EDITORIAL
Published on 09 Sep 2022
Editorial: Chromosome structural variants: Epidemiology, identification and contribution to human diseases
doi 10.3389/fgene.2022.1022918
- 865 views
8,571
Total downloads
36k
Total views and downloads
You will be redirected to our submission process.
EDITORIAL
Published on 09 Sep 2022
ORIGINAL RESEARCH
Published on 02 Jun 2022

ORIGINAL RESEARCH
Published on 14 Apr 2022

ORIGINAL RESEARCH
Published on 23 Mar 2022

CORRECTION
Published on 25 Feb 2022
ORIGINAL RESEARCH
Published on 24 Feb 2022

ORIGINAL RESEARCH
Published on 21 Dec 2021

TECHNOLOGY AND CODE
Published on 01 Dec 2021

ORIGINAL RESEARCH
Published on 04 Nov 2021

ORIGINAL RESEARCH
Published on 20 Sep 2021

ORIGINAL RESEARCH
Published on 26 Aug 2021
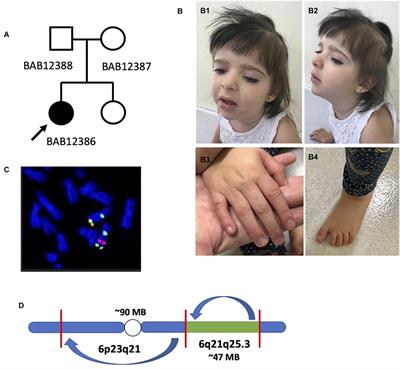
METHODS
Published on 18 Mar 2021

