- 1Institute for Advanced Study of Coastal Ecology, Lu Dong University, Yantai, China
- 2Yantai Institute of Coastal Zone Research, Chinese Academy of Sciences, Yantai, China
- 3Laboratory for Marine Ecology and Environmental Science, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
- 4Key Laboratory of Marine Ecology and Environmental Sciences, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
Although methylmercury (MeHg) has been recognized as a typical heavy metal posing huge damages to various life processes of fish, the response mechanisms of marine fish at early life stages (ELSs) to MeHg is still poorly understood. In this study, non-targeted liquid chromatography-mass spectrometry (LC-MS) based metabolomic and transcriptomic approaches were used to explore response mechanisms of juvenile flounder (Paralichthys olivaceus) to long-term sublethal MeHg exposure (0 and 1.0 μg L-1; 30 d). After exposure, growth parameters of flounder were significantly decreased. Metabolomic and transcriptomic analyses of liver tissue showed obvious difference about biological pathways and identified biomarkers (around 2502 genes and 16 secondary metabolites). Those significantly differentially expressed genes (DEGs) and their enriched pathways were mainly related to immune response, oxidative stress, lipids metabolism, glycometabolism, amino acid and nucleotide metabolism and regulation of protein processes, while those identified secondary metabolites were mainly enriched in tryptophan metabolism, biosynthesis of unsaturated fatty acids, linoleic acid metabolism and glutathione metabolism. Additionally, multi-omic method was used to explore response mechanisms of key pathways under MeHg stress. In this regard, only 57 DEGs and 6 secondary metabolites were significantly enriched in 7 pathways to constitute an integrated regulatory network, including glutathione metabolism, thyroid hormone synthesis, linoleic acid metabolism, biosynthesis of unsaturated fatty acids, tryptophan metabolism pathway, serotonergic synapse and African trypanosomiasis. Above all, we could speculate that antioxidative function, lipids metabolism, nervous system and amino acid metabolism were the more sensitive targets in response to MeHg stress, which were conductive to deeply understand the response mechanisms of fish at ELSs under MeHg exposure. Those identified biomarkers could also be widely used for toxicological studies of pollutants and ecological risks monitoring.
1 Introduction
Mercury (Hg) poses great threats to ecological environment and human health. Generally speaking, Hg compounds in ecosystems are mainly coming from anthropogenic sources, such as industrial emissions, mining, wastes combustion and agricultural materials (Ren et al., 2019b). Once entered into aquatic ecosystems, Hg could be continuously transformed into methylmercury (MeHg) through methylation process by methylated microorganisms (Yuan et al., 2019). Although MeHg concentration in seawater (0.02-0.625 ng L-1) or sediments (0.007-45.1 ng g-1 dw) are at a relative low level, total Hg (THg) level in seawater (0.25-2500 ng L-1) or sediments (2.7-4700 ng g-1 dw) are more higher than MeHg (Cao et al., 2020). Additionally, absorbable MeHg can be easily taken up by aquatic organisms, then transferred along food webs, finally biomagnified to reach a high level in high trophic organisms (especially predatory fish species), with 104-107 times of MeHg in fish tissues than seawater (Stein et al., 1996). As a widely known neurotoxin and endocrine disruptor, MeHg could cause damages to cellular macromolecules, antioxidant systems, immune function, nervous or endocrine systems and many other biological processes, even important life processes of growth, development and reproduction, which have been elaborated in various literatures (Ceccatelli et al., 2010; Fretham et al., 2012; dos Santos et al., 2018; Wu et al., 2018) and our previous studies (Ren et al., 2019a; Ren et al., 2019b; Ren et al., 2020). Up to now, researchers have taken a lot of efforts to explore the response mechanisms of tested animinals especially freshwater fish to MeHg, which had not been quite mastered yet (Brandão et al., 2015; De Marco et al., 2022). Additionally, marine fish at early life stages (ELSs) are more sensitive to various pollutants (Ren et al., 2019b; Ren et al., 2020), which can pose great insights into the response mechanisms of marine fish to MeHg. Meanwhile, laboratory studies about MeHg toxicity for fish have set higher exposure dose (over 10 μg L-1) than that in seawater, which could not reflect true physiological responses of fish in natural aquatic ecosystems. Hence, it is necessary to investigate the regulatory mechanisms of marine fish at ELSs in response to MeHg stress.
With the rapid development of next-generation sequencing technology, omics or multi-omic analyses are widely used in revealing response mechanisms of fish to Hg compounds (especially MeHg). For example, transcriptome analyses of Atlantic cod (Gadus morhua) after MeHg exposure showed that oxidative stress pathways related genes were significantly enriched, and there were disproportionate numbers of genes coding for enzymes involved in metabolism of amino acids, fatty acids and glucose (Yadetie et al., 2013). Furthermore, significantly differentially expressed genes (DEGs) in tusk (Brosme brosme) induced by MeHg were primarily related to protein folding, adipogenesis, notch signaling and lipid metabolism (Olsvik et al., 2021). As for the metabolomic analyses of MeHg-exposed fish, Zhu et al. (2019) found metabolites related to disturbance of tyrosine metabolism, starch and sucrose metabolism, neurotransmitters and oxidative stress were significantly enriched. MeHg had adverse impacts on membrane stabilization/degradation/repair processes, osmoregulation, energy metabolism, gene expression and antioxidant protection of golden grey mullet (Liza aurata) in contaminated wild area (Brandão et al., 2015). Additionally, disturbances in ion osmoregulatory processes were highlighted in gills tissue, whereas differential impairments to energy-producing metabolic pathways, protein catabolism, membrane stabilization processes and alteration of antioxidant defense system were also found in Chelon auratus (De Marco et al., 2022). Moreover, multi-omic techniques had also been used for toxicological research to discuss the complex response mechanisms of tested organisms to different pollutants (Huang et al., 2017; Ortiz-Villanueva et al., 2017; Teng et al., 2018; Pang et al., 2021; Wang et al., 2022).
Flounder (Paralichthys olivaceus) is an important fishery species, widely distributed in the coastal waters of East Asian countries, while its resource declines seriously in the last few decades, with environmental pollution (especially heavy metals) as an important reason. Our recent studies demonstrated waterborne MeHg exposure could pose stress on antioxidant defenses, immune function and many other life processes of flounder, and flounder at ELSs were proved to be sensitive to MeHg. For example, exposure to more than 13 μg L-1, increased morphological deformities and mortality and reduced growth and absorption rate of the embryo-larval flounder (Ren et al., 2019a). After 35 d exposure of MeHg (0-10.0 μg L-1) for larval flounder, the activities of glutathione peroxidase, glutathione reductase and catalase as well as the expression of antioxidant-related genes (gpx, gr and cat) were significantly induced or up-regulation to defense against increased lipid peroxidation level. Additionally, the immune response of larval flounder to MeHg exposure was also active with the activities of lysozyme and immunoglobulin M or their related genes showing opposite trends (Ren et al., 2019b). Moreover, juvenile flounder exerted tissue-specific antioxidant defenses exposed 0-20.0 μg L-1 MeHg exposure (Ren et al., 2020). However, the response mechanisms under these antioxidant defenses and immune responses or other developmental systems were still need to be deeply study. Herein, transcriptomic and metabolomic techniques were performed to excavate the DEGs, changed secondary metabolites and their enriched pathways of juvenile flounder in response to MeHg, finally revealing the response mechanisms to MeHg.
2 Materials and methods
2.1 Fish maintenance and test protocol
About 100 flounder juveniles, which were healthy and uniform in size, were obtained from the Yellow Sea Fishery Station of Shandong Province in China. Before MeHg exposure, these juveniles were acclimated in a flow-through pond (2 m3) with filtered seawater for one week. The primary chemical parameters of seawater were pH 7.98 ± 0.13, salinity 34.0 ± 0.0 psu, dissolved oxygen 7.0 ± 0.4, alkalinity 2.23 ± 0.01 mmol/dm3 and water temperature 19.17 ± 1.53°C, with a light regime of 12L:12D. Fish were fed 5% of the body weight for three times per day with commercial pellets (Santong Central Feed Co. LTD, Anqiu, China). During this period, faces and uneaten feed were removed from the tanks at 8:00 AM per day before the first feeding through circulating water device. Aeration was performed gently by a continuous air-bubbling system.
After acclimation, 15 juveniles of similar size were transferred into each of the experimental tanks (300L, polyethylene), and totally 20 individuals in all the tanks were selected randomly for the measurement of fish length and wet weight respectively, with mean values of them defined as initial length of L0 (18.8 ± 0.44 cm) and initial weight of W0 (95.0 ± 0.77 g). The fish were acclimated for 24 h in these tanks without feeding. Then, the fish were exposed to either a blank control (0 μg L-1) or an MeHg solution of 1.0 μg L-1 with three replicates for each group. Overall, the 1.0 μg L-1 MeHg concentration were verified to be sublethal to flounder at ELSs, while exerted adverse effects to sophisticated life processes of flouner (Ren et al., 2019a; Ren et al., 2019b; Ren et al., 2020). Thus, we set this concentration as the exposre dose to explore the response mechanisms of flounder at ELSs under sublethal MeHg exposure via. Each tank was filled with 200 L of test solution. The analytical reagent CH3ClHg (CAS No: 115-09-3; purity ≥ 99.5%; Sigma-Aldrich Chemical Co., USA) was dissolved in deionized water to obtain a stock solution (10.0 mg L-1), which were diluted in 200 L filtered seawater (the water quality was the same as those in the acclimation period) to produce desired MeHg concentrations (1.0 μg L-1) for each experimental tank. Those tanks in the control group were supplied with 200 L of uncontaminated seawater, the same as those in acclimation period. The seawater in the control group and the solution in the MeHg group were renewed entirely per day with a freshly prepared solution of the same MeHg concentration. During exposure (30 d), seawater and feeding management were identical to those used in fish acclimation.
2.2 Sampling and fish growth determination
At the end of the test, 8 juveniles from each experimental tank were obtained randomly, then they were weighted (WT) and measured (LT) to analyze growth parameters of specific growth rate (SGR) and condition factor (CF), respectively. The former was calculated by the formula: SGRL = (eg - 1) × 100%, where g = (lnLT - lnL0)/t, where t was the experimental period (30 d). Additionally, CF was calculated by other formula: .
After measurements of growth indexes, fish were anesthetized in a metacaine (MS-222) solution of 20 g (m3)-1, then liver tissues were separated and rinsed in ice-cold physiological salt water. After these operation, sampled liver tissues were immediately transferred into RNase-free microtubes and stored in liquid nitrogen until they were analyzed. During sampling period, the tools were all sterilized and the sampling process was generally rapid to avoid contamination by other environmental factors. The test protocol were designed in accordance with the recommendations of the Regulations of the laboratory animal-Guideline for ethical review of animal welfare (Standardization Administration of China, 2018) and was approved by the Committee on the Ethics of Animal Experiments of Lu Dong university in Shandong province of China.
2.3 Transcriptomic analyses
2.3.1 mRNA library construction and sequencing
The liver tissue (approximately 1 g wet weight) of one juvenile from each experimental tank was sampled for high-throughput RNA-seq analyses. Firstly, total RNA was isolated using TRIzol reagent (Invitrogen, USA) following manufacturer’s procedure. Then, RNA amount of each sample were quantified using NanoDrop ND-1000 (NanoDrop, Wilmington, DE, USA), while RNA integrity was assessed by Bioanalyzer 2100 (Agilent, CA, USA) and confirmed by electrophoresis. Then, cleaved RNA fragments were reverse-transcribed to create cDNA by SuperScript™ II Reverse Transcriptase (Invitrogen, cat.1896649, USA). Finally, we performed the 2 × 150bp paired-end sequencing (PE150) on an Illumina Novaseq™ 6000 (LC-Bio Technology CO., Ltd., Hangzhou, China) following vendor’s recommended protocol. Following above steps, 6 sequence libraries (2 groups and each containing 3 replicates) were constructed in this study.
2.3.2 Quantification and differential expression analyses of transcripts
The original data format after sequence analyses was FASTQ. Cutadapt software was used to remove the raw reads that contained adaptor contamination. After removed the low quality bases and undetermined bases, we used HISAT2 software to map reads to the genome. The mapped reads of each sample were assembled using StringTie with default parameters. Then, all transcriptomes from all samples were merged to reconstruct a comprehensive transcriptome using gffcompare software. After the generation of final transcriptomes, StringTie and Ballgown were used to estimate the expression levels of all transcripts by calculating FPKM (Fragments Per Kilobase per Million). The DEGs were selected with fold change > 2 or fold change< 0.5 and p value< 0.05 by R package edgeR, then analyses of GO (Gene ontology) enrichment (p< 0.05) and KEGG (Kyoto Encyclopedia of Genes and Genomes) (p< 0.05) enrichment of the DEGs were performed.
2.3.3 Quantitative real-time PCR of experimental validation
To validate the results of DEGs from transcriptomic analyses, 6 DEGs were selected for quantitative real-time PCR (qRT-PCR) analyses. These genes included pla2g1b, acat2, gpx3, kyat1, gclc and aldh9a1. For qRT-PCR analyses, 18S-rRNA was chosen as the reference gene. Primers used in the qRT-PCR analyses are listed in Table 1. Firstly, liver tissues from each tank were homogenized to extract total RNA using a Trizol kit (Invitrogen, Carlsbad, USA). cDNA synthesis and qRT-PCR were operated followed the handbooks of TUREscript 1st Stand cDNA SYNTHESIS Kit (Aidlab, Beijing, China) and SYBR® Green Supermix (DF, China) Kit. Reactions were carried out on an analytikjena-qTOWER 2.2 qPCR analyzer (Germanty). The relative expression levels of selected genes were calculated according to the 2−ΔΔCT method (Livak and Schmittgen, 2001).

Table 1 The information of forward (F) and reverse (R) primers for target genes used in the qRT-PCR analyses.
2.4 Metabolomic analyses
2.4.1 Metabolite extraction
Approximately 200 mg of liver samples (six samples per group) were selected, which were used for liquid nitrogen grinding. Then collected samples were extracted with 120 μL of precooled 50% methanol by a homogenizer. After centrifugation at 4 000 g for 20 min, the supernatants were transferred into new 96-well plates, which were stored at -80°C prior to the liquid chromatography-mass spectrometry (LC-MS) analyses. Additionally, pooled quality control (QC) samples were also prepared by combining 10 μL of each extraction mixture.
2.4.2 LC-MS analyses
All samples were acquired by the LC-MS system followed machine orders. Firstly, all chromatographic separations were performed using an ExionLC system (SCIEX, Framingham, MA, USA). An ACQUITY UPLC T3 column (100 mm × 2.1 mm, 1.8 μm, Waters, UK) was used for the reversed phase separation. The column oven was maintained at 35°C. The flow rate was 0.4 ml/min and the mobile phase consisted of solvent A (Water, 0.1% formic acid) and solvent B (Acetonitrile, 0.1% formic acid). Gradient elution conditions were set as follows: 0~0.5 min, 5% B; 0.5~7 min, 5% to 100% B; 7~8 min, 100% B; 8~8.1 min, 100% to 5% B; 8.1~10 min, 5% B. The injection volume for each sample was 4 μL. A high-resolution tandem mass spectrometer TripleTOF5600plus (SCIEX, Framingham, MA, USA) was used to detect metabolites eluted form the column. The Q-TOF was operated in both positive and negative ion modes. In order to evaluate the stability of the LC-MS during the whole acquisition, a quality control sample (Pool of all samples) was acquired after every 10 samples.
2.4.3 LC-MS data processing
Acquired MS data pretreatments including peak picking, peak grouping, retention time correction, second peak grouping and annotation of isotopes and adducts was performed using XCMS software. LC-MS raw data files were converted into mzXML format and then processed by the XCMS, CAMERA and metaX toolbox implemented with the R software. Each ion was identified by combining retention time (RT) and m/z data. Intensities of each peak were recorded and a three dimensional matrix containing arbitrarily assigned peak indices (retention time-m/z pairs), sample names (observations) and ion intensity information (variables) was generated. The online KEGG and Human Metabolome Database (HMDB) was used to annotate the metabolites by matching the exact molecular mass data (m/z) of samples with those from database.
The intensity of peak data was further preprocessed by metaX. Those features that were detected in less than 50% of QC samples or 80% of biological samples were removed, the remaining peaks with missing values were imputed with the k-nearest neighbor algorithm to further improve the data quality. Principal component analysis (PCA) was performed for outlier detection and batch effects evaluation using the pre-processed dataset. QC-based robust locally weighted regression (LOESS) signal correction was fitted to the QC data with respect to the order of injection to minimize signal intensity drift over time. In addition, the relative standard deviations of the metabolic features were calculated across all QC samples, and those > 30% were then removed. The p value was adjusted for multiple tests using an false discovery rate (FDR). A VIP (Variable Important for the Projection) cut-off value of 1.0 was used to select important features. The determination of metabolites exerting significant difference among the MeHg treatment group and the control group should simultaneously meet the following requirements: fold-change ≥ 2 or ≤ 0.5, p value ≤ 0.05 and VIP ≥ 1.
2.5 Integrative analyses of metabolome and transcriptome
The DEGs and significantly changed secondary metabolites were used for the integrative analyses. Spearman method was used to analyze the correlation coefficients for the metabolomic and transcriptomic data integration of the control group and the MeHg-treated group. Significantly changed biological pathways with p values less than 0.05 together with at least 1 gene and 1 metabolite included were screened out and identified to reveal the response mechanisms of key biological pathways of juvenile flounder to sublethal MeHg exposure.
2.6 Statistical analyses
One-way ANOVA was used to detected whether there was significant difference among the control and MeHg-treated groups for growth parameters of juvenile flounder. All statistical analyses were carried out in SPSS software (SPSS version 22.0) and the significant difference were set at p< 0.05.
3 Results
3.1 Fish survival and growth
At the end of experiment, no fish died in the control (with surival rate of 100%), while there were 2 flounder died at 1.0 μg L-1 MeHg treatment (96.67%). Additionally, growth parameters including WT (99.0 ± 1.31 g), LT (21.8 ± 0.58 cm) and SGRL (0.49 ± 0.09% d-1) were all significantly reduced than those in the control (WT: 121.0 ± 7.90 g, LT: 23.2 ± 0.17 cm, SGRL: 0.70 ± 0.03% d-1; one-way ANOVA, p< 0.05), although there was no significant difference of CF parameter between MeHg-treated group (0.96 ± 0.08 g/m3) and control group (0.98 ± 0.07 g/m3; ANOVA, p > 0.05).
3.2 Transcriptomic of juvenile flounder under methylmercury exposure
3.2.1 GO enrichment analyses
A total of 3889 GO items were enriched after MeHg exposure (Table S1), among which, 330 items were significantly screened out when the threshold was set as p< 0.05, including 182 items of biological process (BP), 39 items of cellular component (CC) and 109 items of molecular function (MF). As shown in Figure 1, BP items were mainly related to regulation of transcription, DNA template, oxidation-reduction process, phosphorylation, proteolysis, intracellular signal transduction, lipid metabolic processes and immune response. Moreover, most of the DEGs were enriched in membrane, extracellular exosome and endoplasmic reticulum, involved in CC. As for MF, they were closely related to metal ion binding, ATP binding as well as activities of different enzymes, such as transferase, hydrolase, kinase and oxidoreductase.
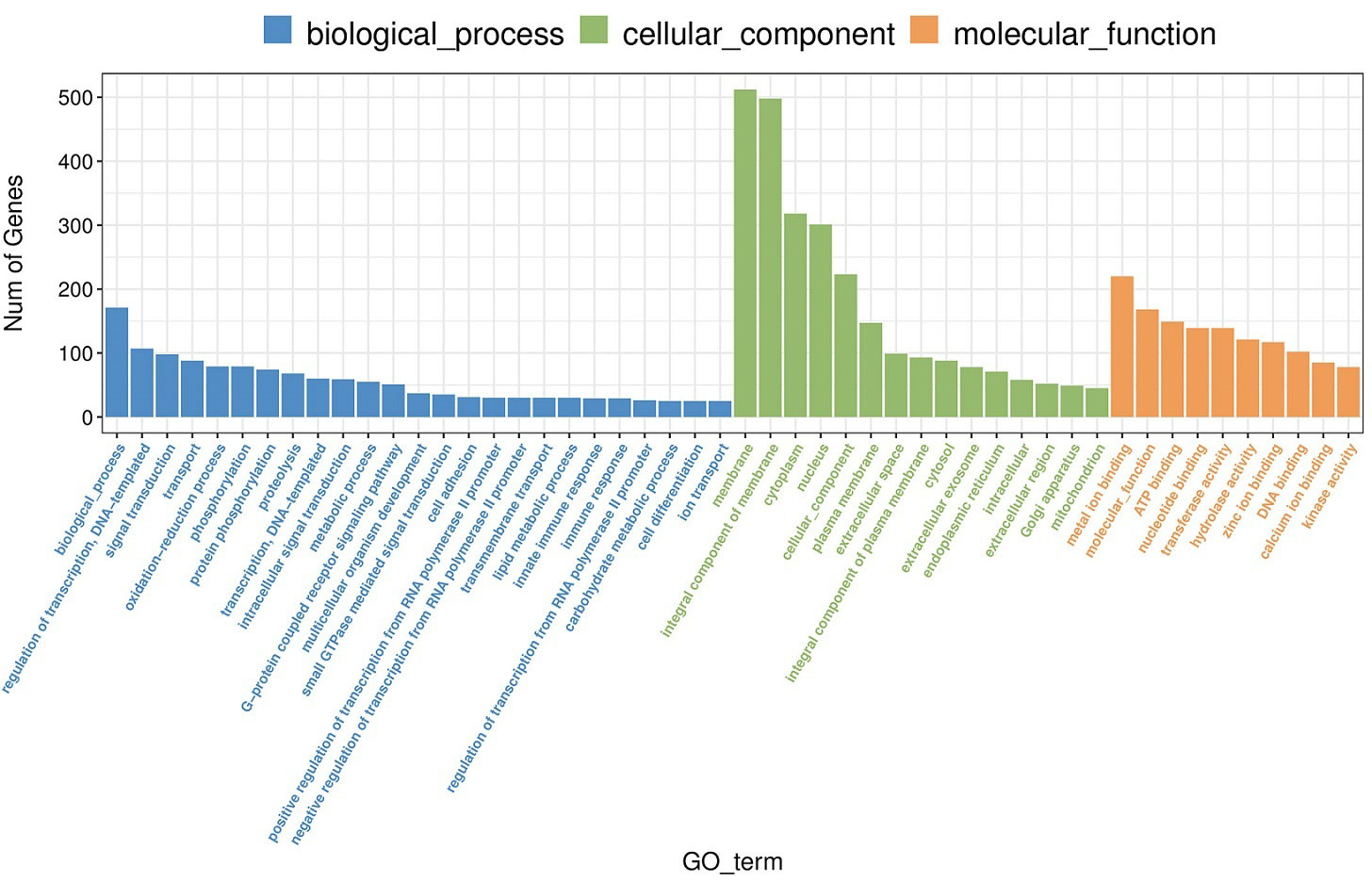
Figure 1 Go enrichement (top 25 of biological process, top 15 of cellular component, top 10 of molecular function) of significantly differentially expressed genes in juvenle flounder under MeHg exposure.
3.2.2 KEGG enrichment analyses
KEGG enrichment analyses showed that 59 pathways were significantly enriched after MeHg exposure, which were mainly related to immune response, oxidative stress, lipids metabolism, glycometabolism, amino acid and nucleotide metabolism and regulation of protein processes (Figure 2). For example, immune response related pathways included phagosome (the number of DEGs: 75), cytokine-cytokine receptor interaction (46), toll-like receptor signaling pathway (34), and so on. However, antioxidative defense related pathways contained protein processing in endoplasmic reticulum (64), peroxisome (27), retinol metabolism (21), metabolism of xenobiotics by cytochrome P450 (10), and so on. Specifically, lipids or glycometabolism related pathways occupied a large part of these KEGG pathways, such as glycerolipid metabolism (23), steroid biosynthesis (22), steroid hormone biosynthesis (21), linoleic acid metabolism (15) for lipids metabolisms, and glycolysis/gluconeogenesis (26), galactose metabolism (19), starch and sucrose metabolism (19), fructose and mannose metabolism (17) for glycometabolism. Moreover, amino acids or nucleotide metabolisms related pathways included biosynthesis of amino acids (28), alanine, aspartate and glutamate metabolism (15), purine metabolism (50) and pyrimidine metabolism (27). Furthermore, several pathways about regulation of protein processes were also observed in KEGG enrichment analyses, such as protein processing in endoplasmic reticulum (64), protein digestion and absorption (31), protein export (6). Additionally, other diseases related pathways were also responding actively to MeHg exposure, including salmonella infection (27), prion diseases (7) and asthma (2) (Table S2).
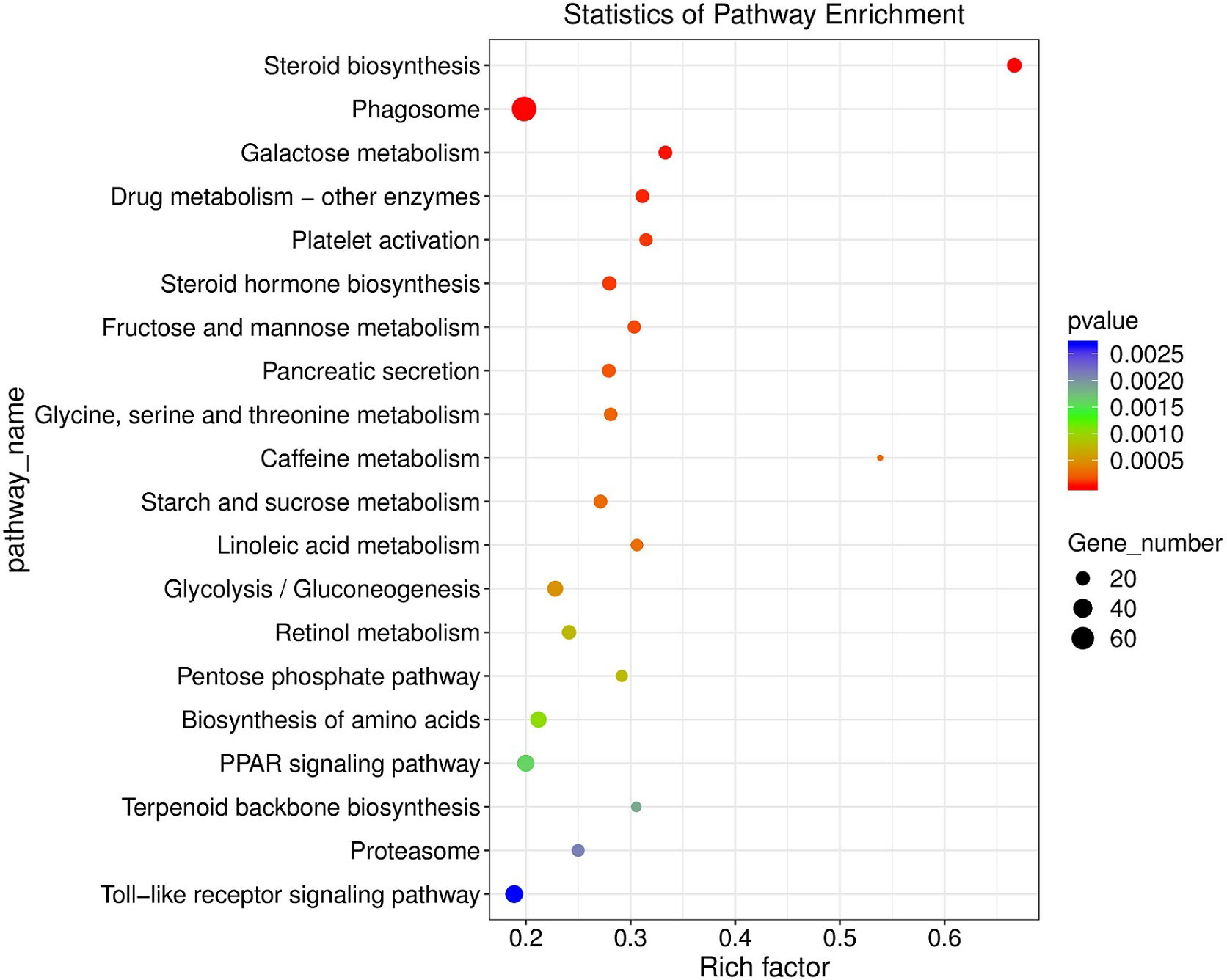
Figure 2 KEGG enriched pathways (top 20, p < 0.05) scatter plot of significantly differentially expressed genes in juvenle flounder under MeHg exposure.
3.2.3 Significantly differentially expressed genes analyses
There were 2502 DEGs including 1139 down-regulation genes and 1363 up-regulation genes between MeHg-treated group and control group (Figure 3). Hierarchical cluster analyses presented visible difference of these DEGs (top 100) (Figure 4), and down-regulation DEGs were significantly enriched in oxidative stress, innate immune system or amino acid metabolism related pathways, such as peroxisome, retinol metabolism, phagosome, arginine, alanine, aspartate, glutamate or histidine metabolisms. However, up-regulation DEGs showed the changes of sorphisticated biological pathways, including glycometabolism or lipids metabolism pathways, such as fructose and mannose metabolism (pfkfb3), PPAR signaling pathway (fabp1), glycerolipid metabolism (pnpla2), steroid biosynthesis (nsdh1, sc5d, ebp, tm7sf2, sqle, lss, msmo1) and so on. Some up-regulation DEGs, including pdia4, pdia6, dnajb11 and hsp90b1, were associated with protein processing in endoplasmic reticulum. Additionally, most of the up-regulation DEGs, such as fdps, hmgcs1, idi1 and hmgcr (enriched in terpenoid backbone biosynthesis), blvra (porphyrin and chlorophyll metabolism), slc4a10 (pancreatic secretion), cda (pyrimidine metabolism) and creld2 (phagosome), were also responding actively to MeHg for the maintainence of general growth or development of flounder.
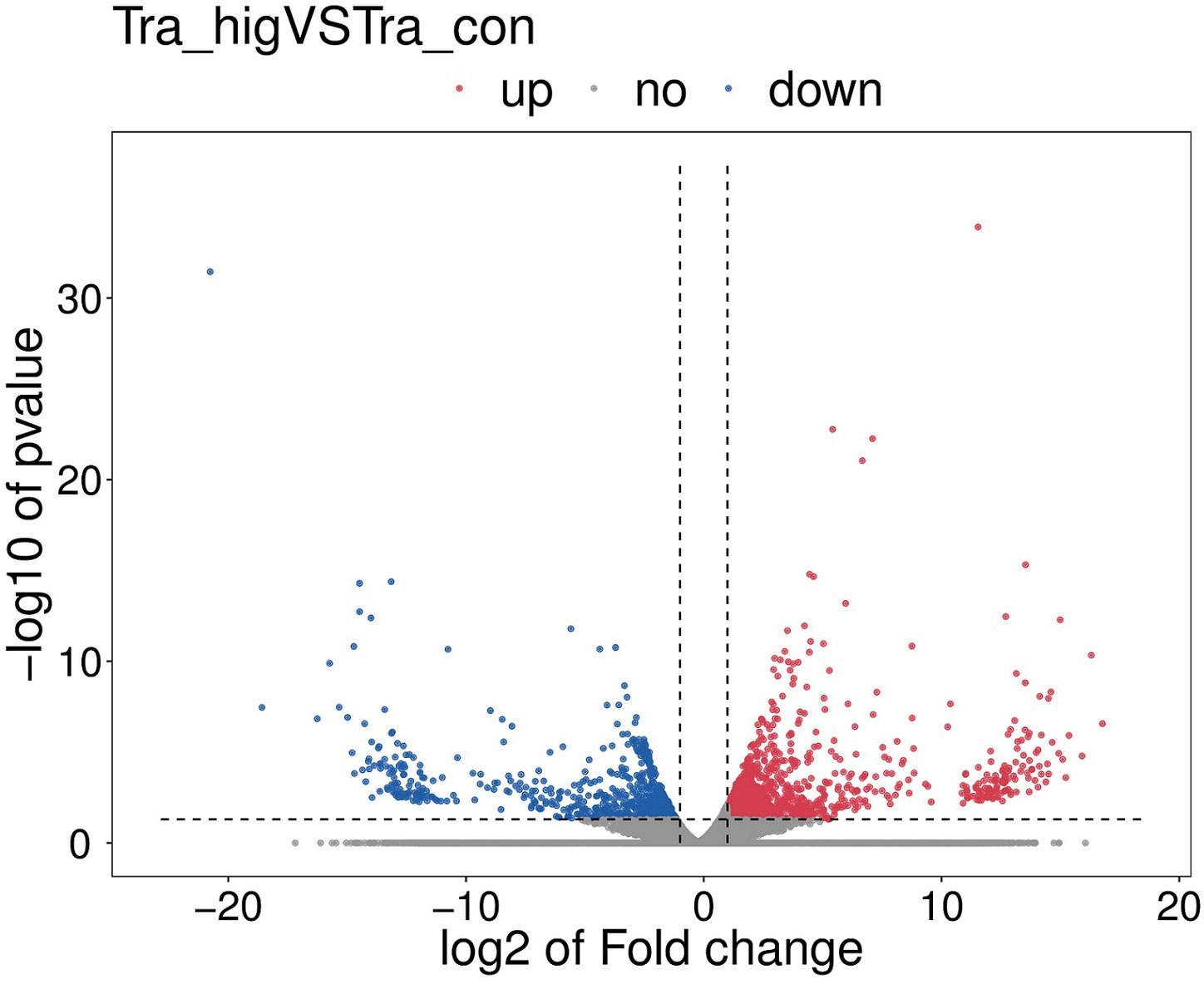
Figure 3 The volcano map of significantly differentially expressed genes (DEGs), including up-regulation DEGs and down-regulation DEGs among the MeHg treatment group and the control group.
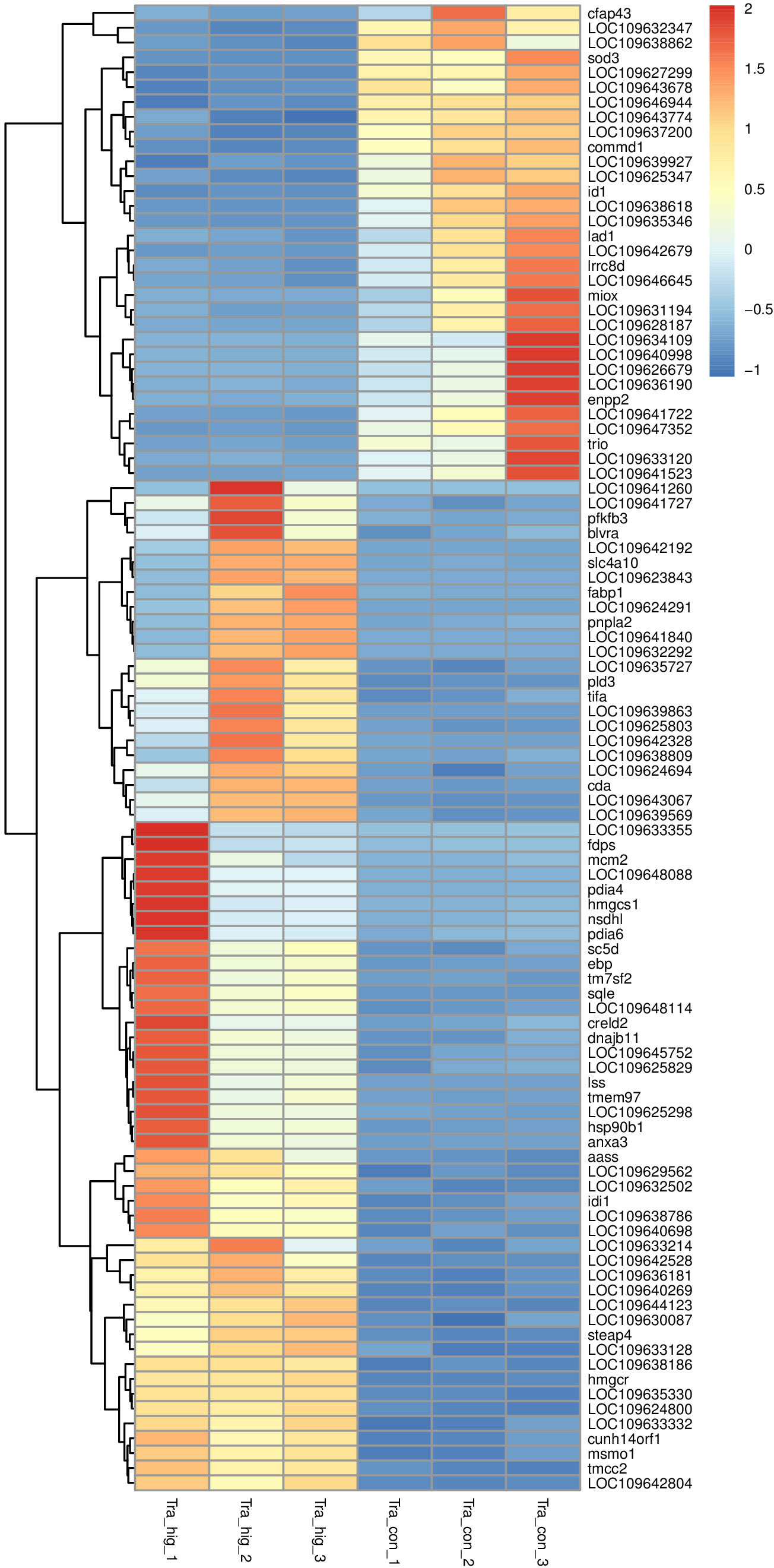
Figure 4 Hierarchical cluster analyses of significantly differentially expressed genes (top 100) in juvenile flounder after MeHg exposure.
To further validate the results of DEGs from transcriptomic analyses, 6 DEGs were selected to quantify their expression tendence patterns by qRT-PCR analyses. The effciency of various primers used in this study were tested no less than twice. These tested genes (acat2, pla2g1b, gpx3, gclc, kyat1, aldh9a1) showed similar expression trends compared to those results from RNA-Seq analyses (Figure S1). Additionally, the expression of acat2, gpx3, kyat1 (up-regulation) and pla2g1b, gclc, aldh9a1 (down-regulation) were sensitive to MeHg, but showed opposite expression trendency.
3.3 Changes of secondary metabolites in juvenile flounder under methylmercury exposure after metabolomic analyses
A total of 10251 ions were accurately detected after metabolomic analyses, among which 233 primary metabolites (ESI+: 184; ESI-: 49) showed significant difference between MeHg-treated group and control group (Table S3). As for the secondary metabolites annoted from these 233 primary metabolites, only 16 metabolite compounds (ESI+: 12; ESI-: 4) changed significantly between MeHg-treated group and control group (Table S4), which were attached to those superclass of lipids and lipid-like molecules, organic acids and derivatives, organoheterocyclic compounds, organic oxygen compounds and benzenoids (Figure 5). Meanwhile, gly-phe, phe-thr and quetiapine were increased significantly, whereas pyroglutamic acid, arachidic acid and taurodeoxycholic acid were decreased significantly. However, only 6 secondary metabolites were significantly enriched in 7 metabolism pathways, mainly including tryptophan metabolism (L-kynurenine, 5-hydroxyindoleacetic acid), glutathione metabolism (oxidized glutathione, pyroglutamic acid), biosynthesis of unsaturated fatty acids (linoleic acid, arachidic acid) and linoleic acid metabolism (linoleic acid) (Figure 6).
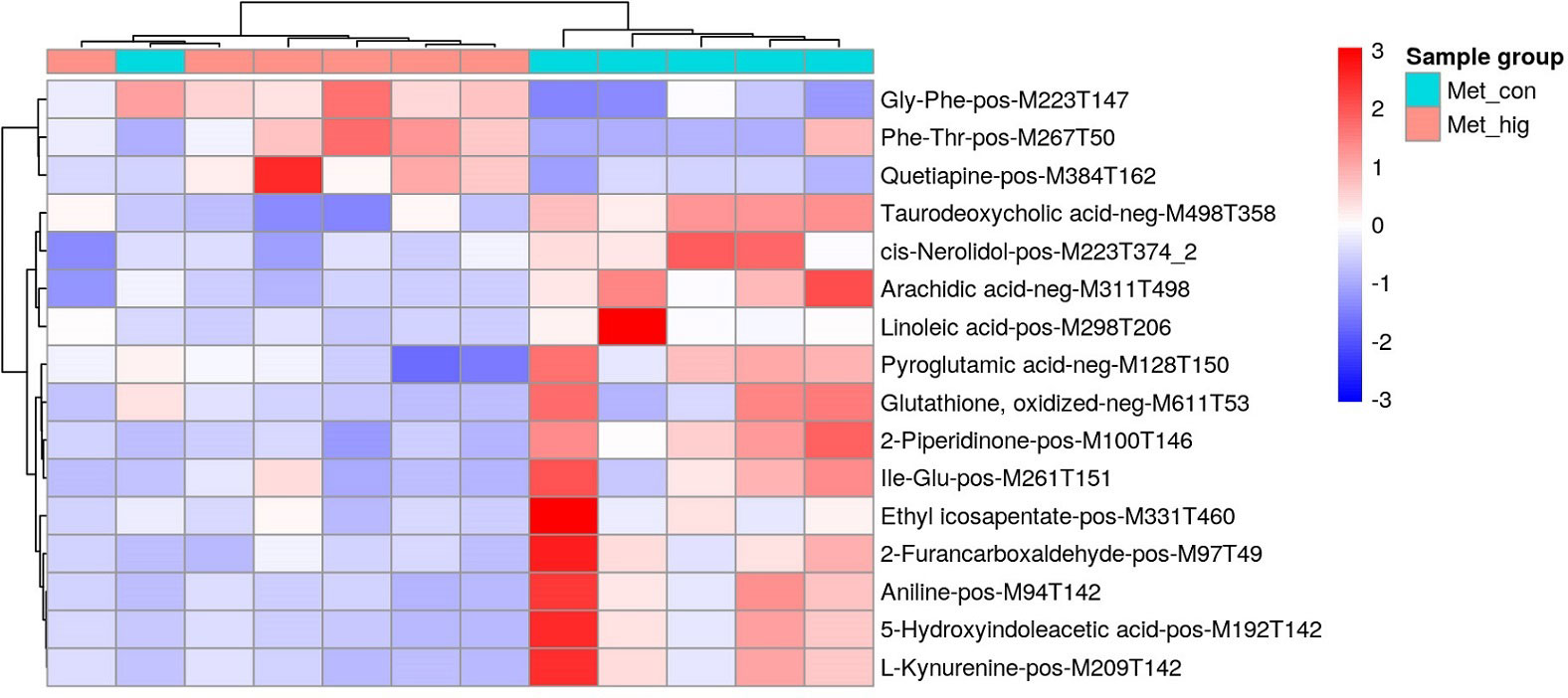
Figure 5 Heatmap of identified secondary metabolites showing significant difference between MeHg treatment and the control groups in the liver tissues of juvenile flounder.
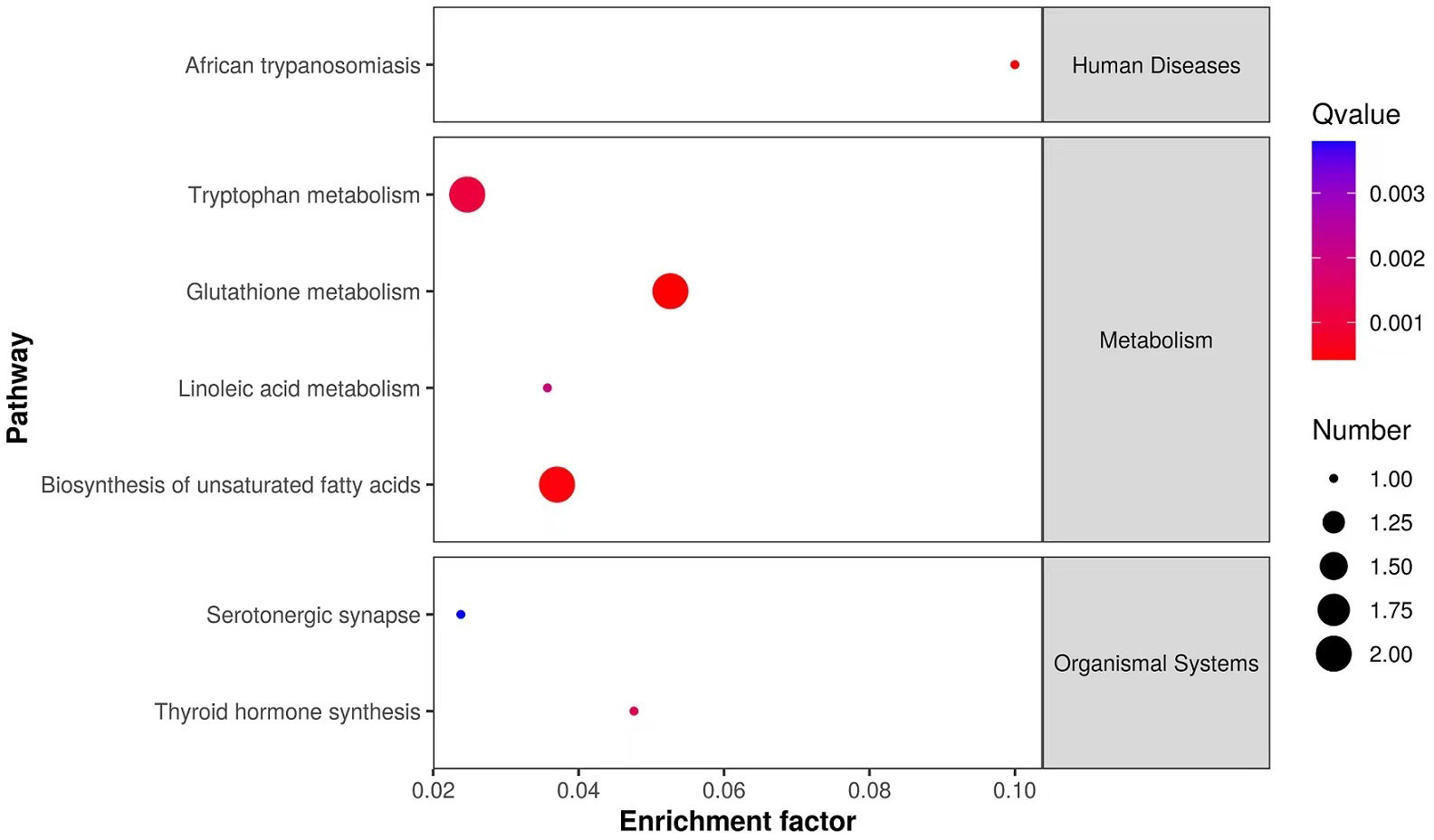
Figure 6 KEGG enriched pathways scatter plot of significantly changed secondary metabolites in liver tissues of juvenile flounder after MeHg exposure.
3.4 Biological pathways in response to methylmercury exposure after multi-omic analyses
After multi-omic analyses, 7 biological pathways including glutathione metabolism, thyroid hormone synthesis, linoleic acid metabolism, biosynthesis of unsaturated fatty acids, tryptophan metabolism pathway, serotonergic synapse and African trypanosomiasis were significantly enriched. In this regard, 6 significantly changed secondary metabolites and 57 DEGs were closely connected through the association among these pathways (Table S5). As shown in Figure 7, the integrated regulatory network was consisted of three clusters, including glutathione metabolism and thyroid hormone synthesis, linoleic acid metabolism and biosynthesis of unsaturated fatty acids, tryptophan metabolism, serotonergic synapse and African trypanosomiasis, respectively.
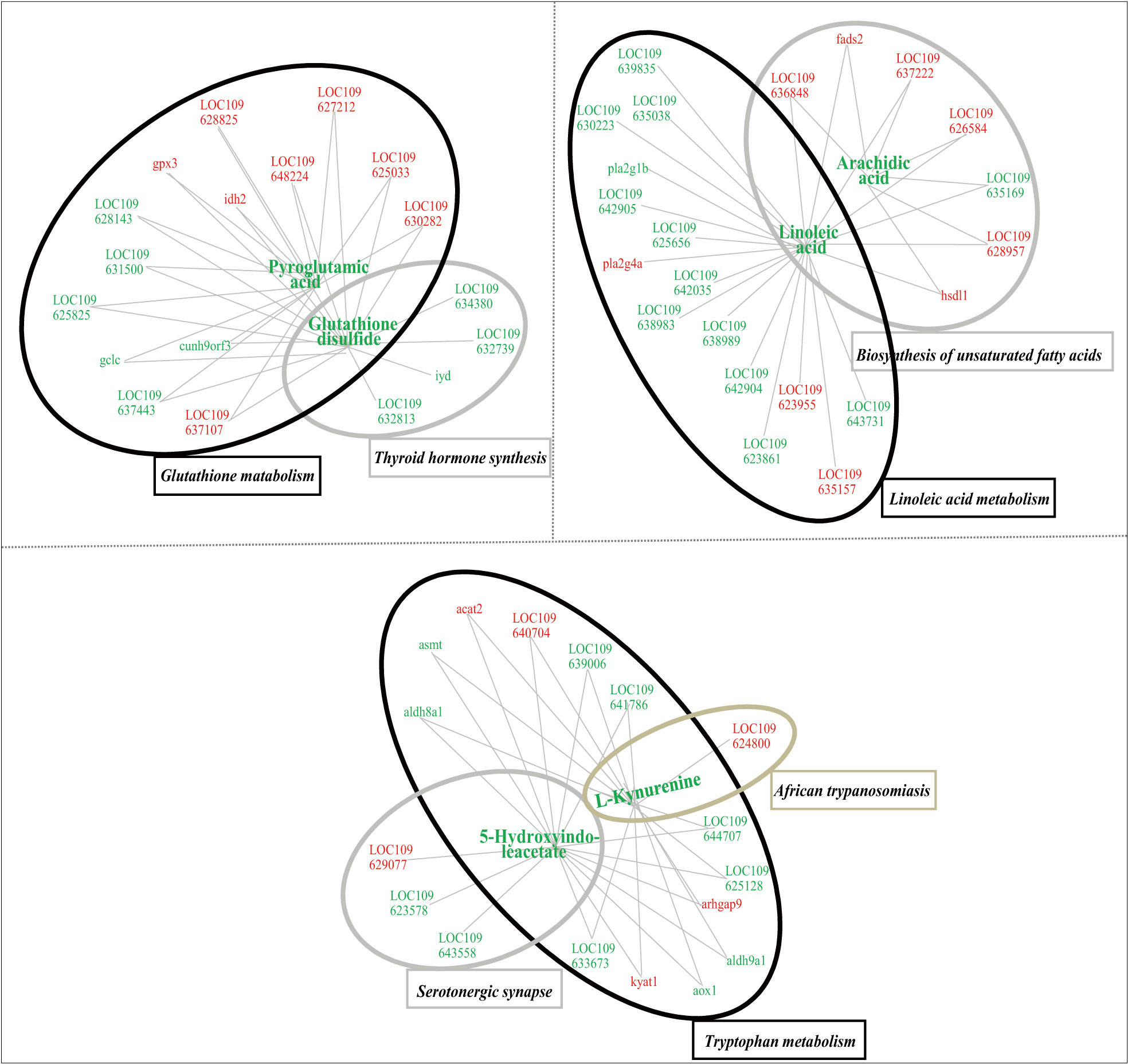
Figure 7 Integrated network of functional interactions between significant changed secondary metabolites and significantly differentially expressed genes, whose levels were significantly affected in the liver tissues of juvenile flounder after MeHg exposure. In this regard, metabolites and genes were connected if they shared at least one common KEGG pathway. Increased or decreased metabolites or genes are indicated by red and blue symbols, respectively.
4 Discussion
4.1 The inhibitory effects of methylmercury on the growth of juvenile flounder
The waterborne MeHg exposure concentrations set in current (last decade) studies of tested fish species at different life stages (Table S6) are generally higher than the environment-relevant dose in seawater (Cao et al., 2020). Therefore, it is difficult to reflect the toxicological effects of MeHg to fish at ELSs under environment-relevant exposure. Hereby, we selected 1.0 μg L-1 as the exposure concentration in this study, closer to the environment-relevant doses of waterborne MeHg, which was also proved to be sublethal to flounder at ELSs in our previous studies. In this study, several growth parameters of juvenile flounder were significantly restrained even under 1.0 μg L-1 exposure, which might be due to the long exposure period compared to the previous study (Ren et al., 2020), where the significantly inhibitory effect for flounder growth was only appeared when MeHg concentration reached 20.0 μg L-1. Lower or higher levels of MeHg exposure could cause huge damages to fish growth, once the MeHg accumulated to a high level in fish body. Similar results were also identified in zebrafish (Danio rerio) larve, suggesting that Hg2+ could alter thyroid hormone levels and related gene expression (Sun et al., 2018). Moreover, available evidence proved MeHg could impact the bioenergetics homeostasis of ATP in fish (Baldissera et al., 2019), which eventually resulted in decreasing energy needed for growth or development. Additionally, Houck and Cech (2004) found MeHg inhibited the growth of juvenile Sacramento blackfish (Orthodon microlepidotus) by disturbing gastrointestinal function, eventually decreasing nutrient absorption. As discussed above, MeHg could inhibit fish growth by a variety of ways, however their regulatory mechanisms under these inhibitory effects are still not deeply mastered. Therefore, we investigated the response mechanisms of juvenile flounder to sublethal MeHg exposure using metabolomic and transcriptomic techniques.
4.2 The transcriptional responses of juvenile flounder to sublethal methylmercury exposure
Comparing those results of current literatures and this study, the number of Hg-induced DEGs varied tested fish species, exposure condition and targeted tissues (Yadetie et al., 2013; Zhang et al., 2020; Zhang et al., 2022). In the present study, most of the DEGs were significantly enriched in cellular components GO items (e.g., membrane, nucleus, cytoplasm), suggesting potential damages of MeHg to cellular structure (especially the membrane) in flounder, which was in agreement with the results identified in common carp (Zhang et al., 2022). In addition, other DEGs were significantly enriched in oxidation-reduction process, phosphorylation, proteolysis, intracellular signal transduction, lipid metabolic processes, immune response and enzymatic activities, suggesting comprehensive acceleration of sophisticated biological processes in fish to avoid disturbance induced by MeHg.
It is well-known that KEGG pathways of immnue response and antioxidative defense are the main targets sensitive to Hg compounds (inorganic or organic mercury) or other pollutants (Wang et al., 2017; Zhang et al., 2020; Olsvik et al., 2021). As shown in this study, phagosome and toll-like receptor signaling pathway were remarkably enriched pathways, which is consistent with the results observed in Chinese rare minnow (Gobiocypris rarus) larvae (Chen et al., 2022) and common carp (Liu et al., 2019). Additionally, immune related pathways including cytokine-cytokine receptor interaction, complement and coagulation cascades were also significantly enriched in flounder. As for antioxidative defense related pathways, protein processing in endoplasmic reticulum, peroxisome, retinol metabolism and metabolism of xenobiotics by cytochrome P450 pathways were significantly enriched in this study, which is helpful to reduce the damage of reactive oxygen species (ROS) induced by MeHg (Jin, 2014; Pedro et al., 2019; Mohamed et al., 2020). Moreover, up-regulation DEGs associated with arachidonic acid metabolism (gpx3, pla2g4a, pla2g1b) and retinol metabolism (rdh10, aldh9a1, aox1) were observed in this study, which was partly in agreement with the findings in zebrafish embryos (Ortiz-Villanueva et al., 2017), suggesting the antioxidant systems were activated to protect juvenile flounder from irreversible MeHg injuries.
Lipids and their derivants are important nutrients for fish health, which are crucial energetic resources and play key roles in cell structure maintenance (Ferain et al., 2018). In this study, steroid biosynthesis and steroid hormone biosynthesis pathways were significantly enriched, which was consistent with the findings in male rare minnow under tributylin exposure (Zhang et al., 2017). Moreover, the DEGs of sqle, lss, msmo1, hsd17b7, ebp, sc5d, soat1, nsdh1 participated in the cholesterol biosynthesis (Gill et al., 2011; Lasunción et al., 2012), which were significantly induced by MeHg, indicating that flounder performed active efforts for maintaining the general development under MeHg stress. Peroxisome proliferator activated receptors (PPARs) could activate transcription of PPAR-target genes (Colliar et al., 2011). For example, the DEG of fabp1, proved to be closely related to PPARα pathway (Laprairie et al., 2016), was significantly upregulated in this study, suggesting lipid metabolism in juvenile flounder were affected under long-term MeHg exposure. Furthermore, as one of the important essential fatty, linoleic acid and its related metabolisms were changed manifestly, which might result in turbulence of nutrient supply for growth and redox state in fish, respectively (Xu et al., 2019; Duan et al., 2020).
Glycometabolism, amino acid and nucleotide metabolisms were also easily to be disturbed in fish under pollutant stress, which had been elucidated in this study and previous literatures (Yadetie et al., 2013; Teng et al., 2018; Pang et al., 2021; Wang et al., 2022). Glycometabolism is of great importance to provide energy and maintain physiological functions in various fish tissues. In the liver tissue of juvenile flounder, most glycometabolism related pathways such as fructose and mannose metabolism and galactose metabolism were responding actively to MeHg, which could change glycometabolism via affecting the electron respiratory chain or tricarboxylic acid cycle (TCA) through the up-regulaton of pfkfb3 and galk genes or the down-regulation of aldh9a1 gene (Yang et al., 2017). In this study, various amino acids, purine and pyrimidine related pathways were significantly affected by MeHg, suggesting MeHg could influence the synthesis and degradation of cellelar macromolecule, and ultimately cause damages to the proteins, DNA or lipids. Similar findings are also described in previous study (Ren et al., 2019b). The upregulation of the DEGs involved in protein processing, digestion or absorption (e.g., pdia4, pdia6, dnajb11, hsp90b1) was also favor the above viewpoint.
4.3 The response mechanisms of key metabolites and pathways in juvenile flounder to sublethal methylmercury exposure
Although 10251 ions were precisely detected in this study, only 16 secondary metabolites were significantly changed. Among which, amino acid and their derivatives, including pyroglutamic acid, oxidized glutathione, gly-phe, ile-glu and phe-thr, were identified as important secondary metabolites of flounder in response to MeHg stress. It is consistent with the results reported in other literatures (Liu et al., 2011; Brandão et al., 2015; Cosio and Renault, 2020; De Marco et al., 2022). For example, glutamic acid, cysteine and glycine are closely related to glutathione biosynthesis, which is essential in fish to avoid oxidative stress or toxic damages (Mohamed et al., 2020). Generally speaking, high attack of MeHg to special ionic groups of amino acids might be an important reason for the decline of intermediates (5-oxoproline, also known as pyroglutamic acid) of glutamic acid metabolism (Abdelmoez et al., 2010). Following the decrease of pyroglutamic acid, the activities of γ-glutamylcysteine synthetase and isocitrate dehydrogenase would be reduced, which might affect the synthesis of glutathione (Figure S2). However, significant up-regulation expression of gpx3 and increased glutathione peroxidase activity caused a reduction of oxidized glutathione contents. The decrease of glutathione peroxidase activity, glutathione levels and gclc expression under MeHg exposure were also favor the above view. Additionally, glutathione metabolism and thyroid hormone synthesis was closely connected by the decrease of oxidized glutathione (Figure S3) in this study. Meanwhile, the expression of creb was also reduced, suggesting the disturbance of thyroid hormones synthesis (Citterio et al., 2019). These results specified the attacks of MeHg for the synthesis or release of thyroid hormones, and eventually affected the growth or development of flounder.
Furthermore, lipids and lipid-like molecules of arachidic acid and linoleic acid and their metabolic pathways of biosynthesis of unsaturated fatty acids and linoleic acid metabolism were significantly changed in this study, which were also used as common biomarkers for studying response mechanisms of tested organisms to pollutants (Huang et al., 2017; Ortiz-Villanueva et al., 2017; Teng et al., 2018; He et al., 2020). Especially, arachidic acid is an essential component of cellular phospholipids that occur mainly in cell membranes and internal membrane structures. Additionally, arachidic acid is also the precursor of prostaglandins, leukotrienes and related compounds, which plays important roles in inflammation and immunity regulation (He et al., 2020). Therefore, significant reduction of arachidic acid in fish would cause adverse influence on cellular membrane structure or function as well as immune systems. As shown in Figure S4, MeHg could inhibit phospholipase A2 activity but induce up-regulation of plag4a, which might affect the conversion from lecithin to linoleate, finally causing disturbance of linoleic acid and arachidonic acid metabolisms. Linoleic acid and arachidonic acid are also essential fatty acids for fish growth and regulation of lipids metabolisms, stress defenses and immune responses (Zhao et al., 2013; Huang et al., 2017; Ortiz-Villanueva et al., 2017; Teng et al., 2018; He et al., 2020). Therefore, these two decreased fatty acids were found in the pathway of biosynthesis of unsaturated fatty acids (Figure S5). Additionally, MeHg induced up-regulation of fads2 and hsdl1, while decreased the activities of CYP2C and CYP2J, which exerted disorders of the biosynthesis of unsaturated fatty acids (Konkel and Schunck, 2011; Kabeya et al., 2017).
L-kynurenine, 5-hydroxyindoleacetate (5-HT) and related metabolism pathway of tryptophan metabolism also showed significant changes in flounder under MeHg exposure. Tryptophan is an essential amino acid for fish growth, which is used for protein synthesis and/or entering catabolism through kynurenine or 5-HT related pathways (Le Floc'h et al., 2011; Hoseini et al., 2019). Generally, increased 5-hydroxyindoleacetic acid (5-HIAA)/5-HT ratio always indicates decreased aggressive behavior or environmental stress in the previous study (Hoseini et al., 2019). Moreover, it has been proposed that tryptophan exhibits antioxidant activity. For example, 5-HT can act as a direct free radical scavenger and decrease lipids oxidative damage (Hoseini et al., 2019). However, due to inhibitory effects of MeHg on the activities or expression of aldehyde dehydrogenase, aldehyde oxidase, monoamine oxidase and aldh9a1, aldh8a1 and aox1, biosynthesis of 5-HT was impeded in this study (Figure S6). Meanwhile, MeHg reduced the activities of hydroxyindole O-methyltransferase and other important oxidoreductases eventually disturbing the biosynthesis of melatonin, which is a rhythmic hormonal signal involved in accelerating the effects of tryptophan metabolism on antioxidant defense, counteracting toxic substances and immune responses (Hoseini et al., 2019). Additionally, SERT protein could be damaged in this study (Figure S7), which could disturb signal transmission of nervous system (Fox et al., 2010). The kynurenine pathway is regarded as dominant route for tryptophan degradation, which is removing excess tryptophan from organisms (Fernstrom, 2016). During this process, kynureninase and acetyl-CoA acetyltransferase are of great importance, whereas their activities were increased with the decrease of L-kynurenine in this study. This phenomenon might be due to that kynurenine pathway will only occur in the presence of tryptophan supplementation (Hoseini et al., 2019). Thus, these results revealed a sign of scarcity of tryptophan contents for flounder growth and development under MeHg stress. Also, the decrease of L-kynurenine might be associated with the reduced activity of tryptophan 2,3-dioxygenase, a key enzyme in the process of catalyzing tryptophan into L-kynurenine (Hoseini et al., 2019). Additionally, the activity of acetyl-CoA acetyltransferase and the expression of acat2 and arhgap9 were significantly induced, which promoted the production of acetyl-CoA, eventually stimulating biological process of glycolysis, suggesting the increased need of energy for fish life.
5 Conclusions
In conclusion, most of the DEGs and significantly changed secondary metabolites in the liver of juvenile flounder were mainly enriched in pathways of immune response, oxidative stress, lipids or amino acid metabolisms under sublethal MeHg exposure.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding authors.
Ethics statement
The animal study was reviewed and approved by Committee on the Ethics of Animal Experiments of Lu Dong University in Shandong province of China.
Author contributions
ZL, JiY, and JuY executed this project and designed the toxicity experiment. ZW and JL conducted the toxicity experiment. ZR, JN, LC and JZ analyzed the experimental results and wrote the manuscript, respectively. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 42176107) and Natural Science Foundation of Shandong Province, China (Grant No.ZR2021QC082).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2022.979357/full#supplementary-material
References
Abdelmoez W., Yoshida H., Nakahasi T. (2010). Pathways of amino acid transformation and decomposition in saturated subcritical water conditions. Int. J. Chem. React. Eng. 8, 21. doi: 10.2202/1542-6580.1903
Baldissera M. D., Souza C. F., Grings M., Descovi S. N., Henn A. S., Flores E. M. M., et al. (2019). Exposure to methylmercury chloride inhibits mitochondrial electron transport chain and phosphotransfer network in liver and gills of grass carp: Protective effects of diphenyl diselenide dietary supplementation as an alternative strategy for mercury toxicity. Aquaculture 509, 85–95. doi: 10.1016/j.aquaculture.2019.05.012
Brandão F., Cappello T., Raimundo J., Santos M. A., Maisano M., Mauceri A., et al. (2015). Unravelling the mechanisms of mercury hepatotoxicity in wild fish (Liza aurata) through a triad approach: bioaccumulation, metabolomic profiles and oxidative stress. Metallomics 7, 1352–1363. doi: 10.1039/c5mt00090d
Cao L., Liu J. H., Dou S. Z., Huang W. (2020). Biomagnification of methylmercury in a marine food web in laizhou bay (North China) and associated potential risks to public health. Mar. pollut. Bull. 150, 12. doi: 10.1016/j.marpolbul.2019.110762
Ceccatelli S., Dare E., Moors M. (2010). Methylmercury-induced neurotoxicity and apoptosis. Chem.-Biol. Interact. 188, 301–308. doi: 10.1016/j.cbi.2010.04.007
Chen C. Z., Li P., Liu L., Li Z. H. (2022). Transcriptomic and proteomic analyses of Chinese rare minnow (Gobiocypris rarus) larvae in response to acute waterborne cadmium or mercury stress. Aquat. Toxicol. 246, 9. doi: 10.1016/j.aquatox.2022.106134
Citterio C. E., Targovnik H. M., Arvan P. (2019). The role of thyroglobulin in thyroid hormonogenesis. Nat. Rev. Endocrinol. 15, 323–338. doi: 10.1038/s41574-019-0184-8
Colliar L., Sturm A., Leaver M. J. (2011). Tributyltin is a potent inhibitor of piscine peroxisome proliferator-activated receptor alpha and beta. Comp. Biochem. Physiol. Part C.: Toxicol. Pharmacol. 153, 168–173. doi: 10.1016/j.cbpc.2010.10.008
Cosio C., Renault D. (2020). Effects of cadmium, inorganic mercury and methyl-mercury on the physiology and metabolomic profiles of shoots of the macrophyte Elodea nuttallii. Environ. pollut. 257, 8. doi: 10.1016/j.envpol.2019.113557
De Marco G., Brandao F., Pereira P., Pacheco M., Cappello T. (2022). Organ-specific metabolome deciphering cell pathways to cope with mercury in wild fish (Golden grey mullet Chelon auratus). Animals 12, 16. doi: 10.3390/ani12010079
dos Santos A. A., Ferrer B., Goncalves F. M., Tsatsakis A. M., Renieri E. A., Skalny A. V., et al. (2018). Oxidative stress in methylmercury-induced cell toxicity. Toxics 6, 15. doi: 10.3390/toxics6030047
Duan Z. H., Duan X. Y., Zhao S., Wang X. L., Wang J., Liu Y. B., et al. (2020). Barrier function of zebrafish embryonic chorions against microplastics and nanoplastics and its impact on embryo development. J. Hazard. Mater. 395, 7. doi: 10.1016/j.jhazmat.2020.122621
Ferain A., Bonnineau C., Neefs I., Das K., Larondelle Y., Rees J. F., et al. (2018). Transcriptional effects of phospholipid fatty acid profile on rainbow trout liver cells exposed to methylmercury. Aquat. Toxicol. 199, 174–187. doi: 10.1016/j.aquatox.2018.03.025
Fernstrom J. D. (2016). A perspective on the safety of supplemental tryptophan based on its metabolic fates. J. Nutr. 146, 2601–2608. doi: 10.3945/jn.115.228643
Fox M. A., Stein A. R., French H. T., Murphy D. L. (2010). Functional interactions between 5-HT2A and presynaptic 5-HT1A receptor-based responses in mice genetically deficient in the serotonin 5-HT transporter (SERT). Br. J. Pharmacol. 159, 879–887. doi: 10.1111/j.1476-5381.2009.00578.x
Fretham S. J. B., Caito S., Martinez-Finley E. J., Aschner M. (2012). Mechanisms and modifiers of methylmercury-induced neurotoxicity. Toxicol. Res. 1, 32–38. doi: 10.1039/c2tx20010d
Gill S., Stevenson J., Kristiana I., Brown A. J. (2011). Cholesterol-dependent degradation of squalene monooxygenase, a control point in cholesterol synthesis beyond HMG-CoA reductase. Cell Metab. 13, 260–273. doi: 10.1016/j.cmet.2011.01.015
He C. Z., Hao R. J., Deng Y. W., Yang C. Y., Du X. D. (2020). Response of pearl oyster Pinctada fucata martensii to allograft-induced stress from lipid metabolism. Fish. Shellfish. Immunol. 98, 1001–1007. doi: 10.1016/j.fsi.2019.11.028
Hoseini S. M., Perez-Jimenez A., Costas B., Azeredo R., Gesto M. (2019). Physiological roles of tryptophan in teleosts: current knowledge and perspectives for future studies. Rev. Aquacult. 11, 3–24. doi: 10.1111/raq.12223
Houck A., Cech J. J.J. (2004). Effects of dietary methylmercury on juvenile Sacramento blackfish bioenergetics. Aquat. Toxicol. 69, 107–123. doi: 10.1016/j.aquatox.2004.04.005
Huang S. S. Y., Benskin J. P., Veldhoen N., Chandramouli B., Butler H., Helbing C. C., et al. (2017). A multi-omic approach to elucidate low-dose effects of xenobiotics in zebrafish (Danio rerio) larvae. Aquat. Toxicol. 182, 102–112. doi: 10.1016/j.aquatox.2016.11.016
Jin L. (2014). Effects and underlying mechaism of vitamin a on syhthesis of selenoprotein and antioxidative function of mammary glands of dairy cows (Inner Mongolia, China: Inner Mongolia Agricultural University).
Kabeya N., Chiba M., Haga Y., Satoh S., Yoshizaki G. (2017). Cloning and functional characterization of fads2 desaturase and elovl5 elongase from Japanese flounder Paralichthys olivaceus. Comp. Biochem. Physiol. Part B.: Biochem. Mol. Biol. 214, 36–46. doi: 10.1016/j.cbpb.2017.09.002
Konkel A., Schunck W. H. (2011). Role of cytochrome P450 enzymes in the bioactivation of polyunsaturated fatty acids. BBA-Proteins. Proteom. 1814, 210–222. doi: 10.1016/j.bbapap.2010.09.009
Laprairie R. B., Denovan-Wright E. M., Wright J. M. (2016). Subfunctionalization of peroxisome proliferator response elements accounts for retention of duplicated fabp1 genes in zebrafish. BMC Evol. Biol. 16, 15. doi: 10.1186/s12862-016-0717-x
Lasunción M. A., Martín-Sánchez C., Canfrán-Duque A., Busto R. (2012). Post-lanosterol biosynthesis of cholesterol and cancer. Curr. Opin. Pharmacol. 12, 717–723. doi: 10.1016/j.coph.2012.07.001
Le Floc'h N., Otten W., Merlot E. (2011). Tryptophan metabolism, from nutrition to potential therapeutic applications. Amino Acids 41, 1195–1205. doi: 10.1007/s00726-010-0752-7
Liu Q., Yang J., Gong Y. F., Cai J. Z., Zhang Z. W. (2019). Role of miR-731 and miR-2188-3p in mediating chlorpyrifos induced head kidney injury in common carp via targeting TLR and apoptosis pathways. Aquat. Toxicol. 215, 9. doi: 10.1016/j.aquatox.2019.105286
Liu X. L., Zhang L. B., You L. P., Yu J. B., Zhao J. M., Li L. Z., et al. (2011). Differential toxicological effects induced by mercury in gills from three pedigrees of Manila clam Ruditapes philippinarum by NMR-based metabolomics. Ecotoxicology 20, 177–186. doi: 10.1007/s10646-010-0569-x
Livak K. J., Schmittgen T. D. (2001). Analyses of relative gene expression data using realtime quantitative PCR and the 2–ΔΔ CT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Mohamed A. A. R., El-Houseiny W., El-Murr A., Ebraheim L. L. M., Ahmed A. I., Abd El-Hakim Y. M. (2020). Effect of hexavalent chromium exposure on the liver and kidney tissues related to the expression of CYP450 and GST genes of Oreochromis niloticus fish: Role of curcumin supplemented diet. Ecotoxicol. Environ. Saf. 188, 12. doi: 10.1016/j.ecoenv.2019.109890
Olsvik P. A., Azad A. M., Yadetie F. (2021). Bioaccumulation of mercury and transcriptional responses in tusk (Brosme brosme), a deep-water fish from a Norwegian fjord. Chemosphere 279, 14. doi: 10.1016/j.chemosphere.2021.130588
Ortiz-Villanueva E., Navarro-Martin L., Jaumot J., Benavente F., Sanz-Nebot V., Pina B., et al. (2017). Metabolic disruption of zebrafish (Danio rerio) embryos by bisphenol a. an integrated metabolomic and transcriptomic approach. Environ. pollut. 231, 22–36. doi: 10.1016/j.envpol.2017.07.095
Pang M. X., Wang Y., Tang Y. J., Dai J. G., Tong J. G., Jin G. (2021). Transcriptome sequencing and metabolite analyses reveal the toxic effects of nanoplastics on tilapia after exposure to polystyrene. Environ. pollut. 277, 12. doi: 10.1016/j.envpol.2021.116860
Pedro S., Dietz R., Sonne C., Rosing-Asvid A., Hansen M., McKinney M. A. (2019). Are vitamins a and e associated with persistent organic pollutants and fatty acids in the blubber of highly contaminated killer whales (Orcinus orca) from Greenland? Environ. Res. 177, 9. doi: 10.1016/j.envres.2019.108602
Ren Z. H., Cao L., Huang W., Liu J. H., Cui W. T., Dou S. Z. (2019a). Toxicity test assay of waterborne methylmercury on the Japanese flounder (Paralichthys olivaceus) at embryonic-larval stages. Bull. Environ. Contam. Toxicol. 102, 770–777. doi: 10.1007/s00128-019-02619-9
Ren Z. H., Liu J. H., Dou S. Z., Zhou D. Y., Cui W. T., Lv Z. B., et al. (2020). Tissue-specific accumulation and antioxidant defenses in flounder (Paralichthys olivaceus) juveniles experimentally exposed to methylmercury. Arch. Environ. Contam. Toxicol. 79, 406–420. doi: 10.1007/s00244-020-00775-2
Ren Z. H., Liu J. H., Huang W., Cao L., Cui W. T., Dou S. Z. (2019b). Antioxidant defenses and immune responses of flounder Paralichthys olivaceus larvae under methylmercury exposure. Comp. Biochem. Physiol. Part C.: Toxicol. Pharmacol. 225, 11. doi: 10.1016/j.cbpc.2019.108589
Standardization Administration of China (2018). Laboratory animal-guideline for ethical review of animal welfare (Beijing, China: General Administration of Quality Supervision, Inspection and Quarantine, and Standardization Administration of China).
Stein E. D., Cohen Y., Winer A. M. (1996). Environmental distribution and transformation of mercury compounds. Crit. Rev. Environ. Sci. Technol. 26, 1–43. doi: 10.1080/10643389609388485
Sun Y. L., Li Y. W., Liu Z. H., Chen Q. L. (2018). Environmentally relevant concentrations of mercury exposure alter thyroid hormone levels and gene expression in the hypothalamic-pituitary-thyroid axis of zebrafish larvae. Fish. Physiol. Biochem. 44, 1175–1183. doi: 10.1007/s10695-018-0504-2
Teng M. M., Zhu W. T., Wang D. Z., Qi S. Z., Wang Y., Yan J., et al. (2018). Metabolomics and transcriptomics reveal the toxicity of difenoconazole to the early life stages of zebrafish (Danio rerio). Aquat. Toxicol. 194, 112–120. doi: 10.1016/j.aquatox.2017.11.009
Wang C. L., Hou M. M., Shang K. Y., Wang H. S., Wang J. W. (2022). Microplastics (Polystyrene) exposure induces metabolic changes in the liver of rare minnow (Gobiocypris rarus). Molecules 27, 16. doi: 10.3390/molecules27030584
Wang M. H., Jeong C. B., Li Y., Lee J. S. (2017). Different transcriptomic responses of two marine copepods, Tigriopus japonicus and Pseudodiaptomus annandalei, to a low dose of mercury chloride (HgCl2). Aquat. Toxicol. 187, 124–131. doi: 10.1016/j.aquatox.2017.03.018
Wu F. Z., Huang W., Liu Q., Xu X. Q., Zeng J. N., Cao L., et al. (2018). Responses of antioxidant defense and immune gene expression in early life stages of large yellow croaker (Pseudosciaena crocea) under methyl mercury exposure. Front. Physiol. 9. doi: 10.3389/fphys.2018.01436
Xu H. G., Liao Z. B., Wang C. Q., Wei Y. L., Liang M. Q. (2019). Hepatic transcriptome of the euryhaline teleost Japanese seabass (Lateolabrax japonicus) fed diets characterized by alpha-linolenic acid or linoleic acid. Comp. Biochem. Physiol. Part D.: Genomics Proteomics 29, 106–116. doi: 10.1016/j.cbd.2018.11.005
Yadetie F., Karlsen O. A., Lanzen A., Berg K., Olsvik P., Hogstrand C., et al. (2013). Global transcriptome analyses of Atlantic cod (Gadus morhua) liver after in vivo methylmercury exposure suggests effects on energy metabolism pathways. Aquat. Toxicol. 126, 314–325. doi: 10.1016/j.aquatox.2012.09.013
Yang Y., Liu W. X., Li D. Z., Qian L., Fu B., Wang C. J. (2017). Altered glycometabolism in zebrafish exposed to thifluzamide. Chemosphere 183, 89–96. doi: 10.1016/j.chemosphere.2017.05.055
Yuan K., Chen X., Chen P., Huang Y. S., Jiang J., Luan T. G., et al. (2019). Mercury methylation-related microbes and genes in the sediments of the pearl river estuary and the south China Sea. Ecotoxicol. Environ. Saf. 185, 7. doi: 10.1016/j.ecoenv.2019.109722
Zhang Q. L., Dong Z. X., Luo Z. W., Zhang M., Deng X. Y., Guo J., et al. (2020). The impact of mercury on the genome-wide transcription profile of zebrafish intestine. J. Hazard. Mater. 389, 9. doi: 10.1016/j.jhazmat.2019.121842
Zhang Y., Lu Y. T., Zhang P. J., Shang X. C., Li Y. H. (2022). Brain injury induced by mercury in common carp: novel insight from transcriptome analyses. Biol. Trace Elem. Res. 1–9. doi: 10.1007/s12011-022-03161-2
Zhang J. L., Zhang C. N., Sun P., Huang M. X., Fan M. Z., Liu M. (2017). RNA-Sequencing and pathway analyses reveal alteration of hepatic steroid biosynthesis and retinol metabolism by tributyltin exposure in male rare minnow (Gobiocypris rarus). Aquat. Toxicol. 188, 109–118. doi: 10.1016/j.aquatox.2017.03.015
Zhao J. Z., Ai Q. H., Mai K. S., Zuo R. T., Luo Y. W. (2013). Effects of dietary phospholipids on survival, growth, digestive enzymes and stress resistance of large yellow croaker, Larmichthys crocea larvae. Aquaculture 410, 122–128. doi: 10.1016/j.aquaculture.2013.05.018
Keywords: methylmercury, paralichthys olivaceus, growth, metabolomic, transcriptomic, response mechanism
Citation: Ren Z, Ning J, Cao L, Liu J, Zhan J, Wang Z, Yu J, Yang J and Lv Z (2022) Metabolomic and transcriptomic analyses reveal response mechanisms of juvenile flounder (Paralichthys olivaceus) to sublethal methylmercury. Front. Mar. Sci. 9:979357. doi: 10.3389/fmars.2022.979357
Received: 27 June 2022; Accepted: 26 September 2022;
Published: 11 October 2022.
Edited by:
Seyyed Morteza Hoseini, Iranian Fisheries Science Research Institute (IFSRI), IranReviewed by:
Patrícia Pereira, University of Aveiro, PortugalMelika Ghelichpour, University of Tehran, Iran
Copyright © 2022 Ren, Ning, Cao, Liu, Zhan, Wang, Yu, Yang and Lv. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhonghua Ren, cmVuemhvbmdodWExNkBtYWlscy51Y2FzLmFjLmNu; Zhenbo Lv, eXRsdnpoZW5ib0AxNjMuY29t
 Zhonghua Ren
Zhonghua Ren Junhao Ning2
Junhao Ning2 Liang Cao
Liang Cao Zhikang Wang
Zhikang Wang Junbao Yu
Junbao Yu Jisong Yang
Jisong Yang