- 1The ithree institute, University of Technology Sydney, Sydney, NSW, Australia
- 2Elizabeth Macarthur Agricultural Institute, NSW Department of Primary Industries, Sydney, NSW, Australia
IncHI2 ST3 plasmids are known carriers of multiple antimicrobial resistance genes. Complete plasmid sequences from multiple drug resistant Escherichia coli circulating in Australian swine is however limited. Here we sequenced two related IncHI2 ST3 plasmids, pSDE-SvHI2, and pSDC-F2_12BHI2, from phylogenetically unrelated multiple-drug resistant Escherichia coli strains SvETEC (CC23:O157:H19) and F2_12B (ST93:O7:H4) from geographically disparate pig production operations in New South Wales, Australia. Unicycler was used to co-assemble short read (Illumina) and long read (PacBio SMRT) nucleotide sequence data. The plasmids encoded three drug-resistance loci, two of which carried class 1 integrons. One integron, hosting drfA12-orfF-aadA2, was within a hybrid Tn1721/Tn21, with the second residing within a copper/silver resistance transposon, comprising part of an atypical sul3-associated structure. The third resistance locus was flanked by IS15DI and encoded neomycin resistance (neoR). An oqx-encoding transposon (quinolone resistance), similar in structure to Tn6010, was identified only in pSDC-F2_12BHI2. Both plasmids showed high sequence identity to plasmid pSTM6-275, recently described in Salmonella enterica serotype 1,4,[5],12:i:- that has risen to prominence and become endemic in Australia. IncHI2 ST3 plasmids circulating in commensal and pathogenic E. coli from Australian swine belong to a lineage of plasmids often in association with sul3 and host multiple complex antibiotic and metal resistance structures, formed in part by IS26.
Introduction
In Australia, restrictions on live animal imports, geographic isolation, and sound antibiotic stewardship have limited the incorporation and spread of genes encoding resistance to antibiotics used to treat serious human infections among Enterobacteriaceae circulating in food animals (Turner, 2011; Abraham et al., 2015; Reid et al., 2017; Kidsley et al., 2018). Australian porcine Escherichia coli are, albeit at low frequency, known to carry blaCMY−2, blaCTX−M−14, and blaCTX−M−9, possibly because of the use of ceftiofur as an off-label, last line antibiotic to treat serious disease (Abraham et al., 2015). First generation antibiotics; commonly tetracyclines, penicillins, and sulphonamides are otherwise used for the therapeutic treatment of bacterial infections in swine (Jordan et al., 2009). Both pathogenic and commensal E. coli sourced from the feces of pig carry class 1 integrons and are often multiple drug resistant (MDR) (Abraham et al., 2015; Wyrsch et al., 2015; Reid et al., 2017). Notably, the insertion element IS26 has infiltrated the genomes of commensal E. coli of porcine origin, where it has played a role in altering the genetic context of clinical class 1 integrons and facilitated the acquisition of further resistances (Reid et al., 2017). Furthermore, it is important to identify and characterize the genetic vehicles that can carry class 1 integrons and genes encoding resistance to first generation antibiotics which are widespread in commensal E. coli populations in Australian pigs (Abraham et al., 2015; Reid et al., 2017; Kidsley et al., 2018) because they are likely to acquire genes encoding resistance to clinically relevant antibiotics when resident in commensal enterobacterial populations in the gut of humans and companion animals.
Plasmids of the incompatibility group HI2 (IncHI2) have been linked with the carriage of tellurium resistance, plus a broad range of antimicrobial resistance genes, including blaIMP, blaNDM, and blaVIM (carbapenemase resistance) (Abraham et al., 2016; Dolejska et al., 2016; Falgenhauer et al., 2017), blaCTX−M (Garcia Fernandez et al., 2007), mcr-1 (plasmid-mediated colistin resistance) (Gilrane et al., 2017; Li et al., 2017; Zheng et al., 2017), oqxAB and aac(6')-Ib-cr (plasmid-mediated quinolone resistance) (Fang et al., 2016), and resistance genes effective against first generation antibiotics and disinfectants (Cain et al., 2010; Cain and Hall, 2012; Abraham et al., 2016; Dolejska et al., 2016; Gilrane et al., 2017). Notably, clinically-important resistance genes have been associated with IncHI2 plasmids that also encode resistance to copper, zinc and, arsenic residues (Fang et al., 2016). In Australia, IncHI2 plasmids carrying a diverse range of antimicrobial resistance genes have been found in MDR E. coli (Dolejska et al., 2016; Saputra et al., 2017), Salmonella enterica serovar Typhimurium (Cain et al., 2010; Cain and Hall, 2012; Billman-Jacobe et al., 2018), Enterobacter cloacae (Sidjabat et al., 2015), and Enterobacter hormaechei susbp. Oharae (Monahan et al., 2019).
The insertion sequence IS26 of the IS6 family plays a significant role in the capture, assembly and mobilization of drug resistance genes found on plasmids (Dionisi et al., 2009; Cain et al., 2010; Shahid, 2010; Venturini et al., 2010, 2013; Lai et al., 2013; Abbo and Hooton, 2014; Chavda et al., 2015; Reid et al., 2015; Garcia et al., 2016) and in the chromosome (Roy Chowdhury et al., 2015, 2018). IS26 may promote plasmid stability and persistence by mediating deletions of plasmid backbone sequence, the expression of which incurs a burden to the host (Porse et al., 2016). IS26 can also facilitate the generation of hybrid virulence/resistance plasmids, formed by co-integration of plasmids separately carrying resistance and virulence gene cargo (Mangat et al., 2017; Wong et al., 2017). Although IS26 has no target site specificity (Harmer et al., 2014), it is often observed localizing next to or within class 1 integrons in multiple drug resistant E. coli recovered from the feces of both healthy pigs, poultry and cattle with gastrointestinal disease (Dawes et al., 2010; Reid et al., 2017). IS26 can shape the structure of class 1 integrons by facilitating the addition of foreign DNA flanked by IS26, and by generating inversion and deletions of sequence within complex resistance structures. Consequently, many class 1 integrons have lost genes that reside within the often observed 3′-conserved sequence (3′-CS), particularly the sulphonamide resistance gene sul1 (Reid et al., 2017). However, sulphonamide resistance persists globally (Grape et al., 2003; Bean et al., 2005; Suhartono et al., 2017), despite restrictions on its use (Enne et al., 2001), with sul2 (Bean et al., 2005) and sul3 genes (Grape et al., 2003; Perreten and Boerlin, 2003; Zhou et al., 2014; Reid et al., 2017) found frequently in close association with class 1 integrons. A sul4 gene associated with a chromosomal locus containing the folate synthesis gene folK and a copy of ISCR20 has recently been described (Razavi et al., 2017). Emerging trends show that genes encoding resistance to last line drugs, such as mcr-1, blaCTX−M, and carbapenemases are captured on plasmids already carrying genes encoding resistances to first generation antibiotics (Alonso et al., 2017; Botts et al., 2017; Delannoy et al., 2017; Poirel et al., 2017). Any one of a number of selection pressures including heavy metals and biocides may be sufficient to then facilitate the persistence and spread of multiple drug resistance plasmids (Argudín et al., 2019). This is further compounded by reports that agrichemicals can alter selection for drug resistant bacteria (Kurenbach et al., 2015, 2018). The potential for complex resistance structures to persist under multiple different selective pressures and across diverse environments underpin the importance of adapting a One Health approach to antimicrobial resistance gene surveillance in humans, food and companion animals, agriculture, effluent (municipal, hospital, and agricultural), and the environments impacted by effluent from diverse sources (Djordjevic et al., 2013; Wyrsch et al., 2016; Huang et al., 2017; Mir et al., 2018).
Here we used a combination of Illumina and PacBio SMRT sequencing to completely close two IncHI2:ST3 MDR plasmids from E. coli recovered from the feces of Australian swine. One of these, pSDE-SvHI2 is from a severe ETEC/ExPEC pathogen (E. coli O157 SvETEC) (Wyrsch et al., 2015), and the other, pSDC-F2_12BHI2, is from Escherichia coli F2_12B, a commensal E. coli ST93 (Reid et al., 2017). Phylogenetic and genomic comparisons were undertaken between these two plasmid sequences and pSTM6-275, recently isolated from a monophasic variant of Salmonella enterica in Australian pigs (Dyall-Smith et al., 2017), as well as all available IncHI2 ST3 plasmids on GenBank.
Materials and Methods
Strains and DNA Preparation
Plasmid pSDE-SvHI2 was resolved from Escherichia coli O157 SvETEC (CC23:O157:H19), a severe pathogen that caused intractable disease within an Australian commercial piggery in 2007 (Wyrsch et al., 2015). Likewise, pSDC-F2_12BHI2 was resolved from Escherichia coli F2_12B, a commensal ST93:O7:H4 strain isolated from a rectal swab of a commercially raised pig, also sourced in 2007 (Reid et al., 2017). Full descriptions of these strains plus information on Illumina short read sequencing and assembly have been published previously (Wyrsch et al., 2015; Reid et al., 2017). Un-sheared genomic DNA suitable for SMRT sequencing was prepared from mid-log phase sub-cultures of strains grown overnight in LB broth using a gentle phenol-chloroform extraction protocol. Genomic DNA samples were checked for appropriate DNA concentrations and integrity using a Qubit dsDNA HS (high sensitivity, 0.2 to 100 ng) Assay Kit on Qubit 2.0 fluorometer (Life Technologies) and for shearing by agarose gel electrophoresis.
Genome Sequencing and Assembly
Long read sequencing was performed by the Ramaciotti Center for Genomics using a Pacific Biosciences RSII sequencer with P6-C4 chemistry. One Single Molecule Real-Time (SMRT) Cell was used for each strain. Plasmid sequences were identified from whole-genome assemblies produced from hybrid read sets (both Illumina and SMRT reads) using the Unicycler pipeline v0.3.1 (Wick et al., 2017), which internally relied upon SPAdes v3.10.1 (Nurk et al., 2013), Bowtie2 v2.3.0 (Langmead and Salzberg, 2012), samtools v1.4.1 (Li et al., 2009), and Pilon v1.22 (Walker et al., 2014).
Annotated sequence for pSDE-SvHI2 and pSDC-F2_12BHI2 have been deposited in GenBank under accession numbers MH287084 and MH287085, respectively.
Plasmid Typing
Plasmids, particularly those that spread amongst the Enterobacteriaceae, have been typed by their in vivo incompatibility and cell exclusion patterns. Many of these incompatibility groups can now be subtyped by allelic variations in select conserved genes, forming plasmid multi-locus sequence typing schemes. Incompatibility typing and plasmid multi-locus sequence typing was performed through the Center of Genomic Epidemiology website (http://www.genomicepidemiology.org/) using PlasmidFinder (Carattoli et al., 2014) and the IncHI2 pDLST scheme (Garcia-Fernandez and Carattoli, 2010).
Phylogeny and Alignment Analyses
Analyses of conserved core single nucleotide polymorphisms (SNPs) was performed using the Harvest suite (Parsnp v1.2, utilizing the Phipack recombination filter, and gingr v1.2) (Treangen et al., 2014). An analysis was run utilizing all available IncHI2 ST3 plasmids from GenBank (Supplementary Table 1), plus two IncHI2 ST1 plasmids, reference pR478 (NC_005211) and pIMP4-SEM1 (KX810825). A second tree was then generated with the ST3 plasmids only, with pSDC_F2_12BHI2 as reference. Gene identification and genomic comparisons were performed using a combination of BLASTn (Camacho et al., 2009) and progressiveMauve (Darling et al., 2010) alignments. Figures were generated from sequence data using SnapGene v3.3.4, BRIG v0.95 (Alikhan et al., 2011), and Easyfig v2.2.2 (Sullivan et al., 2011).
Annotations were managed using SnapGene v3.3.4. Automated annotations were generated by RASTtk (Brettin et al., 2015). Insertion sequences were identified and annotated manually with the aid of ISfinder (Siguier et al., 2006). Remaining annotations were performed manually utilizing BLASTn and publicly available databases, including The Repository of Antibiotic-Resistance Cassettes (Tsafnat et al., 2011) (http://rac.aihi.mq.edu.au/rac/), The Comprehensive Antibiotic Resistance Database (Jia et al., 2017) (https://card.mcmaster.ca/), and the GenBank nucleotide database (https://www.ncbi.nlm.nih.gov/nuccore/).
Results
Unicycler hybrid assemblies resolved pSDE-SvHI2 as a 275,402 bp circular sequence and pSDC-F2_12BHI2 as a 288,288 bp circular sequence with 344 (140 hypotheticals) and 357 (134 hypotheticals) coding sequences (CDS) respectively. Both sequences typed as incompatibility group HI2, sequence type 3 (IncHI2 ST3) plasmids. The full map of each plasmid is presented in Figure 1, along with content comparisons to the most related ST3 plasmids, most of which are from Australia and China. The plasmids had a typical IncHI2 structure, including the RepHIA and RepHI2 replication operons, trh and tra transfer operons and tellurite resistance (ter operon) (Gilmour et al., 2004), plus each carried three variants of complex resistance loci, two of which are class 1 integron associated, one encoding sul1 in a Tn1721/Tn21 hybrid transposon, and one encoding sul3 from within a Tn7-like copper/silver resistance transposon.
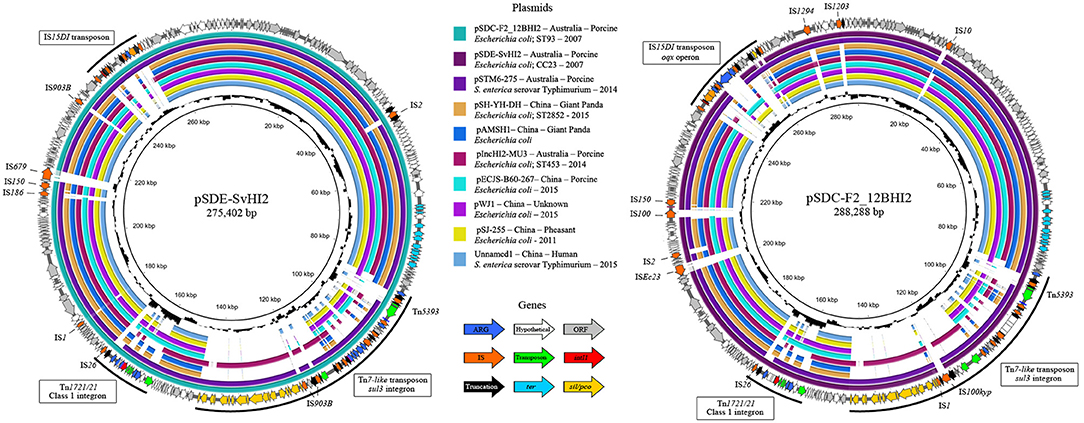
Figure 1. Plasmid maps and alignments of pSDE-SvHI2 (Left) and pSDC-F2_12BHI2 (Right). Maps display plasmid annotations, with genes color coded by function. Annotations highlight important complex resistance structures and insertions on each plasmid. Inner rings show BLASTn alignments against related IncHI2 ST3 plasmid sequences. Center histograms show GC content.
Single nucleotide polymorphism analysis was performed on the set of all ST3 plasmids (Figure 2, Supplementary Table 2), and on this same set plus IncHI2 ST1 plasmids as reference to confirm tree topology (Supplementary Figure 1). Forty-three plasmid sequences available from 2006 to 2017 were included in the ST3-only analysis. Based on the availability of metadata, plasmid sequences were from Enterobacteriaceae of different sources in the pacific region, including Australian porcine production operations and multiple human, agricultural and environmental sources in China. The ST3 sequences formed a single clade with one exception, MH715960 from Taiwan, which separated with 125 core SNPs compared to the Australian reference sequence. Of these 125 core SNPs, 101 are within an ~1.5 kb region of the ter operon. The remaining sequences formed four major subclades, with closest relatives to the Australian reference ranging from two to 14 SNPs. One subclade, highlighted red in Figure 2, demonstrates relatedness between three Australian plasmids from the feces of pigs, pSDE-SvHI2, pSDC-F2_12BHI2, and pSTM-275, an apparently separate Australian plasmid lineage (pIncHI2-MU3) also from pig feces, and six other plasmid sequences from diverse sources in China. Content comparisons of this clade can be seen in Figure 1. Of the remaining plasmids, the most distant relative had 25 SNPs identified from conserved core sequence, suggesting a close evolutionary relationship between plasmid sequences reported from Australia and China.
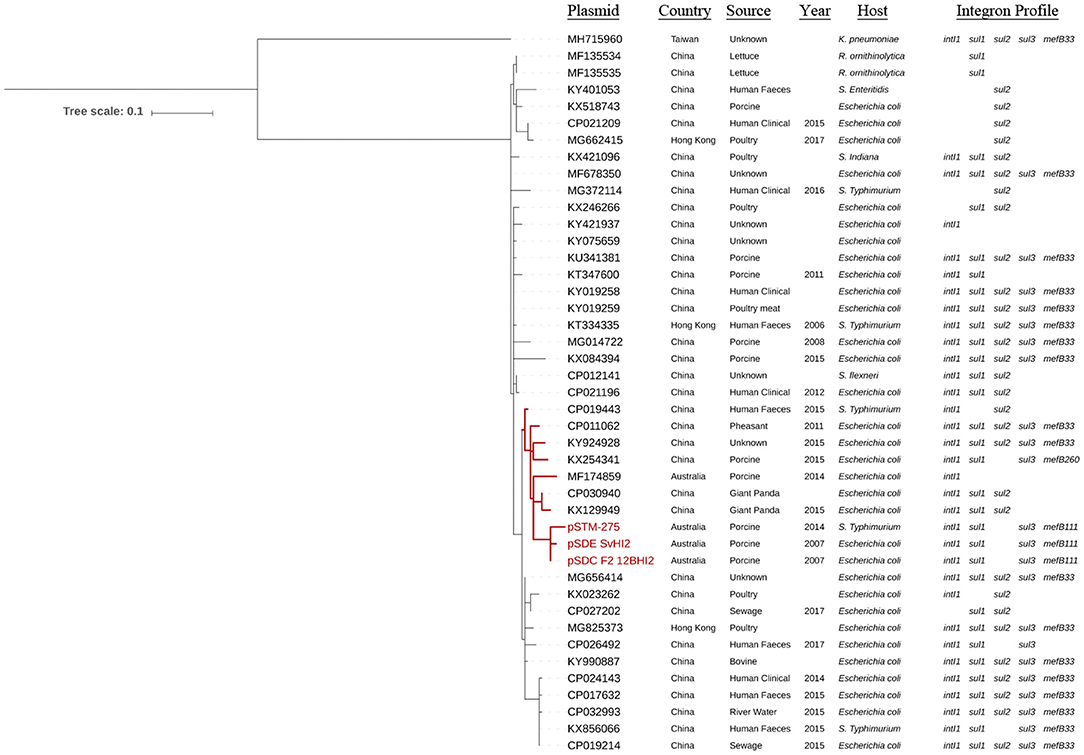
Figure 2. Core phylogeny of IncHI2:ST3 plasmid sequences. Mid-point rooted phylogram constructed using Parsnp, using a conserved core SNP alignment of IncHI2 ST3 sequences, with pSDC-F2_12BHI2 as reference. Sequence metadata and the presence of integron marker genes is also included. Scale shows number of SNPs per site.
The ST3 plasmids carried multiple class 1 integron structures with different sul genes including sul1, sul2, sul3, and a sul3-associated mefB, encoding a macrolide efflux pump (Figure 2). One sequence (KX254341) of Chinese origin was observed with only 260 bp remaining of mefB (ΔmefB260). The Australian IncHI2 ST3 plasmids explored here carry ΔmefB111. The remaining sul3-positive plasmids carry a ΔmefB33 signature, aside from CP026492 from China which completely lacks mefB. This includes both the pseudo-phylogenetically distant MH715960 from Taiwan and the earliest Chinese plasmid sourced from 2006. Interestingly, one single subclade of sequences from China and Hong Kong was universally negative for intI1 and sul3, but not for either sul1 or sul2. Many of these plasmids are also associated with various globally important resistance genes, including blaCTX−M, oqxAB and mcr-1 (Supplementary Table 3).
pSDE-SvHI2, pSDC-F2_12BHI2, and pSTM-275 host a class 1 integron located within a Tn1721/Tn21 hybrid tetracycline resistance transposon (Figure 3). This integron structure is host to the only complete intI1 gene on the plasmids, and has acquired trimethoprim (dfrA12), orfF and streptomycin/spectinomycin (aadA2) resistance gene cassettes. Comparisons to similar Tn1721/Tn21 hybrid transposons show identical sequence across the hybridization point between tnpR1721and tnpM21, however these sequences also show a modified transposition module, with a loss of Tn1721 tnpA sequence between two homologous 8 bp regions (CCAGGGCG), between the second ΔtnpA1721 and its neighboring predicted relaxase. The In2-like class 1 integron tniA gene is truncated to 436 bp by insertion of an IS26 element with no observable associated repeats, suggesting a complex evolutionary path to this final structure. As part of the class 1 integron 3′-CS, the structure encodes sulphonamide resistance (sul1), however in pSTM6-275 the terminal IS26 truncation lies within the sul1 gene, giving a distinguishable gene marker.
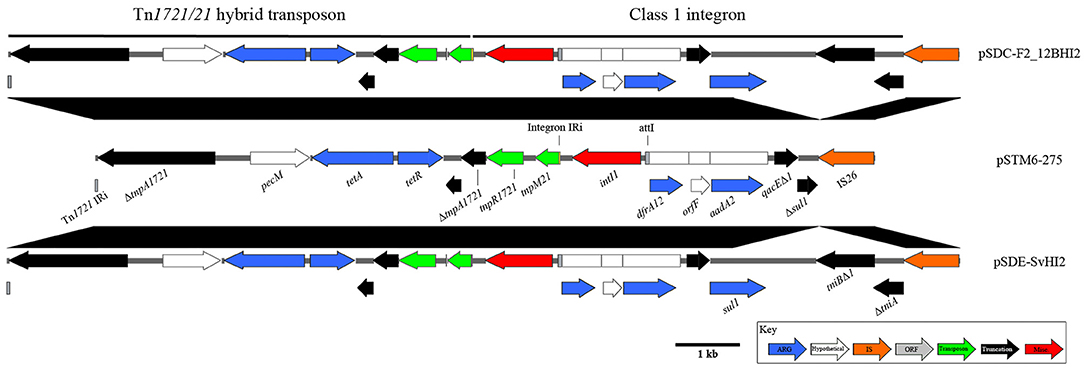
Figure 3. Diagrams and comparisons of Tn1721/Tn21 hybrid transposons carrying a class 1 integron. Clinical class 1 integron-based locus, with comparisons to the highly similar structure found in pSTM6-275 (center). Genes are colored by function, with black connecting regions indicating homology between loci.
A large (58,759 bp), mosaic combination of a globally observed Tn7-like transposon (copper and silver resistance, sil/pco), and transposons mobilizing antimicrobial resistance genes (Figure 1) was also observed in this Australian plasmid lineage. An IS26-associated, class 1 integron-encoding structure has inserted near the Tn7-like transposition module (Figure 4). From the pSTM6-275 sequence, it appears this insertion was in a Tn3 associated gene, however in pSDE-SvHI2 and pSDC-F2_12BHI2 IS26 activity has altered sequences around the insertion site. The integron is a variant of a sul3-associated structure that has been described previously in Australian plasmid pCERC3 (KR827684), now including signature IS26-meditated deletions in intI1 (ΔintI1705) and mefB (ΔmefB111). The integron cassette array encodes estX, psp, aadA2, cmlA, and aadA1. The crossover site described near qacH reported in pCERC3 was identical, confirming it is a derivative of the same integron sequence encoding sul3-qacH-mefB, previously seen within a Tn21 background (Moran et al., 2016). We also noted an IS1203* insertion into the qacH ORF, which may prove epidemiologically useful. The integron structure is followed by a short intermediate sequence encoding rop, then by a Tn5393 variant (encoding streptomycin resistance) with a novel IS903B* insertion into IS1133.
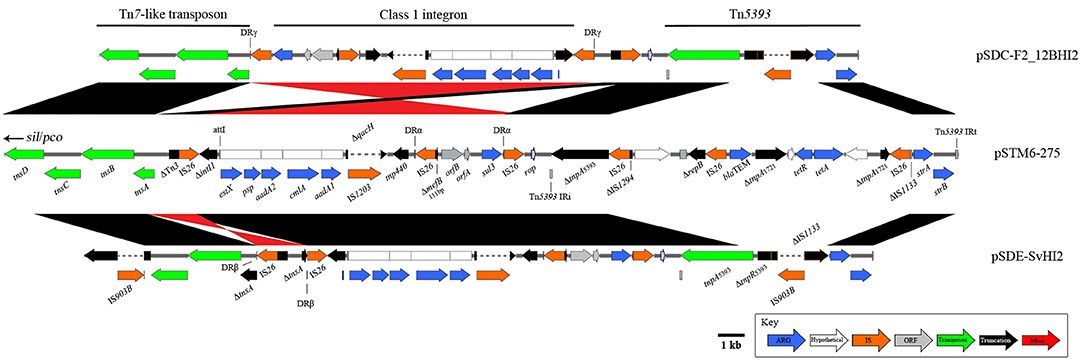
Figure 4. Diagrams and comparisons of sul3-associated resistance structures carrying an IS26 truncated intI1 Structural comparisons between related sul3 encoding regions, comprising a putative transposon, sul3-CS integron and Tn5393. Alignments in black show homology, with inversions indicated in red. Plasmids pSDE-SvHI2 (bottom) and pSDC-F2_12BHI2 (top) both show further rearrangement compared to the more recent pSTM6-275 structure.
From these sequences it is difficult to ascertain what originally mobilized the ΔintI1 module into the Tn7-like transposon. While IS26 flanks the captured sequence (ΔintI1 to sul3), no repeats have been identified to indicate a clean insertion location near the Tn7-like transposon, nor the bordering sequence near rop.
Of the 1,030 bp sequence by rop, 257 bases nearest the Tn5393 repeat are associated with the tetracycline resistance transposon Tn1721 [AJ634602.1] while the remaining 773 bp nearest the IS26/sul3 end matched IncN plasmid backbone [HF545433.1]. These remnant sequences may give indications as to the sources of these structures should more pertinent references become available. Further, the structures we have described have undergone individual insertions and inversion events. In pSDE-SvHI2, an IS26 insertion into tnsA has led to the generation of new direct repeats (DRβ) and has subsequently led to an IS26 mediated inversion. In pSDC-F2_12BHI2, an IS26 insertion near the tnsA ORF has generated DRγ, and an inversion event has again occurred, flipping the structure between ΔintI1 and sul3. A separate inversion event has then occurred between the IS26 elements nearest to rop, re-inverting this sequence to match the original orientation. Importantly, the insertion of a complex Tn1721/Tn2/IS26 structure into Tn5393 within pSTM6-275 demonstrates further resistance consolidation occurring in the 7-year gap between pSDE-SvHI2/pSDC-F2_12BHI2 and pSTM-275 isolations. This insert encoded tetAB and blaTEM. Unfortunately, this insertion has removed most of IS1133, and we cannot determine if pSTM6-275 carried the ΔIS1133-IS903B* insertion described above. The other Australian plasmid, pIncHI2-MU3, encodes the Tn7-like sil/pco heavy metal resistance transposon and is positive for Tn3 and intI1, but lacks any sul gene.
Finally, an IS15DI (IS6-family element; 3 SNPs compared to IS26) mobilized resistance region with a highly recombined and varied structure (Figure 5) has also been identified in pSDE-SvHI2 and pSDC-F2_12BHI2. The original IS15DI insertion into the plasmid backbone has generated 8 bp direct repeats (AACAGCGT) that remain flanking the structure. This composite transposon appears to mobilize neomycin resistance (neoR), and a truncated bleomycin resistance (ble) gene. It has also acquired IncN backbone (tra genes) and the relBE toxin/anti-toxin system, alongside various other whole and truncated IS elements. An internal IS15DI element is present in both pSDE-SvHI2 and pSTM6-275 and is replaced by a Tn6010-like element (similar to KT716391.1, mobilized by IS15DI) carrying oqxABR in pSDC-F2_12BHI2. there is an IS15DI mediated inversion of the tra associated region in pSDC-F2_12BHI2. Plasmid pSDE-SvHI2 has an IS15DI insertion into IS1294, generating DRδ. This has been followed by a rearrangement event leading to the movement of DRδ and neoR toward the terminal IS element, including an inversion, and a loss of sequence through to the ΔtnpATn3.
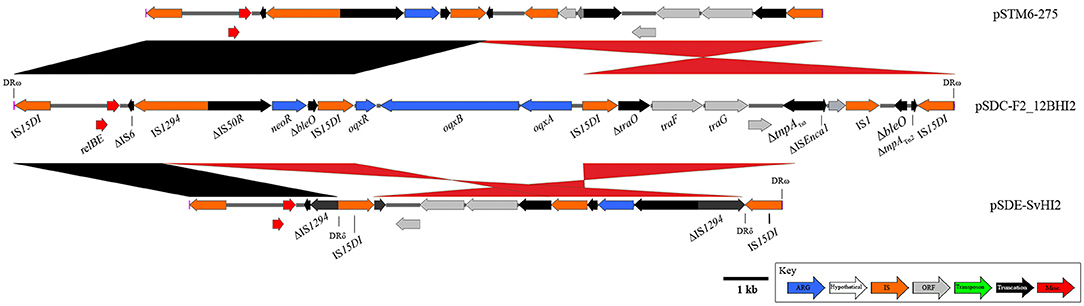
Figure 5. Diagrams and comparisons of an IS15DI-mobilized composite transposon. Composite transposon captured by IS26 variant IS15DI, mobilizing the neomycin resistance gene neoR. pSDC-F2_12BHI2 (center) also hosts a Tn6010-like element, inserted at the central IS15DI.
Discussion
Plasmids belonging to the incompatibility group HI2 are carriers of antimicrobial resistance genes globally. With the increased availability of IncHI2 sequences, a di-locus sequence typing scheme was established (Garcia-Fernandez and Carattoli, 2010) to aid in tracking plasmid lineage development and dissemination. In Australia, most IncHI2 plasmids have been observed mobilizing blaIMP−4, the predominant carbapenemase-encoding gene within Enterobacteriaceae on the eastern seaboard of Australia (Sidjabat et al., 2015). Here we report a comprehensive analysis of two Australian IncHI2 ST3 plasmids from E. coli, and make comparisons to plasmid pSTM6-275 from S. enterica serotype 1,4,[5],12:i:- (Dyall-Smith et al., 2017). Phylogenetic analyses of the current available IncHI2 ST3 dataset showed these three Australian porcine IncHI2 ST3 plasmids are nested within a subclade alongside another Australian porcine ST3 plasmid sequence, pIncHI2-MU3 (Abraham et al., 2018), which hosts neither of the class 1 integrons identified here, but does host the metal resistance genes as part of the Tn7-like transposon. This suggests various lineages of ST3 plasmid may be circulating within Australian porcine agricultural operations.
The IncHI2 ST3 plasmids sequenced to date have been resolved from multiple Enterobacteriaceae including Escherichia coli, various Salmonella enterica serovars, Shigella flexneri, Klebsiella pneumonia, and Raoultella ornithinolytica. Additionally, these Enterobacteriaceae isolates were taken from different sources, including pathogens and non-pathogens. As these ST3 plasmids carry diverse resistance gene cargo and may be found in numerous members of the Enterobacteriaceae family they will likely be important targets for future antimicrobial resistance surveillance (Fang et al., 2018). Our analyses also demonstrate evidence of differential gene acquisition by these plasmids, continuing to expand and alter the antimicrobial resistance gene repertoire which they are associated with internationally. Particularly, sul3-associated integrons carrying different ΔmefB fragments were identified here.
The sul3 gene was first described in pigs (Perreten and Boerlin, 2003) and has since been reported widely in association with Enterobacteriaceae in humans, farm animals and their waste—particularly swine (Antunes et al., 2007; Phuong Hoa et al., 2008; Byrne-Bailey et al., 2009; Curiao et al., 2011; Moran et al., 2016). In Australia and elsewhere, sul3 is identified frequently in commensal E. coli from the feces of swine (Guerra et al., 2004; Bischoff et al., 2005; Reid et al., 2017) but less frequently in avian pathogenic E. coli (APEC) from diverse poultry production systems in Australia (Cummins et al., 2019). These observations suggest that swine production, and food animal production more broadly, plays a major role in the evolution of IncHI2 ST3 plasmids that carry sul3. Plasmid pSTM6-275 was shown to be thermostable at 44°C, and would readily conjugate at 27°C but not at 37°C, highlighting the IncHI2 plasmid families propensity to transfer under environmental conditions (Garcia-Fernandez and Carattoli, 2010; Billman-Jacobe et al., 2018).
Genetic signatures noted in this study; a combination of the IncHI2 ST3 di-locus typing alleles and mefB111 have been seen in commensal E. coli short read assemblies from both the farm were strain F2_12B was sourced, and a separate Australian porcine operation circa 2007 (Reid et al., 2017). Combined with the isolation of pSTM6-275 in 2014, our data suggests these plasmids are purveyors of drug resistance in porcine agricultural settings, likely aided by the use of copper feed additives and the widespread use of first generation antibiotics (Jordan et al., 2009). It is notable that the IncHI2 plasmids sequenced in our study carry a unique IS6 family mobilized transposon encoding neomycin resistance, and additional fluoroquinolone resistance in pSDC-F2_12B. Also, they are host to a globally observed Tn7-like heavy metal resistance transposon (Fang et al., 2016), that has in turn become host to a variant of a sul3-mefB class 1 integron, which appears unique to Australian samples at this time. The ΔmefB111-associated class 1 integron was carried only by the Australian plasmids pSDE-SvHI2, pSDC-F2_12BHI2 and pSTM-275, while ΔmefB33 was observed throughout the remainder of the ST3 plasmid clade. Notably, ΔmefB33 variants are not associated with the presence of the Tn7-like metal resistance transposon, so characterizing their distribution and methods of mobilization will be critical to monitoring complex resistance locus diversification.
Genomic sequence data has shown that IS26 is playing an important role in shaping the context of MDR islands in MDR fecal E. coli in Australian commercial pigs. Recently, we reported high carriage rates (101/103 isolates; 98%) of IS26 among commensal E. coli carrying class 1 integrons (Reid et al., 2017), however we were unable to determine the genetic context of some class 1 integrons. Nonetheless, the frequency of carriage of sul3 and IncHI2 plasmids in our earlier study was significant in two separate commercial swine production facilities. Here we show that IS26 (and the IS15DI variant) has played a pivotal role in the evolution of several porcine IncHI2 ST3 plasmids both as a means of creating deletions and inversions and altering antibiotic resistance gene content.
Finally, the epidemiological analysis of Enterobacteriaceae has been heavily influenced by the spread of antimicrobial resistance and the development of MDR pathogens, leading to a focus on the detection of clinically-relevant resistance genes (blaIMP, mcr-1, and blaCTX−M are examples of this). To further understand the true variety and dissemination of multiple drug resistance, sampling from agricultural and environmental sources impacted by antimicrobials will be important.
Data Availability
The datasets generated for this study can be found in GenBank, MH287084 and MH287085.
Author Contributions
EW performed genomic analyses, generated figures, and drafted the manuscript. TC provided curated E. coli collections for the study. PRC assisted with data interpretation. ML, EW, and CR prepared sequencing samples. MD performed genome assemblies. SD conceived the study and together with EW and PRC wrote the manuscript. All authors provided edits, read, and approved the manuscript.
Funding
This work was in part supported by the Australian Research Council, linkage grants LP150100912 and LE150100031. This project was partly funded by the Australian Center for Genomic Epidemiological Microbiology (Ausgem), a collaborative partnership between the NSW Department of Primary Industries and the ithree institute at the University of Technology Sydney. EW and CR are recipients of Australian Government Research Training Program Scholarships.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful for the helpful discussions with Helen Jacobe-Billman.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2019.00018/full#supplementary-material
References
Abbo, L. M., and Hooton, T. M. (2014). Antimicrobial stewardship and urinary tract infections. Antibiotics 3, 174–192. doi: 10.3390/antibiotics3020174
Abraham, S., Jordan, D., Wong, H. S., Johnson, J. R., Toleman, M. A., Wakeham, D. L., et al. (2015). First detection of extended-spectrum cephalosporin- and fluoroquinolone-resistant Escherichia coli in Australian food-producing animals. J. Global Antimicrob. Resist. 3, 273–277. doi: 10.1016/j.jgar.2015.08.002
Abraham, S., Kirkwood, R. N., Laird, T., Saputra, S., Mitchell, T., Singh, M., et al. (2018). Dissemination and persistence of extended-spectrum cephalosporin-resistance encoding IncI1-blaCTXM-1 plasmid among Escherichia coli in pigs. ISME J. 12, 2352–2362. doi: 10.1038/s41396-018-0200-3
Abraham, S., O'dea, M., Trott, D. J., Abraham, R. J., Hughes, D., Pang, S., et al. (2016). Isolation and plasmid characterization of carbapenemase (IMP-4) producing Salmonella enterica Typhimurium from cats. Sci. Rep. 6:35527. doi: 10.1038/srep35527
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Alonso, C. A., Michael, G. B., Li, J., Somalo, S., Simon, C., Wang, Y., et al. (2017). Analysis of blaSHV-12-carrying Escherichia coli clones and plasmids from human, animal and food sources. J. Antimicrob. Chemother. 72, 1589–1596. doi: 10.1093/jac/dkx024
Antunes, P., Machado, J., and Peixe, L. (2007). Dissemination of sul3-containing elements linked to class 1 integrons with an unusual 3' conserved sequence region among Salmonella isolates. Antimicrob. Agents Chemother. 51, 1545–1548. doi: 10.1128/AAC.01275-06
Argudín, M. A., Hoefer, A., and Butaye, P. (2019). Heavy metal resistance in bacteria from animals. Res. Vet. Sci. 122, 132–147. doi: 10.1016/j.rvsc.2018.11.007
Bean, D. C., Livermore, D. M., Papa, I., and Hall, L. M. (2005). Resistance among Escherichia coli to sulphonamides and other antimicrobials now little used in man. J. Antimicrob. Chemother. 56, 962–964. doi: 10.1093/jac/dki332
Billman-Jacobe, H., Liu, Y., Haites, R., Weaver, T., Robinson, L., Marenda, M., et al. (2018). pSTM6-275, a conjugative IncHI2 plasmid of Salmonella that confers antibiotic and heavy metal resistance under changing physiological conditions. Antimicrob. Agents Chemother. 62:e02357–17. doi: 10.1128/AAC.02357-17
Bischoff, K. M., White, D. G., Hume, M. E., Poole, T. L., and Nisbet, D. J. (2005). The chloramphenicol resistance gene cmlA is disseminated on transferable plasmids that confer multiple-drug resistance in swine Escherichia coli. FEMS Microbiol. Lett. 243, 285–291. doi: 10.1016/j.femsle.2004.12.017
Botts, R. T., Apffel, B. A., Walters, C. J., Davidson, K. E., Echols, R. S., Geiger, M. R., et al. (2017). Characterization of four multidrug resistance plasmids captured from the sediments of an urban Coastal Wetland. Front. Microbiol. 8:1922. doi: 10.3389/fmicb.2017.01922
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Byrne-Bailey, K. G., Gaze, W. H., Kay, P., Boxall, A. B., Hawkey, P. M., and Wellington, E. M. (2009). Prevalence of sulfonamide resistance genes in bacterial isolates from manured agricultural soils and pig slurry in the United Kingdom. Antimicrob. Agents Chemother. 53, 696–702. doi: 10.1128/AAC.00652-07
Cain, A. K., and Hall, R. M. (2012). Evolution of IncHI2 plasmids via acquisition of transposons carrying antibiotic resistance determinants. J. Antimicrob. Chemother. 67, 1121–1127. doi: 10.1093/jac/dks004
Cain, A. K., Liu, X., Djordjevic, S. P., and Hall, R. M. (2010). Transposons related to Tn1696 in IncHI2 plasmids in multiply antibiotic resistant Salmonella enterica serovar Typhimurium from Australian animals. Microb. Drug Resist. 16, 197–202. doi: 10.1089/mdr.2010.0042
Camacho, C., Coulouris, G., Avagyan, V., Ma, N., Papadopoulos, J., Bealer, K., et al. (2009). BLAST+: architecture and applications. BMC Bioinformatics 10:421. doi: 10.1186/1471-2105-10-421
Carattoli, A., Zankari, E., Garcia-Fernandez, A., Voldby Larsen, M., Lund, O., Villa, L., et al. (2014). In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob. Agents Chemother. 58, 3895–3903. doi: 10.1128/AAC.02412-14
Chavda, K. D., Chen, L., Jacobs, M. R., Rojtman, A. D., Bonomo, R. A., and Kreiswirth, B. N. (2015). Complete sequence of a bla(KPC)-harboring cointegrate plasmid isolated from Escherichia coli. Antimicrob. Agents Chemother. 59, 2956–2959. doi: 10.1128/AAC.00041-15
Cummins, M. L., Reid, C. J., Roy Chowdhury, P., Bushell, R. N., Esbert, N., Tivendale, K. A., et al. (2019). Whole genome sequence analysis of Australian avian pathogenic Escherichia coli that carry the class 1 integrase gene. Microb. Genomics 5, 1–13. doi: 10.1099/mgen.0.000250
Curiao, T., Canton, R., Garcillan-Barcia, M. P., De La Cruz, F., Baquero, F., and Coque, T. M. (2011). Association of composite IS26-sul3 elements with highly transmissible IncI1 plasmids in extended-spectrum-beta-lactamase-producing Escherichia coli clones from humans. Antimicrob. Agents Chemother. 55, 2451–2457. doi: 10.1128/AAC.01448-10
Darling, A. E., Mau, B., and Perna, N. T. (2010). progressiveMauve: multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 5:e11147. doi: 10.1371/journal.pone.0011147
Dawes, F. E., Kuzevski, A., Bettelheim, K. A., Hornitzky, M. A., Djordjevic, S. P., and Walker, M. J. (2010). Distribution of class 1 integrons with IS26-mediated deletions in their 3'-conserved segments in Escherichia coli of human and animal origin. PLoS ONE 5:e12754. doi: 10.1371/journal.pone.0012754
Delannoy, S., Le Devendec, L., Jouy, E., Fach, P., Drider, D., and Kempf, I. (2017). Characterization of colistin-resistant Escherichia coli isolated from diseased pigs in France. Front. Microbiol. 8:2278. doi: 10.3389/fmicb.2017.02278
Dionisi, A. M., Lucarelli, C., Owczarek, S., Luzzi, I., and Villa, L. (2009). Characterization of the plasmid-borne quinolone resistance gene qnrB19 in Salmonella enterica serovar Typhimurium. Antimicrob. Agents Chemother. 53, 4019–4021. doi: 10.1128/AAC.00294-09
Djordjevic, S. P., Stokes, H. W., and Roy Chowdhury, P. (2013). Mobile elements, zoonotic pathogens and commensal bacteria: conduits for the delivery of resistance genes into humans, production animals and soil microbiota. Front. Microbiol. 4:86. doi: 10.3389/fmicb.2013.00086
Dolejska, M., Masarikova, M., Dobiasova, H., Jamborova, I., Karpiskova, R., Havlicek, M., et al. (2016). High prevalence of Salmonella and IMP-4-producing Enterobacteriaceae in the silver gull on Five Islands, Australia. J. Antimicrob. Chemother. 71, 63–70. doi: 10.1093/jac/dkv306
Dyall-Smith, M. L., Liu, Y., and Billman-Jacobe, H. (2017). Genome sequence of an australian monophasic Salmonella enterica subsp. enterica Typhimurium Isolate (TW-Stm6) carrying a large plasmid with multiple antimicrobial resistance genes. Genome Announc. 5:e00793–17. doi: 10.1128/genomeA.00793-17
Enne, V. I., Livermore, D. M., Stephens, P., and Hall, L. M. (2001). Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet 357, 1325–1328. doi: 10.1016/S0140-6736(00)04519-0
Falgenhauer, L., Ghosh, H., Guerra, B., Yao, Y., Fritzenwanker, M., Fischer, J., et al. (2017). Comparative genome analysis of IncHI2 VIM-1 carbapenemase-encoding plasmids of Escherichia coli and Salmonella enterica isolated from a livestock farm in Germany. Vet. Microbiol. 200, 114–117. doi: 10.1016/j.vetmic.2015.09.001
Fang, L., Li, X., Li, L., Li, S., Liao, X., Sun, J., et al. (2016). Co-spread of metal and antibiotic resistance within ST3-IncHI2 plasmids from E. coli isolates of food-producing animals. Sci. Rep. 6:25312. doi: 10.1038/srep25312
Fang, L.-X., Li, X.-P., Deng, G.-H., Li, S.-M., Yang, R.-S., Wu, Z.-W., et al. (2018). High genetic plasticity in multidrug resistant ST3-IncHI2 plasmids revealed by sequence comparison and phylogenetic analysis. Antimicrob. Agents Chemother. 62:e02068–17. doi: 10.1128/AAC.02068-17
Garcia Fernandez, A., Cloeckaert, A., Bertini, A., Praud, K., Doublet, B., Weill, F. X., et al. (2007). Comparative analysis of IncHI2 plasmids carrying blaCTX-M-2 or blaCTX-M-9 from Escherichia coli and Salmonella enterica strains isolated from poultry and humans. Antimicrob. Agents Chemother. 51, 4177–4180. doi: 10.1128/AAC.00603-07
Garcia, V., Garcia, P., Rodriguez, I., Rodicio, R., and Rodicio, M. R. (2016). The role of IS26 in evolution of a derivative of the virulence plasmid of Salmonella enterica serovar Enteritidis which confers multiple drug resistance. Infect. Genet. Evol. 45, 246–249. doi: 10.1016/j.meegid.2016.09.008
Garcia-Fernandez, A., and Carattoli, A. (2010). Plasmid double locus sequence typing for IncHI2 plasmids, a subtyping scheme for the characterization of IncHI2 plasmids carrying extended-spectrum beta-lactamase and quinolone resistance genes. J. Antimicrob. Chemother. 65, 1155–1161. doi: 10.1093/jac/dkq101
Gilmour, M. W., Thomson, N. R., Sanders, M., Parkhill, J., and Taylor, D. E. (2004). The complete nucleotide sequence of the resistance plasmid R478: defining the backbone components of incompatibility group H conjugative plasmids through comparative genomics. Plasmid 52, 182–202. doi: 10.1016/j.plasmid.2004.06.006
Gilrane, V. L., Lobo, S., Huang, W., Zhuge, J., Yin, C., Chen, D., et al. (2017). Complete genome sequence of a colistin-resistant Escherichia coli strain harboring mcr-1 on an IncHI2 plasmid in the United States. Genome Announc. 5:e01095–17. doi: 10.1128/genomeA.01095-17
Grape, M., Sundstrom, L., and Kronvall, G. (2003). Sulphonamide resistance gene sul3 found in Escherichia coli isolates from human sources. J. Antimicrob. Chemother. 52, 1022–1024. doi: 10.1093/jac/dkg473
Guerra, B., Junker, E., and Helmuth, R. (2004). Incidence of the recently described sulfonamide resistance gene sul3 among German Salmonella enterica strains isolated from livestock and food. Antimicrob. Agents Chemother. 48, 2712–2715. doi: 10.1128/AAC.48.7.2712-2715.2004
Harmer, C. J., Moran, R. A., and Hall, R. M. (2014). Movement of IS26-associated antibiotic resistance genes occurs via a translocatable unit that includes a single IS26 and preferentially inserts adjacent to another IS26. MBio 5:e01801–01814. doi: 10.1128/mBio.01801-14
Huang, X., Yu, L., Chen, X., Zhi, C., Yao, X., Liu, Y., et al. (2017). High prevalence of colistin resistance and mcr-1 gene in Escherichia coli isolated from food animals in China. Front. Microbiol. 8:562. doi: 10.3389/fmicb.2017.00562
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–d573. doi: 10.1093/nar/gkw1004
Jordan, D., Chin, J. J., Fahy, V. A., Barton, M. D., Smith, M. G., and Trott, D. J. (2009). Antimicrobial use in the Australian pig industry: results of a national survey. Aust. Vet. J. 87, 222–229. doi: 10.1111/j.1751-0813.2009.00430.x
Kidsley, A. K., Abraham, S., Bell, J. M., O'dea, M., Laird, T. J., Jordan, D., et al. (2018). Antimicrobial susceptibility of Escherichia coli and Salmonella spp. isolates from healthy pigs in Australia: results of a Pilot National Survey. Front. Microbiol. 9:1207. doi: 10.3389/fmicb.2018.01207
Kurenbach, B., Hill, A. M., Godsoe, W., Van Hamelsveld, S., and Heinemann, J. A. (2018). Agrichemicals and antibiotics in combination increase antibiotic resistance evolution. PeerJ 6:e5801. doi: 10.7717/peerj.5801
Kurenbach, B., Marjoshi, D., Amabile-Cuevas, C. F., Ferguson, G. C., Godsoe, W., Gibson, P., et al. (2015). Sublethal exposure to commercial formulations of the herbicides dicamba, 2,4-dichlorophenoxyacetic acid, and glyphosate cause changes in antibiotic susceptibility in Escherichia coli and Salmonella enterica serovar Typhimurium. MBio 6:e00009–15. doi: 10.1128/mBio.00009-15
Lai, J., Wang, Y., Shen, J., Li, R., Han, J., Foley, S. L., et al. (2013). Unique class 1 integron and multiple resistance genes co-located on IncHI2 plasmid is associated with the emerging multidrug resistance of Salmonella Indiana isolated from chicken in China. Foodborne Pathog. Dis. 10, 581–588. doi: 10.1089/fpd.2012.1455
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., et al. (2009). The Sequence Alignment/Map format and SAMtools. Bioinformatics 25, 2078–2079. doi: 10.1093/bioinformatics/btp352
Li, R., Xie, M., Zhang, J., Yang, Z., Liu, L., Liu, X., et al. (2017). Genetic characterization of mcr-1-bearing plasmids to depict molecular mechanisms underlying dissemination of the colistin resistance determinant. J. Antimicrob. Chemother. 72, 393–401. doi: 10.1093/jac/dkw411
Mangat, C. S., Bekal, S., Irwin, R. J., and Mulvey, M. R. (2017). A Novel hybrid plasmid carrying multiple antimicrobial resistance and virulence genes in Salmonella enterica Serovar Dublin. Antimicrob. Agents Chemother. 61:e02601–16. doi: 10.1128/AAC.02601-16
Mir, R. A., Weppelmann, T. A., Teng, L., Kirpich, A., Elzo, M. A., Driver, J. D., et al. (2018). Colonization dynamics of cefotaxime resistant bacteria in beef cattle raised without cephalosporin antibiotics. Front. Microbiol. 9:500. doi: 10.3389/fmicb.2018.00500
Monahan, L. G., Demaere, M. Z., Cummins, M. L., Djordjevic, S. P., Roy Chowdhury, P., and Darling, A. E. (2019). High contiguity genome sequence of a multidrug-resistant hospital isolate of Enterobacter hormaechei. Gut Pathog. 11, 3. doi: 10.1186/s13099-019-0288-7
Moran, R. A., Holt, K. E., and Hall, R. M. (2016). pCERC3 from a commensal ST95 Escherichia coli: a ColV virulence-multiresistance plasmid carrying a sul3-associated class 1 integron. Plasmid 84–85, 11–19. doi: 10.1016/j.plasmid.2016.02.002
Nurk, S., Bankevich, A., Antipov, D., Gurevich, A., Korobeynikov, A., Lapidus, A., et al. (2013). Assembling Genomes and Mini-metagenomes from Highly Chimeric Reads. Berlin; Heidelberg: Springer, 158–170.
Perreten, V., and Boerlin, P. (2003). A new sulfonamide resistance gene (sul3) in Escherichia coli is widespread in the pig population of Switzerland. Antimicrob. Agents Chemother. 47, 1169–1172. doi: 10.1128/AAC.47.3.1169-1172.2003
Phuong Hoa, P. T., Nonaka, L., Hung Viet, P., and Suzuki, S. (2008). Detection of the sul1, sul2, and sul3 genes in sulfonamide-resistant bacteria from wastewater and shrimp ponds of north Vietnam. Sci. Total Environ. 405, 377–384. doi: 10.1016/j.scitotenv.2008.06.023
Poirel, L., Jayol, A., and Nordmann, P. (2017). Polymyxins: antibacterial activity, susceptibility testing, and resistance mechanisms encoded by plasmids or chromosomes. Clin. Microbiol. Rev. 30, 557–596. doi: 10.1128/CMR.00064-16
Porse, A., Schonning, K., Munck, C., and Sommer, M. O. (2016). Survival and evolution of a large multidrug resistance plasmid in new clinical bacterial hosts. Mol. Biol. Evol. 33, 2860–2873. doi: 10.1093/molbev/msw163
Razavi, M., Marathe, N. P., Gillings, M. R., Flach, C. F., Kristiansson, E., and Joakim Larsson, D. G. (2017). Discovery of the fourth mobile sulfonamide resistance gene. Microbiome 5, 160. doi: 10.1186/s40168-017-0379-y
Reid, C. J., Roy Chowdhury, P., and Djordjevic, S. P. (2015). Tn6026 and Tn6029 are found in complex resistance regions mobilised by diverse plasmids and chromosomal islands in multiple antibiotic resistant Enterobacteriaceae. Plasmid 80, 127–137. doi: 10.1016/j.plasmid.2015.04.005
Reid, C. J., Wyrsch, E. R., Roy Chowdhury, P., Zingali, T., Liu, M., Darling, A. E., et al. (2017). Porcine commensal Escherichia coli: a reservoir for class 1 integrons associated with IS26. Microb. Genomics 3, 1–42. doi: 10.1099/mgen.0.000143
Roy Chowdhury, P., Charles, I. G., and Djordjevic, S. P. (2015). A role for Tn6029 in the evolution of the complex antibiotic resistance gene loci in genomic island 3 in enteroaggregative hemorrhagic Escherichia coli O104:H4. PLoS ONE 10:e0115781. doi: 10.1371/journal.pone.0115781
Roy Chowdhury, P., Mckinnon, J. M., Liu, M. Y., and Djordjevic, S. P. (2018). Multidrug resistant uropathogenic Escherichia coli ST405 with a novel, composite IS26 transposon in a unique chromosomal location. Front. Microbiol. 9:3212. doi: 10.3389/fmicb.2018.03212
Saputra, S., Jordan, D., Mitchell, T., Wong, H. S., Abraham, R. J., Kidsley, A., et al. (2017). Antimicrobial resistance in clinical Escherichia coli isolated from companion animals in Australia. Vet. Microbiol. 211, 43–50. doi: 10.1016/j.vetmic.2017.09.014
Shahid, M. (2010). Citrobacter spp. simultaneously harboring blaCTX-M, blaTEM, blaSHV, blaampC, and insertion sequences IS26 and orf513: an evolutionary phenomenon of recent concern for antibiotic resistance. J. Clin. Microbiol. 48, 1833–1838. doi: 10.1128/JCM.01467-09
Sidjabat, H. E., Townell, N., Nimmo, G. R., George, N. M., Robson, J., Vohra, R., et al. (2015). Dominance of IMP-4-producing Enterobacter cloacae among carbapenemase-producing Enterobacteriaceae in Australia. Antimicrob. Agents Chemother. 59, 4059–4066. doi: 10.1128/AAC.04378-14
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–36. doi: 10.1093/nar/gkj014
Suhartono, S., Savin, M. C., and Gbur, E. E. (2017). Transmissible plasmids and integrons shift Escherichia coli population toward larger multiple drug resistance numbers. Microb Drug Resist. 24, 244–252. doi: 10.1089/mdr.2016.0329
Sullivan, M. J., Petty, N. K., and Beatson, S. A. (2011). Easyfig: a genome comparison visualizer. Bioinformatics 27, 1009–1010. doi: 10.1093/bioinformatics/btr039
Treangen, T. J., Ondov, B. D., Koren, S., and Phillippy, A. M. (2014). The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 15, 524. doi: 10.1186/s13059-014-0524-x
Tsafnat, G., Copty, J., and Partridge, S. R. (2011). RAC: Repository of Antibiotic resistance Cassettes. Database 2011:bar054. doi: 10.1093/database/bar054
Turner, A. (2011). Quarantine, exports and animal disease in Australia 1901-2010. Aust. Vet. J. 89, 366–371. doi: 10.1111/j.1751-0813.2011.00822.x
Venturini, C., Beatson, S. A., Djordjevic, S. P., and Walker, M. J. (2010). Multiple antibiotic resistance gene recruitment onto the enterohemorrhagic Escherichia coli virulence plasmid. FASEB J. 24, 1160–1166. doi: 10.1096/fj.09-144972
Venturini, C., Hassan, K. A., Roy Chowdhury, P., Paulsen, I. T., Walker, M. J., and Djordjevic, S. P. (2013). Sequences of two related multiple antibiotic resistance virulence plasmids sharing a unique IS26-related molecular signature isolated from different Escherichia coli pathotypes from different hosts. PLoS ONE 8:e78862. doi: 10.1371/journal.pone.0078862
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS ONE 9:e112963. doi: 10.1371/journal.pone.0112963
Wick, R. R., Judd, L. M., Gorrie, C. L., and Holt, K. E. (2017). Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput. Biol. 13:e1005595. doi: 10.1371/journal.pcbi.1005595
Wong, M. H., Chan, E. W., and Chen, S. (2017). IS26-mediated formation of a virulence and resistance plasmid in Salmonella Enteritidis. J. Antimicrob. Chemother. 72, 2750–2754. doi: 10.1093/jac/dkx238
Wyrsch, E., Roy Chowdhury, P., Abraham, S., Santos, J., Darling, A. E., Charles, I. G., et al. (2015). Comparative genomic analysis of a multiple antimicrobial resistant enterotoxigenic E. coli O157 lineage from Australian pigs. BMC Genomics 16:165. doi: 10.1186/s12864-015-1382-y
Wyrsch, E. R., Roy Chowdhury, P., Chapman, T. A., Charles, I. G., Hammond, J. M., and Djordjevic, S. P. (2016). Genomic microbial epidemiology is needed to comprehend the global problem of antibiotic resistance and to improve pathogen diagnosis. Front. Microbiol. 7:843. doi: 10.3389/fmicb.2016.00843
Zheng, B., Huang, C., Xu, H., Guo, L., Zhang, J., Wang, X., et al. (2017). Occurrence and genomic characterization of ESBL-producing, MCR-1-harboring Escherichia coli in farming soil. Front. Microbiol. 8:2510. doi: 10.3389/fmicb.2017.02510
Keywords: Escherichia coli, antimicrobial resistance (AMR), plasmid, genomics, epidemiology
Citation: Wyrsch ER, Reid CJ, DeMaere MZ, Liu MY, Chapman TA, Roy Chowdhury P and Djordjevic SP (2019) Complete Sequences of Multiple-Drug Resistant IncHI2 ST3 Plasmids in Escherichia coli of Porcine Origin in Australia. Front. Sustain. Food Syst. 3:18. doi: 10.3389/fsufs.2019.00018
Received: 14 January 2019; Accepted: 06 March 2019;
Published: 26 March 2019.
Edited by:
Ömer Akineden, University of Giessen, GermanyReviewed by:
Aurora García-Fernández, Istituto Superiore di Sanità (ISS), ItalyCostas C. Papagiannitsis, University of Thessaly, Greece
Copyright © 2019 Wyrsch, Reid, DeMaere, Liu, Chapman, Roy Chowdhury and Djordjevic. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Steven P. Djordjevic, c3RldmVuLmRqb3JkamV2aWNAdXRzLmVkdS5hdQ==
 Ethan R. Wyrsch
Ethan R. Wyrsch Cameron J. Reid1
Cameron J. Reid1 Matthew Z. DeMaere
Matthew Z. DeMaere Michael Y. Liu
Michael Y. Liu Toni A. Chapman
Toni A. Chapman Piklu Roy Chowdhury
Piklu Roy Chowdhury Steven P. Djordjevic
Steven P. Djordjevic