EDITORIAL
Published on 21 Apr 2020
Editorial: Precise Genome Editing Techniques and Applications
doi 10.3389/fgene.2020.00412
- 3,429 views
- 5 citations
38k
Total downloads
222k
Total views and downloads
EDITORIAL
Published on 21 Apr 2020
ORIGINAL RESEARCH
Published on 02 Jul 2019
ORIGINAL RESEARCH
Published on 30 Apr 2019
ORIGINAL RESEARCH
Published on 30 Apr 2019
ORIGINAL RESEARCH
Published on 26 Apr 2019

ORIGINAL RESEARCH
Published on 18 Apr 2019
ORIGINAL RESEARCH
Published on 15 Mar 2019
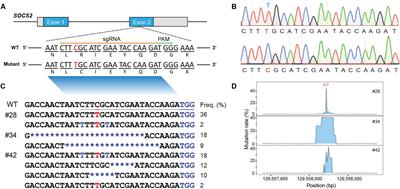
PROTOCOLS
Published on 13 Mar 2019
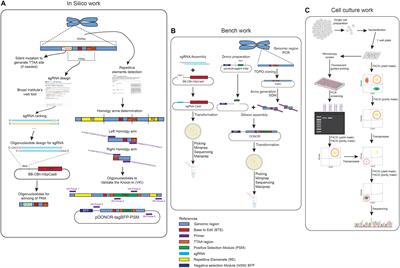
ORIGINAL RESEARCH
Published on 19 Feb 2019

ORIGINAL RESEARCH
Published on 21 Jan 2019
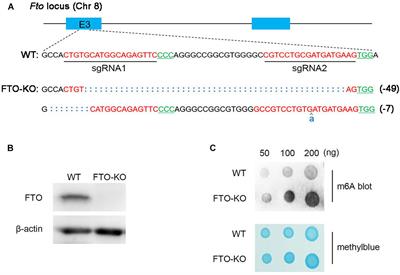
REVIEW
Published on 07 Jan 2019
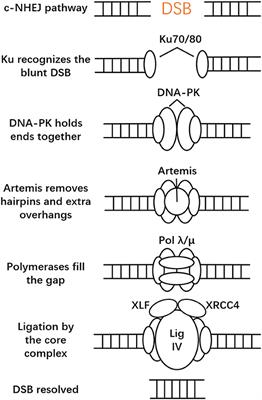
ORIGINAL RESEARCH
Published on 04 Oct 2018