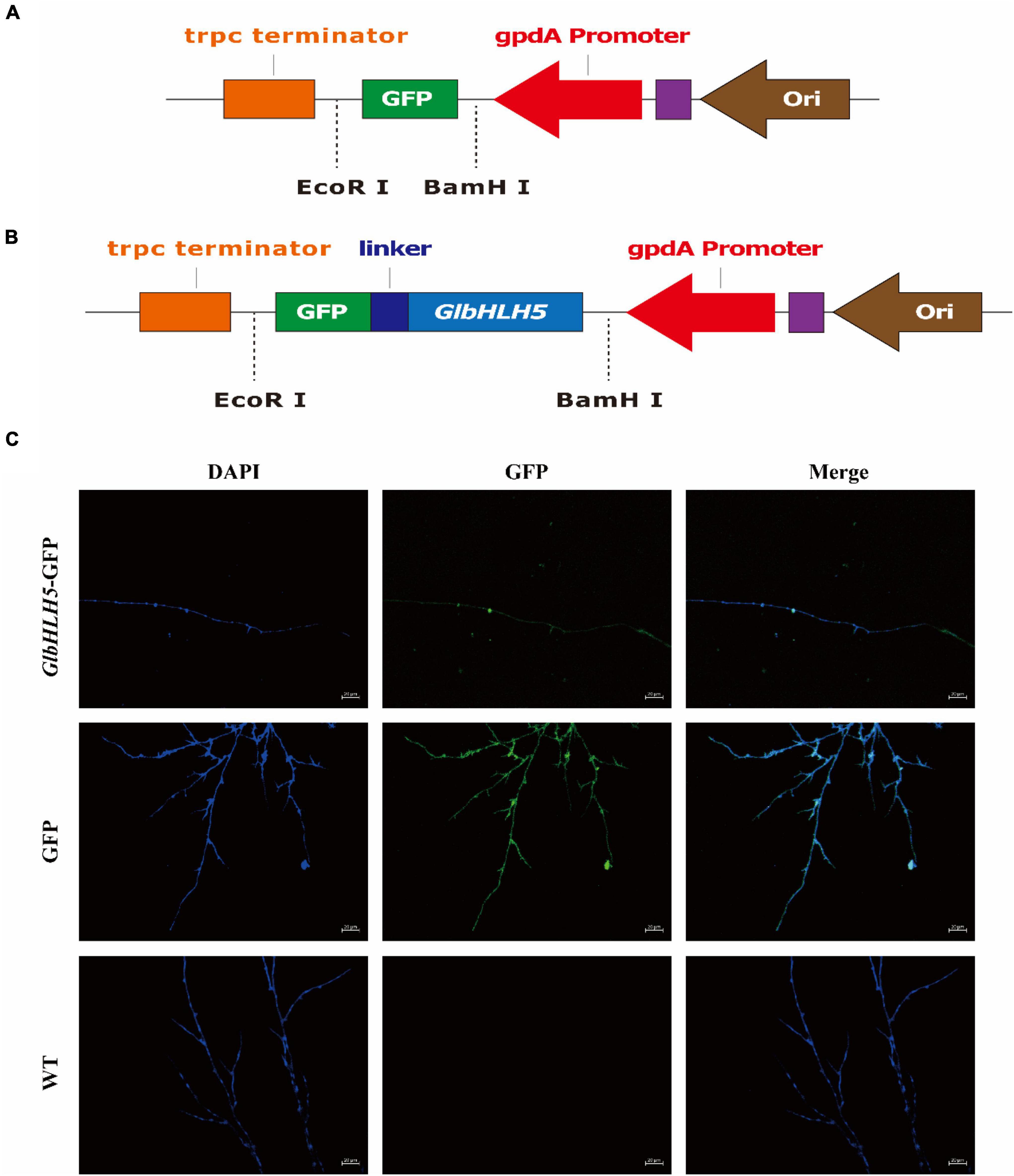
Figure 5. Subcellular localization analysis of GlbHLH5. (A) Construction of the vector pBARGPEI-GPF. (B) Construction of the vector pBARGPEI-GlbHLH5-GPF. (C) Subcellular localization of GlbHLH5 protein in Ganoderma lucidum. DAPI, DAPI nuclear staining; GFP, the field of view under green excitation fluorescence; Merge: the overlapping field of view of DAPI and GFP.
Transgenic Ganoderma lucidum strains overexpressing GlbHLH5 were generated by a PEG-mediated genetic transformation system. The overexpression lines were selected by hygromycin resistance. PCR amplification was also conducted to verify the positive transformants. A 580 bp gpd promoter of the vector was amplified in the GlbHLH5-overexpressed positive transformants but not in the wild strain. It was demonstrated in Figure 6A that GlbHLH5 overexpressed G. lucidum strains could grow well on the hygromycin plate. As for the biosynthesis of Ganoderma triterpenoids, it was shown that the color developing of three overexpression lines (OE-GlbHLH5-1, OE-GlbHLH5-2, and OE-GlbHLH5-3) was deeper than that of the wild type in Salkowski qualitative reaction, visually manifesting the triterpenoid content of the three overexpression lines might be higher than that of the wild type. Further quantitative analysis by the vanillin-perchloric acid chromogenic method revealed that the triterpenoid content of OE-GlbHLH5-1, OE-GlbHLH5-2, and OE-GlbHLH5-3 lines increased by 45, 37, and 21%, respectively (Figure 6B) compared with the wild type. qRT-PCR confirmed that much higher expression levels of GlbHLH5 were observed in the three overexpression lines. Moreover, the transcript levels of the key genes HMGR, SQS, and LS in the biosynthetic pathways of Ganoderma triterpenoids were all upregulated in GlbHLH5 overexpression lines compared with those in the wild type. As shown in Figure 6B, the relative expressions of HMGR, SQS, and LS reached the highest in line OE-GlbHLH5-1, which were 3.16, 3.23, and 2.62 fold of the wild type, respectively. Last but not the least, the variations of the pathway genes transcription were consistent with the accumulation of Ganoderma triterpenoids, and the expression levels of GlbHLH5 as well, indicating that overexpression of GlbHLH5 transcription factor could increase the triterpenoid content by activating the biosynthetic pathways of triterpenoids in G. lucidum.
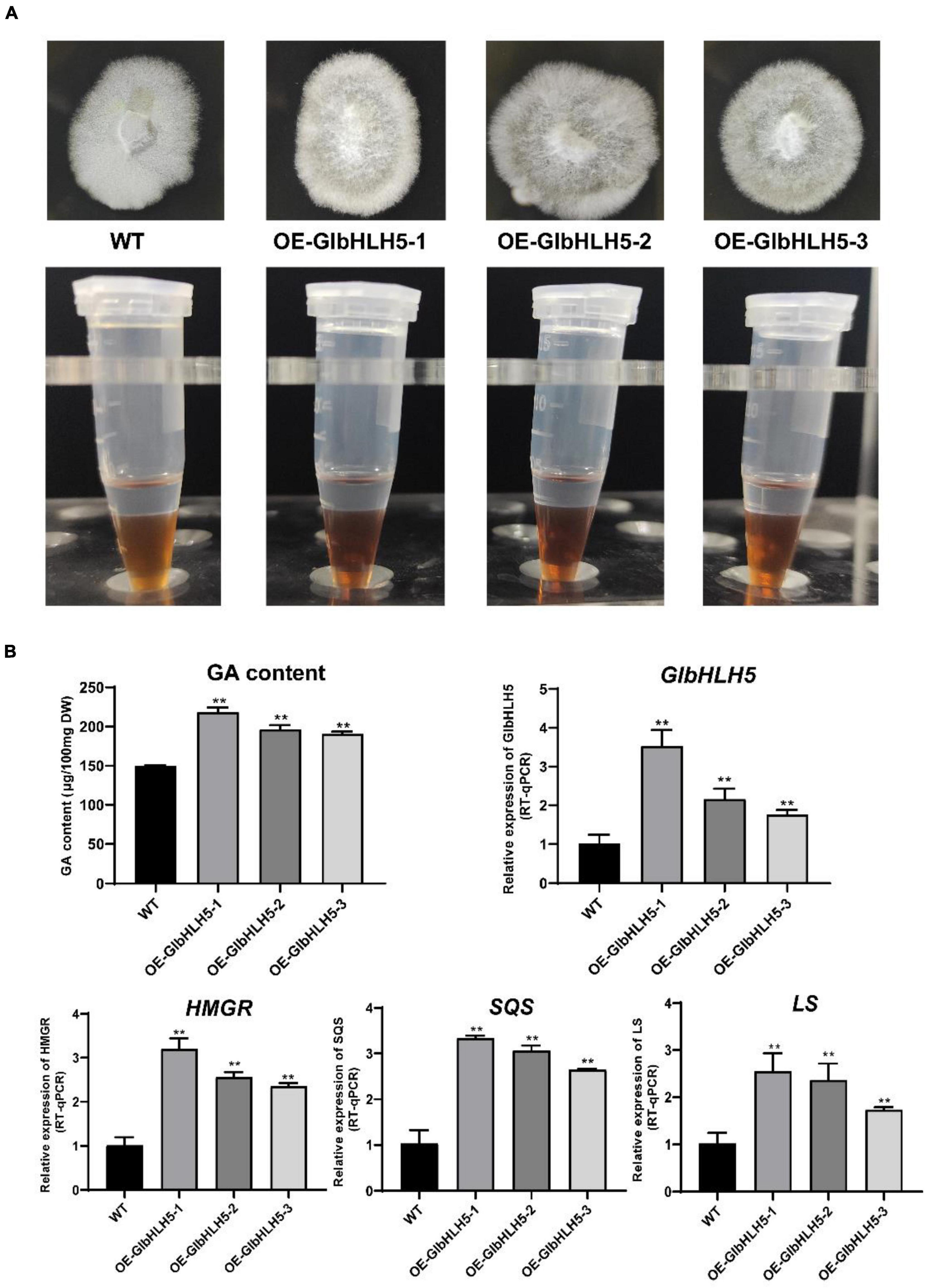
Figure 6. Phenotypic and relative gene expressions analysis of the GlbHLH5 overexpression strains. (A) The phenotypes of the three GlbHLH5 overexpression and the control lines of Ganoderma lucidum and the coloration differences between extracts of different lines. (B) The quantitative analysis of triterpenoids and the key genes transcriptions of the biosynthetic pathway in the GlbHLH5 overexpression and the control lines. “**” means the difference is extremely significant (P < 0.01).
Three stably inherited GlbHLH5 silenced strains were obtained through PEG-mediated multiple-resistance screening on Hyg plates, and the Hyg gene of the vector was amplified from the GlbHLH5-silenced positive transformants, yet not from the wild strain. As shown in Figure 6B, the relative expression levels of GlbHLH5 gene in the three selected GlbHLH5 silencing lines (Si-GlbHLH5-1, Si-GlbHLH5-2, and Si-GlbHLH5-3) were significantly lower than those in wild type (P < 0.05). The effect of silencing GlbHLH5 on the Ganoderma triterpenoids synthesis and the key genes expressions of biosynthetic pathway was completely opposite to that of GlbHLH5 overexpression. Compared with the wild type, the accumulation of Ganoderma triterpenoids decreased in GlbHLH5-silenced lines, as the Salkowski qualitative reaction showed that the color developing of the three silencing strains was shallower than that of the wild type (Figure 7A). Further quantitative analysis by the vanillin-perchloric acid chromogenic method showed that the triterpenoids content of Si-GlbHLH5-1, Si-GlbHLH5-2, and Si-GlbHLH5-3 decreased by 24, 31, and 19%, respectively. To further verify the function of GlbHLH5 on regulating Ganoderma triterpenoids synthesis, the key genes expressions of their biosynthetic pathways were determined by quantitative RT-PCR. It was revealed that silencing of GlbHLH5 gene leaded to down-regulation of the biosynthetic pathways of Ganoderma triterpenoids. The relative expression levels of HMGR, SQS, and LS genes in lines Si-GlbHLH5-1, Si-GlbHLH5-2 and Si-GlbHLH5-3 were all significantly lower than those in wild type (P < 0.01) (Figure 7B), and the variation in triterpenoids biosynthetic pathway gene transcription was generally consistent with GlbHLH5. The results showed that silencing GlbHLH5 transcription factor could reduce the biosynthesis of triterpenoids in G. lucidum, which further proved that GlbHLH5 transcription factor was a transcription factor involved in the regulation of Ganoderma triterpenoids biosynthesis.
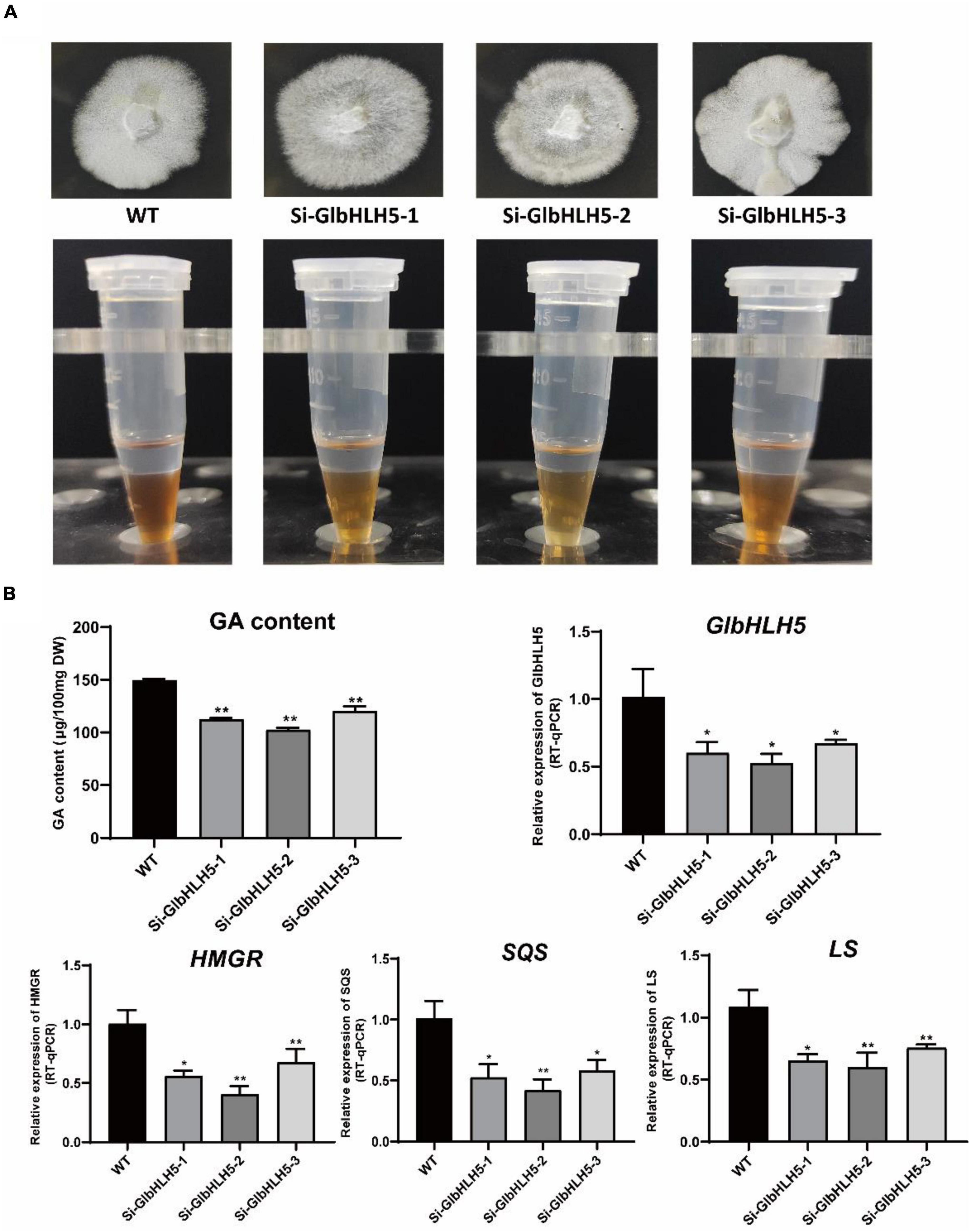
Figure 7. Phenotypic and gene expressions analysis of the GlbHLH5 silencing strains. (A) The phenotypes of the three GlbHLH5 silencing and the control lines of Ganoderma lucidum and the coloration differences between extracts of different lines. (B) The quantitative analysis of triterpenoids and the key genes transcriptions of the biosynthetic pathway in the GlbHLH5 silencing and the control lines. “*” means the difference is significant (P < 0.05) and “**” means the difference is extremely significant (P < 0.01).
The bHLH transcription factor family has many members and diverse functions. It is the second-largest transcription factor in plants (Carretero-Paulet et al., 2010). It plays an important regulatory role in plant growth and development (Verma et al., 2020), morphogenesis (Conway et al., 2010), stress resistance (Zhao et al., 2020), and secondary metabolism (Chezem and Clay, 2016). In the regulation of secondary metabolism, bHLH combines with the cis-acting elements in the promoter region of the target gene at the transcription level to activate the expression of the related genes. Its important role is to regulate the coordinated expression of multiple genes in the biosynthetic pathways of secondary metabolism, so as to globally activate secondary metabolic pathways and promote the biosynthesis of secondary metabolites (Zhou and Memelink, 2016). Studies have shown that bHLH is an important kind of transcription factor regulating terpenoid metabolism. AaTAR2 (Zhou et al., 2020), AaMYC2 (Chen et al., 2017), and AaDELLA (Shen et al., 2015) in Artemisia annua can promote the biosynthesis of artemisinin. The bHLH transcription factors BIS1 (Pan et al., 2018) and CrMYC2 (Zhang et al., 2011) in Catharanthus roseus regulate the synthesis of terpenoid alkaloids in response to the induction of MeJA. SmbHLH51 (Wu et al., 2018), R2R3-MYB (Li and Lu, 2014), and SmMYC2 (Yang et al., 2017) in Salvia can positively regulate the metabolism of tanshinone substances. Recently, an increasing number of bHLH proteins have been identified from diverse fungi, and they involve in a variety of biological processes, such as mycelial growth, cellular differentiation, sporulation, virulence, and secondary metabolism (Sailsbery et al., 2012), but bHLH family in G. lucidum are rarely studied. Whether Ganoderma bHLH can regulate the synthesis of G. lucidum triterpenoids is still unclear.
In this study, the GlbHLH5 gene was cloned and proved to contain the bHLH conserved domains and DNA binding sites (Figure 1A), which was basically consistent with the sequences reported by Chen S. et al. (2012). Phylogenetic tree analysis found that GlbHLH5 and many homologous proteins in Basidiomycota were clustered on the same phylogenetic branch except for Boletus edulis and Pleurotus ostreatus (Figure 1B), indicating that the Ganoderma bHLH transcription factors have species specificity.
MeJA can significantly induce the synthesis of Ganoderma lucidum triterpenoid but the mechanism of its transcriptional regulation remains unclear. Using qRT-PCR, we analyzed the expression patterns of GlbHLH5 and the three key genes of Ganoderma triterpenoid biosynthetic pathway, HMGR, SQS and LS, under the induction of MeJA. As shown in Figure 4, the expressions of GlbHLH5, HMGR, SQS, and LS were all significantly induced by MeJA, and the consistent induced expression pattern between GlbHLH5 and the three key genes of triterpenoids biosynthetic pathway indicated that GlbHLH5 may regulate the transcription of the key genes HMGR, SQS, and LS in the triterpenoid metabolism pathway to promote the biosynthesis of Ganoderma triterpenoids.
We further used transgenic technology to verify the regulatory effect of GlbHLH5, and found that the overexpression of GlbHLH5 in G. lucidum significantly increased the accumulation of Ganoderma triterpenoids and expressions of the key enzyme genes in the biosynthetic pathway of Ganoderma triterpenoids (Figure 6). On the contrary, in GlbHLH5-silenced G. lucidum by RNA interference, the triterpenoids accumulation along with the expression levels of key enzyme genes in the biosynthetic pathway were significantly and synchronously reduced (Figure 7). What’s more, the variations of the pathway genes transcription were consistent with the accumulation of Ganoderma triterpenoids, and the transcription of GlbHLH5 as well, indicating GlbHLH5 transcription factor is a positive regulator of triterpenoids biosynthesis in G. lucidum by activating the synergistic expression of key enzyme genes in the triterpenoid metabolism pathway, hereby a schematic of the predicted regulatory effect of GlbHLH5 in Ganoderma triterpenoids biosynthesis was illustrated in Figure 8. To sum up, GlbHLH5 is a bHLH transcription factor that positively regulates the biosynthesis of Ganoderma triterpenoids in response to JA, the function is similar to that of Salvia SmMYB1 (Zhou et al., 2021), SmbHLH53 (Peng et al., 2020), Catharanthus BIS1 (Singh et al., 2021), CrMYC2 (Hedhili et al., 2010), etc. It is consistent that multiple bHLH transcription factors regulate terpenoid synthesis in response to JA signals.
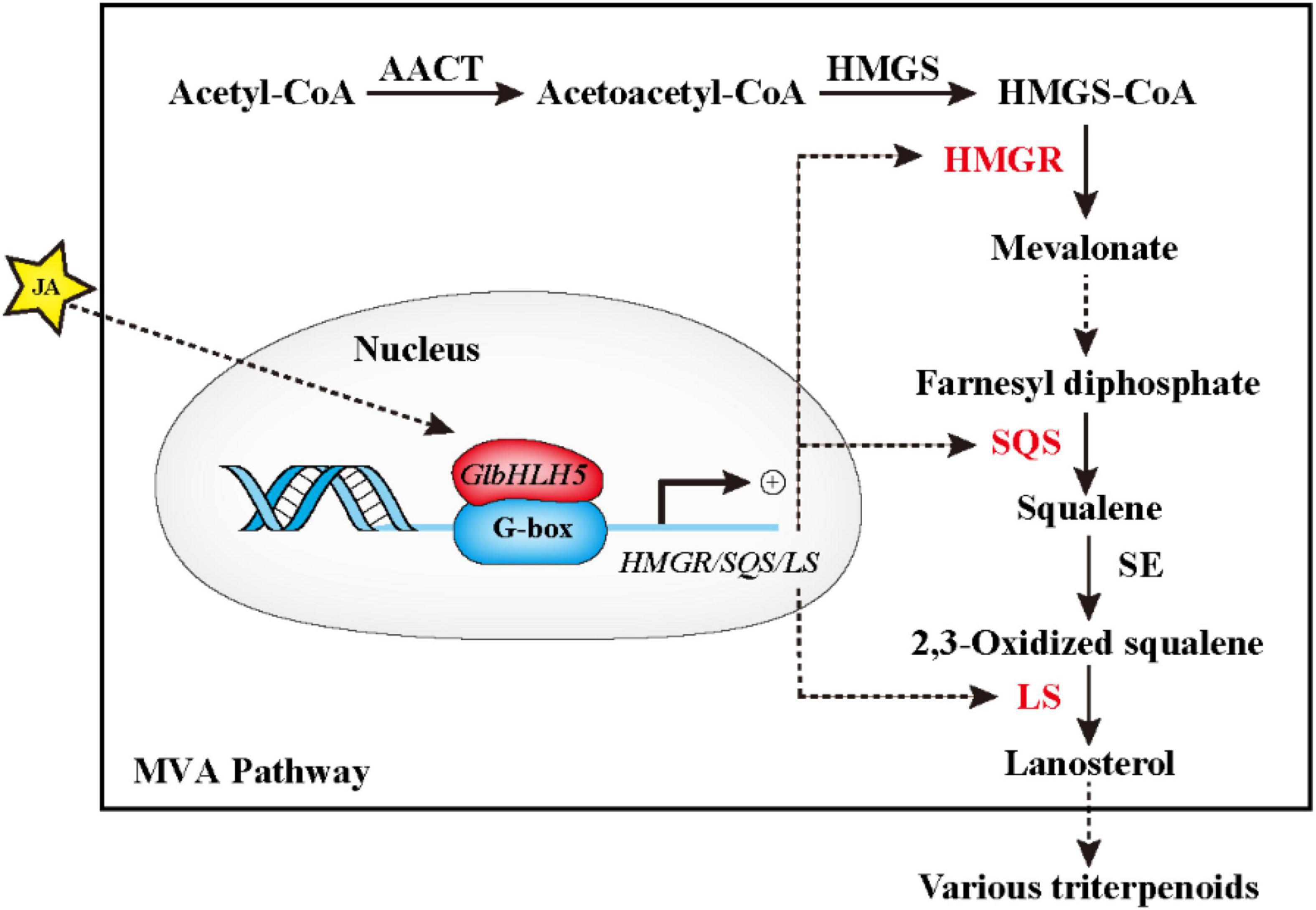
Figure 8. Schematic of the predicted regulatory effect of GlbHLH5 in Ganoderma triterpenoids biosynthesis. The solid box represents the main pathway (MVA pathway) of Ganoderma triterpenoids. AACT, acetyl-CoA acetyltransferase; HMGS, 3-hydroxy-3-methylglutaryl-CoA synthase; HMGR, 3-hydroxy-3-methylglutaryl-CoA reductase, SQS, squalene synthase; SE, squalene monooxygenase; LS, lanosterol synthase.
In this study, the gene of the bHLH transcription factor GlbHLH5 that regulates the biosynthesis of Ganoderma triterpenoids was cloned for the first time. The gene and protein sequence, transcription activation activity and subcellular location were systematically analyzed. Its regulation of triterpenoids synthesis was further confirmed by genetic transformation. It has been shown that GlbHLH5 is a bHLH transcription factor which could positively regulate Ganoderma triterpenoids synthesis by responding to JA signal. It lays the foundation for further exploration of the molecular mechanism of metabolism regulation by GlbHLH5 transcription factor.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
JX performed a part of experiments, designed the experiments and technical route, and mainly revised the manuscript. YW performed the majority of experiments, data analysis, and drafted the manuscript. SR and XY were mainly responsible for the revision of manuscripts. YZ and KX participated in a part of experiments and helped to collect samples. XW conceived the project, revised the manuscript, and provided final approval of the manuscript. All authors read and approved the final manuscript.
This work was supported by the Natural Science Foundation of Zhejiang Province (LQ19C010004) and School Research and Development Fund Project (2016FR010).
KX was employed by Zhejiang Wuyangtang Pharmaceutical Co., Ltd.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.878110/full#supplementary-material
Carretero-Paulet, L., Galstyan, A., Roig-Villanova, I., Martinez-Garcia, J. F., Bilbao-Castro, J. R., and Robertson, D. L. (2010). Genome-wide classification and evolutionary analysis of the bHLH family of transcription factors in Arabidopsis, poplar, rice, moss, and algae. Plant Physiol. 153, 1398–1412. doi: 10.1104/pp.110.153593
Chen, M., Yan, T., Shen, Q., Lu, X., Pan, Q., Huang, Y., et al. (2017). GLANDULAR TRICHOME-SPECIFIC WRKY 1 promotes artemisinin biosynthesis in Artemisia annua. New Phytol. 214, 304–316. doi: 10.1111/nph.14373
Chen, S., Xu, J., Liu, C., Zhu, Y., Nelson, D. R., Zhou, S., et al. (2012). Genome sequence of the model medicinal mushroom Ganoderma lucidum. Nat. Commun. 3:913. doi: 10.1038/ncomms1923
Chen, X., Pei, Z., Peng, L., Qin, Q., Duan, Y., Liu, H., et al. (2021). Genome-Wide identification and functional characterization of CCHC-Type zinc finger genes in ustilaginoidea virens. J. Fungi. 7:947. doi: 10.3390/jof7110947
Chen, Y., Bicker, W., Wu, J., Xie, M., and Lindner, W. (2012). Simultaneous determination of 16 nucleosides and nucleobases by hydrophilic interaction chromatography and its application to the quality evaluation of Ganoderma. J. Agric. Food Chem. 60, 4243–4252. doi: 10.1021/jf300076j
Chen, Z., Xu, J., Xu, H., Li, S., Li, C., Wang, Q., et al. (2021). Effect of continuous cropping soil on the biosynthesis of bioactive compounds and the antioxidant activity of lingzhi or reishi medicinal mushroom, ganoderma lucidum (Agaricomycetes). Int. J. Med. Mushr. 23, 25–37. doi: 10.1615/IntJMedMushrooms.2021039536
Chezem, W. R., and Clay, N. K. (2016). Regulation of plant secondary metabolism and associated specialized cell development by MYBs and bHLHs. Phytochemistry 131, 26–43. doi: 10.1016/j.phytochem.2016.08.006
Conway, S. J., Firulli, B., and Firulli, A. B. (2010). A bHLH code for cardiac morphogenesis. Pediatr. Cardiol. 31, 318–324. doi: 10.1007/s00246-009-9608-x
Dhandapani, S., Tjhang, J. G., and Jang, I. C. (2020). Production of multiple terpenes of different chain lengths by subcellular targeting of multi-substrate terpene synthase in plants. Metab. Eng. 61, 397–405. doi: 10.1016/j.ymben.2020.08.002
Di, P., Zhang, L., Chen, J., Tan, H., Xiao, Y., Dong, X., et al. (2013). (1)(3)C tracer reveals phenolic acids biosynthesis in hairy root cultures of Salvia miltiorrhiza. ACS Chem. Biol. 8, 1537–1548. doi: 10.1021/cb3006962
Finn, R. D., Coggill, P., Eberhardt, R. Y., Eddy, S. R., Mistry, J., Mitchell, A. L., et al. (2016). The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. doi: 10.1093/nar/gkv1344
Fu, X., Peng, B., Hassani, D., Xie, L., Liu, H., Li, Y., et al. (2021). AaWRKY9 contributes to light- and jasmonate-mediated to regulate the biosynthesis of artemisinin in Artemisia annua. New Phytol. 231, 1858–1874. doi: 10.1111/nph.17453
Gharari, Z., Bagheri, K., Danafar, H., and Sharafi, A. (2020). Enhanced flavonoid production in hairy root cultures of Scutellaria bornmuelleri by elicitor induced over-expression of MYB7 and FNScapital PE, Cyrillic2 genes. Plant Physiol. Biochem. 148, 35–44. doi: 10.1016/j.plaphy.2020.01.002
Grienke, U., Mihaly-Bison, J., Schuster, D., Afonyushkin, T., Binder, M., Guan, S. H., et al. (2011). Pharmacophore-based discovery of FXR-agonists. Part II: identification of bioactive triterpenes from Ganoderma lucidum. Bioorg. Med. Chem. 19, 6779–6791. doi: 10.1016/j.bmc.2011.09.039
He, J., Sun, Y., Jia, Y., Geng, X., Chen, R., Zhou, H., et al. (2019). Ganoderma triterpenes protect against hyperhomocysteinemia induced endothelial-mesenchymal transition via TGF-beta signaling inhibition. Front. Physiol. 10:192. doi: 10.3389/fphys.2019.00192
Hedhili, S., De Mattei, M. V., Coudert, Y., Bourrie, I., Bigot, Y., and Gantet, P. (2010). Three non-autonomous signals collaborate for nuclear targeting of CrMYC2, a Catharanthus roseus bHLH transcription factor. BMC Res. Notes 3:301. doi: 10.1186/1756-0500-3-301
Hu, S., Huang, J., Pei, S., Ouyang, Y., Ding, Y., Jiang, L., et al. (2019). Ganoderma lucidum polysaccharide inhibits UVB-induced melanogenesis by antagonizing cAMP/PKA and ROS/MAPK signaling pathways. J. Cell Physiol. 234, 7330–7340. doi: 10.1002/jcp.27492
Hu, Z., Du, R., Xiu, L., Bian, Z., Ma, C., Sato, N., et al. (2020). Protective effect of triterpenes of Ganoderma lucidum on lipopolysaccharide-induced inflammatory responses and acute liver injury. Cytokine 127:154917. doi: 10.1016/j.cyto.2019.154917
Li, C., and Lu, S. (2014). Genome-wide characterization and comparative analysis of R2R3-MYB transcription factors shows the complexity of MYB-associated regulatory networks in Salvia miltiorrhiza. BMC Genomics 15:277. doi: 10.1186/1471-2164-15-277
Li, L. D., Mao, P. W., Shao, K. D., Bai, X. H., and Zhou, X. W. (2019a). Ganoderma proteins and their potential applications in cosmetics. Appl. Microbiol. Biotechnol. 103, 9239–9250. doi: 10.1007/s00253-019-10171-z
Li, L., Hao, X., Liu, H., Wang, W., Fu, X., Ma, Y., et al. (2019b). Jasmonic acid-responsive AabHLH1 positively regulates artemisinin biosynthesis in Artemisia annua. Biotechnol. Appl. Biochem. 66, 369–375. doi: 10.1002/bab.1733
Li, X., Ren, Y., Jing, J., Jiang, Y., Yang, Q., Luo, S., et al. (2021). The inhibitory mechanism of methyl jasmonate on Aspergillus flavus growth and aflatoxin biosynthesis and two novel transcription factors are involved in this action. Food Res. Int. 140:110051. doi: 10.1016/j.foodres.2020.110051
Liu, Z., and Friesen, T. L. (2012). Polyethylene glycol (PEG)-mediated transformation in filamentous fungal pathogens. Methods Mol. Biol. 835, 365–375. doi: 10.1007/978-1-61779-501-5_21
Niego, A. G., Rapior, S., Thongklang, N., Raspe, O., Jaidee, W., Lumyong, S., et al. (2021). Macrofungi as a nutraceutical source: promising bioactive compounds and market value. J. Fungi. 7:397. doi: 10.3390/jof7050397
Pan, Y. J., Lin, Y. C., Yu, B. F., Zu, Y. G., Yu, F., and Tang, Z. H. (2018). Transcriptomics comparison reveals the diversity of ethylene and methyl-jasmonate in roles of TIA metabolism in Catharanthus roseus. BMC Genomics 19:508. doi: 10.1186/s12864-018-4879-3
Peng, J. J., Wu, Y. C., Wang, S. Q., Niu, J. F., and Cao, X. Y. (2020). SmbHLH53 is relevant to jasmonate signaling and plays dual roles in regulating the genes for enzymes in the pathway for salvianolic acid B biosynthesis in Salvia miltiorrhiza. Gene 756:144920. doi: 10.1016/j.gene.2020.144920
Qian, J., Xu, H., Song, J., Xu, J., Zhu, Y., and Chen, S. (2013). Genome-wide analysis of simple sequence repeats in the model medicinal mushroom Ganoderma lucidum. Gene 512, 331–336. doi: 10.1016/j.gene.2012.09.127
Ren, A., Qin, L., Shi, L., Dong, X. Mu da, S., Li, Y. X., et al. (2010). Methyl jasmonate induces ganoderic acid biosynthesis in the basidiomycetous fungus Ganoderma lucidum. Bioresour. Technol. 101, 6785–6790. doi: 10.1016/j.biortech.2010.03.118
Sailsbery, J. K., Atchley, W. R., and Dean, R. A. (2012). Phylogenetic analysis and classification of the fungal bHLH domain. Mol. Biol. Evol. 29, 1301–1318. doi: 10.1093/molbev/msr288
Shang, C. H., Zhu, F., Li, N. Ou-Yang, X., Shi, L., Zhao, M. W., et al. (2008). Cloning and characterization of a gene encoding HMG-CoA reductase from Ganoderma lucidum and its functional identification in yeast. Biosci. Biotechnol. Biochem. 72, 1333–1339. doi: 10.1271/bbb.80011
Shang, C. H., Shi, L., Ren, A., Qin, L., and Zhao, M. W. (2010). Molecular cloning, characterization, and differential expression of a lanosterol synthase gene from Ganoderma lucidum. Biosci. Biotechnol. Biochem. 74, 974–978. doi: 10.1271/bbb.90833
Shen, Q., Cui, J., Fu, X. Q., Yan, T. X., and Tang, K. X. (2015). Cloning and characterization of DELLA genes in Artemisia annua. Genet. Mol. Res. 14, 10037–10049. doi: 10.4238/2015.August.21.10
Shi, L., Yue, S., Gao, T., Zhu, J., Ren, A., Yu, H., et al. (2020). Nitrate reductase-dependent nitric oxide plays a key role on MeJA-induced ganoderic acid biosynthesis in Ganoderma lucidum. Appl. Microbiol. Biotechnol. 104, 10737–10753. doi: 10.1007/s00253-020-10951-y
Singh, S. K., Patra, B., Paul, P., Liu, Y., Pattanaik, S., and Yuan, L. (2021). BHLH IRIDOID SYNTHESIS 3 is a member of a bHLH gene cluster regulating terpenoid indole alkaloid biosynthesis in Catharanthus roseus. Plant Direct. 5:e305. doi: 10.1002/pld3.305
Song, D., Cao, Y., and Xia, Y. (2021). Transcription factor MaMsn2 regulates conidiation pattern shift under the control of MaH1 through homeobox domain in metarhizium acridum. J. Fungi. 7:840. doi: 10.3390/jof7100840
Verma, D., Jalmi, S. K., Bhagat, P. K., Verma, N., and Sinha, A. K. (2020). A bHLH transcription factor, MYC2, imparts salt intolerance by regulating proline biosynthesis in Arabidopsis. FEBS J. 287, 2560–2576. doi: 10.1111/febs.15157
Wang, X., Pereira, J. H., Tsutakawa, S., Fang, X., Adams, P. D., Mukhopadhyay, A., et al. (2021). Efficient production of oxidized terpenoids via engineering fusion proteins of terpene synthase and cytochrome P450. Metab. Eng. 64, 41–51. doi: 10.1016/j.ymben.2021.01.004
Wu, Y., Zhang, Y., Li, L., Guo, X., Wang, B., Cao, X., et al. (2018). AtPAP1 interacts with and activates SmbHLH51, a positive regulator to phenolic acids biosynthesis in salvia miltiorrhiza. Front. Plant Sci. 9:1687. doi: 10.3389/fpls.2018.01687
Xu, J., Xiao, C., Xu, H., Yang, S., Chen, Z., Wang, H., et al. (2021). Anti-inflammatory effects of Ganoderma lucidum sterols via attenuation of the p38 MAPK and NF-kappaB pathways in LPS-induced RAW 264.7 macrophages. Food Chem. Toxicol. 150:112073. doi: 10.1016/j.fct.2021.112073
Yang, N., Zhou, W., Su, J., Wang, X., Li, L., Wang, L., et al. (2017). Overexpression of SmMYC2 increases the production of phenolic acids in salvia miltiorrhiza. Front. Plant Sci. 8:1804. doi: 10.3389/fpls.2017.01804
Yao, C., Wang, Z., Jiang, H., Yan, R., Huang, Q., Wang, Y., et al. (2021). Ganoderma lucidum promotes sleep through a gut microbiota-dependent and serotonin-involved pathway in mice. Sci. Rep. 11:13660. doi: 10.1038/s41598-021-92913-6
Zhang, H., Hedhili, S., Montiel, G., Zhang, Y., Chatel, G., Pre, M., et al. (2011). The basic helix-loop-helix transcription factor CrMYC2 controls the jasmonate-responsive expression of the ORCA genes that regulate alkaloid biosynthesis in Catharanthus roseus. Plant J. 67, 61–71. doi: 10.1111/j.1365-313X.2011.04575.x
Zhao, C., Zhang, C., Xing, Z., Ahmad, Z., Li, J. S., and Chang, M. W. (2019). Pharmacological effects of natural Ganoderma and its extracts on neurological diseases: a comprehensive review. Int. J. Biol. Macromol. 121, 1160–1178. doi: 10.1016/j.ijbiomac.2018.10.076
Zhao, H., Zhang, Q., Zhao, L., Huang, X., Wang, J., and Kang, X. (2012). Spore powder of ganoderma lucidum improves Cancer-Related fatigue in breast cancer patients undergoing endocrine therapy: a pilot clinical trial. Evid. Based Compl. Altern. Med. 2012:809614. doi: 10.1155/2012/809614
Zhao, M. W., Liang, W. Q., Zhang, D. B., Wang, N., Wang, C. G., and Pan, Y. J. (2007). Cloning and characterization of squalene synthase (SQS) gene from Ganoderma lucidum. J. Microbiol. Biotechnol. 17, 1106–1112.
Zhao, Q., Fan, Z., Qiu, L., Che, Q., Wang, T., Li, Y., et al. (2020). MdbHLH130, an apple bHLH transcription factor, confers water stress resistance by regulating stomatal closure and ROS homeostasis in transgenic tobacco. Front. Plant Sci. 11:543696. doi: 10.3389/fpls.2020.543696
Zhou, J., Ji, S., Ren, M., He, Y., Jing, X., and Xu, J. (2014). Enhanced accumulation of individual ganoderic acids in a submerged culture of Ganoderma lucidum by the overexpression of squalene synthase gene. Biochem. Eng. J. 90, 178–183. doi: 10.1016/j.bej.2014.06.008
Zhou, M., and Memelink, J. (2016). Jasmonate-responsive transcription factors regulating plant secondary metabolism. Biotechnol. Adv. 34, 441–449. doi: 10.1016/j.biotechadv.2016.02.004
Zhou, W., Shi, M., Deng, C., Lu, S., Huang, F., Wang, Y., et al. (2021). The methyl jasmonate-responsive transcription factor SmMYB1 promotes phenolic acid biosynthesis in Salvia miltiorrhiza. Hortic. Res. 8:10. doi: 10.1038/s41438-020-00443-5
Keywords: GlbHLH5 transcription factor, positive transcriptional regulator, Ganoderma triterpenoids, overexpression, RNA interference
Citation: Xu J, Wang YY, Zhang Y, Xiong KH, Yan XY, Ruan SY and Wu XQ (2022) Identification of a Novel Metabolic Target for Bioactive Triterpenoids Biosynthesis in Ganoderma lucidum. Front. Microbiol. 13:878110. doi: 10.3389/fmicb.2022.878110
Received: 17 February 2022; Accepted: 29 March 2022;
Published: 09 May 2022.
Edited by:
Dipesh Dhakal, University of Florida, United StatesReviewed by:
Fangcheng Bi, Guangdong Academy of Agricultural Sciences, ChinaCopyright © 2022 Xu, Wang, Zhang, Xiong, Yan, Ruan and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xueqian Wu, d3hxQHphZnUuZWR1LmNu
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.