- 1Department of Pharmacy, The First Affiliated Hospital of University of Science and Technology, China (USTC), Division of Life Sciences and Medicine, University of Science and Technology of China, Hefei, Anhui, China
- 2School of Life Sciences, University of Science and Technology of China, Hefei, Anhui, China
- 3Bloomington High School North, Bloomington, IN, United States
Cyanophages play a pivotal role in controlling cyanobacterial populations in aquatic environments. These dsDNA viruses harbor auxiliary metabolic genes (AMGs) that modulate the key metabolic processes of their cyanobacterial hosts, such as Photosynthesis, nutrient uptake for the optimization of viral replication. Recently, pan1~pan5 and pam1~pam5 cyanophages have been isolated from the fifth largest water resource in China; Lake Chaohu. Detailed genomic analysis of these phages revealed that these isolated cyanophages especially Pan1, Pam2 and Pam3 possess unique AMGs that significantly enhance the metabolic activities of their hosts, potentially leading to the suppression of bloom formation and stabilization of the ecological dynamics of Lake Chaohu. Our findings provide concrete evidence that cyanophages encoding AMGs could serve as effective biocontrol agents against harmful algal blooms, offering a targeted approach to manage these environmental threats. The integration of cyanophage-based management therapies with traditional methods could advance the efficiency and sustainability of controlling cyanobacterial outbreaks, paving the way for novel applications in water resource management. This review emphasizes the importance and critical need for further exploration of phage-host dynamics to fully harness the potential of cyanophages in ecosystem regulation.
1 Introduction
Cyanobacteria are the first known oxygenic photoautotrophs on Earth, more commonly known as “blue-green algae” (1). The fast proliferation and accumulation of cyanobacteria in fresh water bodies can cause Harmful Algal Blooms (HABs) (2), which release chemicals resulting in bad taste, bad odor, and turbidity of water, posing a serious threat to aquatic ecosystem, human health and water sustainability thus disrupting the food chain dynamics (3, 4). Among the various mechanisms influencing cyanobacterial blooms, the role of cyanophages; dsDNA viruses that infect cyanobacteria, has garnered significant attention. Cyanophages play a critical role in controlling cyanobacterial populations in aquatic ecosystems (5–9).
Cyanophages inhabit freshwater lakes, rivers, and marine habitats depending on their cyanobacterial hosts availability. They regulate cyanobacterial populations in freshwater, especially during HABs (5, 10). Temperature, light, and nutrient availability majorly affect cyanophage abundance and activity. Viral proliferation and infection efficiency enhances at optimal temperature, light and nutrition (11, 12). The change in these environmental factors may disrupt the activity of cyanophages. Cyanobacterial hosts multiply in nutrient-rich environments, increasing the number of possible cyanophage hosts. While cyanophages have been proposed as tools for managing cyanobacterial populations and cycling nutrients to promote ecological balance, their role in ecosystems is complex and multifaceted. For instance, phage-induced cyanobacterial lysis might transfer intracellular cyanotoxins into the dissolved phase, worsening algal blooms. Environmental variables that affect cyanophage activity might disturb these dynamics, causing ecological uncertainty. Thus, cyanophages may be a promising biocontrol method, but their advantages, hazards, and ecological impacts must be considered (4, 13).
Cyanophage abundance may vary by season and geography. Because of higher temperatures and more nourishment, cyanobacterial blooms are more prevalent in summer (14). Cyanophage diversity and abundance is greater in nutrient-rich coastal and estuary settings than in open ocean waters (15). Cyanobacterial Harmful Algal Blooms (CyanoHABs) are expanding geographically and now pose a threat to the ecological integrity and sustainability of some of the world’s largest and most resourceful water bodies. These bodies of water include Lakes Victoria in Africa (16), Erie in the United States and Canada (17), Okeechobee in Florida, USA (18), Taihu and Chaohu in China (19, 20), Kasumigaura in Japan (18), the Baltic Sea in Northern Europe (21), and the Caspian Sea in West Asia (4).
The fifth biggest freshwater lake in China, Lake Chaohu, serves as an essential resource, providing drinking and irrigation water to millions. Eutrophication and cyanoHABs have drawn attention to this significant lake. Chaohu’s annual cyanoHAB is dominated by Microcystis and starts in late spring, elevates in summer, and decreases in late autumn (22). The species Microcystis is particularly dominant during the cyanobacterial blooms that occur periodically in Lake Chaohu. Nutrient levels, particularly nitrogen and phosphorus, are one of several environmental variables that affect these blooms. Cyanophyta, Chlorophyta, and Bacillariophyta make up the bulk of the lake’s cyanobacterial ecosystem (20).
Based on morphology and genetics, Myoviridae (contractile tails), Siphoviridae (long, non-contractile tails), and Podoviridae (short, stubby tails) are the three main Cyanophage families (23–25). The fundamental genome structure and function of each family is significantly similar consisting of conserved set of genes that govern the essential viral functions, including replication, assembly, and cell lysis (26). Despite having genetic similarities, these phages employs diverse array of strategies to infect a broad spectrum of cyanobacterial hosts, each with distinct cellular defenses and living conditions (27, 28). These strategies are may or may not influenced by genetic variations occur as a result of horizontal gene transfer such as variations in tail fibers; needed for host recognition and attachment (29, 30), regulatory and auxiliary genes; that enhance the phages’ ability to circumvent host defenses (31). Phages may also optimize their pathogenic potential across environmental and temporal niches due to these or other genetic adaptations (32, 33). Thus, whereas these cyanophages share a common genetic foundation, their capacity to adapt and prosper in a variety of ecological situations is mostly determined by genetic variants, emphasizing the dynamic nature of phage-host interactions (28).
Cyanophages are a distinct group of viruses that have a diverse variety of genetic material, and their genomes include a wide range of genes, including those involved in DNA replication, structural proteins, and host interaction (34). Commonly, cyanophages acquire genes from their hosts or other viruses via horizontal gene transfer to adapt to varied environmental conditions (35). In addition, cyanophages have auxiliary metabolic genes (AMGs) that have an even greater impact on the metabolism of infected cyanobacteria. Despite not being strictly required for viral replication; these genes significantly alter the host’s metabolic processes in a way that promotes viral propagation, particularly in challenging environmental conditions (12). The ability of the host metabolic system to withstand nutritional scarcity and sustain viral replication is aided by AMGs that are engaged in nitrogen fixing and phosphorus acquisition. The significance of AMGs in cyanophages to control HABs is a testament to the complex evolutionary adaptations of the cyanophages and the profound impact they have on the ecology and physiology of cyanobacterial hosts. While cyanophages possess the potential to modulate harmful algal blooms (HABs), it is essential to recognize that their influence extends far beyond mere control mechanisms. The relationship between cyanophages and their cyanobacterial hosts is characterized by a complex interplay that affects both host metabolism and broader ecological dynamics. This intricate interaction demands a more nuanced understanding of cyanophage roles, acknowledging the depth and diversity of their impact on cyanobacterial populations (36–38).
2 Mechanisms of cyanophage infection and lysis
Cyanophages can undergo either a lytic or lysogenic cycle. In every scenario, cyanophages reproduce via the host DNA machinery, which includes the following stages: attachment, penetration, replication and maturation, and then release of viral particles (lytic phase) and cellular content in the environment (26). The complete mechanism of phage infection and the release of viral bodies from the host bacterium is shown in Figure 1. An intricate infection process is initiated by the phage’s specialized attachment to the host cell, which is accomplished by means of tail fibers or surface spikes that identify and bind to surface receptors of the host bacterium. This specificity ensures that phages only infect compatible host cells (39, 40).
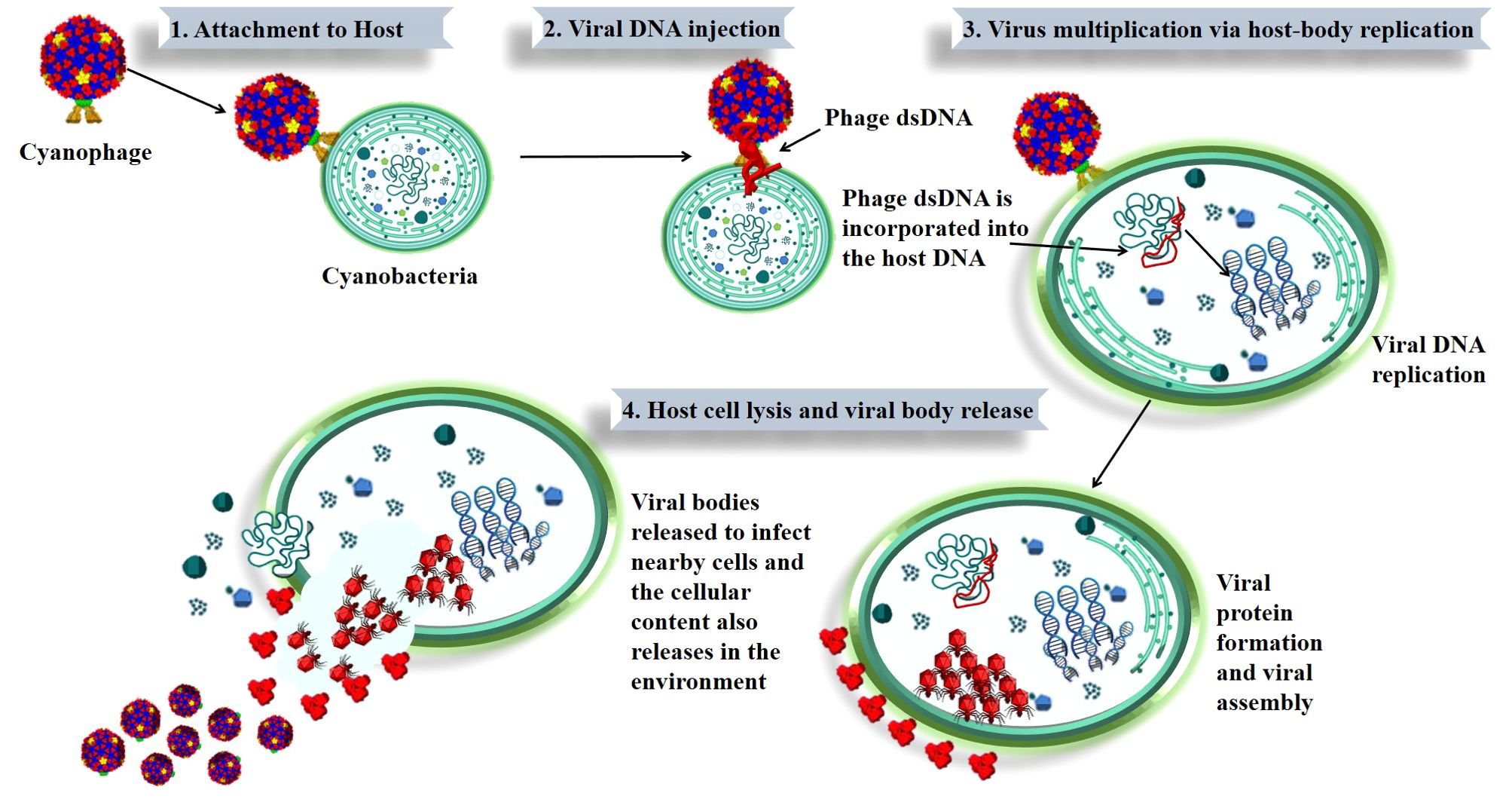
Figure 1. Stages of cyanophage infection and lysis in cyanobacteria. The figure shows how a cyanophage infects a cyanobacterial cell. It starts with (1) the cyanophage attaching to the cyanobacterium, followed by (2) injection of viral DNA into the host. The viral DNA is (3) incorporated into the host, leading to viral replication and protein production. Finally, (4) the host cell breaks open (lysis), releasing new viruses to infect other cells. This process is key to how cyanophages can affect cyanobacterial populations and potentially impact harmful algal blooms (HABs).
After attaching to a cyanobacterium, phages often undergo structural modifications and release enzymes such as lysozymes that penetrate the host cell wall, allowing the phage to inject its DNA into the host cell. Although different cyanophages use different methods for injecting DNA into their hosts, the process usually involves a complex chain reaction of phage structural modifications (41). Once inside the host cell, the phage DNA takes over the cell’s replication machinery and starts making phage components. Normal cellular processes are diminished when the host cell’s resources are diverted towards the generation of viral components (12, 36). By changing the host’s metabolic pathways, the replication process promotes viral production rather than normal cellular processes. At last, enzymes encoded by the phage lyse the host cell, releasing additional infective phage particles. By discharging their contents, phages participate in nutrient cycle in aquatic environments (13, 39). The lytic cycle is often quick and ends with the host cell’s death.
During the lysogenic cycle, the cyanophage’s DNA integrates into the host genome and replicates passively alongside the host’s DNA, producing no immediate damage to the host. This integrated DNA, referred to as a prophage, may stay inactive inside the host genome for an extended period of time until it is reactivated by environmental stimuli. The ability of cyanophages to switch between lysogenic and lytic cycles allows them to adapt to the fluctuating environmental conditions, which can influence the stability and health of cyanobacterial populations. While the lysogenic cycle allows cyanophages to persist in the environment during unfavorable conditions, their reactivation into the lytic cycle under certain stimuli can lead to the rapid destruction of cyanobacterial cells. This dual life cycle contributes to the regulation of cyanobacterial communities and can play a role in mitigating toxic algal blooms, although this connection is complex and influenced by multiple ecological factors. Understanding these dynamics is crucial for appreciating the potential of cyanophages in influencing biogeochemical cycles and the broader biological dynamics of aquatic ecosystems (42).
3 Auxiliary metabolic genes in cyanophages
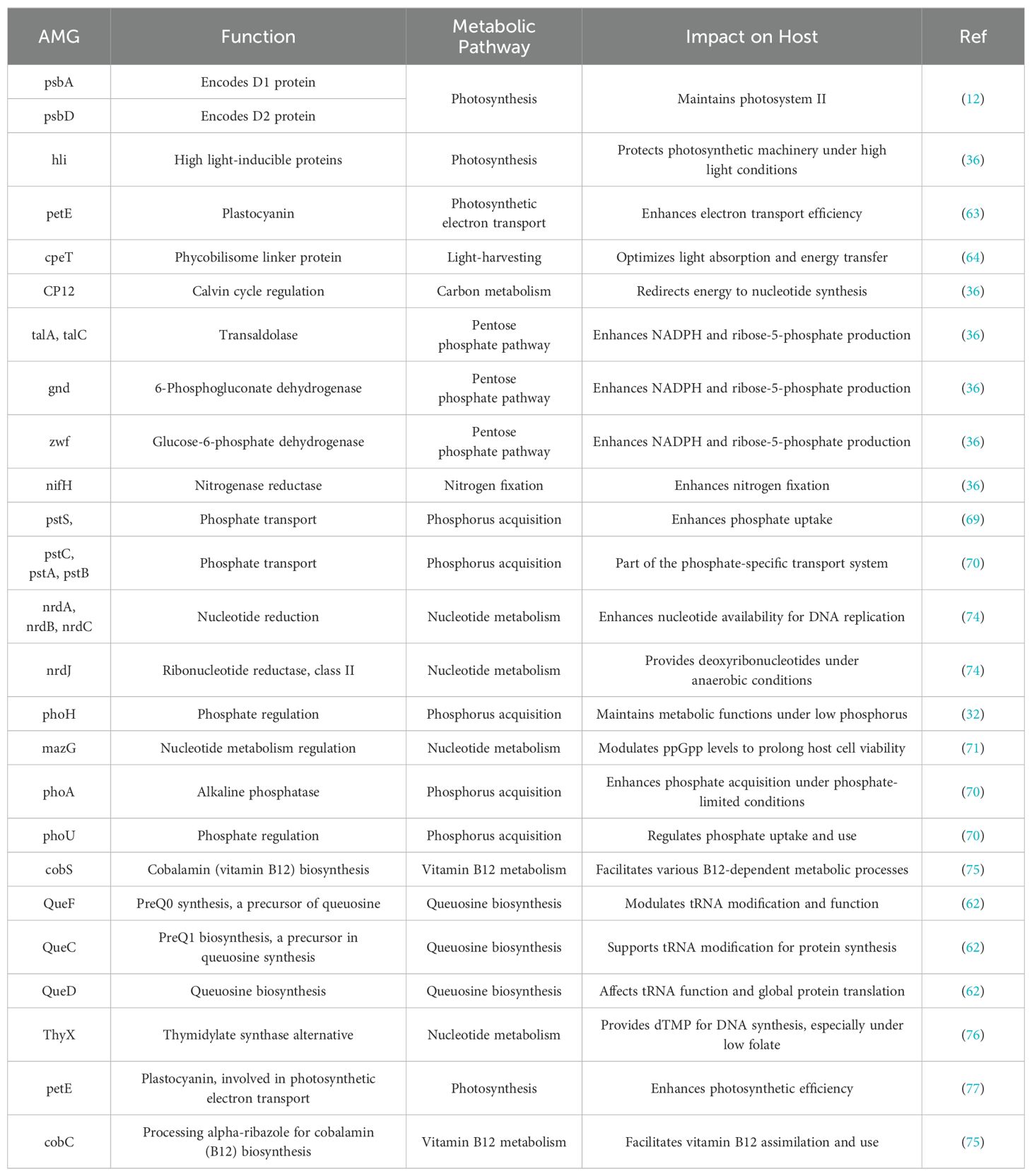
Cyanophages and other viruses carry AMGs that increase the host cell’s metabolic capacity during infection. Rather of relying on core genes for viral propagation, AMGs influence the host’s metabolism in a way that helps the virus replication. Although these genes do not play a crucial role in the viral life cycle, they do have a selection advantage that makes viral replication and assembly more efficient by enhancing the host’s metabolic activities (36, 37).
AMGs are classified into two classes; Class I and Class II. Class I AMGs encode cell metabolic processes and are listed in Kyoto Encyclopedia of Genes and Genomes (KEGG) database (43). These genes are mainly involved in photosynthesis and carbon metabolism (44). While class II AMGs encode peripheral functionalities and are not found in KEGG metabolic pathways. This class contains transport and assembly genes (44), such as those that help balance intermediates of tricarboxylic acid (TCA) cycle (43). This class also acquires biogenic components like phosphate, regulated by pstS in cyanobacterial hosts (32). However, the interpretation of Class II AMGs is often challenging due to the lack of a comprehensive reference database, leading to incomplete annotations and an under-appreciation of their metabolic significance (45). Recently, it has been proposed that Class II AMGs, which are either absent from the KEGG database or have incomplete annotations of metabolic pathways, have a significant role in adaptive capabilities of cyanophages, contributing to their overall function and ecological impact (43).
AMGs such as petE, hli, psbA, psbD, and cpeT are believed to have a significant impact on the photosynthetic activity of infected host cells (12). These genes provide energy to facilitate phage replication and enhance the viability of the phage. The genes associated with carbon metabolism include CP12, gnd, zwf, and talA (36). Previous research has shown that phage-encoded CP12 and talA have the ability to reroute the energy generated during photosynthesis, namely ATP and NADPH, away from the Calvin cycle. Subsequently, this energy is used for the production of dNTPs, which are necessary for phage replication during host infection (36). These genes enable cyanophages to control the host’s metabolic pathways, while ensuring that the infected cyanobacteria can maintain elevated levels of metabolic activity even under unfavorable environmental circumstances (12, 36).
Aside from aiding in viral replication, the presence of AMGs in cyanophages significantly influences the ecology and metabolism of cyanobacterial communities (46). AMGs are found across various families, genera, and species of cyanophages, with some being highly conserved while others exhibit variability in frequency and function depending on the specific cyanophage species (42, 47). This diversity allows different cyanophages to modulate host metabolic pathways in distinct ways, enhancing processes like photosynthesis, nutrient acquisition, and other key metabolic functions (46, 48). This functional integration enables cyanophages to exert a broader ecological impact by not only reproducing but also shaping the metabolic landscape of their cyanobacterial hosts in a species-specific manner, contributing to the overall dynamics of microbial ecosystems (36, 43, 46).
3.1 Mechanisms of AMG integration and expression
AMGs are incorporated into the cyanophage genome through the process of horizontal gene transfer; a process where genetic material is exchanged between different organisms across various species. This transmission of genetic material may take place either between cyanophages or between cyanophages and their host cyanobacteria. As part of the infection cycle, AMGs are expressed after integration to alter the host’s metabolic functions. To help the host cell make the most of its enhanced metabolic pathways, AMG expression is carefully controlled and often coordinated with host cellular machinery (12, 49).
Cyanophages orchestrate the expression of auxiliary metabolic genes (AMGs) by manipulating the host cyanobacteria’s transcriptional and translational machinery during infection. This targeted expression ensures that AMGs are efficiently translated into functional proteins, optimizing the host’s metabolic pathways to facilitate viral replication. The complex interplay involving the integration, expression, and functional impact of AMGs underscores the sophisticated symbiosis between cyanophages and their hosts. Recent studies suggest that the expression of AMGs linked to photosynthesis can be enhanced under conditions of high light availability, which naturally stimulates photosynthesis. This adjustment does not increase the host’s energy burden; rather, it optimizes energy utilization to meet the demands of efficient phage assembly and replication (36, 50).
3.2 Significant cyanophage species with AMGs
Cyanophages, equipped with auxiliary metabolic genes (AMGs), play a pivotal role in the metabolism of diverse cyanobacterial hosts. These cyanophages infect multiple species of cyanobacteria, notably Planktothrix, Pseudanabaena, Dolichospermum, Phormidium foveolarum, Prochlorococcus and Synechococcus; some of the cyanophages infecting these cyanobacterial hosts species are listed in Supplementary Table 1 and also the diversity of these phages and a wide range of metabolism they are manipulating their hosts is illustrated in Figure 2. Prochlorococcus and Synechococcus, which together account for about one-third of the ocean’s primary production and are important contributors to the world nitrogen and carbon cycles; primarily affected by cyanophages in freshwater and marine environments (32, 51). For instance, Cyanophages such as P-SSP7 and S-PM2, found in Prochlorococcus and Synechococcus respectively, are illustrated in Figure 2C, exemplify the complex interplay between viruses and their cyanobacterial hosts. These phages contain AMGs that boost the efficiency of photosynthesis by producing parts of Photosystem II. This process is crucial during infection because the energy from photosynthesis is used to produce nucleotides, which are essential for viral replication. These adaptive strategies allow cyanophages to optimize their propagation by ensuring that their hosts remain metabolically active, even under varying environmental conditions. This interplay between phage-encoded photosystem genes and host metabolism exemplifies how cyanophages are finely tuned to their ecological niches, ensuring their survival and proliferation in diverse aquatic environments (52–54). Some circular cyanophages infecting Prochlorococcus, are illustrated in Figure 2A, have diverse array of genes involved in critical processes such as nucleotide synthesis, photosynthetic efficiency, and nutrient acquisition. Notably genes like talC and hli, which are crucial for adapting to environmental stressors by modulating carbon metabolism and providing protection against photodamage (55). Phages like Syn9, Syn5, and S-WHM1—all infecting Synechococcus, as shown in Figure 2C —harbor genes that safeguard the photosynthetic apparatus and regulate both nucleotide and carbohydrate metabolism which not only enhance nutrient acquisition but also enhances light harvesting capabilities, critical for their proliferation in nutrient-variable aquatic environments. (56–59).
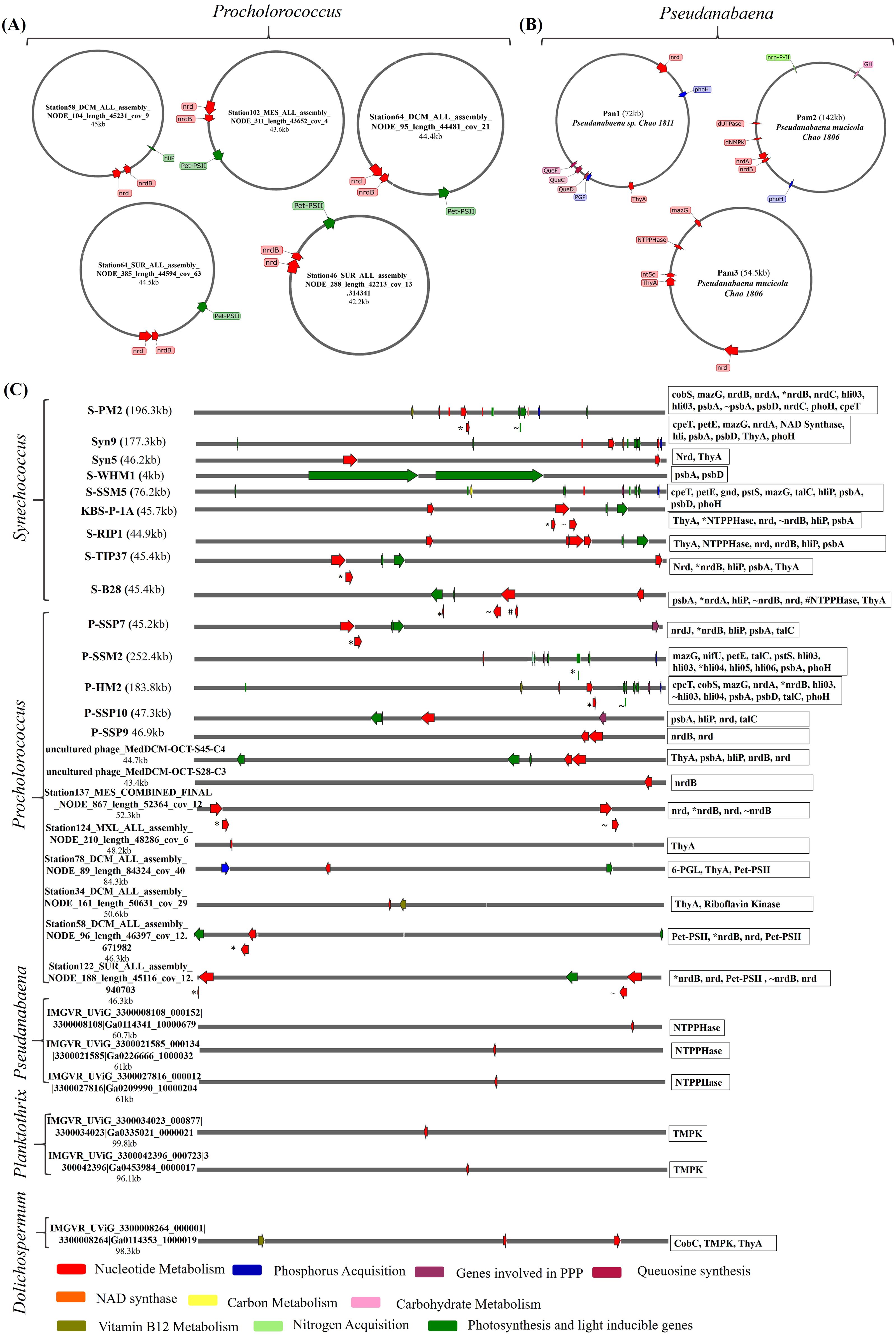
Figure 2. The genomic maps and functional annotations of auxiliary metabolic genes (AMGs) in different cyanobacterial hosts of cyanophages. Each panel (A–C) illustrates the genomic integration and specific roles of AMGs in influencing host cyanobacterial metabolism: (A) shows the circular cyanophages infecting Procholorococcus sp. highlighting their role in nucleotide metabolism, photosynthesis and nutrient acquisition. (B) depicts AMGs in circular phages isolated from lake Chaohu which infect Psuedanabaena sp. with a focus on nutrient metabolism and photosynthetic efficiency (C) presents a detailed mapping of AMGs in different phages having linear genomes infecting different host species- the genes represented in the chart are written from left to right, emphasizing their impact on carbohydrate metabolism, photosystem functionality, and nutrient utilization. Each gene is color-coded based on its metabolic function, providing a visual correlation between phage-encoded genes and their potential metabolic impacts on cyanobacterial hosts. Panels (A–C) not only underscores the genetic complexity and strategic placement of these genes within the phage genomes but also illustrates the intricate interactions between cyanophages and their cyanobacterial hosts in marine ecosystems. These genomic maps serve as a foundation for understanding how cyanophages influence the ecological dynamics of their cyanobacterial hosts in the ocean’s biogeochemical cycles.
In contrast, the larger genome phages like P-SSM2 and P-HM2, infecting Prochlorococcus, feature a diverse array of genes that boost phosphorus acquisition along with enhancing photosynthetic efficiency and respond to environmental stress when nutrients are scarce, as illustrated in Figure 2C (51, 60).
The cyanophages, namely Pam1~Pam5 and Pan1~Pan5, are the primary isolates documented from Lake Chaohu (20, 61). Each has been characterized to various extents, but detailed information on their auxiliary metabolic genes (AMGs) and full genomic data is limited in the accessible public resources (20, 61). On top, phages like Pan1, Pam2 and Pam3 as shown in Figure 2B which infect Pseudanabaena sp., are engaged extensively in nucleotide metabolism through nrd genes and supports queuosine biosynthesis with QueF, QueC, and QueD. This ability to manipulate host metabolic pathways underscores the evolutionary sophistication of these phages and their potential impact on the cyanobacterial communities within aquatic ecosystems. These characteristics reflect the cyanophages’ crucial roles in influencing cyanobacterial population dynamics, potentially affecting global biogeochemical cycles and the ecological balance within their habitats (20, 61, 62).
4 Key metabolic pathways influenced by AMGs
The activation of viral AMGs typically occurs when viruses replicate independently inside host cells, working in acquiescence with the host’s machinery. These AMGs are associated with several physiological activities in hosts infected with viruses, such as photosynthesis (39), carbon metabolism (36), and nucleotide metabolism (53). These AMGs when incorporated into the bacterial host, they enhance the host’s ability to cope with the stress conditions, ensuring continued metabolic function and facilitate efficient phage replication, as illustrated in Figure 3, and the cyanophages having these AMGs infecting their respective hosts and altering their metabolic pathways are illustrated in Figure 2 and also presented in Supplementary Table 1.
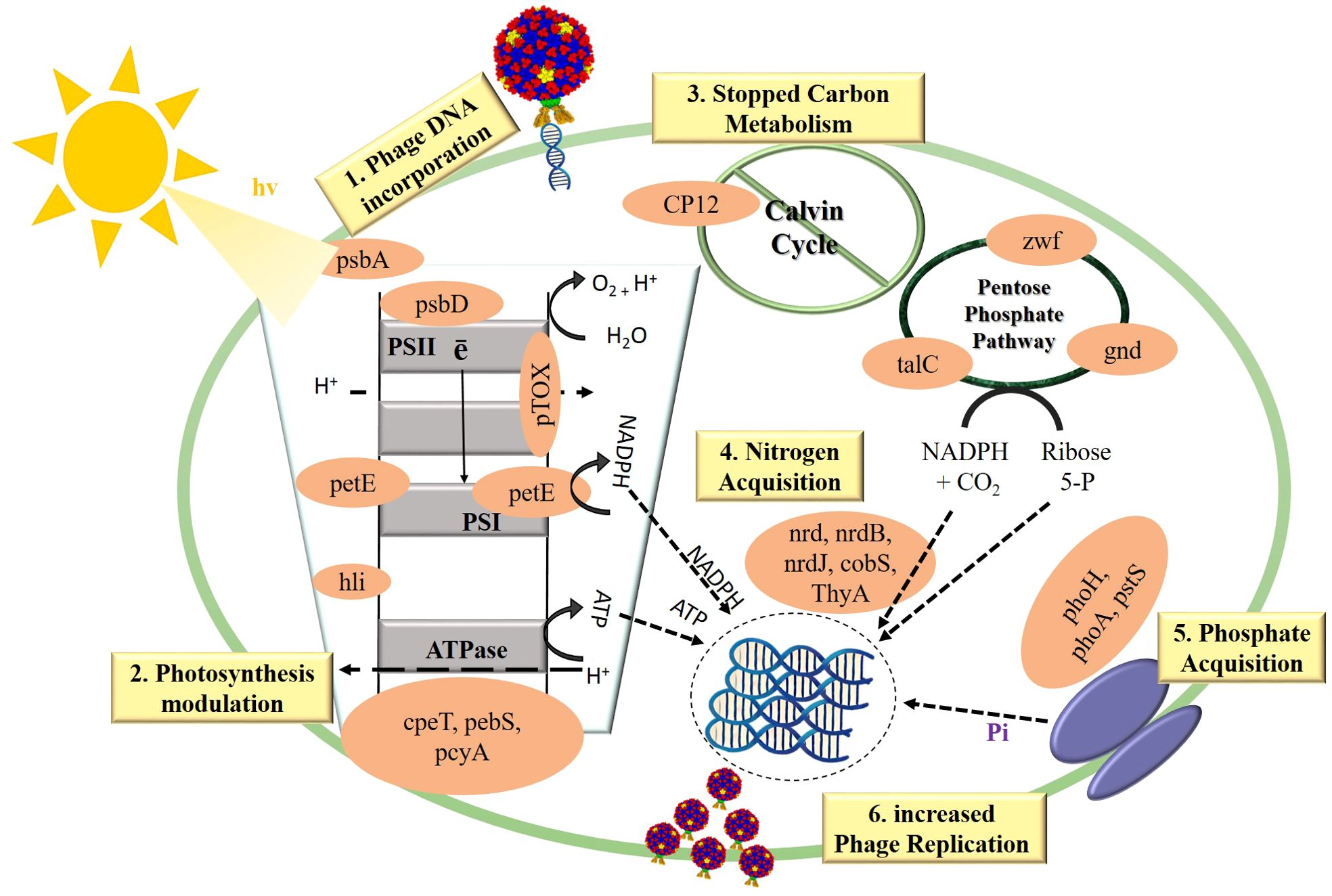
Figure 3. The modulation of key metabolic pathways i.e.: photosynthesis, carbon metabolism, and nitrogen and phosphorus acquisition by the incorporation of AMGs in cyanobacteria. This figure illustrates how cyanophage-encoded AMGs alter cyanobacterial metabolism to favor phage replication. Upon infection, cyanophages integrate their DNA, containing AMGs, into the host cell. 1) Phage DNA Incorporation: Introduction of phage DNA into cyanobacteria initiates metabolic reprogramming. 2) Photosynthesis Modulation: AMGs (psbA, psbD, petE, hli, cpeT, pebS, pcyA) optimize the photosynthetic electron transport chain and light-harvesting systems, ensuring energy production favors phage needs. 3) Carbon Metabolism Inhibition: Disruption of the Calvin Cycle by AMGs (CP12) reroutes carbon flux into the pentose phosphate pathway (zwf, gnd, talC) to generate NADPH for viral replication. 4) Enhanced Nitrogen and Phosphate Acquisition: AMGs (nrdB, nrdJ, cobS, ThyA, phoH, phoA, pstS) enhance nutrient uptake and assimilation, providing essential resources for phage nucleic acid and protein synthesis. 5) Increased Phage Replication: The host’s reprogrammed metabolism supports optimal conditions for phage reproduction.
4.1 Impact on photosynthesis efficiency
Cyanophages have been the subject of substantial study on AMGs, which are linked to photosynthesis. Photosystem II proteins D1 and D2 are encoded by the psbA and psbD genes, respectively. These proteins play an essential role in the light-dependent photosynthesis processes and are essential components of the photosynthetic apparatus. These AMGs maintain photosystem II functionality, which is crucial for viral replication by ensuring the production of ATP and NADPH by the host cyanobacterium (12, 32).
In addition to psbA and psbD genes, cyanophages also carry AMGs such as hli (high light-inducible) genes. These genes ensure uninterrupted photosynthetic performance and energy production during viral infection. Proteins encoded by the hli genes help safeguard photosynthetic membranes from photo-damage and excess light energy by stabilizing and dispersing these rays (36). Additionally, genes like petE and cpeT are essential to photosynthetic processes. The photosynthetic electron transport (petE) chain uses plastocyanin, a copper-containing protein, to move electrons between cytochrome b6f and photosystem I (63). Thus optimizing photosynthetic energy generation for the host and replicating virus by enhancing electron transport efficiency. The cpeT gene is involved in the synthesis of Phycobilisomes which are light-harvesting complexes that enhance light absorption and energy transmission to photosystems particularly under different light conditions (64). By incorporating these genes, cyanophages enhance their host’s light-harvesting abilities, ensuring efficient energy uptake and utilization during infection (65).
These photosynthesis-related AMGs in cyanophages influence the metabolic state of their host’s as illustrated in Figure 3. By affecting host photosynthesis, cyanophages ensure a steady energy supply even under suboptimal conditions, supporting both cyanobacterial and viral proliferation (12, 36). The impact of these genes on the host’s metabolism is significant, but it is important to note that their effects are context-dependent and may vary based on environmental conditions and specific interactions between the phage and host.
4.2 Impact on carbon metabolism
The genes associated with carbon metabolism in cyanophages, including CP12, gnd, zwf, and talA, are instrumental in this metabolic reprogramming. CP12 is a diminutive regulatory protein that plays a crucial role in regulating the Calvin cycle which serves as the principal route for carbon assimilation in photosynthetic organisms. CP12 produced by phages, may block important enzymes in the Calvin cycle, including phosphoribulokinase and glyceraldehyde-3-phosphate dehydrogenase (66). Consequently, redirecting the flow of carbon away from sugar synthesis while towards the pentose phosphate pathway (PPP). This redirection guarantees that the energy and reducing power production during photosynthesis, namely ATP and NADPH, aren’t utilized for the formation of biomass, but rather directed towards the creation of deoxyribonucleotides (dNTPs), which are crucial for phage DNA replication (36).
Furthermore, talA (transaldolase), gnd (6-phosphogluconate dehydrogenase), talC (transaldolase) and zwf (glucose-6-phosphate dehydrogenase) are crucial enzymes in the PPP. This metabolic cycle produces NADPH and ribose-5-phosphate, which serves as a precursor for the production of nucleotides. To enhance viral replication, phages enhance PPP expression, which in turn increases the synthesis of dNTPs. By enhancing the circulation through the PPP, cyanophages deliver ribose-5-phosphate and NADPH consistently. As a result, viral DNA and other important macromolecules may be produced (12, 67).
According to research, phage-encoded CP12 and talA are able to effectively divert reducing power and energy from the Calvin cycle and use it to produce nucleotides (36). In order to satisfy their own reproductive needs, cyanophages use a sophisticated mechanism called rerouting to alter the host’s metabolism. The metabolic flow of the host organism reportedly undergoes a dramatic shift upon infection with cyanophages. The outcome is an increase in PPP flow and a decrease in Calvin cycle flow (32, 36). As shown in Figure 3 this metabolic shift highlights the intricate evolutionary adaptations of cyanophages to control host metabolism for their advantage and ensures the efficient production of viral components.
4.3 Impact on nutrient acquisition
Nutrient acquisition involves the uptake of multiple nutrients like nitrogen, phosphorus, and vitamins in the nutrient scarce environment. Using AMGs that are related with nitrogen fixation such as nrdA, nrdB, nrdC, nrdJ etc., cyanophages may enhance the host’s ability to retain and incorporate nitrogen (68). Nitrogenase reductase; encoded by genes like nifH, is essential for cyanobacteria to absorb because it converts atmospheric nitrogen to ammonia. These AMGs enhance the host’s ability to fix nitrogen, which is a crucial nutrient for both the host and the virus (48). Cyanobacteria have a considerable competitive advantage in many aquatic settings because they are able to fix atmospheric nitrogen, which is a limiting nutrient in some ecosystems. Cyanophages augment the metabolic capacity of their hosts by harboring nitrogen fixation genes, hence promoting more resilient and enduring infections (26).
In addition to nitrogen, phosphorus acquisition is crucial for cyanobacteria, particularly in freshwater environments where phosphorus is typically a limiting resource (36). Cyanophages encode AMGs like pstS and phoH to enhance their host’s phosphate absorption. These genes assist cyanobacteria to absorb and retain phosphate regardless of phosphorus abundance. Managing phosphorus allows cyanobacteria to develop and bloom in nutrient-limited environments (59, 69). The pst system in cyanobacteria is the high affinity phosphate-specific transport system which is involved in the production of permease proteins that microbial cells use to absorb orthophosphate. This transport system is encoded by pst (S, C, A, and B) genes. Cyanobacteria also utilize a low-affinity inorganic phosphate transport system encoded by the pit gene and a glycerol phosphate (Ugp) system involving ugpB, ugpA, ugpE, and ugpC for diverse phosphorus sources (69). Genes like phoH, although its exact function remains unclear, is associated with phosphate stress responses and is thought to play a role in maintaining phosphorus homeostasis under limiting conditions (68). Regulation of these systems is controlled by the Pho regulon, particularly through the phoR and phoB genes, with PhoU acting as a repressor to balance phosphate uptake and utilization. By integrating these phosphorus acquisition genes, cyanophages ensure a sustained supply of this vital nutrient, facilitating effective viral genome replication and assembly (70). In addition to phosphorus absorption, cyanophages encode nucleotide metabolism genes (MazG, nrdA, nrdB, thyX, pyrBI, and cobS) needed for genome replication and transcription. Nucleotide synthesis and regulation by these genes ensure that the viral replication machinery has a consistent supply of DNA building blocks. MazG is thought to regulate host transcription and translation by regulating ppGpp levels. Phage-encoded MazG modulates ppGpp levels to prolong host cell viability under phage infection-induced stress, prolonging viral replication (50, 71).
The collective action of these AMGs illustrates the intricate strategies employed by cyanophages to reprogram host metabolic pathways, ensuring optimal conditions for their replication. While traditionally seen as biological controls that mitigate harmful algal blooms (HABs), cyanophages can also play a more complex role. In certain conditions, phage-induced lysis can release nutrients that fuel the growth of surviving cyanobacteria, potentially intensifying and prolonging HABs. This dual influence highlights the intricate and sometimes paradoxical relationship between cyanophages and their cyanobacterial hosts, affecting the dynamics of aquatic ecosystems (64, 72, 73). AMGs are deliberately placed into cyanophage genomes, typically interspersed with structural and replication genes, to ensure proper expression during infection. These genes serve critical roles in viral replication and host metabolic processes (12, 36, 46). Table 1 highlights major AMGs and their influence on host metabolism. These AMGs demonstrating the complex methods used by cyanophages to exploit their hosts metabolisms and contribute to the viral replication and overall ecological implications of cyanobacterial blooms.
5 Consequences for evolution and ecology
The adaptive advantages provided by auxiliary metabolic genes (AMGs) have proven crucial in the evolution of cyanophages by increasing viral fitness. These genes may be incorporated into phage genomes from other microorganisms, such as host cyanobacteria, via horizontal gene transfer (36, 78, 79). Cyanophages improve their control of host metabolism by using AMGs, which fosters a favorable setting for viral replication and survival (40, 46). Through a process of co-evolution, cyanophages have become more diverse and highly specific, allowing them to infect a vast array of cyanobacterial hosts (51, 80) and significantly shape the evolution and genetic diversity of cyanobacterial genomes.
The recent identification of cyanophages Pan1-Pan5 (61) and Pam1-Pam5 (20) in the Lake Chaohu exemplifies this dynamic interaction. These phages, characterized by their genomic and morphological analyses, interact particularly with Microcystis aeruginosa; a dominant cyanobacterium during bloom conditions in Lake Chaohu (20, 81). They are reported to have diverse AMGs in their genomes that potentially modulate the cyanobacterial metabolism, influencing bloom dynamics and enhancing the resilience of cyanobacterial communities to nutrient-rich conditions (82). Such interactions could lead to novel approaches in managing cyanobacterial blooms, potentially mitigating their intensity or duration through targeted phage applications. Additionally, by facilitating genetic exchanges among cyanobacteria via transduction, these phages contribute to the genetic diversity within these communities, influencing their adaptive responses to environmental fluctuations such as nutrient availability and temperature shifts (20). This role underscores the significant ecological function of cyanophages in shaping microbial community structures and offers promising pathways for future research and biotechnological applications.
One possible mechanism for the introduction of AMGs into cyanobacterial genomes is the lysogenic cycle, which enables phage DNA to integrate into the host genome. Under certain environmental circumstances, these integrated components, known as prophages, may influence the host’s metabolism and stress responses, providing them with an advantage. Sometimes cyanophages may incorporate their genetic material into the genomes of cyanobacterial host as endogenous viral elements (EVEs). Several environmental variables may trigger the reactivation of these EVEs that have been dormant inside the host genome. Host gene expression, stress reactions, and viability may all be affected by EVE presence. Past interactions between cyanobacteria and cyanophages serve as a kind of genetic memory that shapes how cyanobacterial populations develop (5, 43, 83). Ultimately, AMGs have the potential to affect the metabolic pathway development in cyanobacteria, which might have consequences for their ecological roles and competitive dynamics within microbial communities (34, 58, 80).
AMGs have long-term effects on both nutrient cycle and microbial communities. AMGs affect the flux of nutrients and energy in aquatic environments by regulating critical cyanobacterial metabolic pathways. This has the potential to change the make-up and role of microbial communities, which in turn affects nitrogen fixation (48), primary production, and other vital ecosystem functions (12, 69). The variety and number of organisms that rely on cyanobacteria as a food source and habitat may be impacted by these interactions, which in turn can affect higher trophic levels (84).
6 Conclusion and future perspective
This review article highlights the importance of cyanophage-expressed auxiliary metabolic genes (AMGs) in controlling cyanobacterial physiology, particularly in the context of HABs. In order to alter their hosts’ photosynthesis, carbon metabolism, and food absorption, cyanophages have AMGs like psbA, psbD, CP12, talA, pstS, and phoH ultimately ensuring efficient viral replication and spread by enhancing host metabolic pathways. This study highlights the sophisticated evolutionary modifications that cyanophages possess, enabling them to manipulate host metabolism for their own advantage (16, 85).
In order to enhance the metabolic capabilities of cyanobacterial hosts during cyanophage infection, AMGs (36) are essential, and this has a profound effect on how HABs behave. Infected cyanobacteria rely on AMGs for growth and function, which in turn facilitates viral multiplication. This affects the overall structure and function of aquatic microbial communities and also encourages the fast growth of cyanophages. The evolution of cyanophages and their potential regulation of HABs (2) may be better understood by learning more about the functions and processes of AMGs. However, while the importance of AMGs is well-established, there is a need to more specifically address how these genes function within the context of HABs. The existing literature often focuses on marine environments, leaving a gap in understanding their roles in freshwater systems where HABs pose significant environmental and health risks.
Future research on cyanophages should focus on unraveling the comprehensive mechanisms through which AMGs influence cyanobacterial metabolism and bloom dynamics. Advanced genomic and metagenomic approaches are essential to identify novel AMGs and understand their regulatory networks within host cells. Additionally, exploring the ecological interactions between cyanophages with broader microbial communities will provide deeper insights into their role in nutrient cycling and ecosystem functioning. The development of biotechnological applications, or the integration of cyanophage-based therapies; such as phage therapy, with existing HAB management practices offers promising avenues for the targeted control of HABs.
To fully comprehend the impact of cyanophages on HABs, future research must prioritize studies that directly link AMGs to bloom dynamics in these contexts. Furthermore, continuing research and surveillance of cyanophages particularly in freshwater systems like Lake Chaohu are crucial for the development of biologically-oriented management measures to reduce cyanoHABs. In conclusion, gaining a more profound comprehension of the interactions between phages and their hosts especially in Lake Chaohu will provide vital insights into the capacity of phages to uphold ecological equilibrium and guarantee water quality in freshwater ecosystems.
Author contributions
QA: Writing – original draft, Writing – review & editing. KW: Writing – original draft, Writing – review & editing. XW: Writing – original draft. QB: Writing – original draft. QL: Writing – original draft. C-ZZ: Supervision, Writing – original draft. QW: Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. We acknowledge funding from the Key Research and Development Projects of Anhui Province (Grant No. 202104i07020004).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fviro.2024.1461375/full#supplementary-material
Supplementary Table 1 | Comprehensive overview of cyanophages, their hosts, genomic characteristics, and key auxiliary metabolic genes (AMGs).
References
1. Planavsky NJ, Asael D, Hofmann A, Reinhard CT, Lalonde SV, Knudsen A, et al. Evidence for oxygenic photosynthesis half a billion years before the Great Oxidation Event. Nat Geosci. (2014) 7:283–6. doi: 10.1038/ngeo2122
2. Zohdi E, Abbaspour M. Harmful algal blooms (red tide): a review of causes, impacts and approaches to monitoring and prediction. Int J Environ Sci Technol. (2019) 16:1789–806. doi: 10.1007/s13762-018-2108-x
3. Huisman JM, Matthijs HC, Visser PM. Harmful Cyanobacteria Springer Aquatic Ecology Series 3. Dordrecht, The Netheralands. (2005). Vol. 3. doi: 10.1007/1-4020-3022-3
4. Paerl HW, Otten TG. Harmful cyanobacterial blooms: causes, consequences, and controls. Microbial Ecol. (2013) 65:995–1010. doi: 10.1007/s00248-012-0159-y
5. Suttle CA. Marine viruses—major players in the global ecosystem. Nat Rev Microbiol. (2007) 5:801–12. doi: 10.1038/nrmicro1750
6. Wilhelm SW, Suttle CA. Viruses and nutrient cycles in the sea: viruses play critical roles in the structure and function of aquatic food webs. Bioscience. (1999) 49:781–8. doi: 10.2307/1313569
7. Zhang D, He Y, Gin KY-H. Novel freshwater cyanophages provide new insights into evolutionary relationships between freshwater and marine cyanophages. Microbiol Spectr. (2021) 9:e00593–21. doi: 10.1128/Spectrum.00593-21
8. Aranda YN, Bhatt P, Ates N, Engel BA, Simsek H. Cyanophage-cyanobacterial interactions for sustainable aquatic environment. Environ Res. (2023) 229:115728. doi: 10.1016/j.envres.2023.115728
9. Dantas CWD, Martins DT, Nogueira WG, Alegria OVC, Ramos RTJ. Tools and methodology to in silico phage discovery in freshwater environments. Front Microbiol. (2024) 15:1390726. doi: 10.3389/fmicb.2024.1390726
10. Deng L, Hayes PK. Evidence for cyanophages active against bloom-forming freshwater cyanobacteria. Freshw Biol. (2008) 53:1240–52. doi: 10.1111/j.1365-2427.2007.01947.x
11. Mojica KD, Brussaard CP. Factors affecting virus dynamics and microbial host–virus interactions in marine environments. FEMS Microbiol Ecol. (2014) 89:495–515. doi: 10.1111/fem.2014.89.issue-3
12. Lindell D, Jaffe JD, Johnson ZI, Church GM, Chisholm SW. Photosynthesis genes in marine viruses yield proteins during host infection. Nature. (2005) 438:86–9. doi: 10.1038/nature04111
13. Weitz JS, Wilhelm SW. Ocean viruses and their effects on microbial communities and biogeochemical cycles. F1000 Biol Rep. (2012) 4:17. doi: 10.3410/B4-17
14. Wood SA, Borges H, Puddick J, Biessy L, Atalah J, Hawes I, et al. Contrasting cyanobacterial communities and microcystin concentrations in summers with extreme weather events: insights into potential effects of climate change. Hydrobiologia. (2017) 785:71–89. doi: 10.1007/s10750-016-2904-6
15. Comeau AM, Chan AM, Suttle CA. Genetic richness of vibriophages isolated in a coastal environment. Environ Microbiol. (2006) 8:1164–76. doi: 10.1111/j.1462-2920.2006.01006.x
16. Bhatt P, Engel BA, Reuhs M, Simsek H. Cyanophage technology in removal of cyanobacteria mediated harmful algal blooms: A novel and eco-friendly method. Chemosphere. (2023) 315:137769. doi: 10.1016/j.chemosphere.2023.137769
17. Bothe H, Schmitz O, Yates MG, Newton WE. Nitrogen fixation and hydrogen metabolism in cyanobacteria. Microbiol Mol Biol Rev. (2010) 74:529–51. doi: 10.1128/MMBR.00033-10
18. Havens K, Fukushima T, Xie P, Iwakuma T, James R, Takamura N, et al. Nutrient dynamics and the eutrophication of shallow lakes Kasumigaura (Japan), Donghu (PR China), and Okeechobee (USA). Environ pollut. (2001) 111:263–72. doi: 10.1016/S0269-7491(00)00074-9
19. Qin B. The changing environment of Lake Taihu and its ecosystem responses. J Freshwater Ecol. (2015) 30(1):1–3.
20. Zhu J, Yang F, Du K, Wei Z-L, Wu Q-F, Chen Y, et al. Phylogenomics of five Pseudanabaena cyanophages and evolutionary traces of horizontal gene transfer. Environ Microbiome. (2023) 18:3. doi: 10.1186/s40793-023-00461-5
21. Carstensen J, Andersen JH, Gustafsson BG, Conley DJ. Deoxygenation of the Baltic Sea during the last century. Proc Natl Acad Sci. (2014) 111:5628–33. doi: 10.1073/pnas.1323156111
22. Yuan L, Yu P, Huang X, Zhao Z, Chen L, Ju F. Seasonal succession, host associations and biochemical roles of aquatic viruses in a eutrophic lake plagued by cyanobacterial blooms. Research Square [PREPRINT]. (2023). doi: 10.21203/rs.3.rs-3510205/v1
23. Zhou Y, Lin J, Li N, Hu Z, Deng F. Characterization and genomic analysis of a plaque purified strain of cyanophage PP. Virologica Sin. (2013) 28:272–9. doi: 10.1007/s12250-013-3363-0
24. Xia H, Li T, Deng F, Hu Z. Freshwater cyanophages. Virologica Sin. (2013) 28:253–9. doi: 10.1007/s12250-013-3370-1
25. Jaskulska A, Mankiewicz-Boczek J. Cyanophages specific to cyanobacteria from the genus Microcystis. Ecohydrology Hydrobiology. (2020) 20:83–90. doi: 10.1016/j.ecohyd.2019.06.001
26. Grasso CR, Pokrzywinski KL, Waechter C, Rycroft T, Zhang Y, Aligata A, et al. A review of cyanophage-host relationships: highlighting cyanophages as a potential cyanobacteria control strategy. Toxins (Basel). (2022) 14(6):385. doi: 10.3390/toxins14060385
27. Mavrich TN, Hatfull GF. Bacteriophage evolution differs by host, lifestyle and genome. Nat Microbiol. (2017) 2:17112. doi: 10.1038/nmicrobiol.2017.112
28. Dion MB, Oechslin F, Moineau S. Phage diversity, genomics and phylogeny. Nat Rev Microbiol. (2020) 18(3):125–38. doi: 10.1038/s41579-019-0311-5
29. Latka A, Leiman PG, Drulis-Kawa Z, Briers Y. Modeling the architecture of depolymerase-containing receptor binding proteins in Klebsiella phages. Front Microbiol. (2019) 10:2649. doi: 10.3389/fmicb.2019.02649
30. De Jonge PA, Nobrega FL, Brouns SJ, Dutilh BE. Molecular and evolutionary determinants of bacteriophage host range. Trends Microbiol. (2019) 27:51–63. doi: 10.1016/j.tim.2018.08.006
31. Deng L, Gregory A, Yilmaz S, Poulos BT, Hugenholtz P, Sullivan MB. Contrasting life strategies of viruses that infect photo-and heterotrophic bacteria, as revealed by viral tagging. MBio. (2012) 3(6):10–128. doi: 10.1128/mbio.00373-12
32. Sullivan MB, Waterbury JB, Chisholm SW. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature. (2003) 424:1047–51. doi: 10.1038/nature01929
33. Koskella B, Brockhurst MA. Bacteria–phage coevolution as a driver of ecological and evolutionary processes in microbial communities. FEMS Microbiol Rev. (2014) 38:916–31. doi: 10.1111/1574-6976.12072
34. Roux S, Hallam SJ, Woyke T, Sullivan MB. Viral dark matter and virus-host interactions resolved from publicly available microbial genomes. Elife. (2015) 4:e08490. doi: 10.7554/eLife.08490
35. Millard A, Clokie MR, Shub DA, Mann NH. Genetic organization of the psbAD region in phages infecting marine Synechococcus strains. Proc Natl Acad Sci. (2004) 101:11007–12. doi: 10.1073/pnas.0401478101
36. Thompson LR, Zeng Q, Kelly L, Huang KH, Singer AU, Stubbe J, et al. Phage auxiliary metabolic genes and the redirection of cyanobacterial host carbon metabolism. Proc Natl Acad Sci U.S.A. (2011) 108:E757–64. doi: 10.1073/pnas.1102164108
37. Breitbart M, Rohwer F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. (2005) 13:278–84. doi: 10.1016/j.tim.2005.04.003
38. Lin W, Li D, Pan L, Li M, Tong Y. Cyanobacteria-cyanophage interactions between freshwater and marine ecosystems based on large-scale cyanophage genomic analysis. Sci Total Environ. (2024) 950:175201. doi: 10.1016/j.scitotenv.2024.175201
39. Mann NH, Cook A, Millard A, Bailey S, Clokie M. Bacterial photosynthesis genes in a virus. Nature. (2003) 424:741–1. doi: 10.1038/424741a
40. Waterbury JB, Valois FW. Resistance to co-occurring phages enables marine Synechococcus communities to coexist with cyanophages abundant in seawater. Appl Environ Microbiol. (1993) 59:3393–9. doi: 10.1128/aem.59.10.3393-3399.1993
41. Kutter E, Sulakvelidze A. Bacteriophages: biology and applications. Crc press (2004). Available at: taylorfrancis.com.
42. Grasso CR, Pokrzywinski KL, Waechter C, Rycroft T, Zhang Y, Aligata A, et al. A review of cyanophage–host relationships: Highlighting cyanophages as a potential cyanobacteria control strategy. Toxins. (2022) 14:385. doi: 10.3390/toxins14060385
43. Hurwitz BL, U’ren JM. Viral metabolic reprogramming in marine ecosystems. Curr Opin Microbiol. (2016) 31:161–8. doi: 10.1016/j.mib.2016.04.002
44. Brum JR, Sullivan MB. Rising to the challenge: accelerated pace of discovery transforms marine virology. Nat Rev Microbiol. (2015) 13:147–59. doi: 10.1038/nrmicro3404
45. Hurwitz BL, Brum JR, Sullivan MB. Depth-stratified functional and taxonomic niche specialization in the ‘core’and ‘flexible’Pacific Ocean Virome. ISME J. (2015) 9:472–84. doi: 10.1038/ismej.2014.143
46. Gao EB, Huang Y, Ning D. Metabolic genes within cyanophage genomes: implications for diversity and evolution. Genes (Basel). (2016) 7(10):80. doi: 10.3390/genes7100080
47. Heyerhoff B, Engelen B, Bunse C. Auxiliary metabolic gene functions in pelagic and benthic viruses of the baltic sea. Front Microbiol. (2022) 13:863620. doi: 10.3389/fmicb.2022.863620
48. Bothe H, Schmitz O, Yates MG, Newton WE. Nitrogen fixation and hydrogen metabolism in cyanobacteria. Microbiol Mol Biol Rev. (2010) 74:529–51. doi: 10.1128/MMBR.00033-10
49. Bragg JG, Chisholm SW. Modeling the fitness consequences of a cyanophage-encoded photosynthesis gene. PLoS One. (2008) 3:e3550. doi: 10.1371/journal.pone.0003550
50. Lindell D, Jaffe JD, Coleman ML, Futschik ME, Axmann IM, Rector T, et al. Genome-wide expression dynamics of a marine virus and host reveal features of co-evolution. Nature. (2007) 449:83–6. doi: 10.1038/nature06130
51. Sullivan MB, Huang KH, Ignacio-Espinoza JC, Berlin AM, Kelly L, Weigele PR, et al. Genomic analysis of oceanic cyanobacterial myoviruses compared with T4-like myoviruses from diverse hosts and environments. Environ Microbiol. (2010) 12:3035–56. doi: 10.1111/j.1462-2920.2010.02280.x
52. Sabehi G, Lindell D. The P-SSP7 cyanophage has a linear genome with direct terminal repeats. PLoS One. (2012) 7:e36710. doi: 10.1371/journal.pone.0036710
53. Dwivedi B, Xue B, Lundin D, Edwards RA, Breitbart M. A bioinformatic analysis of ribonucleotide reductase genes in phage genomes and metagenomes. BMC evolutionary Biol. (2013) 13:1–17. doi: 10.1186/1471-2148-13-33
54. Mann NH, Clokie MR, Millard A, Cook A, Wilson WH, Wheatley PJ, et al. The genome of S-PM2, a “photosynthetic“ T4-type bacteriophage that infects marine Synechococcus strains. J bacteriology. (2005) 187:3188–200. doi: 10.1128/JB.187.9.3188-3200.2005
55. Wang RH, Yang S, Liu Z, Zhang Y, Wang X, Xu Z, et al. Phagescope: a well-annotated bacteriophage database with automatic analyses and visualizations. Nucleic Acids Res. (2024) 52(D1):D756–61. doi: 10.1093/nar/gkad979
56. Weigele PR, Pope WH, Pedulla ML, Houtz JM, Smith AL, Conway JF, et al. Genomic and structural analysis of Syn9, a cyanophage infecting marine Prochlorococcus and Synechococcus. Environ Microbiol. (2007) 9:1675–95. doi: 10.1111/j.1462-2920.2007.01285.x
57. Pope WH, Weigele PR, Chang J, Pedulla ML, Ford ME, Houtz JM, et al. Genome sequence, structural proteins, and capsid organization of the cyanophage Syn5: a “horned“ bacteriophage of marine synechococcus. J Mol Biol. (2007) 368:966–81. doi: 10.1016/j.jmb.2007.02.046
58. Pope WH, Bowman CA, Russell DA, Jacobs-Sera D, Asai DJ, Cresawn SG, et al. Whole genome comparison of a large collection of mycobacteriophages reveals a continuum of phage genetic diversity. eLife. (2015) 4:e06416. doi: 10.7554/eLife.06416.028
59. Clokie MR, Mann NH. Marine cyanophages and light. Environ Microbiol. (2006) 8:2074–82. doi: 10.1111/j.1462-2920.2006.01171.x
60. Huang YM, Straub D, Blackwell N, Kappler A, Kleindienst S. Meta-omics reveal gallionellaceae and rhodanobacter species as interdependent key players for fe(II) oxidation and nitrate reduction in the autotrophic enrichment culture KS. Appl Environ Microbiol. (2021) 87:e0049621. doi: 10.1128/AEM.00496-21
61. Du K, Yang F, Zhang JT, Yu RC, Deng Z, Li WF, et al. Comparative genomic analysis of five freshwater cyanophages and reference-guided metagenomic data mining. Microbiome. (2022) 10:128. doi: 10.1186/s40168-022-01324-w
62. Mohammad A, Bon Ramos A, Lee BW, Cohen SW, Kiani MK, Iwata-Reuyl D, et al. Protection of the queuosine biosynthesis enzyme queF from irreversible oxidation by a conserved intramolecular disulfide. Biomolecules. (2017) 7(1):30. doi: 10.3390/biom7010030
63. Scanlan DJ, Ostrowski M, Mazard S, Dufresne A, Garczarek L, Hess WR, et al. Ecological genomics of marine picocyanobacteria. Microbiol Mol Biol Rev. (2009) 73:249–99. doi: 10.1128/MMBR.00035-08
64. Wiethaus J, Busch AW, Dammeyer T, Frankenberg-Dinkel N. Phycobiliproteins in Prochlorococcus marinus: biosynthesis of pigments and their assembly into proteins. Eur J Cell Biol. (2010) 89:1005–10. doi: 10.1016/j.ejcb.2010.06.017
65. Ledermann B, Aras M, Frankenberg-Dinkel N. Biosynthesis of cyanobacterial light-harvesting pigments and their assembly into phycobiliproteins. In: Hallenbeck PC, editor. Modern Topics in the Phototrophic Prokaryotes: Metabolism, Bioenergetics, and Omics. Springer International Publishing, Cham (2017).
66. Marri L, Thieulin-Pardo G, Lebrun R, Puppo R, Zaffagnini M, Trost P, et al. CP12-mediated protection of Calvin-Benson cycle enzymes from oxidative stress. Biochimie. (2014) 97:228–37. doi: 10.1016/j.biochi.2013.10.018
67. Sharkey TD. Pentose phosphate pathway reactions in photosynthesizing cells. Cells. (2021) 10:1547. doi: 10.3390/cells10061547
68. Lu J, Zhu B, Struewing I, Xu N, Duan S. Nitrogen-phosphorus-associated metabolic activities during the development of a cyanobacterial bloom revealed by metatranscriptomics. Sci Rep. (2019) 9:2480. doi: 10.1038/s41598-019-38481-2
69. Martiny AC, Huang Y, Li W. Occurrence of phosphate acquisition genes in Prochlorococcus cells from different ocean regions. Environ Microbiol. (2009) 11:1340–7. doi: 10.1111/j.1462-2920.2009.01860.x
70. Hsieh Y-J, Wanner BL. Global regulation by the seven-component Pi signaling system. Curr Opin Microbiol. (2010) 13:198–203. doi: 10.1016/j.mib.2010.01.014
71. Bryan MJ, Burroughs NJ, Spence EM, Clokie MR, Mann NH, Bryan SJ. Evidence for the intense exchange of MazG in marine cyanophages by horizontal gene transfer. PLoS One. (2008) 3:e2048. doi: 10.1371/journal.pone.0002048
72. Brussaard CP. Viral control of phytoplankton populations–a review. J Eukaryot Microbiol. (2004) 51:125–38. doi: 10.1111/j.1550-7408.2004.tb00537.x
73. Bullerjahn GS, Mckay RM, Davis TW, Baker DB, Boyer GL, D'anglada LV, et al. Global solutions to regional problems: Collecting global expertise to address the problem of harmful cyanobacterial blooms. A Lake Erie case study. Harmful Algae. (2016) 54:223–38. doi: 10.1016/j.hal.2016.01.003
74. Rozman Grinberg I, Martínez-Carranza M, Bimai O, Nouaïria G, Shahid S, Lundin D, et al. A nucleotide-sensing oligomerization mechanism that controls NrdR-dependent transcription of ribonucleotide reductases. Nat Commun. (2022) 13:2700. doi: 10.1038/s41467-022-30328-1
75. Helliwell KE, Lawrence AD, Holzer A, Kudahl UJ, Sasso S, Kräutler B, et al. Cyanobacteria and eukaryotic algae use different chemical variants of vitamin B12. Curr Biol. (2016) 26:999–1008. doi: 10.1016/j.cub.2016.02.041
76. Sullivan MB, Coleman ML, Weigele P, Rohwer F, Chisholm SW. Three Prochlorococcus cyanophage genomes: signature features and ecological interpretations. PLoS Biol. (2005) 3:e144. doi: 10.1371/journal.pbio.0030144
77. Kuntz M. Plastid terminal oxidase and its biological significance. Planta. (2004) 218:896–9. doi: 10.1007/s00425-004-1217-6
78. Suttle CA, Chan AM. Dynamics and distribution of cyanophages and their effect on marine synechococcus spp. Appl Environ Microbiol. (1994) 60:3167–74. doi: 10.1128/aem.60.9.3167-3174.1994
79. Huang X, Jiao N, Zhang R. The genomic content and context of auxiliary metabolic genes in roseophages. Environ Microbiol. (2021) 23:3743–57. doi: 10.1111/1462-2920.15412
80. Samson JE, Magadán AH, Sabri M, Moineau S. Revenge of the phages: defeating bacterial defences. Nat Rev Microbiol. (2013) 11:675–87. doi: 10.1038/nrmicro3096
81. Chen Z, Zhang J, Li R, Tian F, Shen Y, Xie X, et al. Metatranscriptomics analysis of cyanobacterial aggregates during cyanobacterial bloom period in Lake Taihu, China. Environ Sci pollut Res. (2018) 25:4811–25. doi: 10.1007/s11356-017-0733-4
82. Yan L, Xu Z, Hu Y, Wang Y, Zhou F, Gao X, et al. Cyanobacteria bloom hazard function and preliminary application in lake taihu, China. Chemosphere. (2022) 307:136122. doi: 10.1016/j.chemosphere.2022.136122
83. Six C, Thomas J-C, Garczarek L, Ostrowski M, Dufresne A, Blot N, et al. Diversity and evolution of phycobilisomes in marine Synechococcus spp.: a comparative genomics study. Genome Biol. (2007) 8:1–22. doi: 10.1186/gb-2007-8-12-r259
84. Weitz JS, Mileyko Y, Joh RI, Voit EO. Collective decision making in bacterial viruses. Biophys J. (2008) 95:2673–80. doi: 10.1529/biophysj.108.133694
Keywords: algal blooms, cyanobacteria, cyanophages, auxiliary metabolic genes, viruses
Citation: Ain Qu, Wu K, Wu X, Bai Q, Li Q, Zhou C-Z and Wu Q (2024) Cyanophage-encoded auxiliary metabolic genes in modulating cyanobacterial metabolism and algal bloom dynamics. Front. Virol. 4:1461375. doi: 10.3389/fviro.2024.1461375
Received: 08 July 2024; Accepted: 09 September 2024;
Published: 01 October 2024.
Edited by:
Jessica Labonté, Texas A&M University at Galveston, United StatesReviewed by:
Marcos Godoy, San Sebastián University, ChileJordan Walker, Texas A&M University at Galveston, United States
Copyright © 2024 Ain, Wu, Wu, Bai, Li, Zhou and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qingfa Wu, wuqf@ustc.edu.cn
†These authors have contributed equally to this work
 Qurat ul Ain
Qurat ul Ain