
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Plant Sci. , 27 June 2019
Sec. Plant Physiology
Volume 10 - 2019 | https://doi.org/10.3389/fpls.2019.00841
 Dachuan Gu1,2
Dachuan Gu1,2 Rujun Ji1,2
Rujun Ji1,2 Chunmei He1,2
Chunmei He1,2 Tao Peng1,2
Tao Peng1,2 Mingyong Zhang1,2
Mingyong Zhang1,2 Jun Duan1,2
Jun Duan1,2 Changyun Xiong3*
Changyun Xiong3* Xuncheng Liu1,2*
Xuncheng Liu1,2*Plant lifecycle starts from seed germination, which is regulated by various environmental cues and endogenous hormones. Light promotes seed germination mainly by phytochrome B (PHYB) during the initial phase of imbibition, which involves genome-wide light-responsive transcription changes. Recent studies indicated an involvement of multiple epigenetic factors in the control of seed germination. However, few studies have been reported about the role of a histone methyltransferase in light-mediated seed germination process. Here, we identified SUVH5, a histone H3 lysine 9 methyltransferase, as a positive regulator in light-mediated seed germination in Arabidopsis. Loss of function of SUVH5 leads to decreased PHYB-dependent seed germination. RNA-sequencing analysis displayed that SUVH5 regulates 24.6% of light-responsive transcriptome in imbibed seeds, which mainly related to hormonal signaling pathways and developmental processes. Furthermore, SUVH5 represses the transcription of ABA biosynthesis and signal transduction-related genes, as well as a family of DELAY OF GERMINATION (DOG) genes via dimethylation of histone H3 at lysine 9 (H3K9me2) in imbibed seeds. Taken together, our findings revealed that SUVH5 is a novel positive regulator of light-mediated seed germination in Arabidopsis.
Plant life cycle initiates from seed germination, which is of both economic and ecologic importance (Rajjou et al., 2012). Arabidopsis seeds consist of embryo, single cell endosperm and testa from inside to outside (Finch-Savage and Leubner-Metzger, 2006). Arabidopsis seed germination includes two-step process, which is testa rupture followed by endosperm rupture (Yamaguchi, 2008; Weitbrecht et al., 2011). It is well known that seed germination is regulated by endogenous and exogenous factors, such as light, temperature, moisture, oxygen, nutrients, and multiple plant hormones (Finch-Savage and Leubner-Metzger, 2006; De Wit et al., 2016; Shu et al., 2016).
Light is a key environmental factor in the control of seed germination. Plants perceive different parts of the light spectrum by distinct sets of photoreceptors, such as phytochromes, cryptochromes, phototropins, ZEITLUPE family, and UVR8 (Briggs and Christie, 2002; Chaves et al., 2011; Rizzini et al., 2011; Wang and Wang, 2015). Phytochromes are red and far-red light photoreceptors that play critical role in regulating seed germination in various plants species (Borthwick et al., 1952; Shinomura et al., 1994, 1996). Dark-imbibed lettuce seeds (Lactuca sativa L.) irradiated with red light (R) will induce germination, whereas subsequent exposure to far-red light (FR) can reverse this process (Borthwick et al., 1952). The light signaling mechanism relies on conformational conversion between inactive state (Pr) and active state (Pfr), which are photo-convertible isoforms of phytochromes (De Wit et al., 2016). In Arabidopsis, there are five phytochromes, designated phytochrome A (PHYA) to phytochrome E (PHYE) (Jiao et al., 2007). PHYB plays a fundamental role in the promotion of seed germination during the initiate phase of seed imbibition (Shinomura et al., 1994; Seo et al., 2008).
It’s well known that plant hormone abscisic acid (ABA) plays a predominate role in the repression of seed germination (Seo et al., 2008; Shu et al., 2016). Endogenous ABA levels are regulated by a balance of between its biosynthesis and catabolism. The major ABA biosynthesis pathway is regulated by multiple factors, including the rate limiting enzymes 9-cis-epoxycarotenoid dioxygenases (NCEDs), zeaxanthin epoxidase (ZEP) ABA1, cytosolic short-chain dehydrogenase (SDR) ABA2, molybdenumcofactor sulfurase ABA3, as well as aldehyde oxidase AAO3 (Seo and Koshiba, 2002). Seeds of triple mutant nced5 nced6 nced9 germinate faster than the wild-type (Frey et al., 2012), whereas transgenic plants constitutively expressing NCED6 increase ABA levels and prevent seed germination (Martinez-Andujar et al., 2011). The core ABA signaling network is composed of PYR/PYL/RCAR receptors, PP2C phosphatases, SnRK2 kinases, bZIP-type transcription factors known as ABA-responsive element (ABRE) binding factors (Raghavendra et al., 2010; Antoni et al., 2011; Hauser et al., 2011). ABI3 (ABA insensitive 3) and ABI5, two key transcription factors in ABA signal transduction, have been reported to play crucial roles in maintaining seed dormancy, and repressing seed germination (Koornneef et al., 2002; Piskurewicz et al., 2008; Kanai et al., 2010). Moreover, seed dormancy can prevent germination when environmental conditions are suitable for germination. DELAY OF GERMINATION 1 (DOG1) is a master regulator in control of seed dormancy, which belongs to a plant-specific gene family with other four additional members (Bentsink et al., 2006). Loss of function of DOG1 in Arabidopsis results in abolished seed dormancy and fast germination even under unfavorable conditions (Nakabayashi et al., 2012; Graeber et al., 2014).
Previous studies displayed that multiple epigenetic factors, including chromatin-remodeling factors, histone deacetylases, histone demethylases and histone methyltransferases, play diverse roles in the regulation of seed germination and dormancy (Dean Rider et al., 2003; Perruc et al., 2007; Saez et al., 2008; Cho et al., 2012; Luo et al., 2012; Zheng et al., 2012; Zhou et al., 2013; Lee et al., 2014; Liu et al., 2014; Zhao et al., 2015; Gu et al., 2017). The chromatin-remodeling factor PICKLE selectively regulates a number of genes to repress embryonic identity during germination (Dean Rider et al., 2003; Perruc et al., 2007). SWI3B, an Arabidopsis homolog of the yeast SWI3 subunit of SWI/SNF chromatin-remodeling complexes, plays a negative role in ABA-repressed seed germination (Saez et al., 2008). Loss of function of histone deacetylases HDA6 and HD2C result in increased sensitivity to ABA and NaCl stresses during germination (Luo et al., 2012). Moreover, a recent study demonstrated histone deacetylase HDA15 interacts with Phytochrome Interacting Factor 1 (PIF1), a key negative transcription factor in light signaling pathway, in repressing light-mediated seed germination. HDA15 and PIF1 co-repress the genes associate with multiple hormonal signaling pathways and cellular processes by decreasing the histone H3 acetylation levels in the dark conditions (Gu et al., 2017). Furthermore, two histone arginine demethylases, JMJ20 and JMJ22, were found to be positive regulators in PHYB-dependent seed germination (Cho et al., 2012). JMJ20/JMJ22 increase gibberellic acid (GA) levels via removal of histone arginine methylations of GA biosynthesis genes, GA3ox1/GA3ox2, and ultimately promote seed germination (Cho et al., 2012). Whereas, another two histone demethylases (LDL1/LDL2) have been reported that function redundantly in repressing seed dormancy (Zhao et al., 2015). Furthermore, histone methyltransferase KYP/SUVH4 also controlled Arabidopsis primary seed dormancy, while another methyltransferase EFS inhibited seed germination (Zheng et al., 2012; Lee et al., 2014). However, few studies have been reported the function of a histone methyltransferase in light-mediated seed germination process.
SUVH5, a histone H3 lysine 9 methyltransferase, belonging to the SUV(R) group of SET domain proteins, has been reported to maintain transposon elements and inverted repeats silencing via histone H3K9 dimethylation (Ebbs and Bender, 2006; Rajakumara et al., 2011; Yu et al., 2017). In the present study, we identified SUVH5 as a novel component of light-mediated transcriptional regulatory network in seed germination. SUVH5 represses the expression of key seed germination-related genes, such as ABA biosynthesis and signal transduction-related genes, as well as a group of DOG genes by H3K9 dimethylation in imbibed seeds.
All Arabidopsis plants used in this study are in Col-0 background. The suvh5-2 (SALK_074957) and suvh4/5/6 mutant was a kind gift from Professor Judith Bender at the University of Brown. suvh5-2 allele was backcrossed to Col-0 for three times. suvh4 mutant kyp-6 (SALK_041474) was obtained from the Arabidopsis Information Resource Center1. The seeds used for germination comparison were harvested in the same batch of plants grown at 22°C under long days (16 h WL/8 h dark). Following seeds harvesting, seeds were kept in an incubator at 22°C for about 1 month to break dormancy prior to germination assays.
The PHYB-dependent seed germination assays were performed as described previously (Oh et al., 2007). Briefly, seeds were surface-sterilized and plated on half-strength Murashige-Skoog (Sigma-Aldrich) agar plates containing 0.3% sucrose and 1% phytoagar (pH 5.7). The plates were placed in an illuminated incubator with white light (80 μmol⋅m–2⋅s–1) at 22°C. 1 h after imbibition and sterilization, seeds were irradiated with far-red light (3.82 μmol⋅m–2⋅s–1) for 5 min (indicated as FR or dark conditions), or exposure to far-red light (3.82 μmol⋅m–2⋅s–1) for 5 min following irradiation with red light (13.12 μmol⋅m–2⋅s–1) for 5 min (referred as R or light conditions). The seeds were kept in the dark to calculate the germination rates at the indicated time. At least 60 seeds were used for each experimental point, and seeds harvested from three independent batches were performed for statistical analysis.
After FR or R treatment, the seeds were incubated in the dark at 22°C for the indicated time. The imbibed seeds were ground to powder in liquid nitrogen and total RNA was extracted with TRIZOL Reagent (Invitrogen) according to the manufacture’s protocol. After DNase I treatment, the first strand cDNA was synthesized using 2 μg total RNA according to the manufacturer’s instruction of TransScript One-Step gDNA Removal and cDNA Synthesis Super Mix Kit (TransGen, Beijing). Quantitative RT-PCR was performed by using SYBR Green Mix (Bio-Rad) in an ABI7500 Real-Time PCR System (Applied Biosystems). Three biological replicates were performed, and three technical repeats were carried out for each biological replicate. PP2A was used as an internal control (Czechowski et al., 2005). The primer pairs for quantitative RT-PCR are listed in Supplementary Table S4.
For whole genomic transcriptome analysis, the seeds after R light treatment were incubated in the dark at 22°C for 24 h prior to RNA extraction. Total RNA was extracted as described above and an mRNA-seq library was prepared by using an mRNA Seq Kit (Illumina). RNA-seq were performed by Genepioneer Biotechnologies (Nanjing, China) with triplicate biological samples. High-quality clean reads were obtained by removing the adaptor sequences, ambiguous reads (“N” > 10%), and low-quality reads (i.e., more than 50% of bases in a read had a quality value Q ≤ 5). Then the clean reads were mapped to Arabidopsis genome TAIR10 using HISAT2 software with default parameters (Pertea et al., 2016). Cuffdiff2 was applied to detect differentially expressed genes (DEGs). Genes with more than 1.5-fold changes with statistically significance (adjusted P-value < 0.05) were selected. GO (gene ontology) analyses of DEGs were performed with Metascape software3 with a cutoff of P ¡ 0.05 and a minimum overlap of 3. The regulated trends of DEGs were visualized by use of heat-map made by HemI (version 1.0.1) (Deng et al., 2014). Hierarchical clustering analysis was done with the average linkage method using the HemI software. These raw sequencing data sets were deposited in NCBI-SRA database (BioProject accession number: PRJNA489162).
Equal amount of Col-0 and suvh5-2 mutant seeds were treated with R light pulse and subsequently incubated in dark at 22°C for 24 h before ChIP-qPCR analyses. ChIP-qPCR assays were performed as previously described (Gendrel et al., 2005). After fixation with formaldehyde, the chromatin was extracted and then sheared to an average length of 500 bp by sonication. The chromatin was immunoprecipitated with anti-di-methylated histone H3K9 (catalog no. 39753; Active Motif). After cross-linking reversed, the amount of each precipitated DNA fragment was detected by quantitative PCR using specific primers listed in Supplementary Table S4. The amounts of DNA after ChIP were quantified and normalized to TA3, the relative enrichment refers to the H3K9me2 enrichment vs. the histone H3 occupancy. Three biological replicates were performed, and three technical repeats were carried out for each biological replicate.
After R treatment, the seeds were incubated in the dark at 22°C for 24 h. Samples were harvested and extracted for ABA as described previously (Wu et al., 2007). Finely powdered sample (30 mg, fresh weight) was extracted with 3 mL ethyl acetate by vortexing for 30 s followed by ultrasonic extraction in ice-cold water for 20 min. Before ultrasonic extraction, 1 ng [2H6] ABA was added to the mixture as an internal standard. After centrifuging at 10000 × g for 5 min at 4°C, 2.9 mL supernatants were collected, and then dried under a stream of nitrogen. The residue was re-dissolved in 100 μL methanol. The supernatants were filtered through a 0.22 μm membrane, and subjected to an ultra–performance liquid chromatography/quadrupole time–of–flight mass spectrometry (UPLC–QTOF–MS) (Acquity UPLC I-Class/ Xevo® G2-XS QTOF, Waters Corporation, MA, United States). Each sample (5 μL) was injected onto a Waters ACQUITY UPLC HSS T3 C18 column (2.1 mm × 100 mm, 1.8 μm). Solvent A was Milli-Q water with 0.1% (v/v) formic acid. Solvent B was acetonitrile with 0.1% (v/v) formic acid. The solvent gradient was started at 20% B, then linearly increased to 35% within 10 min, later increased to 95% B in 0.1 min and kept for 3 min. In that moment, it suddenly dropped to 20% in 0.1 min and maintain for 3 min. The flow rate was 0.4 mL/min. The column temperature was 30°C. The electrospray ionization operated on negative mode. The MS conditions were capillary voltage: 1.5 kV; source temperature: 100°C; desolvation temperature: 300°C; cone gas flow: 50 L/h; and desolvation gas flow: 600 L/h. The quantitative analysis of ABA was based on calibration curve, which was constructed by plotting the concentration of ABA standard against the peak area of [2H6] ABA. The ABA content was determined three times for each sample. Three biological replicates were performed.
To investigate whether histone methyltransferase SUVH5 plays a role in light-regulated seed germination, we examined the germination trait of a previously reported loss of function of SUVH5 mutant suvh5-2 (Ebbs and Bender, 2006) by PHYB-dependent germination protocol (Oh et al., 2004; Oh et al., 2006). 1 h after white light (WL) irradiation and surface sterilization, the seeds were exposed to 5 min far-red (FR) light (PHYB inactive, referred as FR or dark conditions) or followed by illumination with 5 min red light (PHYB activation, indicated as R or light conditions), and subsequently kept in the dark for 2 days (Figure 1A). Upon FR conditions, both wild-type Col-0 and suvh5-2 seeds failed to germinate (the germination rates were 0) (Figure 1B). Next, we examined the germination rates of Col-0 and suvh5-2 after R treatment. 66.6% of the wild-type seeds germinated 48 h after treatment, while suvh5-2 seeds displayed relatively lower germination rates (46.6%) compared to the wild-type at the indicated time point (Figure 1B). As a control, seeds were kept under continuous white light (WL) and scored for germination (Figure 1B). Moreover, we also tested dynamics of germination rates of Col-0 and suvh5-2 on PHYB activated (R) conditions after a long period. As shown in Supplementary Figure S1, except at 84 h, there were no significant germination rates difference in Col-0 and suvh5-2 48 h after treatment. The results indicated that SUVH5 may mainly modulate the germination kinetic in the initial phases and slightly affect the final rate of germination.
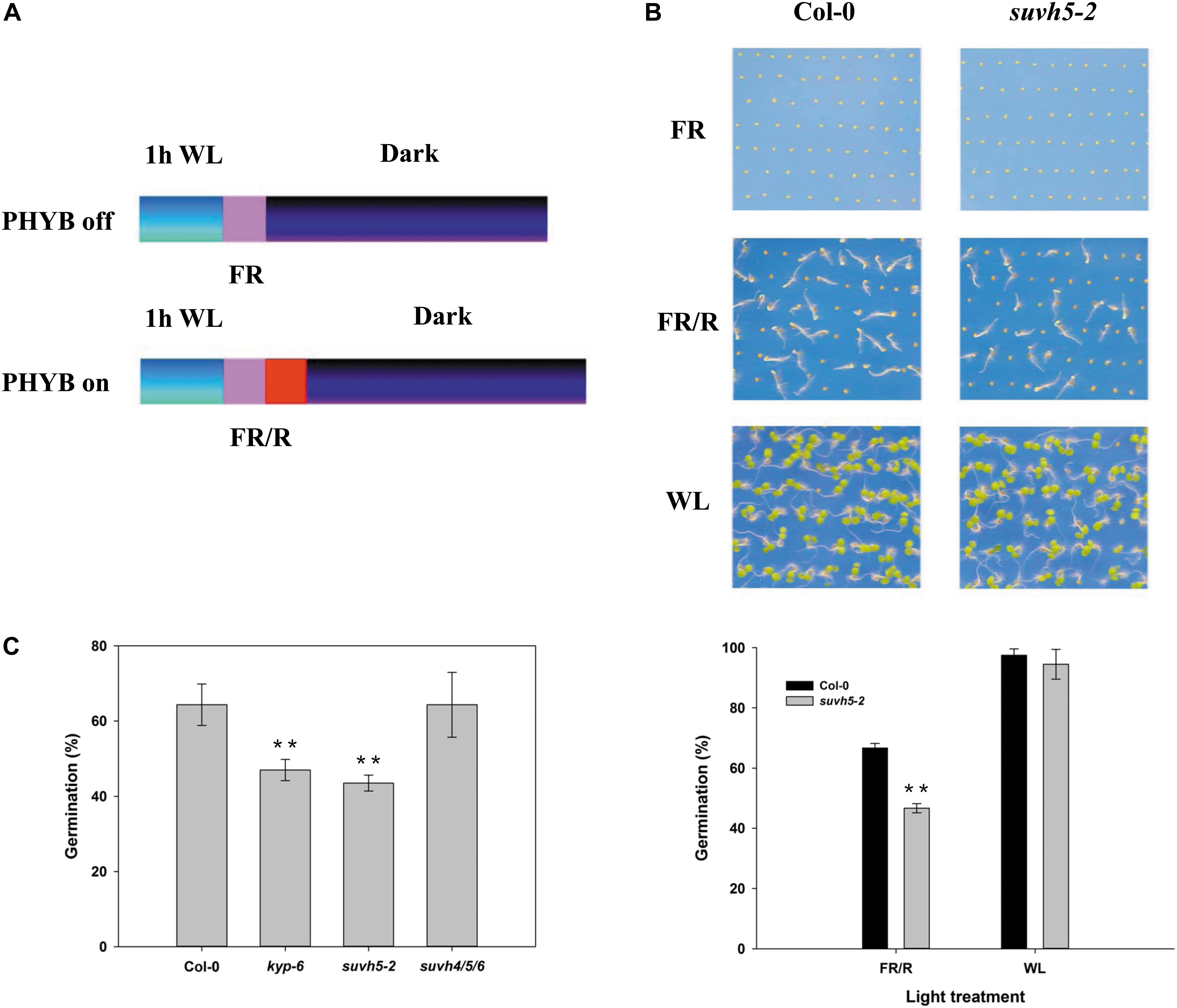
Figure 1. Analysis of the germination rate of kyp-6, suvh5-2, and suvh4/5/6 mutant using PHYB-dependent seed germination assays. (A) Germination protocols of PHYB inactivated (PHYB-off, FR) and activated (PHYB-on, R) assays. (B) Germination patterns of Col-0 and suvh5-2 under PHYB inactivated (FR), PHYB activated (R), and continuous white light (WL) conditions. Profiles (top) and germination rates (bottom) of Col-0 and suvh5-2 on PHYB inactivated (FR), PHYB activated (R), and continuous white light (WL) conditions. After FR or R treatment, the seeds were kept in the dark for 48 h. Seeds were kept under continuous white light (WL) as a control. (C) Germination rates of Col-0, kyp-6, suvh5-2, and suvh4/5/6 on PHYB activated (R) conditions. Germination frequencies were recorded at 48h after treatment. Values are shown as means ± SD (n = 3) (t-test, ∗∗P < 0.01, difference from Col-0).
Previous study displayed that the plant SUPPRESSOR OF VARIEGATION 3-9 HOMOLOG (SUVH) family members, SUVH4, SUVH5, and SUVH6 act redundantly in regulating methylation of H3K9 and transposon silencing (Ebbs and Bender, 2006; Yu et al., 2017). We further examined the phenotype of the suvh4 single mutant, kyp-6 and the suvh4/5/6 triple mutant (Ebbs and Bender, 2006). Similar to suvh5-2, kyp-6 showed relatively lower germination rates compared to Col-0, which indicated that SUVH4 and SUVH5 may act redundantly in the control of seed germination. Surprisingly, suvh4/5/6 triple mutant displayed a similar germination phenotype with wild-type (Figure 1C), which suggested that SUVH6 may act oppositely to SUVH4 and SUVH5 in regulating light-mediated seed germination. Together, these data indicated that SUVH5 may act as a positive regulator of PHYB-dependent seed germination.
We further examined the expression patterns of SUVH5 and its close homologs, SUVH4 and SUVH6 under FR and R conditions in germinating seeds. Seeds of Col-0 were treated with FR and FR/R exposure separately, after imbibed for indicated time (3, 6, 12, and 24 h), the seeds were harvested for gene expression analysis. SUVH4/5/6 demonstrated a similar expression profile during light-mediated seed germination process (Figure 2). Relatively higher expression levels of SUVH4/5/6 were detected in dry seeds (0 h as indicated). 1 h after imbibition, the transcripts of SUVH4/5/6 were significantly decreased. Except for a recover of SUVH4 expression 12 and 24 h after R treatment, both FR and R treatments decreased the expression of SUVH4/5/6 in imbibed seeds compared to 0 h (Figure 2). Moreover, we also analyzed the expression of SUVH4/5/6 in imbibed seeds under continuous FR and R irradiation. Except for continuous R light promoted SUVH4 expression and a recover of SUVH4/SUVH6 expression 12 h after continuous FR treatment, both continuous FR and R impulse repressed the expression of SUVH4/5/6 in imbibed seeds compared to 0 h (Supplementary Figure S2). Collectively, these data suggested that the expression of SUVH4/5/6 was regulated by light during the initial phase of seed germination.
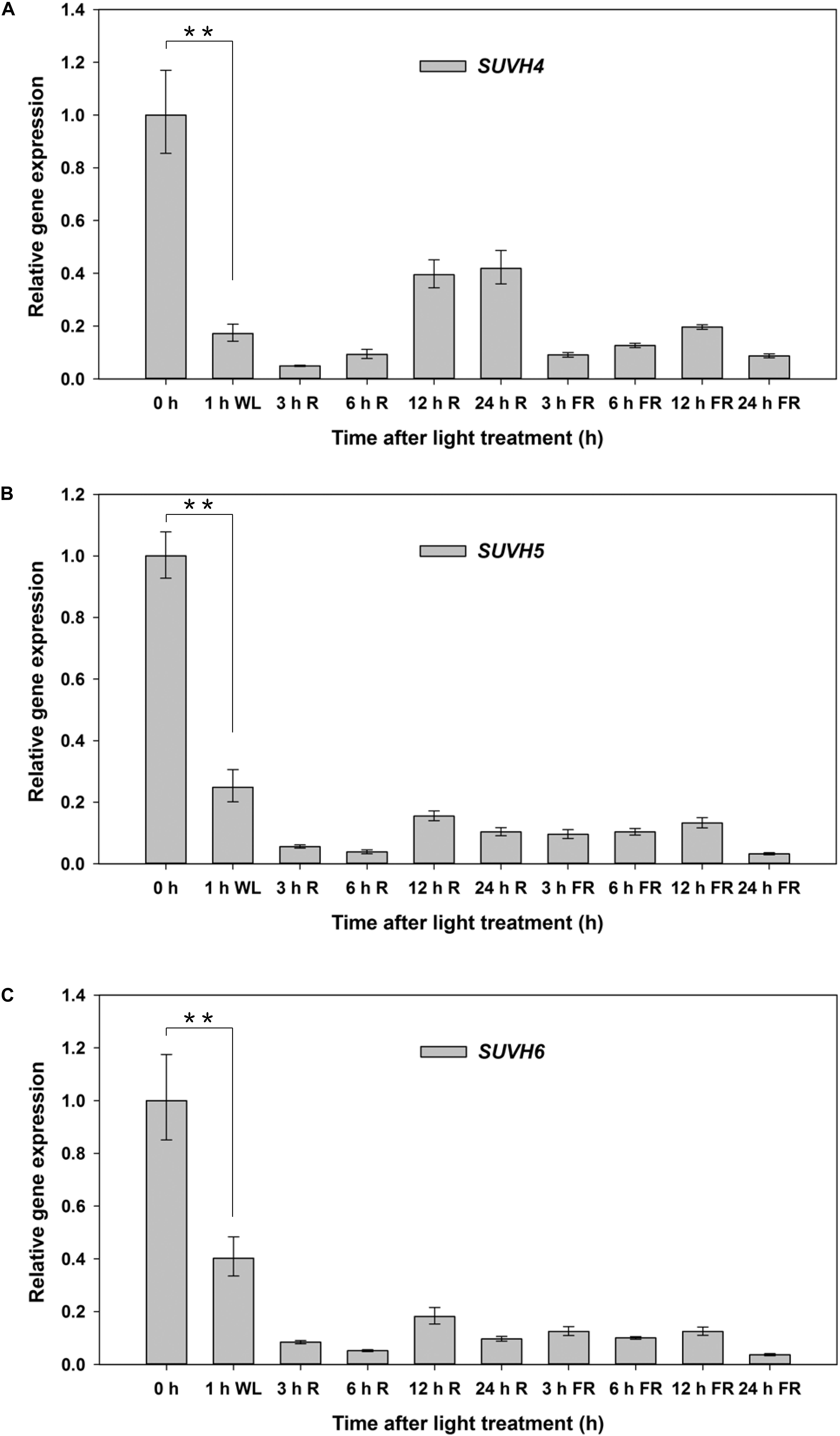
Figure 2. Expression patterns of SUVH4/5/6 under R and FR conditions. Equal amount of Col-0 seeds were treated with FR or R light pulse and subsequently incubated in dark for indicated times before extracting mRNA. 0 h indicates dry seeds. PP2A was used as an internal control. Values are shown as means ± SD (n = 3) (t-test, ∗∗P < 0.01, difference from 0 h). (A) Expression patterns of SUVH4 under R and FR conditions. (B) Expression patterns of SUVH5 under R and FR conditions. (C) Expression patterns of SUVH6 under R and FR conditions.
To further study the function of SUVH5 in light-mediated seed germination, we examined the SUVH5-regulated transcriptome changes under R conditions by RNA-sequencing (RNA-seq) assays. Seeds of Col-0 and suvh5-2 were treated under R conditions and kept in the dark, 24 h after imbibition, the seeds were harvested for RNA extraction, library construction, and high-throughput sequencing. To get reliable RNA-seq results, three independent biological replicated samples were harvest for analysis. Genes with more than 1.5-fold changes with statistical significance (adjusted P value < 0.05) were selected. Compared with the wild-type, 982 genes were up-regulated whereas only 101 genes were down-regulated in suvh5-2 mutant, which suggested that SUVH5 may act mainly as a transcription repressor in light-mediated seed germination process (Supplementary Tables S1, S2).
Next, GO and functional clustering analysis of SUVH5-regulated genes were performed by Metascape software. We showed that the genes up-regulated in suvh5-2 are mainly associated with the biological processes including response to toxic substance, response to temperature stimulus, secondary metabolic process, response to drug, response to abscisic acid, lipid storage, toxin catabolic processes, and response to karrikin (Figure 3A). In contrast, the genes down-regulated in suvh5-2 are preferentially enriched in syncytium formation, tissue development, response to light stimulus, xylem and phloem pattern formation, phloem or xylem histogenesis, and DNA metabolic process (Figure 3B). Collectively, these results suggested that SUVH5 may integrate multiple internal and external factors to regulate many developmental processes, including light-regulated seed germination.
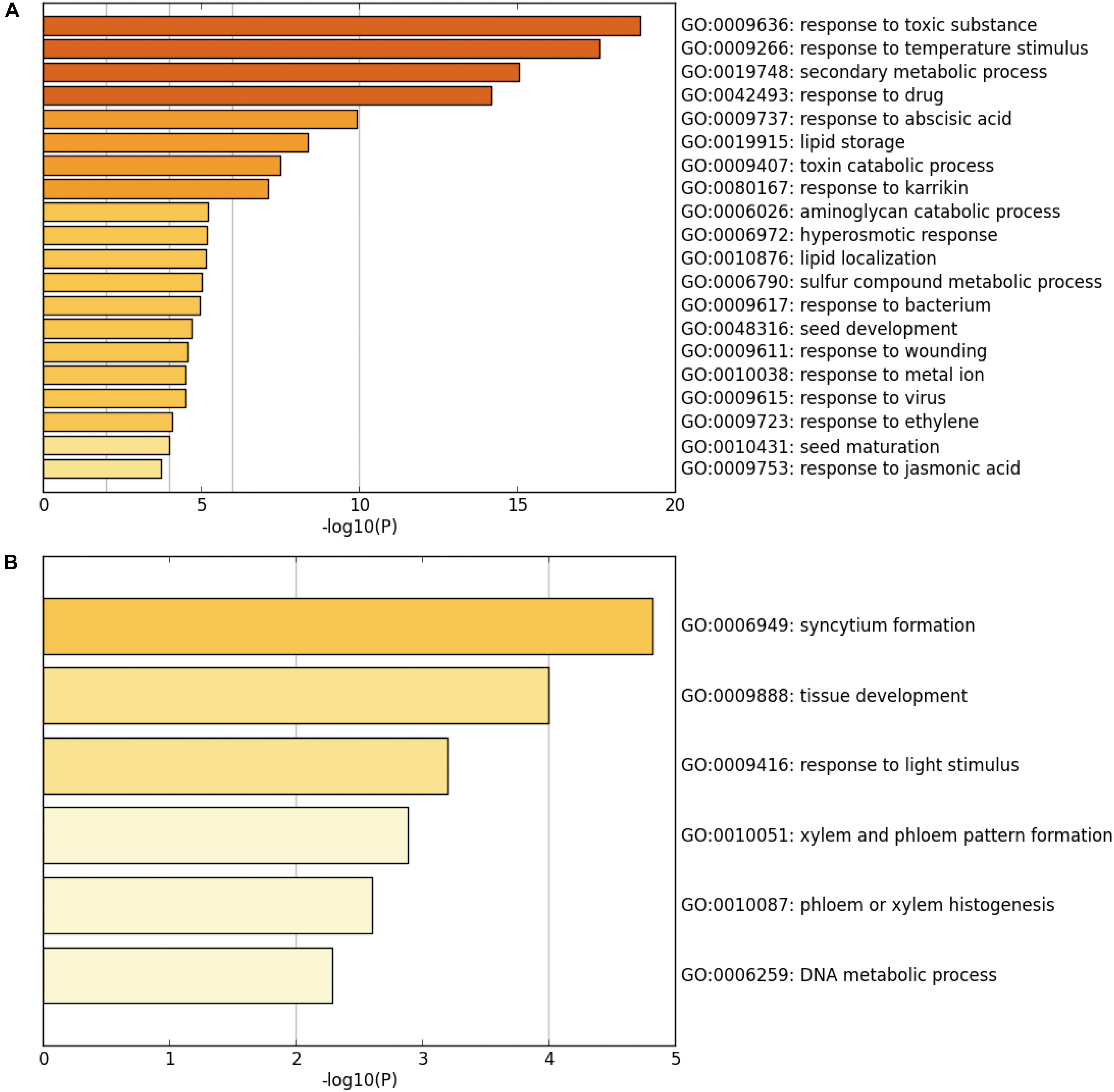
Figure 3. Chart of enriched ontology clusters of significantly expressed genes, which is repressed (A) or activated (B) by SUVH5 (P < 0.05).
Previous transcriptome analysis displayed that 2069 genes are regulated by light in imbibed seeds in Arabidopsis (Col-0 R vs. Col-0 FR) (Shi et al., 2013). Analysis in combination with SUVH5 and light-regulated transcriptomes demonstrated that the expression of about 24.6% (510) light-regulated genes was altered in suvh5 mutant (Figure 4A and Supplementary Table S3). Interestingly, most of these genes (97.6%, 498) were down-regulated by light whereas up-regulated in suvh5-2 (Figure 4A). Consistently, heatmap displayed that suvh5-2 and light modulated the transcriptome changes in an opposite manner (Figure 4B). These data indicated that SUVH5 may act as a key positive regulator of light-regulated transcriptome in imbibed seeds.
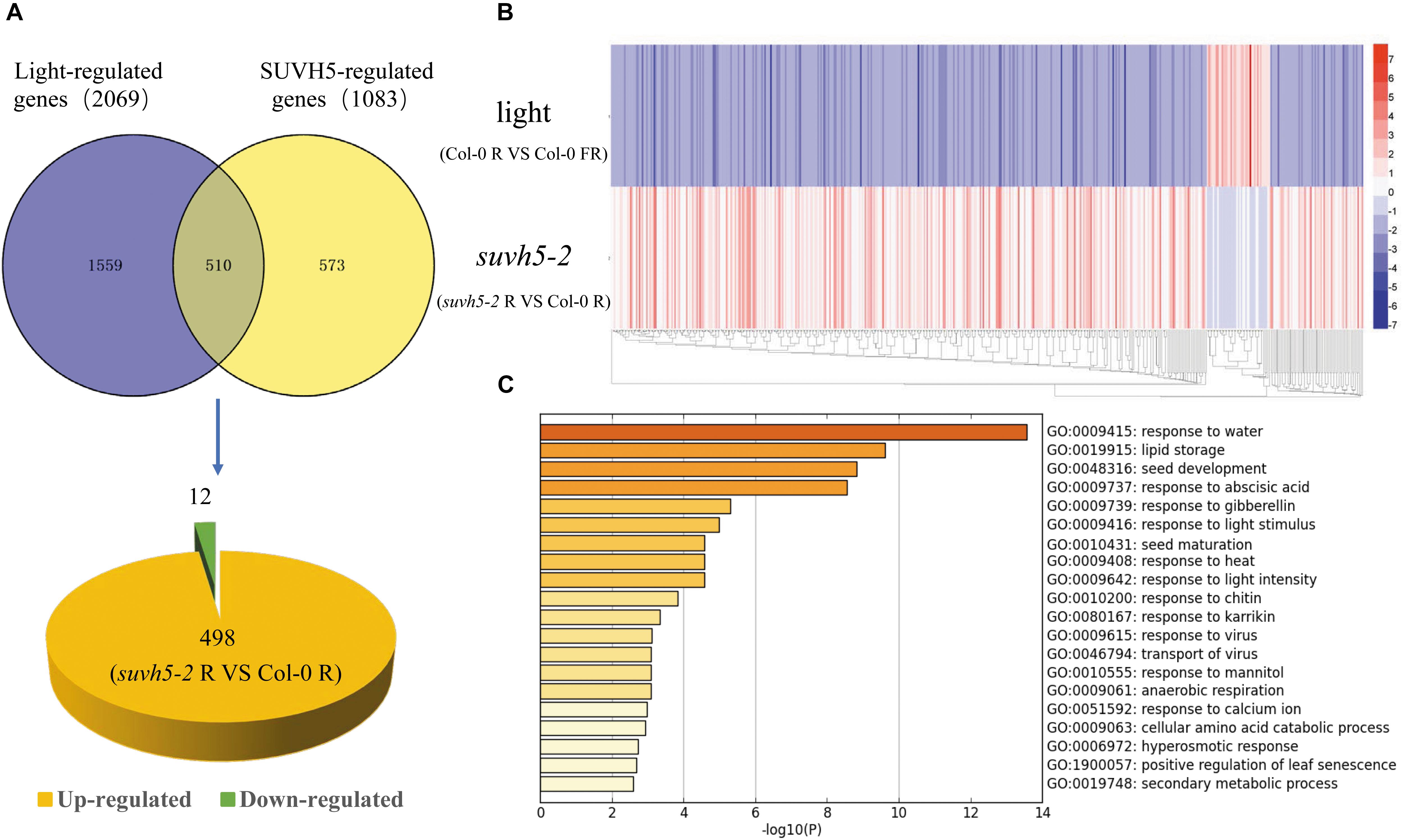
Figure 4. Genome-wide analysis of SUVH5-regulated transcriptome in light-mediated seed germination process. (A) Venn diagram shows the overlap of SUVH5- and light-regulated genes. Among these co-regulated genes, 498 were up-regulated whereas 12 were down-regulated in suvh5. (B) Heatmap of light and SUVH5 co-regulated genes. The bar indicates the fold change (suvh5-2 R vs. Col-0 R and Col-0 R vs. Col-0 FR). (C) Chart of enriched ontology clusters of significantly expressed genes co-regulated by SUVH5 and light (P < 0.05).
Gene ontology analysis and functional clustering analysis revealed that SUVH5 and light co-regulated genes are mainly related to the processes including response to water, lipid storage, seed development, response to abscisic acid, response to gibberellin, response to light stimulus, seed maturation, response to heat, response to light intensity, response to chitin, and response to karrikin (Figure 4C). Collectively, these findings suggested that SUVH5 and light co-regulated many developmental processes by affecting the expression of the light-responsive genes related to multiple hormonal signaling pathways and development processes.
It’s well known that ABA plays a predominant negative role in the regulation of seed germination. Our RNA-seq analysis demonstrated that the expression of a large subset of ABA signaling-related genes, including ABA biosynthesis genes (ABA1, ABA3, NCED6, and AAO3), ABA signal transduction genes (ABI5, EEL, ABF4, HAI2, and PYL13) and ABA-responsive genes (ABR, ABR1, EM1, USP, RAB18, and et al.) were up-regulated in suvh5 mutant compared with wild-type upon R conditions (Supplementary Table S1, S5). Further quantitative RT-PCR (qRT-PCR) analysis displayed that relatively higher expression levels of these genes were detected in imbibed suvh5-2 seeds compared with wild-type (Figures 5A–C). Consistently, we showed that the ABA content in suvh5-2 seeds was 6.5-fold higher than that in wild-type (Figure 5F). GA is another critical plant hormone in the regulation of seed germination, we further analyzed the expression of genes relate to GA signaling and metabolism processes. DELLA (GAI, RGA, RGL1, RGL2, and RGL3) proteins are master negative components in GA signaling, and RGL2 is the main repressor of seed germination (Nelson and Steber, 2016). GA2ox is crucial catabolic enzyme of bioactive GAs and negatively regulated GA metabolism (Liu and Hou, 2018). Increased expression level of GAI, RGL2, RGL3, GA2ox2, and GA2ox4 was examined in imbibed suvh5 seeds compared with wild-type under R condition (Figure 5E), which suggested that SUVH5 may also regulate GA signaling and GA catabolism in the control of seed germination. Together, these data suggested that SUVH5 may promote light-mediated seed germination by modulating the balance of ABA and GA in imbibed seeds.
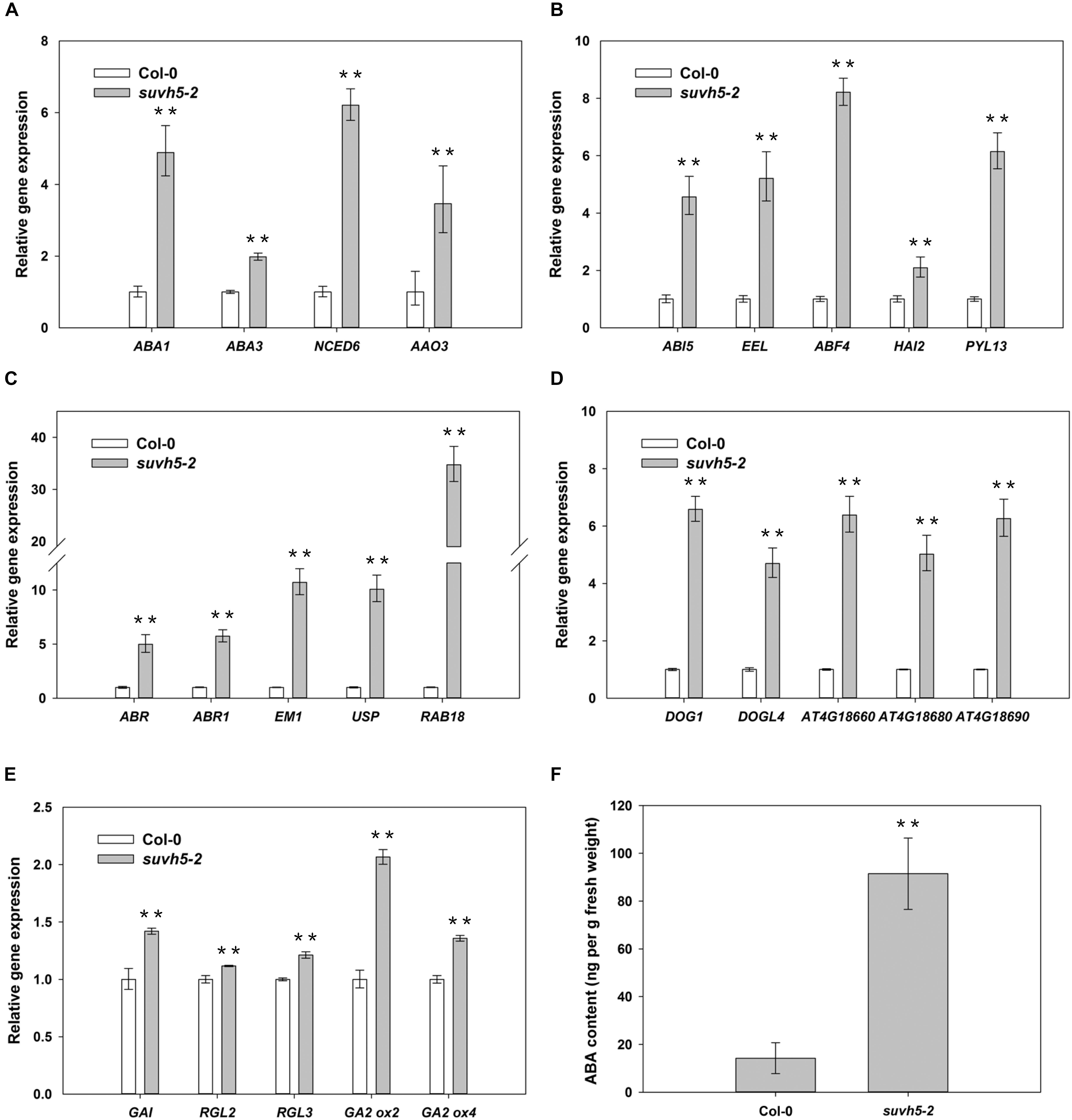
Figure 5. qRT-PCR analyses of the expression levels of ABA biosynthesis genes (A), ABA signaling transduction genes (B), ABA responsive genes (C), DOG family genes (D), GA signaling transduction/deactivating genes (E), and analyses endogenous abscisic acid (ABA) contents (F) in imbibed Col-0 and suvh5-2 seeds under R conditions. Equal amount of Col-0 and suvh5-2 mutant seeds were treated with R light pulse and subsequently incubated in dark for 24 h before extracting mRNA and ABA. PP2A was used as an internal control of qRT-PCR analyses. Values are shown as means ± SD (n = 3) (t-test, ∗∗P < 0.01, difference from Col-0).
DELAY OF GERMINATION 1 is a master regulator of seed dormancy and belongs to a plant-specific gene family with other four DOG1-like genes, At4g18660, At4g18680, At4g18690, and DOGL4 (Bentsink et al., 2006). RNA-seq analysis revealed that the expression of some DOG1-like genes was up-regulated in suvh5-2 mutant (Supplementary Table S1). We further examined the transcripts of DOG1 and DOG1-like genes by qRT-PCR assays. The levels of the expression of DOG1, At4g18660, At4g18680, At4g18690, and DOGL4 were significantly up-regulated in suvh5-2 compared with wild-type seeds under R conditions (Figure 5D), which indicated that SUVH5 may increase seed germination by repressing the expression of the DOG genes.
Previous studies suggested that SUVH5 modulates transcriptional gene silencing through histone H3K9 methyltransferase activity (Ebbs and Bender, 2006; Rajakumara et al., 2011; Yu et al., 2017), and the SRA domain of SUVH5 is required for the accumulation of the H3K9 dimethylation (Rajakumara et al., 2011; Yu et al., 2017). We then analyzed the levels of histone H3K9me2 of the ABA signaling and DOG genes in wild-type and suvh5-2 upon R conditions by chromatin immunoprecipitation in combination with quantitative PCR (ChIP-qPCR) assays. Since previously genome-wide profiling indicated that H3K9me2 modification occurred in target gene promoters or in gene bodies (Zhou et al., 2010), then the regions proximal to the transcriptional starting sites (P), and the first exon regions (E) of these genes were selected for analysis (Figure 6A).
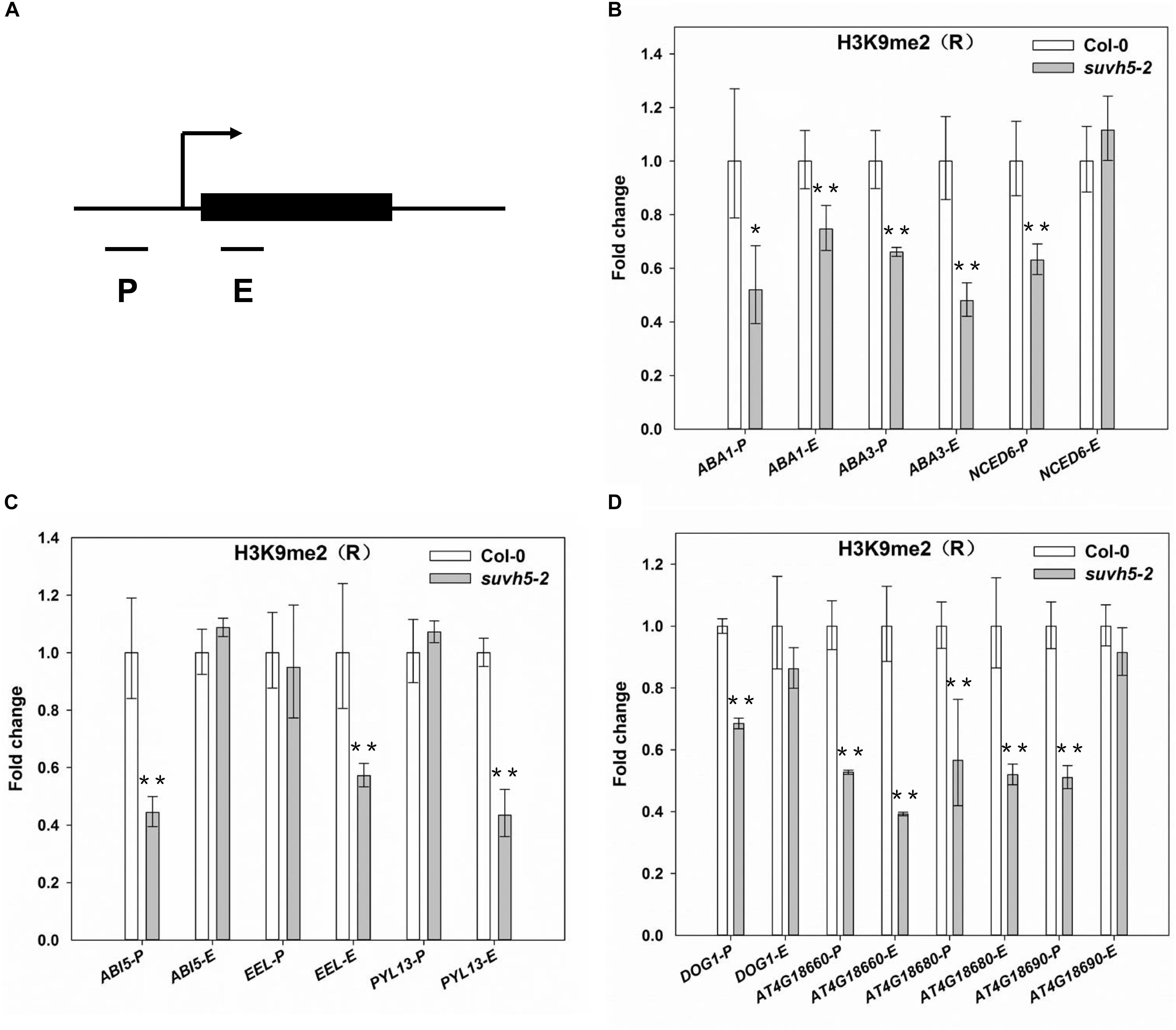
Figure 6. ChIP-qPCR analyses of H3K9me2 levels of SUVH5-regulated genes. (A) Schematic diagram of the regions for ChIP analysis. P and E indicate proximal promoter and first exon regions, respectively. ChIP-qPCR analyses of the histone H3K9me2 levels at the promoter and first exon regions of ABA biosynthesis genes (B), ABA signaling transduction genes (C), and DOG family genes (D) in imbibed Col-0 and suvh5-2 seeds under R conditions. Equal amount of Col-0 and suvh5-2 mutant seeds were treated with R light pulse and subsequently incubated in dark for 24 h before ChIP-qPCR analyses. The amounts of DNA after ChIP were quantified and normalized to TA3, the relative enrichment refers to the H3K9me2 enrichment vs. the histone H3 occupancy. Values are shown as means ± SD (n = 3) (t-test, *P < 0.05, ∗∗P < 0.01, difference from Col-0).
For the ABA biosynthesis genes, the levels of H3K9me2 were significantly decreased at the promoter and first exon regions of ABA1 and ABA3 as well as the promoter region of NCED6 in imbibed suvh5 seeds compared with wild-type (Figure 6B). For the ABA signal transduction-related genes, a significant decrease of H3K9me2 level was detected at the promoter of ABI5 and the exon regions of EEL and PYL13 in suvh5 mutant (Figure 6C). Furthermore, relatively lower levels of H3K9me2 were detected at the promoters of DOG1, At4g18660, At4g18680, and At4g18690, as well as at the exons of At4g18660 and At4g18680 in suvh5 mutant compared with wild-type (Figure 6D). Together, these data suggested that SUVH5 represses the expression of ABA signaling and DOG genes by histone H3K9 dimethylation.
In this work, we present evidence indicating that SUVH5 is a positive component of light-mediated seed germination process. Loss of function of SUVH5 results in decreased germination and leads to 24.6% of light-responsive transcriptome changes. Moreover, SUVH5 represses the expression of ABA signaling and DOG genes via dimethylation of histone H3 at lysine 9.
Abscisic acid is a critical plant hormone in regulating seed germination and dormancy. Previous studies demonstrated that loss of function of ABA biosynthesis mutant aba1 displayed reduced seed dormancy and faster germination rate both in Arabidopsis and tobacco (Koornneef, 1982; Grappin et al., 2000). Mutation of aba3 impaired in ABA biosynthesis and resulted in reduced seed dormancy (Léon-Kloosterziel et al., 1996). NCED6, another key regulator of ABA biosynthesis, also functions in the induction of seed dormancy (Lefebvre et al., 2006). In present work, we showed that SUVH5 represses the expression of these ABA biosynthesis genes by dimethylation of histone H3K9, which indicated that SUVH5 may promote light-mediated seed germination by decreasing ABA content in imbibed seeds. Furthermore, bZIP-type transcription factor ABI5 maintains seed dormancy by activating the expression of genes including seed storage protein genes (Piskurewicz et al., 2008; Kanai et al., 2010; Wang et al., 2011). EEL, a transcription factor homologous to ABI5, is also able to bind to the ABA-responsive elements (ABRE) of seed storage protein genes during late embryogenesis (Bensmihen et al., 2005). Repression of ABI5 and EEL transcripts by SUVH5 indicated that SUVH5 may act to accelerate the launch of germination in imbibed seeds. Moreover, PYR/PYL/RCAR proteins are intracellular ABA receptors regulating ABA-dependent gene expression. Recent works demonstrated that pyl duodecuple mutant, pyr1pyl1/2/3/4/5/7/8/9/10/11/12 is extremely insensitive to ABA effects on seed germination, whereas transgenic plants overexpressing PYL13 show increased ABA sensitivity in seed germination (Zhao et al., 2013; Fuchs et al., 2014). Repressing of PYL13 expression by SUVH5 indicated that SUVH5 may decrease ABA perception thus restrain ABA signal transduction in imbibed seeds. In addition, a number of ABA-responsive genes, such as ABR, ABR1, EM1, USP, and RAB18 were up-regulated in suvh5 mutant, which confirmed a negative role of SUVH5 in ABA signal transduction in imbibed seeds. Together, these findings revealed that SUVH5 may promote seed germination via inhibiting both ABA biosynthesis and ABA signal transduction pathways in imbibed seeds.
Diverse epigenetic modifications, such as DNA methylation, histone modification and chromatin-remodeling, have been reported to play critical roles in regulating seed germination (Dean Rider et al., 2003; Perruc et al., 2007; Saez et al., 2008; Cho et al., 2012; Luo et al., 2012; Zhou et al., 2013; Liu et al., 2014; Gu et al., 2017; Kawakatsu et al., 2017). Recent work displayed that the expression of DOGL4, a paralogous gene of DOG1, is regulated by DNA demethylase ROS1-mediated DNA demethylation (Zhu et al., 2007, 2018). In present work, we showed that the expression of DOGL4 is also repressed by SUVH5-mediated histone H3K9 dimethylation. Interestingly, structure-based studies indicated that a functional SUVH5 SRA domain is required for both DNA methylation and accumulation of the H3K9me2 (Rajakumara et al., 2011). These findings suggested that SUVH5 may repress the expression of DOG genes through both histone H3K9 dimethylation and DNA methylation manners. It will be meaningful to study interplay of SUVH5 and ROS1 in the regulation of DOG genes expression in light-mediated process. Moreover, it’s well known that the epigetic factors usually act in multi-protein complexes in regulating gene expression (Liu et al., 2014). Previous studies demonstrated that RPD3-HDA1-type histone deacetylase HDA6 represses the expression of the ABA pathway genes by regulating the levels of H3ac, H3K4me3 and H3K9me2 (Chen et al., 2010; Chen and Wu, 2010; Luo et al., 2012). In present work, we also showed that SUVH5 represses the expression of ABA signaling-related genes via H3K9me2. A recent study reported that SUVH4/5/6 and HDA6 act in a same protein complex (Yu et al., 2017). These findings strongly suggested that SUVH5 may associate with HDA6 in the regulation of light-mediated seed germination. Further analysis of SUVH5-containing protein complexes will help to elucidate its role of in light-regulated seed germination process.
In imbibed seeds, SUVH5 and SUVH6 displayed similar expression patterns after R and FR irradiation, which indicated that their transcripts might be regulated by the same upstream light-responsive transcription regulators. However, the similar gene expression profiles do not mean they play same roles in light-mediated seed germination. Phenotypic analysis of suvh4, suvht5 and suvh4/5/6 triple mutant suggested that SUVH6 may act oppositely to SUVH4 and SUVH5 and negatively regulate light-mediated seed germination. SUVH proteins generally functions as repressors of gene expression via histone H3K9 dimethylation (Ebbs and Bender, 2006). A recent study demonstrated that KPY/SUVH5/SUVH6 proteins have distinct methylated DNA binding preference, which suggested that these proteins may target different downstream genes (Li et al., 2018). In present work, we showed that SUVH5 represses the expression of ABA biosynthesis, ABA signal transduction as well as DOG genes via histone H3K9 dimethylation. SUVH6 may repress light-mediated seed germination by repressing the expression of some other target genes, such as GA biosynthesis and GA signal transduction related genes in imbibed seeds. Further genome-wide analysis of the downstream genes of SUVH6 will help to explore its role in light-mediated seed germination.
In summary, we identified histone methyltransferase SUVH5 as a positive regulator in light-mediated seed germination. Upon R or strong light conditions, SUVH5 depresses the expression of ABA signaling and DOG genes via dimethylation of H3K9, resulting in reduced levels of ABA contents and increased germination kinetic in imbibed seeds. Moreover, SUVH5 may increase GA levels in imbibed seeds by repress the expression of GA catabolic genes. Ultimately, the changed balance between ABA and GA by SUVH5 leads to promotion of seed germination.
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
XL designed the study. DG conducted most of the experiments. RJ, CH, and TP conducted some of the experiments. MZ, JD, and CX contributed with some materials and reagents. DG and XL analyzed the data and wrote the manuscript.
This work was supported by grants from the National Natural Science Foundation of China (Nos. 31801091 and 31771366), the Youth Innovation Promotion Association of Chinese Academy of Sciences (No. 2014319), the China Postdoctoral Science Foundation (No. 2017M622819), and the Science and Technology Foundation of Guangzhou City (No. 201707010335).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
We thank TAIR for providing the information.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00841/full#supplementary-material
FIGURE S1 | Dynamics of germination rates of Col-0 and suvh5-2 on PHYB activated (R) conditions. Germination frequencies were recorded at indicated time points after treatment. Values are shown as means ± SD (n = 3) (t-test, *P < 0.05, ∗∗P < 0.01, difference from Col-0).
FIGURE S2 | Expression patterns of SUVH4/5/6 under continuous R light and FR light conditions. Equal amount of Col-0 seeds were treated with continuous FR or R light pulse at 22°C for indicated times before extracting mRNA. 0 h indicates dry seeds. PP2A was used as an internal control. Values are shown as means ± SD (n = 3). (A) Expression patterns of SUVH4 under continuous R and FR conditions. (B) Expression patterns of SUVH5 under continuous R and FR conditions. (C) Expression patterns of SUVH6 under continuous R and FR conditions.
TABLE S1 | Genes up-regulated in suvh5-2 in R conditions.
TABLE S2 | Genes down-regulated in suvh5-2 in R conditions.
TABLE S3 | SUVH5 and light co-regulated genes.
TABLE S4 | Primer pairs used in this study.
TABLE S5 | List of ABA signaling-related genes repressed by SUVH5 in imbibed seeds.
Antoni, R., Rodriguez, L., Gonzalez-Guzman, M., Pizzio, G. A., and Rodriguez, P. L. (2011). News on ABA transport, protein degradation, and ABFs/WRKYs in ABA signaling. Curr. Opin. Plant Biol. 14, 547–553. doi: 10.1016/j.pbi.2011.06.004
Bensmihen, S., Giraudat, J., and Parcy, F. (2005). Characterization of three homologous basic leucine zipper transcription factors (bZIP) of the ABI5 family during Arabidopsis thaliana embryo maturation. J. Exp. Bot. 56, 597–603. doi: 10.1093/jxb/eri050
Bentsink, L., Jowett, J., Hanhart, C. J., and Koornneef, M. (2006). Cloning of DOG1, a quantitative trait locus controlling seed dormancy in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 103, 17042–17047. doi: 10.1073/pnas.0607877103
Borthwick, H. A., Hendricks, S., Parker, M., Toole, E., and Toole, V. K. (1952). A reversible photoreaction controlling seed germination. Proc. Natl. Acad. Sci. U.S.A. 38, 662–666. doi: 10.1073/pnas.38.8.662
Briggs, W. R., and Christie, J. M. (2002). Phototropins 1 and 2: versatile plant blue-light receptors. Trends Plant Sci. 7, 204–210. doi: 10.1016/s1360-1385(02)02245-8
Chaves, I., Pokorny, R., Byrdin, M., Hoang, N., Ritz, T., Brettel, K., et al. (2011). The cryptochromes: blue light photoreceptors in plants and animals. Annu. Rev. Plant Biol. 62, 335–364. doi: 10.1146/annurev-arplant-042110-103759
Chen, L. T., Luo, M., Wang, Y. Y., and Wu, K. (2010). Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 61, 3345–3353. doi: 10.1093/jxb/erq154
Chen, L. T., and Wu, K. (2010). Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal. Behavior. 5, 1318–1320. doi: 10.4161/psb.5.10.13168
Cho, J. N., Ryu, J. Y., Jeong, Y. M., Park, J., Song, J. J., Amasino, RM., et al. (2012). Control of seed germination by light-induced histone arginine demethylation activity. Dev. Cell 22, 736–748. doi: 10.1016/j.devcel.2012.01.024
Czechowski, T., Stitt, M., Altmann, T., Udvardi, M. K., and Scheible, W. R. (2005). Genome-wide identification and testing of superior reference genes for transcript normalization in arabidopsis. Plant Physiol. 139, 5–17. doi: 10.1104/pp.105.063743
Grappin, P., Bouinot, D., Sotta, B., Miginiac, E., and Jullien, M. (2000). Control of seed dormancy in Nicotiana plumbaginifolia: post-imbibition abscisic acid synthesis imposes dormancy maintenance. Planta. 210, 279–285. doi: 10.1007/PL00008135
De Wit, M., Galvao, V. C., and Fankhauser, C. (2016). Light-mediated hormonal regulation of plant growth and development. Annu. Rev. Plant Biol. 67, 513–537. doi: 10.1146/annurev-arplant-043015-112252
Dean Rider, S., Henderson, J. T., Jerome, R. E., Edenberg, H. J., Romero-Severson, J., Ogas, J., et al. (2003). Coordinate repression of regulators of embryonic identity by PICKLE during germination in Arabidopsis. Plant J. 35, 33–43. doi: 10.1046/j.1365-313X.2003.01783.x
Deng, W., Wang, Y., Liu, Z., Cheng, H., and Xue, Y. (2014). HemI: a toolkit for illustrating heatmaps. PLoS One 9:e111988. doi: 10.1371/journal.pone.0111988
Ebbs, M. L., and Bender, J. (2006). Locus-specific control of DNA methylation by the Arabidopsis SUVH5 histone methyltransferase. Plant Cell 18, 1166–1176. doi: 10.1105/tpc.106.041400
Finch-Savage, W. E., and Leubner-Metzger, G. (2006). Seed dormancy and the control of germination. New Phytol. 171, 501–523. doi: 10.1111/j.1469-8137.2006.01787.x
Frey, A., Effroy, D., Lefebvre, V., Seo, M., Perreau, F., Berger, A., et al. (2012). Epoxycarotenoid cleavage by NCED5 fine-tunes ABA accumulation and affects seed dormancy and drought tolerance with other NCED family members. Plant J. 70, 501–512. doi: 10.1111/j.1365-313X.2011.04887.x
Fuchs, S., Tischer, S. V., Wunschel, C., Christmann, A., and Grill, E. (2014). Abscisic acid sensor RCAR7/PYL13, specific regulator of protein phosphatase coreceptors. Proc. Natl. Acad. Sci. U.S.A. 111, 5741–5746. doi: 10.1073/pnas.1322085111
Gendrel, A.-V., Lippman, Z., Martienssen, R., and Colot, V. (2005). Profiling histone modification patterns in plants using genomic tiling microarrays. Nat. Methods 2:213. doi: 10.1038/nmeth0305-213
Graeber, K., Linkies, A., Steinbrecher, T., Mummenhoff, K., Tarkowska, D., Tureèková, V. et al. (2014). Delay of germination 1 mediates a conserved coat-dormancy mechanism for the temperature- and gibberellin-dependent control of seed germination. Proc. Natl. Acad. Sci. U.S.A. 111, E3571–E3580. doi: 10.1073/pnas.1403851111
Gu, D., Chen, C. Y., Zhao, M., Zhao, L., Duan, X., Duan, J., et al. (2017). Identification of HDA15-PIF1 as a key repression module directing the transcriptional network of seed germination in the dark. Nucleic Acids Res. 45, 7137–7150. doi: 10.1093/nar/gkx283
Hauser, F., Waadt, R., and Schroeder, J. I. (2011). Evolution of abscisic acid synthesis and signaling mechanisms. Curr. Biol. 21, R346–R355. doi: 10.1016/j.cub.2011.03.015
Jiao, Y., Lau, O. S., and Deng, X. W. (2007). Light-regulated transcriptional networks in higher plants. Nat. Rev. Genet. 8, 217–230. doi: 10.1038/nrg2049
Kanai, M., Nishimura, M., and Hayashi, M. (2010). A peroxisomal ABC transporter promotes seed germination by inducing pectin degradation under the control of ABI5. Plant J. 62, 936–947. doi: 10.1111/j.1365-313X.2010.04205.x
Kawakatsu, T., Nery, J. R., Castanon, R., and Ecker, J. R. (2017). Dynamic DNA methylation reconfiguration during seed development and germination. Genome Biol. 18:171. doi: 10.1186/s13059-017-1251-x
Koornneef, M. (1982). The Genetics of Some Planthormones and Photoreceptors in Arabidopsis thaliana (L.) Heynh. Ph.D. thesis, Wageningen University & Research, Wageningen
Koornneef, M., Bentsink, L., and Hilhorst, H. (2002). Seed dormancy and germination. Curr. Opin. Plant Biol. 5, 33–36. doi: 10.1016/S1369-5266(01)00219-9
Lee, N., Kang, H., Lee, D., and Choi, G. (2014). A histone methyltransferase inhibits seed germination by increasing PIF1 mRNA expression in imbibed seeds. Plant J. 78, 282–293. doi: 10.1111/tpj.12467
Lefebvre, V., North, H., Frey, A., Sotta, B., Seo, M., Okamoto, M., et al. (2006). Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 45, 309–319. doi: 10.1111/j.1365-313X.2005.02622.x
Li, X., Harris, C. J., Zhong, Z., Chen, W., Liu, R., Jia, B., et al. (2018). Mechanistic insights into plant SUVH family H3K9 methyltransferases and their binding to context-biased non-CG DNA methylation. Proc. Natl. Acad. Sci. U.S.A. 115, E8793–E8802. doi: 10.1073/pnas.1809841115
Liu, X., and Hou, X. (2018). Antagonistic regulation of aba and ga in metabolism and signaling pathways. Front. Plant Sci. 9:251. doi: 10.3389/fpls.2018.00251
Liu, X., Yang, S., Zhao, M., Luo, M., Yu, C. W., Chen, C. Y., et al. (2014). Transcriptional repression by histone deacetylases in plants. Mol Plant. 7, 764–772. doi: 10.1093/mp/ssu033
Luo, M., Wang, Y. Y., Liu, X., Yang, S., Lu, Q., Cui, Y., et al. (2012). HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 63, 3297–3306. doi: 10.1093/jxb/ers059
Léon-Kloosterziel, K. M., Gil, M. A., Ruijs, G. J., Jacobsen, S. E., Olszewski, N. E., Schwartz, S. H., et al. (1996). Isolation and characterization of abscisic acid-deficient Arabidopsis mutants at two new loci. Plant J. 10, 655–661. doi: 10.1046/j.1365-313X.1996.10040655.x
Martinez-Andujar, C., Ordiz, M. I., Huang, Z., Nonogaki, M., Beachy, R. N., Nonogaki, H., et al. (2011). Induction of 9-cis-epoxycarotenoid dioxygenase in Arabidopsis thaliana seeds enhances seed dormancy. Proc. Natl. Acad. Sci. U.S.A. 108, 17225–17229. doi: 10.1073/pnas.1112151108
Nakabayashi, K., Bartsch, M., Xiang, Y., Miatton, E., Pellengahr, S., Yano, R., et al. (2012). The time required for dormancy release in arabidopsis is determined by delay of germination1 protein levels in freshly harvested seeds. Plant Cell 24, 2826–2838. doi: 10.1105/tpc.112.100214
Nelson, S. K., and Steber, C. M. (2016). “Gibberellin hormone signal perception: down-regulating DELLA repressors of plant growth and development,” in Annual Plant Reviews, eds P. Hedden and S. G. Thomas (Hoboken, NJ: JohnWiley & Sons, Ltd.) 153–188.
Oh, E., Kim, J., Park, E., Kim, J. I., Kang, C., Choi, G., et al. (2004). PIL5, a phytochrome-interacting basic helix-loop-helix protein, is a key negative regulator of seed germination in Arabidopsis thaliana. Plant Cell 16, 3045–3058. doi: 10.1105/tpc.104.025163
Oh, E., Yamaguchi, S., Hu, J., Yusuke, J., Jung, B., Paik, I., et al. (2007). PIL5, a phytochrome-interacting bHLH protein, regulates gibberellin responsiveness by binding directly to the GAI and RGA promoters in Arabidopsis seeds. Plant Cell 19, 1192–1208. doi: 10.1105/tpc.107.050153
Oh, E., Yamaguchi, S., Kamiya, Y., Bae, G., Chung, W. I., Choi, G., et al. (2006). Light activates the degradation of PIL5 protein to promote seed germination through gibberellin in Arabidopsis. Plant J. 47, 124–139. doi: 10.1111/j.1365-313X.2006.02773.x
Perruc, E., Kinoshita, N., and Lopez-Molina, L. (2007). The role of chromatin-remodeling factor PKL in balancing osmotic stress responses during Arabidopsis seed germination. Plant J. 52, 927–936. doi: 10.1111/j.1365-313X.2007.03288.x
Pertea, M., Kim, D., Pertea, G. M., Leek, J. T., and Salzberg, S. L. (2016). Transcript-level expression analysis of RNA-seq experiments with HISAT, stringtie and ballgown. Nat. Protoc. 11:1650. doi: 10.1038/nprot.2016.095
Piskurewicz, U., Jikumaru, Y., Kinoshita, N., Nambara, E., Kamiya, Y., Lopez-Molina, L., et al. (2008). The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20, 2729–2745. doi: 10.1105/tpc.108.061515
Raghavendra, A. S., Gonugunta, V. K., Christmann, A., and Grill, E. (2010). ABA perception and signalling. Trends Plant Sci. 15, 395–401. doi: 10.1016/j.tplants.2010.04.006
Rajakumara, E., Law, J. A., Simanshu, D. K., Voigt, P., Johnson, L. M., Reinberg, D., et al. (2011). A dual flip-out mechanism for 5mC recognition by the Arabidopsis SUVH5 SRA domain and its impact on DNA methylation and H3K9 dimethylation in vivo. Genes Dev. 25, 137–152. doi: 10.1101/gad.1980311
Rajjou, L., Duval, M., Gallardo, K., Catusse, J., Bally, J., Job, C., et al. (2012). Seed germination and vigor. Annu. Rev. Plant Biol. 63, 507–533. doi: 10.1146/annurev-arplant-042811-105550
Rizzini, L., Favory, J.-J., Cloix, C., Faggionato, D., O’hara, A., Kaiserli, E., et al. (2011). Perception of UV-B by the Arabidopsis UVR8 protein. Science 332, 103–106. doi: 10.1126/science.1200660
Saez, A., Rodrigues, A., Santiago, J., Rubio, S., and Rodriguez, P. L. (2008). HAB1-SWI3B interaction reveals a link between abscisic acid signaling and putative SWI/SNF chromatin-remodeling complexes in Arabidopsis. Plant Cell 20, 2972–2988. doi: 10.1105/tpc.107.056705
Seo, M., and Koshiba, T. (2002). Complex regulation of ABA biosynthesis in plants. Trends Plant Sci. 7, 41–48. doi: 10.1016/S1360-1385(01)02187-2
Seo, M., Nambara, E., Choi, G., and Yamaguchi, S. (2008). Interaction of light and hormone signals in germinating seeds. Plant Mol. Biol. 69, 463–472. doi: 10.1007/s11103-008-9429-y
Shi, H., Zhong, S., Mo, X., Liu, N., Nezames, C. D., Deng, X. W., et al. (2013). HFR1 sequesters pif1 to govern the transcriptional network underlying light-initiated seed germination in Arabidopsis. Plant Cell 25, 3770–3784. doi: 10.1105/tpc.113.117424
Shinomura, T., Nagatani, A., Chory, J., and Furuya, M. (1994). The induction of seed germination in Arabidopsis thaliana is regulated principally by phytochrome b and secondarily by phytochrome A. Plant Physiol. 104, 363–371. doi: 10.1104/pp.104.2.363
Shinomura, T., Nagatani, A., Hanzawa, H., Kubota, M., Watanabe, M., Furuya, M., et al. (1996). Action spectra for phytochrome A-and B-specific photoinduction of seed germination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 93, 8129–8133. doi: 10.1073/pnas.93.15.8129
Shu, K., Liu, X. D., Xie, Q., and He, Z. H. (2016). Two faces of one seed: hormonal regulation of dormancy and germination. Mol. Plant 9, 34–45. doi: 10.1016/j.molp.2015.08.010
Wang, H., and Wang, H. (2015). Phytochrome signaling: time to tighten up the loose ends. Mol. Plant 8, 540–551. doi: 10.1016/j.molp.2014.11.021
Wang, Y., Li, L., Ye, T., Zhao, S., Liu, Z., Feng, Y. Q., et al. (2011). Cytokinin antagonizes ABA suppression to seed germination of Arabidopsis by downregulating ABI5 expression. Plant J. 68, 249–261. doi: 10.1111/j.1365-313X.2011.04683.x
Weitbrecht, K., Müller, K., and Leubner-Metzger, G. (2011). First off the mark: early seed germination. J. Exp. Bot. 62, 3289–3309. doi: 10.1093/jxb/err030
Wu, J., Hettenhausen, C., Meldau, S., and Baldwin, I. T. (2007). Herbivory rapidly activates MAPK signaling in attacked and unattacked leaf regions but not between leaves of Nicotiana attenuata. Plant Cell 19, 1096–1122. doi: 10.1105/tpc.106.049353
Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. doi: 10.1146/annurev.arplant.59.032607.092804
Yu, C. W., Tai, R., Wang, S. C., Yang, P., Luo, M., Yang, S., et al. (2017). Histone deacetylase6 acts in concert with histone methyltransferases suvh4, suvh5, and suvh6 to regulate transposon silencing. Plant Cell 29, 1970–1983. doi: 10.1105/tpc.16.00570
Zhao, M., Yang, S., Liu, X., and Wu, K. (2015). Arabidopsis histone demethylases LDL1 and LDL2 control primary seed dormancy by regulating delay of germination 1 and ABA signaling-related genes. Front. Plant Sci. 6:159. doi: 10.3389/fpls.2015.00159
Zhao, Y., Chan, Z., Xing, L., Liu, X., Hou, Y. J., Chinnusamy, V., et al. (2013). The unique mode of action of a divergent member of the ABA-receptor protein family in ABA and stress signaling. Cell Res. 23, 1380–1395. doi: 10.1038/cr.2013.149
Zheng, J., Chen, F., Wang, Z., Cao, H., Li, X., Deng, X., et al. (2012). A novel role for histone methyltransferase KYP/SUVH4 in the control of Arabidopsis primary seed dormancy. New Phytol. 193, 605–616. doi: 10.1111/j.1469-8137.2011.03969.x
Zhou, J., Wang, X., He, K., Charron, J. B., Elling, A. A., Deng, X. W., et al. (2010). Genome-wide profiling of histone H3 lysine 9 acetylation and dimethylation in Arabidopsis reveals correlation between multiple histone marks and gene expression. Plant Mol Biol. 72, 585–595. doi: 10.1007/s11103-009-9594-7
Zhou, Y., Tan, B., Luo, M., Li, Y., Liu, C., Chen, C., et al. (2013). Histone deacetylase19 interacts with hsl1 and participates in the repression of seed maturation genes in Arabidopsis seedlings. Plant Cell 25, 134–148. doi: 10.1105/tpc.112.096313
Zhu, H., Xie, W., Xu, D., Miki, D., Tang, K., Huang, C. F., et al. (2018). DNA demethylase ROS1 negatively regulates the imprinting of DOGL4 and seed dormancy in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 115, E9962–E9970. doi: 10.1073/pnas.1812847115
Keywords: histone methylation, SUVH5, histone methyltransferase, seed germination, Arabidopsis
Citation: Gu D, Ji R, He C, Peng T, Zhang M, Duan J, Xiong C and Liu X (2019) Arabidopsis Histone Methyltransferase SUVH5 Is a Positive Regulator of Light-Mediated Seed Germination. Front. Plant Sci. 10:841. doi: 10.3389/fpls.2019.00841
Received: 08 March 2019; Accepted: 12 June 2019;
Published: 27 June 2019.
Edited by:
Catherine Bellini, Umeå University, SwedenReviewed by:
Paola Vittorioso, Sapienza University of Rome, ItalyCopyright © 2019 Gu, Ji, He, Peng, Zhang, Duan, Xiong and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Changyun Xiong, spandax@163.com; Xuncheng Liu, xunchengliu@scbg.ac.cn
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.