ORIGINAL RESEARCH
Published on 08 May 2024
IL-38 promotes the development of prostate cancer
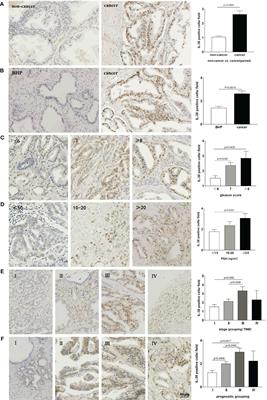
doi 10.3389/fimmu.2024.1384416
- 807 views
- 2 citations
20k
Total downloads
57k
Total views and downloads
Select the journal/section where you want your idea to be submitted:
ORIGINAL RESEARCH
Published on 08 May 2024

REVIEW
Published on 14 Mar 2024
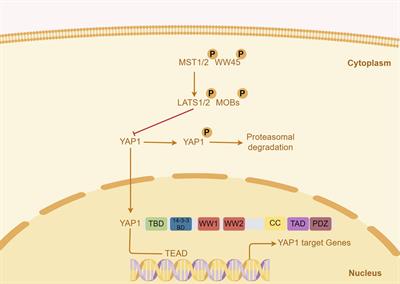
ORIGINAL RESEARCH
Published on 12 Mar 2024

ORIGINAL RESEARCH
Published on 29 Feb 2024
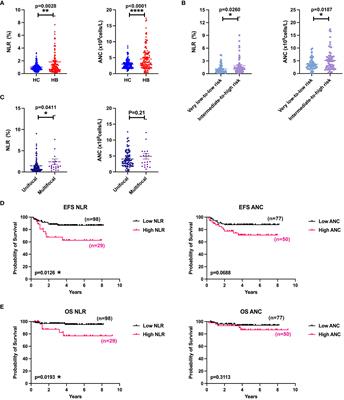
REVIEW
Published on 22 Feb 2024
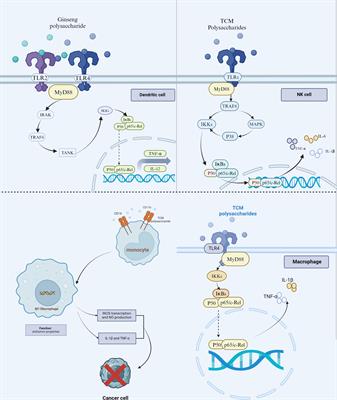
REVIEW
Published on 15 Feb 2024

STUDY PROTOCOL
Published on 12 Feb 2024
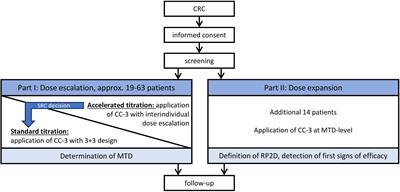
CASE REPORT
Published on 06 Feb 2024

ORIGINAL RESEARCH
Published on 19 Jan 2024
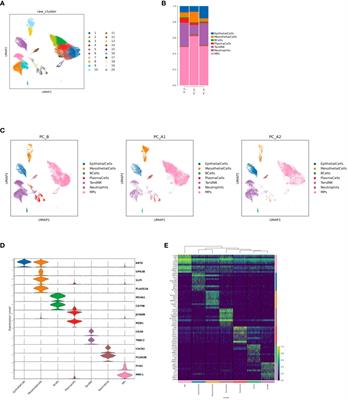
ORIGINAL RESEARCH
Published on 08 Jan 2024

ORIGINAL RESEARCH
Published on 22 Dec 2023
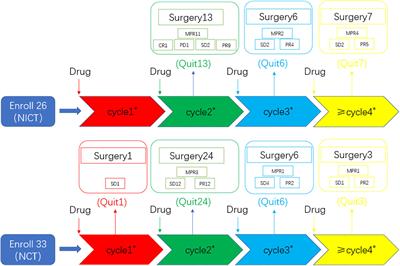
ORIGINAL RESEARCH
Published on 20 Dec 2023

Frontiers in Oncology