
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Genet. , 04 December 2018
Sec. Behavioral and Psychiatric Genetics
Volume 9 - 2018 | https://doi.org/10.3389/fgene.2018.00602
This article is part of the Research Topic Genomics and Epigenomics of Alcoholism View all 12 articles
 Diana Zhou1†
Diana Zhou1† Yinghong Zhao2†
Yinghong Zhao2† Michael Hook1†
Michael Hook1† Wenyuan Zhao1
Wenyuan Zhao1 Athena Starlard-Davenport1
Athena Starlard-Davenport1 Melloni N. Cook1,3
Melloni N. Cook1,3 Byron C. Jones1
Byron C. Jones1 Kristin M. Hamre4
Kristin M. Hamre4 Lu Lu1*
Lu Lu1*Coenzyme Q (CoQ) is a well-studied molecule, present in every cell membrane in the body, best known for its roles as a mitochondrial electron transporter and a potent membrane anti-oxidant. Much of the previous work was done in vitro in yeast and more recent work has suggested that CoQ may have additional roles prompting calls for a re-assessment of its role using in vivo systems in mammals. Here we investigated the putative role of Coenzyme Q in ethanol-induced effects in vivo using BXD RI mice. We examined hippocampal expression of Coq7 in saline controls and after an acute ethanol treatment, noting enriched biologic processes and pathways following ethanol administration. We also identified 45 ethanol-related phenotypes that were significantly correlated with Coq7 expression, including six phenotypes related to conditioned taste aversion and ethanol preference. This analysis highlights the need for further investigation of Coq7 and related genes in vivo as well as previously unrecognized roles that it may play in the hippocampus.
Coenzyme Q (CoQ or ubiquinol) is a lipophilic molecule present in every cell membrane in the body (Crane, 2001; Turunen et al., 2004). It is best known for its roles as a mitochondrial electron transporter and a potent membrane anti-oxidant (Ernster and Dallner, 1995; Bentinger et al., 2007). CoQ is made up of a benzoquinone ring with an isoprenoid side chain (containing 6–10 units) conserved across species from yeast (as CoQ6), to mice (as CoQ7), to humans (as CoQ10) (Lenaz, 1985). CoQ production in any species is the result of a complex biosynthesis process involving 10 to 15 or more genes (depending on the species) encoding a series of enzymes and non-enzymatic proteins, many of which belong to the Coq family of genes (Coq 1 – Coq 10A/B) (Acosta et al., 2016, see their Table 1 and Figure 1 for a complete list). Despite this molecule being characterized and isolated nearly 60 years ago (Festenstein et al., 1955; Wolf et al., 1958; Crane et al., 1989), it continues to remain relevant through ongoing investigations that are fine-tuning its roles in bioenergetics and anti-oxidant defense.
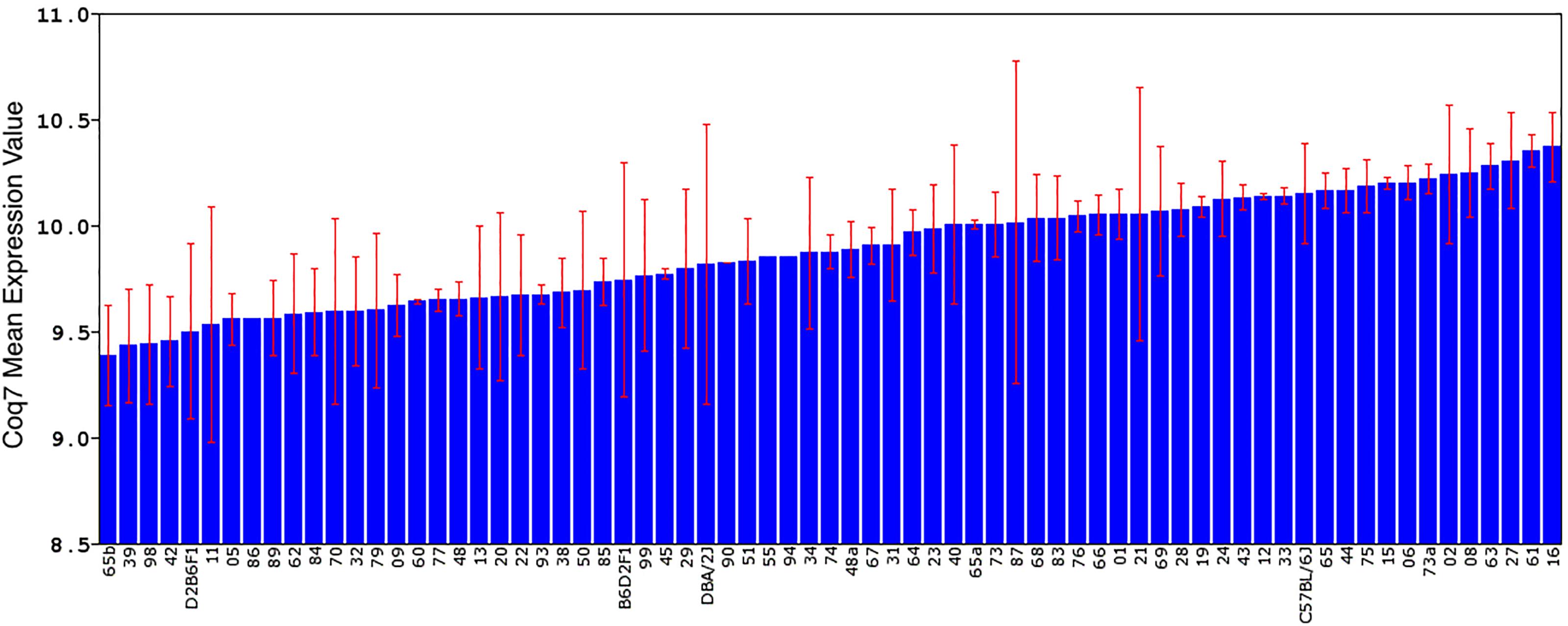
FIGURE 1. Differential expression of Coq7 across BXD RI strains rank ordered by expression level. The standard deviation and mean expression of Coq7 in each strain is shown across the parental DBA/2J and C57BL/6J strains, F1 hybrids, and 67 BXD strains. The x-axis represents the mouse strain, while the y-axis shows the mean gene expression using the log2 scale.
Much of the early genetic work regarding CoQ stemmed from submitochondrial fraction studies (Mellors and Tappel, 1966; Landi et al., 1984) and yeast Saccharomyces cerevisiae (González-Mariscal et al., 2014), which highlighted CoQ biosynthesis as necessary for mitochondrial antioxidant defense, with less CoQ production resulting in impaired defenses and increased presence of anti-oxidant molecules. But ongoing work has revealed CoQ’s role as an anti-oxidant to be more complex than previously thought. In vitro studies using CoQ deficient skin fibroblasts showed severe (<20% of normal) and mild (>60%) CoQ deficiency did not increase reactive oxygen species (ROS) production while moderate deficiency (30–50%) markedly increased ROS production (Quinzii et al., 2008, 2010). Recent in vivo studies in CoQ knockout mouse models have been more equivocal, finding that CoQ deficiency does not always directly correspond with ROS production or tissue dysfunction (Quinzii et al., 2013; Licitra and Puccio, 2014; Wang et al., 2015; Luna-Sánchez et al., 2015). This has led some researchers to emphasize the need for better characterization of CoQ’s roles in vivo using mammalian models vs. studies in vitro or using yeast (Wang and Hekimi, 2016).
In the current study, we use a systems genetics approach to examine the relationship between acute ethanol effects and hippocampal Coq7 expression in a well characterized genetic population of BXD Recombinant Inbred (RI) mice derived from the C57BL/6J (B6) and DBA/2J (D2) inbred strains, furthering our understanding of CoQ biosynthesis regulation in vivo. Currently, there is little evidence connecting Coq7 and regulation of ethanol responses. However, ethanol metabolism into acetaldehyde is a well understood source of oxidative stress in the brain, causing lipid peroxidation and other oxidative damage to brain tissue (Hipolito et al., 2007; Hernández et al., 2016). This metabolic damage is thought to be warded off by increased gene expression of endogenous anti-oxidants, such as superoxide dismutase (Reddy et al., 1999; Enache et al., 2008). Coq7 (also known as mclk-1) encodes a hydroxylase (coq7p) involved in one of the final steps of CoQ synthesis, conversion of demethoxyubiquinone (DMQ) to CoQ (Acosta et al., 2016). This step in biosynthesis is thought to be the regulated step in CoQ biosynthesis (Marbois and Clarke, 1996; Padilla et al., 2009; Martín-Montalvo et al., 2013; Lohman et al., 2014), making it a prime target to explore how endogenous CoQ production changes in response to acute oxidative stress. Additional evidence suggesting that Coq7 is important in alcohol responses comes from a study in HXB/BXH RI rats where Coq7 has been proposed as a candidate gene for alcohol dependency and consumption (Tabakoff et al., 2009).
Previous studies from our lab have shown that gene expression in the hippocampus is particularly sensitive to the effects of acute ethanol (1.8 g/kg) (Urquhart et al., 2016; Baker et al., 2017). Others have also shown that acute ethanol (2.0 g/kg) produces brain region-specific changes in gene expression, including in the hippocampus (Kerns et al., 2005). Here, we demonstrate a positive relationship between acute ethanol ingestion and Coq7 expression in hippocampus. We also map an expression quantitative trait locus (eQTL) for Coq7 in BXD RI mice as well as identify pathways associated with Coq7 and its correlated genes. Through this, we aim to better characterize Coq7 as a vital gene in the oxidative stress response caused by brain ethanol metabolism.
The BXD RI mouse strains were derived by crossing the parental strains B6 and D2. The F1 progeny were subsequently intercrossed followed by inbreeding to fix parental genotypes at each locus. The BXD mice are a densely phenotyped and genotyped family and have been used as a genetic reference panel for identifying the genetic basis of phenotypes and diseases, including molecular expression phenotypes, as well as for identifying pathways regulating gene expression. For this study, the dataset of Hippocampus Consortium M430v2 (Jun06) RMA that we generated previously (Overall et al., 2009) was used for genetic mapping, transcript measurement of which was taken from the hippocampus of 67 BXD strains, the parental B6 and D2 strains, and reciprocal F1 hybrids (B6D2F1 and D2B6F1). Detailed information on the strain, sex, age of each animal can be accessed from http://genenetwork.org/webqtl/main.py?FormID=sharinginfo&GN_AccessionId=110. This data set has been uploaded into Gene Expression Omnibus (GEO) with accession number GSE84767 where the data can be downloaded.
The BXD parental strains B6 and D2 mice were used for acute ethanol treatment. Ten mice per strain (2∼3 mice per sex) including both males and females at 2–4 months old were divided into two groups: (1) saline group: given an isovolumetric IP injection of saline and (2) ethanol group: treated with an IP injection of 2.0 g/kg i.p., ethanol (12.5% v/v). The dose of 2.0 g/kg of ethanol was chosen to allow for comparisons with other studies in mice seeking to produce genetically based ethanol-sensitive behaviors (Cunningham and Noble, 1992; Cunningham and Prather, 1992; Cunningham, 1995; Risinger and Cunningham, 1995, 1998). Twenty-four hours after treatment, these mice were sacrificed for tissue harvest (Cook et al., 2015).
The B6 and D2 mice treated with ethanol or saline were anesthetized with an overdose of avertin (1.25% 2,2,2- tribromoethanol and 0.8% tert-pentyl alcohol in water; 0.8–1.0 ml, i.p.) until they were immobile, a period of less than 2 min. After this time mice were sacrificed by cervical dislocation. The hippocampi were harvested according to previously described methods (Lu et al., 2001). The left and right hippocampi were pooled and stored in RNAlater overnight at 4°C, then kept at -80°C until RNA extraction.
RNA was extracted from the hippocampus using RNA STAT-60 (protocols can be found at Tel-Test1) as per the manufacturer’s instructions. A spectrophotometer (Nanodrop Technologies2) was used to measure RNA concentration and purity, and the Agilent 2100 Bioanalyzer was used to evaluate RNA integrity. To pass quality control, the RNA integrity values needed to be greater than 8. The majority of samples had values between 8 and 10.
The Affymetrix GeneChipTM Mouse Transcriptome Array 1.0 (MTA 1.0) was used to generate gene expression data, from B6 and D2 mice treated with ethanol or saline, according to the manufacturers’ protocol. Affymetrix Expression Console Software was used to identify and remove outlier arrays, and normalize raw data in CEL files using the Robust Multichip Array (RMA) method (Pan et al., 2011). The expression data were then re-normalized using a modified Z score described in a previous publication (Bolstad et al., 2003). We calculated the log base 2 of the normalized values, computed Z scores for each array, multiplied the Z scores by 2, and added an offset of 8 units to each value. This transformation yields a set of Z-like scores for each array that have a mean of 8, a variance of 4, and standard deviation of 2. The advantage of this modified Z score is that a twofold difference in expression corresponds approximately to a 1 unit change.
Total RNA from 10 hippocampi per treatment group (both B6 and D2 mice) was used for a quantitative RT-PCR experiment. The gene-specific probe and primer sets for Coq7 (upstream 5′-tttggaccatagctgcattg-3′, downstream 5′-tgaggcctcttccatactctg-3′) were deduced using Universal Probe Library Assay Design software3. Coq7 mRNA levels were detected and analyzed on a LightCycler 480 System (Roche, Indianapolis, IN, Unites States3) under the following cycling conditions: 1 cycle at 95°C for 5 min and then 40 cycles at 95°C for 10 s, 60°C for 30 s, and 72°C for 10 s. The PCR mix contained 0.2 μl of 10 μM primers, 0.1 μl of 10 μM Universal library probe, 5 μl of LC 480 master mix (2×), 2 μl of template cDNA, and RNase-free water to 10 μl. TATA box-binding protein (TBP) was selected as the endogenous quantity control. The relative gene expression of Coq7 was analyzed with the ΔΔCT method with TBP used as the reference gene for normalization. Coq7 expression (fold change) in each ethanol treated mouse relative to average of the corresponding control mice was calculated as: Fold Change = 2ˆ-[ΔCT(Coq7 in each ethanol treated mouse) - (Mean of ΔCT of control mice)].
The Coq7 gene expression data from saline or alcohol treatment in B6 and D2 mice were evaluated using the analysis of covariance (ANCOVA) with treatment, strain, and sex as factors, and adjusted for age and body weight.
We performed phenotype QTL and eQTL analyses using the WebQTL module on GeneNetwork4 according to our published methods (Chesler et al., 2005). The genome-wide efficient mixed model association algorithm (GEMMA) was used to identify potential eQTLs regulating Coq7 expression levels and phenotype QTLs near the Coq7 locus, and to estimate the significance at each location using known genotypic data for those sites. Each of these analyses produced a likelihood ratio statistic (LRS) score, providing us with a quantitative measure of confidence of linkage between the observed phenotypes or expression level of Coq7 and known genetic markers. The significance of the QTL and eQTLs were calculated using more than 2000 permutations tests. Loci were considered statistically significant if genome-wide p < 0.05. Sequence variability between B6 and D2 was then determined using the Sanger mouse SNP database5.
Prior to gene function analysis, co-expression and literature correlation were performed to filter a list of transcripts correlated with Coq7 gene expression in the ethanol and saline groups. Co-expression analysis was performed on GeneNetwork. Coq7 probe set expression was compared to all probe sets on the MTA 1.0 array. Criterion for significant co-expression included average log2 probe set expression greater than 7.0, as well as a significant correlation with Coq7, indicated by a Pearson product correlation value (p < 0.05). We further filtered Coq7 co-expressed probe sets in the ethanol and saline groups by performing literature correlations using the Semantic Gene Organizer to find the potential biological correlation between Coq7 and other genes (Homayouni et al., 2004). Genes with higher correlation values (r > 0.3) were selected for further analysis. The top 500 genes with significant co-expression (p < 0.05) and literature correlations (r > 0.45) for the ethanol and saline data sets were then selected and uploaded to Webgestalt6 for gene function analyses (Zhang et al., 2005). Enrichment of biological function in the top 500 Coq7 ethanol and saline co-expression data sets was determined using the hypergeometric test. The p-values from the hypergeometric test were automatically adjusted to account for multiple comparisons using the Benjamini and Hochberg correction (Benjamini and Hochberg, 1995). Categories with an adjusted p-value of less than 0.05 indicated that the set of submitted genes was significantly over-represented in those categories.
We used the BXD phenotype database in our GeneNetwork website4 to find alcohol phenotypes highly correlated (Pearson product correlation, p < 0.05) with expression of the Coq7 probe set in hippocampus from naïve BXD mice (Hippocampus Consortium M430v2 (Jun06) RMA data set).
Only one probe set in the Affymetrix M430 dataset represents the Coq7 gene (1415556_at), which targets the last five coding exons. Coq7 expression (log2 scale) varied widely between BXD strains, with a fold-change of 2.57 (Figure 1). BXD65b had the lowest expression (9.38 ± 0.24), and BXD16 had the highest expression (10.70 ± 0.16). There is also a 1.26-fold difference in Coq7 transcript abundance between B6 (10.15 ± 0.23) and D2 (9.81 ± 0.66).
The Affymetrix M430 database was used for eQTL mapping. Coq7 is located on chromosome 7 at 118.53 Mb, and a significant eQTL modulating the expression of this gene with a likelihood ratio statistics (LRS) of 81 was mapped to the Coq7 gene locus (Figure 2). This indicates that Coq7 is cis-regulated, meaning one or more sequence variants affecting its expression is located within or near the gene itself. Using the open access sequence data resources at Sanger5, we identified 140 SNPs in Coq7 between the BXD parental strains B6 and D2. Eleven SNPs are located at 3′-UTR and coding area (Table 1) including one stop gain variance that could have strong downstream effect. The rest of them are located at non-coding area. All SNPs are listed in Supplementary Table 1. In addition, we identified 42 indels in Coq7 between the BXD parental strains (Supplementary Table 2), with two of them being frameshift variants that could also have a strong downstream effect.
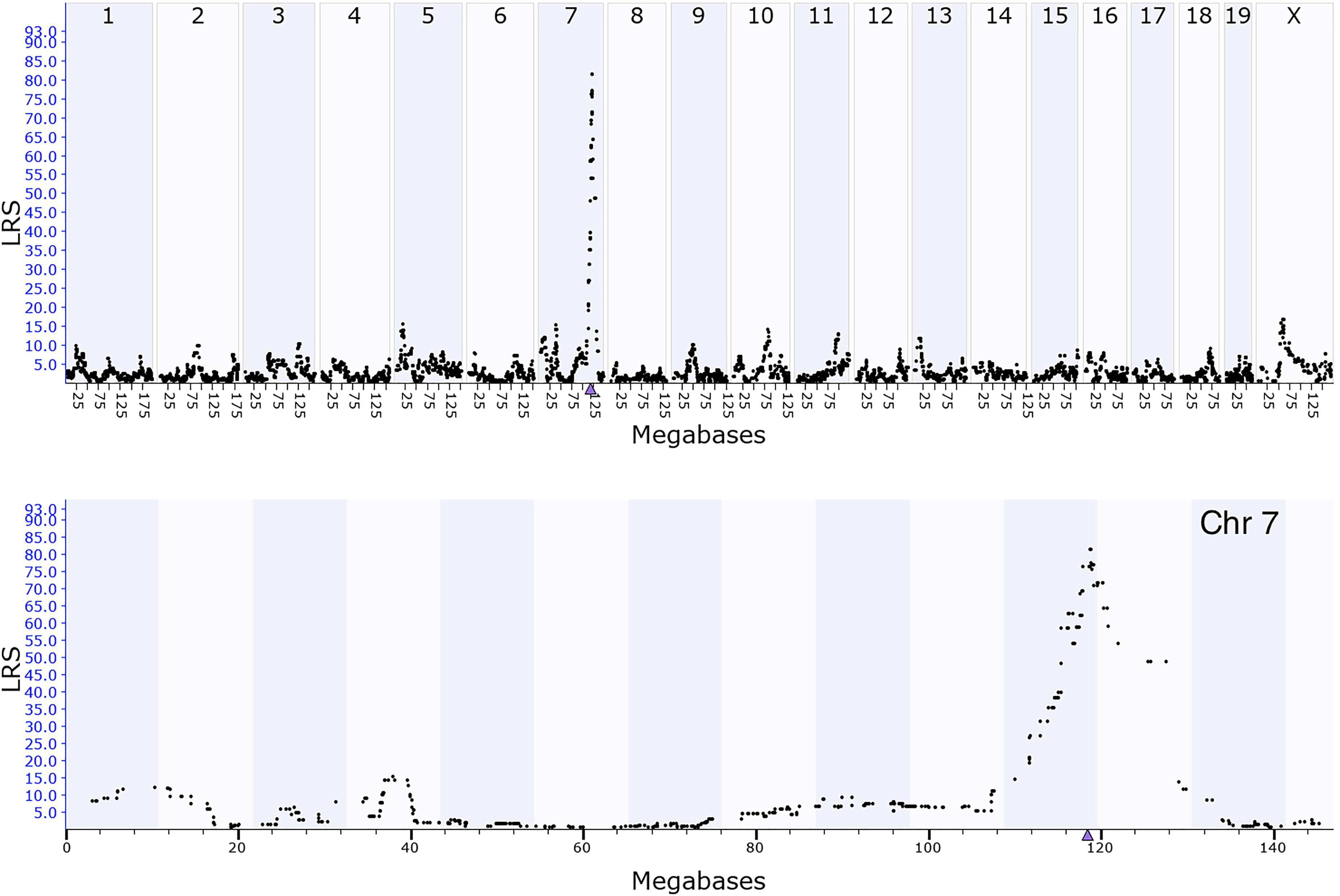
FIGURE 2. GEMMA eQTL mapping for Coq7 expression in hippocampus. The x-axis represents positions on the mouse genome in megabases (Mb), while the y-axis represents the likelihood ratio statistic scores (LRS), a measurement of the linkage between expression of Coq7 and inheritance of B6 and D2 alleles at different regions of the genome. Top panel shows interval mapping for the whole mouse genome, indicating a significant eQTL at chromosome 7. Bottom panel shows a close-up view of chromosome 7 in which the peak of eQTL was around 119 Mb of chromosome 7 where Coq7 is located (triangle).
We used the Affymetrix MTA datasets to analyze the effect of alcohol on the expression of Coq7 in the hippocampal tissue of B6 and D2 mice using an ANOVA with treatment, strain and sex as the between subject factors. The average of Coq7 expression in the saline group was 7.46 ± 0.01 (log2 scale), while the average for the ethanol group was 7.52 ± 0.01 (log2 scale), indicating an increase in expression after ethanol treatment. ANCOVA analysis showed a significant effect of ethanol treatment on Coq7 transcript abundance (P = 0.029) and also interaction of ethanol treatment and sex significantly effects on Coq7 transcript abundance (P = 0.002) (Table 2). For B6 strain, the average of Coq7 expression in the ethanol group was 7.53 ± 0.04 (log2 scale), while the average for the saline group was 7.46 ± 0.03 (log2 scale). For D2 strain, the average of Coq7 expression in the ethanol group was 7.51 ± 0.05 (log2 scale), while the average for the saline group was 7.47 ± 0.03 (log2 scale). The T-test analysis showed a significant effect of ethanol treatment on Coq7 transcript abundance in the B6 strain (P = 0.0234), not in the D2 strain (P = 0.2454).
Coq7 expression was validated using RT-PCR. The RT-PCR results showed significantly increased expression of Coq7 (F = 11.77, P = 0.027) after ethanol treatment. This was consistent with the results from microarray analysis. In addition, we also found significant strain differences between B6 and D2 (F = 9.326, P = 0.038), and sex difference between males and females (F = 11.788, P = 0.027).
We used the top 500 transcripts co-expressed with Coq7 from both saline and ethanol group to perform gene ontology (GO) analysis. For the saline-treated group, significant biological processes included “ATP metabolic process,” “mitochondrion organization,” “electron transport chain,” and “ubiquinone biosynthetic process” (Supplementary Table 3). For the ethanol treated group, 54 enriched GO biological processes categories reached significance (Supplementary Table 4). Many of the categories in the ethanol group overlapped with those in the saline group, but all of the ATP metabolic and mitochondrial function related categories were no longer significant. Instead, several new enriched categories including serine metabolic process, behavior and cognition show up.
Gene pathway enrichment analysis for the saline-treated group revealed 21 related pathways (adj P < 0.05, Table 3), while the ethanol-treated group revealed six related pathways (adj P < 0.05, Table 4). The top enriched pathway in the saline group including “Electron Transport Chain” and “Oxidative phosphorylation” are not shared with the ethanol group. A pathway of interest unique to the ethanol group is “Oxidative stress.”
We performed correlational analyses with phenotypes archived in the GeneNetwork database to identify ethanol-related behaviors correlated with Coq7 (p < 0.05) expression in the hippocampus. We found 45 ethanol-related phenotypes significantly correlated with Coq7 expression (Supplementary Table 5), most of which are related to ethanol consumption, ethanol response, ethanol preference, and body temperature after ethanol treatment; some of which are mapped near Coq7 location. For example, one phenotype (record ID 10496, ethanol response) has a significant QTL with a LRS of 13.8 (P < 0.05) on chromosome 7 at 114∼119 Mb (Figure 3) where Coq7 is located.
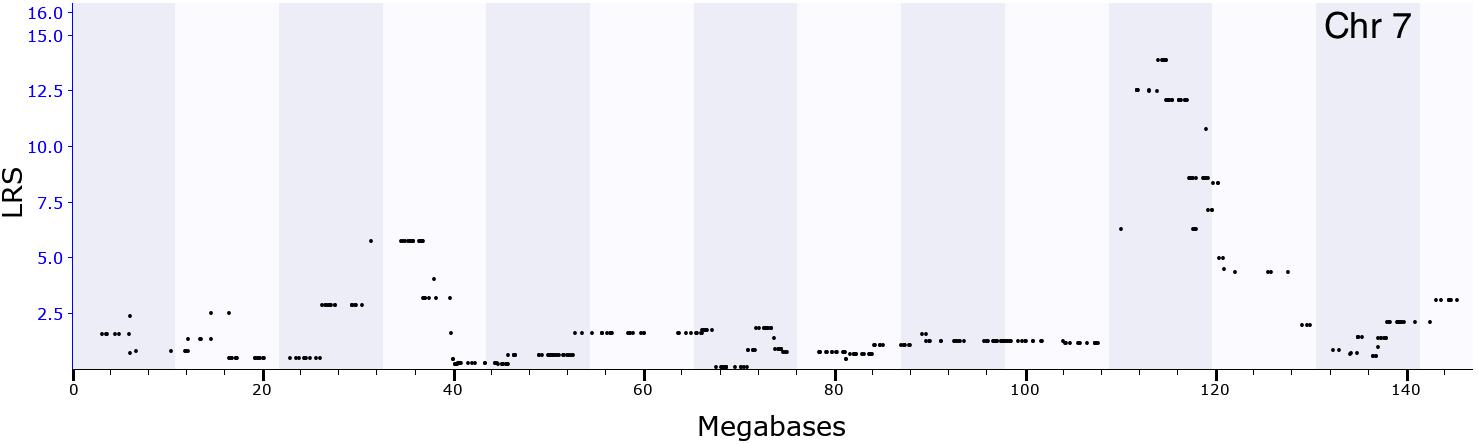
FIGURE 3. GEMMA QTL mapping for locomotion behavior after ethanol injection (2 g/kg). The x-axis represents a position on the mouse genome in megabases (Mb), while the y-axis gives the likelihood ratio statistic scores (LRS). The mapping result showed that the peak of QTL was around 114∼119 Mb of chromosome 7.
Currently, mechanisms through which ethanol affects Coq7 and ubiquinone are unknown and previous studies analyzing Coq7 and ethanol are lacking. To this end, we attempted to elucidate the relationship between ethanol and expression of Coq7 in the hippocampus of BXD mice by identifying genes and biological pathways that may link the two. Coq7 is variably expressed in naïve hippocampal tissue from BXD RI strains and is cis-regulated, making it an excellent candidate for study as a modifier of gene expression or biological phenotypes (Ciobanu et al., 2010). We found that there are multiple SNPs and Indels in Coq7, which may be responsible for its differential expression in the hippocampal tissues across BXD strains. This also suggests that one or more polymorphism may affect Coq7 expression and play a role in gene regulation (Kunugi et al., 2001; Jablonski et al., 2005). Furthermore, we found increased expression of Coq7 following acute ethanol injection and identified 45 ethanol-related phenotypes correlated with Coq7, supporting its likely involvement in ethanol responses (Supplementary Table 5). These findings included six phenotypes related to conditioned taste aversion and ethanol preference. It has been previously proposed that Coq7 may play a role in alcohol consumption (Tabakoff et al., 2009), aligning with the result of our phenotypic analysis. While this phenotypic alignment is interesting, it should be noted that our experiment did not directly test ethanol consumption behaviors before and after ethanol injection, limiting how generalizable this result may be. Future direct observation of changes in ethanol preferences following ethanol injection or a period of oral consumption would likely be worthwhile.
Comparison of functional enrichment analysis results between our treatment groups may hold insight into why Coq7 was upregulated following ethanol injection. In the saline treatment group, we found processes related to cellular metabolism and ATP generation (Supplementary Table 3). Presumably, when proteins needed to break down energy substrates are expressed, Coq7 is also expressed. This is unsurprising considering CoQ’s role in the electron transport chain as an electron shuttle, representing a lynchpin of the final step of ATP generation following metabolism. Conversely, these metabolic functions were largely insignificant in the ethanol treatment group’s analysis, suggesting a less prominent role. This may well be representative of ethanol’s effect on the mitochondria, which has been shown to decrease respiratory rates and the rate of ATP synthesis following chronic exposure (Thayer and Rubin, 1979; Manzo-Avalos and Saavedra-Molina, 2010). This notion was further corroborated by pathway enrichment analysis, which found pathways such “Electron Transport Chain,” “TCA cycle,” “Amino Acid Cycle,” “Oxidative phosphorylation,” and “Glycolysis and Gluconeogenesis” associated with the saline treated group (Table 3), but not the ethanol treatment group.
The ethanol group uniquely had “Oxidative Stress” as a significantly enriched gene pathway (Table 4). Notably, three of the four genes in this gene pathway encode proteins contributing to anti-oxidant enzymes: Sod2 (superoxide dismutase, Azadmanesh and Borgstahl, 2018), Gclc (glutamate-cysteine ligase, a catalyst for glutathione synthesis; Lu, 2009, 2013), and Txnrd1 (thioredoxin reductase 1; Turanov et al., 2010). While all three are well-established anti-oxidants, previous work on their interactions with ethanol vary greatly. Superoxide dismutase (SOD) is perhaps best studied but also the most controversial. SOD activity in the murine brain following ethanol exposure has been shown to increase (Somani et al., 1996; Enache et al., 2008; Reis et al., 2017), remain unchanged (Gönenç et al., 2005) and decrease (Ledig et al., 1981) under various conditions. The other two genes are less well characterized. Gclc mRNA expression shown to be induced by ethanol in rat liver and brain (Lu et al., 1999; Narasimhan et al., 2011) and Txnrd1 has not been as well studied in terms of ethanol-induced stress.
However, a recent paper by Casañas-Sánchez et al. (2016) found all three of these genes, as well as many other anti-oxidant genes, to have increased expression in hippocampal derived HT22 cells following acute, sub-toxic ethanol exposure. To explain its result, this paper emphasized the growing understanding of ROS as second messengers, capable of influencing gene expression (D’Autréaux and Toledano, 2007; Kaspar et al., 2009; Schieber and Chandel, 2014). These findings have been pioneered in the growing field of “redox biology,” which heavily emphasizes a more nuanced understanding of the impact of ROS. Current work suggests that varying levels of ROS production (so called “basal,” “low,” “intermediate,” and “high” oxidative stress) cause different cellular reactions (Lushchak, 2014; Sies, 2015). Casañas-Sánchez et al. (2016) reasoned that low levels of ethanol increased the level of oxidative stress, as ethanol is a well-documented potent producer of ROS by virtue of its metabolism (Lieber, 1976; Zakhari, 2006; Das and Vasudevan, 2007), resulting in beneficial mitohormetic anti-oxidant gene expression.
This phenomenon may similarly be occurring in our study, as evidenced by the induction of Coq7 and the phenotypic shift to oxidative stress protection in the ethanol treatment group. It may be that CoQ is unimportant as an anti-oxidant under basal or low-level oxidative stress, as seen in CoQ knockout mouse studies, but is induced under heightened oxidative stress levels alongside other well-known anti-oxidants. The variation of Coq7 expression in BXD RI mice could result in variation in this inducible response, accounting for differences in how individual strains of BXDs react to ethanol ingestion. Tabakoff et al. (2009) argue that several of their identified “alcohol consumption genes” could theoretically modulate GABA release from the hypothalamus to areas like the ventral tegmental area, ultimately affecting downstream reward behaviors through a variety of molecular mechanisms (see their Figure 3). Logically, the variation in genetic expression among individuals or under different conditions may end up modulating behavior. While Tabakoff mentions Coq7 as an identified “ethanol consumption gene,” they do not outline how it might affect GABA release in this way. With the idea of ROS as signaling molecules in mind, variation in Coq7 expression (or other anti-oxidant defenses) could limit or permit ROS signaling, potentially altering critical neurologic signaling pathways related to reward-seeking behavior, similar to those proposed by Tabakoff et al. (2009). How these alterations would occur is still a matter of investigation. Work has examined some of the intracellular mechanisms by which ROS can affect plasticity and signaling in the hippocampus, mainly through induction of long-term potentiation (LTP). This induction seems to be through ROS acting as a second messenger, inducing phosphorylation of established LTP effectors such as PKC, ERK, and CamKII (Malinow et al., 1989; Klann et al., 1998; Kishida et al., 2005). For recent reviews on the topic, please see Beckhauser et al. (2016) and Oswald et al. (2018). However, there has been little investigation into ROS’s effect on motivational circuitry. While further work is needed, ethanol-induced oxidative stress may represent a worthwhile line of inquiry to better understand hippopcampal cellular responses to heightened oxidative stress in vivo and their downstream effects on neurotransmission and behavior.
In sum, we analyzed the effect of ethanol on the expression of Coq7 using a systems genetic approach. We identified an eQTL showing variability in Coq7 expression in BXD RI mice and found its expression to be increased following ethanol treatment. We also uncovered several pathways and genes which may interact with Coq7 to regulate the ethanol response. Based on this and previous reports, Coq7 may act as an inducible anti-oxidant at heightened levels of oxidative stress, aligning both with previous work and current questions on this ubiquitous molecule. Further research into the specific interactions between Coq7 and identified genes may elucidate their relationships and shed light on how Coq7 affects mitochondria following acute ethanol consumption.
All animal work was conducted in accordance with of and procedures approved by the Institutional Animal Care and Use Committees at The University of Tennessee Health Science Center and University of Memphis following NIH guidelines.
DZ provided the primary analysis and primary writing for the manuscript. YZ provided the primary interpretation of experiment and primary writing for the manuscript. MH provided the primary writing, editing, literature review, and for the manuscript and data interpretation of experiment. WZ provided the analysis and RT-PCR experiment. AS-D, MC, AD, and BJ provided the final editing and critique of manuscript. KH provided the primary experiment, final editing and critique of the manuscript. LL provided the procedure, planning, funding, and oversight for experiment as well as final editing and critique of manuscript.
This study was supported by National Institutes of Health grants R01AA021951 (LL, BJ) and U01AA014425 (LL), and Internal New Grant Support Funding (E073239005) from the University of Tennessee Health Science Center (LL).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors thank Dr. Robert Williams for his financial support, bioinformatics support, and experiment design.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fgene.2018.00602/full#supplementary-material
Acosta, M. J., Fonseca, L. V., Desbats, M. A., Cerqua, C., Zordan, R., Trevisson, E., et al. (2016). Coenzyme Q biosynthesis in health and disease. Biochim. Biophys. Acta 1857, 1079–1085. doi: 10.1016/j.bbabio.2016.03.036
Azadmanesh, J., and Borgstahl, G. E. (2018). A review of the catalytic mechanism of human manganese superoxide dismutase. Antioxidants 7:E25. doi: 10.3390/antiox7020025
Baker, J. A., Li, J., Zhou, D., Yang, M., Cook, M. N., Jones, B. C., et al. (2017). Analyses of differentially expressed genes after exposure to acute stress, acute ethanol, or a combination of both in mice. Alcohol 58, 139–151. doi: 10.1016/j.alcohol.2016.08.008
Beckhauser, T. F., Francis-Oliveira, J., and De Pasquale, R. (2016). Reactive oxygen species: physiological and physiopathological effects on synaptic plasticity: supplementary issue: brain plasticity and repair. J. Exp. Neurosc. 10(Suppl. 1), 23–48. doi: 10.4137/JEN.S39887
Benjamini, Y., and Hochberg, Y. (1995). Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300.
Bentinger, M., Brismar, K., and Dallner, G. (2007). The antioxidant role of coenzyme Q. Mitochondrion 7, S41–S50. doi: 10.1016/j.mito.2007.02.006
Bolstad, B. M., Irizarry, R. A., Åstrand, M., and Speed, T. P. (2003). A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19, 185–193. doi: 10.1093/bioinformatics/19.2.185
Casañas-Sánchez, V., Pérez, J. A., Quinto-Alemany, D., and Díaz, M. (2016). Sub-toxic ethanol exposure modulates gene expression and enzyme activity of antioxidant systems to provide neuroprotection in hippocampal HT22 cells. Front. Physiol. 7:312. doi: 10.3389/fphys.2016.00312
Chesler, E. J., Lu, L., Shou, S., Qu, Y., Gu, J., Wang, J., et al. (2005). Complex trait analysis of gene expression uncovers polygenic and pleiotropic networks that modulate nervous system function. Nat. Genet. 37, 233–242. doi: 10.1038/ng1518
Ciobanu, D. C., Lu, L., Mozhui, K., Wang, X., Jagalur, M., Morris, J. A., et al. (2010). Detection, validation, and downstream analysis of allelic variation in gene expression. Genetics 184, 119–128. doi: 10.1534/genetics.109.107474
Cook, M. N., Baker, J. A., Heldt, S. A., Williams, R. W., Hamre, K. M., and Lu, L. (2015). Identification of candidate genes that underlie the QTL on chromosome 1 that mediates genetic differences in stress-ethanol interactions. Physiol. Genom. 47, 308–317. doi: 10.1152/physiolgenomics.00114.2014
Crane, F., Hatefi, Y., Lester, R., and Widmer, C. (1989). Isolation of a quinone from beef heart mitochondria. 1957. Biochim. Biophys. Acta 1000, 362–363.
Crane, F. L. (2001). Biochemical functions of coenzyme Q10. J. Am. Coll. Nutr. 20, 591–598. doi: 10.1080/07315724.2001.10719063
Cunningham, C. L. (1995). Localization of genes influencing ethanol-induced conditioned place preference and locomotor activity in BXD recombinant inbred mice. Psychopharmacology 120, 28–41. doi: 10.1007/BF02246142
Cunningham, C. L., and Noble, D. (1992). Conditioned activation induced by ethanol: role in sensitization and conditioned place preference. Pharmacol. Biochem. Behav. 43, 307–313. doi: 10.1016/0091-3057(92)90673-4
Cunningham, C. L., and Prather, L. K. (1992). Conditioning trial duration affects ethanol-induced conditioned place preference in mice. Anim. Learn. Behav. 20, 187–194. doi: 10.3758/BF03200416
Das, S. K., and Vasudevan, D. (2007). Alcohol-induced oxidative stress. Life Sci. 81, 177–187. doi: 10.1016/j.lfs.2007.05.005
D’Autréaux, B., and Toledano, M. B. (2007). ROS as signalling molecules: mechanisms that generate specificity in ROS homeostasis. Nat. Rev. Mol. Cell Biol. 8, 813–824. doi: 10.1038/nrm2256
Enache, M., Van Waes, V., Vinner, E., Lhermitte, M., Maccari, S., and Darnaudéry, M. (2008). Impact of an acute exposure to ethanol on the oxidative stress status in the hippocampus of prenatal restraint stress adolescent male rats. Brain Res. 1191, 55–62. doi: 10.1016/j.brainres.2007.11.031
Ernster, L., and Dallner, G. (1995). Biochemical, physiological and medical aspects of ubiquinone function. Biochim. Biophys. Acta 1271, 195–204. doi: 10.1016/0925-4439(95)00028-3
Festenstein, G., Heaton, F., Lowe, J., and Morton, R. (1955). A constituent of the unsaponifiable portion of animal tissue lipids (λmax. mμ.). Biochem. J. 59, 558–566. doi: 10.1042/bj0590558
Gönenç, S., Uysal, N., Açikgöz, O., and Kayatekin, B. (2005). Effects of melatonin on oxidative stress and spatial memory impairment induced by acute ethanol treatment in rats. Physiol. Res. 54, 341–348.
González-Mariscal, I., García-Testón, E., Padilla, S., Martín-Montalvo, A., Viciana, T. P., Vazquez-Fonseca, L., et al. (2014). The regulation of coenzyme q biosynthesis in eukaryotic cells: all that yeast can tell us. Mol. Syndromol. 5, 107–118. doi: 10.1159/000362897
Hernández, J. A., López-Sánchez, R. C., and Rendón-Ramírez, A. (2016). Lipids and oxidative stress associated with ethanol-induced neurological damage. Oxid. Med. Cell. Longev. 2016:1543809. doi: 10.1155/2016/1543809
Hipolito, L., Sanchez, M., Polache, A., and Granero, L. (2007). Brain metabolism of ethanol and alcoholism: an update. Curr. Drug Metab. 8, 716–727. doi: 10.2174/138920007782109797
Homayouni, R., Heinrich, K., Wei, L., and Berry, M. W. (2004). Gene clustering by latent semantic indexing of MEDLINE abstracts. Bioinformatics 21, 104–115. doi: 10.1093/bioinformatics/bth464
Jablonski, M. M., Dalke, C., Wang, X., Lu, L., Manly, K. F., Pretsch, W., et al. (2005). An ENU-induced mutation in Rs1h causes disruption of retinal structure and function. Mol. Vis. 11, 569–581.
Kaspar, J. W., Niture, S. K., and Jaiswal, A. K. (2009). Nrf2: INrf2 (Keap1) signaling in oxidative stress. Free Radic. Biol. Med. 47, 1304–1309. doi: 10.1016/j.freeradbiomed.2009.07.035
Kerns, R. T., Ravindranathan, A., Hassan, S., Cage, M. P., York, T., Sikela, J. M., et al. (2005). Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. J. Neurosci. 25, 2255–2266. doi: 10.1523/JNEUROSCI.4372-04.2005
Kishida, K. T., Pao, M., Holland, S. M., and Klann, E. (2005). NADPH oxidase is required for NMDA receptor-dependent activation of ERK in hippocampal area CA1. J. Neurochem. 94, 299–306. doi: 10.1111/j.1471-4159.2005.03189.x
Klann, E., Roberson, E. D., Knapp, L. T., and Sweatt, J. D. (1998). A role for superoxide in protein kinase C activation and induction of long-term potentiation. J. Biol. Chem. 273, 4516–4522. doi: 10.1074/jbc.273.8.4516
Kunugi, H., Ishida, S., Akahane, A., and Nanko, S. (2001). Exon/intron boundaries, novel polymorphisms, and association analysis with schizophrenia of the human synaptic vesicle monoamine transporter (SVMT) gene. Mol. Psychiatry 6, 456–460. doi: 10.1038/sj.mp.4000895
Landi, L., Cabrini, L., Sechi, A. M., and Pasquali, P. (1984). Antioxidative effect of ubiquinones on mitochondrial membranes. Biochem. J. 222, 463–466. doi: 10.1042/bj2220463
Ledig, M., M’Paria, J.-R., and Mandel, P. (1981). Superoxide dismutase activity in rat brain during acute and chronic alcohol intoxication. Neurochem. Res. 6, 385–390. doi: 10.1007/BF00963853
Lenaz, G. (1985). Coenzyme Q: Biochemistry, Bioenergetics, and Clinical Applications of Ubiquinone. Hoboken, NJ: John Wiley & Sons Incorporated.
Licitra, F., and Puccio, H. (2014). An overview of current mouse models recapitulating coenzyme q10 deficiency syndrome. Mol. Syndromol. 5, 180–186. doi: 10.1159/000362942
Lieber, C. S. (1976). The metabolism of alcohol. Sci. Am. 234, 25–33. doi: 10.1038/scientificamerican0376-25
Lohman, D. C., Forouhar, F., Beebe, E. T., Stefely, M. S., Minogue, C. E., Ulbrich, A., et al. (2014). Mitochondrial COQ9 is a lipid-binding protein that associates with COQ7 to enable coenzyme Q biosynthesis. Proc. Natl. Acad. Sci. U.S.A. 111, E4697–E4705. doi: 10.1073/pnas.1413128111
Lu, L., Airey, D. C., and Williams, R. W. (2001). Complex trait analysis of the hippocampus: mapping and biometric analysis of two novel gene loci with specific effects on hippocampal structure in mice. J. Neurosci. 21, 3503–3514. doi: 10.1523/JNEUROSCI.21-10-03503.2001
Lu, S. C. (2009). Regulation of glutathione synthesis. Mol. Aspects Med. 30, 42–59. doi: 10.1016/j.mam.2008.05.005
Lu, S. C. (2013). Glutathione synthesis. Biochim. Biophys. Acta 1830, 3143–3153. doi: 10.1016/j.bbagen.2012.09.008
Lu, S. C., Huang, Z. Z., Yang, J. M., and Tsukamoto, H. (1999). Effect of ethanol and high-fat feeding on hepatic γ-glutamylcysteine synthetase subunit expression in the rat. Hepatology 30, 209–214. doi: 10.1002/hep.510300134
Luna-Sánchez, M., Díaz-Casado, E., Barca, E., Tejada, M. Á., Montilla-García,Á., Cobos, E. J., et al. (2015). The clinical heterogeneity of coenzyme Q10 deficiency results from genotypic differences in the Coq9 gene. EMBO Mol. Med. 7, 670–687. doi: 10.15252/emmm.201404632
Lushchak, V. I. (2014). Free radicals, reactive oxygen species, oxidative stress and its classification. Chem. Biol. Interact. 224, 164–175. doi: 10.1016/j.cbi.2014.10.016
Malinow, R., Schulman, H., and Tsien, R. W. (1989). Inhibition of postsynaptic PKC or CaMKII blocks induction but not expression of LTP. Science 245, 862–866. doi: 10.1126/science.2549638
Manzo-Avalos, S., and Saavedra-Molina, A. (2010). Cellular and mitochondrial effects of alcohol consumption. Int. J. Environ. Res. Public Health 7, 4281–4304. doi: 10.3390/ijerph7124281
Marbois, B. N., and Clarke, C. F. (1996). The COQ7 gene encodes a protein in Saccharomyces cerevisiae necessary for ubiquinone biosynthesis. J. Biol. Chem. 271, 2995–3004. doi: 10.1074/jbc.271.6.2995
Martín-Montalvo, A., González-Mariscal, I., Pomares-Viciana, T., Padilla-López, S., Ballesteros, M., Vazquez-Fonseca, L., et al. (2013). The phosphatase Ptc7 induces coenzyme Q biosynthesis by activating the hydroxylase Coq7 in yeast. J. Biol. Chem. 288, 28126–28137. doi: 10.1074/jbc.M113.474494
Mellors, A., and Tappel, A. L. (1966). The inhibition of mitochondrial peroxidation by ubiquinone and ubiquinol. J. Biol. Chem. 241, 4353–4356.
Narasimhan, M., Mahimainathan, L., Rathinam, M. L., Riar, A. K., and Henderson, G. I. (2011). Overexpression of Nrf2 protects cerebral cortical neurons from ethanol-induced apoptotic death. Mol. Pharmacol. 80, 988–999. doi: 10.1124/mol.111.073262
Oswald, M. C., Garnham, N., Sweeney, S. T., and Landgraf, M. (2018). Regulation of neuronal development and function by ROS. FEBS Lett. 592, 679–691. doi: 10.1002/1873-3468.12972
Overall, R. W., Kempermann, G., Peirce, J., Lu, L., Goldowitz, D., Gage, F. H., et al. (2009). Genetics of the hippocampal transcriptome in mouse: a systematic survey and online neurogenomics resource. Front. Neurosci. 3:55. doi: 10.3389/neuro.15.003.2009
Padilla, S., Tran, U., Jimenez-Hidalgo, M., Lopez-Martin, J., Martin-Montalvo, A., Clarke, C., et al. (2009). Hydroxylation of demethoxy-Q6 constitutes a control point in yeast coenzyme Q6 biosynthesis. Cell. Mol. Life Sci. 66, 173–186. doi: 10.1007/s00018-008-8547-7
Pan, Y., Balazs, L., Tigyi, G., and Yue, J. (2011). Conditional deletion of Dicer in vascular smooth muscle cells leads to the developmental delay and embryonic mortality. Biochem. Biophys. Res. Commun. 408, 369–374. doi: 10.1016/j.bbrc.2011.02.119
Quinzii, C. M., Garone, C., Emmanuele, V., Tadesse, S., Krishna, S., Dorado, B., et al. (2013). Tissue-specific oxidative stress and loss of mitochondria in CoQ-deficient Pdss2 mutant mice. FASEB J. 27, 612–621. doi: 10.1096/fj.12-209361
Quinzii, C. M., López, L. C., Gilkerson, R. W., Dorado, B., Coku, J., Naini, A. B., et al. (2010). Reactive oxygen species, oxidative stress, and cell death correlate with level of CoQ10 deficiency. FASEB J. 24, 3733–3743. doi: 10.1096/fj.09-152728
Quinzii, C. M., López, L. C., Von-Moltke, J., Naini, A., Krishna, S., Schuelke, M., et al. (2008). Respiratory chain dysfunction and oxidative stress correlate with severity of primary CoQ10 deficiency. FASEB J. 22, 1874–1885. doi: 10.1096/fj.07-100149
Reddy, S., Husain, K., Schlorff, E., Scott, R., and Somani, S. (1999). Dose response of ethanol ingestion on antioxidant defense system in rat brain subcellular fractions. Neurotoxicology 20, 977–987.
Reis, R., Charehsaz, M., Sipahi, H., Ekici, A. I. D., Macit,Ç, Akkaya, H., et al. (2017). Energy drink induced lipid peroxidation and oxidative damage in rat liver and brain when used alone or combined with alcohol. J. Food Sci. 82, 1037–1043. doi: 10.1111/1750-3841.13662
Risinger, F. O., and Cunningham, C. L. (1995). Genetic differences in ethanol-induced conditioned taste aversion after ethanol preexposure. Alcohol 12, 535–539. doi: 10.1016/0741-8329(95)00040-2
Risinger, F. O., and Cunningham, C. L. (1998). Ethanol-induced conditioned taste aversion in BXD recombinant inbred mice. Alcoholism 22, 1234–1244. doi: 10.1111/j.1530-0277.1998.tb03904.x
Schieber, M., and Chandel, N. S. (2014). ROS function in redox signaling and oxidative stress. Curr. Biol. 24, R453–R462. doi: 10.1016/j.cub.2014.03.034
Sies, H. (2015). Oxidative stress: a concept in redox biology and medicine. Redox Biol. 4, 180–183. doi: 10.1016/j.redox.2015.01.002
Somani, S., Husain, K., Diaz-Phillips, L., Lanzotti, D., Kareti, K., and Trammell, G. (1996). Interaction of exercise and ethanol on antioxidant enzymes in brain regions of the rat. Alcohol 13, 603–610. doi: 10.1016/S0741-8329(96)00075-4
Tabakoff, B., Saba, L., Printz, M., Flodman, P., Hodgkinson, C., Goldman, D., et al. (2009). Genetical genomic determinants of alcohol consumption in rats and humans. BMC Biol. 7:70. doi: 10.1186/1741-7007-7-70
Thayer, W. S., and Rubin, E. (1979). Effects of chronic ethanol intoxication on oxidative phosphorylation in rat liver submitochondrial particles. J. Biol. Chem. 254, 7717–7723.
Turanov, A. A., Kehr, S., Marino, S. M., Yoo, M.-H., Carlson, B. A., Hatfield, D. L., et al. (2010). Mammalian thioredoxin reductase 1: roles in redox homoeostasis and characterization of cellular targets. Biochem. J. 430, 285–293. doi: 10.1042/BJ20091378
Turunen, M., Olsson, J., and Dallner, G. (2004). Metabolism and function of coenzyme Q. Biochim. Biophys. Acta 1660, 171–199. doi: 10.1016/j.bbamem.2003.11.012
Urquhart, K. R., Zhao, Y., Baker, J. A., Lu, Y., Yan, L., Cook, M. N., et al. (2016). A novel heat shock protein alpha 8 (Hspa8) molecular network mediating responses to stress-and ethanol-related behaviors. Neurogenetics 17, 91–105. doi: 10.1007/s10048-015-0470-0
Wang, Y., and Hekimi, S. (2016). Understanding ubiquinone. Trends Cell Biol. 26, 367–378. doi: 10.1016/j.tcb.2015.12.007
Wang, Y., Oxer, D., and Hekimi, S. (2015). Mitochondrial function and lifespan of mice with controlled ubiquinone biosynthesis. Nat. Commun. 6:6393. doi: 10.1038/ncomms7393
Wolf, D. E., Hoffman, C. H., Trenner, N. R., Arison, B. H., Shunk, C. H., Linn, B. O., et al. (1958). Coenzyme QI Structure studies on the coenzyme Q group. J. Am. Chem. Soc. 80:4752.
Zakhari, S. (2006). Overview: How is alcohol metabolized by the body? Alcohol Res. Health 29, 245–255.
Keywords: ethanol, Coenzyme Q, oxidative stress, hippocampus, mouse models, genetics, genomics
Citation: Zhou D, Zhao Y, Hook M, Zhao W, Starlard-Davenport A, Cook MN, Jones BC, Hamre KM and Lu L (2018) Ethanol’s Effect on Coq7 Expression in the Hippocampus of Mice. Front. Genet. 9:602. doi: 10.3389/fgene.2018.00602
Received: 04 June 2018; Accepted: 16 November 2018;
Published: 04 December 2018.
Edited by:
Elizabeth A. Thomas, The Scripps Research Institute, United StatesReviewed by:
Camron D. Bryant, Boston University, United StatesCopyright © 2018 Zhou, Zhao, Hook, Zhao, Starlard-Davenport, Cook, Jones, Hamre and Lu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lu Lu, lulu@uthsc.edu
†These authors have contributed equally to this work as co-first authors
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.