- 1Soil Science and Plant Nutrition, Faculty of Life Sciences, Rhine-Waal University of Applied Sciences, Kleve, Germany
- 2Department of Soil Science, Bangladesh Agricultural University, Mymensingh, Bangladesh
- 3Soil Biology and Plant Nutrition, University of Kassel, Witzenhausen, Germany
Sea-water level rise leads to increased saltwater intrusion causing soil salinity on arable land with negative effects on soil microbial processes. Organic amendments are known to reduce the effects of salinity on soil microorganisms, therefore positively influencing microbial activity and nutrient cycling. However, the extent of this effect in paddy rice soils under aerobic compared to anaerobic conditions is unknown. Consequently, benefits of organic matter addition on carbon (C) and nitrogen (N) mineralisation under saline conditions were evaluated in a short-term laboratory incubation experiment. Two soils from Bangladesh were incubated with rice straw, manure or a manure-rice straw mixture at 50 and 100% water holding capacity (25°C, 27 days). In addition, NaCl was added to half of the samples, which resulted in a set of non-saline (ECe = 1.1–1.3 dS m−1) and saline (ECe = 24.0–32.4 dS m−1) soils. Soil respiration (CO2-release) was measured throughout the experiment. At the end of the experiment, dissolved organic C, inorganic N, microbial biomass as well as bacterial, archaeal, and fungal domains were determined. Overall, effects of substrate addition overruled effects caused by salinity and water content. Microbial activity and biomass in particular fungi increased most strongly after rice straw addition and resulted in N immobilisation independent of moisture level. Rice straw and manure alleviated the effects of salinity on microorganisms; these were therefore mainly detectable in the non-amended soils. The reason for this is likely a higher C availability for soil microorganisms after amendment of organic materials, which allows them to produce osmolytes, counteracting the osmotic effects of increased salinity. However, the microbial communities of the two soils under investigation showed different response patterns to salinity reflected by a substantially higher fungi-bacteria ratio in soil B prone to salinity at 50% WHC as compared to soil A. Likewise, the metabolic quotient was higher in soil B and not affected by any of the short term treatments, revealing a soil legacy. Our results highlight the importance of organic amendments, such as rice straw to paddy soils under saline conditions to reduce negative effects on soil microbial processes, allowing them to maintain their major functions.
Introduction
Soil salinization is a regionally or locally important process of soil degradation resulting in temporal or permanent loss of arable land, mainly because increasing salt content results in reduced plant growth of crops. Even though there is considerable variation in the extent of soil salinity (Luedeling and Wichern, 2007), about 20% of the world's cultivated land and 50% of the arable land are affected to a certain extent by salinity (Flowers and Yeo, 1995). Soil salinity is caused by natural processes, i.e., primary salinization or is the result of human activities, i.e., secondary salinization, occurring in irrigated as well as in rain-fed dryland agriculture.
Many of the future challenges of food production caused by climate change, such as a rising sea-water level leading to increased saltwater intrusion causing soil salinity can be found in Bangladesh. Especially the coastal area is largely affected (about 50%) by different degrees of salinity (Haque, 2006). The pronounced dry and rainy seasons, which result in temporal accumulation of salts in the soils, contribute to this problem, even further away from the coast.
Rice is the major staple food in Bangladesh and cultivated on large areas in paddy systems, often affected by soil salinity. Solutions to maintain rice production under salt stress conditions are necessary, where cropping of salt tolerant rice varieties is one option. However, soil salinity may affect not only plants but also the soil microorganisms responsible for nutrient cycling. It is known that soil salinity is a severe stress to the living components of ecosystems by increasing the osmotic pressure on cells and reducing water availability (Killham and Firestone, 1984; Schimel et al., 1989; Borken and Matzner, 2009; Mitran et al., 2017). Paddy systems are characterised by periods of water saturation and periods of aerobic conditions, which can be both affected by salinization. However, after flooding the paddy field, water saturation might result in a dilution of accumulated salts. Later, when water is lost from the system, salt accumulation most likely also increases in a paddy system. It is known that soil microorganisms are affected by both, increased soil salinity and reduced oxygen availability.
Soil salinity is a major threat to soil microorganisms, strongly altering soil organic matter turnover and nutrient recycling (Wichern et al., 2006; Chowdhury et al., 2011; Setia et al., 2011; Rath and Rousk, 2015; Leogrande and Vitti, 2019). Salt effects on soil microorganisms and biogeochemical processes have been repeatedly studied for more than 40 years (Laura, 1974; Rietz and Haynes, 2003; Sardinha et al., 2003; Wichern et al., 2006; Khan et al., 2008). The clearest effects were observed for microbial activity parameters, such as respiration, N mineralization, and enzyme activity, which usually decreased with increasing salinity (Laura, 1974; Pathak and Rao, 1998; Rietz and Haynes, 2003; Wichern et al., 2006; Rath and Rousk, 2015). Less consistent were the salinity effects on the soil microbial biomass and microbial community composition (Wichern et al., 2006; Rath and Rousk, 2015; Iqbal et al., 2016). Often microbial biomass is not altered in the short term (Rath and Rousk, 2015). Inconsistent effects were observed on the response of fungi to salinity (Wichern et al., 2006; Rath and Rousk, 2015; Iqbal et al., 2016; Leogrande and Vitti, 2019).
One method for decreasing negative salinity effects is the incorporation of organic materials into soil (Luedeling et al., 2005; Wichern et al., 2006), due to their beneficial effects on soil physical, chemical, and biological properties (Wichern et al., 2006; Iqbal et al., 2016; Chahal et al., 2017; Leogrande and Vitti, 2019). The positive biological effects are most likely caused by the positive effect of available carbon (C) derived from the added organic matter to microbial cells allowing their adjustment to osmotic stress by producing osmolytes, which counteract osmotic stress (Killham and Firestone, 1984; Schimel et al., 1989; Wichern et al., 2006). Moreover, crop residues are decomposed by soil microorganisms and thus contribute substantially to nutrient availability in the soil, especially, when external input is low. The decomposition of crop residues and organic amendments is largely determined by the presence of soil microorganisms and optimal conditions for their activity, such as temperature and sufficient oxygen for heterotrophs.
Many short-term experiments investigating the effect of elevated salinity on soil microorganisms have been carried out under aerobic conditions. Here plant residues were added as a means to provide available substrate to the soil microorganisms. The added substrates often resulted in alleviation of the negative effects of salinity, when providing easily available energy and nutrient sources for the soil microorganisms (Wichern et al., 2006). When substrates of lower availability to microorganisms were added, the effects of salinity remained more pronounced (Iqbal et al., 2016). Consequently, the question arises, if addition of organic amendments and crop residues are means to reduce the negative effects of soil salinity on the microbial driven mineralisation processes in paddy rice soils, where conditions are temporarily anaerobic and thus potentially adding additional stress to part of the microbial community. Therefore, our objectives were to evaluate the effects of organic matter addition of different inherent quality (manure, rice straw) on C and N fluxes under saline conditions, and to evaluate the effect of anaerobic vs. aerobic soil conditions on salinity effects on soil microbial activity, microbial biomass and microbial community composition.
We hypothesized, that (i) rice straw as a source of easily available C allows soil microorganisms to better respond to osmotic stress caused by soil salinity addition, thus maintaining their functionality, that (ii) rice straw supports fungi also under saline conditions, and that (iii) anaerobic conditions add another stress to soil microbial communities in paddy rice soils, which results in reduced microbial activity and biomass in particular under saline conditions.
Materials and Methods
Properties of Soils and Organic Amendments
Composite field moist paddy rice soil samples (0–15 cm depth) were collected in May 2015 at 15 locations from two farmers' fields in Bangladesh in Mymensingh (soil A) and Nalitabari (soil B), which are situated in an area characterized by rice-based arable farming systems. Mymensingh is situated in the “Old Brahmaputra Floodplain” and Nalitabari in the “Northern and Eastern Piedmont Plains.” To the best of our knowledge, both sites have not experienced soil salinization before. The climate in the region is a typical tropical monsoonal climate with temperature and rainfall peaking during the Kharif season (April until September) and scanty rainfall with substantially lower average temperature during the Rabi season (October until March). Average annual rainfall is 2,220 mm with a peak in July and August. The average annual temperature in the region is 25°C. At the time of sampling soils were non-saline with an electrical conductivity in the extract of the water saturated soil (ECe) of 1.0 dS m−1 in soil A and 0.5 dS m−1 in soil B. Field-moist soil was gently mixed, air-dried and ground to pass a 2 mm sieve prior to pre-incubation and soil analyses. Soil A had a higher clay content (24%) and a higher pH (7.1) compared to soil B (14% clay, pH = 5.0) with comparable sand content (28 and 22%). Salt addition significantly reduced each soil pH, which was especially pronounced in soil A and substrate addition induced a slight pH increase, in particular in soil B (Table S1). Increasing the soil water content from 50 to 100% of the water holding capacity (WHC) resulted in a slight but significant increase in pH in all soils. Soil organic carbon (SOC) content was 12.3 mg g−1 soil in soil A and 12.7 mg g−1 in soil B. Soil microbial biomass C content was 543 μg g−1 soil in soil A and 226 in soil B μg g−1 soil.
Rice straw samples were collected from the rice field in Mymensingh during harvest, dried at 60°C until constant weight and then ground in a grinding mill to pass through a 2 mm sieve. The straw sample was stored in paper bags until analyses and use in the incubation experiment. Its dry matter had 40% C, 0.5% N (C:N ratio = 80), 0.12% phosphorus (P), 1.5% potassium (K) and 0.04% sulphur S. Well–decomposed manure (cow dung) with a C:N ratio of 24 (25.4% C, 1.05% N), a total P, K, and S content of 0.47, 0.75, and 0.25, respectively, was collected from the same farm, dried, mixed and sieved (2 mm) for further analyses and use in the incubation study.
Experimental Set-Up
To investigate the salinity effects on C and N mineralisation, an incubation experiment was set up using the two soils. Briefly, soil samples (95 g dry weight basis) were weighed into 500 ml stopper Pyrex bottles and treated with or without 15 mg NaCl g−1 soil. Median values of ECe in the control treatments were 1.3 dS m−1 for soil A and 1.1 dS m−1 for soil B. In soil A, after salt addition the median was 25.8 dS m−1 at 100% WHC compared to 32.4 dS m−1 at 50% WHC. In soil B after salt addition the median was 24.0 dS m−1 at 100% WHC compared to 26.4 dS m−1 at 50% WHC. The requisite amount of salt was added dissolved in the water required to raise and maintain the gravimetric water content either to aerobic conditions (50% WHC) or to anaerobic conditions (100% WHC). As a consequence of the different water contents, ECe was lower in the 100% WHC treatments compared to 50% WHC especially where salt was added. In addition to salt addition and the two different soil moisture levels, organic amendments were added to half of the samples. Rice straw and manure, both were added at 2.8 mg g−1 soil. A third organic matter treatment consistent of both, rice and manure applied at the rates given above. The organic amendments were thoroughly mixed into the soil. After that, samples were pre-incubated in a thermostatic cabinet for 7 days at 25°C in the dark to re-activate the microbial community as the soil was very dry at times of sampling. All treatments consisted of four replicates and were randomly distributed in the incubation cabinet. After pre-incubation for seven days, all samples were aerated and incubated again for 27 days. At the end of the incubation experiment, subsamples of each treatment were analysed for soil microbial biomass C and N, dissolved organic C and N, and inorganic N.
Determination of Soil Chemical and Physical Properties
Particle size distribution of the soils was determined using a pipette method (Gee and Or, 2002). The pH of the soil was determined before and after the incubation experiment using a soil-to-water ratio of 1:5. Electrical conductivity (ECe) of the original soils was measured in the extract of a water saturated soil paste according to Ryan et al. (1996) and using a 1:5 soil-to-water ratio (EC1:5). After incubation, the EC1:5 was determined in all samples using a 1:5 soil-to-water ratio and the ECe was calculated based on the EC1:5/ECe ratio of the original soils. Total soil C was calculated based on the organic matter content determined from loss on ignition after combustion at 550°C for at least 3 h. The K2SO4 extracts of the non-fumigated samples from microbial biomass determination (see below) were analysed for extractable organic C, which was defined as dissolved organic C (DOC). Additionally, nitrate-N (-N), nitrite-N (-N) and ammonium-N (-N) were determined in these extracts using an Autoanalyser 3 continuous flow system (SEAL Analytical GmbH, Norderstedt, Germany).
Determination of Soil Microbial Properties
To determine microbial activity, on days 3, 6, 15, 21, and 27 of the incubation experiment, carbon mineralisation was determined as soil respiration, which was measured using alkaline traps (Anderson and Domsch, 1990). The evolved CO2 was captured in 0.5 M NaOH and titrated with 0.1 M HCl after addition of 5 ml M BaCl2 using a Titroline® 6,000 automatic titrator (SI Analytics, Mainz, Germany). The CO2-C (μg g−1 soil) produced was calculated by subtracting the values of the samples from the mean blank values. The organic amendment (manure, rice straw, rice straw-manure) derived CO2-C (% of substrate-C) was estimated by subtracting the soil respiration of the control treatments from the respiration in the respective treatment with organic amendments and dividing it by the amount of C added with the substrate. Further, net microbial N mineralisation or immobilisation was determined by subtracting inorganic N values of the original soils before incubation from the inorganic N values at the end of the incubation. Positive values indicated net N mineralisation, whereas negative values showed net N immobilisation or N losses, the latter especially under anaerobic conditions.
Soil microbial biomass C (MBC) and N (MBN) were determined in moist subsamples at the end of the incubation experiment using the chloroform fumigation extraction method (Brookes et al., 1985; Vance et al., 1987). About 10 g (on an oven-dry basis) of soil were fumigated for 24 h at 25°C with ethanol-free chloroform (CHCl3). Following the removal of CHCl3, the sample was extracted with 40 ml 0.5 M K2SO4 for 30 min by horizontal shaking at 200 rev min−1 and filtered through a folded filter paper (VWR 305, particle retention: 2–3 μm). At the same time an identical non-fumigated (10 g) soil sample was extracted similarly. Organic C and total N in the extracts were measured using a multi N/C 2100S analyser (Analytik Jena AG, Jena, Germany). MBC was calculated as EC/kEC, where EC = (organic C extracted from fumigated soils)—(organic C extracted from non-fumigated soils) and kEC = 0.45 (Wu et al., 1990). MBN was calculated as EN/kEN, where EN = (organic N extracted from fumigated soils)—(organic N extracted from non-fumigated soils) and kEN = 0.54 (Brookes et al., 1985).
The ergosterol content after incubation as an indicator of saprotrophic fungi was measured according to Djajakirana et al. (1996). Briefly, 2 g moist soil were extracted with 100 ml ethanol for 30 min by oscillating shaking at 250 rev min−1 and then filtered (Whatman GF/A). Ergosterol determination was carried out using a reversed-phase HPLC analysis at 26°C, using a 125 mm × 4 mm Sphereclone 5 μm ODS II column with a Phenomenex guard column (4 mm × 3 mm). Chromatography was performed isocratically with methanol (100%) and a resolution of detection of 282 nm (Dionex UVD 170 S).
Total genomic dsDNA (from now on DNA) was extracted from 0.5 g of soil sample using FastDNATM SPIN Kit for Soil and FastPrep®-24 bead-based homogenizer (both MP Bio, Santa Ana, CA) according to Hemkemeyer et al. (2014). The concentration of extracted DNA was measured based on the intercalating dye QuantiFluor® dsDNA System (Promega GmbH, Mannheim, Germany) in the microplate reader FLUOstar® Omega (BMG Labtech, Ortenberg, Germany) at 485 nm excitation and 520 nm emission. Quantitative PCR of archaeal as well as bacterial 16S rRNA genes and fungal ITS1 sequences was conducted in a LightCycler® 480 II using LightCycler® 480 Probes Master and LightCycler® 480 SYBR Green I Master, respectively (all Roche, Penzberg, Germany). A total volume of 20 μL contained 10 μL master. In the case of prokaryotes, the reaction mix contained 0.5 μM forward primer, 0.5 μM reverse primer, and 0.2 μM probe, while for fungi the end concentration of each primer was 0.4 μM. For duplicate determinations, half of the reactions were supplied with 10-fold and the other half with 50-fold dilutions of template DNA (2 μL). According DNA fragments of Methanobacterium oryzae, Bacillus subtilis, and Fusarium graminearum cloned into pGEM®-T vector (Promega), respectively, served as standards. For primers, probes, and qPCR reaction conditions see Table S2 (Martin and Rygiewicz, 2005; Yu et al., 2005).
Statistical Analyses
The results presented in tables and figures are median values and presented on an oven-dry basis (about 24 h at 105°C). Due to pronounced differences and different responses of the two soils, soils were analysed separately instead of using a 4-factorial statistical model. For the separate statistical analysis of each soil, a three-way Analysis of Variance based on Type II sums of squares (Langsrud, 2003) using R (R Core Team, 2018) and the R package CAR (Fox and Weisberg, 2011) was applied with subsequent model simplification. If necessary, data were box-cox transformed using MASS (Venables and Ripley, 2002) and if residuals were still nonparametric or heteroscedastic, the rank-based ANOVA from the package RFIT (Kloke and McKean, 2012) was employed. The application of the post hoc-test, i.e., estimated marginal means using EMMEANS (Lenth, 2019) and Dunn's test using FSA (Ogle et al., 2018), respectively, was supported by the CLD- and cldList-command, respectively, the latter deriving from the package RCOMPANION (Mangiafico, 2018). F- and p-value tables are given in the supplementary materials part. To elucidate the effect of salt addition for each substrate under each oxygen condition separately, Wilcoxon rank sum test was employed. Correlations were calculated in dependence of normal distribution and homoscedasticity either as Pearson's r or Kendall's τ. Figures were prepared with GGPLOT2 (Wickham, 2016) in combination with SCALES (Wickham, 2018).
Results
Salinity Effects on Soil Microorganisms and Microbial Processes
Salt addition reduced respiration in the control (CON) and manure (M) treatments of both soils at 50 and 100% WHC (Table 1; for full model F- and p-values see, Table S3). In soil A at 50% WHC, rice straw addition (R) and manure plus rice straw addition (RM) even showed no reduction in respiration with increased salt level. Overall, the reducing effect of soil salinity on soil respiration as an indicator of C mineralisation was more pronounced in soil B and was alleviated by organic amendments especially in those treatments receiving rice straw. Salinity reduced soil respiration by 16 to 53% in CON and M treatments, with a tendency of larger reduction at 100% WHC and a slightly stronger response in soil A as compared to soil B. When rice straw was added, salinity did not affect soil respiration in soil A at 50% WHC, whereas it decreased respiration at 100% WHC by 20% and in soil B at 50 and 100% WHC by 32 and 20%, respectively. The same pattern was observed after combined application of rice straw and manure, with lower reduction of respiration in the saline treatment at 100% WHC in soil A (11%) and a reduction in soil B by 26%. Salinity showed inconsistent effects on DOC changes in the various treatments (Table 1, Table S5). At 50% WHC, salt addition increased DOC. Also at 100% WHC, DOC increased after salt addition but this was mainly in the rice straw treatments in soil B. Opposite to the effects of soil salinity on soil respiration described above, N mineralisation was increased after salt addition in the control and manure treatments at 50 and 100% WHC likewise (Figure 1, Table S6). The effect was stronger for the manure treatments and at 100% WHC. On the other hand, the immobilisation effect of rice straw and manure plus rice straw addition on inorganic N was diminished in the saline treatments. Salinity is known not only to reduce microbial respiration and N mineralisation-immobilisation but also nitrification. In soil B, salinity consistently resulted in large contribution of to inorganic N (84 to 100%) as compared to , whereas in soil A always had a higher share (57 to 100% of inorganic N) in particular in CON and M treatments (Table 1, Tables S7, S8). In the R and RM treatments inorganic N was on an overall smaller level and salinity supported accumulation of in both soils with larger contribution in soil B. However, in soil A at 100% WHC there was no salinity effect on nitrification observed and inorganic N was very low.
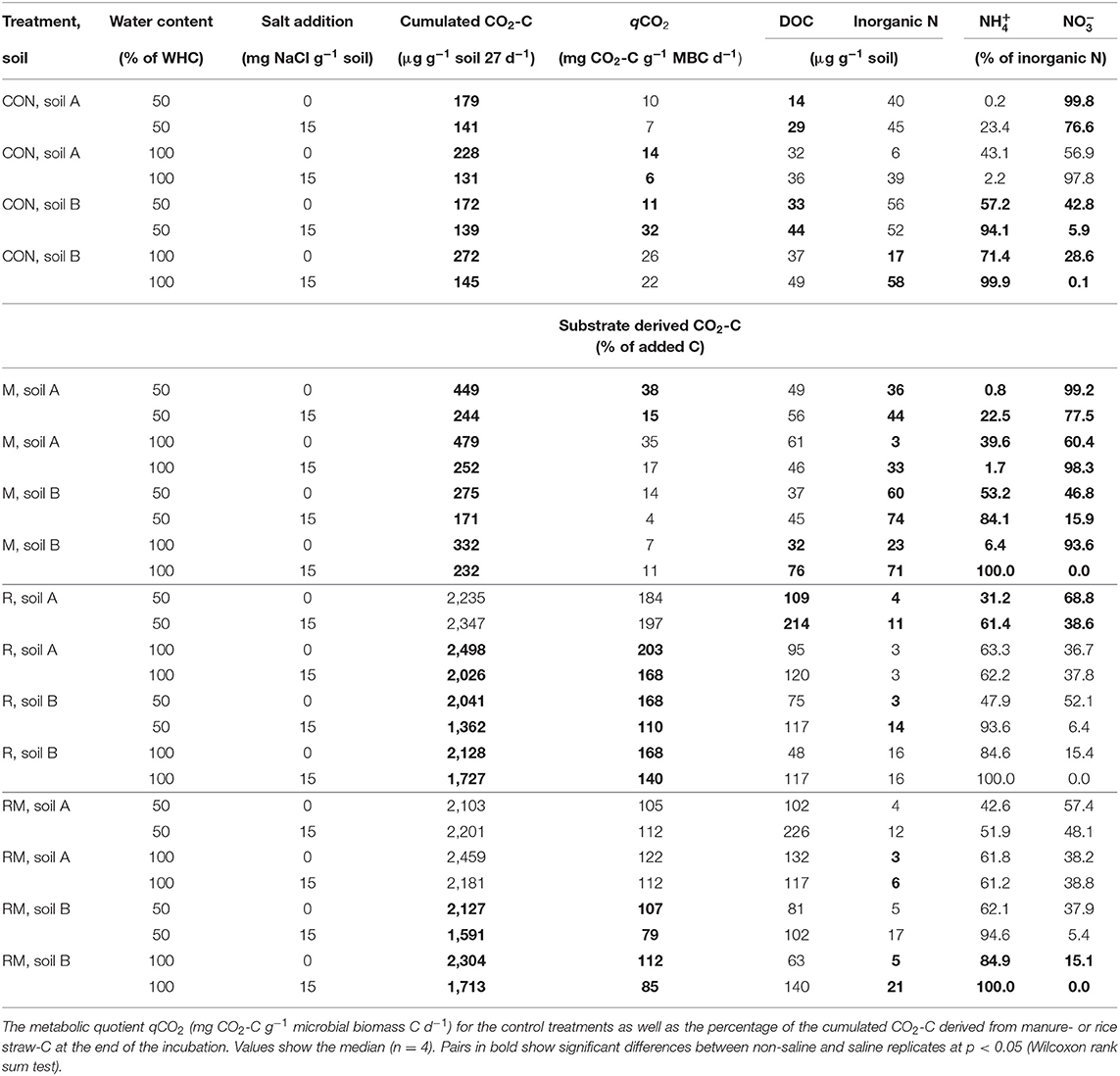
Table 1. Cumulated respiration, dissolved organic C (DOC), inorganic N, the proportion of ammonium () and nitrate () on inorganic N for the unamended treatments (CON), the manure (M), the rice straw (R) and the rice-straw plus manure (RM) treatments.
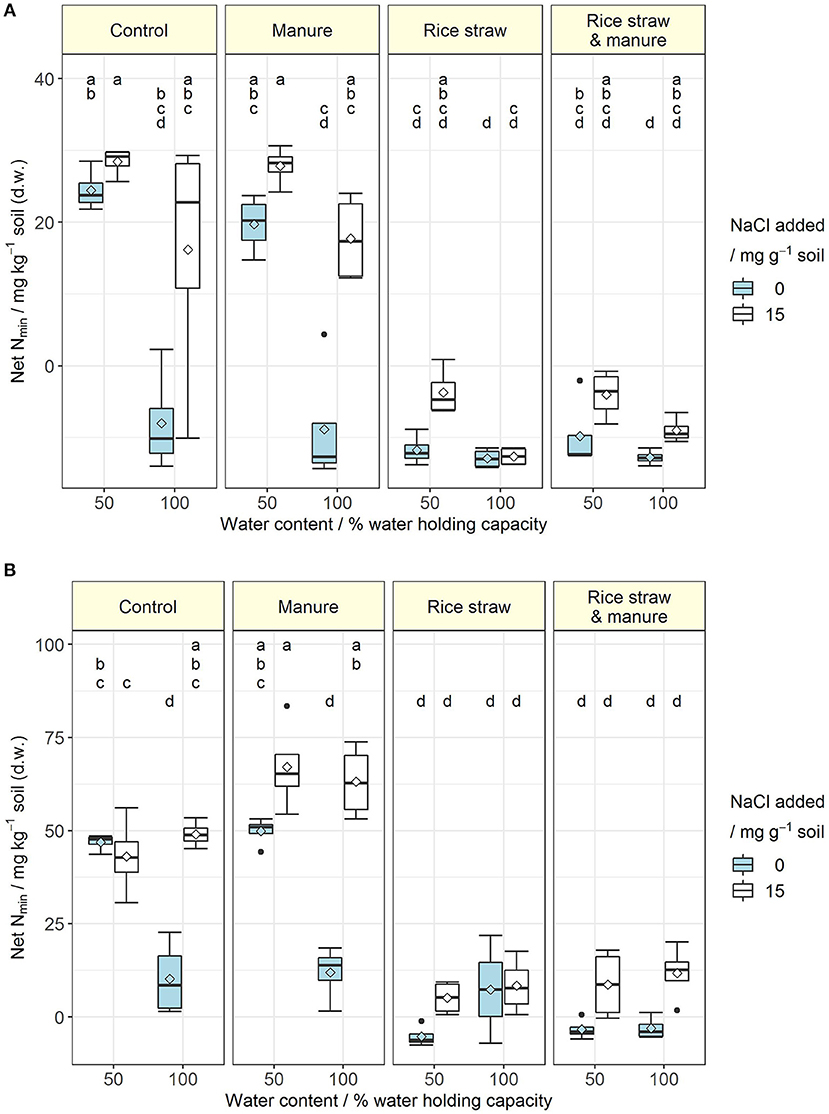
Figure 1. (A,B) Net N mineralisation (mg inorganic N kg−1 soil) at the end of the incubation for soil A (A) and soil B (B) at 50 and 100% water holding capacity and with (15 mg NaCl g−1 soil) or without salt addition after manure, rice straw or manure plus rice straw addition. Diamonds indicate the mean and letters indicate significant differences at p < 0.05 (Rank-based ANOVA for soil A and ANOVA for soil B; n = 4).
MBC was not affected by salinity in soil A, while in soil B it was significantly reduced at 50% WHC (Figure 2, Table 2, Table S9). MBN showed inconsistent responses to salinity in both soils (Figure 2, Table 2, Table S10). In contrast, total extractable DNA (μg g−1 soil) was significantly reduced in relation to salinity in both soils, except for soil A at 100% WHC (Table 2, Table S11). Bacterial and archaeal 16S rRNA gene copy numbers g−1 soil were slightly but significantly reduced by salinity in soil B but not in soil A, where they remained on the same level (Table 2, Tables S12, S13. In contrast, salinity significantly increased ergosterol content in R and RM-amended replicates of soil B at 100% WHC, whereas there was no such effect of salinity in soil A (Table 2, Table S14). This was partly reflected by the content of fungal ITS1 sequences; for example, in soil A there was an increase at 100% WHC, while for soil B this effect depended on water content and substrate amendment (Table 2, Table S15). Based on this, the fungi-to-bacteria and fungi-to-archaea ratios also increased in these treatments (data not shown). The biomass specific respiration of the CON treatments, also referred to as metabolic quotient qCO2, was not significantly affected by short term salinity but varied significantly between the two soils, with higher values in soil B (Table 1). Moreover, it increased with increasing ergosterol-to-MBC ratio (τ = 0.500, p = 006, Figure 3).
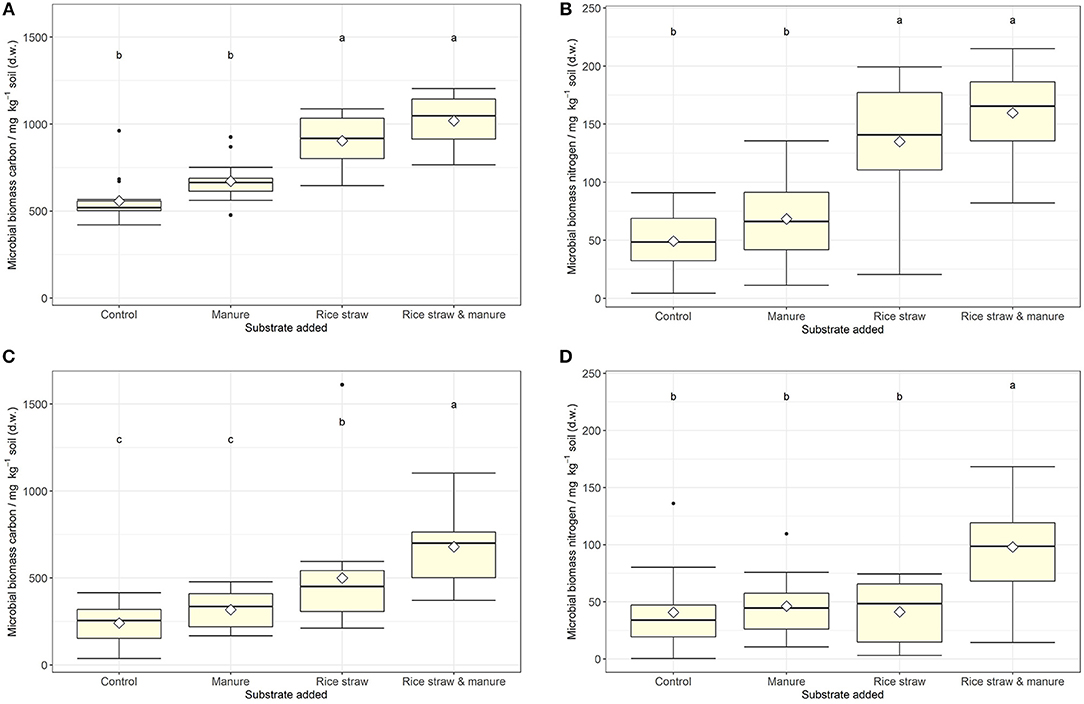
Figure 2. (A–D) Microbial biomass C and N (mg kg−1 soil) at the end of the incubation for soil A (upper panel, A,B) and soil B (lower panel, C,D) after manure, rice straw or manure plus rice straw addition. Diamonds indicate the mean and different letters indicate significant differences at p < 0.05 (ANOVA with model simplification; n = 4). Substrate addition was the only consistent significant effect on microbial biomass C and N in both soils. Salt addition had a significant reducing effect on soil microbial C in soil A at 50% water holding capacity only.
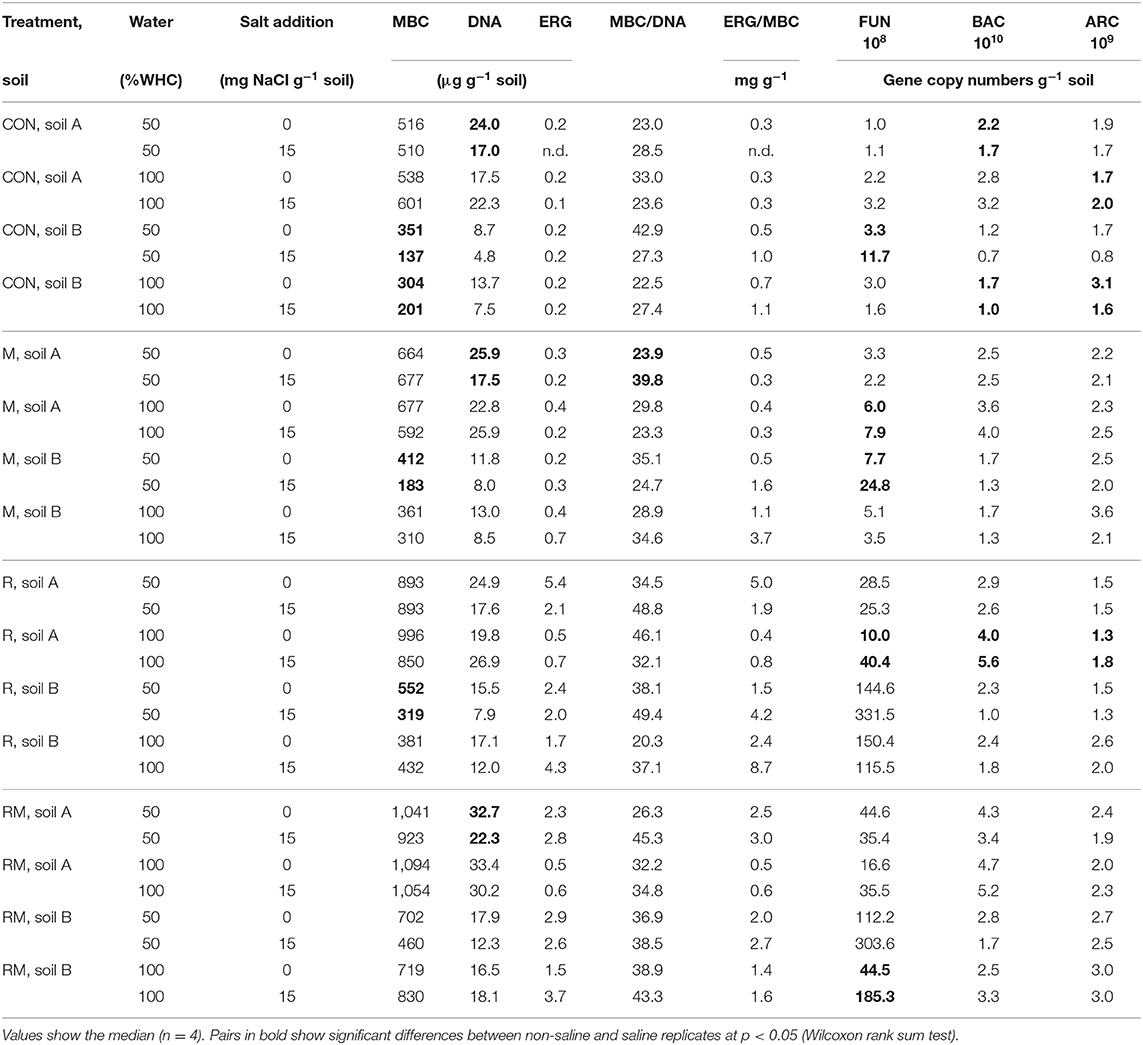
Table 2. Microbial biomass C (MBC), total extractable DNA, ergosterol (ERG), the MBC/DNA ratio, the ERG/MBC ratio, fungal ITS1 sequences g−1 soil (FUN), bacterial (BAC) and archaeal (ARC) 16 S rRNA gene copies g−1 soil in control (CON), manure (M), rice straw (R) and rice straw with manure (RM) amended soils.
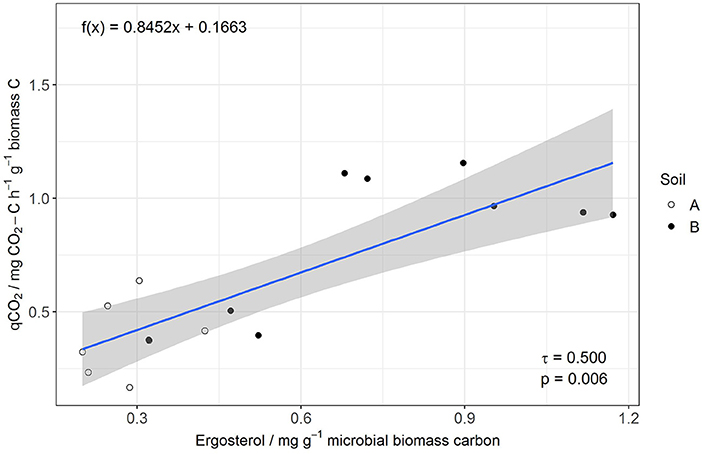
Figure 3. Linear regression of qCO2 (y) and Erg-to-MBC (x) for the control without manure and rice straw amendment.
Effects of Organic Amendments on C and N Mineralisation and Soil Microorganisms
Addition of organic amendments increased soil respiration (Table 1, Table S3). Manure (M) resulted in a 2-fold increase in respiration compared to the CON treatment. The R treatment on the other hand showed an approximately 10-fold increase of soil respiration. The RM treatment was in the same range as R. While the cumulated CO2-C derived from the respective substrates was between 4 and 38% in the M treatments, it was between 110 and 203% in the R and between 79 and 122% in the RM treatments (Table S4) with larger values in soil A as compared to soil B caused by a higher microbial biomass (Table 1, Tables S9, S10). Along with that, the DOC contents were generally increased in the R and RM treatments, but only in the M treatments of soil A (Table 1, Table S5). Further, a net N mineralisation was always observed at 50% WHC in the CON and M treatments (Figure 1, Table S6), whereas a net N immobilisation occurred in the R and RM treatments in most cases. However, at 100% WHC, all treatments without salt addition showed a lower inorganic N content compared to the CON treatment at 50% WHC (Tables S7, S8). On the other hand, salt addition at 100% WHC enhanced the inorganic N content in almost all cases, and was particularly pronounced in CON and M treatments.
MBC increased in all treatments with organic amendments, independent of water content and salt addition (Figure 2, Table 2, Table S9) and reached very high levels of above 1,000 μg MBC g−1 soil in the RM treatment of soil A. Values were generally higher in soil A as compared to soil B. Also MBN increased after addition of organic amendments (Figure 2, Table S10). Even though MBC and MBN were closely correlated (τSoilA = 0.670, τSoilB = 0.544, both p < 0.001), a stronger variability of this relationship was observed especially at lower MBN values. Total extractable DNA followed the same pattern as MBC (Table S11), even though the relationship between DNA and MBC was weak or moderate (τSoilA = 0.384, p = 0.002, τSoilB = 0.579, p < 0.001). The ratio between MBC and DNA (MBC-to-DNA) or the conversion factor FDNA derived from total extractable dsDNA and microbial biomass C data was 35 on average for the two soils. Treatment average values ranged from 20 to 50 and individual values varied between 3.5 and 97.4. Bacterial 16S rRNA gene copy numbers g−1 soil were also increased in the order RM > R > M > CON, whereas it was only significant for the R and RM treatments as compared to CON (Table S12). In contrast, archaeal 16S rRNA gene copy numbers g−1 soil, were increased in manure containing treatments (M and RM) in both soils (Table S13), whereas the ergosterol content generally increased in both soils with rice addition in the R and RM treatments (Table S14). This increase in ergosterol was also reflected by increased fungal ITS1 sequences g−1 soil (Table S15), which showed a much stronger response than ergosterol. However, ergosterol content and fungal ITS1 were only weakly correlated (τSoilA = 0.353, τSoilB = 0.460, both p < 0.001).
Effects of Anaerobic Conditions on C and N Fluxes and Soil Microorganisms
Anaerobic conditions (100% WHC) resulted in increased respiration for some of the treatments compared to 50% WHC under non-saline conditions (Table 1, Table S3). When salt was added, this effect was far less apparent, especially in the control treatments. DOC was not consistently affected by water content. Net N mineralisation, on the other hand, was significantly reduced in CON and M treatments at 100% WHC in soil A with the same trend observed in soil A, where net N mineralisation reached the same level as in the rice straw treatments with net N immobilisation in soil A and very little N mineralisation in soil B (Figure 1, Table S6).
MBC was not affected by water content (Table 2, Table S9). However, total extractable DNA was slightly enhanced at higher water content (Table S11). Furthermore, whereas archaeal 16S rRNA gene copy numbers g−1 soil were slightly and significantly enhanced at higher water content in soil B there was no change in soil A (Table S13). Bacterial 16S rRNA counts, however, where significantly increased in soil A and B at 100% WHC, with a stronger response in soil A (Table S12). On the other hand, water content had no effect on the ergosterol content (Table S14) or the relative contribution of ergosterol to microbial biomass C. In contrast, fungal ITS1 was significantly affected by water content in soil B (Table S15), with a tendency of reduced contents at higher water levels and a lower fungi-to-bacteria ratio (data not shown). Also in soil A there were some trends with reduced ITS1 in soils at 100% WHC not treated with salt.
Discussion
Effects of Salinity and Organic Amendments on C and N Processes
Our results show some expected outcomes on the effect of salinity and organic amendments on soil respiration. However, as shown below, the microbial responses substantially vary between the two soils. Firstly, like in many previous studies (Laura, 1974; Pathak and Rao, 1998; Wichern et al., 2006; Setia et al., 2011; Iqbal et al., 2016) and recently qualitatively summarized by Leogrande and Vitti (2019), soil respiration assessed by CO2-evolution, was reduced by soil salinity in our study for most of the treatments with and without organic matter addition. However, addition of organic matter increased soil respiration substantially, even under saline conditions, which was observed in earlier studies as well (Wichern et al., 2006; Sall et al., 2015; Iqbal et al., 2016). Organic matter thus provides a means to reduce the negative effects of soil salinity on soil microorganisms (Chahal et al., 2017; Leogrande and Vitti, 2019). One reason for this is the enhanced availability of energy rich C-containing compounds in the organic amendments, which allow soil microorganisms to synthesise osmolytes counteracting the osmotic pressure from elevated salinity or to invest into metabolic processes for detoxification and cell repair (Killham and Firestone, 1984; Schimel et al., 1989; Wichern et al., 2006).
Along with the reduced soil respiration and thus lower C mineralisation, salinity increased DOC content in some treatments, in particular after rice straw addition, which was observed earlier (Mavi et al., 2012; Chahal et al., 2017). This increase is probably partly caused by high sodium loads at the exchange complex leading to soil dispersion and C release from destroyed aggregates. This is in line with findings from Wang et al. (2015), who investigated rice straw decomposition in paddy systems, and with one of our earlier studies (Iqbal et al., 2016), where DOC values increased with increasing salinity, especially after rice straw addition. Water extractable C was proposed as an indicator of management effects on SOC by Benbi et al. (2015), where long term manure application resulted in increased labile C and stable C and therefore may contribute to long-term C stability in paddy soils.
In our study, rice straw addition with or without manure increased MBC and the fungal contribution significantly in the short term, which is in contrast to an earlier study where rice straw did not substantially increase microbial biomass in saline paddy soils (Iqbal et al., 2016). Manure addition alone showed only a trend towards an increase of the MBC, which highlights the importance of rice plant residues for soil fertility maintenance, in particular under saline conditions (Iqbal, 2018). Enhanced microbial biomass will ultimately contribute to increased microbial necromass, which accounts for at least half of soil organic matter (Miltner et al., 2012; Kallenbach et al., 2016; Khan et al., 2016) and is thus a prerequisite for SOC maintenance. Even though salinity decreased microbial respiration, MBC was not consistently reduced by salinity in our study as observed earlier (Rath and Rousk, 2015) contradicting findings of others (Rietz and Haynes, 2003; Sardinha et al., 2003; Wichern et al., 2006). However, it is worth mentioning that the microbial communities of the two soils responded differently to salinity as discussed below.
Along with an increase of MBC, N immobilisation in paddy rice soils was enhanced after rice straw addition, as expected. However, microbial N immobilisation was enhanced in the control and manure treatments under anaerobic conditions (100% WHC) as compared to aerobic conditions (50% WHC), where a net N mineralisation occurred during the period of the incubation experiment. This was also observed by Said-Pullicino et al. (2014) and reflects higher C and N availability to soil microorganisms due to higher mobility of C and nutrients in the saturated soil. However, at saline and anaerobic conditions, a net N release occurred in the control and manure treatments, which is most likely caused by microbial death and inorganic N release as shown by slightly reduced microbial C and N, and total DNA in these treatments. Inorganic N increase along with a reduction of MBC under saline conditions has also been shown in earlier studies (Wichern et al., 2006; Sall et al., 2015). Addition of rice straw as a source of labile C is thus a good option to support microbial N immobilisation under aerobic and anaerobic conditions as compared to manure alone and may better contribute to N conservation under saline conditions in paddy rice soils.
It should be noted that extracted DNA in the investigated soils was not related to MBC determined by fumigation extraction as shown in earlier work (Leckie et al., 2004; Calbrix et al., 2007), but in contrast to a variety of more positive studies (Marstorp et al., 2000; Gangneux et al., 2011; Semenov et al., 2018). In addition, the MBC-to-DNA ratio was substantially higher than reported previously (Joergensen and Emmerling, 2006; Anderson and Martens, 2013). In contrast to microbial biomass and microbial residues (Joergensen and Wichern, 2008), the extractable microbial DNA is dominated by bacterial DNA as in the current study. One reason could be the stronger packing in dense protein-DNA complexes in the nucleus of eukaryotic fungi. Another reason is the variation in ribosomal gene copy numbers and cellular nucleic acid concentrations between fungi and bacteria so that they do not provide an estimate of biomass ratios (Rousk et al., 2010). Consequently, small fluctuations in bacterial DNA may lead to strong shifts in the MBC-to-DNA ratio. Such shifts could be caused by straw and manure addition in the current experiment, particular in soil B. In addition, the extractable DNA fraction varies more in fungi (Leckie et al., 2004), due to differences in the ratio of cytoplasm to genome. However, the decreasing ratio of the cell-membrane component ergosterol to fungal DNA after rice straw addition points to the reverse direction. More DNA seems to be extractable from actively straw-decomposing fungi. It should be noted that the DNA from arbuscular-mycorrhizal fungi is hardly extractable (Hartmann et al., 2015), despite the probably high contribution of this fungal phylum to the microbial biomass (Faust et al., 2017). Another reason for the rather low correlation between MBC and extractable DNA might be the presence of residual DNA of soil microorganisms and plants in soil or interference of DNA with humic compounds during extraction (Pietramellara et al., 2009; Carini et al., 2016). These observations question the applicability of a general conversion factor to determine MBC from DNA extraction (Joergensen and Emmerling, 2006). However, the variation in the MBC-to-DNA ratio opens the possibility to gain additional information, e.g., on the contribution of bacteria to the MBC if the knowledge for the reasons of the aforementioned variability will be improved in the near future.
It is well-known that organic amendments can increase methane (CH4) emissions in paddy rice soils, in particular when applied before transplanting of rice (Yan et al., 2005; Sanchis et al., 2012). However, as we did not measure CH4 emissions and other trace gases, such as N2O and NH3, we cannot exclude that gaseous losses of C and N may have occurred, which needs to be considered in a full C and N balance, which was not in the scope of the present work.
Distinct Microbial Response of Different Soils Indicate Soil Legacy
Even though -to our knowledge- the two investigated soils where not prone to salinity before, they, along with their pronounced differences in pH, showed distinct responses to salinity, probably caused by differences in the microbial community composition. For example, most of the inorganic N was in soil A at non-saline conditions, which remained substantially higher also at saline conditions as compared to soil B, where N was almost exclusively . Here, microbial nitrifiers seem to be present only in lower abundance, as also under non-saline and aerobic conditions, a large proportion of the inorganic N was present as . The proportion of increased under saline conditions to approximately 84 to 100% in soil B, indicating inhibition of nitrification, as observed before (Laura, 1974; Pathak and Rao, 1998; Wichern et al., 2006). However, soil A showed microbial nitrifiers better adapted to salinity, with ~40% (R and RM) to more than 95% (CON and M) of the inorganic N at the end of the incubation being present as even under saline conditions. This is in line with earlier findings, where paddy rice soils from Bangladesh also showed pronounced differences in the response of microbial nitrifiers to enhanced salinity (Iqbal et al., 2016), most likely inhabited by adapted microbial consortia (Moussa et al., 2006). The higher tolerance of the microbial community to salinity in soil A is also reflected by an overall lower response of MBC, in contrast to a reduction of the already lower MBC contents in soil B as also observed by Wichern et al. (2006).
Ergosterol is often used as an indicator of saprotrophic fungi (Joergensen and Wichern, 2008; Faust et al., 2017). Interestingly, in soil B after rice straw addition, salinity in anaerobic conditions enhanced fungi as indicated by increased ergosterol content and larger amounts of extractable ITS1 sequences. The extremely low contribution of ergosterol to MBC (Djajakirana et al., 1996; Joergensen and Wichern, 2008) indicates that the concentration of saprotrophic fungi is probably very low in the current paddy soils. Other studies also showed fungal growth rates are less sensitive to salinity than bacterial growth rates after application of fresh substrate (Rath et al., 2016, 2019; Dang et al., 2019). This might be different for the contribution of fungal residues to SOC under saline conditions (Khan et al., 2016) as the biomass of many soil fungi declines when exposed to salinity (Sardinha et al., 2003). However, Roth et al. (2011) and Murugan et al. (2014) observed high contributions of fungi to microbial necromass in paddy fields. One reason for these discrepancies observed could be differences in the length of the aerobic period between different rice cultivation seasons, which probably stronger support saprotrophic aerobic fungi.
The relative increase of saprotrophic fungi as indicated by the ergosterol-to-microbial biomass C ratio was positively related to an increase of the metabolic quotient qCO2, a simple indicator of the demand of microorganisms to maintain their biomass or a simplified carbon use efficiency (CUE) (Joergensen and Wichern, 2018) Consequently, increased fungal dominance will result in lower C use efficiency as shown earlier (Nannipieri et al., 2003; Iqbal et al., 2016) and may result in organic matter loss in the long run. A higher specific metabolic activity may reflect stronger catabolic activity and production of osmolytes, cell repair or detoxification (Jones et al., 2019) and thus reflects an adaptation of the soil microbial community to abiotic stress, such as salinity (Wichern et al., 2006).
Saline conditions usually increase metabolic activity to avoid cell damage by the enhanced osmotic pressure and toxicity of the salts present. However, the metabolic quotient qCO2 was not affected by salinity in or study in the short term but showed differences between the two investigated soils, indicating that the microbial communities adjusted their metabolism over a longer period of time. It was higher in soil B with a lower pH as compared to soil A, which is in line with Jones et al. (2019), who observed a lower CUE in soils with lower pH (below 5.5). Soil B had a higher qCO2 and therefore more C was used for maintenance of microbial cells, resulting in a less efficient C use, which in the long run yield a lower CUE and consequently may result in stronger organic matter decomposition. However, this was not represented by lower SOM accumulation in soil B as SOC content in both soils was the same, probably because the qCO2 was on an overall low level as compared to other studies (Wichern et al., 2003, 2006; Hartman and Richardson, 2013; Iqbal et al., 2016). Further, next to C lost as CO2 for a full microbial CUE assessment also microbial necromass and other microbial metabolites need to be considered (Joergensen and Wichern, 2018), which we were not able to do in the present study. However, it can be concluded that short term abiotic stress, such as salinity or anoxic conditions, does not directly trigger metabolic adaptation of the whole microbial community, but rather results in survival of the microbes better adapted to stress and likely having adapted energy intensive metabolic mechanisms as explained above. Consequently, the microbial community changes and along with that the microbial CUE decreases in the long run.
Conclusions
Organic amendments, such as rice straw or manure alleviate some of the negative effects of salinity on soil microorganisms and microbial processes. As hypothesized, addition of rice straw allows soil microbial communities, in particular fungi to better adapt to the osmotic stress making use of the added energy to maintain their metabolism or even forming additional biomass, which, as expected, results in increased N immobilisation. Contrary to our hypothesis, enhanced water content in the paddy soils did not add additional stress to the microbial community, but rather improved C availability for soil microorganisms as reflected by enhanced C mineralisation and N immobilisation allowing the microorganisms to counteract the negative salinity effects. However, microbial communities in different soils show a distinct response to salinity, to anoxic conditions and organic amendments with a pronounced legacy from previous abiotic and biotic experiences. Future investigations should focus on plant-soil microbial interactions under saline conditions after addition of organic amendments. Overall, to maintain soil microbial functions under saline conditions, plant residue and nutrient management needs to consider application of sufficient amounts of available C and nutrients by combining C and nutrient sources, such as rice straw and manure. Maintaining microbial life under salt stress will ultimately also support maintenance of soil organic matter, as it derives to a large extent from microbial necromass.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
FW designed the experiment, analysed the data, and wrote the manuscript. MI and CW co-designed the experiment, conducted the research, and contributed to writing the manuscript. MH analysed the data, designed figures, and contributed to writing the manuscript. RJ contributed to writing the manuscript.
Funding
The research was funded by Rhine-Waal University of Applied Sciences. MI received a short term scholarship from the German Academic Exchange Programme (DAAD). Open access publication was facilitated by Frontiers.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We highly appreciate the support of Linda Claaßen and Anne-Sophie Neumann in DNA-extraction, qPCR, and ergosterol extraction. The support of Dr. Stefan Weber for ergosterol determination is acknowledged. Furthermore, the technical assistance during lab work by various student assistants is highly appreciated.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2020.00030/full#supplementary-material
References
Anderson, T.-H., and Domsch, K. H. (1990). Application of eco-physiological quotients (qCO2 and qD) on microbial biomasses from soils of different cropping histories. Soil Biol. Biochem. 22, 251–255. doi: 10.1016/0038-0717(90)90094-G
Anderson, T.-H., and Martens, R. (2013). DNA determinations during growth of soil microbial biomasses. Soil Biol. Biochem. 57, 487–495. doi: 10.1016/j.soilbio.2012.09.031
Benbi, D. K., Brar, K., Toor, A. S., and Sharma, S. (2015). Sensitivity of labile soil organic carbon pools to long-term fertilizer, straw and manure management in rice-wheat system. Pedosphere 25, 534–545. doi: 10.1016/S1002-0160(15)30034-5
Borken, W., and Matzner, E. (2009). Reappraisal of drying and wetting effects on C and N mineralization and fluxes in soils. Glob. Chang. Biol. 15, 808–824. doi: 10.1111/j.1365-2486.2008.01681.x
Brookes, P. C., Landman, A., Pruden, G., and Jenkinson, D. S. (1985). Chloroform fumigation and the release of soil nitrogen: a rapid direct extraction method to measure microbial biomass nitrogen in soil. Soil Biol. Biochem. 17, 837–842. doi: 10.1016/0038-0717(85)90144-0
Calbrix, R., Barray, S., Chabrerie, O., Fourrie, L., and Laval, K. (2007). Impact of organic amendments on the dynamics of soil microbial biomass and bacterial communities in cultivated land. Appl. Soil Ecol. 35, 511–522. doi: 10.1016/j.apsoil.2006.10.007
Carini, P., Marsden, P. J., Leff, J. W., Morgan, E. E., Strickland, M. S., and Fierer, N. (2016). Relic DNA is abundant in soil and obscures estimates of soil microbial diversity. Nat. Microbiol. 2:16242. doi: 10.1038/nmicrobiol.2016.242
Chahal, S. S., Choudhary, O. P., and Mavi, M. S. (2017). Organic amendments decomposability influences microbial activity in saline soils. Arch. Agron. Soil. Sci. 63, 1875–1888. doi: 10.1080/03650340.2017.1308491
Chowdhury, N., Marschner, P., and Burns, R. G. (2011). Soil microbial activity and community composition: impact of changes in matric and osmotic potential. Soil Biol. Biochem. 43, 1229–1236. doi: 10.1016/j.soilbio.2011.02.012
Dang, C., Morrissey, E. M., Neubauer, S. C., and Franklin, R. B. (2019). Novel microbial community composition and carbon biogeochemistry emerge over time following saltwater intrusion in wetlands. Glob. Chang. Biol. 25, 549–561. doi: 10.1111/gcb.14486
Djajakirana, G., Joergensen, R. G., and Meyer, B. (1996). Ergosterol and microbial biomass relationship in soil. Biol. Fertil. Soils 22, 299–304. doi: 10.1007/BF00334573
Faust, S., Heinze, S., Ngosong, C., Sradnick, A., Oltmanns, M., Raupp, J., et al. (2017). Effect of biodynamic soil amendments on microbial communities in comparison with inorganic fertilization. Appl. Soil Ecol. 114, 82–89. doi: 10.1016/j.apsoil.2017.03.006
Flowers, T. J., and Yeo, A. R. (1995). Breeding for salinity resistance in crop plants: where next? Funct. Plant Biol. 22, 875–884. doi: 10.1071/PP9950875
Fox, J., and Weisberg, S. (2011). An R Companion to Applied Regression, 2nd Edn. Thousand Oaks, CA: Sage.
Gangneux, C., Akpa-Vinceslas, M., Sauvage, H., Desaire, S., Houot, S., and Laval, K. (2011). Fungal, bacterial and plant dsDNA contributions to soil total DNA extracted from silty soils under different farming practices: relationships with chloroform-labile carbon. Soil Biol. Biochem. 43, 431–437. doi: 10.1016/j.soilbio.2010.11.012
Gee, G. W., and Or, D. (2002). “Particle size analysis,” in Methods of Soil Analysis, Part 4—Physical Methods. eds J. H. Dane and G. C. Topp. (Madison, WI: Soil Science Society of America), 255–293.
Haque, S. (2006). Salinity problems and crop production in coastal regions of Bangladesh. Pak. J. Bot. 38, 1359–1365.
Hartman, W. H., and Richardson, C. J. (2013). Differential nutrient limitation of soil microbial biomass and metabolic quotients (qCO2): is there a biological stoichiometry of soil microbes? PLoS ONE 8:e57127. doi: 10.1371/journal.pone.0057127
Hartmann, M., Frey, B., Mayer, J., Mäder, P., and Widmer, F. (2015). Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 9, 1177–1194. doi: 10.1038/ismej.2014.210
Hemkemeyer, M., Pronk, G. J., Heister, K., Kögel-Knabner, I., Martens, R., and Tebbe, C. C. (2014). Artificial soil studies reveal domain-specific preferences of microorganisms for the colonisation of different soil minerals and particle size fractions. FEMS Microbiol. Ecol. 90, 770–782. doi: 10.1111/1574-6941.12436
Iqbal, M. T., Joergensen, R. G., Knoblauch, C., Lucassen, R., Singh, Y., Watson, C., et al. (2016). Rice straw addition does not substantially alter microbial properties under hypersaline soil conditions. Biol. Fertil. Soils 52, 867–877. doi: 10.1007/s00374-016-1126-4
Iqbal, T. (2018). Rice straw amendment ameliorates harmful effect of salinity and increases nitrogen availability in a saline paddy soil. J. Saudi Soc. Agri. Sci. 17, 445–453. doi: 10.1016/j.jssas.2016.11.002
Joergensen, R. G., and Emmerling, C. (2006). Methods for evaluating human impact on soil microorganisms based on their activity, biomass, and diversity in agricultural soils. J. Plant Nutr. Soil Sci. 169, 295–309. doi: 10.1002/jpln.200521941
Joergensen, R. G., and Wichern, F. (2008). Quantitative assessment of the fungal contribution to microbial tissue in soil. Soil Biol. Biochem. 40, 2977–2991. doi: 10.1016/j.soilbio.2008.08.017
Joergensen, R. G., and Wichern, F. (2018). Alive and kicking: why dormant soil microorganisms matter. Soil Biol. Biochem. 116, 419–430. doi: 10.1016/j.soilbio.2017.10.022
Jones, D. L., Cooledge, E. C., Hoyle, F. C., Griffiths, R. I., and Murphy, D. V. (2019). pH and exchangeable aluminum are major regulators of microbial energy flow and carbon use efficiency in soil microbial communities. Soil Biol. Biochem. 138:107584. doi: 10.1016/j.soilbio.2019.107584
Kallenbach, C. M., Frey, S. D., and Grandy, A. S. (2016). Direct evidence for microbial-derived soil organic matter formation and its ecophysiological controls. Nat. Commun. 7:13630. doi: 10.1038/ncomms13630
Khan, K. S., Gattinger, A., Buegger, F., Schloter, M., and Joergensen, R. G. (2008). Microbial use of organic amendments in saline soils monitored by changes in the 13C/12 C ratio. Soil Biol. Biochem. 40, 1217–1224. doi: 10.1016/j.soilbio.2007.12.016
Khan, K. S., Mack, R., Castillo, X., Kaiser, M., and Joergensen, R. G. (2016). Microbial biomass, fungal and bacterial residues, and their relationships to the soil organic matter C/N/P/S ratios. Geoderma 271, 115–123. doi: 10.1016/j.geoderma.2016.02.019
Killham, K., and Firestone, M. K. (1984). Salt stress control of intracellular solutes in streptomycetes indigenous to saline soils. Appl. Environ. Microbiol. 47, 301–306. doi: 10.1128/AEM.47.2.301-306.1984
Kloke, J. D., and McKean, J. W. (2012). Rfit: rank-based estimation for linear models. R. J. 4, 57–64. doi: 10.32614/RJ-2012-014
Langsrud, Y. (2003). ANOVA for unbalanced data: use type II instead of type III sums of squares. Stat. Comput. 13, 163–167. doi: 10.1023/A:1023260610025
Laura, R. D. (1974). Effects of neutral salts on carbon and nitrogen mineralisation of organic matter in soil. Plant Soil 41, 113–127. doi: 10.1007/BF00017949
Leckie, S. E., Prescott, C. E., Grayston, S. J., Neufeld, J. D., and Mohn, W. W. (2004). Comparison of chloroform fumigation-extraction, phospholipid fatty acid, and DNA methods to determine microbial biomass in forest humus. Soil Biol. Biochem. 36, 529–532. doi: 10.1016/j.soilbio.2003.10.014
Lenth, R. (2019). Emmeans: Estimated Marginal Means, Aka Least-Squares Means. R package version 1.3.2. Available online at: https://CRAN.R-project.org/package=emmeans.
Leogrande, R., and Vitti, C. (2019). Use of organic amendments to reclaim saline and sodic soils: a review. Arid Land Res. Manage. 33, 1–21. doi: 10.1080/15324982.2018.1498038
Luedeling, E., Nagieb, M., Wichern, F., Brandt, M., Deurer, M., and Buerkert, A. (2005). Drainage, salt leaching and physico-chemical properties of irrigated man-made terrace soils in a mountain oasis of northern Oman. Geoderma 125, 273–285. doi: 10.1016/j.geoderma.2004.09.003
Luedeling, E., and Wichern, F. (2007). The need for site-specific adaptation of organic standards: The example of dryland salinity in Australia. J. Agri. Rural Dev. Tropics Subtrop. Suppl. 89, 121–142.
Mangiafico, S. (2018). Rcompanion: Functions to Support Extension Education Program Evaluation. R package version 2.0.10. available online at: https://CRAN.R-project.org/package=rcompanion.
Marstorp, H., Guan, X., and Gong, P. (2000). Relationship between dsDNA, chloroform labile C and ergosterol in soils of different organic matter contents and pH. Soil Biol. Biochem. 32, 879–882. doi: 10.1016/S0038-0717(99)00210-2
Martin, K. J., and Rygiewicz, P. T. (2005). Fungal-specific PCR primers developed for analysis of the ITS region of environmental DNA extracts. BMC Microbiol. 5:28. doi: 10.1186/1471-2180-5-28
Mavi, M. S., Sanderman, J., Chittleborough, D. J., Cox, J. W., and Marschner, P. (2012). Sorption of dissolved organic matter in salt-affected soils: effect of salinity, sodicity and texture. Sci. Total Environ. 435–436, 337–344. doi: 10.1016/j.scitotenv.2012.07.009
Miltner, A., Bombach, P., Schmidt-Brücken, B., and Kästner, M. (2012). SOM genesis: microbial biomass as a significant source. Biogeochemistry 111, 41–55. doi: 10.1007/s10533-011-9658-z
Mitran, T., Mani, P. K., Basak, N., Biswas, S., and Mandal, B. (2017). Organic amendments influence on soil biological indices and yield in Rice-Based cropping system in coastal sundarbans of India. Commun. Soil Sci. Plant Anal. 48, 170–185. doi: 10.1080/00103624.2016.1254229
Moussa, M. S., Sumanasekera, D. U., Ibrahim, S. H., Lubberding, H. J., Hooijmans, C. M., Gijzen, H. J., et al. (2006). Long term effects of salt on activity, population structure and floc characteristics in enriched bacterial cultures of nitrifiers. Water Res. 40, 1377–1388. doi: 10.1016/j.watres.2006.01.029
Murugan, R., Loges, R., Taube, F., Sradnick, A., and Joergensen, R. G. (2014). Changes in Soil microbial biomass and residual indices as ecological indicators of land use change in temperate permanent grassland. Microb. Ecol. 67, 907–918. doi: 10.1007/s00248-014-0383-8
Nannipieri, P., Ascher-Jenull, J., Ceccherini, M. T., Landi, L., Pietramellara, G., and Renella, G. (2003). Microbial diversity and soil functions. Eur. J. Soil Sci. 54, 655–670. doi: 10.1046/j.1351-0754.2003.0556.x
Ogle, D. H., Wheeler, P., and Dinno, A. (2018). FSA: Fisheries Stock Analysis. R package version 0.8.22. Available online at: https://github.com/droglenc/FSA.
Pathak, H., and Rao, D. (1998). Carbon and nitrogen mineralization from added organic matter in saline and alkali soils. Soil Biol. Biochem. 30, 695–702. doi: 10.1016/S0038-0717(97)00208-3
Pietramellara, G., Ascher, J., Borgogni, F., Ceccherini, M. T., Guerri, G., and Nannipieri, P. (2009). Extracellular DNA in soil and sediment: fate and ecological relevance. Biol. Fertil. Soils 45, 219–235. doi: 10.1007/s00374-008-0345-8
R Core Team (2018). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna. Available online at: http://www.R-project.org/
Rath, K. M., Maheshwari, A., Bengtson, P., and Rousk, J. (2016). Comparative toxicities of salts on microbial processes in soil. Appl. Environ. Microbiol. 82:2012. doi: 10.1128/AEM.04052-15
Rath, K. M., Murphy, D. N., and Rousk, J. (2019). The microbial community size, structure, and process rates along natural gradients of soil salinity. Soil Biol. Biochem. 138:107607. doi: 10.1016/j.soilbio.2019.107607
Rath, K. M., and Rousk, J. (2015). Salt effects on the soil microbial decomposer community and their role in organic carbon cycling: a review. Soil Biol. Biochem. 81, 108–123. doi: 10.1016/j.soilbio.2014.11.001
Rietz, D. N., and Haynes, R. J. (2003). Effects of irrigation-induced salinity and sodicity on soil microbial activity. Soil Biol. Biochem. 35, 845–854. doi: 10.1016/S0038-0717(03)00125-1
Roth, P. J., Lehndorff, E., Cao, Z. H., Zhuang, S., and Bannert, A. (2011). Accumulation of nitrogen and microbial residues during 2000 years of rice paddy and non-paddy soil development in the Yangtze River Delta, China. Glob. Chang. Biol. 17, 3405–3417. doi: 10.1111/j.1365-2486.2011.02500.x
Rousk, J., Bååth, E., Brookes, P. C., Lauber, C. L., Lozupone, C., Caporaso, J. G., et al. (2010). Soil bacterial and fungal communities across a pH gradient in an arable soil. ISME J. 4, 1340–1351. doi: 10.1038/ismej.2010.58
Ryan, J., Garabet, S., Harmsen, K., and Rashid, A. (1996). A soil and plant analysis manual adapted for the West Asia and North Africa Region. Aleppo: ICARDA.
Said-Pullicino, D., Cucu, M. A., Sodano, M., Birk, J. J., Glaser, B., and Celi, L. (2014). Nitrogen immobilization in paddy soils as affected by redox conditions and rice straw incorporation. Geoderma 228–229, 44–53. doi: 10.1016/j.geoderma.2013.06.020
Sall, S. N., Ndour, N. Y. B., Diédhiou-Sall, S., Dick, R., and Chotte, J.-L. (2015). Microbial response to salinity stress in a tropical sandy soil amended with native shrub residues or inorganic fertilizer. J. Environ. Manage. 161, 30–37. doi: 10.1016/j.jenvman.2015.06.017
Sanchis, E., Ferrer, M., Torres, A. G., Cambra-López, M., and Calvet, S. (2012). Effect of water and straw management practices on methane emissions from rice fields: a review through a meta-analysis. Environ. Eng. Sci. 29, 1053–1062. doi: 10.1089/ees.2012.0006
Sardinha, M., Müller, T., Schmeisky, H., and Joergensen, R. G. (2003). Microbial performance in soils along a salinity gradient under acidic conditions. Appl. Soil Ecol. 23, 237–244. doi: 10.1016/S0929-1393(03)00027-1
Schimel, J. P., Scott, W. J., and Killham, K. (1989). Changes in cytoplasmic carbon and nitrogen pools in a soil bacterium and a fungus in response to salt stress. Appl. Environ. Microbiol. 55, 1635–1637. doi: 10.1128/AEM.55.6.1635-1637.1989
Semenov, M., Blagodatskaya, E., Stepanov, A., and Kuzyakov, Y. (2018). DNA-based determination of soil microbial biomass in alkaline and carbonaceous soils of semi-arid climate. J. Arid Environ. 150, 54–61. doi: 10.1016/j.jaridenv.2017.11.013
Setia, R., Marschner, P., Baldock, J., Chittleborough, D., Smith, P., and Smith, J. (2011). Salinity effects on carbon mineralization in soils of varying texture. Soil Biol. Biochem. 43, 1908–1916. doi: 10.1016/j.soilbio.2011.05.013
Vance, E. D., Brookes, P. C., and Jenkinson, D. S. (1987). An extraction method for measuring soil microbial biomass C. Soil Biol. Biochem. 19, 703–707. doi: 10.1016/0038-0717(87)90052-6
Venables, W. N., and Ripley, B. D. (2002). Modern Applied Statistics with S. 4th Edn. New York, NY: Springer.
Wang, W., Lai, D. Y. F., Wang, C., Pan, T., and Zeng, C. (2015). Effects of rice straw incorporation on active soil organic carbon pools in a subtropical paddy field. Soil Tillage Res. 152, 8–16. doi: 10.1016/j.still.2015.03.011
Wichern, F., Richter, C., and Joergensen, R. G. (2003). Soil fertility breakdown in a subtropical South African vertisol site used as a home garden. Biol. Fertil. Soils 37, 288–294. doi: 10.1007/s00374-003-0596-3
Wichern, J., Wichern, F., and Joergensen, R. G. (2006). Impact of salinity on soil microbial communities and the decomposition of maize in acidic soils. Geoderma 137, 100–108. doi: 10.1016/j.geoderma.2006.08.001
Wickham, H. (2016). ggplot2: Elegant Graphics for Data Analysis. New York, NY: Springer. doi: 10.1007/978-3-319-24277-4
Wickham, H. (2018). Scales: Scale Functions for Visualization. R package version 1.0.0. Available at online: https://CRAN.R-project.org/package=scales.
Wu, J., Joergensen, R. G., Pommerening, B., Chaussod, R., and Brookes, P. C. (1990). Measurement of soil microbial biomass C by fumigation-extraction—an automated procedure. Soil Biol. Biochem. 22, 1167–1169. doi: 10.1016/0038-0717(90)90046-3
Yan, X., Yagi, K., Akiyama, H., and Akimoto, H. (2005). Statistical analysis of the major variables controlling methane emission from rice fields. Glob. Chang. Biol. 11, 1131–1141. doi: 10.1111/j.1365-2486.2005.00976.x
Keywords: microbial activity, microbial community composition, mineralisation, nitrification, organic amendments, osmotic stress, residue management, salinity
Citation: Wichern F, Islam MR, Hemkemeyer M, Watson C and Joergensen RG (2020) Organic Amendments Alleviate Salinity Effects on Soil Microorganisms and Mineralisation Processes in Aerobic and Anaerobic Paddy Rice Soils. Front. Sustain. Food Syst. 4:30. doi: 10.3389/fsufs.2020.00030
Received: 18 December 2019; Accepted: 27 February 2020;
Published: 17 March 2020.
Edited by:
Somasundaram Jayaraman, Indian Institute of Soil Science (ICAR), IndiaReviewed by:
Nirmalendu Basak, Central Soil Salinity Research Institute (ICAR), IndiaSuprasanna Penna, Bhabha Atomic Research Centre (BARC), India
Copyright © 2020 Wichern, Islam, Hemkemeyer, Watson and Joergensen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Florian Wichern, Zmxvcmlhbi53aWNoZXJuJiN4MDAwNDA7aHNydy5ldQ==
 Florian Wichern
Florian Wichern Md. Rafiqul Islam1,2
Md. Rafiqul Islam1,2 Michael Hemkemeyer
Michael Hemkemeyer Conor Watson
Conor Watson Rainer Georg Joergensen
Rainer Georg Joergensen