EDITORIAL
Published on 13 Dec 2021
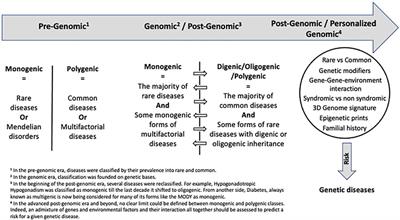
Editorial: Monogenic vs. Oligogenic Reclassification
doi 10.3389/fgene.2021.821591
- 2,683 views
- 1 citation
6,050
Total downloads
29k
Total views and downloads
EDITORIAL
Published on 13 Dec 2021

CASE REPORT
Published on 06 Oct 2021

ORIGINAL RESEARCH
Published on 03 Sep 2021

BRIEF RESEARCH REPORT
Published on 02 Jun 2021

REVIEW
Published on 01 Apr 2021

