- 1Department of Plant Sciences, University of California, Davis, Davis, CA, United States
- 2Center of Agricultural Biochemistry and Biotechnology, University of Agriculture Faisalabad, Faisalabad, Pakistan
- 3Department of Biology, University of Florence, Florence, Italy
- 4Dipartimento di Scienze Agrarie, Alimentari e Forestali, Università degli Studi di Palermo, Palermo, Italy
Walnuts are among the most important nut crops grown in temperate regions of the world. Commercial production in California, and increasingly worldwide, relies on only few clonally grafted scion genotypes, particularly “Chandler,” and more recently clonally propagated disease-resistant rootstocks. Diseases, nematodes, insect pests, abiotic stresses, and other nutritional and environmental factors, can reduce walnut productivity and quality, affecting grower profitability. The California Walnut Breeding Program at UC Davis has developed and released scion cultivars and rootstocks to help address some of these problems. Sequencing of the walnut genome is expected to speed walnut breeding by facilitating development of molecular markers that can be linked to phenotypic traits, and by reducing the time needed to screen new genotypes. Nonetheless, conventional breeding of tree crops is still a long-term proposition. Here we describe a toolkit that utilizes the CRISPR-Cas9 system to enable rapid and precise editing of the genomes of currently grown commercial clonal walnut genotypes. A computational tool was developed to aid selection of guide RNAs targeting specific genomic sites of this crop and to identify potential off-target sites. As a proof-of-concept, the gene encoding phytoene desaturase (PDS) of walnut was targeted, disrupting its expression. A dominant visual phenotype associated with the successful editing of this gene was observed in non-pigmented “albino” shoots obtained from in vitro cultures. This toolkit can now be adapted to other important tree crop members of the order Fagales, which includes other valuable nut and tree species such as pecans, butternuts, chestnuts, hickory nuts, hazelnut, wingnut, and oaks.
Introduction
Persian (English) walnut (Juglans regia) is a widely grown temperate-region nut crop that provides an excellent dietary source of plant-based protein and fiber, contains a rich supply of healthy oils including in particular omega-3 fatty acids, and is an important component of a heart-healthy Mediterranean diet (Vinson and Cai, 2012). It also ranks 7th in production and 5th in export among California's top agricultural commodities, grown on 335,000 acres with a production of around 630,000 tons valued at U$1.6 billion, of which 411,000 tons are exported (CA Stat Review, 2018). Sustainability of this industry in California has been dependent on F1 Juglans hindsii × J. regia hybrid rootstock, referred to commercially as Paradox, and on improved J. regia scion varieties such as “Chandler” that have preferred nut quality and production traits. Walnut production faces a number of agronomic issues, including diseases, soil-borne pests, insects, abiotic stresses (low water availability, salt, heat), and other environmental factors. Combined these can drastically reduce productivity and quality thus affecting exports and consequently grower profitability. The most secure and sustainable solutions to these problems can be gained genetically by the modification of useful phenotypic traits. The walnut breeding program in CA has developed both sexual and asexual approaches to introduce new genetic information into walnut scion and rootstock genotypes (Brown et al., 2019). Traditional breeding methods create a vast number of recombinant progeny that need to be evaluated by intensive and resource-consuming phenotyping to identify the few useful progeny. This is a cumbersome and time-consuming process, requiring large investments, and many years/decades. Efforts are underway to develop genome based genetic markers that are linked to phenotypic traits of interest to enhance early selection and to reduce the inordinate amount of time required to generate improved cultivars (Aradhya et al., 2019). However, relating accurate and reliable phenotyping of traits to the genetic information necessary for marker development is a limiting step. It is still a huge challenge for precision breeding, as the molecular mechanisms that underlie most phenotypic traits of interest in walnuts are not well-understood. Developing mutants/knock-outs in specific genes can improve the interpretation of relationships between genes and phenotypic traits.
Sequencing of the walnut genome was a major breakthrough in walnut breeding, providing an initial navigation platform for molecular breeding (Martinez-Garcia et al., 2016; Stevens et al., 2018). In addition to developing resources for identifying potential molecular markers, the genome sequence also revealed genetic similarities between walnut and other plant species providing a reference sequence essential for detailed transcriptomic and proteomic studies. With current genome sequence versions achieving chromosome-level assemblies of several walnut varieties and species (Zhu et al., 2019), precise genetic modifications can finally be rationally planned, paving the way for expedited molecular breeding. A new tool, CRISPR-Cas9, has been developed recently enabling precise genome engineering for gene silencing, gene activation, transcriptional regulation, and genetic base edits (Doudna and Charpentier, 2014; Noman et al., 2016). This two component system is composed of a Cas9 endonuclease from Streptococcus pyrogenes, and a single guide RNA (gRNA) that is able to target a DNA sequence of 20 nt before every NGG (protospacer-adjacent motif (PAM) site). This editing system was discovered in bacteria that use it in nature to defend themselves from attacks by bacterial viruses (Doudna and Charpentier, 2014). This discovery has been exploited to create a revolutionary biotechnological tool that can be used for plant genetic improvement. CRISPR-Cas9 has been widely used for targeted gene editing in many plant species (Brooks et al., 2014; Nishitani et al., 2016; Chen et al., 2019; Zhang et al., 2019) and the availability of a complete walnut genome sequence now allows a precise guide RNA selection and avoidance of off-target sequences.
Phytoene is a central precursor of carotenoids, affecting plant coloration, photosynthesis, photoprotection, and cell signaling (Meléndez-Martínez et al., 2015; Wang and Fu, 2016). The first committed step in carotenoid biosynthesis is carried out by phytoene synthase, followed by phytoene desaturase (PDS) (Foudree et al., 2010; Koschmieder et al., 2017). PDS can be inhibited by norflurazon-containing herbicides that disrupt chloroplast function leading to a bleached phenotype (Arias et al., 2006). For this reason PDS is commonly used as an endogenous reporter gene for proof of concept gene-editing in plants (Odipio et al., 2017; Shan et al., 2018; Bernard et al., 2019; Charrier et al., 2019; Ma et al., 2019; Wilson et al., 2019). In this study, we integrated knowledge of the complete walnut genomic sequence with Cas9-based gene-editing to efficiently select guide RNAs specifically targeting JrPDS. Edited lines displayed stunting and/or albino shoots in tissue culture. We have entered the era of precise genome-editing of walnuts, paving the way for its application in other nut tree crops.
Materials and Methods
Selection of sgRNA and Assembly of CRISPR-Cas9 Construct
The fasta and gene model (gff) files of J. regia were downloaded from NCBI (https://www.ncbi.nlm.nih.gov/assembly/GCF_001411555.1/). To generate a search database, we extracted the gene, mRNA, CDS, exon, and intron information from the gene model file and fetched the gene sequences from the Fasta file using in-house Perl script (Figure 2). We developed another Perl script, which extracts 23 length nucleotide sequences (gRNA + PAM) from the given gene sequence. In addition, the script will report the “TTTT” count, GC percentage, Melting temperature, and the presence of restriction enzyme cut sites. The positon of the gRNA and the gene coordinates were calculated using the generated database. The columns “20 mer Total” and “20 mer inside Gene” provide the number of occurrences of gRNA (without any mismatch) inside the entire walnut genome and inside the gene of interest. In addition, our program will report the occurrence of the “12-Mer” seed sequences (without any mismatch) in the entire genome and inside the gene, which helps the user select specific gRNAs by avoiding the potential off target candidates. The computational workflow used for the prediction of gRNA is shown in Figure S1. The complete processing pipeline is available at our GitHub repository (https://github.com/dandekarlab/SoggyCas). The complete search results for gRNA targeting JrPDS using our workflow is given in Table S1.
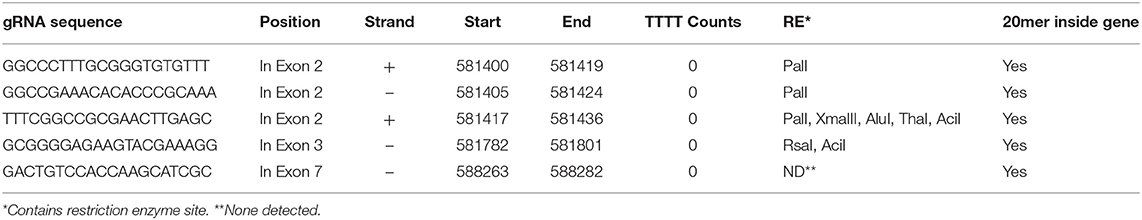
Five sets of complementary DNA oligos containing selected gRNA and adapter sequences for BsaI restriction site at 5′ ends were commercially synthesized (IDT Technologies) (Table 1). The complementary oligos were annealed in a thermocycler leaving single stranded overhangs of adapter sequences. Annealing reaction consisted of 50 mM of each oligo in 10 μL total reaction volume. After an initial incubation at 37°C for 30 min, the reaction was heated to 94°C for 5 min and then cooled to room temperature on a ramp of −5°C/s. Plant expression vector pHSE401 (Addgene) having plant codon optimized Cas9 was digested with BsaI, a 1213-bp fragment of SmR gene was removed and vector backbone was gel purified for further cloning. Annealed complementary oligos were ligated with BsaI-digested fragment of the plant expression vector. The integration of distinct 20-bp DNA targeting JrPDS was confirmed by sequencing using M13 forward primer. This plasmid (Figure 1) was electroporated into the disarmed Agrobacterium tumefaciens strain EHA105 and vector was designated as pDM17.0101.01.
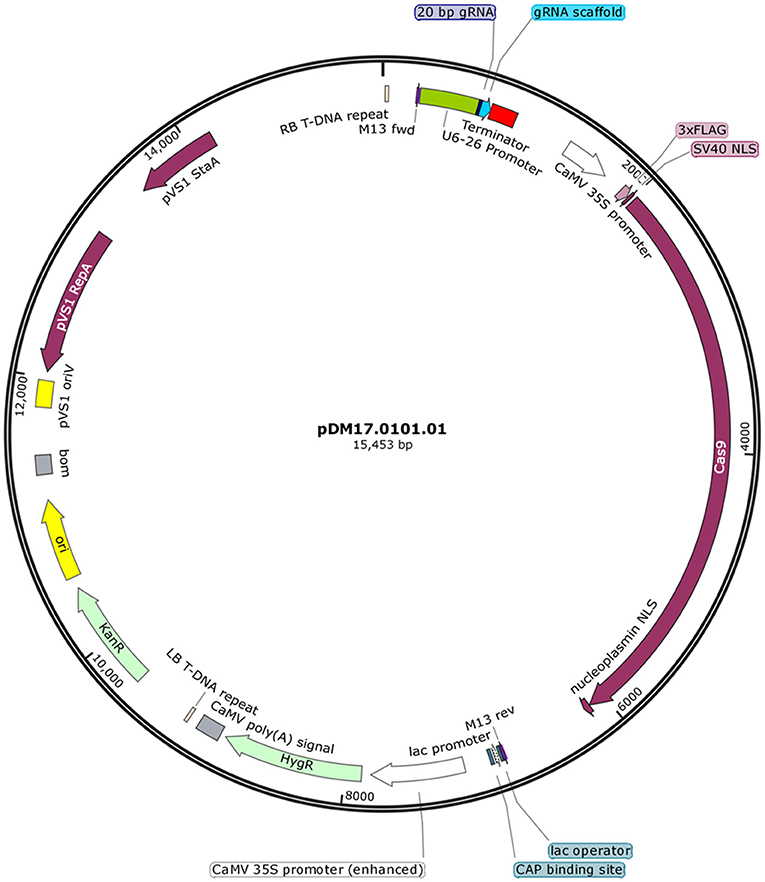
Figure 1. DNA vector used to deliver Cas9 and sgRNA targeting JrPDS. Binary vector for generation of mutant plant lines carrying a construct expressing the sgRNA controlled by the U6-26 promoter and another expressing SpCas9 containing a nuclear localization signal and a 3xFlag-tag.
Transformation of Walnut Embryos and Maintenance of Transformants
Agrobacterium-mediated transformation of the somatic embryo line CR1 from the walnut cultivar Chandler was performed as previously described (Walawage et al., 2014). About 40 small (2–5 mm) white, intact walnut somatic embryos were transformed using binary vector pDM17.0101.01 as previously described (Walawage et al., 2014). Transformants were selected on 25 mg/l hygromycin. The inoculated embryos were designated E0 embryos, the initial secondary embryos were E1 embryos, and subsequent generations of embryos were called E2, E3, etc. E1 embryos were transferred into separate plates containing basal Driver/Kuniyuki walnut medium (DKW) (Driver and Kuniyuki, 1984) containing hygromycin and allowed to grow in the dark. Independent embryo lines surviving on hygromycin after 8 weeks producing E2 generations were desiccated over saturated ammonium nitrate (Walawage et al., 2014) and germinated on DKW supplemented with 1 mg/l 6-Benzylaminopurine (BAP) and 0.01 mg/l Indole-3-butyric acid (IBA). Desiccated embryos were cultured at ambient temperature under cool white fluorescent lights (16 h day length, ~100 μE) for 4 weeks to induce shoot formation. Similarly, untransformed wild type Chandler embryos were germinated and used as controls. Transformed and untransformed embryos, shoots, and roots were evaluated for endogenous phytoene desaturase (PDS) gene disruption. Molecular analysis such as PCR, Sanger sequencing, and phenotypic characters of germinating embryos were taken into consideration when analyzing the transformants.
Analysis of Transformants
Transgenic and non-transgenic plantlets were grown in vitro as described above. Leaf, root, and somatic embryo tissues were collected for molecular analysis. Transgenic line 2-1-1 was analyzed in detail, and sampling included leaves from albino shoots, variegated shoots, green shoots, and geminated embryos with only roots. Non-transgenic line samples included leaves from green tissues, roots and embryos. Approximately 0.1 g of leaf tissue from each sample was placed in microcentrifuge tubes and simultaneously disrupted and homogenized by high-speed shaking with a bead in a sealed tube (Tissuelyser) set at 30 Hz for 1 min. Genomic DNA was isolated using a DNeasy Plant Mini Kit (Qiagen) according to the manufacturer's protocols. A DNA fragment of 482 bp spanning the Cas9/gRNA target sequences were amplified by PCR using LongAmp Taq DNA Polymerase (New England Biolabs). Oligonucleotide primers used were Forward: 5′-CCCGGTATCTTGGTTTCCGCGATGGGGC and Reverse: 5′-GGTCTTGGGAAGTCCATGCATACTACCTGCG. As the template 200 ng of genomic DNA was used from each sample. The thermocyler conditions were 94°C for 30 s for initial denaturation, followed by 30 cycles at 94°C for 30 s, 65°C for 1 min for annealing, 65°C for 25 s for extension, and 65°C 10 min for the final extension. After purification, the DNA PCR product was sequenced by the Sanger method (Quintara Biosciences). DNA sequences obtained from each sample were aligned with the reference sequence to verify mutations that occurred in the different DNA samples (SnapGene v4.3.4, GSL Biotech LLC). Phenotypic characters of PDS edited plants and embryos were observed and recorded.
Results
PDS Is a Single Copy Gene in Juglans regia
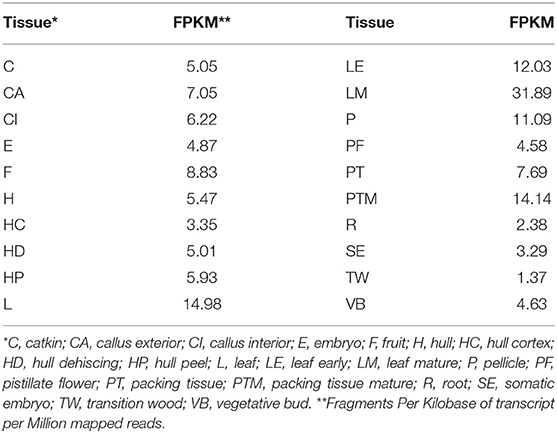
Phytoene desaturase in J. regia (JrPDS) was predicted to be encoded by a single copy gene in the initial fragmented genome assembly. This was confirmed with the new complete Chandler assembly (Dr. Annarita Marrano, unpublished results), locating it in chromosome 7 and identified as Jr07_15220 (Figure 2). Both versions of the gene predictions are 99% identical, with all 15 exons present in both. Transcripts mapping to JrPDS were detected in 20 different walnut tissues (Chakraborty et al., 2016), with greater abundance in green tissues (Table 1).
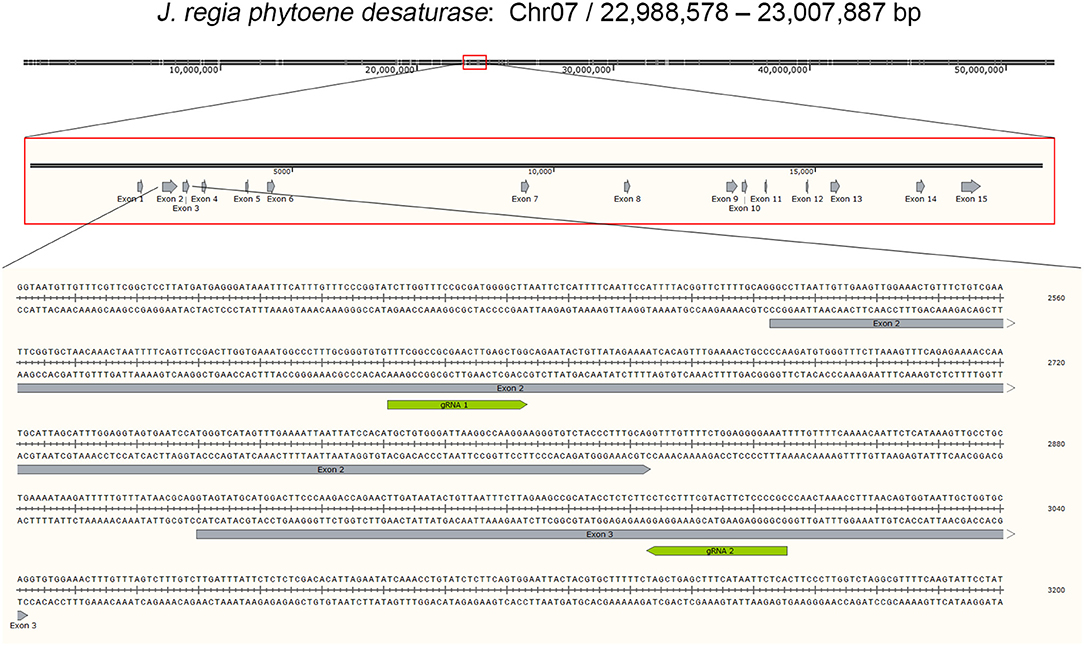
Figure 2. Gene model of JrPDS. Representation of chromosome 7 from Juglans regia cv. Chandler, highlighting the genome vicinity containing CDS Jr07_15220 encoding Phytoene desaturase. The zoomed region shows the relative size and location of the 15 exons. The bottom panel shows in further detail the segment comprising exons 2 and 3, and the location of 2 selected gRNAs.
Selection of gRNAs Targeting JrPDS
Based on the availability of the complete genome sequence and JrPDS being single copy, a strategy was devised to disrupt its functionality by introducing frame shift mutations using Cas9. In order to direct the endonuclease to the proper genomic site, a computational pipeline was developed to assist in guide RNAs selection (Figure S1). With this search strategy 1,303 possible gRNA sequences were retrieved, and overall search metrics are shown in Table 2. The complete information of the 1,303 predicted gRNAs using our pipeline is listed in Table S1. Our pipeline reported that 223 of the predicted gRNAs contain “TTTT”s, which may trigger PolIII transcriptional termination (Ui-Tei et al., 2017), reducing their score in our ranking of preference. In addition, our pipeline calculated the GC content, melting temperature, and the off-target sites of the gRNA sequences which are the major factors in the specificity of gRNAs. We selected the gRNAs which were falling inside exons, had GC percentage of 60 and had no off-target sites in the total 20 nt sequence if the gRNA or just the 12 mer “seed sequence” (“20 mer Total” = 1, “20 mer inside Gene” = 1, “12 mer Total” = 1, “12 mer inside Gene” = 1) and no “TTTT” counts. Five candidate gRNA sequences were selected based on these filter criteria (Table 3). Since 3 were partially overlapped in exon 2, one was chosen along with other 2 (annealing in exons 3 and 7, Table 3). These were used to build the vectors for plant transformation. The construct targeting exon 2 was used for plant transformation and described below.
Phenotypic Characters of Germinated Shoots With Functional Inactivation of JrPDS
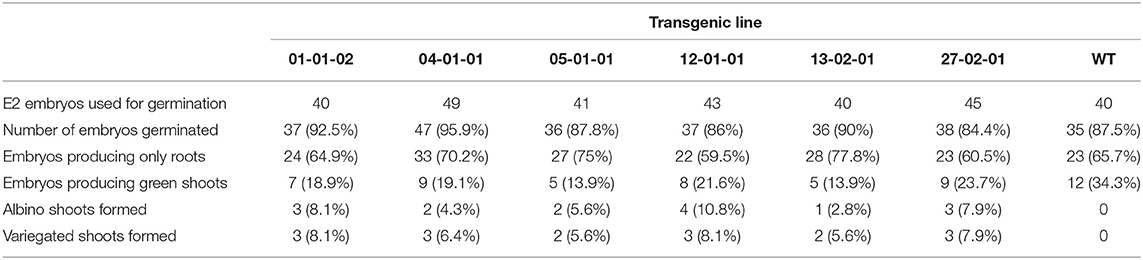
After transforming ~40 small white embryos, 20 transgenic lines were selected and maintained. Embryo lines producing white, healthy E2 generations were maintained and used for further analysis (Figure 3A). Embryos from six transgenic lines were germinated to study phenotypic characters (Table 4). Despite transformants exhibiting normal phenotype at the embryo stage, when transferred to media containing IBA and BAP to induce shoot formation, functional inactivation of PDS became more visually evident. Compared to untransformed controls, lines targeted for PDS inactivation displayed smaller/stunted shoots and the bleached phenotype (reduced photosynthetic capacity) (Figure 3B).
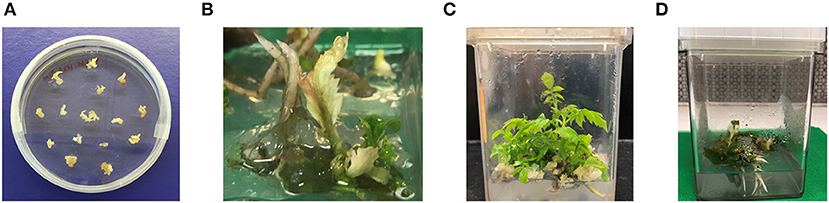
Figure 3. Phenotype of edited lines. (A) Walnut somatic embryos maintained in vitro were transformed by Agroinfection and kept in selective DKW medium containing hygromycin. Surviving embryo lines were transferred to shoot-inducing medium and phenotypes of resulting (B) stunted/albino, and (C) normal shoots were observed. (D) PDS inactivated lines also displayed vigorous root formation.
From the six transgenic lines tested, 18.5% of geminated embryos produced green shoots and were phenotypically similar to the non-transformed parental line, while 6.5% produced albino shoots, and 7% had variegated leaves (Figures 3B,C). All the albino or variegated shoots displayed stunted phenotype with shorter internodes. Interestingly, the PDS-edited shoots also displayed healthy vigorous root system (Figure 3D). In transgenic lines about 65.7% embryos only developed roots and did not develop shoots (Table 4). Embryos developed into variegated or albino shoots are due to disruption of the PDS gene. This confirms that not only can we target a specific site in the walnut genome for genetic mutation, but that PDS is essential for proper plant growth. We chose shoots and embryos of transgenic line 2-1-1 for molecular analysis. DNA was isolated from independent green leaves, albino leaves, variegated leaves, roots, and transgenic embryos of line 2-1-1 and analyzed for the mutations, comparing to untransformed embryos and shoots.
Mutations in Transgenic Lines
In order to determine the exact mutations generated by Cas9 targeting JrPDS, DNA sequencing of selected lines was performed. A common target site was 3 bases upstream of the PAM site (Figure 4A), where both single nucleotide insertions and deletions resulting in frameshifts were detected in the clones analyzed. Besides these single nucleotide mutations, the sequence alignment also shows an example in which many other mutations were detected. A closer inspection of the sequence chromatogram however reveals the sequencing artifact (Figure 4B). This is observed in cases in which a single allele is edited, generating a polymorphic region in the alignment. When this genomic locus is PCR-amplified and sequenced, the polymorphic region due to the frameshift results in ambiguous base-calling annotated as apparent multiple errors in sequence. This illustrates the necessity to verify each selected line individually in order to understand the precise mutation generated in each line.
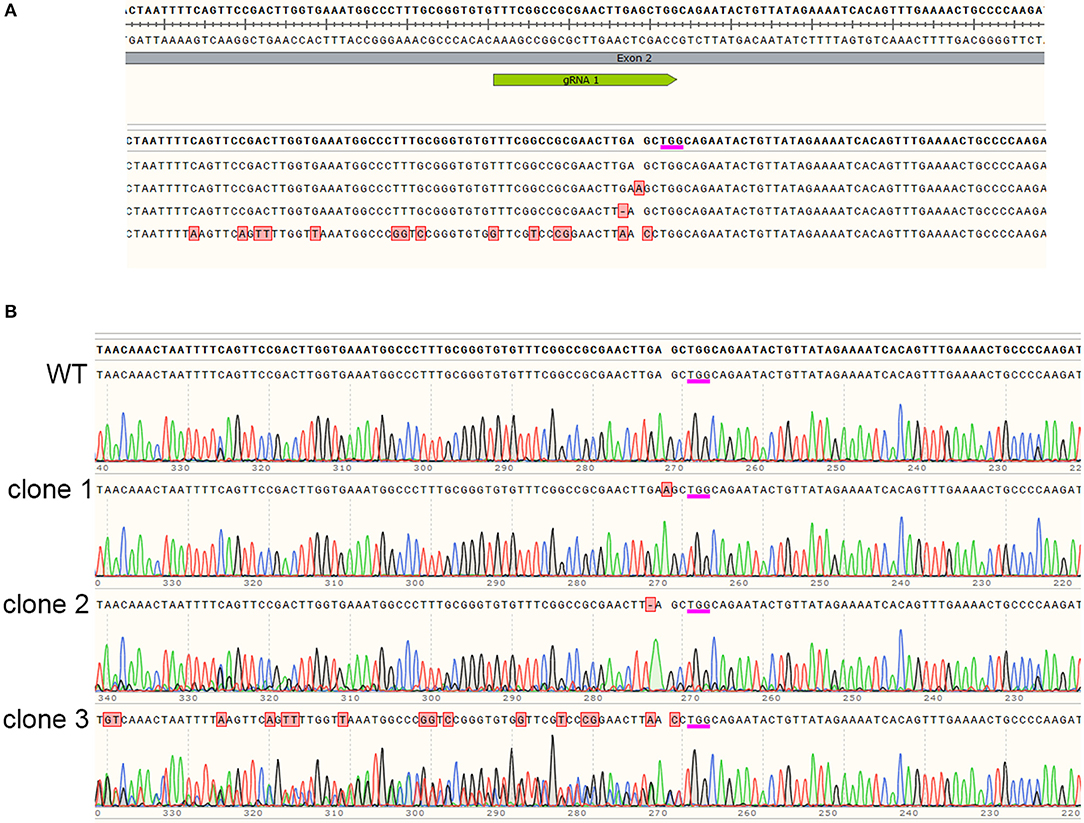
Figure 4. Sequence analysis of edited lines. Mutations in JrPDS of selected lines displaying stunted or albino phenotypes were investigated by DNA sequencing. (A) Alignment of sequencing reads obtained from selected edited lines. Sequence on top shows site of gRNA annealing and PAM site (underlined in magenta) with reads showing detected mutations in red, or the lack of mutations in a wild type clone. (B) Sequence chromatograms of the reads shown in the top panel, revealing the original sequence, a mutated line with a adenosine insertion, a mutated line with a guanosine deletion, and the bottom read which seemed to contain multiple mutations, but revealed edition of only one of the alleles, generating an heterozygotic region during the sequencing reaction.
Discussion
Nut crop breeding faces numerous challenges, including long generation times and space requirements that considerably limit the development of new cultivars. In addition, breeding for desired traits can also carry undesired closely linked traits. Recent availability of vastly improved complete genome sequences for Juglans microcarpa and Juglans regia cultivars “Chandler,” and “Serr” provide an enormous advancement in our understanding of the chromosomal arrangement and gene/marker repertoire from an ordered point of view, bringing order to the previous map composed of over 105 thousand scaffolds. With more cultivars being sequenced, the path from genome to phenome becomes clearer with tremendous impact in marker selection and validation. Synergistically, with the recent introduction of the CRISPR-Cas9 toolbox in plants of agronomical importance, not only can we understand and read genome structure like never before but also modify it with previously unattainable precision (Chen et al., 2019). It has become a powerful tool to learn more about gene function (Arora and Narula, 2017). Here we presented the first report to our knowledge of gene-editing using Cas9 in a nut crop.
The versatility of CRISPR-Cas9 tools goes beyond point mutations adjacent to PAM sites, with options in base-editing variations already employed in some crops (Bastet et al., 2019; Wu et al., 2019). When used in combination, multiple sgRNAs targeting the same exon or different exons within the same gene provide a higher chance of a functional knockout (Xie et al., 2015). Whole gene deletions are also possible if two events of double strand break are simultaneous (Xie et al., 2015; Durr et al., 2018), and if a donor strand is delivered with the sgRNAs, incorporation of a new sequence (allele substitution) may occur (Collonnier et al., 2017). To achieve optimal functioning of these ribonucleoproteins (RNPs) one may also optimize the vectors used in agroinfiltration with alternative promoter sequences and components such as self-cleaving tRNAs for multi-targeting, for example (Xie et al., 2015). Other delivery methods of RNPs such as transfection (Cao et al., 2019), biolistics (Liang et al., 2018), synthetic vesicles (Lin et al., 2018), and functionalized carbon nanotubes (Demirer et al., 2019), are quickly becoming available as well. In the case reported here, single nucleotide insertions and deletions were detected in JrPDS, in both or in one of the alleles. In addition, despite our gRNA selection pipeline being designed to minimize the chance of off-target editing, we cannot rule out this possibility with the current data. This is currently a concern with CRISPR-related technologies and advancements to minimize off-targets are being pursued (Doench et al., 2016; Kleinstiver et al., 2016; Hajiahmadi et al., 2019). In this work, we made a holistic effort to combine filters to minimize the off-target effects for the gRNA selection, a conservative approach that limits the options for gRNA selection but reduces even further the chance of off-targets. Still, we cannot exclude the possibility of off-targets, and advocate for any plant line generated with the current CRISPR toolbox to be analyzed carefully before being adopted by breeding programs. One possible way of doing this is PCR-amplifying and sequencing all regions with similarity to the chosen target site, which could still be not enough to detect off-targets. However, absolute certainty could only be achieved by sequencing the entire genome of the edited line, something beyond the scope of our proof-of-concept presented here. Still regarding CRISPR technologies, another point to consider is the persistence of Cas9 in edited lines intended for commercial use. Besides trying to segregate out the Cas9 in future generations, efforts are underway in our group to develop a Cas9 removal procedure once the desired mutations are detected. An alternative is the adoption of carbon nanotubes and other polymers or procedures to deliver the gRNA-Cas9 ribonucleo-protein complex directly inside cells.
Phytoene desaturase is a single-copy gene in walnuts and is an interesting target for demonstrating Cas9 functionality. PDS inactivation results in an easily scored visual (bleached) phenotype and alteration of function of this key enzyme in carotene synthesis can provide insight into the essential role of carotenes in plastid morphogenesis and intimately regulate health. Although expressed in all 20 tissue types investigated, PDS is greatly enriched in green tissues, confirming its importance for the development, and functioning of the photosynthetic machinery. Besides accessory light-harvesting photosynthetic pigments, they play essential roles as anti-oxidants (Stahl and Sies, 2003) aiding the maintenance of membranes and other plastid components. This has implications in general plant health, as the redox state and integrity of plastids influence many cellular processes (Foyer, 2018) and also serve as a reliable abiotic and biotic stress marker (Zechmann, 2019).
Conclusion
In conclusion, the combination of several technologies is creating the necessary framework for streamlining the generation of high throughput tailored variants of walnut cultivars commonly used as rootstocks and scions. This holds great promise for the study of gene function and plant breeding of nut crops. This is timely important given the increase in access and interest in diets rich in polyunsaturated fatty acids and polyphenols (Vinson and Cai, 2012), plant-based proteins and therapeutics. As new advancements are added to the CRISPR toolbox, precise gene-editing can be increasingly adopted by breeding programs.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
AD and CL obtained funding. AD, CL, SW, and MM planned the experiments. BB, FM, MM, and SW performed the experiments. AD, BB, FM, MM, PZ, and SW analyzed the data. All authors contributed to the manuscript preparation and review.
Funding
The authors wish to acknowledge the funding from the California Walnut Board to support the research described in this paper.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Sandeep Chakraborty for assistance in the early steps of gRNA analysis and Steven H. Lee for assistance in maintaining the walnut tissue cultures.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2019.00100/full#supplementary-material
Table S1. Complete list of retrieved gRNA sequences targeting Phytoene desaturase of Juglans regia.
References
Aradhya, M. K., Velasco, D., Wang, J. R., Ramasamy, R., You, F. M., Leslie, C. A., et al. (2019). A fine-scale genetic linkage map reveals genomic regions associated with economic traits in walnut (Juglans regia L.). Plant Breed. 138, 635–646. doi: 10.1111/pbr.12703
Arias, R. S., Dayan, F. E., Michel, A., Howell, J., and Scheffler, B. E. (2006). Characterization of a higher plant herbicide-resistant phytoene desaturase and its use as a selectable marker. Plant Biotechnol. J. 4, 263–273. doi: 10.1111/j.1467-7652.2006.00179.x
Arora, L., and Narula, A. (2017). Gene editing and crop improvement using CRISPR-Cas9 system. Front. Plant Sci. 8:1932. doi: 10.3389/fpls.2017.01932
Bastet, A., Zafirov, D., Giovinazzo, N., Guyon-Debast, A., Nogué, F., Robaglia, C., et al. (2019). Mimicking natural polymorphism in eIF4E by CRISPR-Cas9 base editing is associated with resistance to potyviruses. Plant Biotechnol. J. 17, 1736–1750. doi: 10.1111/pbi.13096
Bernard, G., Gagneul, D., Alves Dos Santos, H., Etienne, A., Hilbert, J. L., and Rambaud, C. (2019). Efficient genome editing using CRISPR/Cas9 technology in chicory. Int. J. Mol. Sci. 20:E1155. doi: 10.3390/ijms20051155
Brooks, C., Nekrasov, V., Lippman, Z. B., and Van Eck, J. (2014). Efficient gene editing in tomato in the first generation using the clustered regularly interspaced short palindromic repeats/CRISPR-associated9 system. Plant Physiol. 166, 1292–1297. doi: 10.1104/pp.114.247577
Brown, P. J. C, Leslie, A., and Dandekar (2019). “Juglans regia L. Walnut,” in Biotechnology of Fruit and Nut Crops, 2nd Edn., eds R. E. Litz, F. Pliego-Alfaro, and J. I. Hormaza (Cambridge, MA: C.A.B. International), 307–323.
California Agricultural Statistics Review 2017-2018 (2018). California Department of Food and Agriculture. Available online at: https://www.cdfa.ca.gov/statistics/PDFs/2017-18AgReport.pdf (accessed July 22, 2019).
Cao, Y., Ma, E., Cestellos-Blanco, S., Zhang, B., Qiu, R., Su, Y., et al. (2019). Nontoxic nanopore electroporation for effective intracellular delivery of biological macromolecules. PNAS 116, 7899–7904 doi: 10.1073/pnas.1818553116
Chakraborty, S., Britton, M., Martínez-García, P. J., and Dandekar, A. M. (2016). Deep RNA-Seq profile reveals biodiversity, plant-microbe interactions and a large family of NBS-LRR resistance genes in walnut (Juglans regia) tissues. AMB Express 6:12. doi: 10.1186/s13568-016-0182-3
Charrier, A., Vergne, E., Dousset, N., Richer, A., Petiteau, A., and Chevreau, E. (2019). Efficient targeted mutagenesis in apple and first time edition of pear using the CRISPR-Cas9 system. Front. Plant Sci. 10:40. doi: 10.3389/fpls.2019.00040
Chen, K., Wang, Y., Zhang, R., Zhang, H., and Gao, C. (2019). CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 70, 667–697. doi: 10.1146/annurev-arplant-050718-100049
Collonnier, C., Guyon-Debast, A., Maclot, F., Mara, K., Charlot, F., and Nogué, F. (2017). Towards mastering CRISPR-induced gene knock-in in plants: survey of key features and focus on the model Physcomitrella patens. Methods 121, 103–117. doi: 10.1016/j.ymeth.2017.04.024
Demirer, G. S., Zhang, H., Matos, J. L., Goh, N. S., Cunningham, F. J., Sung, Y., et al. (2019). High aspect ratio nanomaterials enable delivery of functional genetic material without DNA integration in mature plants. Nat. Nanotechnol. 14, 456–464. doi: 10.1038/s41565-019-0382-5
Doench, J. G., Fusi, N., Sullender, M., Hegde, M., Vaimberg, E. W., Donovan, K. F., et al. (2016). Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat. Biotechnol. 34, 184–191. doi: 10.1038/nbt.3437
Doudna, J. A., and Charpentier, E. (2014). The new frontier of genome engineering with CRISPR-Cas9. Science 346:1258096. doi: 10.1126/science.1258096
Driver, J. A., and Kuniyuki, A. H. (1984). In vitro propagation of Paradox walnut rootstock [Juglans hindsii X Juglans regia, tissue culture]. AGRIS 19, 507–509.
Durr, J., Papareddy, R., Nakajima, K., and Gutierrez-Marcos, J. (2018). Highly efficient heritable targeted deletions of gene clusters and non-coding regulatory regions in Arabidopsis using CRISPR/Cas9. Sci. Rep. 8:4443. doi: 10.1038/s41598-018-22667-1
Foudree, A., Aluru, M., and Rodermel, S. (2010). PDS activity acts as a rheostat of retrograde signaling during early chloroplast biogenesis. Plant Signal Behav. 5, 1629–1632. doi: 10.4161/psb.5.12.13773
Foyer, C. H. (2018). Reactive oxygen species, oxidative signaling and the regulation of photosynthesis. Environ. Exp. Bot. 154, 134–142. doi: 10.1016/j.envexpbot.2018.05.003
Hajiahmadi, Z., Movahedi, A., Wei, H., Li, D., Orooji, Y., Ruan, H., et al. (2019). Strategies to increase on-target and reduce off-target effects of the CRISPR/Cas9 system in plants. Int. J. Mol. Sci. 20:E3719. doi: 10.3390/ijms20153719
Kleinstiver, B. P., Pattanayak, V., Prew, M. S., Tsai, S. Q., Nguyen, N. T., Zheng, Z., et al. (2016). High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature 529, 490–495. doi: 10.1038/nature16526
Koschmieder, J., Fehling-Kaschek, M., Schaub, P., Ghisla, S., Brausemann, A., Timmer, J., et al. (2017). Plant-type phytoene desaturase: functional evaluation of structural implications. PLoS ONE 12:e0187628. doi: 10.1371/journal.pone.0187628
Liang, Z., Chen, K., Zhang, Y., Liu, J., Yin, K., Qiu, J. L., et al. (2018). Genome editing of bread wheat using biolistic delivery of CRISPR/Cas9 in vitro transcripts or ribonucleoproteins. Nat. Protoc. 13, 413–430. doi: 10.1038/nprot.2017.145
Lin, C. S., Hsu, C. T., Yang, L. H., Lee, L. Y., Fu, J. Y., Cheng, Q. W., et al. (2018). Application of protoplast technology to CRISPR/Cas9 mutagenesis: from single-cell mutation detection to mutant plant regeneration. Plant Biotechnol. J. 16, 1295–1310. doi: 10.1111/pbi.12870
Ma, C., Zhu, C., Zheng, M., Liu, M., Zhang, D., Liu, B., et al. (2019). CRISPR/Cas9-mediated multiple gene editing in Brassica oleracea var. capitata using the endogenous tRNA-processing system. Hortic. Res. 6:20. doi: 10.1038/s41438-018-0107-1
Martinez-Garcia, P. J., Crepeau, M. W., Puiu, D., Gonzalez-Ibeas, D., Whalen, J., Stevens, K. A., et al. (2016). The walnut (Juglans regia) genome sequence reveals rich diversity in genes coding for the biosynthesis of nonstructural polyphenols. Plant J. 87, 507–532. doi: 10.1111/tpj.13207
Meléndez-Martínez, A. J., Mapelli-Brahm, P., Benítez-González, A., and Stinco, C. M. (2015). A comprehensive review on the colorless carotenoids phytoene and phytofluene. Arch. Biochem. Biophys. 572, 188–200. doi: 10.1016/j.abb.2015.01.003
Nishitani, C., Hirai, N., Komori, S., Wada, M., Okada, K., Osakabe, K., et al. (2016). Efficient genome editing in apple using a CRISPR/Cas9 system. Sci. Rep. 6:31481. doi: 10.1038/srep31481
Noman, A., Aqeel, M., and He, S. (2016). CRISPR-Cas9: tool for qualitative and quantitative plant genome editing. Front. Plant Sci. 7:1740. doi: 10.3389/fpls.2016.01740
Odipio, J., Alicai, T., Ingelbrecht, I., Nusinow, D. A., Bart, R., and Taylor, N. J. (2017). Efficient CRISPR/Cas9 genome editing of phytoene desaturase in Cassava. Front. Plant Sci. 8:1780. doi: 10.3389/fpls.2017.01780
Shan, S., Mavrodiev, E. V., Li, R., Zhang, Z., Hauser, B. A., Soltis, P. S., et al. (2018). Application of CRISPR/Cas9 to Tragopogon (Asteraceae), an evolutionary model for the study of polyploidy. Mol. Ecol. Resour. 18, 1427–1443. doi: 10.1111/1755-0998.12935
Stahl, W., and Sies, H. (2003). Antioxidant activity of carotenoids. Mol. Aspects Med. 24, 345–351. doi: 10.1016/S0098-2997(03)00030-X
Stevens, K. A., Woeste, K., Chakraborty, S., Crepeau, M. W., Leslie, C. A., Martínez-García, P. J., et al. (2018). Genomic variation among and within six Juglans species. G3 8, 2153–2165. doi: 10.1534/g3.118.200030
Ui-Tei, K., Maruyama, S., and Nakano, Y. (2017). Enhancement of single guide RNA transcription for efficient CRISPR/Cas-based genomic engineering. Genome 60, 537–545. doi: 10.1139/gen-2016-0127
Vinson, J. A., and Cai, Y. (2012). Nuts, especially walnuts, have both antioxidant quantity and efficacy and exhibit significant potential health benefits. Food Funct. 3, 134–140. doi: 10.1039/C2FO10152A
Walawage, S. L., Leslie, C. A., Escobar, M. A., and Dandekar, A. M. (2014). Agrobacterium tumefaciens-mediated transformation of walnut (Juglans regia). Bio-protocol 4:e1258. doi: 10.21769/BioProtoc.1258
Wang, D., and Fu, A. (2016). The plastid terminal oxidase is a key factor balancing the redox state of thylakoid membrane. Enzymes 40, 143–171. doi: 10.1016/bs.enz.2016.09.002
Wilson, F. M., Harrison, K., Armitage, A. D., Simkin, A. J., and Harrison, R. J. (2019). CRISPR/Cas9-mediated mutagenesis of phytoene desaturase in diploid and octoploid strawberry. Plant Methods 15:45. doi: 10.1186/s13007-019-0428-6
Wu, Y., Xu, W., Wang, F., Zhao, S., Feng, F., Song, J., et al. (2019). Increasing cytosine base editing scope and efficiency with engineered Cas9-PmCDA1 fusions and the modified sgRNA in rice. Front. Genet. 10:379. doi: 10.3389/fgene.2019.00379
Xie, K., Minkenberg, B., and Yang, Y. (2015). Boosting CRISPR/Cas9 multiplex editing capability with the endogenous tRNA-processing system. PNAS 112, 3570–3575. doi: 10.1073/pnas.1420294112
Zechmann, B. (2019). Ultrastructure of plastids serves as reliable abiotic and biotic stress marker. PLoS ONE 14:e0214811. doi: 10.1371/journal.pone.0214811
Zhang, Y., Malzahn, A. A., Sretenovic, S., and Qi, Y. (2019). The emerging and uncultivated potential of CRISPR technology in plant science. Nat. Plants 5, 778–794. doi: 10.1038/s41477-019-0461-5
Keywords: phytoene desaturase, nut crops, gene-editing, CRISPR-Cas9, gRNA, oxidative stress, plastid health
Citation: Walawage SL, Zaini PA, Mubarik MS, Martinelli F, Balan B, Caruso T, Leslie CA and Dandekar AM (2019) Deploying Genome Editing Tools for Dissecting the Biology of Nut Trees. Front. Sustain. Food Syst. 3:100. doi: 10.3389/fsufs.2019.00100
Received: 12 August 2019; Accepted: 18 October 2019;
Published: 05 November 2019.
Edited by:
Avtar Krishan Handa, Purdue University, United StatesReviewed by:
Tahira Fatima, Purdue University, United StatesJas Singh, Agriculture and Agri-Food Canada (AAFC), Canada
Copyright © 2019 Walawage, Zaini, Mubarik, Martinelli, Balan, Caruso, Leslie and Dandekar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Abhaya M. Dandekar, amdandekar@ucdavis.edu
 Sriema L. Walawage
Sriema L. Walawage Paulo A. Zaini
Paulo A. Zaini Muhammad S. Mubarik
Muhammad S. Mubarik Federico Martinelli1,3
Federico Martinelli1,3 Bipin Balan
Bipin Balan Tiziano Caruso
Tiziano Caruso Charles A. Leslie
Charles A. Leslie Abhaya M. Dandekar
Abhaya M. Dandekar