- 1School of Sciences, Bath Spa University, Bath, United Kingdom
- 2School of Physiology, Pharmacology and Neuroscience, University of Bristol, Bristol, United Kingdom
Coding proficiency is becoming an essential skill for future life sciences graduates. Not only must researchers be able to handle and scrutinise increasingly large datasets, but alternative career paths often list coding as a desirable skill for applicants. With the additional advantage of promoting the development of key skills such as problem solving, critical thinking, and group work, there is an increasing need to expose students to principles of coding in a structured and paced way. Despite this, computational tools are often missing from Life Sciences undergraduate curricula. We suggest the reasons for this are barriers faced by both students and staff, which coalesce into a general fear surrounding the topic. From reservations surrounding the complexity of the topic and time input required, to the increased staffing load, there are several factors which contribute to a lack of coding proficiency in Life Sciences curricula. To overcome these barriers, we propose a scaffolding approach using RStudio for data analysis, that has been successfully implemented to teach basic coding skills to students in their first and second years of Neuroscience and Physiology programmes at University of Bristol (UK). We believe that this approach has been a successful route to lower students’ anxiety around coding and ease them into more complex tasks. In this perspective we will reflect on the current barriers, discuss our experience with overcoming them and consider implementations for future iterations.
1 Introduction
With the advancement of experimental techniques, big data is becoming increasingly prevalent across the Life Sciences (Juavinett, 2022). In response, there has been widespread development of computational tools to conduct experiments, test hypotheses and to build simulations and models (Pevzner and Shamir, 2009; Markowetz, 2017). However, to benefit from these tools, it is necessary to have at least an elementary knowledge of coding (defined as writing instructions in a format that a computer can understand and act on to complete a task).
It can be argued, therefore, that coding is rapidly becoming an essential employability skill for new Life Sciences graduates entering the workplace (World Economic Forum, 2016, 2023; Brynjolfsson et al., 2019; Juavinett, 2022; QAA, 2023). Notably, jobs that require coding are often associated with higher salaries, with data scientist roles typically boasting starting salaries above the national average both in the US and UK (National Careers Service, n.d.; U.S. Bureau of Labor Statistics, n.d.). For these reasons, we believe some principles of coding (e.g., understanding the structure of a script, using a script to perform analysis, write small and easy piece of code to perform a task) must be successfully integrated into Life Sciences curricula, to provide opportunities for our students to develop the skills necessary to access desirable jobs post-graduation (Glass, 2016).
Employability aside, we have found that coding-based activities promote the development of skills such as analytical and critical thinking, problem solving and creativity (Tuomi et al., 2018; Psycharis and Kallia, 2017; Romero et al., 2017). Creating a setting in which students often enjoy and seek peer support, coding-based activities also promote successful group work and encourage active student–student and student-teacher discussions.
To summarise, we believe that coding content allows students to develop key skills during their degree that will be invaluable for any career they choose to pursue. However, as coding is not a staple of all current curricula, we have attempted to develop a framework around which coding can be effectively embedded within Life Sciences degree programmes in a way that is both accessible and engaging to students, and realistic in terms of the commitment required on behalf of staff. In this article we will discuss some of the existing barriers to teaching coding skills and describe our approach to overcoming them.
2 The challenges of teaching coding
There are, as such, many benefits to coding-based activities for Life Sciences undergraduates but it would be naïve to suggest that the introduction of such activities to the core curriculum is not without its challenges for both students and staff (Figure 1). We have observed that students often have negative preconceptions of coding, anxieties around already challenging courseloads, and inequalities surrounding digital literacy. On the other hand, staff often face a lack of training, and often overburdened workloads in addition to the challenge of identifying relevant sessions into which coding can be integrated.
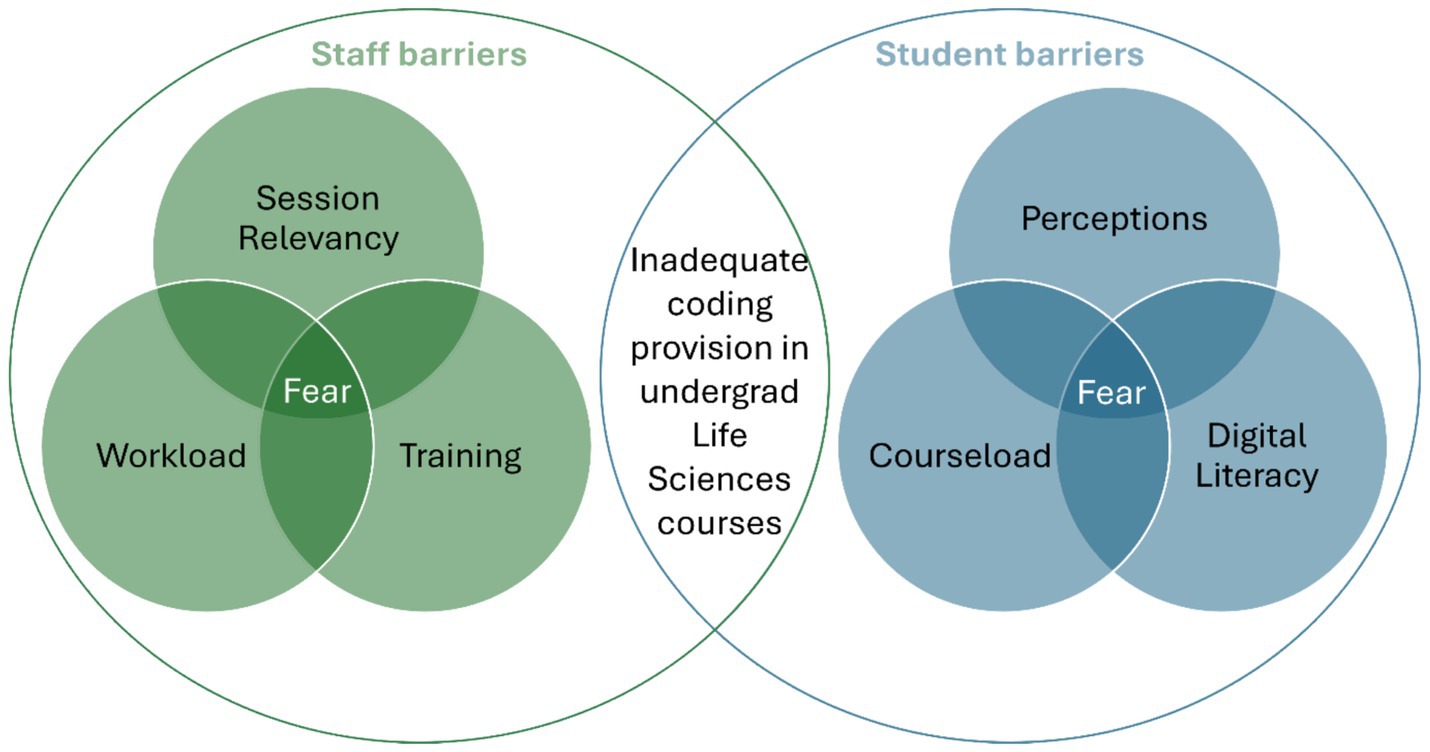
Figure 1. Barriers and challenges to teaching coding from a student and staff perspective. The diagram shows the key barriers that we feel both staff and students face when teaching and learning coding. We have found these factors accumulate into a sense of fear around coding, which fuels the lack of coding provision in Life Sciences courses.
To first consider the student-centred challenges, we have found that perception is often the largest issue to overcome. Many of our students approach coding with a sense of foreboding that we do not see in other sessions, such as lab practicals. When asked about their concerns, students have explained that they worry coding will be too hard, or that it will require high-level mathematics. Others think coding is irrelevant to them and simply refuse to engage. These perceptions are further compounded by the second challenge outlined above, that first-year undergraduates tend towards low digital literacy and can struggle with tasks that form the basis of coding, such as the production, use and manipulation of tables in widely used programmes such as Microsoft Excel. Students often require a considerable level of foundation teaching on the use of basic computer commands to be able to tackle coding-based activities. Both challenges are compounded by the fact that many of our students feel overwhelmed by their courseload and often decide to set aside coding to prioritise other subjects.
To overcome these interlinked challenges, we introduced coding in a series of workshops using simple language, accessible software and crucially, in sessions that have direct relevancy to the core curriculum. Using data taken from recently completed practical sessions, these workshops allowed students to see the applicability of coding, only after they have had the time to develop their digital literacy skills to a baseline standard. This approach ensured that students understood the relevancy of the activity being attempted, and that coding-based sessions were not simply viewed as optional “add ons” to their courseload. This, we feel, was key to our approach. While optional modules could offer coding provision to engaged students, many students described above would choose not to engage, leaving them with a problematic hole in their skill set. Optional modules, we suggest, are better placed at third-year level and above, once all students have had the opportunity to learn some principles of coding within the core curriculum, to ensure equal opportunities amongst all students.
However, it would be dishonest to suggest that the approaches described above were not implemented without considerable investment on behalf of the staff teaching on our undergraduate programmes. Initially, staff invested time and effort in appropriate training, which for many is self-led and not always compatible with high academic workloads. While there is little that can be done to circumvent this issue, the provision of in-person or online training modules by institutions, and the time to engage with them would allow staff to develop their coding skills. However, even when appropriately trained, staff must find the time to dedicate to coding within the classroom and the curriculum, which with an already extensive amount of information and skill development to include in the core content, there is often little to no space for educators to expand on coding skills. Identifying opportunities for coding to be inserted into existing sessions is one way we have attempted to overcome this. For example, where data analysis was previously completed on Excel, we have modified the activity so that students must complete the same analysis in RStudio, a coding-based programme. This enabled us to highlight the applications and relevance of coding to the course, without having to design and develop additional sessions.
A final challenge we encountered was that students often require a high level of support to complete coding-centred activities. This creates a high staffing demand or need for smaller classes (with multiple repetitions of the same session), without which, we have found that students’ questions and confusion mount, and very simple issues including not being able to locate and open files can form a barrier to progression, and ultimately, student disengagement. However, such intensive staff support is not always possible, for the reasons outlined above. In these instances, we have successfully employed asynchronous approaches to provide “hands-off” support. For example, providing video tutorials, additional examples and explanation documents that students can refer to during and after the session can free up staff to tackle more complex questions. Additionally, we have introduced AI technology to sessions, so that students can answer their own questions and solve syntax errors, furthering their engagement with the task, as well as their critical thinking and problem-solving skills.
3 Our approach to overcoming these challenges
In line with the above, we designed and implemented a scaffolding approach to integrate coding-based activities into the first- and second-year curricula of our Neuroscience and Psychology undergraduate programmes. While still under development, we have identified methods to overcome many of the challenges outlined above and have received positive feedback from our students accordingly.
We adopted scaffolding theory, wherein student expertise and knowledge are slowly built, and a high level of support is initially provided but then gradually stripped away, leading to independence and autonomy (Gonulal and Loewen, 2018). This approach allowed us to gently introduce students to the basic concepts of coding and to the programming software without overwhelming them. In addition, we have found that scaffolding fosters key skills, including but not limited to, critical thinking, problem solving, and mathematical thinking, as well as encouraging students to engage with content and take responsibility for their learning (Lin et al., 2012; Bakker et al., 2015). Scaffolding is a good choice for teaching coding, as it allows students to perform tasks that they would not normally be able to do without a high level of support, furthering their confidence in their abilities and combatting one of the most striking barriers identified. From our experience, introducing multiple sessions throughout the degree with increasing demands placed on independence, autonomy and level of content, exposed students little and often to elements of coding, to counter the barriers our students face as outlined above (Figure 2). We have found an effective way of doing this is to initially ask students to work through an already-completed script (full scripts available1; Swainson et al., 2024), then in a following session prompt them to start editing some key lines of code and finally, to instruct them to begin inserting their own pieces of code. Building on this, we hope in future to get students to the stage in which they can write their own script from start to finish.

Figure 2. Example of our scaffolding approach to coding. Diagram showing the structure of workshops that we embedded in the curriculum. Three initial workshops including elements of coding were inserted in year one and two, with tasks set at increasing difficulty based on how much independence, level of content and autonomy was required in the session. ILOs, intended learning outcomes.
There are many programming languages used in the context of Life Sciences, such as MATLAB, Python, SQL and R. We suggest adopting R programming language as it is one of the most popular languages used in biostatistics to compute statistics and graphics, it is open-source and free to use, and it has some easy-to-understand environments (R Foundation, n.d.). In particular, we suggest adopting RStudio, an integrated development environment (IDE) for R, as we have found RStudio to be remarkably user-friendly and therefore less intimidating for students upon first use. To increase accessibility, we recommend using RStudio hosted on Posit Cloud (Posit Software, n.d.), which allows the software to be used online without download. This simplifies things for students, as once projects are set up, they can be saved and accessed from any device. In addition, the use of Posit Cloud means commands are the same across Windows and Mac machines in a way that is not the case on desktop versions, further simplifying matters for staff and students. Another benefit to RStudio is RMarkdown (RStudio, n.d.), which allows for the creation of neat and intuitive documents that alternate between paragraphs of guided explanation, and lines of code, which are directly followed by an output section (Finch et al., 2021; Grayson et al., 2022; see Footnote 1 for complete script files; Swainson et al., 2024). This is in comparison to the basic, unembellished, and often intimidating basic text of many coding software options, making the user experience easier and more relaxed. Once completed, R Markdown can be exported and saved as a revision document in which students can see the code, graphs and analysis generated. We have found that this final format helps to consolidate the statistical and analytical part of the workshop, together with reinforcing the coding elements. This also provides the advantage of highlighting the importance of reproducibility in science to students, and good practice associated with it (Baumer and Udwin, 2015).
To ease student pressure and workload during coding-based activities, we recommend that the intended learning outcomes (ILOs) complement course-specific content, such as investigating data describing recently taught physiological mechanisms. This allows students to review and consolidate previously taught teaching, and removes the initial expectation students may have, of needing to achieve a thorough understanding of coding in a single session. We have observed that by focusing ILOs on the understanding and application of appropriate statistical tests and identification of the best graphical representation of data they have previously collected in experimental sessions, students naturally familiarised themselves with the RStudio environment and basic coding commands without feeling they had to get it right from the first exposure. Once students had a few sessions of working with RStudio, we introduced coding proficiency elements into ILOs and associated assessments. The assessment (multiple choice answer type questions) highlighted that although students mostly get the answers right immediately after the workshop, their performance decreases in the final exam, perhaps highlighting the need for further sessions to maintain students’ coding proficiency. Such sessions need not necessarily take the form of workshops, however, as we have found a complementary lecture and asynchronous resources helpful to provide support and a theoretical understanding of coding, to support students in their learning.
Finally, in line with current conversations in education, we recommend the inclusion of the use of ChatGPT or similar, (OpenAI, n.d.) as an additional source of support during coding-based activities. We want students to be aware of the flaws and limitations of using such technology, but also the opportunities it can offer. To help students in the use of generative AI, we provided an example prompt that they could input into ChatGPT to request an explanation of a specific line of code. ChatGPT has been used in other educational contexts, and it has been shown to be useful in reviewing code and explaining functions, enabling students to appreciate such tools and reinforce the content covered in in-person sessions (Adeshola and Adepoju, 2023; Popovici, 2023; da Silva et al., 2024). There is now a specific R Tutor tool (using GPT 4 model; Orditus, n.d.) available, and although we have not tested it directly it looks to be a promising addition to future iterations of this coding series.
4 Lessons learned
Throughout this process, we have identified several lessons learned and solutions to these. One of the first things that emerged was the importance of including very clear and defined explanations of every single step, including instructions on how to download and import different file formats. Without these, students became quite confused at the very beginning of sessions, and problems with the simple task of setting up their RStudio project often demotivated them to continue engaging with the workshop. Another important thing to consider is that students might be using different software versions and operating systems and therefore alternative instructions need to be included for all possible options. During one of the initial sessions, for example, there were some issues with the Excel data file format leading to Mac users not being able to download the file. This slowed down the progress of some students and discouraged them. An easy solution to overcome this is to use Posit Cloud, as it ensures that the instructions are uniform across all devices.
Additionally, preliminary staff numbers were based on the support previously needed for other workshops and labs, but it became clear very quickly that students require additional support during these sessions. This issue was also identified by students, with most requesting either smaller sessions or more staff members present to support them. Increasing staff numbers was not possible, however, we had success in implementing alternative support for learners during coding sessions, such as asynchronous instructions and video guides. In future, we aim to provide further supporting resources, such as sheets containing most common errors and tips for troubleshooting, a syllabus of key functions, documentation on R, as well as encouraging further use of ChatGPT (or R Tutor).
Three sessions across years one and two were a good starting point to introduce coding, but to enable our students to develop a satisfactory understanding of coding, further sessions will be required. To further embed coding in the curriculum, more staff members need to be on board and given that many do not feel confident on the subject, it is important for universities to provide training for staff on coding and programming skills so that more aspects of coding can be embedded in current teaching. While this is an issue that must be tackled at a faculty or institutional level, the more call for such training by staff, the higher the likelihood of such training being made available.
Encouragingly, after completing the workshops, many students appreciated the use of coding, and some have explored further courses and opportunities (such as third year projects requiring coding) to advance their knowledge, recognising the positive impact this might have on their career. An aspect that they enjoyed was problem solving in groups. Although they found it frustrating when they could not solve an error, especially when a member of staff was not readily available to help, when they did overcome the challenge and produce an output, they found it very rewarding. Certainly, by the end of the sessions most students felt less anxious about approaching coding in the future, a key aim when establishing the coding workshops.
5 Future directions
The scaffolding approach we have described above has been a positive experience overall and an effective direction that we would recommend when teaching coding to students in Life Sciences degrees. However, as mentioned above, there are still some aspects that we feel require further improvements. There are three areas that we wish to alter over future iterations:
i. Find further opportunities to include more sessions throughout the degree and have a consistent use of R in multiple modules.
ii. Produce supporting resources such as interactive videos to practice, sheets to revise key functions and additional examples of code to allow students to facilitate independent learning.
iii. Introduce the use of AI earlier on, to also allow students to expand on their independent learning. However, this should be accompanied by clear guidance on advantages and pitfalls of tools such as ChatGPT, as well as institutional policies on the use of AI.
We hope that by expanding on these areas, students will reach their final year with greater confidence in coding and able to access further career opportunities. Of course, there will always be students that struggle and will choose not to engage with coding. We therefore need to be mindful about how to engage these students, and ensure they are not left behind. So far, however, with the current approach, most students have identified positive aspects of coding and have largely overcome their fear of the subject. With additional support and opportunities for practice, alongside training and support to empower staff to teach coding, we expect even more students will engage with and embrace the benefits coding skills can bring to a degree and career in the Life Sciences.
Data availability statement
Publicly available code were used in this study. This data can be found at: https://osf.io/j6rfg/?view_only=a4bfbf7483a74b90a4650c43a1c8c135.
Author contributions
EP: Writing – original draft, Writing – review & editing. AS: Writing – original draft, Writing – review & editing. NC: Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/feduc.2024.1504877/full#supplementary-material
Footnotes
References
Adeshola, I., and Adepoju, A. P. (2023). The opportunities and challenges of ChatGPT in education. Interact. Learn. Environ, 32, 1–14. doi: 10.1080/10494820.2023.2253858
Bakker, A., Smit, J., and Wegerif, R. (2015). Scaffolding and dialogic teaching in mathematics education: introduction and review. ZDM 47, 1047–1065. doi: 10.1007/s11858-015-0738-8
Baumer, B., and Udwin, D. (2015). R Markdown. Wiley Interdiscip Rev Comput Stat 7, 167–177. doi: 10.1002/WICS.1348
Brynjolfsson, E., Rock, D., and Tambe, P. (2019). How will machine learning transform the labor market? |. Hoover Institution. Available at: https://www.hoover.org/research/how-will-machine-learning-transform-labor-market (Accessed August 28, 2024).
Finch, S., Gordon, I., and Patrick, C. (2021). Taking the aRghhhh out of teaching statistics with R: using R markdown. Teach. Stat. 43, S143–S147. doi: 10.1111/TEST.12251
Glass, Burning (2016). Beyond point and click: the expanding demand for coding skills. Available at: www.burning-glass.com (Accessed August 28, 2024).
Gonulal, T., and Loewen, S. (2018). “Scaffolding technique” in The TESOL encyclopedia of English language teaching. ed J. I. Liontas. (Netherland: Wiley), 1–5.
Grayson, K. L., Hilliker, A. K., and Wares, J. R. (2022). R markdown as a dynamic interface for teaching: modules from math and biology classrooms. Math. Biosci. 349:108844. doi: 10.1016/j.mbs.2022.108844
Juavinett, A. L. (2022). The next generation of neuroscientists needs to learn how to code, and we need new ways to teach them. Neuron 110, 576–578. doi: 10.1016/j.neuron.2021.12.001
Lin, T.-C., Hsu, Y.-S., Lin, S.-S., Changlai, M.-L., Yang, K.-Y., and Lai, T.-L. (2012). A review of empirical evidence on scaffolding for science education. Int. J. Sci. Math. Educ. 10, 437–455. doi: 10.1007/s10763-011-9322-z
Markowetz, F. (2017). All biology is computational biology. PLoS Biol. 15:e2002050. doi: 10.1371/journal.pbio.2002050
National Careers Service. (n.d.). Explore careers. Available at: https://nationalcareers.service.gov.uk/explore-careers (Accessed August 28, 2024).
OpenAI (n.d.). Introducing ChatGPT. Available at: https://openai.com/index/chatgpt/ (Accessed August 28, 2024).
Orditus, LLC (n.d.). RTutor 0.99. Available at: https://rtutor.ai/ (Accessed August 28, 2024).
Pevzner, P., and Shamir, R. (2009). Computing has changed biology—biology education must catch up. Science 1979, 541–542. doi: 10.1126/SCIENCE.1173876
Popovici, M. D. (2023). ChatGPT in the classroom. Exploring its potential and limitations in a functional programming course. Int. J. Hum. Comput. Interact. 40, 7743–7754. doi: 10.1080/10447318.2023.2269006
Posit Software (n.d.). Posit cloud | take your data science to the cloud. Available at: https://posit.co/products/cloud/cloud/ (Accessed August 28, 2024).
Psycharis, S., and Kallia, M. (2017). The effects of computer programming on high school students’ reasoning skills and mathematical self-efficacy and problem solving. Instr. Sci. 45, 583–602. doi: 10.1007/s11251-017-9421-5
R Foundation (n.d.). R: the R project for statistical computing. Available at: https://www.r-project.org/ (Accessed August 28, 2024).
Romero, M., Lepage, A., and Lille, B. (2017). Computational thinking development through creative programming in higher education. Int. J. Educ. Technol. High. Educ. 14, 42–57. doi: 10.1186/s41239-017-0080-z
RStudio (n.d.). R Markdown. Available at: https://rmarkdown.rstudio.com/ (Accessed August 28, 2024).
Silva, C. A. G.da, Ramos, F. N., de Moraes, R. V., and dos Santos, E. L. (2024). ChatGPT: challenges and benefits in software programming for higher education. Sustain. For., Vol. 16, Page:1245. doi: 10.3390/SU16031245
Swainson, A., Paci, E., and Cerminara, N. L. (2024). Facing the fear: a scaffolding approach to teaching life sciences undergraduates R coding. Available at: osf.io/j6rfg.
Tuomi, P., Multisilta, J., Saarikoski, P., and Suominen, J. (2018). Coding skills as a success factor for a society. Educ Inf Technol (Dordr) 23, 419–434. doi: 10.1007/s10639-017-9611-4
U.S. Bureau of Labor Statistics (n.d.). Data scientists: occupational outlook handbook. Available at: https://www.bls.gov/ooh/math/data-scientists.htm (Accessed November 08, 2024).
World Economic Forum (2016). Five million jobs by 2020: the real challenge of the fourth industrial revolution. Available at: https://www.weforum.org/press/2016/01/five-million-jobs-by-2020-the-real-challenge-of-the-fourth-industrial-revolution/ (Accessed August 28, 2024).
World Economic Forum (2023). The future of jobs report 2023. Available at: www.weforum.org (Accessed August 28, 2024).
Keywords: coding, life sciences, R, undergraduates, neuroscience, physiology
Citation: Paci E, Swainson A and Cerminara NL (2025) Facing the fear: a scaffolding approach to teaching life sciences undergraduates R coding. Front. Educ. 9:1504877. doi: 10.3389/feduc.2024.1504877
Edited by:
Sorinel A. Oprisan, College of Charleston, United StatesReviewed by:
Michael J. Wolyniak, Hampden–Sydney College, United StatesCopyright © 2025 Paci, Swainson and Cerminara. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Nadia L. Cerminara, bi5jZXJtaW5hcmFAYnJpc3RvbC5hYy51aw==
†These authors share first authorship
 Elena Paci
Elena Paci Alex Swainson
Alex Swainson Nadia L. Cerminara
Nadia L. Cerminara