- 1Department of Paediatrics, Fribourg Hospital, Fribourg, Switzerland
- 2Department of Health Science and Medicine, University Lucerne, Lucerne, Switzerland
- 3Department of Paediatrics, The University of Melbourne, Parkville, VIC, Australia
- 4Infectious Diseases Research Group, Murdoch Children’s Research Institute, Parkville, VIC, Australia
- 5Infectious Diseases Unit, The Royal Children’s Hospital Melbourne, Parkville, VIC, Australia
- 6Department for Community Health, Faculty of Science and Medicine, University of Fribourg, Fribourg, Switzerland
Background: Children are the age group with the highest exposure to antibiotics (ABX). ABX treatment changes the composition of the intestinal microbiota. The first few years of life are crucial for the establishment of a healthy microbiota and consequently, disturbance of the microbiota during this critical period may have far-reaching consequences. In this review, we summarise studies that have investigated the effect of ABX on the composition of the intestinal microbiota in children.
Methods: According to the PRISMA guidelines, a systematic search was done using MEDLINE and Embase to identify original studies that have investigated the effect of systemic ABX on the composition of the intestinal microbiota in children.
Results: We identified 89 studies investigating a total of 9,712 children (including 4,574 controls) and 14,845 samples. All ABX investigated resulted in a reduction in alpha diversity, either when comparing samples before and after ABX or children with ABX and controls. Following treatment with penicillins, the decrease in alpha diversity persisted for up to 6–12 months and with macrolides, up to the latest follow-up at 12–24 months. After ABX in the neonatal period, a decrease in alpha diversity was still found at 36 months. Treatment with penicillins, penicillins plus gentamicin, cephalosporins, carbapenems, macrolides, and aminoglycosides, but not trimethoprim/sulfamethoxazole, was associated with decreased abundances of beneficial bacteria including Actinobacteria, Bifidobacteriales, Bifidobacteriaceae, and/or Bifidobacterium, and Lactobacillus. The direction of change in the abundance of Enterobacteriaceae varied with ABX classes, but an increase in Enterobacteriaceae other than Escherichia coli was frequently observed.
Conclusion: ABX have profound effects on the intestinal microbiota of children, with notable differences between ABX classes. Macrolides have the most substantial impact while trimethoprim/sulfamethoxazole has the least pronounced effect.
Introduction
Children are the age group with the highest exposure to antibiotics (ABX). ABX are the second most prescribed drugs for children, surpassed only by analgesics (1–5). More than two-thirds of children receive ABX before reaching the age of two years (6) with exposure to an average of almost three ABX in the first year of life (7). Approximately one third of hospitalised children (8) and nearly half of acutely ill children in outpatient settings receive ABX (9), often with inappropriate indications or drugs (10). While the widespread use of ABX has significantly reduced childhood morbidity and mortality during the last century (11), exposure to ABX is also associated with adverse long-term health effects. These include an increased risk for atopic dermatitis, allergies, wheezing and asthma, obesity, arthritis idiopathic disorder, psoriasis, and neurodevelopmental disorders (12). These adverse effects likely result from changes in the microbiota, particularly the intestinal microbiota, which undergoes significant development during the first two to three years of life (13–15). Children are highly susceptible to ABX-induced dysbiosis (an imbalance in the microbiota), whereas adults typically have a more stable and resilient microbiota that recovers more easily from such disturbances (16). The first years of life are critical for the development and stabilisation of the intestinal microbiota, coinciding with key milestones in the development of the immune system, metabolism and neurodevelopment (17–19). Consequently, any disturbances of the microbiota during this critical period may have long-lasting and far-reaching consequences.
In this review, we systematically summarise studies that have investigated the effect of ABX on the composition of the intestinal microbiota in children of all age groups.
Methods
In May 2024, MEDLINE (1946 to present) and Embase (1972 to present) were searched. Embase was searched using the OVID interface. The detailed search terms can be found in the supplementary data. No geographical limitations were used. References of retrieved articles were searched for additional publications.
Original studies which investigated effect of ABX on the composition of the bacterial intestinal microbiota in children less than 18 years of age were included, as well as studies involving mixed-age populations that provided separate data specifically for this age group. Exclusion criteria were studies which (i) included children with underlying diseases (e.g., oncological diseases, cystic fibrosis) and (ii) studies not published in English, German, French, Spanish, Portuguese or Italian.
The following variables were extracted from included studies: year of publication, country, study design, number and characteristics of included children, number of samples, ABX treatment (drug, dose, frequency, route of administration, duration), previous ABX, microbiota analysis method, timing of stool analysis and key findings (including changes in diversity, abundance of microbes, antibiotic resistance genes, ARGs).
Studies were identified, selected, appraised, and synthesised following the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) guidelines for systematic reviews (20, 21). The level of evidence of each study was classified according to the 2011 Oxford Centre for Evidence-Based Medicine (OCEBM) Levels of Evidence (22). Risk of bias was assessed using the 2017 Joanna Briggs Institution (JBI) standardised critical appraisal checklist for case-control and cohort studies (23).
Results
The search identified 6,146 and 11,470 studies in MEDLINE and Embase, respectively. From the 17,616 studies, 2,183 duplicates were removed. 89 studies met the inclusion criteria (24–112). No additional relevant studies were found through citation searching. The selection of included studies is summarized in Figure 1.
The 89 studies investigated a total of 9,712 children (of these 4,574 were controls; mean 111 children per study, range 9 to 1,023) and 14,845 samples (mean 322 samples per study, range 20 to 1,247). Of the included studies, 47 were done in Europe (Denmark, Netherlands, Finland, Spain, France, Italy, Germany, Ireland, Estonia, United Kingdom, Sweden, Norway, Austria) (24, 26, 28, 29, 31, 32, 34, 35, 37–39, 43, 45, 48–52, 60, 61, 63, 64, 66, 67, 70, 71, 74, 75, 81–83, 88, 92, 93, 95, 98, 101, 102, 104–112), 21 on the American continent (USA, Chile, Canada) (25, 30, 41, 42, 44, 54–56, 58, 59, 69, 73, 76, 78, 84–86, 94, 96, 97, 99), 13 in Asia (China, India, Japan, Korea, Taiwan, Lebanon) (27, 40, 53, 57, 65, 68, 72, 77, 80, 87, 90, 91, 100), seven in Africa (Niger, Burkina Faso, Zimbabwe, South Africa) (36, 46, 47, 62, 79, 89, 103) and one in Australia (33). The age of participants ranged from preterm birth to 15 years. Of the included studies, 75 were observational studies, including 60 cohort studies (24, 25, 27, 28, 30–35, 37–42, 44, 45, 48–58, 61, 64–68, 70, 71, 73, 74, 76–78, 84, 87–92, 94, 97–102, 109–112), 12 cross-sectional studies (26, 29, 43, 72, 75, 82, 85, 93, 95, 99, 108, 113), three pre-post-intervention studies (35, 60, 96); and 13 (cluster) randomised controlled trials (36, 46, 47, 59, 62, 63, 69, 79–81, 83, 86, 103). In total, 42 studies used 16S rRNA gene sequencing (25–28, 40–46, 48, 51, 52, 55, 57, 58, 62, 64, 66, 68, 69, 71–73, 76, 77, 79, 80, 83–86, 91, 92, 98–103, 113), ten shotgun metagenomic sequencing (29, 30, 36, 47, 54, 75, 83, 89, 94, 98), seven polymerase chain reaction (PCR) or PCR-temperature gradient gel electrophoresis (PCR-TGGE) (39, 49, 50, 65, 67, 78, 82), one each fluorescence in situ hybridization (FISH) (104) and 16S-23S IS profiling (48), and 30 cultures (24, 31–35, 37–39, 49, 53, 56, 59–61, 63, 74, 81, 87, 88, 90, 93, 96, 105, 107–112).
Supplementary Table S1 provides a summary of the main findings of the 89 studies included in this review.
All included studies had an overall risk of bias score (JBI standardised critical appraisal checklist, yes%) over 60% (acceptable quality) and 49% (44/89) of studies had an overall score ≥80% (good quality) (Supplementary Table S3). The most frequent risk of bias was attrition bias [present in 24% (21/89) studies].
Penicillins
The effect of penicillins on the intestinal microbiota was investigated in 13 studies including 1,311 children [one study did not specify the number of children investigated (70); for detailed information see Supplementary Table S3] (35, 55, 70, 71, 75, 78, 79, 87, 89, 90, 100, 107, 112).
Diversity
Two studies found a lower alpha diversity (a measure of the variety and abundance of bacterial taxa within a sample) of the intestinal microbiota during the treatment with amoxicillin compared to controls with recovery either immediately after stopping ABX (89) or six months after (71), respectively. However, compared to controls, the first study found a higher alpha diversity at the latest follow-up time point 24 months after ABX (89). In other studies, compared to controls, a lower richness was found to persist for less than two weeks after treatment with ampicillin and penicillin (75), and for up to 6–12 months (latest follow-up being after 12–24 months) in a study (which did not report separate results for treatment with amoxicillin with or without clavulanate and penicillin V) (70) (Figure 2). One study found a lower alpha diversity at day seven of life compared to day three in preterm neonates after treatment with penicillin plus moxalactam or piperacillin-tazobactam, but no difference in diversity after ABX with the two treatment regimens (100). Another study found no difference in alpha diversity, at the latest follow-up point of the study, five days after treatment with amoxicillin (79). Three studies investigated beta diversity (a measure of the differences in bacterial taxa between different samples) by comparing the ABX group and controls and found a higher beta diversity after treatment with penicillin, amoxicillin/ampicillin, penicillin plus moxalactam and piperacillin-tazobactam, respectively (75, 89, 100).
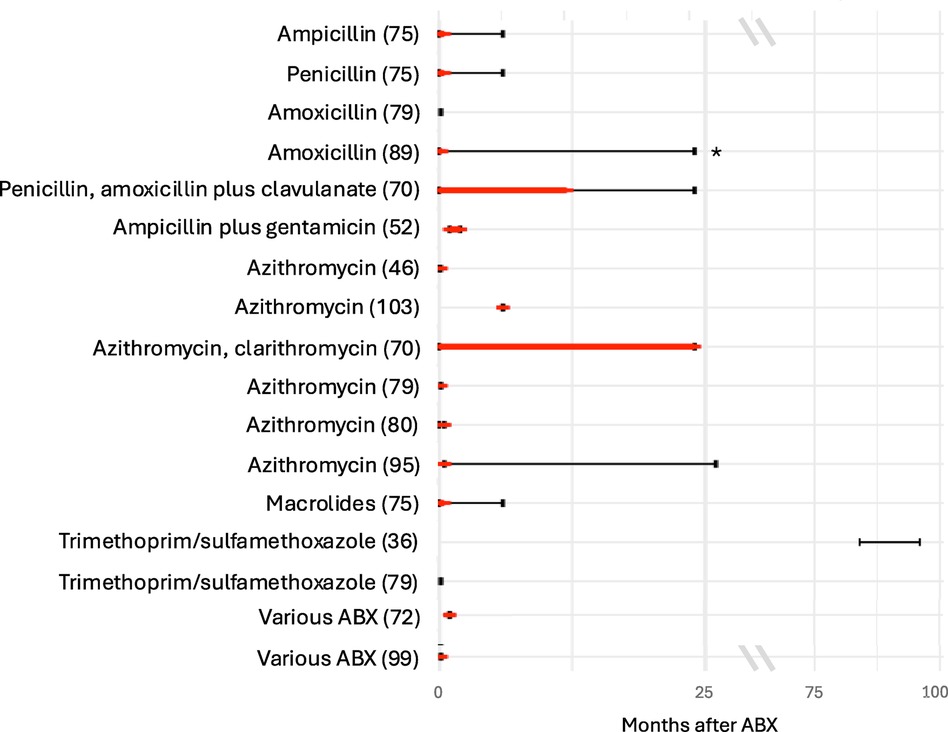
Figure 2. Duration of decreased alpha diversity after ABX reported in different studies. Red bars depict duration of decreased alpha diversity in ABX group compared to controls (red bars); black bars depict the duration of follow-up after ABX. Studies specifying Shannon index, inverse Simpson index or richness for the ABX and control group were included. Studies which did not specify the time points when the alpha diversity was investigated were excluded.
Composition
When comparing the composition of the intestinal microbiota during or after oral administration of amoxicillin, compared to controls, increased abundances of Firmicutes (71), Enterobacteriales (71), Ruminococcaceae (71), Lachnospiraceae (71), Megasphaera (71), Coprococcus (71), Escherichia (89), Dialister (71), Weissela confusa (89), Prevotella sp. 885 (89), Prevotella stercorea (89), Holdemanella biformis (89), Lactobacillus animalis (89), Fusicatenibacter saccharivorans (89), Catenibacterium mitsuokai (89), Slackia isoflavoniconvertens (89), Weissella cibaria (89), Streptococcus macedonicus (89), Gemmiger formicilis (89), and Actinomyces odontolyticus (89), and decreased abundances of Coriobacteriaceae (71), Bacteroidaceae (71), Streptococcaceae (71), Lactobacillus (89), Bifidobacterium (71), Enterococcus (71), Streptococcus (87, 89), Klebsiella (89), Holdemanella (89), Dorea (89), Bifidobacterium bifidum (89), and Bifidobacterium longum (89) were observed.
When comparing the composition before and during treatment with penicillin V, ampicillin and methicillin, respectively, decreased bacterial counts of Lactobacillus (87), Streptococcus (87), and Bifidobacterium (87) were found. One study found increased abundances of Bacteroidetes, Rikenellaceae, and Dialister and decreased abundances of Actinobacteria, Gemellales, Gemellaceae, Lactobacillus, and Collinsella in children who were given penicillin or amoxicillin with or without clavulanate, without reporting results for these ABX separately and without reporting the route of administration (70). Fluctuations in abundance over time were found for Veillonellaceae (71), Clostridiaceae (both being lower during treatment with amoxicillin and higher than in controls after treatment) (71) and Parabacteroides (being higher within less than 6 months after treatment with amoxicillin with or without clavulanic acid or penicillin (without separate analysis) compared with controls and then being lower 6 to 12 months after treatment compared with controls (70) (Figure 3). One study found no difference in bacterial abundance after treatment with penicillin (75).
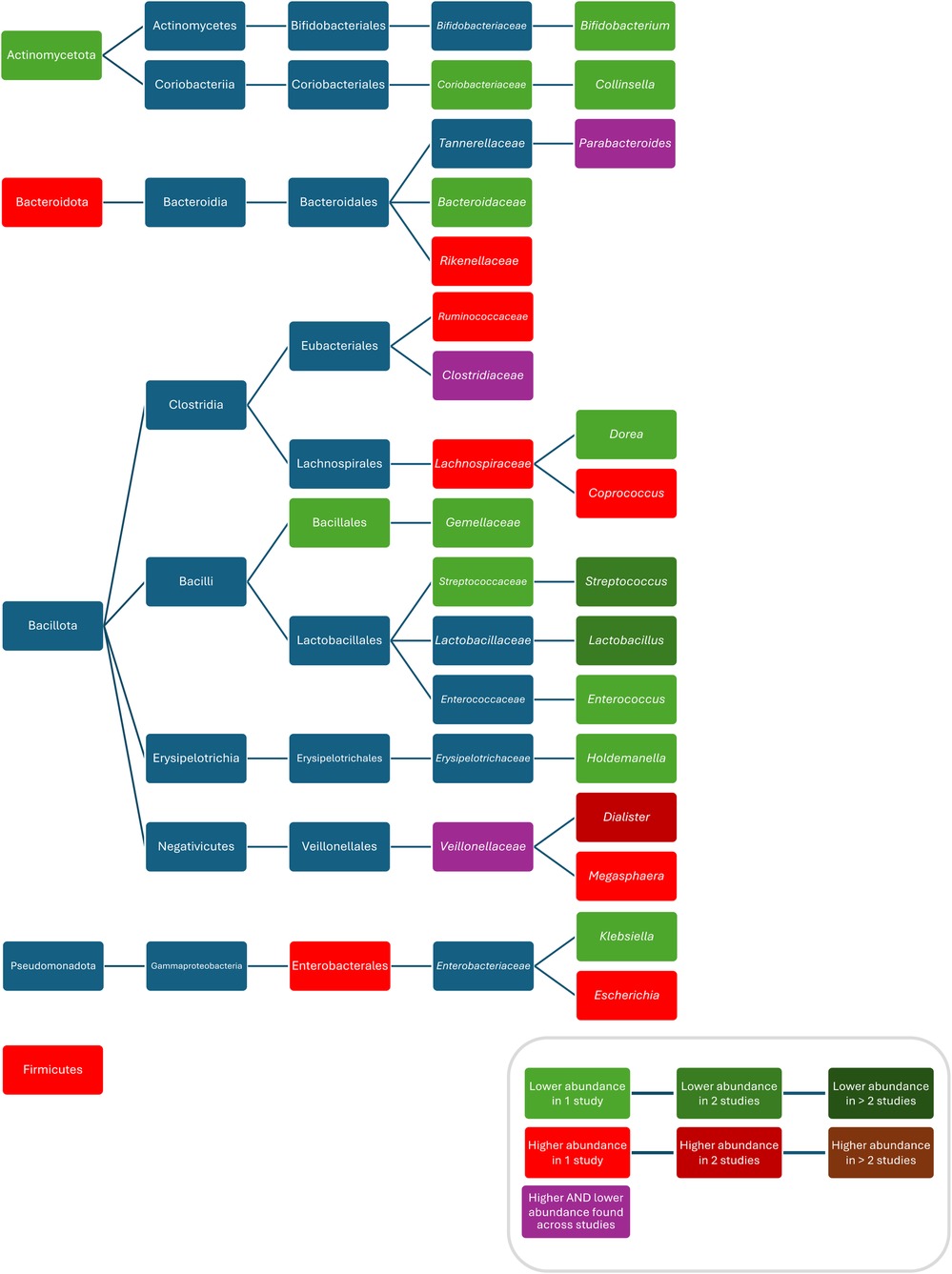
Figure 3. Differences in bacterial abundance between children treated with penicillins and controls. Included studies investigated amoxicillin (71, 89) and amoxicillin with or without clavulanate and penicillin V (70). Studies which did not compare ABX group to controls and studies not providing p-values were excluded.
ARGs
The effect of penicillins on the abundance of ARGs was studied in three studies. Two studies found an increased abundance of ARGs after treatment with penicillin and ampicillin/amoxicillin, respectively, which returned to normal three to four weeks after treatment (75, 89). The other study found increased abundances of multiple ARGs, e.g., for β-lactamases (unknown), ABC efflux pumps, araC, and emrD (55). A higher abundance of plasmids after treatment with penicillin or ampicillin (without separate analysis) was reported in another study (75).
Penicillins plus aminoglycosides
13 studies investigated combinations of different penicillins plus gentamicin in 3,141 children (for detailed information see Supplementary Table S3) (26, 37, 40, 52, 58, 63, 69, 81, 83, 85, 93, 94, 110).
Diversity
Compared with controls, three studies found a lower alpha diversity in children treated with penicillin, amoxicillin, or ampicillin plus gentamicin (52, 58, 83). The duration of treatment with ampicillin plus gentamicin positively correlated with the decrease in alpha diversity (58). One study found a lower alpha diversity in children treated for more than seven days with ampicillin plus gentamicin compared with these treated for a shorter duration (94). Another study found a lower richness in children treated with ampicillin plus gentamicin compared with controls two months after treatment (52) (Figure 2). In contrast, one study found no difference in alpha diversity at two weeks of life after treatment with ampicillin plus gentamicin (69), and another one, one year after treatment with penicillin plus tobramycin (26). Yet another study compared the alpha diversity one week after treatment with ampicillin plus tobramycin, ampicillin plus tobramycin plus metronidazole, and ampicillin plus cefotaxime and found no difference between these groups (85). Another study compared alpha and beta diversity after treatment with ampicillin plus gentamicin and ampicillin plus cefotaxime and found no differences at the latest follow up at 30 days of life (40).
Composition
Compared to controls, after treatment with ampicillin plus gentamicin increased abundances/colonisation rates of Proteobacteria (52), Bacilli (94), Clostridiales (94), Bacteroidales (94), Enterobacteriaceae (52), Peptostreptococcaceae (52), Clostridium (52), Klebsiella (37, 83, 94), Enterobacter (58, 93), Klebsiella/Enterobacter (93), Veillonella (80), Streptococcus (80), and Enterococcus faecalis (37), and decreased abundances of Bifidobacteriacea (52), Bifidobacterium (52, 83), Escherichia (83), Staphylococcus (58, 83), and Lactobacillus (52); after treatment with penicillin plus gentamicin increased abundances of Acinetobacter (83), and Klebsiella (83), and decreased abundances of Bifidobacterium (83), Escherichia (83), Staphylococcus (83), and Escherichia coli (83); after treatment with penicillin plus netilmicin decreased abundance of Clostridum difficile (63); and after treatment with penicillin plus tobramycin decreased abundances of Bacteroidetes were found (26). Conflicting results were reported for the abundance of Actinobacteria (52, 69), Enterococcus (52, 58, 83) and Bacteroides (83, 94), and Escherichia coli (37, 93, 94) (Figure 4).
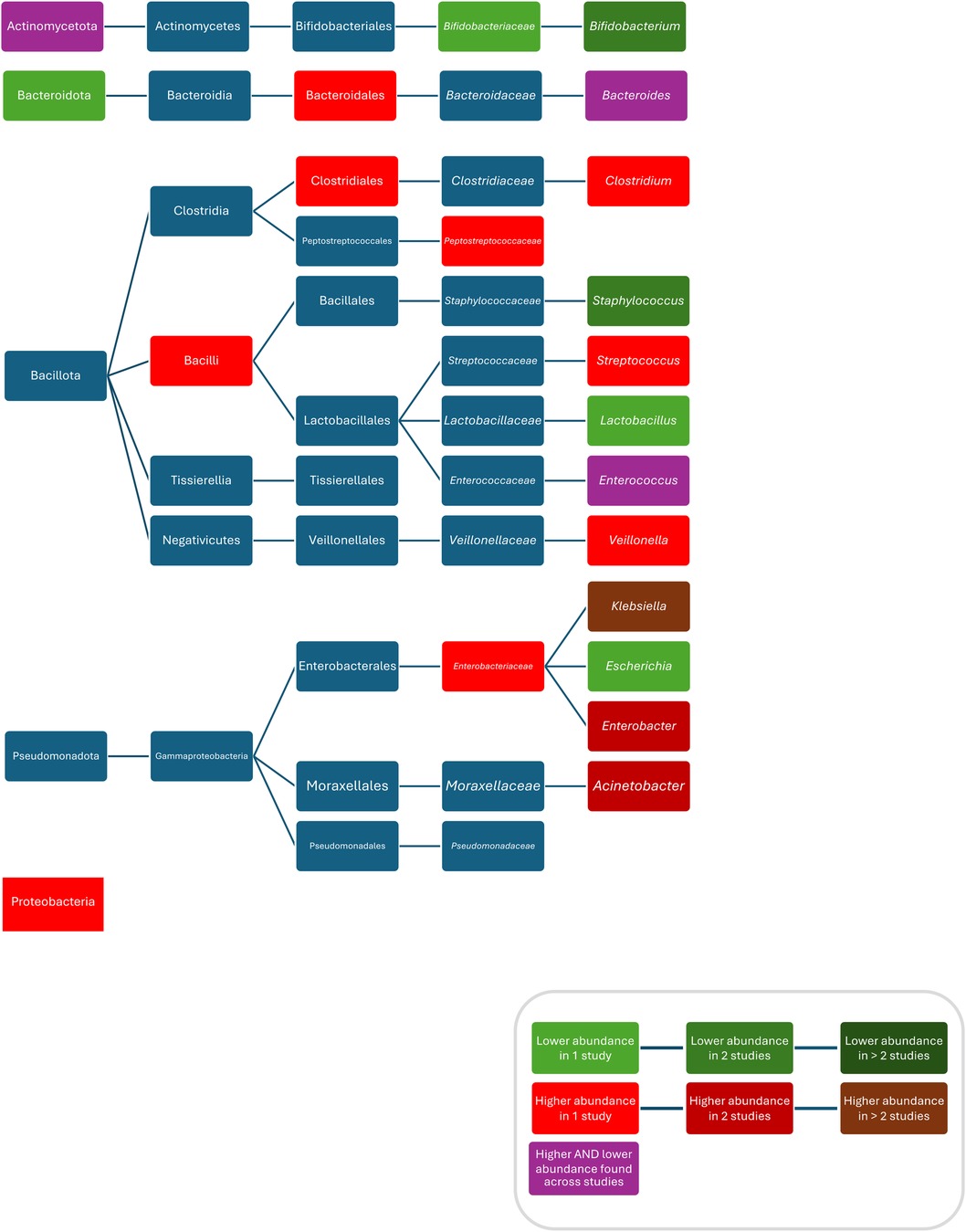
Figure 4. Differences in bacterial abundance or colonization rate between children treated with penicillins plus aminoglycosides and controls. Studies included investigated ampicillin plus gentamicin (52, 58, 69, 81, 93, 94), penicillin plus gentamicin (81, 83), and penicillin plus tobramycin (26). Studies which did not compare ABX group to controls and studies not providing p-values were excluded.
ARGs
One study reported that changes in the ARG profile persisted for up to four months after treatment with amoxicillin/clavulanate plus gentamicin with 10 of 31 ARGs being more abundant, while after penicillin plus gentamicin five of 10 ARGs were found to be more abundant (83). One study found resistance to ampicillin in E. coli, Klebsiella, and Enterobacter after ampicillin plus gentamicin treatment (93).
Penicillins plus cephalosporins
Five studies investigated combinations of penicillins plus cephalosporins, including 1,123 children (for detailed information see Supplementary Table S3) (34, 40, 83, 85, 101).
Diversity
One study found a lower alpha diversity in children treated with amoxicillin plus cefotaxime compared with controls (83). The same study also found a high beta diversity with a dissimilar composition between children treated with amoxicillin plus cefotaxime and controls, which was still present four months after stopping ABX (83). One study compared alpha diversity one week after treatment with ampicillin plus tobramycin, ampicillin plus tobramycin plus metronidazole, and ampicillin plus cefotaxime and found no difference between these groups (85). Another study compared alpha and beta diversity after treatment with ampicillin plus gentamicin and ampicillin plus cefotaxime and found no differences at the latest follow-up at 30 days of life (40).
Composition
Increased abundances of Enterococcus (101), Clostridium (101), and Acinetobacter (83), and decreased abundances of Bifidobacterium (101), Akkermansia (83), and Escherichia coli (83) were found in children after treatment amoxicillin plus cefotaxime or ceftazidime compared to controls. One study compared abundances after treatment with ampicillin plus tobramycin, ampicillin plus tobramycin plus metronidazole, and ampicillin plus cefotaxime and found no difference between groups (85). One study did not provide p-values for their analysis of colonisation rates (34).
ARGs
One study reported changes in the ARG profile with 10 of 31 ARGs being more abundant after treatment with amoxicillin plus cefotaxime. Compared to penicillin plus gentamicin (five of 10 ARGs being more abundant), amoxicillin plus cefotaxime was found to have a higher impact on ARG abundance (83).
Cephalosporins
The effect of cephalosporins on the intestinal microbiota in children was investigated in nine studies including 916 children (for detailed information see Supplementary Table S3) (55, 57, 60, 63, 87, 88, 90, 91, 93).
Diversity
One study found a lower alpha diversity in infants treated with cefalexin compared with controls at two months of life (91). Another study found a lower richness immediately after treatment with cefotaxime and cefazoline compared to before ABX (55). One study found no difference in alpha diversity in neonates at ten days of life between the ABX group and controls after treatment with cefotaxime (57). Three studies did not analyse alpha diversity or richness (60, 87, 93).
Composition
The administration of cefalexin was associated with increased abundances of Enterobacteriaceae (91), Enterococcus (91), and decreased abundances of Bifidobacterium (91) compared to controls. The administration of cefuroxime was associated with a decreased abundance of Escherichia coli (93). When comparing before and after treatment with cefaclor decreased bacterial counts of Enterobacteriaceae (87), and Bifidobacterium (87), were found, while when comparing before and after treatment with ceftazidime decreased bacterial counts of Enterobacteriaceae (87), Lactobacillus (87), and Bifidobacterium (87), were found. After treatment with cefotaxime, compared to controls, increased abundances of Enterobacteriaceae (57) and Parabacteroides (57), and decreased abundances of Bifidobacterium (57), Clostridium difficile (63) and Escherichia coli (55) were found. After treatment with ceftriaxone, abundances of Enterobacteriaceae and Lactobacillus increased compared to before ABX (88). When comparing before and after treatment with cefpiramide decreased counts of Enterobacteriaceae (87), Bacteroidaceae (87), Bifidobacterium (87), Lactobacillus (87), and Staphylococcus (87) were found.
ARGs
After treatment with ceftriaxone, one study found Klebsiella/Enterobacter, Citrobacter, Serratia and E. coli to be resistant to cefoperazone and ceftriaxone and Pseudomonas aeruginosa to ceftriaxone (60). One study found increased abundances of multiple ARGs after treatment with cefotaxime, e.g., β-lactamase (CMY-LAT-MOX), MFS efflux, ABC efflux, and robA (55).
Carbapenems
The effect of carbapenems on the intestinal microbiota was investigated in three studies including 67 children (for detailed information see Supplementary Table S3) (55, 96, 105).
Diversity
One study found a reduced richness comparing before and two days after treatment with meropenem (55). Two studies did not analyse alpha or beta diversity (96, 105).
Composition
The studies reported an increased bacterial count of Enterococcus (96), Proteus (96), Pseudomonas (96), Enterobacter (96), and Staphylococcus epidermidis (55), and a decreased abundance of Klebsiella (96), Lactobacillus (96), and Streptococcus (96) comparing before and after treatment with imipenem-cilastatin, while the second study did not find changes in the abundance of different bacteria (105).
ARGs
The first study did not identify bacteria with resistance to imipenem (96), while in the second study, in one child P. aeruginosa resistant to imipenem was found after treatment. This child was previously also treated with aztreonam (105). One study found increased abundances of multiple ARGs after treatment with meropenem, e.g., mecA, norA, dfrC, gyrA, and qacA (55).
Macrolides
The effect of macrolides on the intestinal microbiota was investigated in 13 studies including 802 children (for detailed information see Supplementary Table S3) (38, 46, 62, 70, 71, 75, 79, 80, 87, 95, 103, 111, 112).
Diversity
Four studies found a lower alpha diversity (46, 79, 95, 103) and three a lower richness (70, 75, 80) between five days and 12–24 months after treatment with macrolides. One study reported a decrease in alpha diversity after 14 days but no further changes between 13 and 39 months after treatment with azithromycin (95). After treatment with azithromycin, one study found a difference in richness between children with ABX and controls, but no difference in alpha diversity (80) (Figure 2). Three studies analysed beta diversity and found a high beta diversity between ABX group and controls up to 14 days after treatment with azithromycin (80, 95) and a distinct composition on phylum and genus level in the ABX group up until six months after treatment with either azithromycin or clarithromycin without reporting results separately (70). One study did not detect a difference in beta diversity five days after treatment with azithromycin (46).
Composition
Compared to controls, after treatment with azithromycin increased abundances/bacterial counts of Clostridium (95), and Blautia (46), and decreased abundances/bacterial counts of Actinobacteria (95), Verrucomicrobia (80), Betaproteobacteria (80), Verrucomicrobiae (80), Bifidobacteriales (95), Bifidobacteriaceae (95), Clostridiacea (95), Bifidobacterium (95), Anaerovibrio (46), Peptoniphilus (46), Succinivibrio (46), Megasphaera (46), Escherichia (80), Akkermansia (80), Peptostreptococcus (80), Campylobacter hominis (62), Campylobacter ureolyticus (62), and Campylobacter jejuni (62) were found. Two studies did not report separate results for changes in abundances after treatment with different macrolides and found compared to controls, increased abundances of Bacteroidetes (70), Alphaproteobacteria (71), Bacteroidales (70), Lactobacillales (70), Rikenellaceae (70), Subdoligranulum (71), Salmonella (71), Eggerthella (70), Bacteroides (70), Parabacteroides (70), Eubacterium (70), and Clostridium (70), and decreased abundances of Actinobacteria (70), Bifidobacteriales (70), Coriobacteriales (70), Clostridiales (70), Gemellaceae (70), Collinsella (70), and Bifidobacterium (70, 71). Conflicting results were reported for abundances of Proteobacteria (70, 80), Bacillales/Gemellales (70, 71), and Dialister (70, 95) (Figure 3). Two studies found no difference in bacterial abundance between children with ABX and controls (38, 75). Three studies did report p-values for abundances (38, 79, 111). One study did not investigate bacterial abundances (103) (Figure 5).
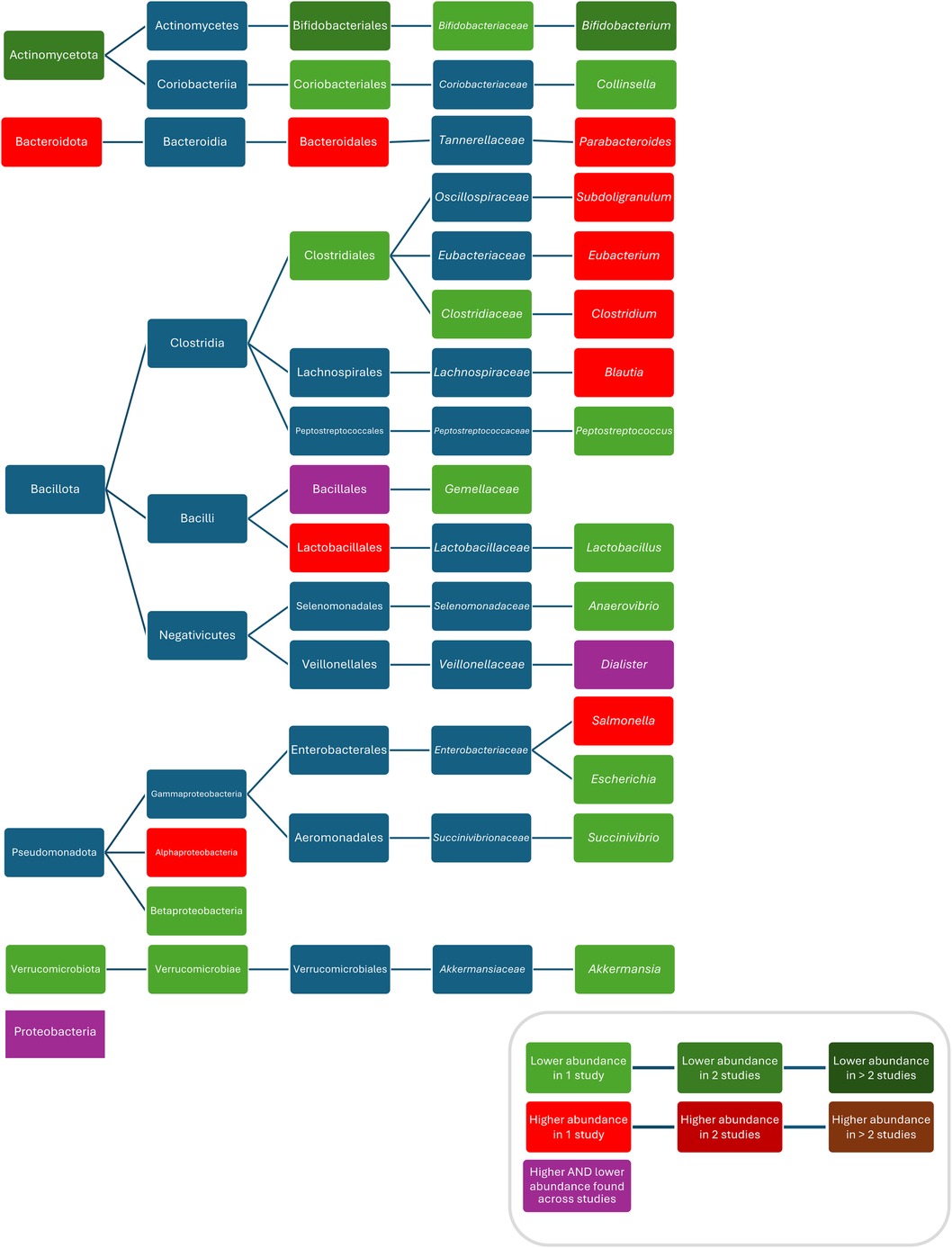
Figure 5. Differences in bacterial abundance or total bacterial count between children treated with macrolides and controls. Included studies investigated azithromycin (80, 95, 103), azithromycin and clarithromycin (70) or did not separately analyse different macrolides (71). Studies which did not compare ABX group to controls and studies not providing p-values were excluded.
ARGs
ARGs were investigated in two studies (70, 114). One study found increased macrolide resistance after treatment with azithromycin or clarithromycin (without separate analysis), which declined linearly until going back to baseline 6 to 12 months after ABX. The study also found that the abundance of ermF and ermB genes correlated negatively with time since the last macrolide treatment, while the abundance of the bsh gene correlated positively with time since the last macrolide treatment (70). One study found a higher abundance of ARGs and plasmids in the ABX group compared with controls, without performing a separate analysis for different macrolides (75).
Trimethoprim/sulfamethoxazole
The effect of trimethoprim/sulfamethoxazole on the intestinal microbiota was investigated in six studies, including 254 children (27, 36, 47, 53, 55, 79).
Diversity
Two studies detected no difference in alpha diversity between the ABX group and the control group within the first week after ABX and after seven and eight years of continuous prophylactic treatment (Figure 2) (36, 79). Another study found a longitudinal increase in alpha diversity between six weeks and six months after the ABX in the ABX group (47). Another study found a decrease in diversity two weeks after the start of prophylactic treatment compared to before treatment, which recovered within one to two months (27). Another study found a decreased richness comparing before and two days after ABX (55). One study found no difference in bacterial counts between the two groups (53). Two studies analysed beta diversity, of which one did not detect a difference in between the ABX group and controls (36) and the other one found a lower beta diversity in the ABX group compared to controls (47). Comparing before to after treatment with trimethoprim/sulfamethoxazole decreased bacterial counts of Enterobacter (53) and Veillonella (53) were found.
Composition
Comparing the ABX group to controls increased abundances of Enterobacteriales (27), Alistipes onderdonkii (36), Eggerthella lenta (36), Clostridium bartlettii (36). Haemophilus parainfluenzae (36), Streptococcus mutans (36), Streptococcus parasanguinis (36), and Streptococcus vestibularis (36), and decreased abundances of Enterobacteriaceae (36) were found after ABX. One study detected no differences in bacterial abundance after treatment with trimethoprim/sulfamethoxazole (47) and another study did not provide p-values for changes in abundances (79).
ARGs
ARGs were studied in one study, dfr and sul genes were found to be more abundant in the ABX group after the ABX compared to before (47).
Aminoglycosides
The effect of aminoglycosides on the intestinal microbiota was investigated in three studies, including 52 children (for detailed information see Supplementary Table S3) (55, 87, 112).
Diversity
The first study found no difference in alpha diversity three days after treatment with gentamicin (55) and the second did not analyse alpha diversity (87). Both studies did not analyse beta diversity.
Composition
One study observed decreased bacterial counts of Enterobacteriaceae (87), Streptococcus (87), Clostridium (87), and Lactobacillus (87) comparing before and three to six days after treatment with gentamicin.
ARGs
One study found increased abundances of multiple ARGs after treatment with gentamicin, e.g., evgA and emrK (55).
Glycopeptides
The effect of glycopeptides on the intestinal microbiota was investigated in three studies, including 48 children (for detailed information see Supplementary Table S3) (45, 55, 84).
Diversity
One study found a lower alpha diversity seven days after treatment with vancomycin compared to before ABX, but no difference between the ABX group and controls (45). Another study found a lower richness two days after treatment with vancomycin compared to before ABX (55). One study found no difference in alpha diversity after treatment with vancomycin at 25 days of life (84).
Composition
One study found increased abundances of Staphylococcus and decreased abundances of Commamonadaceae, Pseudomonas, Bifidobacterium when comparing before and after ABX (45). Another study reported increased abundances of Staphylococcus (in intestinal tissue but not in stool) when comparing children treated with vancomycin to other ABX (84).
ARGs
One study found an increase in ARGs after treatment with vancomycin, e.g., evgA, emrK (55).
Mixed and not specified ABX
Various ABX without reporting separate results were investigated in 19 studies (24, 31–33, 44, 48, 49, 54, 59, 64–66, 72–74, 77, 86, 92, 97), while another 20 did not specify which ABX were investigated (28–30, 38, 42, 43, 50, 51, 56, 61, 67, 68, 76, 82, 98, 99, 104, 106, 109, 115). These studies included a total of 5,139 children (for detailed information see Supplementary Table S3).
Diversity
A decreased alpha diversity in ABX compared to controls was found in seven studies (29, 54, 72, 74, 77, 97, 98) and lower richness in two studies (66, 99). One study found a decrease in alpha diversity during ABX and an increase afterwards (64). One study found a lower alpha diversity in children treated for two days compared to seven days or more (44). Two studies reported that alpha diversity inversely correlated with increase in number of ABX courses (54, 74). One study found a decrease in diversity and in richness with each additional day of ABX (76) and another study found a lower diversity within the Bacteroidetes phylum in the ABX group than in controls (48). Other studies found no difference in alpha diversity between birth and hospital discharge (86), at ten days of life (43), or up to six month of life, when comparing children after ABX to controls (92). One study found no difference in alpha diversity comparing before, during, and after ABX (25). Twelve studies did not analyse alpha diversity or richness (28, 31–33, 39, 42, 49, 56, 61, 67, 73, 82, 104). Five studies found a high beta diversity with a dissimilar bacterial composition between the ABX group and controls (54, 72, 92, 98, 25). One study found a dissimilar composition in children with two days of ABX compared to seven or more days (44). One study found a dissimilar composition between neonates receiving ABX in the first week of life only and neonates receiving ABX in the first week of life plus after the first week of life (54). Samples collected in controls showed more similarity to each other than samples from the period when children were starting or stopping ABX (30). One study found no effect of ABX on beta diversity at one year of life (51). Another study found a decrease in beta diversity in neonates from week one to week three of life in the ABX group compared to controls (97).
Composition
When comparing ABX groups and controls, after ABX, increased abundances/colonisation rates/bacterial counts for Proteobacteria (28), Gammaproteobacteria (73), Enterobacteriaceae (29, 54), Veillonella (54), Klebsiella (56), Escherichia/Shigella (72), Enterobacter (56), Klebsiella/Enterobacter (31), Sphingomonas (97), Acidovorax (97), Proteus (42), Bacteroides vulgatus (74), Bifidobacterium bifidum (74), Staphylococcus epidermidis (54), Veillonella parvula (54), Veillonella unspecified (54), Klebsiella oxytoca (54), Escherichia coli (54), Bifidobacterium breve (54), and decreased abundances for Bacteroidetes (48, 72, 76, 92), Clostridiales (54), Bifidobacteraceae (54), Prevotella (54), Bacteroides (29, 65, 72, 77, 82), Parabacteroides (29), Ruminococcus (25), Haemophilus (97), Blautia (97), Erysipelatoclostridium (97), Gemella (97), Rothia (97), Streptococcus (97), Clostridium perfringens (33), Eubacterium rectale (98), Akkermansia muciniphila (80), Lactobacillus mucosae (80), Bacteroides fragilis (80), Actinomyces odontolyticus (74), Bifidobacterium longum (50, 54, 74), Bifidobacterium bifidum (50, 54) Bifidobacterium lactis (50), Escherichia coli (30, 31) Escherichia coli 8711 (80), and Escherichia coli 17709 (80) were found. Conflicting results were found for Actinobacteria (28, 72), Firmicutes (28, 72, 92), Clostridium (29, 65, 73, 76, 98), Enterococcus (54, 77, 97), Lactobacillus (32, 33, 61, 65, 92), Bifidobacterium (29, 32, 54, 72, 82), Citrobacter (42, 56), Staphylococcus (54, 97), Clostridium difficile (33, 67), Clostridium butyricum (33, 49), Enterococcus faecialis (31, 54), and Bifidobacterium breve (50, 54, 74) (Figure 6). One study found Clostridium perfringens abundance decreased compared to controls 3–4 weeks after ABX and increased 5–6 weeks after ABX (42). One study found a lower abundance of Bacteroides and a higher abundance of Actinobacteria, Proteobacteria, Bacteroidetes in children treated for more than seven days compared to these treated for two days (44). Another study found lower abundances of Lactobacillus and Enterococcus after more than seven days of treatment compared to less than seven days of treatment (68). Three studies did not compare bacterial abundances (66, 99, 104). One study, only investigating Bifidobacterium abundance, found no difference in abundance between the ABX group and controls (39).
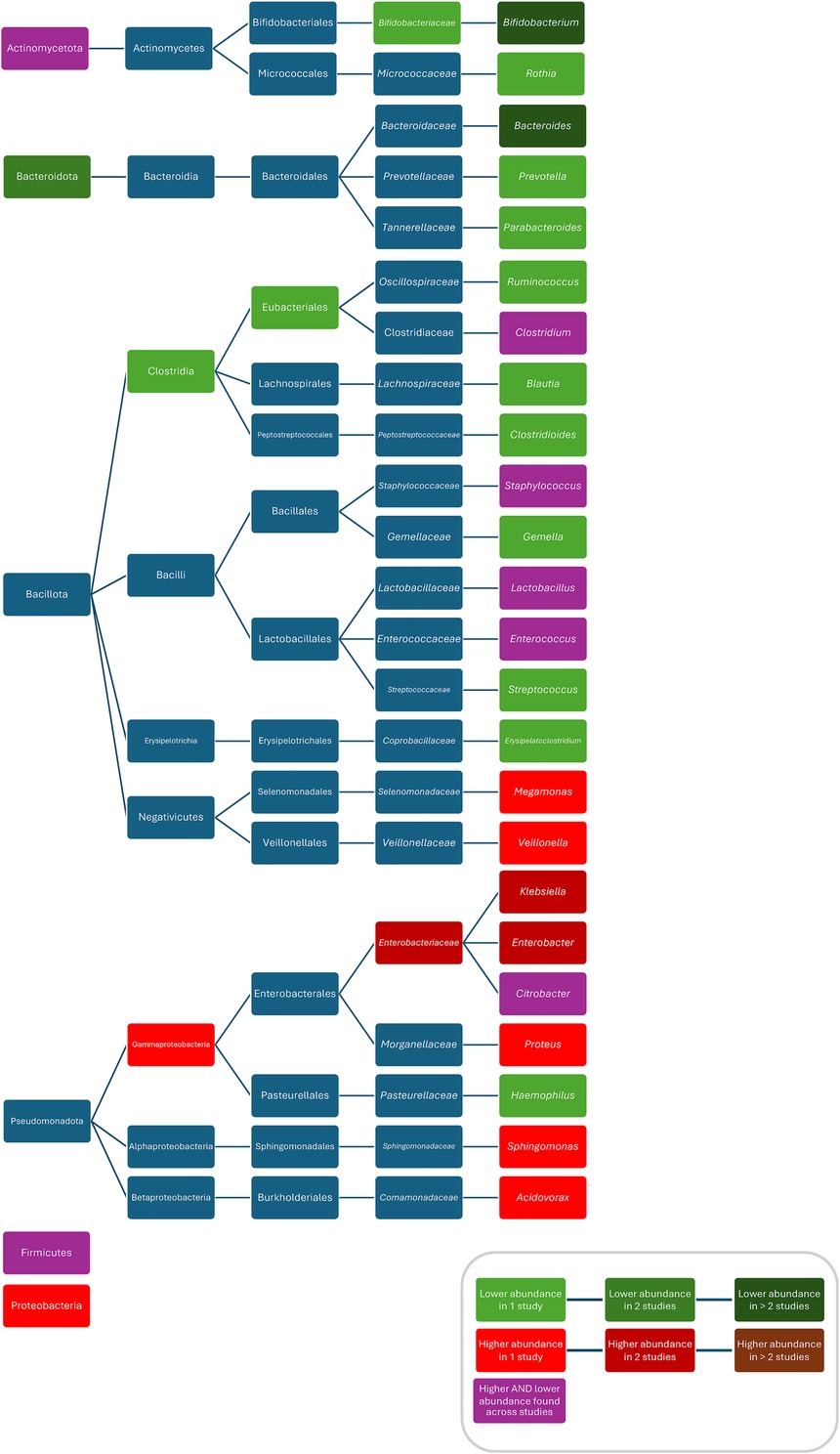
Figure 6. Differences in bacterial abundance or colonisation rate between ABX groups and controls of studies investigating various ABX without separate analysis of individual ABX. Included studies investigated: ampicillin, nafcillin, gentamicin, tobramycin, others (97), penicillin (plus aminoglycoside), others (92), ampicillin/sulbactam, cefotaxime (72), penicillin plus gentamicin, amoxicillin plus gentamicin, amoxicillin plus ceftazidime, others (48), ampicillin and cefotaxime, amikacin, vancomycin, others (65), ampicillin, cefotaxime, gentamicin, others (54), cephalosporin, penicillin, others (113), penicillins, macrolides, cephalosporins, others (115), beta-lactams, aminoglycosides, vancomycin, others (73), benzylpenicillin, cloxacillin, flucloxacillin, others (32), benzylpenicillin, cloxacillin, flucloxacillin, others (31), penicillin, penicillin plus gentamicin, others (33), or did not specify which ABX they investigated (28, 29, 42, 56, 61, 82, 98). Studies which did not compare ABX group to controls and studies not providing p-values were excluded.
ARGs
ARGs were studied in two studies, which found a higher abundance of ARGs in ABX group including resistance to ABX rarely/never used in neonates and to multidrug-resistant organisms (MDROs) (54), higher abundance of ARGs and episomally encoded genes in the ABX group (98).
Discussion
The first three years of life are pivotal for the development of the intestinal microbiota (14, 25), which, in turn, plays a crucial role in shaping the immune system. Moreover, early colonization of the intestine significantly impacts later microbial communities, so changes in this early period will have long-lasting consequences (115). A lower diversity of the intestinal microbiota has been associated with an increased risk of developing allergic diseases, type 1 diabetes, and rheumatic diseases (116–118). It is therefore concerning that all the ABX investigated in the studies in our review had profound effects on the intestinal microbiota in children associated with a decrease in alpha diversity. In some studies, this decrease was positively associated with duration of ABX. This effect was prolonged, persisting up to 12–24 months after stopping ABX for macrolides, and up to 36 months for ABX in the neonatal period.
In our review, we found that exposure to certain ABX (penicillins, penicillins plus gentamicin, cephalosporins, carbapenems, macrolides, and aminoglycosides, but not trimethoprim/sulfamethoxazole) is associated with decreased abundances of Actinobacteria (52, 70, 95), Bifidobacteriales (70, 95), Bifidobacteriaceae (52, 54, 95), and/or Bifidobacterium (52, 57, 70, 83, 87), and Lactobacillus (52, 70, 87, 88, 96). The direction of change in the abundance of Enterobacteriaceae depends on the ABX class but often an increase in Enterobacteriaceae other than E. coli is observed. These findings are in accordance with findings from a similar review in adults, which, after ABX, along with a decrease in alpha diversity, also found decreased abundances of Bifidobacterium and Lactobacillus and increased abundances of Enterobacteriaceae, other than E. coli (e.g., Klebsiella, Citrobacter and Enterobacter) (16). The results from our review, also align with results from a large in vitro study, which tested the effect of 144 ABX on 38 common human intestinal microbiota species (119). The study found that macrolides strongly inhibit the growth of most tested intestinal microbes, while beta-lactams have strain specific effects. This strain-specific effect of ABX might lead to large community composition disturbances with “killing-sensitive” strains more readily being eliminated from communities.
As mentioned above, almost all ABX are associated with a reduction in bacteria which have been identified as being beneficial. Bifidobacterium and Lactobacillus contribute to maintaining the gut barrier by producing high concentrations of short chain fatty acids (SCFA) such as acetate, propionate, and butyrate (120). However, SCFA not only serve as energy sources for the interstitial epithelium, but also have diverse effects on host physiology and immunity. In children, a decreased abundance of Actinobacteria has been associated with type 1 diabetes (121), while lower abundances of Lactobacillaceae and Bifidobacteriaceae have been associated with the development of allergic sensitization, eczema, or asthma (116). Reduced Bifidobacterium abundance has also been linked to childhood obesity (122, 123). Furthermore, it has been shown that peptidoglycans from Bifidobacterium can cross the blood-brain barrier and enhance neuronal maturation by influencing cytokine production by microglia (124). However, it's important to note that most of the studies investigating associations between variations in the intestinal microbiota composition and diseases are cross-sectional and therefore, it cannot be determined whether factors like ABX use, which may transiently decrease the abundance of different beneficial taxa, impact the development of these diseases. Nevertheless, a large meta-analysis has shown that ABX exposure in the first years of life is associated with an increased risk of developing atopic dermatitis, allergies, wheezing and asthma, obesity, rheumatological and neurodevelopmental diseases (12) and it is very likely that the mechanism behind this are changes in the microbiota (125).
The exact mechanism by which different intestinal bacteria influence the developing immune system is not yet clear but dysbiosis has been associated with a pro-inflammatory state (e.g., increased T-helper 17 cells (126). A decreased abundance of Bacteroides uniformis has been associated with increased production of interleukin (IL)-17, which is associated with increased neutrophil extracellular trap (NET) formation (114). These web-like structures are composed of DNA, histones, and antimicrobial proteins and released by activated neutrophils in response to infection or inflammation. NETs play an important role in fighting pathogens. In critically ill patients, progressive intestinal dysbiosis characterised by a high abundance of Enterobacteriaceae has been associated with a shift towards immature neutrophil populations with reduced NET formation (127). In contrast, dysregulated NET formation has been associated with various inflammatory and autoimmune diseases, e.g., rheumatological diseases (128). Other studies showed that the abundance of Klebsiella pneumoniae and Streptococcus mitis in the intestine positively correlates with the amount of natural killer cells in blood, the abundance of B. uniformis with immunoglobulin M levels as well as the erythrocyte sedimentation rate, and the abundance Eubacterium eligens with IL-4 and CD3 + CD8+ T cells levels (129).
Antimicrobial resistance is an increasing problem; it is estimated that by the year 2050, 10 million people will die annually because of infections with ABX-resistant bacteria (130). Therefore, another important finding in our review is the increase in ARG following ABX with all ABX classes, which persisted for as short as three weeks for some ABX, but up to four months after treatment for others. This is particularly important for ABX, such as amoxicillin and trimethoprim/sulfamethoxazole, which are often given for long-term prophylaxis. Even a transient increase in ARGs can have significant clinical implications, especially if a child develops a new infection during this period, as it may lead to infections that are harder to treat, requiring ABX with more side effects or which have broader spectrum activity, further exacerbating the problem of antimicrobial resistance.
The impact of ABX on the intestinal microbiota is multifaceted, influenced by both their spectrum of activity, and whether ABX are administered orally or intravenously. Formulation is also a factor: for example bacampicillin syrup alters bacterial abundance but tablets do not (131). ABX with a broad spectrum of activity and selective killing are the most dangerous to the intestinal microbiota and it is important to find drugs with a narrow spectrum of activity that inhibit pathogens but not intestinal commensals.
Other factors can mitigate ABX-induced dysbiosis. In infants, this includes breastfeeding and administration of pre-, pro- and postbiotics (132). Breastfeeding has been associated with an increased abundance of Bifidobacterium, Staphylococcus, and Streptococcus and lower abundances of Enterococcus and Enterobacteriaceae (133). Breast milk itself is an important source of pre-, pro- and postbiotics. A recent study showed that breastfeeding reduced the decrease in B. infantis which was seen after ABX and protected from ABX-associated increased asthma risk (134). The European Society for Paediatric Gastroenterology Hepatology and Nutrition recommends probiotics for the prevention of ABX-associated diarrhoea (132, 135), particularly Saccharomyces boulardii and Lactobacillus rhamnosus GG (136). In an RCT involving healthy children, the administration of Bacillus subtilis DE111 increased alpha diversity (137). Similarly, an observational study in preterm neonates showed an increase in alpha diversity, higher abundances of Bifidobacterium and Lactobacillus, and lower abundances of Streptococcus after the administration of a probiotic containing Lactobacillus acidophilus and Bifidobacterium bifidum (138). These findings, which contrast with the ABX-induced microbial disturbances observed in our systematic review, suggest that probiotics might help restore gut microbiota balance after ABX. A Cochrane review including 33 RCTs concluded that the administration of probiotics led to a reduction of ABX-associated diarrhoea from 19 to 8% with a number needed to treat of nine (132). However, many important questions remain open, such as the ideal timing, dosage, duration and strain selection for probiotics, as well as the benefit of co-administration with pre- and postbiotics.
Strengths and limitations
A strength of this study is the comprehensive literature search, including children of all age groups and various ABX classes. However, the study is also subject to some limitations: First, most of the included studies were observational studies and only few RCTs were identified. Second, although most studies used longitudinal designs, the variability in follow-up periods, ranging from days to years, complicates inter-study comparisons. The timing of stool sampling is crucial, as changes in the bacterial composition of the intestinal microbiota can be transient or fluctuant. Third, a significant portion of the studies did not differentiate between various ABX or did not specify ABX. This may introduce bias into the results, as different ABX classes have been shown to have different effects. Therefore, the results of these studies have been pooled for this review. Even within an ABX class, the spectrum of activity differs and the effect on the intestinal microbiota will therefore be different. Fourth, except for five studies, all included participants had (suspected) infections, potentially introducing infections as a confounding factor. Furthermore, the analysis encompassed diverse age groups, though the impact of ABX on the microbiota is likely most pronounced in younger children, particularly neonates or infants. Lastly, microbiota research is largely influenced by the used analysis techniques. Molecular diagnostics are influenced by DNA extraction and library preparation method, used sequencing platforms and protocols and bioinformatic pipelines and tools. Culture-based diagnostics are influenced by the choice of culture media and incubation conditions and techniques for assessing ABX resistance.
In summary, ABX have profound effects on the intestinal microbiota, with notable differences between ABX classes. The duration of ABX likely influences the magnitude of these changes. Among those studied, macrolides have the most substantial impact while trimethoprim/sulfamethoxazole has the least pronounced effect. Important remaining questions include how long ABX-induced changes in the composition of the intestinal microbiota persist and the long-term effect of transient changes on health outcomes, particularly if they are given beyond the critical period of microbiota development in the first two to three years of life. Additionally, it is crucial to investigate whether ABX-resistant strains persist in the absence of selective pressure from ABX. Another area for further research is the development of remedies which can selectively protect intestinal microbiomes.
Author contributions
JW: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. NC: Writing – review & editing. PZ: Conceptualization, Data curation, Formal Analysis, Investigation, Methodology, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. PZ is supported by an Ambizione Grant from the Swiss National Science Foundation (PZPGP3_193140).
Acknowledgments
Poh Chua provided valuable assistance in defining the search terms.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/falgy.2024.1458688/full#supplementary-material
References
1. Bénard-Laribière A, Jové J, Lassalle R, Robinson P, Droz-Perroteau C, Noize P. Drug use in French children: a population-based study. Arch Dis Child. (2015) 100(10):960–5. doi: 10.1136/archdischild-2014-307224
2. Hellman J, Grape M, Ternhag A. Antibiotic consumption among a Swedish cohort of children born in 2006. Acta Paediatr. (2015) 104(10):1035–8. doi: 10.1111/apa.13097
3. Masarwa R, Lefebvre C, Platt RW, Filion KB. General practitioner prescribing trends among pediatric patients in the United Kingdom: 1998–2018. Pharmacoepidemiol Drug Saf. (2022) 31(3):302–13. doi: 10.1002/pds.5377
4. Tomlin AM, Woods DJ, Lloyd HS, Tilyard MW. Trends in outpatient prescription medicine use in New Zealand children 2010–2015: a national population-based study. Pediatric Drugs. (2018) 20:465–74. doi: 10.1007/s40272-018-0303-3
5. Vernacchio L, Kelly JP, Kaufman DW, Mitchell AA. Medication use among children <12 years of age in the United States: results from the Slone survey. Pediatrics. (2009) 124(2):446–54. doi: 10.1542/peds.2008-2869
6. Aversa Z, Atkinson EJ, Schafer MJ, Theiler RN, Rocca WA, Blaser MJ, et al. Association of infant antibiotic exposure with childhood health outcomes. In: Mayo Clinic Proceedings. Elsevier (2021). p. 66–77.
7. Hicks LA, Jr THT, Hunkler RJ. US outpatient antibiotic prescribing, 2010. N Engl J Med. (2013) 368(15):1461–2. doi: 10.1056/NEJMc1212055
8. Tribble AC, Lee BR, Flett KB, Handy LK, Gerber JS, Hersh AL, et al. Appropriateness of antibiotic prescribing in United States children’s hospitals: a national point prevalence survey. Clin Infect Dis. (2020) 71(8):e226–34. doi: 10.1093/cid/ciaa036
9. Burvenich R, Dillen H, Trinh NTH, Freer J, Wynants L, Heytens S, et al. Antibiotic use in ambulatory care for acutely ill children in high-income countries: a systematic review and meta-analysis. Arch Dis Child. (2022) 107(12):1088–94. doi: 10.1136/archdischild-2022-324227
10. Nasso C, Scarfone A, Pirrotta I, Rottura M, Giorgi DA, Pallio G, et al. Appropriateness of antibiotic prescribing in hospitalized children: a focus on the real-world scenario of the different paediatric subspecialties. Front Pharmacol. (2022) 13:890398. doi: 10.3389/fphar.2022.890398
11. Shulman ST. The history of pediatric infectious diseases. Pediatr Res. (2004) 55(1):163–76. doi: 10.1203/01.PDR.0000101756.93542.09
12. Duong QA, Pittet LF, Curtis N, Zimmermann P. Antibiotic exposure and adverse long-term health outcomes in children: a systematic review and meta-analysis. J Infect. (2022) 85(3):213–300. doi: 10.1016/j.jinf.2022.01.005
13. Bergström A, Skov TH, Bahl MI, Roager HM, Christensen LB, Ejlerskov KT, et al. Establishment of intestinal microbiota during early life: a longitudinal, explorative study of a large cohort of Danish infants. Appl Environ Microbiol. (2014) 80(9):2889–900. doi: 10.1128/AEM.00342-14
14. Zimmermann P, Curtis N. Factors influencing the intestinal microbiome during the first year of life. Pediatr Infect Dis J. (2018) 37(12):e315–35. doi: 10.1097/INF.0000000000002103
15. Park H, Park NY, Koh A. Scarring the early-life microbiome: its potential life-long effects on human health and diseases. BMB Rep. (2023) 56(9):469. doi: 10.5483/BMBRep.2023-0114
16. Zimmermann P, Curtis N. The effect of antibiotics on the composition of the intestinal microbiota-a systematic review. J Infect. (2019) 79(6):471–89. doi: 10.1016/j.jinf.2019.10.008
17. Jain N. The early life education of the immune system: moms, microbes and (missed) opportunities. Gut Microbes. (2020) 12(1):1824564. doi: 10.1080/19490976.2020.1824564
18. van Beijsterveldt IA, Snowden SG, Myers PN, de Fluiter KS, van de Heijning B, Brix S, et al. Metabolomics in early life and the association with body composition at age 2 years. Pediatr Obes. (2022) 17(3):e12859. doi: 10.1111/ijpo.12859
19. Gilmore JH, Shi F, Woolson SL, Knickmeyer RC, Short SJ, Lin W, et al. Longitudinal development of cortical and subcortical gray matter from birth to 2 years. Cerebral cortex. (2012) 22(11):2478–85. doi: 10.1093/cercor/bhr327
20. Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. (2009) 6(7):e1000097. doi: 10.1371/journal.pmed.1000097
21. Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. Int J Surg. (2021) 88:105906. doi: 10.1016/j.ijsu.2021.105906
22. Phillips B. Oxford Center for Evidence-based Medicine. Levels of Evidence. Available online at: http://www.cebm.net/index.aspx?o=1025.2009
23. Institute JB. JBI Critical appraisal checklist for analytical cross sectional studies. (2017) Available online at: https://jbi.global/sites/default/files/2019-05/JBI_Critical_Appraisal-Checklist_for_Analytical_Cross_Sectional_Studies2017_0.pdf (Accessed April 1, 2024).
24. Mathieu H, Lambert-Zechovsky N, Beaufils F, Blum C, Bingen E, Guillot M. Amikacine chez l'enfant: efficacité, tolérance, effets sur l'écosystème microbien intestinal. (1978).
25. Bokulich NA, Chung J, Battaglia T, Henderson N, Jay M, Li H, et al. Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci Transl Med. (2016) 8(343):343ra82. doi: 10.1126/scitranslmed.aad7121
26. Ainonen S, Tejesvi MV, Mahmud MR, Paalanne N, Pokka T, Li W, et al. Antibiotics at birth and later antibiotic courses: effects on gut microbiota. Pediatr Res. (2022) 91(1):154–62. doi: 10.1038/s41390-021-01494-7
27. Akagawa Y, Kimata T, Akagawa S, Yamaguchi T, Kato S, Yamanouchi S, et al. Impact of long-term low dose antibiotic prophylaxis on gut microbiota in children. J Urol. (2020) 204(6):1320–5. doi: 10.1097/JU.0000000000001227
28. Arboleya S, Sánchez B, Solís G, Fernández N, Suárez M, Hernández-Barranco AM, et al. Impact of prematurity and perinatal antibiotics on the developing intestinal microbiota: a functional inference study. Int J Mol Sci. (2016) 17(5):649. doi: 10.3390/ijms17050649
29. Barnett DJM, Endika MF, Klostermann CE, Gu F, Thijs C, Nauta A, et al. Human milk oligosaccharides, antimicrobial drugs, and the gut microbiota of term neonates: observations from the KOALA birth cohort study. Gut Microbes. (2023) 15(1). doi: 10.1080/19490976.2022.2164152
30. Bender JM, Li F, Purswani H, Capretz T, Cerini C, Zabih S, et al. Early exposure to antibiotics in the neonatal intensive care unit alters the taxonomic and functional infant gut microbiome. J Matern Fetal Neonatal Med. (2021) 34(20):3335–43. doi: 10.1080/14767058.2019.1684466
31. Bennet R, Eriksson M, Nord C, Zetterström R. Fecal bacterial microflora of newborn infants during intensive care management and treatment with five antibiotic regimens. Pediatr Infect Dis. (1986) 5(5):533–9. doi: 10.1097/00006454-198609000-00009
32. Bennet R, Nord CE. Development of the faecal anaerobic microflora after caesarean section and treatment with antibiotics in newborn infants. Infection. (1987) 15(5):332–6. doi: 10.1007/BF01647733
33. Blakey J, Lubitz L, Barnes GL, Bishop RF, Campbell NT, Gillam GL. Development of gut colonisation in pre-term neonates. J Med Microbiol. (1982) 15(4):519–29. doi: 10.1099/00222615-15-4-519
34. Bonnemaison E, Lanotte PH, Cantagrel S, Thionois S, Quentin R, Chamboux C, et al. Comparison of fecal flora following administration of two antibiotic protocols for suspected maternofetal infection. Biol Neonate. (2003) 84(4):304–10. doi: 10.1159/000073639
35. Borderon JC, Rastegar A, Rolland JC, Laugier J. Effect of an amoxicillin-clavulanic acid combination on the aerobic fecal flora in children. Pathol Biol. (1985) 33(5 Pt 2):569–72.3937129
36. Bourke CD, Gough EK, Pimundu G, Shonhai A, Berejena C, Terry L, et al. Cotrimoxazole reduces systemic inflammation in HIV infection by altering the gut microbiome and immune activation. Sci Transl Med. (2019) 11(486):eaav0537. doi: 10.1126/scitranslmed.aav0537
37. Bourrillon A, Lambert-Zechovsky N, Beaufils F, Lejeune C, Bingen E, Blum C, et al. Antibiotic therapy, intestinal microbial pullulation and risk of infection in children. Arch Fr Pediatr. (1978) 35(10 Suppl):23–37.255089
38. Butel MJ, Boussougnant Y. Influence de ĺérythromycine sur la flore fécale de ĺenfant de moins de un an. Pathol Biol. (1986) 34(5):591–5.3534749
39. Butel MJ, Suau A, Campeotto F, Magne F, Aires J, Ferraris L, et al. Conditions of Bifidobacterial Colonization in Preterm Infants: A Prospective Analysis (2007). Available online at: http://journals.lww.com/jpgn
40. Chang HY, Chiau JSC, Ho YH, Chang JH, Tsai KN, Liu CY, et al. Impact of early empiric antibiotic regimens on the gut microbiota in very low birth weight preterm infants: an observational study. Front Pediatr. (2021) 9:651713. doi: 10.3389/fped.2021.651713
41. Chernikova DA, Koestler DC, Hoen AG, Housman ML, Hibberd PL, Moore JH, et al. Fetal exposures and perinatal influences on the stool microbiota of premature infants. J Matern Fetal Neonatal Med. (2016) 29(1):99–105. doi: 10.3109/14767058.2014.987748
42. D'Agata AL, Wu J, Welandawe MKV, Dutra SVO, Kane B, Groer MW. Effects of early life NICU stress on the developing gut microbiome. In: Developmental Psychobiology. John Wiley and Sons Inc. (2019) p. 650–60.
43. Dahl C, Stigum H, Valeur J, Iszatt N, Lenters V, Peddada S, et al. Preterm infants have distinct microbiomes not explained by mode of delivery, breastfeeding duration or antibiotic exposure. Int J Epidemiol. (2018) 47(5):1658–69. doi: 10.1093/ije/dyy064
44. Dardas M, Gill SR, Grier A, Pryhuber GS, Gill AL, Lee YH, et al. The impact of postnatal antibiotics on the preterm intestinal microbiome. Pediatr Res. (2014) 76(2):150–8. doi: 10.1038/pr.2014.69
45. d'Haens EJ, Zwittink RD, Belzer C, Hemels MAC, van Lingen RA, Renes IB, et al. Short-term changes of intestinal microbiota composition in preterm infants after two prophylactic doses of vancomycin. Acta Paediatr Int J Paediatr. (2019) 108(10):1919–20. doi: 10.1111/apa.14915
46. Doan T, Arzika AM, Ray KJ, Cotter SY, Kim J, Maliki R, et al. Gut microbial diversity in antibiotic-naive children after systemic antibiotic exposure: a randomized controlled trial. Clin Infect Dis. (2017) 64(9):1147–53. doi: 10.1093/cid/cix141
47. D’Souza AW, Moodley-Govender E, Berla B, Kelkar T, Wang B, Sun X, et al. Cotrimoxazole prophylaxis increases resistance gene prevalence and α-diversity but decreases β-diversity in the gut microbiome of human immunodeficiency virus–exposed, uninfected infants. Clin Infect Dis. (2020) 71(11):2858–68. doi: 10.1093/cid/ciz1186
48. Eck A, Rutten NBMM, Singendonk MMJ, Rijkers GT, Savelkoul PHM, Meijssen CB, et al. Neonatal microbiota development and the effect of early life antibiotics are determined by two distinct settler types. PLoS One. (2020) 15(2):e0228133. doi: 10.1371/journal.pone.0228133
49. Ferraris L, Butel MJ, Campeotto F, Vodovar M, Rozé JC, Aires J. Clostridia in premature neonates’ gut: incidence, antibiotic susceptibility, and perinatal determinants influencing colonization. PLoS One. (2012) 7(1):e30594. doi: 10.1371/journal.pone.0030594
50. Forsgren M, Isolauri E, Salminen S, Rautava S. Late preterm birth has direct and indirect effects on infant gut microbiota development during the first six months of life. Acta Paediatr Int J Paediatr (2017) 106(7):1103–9. doi: 10.1111/apa.13837
51. Fouhy F, Watkins C, Hill CJ, O’Shea CA, Nagle B, Dempsey EM, et al. Perinatal factors affect the gut microbiota up to four years after birth. Nat Commun. (2019) 10(1):1517. doi: 10.1038/s41467-019-09252-4
52. Fouhy F, Guinane CM, Hussey S, Wall R, Ryan CA, Dempsey EM, et al. High-throughput sequencing reveals the incomplete, short-term recovery of infant gut microbiota following parenteral antibiotic treatment with ampicillin and gentamicin. Antimicrob Agents Chemother. (2012) 56(11):5811–20. doi: 10.1128/AAC.00789-12
53. Fujita K, Sakata H, Yoshioka H. Selective inhibition of enterobacteriaceae in fecal flora of children by oral trimethoprim-sulfamethoxazole. Pediatr Infect Dis. (1984) 3(6):530–3. doi: 10.1097/00006454-198411000-00010
54. Gasparrini AJ, Wang B, Sun X, Kennedy EA, Hernandez-Leyva A, Ndao IM, et al. Persistent metagenomic signatures of early-life hospitalization and antibiotic treatment in the infant gut microbiota and resistome. Nat Microbiol. (2019) 4(12):2285–97. doi: 10.1038/s41564-019-0550-2
55. Gibson MK, Wang B, Ahmadi S, Burnham CAD, Tarr PI, Warner BB, et al. Developmental dynamics of the preterm infant gut microbiota and antibiotic resistome. Nat Microbiol. (2016) 1(4):1–10. doi: 10.1038/nmicrobiol.2016.24
56. Goldmann DA, Leclair J, Macone A. Bacterial colonization of neonates admitted to an intensive care environment. J Pediatr. (1978) 93(2):288–93. doi: 10.1016/S0022-3476(78)80523-X
57. Gong C, Yang L, Liu K, Shen S, Zhang Q, Li H, et al. Effects of antibiotic treatment and probiotics on the gut microbiome of 40 infants delivered before term by cesarean section analysed by using 16s rRNA quantitative polymerase chain reaction sequencing. Med Sci Monit. (2021) 27:e928467-1. doi: 10.12659/MSM.928467
58. Greenwood C, Morrow AL, Lagomarcino AJ, Altaye M, Taft DH, Yu Z, et al. Early empiric antibiotic use in preterm infants is associated with lower bacterial diversity and higher relative abundance of enterobacter. J Pediatr. (2014) 165(1):23–9. doi: 10.1016/j.jpeds.2014.01.010
59. Grylack L, Neugebauer D, Scanlon JW. Effects of oral antibiotics on stool Flora and overall sensitivity patterns in an intensive care nursery. Pediatr Res. (1982) 16:509–51. doi: 10.1203/00006450-198207000-00001
60. Guggenbichler JP, Koflerj J. Influence of third-generation cephalosporins on aerobic intestinal flora. J Antimicrob Chemother. (1984) 14:67–70. doi: 10.1093/jac/14.suppl_B.67
61. Hall MA, Cole CB, Smith SL, Fuller R, Rolles CJ. Factors influencing the presence of faecal lactobacilli in early infancy. Arch Dis Child. (1990) 65(2):185. doi: 10.1136/adc.65.2.185
62. Hinterwirth A, Sié A, Coulibaly B, Ouermi L, Dah C, Tapsoba C, et al. Rapid reduction of campylobacter species in the gut microbiome of preschool children after oral azithromycin: a randomized controlled trial. Am J Trop Med Hyg. (2020) 103(3):1266–9. doi: 10.4269/ajtmh.19-0940
63. Holton AF, Hall MA, Lowes JA. Antibiotic exposure delays intestinal colonization by Clostridium difficile in the newborn. J Antimicrob Chemother. (1989) 24(5):811–7. doi: 10.1093/jac/24.5.811
64. Hutchinson RA, Costeloe KL, Wade WG, Millar MR, Ansbro K, Stacey F, et al. Intravenous antibiotics in preterm infants have a negative effect upon microbiome development throughout preterm life. Gut Pathog. (2023) 15(1):18. doi: 10.1186/s13099-023-00544-1
65. Itani T, Moubareck CA, Melki I, Rousseau C, Mangin I, Butel MJ, et al. Establishment and development of the intestinal microbiota of preterm infants in a Lebanese tertiary hospital. Anaerobe. (2017) 43:4–14. doi: 10.1016/j.anaerobe.2016.11.001
66. Jacquot A, Neveu D, Aujoulat F, Mercier G, Marchandin H, Jumas-Bilak E, et al. Dynamics and clinical evolution of bacterial gut microflora in extremely premature patients. J Pediatr. (2011) 158(3):390–6. doi: 10.1016/j.jpeds.2010.09.007
67. Jenke AC, Postberg J, Mariel B, Hensel K, Foell D, Däbritz J, et al. S100a12 and HBD2 correlate with the composition of the fecal microflora in ELBW infants and expansion of E. COLI is associated with NEC. BioMed Res Int. (2013) 2013:150372. doi: 10.1155/2013/150372
68. Jia J, Xun P, Wang X, He K, Tang Q, Zhang T, et al. Impact of postnatal antibiotics and parenteral nutrition on the gut microbiota in preterm infants during early life. J Parenter Enteral Nutr. (2020) 44(4):639–54. doi: 10.1002/jpen.1695
69. Kim CS, Grady N, Derrick M, Yu Y, Oliphant K, Lu J, et al. Effect of antibiotic use within first 48 hours of life on the preterm infant microbiome: a randomized clinical trial. JAMA Pediatr. (2021) 175:303–5. doi: 10.1001/jamapediatrics.2020.4916
70. Korpela K, Salonen A, Virta LJ, Kekkonen RA, Forslund K, Bork P, et al. Intestinal microbiome is related to lifetime antibiotic use in Finnish pre-school children. Nat Commun. (2016) 7:10410. doi: 10.1038/ncomms10410
71. Korpela K, Salonen A, Saxen H, Nikkonen A, Peltola V, Jaakkola T, et al. Antibiotics in early life associate with specific gut microbiota signatures in a prospective longitudinal infant cohort. Pediatr Res. (2020) 88(3):438–43. doi: 10.1038/s41390-020-0761-5
72. Kwon Y, Cho YS, Lee YM, Kim SJ, Bae J, Jeong SJ. Changes to gut microbiota following systemic antibiotic administration in infants. Antibiotics. (2022) 11(4):470. doi: 10.3390/antibiotics11040470
73. Rosa PSL, Warner BB, Zhou Y, Weinstock GM, Sodergren E, Hall-Moore CM, et al. Patterned progression of bacterial populations in the premature infant gut. Proc Natl Acad Sci U S A. (2014) 111(34):12522–7. doi: 10.1073/pnas.1409497111
74. Lebeaux RM, Madan JC, Nguyen QP, Coker MO, Dade EF, Moroishi Y, et al. Impact of antibiotics on off-target infant gut microbiota and resistance genes in cohort studies. Pediatr Res. (2022) 92(6):1757–66. doi: 10.1038/s41390-022-02104-w
75. Li X, Brejnrod A, Thorsen J, Zachariasen T, Trivedi U, Russel J, et al. Differential responses of the gut microbiome and resistome to antibiotic exposures in infants and adults. Nat Commun. (2023) 14(1):8526. doi: 10.1038/s41467-023-44289-6
76. Lindberg TP, Caimano MJ, Hagadorn JI, Bennett EM, Maas K, Brownell EA, et al. Preterm infant gut microbial patterns related to the development of necrotizing enterocolitis. J Matern Fetal Neonatal Med. (2020) 33(3):349–58. doi: 10.1080/14767058.2018.1490719
77. Lu S, Huang Q, Wei B, Chen Y. Effects of β-lactam antibiotics on gut microbiota colonization and metabolites in late preterm infants. Curr Microbiol. (2020) 77(12):3888–96. doi: 10.1007/s00284-020-02198-7
78. Mangin I, Suau A, Gotteland M, Brunser O, Pochart P. Amoxicillin treatment modifies the composition of bifidobacterium species in infant intestinal microbiota. Anaerobe. (2010) 16(4):433–8. doi: 10.1016/j.anaerobe.2010.06.005
79. Oldenburg CE, Sié A, Coulibaly B, Ouermi L, Dah C, Tapsoba C, et al. Effect of commonly used pediatric antibiotics on gut microbial diversity in preschool children in Burkina Faso: a randomized clinical trial. Open Forum Infect Dis. (2018) 5(11):ofy289. doi: 10.1093/ofid/ofy289
80. Parker EPK, Praharaj I, John J, Kaliappan SP, Kampmann B, Kang G, et al. Changes in the intestinal microbiota following the administration of azithromycin in a randomised placebo-controlled trial among infants in south India. Sci Rep. (2017) 7(1):9168. doi: 10.1038/s41598-017-06862-0
81. Parm Ü, Metsvaht T, Sepp E, Ilmoja ML, Pisarev H, Pauskar M, et al. Impact of empiric antibiotic regimen on bowel colonization in neonates with suspected early onset sepsis. Eur J Clin Microbiol Infect Dis. (2010) 29:807–16. doi: 10.1007/s10096-010-0931-1
82. Penders J, Thijs C, Vink C, Stelma FF, Snijders B, Kummeling I, et al. Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics. (2006) 118(2):511–21. doi: 10.1542/peds.2005-2824
83. Reyman M, van Houten MA, Watson RL, Chu MLJN, Arp K, de Waal WJ, et al. Effects of early-life antibiotics on the developing infant gut microbiome and resistome: a randomized trial. Nat Commun. (2022) 13(1):893. doi: 10.1038/s41467-022-28525-z
84. Romano-Keeler J, Shilts MH, Tovchigrechko A, Wang C, Brucker RM, Moore DJ, et al. Distinct mucosal microbial communities in infants with surgical necrotizing enterocolitis correlate with age and antibiotic exposure. PLoS One. (2018) 13(10):e0206366. doi: 10.1371/journal.pone.0206366
85. Rooney AM, Timberlake K, Brown KA, Bansal S, Tomlinson C, Lee KS, et al. Each additional day of antibiotics is associated with lower gut anaerobes in neonatal intensive care unit patients. Clin Infect Dis. (2020) 70(12):2553–60. doi: 10.1093/cid/ciz698
86. Russell JT, Ruoss JL, de la Cruz D, Li N, Bazacliu C, Patton L, et al. Antibiotics and the developing intestinal microbiome, metabolome and inflammatory environment in a randomized trial of preterm infants. Sci Rep. (2021) 11(1):1943. doi: 10.1038/s41598-021-80982-6
87. Sakata H, Fujiia K, Yoshioka H. The effect of antimicrobial agents on fecal Flora of children. Antimicrob Agents Chemother. (1986) 29:225–9. doi: 10.1128/AAC.29.2.225
88. Savino F, Roana J, Mandras N, Tarasco V, Locatelli E, Tullio V. Faecal microbiota in breast-fed infants after antibiotic therapy. Acta Paediatr Int J Paediatr. (2011) 100(1):75–8. doi: 10.1111/j.1651-2227.2010.01988.x
89. Schwartz DJ, Langdon A, Sun X, Langendorf C, Berthé F, Grais RF, et al. Effect of amoxicillin on the gut microbiome of children with severe acute malnutrition in Madarounfa, Niger: a retrospective metagenomic analysis of a placebo-controlled trial. Lancet Microbe. (2023) 4(11):e931–42. doi: 10.1016/S2666-5247(23)00213-6
90. Sunakawa K, Akitaf H, Iwataf S, Satof Y. The influence of cefotaxime on intestinal flora and bleeding diathesis in infants and neonates, compared with other/mactams. J Antimicrob Chemother. (1984) 14:317–24. doi: 10.1093/jac/14.suppl_B.317
91. Tanaka S, Kobayashi T, Songjinda P, Tateyama A, Tsubouchi M, Kiyohara C, et al. Influence of antibiotic exposure in the early postnatal period on the development of intestinal microbiota. FEMS Immunol Med Microbiol. (2009) 56(1):80–7. doi: 10.1111/j.1574-695X.2009.00553.x
92. Tapiainen T, Koivusaari P, Brinkac L, Lorenzi HA, Salo J, Renko M, et al. Impact of intrapartum and postnatal antibiotics on the gut microbiome and emergence of antimicrobial resistance in infants. Sci Rep. (2019) 9(1):10635. doi: 10.1038/s41598-019-46964-5
93. Tullus K, Berglund B, Fryklund B, Kühn I, Burman LG. Influence of antibiotic therapy on faecal carriage of P-fimbriated Escherichia coli and other gram-negative bacteria in neonates. J Antimicrob Chemother. (1988) 22(4):563–8. doi: 10.1093/jac/22.4.563
94. Ward DV, Scholz M, Zolfo M, Taft DH, Schibler KR, Tett A, et al. Metagenomic sequencing with strain-level resolution implicates uropathogenic E. coli in necrotizing enterocolitis and mortality in preterm infants. Cell Rep. (2016) 14(12):2912–24. doi: 10.1016/j.celrep.2016.03.015
95. Wei S, Mortensen MS, Stokholm J, Brejnrod AD, Thorsen J, Rasmussen MA, et al. Short- and long-term impacts of azithromycin treatment on the gut microbiota in children: a double-blind, randomized, placebo-controlled trial. EBioMedicine. (2018) 38:265–72. doi: 10.1016/j.ebiom.2018.11.035
96. Welkon CJ, Long SS, Gilligan PH. Effect of imipenem-cilastatin therapy on fecal Flora. Antimicrob Agents Chemother. (1986) 29:741–3. doi: 10.1128/AAC.29.5.741
97. Xu Y, Milburn O, Beiersdorfer T, Du L, Akinbi H, Haslam DB. Antibiotic exposure prevents acquisition of beneficial metabolic functions in the preterm infant gut microbiome. Microbiome. (2022) 10(1):103. doi: 10.1186/s40168-022-01300-4
98. Yassour M, Vatanen T, Siljander H, Hämäläinen AM, Härkönen T, Ryhänen SJ, et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Available online at: https://www.ScienceTranslationalMedicine.org (Accessed 15 May, 2024).
99. Zhou Y, Shan G, Sodergren E, Weinstock G, Walker WA, Gregory KE. Longitudinal analysis of the premature infant intestinal microbiome prior to necrotizing enterocolitis: a case-control study. PLoS One. (2015) 10(3):e0118632. doi: 10.1371/journal.pone.0118632
100. Zhu D, Xiao S, Yu J, Ai Q, He Y, Cheng C, et al. Effects of one-week empirical antibiotic therapy on the early development of gut microbiota and metabolites in preterm infants. Sci Rep. (2017) 7(1):8025. doi: 10.1038/s41598-017-08530-9
101. Zwittink RD, Renes IB, van Lingen RA, van Zoeren-Grobben D, Konstanti P, Norbruis OF, et al. Association between duration of intravenous antibiotic administration and early-life microbiota development in late-preterm infants. Eur J Clin Microbiol Infect Dis. (2018) 37(3):475–83. doi: 10.1007/s10096-018-3193-y
102. Zwittink RD, Zoeren-Grobben DV, Renes IB, Lingen RAV, Norbruis OF, Martin R, et al. Dynamics of the bacterial gut microbiota in preterm and term infants after intravenous amoxicillin/ceftazidime treatment. BMC Pediatr. (2020) 20(1):1–10. doi: 10.1186/s12887-020-02067-z
103. Doan T, Hinterwirth A, Arzika AM, Cotter SY, Ray KJ, O'Brien KS, et al. Mass azithromycin distribution and community microbiome: a cluster-randomized trial. Open Forum Infect Dis. (2018) 5(8):ofy182. doi: 10.1093/ofid/ofy182
104. Westerbeek E, Slump R, Lafeber H, Knol J, Georgi G, Fetter W, et al. The effect of enteral supplementation of specific neutral and acidic oligosaccharides on the faecal microbiota and intestinal microenvironment in preterm infants. Eur J Clin Microbiol Infect Dis. (2013) 32:269–76. doi: 10.1007/s10096-012-1739-y
105. Borderon JC, Rastegar A, Laugier J, Gold F. The effect of imipenem/cilastatin on the aerobic faecal flora of children. J Antimicrob Chemother. (1986) 18:121–5. doi: 10.1093/jac/18.Supplement_E.121
106. Lambert-Zechovsky N, Bingen E, Proux M, Aujard Y, Mathieu H. Influence of cefoperazone on the fecal flora in children. Pathol Biol. (1984) 32(5):439–42.6739153
107. Lambert-Zechovsky N, Bingen E, Proux MC, Aujard Y, Mathieu H. Influence de l'amoxicilline associée à l'acide clavulanique sur la flore fécale de l'enfant. Pathol Biol. (1984) 32(5):436–8.6739152
108. Lambert-Zechovsky N, Bingen E, Beaufils F, Bourrilon A, Mathieu H. Etude de ĺécosystème intestinal de ĺenfant - influence de la colistine. Pathol Biol. (1981) 29(5):293–7.6785710
109. Lambert-Zechovsky N, Bingen E, Proux MC, Aufrant C, Mathieu H. Effet du céfotaxime sur ĺécosystème bactérien intestinal de ĺenfant. La Nouvelle Presse Médicale. (1981) 10(8):553–5.
110. Bingen E, Leluan G, Desvignes A. Influence de l'ampicilline associée aux aminosides sur l'ecosystème bactérien intestinal de l'enfant (1982).
111. Brignoli C, Vassena E. Effect of spiramycin on intestinal flora in infants. Minerva Pediatr. (1958) 10(29–30):757–62. doi: 10.1097/NNR.0000000000000133
112. Moustardier G, Bentegeat J, Le Noc P. Changes in aerobic intestinal flora of infants and young children caused by antibiotics. Ann Inst Pasteur. (1956) 10–21.
113. Bai L, Zhou P, Li D, Ju X. Changes in the gastrointestinal microbiota of children with acute lymphoblastic leukaemia and its association with antibiotics in the short term. J Med Microbiol. (2017) 66(9):1297–307. doi: 10.1099/jmm.0.000568
114. Li G, Liu L, Lu T, Sui Y, Zhang C, Wang Y, et al. Gut microbiota aggravates neutrophil extracellular traps-induced pancreatic injury in hypertriglyceridemic pancreatitis. Nat Commun. (2023) 14(1):6179. doi: 10.1038/s41467-023-41950-y
115. Wampach L, Heintz-Buschart A, Hogan A, Muller EEL, Narayanasamy S, Laczny CC, et al. Colonization and succession within the human gut microbiome by archaea, bacteria, and microeukaryotes during the first year of life. Front Microbiol. (2017) 8:738. doi: 10.3389/fmicb.2017.00738
116. Zimmermann P, Messina N, Mohn WW, Finlay BB, Curtis N. Association between the intestinal microbiota and allergic sensitization, eczema, and asthma: a systematic review. J Allergy Clin Immunol. (2019) 143(2):467–85. doi: 10.1016/j.jaci.2018.09.025
117. Mokhtari P, Metos J, Babu PVA. Impact of type 1 diabetes on the composition and functional potential of gut microbiome in children and adolescents: possible mechanisms, current knowledge, and challenges. Gut Microbes. (2021) 13(1):1926841. doi: 10.1080/19490976.2021.1926841
118. Wang Y, Wei J, Zhang W, Doherty M, Zhang Y, Xie H, et al. Gut dysbiosis in rheumatic diseases: a systematic review and meta-analysis of 92 observational studies. EBioMedicine. (2022) 80. doi: 10.1016/j.ebiom.2022.104055
119. Maier L, Goemans CV, Wirbel J, Kuhn M, Eberl C, Pruteanu M, et al. Unravelling the collateral damage of antibiotics on gut bacteria. Nature. (2021) 599(7883):120–4. doi: 10.1038/s41586-021-03986-2
120. LeBlanc JG, Chain F, Martín R, Bermúdez-Humarán LG, Courau S, Langella P. Beneficial effects on host energy metabolism of short-chain fatty acids and vitamins produced by commensal and probiotic bacteria. Microb Cell Fact. (2017) 16:1–10. doi: 10.1186/s12934-017-0691-z
121. Murri M, Leiva I, Gomez-Zumaquero JM, Tinahones FJ, Cardona F, Soriguer F, et al. Gut microbiota in children with type 1 diabetes differs from that in healthy children: a case-control study. BMC Med. (2013) 11:1–12. doi: 10.1186/1741-7015-11-46
122. Gao X, Jia R, Xie L, Kuang L, Feng L, Wan C. Obesity in school-aged children and its correlation with gut E. coli and bifidobacteria: a case–control study. BMC Pediatr. (2015) 15:1–4. doi: 10.1186/s12887-015-0318-7
123. Pihl AF, Fonvig CE, Stjernholm T, Hansen T, Pedersen O, Holm JC. The role of the gut microbiota in childhood obesity. Childhood Obes. (2016) 12(4):292–9. doi: 10.1089/chi.2015.0220
124. Parker A, Fonseca S, Carding SR. Gut microbes and metabolites as modulators of blood-brain barrier integrity and brain health. Gut Microbes. (2020) 11(2):135–57. doi: 10.1080/19490976.2019.1638722
125. Laforest-Lapointe I, Arrieta MC. Patterns of early-life gut microbial colonization during human immune development: an ecological perspective. Front Immunol. (2017) 8:276727. doi: 10.3389/fimmu.2017.00788
126. Shao L, Li M, Zhang B, Chang P. Bacterial dysbiosis incites Th17 cell revolt in irradiated gut. Biomed Pharmacother. (2020) 131:110674. doi: 10.1016/j.biopha.2020.110674
127. Schlechte J, Zucoloto AZ, Yu I, Doig CJ, Dunbar MJ, McCoy KD, et al. Dysbiosis of a microbiota–immune metasystem in critical illness is associated with nosocomial infections. Nat Med. (2023) 29(4):1017–27. doi: 10.1038/s41591-023-02243-5
128. Apel F, Zychlinsky A, Kenny EF. The role of neutrophil extracellular traps in rheumatic diseases. Nat Rev Rheumatol. (2018) 14:467–75. doi: 10.1038/s41584-018-0039-z
129. Lin X, Abdalla M, Yang J, Liu L, Fu Y, Zhang Y, et al. Relationship between gut microbiota dysbiosis and immune indicator in children with sepsis. BMC Pediatr. (2023) 23(1):516. doi: 10.1186/s12887-023-04349-8
130. O'Neill J. Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Review on Antimicrobial Resistance. Wellcome Trust and HM Government. (2016). Available online at: https://amr-review.org/sites/default/files/160525_Final%20paper_with%20cover.pdf (Accessed September 17, 2024).
131. Heimdahl A, Nord CE, Weilander K. Effect of bacampicillin on human mouth, throat and colon flora. Infection. (1979) 7(Suppl 5):S446–51. doi: 10.1007/BF01659768
132. Guo Q, Goldenberg JZ, Humphrey C, El Dib R, Johnston BC. Probiotics for the prevention of pediatric antibiotic-associated diarrhea. Cochrane Database Syst Rev. (2019) 4. doi: 10.1002/14651858.CD004827.pub5
133. Leo S, Cetiner OF, Pittet LF, Messina NL, Jakob W, Falquet L, et al. The association between the composition of the early-life intestinal microbiome and eczema in the first year of life. Front Microbiomes. (2023) 2:1147082. doi: 10.3389/frmbi.2023.1147082
134. Dai DLY, Petersen C, Hoskinson C, Bel KLD, Becker AB, Moraes TJ, et al. Breastfeeding enrichment of B. longum subsp. infantis mitigates the effect of antibiotics on the microbiota and childhood asthma risk. Med (New York, NY). (2023) 4(2):92–112.e5. doi: 10.1016/j.medj.2022.12.002
135. Szajewska H, Canani RB, Domellöf M, Guarino A, Hojsak I, Indrio F, et al. Probiotics for the management of pediatric gastrointestinal disorders: position paper of the ESPGHAN special interest group on gut Microbiota and modifications. J Pediatr Gastroenterol Nutr. (2023) 76(2):232–47. doi: 10.1097/MPG.0000000000003633
136. Szajewska H, Kołodziej M. Systematic review with meta-analysis: saccharomyces boulardii in the prevention of antibiotic-associated diarrhoea. Aliment Pharmacol Ther. (2015) 42(7):793–801. doi: 10.1111/apt.13344
137. Paytuví-Gallart A, Sanseverino W, Winger A. Daily intake of probiotic strain Bacillus subtilis DE111 supports a healthy microbiome in children attending day-care. Benefic Microbes. (2020) 11(7):611–20. doi: 10.3920/BM2020.0022
138. Westaway JA, Huerlimann R, Kandasamy Y, Miller CM, Norton R, Watson D, et al. To probiotic or not to probiotic: a metagenomic comparison of the discharge gut microbiome of infants supplemented with probiotics in NICU and those who are not. Front Pediatr. (2022) 10:838559. doi: 10.3389/fped.2022.838559
Keywords: microbiome, stool, intestine, sequencing, 16S rRNA, penicillin, cephalosporin, macrolide
Citation: Wurm J, Curtis N and Zimmermann P (2024) The effect of antibiotics on the intestinal microbiota in children - a systematic review. Front. Allergy 5:1458688. doi: 10.3389/falgy.2024.1458688
Received: 3 July 2024; Accepted: 6 September 2024;
Published: 7 October 2024.
Edited by:
Lisa A. Miller, University of California, Davis, United StatesReviewed by:
Priya Katari, Weill Cornell Medical Center, United StatesJosé J. Leija-Martínez, Autonomous University of San Luis Potosí, Mexico
Copyright: © 2024 Wurm, Curtis and Zimmermann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Petra Zimmermann, cGV0cmEuemltbWVybWFubkB1bmlmci5jaA==
 Juliane Wurm
Juliane Wurm Nigel Curtis
Nigel Curtis Petra Zimmermann
Petra Zimmermann