- 1Department of Biomedical Engineering, Tufts University, Medford, MA, United States
- 2New Harvest, Brooklyn, NY, United States
- 3Department of Biological Sciences, Chico State University, Chico, CA, United States
Cellular agriculture is defined as the production of agricultural products from cell cultures rather than from whole plants or animals. With growing interest in cellular agriculture as a means to address public health, environmental, and animal welfare challenges of animal agriculture, the concept of producing seafood from fish cell- and tissue-cultures is emerging as an approach to address similar challenges with industrial aquaculture systems and marine capture. Cell-based seafood—as opposed to animal-based seafood—can combine developments in biomedical engineering with modern aquaculture techniques. Biomedical engineering developments such as closed-system bioreactor production of land animal cells create a basis for the large scale production of marine animal cells. Aquaculture techniques such as genetic modification and closed system aquaculture have achieved significant gains in production that can pave the way for innovations in cell-based seafood production. Here, we present the current state of innovation relevant to the development of cell-based seafood across multiple species, as well as specific opportunities and challenges that exist for advancing this science. The authors find that the physiological properties of fish cell- and tissue- culture may be uniquely suited to cultivation in vitro. These physiological properties, including tolerance to hypoxia, high buffering capacity, and low-temperature growth conditions, make marine cell culture an attractive opportunity for scaled production of cell-based seafood; perhaps even more so than mammalian and avian cell cultures for cell-based meats. This opportunity, coupled with the unique capabilities of crustacean tissue-friendly scaffolding such as chitosan, a common seafood waste product and mushroom derivative, presents promise for cell-based seafood production via bioreactor cultivation. To become fully realized, cell-based seafood research will require more understanding of fish muscle cell and tissue cultivation; more investigation into serum-free media formulations optimized for fish cell culture; and bioreactor designs tuned to the needs of fish cells for large scale production.
Introduction
It is estimated that industrialized fisheries and fishing due to marine capture have lowered ocean biomass content by up to 80% (Myers and Worm, 2003). This effect, coupled with the effects of global warming on oceans, threatens to decimate wild fish populations (Funk and Brown, 2009). While some herald the rise of aquaculture as an ecological and economic boon (Tidwell and Allan, 2001), others feel that its benefits have been overstated and that it does not fundamentally solve current strains on wild fish populations (Naylor et al., 2000; Merino et al., 2012). At first glance, aquaculture seems capable of relieving the pressures of wild capture, however, this may not be the case. Carnivorous farmed fish are often fed wild fish, modifying natural habitats through harvest and cultivation (Naylor et al., 2000). While aquaculture was found to be a suitable means of feeding the earth's growing population, this would necessitate a reduction of wild-caught fish used for aquaculture feed, as many aquacultured fish eat fishmeal derived from wild-caught fish (Merino et al., 2012). Thus, even as aquaculture increases, so does wild-capture.
The challenges of feeding the world's growing population in a healthy way and in the context of sustainability has been presented (Lindgren et al., 2018). In this thesis, “Eating for the post-Anthropocene: Alternative proteins, Silicon Valley and the (bio)politics of food security,” novel animal proteins as a new locus for biopolitical disruption are presented as agents that affect culture and geographies (Sexton, 2018).
With oceans at risk, cell-based seafood provides a unique opportunity to transform the sustainability landscape. The concept of producing meat from cell cultures rather than from whole animals as a means to provide nutritional muscle tissue is no longer novel (Datar and Betti, 2010). The conversation around this concept has centered on the growth of mammalian or avian cells and tissues to replace meat, but cellular agriculture could easily be extended to fish, mollusk, and crustacean cells and tissues to replace seafoods. Such an extension could yield unique benefits that have not yet been explored. There are many considerations for cell-based seafood, with significant differences to those of cell-based land animals. These considerations include the scientific considerations of working with these unique cell lines, and human factors including cultural, gustatory, and regulatory considerations, all fall within the broader framework of the necessity of this pursuit from a sustainability standpoint (Figure 1). While cell-based seafood shares some common grounding with its land-based analogs in science and human considerations, sustainability is an even more substantial driver, as cell-based seafood could lead to greater preservation of marine environments.
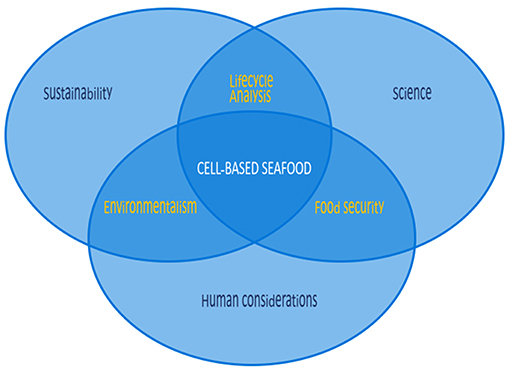
Figure 1. Considerations for cell-based seafood are numerous, differing in many respects from those of cell-based land animals. While cell-based seafood shares some common ground with land-based analogs in science and human considerations, sustainability is a more substantial driver, as cell-based seafood could lead to greater preservation of marine environments.
Preliminary life-cycle analyses indicate that cultured meat production could reduce greenhouse gas emissions by as much as 95% (Tuomisto and Teixeira de Mattos, 2011). However, the cost of these products remains to be seen as these products have not yet reached the market. There is much thought that initial product offerings will be more expensive than conventional meat. It is possible that the costs of cultured meat and fish production might lead them to become the food of the elite—allowing these individuals the opportunity to enjoy the nutritional and food safety benefits, and in some cases bypass the guilt of animal consumption (Stephens et al., 2018).
Certain considerations, such as safety regulation and cultural acceptance, are much harder to quantify, and there is reason to believe that the passage of time and increasing familiarity with cultured foods may support broader acceptance. Cultured meat adoption may prove easier and quicker in countries such as China and India, rather than in the United States (Bryant et al., 2019). There are other reports that indicate cultured fish may have a mixed reception initially; given most individuals prefer a beef burger or a plant-based burger to a cultured meat burger (Slade, 2018). Such preferences may shift over time but do pose short-term challenges for the field.
In addition, safety regulations are not fully established for cellular agriculture products in the United States, United Kingdom, or the EuropeanUnion, though a number of regulation-relevant topics were recently summarized (Stephens et al., 2018). Such regulations have not yet reached cellular aquaculture.
Trends in Seafood Production
While aquaculture is a very old technology—pictorial engravings suggest that Egyptians used aquaculture for fish as early as 2,500 BC (Anon—FAO, 1987)- it was only in the 1970s that aquaculture production became relevant beyond the subsistence and small scale (Asche et al., 2008). Similar to the evolution of agricultural land animal farming, aquaculture has moved from extensive systems that depend on natural environments with minimal human intervention, to semi-intensive and intensive systems, where producers actively control growing conditions through feeding, breeding, disease control and waste removal, resulting in much higher production rates (Asche et al., 2008). With the intensification of aquaculture comes several challenges similar to the intensification of land animal husbandry, including the greater production of nitrogen, and phosphorus waste materials from uneaten feed and metabolic waste products (Yeo et al., 2004). This creates a higher risk of negative environmental impacts (Piedrahita, 2003), such as the damage of waterways and promotion of algal blooms (Paerl et al., 2014). Intensification also increases the risk of pathogen spread from farmed fish to native species (Naylor et al., 2000). Furthermore, an absence of research on the welfare of most aquaculture species has made it difficult to develop welfare standards for farmed aquatic animals, and thus the ethical implications and appropriate welfare regulations of aquaculture remain an open question (Browman et al., 2018).
Aquaculture has recently surpassed marine capture as the main source of seafood for human consumption (Figure 2) (Food FAO, 2018). Global landings for marine capture have remained constant at around 90 million tons per year since 1994, while global aquaculture of fish and shellfish has nearly doubled over the same time period (Food FAO, 2018), thus aquaculture has not relieved the demand for wild-caught seafood (Naylor et al., 2000). While aquaculture is often seen as mitigating overfishing, many farmed fish depend on feed originating from marine capture. Because several marine captured fish are used to feed a single carnivorous farmed fish; aquaculture may be decreasing the global fish supply, rather than increasing it (Naylor and Burke, 2005). While fully-controlled aquaculture environments are more efficient production systems than the use of natural conditions (Tacon and Metian, 2008), to date there is no scientific literature about fully-controlled environments for producing seafood from cell cultures rather than from marine animal husbandry.
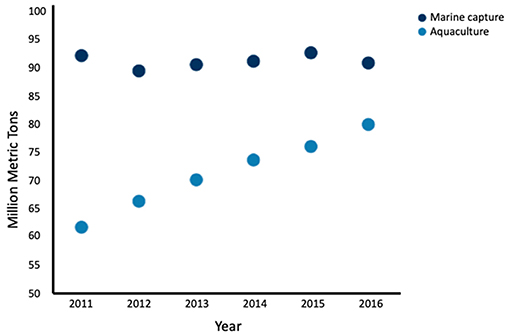
Figure 2. Global seafood production by marine capture and aquaculture by year from 2011 to 2016. Aquaculture production has increased while marine capture has remained fairly constant. Fish, crustaceans, mollusks and other aquatic animals are included, and aquatic mammals, crocodiles, caimans, seaweeds, and other aquatic plants are excluded Data adapted from the FAO (2018).
Engineering Biology for Seafood Production
Genetic modification has resulted in spectacular production increases, feed conversion rates, and reduced development times for fish in closed systems (Muir, 2004). Genetically-modified rohu develop over four times faster than their traditionally bred counterparts (Venugopal et al., 2004), while genetically-modified mud loaches are able to grow 35 times faster and substantially larger (Nam et al., 2001). Like the mud loach, genetically-modified tilapia containing a salmon growth hormone spliced to an ocean pout antifreeze promoter gene can grow over 320% larger than similarly reared non-transgenic fish (Rahman et al., 1998). The OPAFPcsGH gene, which encodes for regulatory elements from ocean pout followed by growth hormone, forms the “all fish” chimera gene sequence used in Aquabounty salmon (Hew and Fletcher, 1996). Aquabounty salmon, approved for manufacture and sale in Canada, develop twice as fast as their wild type counterparts with an impressive feed conversion rate 10% higher than farmed salmon (Bisson, 2015). These fish are formidable food sources and an ecosystem threat—and thus are produced only in closed, in-land systems as triploid, and therefore infertile females (Reichhardt, 2000). Despite these safety measures, significant regulatory hurdles, coupled with legislative pressure, have prevented the fish from entering the US market (Waltz, 2017). These fish are a good example of the power of engineering biology to improve closed-system seafood production. These genome-level advances for closed aquaculture systems may lay important groundwork for cell-based fish production research and development.
Cell-Based Seafood Production
The Progression of Cellular Agriculture
In the past 5 years, research focused on cell-based meats has accelerated from the tasting of the cell-cultured hamburger in 2013 (Zaraska, 2013) and early publications, including life-cycle assessments and basic research (Tuomisto and Teixeira de Mattos, 2011; Post, 2012, 2014), to a growing start-up community, updated life cycle assessments, and publications focused on refining the technologies required to accelerate cell-based meat production (Tuomisto et al., 2014; Krieger et al., 2018; Rubio et al., 2019). To date, research decisions in cell-based meat production, such as selection of cell species and cell type have been largely driven by market size and environmental impact (Rodriguez-Fernandez, 2019), rather than suitability of cells species and types suitable for large scale bioreactor cultivation. This is the result of a general lack of basic research on the cell cultures of commonly-consumed animals.
Cellular agriculture has recently become a topic of interest for regulators in the United States. Given the regulatory jurisdiction of the Food and Drug Administration (FDA) over products of biotechnology and the United States Department of Agriculture over livestock, there is interest in the regulations necessary for potentially jurisdiction over cell-based meat products (Stephens et al., 2018). Despite relative clarity over seafood, which is regulated entirely by the FDA (except for animals in the order Siluriformes) (Kobbeman, 2004), cell-based seafood production, and its potential to address a growing demand for seafood while avoiding the challenges of industrial aquaculture, has remained relatively uninvestigated from a regulatory standpoint.
Basic Methodology for Cell-Based Seafood Production
As with cell-based meat production, cell-based seafood relies on advanced technological developments in the optimization of cell lines, media formulation, and bioreactor design (Figure 3) (Datar and Betti, 2010). Like any closed cell- or tissue-culture system, a cell-based seafood production system would consist of the following integrated elements: appropriate cell type(s) from the tissue of interest; a growth media to provide nutrients to proliferating and differentiating cells, and a bioreactor to provide the closed environment to support the growth. For three-dimensional tissues, a biocompatible scaffold would be needed to provide structure for cell growth and maturation. A muscle protein production system (mpps) for goldfish was developed in 2002 with NASA funding (Benjaminson et al., 2002), however, there are no other examples of cell-based seafood production published during this time period.
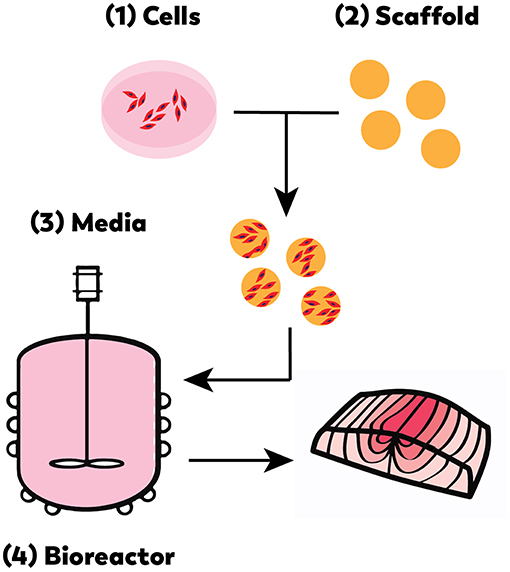
Figure 3. Cell-based seafood production will require the appropriate: (1) cell line(s), (2) compatible scaffold, (3) medium, and (4) bioreactor (Edelman et al., 2005).
Bioreactors are complex closed-system environments for producing biomass that require the constant monitoring (feedback loops for process control), maintenance, and optimization of several parameters. Marine cell cultures may be more forgiving in terms of temperature, pH and oxygen requirements compared to mammalian cell cultures, based on the unique characteristics of native fish muscle tissue. This difference has significant implications for energy use and thus cost considerations for mass production of cell-based seafood. These differences also present a new set of research opportunities for cellular agriculture and toward large-scale tissue engineering in general; most medically-motivated tissue engineering is focused on generating smaller scale tissues, usually no larger than organs (Griffith and Naughton, 2002).
Characteristics of Native Fish Muscle Tissue Compared to Mammalian Cell Culture
Disciplines such as bioengineering, cell biology, and genetics have made significant gains in the understanding of cell and tissue culture over the past 20 years—however these advances have been mostly focused on mammalian systems. Cells and tissues from fish and other marine species have not yet been widely investigated in vitro. In contrast, native fish muscle physiology has been relatively well-researched because of the implications for aquaculture. Some experimentation and assays have been carried out on harvested native muscle tissues from fresh water and marine animals (Castellini and Somero, 1981; Eberlee and Storey, 1984; Anchelin et al., 2011), and these studies provide interesting insights into the potential capabilities of fish muscle in culture.
Native fish muscle consists of three muscle types—each of which differs between species depending on fish type, location, and feed (Kiessling et al., 2006). In teleost and elasmobranch fish, there are three muscle types: red, white, and pink. Red muscle is highly vascularized and is comprised of slow twitch fibers with a high density of mitochondria and a rich supply of capillaries (Johnston, 2001). This muscle is for slow, sustainable swimming speeds, and relies on aerobic metabolic pathways. White muscles are fast twitch, tightly packed with myofibrils, and primarily utilize anaerobic metabolic pathways (Kiessling et al., 2006). White muscle is used for burst-swimming and fast starts. Pink muscle shares some of the characteristics of both white and red muscle (Johnston, 2001).
The variety of fish muscle cell types, which vary in physiology and metabolic pathway, may offer a range of options when designing closed cultivation systems for cell-based seafood production. Native fish muscle also has characteristics related to metabolism, as described below, that, if exploited in ex vivo tissue culture, would make this tissue source uniquely suited for cell-based production.
Oxygen Requirements
Mammalian Cell Culture
Low oxygen concentrations are detrimental to mammalian cell growth and recombinant protein production (Wang et al., 1994), and thus dissolved oxygen concentration and air saturation are important variables that must be monitored and maintained in bioreactor systems. Air saturation levels of 40–60% are often required for mammalian cell culture (Furukawa and Ohsuye, 1998; Link et al., 2004; Zhu et al., 2005). Air saturation and oxygen levels become even more critical in three-dimensional, thick vascularized tissues where oxygen diffusion limits the development of tissues past about 1.8 mm (Griffith et al., 2005).
Fish Cell Culture
Fish are well-adapted to tolerate low oxygen environments as aquatic animals, as they are frequently subjected to low oxygen levels that result in hypoxic conditions (Vaquer-Sunyer and Duarte, 2008). Interestingly, many coral reef fish are tolerant of hypoxic conditions as low as 2.8–0.5% air saturation for the goby Gobiodon histrio and 1.6–0.7% air saturation for the blenny Atrosalarias fuscus before showing signs of distress (Nilsson and Ostlund-Nilsson, 2004). Using a microarray- and bioinformatics-based approach, over two hundred hypoxia-responsive genes were identified in the Japanese medaka brain, gill, and liver ranging from cell metabolism to RNA processing to protein degradation through the ubiquitin system (Zhang et al., 2009). For instance, Hif-1-alpha is a transcription factor that is present under hypoxic conditions–when molecular oxygen is present the protein becomes hydroxylated and subsequently degraded by the ubiquitin system (Ivan et al., 2001). This highly tuned system, discovered in mammals, plays a notable role in the hypoxia response of fish. Analysis of icefish mRNA revealed that the functional domains of Hif-1-alpha are highly conserved, indicating continued selective pressure exerted by hypoxia (Rix et al., 2017). These and other hypoxia response gene relationships have been characterized in detail in the hypoxic fish database, HRGFish, which provides a variety of prediction tools and validation methods to further evaluate the conservation of hypoxia genes in diverse fish species (Rashid et al., 2017).
In the absence of research on the oxygen requirements of fish cell culture in bioreactor systems, the authors speculated that the genetic adaptations of fish to the levels of hypoxia suggest that fish tissues may be uniquely suited to the oxygen-limited environments found in tissue culture and provided via bioreactor conditions. Further, the presence of hypoxia response genes that are conserved in mammals suggest potential targets to increase the ability of mammalian cells to tolerate low oxygen environments.
pH Considerations
Mammalian Cell Culture
Extracellular pH correlates with intracellular pH in mammalian cell culture (Lagadic-Gossmann et al., 2004), while lactic acid, which is formed by cells in anaerobic conditions (Brooks, 2009), accumulates over extended periods of culture in bioreactor systems (Zhao and Ma, 2005). This accumulation contributes to acidification of the bioreactor environment. The well-documented relationship between intracellular acidification and cell death (Lagadic-Gossmann et al., 2004), and because reduction of extracellular pH from 6.8 to 6.3 can render cells quiescent (Taylor and Hodson, 1984), pH control of bioreactor systems requires fine tuning to optimize cell growth. Shifting the pH can also be used as an optimization tool, as changes in pH can affect protein production and glycosylation (Borys et al., 1993), however altering pH to increase protein production and process performance usually reduces cell growth and metabolism (Trummer et al., 2006). For mammalian cells cultures, the pH of a bioreactor system must be actively maintained to counter the build-up of acidic metabolites like lactic acid, for either cell proliferation or protein production, depending on the goals of the culture system.
Fish Cell Culture
A key physiological characteristic of in vivo muscle across species is intracellular buffering capacity (Burton, 1978), the ability of a cell to maintain a neutral pH in the presence of metabolic end products such as lactic acid (Rogatzki et al., 2015). Intracellular buffering capacity is measured in Slykes, or micromoles of base required to change the pH of homogenized tissue by one unit per gram of wet weight (Slyke, 1922).
Land mammals such as pigs have buffering capacities of 49.7 Slykes, and cows 51.9 Slykes, while sea mammals such as the fur seal have buffering capacities of 79.2 Slykes, and spotted dolphins 84.1 Slykes (Castellini and Somero, 1981). White muscle in warm-bodied fishes have particularly high buffering capacities; 102 Slykes in the black skipjack, and 107 Slykes in the albacore or double that of many land mammals (Castellini and Somero, 1981). Further, muscle lactate dehydrogenase (LDH), the enzyme responsible for breaking down lactic acid in muscle tissue, exhibits up to 2-fold higher activity in warm-bodied fish such as the albacore when compared to land animals such as the cow (Castellini and Somero, 1981).
The higher intracellular buffering capacity and the higher muscle lactate dehydrogenase activity unique to warm bodied white fish muscle in vivo suggest that these muscle tissues may be more resilient than mammalian cells in terms of in vitro tissue culture, with an ability to grow well over a wider pH range. More research is required to determine if buffering capacity of native muscle tissue correlates to the buffering capacity of these tissues when cultured in vitro.
Temperature Requirements
Mammalian Cell Culture
Mammalian cells in culture are most often maintained at 37°C, human body temperature, a key factor in mammalian bioreactor process design. However, the viability of mutant Chinese hamster ovary cells was improved by alternating culture temperatures between 39 and 34°C (Jenkins and Hovey, 1993), while cultures grown at 30°C can live longer and produce more protein (Moore et al., 1997) than those cultivated at 37°C.
Fish Cell Culture
Fish vary widely in their adaptation strategies to frigid arctic and Antarctic environments, speciating at a higher rate near the earth's poles than near the equator (Rabosky et al., 2018). Some of these adaptations have resulted in the development of antifreeze defense systems that are optimized for temperatures of 0° to 10°C (Abele and Puntarulo, 2004) and an increased production of antioxidants, such as marine-derived tocopherol, which is found in greater quantities in salmon from cold waters than fish from equatorial waters (Yamamoto et al., 2001). Other adaptations include unique oxygen carrying and metabolic capabilities (Verde et al., 2008). In some cases, these adaptations manifest as even the lack of hemoglobin, as is the case in the demersal icefish, Chaenocephalus aceratus; these fish compensate for their lack of hemoglobin with increased numbers of mitochondria (Johnston, 1987). Adaptations in arctic species exhibit muscle design and mitochondrial density similar to high-performance adaptations in more equatorial species (Pörtner, 2002). Fish cell culture conditions typically mirror those of their typical habitats, with culture temperatures varying from 15 to 30°C. Some fish cell lines can vary their rate of metabolism within a 5°C temperature range, with the cells metabolizing more quickly at higher temperatures (Courtenay and Williams, 2004). Since fish cells can be cultured at cooler temperatures, this reduces the energy required to maintain a constant temperature of a culture system, and costs associated with producing cell-based seafood at scale. Thus, cell-based seafood may offer cost benefits at scale compared to cell-based meats.
Fish Cell Lines
Until recently, fish cell lines have found limited applications, mainly to test viral load, to examine water toxicology, and to generate vaccines for farmed fish (Wolf and Mann, 1980; Lai et al., 2008). Few of these cell lines have been utilized to produce edible fish; except for a study focused on producing fish-based proteins for astronauts (Benjaminson et al., 2002). The American Type Culture Collection (ATCC) contains over 4,000 cell lines, however, it does not currently contain any fish muscle cell lines (Hay et al., 1992).
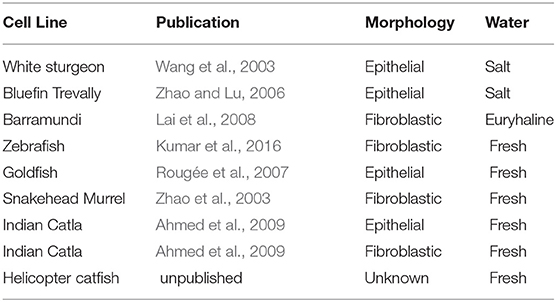
Few cell line databases include fish cell lines, while two (FICELdb and Cellosaurus) do so. FICELdb is a fish cell database that refers to many legacy lines dating from 1962 to 1999 (Braga et al., 2006). Cellosaurus is a cell line compendium created in 2012 and published in 2018, housed within ExPASy, a database maintained by the Swiss Institute for Bioinformatics (Bairoch, 2018). It consists of cell line repositories from around the world, categorizing over 100,000 known cell lines (Artimo et al., 2012). Interestingly, only 558 are immortalized fish cell lines, only nine are lines of fish cells isolated from fish muscle, none of which are myoblastic, all of which have spontaneously immortalized (Bairoch, 2018). While it is likely that not all immortalized fish cell lines are represented in these databases, the nine fish cell lines identified in Cellosaurus are displayed in Table 1.
Two of the nine fish cells lines originated from salt water species, both of which are epithelial in nature: a white sturgeon line spontaneously immortalized in 2003, and a bluefin trevally line from 2006. The cell line originating from barramundi, a euryhaline fish, is fibroblastic. The remaining six fish cell lines originate from fresh water species. Zebrafish Brachydanio rerio and Goldfish Carassius auratus are commonly-known model organisms and pets; the remaining cell lines originate from commonly cultivated species in Southeast Asia; the snakehead murrel, a common food fish in Thailand; the Sahul Indian catla, a farmed fish that is often grown with other carp species, and the helicopter catfish, a species farmed commercially in Malaysia. Of interest, a goldfish cell line, while fibroblastic in nature, was successfully utilized to grow edible fish filets (Benjaminson et al., 2002).
While these nine cell lines have been useful for culturing a number of fish pathogens, none have yet been used for bioengineering or tissue engineering applications, and none are myoblastic lines. Derivation and characterization of myoblastic cell lines from various fish would be an important resource for researchers advancing cell-based seafood production. The availability of fish cell lines that are well-characterized could be invaluable for cell-based seafood research; similar to the availability of C2C12 cells and their impact on mammalian muscle tissue engineering (Burattini et al., 2004).
Cell Isolation Methods
Lack of fish cell lines means that most fish tissue culture work is conducted on primary cells isolated from fish. Fish cell isolation methods are similar to mammalian cell isolations (An Introduction to Mammalian Cell Culture, 2016). In general, the fish is initially sterilized in ethanol, anesthetized, and a tissue sample is removed with a biopsy. The tissue sample is then either explanted or enzymatically-digested with collagenase or trypsin. Explanted tissues are allowed to attach to the culture plate, and cells migrate from the tissue to the culture surface. Culture plates can be coated with proteins to enhance cell adhesion (e.g., gelatin, collagen, laminin). Enzymatically-digested tissues are in prepared aqueous solutions, where digestion by trypsin or collagenase releases the cells into the liquid medium. The cells are then rinsed with buffer to remove contaminants and are often filtered to liberate the cells from the residual debris (Oestbye and Ytteborg, 2018).
While these techniques work well for isolating tissue from adult fish, recent research has begun investigating the isolation of cells from embryonic teleosts (Anchelin et al., 2011). Embryonic cells from many wild species and wild fish that are artificially fertilized at fish hatcheries are challenging to isolate. Additionally, many species of fish have high telomerase activity, so primary embryonic cells do not senesce as quickly as in mammalian systems (Anchelin et al., 2011). Importantly, while fish do not have embryonic stem cells per se, they do have embryonic progenitors that may be coaxed down different differentiation pathways (Ghosh et al., 1997; Ma et al., 2001; Ciarlo et al., 2017).
As more research is carried out on fish muscle cell types, it should be possible to isolate cell types and populations of interest. Magnetic beads coupled to muscle-progenitor-specific antibodies can facilitate rapid isolation of these cells with minimal damage (Plouffe et al., 2015). Additionally, fluorescence-activated cell sorting (FACS) is an effective technique for cell isolation. The main challenge is identifying cell-surface proteins that distinguish these progenitor and muscle cells; particularly because the conservation in sequence with mammalian extracellular proteins is very low (Liongue and Ward, 2007), so most mammalian antibodies do not cross-react with fish-derived cells, thus limiting isolation and identification options.
Culture Conditions
Extracellular Environments and Scaffolds
The extracellular matrix (ECM) that fish cells need to survive and proliferate in vitro is a key area of research interest; most mammalian cell lines are plated on tissue-culture plates coated with vacuum gas plasma, which makes the polystyrene surface more hydrophilic. Atlantic salmon primary muscle cells have been successfully cultured and differentiated on laminin-coated tissue culture plates (Oestbye and Ytteborg, 2018). However, fish cells may require surfaces or scaffolds of different ECM proteins such as elastins, collagens, fibronectin, and laminin to adhere and grow efficiently, and may also require fish glycoaminoglycans (Frantz et al., 2010; Arima et al., 2013). Optimizing these ECM proteins would also likely be critical to efficient fish culture in vitro, especially since fish protein glycosylation patterns are different from mammals.
Growth Media
Variables to consider for fish cell culture growth media include salt concentration, buffer, temperature, carbon source, and pH. Past growth media formula suggest a trial and error approach that may not be optimal, including the use of mammalian or insect cell formulations such as Eagle's Medium, Modified Eagle's Medium (MEM), Medium 199 (M199) and Leibowitz's 15 (L-15) medium (Fernandez et al., 1993). These media are fully defined and easily replicated, including the content of inorganic salts, amino acids, and basic nutrients for culture growth. Additional salts are also added for some marine species such as the grunt (Clem et al., 1961). Fetal bovine serum (FBS) and fetal calf serum (FCS) are common additives (Arora, 2013). The use of bicarbonate in media formulations aids cell growth and buffering capabilities as well (Fernandez et al., 1993). The ability to control the buffering capacity of the media is important; some fish cells require growth at 5% carbon dioxide, but others utilize anoxic or standard oxygen tension (Fernandez et al., 1993). These conditions vary between species (and even between tissues from the same fish) but can be controlled for cell-based seafood production. The addition of growth factors, such as fibroblast growth factor (FGF2) has proven to be helpful for the growth of some muscle cell cultures, including epithelial tuna cells in L-15 media with 10–20% FBS (Bain et al., 2013). Attempts to lower the FBS content resulted in reduced cell proliferation, while the addition of vitamin E and fatty acids improved proliferation (Scholefield and Schuller, 2014).
Removing FBS or FCS from the media for cell-based seafood production would be advantageous due to the high cost, their animal sources, and their potential for carrying mammalian viruses and prions (Van der Valk et al., 2010). Investigations into the molecular components of fish serum should be useful to identify and then generate in vitro the required components to keep fish cells active in culture. Importantly, fats are nutritionally-relevant in seafood, especially omega-3 long-chain polyunsaturated fatty acids (PUFAs), eicosapentaenoic acid (EPA) and docosahexaenoic acid (DHA) (Wall et al., 2010). More research is necessary to understand how media composition affects the nutritional quality and taste of cell-based seafood. EPA and DHA are normally produced by marine algae, and studies have focused on large-scale production of omega-3 fatty acids from algal biomass; these studies indicate that optimization of algal growth and EPA and DHA production has strong potential for optimizing the industrial production of PUFAs (Chauton et al., 2015; Hamilton et al., 2015) and their inclusion into the media and cells of in vitro seafood cultures. The potential for non-animal sourcing of these PUFAs is environmentally attractive, avoiding ethical considerations, reducing potential contamination of cell-based seafood with animal pathogens, and also significantly reducing costs.
Crustacean Cell Culture
Cell Lines
Crustacean cell culture research has been pursued for use in aquaculture and pharmaceutical industries for studying diseases affecting farmed seafood and to identify biologics with human clinical relevance (Zhao et al., 2003). Attempts have been made to establish cell lines from crustacean tissues to improve isolation and maintenance methodology, including the use of short-term cultures obtained from repeated primary cell isolations (Rinkevich, 2005). Short-term cultures are sufficient for bench-scale research but not sufficient for cell-based seafood applications with long-term goals of mass production and commercialization; thus studies into the long-term culture of these cells are essential.
To advance crustacean tissue culture for food purposes, new strategies for developing immortalized or long-term cell lines of relevant lineages such as muscle and fat are needed. Previous crustacean tissue culture studies have instead largely focused on ovarian epithelial and fibroblast cells (Luedeman and Lightner, 1992; Fan and Wang, 2002; Maeda et al., 2003). Studies that examined muscle, eye stalk, and hepatopancreas cells in crustaceans found poor survival and slow growth compared to the ovary-derived epithelial and fibroblast cells (George and Dhar, 2010). The most frequently studied crustacean genus is Penaeus which includes shrimp and prawn species. Studies with this species have derived primary cell cultures from ovary, hepatopancreas, nerve, lymphoid and hematopoietic tissue (Rinkevich, 2005). However, to date no continuous crustacean cell lines have been established (Rinkevich, 2005).
Significant effort is needed to derive and characterize crustacean cells for use in cell-based seafood research and development. It is possible that establishment of such cell lines could also be relevant to the aquaculture and pharmaceutical sectors.
Cell Isolation Methods
Cell isolation procedures for crustaceans and vertebrates share some similarities in terms of animal age and digestion procedures. For example, cells isolated from younger “pre-molting” prawns have a higher success rate for survival and growth (Tong and Miao, 1996). This is comparable to how more proliferative cells are correlated with younger mammal and avian donors (Doumit et al., 1990; Mesires and Doumit, 2002). Cell cultivation studies often attempt to isolate from both explanted and enzyme-digested tissues. For the explant approach, which is better suited for less dense tissues, the target tissue is harvested from the animal, rinsed in buffer solution, dissected into small sections and allowed to attach to a tissue culture surface. The cells within the explant then proliferate, migrate, and adhere to the substrate. For the enzyme digestion method, digestive enzymes (e.g., trypsin, collagenase) are incubated with the dissected explants to degrade the tissue into single cells in suspension. The slurry is often filtered to remove undigested tissue and the enzymes are inhibited after a certain amount of incubation time to prevent cell damage. For cell isolations from most shrimp tissues, the explant method tends to be more successful than enzyme-digestion method in terms of cell proliferation, although enzyme digestion is preferred for hepatopancreas tissues (Ma et al., 2017).
Culture Conditions
Variables such as temperature, pH and osmolality differ between vertebrate and invertebrate cell culture. The incubation temperature for invertebrates is lower than for vertebrates and typically falls within the range of 25–30°C (Hink, 1979). The pH of shrimp and prawn cell cultures is typically within pH 7.0-7.6 (Ma et al., 2017). Osmolality can range between 472 and 760 mmol/kg depending on the hemolymph osmolality of the origin animal (Chen et al., 1986; Rinkevich, 2005). Carbon dioxide exchange is generally not necessary for invertebrate tissue culture because the basal media tends to be buffered by phosphates rather than sodium bicarbonate. While some of the reported crustacean cell cultures (e.g., Penaeus monodon gonad, heart, and epidermis cells) reach confluency and survive for extended periods of time with routine media changes, the cultures often degenerate after a few rounds of subculture (Chen et al., 1986; Owens and Smith, 1999). This loss of vitality in culture during passaging can be due to many factors, but may be in part due to sensitivity of the cells to the enzymes employed during passaging.
Like fish cells, crustacean cells can be cultured at lower temperatures (Toullec, 1999) suggesting an energy savings opportunity for large scale bioreactor cultures compared to mammalian cells. Similarly, crustacean cell culture does not require carbon dioxide exchange (Ma et al., 2017) obviating the need for a common mammalian cell culture bioreactor parameter control system.
Growth Media
The basal medium for invertebrate cell culture commonly consists of Grace's medium or L-15 medium, though formulations such as M199 have exhibited superior effects in some studies (Ma et al., 2017). The standard for mammalian cell culture is basal media (e.g., Dulbecco's Modified Eagle Medium, RPMI 1640, DMEM/F-12) supplemented with animal serum from domestic species like cows, horses or chickens. Fetal bovine serum is also important for shrimp cell culture at a concentration of 10–20% (Ma et al., 2017). However, this strategy has not been sufficient to maintain crustacean cells in culture long-term, so supplemented media with marine-relevant factors such as lipid solutions or hemolymph extracted from various sea creatures has been explored (Chen et al., 1986; Rinkevich, 2005). Common supplements include shrimp or crab muscle extract and hemolymph from Penaeus species (Ma et al., 2017). These additions likely provide trace and critical growth factors not supplied by mammalian serum. Some growth factors have been identified to improve shrimp lymphoid and ovarian cultures, including epidermal growth factor and transforming growth factor beta (Ma et al., 2017).
More study is needed in the development of a sustainable, animal-free media for the growth of crustacean cells. The emphasis on animal-based media likely reflects the lack of understanding of the fundamental metabolic and growth requirements for crustacean cells and tissues.
Mollusk Considerations
Buffering Capacity
Buffering capacity varies widely in mollusks with some species exhibiting particularly high buffering capacity (Eberlee and Storey, 1984). The channeled whelk, Busycotypus canaliculatus, has an aerobic buffering capacity in the hepatopancreas of 79.4 plus or minus 17.2 Slykes, and a remarkable buffering capacity of almost 120 Slykes following 24 h of anoxic stress (Hetrick et al., 1981). While heart muscle tissues appear to have a lower buffering capacity, anoxic stress increases the buffering capacity to over 60 Slykes (Eberlee and Storey, 1984), a value that is still higher than that of many land animals. The high buffering capacity of mollusks suggests an intrinsic ability of mollusk cell cultures to tolerate a wider pH range than mammalian cell cultures; assumptions that will require additional research.
Cell Culture
Mollusk culture has been even more elusive than fish or crustacean cell cultures. While primary cell cultures have been attained, continuous cultures of mollusk cell lines have yet to achieve sustained proliferation in vitro (Chen and Wang, 1999). This includes attempts at long-term cultures of clam and oyster lines with 138 unique different media formulations (Hetrick et al., 1981). Some mitotic activity was observed in scallop cell cultures over 4 months (Odintsova and Khomenko, 1991), and in the surf clam, Spisula soldissima, although the media in the later study contained fetal calf serum and whole egg extract (Cecil, 1969). An exception to the challenge of proliferative cultures are from abalone species cultivated for pearl production and hemocyte (blood cell) lines used for toxicological studies. The mantle cells derived from tissue explants of Haliotis varia has been successfully cultured for 370 days in Medium 199 containing additional salts, lactalbumin, kanamycin, sodium bicarbonate and fetal bovine serum (Suja and Dharmaraj, 2005). The mantle of H. varia was grown in serum free media containing whole-body extract (Suja et al., 2007). Even fewer mollusk muscle cells have been isolated compared to fish. Clam primary cardiomyocytes were grown up to a month (Hanana et al., 2011) and primary cultures of clam heart tissue originating from Meretrix luxoria was successfully cultured for over 5 months (Chen and Wang, 1999).
Mollusk cell culture has been understudied, with demonstrated challenges in establishing long term, proliferative, food-relevant cell lines. The success in the abalone industry suggests the opportunities are there with sufficient focus on the cell sources and cultivation conditions.
Scaffolds for Three-Dimensional Tissue Cultivation
Cultivating three-dimensional tissues relies on the presence of a scaffold, a biocompatible material capable of supporting cell growth and differentiation by providing a suitable morphology, chemical and structural template. Numerous scaffolding materials that are employed in medical tissue engineering could be relevant to biofabricated food such as cellulose, alginate and chitosan. Chitosan is of particular interest because it is edible, inexpensive, accessible and well-referenced in tissue engineering literature. Chitosan is a food-relevant material that is derived from chitin; a primary component of insect and crustacean exoskeletons and one of the most prevalent biopolymers on earth (along with cellulose). Within exoskeletons, chitin exists as nanofibril structures, providing mechanical strength to the cuticle (Ifuku, 2014). Chitosan can also be derived from non-animal sources like fungi, algae, and yeast (Croisier and Jérôme, 2013). Chitosan powder can be prepared from crustacean, mushroom or microbial sources after the isolation of chitin and the chemical deacetylation to generate chitosan. This material is then dissolved into an aqueous solution which can be cast into a variety of formats such as membranes, hydrogels, and sponges (Croisier and Jérôme, 2013). Fungal chitosan is a preferred source as it is: (1) non-allergenic, (2) customizable in terms of molecular weight, and (3) approved for human consumption as a food additive or nutrition supplement (Pochanavanich and Suntornsuk, 2002; Wu et al., 2004; Nitschke et al., 2011).
Scaffold Fabrication
Chitosan can be dissolved in acidic solution, such as 1–2% acetic acid in distilled water. The viscosity of the solution increases rapidly with an increase in concentration (Jana et al., 2013). Many scaffolds that support various cell cultures use chitosan solutions as low as 0.5% (Katalinich, 2001). To create films to support cell culture, chitosan solutions are cast on glass substrates and allowed to dry, then rinsed with buffer to neutralize the acetyl groups, washed with water or PBS and sterilized with 70% ethanol and UV light prior to cell seeding (Rubio et al., 2019). Chitosan can also be manipulated to produce physically associated or cross-linked hydrogels (Drury and Mooney, 2003), porous sponges with tunable mechanical properties and pore size distributions (Jana et al., 2013; Rubio et al., 2019) and dry or wet spun fibers (Croisier and Jérôme, 2013).
Discussion
Given the progression of aquaculture toward more intensive, controlled and efficient systems, cell-based seafood production offers a new option to potentially avert the challenges associated with industrial aquaculture and marine capture. The opportunities and challenges in the field of cell-based seafood production were reviewed, covering aspects of marine cell culture, native marine muscle tissue, and marine animal considerations. Because of the minimal literature on marine cell cultures, some insights and speculations on the characteristics of marine cell cultures are provided, based on the unique properties of native muscle tissue in fish.
Overall, fish muscle tissue may be well-suited for bioreactor cultivation relative to mammalian muscle tissues, given the ability of fish tissues to: (1) endure hypoxic conditions, reducing the need for active oxygenation in oxygen-limited bioreactor environments; (2) tolerate pH ranges, potentially establishing a wider range of pH in which cell growth is feasible; and (3) growth at lower temperatures, potentially reducing heat transfer needs of bioreactor cultivation at scale and reducing production costs.
A review of the literature on crustacean cell culture indicates that there is very little research that is directly relevant to cell-based food production. A review of the literature on mollusk is even more sparse. This dearth in direct insight into marine organisms related to cell isolations, cultivation conditions and related needs provides a unique opportunity for scientific activity to help propel the field forward, not only for the field of cellular agriculture, but also for novel marine bioproducts and unique metabolic insights with potential translation to benefit human health and the environment.
Approaches to cell-based seafood production can range from large scale cell cultivation, resulting in large masses of seafood-relevant cells and tissues for applications in processed seafoods like surimi; to three-dimensional tissue cultivation, resulting in structured products more akin to filets. Chitosan is a promising, marine-relevant biomaterial for three-dimensional tissue culture applications. It is customizable, edible, and widely available; making it a suitable material to explore for cell-based seafood production, as well as for other tissue engineering and cellular agriculture applications.
There could be several advantages to producing seafood from cell cultures rather than whole marine animals. The production of cell-based seafood has the potential to alter many fundamental parameters considered immutable in food cultivation, including the production of inedible excess tissues such as bones, skin, shells, and scales, that are often discarded and can negatively impact the environment. Cell-based seafood may also shorten cycle time; cell cultures may require weeks to months to generate functional foods; while by comparison, genetically-modified Aquabounty salmon require 18 months to grow to market size, roughly half the time of a normal salmon (Waltz, 2017).
Conclusions
Producing seafood from marine cell cultures is a novel seafood production method and an intriguing opportunity for cellular agriculture. A survey of relevant literature reveals that marine cell and tissue culture is an enormously neglected field of research. Few cell lines are available from marine species and none are directly relevant to seafood production. Despite the properties of marine muscle tissue that suggest this as a promising opportunity for bioreactor cultivation, the behavior of marine cells in large scale culture environments remains speculative. There are several research gaps that exist for marine cell culture, alongside several opportunities that make these research gaps worth addressing. With growing interest in cellular agriculture as a means to produce meat, milk, eggs, and other animal proteins from cell cultures, and with the rapid intensification of aquaculture systems, the time is right to investigate the production of seafood without marine animals.
Author Contributions
KK oversaw the preparation and writing of the manuscript and themes, contributing sections on mollusk cell culture, and oxygen and pH considerations. NR contributed material on crustacean cell culture and chitosan biomaterials, and DS contributed to fish tissue isolation, embryonic cells, and muscle cell type sections. ID and DK contributed to the organization, writing and editing of the manuscript.
Funding
New Harvest is a 501(c)(3) research institute that supports basic research in cellular agriculture The research for this review was performed using funding to New Harvest from an anonymous donor.
Conflict of Interest Statement
DS is a scientific advisor for Finless Foods.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge Bradley Silverman for his insights on process engineering considerations, Arif Malik for his insights on the fish musculature section of this review, Andrew Stout for his insights on the Warburg effect, Kevin Cadena for his cultured meat illustrations, Mike Selden for his introductions.
References
Abele, D., and Puntarulo, S. (2004). Formation of reactive species and induction of antioxidant defence systems in polar and temperate marine invertebrates and fish. Comp. Biochem. Physiol. 138, 405–415. doi: 10.1016/j.cbpb.2004.05.013
Ahmed, V. I., Babu, V. S., Chandra, V., Nambi, K. S., Thomas, J., Bhonde, R., et al. (2009). A new fibroblastic-like cell line from heart muscle of the Indian major carp (Catla catla): development and characterization. Aquaculture 293, 180–186. doi: 10.1016/j.aquaculture.2009.05.012
An Introduction to Mammalian Cell Culture (2016). An Introduction to Mammalian Cell Culture. Available online at: https://www.aiche.org/resources/publications/cep/2016/april/introduction-mammalian-cell-culture
Anchelin, M., Murcia, L., Alcaraz-Pérez, F., García-Navarro, E. M., and Cayuela, M. L. (2011). Behaviour of telomere and telomerase during aging and regeneration in Zebrafish. PLoS ONE 6:16955. doi: 10.1371/journal.pone.0016955
Arima, K., Fujita, H., Toita, R., Imazu-Okada, A., Tsutsumishita-Nakai, N., Takeda, N., et al. (2013). Amounts and compositional analysis of glycosaminoglycans in the tissue of fish. Carbohydrate Res. 366, 25–32. doi: 10.1016/j.carres.2012.11.010
Artimo, P., Jonnalagedda, M., Arnold, K., Baratin, D., Csardi, G., De Castro, E., et al. (2012). ExPASy: SIB bioinformatics resource portal. Nucl. Acids Res. 40, W597–W603. doi: 10.1093/nar/gks400
Asche, F., Roll, K. H., and Tveterås, S. (2008). “Future trends in aquaculture: productivity growth and increased production,” in Aquaculture in the Ecosystem, eds M. Holmer, K. Black, C. M. Duarte, N. Marbà, and I. Karakassis (Dordrecht: Springer Netherlands), 271–292. doi: 10.1007/978-1-4020-6810-2_9
Bain, P. A., Hutchinson, R. G., Marks, A. B., Crane, M. S. J., and Schuller, K. A. (2013). Establishment of a continuous cell line from southern bluefin tuna (Thunnus maccoyii). Aquaculture 376, 59–63. doi: 10.1016/j.aquaculture.2012.11.008
Bairoch, A. (2018). The cellosaurus, a cell-line knowledge resource. J. Biomol. Tech. 29, 25–38. doi: 10.7171/jbt.18-2902-002
Benjaminson, M. A., Gilchriest, J. A., and Lorenz, M. (2002). In vitro edible muscle protein production system (MPPS): Stage 1, fish. Acta astronautica 51, 879–889. doi: 10.1016/S0094-5765(02)00033-4
Bisson, R. (2015). AquaBounty: Label The Salmon, for Everyone's Sake. The Guardian. Available online at: https://www.theguardian.com/sustainable-business/2015/dec/21/aquabounty-salmon-fda-gmo-foods-labeling (accessed September 7, 2018).
Borys, M. C., Linzer, D. I. H., and Papoutsakis, E. T. (1993). Culture pH affects expression rates and glycosylation of recombinant mouse placental lactogen proteins by chinese hamster ovary (CHO) Cells. Nat. Biotechnol. 11, 720–724. doi: 10.1038/nbt0693-720
Braga, D., Laizé, V., Tiago, D. M., and Cancela, M. L. (2006). Enhanced DNA transfer into fish bone cells using polyethylenimine. Mol. Biotechnol. 34, 51–54. doi: 10.1385/MB:34:1:51
Brooks, G. A. (2009). Cell–cell and intracellular lactate shuttles. J. Physiol. 587, 5591–5600. doi: 10.1113/jphysiol.2009.178350
Browman, H. I., Cooke, S. J., Cowx, I. G., Derbyshire, S. W. G., Kasumyan, A., Key, B., et al. (2018). Welfare of aquatic animals: where things are, where they are going, and what it means for research, aquaculture, recreational angling, and commercial fishing. ICES J. Mar. Sci. 76, 82–92. doi: 10.1093/icesjms/fsy067
Bryant, C., Szejda, K., Parekh, N., Desphande, V., and Tse, B. (2019). A survey of consumer perceptions of plant-based and clean meat in the USA, India, and China. Front. Sustain. Food Syst. 3:11. doi: 10.3389/fsufs.2019.00011
Burattini, S., Ferri, P., Battistelli, M., Curci, R., Luchetti, F., and Falcieri, E. (2004). C2C12 murine myoblasts as a model of skeletal muscle development: morpho-functional characterization. Eur. J. Histochem. 48, 223–234. doi: 10.4081/891
Burton, R. F. (1978). Intracellular buffering. Respir. Physiol. 33, 51–58. doi: 10.1016/0034-5687(78)90083-X
Castellini, M. A., and Somero, G. N. (1981). Buffering capacity of vertebrate muscle: correlations with potentials for anaerobic function. J. Comp. Physiol. B 143, 191–198. doi: 10.1007/BF00797698
Cecil, J. T. (1969). Mitoses in cell cultures from cardiac tissue of the surf clam Spisula solidissima. J. Invertebr. Pathol. 14, 407–410. doi: 10.1016/0022-2011(69)90170-0
Chauton, M. S., Reitan, K. I., Norsker, N. H., Tveter, R., and Kleivdal, H. T. (2015). A techno-economic analysis of industrial production of marine microalgae as a source of EPA and DHA-rich raw material for aquafeed: Research challenges and possibilities. Aquaculture 436, 95–103. doi: 10.1016/j.aquaculture.2014.10.038
Chen, S. N., CHI, S.-C., Kou, G. H., and Liao, I. C. (1986). Cell culture from tissues of grass prawn, Penaeus monodon. Fish Pathol. 21, 161–166. doi: 10.3147/jsfp.21.161
Chen, S. N., and Wang, C. S. (1999). Establishment of cell lines derived from oyster, crassostrea gigas thunberg and hard clam, meretrix lusoria röding. Methods Cell Sci. 21, 183–192. doi: 10.1023/A:1009829807954
Ciarlo, C., Kaufman, C. K., Kinikoglu, B., Michael, J., Yang, S., D′Amato, C., et al. (2017). A chemical screen in zebrafish embryonic cells establishes that Akt activation is required for neural crest development. eLife 6:e29145. doi: 10.7554/eLife.29145
Clem, L. W., Moewus, L., and Michael Sigel, M. (1961). Studies with cells from marine fish in tissue culture. Proc. Soc. Exp. Biol. Med. 108, 762–766. doi: 10.3181/00379727-108-27059
Courtenay, P. W. R., and Williams, J. D. (2004). Snakeheads (Pisces, Channidae): A Biological Synopsis and Risk Assessment. Reston, VA: United States Geological Survey.
Croisier, F., and Jérôme, C. (2013). Chitosan-based biomaterials for tissue engineering. Eur. Poly. J. 49, 780–792. doi: 10.1016/j.eurpolymj.2012.12.009
Datar, I., and Betti, M. (2010). Possibilities for an in vitro meat production system. Innov. Food Sci. Emerg. Technol. 11, 13–22. doi: 10.1016/j.ifset.2009.10.007
Doumit, M. E., McFarland, D. C., and Minshall, R. D. (1990). Satellite cells of growing turkeys: influence of donor age and sex on proliferation and differentiation in vitro. Exper. Cell Res. 189, 81–86. doi: 10.1016/0014-4827(90)90259-D
Drury, J. L., and Mooney, D. J. (2003). Hydrogels for tissue engineering: scaffold design variables and applications. Biomaterials 24, 4337–4351. doi: 10.1016/S0142-9612(03)00340-5
Eberlee, J. C., and Storey, K. B. (1984). Buffering capacities of the tissues of marine molluscs. Physiol. Zool. 57, 567–572. doi: 10.1086/physzool.57.5.30163950
Edelman, P. D., McFarland, D. C., Mironov, V. A., and Matheny, J. G. (2005). Commentary: in vitro-cultured meat production. Tissue Eng. 11, 659–662. doi: 10.1089/ten.2005.11.659
Fan, T.-J., and Wang, X.-F. (2002). In vitro culture of embryonic cells from the shrimp, Penaeus chinensis. J. Exp. Marine Biol. Ecol. 267, 175–184. doi: 10.1016/S0022-0981(01)00364-1
FAO (1987). FAO Fisheries & Aquaculture-National Aquaculture Sector Overview-Egypt. Available online at: http://www.fao.org/fishery/countrysector/naso_egypt/en
FAO (2018). The State of World Fisheries and Aquaculture 2018 - Meeting the Sustainable Development Goals. Rome: FAO. Licence: CC BY-NC-SA 3.0 IGO.
Fernandez, R. D., Yoshimizu, M., Ezura, Y., and Kimura, T. (1993). Comparative growth response of fish cell lines in different media, temperatures, and sodium chloride concentrations. Fish Pathol. 28, 27–34. doi: 10.3147/jsfp.28.27
Food FAO (2018). The State of World Fisheries and Aquaculture 2018. Rome: Contributing to Food Security and Nutrition for All.
Frantz, C., Stewart, K. M., and Weaver, V. M. (2010). The extracellular matrix at a glance. J. Cell. Sci. 123, 4195–4200. doi: 10.1242/jcs.023820
Funk, C. C., and Brown, M. E. (2009). Declining global per capita agricultural production and warming oceans threaten food security. Food Security 1, 271–289. doi: 10.1007/s12571-009-0026-y
Furukawa, K., and Ohsuye, K. (1998). Effect of culture temperature on a recombinant CHO cell line producing a C-terminal α-amidating enzyme. Cytotechnology 26, 153–164. doi: 10.1023/A:1007934216507
George, S. K., and Dhar, A. K. (2010). An improved method of cell culture system from eye stalk, hepatopancreas, muscle, ovary, and hemocytes of Penaeus vannamei. In Vitro Cell. Dev. Biol. Anim. 46, 801–810. doi: 10.1007/s11626-010-9343-x
Ghosh, C., Liu, Y., Ma, C., and Collodi, P. (1997). Cell cultures derived from early zebrafish embryos differentiate in vitro into neurons and astrocytes. Cytotechnology 23, 221–230. doi: 10.1023/A:1007915618413
Griffith, C. K., Miller, C., Sainson, R. C. A., Calvert, J. W., Jeon, N. L., Hughes, C. C. W., et al. (2005). Diffusion limits of an in vitro thick prevascularized tissue. Tissue Eng. 11, 257–266. doi: 10.1089/ten.2005.11.257
Griffith, L. G., and Naughton, G. (2002). Tissue engineering–current challenges and expanding opportunities. Science 295, 1009–1014. doi: 10.1126/science.1069210
Hamilton, M. L., Warwick, J., Terry, A., Allen, M. J., Napier, J. A., and Sayanova, O. (2015). Towards the industrial production of omega-3 long chain polyunsaturated fatty acids from a genetically modified diatom Phaeodactylum tricornutum. PLoS ONE 10:e0144054. doi: 10.1371/journal.pone.0144054
Hanana, H., Talarmin, H., Pennec, J. P., Droguet, M., Gobin, E., Marcorelle, P., et al. (2011). Establishment of functional primary cultures of heart cells from the clam Ruditapes decussatus. Cytotechnology 63, 295–305. doi: 10.1007/s10616-011-9347-8
Hay, R. J., Caputo, J., and Macy, M. L. (1992). ATCC Quality Control Methods for Cell Lines. Manassas, VA: Amer Type Culture Collection.
Hetrick, F. M., Stephens, E., Lomax, N., and Lutrell, K. (1981). “Attempts to develop a marine molluscan cell line,” in College Park, University of Maryland. Sea Grant College Program Technical Report UM-SG-TS-81–S-06.
Hew, C. L., and Fletcher, G. L. (1996). Transgenic Salmonid Fish Expressing Exogenous Salmonid Growth Hormone. Introduction To Aquaculture. Available online at: http://www.fao.org/docrep/field/003/ac169e/ac169e00.htm (accessed October 4, 2018).
Hink, W. F. (1979). “[39] Cell lines from invertebrates,” in Methods in Enzymology, eds W. B. Jakoby and I. H. Pastan (Elsevier), 450–466. doi: 10.1016/S0076-6879(79)58160-9
Ifuku, S. (2014). Chitin and chitosan nanofibers: Preparation and chemical modifications. Molecules 19, 18367–18380. doi: 10.3390/molecules191118367
Ivan, M., Kondo, K., Yang, H., Kim, W., Valiando, J., Ohh, M., et al. (2001). HIFalpha targeted for VHL-mediated destruction by proline hydroxylation: implications for O2 sensing. Science 292, 464–468. doi: 10.1126/science.1059817
Jana, S., Cooper, A., and Zhang, M. (2013). Chitosan scaffolds with unidirectional microtubular pores for large skeletal myotube generation. Adv. Healthcare Mater. 2, 557–561. doi: 10.1002/adhm.201200177
Jenkins, N., and Hovey, A. (1993). Temperature control of growth and productivity in mutant Chinese hamster ovary cells synthesizing a recombinant protein. Biotechnol. Bioeng. 42, 1029–1036. doi: 10.1002/bit.260420903
Johnston, I. A. (1987). Respiratory characteristics of muscle fibres in a fish (Chaenocephalus aceratus) that lacks haem pigments. J. Experi. Biol. 133, 415–428.
Johnston, I. A. (2001). Fish Physiology: Muscle Development and Growth. Houston, TX: Gulf Professional Publishing.
Katalinich, M. (2001). Characterization of Chitosan Films for Cell Culture Applications. Orono, ME: University of Maine Digital Library.
Kiessling, A., Ruohonen, K., and Bjørnevik, M. (2006). Muscle fibre growth and quality in fish. Archiv fur Tierzucht. 49, 137–141.
Kobbeman, K. E. (2004). Hook, line and sinker: how congress swallowed the domestic catfish industry's narrow definition of this ubiquitous bottomfeeder. Ark. L. Rev. 57:407.
Krieger, J., Park, B.-W., Lambert, C. R., and Malcuit, C. (2018). 3D skeletal muscle fascicle engineering is improved with TGF-β1 treatment of myogenic cells and their co-culture with myofibroblasts. PeerJ. 6:e4939. doi: 10.7717/peerj.4939
Kumar, A., Singh, N., Goswami, M., Srivastava, J. K., Mishra, A. K., and Lakra, W. S. (2016). Establishment and characterization of a new muscle cell line of Zebrafish (Danio rerio) as an in vitro model for gene expression studies. Anim. Biotechnol. 27, 166–173. doi: 10.1080/10495398.2016.1147455
Lagadic-Gossmann, D., Huc, L., and Lecureur, V. (2004). Alterations of intracellular pH homeostasis in apoptosis: origins and roles. Cell Death Different. 11:953. doi: 10.1038/sj.cdd.4401466
Lai, Y.-S., Chiou, P. P., Chen, W.-J., Chen, Y.-C., Chen, C.-W., Chiu, I.-S., et al. (2008). Characterization of apoptosis induced by grouper iridovirus in two newly established cell lines from barramundi, Lates calcarifer (Bloch). J. Fish Dis. 31, 825–834. doi: 10.1111/j.1365-2761.2008.00957.x
Lindgren, E., Harris, F., Dangour, A. D., Gasparatos, A., Hiramatsu, M., Javadi, F., et al. (2018). Sustainable food systems—a health perspective. Sustainab. Sci. 13, 1505–1517. doi: 10.1007/s11625-018-0586-x
Link, T., Bäckström, M., Graham, R., Essers, R., Zörner, K., Gätgens, J., et al. (2004). Bioprocess development for the production of a recombinant MUC1 fusion protein expressed by CHO-K1 cells in protein-free medium. J. Biotechnol. 110, 51–62. doi: 10.1016/j.jbiotec.2003.12.008
Liongue, C., and Ward, A. C. (2007). Evolution of class I cytokine receptors. BMC Evolut. Biol. 7:120. doi: 10.1186/1471-2148-7-120
Luedeman, R., and Lightner, D. V. (1992). Development of an in vitro primary cell culture system from the penaeid shrimp, Penaeus stylirostris and Penaeus vannamei. Aquaculture 101, 205–211. doi: 10.1016/0044-8486(92)90024-F
Ma, C., Fan, L., Ganassin, R., Bols, N., and Collodi, P. (2001). Production of zebrafish germ-line chimeras from embryo cell cultures. Proc. Natl. Acad. Sci. U.S.A. 98, 2461–2466. doi: 10.1073/pnas.041449398
Ma, J., Zeng, L., and Lu, Y. (2017). Penaeid shrimp cell culture and its applications. Rev. Aquaculture 9, 88–98. doi: 10.1111/raq.12106
Maeda, M., Mizuki, E., Itami, T., and Ohba, M. (2003). Ovarian primary tissue culture of the kuruma shrimp Marsupenaeus japonicus. In Vitro Cell. Dev. Biol. Anim. 39, 208–212. doi: 10.1290/1543-706X(2003)039<0208:OPTCOT>2.0.CO;2
Merino, G., Barange, M., Blanchard, J. L., Harle, J., Holmes, R., Allen, I., et al. (2012). Can marine fisheries and aquaculture meet fish demand from a growing human population in a changing climate? Global Environ. Change 22, 795–806. doi: 10.1016/j.gloenvcha.2012.03.003
Mesires, N. T., and Doumit, M. E. (2002). Satellite cell proliferation and differentiation during postnatal growth of porcine skeletal muscle. Am. J. Physiol. Cell Physiol. 282, C899–C906. doi: 10.1152/ajpcell.00341.2001
Moore, A., Mercer, J., Dutina, G., Donahue, C. J., Bauer, K. D., Mather, J. P., et al. (1997). Effects of temperature shift on cell cycle, apoptosis and nucleotide pools in CHO cell batch cultues. Cytotechnology 23, 47–54. doi: 10.1023/A:1007919921991
Muir, W. M. (2004). The threats and benefits of GM fish. EMBO Rep. 5, 654–659. doi: 10.1038/sj.embor.7400197
Myers, R. A., and Worm, B. (2003). Rapid worldwide depletion of predatory fish communities. Nature 423:280. doi: 10.1038/nature01610
Nam, Y. K., Noh, J. K., Cho, Y. S., Cho, H. J., Cho, K. N., Kim, C. G., et al. (2001). Dramatically accelerated growth and extraordinary gigantism of transgenic mud loach Misgurnus mizolepis. Transgenic Res. 10, 353–362. doi: 10.1023/A:1016696104185
Naylor, R., and Burke, M. (2005). Aquaculture and ocean resources: raising tigers of the sea. Annu. Rev. Environ. Resour. 30, 185–218. doi: 10.1146/annurev.energy.30.081804.121034
Naylor, R. L., Goldburg, R. J., Primavera, J. H., Kautsky, N., Beveridge, M. C., Clay, J., et al. (2000). Effect of aquaculture on world fish supplies. Nature 405:1017. doi: 10.1038/35016500
Nilsson, G. E., and Ostlund-Nilsson, S. (2004). Hypoxia in paradise: widespread hypoxia tolerance in coral reef fishes. Proc. Biol. Sci. 271 (Suppl. 3), S30–S33. doi: 10.1098/rsbl.2003.0087
Nitschke, J., Altenbach, H.-J., Malolepszy, T., and Mölleken, H. (2011). A new method for the quantification of chitin and chitosan in edible mushrooms. Carbohydrate Res. 346, 1307–1310. doi: 10.1016/j.carres.2011.03.040
Odintsova, N. A., and Khomenko, A. V. (1991). Primary cell culture from embryos of the Japanese scallop Mizuchopecten yessoensis (Bivalvia). Cytotechnology 6, 49–54. doi: 10.1007/BF00353702
Oestbye, T. K., and Ytteborg, E. (2018). Preparation and culturing of atlantic salmon muscle cells for in vitro studies. Myogenesis 1889, 319–330. doi: 10.1007/978-1-4939-8897-6_19
Owens, L., and Smith, J. (1999). Early attempts at production of prawn cell lines. Methods Cell Sci. 21, 207–212. doi: 10.1023/A:1009806606562
Paerl, H. W., Gardner, W. S., McCarthy, M. J., Peierls, B. L., and Wilhelm, S. W. (2014). Algal blooms: noteworthy nitrogen. Science 346, 175–175. doi: 10.1126/science.346.6206.175-a
Piedrahita, R. H. (2003). Reducing the potential environmental impact of tank aquaculture effluents through intensification and recirculation. Aquaculture 226, 35–44. doi: 10.1016/S0044-8486(03)00465-4
Plouffe, B. D., Murthy, S. K., and Lewis, L. H. (2015). Fundamentals and application of magnetic particles in cell isolation and enrichment. Rep. Prog. Phys. 78:016601. doi: 10.1088/0034-4885/78/1/016601
Pochanavanich, P., and Suntornsuk, W. (2002). Fungal chitosan production and its characterization. Lett. Appl. Microbiol. 35, 17–21. doi: 10.1046/j.1472-765X.2002.01118.x
Pörtner, H.-O. (2002). Climate variations and the physiological basis of temperature dependent biogeography: systemic to molecular hierarchy of thermal tolerance in animals. Compa. Biochem. Physiol. Part A 132, 739–761. doi: 10.1016/S1095-6433(02)00045-4
Post, M. J. (2012). Cultured meat from stem cells: challenges and prospects. Meat Sci. 92, 297–301. doi: 10.1016/j.meatsci.2012.04.008
Post, M. J. (2014). An alternative animal protein source: cultured beef. Anna. N. York Acad. Sci. 1328, 29–33. doi: 10.1111/nyas.12569
Rabosky, D. L., Chang, J., Title, P. O., Cowman, P. F., Sallan, L., Friedman, M., et al. (2018). An inverse latitudinal gradient in speciation rate for marine fishes. Nature 559, 392–395. doi: 10.1038/s41586-018-0273-1
Rahman, M. A., Mak, R., Ayad, H., Smith, A., and Maclean, N. (1998). Expression of a novel piscine growth hormone gene results in growth enhancement in transgenic tilapia (Oreochromis niloticus). Transgenic Res. 7, 357–369. doi: 10.1023/A:1008837105299
Rashid, I., Nagpure, N. S., Srivastava, P., Kumar, R., Pathak, A. K., Singh, M., et al. (2017). HRGFish: a database of hypoxia responsive genes in fishes. Sci. Rep. 7:42346. doi: 10.1038/srep42346
Reichhardt, T. (2000). Will Souped Up Salmon Sink or Swim? London, UK: Nature Publishing Group. doi: 10.1038/35017657
Rinkevich, B. (2005). Marine invertebrate cell cultures: new millennium trends. Marine Biotechnol. 7, 429–439. doi: 10.1007/s10126-004-0108-y
Rix, A. S., Grove, T. J., and O'Brien, K. M. (2017). Hypoxia-inducible factor-1α in Antarctic notothenioids contains a polyglutamine and glutamic acid insert that varies in length with phylogeny. Polar Biol. 40, 2537–2545. doi: 10.1007/s00300-017-2164-6
Rodriguez-Fernandez, C. (2019). Beyond the lab-grown burger: “cellular agriculture” is taking over the food industry. Labiotech.eu. Available online at: https://labiotech.eu/features/cellular-agriculture-food-industry/
Rogatzki, M. J., Ferguson, B. S., Goodwin, M. L., and Gladden, L. B. (2015). Lactate is always the end product of glycolysis. Front. Neurosci. 9:00022. doi: 10.3389/fnins.2015.00022
Rougée, L., Ostrander, G. K., Richmond, R. H., and Lu, Y. (2007). Establishment, characterization, and viral susceptibility of two cell lines derived from goldfish Carassius auratus muscle and swim bladder. Dis. Aquat. Organ. 77, 127–135. doi: 10.3354/dao01802
Rubio, N. R., Fish, K. F., Trimmer, B. A., and Kaplan, D. L. (2019). In vitro insect muscle for tissue engineering applications. ACS Biomater. Sci. Eng. 5, 1071–1082. doi: 10.1021/acsbiomaterials.8b01261
Scholefield, A. M., and Schuller, K. A. (2014). Cell proliferation and long chain polyunsaturated fatty acid metabolism in a cell line from southern bluefin tuna (Thunnus maccoyii). Lipids 49, 703–714. doi: 10.1007/s11745-014-3910-y
Sexton, A. E. (2018). Eating for the post-Anthropocene: alternative proteins and the biopolitics of edibility. Transac. Institute Br. Geogr. 43, 586–600. doi: 10.1111/tran.12253
Slade, P. (2018). If you build it, will they eat it? Consumer preferences for plant-based and cultured meat burgers. Appetite 125, 428–437. doi: 10.1016/j.appet.2018.02.030
Slyke, D. D. V. (1922). On the measurement of buffer values and on the relationship of buffer value to the dissociation constant of the buffer and the concentration and reaction of the buffer solution. J. Biol. Chem. 52, 525–570.
Stephens, N., Di Silvio, L., Dunsford, I., Ellis, M., Glencross, A., and Sexton, A. (2018). Bringing cultured meat to market: technical, socio-political, and regulatory challenges in cellular agriculture. Trends Food Sci. Technol. 78, 155–166. doi: 10.1016/j.tifs.2018.04.010
Suja, C. P., and Dharmaraj, S. (2005). In vitro culture of mantle tissue of the abalone Haliotis varia Linnaeus. Tissue Cell 37, 1–10. doi: 10.1016/j.tice.2004.08.002
Suja, C. P., Sukumaran, N., and Dharmaraj, S. (2007). Effect of culture media and tissue extracts in the mantle explant culture of abalone, Haliotis varia Linnaeus. Aquaculture 271, 516–522. doi: 10.1016/j.aquaculture.2007.04.086
Tacon, A. G. J., and Metian, M. (2008). Global overview on the use of fish meal and fish oil in industrially compounded aquafeeds: trends and future prospects. Aquaculture 285, 146–158. doi: 10.1016/j.aquaculture.2008.08.015
Taylor, I. W., and Hodson, P. J. (1984). Cell cycle regulation by environmental pH. J. Cell. Physiol. 121, 517–525. doi: 10.1002/jcp.1041210310
Tidwell, J. H., and Allan, G. L. (2001). Fish as food: aquaculture's contribution. EMBO Rep. 2, 958–963. doi: 10.1093/embo-reports/kve236
Tong, S., and Miao, H.-Z. (1996). Attempts to initiate cell cultures from Penaeus chinensis tissues. Aquaculture 147, 151–157. doi: 10.1016/S0044-8486(96)01386-5
Toullec, J.-Y. (1999). Crustacean primary cell culture: a technical approach. Methods Cell Sci. 21, 193–198. doi: 10.1023/A:1009833924792
Trummer, E., Fauland, K., Seidinger, S., Schriebl, K., Lattenmayer, C., Kunert, R., et al. (2006). Process parameter shifting: Part I. Effect of DOT, pH, and temperature on the performance of Epo-Fc expressing CHO cells cultivated in controlled batch bioreactors. Biotechnol. Bioeng. 94, 1033–1044. doi: 10.1002/bit.21013
Tuomisto, H. L., Ellis, M. J., and Haastrup, P. (2014). “Environmental impacts of cultured meat: alternative production scenarios,” in Proceedings of the 9th International Conference on Life Cycle Assessment in the Agri-Food Sector (San Francisco, CA), 8–10.
Tuomisto, H. L., and Teixeira de Mattos, M. J. (2011). Environmental impacts of cultured meat production. Environ. Sci. Technol. 45, 6117–6123. doi: 10.1021/es200130u
Van der Valk, J., Brunner, D., De Smet, K., Fex Svenningsen, A. A., Honegger, P., Knudsen, L. E., et al. (2010). Optimization of chemically defined cell culture media–replacing fetal bovine serum in mammalian in vitro methods. Toxicol. In Vitro 24, 1053–1063. doi: 10.1016/j.tiv.2010.03.016
Vaquer-Sunyer, R., and Duarte, C. M. (2008). Thresholds of hypoxia for marine biodiversity. PNAS 105, 15452–15457. doi: 10.1073/pnas.0803833105
Venugopal, T., Anathy, V., Kirankumar, S., and Pandian, T. J. (2004). Growth enhancement and food conversion efficiency of transgenic fish Labeo rohita. J. Exper. Zool. Part A. 301A, 477–490. doi: 10.1002/jez.a.78
Verde, C., Giordano, D., and di Prisco, G. (2008). The adaptation of polar fishes to climatic changes: structure, function and phylogeny of haemoglobin. IUBMB Life 60, 29–40. doi: 10.1002/iub.1
Wall, R., Ross, R. P., Fitzgerald, G. F., and Stanton, C. (2010). Fatty acids from fish: the anti-inflammatory potential of long-chain omega-3 fatty acids. Nutri. Rev. 68, 280–289. doi: 10.1111/j.1753-4887.2010.00287.x
Waltz, E. (2017). First genetically engineered salmon sold in Canada. Nat. News 548:148. doi: 10.1038/nature.2017.22116
Wang, G., LaPatra, S., Zeng, L., Zhao, Z., and Lu, Y. (2003). Establishment, growth, cryopreservation and species of origin identification of three cell lines from white sturgeon, Acipenser transmontanus. Methods Cell Sci. 25, 211–220. doi: 10.1007/s11022-004-9120-x
Wang, J., Honda, H., Park, Y. S., Iijima, S., and Kobayashi, T. (1994). “Effect of Dissolved Oxygen Concentration on Growth and Production of Biomaterials by Animal Cell Culture,” in Animal Cell Technology: Basic and Applied Aspects: Proceedings of the Sixth International Meeting of the Japanese Association for Animal Cell Technology, Nagoya, Japan, November 9–12, 1993 The Sixth International Meeting of Japanese Association for Animal Cell Technology JAACT'93, eds T. Kobayashi, Y. Kitagawa, and K. Okumura (Dordrecht: Springer Netherlands, 191–195.
Wolf, K., and Mann, J. A. (1980). Poikilotherm vertebrate cell lines and viruses: a current listing for fishes. In Vitro 16, 168–179. doi: 10.1007/BF02831507
Wu, T., Zivanovic, S., Draughon, F. A., and Sams, C. E. (2004). Chitin and chitosan value-added products from mushroom waste. J. Agri. Food Chem. 52, 7905–7910. doi: 10.1021/jf0492565
Yamamoto, Y., Fujisawa, A., Hara, A., and Dunlap, W. C. (2001). An unusual vitamin E constituent (α-tocomonoenol) provides enhanced antioxidant protection in marine organisms adapted to cold-water environments. Proc. Natl. Acad. Sci. U.S.A. 98, 13144–13148. doi: 10.1073/pnas.241024298
Yeo, S. E., Binkowski, F. P., and Morris, J. E. (2004). Aquaculture Effluents and Waste By-Products Characteristics, Potential Recovery, and Beneficial Reuse. Ames, IA: NCRAC Technical Bulletins.
Zhang, Z., Ju, Z., Wells, M. C., and Walter, R. B. (2009). Genomic approaches in the identification of hypoxia biomarkers in model fish species. J. Experi. Marine Biol. Ecol. 381, S180–S187. doi: 10.1016/j.jembe.2009.07.021
Zhao, F., and Ma, T. (2005). Perfusion bioreactor system for human mesenchymal stem cell tissue engineering: dynamic cell seeding and construct development. Biotechnol. Bioeng. 91, 482–493. doi: 10.1002/bit.20532
Zhao, Z., and Lu, Y. (2006). Establishment and characterization of two cell lines from bluefin trevally Caranx melampygus. Dis. Aquat. Organ. 68, 91–100. doi: 10.3354/dao068091
Zhao, Z., Montgomery-Brock, D., Lee, C.-S., and Lu, Y. (2003). Establishment, characterization and viral susceptibility of 3 new cell lines from snakehead, Channa striatus (Blooch). Methods Cell Sci. 25, 155–166. doi: 10.1007/s11022-004-3804-0
Keywords: cellular agriculture, cell-based seafood, fish tissue culture, bioreactor, serum-free media, ocean conservation, marine cell culture, aquaculture
Citation: Rubio N, Datar I, Stachura D, Kaplan D and Krueger K (2019) Cell-Based Fish: A Novel Approach to Seafood Production and an Opportunity for Cellular Agriculture. Front. Sustain. Food Syst. 3:43. doi: 10.3389/fsufs.2019.00043
Received: 08 November 2018; Accepted: 21 May 2019;
Published: 11 June 2019.
Edited by:
Selena Ahmed, Montana State University, United StatesReviewed by:
Peter Neubauer, Technische Universität Berlin, GermanySteve Oh, Bioprocessing Technology Institute (A*STAR), Singapore
Copyright © 2019 Rubio, Datar, Stachura, Kaplan and Krueger. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kate Krueger, a2F0ZUBuZXctaGFydmVzdC5vcmc=
 Natalie Rubio
Natalie Rubio Isha Datar2
Isha Datar2 Kate Krueger
Kate Krueger