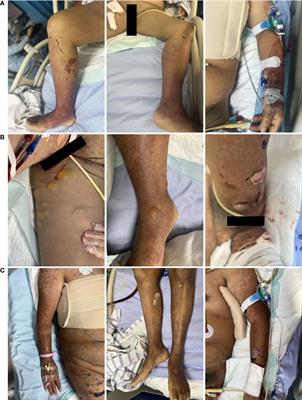
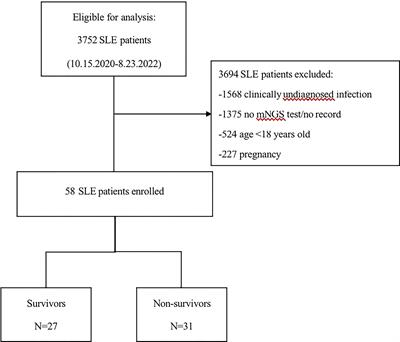
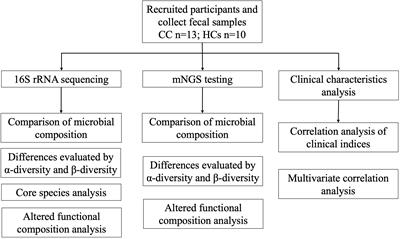
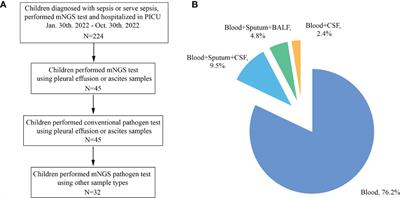
ORIGINAL RESEARCH
Published on 10 Nov 2023
A preliminary evaluation of targeted nanopore sequencing technology for the detection of Mycobacterium tuberculosis in bronchoalveolar lavage fluid specimens

doi 10.3389/fcimb.2023.1107990
- 2,697 views
- 8 citations