
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Mar. Sci. , 28 August 2018
Sec. Marine Molecular Biology and Ecology
Volume 5 - 2018 | https://doi.org/10.3389/fmars.2018.00309
This article is part of the Research Topic The Cellular Biomineralization Pathways of Marine Organisms View all 12 articles
 Anna V. Ivanina1
Anna V. Ivanina1 Ballav Borah2
Ballav Borah2 Tadas Rimkevicius1
Tadas Rimkevicius1 Jason Macrander1
Jason Macrander1 Helen Piontkivska3
Helen Piontkivska3 Inna M. Sokolova4,5*
Inna M. Sokolova4,5* Elia Beniash2*
Elia Beniash2*Vascular endothelial growth factor (VEGF) and vascular endothelial growth factor receptor (VEGFR) play a role in early development, organogenesis, and regeneration, as well as biomineralization of invertebrates. The involvement of VEGF and VEGFR in biomineralization was demonstrated in sea urchin larvae but its role in the biomineralization of other invertebrate groups such as mollusks is not known. We assessed the potential role of VEGF signaling on biomineralization of a model marine bivalve, the Pacific oyster Crassostrea gigas, by analyzing the effects of a VEGFR inhibitor (50 nM axitinib) on shell growth, shell hardness, and expression profiles of biomineralization-related genes. Bioinformatics analysis identified a wide range of biomineralization-related genes potentially activated by VEGF including carbonic anhydrases (CAs), sodium/proton and sodium/calcium exchangers, calmodulins, and genes involved in chitin metabolism. Exposure to a VEGFR inhibitor axitinib led to upregulation of CAs (CA7 and CA12), sodium/proton exchanger 10, sodium/calcium exchanger 1 and 3, as well as calmodulin mRNA in hemocytes (HCs). In the mantle tissue, axitinib exposure led to a compensatory upregulation of VEGFR mRNA but did not affect the expression of other studied biomineralization genes. Expression of VEGF, VEGFR mRNA, or that of the downstream biomineralization-related genes in a non-biomineralizing tissue (the gill) was not affected by axitinib. The shell mass was higher in the axitinib treatment group. Inhibition of VEGFR led to significant increase in shell microhardness, and to a small but statistically significant decrease in crystallinity. Our results therefore indicate that, although VEGF signaling might not play a crucial role in shell biomineralization of C. gigas as it does in mammals and echinoderms, it is involved in regulation of shell formation in bivalves and that effects of VEGF on biomineralization are likely mediated by HCs.
Mollusk shells are composite organomineral structures made of calcium carbonate mineral (primarily calcite and aragonite), and a minor but structurally important organic fraction. Shell formation is a complex biologically regulated process (Zhang et al., 2012) which often occurs under the physicochemical conditions of low or under-saturation with respect to CaCO3 precipitation (Beniash et al., 2010; Dickinson et al., 2013; Ivanina et al., 2013b). Two major tissue types – the mantle and the blood cells [hemocytes (HCs)] – have been identified as the major players in molluscan biomineralization. Epithelial cells of the mantle edge (ME) are traditionally associated with the shell formation as they produce the extracellular matrix (ECM) proteins directing CaCO3 deposition and controlling the structure, orientation and size of CaCO3 crystals (Falini et al., 1996; Nudelman et al., 2006). Transcriptomic studies of the cells of the ME revealed high expression levels of genes encoding ECM proteins thereby supporting earlier reports of the role of the ME in the regulation of shell biomineralization (Marie et al., 2012; Ivanina et al., 2017). Recent studies also implicate mollusk HCs in the biomineralization process. HCs are multifunctional cells involved in the immune defense, wound repair and biomineralization (Roch, 1999; Sokolova, 2009). Different HC populations are specialized on either immune defense or biomineralization (O’Neill et al., 2013; Li et al., 2016a; Ivanina et al., 2017). Biomineralizing HCs accumulate calcium carbonate mineral particles and can move by diapedesis from the blood stream to the extrapallial space (between the mollusk mantle and the shell) depositing CaCO3 onto the shell surface (Mount et al., 2004; Li et al., 2016b). The transcriptomic profiles of the putative biomineralization HCs indicate that they play a role in the mineral transport, and potentially contribute to the shell matrix formation (Ivanina et al., 2017). To date, the molecular mechanisms controlling molluscan biomineralization and regulating the interplay between different biomineralizing cells are not well understood. Knowledge of such mechanisms is essential for understanding the impacts of environmental change on the shell, the main protective structure of mollusks, and thus on the survival and performance of mollusks, many of whom are key aquaculture species and ecosystem engineers (Beniash et al., 2010; Dickinson et al., 2012; Ivanina et al., 2013a).
Vascular endothelial growth factor (VEGF) stimulates angiogenesis and osteogenesis in mammals (Street et al., 2002). Angiogenesis and osteogenesis are closely associated (Schipani et al., 2009), and bone mineralization initiates around blood vessels and potentially involves blood cells (Akiva et al., 2015; Kerschnitzki et al., 2016). In mammals, VEGF induces differentiation of osteoblasts and mineralizing chondrocytes and plays a critical role in bone formation, healing and regeneration (d’Aquino et al., 2007; Wang et al., 2007; Liu et al., 2012; Chan et al., 2015; Hu and Olsen, 2016). Recently, VEGF was found to play a critical role in the regulation of mineralization in echinoderms (sea urchin larvae) where it controls skeleton morphogenesis (Duloquin et al., 2007; Sun and Ettensohn, 2014). VEGF signaling is essential for regulation of morphogenetic patterning of the primary mesenchyme cells (a major biomineralizing cell type of echinoderms) and for activation of the skeletogenic gene regulatory network (Duloquin et al., 2007; Adomako-Ankomah and Ettensohn, 2013, 2014; Sun and Ettensohn, 2014), so that inhibition of VEGF signaling causes significant defects in skeletogenesis (Adomako-Ankomah and Ettensohn, 2013). Furthermore, addition of VEGF to isolated sea urchin micromeres in vitro induces formation of calcitic spicules (Knapp et al., 2012). These data indicate that VEGF is an evolutionary conserved and potent regulator of skeletal morphogenesis. Based on these findings, we hypothesized that VEGF may be involved in the control of mollusk shell formation by regulating the biomineralizing functions of the circulatory HCs and/or those of the biomineralizing mantle cells. To test these hypotheses, we used a specific VEGF receptor (VEGFR) inhibitor axitinib to disrupt VEGF signaling in juvenile and adult C. gigas, and tested the effects of this disruption on transcriptomic profile of key biomineralization genes in the HCs and the mantle as well as on the mechanical and mineralogical properties of the shells.
Juvenile (5–10 mm shell length) and adult Crassostrea gigas (6–7 cm shell length) were obtained from Ocean Alaska Hatchery (Alaska, United States) and Fanny Bay (British Columbia, Canada), respectively. Prior to the experiments, the oysters were acclimated for a week at 20°C and salinity 30 practical salinity units (PSU) in re-circulating water tanks with artificial sea water (ASW; Instant Ocean®, Kent Marine) prior to experiments.
A stock solution of axitinib (ApexBio Technology, Houston, TX, United States) was prepared in DMSO and stored at -20°C. Axitinib is a potent and highly selective inhibitor of VEGFR tyrosine kinase 1, 2, and 3 that binds to the intracellular ATP site domain of the VEGFR thereby inhibiting signal transduction by VEGF at low nanomolar concentrations of axitinib (Gross-Goupil et al., 2013). Oysters were cultured in ASW containing 50 nM axitinib (VEGF-inhibited) or the equivalent concentration of DMSO (control). The working concentration of axitinib was chosen based on the previous published research (Adomako-Ankomah and Ettensohn, 2013; Sun and Ettensohn, 2014) and tested for non-specific toxicity by measuring respiration of juvenile oysters in the presence of different concentrations of axitinib. These pilot studies showed that 50 nM axitinib did not inhibit oyster respiration indicating the lack of non-specific toxicity.
Oysters were divided into eight batches and randomly assigned to either axitinib or control (DMSO) treatment. For each treatment, four replicate tanks for juveniles or two tanks for adults were set up, each tank containing 5 L of ASW with ∼50 juveniles or 5 adult oysters per tank. Water temperature was maintained at 21 ± 1°C and salinity at 30 ± 0.5 PSU in all tanks. Water was changed daily, and axitinib or DMSO was added after each water change. Juvenile oysters were exposed for 14 weeks, and adults for 2 weeks. Oysters were fed ad libitum every day with a commercial algal blend (2 ml tank-1) containing Nannochloropsis oculata, Phaeodactylum tricornutum, and Chlorella spp. (DT’s Live Marine Phytoplankton, Sycamore, IL, United States). Algae were added to the tanks immediately following the water change. No mortality was detected throughout the experiments.
The O2 consumption rates (MO2) were measured in juvenile oysters every week for 14 weeks by closed-chamber respirometry. MO2 was measured in 3 ml water-jacketed chambers using Clarke-type oxygen electrodes (OX1LP-3 ml, Qubit Systems, Kingston, ON, Canada) in ASW at 20°C and salinity 30. For each measurement, two similar-sized juveniles were placed into the chamber and allowed to recover for 30 min from the handling stress. The chambers were then closed, and O2 consumption was monitored for 20–30 min. After the MO2 determinations, all juvenile oysters were collected and stored in 70% ethanol for body and shell mass measurements.
Juveniles of C. gigas from control and axitinib-treated groups were collected at time 0 (prior to exposures) and after 14 weeks of exposure. Total wet body mass of specimens was determined using a microbalance XP 56 (Metler-Toledo, Toledo, OH, United States) with a precision of 0.01 mg or better, and dry tissue mass was calculated from the total dry mass by subtracting the mass of the shell. To obtain the shell mass, juveniles were crushed and incubated in NaOCl overnight to digest the tissues; the remaining shell pieces were then washed in ultrapure water followed by 70% ethanol, freeze dried, and their mass determined using a microbalance.
To ascertain whether sequences annotated as predicted VEGF-A (XP_011449745.1), VEGF-D (EKC18709.1), and VEGFR1 (EKC41995) loci in the C. gigas genome belong to the VEGF and VEGFR gene families in invertebrates, we conducted phylogenetic analysis. Protein sequences were identified using BLASTp at the NCBI, using C. gigas sequences EKC18709 and EKC41995 as queries. Described Pvf1/VEGF (McDonald et al., 2003) and PDGF/VEGF-receptor-related (Pvr) (Parsons and Foley, 2013) homologs from two Drosophila species and representative molluscan genomes were collected. Representative VEGF-A, -B, -C, and -D, and VEGFR1 and VEGFR2 (Holmes and Zachary, 2005) family members from four mammalian genomes (human, chimpanzee, mouse, and rat), collected from Ensembl1, were also included in the analysis. Amino acid sequences were aligned using MUSCLE (Edgar, 2004) as implemented in MEGA7 (Kumar et al., 2016). The resultant multiple sequence alignments of VEGF and VEGFR homologs, respectively, were used to reconstruct the maximum-likelihood trees based on the JTT (Jones et al., 1992) substitution model using MEGA7 (Kumar et al., 2016). Gamma distribution was used to account for the rate of heterogeneity across sites, estimated as ∼7.4 and ∼4 for VEGF and VEGFR datasets, respectively. To maximize the number of shared sites used in the analysis, we used a partial deletion option with a cut-off of 90% coverage to eliminate positions with multiple gaps. In other words, only positions that had less than 10% of gaps across all sequences were included in the tree reconstruction, resulting in 166 and 1027 shared positions used in VEGF and VEGFR tree reconstructions. About 100 bootstrap replications were used to evaluate the reliability of internal branches. Pvf2 and Pvf3 sequences from Drosophila melanogaster and D. simulans were also included in the VEGF phylogenetic tree (Holmes and Zachary, 2005; Tarsitano et al., 2006). Pvf-1 sequences, representing Pvr genes from nematodes Caenorhabditis elegans (Tarsitano et al., 2006) and C. briggsae were used to root the VEGF tree. The VEGFR tree was rooted with human and chimpanzee sequences of PTK7 (tyrosine kinase receptor with seven Ig-like domains), a distant homolog of VEGF (Seipel et al., 2004).
To identify the potential biomineralization genes regulated by the VEGF-dependent signaling cascade, we used a custom bioinformatic pipeline to identify genes that were adjacent to or contained STAT3, AP-1, and CREB binding sites in their promoter regions using annotated genome of the oyster (Crassostrea gigas) (version 9). These transcription factors are key signal transducers activated by the VEGF-VEGFR signaling (Abdollahi et al., 2007; Cao, 2010; Gao et al., 2017). Briefly, the genome was screened for the consensus binding sites for STAT 3 (5′-TTCC C/G T/G AA-3′), AP-1 (5′-TGA G/C TCA-3′), and CREB (5′-TGACGTCA-3′). About 70 base-pair regions containing each potential binding site were obtained from the genome and used in a subsequent BLASTn search to identify its position within the C. gigas genome scaffolds. Position information was then used in combination with a custom Python script (Supplementary Table 1) to isolate 1000 bases upstream within each genome scaffold. For each potential binding site, an approximately 2-kb downstream region of the genome was used to search for annotated genes from the C. gigas genome project, using predicted proteins as query sequences in a BLASTx search. Predicted proteins with e-values less than 1e-10 and an identity score greater than 95% were assessed for their possible function through gene ontology (GO) analysis. GO terms were clustered in the program REVIGO with the whole UniProt database used to reference the size of each term and semantic similarities were determined using SimRel medium (0.7) similarity. Among 12,436 genes that have binding sites for STAT3 (1528 genes), AP-1 (7976 genes), and CREB (3932 genes) in the promoter, we identified five isoforms of carbonic anhydrase (CA2, CA3, CA7, CA12, and CA13), two isoforms of sodium-calcium exchangers (NaHE1 and NaHE3), two isoforms of sodium-calcium exchangers (NaCaE1 and NaCaE3), a sarcoplasmic Ca binding protein, two isoforms of calmodulin (1 and 12) as well as four isoforms of CSs (CSC, CS1, CS6, and CS8), which were further used for transcription analyses (Supplementary Table 2).
The mRNA expression of selected biomineralization-related genes (Table 1) was determined in the ME, central mantle (CM), gills, and HCs of adult C. gigas exposed to axitinib or DMSO for 2 weeks. Total RNA was extracted from 50 to 100 mg of the mantle or gill tissues using TRI Reagent (Sigma-Aldrich, St. Louis, MO, United States) according to the manufacturer’s protocol. RNA from HCs was isolated using mini ZR RNA MiniPrep™ kit (Zymo Research, Irvine, CA, United States) according to the manufacturer’s instructions. RNA concentration was determined using NanoDrop 2000 spectrophotometer (Thermo Scientific, Pittsburg, CA, United States). Single-stranded cDNA was obtained from 0.2 μg of the total RNA using 50 U μl-1 SMARTScribe™ reverse Transcriptase (Clontech, Mountain View, CA, United States) and 20 μmol l-1 of oligo(dT)18 primers. Transcript expression of the target genes was determined by qRT-PCR using a 7500 Fast Real-Time PCR System (Applied BioSystems/Life Technologies, Carlsbad, CA, United States) and SYBR® Green PCR kit (Life Technologies, Bedford, MA, United States) according to the manufacturer’s instructions. The qRT-PCR reaction mixture consisted of 7.5 μl of 2X SYBR® Green master mix, 0.3 μmol l-1 of each forward and reverse gene-specific primers (Table 1), and 1.5 μl of 10X diluted cDNA template and water to adjust to 15 μl. The reaction mixture was subjected to the following cycling: 10 min at 95°C to denature DNA and activate Taq polymerase, and 40 cycles each of 15 s at 94°C, 30 s at 60°C, and 30 s at 72°C. Serial dilutions of a single cDNA sample (from the ME) were amplified during each run to determine amplification efficiency and to test for potential run-to-run amplification variability (Pfaffl, 2001). The target gene mRNA expression was standardized relative to β-actin mRNA and the internal cDNA standard (Pfaffl, 2001).
Juveniles of C. gigas from control and axitinib-treated groups were collected at time 0 (prior to exposures), 2 days, 6 weeks, and 14 weeks of exposure and fixed in 70% ethanol. Juveniles were placed overnight in 4% NaOCl solution to remove organic material, washed two times 30 min each with deionized water followed by a wash in 70% ethanol. Isolated shells were lyophilized for 24 h, and five specimens per group were ground and pressed in KBr pellets. Pellets were analyzed using a FTIR spectrometer (Bruker Vertex 70, Bruker, Billerica, MA, United States) in a transmittance mode with the resolution of 4 cm-1 and 128 scans per sample. Absorbance spectra were processed using Spectrum 5.1 software (Perkin Elmer, Shelton, CN, United States). The 600–1000 cm-1 regions of the spectra were isolated, baseline corrected and normalized, and the positions and heights of absorption peaks of ν4 and ν2 vibrational bands were measured. The ν4/ν2 band intensity ratio is inversely proportional to calcite crystal and is commonly used as a proxy for the crystallinity of the mineral phase (Beniash et al., 1997; Gueta et al., 2007).
The lyophilized right valves of the juvenile oysters were mounted and polymerized in Epofix epoxy resin (EMS, Hatfield, PA, United States) at room temperature. Five individuals from the axitinib-treated and control groups collected at time 0, 2 days, 6 weeks, and 14 weeks of exposure were used for analyses. To expose shells, the blocks were manually polished with 320 grit paper in the plane normal to the surface and transecting the shell from acute apical tip to its most distal edge until the midline of the shell was exposed. The samples were further polished with 400, 600, and 800 grit paper using Buehler Minimet 1000 polisher, followed by fine polishing with Metadi diamond suspensions down to 0.25 μm. All polishing was carried out in CaCO3-saturated water. The microhardness tests were performed using Identamet (Buehler, Lake Bluff, IL, United States) microindenter at 0.25 N load and 5 s dwelling time with Vickers tip. Five indentations were conducted per each sample. To avoid edge effects, each indentation was positioned at least two indentation diagonals away from the neighboring indentations and the edges of the shell. Microhardness values were averaged for five indentations from each shell, and the average value used as a biological replicate.
Two-way ANOVA was used to test the effects of the tissues and presence/absence of VEGF inhibitor on the studied traits (Table 2). For shell traits, one-way ANOVA and/or Student’s t-test were used. Prior to analyses, data were tested for normality and homogeneity of variance by Kolmogorov-Smirnoff and Levine’s tests, respectively, and normalized as needed using Box-Cox common transforming method. Fisher’s Least Significant Differences (LSD) tests were used for planned posthoc comparisons of the differences between the pairs of means of interest. All statistical analyses were performed with JMP Pro 12 (Cary, NC, United States) or OriginPro 2015 software package (Northampton, MA, United States). Differences were considered significant if the probability of Type I error was less than 0.05. The data are presented as means ± standard errors of means (SEM) unless indicated otherwise.
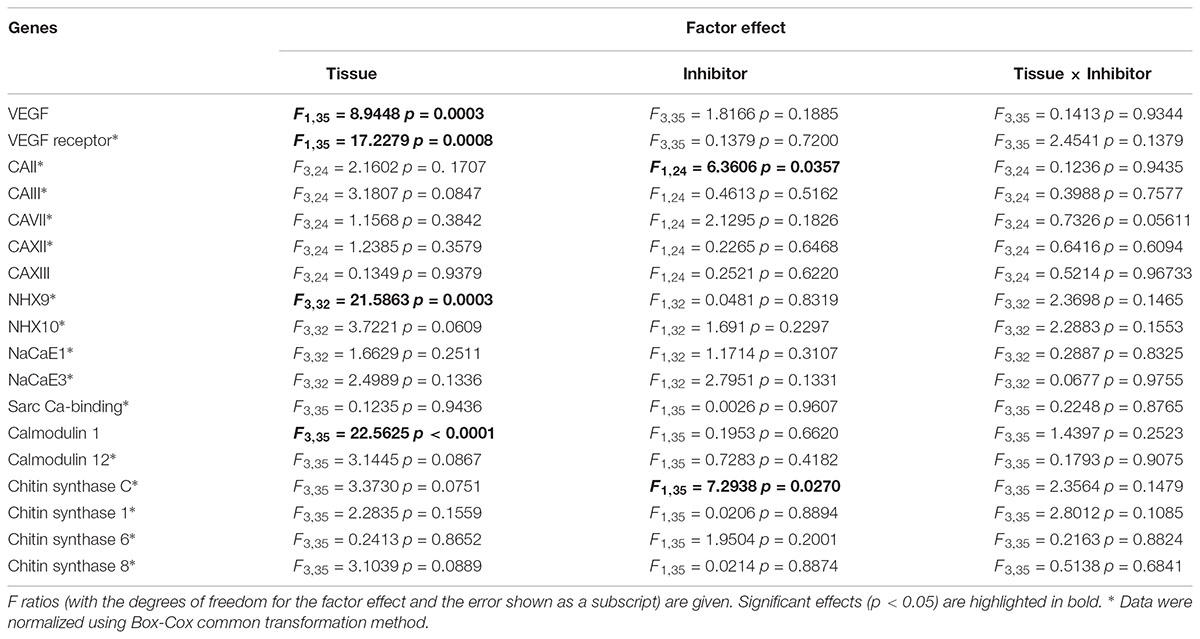
TABLE 2. ANOVA: Effects of the axitinib on the expression of biomineralization mRNAs in Crassostrea gigas.
Dry mass of the soft tissues of juveniles prior to experiment was 0.32 ± 0.05 mg (N = 10); after 14 weeks of exposure it increased to 0.42 ± 0.06 mg (N = 15) and 0.47 ± 0.11 mg (N = 15) in the control and axitinib-exposed group, respectively (p > 0.05 for the differences between the groups). There was no significant difference in shell mass between 0 and 14 weeks time-points in the control group (5.31 ± 0.32 mg, N = 10 and 5.27 ± 0.25 mg, N = 15), respectively. A difference approaching significance in the shell mass was found between the time 0 and 14 weeks of exposure to axitinib-group (p = 0.074), with masses of 5.31 ± 0.32 mg (N = 10) and 6.28 ± 0.39 mg (N = 15), respectively. Importantly, significant difference in the shell mass was found after 14 weeks of exposure between the axitinib-treated and control juveniles (p = 0.048). These data indicate that inhibition of VEGF signaling pathway lead to a small but significant increase in the shell mass while the soft tissue mass was not affected.
The duration of experimental exposure had no significant effect on the respiration of control or axitinib-exposed juvenile oysters (F165,13 = 1.56, p = 0.149); therefore, data for different time points within each experimental group were pooled for subsequent analyses. Exposure to axitinib had no effect on O2 consumption rates of juveniles (24.17 ± 2.8 μmol O2 animal-1 h-1 and 28.47 ± 3.09 μmol O2 animal-1 h-1 for control and axitinib-exposed group, respectively, N = 82–84, p = 0.307).
The FTIR spectroscopic analysis showed a significantly higher ν4/ν2 intensity ratio in the shells of juveniles from the axitinib-treated group compared with those from the control group (3.24 ± 0.28 and 2.91 ± 0.06, respectively, p = 0.046) after 14 weeks exposure. This indicates that the mineral of the shells in the axitinib-treated group is less crystalline than in the controls (Beniash et al., 1997; Gueta et al., 2007).
The Vickers microhardness of shells of juveniles in the axitinib-treated group was higher than in the control (ANOVA for the effect of treatment: F1,26 = 23.48, p < 0.001) and increased with the exposure duration (i.e., the age of oysters) in both groups (ANOVA for the effect of exposure time: F2,26 = 8.08, p = 0.002). After 14 weeks of exposure, the Vickers microhardness was ∼20% higher (202.74 ± 21.72) in the axitinib-treated group compared to the controls (166.75 ± 24.17) (p = 0.04).
Phylogenetic analysis confirmed presence of VEGF and VEGFR homologs in the oyster genome. As Figure 1A shows, both C. gigas sequences annotated as VEGF (XP_011449745.1, annotated as VEGF-A, and EKC18709.1, annotated as VEGF-D) cluster with other molluscan VEGF-like sequences, including XP_022327278.1 sequence from C. virginica annotated as VEGF C-like. Oyster VEGF-like sequences form a cluster with a pair of sequences, one of which is annotated as a platelet-derived growth factor subunit B (PDGF-B) from Yesso scallop grouped with a recently described PDGF/VEGF homolog from the pearl oyster Pinctada fucata (Huang et al., 2017), although the internal branch lacks significant bootstrap support. Jointly, these bivalve sequences, together with representatives of gastropods, form a clade with Pvf1 sequences from Drosophila genomes. Mammalian representatives of VEGF gene family form a separate clade from the invertebrate Pvf1-like group, subdivided into VEGF-A and -B, and VEGF-C and -D clusters, supported by the bootstrap values of 89, 94, and 100%, respectively. Pvf2 and Pvf3 sequences from Drosophila cluster together outside of this mammalian clade, with the bootstrap support of 85%, thus, offering (albeit weak) support for the hypothesis that oyster VEGF-like sequences are more closely related to Pvf1 genes. The lack of strong bootstrap support for internal branches representing deeper divergences within this gene superfamily likely reflects ancient divergence times between taxa, for example, the estimated divergence time between oysters and Drosophila is ∼753 MYA, per TimeTree.org (Hedges et al., 2015) as well as a relatively small number of shared sites available in this alignment (166 amino acid sites were used in tree reconstruction, out of total 328 amino acids in C. gigas protein).
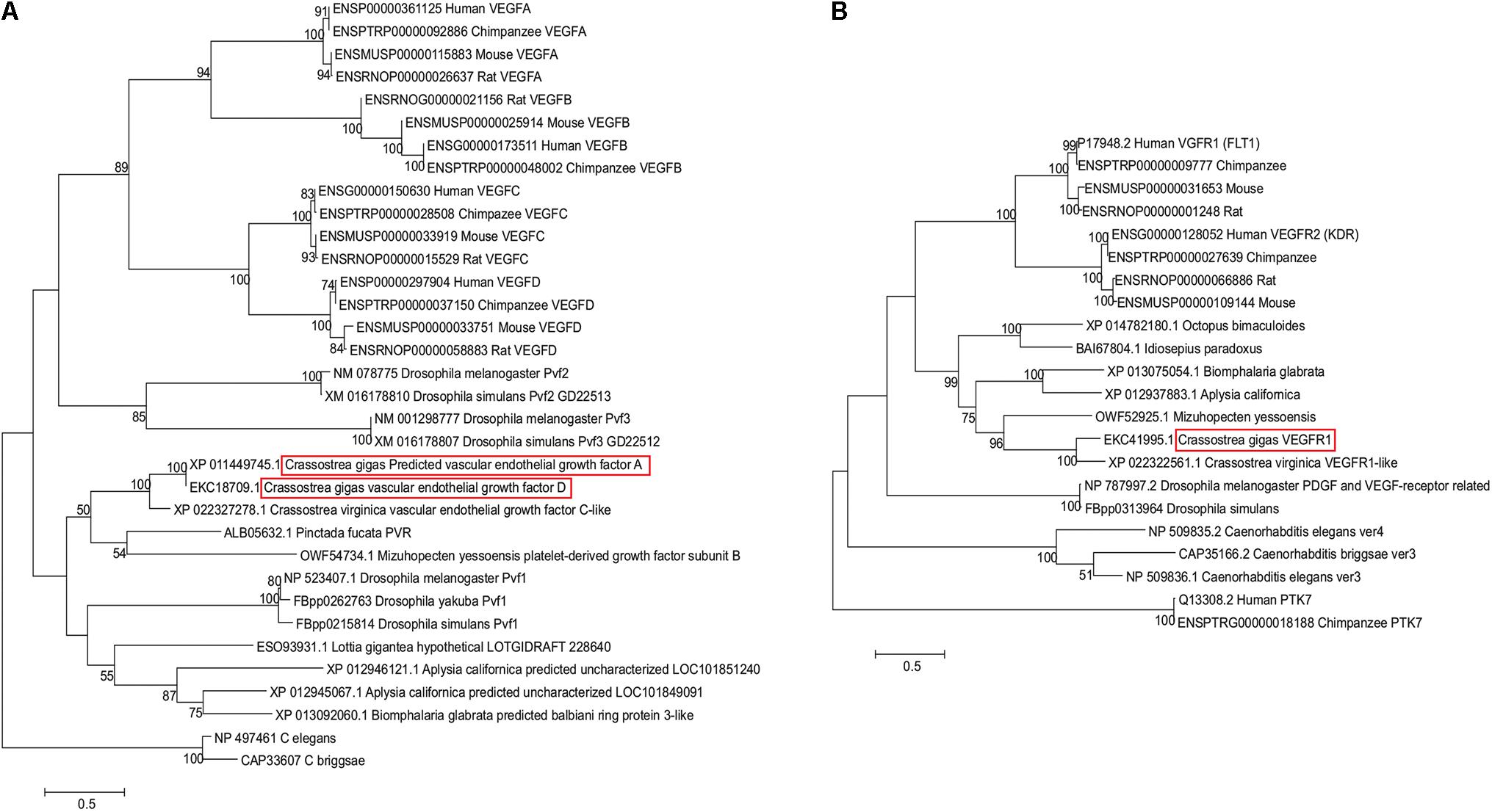
FIGURE 1. Maximum-likelihood-based phylogenetic trees of VEGF (A) and VEGFR (B) sequences reconstructed using JTT+gamma substitution model. Each sequence is designated by its GenBank or Ensembl accession number and species name. Sequences from C. gigas are highlighted with red boxes. Numbers above branches represent percent bootstrap support.
Likewise, predicted VEGFR sequence from C. gigas is clustered with the VEGFR-like sequence from C. virginica, joined by a VEGFR sequence from Yesso scallop (Figure 1B). These bivalve sequences form a clade with representatives of gastropods, supported by a 82% bootstrap, and together they form a molluscan clade with cephalopod representatives (99% support). Representative mammalian VEGFR1 (FLT1) and VEGFR2 (KDR) sequences form a separate sister clade, split into two clusters, supported by 100% bootstrap values, with Drosophila representatives clustered outside of this grouping, with 61% support for the joint cluster. This indicates that oyster VEGFR-like sequences are likely related to mammalian VEGFR1 and -R2 sequences. Interestingly, according to the multiple sequence alignment, the binding region of axitinib at human VEGFR2 protein [residues 801–826 (McTigue et al., 2012)] appears to share multiple sites with amino acid residues that have the same physicochemical properties not only with mammalian VEGFR1 sequences, but also with invertebrate sequences (Figure 2). Such conservation at the level of physicochemical amino acid properties suggests that similar to interactions with the mammalian VEGFR2 proteins, axitinib likely has an inhibitory effect on oyster VEGF receptors.
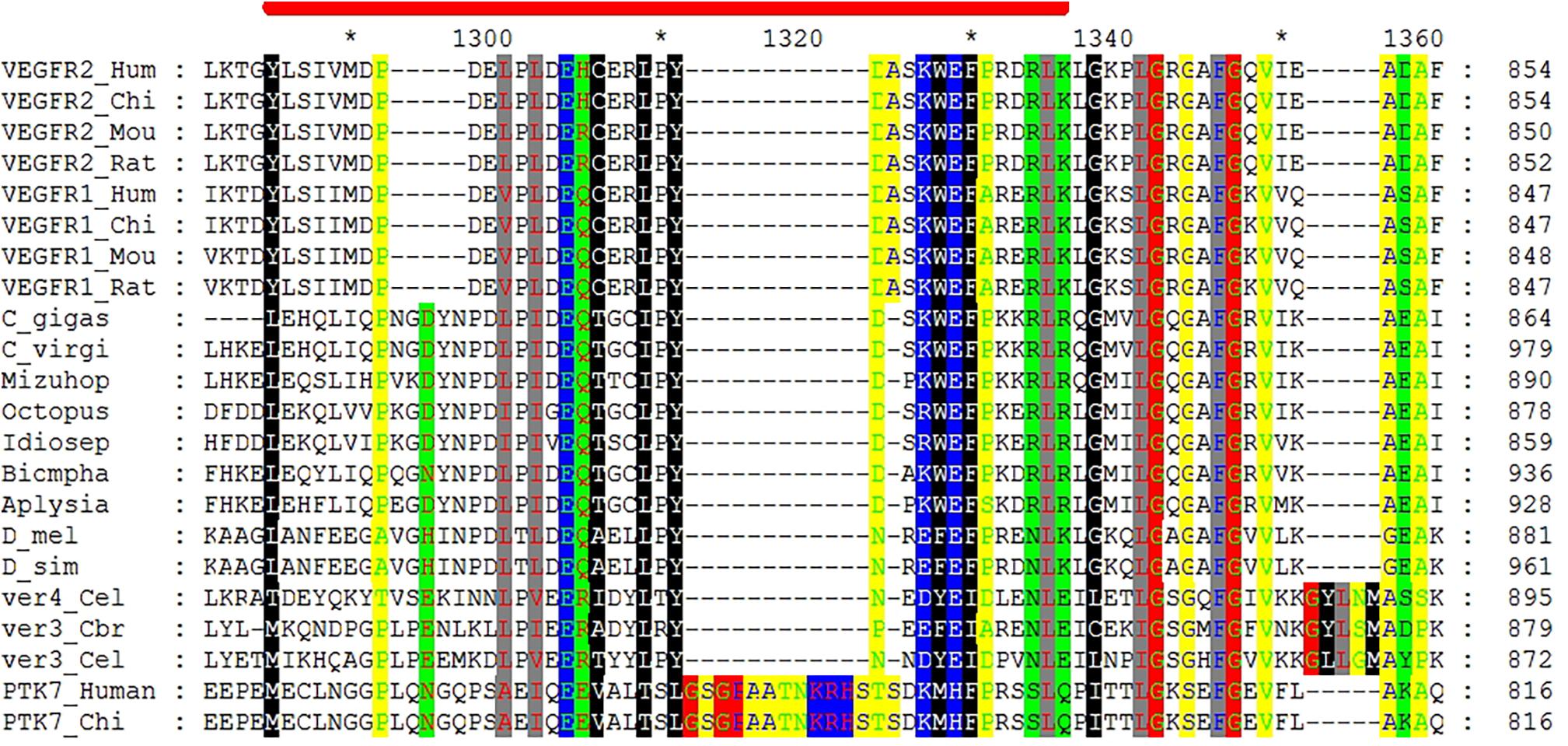
FIGURE 2. Section of the multiple sequence alignment of VEGFR proteins sequences that shows conservation of physicochemical properties of residues involved in interactions with axitinib. The coordinates on right refer to the absolute amino acid position number for each sequence, while coordinates above the alignment refer to the position in the alignment. Red stripe marks the span between Y801 and K826 residues of human VEGFR2 involved in axitinib binding according to McTigue et al. (2012). The species name abbreviations, top to bottom, are Hum (human), Chi (chimpanzee), Mou (mouse), Rat (rat), C. gigas, C. virgi (C. virginica), Mizuhop (Mizuhopecten yessoensis), Octopus (Octopus bimaculoides), Idiosep (Idiosepius paradoxus), Biompha (Biomphalaria glabrata), Aplysia (Aplysia californica), D_mel (Drosophila melanogaster), D_sim (Drosophila simulans), Cel (Caenorhabditis elegans), and Cbr (Caenorhabditis briggsae). The following sequences (GenBank, FlyBase, or Ensembl accession numbers) were used in the alignment (top to bottom): ENSG00000128052, ENSPTRP00000027639, ENSMUSP00000109144, ENSRNOP00000066886, P17948.2, ENSPTRP00000009777, ENSMUSP00000031653, ENSRNOP00000001248, EKC41995.1, XP_022322561.1, OWF52925.1, XP_014782180.1, BAI67804.1, XP_013075054.1, XP_012937883.1, NP_787997.2, FBpp0313964, NP_509835.2, CAP35166.2, NP_509836.1, Q13308.2, and ENSPTRG00000018188. Conserved sites containing amino acid residues with identical physicochemical properties show polar (black lettering, green background), hydrophobic (white lettering, black background), positively charged (pink lettering, blue background), amphoteric (red lettering, green background), small (green lettering, yellow background), aliphatic (red lettering, gray background), negatively charged (green lettering, blue background), and charged (white lettering, blue background) residues.
Our bioinformatic pipeline recovered 79859 predicted total sites for the studied transcription factors, including 6040 sites for STAT3, 42967 sites for AP1, and 30852 sites for CREB. Of these, there were 12436 genes found adjacent to the transcription factors, specifically with 1528 adjacent to STAT3; 7976 to AP1 and 3932 to CREB. Of these, 996, 5152, and 2522 genes contained GO information for STAT3, AP1, and CREB, respectively (Supplementary Table 1). In total 526, 1663, and 897 GO groups were used in the program REVIGO for STAT3, AP1, and CREB, respectively.
Across all three transcription factors, we found similar REVIGO profiles associated with different GO groups in the Biological Process and Molecular Function domains. Some of the most abundant GO groups for the Biological Process domain were relatively large and encompassing GO groups, including metabolism [GO:0008152], regulation of DNA-templated transcription [GO: 0006355], carbohydrate metabolism [GO:0005975], intracellular signal transduction [GO:0035556], and transmembrane transport [GO:0055085]. Some of the smaller GO categories found in high abundance associated with the transcription factors included chitin metabolism [GO:0006030], hemophilic cell adhesion via plasma membrane adhesion molecules [GO:0007156], and integrin-mediated signaling pathway [GO:0007229] (Supplementary Figure 1).
In the Molecular Function domain, the most abundant GO groups corresponded to large encompassing groups, such as ATP binding [GO:0005524] and zinc ion binding [GO:0008270]. There were also abundant groups associated with intermediate GO encompassing groups, such as calcium ion binding [GO:0005509] and G-protein coupled receptor activity [GO:0004930]. Finally, among the lower abundant GO groups, there was proportionately larger representation of chitin binding [GO:0008061] and scavenger receptor activity [GO:0005044] compared to other GO categories occurring in similar frequencies (Supplementary Figure 1). Among these, one of the G-protein coupled receptors was linked back to a kisspeptin protein (K1PWK8_CRAGI) which has putative regulatory role in the mantle function (Cardoso et al., 2016). Additionally, there were numerous proteins associated with calcium ion binding, chitin binding, and other GO groups relevant to mantle function and formation selected for further investigation of the transcript levels (as described below).
Under the control conditions, VEGF mRNA expression varied among the tissues of adult oysters, with significantly lower expression levels in HCs than in the ME or the gills, and intermediate values in the CM (Figure 3A). The exposure to axitinib led to a modest increase in the VEGF expression in HCs, which eliminated the tissue-specific differences in VEGF mRNA levels (Figure 3A). Expression of VEGFR in the control group was significantly higher in gills than in the tissues involved in biomineralization (Figure 3B). Exposure to axitinib led to an increase of VEGFR mRNA expression in the mantle and a decrease in the gills (Figure 3B).
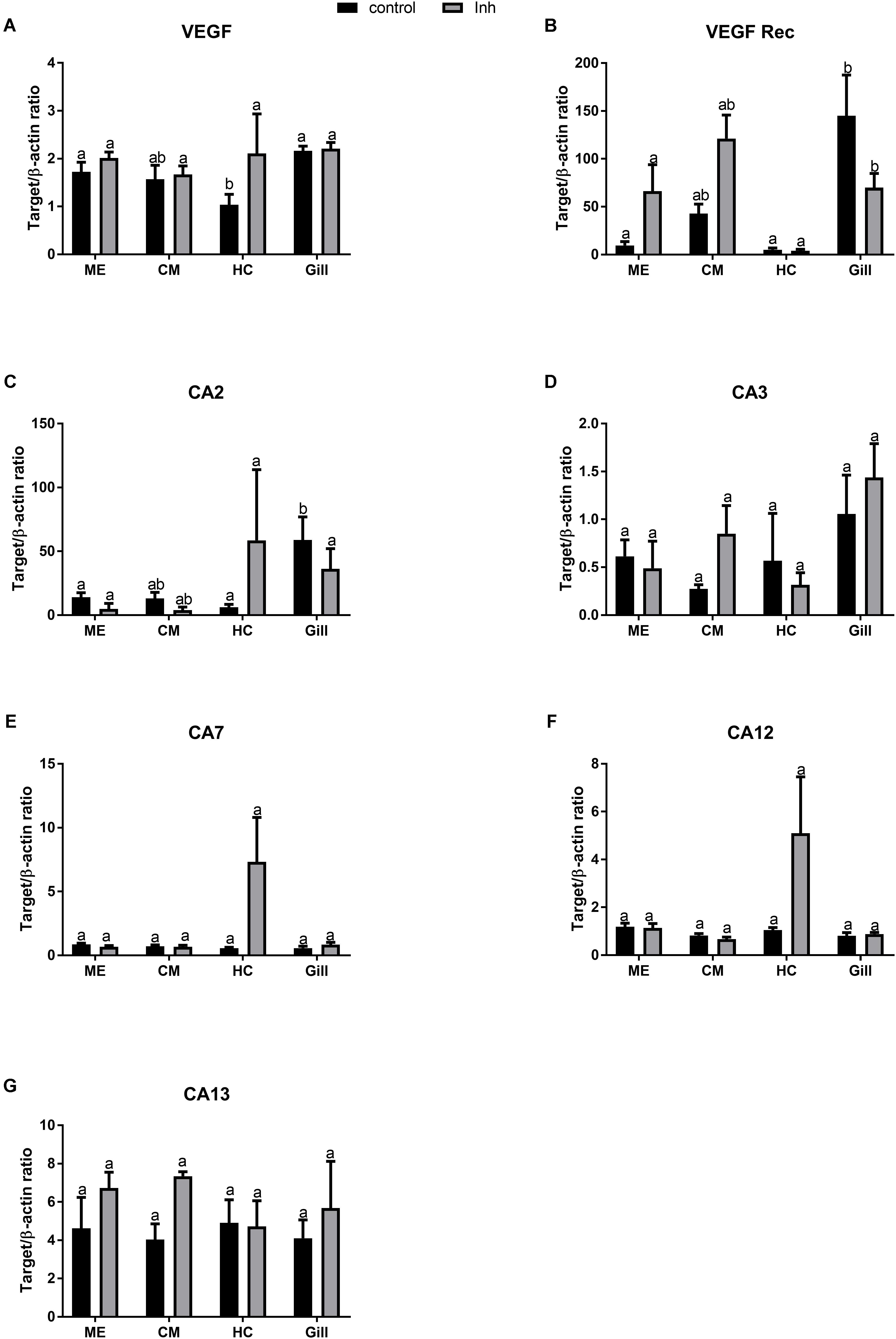
FIGURE 3. Levels of mRNA expression of VEGF, VEGF receptor, and different isoforms of carbonic anhydrase in tissues of adult C. gigas in response to a 2-week axitinib exposure. Genes: (A) VEGF – vascular endothelial growth factor, (B) VEGF Rec – vascular endothelial growth factor receptor; (C–G) CA2, CA3, CA7, CA12, and CA13 – carbonic anhydrases 2, 3, 7, 12, and 13, respectively. Control – control oysters and Inh – axitinib-exposed oysters. Expression levels in the mantle edge (ME), central mantle (CM), hemocytes (HC), and the gills (Gill) are shown. Different letters indicate significant differences between different tissues under the same treatment conditions (i.e., control or axitinib exposure); columns that do not share a letter are significantly different (p < 0.05). N = 3–5.
Out of five studied CA isoforms (CA2, CA3, CA7, CA12, and CA134), only CA2 was differentially expressed in the studied tissues with higher mRNA level in the gills compared with the mantle or HCs (Figures 3C–G). Exposure to VEGFR inhibitor significantly affected expression profile of CA2 but not that of other studied CA isoforms in oyster tissues (Figures 3C–G and Table 2). Notably, exposure to axitinib led to a strong increase in mRNA expression of CA2, CA7, and CA12 in HCs (by ∼9, 12- and 5-fold, respectively); however, this increase was not statistically significant due to a large variation and relatively small sample size (N = 3–5).
Under the control conditions, expression of NaHE9 transporter was higher in the gills than in the other studied tissues (Figure 4A and Table 2), while mRNA expression of NaHE10 was significantly lower in the gills than in the ME cells (Figure 4B). Expression of NaCaE1 was significantly elevated in HCs (Figure 4C), where expression of NaCaE3 was similar in all studied tissues (Figure 4D). Exposure to axitinib led to a notable albeit statistically non-significant increase of NaHE10, NaCaE1 and NaCaE3 mRNA expression in HCs (by ∼4.5, 2- and 4-fold, respectively), whereas NaHE9 expression remained low (Figure 4). Axitinib exposure also significantly stimulated NaHE10 mRNA expression in the gills by 85% (Figure 4B).
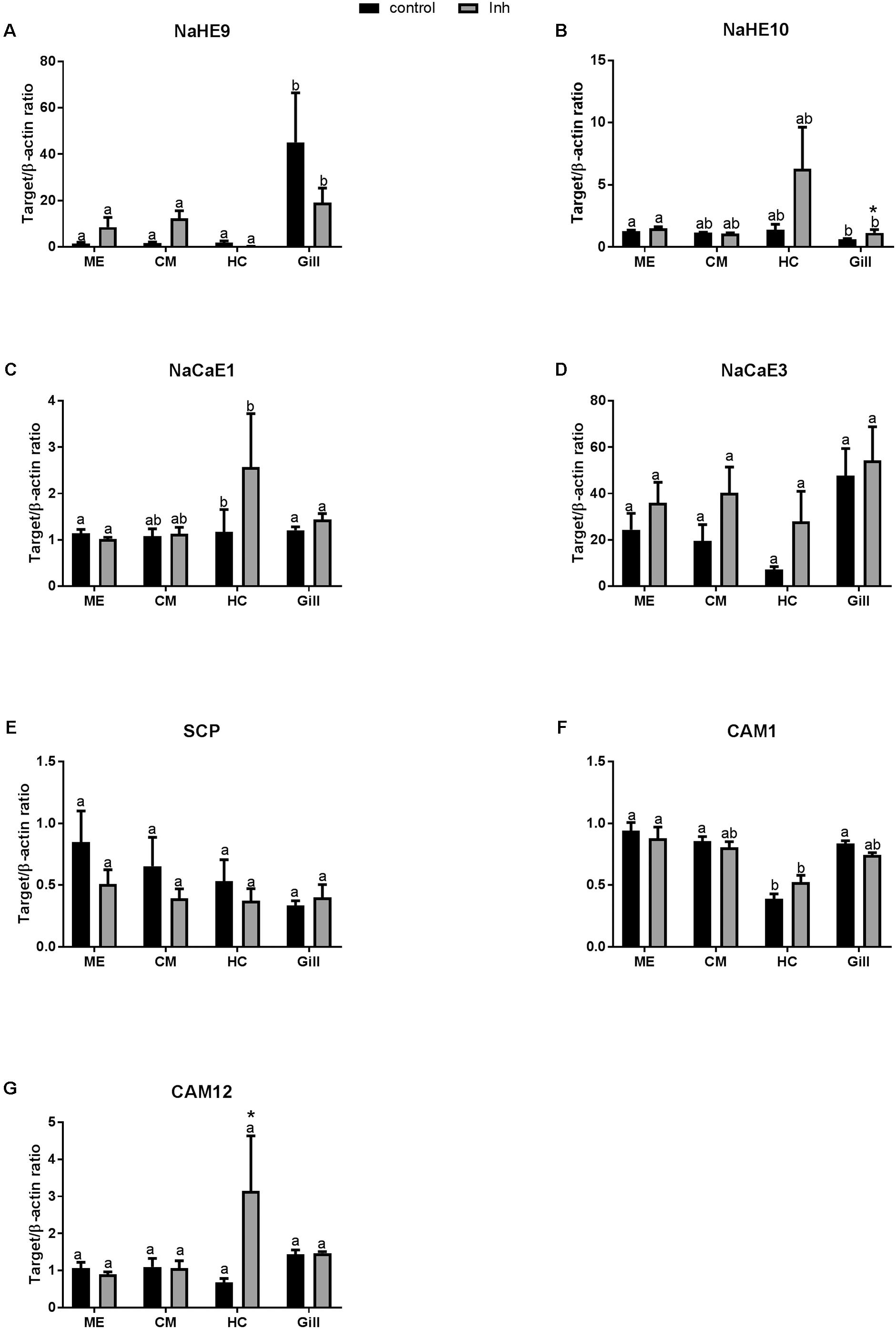
FIGURE 4. Levels of mRNA expression of ion transporters and Ca binding proteins in tissues of adult C. gigas in response to a 2-week axitinib exposure. Genes: (A,B) NaHE9 and NaHE10 – Na+/H+ exchangers 9 and 10, respectively; (C,D) NaCaE1 and NaCaE3 – Na+/Ca2+ exchangers 1 and 3, respectively; (E) SCP – sarcoplasmic calcium-binding protein; (F,G) CAM 1 and CAM 12 – calmodulin 1 and 12, respectively. Control – control oysters and Inh – axitinib-exposed oysters. Expression levels in the mantle edge (ME), central mantle (CM), hemocytes (HC), and the gills (Gill) are shown. Different letters indicate significant differences between different tissues under the same treatment conditions (i.e., control or axitinib exposure); columns that do not share a letter are significantly different (p < 0.05). Asterisks indicate significant differences between the control and axitinib-exposed oysters in the same tissue. N = 3–5.
Among the three studied Ca2+-binding genes [calmodulin 1 (CAM1), calmodulin 12 (CAM12) and sarcoplasmic calcium-binding protein (SCP)], mRNA expression or CAM1 and CAM12 were lower in HCs than in other tissues of the control oysters (Figures 4E–G and Table 2). Expression levels of CAM1 and SCP mRNA were not significantly affected by axitinib exposure (Figures 4E,F). In contrast, mRNA expression of CAM12 was significantly higher in HC of animals exposed to the VEGFR inhibitor compared with the respective controls (Figure 4G).
In control oysters, mRNA expression of chitin synthase C (CSC) and chitin synthase 6 (CS6) were similar in all studied tissues (Figures 5A,C). Expression of CS1 mRNA was lower in the gills than in the other tissues, although the differences were only significant between the gills and the CM cells (Figure 5B). Expression of CS8 mRNA was the lowest in HCs compared with other tissues (Figure 5D). Axitinib treatment led to a statistically significant increase in CSC expression in the gills (Figure 5A and Table 2). The expression of CS1 was higher in CM than in other tissues, and this difference was significant between CM and gills (Figure 3B). The exposure to the VEGFR inhibitor led to a decrease in CS1 expression in CM, yet significant differences remained between CM and gills (Figure 5B). Notably, axitinib exposure led to a strong upregulation of CS6 and CS8 mRNA expression in HCs (by ∼15 and 45-fold, respectively) albeit these differences were not statistically significant (Figures 5C,D).
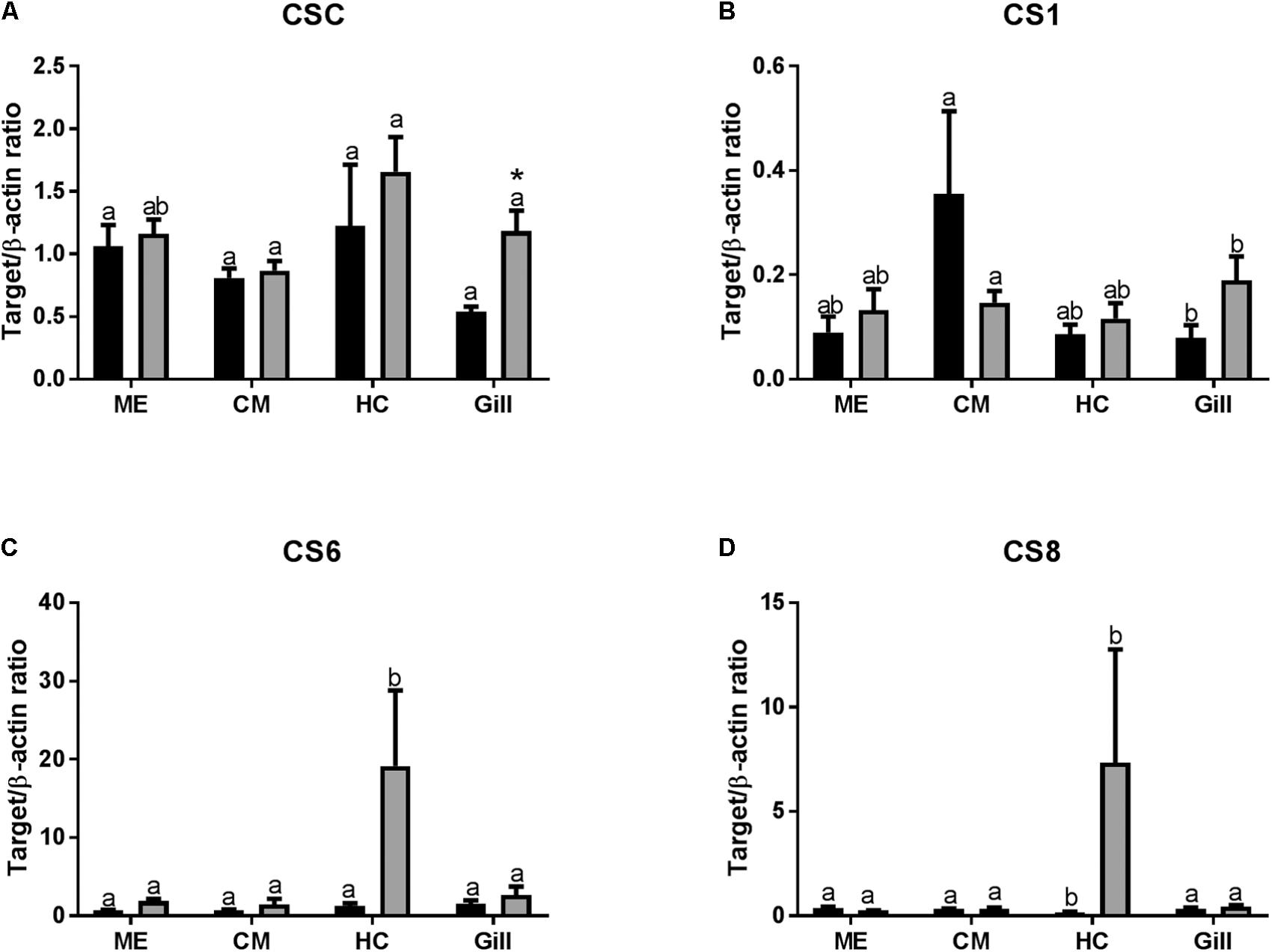
FIGURE 5. Levels of mRNA expression of chitin synthases in tissues of adult C. gigas in response to a 2-week axitinib exposure. Genes: (A–D) CSC, CS1, CS6, and CS8 – chitin synthases C, 1, 6, and 8, respectively. Control – control oysters and Inh – axitinib-exposed oysters. Expression levels in the mantle edge (ME), central mantle (CM), hemocytes (HC), and the gills (Gill) are shown Different letters indicate significant differences between different tissues under the same treatment conditions (i.e., control or axitinib exposure); columns that do not share a letter are significantly different (p < 0.05). Asterisks indicate significant differences between the control and axitinib-exposed oysters in the same tissue. N = 3–5.
Disruption of VEGF signaling did not result in the detrimental effects on biomineralization in juvenile C. gigas. The shell mass after 14 weeks of axitinib exposure was significantly higher than in the controls, suggesting that VEGF signaling might act as a suppressor of shell growth. Notably, calcite crystallinity slightly but significantly decreased, while there was a significant 20% increase in the microhardness of the shells from the specimens exposed to axitinib, suggesting a complex effects of this inhibitor on shell formation. However, the body growth (assessed by the soft body mass) and metabolic rates were similar in the control and axitinib-exposed juveniles indicating that the disruption of VEGF signaling does not lead to major negative systemic effects in oysters. This contrasts findings in sea urchin larvae where axitinib exposure disrupts the skeletogenic gene regulatory network, resulting in a dramatic disruption of skeletogenesis (Adomako-Ankomah and Ettensohn, 2013, 2014; Sun and Ettensohn, 2014) and the fact that VEGF activates spiculogenesis in the in vitro sea urchin PMC culture (Knapp et al., 2012). Lack of the major disruption of shell formation in C. gigas juveniles during prolonged (14 weeks) exposure to axitinib may indicate that VEGF signaling plays a less prominent and potentially different role in biomineralization in bivalves than in echinoderms and vertebrates (d’Aquino et al., 2007; Wang et al., 2007; Liu et al., 2012; Adomako-Ankomah and Ettensohn, 2013, 2014; Sun and Ettensohn, 2014; Chan et al., 2015; Hu and Olsen, 2016). Alternatively, this may indicate lower sensitivity of the bivalve VEGFR to axitinib. Although the axitinib-binding properties of the bivalve VEGFR have not been studied, the latter explanation appears less probable, given similarity of the putative axitinib-binding sites of oyster and mammalian VEGFR (Figure 2) and consistent responses of the shell mass and mechanical properties as well as the transcription profiles of VEGF-responsive genes to axitinib exposure in oysters.
It is worth noting that mineral crystallinity of the shells of juvenile C. gigas under control conditions was higher than the reported crystallinity values for the shells of the juveniles of a closely related eastern oyster Crassostrea virginica (Beniash et al., 2010), with ν4/ν2 ratios 2.9 vs. 3.9, respectively. The microhardness of the shell was lower in the juveniles of C. gigas than in the juveniles of C. virginica, 170 VH vs. 250 VH (Beniash et al., 2010). The reasons for these differences in shell properties between two morphologically and ecologically similar species are presently unknown, but emphasize the notion that shell formation in bivalves is under the tight and potentially species-specific biological control (Addadi et al., 2006; Checa et al., 2009; Marin et al., 2012).
Our bioinformatic analyses identified three closely related isoforms of VEGF (VEGF-A, -C, and -D) and two isoforms of VEGF receptor (similar to the vertebrate VEGFR1) in the genome of C. gigas. Transcriptomic profiles of biomineralization-related genes showed modest change in response to a 2-week exposure to a potent VEGFR inhibitor, axitinib, in adult C. gigas. Expression patterns of the studied CAs, ion transporters, Ca2+ binding proteins and shell matrix producing enzymes (CSs) did not strongly respond to VEGF inhibition in a major biomineralizing tissue, the mantle. This was similar to the general lack of a transcriptomic response of these genes in a non-biomineralizing tissue (the gills), except for the slight but significant upregulation of mRNA level of Na+/H+ exchanger 10 and CSC in the gills of axitinib-exposed oysters. Interestingly, HCs showed the strongest transcriptomic response to VEGFR inhibition of all the studied tissues, with upregulation of multiple genes by 2- to 45-fold in axitinib treatments. Albeit many of these increases were statistically not significant due to the large individual variation and a limited sample size, the consistent trend shows an increase in VEGF expression in HCs of axitinib-treated oysters, as well as a strong upregulation of mRNA for CAs (CA2, CA7, and CA12), Na+-dependent ion transporters (NaHE10, NaCaE1), calmodulin 12, and CSs (CS6 and CS8). This indicates that HCs are more sensitive to the disruption of VEGF signaling and may potentially compensate for it by increased production of VEGF. Furthermore, this indicates that transcriptional regulation of the same genes differs in different tissues of oysters potentially reflecting tissue-specific differences in the densities and/or properties of VEGFR and/or epigenetic changes in the gene promoters that alter sensitivity to VEGF-derived signals. It is intriguing to speculate that the relatively high sensitivity of oyster HCs to VEGF-related signals may be related to the mesodermal origin of these cells (Smith et al., 2016) similar to the primary mesenchyme cells of echinoderms, as well as the mineralizing odontoblasts and chondrocytes of vertebrates, and opposed to the less VEGF-sensitive ectoderm-originated tissues such as the mantle and the gills. This hypothesis as well as the mechanisms underlying the differential sensitivity of tissue types and biomineralization processes to VEGF signaling in different invertebrates require further investigation.
As a corollary, our present study shows that although VEGF might play a role in some aspects of shell formation in mollusks, it is not a key regulator of the mineralization processes as in vertebrates and echinoderms (Mayer et al., 2005; Duloquin et al., 2007; Knapp et al., 2012; Liu et al., 2012) hinting toward a possibility that the involvement of VEGF in biomineralization may have developed in deuterostomata. Future studies are needed to determine the physiological roles of VEGF-VEGFR signaling in oysters and other mollusks such as involvement in the regulation of energy and carbohydrate metabolism, DNA transcription and G-protein coupled receptor signaling identified as the candidate processes for VEGF regulation in our bioinformatic pipeline.
EB and IS conceived the study, developed the research design, analyzed the data, and wrote the manuscript. AI participated in the development of the experimental design, conducted the axitinib exposure experiments, performed the gene expression and respiration studies, and participated in the manuscript writing. BB conducted the microhardness tests and FTIR spectroscopy of the shells and performed the mass measurements. TR assisted AI and BB in conducting the experiments, sample preparation, and data collection. HP conducted the phylogenetic analysis and participated in the manuscript preparation. JM conducted the bioinformatic analysis of gene networks and participated in the manuscript preparation.
This work was supported by the United States National Science Foundation (awards IOS-1557870 and IOS-1557551 to IS and EB). JM was supported by the United States National Science Foundation award OCE-1536530. TR is a participant in the REU program supported by NSF awards IOS-1557870 and IOS-1557551.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmars.2018.00309/full#supplementary-material
Abdollahi, A., Schwager, C., Kleeff, J., Esposito, I., Domhan, S., Peschke, P., et al. (2007). Transcriptional network governing the angiogenic switch in human pancreatic cancer. Proc. Natl. Acad. Sci. U.S.A. 104, 12890–12895. doi: 10.1073/pnas.0705505104
Addadi, L., Joester, D., Nudelman, F., and Weiner, S. (2006). Mollusk shell formation: a source of new concepts for understanding biomineralization processes. Chem. Eur. J. 12, 981–987. doi: 10.1002/chem.200500980
Adomako-Ankomah, A., and Ettensohn, C. A. (2013). Growth factor-mediated mesodermal cell guidance and skeletogenesis during sea urchin gastrulation. Development 140, 4214–4225. doi: 10.1242/dev.100479
Adomako-Ankomah, A., and Ettensohn, C. A. (2014). Growth factors and early mesoderm morphogenesis: insights from the sea urchin embryo. Genesis 52, 158–172. doi: 10.1002/dvg.22746
Akiva, A., Malkinson, G., Masic, A., Kerschnitzki, M., Bennet, M., Fratzl, P., et al. (2015). On the pathway of mineral deposition in larval zebrafish caudal fin bone. Bone 75, 192–200. doi: 10.1016/j.bone.2015.02.020
Beniash, E., Aizenberg, J., Addadi, L., and Weiner, S. (1997). Amorphous calcium carbonate transforms into calcite during sea urchin larval spicule growth. Proc. R. Soc. Lond. B 264, 461–465. doi: 10.1098/rspb.1997.0066
Beniash, E., Ivanina, A., Lieb, N. S., Kurochkin, I., and Sokolova, I. M. (2010). Elevated levels of carbon dioxide affect metabolism and shell formation in oysters Crassostrea virginica. Mar. Ecol. Prog. Ser. 419, 95–108. doi: 10.3354/meps08841
Cao, Y. (2010). Wake-up call for endothelial cells. Blood 115, 2336–2337. doi: 10.1182/blood-2009-12-256933
Cardoso, J. C. R., Félix, R. C., Bjärnmark, N., and Power, D. M. (2016). Allatostatin-type A, kisspeptin and galanin GPCRs and putative ligands as candidate regulatory factors of mantle function. Mar. Genomics 27, 25–35. doi: 10.1016/j.margen.2015.12.003
Chan, C. K., Seo, E. Y., Chen, J. Y., Lo, D., Mcardle, A., Sinha, R., et al. (2015). Identification and specification of the mouse skeletal stem cell. Cell 160, 285–298. doi: 10.1016/j.cell.2014.12.002
Checa, A. G., Esteban-Delgado, F. J., Ramírez-Rico, J., and Rodríguez-Navarro, A. B. (2009). Crystallographic reorganization of the calcitic prismatic layer of oysters. J. Struct. Biol. 167, 261–270. doi: 10.1016/j.jsb.2009.06.009
d’Aquino, R., Graziano, A., Sampaolesi, M., Laino, G., Pirozzi, G., De Rosa, A., et al. (2007). Human postnatal dental pulp cells co-differentiate into osteoblasts and endotheliocytes: a pivotal synergy leading to adult bone tissue formation. Cell Death Differ. 14, 1162–1171. doi: 10.1038/sj.cdd.4402121
Dickinson, G. H., Ivanina, A. V., Matoo, O. B., Pörtner, H. O., Lannig, G., Bock, C., et al. (2012). Interactive effects of salinity and elevated CO2 levels on juvenile eastern oysters, Crassostrea virginica. J. Exp. Biol. 215, 29–43. doi: 10.1242/jeb.061481
Dickinson, G. H., Matoo, O. B., Tourek, R. T., Sokolova, I. M., and Beniash, E. (2013). Environmental salinity modulates the effects of elevated CO2 levels on juvenile hard shell clams, Mercenaria mercenaria. J. Exp. Biol. 216, 2607–2618. doi: 10.1242/jeb.082909
Duloquin, L., Lhomond, G., and Gache, C. (2007). Localized VEGF signaling from ectoderm to mesenchyme cells controls morphogenesis of the sea urchin embryo skeleton. Development 134, 2293–2302. doi: 10.1242/dev.005108
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Falini, G., Albeck, S., Weiner, S., and Addadi, L. (1996). Control of aragonite or calcite polymorphism by mollusk shell macromolecules. Science 271, 67–69. doi: 10.1126/science.271.5245.67
Gao, P., Niu, N., Wei, T., Tozawa, H., Chen, X., Zhang, C., et al. (2017). The roles of signal transducer and activator of transcription factor 3 in tumor angiogenesis. Oncotarget 8, 69139–69161. doi: 10.18632/oncotarget.19932
Gross-Goupil, M., François, L., Quivy, A., and Ravaud, A. (2013). Axitinib: a review of its safety and efficacy in the treatment of adults with advanced renal cell Carcinoma. Clin. Med. Insights Oncol. 7, 269–277. doi: 10.4137/CMO.S10594
Gueta, R., Natan, A., Addadi, L., Weiner, S., Refson, K., and Kronik, L. (2007). Local atomic order and infrared spectra of biogenic calcite. Angew. Chem. Int. Ed. Engl. 46, 291–294. doi: 10.1002/anie.200603327
Hedges, S. B., Marin, J., Suleski, M., Paymer, M., and Kumar, S. (2015). Tree of life reveals clock-like speciation and diversification. Mol. Biol. Evol. 32, 835–845. doi: 10.1093/molbev/msv037
Holmes, D. I., and Zachary, I. (2005). The vascular endothelial growth factor (VEGF) family: angiogenic factors in health and disease. Genome Biol. 6:209. doi: 10.1186/gb-2005-6-2-209
Hu, K., and Olsen, B. R. (2016). Osteoblast-derived VEGF regulates osteoblast differentiation and bone formation during bone repair. J. Clin. Investig. 126, 509–526. doi: 10.1172/JCI82585
Huang, X. D., Zhang, H., and He, M. X. (2017). A PDGF/VEGF homologue provides new insights into the nucleus grafting operation and immune response in the pearl oyster Pinctada fucata. Gene 637, 1–8. doi: 10.1016/j.gene.2017.09.019
Ivanina, A. V., Beniash, E., Etzkorn, M., Meyers, T. B., Ringwood, A. H., and Sokolova, I. M. (2013a). Short-term acute hypercapnia affects cellular responses to trace metals in the hard clams Mercenaria mercenaria. Aquat. Toxicol. 14, 123–133. doi: 10.1016/j.aquatox.2013.05.019
Ivanina, A. V., Dickinson, G. H., Matoo, O. B., Bagwe, R., Dickinson, A., Beniash, E., et al. (2013b). Interactive effects of elevated temperature and CO2 levels on energy metabolism and biomineralization of marine bivalves Crassostrea virginica and Mercenaria mercenaria. Comp. Biochem. Physiol. A Mol. Integr. Physiol. 166, 101–111. doi: 10.1016/j.cbpa.2013.05.016
Ivanina, A. V., Falfushynska, H. I., Beniash, E., Piontkivska, H., and Sokolova, I. M. (2017). Biomineralization-related specialization of hemocytes and mantle tissues of the Pacific oyster Crassostrea gigas. J. Exp. Biol. 220, 3209–3221. doi: 10.1242/jeb.160861
Jones, D. T., Taylor, W. R., and Thornton, J. M. (1992). The rapid generation of mutation data matrices from protein sequences. Bioinformatics 8, 275–282. doi: 10.1093/bioinformatics/8.3.275
Kerschnitzki, M., Akiva, A., Ben Shoham, A., Koifman, N., Shimoni, E., Rechav, K., et al. (2016). Transport of membrane-bound mineral particles in blood vessels during chicken embryonic bone development. Bone 83, 65–72. doi: 10.1016/j.bone.2015.10.009
Knapp, R. T., Wu, C. H., Mobilia, K. C., and Joester, D. (2012). Recombinant sea urchin vascular endothelial growth factor directs single-crystal growth and branching in vitro. J. Am. Chem. Soc. 134, 17908–17911. doi: 10.1021/ja309024b
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Li, S., Liu, C., Huang, J., Liu, Y., Zhang, S., Zheng, G., et al. (2016a). Transcriptome and biomineralization responses of the pearl oyster Pinctada fucata to elevated CO2 and temperature. Sci. Rep. 6:18943. doi: 10.1038/srep18943
Li, S. G., Liu, Y. J., Liu, C., Huang, J. L., Zheng, G. L., Xie, L. P., et al. (2016b). Hemocytes participate in calcium carbonate crystal formation, transportation and shell regeneration in the pearl oyster Pinctada fucata. Fish Shellfish Immunol. 51, 263–270. doi: 10.1016/j.fsi.2016.02.027
Liu, Y., Berendsen, A. D., Jia, S., Lotinun, S., Baron, R., Ferrara, N., et al. (2012). Intracellular VEGF regulates the balance between osteoblast and adipocyte differentiation. J. Clin. Invest. 122, 3101–3113. doi: 10.1172/JCI61209
Marie, B., Joubert, C., Tayale, A., Zanella-Cleon, I., Belliard, C., Piquemal, D., et al. (2012). Different secretory repertoires control the biomineralization processes of prism and nacre deposition of the pearl oyster shell. Proc. Natl. Acad. Sci. U.S.A. 109, 20986–20991. doi: 10.1073/pnas.1210552109
Marin, F., Le Roy, N., and Marie, B. (2012). The formation and mineralization of mollusk shell. Front. Biosci. 4:1099–10125. doi: 10.2741/s321
Mayer, H., Bertram, H., Lindenmaier, W., Korff, T., Weber, H., and Weich, H. (2005). Vascular endothelial growth factor (VEGF-A) expression in human mesenchymal stem cells: autocrine and paracrine role on osteoblastic and endothelial differentiation. J. Cell. Biochem. 95, 827–839. doi: 10.1002/jcb.20462
McDonald, J. A., Pinheiro, E. M., and Montell, D. J. (2003). PVF 1, a PDGF/VEGF homolog, is sufficient to guide border cells and interacts genetically with Taiman. Development 130, 3469–3478. doi: 10.1242/dev.00574
McTigue, M., Murray, B. W., Chen, J. H., Deng, Y. L., Solowiej, J., and Kania, R. S. (2012). Molecular conformations, interactions, and properties associated with drug efficiency and clinical performance among VEGFR TK inhibitors. Proc. Natl. Acad. Sci. U.S.A. 109, 18281–18289. doi: 10.1073/pnas.1207759109
Mount, A. S., Wheeler, A. P., Paradkar, R. P., and Snider, D. (2004). Hemocyte-mediated shell mineralization in the eastern oyster. Science 304, 297–300. doi: 10.1126/science.1090506
Nudelman, F., Gotliv, B. A., Addadi, L., and Weiner, S. (2006). Mollusk shell formation: mapping the distribution of organic matrix components underlying a single aragonitic tablet in nacre. J. Struct. Biol. 153, 176–187. doi: 10.1016/j.jsb.2005.09.009
O’Neill, M., Gaume, B., Denis, F., and Auzoux-Bordenave, S. (2013). Expression of biomineralisation genes in tissues and cultured cells of the abalone Haliotistuberculata. Cytotechnology 65, 737–747. doi: 10.1007/s10616-013-9576-0
Parsons, B., and Foley, E. (2013). The drosophila platelet-derived growth factor and vascular endothelial growth factor-receptor related (Pvr) protein ligands Pvf2 and Pvf3 control hemocyte viability and invasive migration. J. Biol. Chem. 288, 20173–20183. doi: 10.1074/jbc.M113.483818
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29, 2002–2007. doi: 10.1093/nar/29.9.e45
Roch, P. (1999). Defense mechanisms and disease prevention in farmed marine invertebrates. Aquaculture 172, 125–145. doi: 10.1016/S0044-8486(98)00439-6
Schipani, E., Maes, C., Carmeliet, G., and Semenza, G. L. (2009). Regulation of osteogenesis-angiogenesis Coupling by HIFs and VEGF. J. Bone Min. Res. 24, 1347–1353. doi: 10.1359/jbmr.090602
Seipel, K., Eberhardt, M., Muller, P., Pescia, E., Yanze, N., and Schmid, V. (2004). Homologs of vascular endothelial growth factor and receptor, VEGF and VEGFR, in the jellyfish Podocoryne carnea. Dev. Dyn. 231, 303–312. doi: 10.1002/dvdy.20139
Smith, V. J., Accorsi, A., and Malagoli, D. (2016). “Chapter 1 - hematopoiesis and hemocytes in pancrustacean and molluscan models,” in The Evolution of the Immune System, ed. D. Malagoli (Cambridge, MA: Academic Press), 1–28.
Street, J., Bao, M., Bunting, S., Peale, F. V., Ferrara, N., Steinmetz, H., et al. (2002). Vascular endothelial growth factor stimulates bone repair by promoting angiogenesis and bone turnover. Proc. Nat. Acad. Sci. U.S.A. 99, 9656–9661. doi: 10.1073/pnas.152324099
Sun, Z., and Ettensohn, C. A. (2014). Signal-dependent regulation of the sea urchin skeletogenic gene regulatory network. Gene Expr. Patterns 16, 93–103. doi: 10.1016/j.gep.2014.10.002
Tarsitano, M., De Falco, S., Colonna, V., Mcghee, J. D., and Persico, M. G. (2006). The C. elegans pvf-1 gene encodes a PDGF/VEGF-like factor able to bind mammalian VEGF receptors and to induce angiogenesis. FASEB J. 20, 227–233. doi: 10.1096/fj.05-4147com
Wang, Y., Wan, C., Deng, L. F., Liu, X. M., Cao, X. M., Gilbert, S. R., et al. (2007). The hypoxia-inducible factor a pathway couples angiogenesis to osteogenesis during skeletal development. J. Clin. Investig. 117, 1616–1626. doi: 10.1172/JCI31581
Keywords: biomineralization, mollusk, VEGF, gene expression regulation, hardness, crystallinity, respiration
Citation: Ivanina AV, Borah B, Rimkevicius T, Macrander J, Piontkivska H, Sokolova IM and Beniash E (2018) The Role of the Vascular Endothelial Growth Factor (VEGF) Signaling in Biomineralization of the Oyster Crassostrea gigas. Front. Mar. Sci. 5:309. doi: 10.3389/fmars.2018.00309
Received: 13 March 2018; Accepted: 10 August 2018;
Published: 28 August 2018.
Edited by:
Vera Bin San Chan, Clemson University, United StatesReviewed by:
Daniele Tibullo, Università degli Studi di Catania, ItalyCopyright © 2018 Ivanina, Borah, Rimkevicius, Macrander, Piontkivska, Sokolova and Beniash. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Inna M. Sokolova, inna.sokolova@uni-rostock.de Elia Beniash, ebeniash@pitt.edu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.