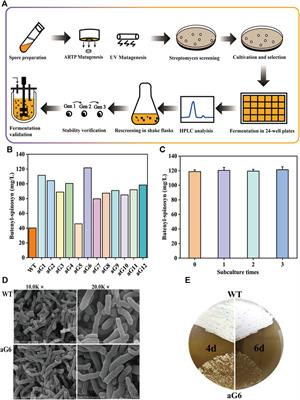
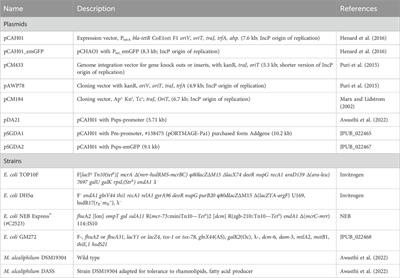
ORIGINAL RESEARCH
Published on 15 May 2024
Optimization of electroporation method and promoter evaluation for type-1 methanotroph, Methylotuvimicrobium alcaliphilum
doi 10.3389/fbioe.2024.1412410
- 1,034 views
- 1 citation
1,269
Total downloads
5,822
Total views and downloads
You will be redirected to our submission process.
ORIGINAL RESEARCH
Published on 15 May 2024

ORIGINAL RESEARCH
Published on 19 Apr 2024

ORIGINAL RESEARCH
Published on 15 Jan 2024