
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst. , 16 April 2019
Sec. Agro-Food Safety
Volume 3 - 2019 | https://doi.org/10.3389/fsufs.2019.00020
This article is part of the Research Topic Mobilization of antimicrobial resistance traits from agriculture and aquaculture to food View all 5 articles
 Tameka N. Bythwood1
Tameka N. Bythwood1 Vivek Soni1
Vivek Soni1 Karen Lyons1
Karen Lyons1 Anne Hurley-Bacon1
Anne Hurley-Bacon1 Margie D. Lee1†
Margie D. Lee1† Charles Hofacre1
Charles Hofacre1 Susan Sanchez2
Susan Sanchez2 John J. Maurer1*†
John J. Maurer1*†The rise in Salmonella resistance to cephalosporins and fluoroquinolones has become a significant threat to public health. At issue, is whether agricultural use of antimicrobials is selecting antibiotic resistance in Salmonella and the degree to which large antimicrobial resistance gene reservoirs, present in animal manures, contribute to this resistance. Two in vivo studies were performed to address these questions. In the first study, chickens were administered Salmonella and commensals, including an Escherichia coli strain with a mobile, ceftiofur-resistance plasmid, in order to determine how antibiotic administration impacted resistance in E. coli and Salmonella. All antibiotics administered to chickens increased streptomycin resistance in E. coli. However, only ceftiofur administration increased resistance in Salmonella and specifically to extended-spectrum β-lactams and cephalosporins (ESBL). There was no significant increase in ESBL-resistant Salmonella in chickens administered a ceftiofur-resistance plasmid donor. In the second study, chickens were administered two different isolates of S. enterica Typhimurium and a chicken resistome to serve as a gene donor. Birds were subsequently administered chlortetracycline or streptomycin. Antimicrobial administration significantly altered aminoglycoside and tetracycline resistance in the Enterobacteriaceae population. However, there was no significant increase in antimicrobial resistant Salmonella. Administration of a chicken resistome had no significant impact on prevalence of resistance in Enterobacteriaceae populations, including Salmonella. Evident, from both studies, was that these treatments had minimal effect on increasing the prevalence of resistance in Salmonella, suggesting that other factors may be more important in dissemination of antimicrobial resistant Salmonella in chickens.
Salmonella enterica is the 3rd leading cause of foodborne illnesses globally; accounting for 78 million illnesses and 59 thousand foodborne-related deaths annually (Havelaar et al., 2015). In the United States, non-typhoidal Salmonella is responsible for 1 million illnesses, 19 thousand hospitalizations, and 378 deaths each year (Scallan et al., 2011). Children (≤5 years old) and the elderly (>65 years old) are most susceptible to Salmonella infection, requiring some medical intervention (Scallan et al., 2013, 2015, 2018). Antibiotic therapeutics can lessen the severity and length of illness (Hu et al., 2014). However, the rise in antimicrobial-resistant Salmonella has become a significant public health concern. Especially problematic is its reduced susceptibility to antimicrobials commonly prescribed to treat salmonellosis (Wasyl et al., 2015; Iwamoto et al., 2017; Tyson et al., 2017; Duong et al., 2018) and documented cases of treatment failure (Collard et al., 2007; Tribble, 2017; Duong et al., 2018).
As most Salmonella infections are associated with consumption of meats, milk, and eggs, the veterinary use of antimicrobials, especially those important for treatment of humans, has long been a contentious issue (Cox and Ricci, 1969). While there is epidemiological data to support the link between antimicrobial use and resistance (Bortolaia et al., 2015; Noyes et al., 2016; Andersen et al., 2017; Caffrey et al., 2017; Shigemura et al., 2018), it is generally difficult to ascertain causal effect depending on the antimicrobial (Fairchild et al., 2005; Smith et al., 2007; Agga et al., 2016), food animal (Agga et al., 2016), level or scope of food animal production system (Idris et al., 2006; Andersen et al., 2017; Liljebjelke et al., 2017), or method of quantifying resistance (Fairchild et al., 2005; Smith et al., 2007; Agga et al., 2016). In poultry production, the antimicrobials available for use in the US are limited to 15 drugs (Singer and Hofacre, 2006). The antimicrobials with Gram-negative spectrum are chlortetracycline, oxytetracycline, streptomycin, gentamicin, and ceftiofur. The likelihood that any of these antimicrobials are used are mostly dependent on economics, weighing the cost of the drug against the level of disease (Singer and Hofacre, 2006). A commercial poultry farm may go a full year without any therapeutic antimicrobial use (Liljebjelke et al., 2017).
Antimicrobial resistance in Salmonella is dependent on the acquisition of resistance genes from its environment. As Salmonella is not naturally transformable (Lorenz and Wackernagel, 1994), resistance gene transfer is therefore reliant on cell-to-cell contact (Sieckmann et al., 1969) with another cell harboring a conjugative genetic element with resistance gene(s). Therefore, the population size of antibiotic resistance donors is an important factor, influencing antimicrobial resistance in Salmonella. If this is an especially small population, antibiotic selection pressure would be expected to play an important role in amplifying the donor population (Schjørring et al., 2008) or the newly acquired resistance in Salmonella.
The Enterobacteriaceae, which includes member species Salmonella and Escherichia coli, are a minor component of the chicken gastrointestinal microbiome (Lu et al., 2003a) and the poultry litter bacterial community (Lu et al., 2003b). However, Salmonella and E. coli isolated from the poultry environment tend to harbor class 1 integrons, which are genetic elements that can capture and integrate antimicrobial resistance genes within the bacterial host genome (Bass et al., 1999; Goldstein et al., 2001). The poultry litter bacterial community contains gram-positive species that have a very high prevalence of class 1 integrons and have the same integron-associated antimicrobial resistance genes as Gram-negatives cultured from the same environment (Nandi et al., 2004). This suggests that the poultry litter community is an important reservoir of resistance elements. There exists a diversity of integron-associated resistance genes in this environment, including resistances to β-lactams, phenicols, sulfonamides, aminoglycosides, and quaternary ammonium (Lu et al., 2003b; Nandi et al., 2004; Smith et al., 2007). In addition, the poultry environment also has a diverse array of tetracycline resistance genes with evidence of horizontal gene transfer among evolutionarily disparate genera, as evident in the presence of tetO in Campylobacter and Enterococcus (Fairchild et al., 2005). It is therefore not surprising that Salmonella isolated from the poultry environment can possess a diverse array of antimicrobial resistances, even on farms where there has been no therapeutic use of antimicrobials (Liljebjelke et al., 2017). However, it is not clear what factors have the highest impact on resistance in Salmonella. One would expect an increase in resistant Salmonella in birds raised in a deep litter system such as is common in the US, regardless of therapeutic antimicrobial treatment. The objective of this study was to determine the impact of resistance reservoirs and antimicrobial administration on prevalence of antimicrobial resistant Salmonella in chickens. The studies revealed that while there were significant changes in resistance in Gram-negative commensals, with the exception of ceftiofur, most antibiotics did not increase prevalence of antibiotic-resistant Salmonella.
Salmonella enterica Typhimurium isolates, 934 and 3147, were used as recipients in antimicrobial resistance transfer experiments. The Salmonella isolates were obtained from a commercial poultry farm in northeast Georgia and represent the same strain as determined by PFGE (pulsed-field gel electrophoresis) (Liljebjelke et al., 2017). Typhimurium isolate 934 is resistant to streptomycin and sulfisoxazole and 3147 is only resistant to sulfisoxazole. Isolate 3147 contains a class 1 integron (intI1) with an empty integration site (Lévesque et al., 1995). Rifampicin and nalidixic acid resistant variants were selected by plating 100 μl of an overnight culture on Luria Bertani (LB) (Murooka and Harada, 1994) agar with increasing concentrations of each antibiotic to obtain resistance to 64 μg/ml (Kruse and Sørum, 1994). Therefore, strains used in subsequent experiments will be referred to as R (rifampicin-resistant) or NR (nalidixic acid and rifampicin-resistant) if the phenotype was important for detection or selection of transconjugants.
The multi-drug resistance plasmid (floR, blaCMY2, strA, tetA) harbored by S. Newport strain 14407 was moved into nalidixic-resistant E. coli strain 1932 (Wooley et al., 1992) through filter mating (Taylor et al., 1981). Escherichia coli strain 1932 harboring the resistance plasmid was used as a plasmid donor in both in vitro and in vivo studies. Ceftiofur and florfenicol-susceptible Salmonella enterica serovars Enteritidis, Heidelberg, Infantis, Kentucky, Montevideo, and Typhimurium isolates were made rifampicin (64 μg/ml) resistant as previously described, and used in in vitro conjugation experiments in order to assess the plasmid transfer rate in multiple strain backgrounds. The florfenicol resistance gene, floR was used as a marker in PCR screening of transconjugants (Keyes et al., 2000).
Plasmid donor and rifampicin-resistant Salmonella recipient strains were grown in LB at 37°C overnight in standing culture. Strains were diluted in 5 ml of fresh LB broth, 1/500, at a donor to recipient ratio of 1:1. Cultures were subsequently incubated at 37°C standing overnight then diluted 10-fold in PBS and plated on LB agar with rifampicin alone (recipients), and LB agar with rifampicin and florfenicol (transconjugants), for enumeration. Plates were incubated overnight at 37°C. Plasmid transfer frequency was calculated by taking the total number of transconjugants divided by the total number of recipients.
Plasmid segregation rate was determined for a Typhimurium 934R transconjugant passaged for 5 serial cultures in LB broth with no antibiotic selection using the following protocol. Transconjugant was streaked onto LB with florfenicol and incubated overnight at 37°C, as a standing culture then a single colony was used to inoculate LB broth with florfenicol and incubated overnight at 37°C. One hundred microliters was used to inoculate 100 ml of LB with no antibiotics, and incubated overnight at 37°C. Following overnight incubation, the broth culture was diluted 10-fold and plated onto LB agar with no antibiotics and LB agar with florfenicol. The overnight culture was used to inoculate fresh LB and repeated for four more passages. Plate counts were recorded daily and plasmid segregation rate was calculated as described by Modi and Adams (Modi and Adams, 1991).
To measure the fitness cost of the multidrug-resistance plasmid on isolate 934R, transconjugant and recipient were grown alone or together in LB broth with no antibiotics. Seed cultures in LB broth with and without antibiotics were incubated overnight at 37°C. The next day, 1 ml of the overnight broth culture was transferred to microfuge tubes and centrifuged at 4,500 × g for 10 min to pellet bacterial cells. The supernatant was decanted and the cells were suspended in 1 ml of deionized H2O. Fifty microliters of each cell suspension was used to inoculate 10 ml of LB with no antibiotics, alone or together, of which 10-fold serial dilutions (10−4 – 10−7) were plated onto LB agar with and without florfenicol (0 h). After incubation at 37°C for 24 h serial 10-fold dilutions were plated onto LB agar with and without florfenicol and plates were incubated at 37°C for 24 h. Fitness cost was calculated as described by Melnyk et al. (2015) and Lenski et al. (1994).
Three hundred and sixty, 1 day-old SPF chicks were separated into 12 groups and placed into individual pens (30 birds per pen) in a research flock house, as described in Table 1. The pen floors had been sanitized with Vircon S (Lanxess Corp.; Pittsburgh, PA) and covered with fresh pine shavings. For ceftiofur administration group, 1 day old, chicks received a subcutaneous injection of ceftiofur (0.1 mg/chick). At 1 day of age, chicks were orally inoculated with Typhimurium 934R (106 CFUs/chick) or Typhimurium 934R with Escherichia coli strain 1932 harboring Newport multidrug-resistance plasmid (106 CFUs/chick). At 14 days of age, some groups were given antibiotics in their drinking water continuously for 3 consecutive days (0.60 g/gallon streptomycin or 1.45 g/gallon oxytetracycline).
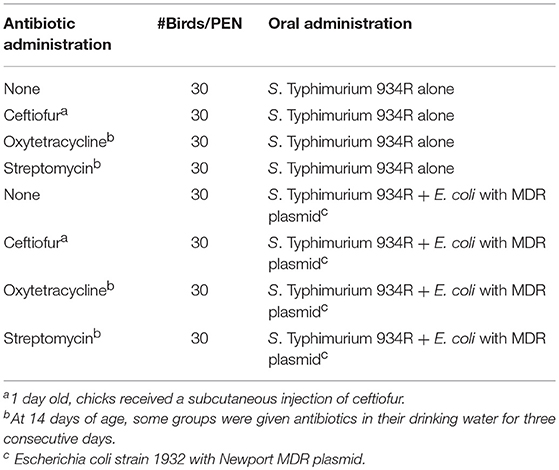
Table 1. Animal Study 1: Does antibiotic administration enhance acquisition of the Newport multidrug-resistance plasmid by Typhimurium 934R in chickens?.
Fresh cecal droppings were collected from the pens on days 13, 15, and 20 by placing 0.75 m × 1 m sheets of paper in each pen. Three pools of 10 cecal samples were recovered from each paper and placed into 50 ml centrifuge tube, weighed and diluted 1/10 ml in freezer stock solution (1% peptone and 15% glycerol in water), and aliquots were placed into three 1 ml Eppendorf tubes for freezer storage at −80°C.
Resistant bacteria were isolated by plating onto selective media as follows. Salmonella were isolated onto Xylose Lysine Tergitol 4 (XLT4) agar (Sigma; St. Louis, MO) plates containing ampicillin (25 μg/ml), florfenicol (32 μg/ml), streptomycin (32 μg/ml), or tetracycline (10 μg/ml). Total Salmonella Typhimurium 934R were enumerated by plating onto XLT containing rifampicin (64 μg/ml). Resistant coliforms were enumerated by plating onto MacConkey agar containing ampicillin (25 μg/ml), florfenicol (32 μg/ml), streptomycin (32 μg/ml), or tetracycline (10 μg/ml). Plates were incubated at 37°C for 18–22 h. Isolates were stocked in freezer stock medium and stored at −80°C.
The chicken resistome was prepared by administering used litter to chicks and collecting the subsequent resistant cecal microbiome. Eighteen days old, embryonating chicken eggs from specific pathogen free (SPF), white leghorn chickens (Charles River Laboratories; Willmington, MA) were cleaned and disinfected by immersing in pre-warmed (100°F) 10% commercial bleach solution (James Austin Co.; Mars, PA) for approximately 3 min as previously described (Wang and Slavik, 1998). Fifty-eight day of hatch, SPF chickens were separated into four groups containing 14 chickens per Horsfall unit. Prior to placement of chicks, HEPA-filtered, Horsfall isolator units were disinfected by washing with bleach solution followed by fumigation with formaldehyde for approximately 24 h (Williams, 1970). Each bird was orally inoculated with 0.5 ml of pooled, poultry litter samples harvested from commercial broiler farms in the Northeast Georgia area; this litter microbiota had been shown previously to harbor a variety of antimicrobial resistance genes (Lu et al., 2003b). At 3 weeks, post-inoculation, chicks were sacrificed by cervical dislocation. The cecal contents were pooled, homogenized for 5 min at maximum speed using a stomacher (Tekmar Company; Cincinnati, OH) with 1 ml of phosphate buffered saline (PBS, pH = 8). Homogenates were filtered through gauze and confirmed to be Salmonella-free by culture as described below. The resistome homogenate was administered to broiler chicks in subsequent experiments.
Experiments were conducted with day of hatch, broiler chicks housed in research flock houses at stocking densities simulating commercial conditions. Broiler eggs, hatching incubators, and floor pen houses at the Poultry Diagnostic and Research Center were disinfected and fumigated with formaldehyde. Five days prior to placing broiler chicks, the floor pens, walls and floors were randomly sampled with drag swabs (Liljebjelke et al., 2005) and confirmed as Salmonella-negative according to procedures described below. Cobb/Cobb broiler chicks (Siloam Springs, AR) were collected from incubators at day of hatch and transferred to an adjacent floor pen house containing 3 × 3 m pens lined with fresh softwood shavings. Chicks were divided into 16 groups of ~50 birds per pen, as described in Table 2.
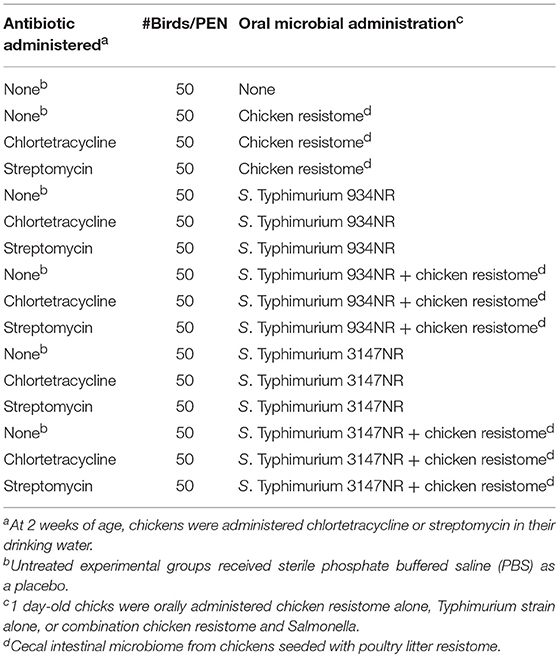
Table 2. Animal Study 2: Does antibiotic administration enhance acquisition of antimicrobial resistance in Salmonella in vivo from the chicken resistome?
Salmonella inoculum was prepared by streaking onto XLT4NR (containing nalidixic acid and rifampicin), incubating at 37°C overnight which was used to inoculate 500 ml Brain Heart Infusion broth (Sigma) which was incubated at 37°C standing for 4 h. Approximately 107 CFU were administered to the broiler chicks on day of hatch. One day following oral inoculation, a 1 ml suspension of litter resistome was added to the chicks' drinking water. All birds were given clean drinking water daily ad libitum and fed commercial feed supplemented with monensin (Elanco Animal Health; Indianapolis, IN) (110 g/ton) and bacitracin methylene disalicylate (Zoetis Inc.; Parippany, NJ) (55 g/ton). At 2 weeks of age, the birds were administered chlortetracycline (55 mg/kg, body weight) or streptomycin (33 mg/kg, body weight) via their water drinkers for 1 week. Control birds received no chlortetracycline or streptomycin. The colony house environment was sampled weekly for Salmonella by drag swabs (Sigma) as previously described (Liljebjelke et al., 2005). To assay for resistant Salmonella, 10 μl of tetrathionate brilliant green broth (TTBG) drag swab enrichment were streaked onto XLT4 agar with no antibiotics or XLT4NR in order to detect 934NR and 3147NR. Additionally, TTBG enrichments were streaked onto XLT4NR and one of the following antibiotics: ampicillin (10 μg/ml), chloramphenicol (25 μg/ml), gentamicin (16 μg/ml), kanamycin (25 μg/ml), streptomycin (25 μg/ml), or tetracycline (10 μg/ml). Plates were incubated for 24 h at 37°C; presumptive positive isolates were stocked in freezer stock medium (1% peptone, 15% glycerol) and stored at −80°C.
Poultry litter, water and feed from each pen (n = 16) were collected and processed on a weekly basis, as follows. Chicken litter samples were randomly collected from five general areas within a pen then pooled (n = 16 pens) in order to enumerate Gram-negative enterics. Five grams of litter was suspended in approximately 30 ml of PBS and vigorously shaken for 10 min with a “wrist-action” shaker set at maximum speed (Burrell Scientific; Pittsburgh, PA). Samples were then filtered through sterile gauze and centrifuged at low speed (50 × g for 15 min at 4°C) to remove litter debris, the bacteria were pelleted by high-speed centrifugation (3,650 × g for 15 min at 4°C); supernatant was discarded and the bacterial pellet suspended in 1 ml PBS. A 0.5 ml aliquot was collected by high-speed centrifugation, and the bacterial pellet was suspended in Superbroth (Murooka and Harada, 1994) with 20% sterile glycerol and stored at −80°C. Enumeration of resistant, Gram-negative enterics was done by serially diluting frozen-glycerol stocks of litter suspensions in PBS (dilution range: 10−1 – 10−5), spread-plating onto MacConkey agar, MacConkey agar supplemented with nalidixic acid and rifampicin to enumerate Salmonella, and MacConkey agar with other antibiotics (ampicillin, chloramphenicol, gentamicin, kanamycin, streptomycin or tetracycline), at same concentrations used to enumerate antimicrobial resistant Salmonella. Plates were incubated for 18–24 h at 37°C.
Water and feed samples were screened for Salmonella by culture as follows. One hundred ml of water was added to 2X TTBG, 25 grams of feed was added to 225 ml TTBG, and both were incubated at 41.5 C overnight. TTBG enrichments were then plated onto XLT4 agar and incubated at 37°C for 18–24 h. Black colonies were confirmed as S. enterica Typhimurium by whole-cell agglutination test using polyvalent Salmonella “O” and Group B antiserum (Sigma).
Whole cells were used as template for PCR as described by Bass et al. (1999) using a RapidcyclerTM, hot-air thermocycler (Wittwer et al., 1989). Escherichia coli K12 strain SK1592 (Tn21::pDU202) and S. enterica Typhimurium strain SR11 served as positive and negative controls, respectively for Tn21 (Liljebjelke et al., 2017) and the class 1 integron associated genes intI1 and sul1 (Lévesque et al., 1995; Bass et al., 1999) (Table 3). DNA products were visualized by gel electrophoresis with 1.5% molecular grade agarose containing 0.2 μg/ml ethidium bromide (57). Antibiotic-resistant Salmonella isolates were also screened by PCR for aadA1 (streptomycin/spectinomycin resistance) (Shahi and Kumar, 2015), aadB (gentamicin resistance) (Shahi and Kumar, 2015), tetA (tetracycline resistance) (Ng et al., 2001), and blaTEM (ampicillin resistance) (Mabilat et al., 1990) (Table 3). Antimicrobial resistance genes were selected based on their abundance in poultry litter or commensal species present in the poultry house environment (Lu et al., 2003b; Nandi et al., 2004; Smith et al., 2007). Plasmids pDU202, pUC8 and pSL18 served as positive controls for aadA1, blaTEM and tetA, respectively (Vieira and Messing, 1982; Lee et al., 1993; Bass et al., 1999). A poultry litter, E. coli isolate 529.3-1, with an aadB integron cassette, served as a positive control for aadB PCR screens. S. Typhimurium SR11 served as a negative control in these antibiotic resistance gene screens.
Salmonella isolates cultured from the chickens were strain typed using PGFE with 2.5 U of restriction endonuclease, Xba I (Roche; Indianapolis, IN) as previously described. DNA fragments were separated with a CHEF DR-II electrophoresis apparatus (Bio-Rad; Hercules, CA) at 200 V for 25 h and pulse time of 2–40 s.(Liljebjelke et al., 2005) To increase the resolution of DNA band patterns for some isolates, 50 μM thiourea (Sigma Aldrich) was added to the running buffer (Koort et al., 2002). Yeast strain Saccharomyces cerevisiae YPH80 served as the molecular weight marker (220- 1100 kb) (BioWhittaker Molecular Applications; Rockland, ME).
Large molecular weight plasmids were isolated and analyzed as follows. Cultures were grown to an OD600 nm of 0.8–1.0 in 3 ml LB broth at 37°C with aeration (shaking: 220 rpm). Plasmid isolation, was carried out as described by Williams et al. (2006) using the CosMC Prep Tube Protocol for High and Low Copy Plasmid Purification (Agencourt Bioscience; Beverly, MA). Plasmids were separated on a 0.5% Seakem Gold agarose (Lonza; Rockland, ME) gel by electrophoresis at 5.14V/cm for 18 h at room temperature. Ten microliter of BAC-Tracker Supercoiled DNA ladder (Epicenter Biotech; Madison, WI) was used as molecular weight markers. Gels were stained with SYBR green I nucleic acid gel stain (Lonza) for 60 min. The DNA band, containing the plasmid of interest, was excised from the agarose gel and washed in cold 1:10 strength TE buffer pH 8.0 (10 mM Tris HCl, 1 mM EDTA) at 4°C. The agarose gel slice was subsequently washed with 100 μl SuRE/Cut Buffer B (10 mM Tris HCl, 10 mM MgCl2, 100 mM NaCl, 1 M 2-mercaptoethanol), replaced with fresh enzyme buffer with Hind III (1U) (Roche), and incubated at 37°C for 2 h. The cut DNA was embedded in 0.75% Seakem Gold agarose gel and fragments were separated by gel electrophoresis at 45V, for 18 h at room temperature. One Kilobase DNA Ladder Plus (Fermentas; Glen Burnie, MD), and Hind III digest of λ DNA were used as the molecular weight markers. Gels were stained with Sybr Green for 60 min. DNA fragment sizes were then determined using Gene Profiler 4.05 by Scanalytics, Inc. (Fairfax, VA).
Salmonella isolates were initially screened for resistance using the Kirby-Bauer disk diffusion method against a panel of eight antimicrobials (CLSI, 2017). This panel included ampicillin (10 μg), chloramphenicol (30 μg), gentamicin (10 μg), kanamycin (30 μg), nalidixic acid (30 μg), rifampicin (5 μg), streptomycin (10 μg), and tetracycline (30 μg) (Sigma). The Salmonella parent strain was run as the negative control in order to inferred acquired resistances. The Sensititre® susceptibility system (Trek Diagnostics Systems, Ltd.; Oakwood Village, OH) was used to c determine the minimum inhibitory concentration (MIC) for select Salmonella isolates (CLSI, 2017). The antimicrobials contained in each Sensititre® plate were: enrofloxacin, gentamicin, ceftiofur, neomycin, oxytetracycline, tetracycline, amoxicillin, spectinomycin, sulphadimethoxine, trimethoprim/sulphamethoxazole, sarafloxacin, sulphathiazole, and streptomycin. Antimicrobial susceptibilities for Salmonella isolated in Animal Study 2 were assessed using a Sensititre® plate that contained 17 antimicrobials including: amikacin, amoxicillin, ampicillin, cefazolin, cefoxitin, cefpodoxime, ceftiofur, cephalothin, chloramphenicol, enrofloxacin, gentamicin, imipenem, marbofloxacin, orbifloxacin, rifampicin, tetracycline, ticarcillin, clavulanic acid, and trimethoprim. Susceptibilities were controlled and interpreted according to guidelines established by the Clinical and Laboratory Standards Institute (CLSI, 2017).
The mean and standard deviations were computed for each experimental group and therapeutic treatment. Paired and unpaired student t-test analyses were used to evaluate the effects of therapeutic treatments over time on the degree of resistance to each antibiotic tested. X2 test was used to determine if there was a random association between administration and antimicrobial resistance.
In vitro, the Newport multidrug-resistance plasmid was readily transmitted to multiple Salmonella serovars at frequencies ranging from 3.8 × 10−7 to 8.0 × 10−6 (Table 4). This plasmid was stably maintained in Typhimurium 934R even after 50 generations in the absence of antibiotic selective pressure (day 5; Table 5) and the plasmid segregation rate was low (v = 8.6 × 10−2). This indicates that plasmid acquisition would not be a barrier to Salmonella acquiring antibiotic resistance. However, the plasmid did impair Typhimurium 934R fitness (S = −0.10, Sd = 3.86; Sl = −0.01) (Lenski et al., 1994; Melnyk et al., 2015), indicating that plasmid maintenance may be a challenge in vivo. Since there was a fitness cost, acquisition, and maintenance of the plasmid in vivo may be enhanced with antimicrobial selection pressure. Therefore, administration of streptomycin, tetracyclines, or the cephalosporins would be expected to increase plasmid-encoded resistance in the microbiome of chickens.
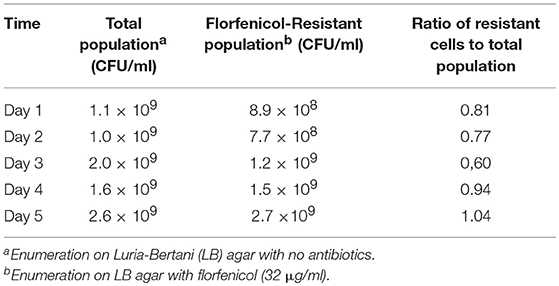
Table 5. In vitro maintenance of Newport MDR plasmid in S. Typhimurium 934R: stability in the absence of antibiotic selection pressure.
In vivo, none of the antibiotics increased abundance of resistant Salmonella (Table 6). In addition, no florfenicol-resistant Salmonella were detected suggesting that the Newport multidrug-resistance plasmid was not transferred and/or maintained in Salmonella in vivo. There was no increase in Salmonella extended-spectrum β-lactam (ESBL) resistance in birds co-infected with E. coli harboring the Newport MDR plasmid compared to group colonized with Salmonella alone (61.4% vs. 67.5%). This was confirmed in that the plasmid genotype of Typhimurium expressing ESBL resistance, gentamicin resistance or antimicrobial susceptible phenotypes were identical to Salmonella Typhimurium LT2 spvB-virulence plasmid control using Hind III restriction enzyme mapping (Supplemental Figure 1). There were only three antimicrobial resistance phenotypes detected: ESBL resistance to amoxicillin, ampicillin, cefazolin, cephalothin, cefoxitin, cefpodoxime, and ceftiofur (59.8%); gentamicin resistance (0.5%); or pan-susceptible (39.7%). Ceftiofur administration was the only antibiotic that significantly increased ESBL resistance abundance in Typhimurium 934R compared to the untreated group (37.5% vs. 88.5%; p = 1.1 × 10−7) (Table 7).
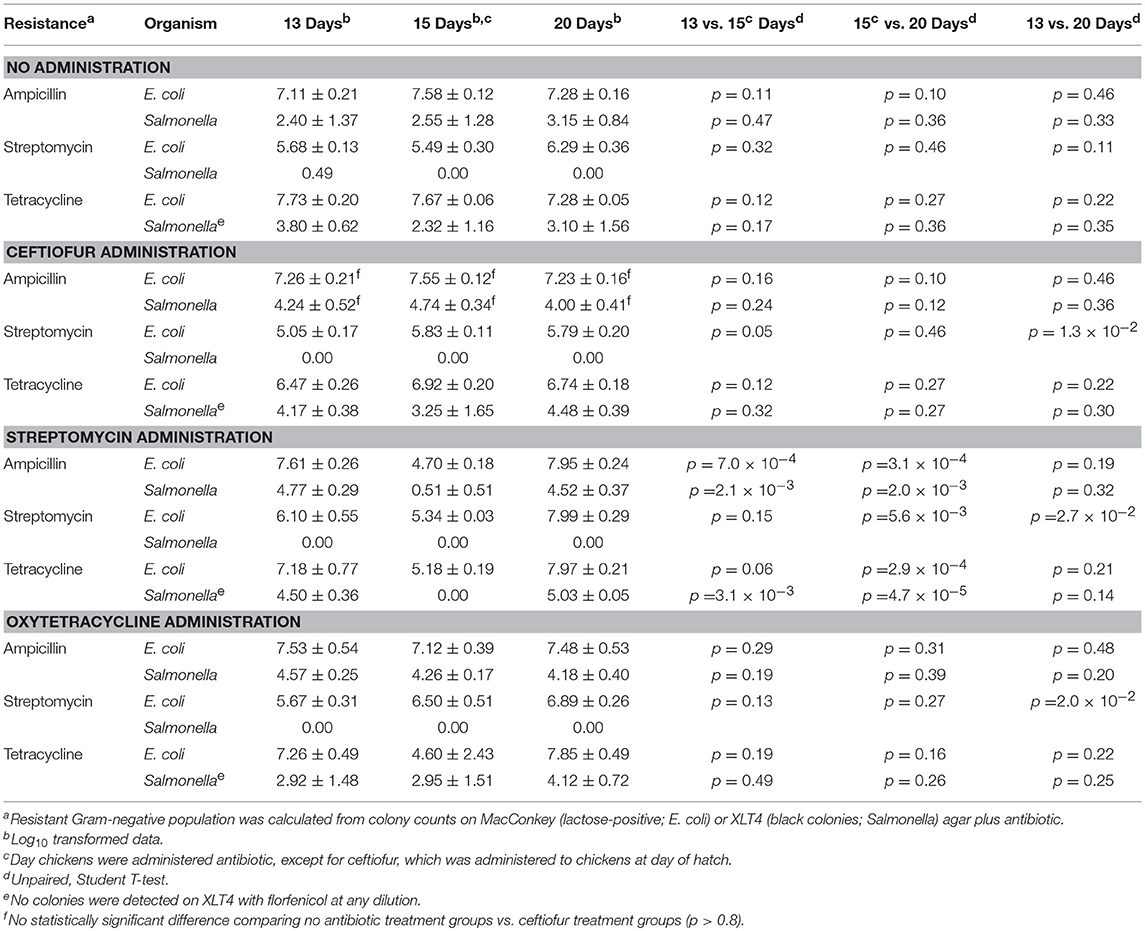
Table 6. Impact of antibiotic administration on antimicrobial resistance in Gram-negative bacteria colonizing chickens.
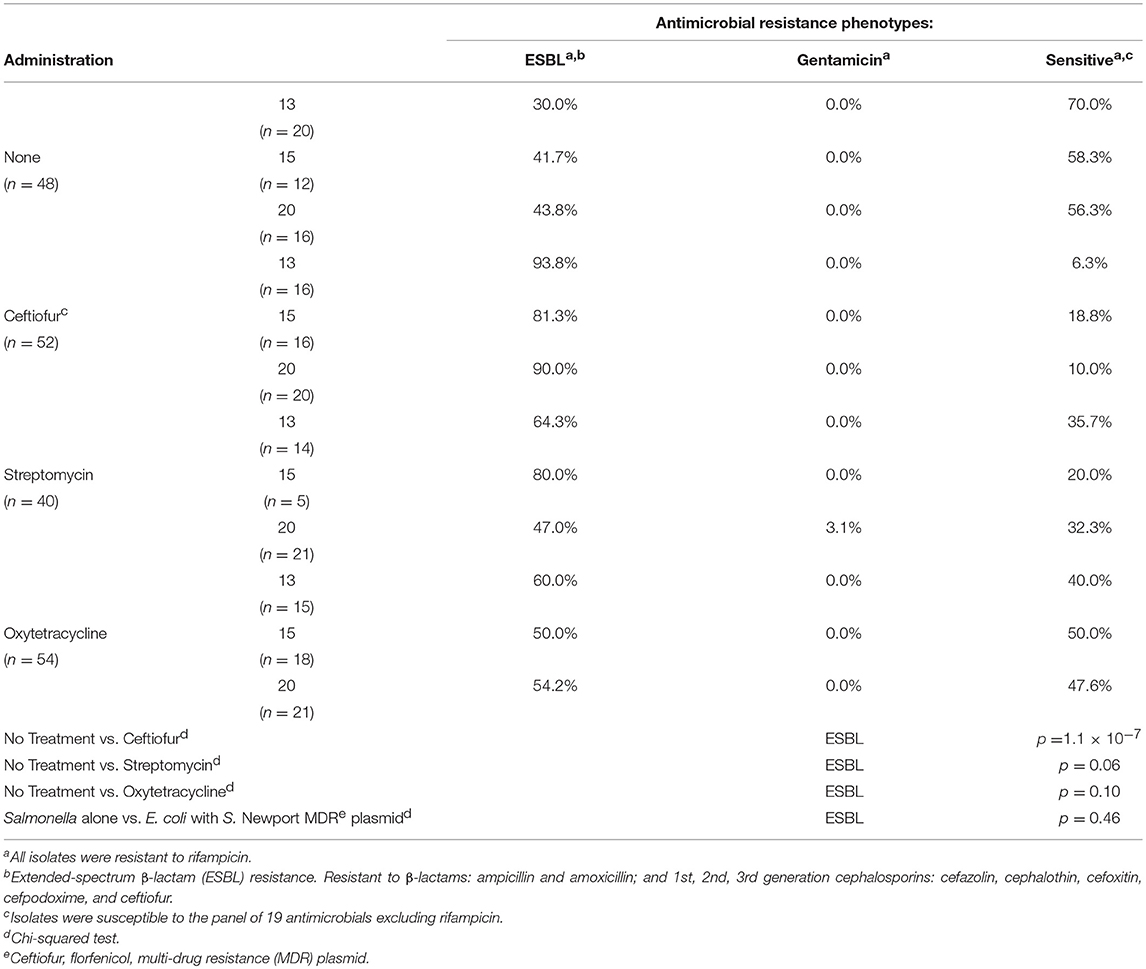
Table 7. Impact of antibiotic administration on S. Typhimurium 934R antimicrobial resistance phenotype.
In this experiment, two S. Typhimurium isolates (934NR and 3147NRintI1) of the same strain (See Supplemental Figure 2) were used in order to determine if antimicrobial resistance genotype influenced the acquisition of new resistances. Isolate 3147NR contained a type 1 integron with an empty integration site enabling easy PCR screening of integron-associated resistance. Because inflammation has been shown to affect the antimicrobial resistance in Salmonella (Stecher et al., 2012), birds were administered an infectious dose high enough to cause clinical symptoms. A difference in mortality (Table 8) and poultry environment abundance (Figure 1) was shown between the isolates. Mortality was reduced in chickens fed litter resistome then administered isolate 3147NRintI1 (31.8% vs. 45.4%; p = 0.035) and the isolate was less abundant in the poultry environment compared to isolate 934NR (Figure 1). Isolate 934NR was also more likely to be ampicillin resistant (91.7% vs. 67.4%; p = 3.7 × 10−3) (Table 9).
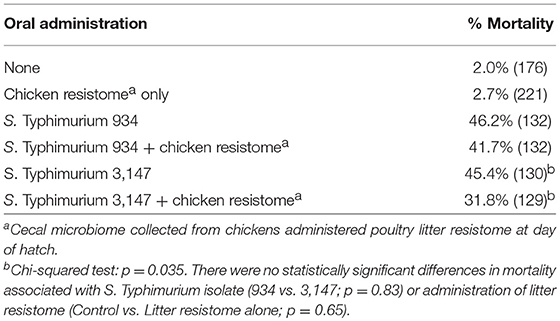
Table 8. Mortality in chickens orally inoculated with nalidixic acid, rifampicin-resistant Salmonella Typhimurium strains.
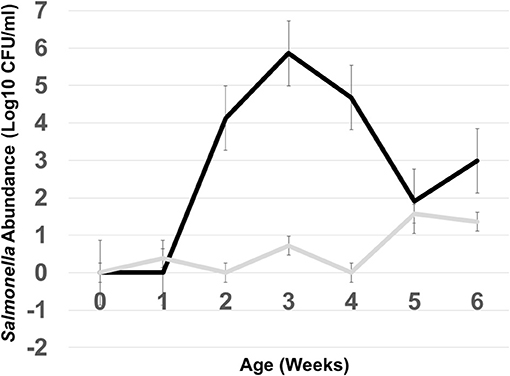
Figure 1. Abundance of Salmonella in the poultry environment following oral administration of S. Typhimurium 934NR (black line) or 3147NR (gray line) to 1 day old chicks. Poultry litter was collected at placement of day of hatch chicks (time 0) and weekly, for 6 weeks. Salmonella abundance was determined from colony counts obtained from 10-fold serial dilutions and plating on MacConkey agar with nalidixic acid and rifampicin.
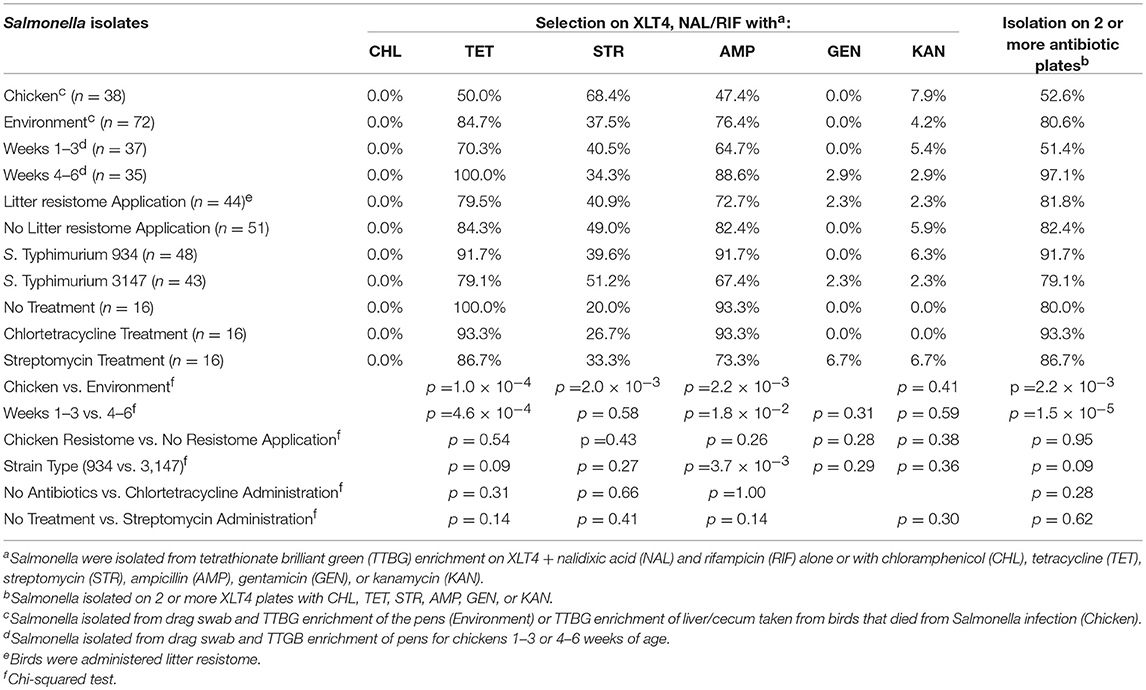
Table 9. Antimicrobial resistance in Salmonella isolated from chickens administered antibiotics and chicken resistome.
Antibiotic administration had a minor effect on Salmonella resistance while it increased resistance in other Gram-negative enterics. The total Gram-negative enteric population peaked at 109 CFU/g in the feces of 2 weeks old birds, and remained high (~108 CFU/g) up to 6 weeks of age (see Figure 2) however for the most part the resistant Gram-negatives did not exceed the abundance of susceptible population. There were also no significant differences in abundance of resistant Gram-negative enteric in the feces of birds colonized with the litter resistome. Nor did administering litter resistome as a gene donor affect resistance in Salmonella.
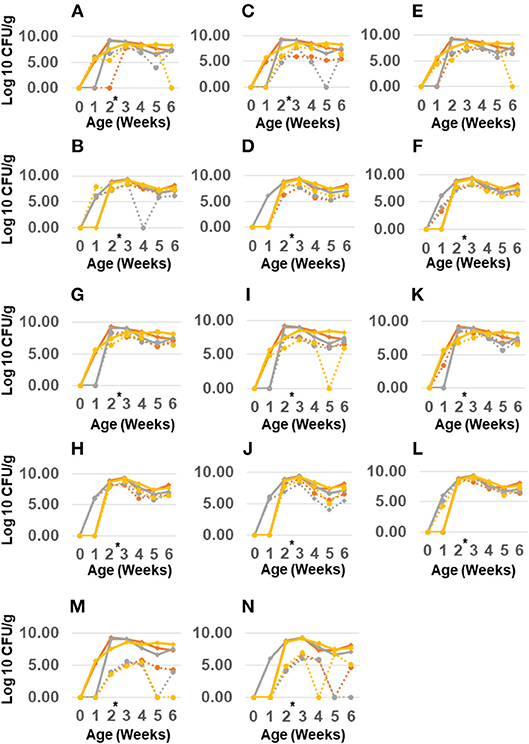
Figure 2. Abundance of antimicrobial resistant Gram-negatives in chickens administered poultry litter resistome and antibiotics. Salmonella Typhimurium 934R was orally administered to 1 day old chicks with (B,D,F,H,J,L,N) or without (A,C,E,G,I,K,M) poultry litter resistome. At 2 weeks of age (black arrow), chlortetracycline (gray line), or streptomycin (yellow line) was administered; control birds received no antibiotic treatment (red line). Abundance of Gram-negatives resistant to streptomycin (A,B), gentamicin (C,D), kanamycin (E,F), tetracycline (G,H), chloramphenicol (I,J), ampicillin (K,L), or nalidixic acid/rifampicin (M,N) was determined from plating samples on MacConkey agar with antibiotic (dashed lines). MacConkey agar without antibiotics was used to enumerate total Gram-negatives (solid line). *Antibiotic treatment at 2 weeks of age.
However, antimicrobial administration had a significant impact on resistance to certain antibiotics (Table 10). Streptomycin administration increased aminoglycoside and tetracycline resistance in Gram-negative enterics cultured from the poultry environment. Chlortetracycline usage also increased tetracycline resistance (p = 1.9 × 10−3) but had no effect on resistance to the other antimicrobials. There was no increase in resistance to chloramphenicol or ampicillin in birds administered antibiotics, nor was there a significant change in Salmonella abundance. The prevalence of resistance in Salmonella was impacted by the source (bird or environment) and the age of the birds (Tables 9, 11). A significantly greater proportion of Salmonella isolated from the poultry environment were resistant to ampicillin, streptomycin, and tetracycline, compared to Salmonella isolated from birds (80.6% vs. 52.6%; p = 2.2 × 10−3). In addition, ampicillin and tetracycline resistance was more likely in isolates cultured from the environment of 4–6 weeks old birds compared to 1–3 weeks old. Resistance to two or more antibiotics was also significantly more likely in isolates from the environment of older birds (97.1% vs. 51.4%; p = 1.5 × 10−5).
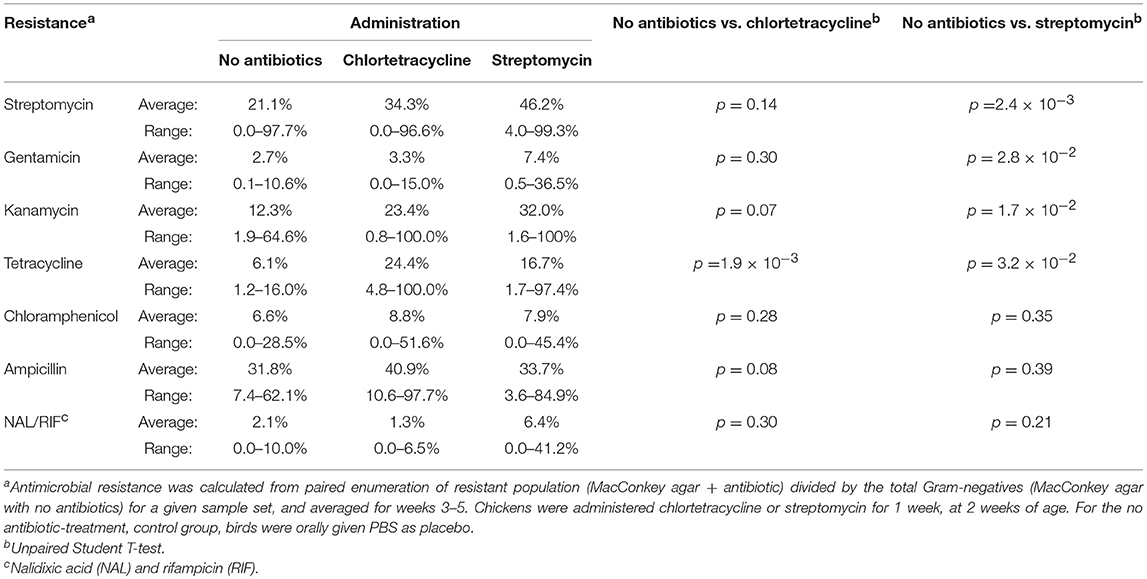
Table 10. Antimicrobial resistance in Gram-negative bacteria isolated from chickens administered antimicrobials and chicken resistome.

Table 11. Antimicrobial resistance genotype and phenotypes of S. Typhimurium recovered from poultry environment of chickens experimentally colonized with Salmonella.
Antimicrobial resistance phenotype and genotype was characterized in Salmonella isolated from the poultry environment in order to verify gene transfer to the Typhimurium strain. Isolates were screened for class 1 integrons and the integron-associated resistance genes sul1, aadA1, and aadB; blaTEM1; and tetA. Isolate 934NR, naturally resistant to streptomycin and sulfisoxazole, acquired resistances to ampicillin, amoxicillin, kanamycin, or tetracycline (Table 11). However, half of the 934NR isolated in this experiment, were sensitive to streptomycin. Seven resistance phenotypes were identified with sulfisoxazole, ampicillin, and amoxicillin resistance as the most common. Eighty-seven percent of isolates were resistant to three or more antimicrobials. All β-lactam resistant 934NR isolates were positive for the blaTEM β-lactamase gene allele. The only other antimicrobial resistance gene detected was aadA1 in one isolate that was resistant to streptomycin and sulfisoxazole, however none of the 934NR isolates were positive for class 1 integrons. Of the three tetracycline-resistant isolates examined, none were positive for tetA.
Similarly, isolate 3147NRintI1, naturally sulfisoxazole resistant, acquired resistances including ampicillin, amoxicillin, gentamicin, kanamycin, streptomycin and tetracycline. However, one third of 3147NRintI1 isolates were sensitive to sulfisoxazole. Eight antimicrobial resistance phenotypes were identified in 3147NRintI1, with resistances to streptomycin and sulfisoxazole the most common. Twenty-percent of these isolates were resistant to three or more antibiotics. However, three 3147NRintI1 isolates were cultured that were resistant to six or more antimicrobials. All β-lactam resistant 3147NRintI1 isolates were positive for blaTEM. Four of five gentamycin-resistant isolates were positive for the aadB; and all tetracycline resistant 3147NRintI1 isolates were positive for tetA. The most common antimicrobial resistance genotype identified in 3147NRintI1 isolates was aadA1, aadB, blaTEM, sul1, tetA. The other antimicrobial resistance genotypes identified included blaTEM; and aadA1, blaTEM, sul1, tetA. All 3147NRintI1 isolates harboring aadA1, aadB, blaTEM, sul1, tetA or aadA1, blaTEM, sul1, tetA were positive for the class 1 integron. While the original Salmonella 3147NRintI1 isolate administered to the birds contained the class 1 integron, only 28% of antimicrobial resistant 3147 isolates cultured from the birds contained an integron. And Tn21, a transposon responsible for dissemination of class 1 integrons in avian E. coli (Bass et al., 1999), was not detected in any antimicrobial-resistant Salmonella isolates indicating that the integron was encoded on a different element.
While acquisition of antimicrobial resistance in Salmonella occurs in nature, how quickly does this occur in vivo and what is the impact of antibiotic usage on the acquisition of resistance? We performed two trials in birds using multiple genotypes of Salmonella and two sources of resistant genes: a defined MDR plasmid and an environmental resistome. This study documented an abundant resistant Enterobacteriaceae population in the birds and the poultry litter in the absence of treatments. Others have reported similar findings; abundant antimicrobial resistance in the absence of antimicrobial usage (Fairchild et al., 2005; Smith et al., 2007; Agga et al., 2016; Liljebjelke et al., 2017). Emergence of antimicrobial-resistant Salmonella in chickens occurred readily despite its existence as a minor population in bird and litter bacterial community outnumbered by gram-positives 100-1,000 to 1 (Lu et al., 2003a,b). Salmonella colonizing chickens easily acquire antimicrobial resistance from the resident bacteria present in chickens, even in the absence of antimicrobial use. For Salmonella to acquire resistance genes there needs to be a sufficient antibiotic resistance reservoir. Poultry litter has a rich resistome with a high abundance of class 1 integrons at 1 copy per 1-100 bacterial genomes (Nandi et al., 2004). The most abundant integron associated antimicrobial resistance genes in poultry Salmonella tend to be the same as those present in litter (Nandi et al., 2004; Liljebjelke et al., 2017). Emergence of extended-spectrum β-lactam/cephalosporin resistance in Salmonella has been attributed to acquisition of conjugative plasmids (Winokur et al., 2000). These plasmids often contain other resistances including chloramphenicol, florfenicol, streptomycin, and tetracycline (Winokur et al., 2000; Doublet et al., 2004; Fernández-Alarcón et al., 2011). In nature, Salmonella acquires MDR plasmids from commensals that inhabit the same environment (Winokur et al., 2001; Fricke et al., 2009; McCollister et al., 2016) and its acquisition of antimicrobial resistance is dependent on an abundant, antimicrobial-resistant donor population (Smith, 1975; Schjørring et al., 2008; Faure et al., 2009; Card et al., 2017). How quickly Salmonella acquires plasmids and associated antimicrobial resistances are dependent on the presence and abundance of the plasmid in the intestinal microbiome, rate of plasmid transmission, plasmid stability and fitness cost (Stewart and Levin, 1977; Ponciano et al., 2007). Antimicrobial usage may also play an important role in the acquisition and maintenance of plasmid-borne resistance in Salmonella by ameliorating the fitness cost.
With the exception of ceftiofur, antibiotic administration did not significantly increase antimicrobial resistance in Salmonella. ESBL-resistance in Salmonella was not due to horizontal transmission of the Newport MDR-plasmid. Subbiah et al. reported an ESBL incA/C plasmid imposed a fitness cost in vitro and in vivo (Subbiah et al., 2011) therefore it is possible that ESBL-resistant Salmonella acquired the resistance gene on a transposon. Given that the ESBL-resistance gene blaCYM2 is adjacent to a transposon in the S. Newport MDR plasmid (Giles et al., 2004) and a recent report of blaCMX in a transposon (Huang et al., 2017), one possible explanation for plasmid-less ESBL resistance is blaCMY transposition on and off a conjugative plasmid that ferried this element between cells. Whole genome sequencing will determine the nature of ESBL resistance in these Salmonella isolates; where the resistance gene resides; and the genetic element that bears it.
In other animal models, the intestinal microbiome can be a formidable barrier to the transfer of antimicrobial resistance (Schjørring et al., 2008; Faure et al., 2009; Stecher et al., 2012). However, this study showed that antimicrobial-resistant Salmonella occurred in the absence of administration of a resistant microbiome. This finding shows that ample reservoirs exist among the normal microbiome of baby chicks, even in a sanitized environment.
Salmonella can easily acquire antimicrobial resistance from the chicken resistome. With the exception of ceftiofur use, antimicrobial resistance in Salmonella occurred even in the absence of antibiotic administration. The animal resistome is one important contributor to antimicrobial resistance in Salmonella. A detailed analysis of the animal resistome will provide insights into important players in the dissemination of antimicrobial resistance within these production systems.
The University of Georgia's, Institutional Animal Care and Use Committee approved all animal care protocols described in this work.
JM, CH, and ML contributed to the conception and design of this study. JM, TB, AH-B, VS, and KL were responsible for the acquisition of data analyzed in this study. JM was involved in the analysis and interpretation associated with this work. SS directed antimicrobial susceptibility testing and interpretation of susceptibility results. All authors were involved in manuscript writing, revisions, and final approval of this manuscript.
This work was supported by grants from the United States Department of Agriculture (99-35212-8680, 2017-05586) and the National Institute of Health (NIH-NAID 1R15AI 54401-1A1).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work, presented here, was part of TB, VS, and KL MS thesis at the University of Georgia (Bythwood, 2003; Soni, 2007; Lyons, 2009).
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2019.00020/full#supplementary-material
Supplemental Figure 1. Detection of high molecular weight plasmids in antimicrobial-resistant Salmonella cultured from the environment of chickens administered S. Typhimurium 934R. (A) Uncut plasmid DNA. Lane 1: BAC-Tracker Supercoiled DNA ladder; Lane 2: RP4 plasmid control; lane 3: S. Typhimurium 934R; lanes 4–12: antimicrobial-resistant isolates: 1543Rc (lane 4), 1546RAa (lane 5), 1546RTb (lane 6), 1546Rc (lane 7), 1549Rb (lane 8), 1549RAb (lane 9), 1552RAa (lane 10), 1552RTb lane 11), and 1552 Rc (lane 12). (B) Hind III restriction digestion of plasmid DNA. Lane 1: λ DNA ladder; lane 2: λ DNA, lane 3: Hind III-digested λ DNA; lane 4: S. Typhimurium LT2 (virulence plasmid control); lane 5: S. Typhimurium 934R; lanes 6–11: antimicrobial-resistant isolates: 1543Rc (lane 6), 1546RAa (lane 7), 1546RTb (lane 8), 1546Rc (lane 9), 1549Rb (lane 10), and 1549RAb (lane 11). DNA was separated on 0.5% agarose gel at 5.15V/cm for 18 h at room temperature and stained with Sybr-Green.
Supplemental Figure 2. Pulsed-field gel electrophoresis (PFGE XbaI) confirms antibiotic-resistant Salmonella recovered from the poultry environment as the S. Typhimurium strain administered to the birds as isolates 934R and 3147. Lanes 1 and 16: MW DNA standard Saccharomyces cerevisiae chromosomes; Lane 2: laboratory strain S. Typhimurium SR11; Lane 3: S. Typhimurium 934R; lane 4: S. Typhimurium 3147R; lanes 5–15: antibiotic resistant Salmonella isolated from poultry environment of chickens administered S. Typhimurium strains 93R4 or 3147R.
AMP, ampicillin; AMX, amoxicillin; ESBL, extended spectrum β-lactams; GEN, gentamicin; KAN, kanamycin; LB, Luria-Bertani medium; MPN, most probable number; MDR, multi-drug resistance; NAL, nalidixic acid; PCR, polymerase chain reaction; PFGE, pulsed-field gel electrophoresis; RIF, rifampicin; SPF, specific pathogen free; STR, streptomycin; SMX, sulfisoxazole; TET, tetracycline; TTBG, tetrathionate brilliant green; XLT4, xylose, lysine, tergitol 4.
Agga, G. E., Schmidt, J. W., and Arthur, T. M. (2016). Antimicrobial-resistant fecal bacteria from ceftiofur-treated and nonantimicrobial-treated comingled beef cows at a cow-calf operation. Microb. Drug Resist. 22, 598–608. doi: 10.1089/mdr.2015.0259
Andersen, V. D., DE Knegt, L. V., Munk, P., Jensen, M. S., Agersø, Y., et al. (2017). The association between measurements of antimicrobial use and resistance in the faeces microbiota of finisher batches. Epidemiol. Infect. 145, 2827–2837. doi: 10.1017/S0950268817001285
Bass, L., Liebert, C. A., Lee, M. D., Summers, A. O., White, D. G., Thayer, S. G., et al. (1999). Incidence and characterization of integrons, genetic elements mediating multiple-drug resistance, in avian Escherichia coli. Antimicrob. Agents Chemother. 43, 2925–2929. doi: 10.1128/AAC.43.12.2925
Bortolaia, V., Mander, M., Jensen, L. B., Olsen, J. E., and Guardabassi, L. (2015). Persistence of vancomycin resistance in multiple clones of Enterococcus faecium isolated from Danish broilers 15 years after the ban of avoparcin. Antimicrob. Agents Chemother. 59, 2926–2929. doi: 10.1128/AAC.05072-14
Bythwood, T. N. (2003). In Vivo Transfer of Antibiotic Resistance Genes Between The Resident Intestinal Microflora Of Broiler Chickens and Salmonella typhimurium. University of Georgia. Available online at: http://purl.galileo.usg.edu/uga_etd/buffington_tameka_n_200312_ms (accessed April 30, 2018). 1–132.
Caffrey, N., Nekouei, O., Gow, S., Agunos, A., and Checkley, S. (2017). Risk factors associated with the A2C resistance pattern among E. coli isolates from broiler flocks in Canada. Prev. Vet. Med. 148, 115–120. doi: 10.1016/j.prevetmed.2017.11.001
Card, R. M., Cawthraw, S. A., Nunez-Garcia, J., Ellis, R. J., Kay, G., Pallen, M. J., et al. (2017). An in vitro chicken gut model demonstrates transfer of a multidrug resistance plasmid from salmonella to commensal Escherichia coli. MBio 8, 1–15. doi: 10.1128/mBio.00777-17
CLSI (2017). M100 Performance Standards for Antimicrobial Suceptibility Testing, 27th Ed. Available online at: https://1pdf.net/m100-performance-standards-for-antimicrobial-susceptibility-clsi_59de3ac3f6065dc81db4fc (accessed April 30, 2018).
Collard, J. M., Place, S., Denis, O., Rodriguez-Villalobos, H., Vrints, M., Weill, F. X., et al. (2007). Travel-acquired salmonellosis due to Salmonella Kentucky resistant to ciprofloxacin, ceftriaxone and co-trimoxazole and associated with treatment failure. J. Antimicrob. Chemother. 60, 190–192. doi: 10.1093/jac/dkm114
Cox, L. A., and Ricci, P. F. (1969). The Joint Commitee on the Use of Antibiotics in Animal Husbandry and Veterinary Medicine. London: Her Majesty's Stationary Office.
Doublet, B., Carattoli, A., Whichard, J. M., White, D. G., Baucheron, S., Chaslus-Dancla, E., et al. (2004). Plasmid-mediated florfenicol and ceftriaxone resistance encoded by the floR and bla(CMY-2). genes in Salmonella enterica serovars typhimurium and Newport isolated in the United States. FEMS Microbiol Lett. 233, 301–305. doi: 10.1111/j.1574-6968.2004.tb09496.x
Duong, V. T., Tuyen, H. T., Van Minh, P., Campbell, J. I., Phuc, H. L., Nhu, T. D. H., et al. (2018). No clinical benefit of empirical antimicrobial therapy for pediatric diarrhea in a high-usage, high-resistance setting. Clin. Infect. Dis. 66, 504–511. doi: 10.1093/cid/cix844
Fairchild, A. S., Smith, J. L., Idris, U., Lu, J., Sanchez, S., Purvis, L. B., et al. (2005). Effects of orally administered tetracycline on the intestinal community structure of chickens and on tet determinant carriage by commensal bacteria and Campylobacter jejuni. Appl. Environ. Microbiol. 71, 5865–5872. doi: 10.1128/AEM.71.10.5865-5872.2005
Faure, S., Perrin-Guyomard, A., Delmas, J. M., and Laurentie, M. (2009). Impact of therapeutic treatment with beta-lactam on transfer of the bla(CTX-M-9) resistance gene from Salmonella enterica serovar Virchow to Escherichia coli in gnotobiotic rats. Appl. Environ. Microbiol. 75, 5523–5528. doi: 10.1128/AEM.00020-09
Fernández-Alarcón, C., Singer, R. S., and Johnson, T. J. (2011). Comparative genomics of multidrug resistance-encoding IncA/C plasmids from commensal and pathogenic Escherichia coli from multiple animal sources. PLoS ONE 6:e23415. doi: 10.1371/journal.pone.0023415
Fricke, W. F., McDermott, P. F., Mammel, M. K., Zhao, S., Johnson, T. J., Rasko, D. A., et al. (2009). Antimicrobial resistance-conferring plasmids with similarity to virulence plasmids from avian pathogenic Escherichia coli strains in Salmonella enterica serovar Kentucky isolates from poultry. Appl. Environ. Microbiol. 75, 5963–5971. doi: 10.1128/AEM.00786-09
Giles, W. P., Benson, A. K., Olson, M. E., Hutkins, R. W., Whichard, J. M., Winokur, P. L., et al. (2004). DNA sequence analysis of regions surrounding blaCMY-2 from multiple Salmonella plasmid backbones. Antimicrob. Agents Chemother. 48, 2845–2852. doi: 10.1128/AAC.48.8.2845-2852.2004
Goldstein, C., Lee, M. D., Sanchez, S., Hudson, C., Phillips, B., Register, B., et al. (2001). Incidence of class 1 and 2 integrases in clinical and commensal bacteria from livestock, companion animals, and exotics. Antimicrob. Agents Chemother. 45, 723–726. doi: 10.1128/AAC.45.3.723-726.2001
Havelaar, A. H., Kirk, M. D., Torgerson, P. R., Gibb, H. J., Hald, T., Lake, R. J., et al. (2015). World Health Organization global estimates and regional comparisons of the burden of foodborne disease in 2010. PLoS Med. 12:e1001923. doi: 10.1371/journal.pmed.1001923
Hu, H. H., Chiou, C. C., Cheng, M. F., Chen, Y. S., Sheu, S. K., Wu, T. C., et al. (2014). The clinical outcomes of antimicrobial therapy in pediatric patients with nontyphoid salmonellosis with different levels of severity. Clin. Pediatr. 53, 967–974. doi: 10.1177/0009922814540792
Huang, W., Wang, G., Sebra, R., Zhuge, J., Yin, C., Aguero-Rosenfeld, M. E., et al. (2017). Emergence and evolution of multidrug-resistant Klebsiella pneumoniae with both blaKPC and blaCTX-M Integrated in the Chromosome. Antimicrob. Agents Chemother. 61:e00076–e00017. doi: 10.1128/AAC.00076-17
Idris, U., Lu, J., Maier, M., Sanchez, S., Hofacre, C. L., Harmon, B. G., et al. (2006). Dissemination of fluoroquinolone-resistant Campylobacter spp. within an integrated commercial poultry production system. Appl. Environ. Microbiol. 72, 3441–3447. doi: 10.1128/AEM.72.5.3441-3447.2006
Iwamoto, M., Reynolds, J., Karp, B. E., Tate, H., Fedorka-Cray, P. J., Plumblee, J. R., et al. (2017). Ceftriaxone-resistant nontyphoidal salmonella from humans, retail meats, and food animals in the United States, 1996-2013. Foodborne Pathog. Dis. 14, 74–83. doi: 10.1089/fpd.2016.2180
Keyes, K., Hudson, C., Maurer, J. J., Thayer, S., White, D. G., and Lee, M. D. (2000). Detection of florfenicol resistance genes in Escherichia coli isolated from sick chickens. Antimicrob. Agents Chemother. 44, 421–424. doi: 10.1128/AAC.44.2.421-424.2000
Koort, J. M., Lukinmaa, S., Rantala, M., Unkila, E., and Siitonen, A. (2002). Technical improvement to prevent DNA degradation of enteric pathogens in pulsed-field gel electrophoresis. J. Clin. Microbiol. 40, 3497–3498. doi: 10.1128/JCM.40.9.3497-3498.2002
Kruse, H., and Sørum, H. (1994). Transfer of multiple drug resistance plasmids between bacteria of diverse origins in natural microenvironments. Appl. Environ. Microbiol. 60, 4015–4021.
Lee, C., Langlois, B. E., and Dawson, K. A. (1993). Detection of tetracycline resistance determinants in pig isolates from three herds with different histories of antimicrobial agent exposure. Appl. Environ. Microbiol. 59, 1467–1472.
Lenski, R. E., Simpson, S. C., and Nguyen, T. T. (1994). Genetic analysis of a plasmid-encoded, host genotype-specific enhancement of bacterial fitness. J. Bacteriol. 176, 3140–3147. doi: 10.1128/jb.176.11.3140-3147.1994
Lévesque, C., Piché, L., Larose, C., and Roy, P. H. (1995). PCR mapping of integrons reveals several novel combinations of resistance genes. Antimicrob. Agents Chemother. 39, 185–191. doi: 10.1128/AAC.39.1.185
Liljebjelke, K. A., Hofacre, C. L., Liu, T., White, D. G., Ayers, S., Young, S., et al. (2005). Vertical and horizontal transmission of salmonella within integrated broiler production system. Foodborne Pathog. Dis. 2, 90–102. doi: 10.1089/fpd.2005.2.90
Liljebjelke, K. A., Hofacre, C. L., White, D. G., Ayers, S., Lee, M. D., and Maurer, J. J. (2017). Diversity of antimicrobial resistance phenotypes in Salmonella isolated from commercial poultry farms. Front. Vet. Sci. 4:96. doi: 10.3389/fvets.2017.00096
Lorenz, M. G., and Wackernagel, W. (1994). Bacterial gene transfer by natural genetic transformation in the environment. Microbiol. Rev. 58, 563–602.
Lu, J., Idris, U., Harmon, B., Hofacre, C., Maurer, J. J., and Lee, M. D. (2003a). Diversity and succession of the intestinal bacterial community of the maturing broiler chicken. Appl. Environ. Microbiol. 69, 6816–6824. doi: 10.1128/AEM.69.11.6816-6824.2003
Lu, J., Sanchez, S., Hofacre, C., Maurer, J. J., Harmon, B. G., and Lee, M. D. (2003b). Evaluation of broiler litter with reference to the microbial composition as assessed by using 16S rRNA and functional gene markers. Appl. Environ. Microbiol. 69, 901–908. doi: 10.1128/AEM.69.2.901-908.2003
Lyons, K. (2009). The Distribution of Large Molecular Weight Plasmids and Plasmid Genomics in Salmonella. University of Georgia. Available online at: http://purl.galileo.usg.edu/uga_etd/lyons_karen_m_200912_ms (accessed April 30, 2018), 1–85.
Mabilat, C., Goussard, S., Sougakoff, W., Spencer, R. C., and Courvalin, P. (1990). Direct sequencing of the amplified structural gene and promoter for the extended-broad-spectrum beta-lactamase TEM-9 (RHH-1) of Klebsiella pneumoniae. Plasmid 23, 27–34. doi: 10.1016/0147-619X(90)90041-A
McCollister, B., Kotter, C. V., Frank, D. N., Washburn, T., and Jobling, M. G. (2016). Whole-genome sequencing identifies in vivo acquisition of a blaCTX-M-27-carrying IncFII transmissible plasmid as the cause of ceftriaxone treatment failure for an invasive Salmonella enterica serovar typhimurium infection. Antimicrob. Agents Chemother. 60, 7224–7235. doi: 10.1128/AAC.01649-16
Melnyk, A. H., Wong, A., and Kassen, R. (2015). The fitness costs of antibiotic resistance mutations. Evol. Appl. 8, 273–283. doi: 10.1111/eva.12196
Modi, R. I., and Adams, J. (1991). Coevolution in bacterial-plasmid populations. Evolution 45, 656–667. doi: 10.1111/j.1558-5646.1991.tb04336.x
Murooka, Y., and Harada, T. III. (1994). “Gene transfer in gram-negative bacteria,” in Methods for General and Molecular Bacteriology, eds P. Gerhardt, R. G. E. Murray, W. A. Wood, and N. R. Krieg (Washington, DC: ASM Press), 317–364.
Nandi, S., Maurer, J. J., Hofacre, C., and Summers, A. O. (2004). Gram-positive bacteria are a major reservoir of Class 1 antibiotic resistance integrons in poultry litter. Proc. Natl. Acad. Sci. U S A. 101, 7118–7122. doi: 10.1073/pnas.0306466101
Ng, L. K., Martin, I., Alfa, M., and Mulvey, M. (2001). Multiplex PCR for the detection of tetracycline resistant genes. Mol. Cell. Probes 15, 209–215. doi: 10.1006/mcpr.2001.0363
Noyes, N. R., Benedict, K. M., Gow, S. P., Waldner, C. L., Reid-Smith, R. J., Booker, C. W., et al. (2016). Modelling considerations in the analysis of associations between antimicrobial use and resistance in beef feedlot cattle. Epidemiol. Infect. 144, 1313–1329. doi: 10.1017/S0950268815002423
Ponciano, J. M., De Gelder, L., Top, E. M., and Joyce, P. (2007). The population biology of bacterial plasmids: a hidden Markov model approach. Genetics 176, 957–968. doi: 10.1534/genetics.106.061937
Scallan, E., Crim, S. M., Runkle, A., Henao, O. L., Mahon, B. E., Hoekstra, R. M., et al. (2015). Bacterial enteric infections among older adults in the United States: foodborne diseases active surveillance network, 1996-2012. Foodborne Pathog. Dis. 12, 492–499. doi: 10.1089/fpd.2014.1915
Scallan, E., Griffin, P. M., McLean, H. Q., and Mahon, B. E. (2018). Hospitalisations due to bacterial gastroenteritis: a comparison of surveillance and hospital discharge data. Epidemiol. Infect. 146, 954–960. doi: 10.1017/S0950268818000882
Scallan, E., Hoekstra, R. M., Angulo, F. J., Tauxe, R. V., Widdowson, M. A., Roy, S. L., et al. (2011). Foodborne illness acquired in the United States–major pathogens. Emerg. Infect. Dis. 17, 7–15. doi: 10.3201/eid1701.P11101
Scallan, E., Mahon, B. E., Hoekstra, R. M., and Griffin, P. M. (2013). Estimates of illnesses, hospitalizations and deaths caused by major bacterial enteric pathogens in young children in the United States. Pediatr. Infect. Dis. J. 32, 217–221. doi: 10.1097/INF.0b013e31827ca763
Schjørring, S., Struve, C., and Krogfelt, K. A. (2008). Transfer of antimicrobial resistance plasmids from Klebsiella pneumoniae to Escherichia coli in the mouse intestine. J. Antimicrob. Chemother. 62, 1086–1093. doi: 10.1093/jac/dkn323
Shahi, S. K., and Kumar, A. (2015). Isolation and genetic analysis of multidrug resistant bacteria from diabetic foot ulcers. Front. Microbiol. 6:1464. doi: 10.3389/fmicb.2015.01464
Shigemura, H., Matsui, M., Sekizuka, T., Onozuka, D., Noda, T., Yamashita, A., et al. (2018). Decrease in the prevalence of extended-spectrum cephalosporin-resistant Salmonella following cessation of ceftiofur use by the Japanese poultry industry. Int. J. Food Microbiol. 274, 45–51. doi: 10.1016/j.ijfoodmicro.2018.03.011
Sieckmann, D. G., Reed, N. D., and Georgi, C. E. (1969). Transferable drug resistance among Enterobacteriaceae isolated from human urinary tract infections. Appl. Microbiol. 17, 701–706.
Singer, R. S., and Hofacre, C. L. (2006). Potential impacts of antibiotic use in poultry production. Avian Dis. 50, 161–172. doi: 10.1637/7569-033106R.1
Smith, J. L., Drum, D. J., Dai, Y., Kim, J. M., Sanchez, S., Maurer, J. J., et al. (2007). Impact of antimicrobial usage on antimicrobial resistance in commensal Escherichia coli strains colonizing broiler chickens. Appl. Environ. Microbiol. 73, 1404–1414. doi: 10.1128/AEM.01193-06
Smith, M. G. (1975). In vivo transfer of R factors between Escherichia coli strains inoculated into the rumen of sheep. J. Hyg. 75, 363–370. doi: 10.1017/S0022172400024426
Soni, V. (2007). Genetic Approach to Understanding the Behavior of Salmonella in Meat and Poultry System. University of Georgia. Available online at: http://purl.galileo.usg.edu/uga_etd/soni_vivek_200708_ms (accessed April 30, 2018), 1–100.
Stecher, B., Denzler, R., Maier, L., Bernet, F., Sanders, M. J., Pickard, D. J., et al. (2012). Gut inflammation can boost horizontal gene transfer between pathogenic and commensal Enterobacteriaceae. Proc. Natl. Acad. Sci. U S A. 109, 1269–1274. doi: 10.1073/pnas.1113246109
Stewart, F. M., and Levin, B. R. (1977). The population biology of bacterial plasmids: a PRIORI conditions for the existence of conjugationally transmitted factors. Genetics 87, 209–228.
Subbiah, M., Top, E. M., Shah, D. H., and Call, D. R. (2011). Selection pressure required for long-term persistence of blaCMY-2-positive IncA/C plasmids. Appl. Environ. Microbiol. 77, 4486–4493. doi: 10.1128/AEM.02788-10
Taylor, D. E., De Grandis, S. A., Karmali, M. A., and Fleming, P. C. (1981). Transmissible plasmids from Campylobacter jejuni. Antimicrob. Agents Chemother. 19, 831–835. doi: 10.1128/AAC.19.5.831
Tribble, D. R. (2017). Resistant pathogens as causes of traveller's diarrhea globally and impact(s) on treatment failure and recommendations. J. Travel Med. 24(Suppl. 1), S6–S12. doi: 10.1093/jtm/taw090
Tyson, G. H., Tate, H. P., Zhao, S., Li, C., Dessai, U., Simmons, M., et al. (2017). Identification of plasmid-mediated quinolone resistance in Salmonella isolated from swine ceca and retail pork chops in the United States. Antimicrob. Agents Chemother. 61, e01318–e01317. doi: 10.1128/AAC.01318-17
Vieira, J., and Messing, J. (1982). The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene 19, 259–268. doi: 10.1016/0378-1119(82)90015-4
Wang, H., and Slavik, M. F. (1998). Bacterial penetration into eggs washed with various chemicals and stored at different temperatures and times. J. Food Prot. 61, 276–279. doi: 10.4315/0362-028X-61.3.276
Wasyl, D., Kern-Zdanowicz, I., Domanska-Blicharz, K., Zajac, M., and Hoszowski, A. (2015). High-level fluoroquinolone resistant Salmonella enterica serovar Kentucky ST198 epidemic clone with IncA/C conjugative plasmid carrying bla(CTX-M-25) gene. Vet. Microbiol. 175, 85–91. doi: 10.1016/j.vetmic.2014.10.014
Williams, J. E. (1970). Effect of high-level formaldehyde fumigation on bacterial populations on the surface of chicken hatching eggs. Avian Dis. 14, 386–392. doi: 10.2307/1588482
Williams, L. E., Detter, C., Barry, K., Lapidus, A., and Summers, A. O. (2006). Facile recovery of individual high-molecular-weight, low-copy-number natural plasmids for genomic sequencing. Appl. Environ. Microbiol. 72, 4899–4906. doi: 10.1128/AEM.00354-06
Winokur, P. L., Brueggemann, A., DeSalvo, D. L., Hoffmann, L., Apley, M. D., Uhlenhopp, E. K., et al. (2000). Animal and human multidrug-resistant, cephalosporin-resistant salmonella isolates expressing a plasmid-mediated CMY-2 AmpC beta-lactamase. Antimicrob. Agents Chemother. 44, 2777–2783. doi: 10.1128/AAC.44.10.2777-2783.2000
Winokur, P. L., Vonstein, D. L., Hoffman, L. J., Uhlenhopp, E. K., and Doern, G. V. (2001). Evidence for transfer of CMY-2 AmpC beta-lactamase plasmids between Escherichia coli and Salmonella isolates from food animals and humans. Antimicrob. Agents Chemother. 45, 2716–2722. doi: 10.1128/AAC.45.10.2716-2722.2001
Wittwer, C. T., Fillmore, G. C., and Hillyard, D. R. (1989). Automated polymerase chain reaction in capillary tubes with hot air. Nucleic Acids Res. 17, 4353–4357. doi: 10.1093/nar/17.11.4353
Keywords: Salmonella, antibiotic usage, antimicrobial resistance, poultry, plasmids
Citation: Bythwood TN, Soni V, Lyons K, Hurley-Bacon A, Lee MD, Hofacre C, Sanchez S and Maurer JJ (2019) Antimicrobial Resistant Salmonella enterica Typhimurium Colonizing Chickens: The Impact of Plasmids, Genotype, Bacterial Communities, and Antibiotic Administration on Resistance. Front. Sustain. Food Syst. 3:20. doi: 10.3389/fsufs.2019.00020
Received: 05 December 2018; Accepted: 14 March 2019;
Published: 16 April 2019.
Edited by:
Walid Alali, Kuwait University, KuwaitReviewed by:
Steven Lee Foley, National Center for Toxicological Research (FDA), United StatesCopyright © 2019 Bythwood, Soni, Lyons, Hurley-Bacon, Lee, Hofacre, Sanchez and Maurer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: John J. Maurer, amptYXVyZXJAdnQuZWR1
†Present Address: Margie D. Lee, Department of Biomedical Sciences and Pathobiology, Virginia-Maryland College of Veterinary Medicine, Blacksburg, VA, United States
John J. Maurer, Department of Animal and Poultry Sciences, College of Agriculture and Life Sciences, and Virginia Tech University, Blacksburg, VA, United States
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.