- 1GeneOne Life Science, Inc., Seoul, Republic of Korea
- 2New York University (NYU) Langone Health, Dermatology, New York, NY, United States
- 3The Wistar Institute, HIV Cure and Viral Disease Center, Philadelphia, PA, United States
- 4NanoBio Diagnostics, West Chester, PA, United States
In the past 25 years, the world has witnessed outbreaks of illnesses in humans from three different coronaviruses. Both the SARS-CoV outbreak of 2003 and the MERS-CoV outbreak of 2013 resulted in overall low fatalities in part due to inefficient human-to-human spread of each virus. In contrast, SARS-CoV-2, which emerged in 2019, was highly efficient at human-to-human spread and caused a global pandemic resulting in millions of casualties. Zoonotic transmission of viruses, including the three coronaviruses, poses an ongoing threat that cannot be ignored. In this review, we have focused on the diagnostics and therapeutics fronts using SARS-CoV-2 as a model. Specifically, we have selected proteins associated with the virus particles as targets and discussed various platform technologies. These insights hold the potential to inform the development of more effective therapeutics and vaccines not only for SARS-CoV-2 but also for future viral pandemics, thus contributing to global health on a broader scale.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), the etiological cause of coronavirus disease 2019 (COVID-19), is highly transmissible, and while most clinical symptoms overlap with many respiratory illnesses, it can lead to adverse effects in certain groups including the elderly and those who suffer from certain chronic diseases (1, 2). In comparison to SARS-CoV-1 and MERS-CoV, the high transmission capability of SARS-CoV-2 has resulted in a much higher incidence and number of deaths with a far greater number of countries being affected (3–5). Disease outcomes from SARS-CoV-2 infection have also demonstrated the importance of early diagnosis; greater time between symptom onset and confirmed diagnosis has been linked with worse prognosis for SARS-CoV-2 patients. The need for accurate diagnosis at early-stage infection remains a persistent challenge.
Various diagnostic assays have been created to detect SARS-CoV-2, including nucleic acid-based assays that detect the viral genome and protein-based assays that detect antibodies specific to various viral antigens. The nucleic acid-based assays (RT-PCR etc.) exhibit high sensitivity and specificity (6–10). However, their availability in resource-poor settings is limited. Antigen assays can detect viral proteins and can be conducted at point-of-care settings providing information about SARS-CoV-2 infection, usually with faster results than for RT-PCR, albeit with a reduced sensitivity. Thus, there is still a need for rapid, sensitive, and accessible diagnostic tests to address this issue.
The development of improved diagnostic antigen assays, antibody therapeutics and vaccines against SARS-CoV-2 depends on a comprehensive understanding of proteins present in viral particles. While technologies such as cryo-electron microscopy and tomography have provided information on the architecture of the spike envelope protein present on the surface of virus particles, there is a relative lack of data regarding other structural proteins such as membrane (M) and envelope (E) proteins, as well as the nucleocapsid (N) protein of SARS-CoV-2 (11–14). The goal of this review is to summarize the available data regarding the quantification and structure of the proteins present in the virus particles. This information is discussed in the context of diagnostic assays currently in the market and potential avenues for enhancing the sensitivity and specificity of such assays. Structural studies showed striking alterations in the spike protein from inactivated viruses in comparison to the wild type virus with respect to the number and the conformation status of viral protein molecules. The efficacy noted in the range of 51-79% for inactivated virus vaccines could be due to these alterations (15, 16). Hence, the information generated on SARS-CoV-2 by investigators will serve as a useful guide for enhancing the sensitivity and specificity of diagnostic antigen assays as well as development of inactivated virus vaccines and other novel therapeutics for combating likely future pandemics.
Overview of SARS-CoV-2 viral proteins
SARS-CoV-2 has a positive-sense single-stranded RNA (ssRNA) genome of around 29.8-29.9 kb, that encodes a total of 27 proteins (Figure 1). These proteins consist of nonstructural proteins (nsp1-16), accessory proteins (ORF 3a, ORF 3b, p6, ORF 7a, ORF 7b, ORF 8b, ORF 9b, and ORF 14) and structural proteins [nucleocapsid (N), membrane (M), Envelope (E) and spike (S)] (14, 17–21). Viral particles and their proteins are subject to various structural alterations during the processes of infection, genomic replication, and the formation of new viral particles (22). The readers may take advantage of several recent reviews available on this topic (14, 23–29). The virus particle exhibits a spherical or oval structure with a diameter ranging from 60 to 140 nm. The assembly of virus particles has been observed in various intracellular locations, including endosomes, the rough endoplasmic reticulum, double-membrane vesicles, intermediate vesicles, and exosomes (30, 31). Five of the SARS-CoV-2 accessory proteins (p3a, p6, p7a, p7b, and p9b) have been shown to be incorporated into mature virions as summarized in Table 1 and Figure 2.

Figure 1. Visual representation of (A) major structural proteins in a SARS-CoV-2 virion and (B) map illustrating layout of SARS-CoV-2 genome. Created with BioRender.com.
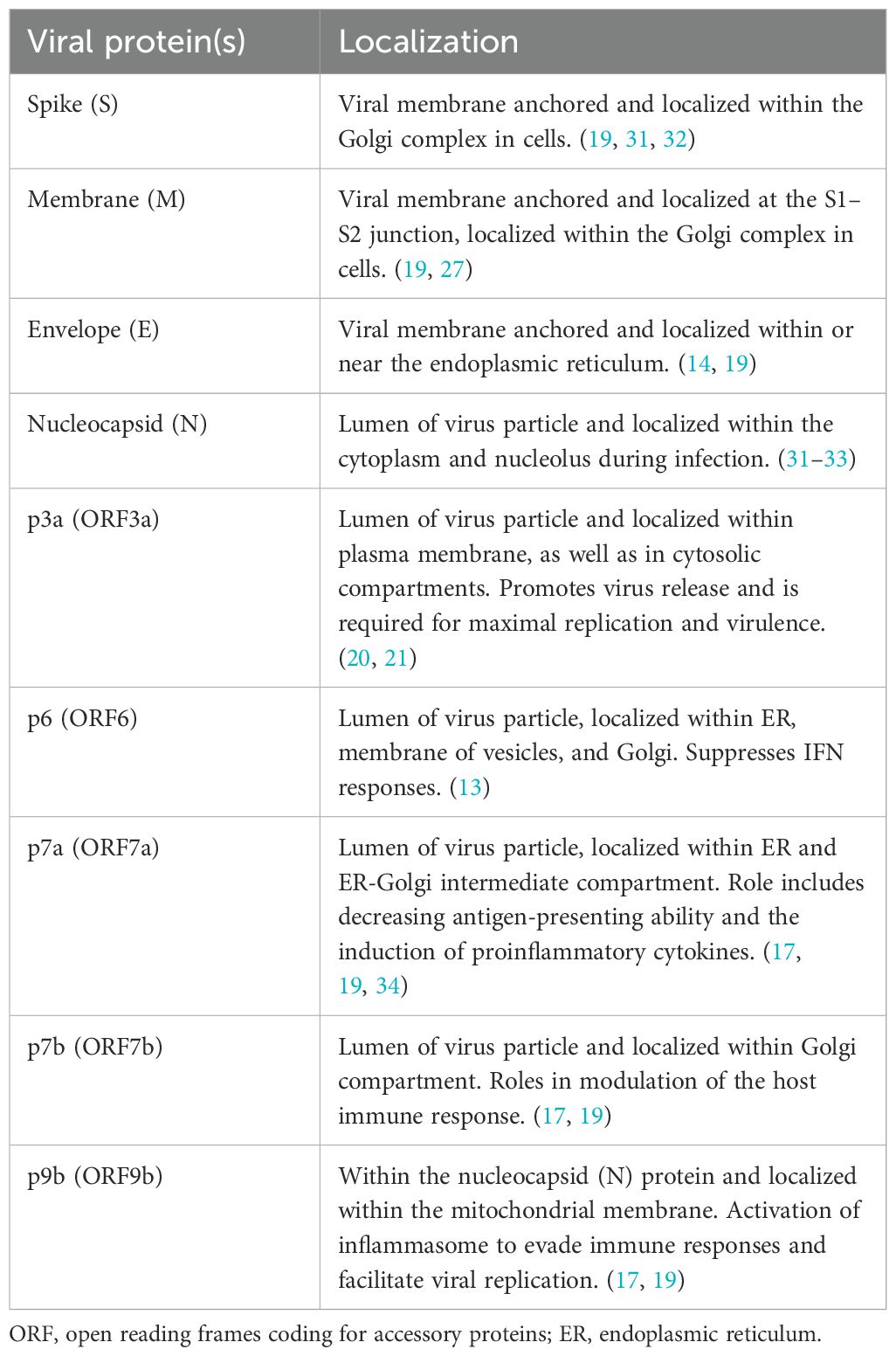
Table 1. Localization of SARS-CoV-2 proteins within the host cell or virus particle and their major functions.
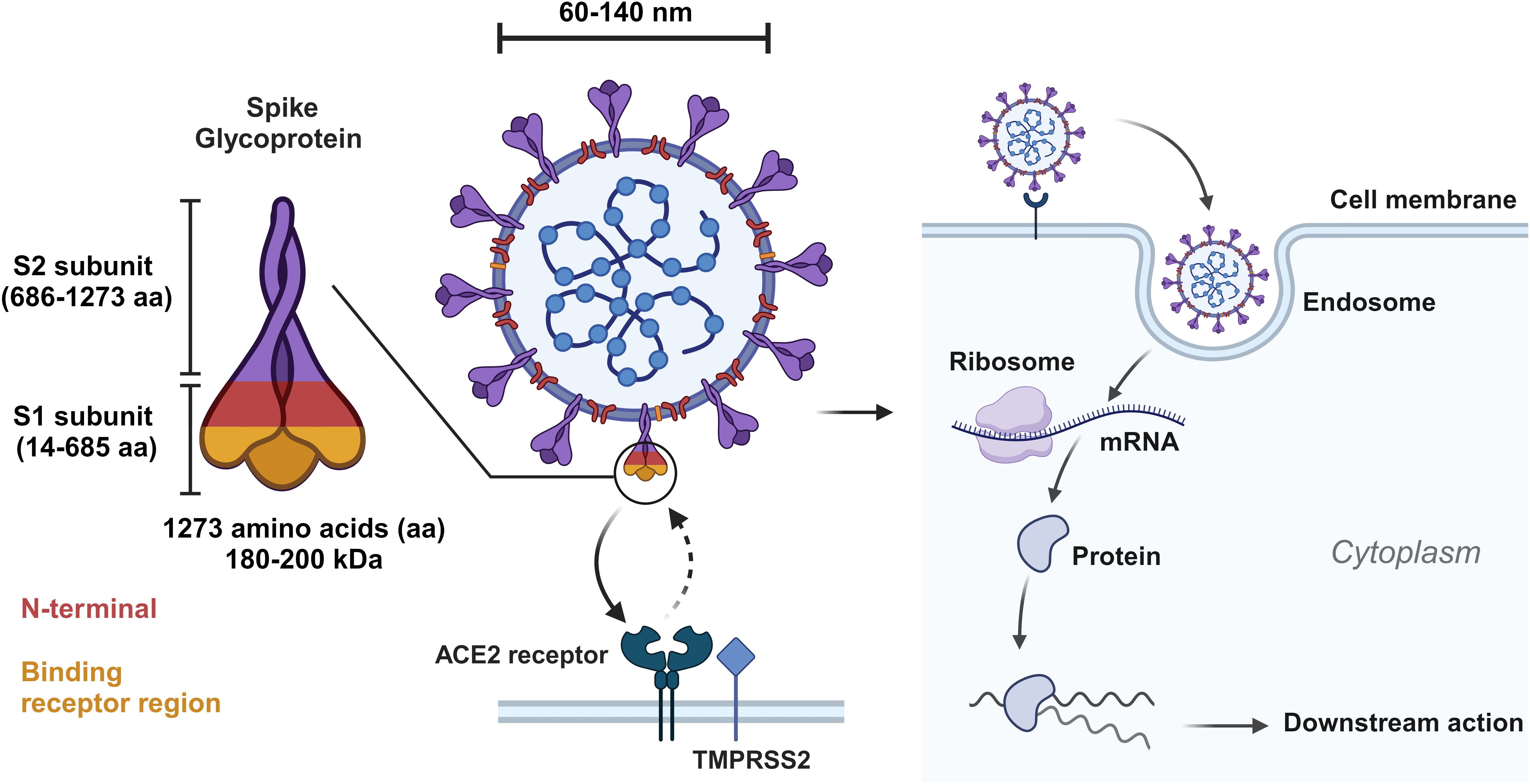
Figure 2. Visual representation of spike glycoprotein of SARS-CoV-2 and depiction of cell entry and early events of SARS-CoV-2 lifecycle. Created with BioRender.com.
Structural features and functions of the spike protein of SARS-CoV-2
The S protein, a critical constituent weighing 180-200 kDa, is integral to the SARS-CoV-2 viral life cycle (35). It consists of an extracellular N-terminus, a transmembrane domain anchored in the viral membrane, and a short intracellular C-terminal segment. Comprising 1273 amino acids, the S protein is divided into two subunits: the S1 subunit (14-685aa) and the S2 subunit (686-1273aa). The S1 subunit contains the N-terminal domain (14-305aa), receptor-binding domain (RBD; 319-541aa), and receptor binding motif (RBM; 437-508aa). The N-terminal domain of the S1 subunit participates in receptor binding, with the receptor-binding domain (RBD) and receptor binding motif contributing to high-affinity interactions with host cell receptors. The S2 subunit includes the fusion peptide domain (788-806 aa), heptapeptide repeat sequences 1 and 2 (912-984 aa and 1163-1213 aa), transmembrane domain (1213-1237aa), and cytoplasmic tail (1238-1273aa) (36). The S2 subunit is responsible for membrane fusion, structural stability, and anchoring the S protein in the viral membrane. Employing cryo-electron microscopy (EM) and X-ray crystallography, the examination of over 300 S protein structures has provided insights into the mechanisms governing S protein receptor binding (37). The metastable prefusion conformation of the S protein undergoes substantial structural rearrangement upon interaction with the host cell. Notably, a permanent transition occurs from the “pre-fusion” state, characterized by noncovalent bonding of S1 and S2 subunits to a “post-fusion” state involving cleavage of the N-terminal S1 subunit, followed by the refolding of the remaining S2 subunit (25, 38–41). The Spike (S) protein RBD exhibits diverse conformations, including both down and up forms. Given that S protein on viral surfaces exists in a homotrimeric form, S trimers conformations may be referred to as one-up, two-up, or three-up. ACE2 receptor binding is facilitated by RBD in up conformations, while the down conformation precludes such binding, in part through shielding from antibody recognition with a bulky glycan coat (41).
As an RNA virus, SARS-CoV-2 evolves rapidly. It is estimated that the mutation rate is in the range of 1x106-2x106 per nucleotide per replication cycle, resulting in the generation of variant viruses (29). The S protein, present in smaller numbers per viral particle, is particularly susceptible to mutations which can give rise to new variants. As mentioned earlier, virulence can be enhanced or diminished based on the specific interaction energies between different spike and ACE2 variants (42, 43). Furthermore, mutations in S protein can result in immune evasion and resistance to treatment (44). Hence, the genetic alterations in the spike protein can impact the efficacy of vaccine and antibody therapeutics targeting spike protein-receptor interactions.
SARS-CoV-2 nucleocapsid protein
The nucleocapsid protein of SARS-CoV-2 plays a crucial role in viral genome packaging and regulation of RNA synthesis during replication (45). N protein is highly conserved among SARS-CoV-2 variants, making it an attractive therapeutic target (33) and it is also considered highly immunogenic. (45) The SARS-CoV-2 N protein consists of 419 amino acids (aa) with five domains: the N-terminal domain (NTD: 1-50 aa), RNA binding domain (51-174aa), linker domain (LINK: 175-246aa), dimerization domain (247-365aa), and C-terminal domain (CTD: 366-419aa). While the NTD, LINK, and CTD regions are highly dynamic and disordered, the RNA binding domain and dimerization domain possess well-defined structures. All five domains are predicted to engage in RNA binding, and higher-ordered oligomerization may also occur in an RNA-independent manner (33, 46).
The disordered regions of the N protein harbor sites for protein-protein and protein-RNA interactions. Notably, intra-protein interactions between the arginine-rich C terminus of the NTD and the basic beta strand from the RNA binding domain have been observed. However, minimal interactions have been reported between the RNA binding domain and dimerization domain. Additionally, potential N-linked and O-linked glycosylation sites within the N protein may influence protein folding and stability (33, 47).
Envelope protein of SARS-CoV-2
The E protein of SARS-CoV-2, the smallest among the major structural proteins, plays diverse roles in the virus life cycle. It is involved in virus particle assembly, budding from the cell membrane, and envelope formation. The E protein consists of a hydrophilic N-terminus, a transmembrane domain, and a long hydrophilic C-terminus. It is primarily located within the intracellular trafficking network, including the endoplasmic reticulum (ER), Golgi apparatus, and ER-Golgi intermediate compartment (ERGIC). Computational and biochemical studies suggest that the E protein can form dimers, trimers, or pentamers, with a native pentameric structure proposed as a voltage-gated channel (14, 31). Moreover, the E protein functions as an ion channeling viroporin, facilitating ion transport across membranes. It interacts with various host proteins, including those associated with the host cell membrane, Golgi apparatus, and endoplasmic reticulum (14, 48). The E protein of SARS-CoV-2, with its distinct structural characteristics and functional roles, holds great promise as a target for antiviral therapy.
SARS-CoV-2 membrane protein
The M protein of SARS-CoV-2 is the most abundant protein found in the viral membrane. It consists of approximately 230 amino acids and possesses a short N-terminal domain, three transmembrane domains, and a carboxy-terminal domain located inside the viral particle. Dimeric complexes of the M protein are present within the virion envelope. The protein further assembles into higher order oligomers (27). Notably, an amphipathic region at the end of the third transmembrane domain is a characteristic feature shared by M proteins (27, 47).
The M protein plays crucial roles in maintaining virion size and shape, as well as facilitating the assembly of other structural proteins, including S, E, and N proteins (49). It also engages in interactions with accessory and non-structural proteins, which are essential for viral structural protein processing, modification, and trafficking contributing to viral particle assembly and egress, impacting the overall virus life cycle (50). Due to its critical functions and interactions, the M protein represents a potential target for therapeutic interventions against SARS-CoV-2.
Non-structural proteins associated with virus particles: ORF3a and ORF7a proteins
The ORF3a protein of SARS-CoV-2 is a viroporin ion channel that exerts various effects on the virus life cycle, playing a significant role in modulating autophagy, viral replication, and viral release (21, 32). ORF3a is a 275-amino acid protein with three transmembrane domains, an N-terminal region, and a C-terminal region. It forms dimers and exhibits a distinctive structure that contributes to its functional properties
The SARS-CoV-2 ORF7a protein, comprising 121 amino acid, is a type-I transmembrane protein involved in immunomodulation, particularly in CD14 monocytes (34, 51). It consists of an N-terminal signaling region, an Ig-like ectodomain, a hydrophobic transmembrane domain, and an ER retention motif. The Ig-like domain, known for its diverse actions in intercellular adhesion, identification, and binding, exhibits a conserved beta-sandwich fold with seven beta-strands, organized into two beta sheets. The beta sheet structure is stabilized by two disulfide bonds (34).
Quantification of protein molecules in SARS-CoV-2 virus particles
While several studies have characterized the biochemical properties of structural and non-structural proteins encoded by SARS-CoV-2, the precise number of each protein in virus particles remains unclear. Bar-On et al. (18) estimated the number of molecules of S, N, M, and E proteins based on data from SARS-CoV and related viruses (18). 100 trimers are estimated of the spike protein per virus particle, translating to 300 monomers of Spike on the surface of each virion. The number of N, M, and E proteins was estimated to be around 1000, 2000, and below 20, respectively. However, cryo-electron microscopy studies by Laue et al. (52) and Ogando et al. (53) revealed differences in the number of spike molecules between SARS-CoV and SARS-CoV-2 (52, 53). The median number of spike proteins was found to be 25 on SARS-CoV-2 virions and 32 on SARS-CoV virions. Previous reports for SARS-CoV indicated a higher number of around 65 spikes per virion though this was with a different isolate.
Bezstarosti et al. (54) employing mass spectrometry approaches proposed that 90% of the SARS-CoV-2 proteome consists of N protein (54). Furthermore, it is known that the N protein forms complexes with genomic RNA, designated as ribonucleoprotein (RNP) complexes, with approximately 30-35 RNPs per virion. However, the exact quantification of RNPs in SARS-CoV-2 virus particles requires further investigation. It should be noted that currently there is no information available regarding the number of molecules of accessory proteins present in the virus particles.
In summary, while several studies have provided estimates of protein molecules within SARS-CoV-2 virus particles, there are discrepancies and uncertainties regarding the precise numbers. Comparative studies with related coronaviruses and advanced techniques such as cryo-electron microscopy and mass spectrometry contribute to our understanding of protein quantification in SARS-CoV-2, providing valuable insights into visual structure and biology.
Enhancing the sensitivity of rapid antigen tests for SARS-CoV-2 detection
The containment of SARS-CoV-2 transmission relies on the timely identification of infection status in the individuals through rapid screening tests. Initially, diagnostic tests predominantly utilized RT-PCR targeting specific genes (N, E, S, and RNA-dependent RNA polymerase (Rd-Rp) of the viral genome (55, 56). However, the requirement for technical expertise, long waiting times, high costs, and necessary infrastructure limited the accessibility of these nucleic acid-based molecular tests. To address this gap, point-of-care (POC) rapid antigen tests (RATs) targeting viral antigens were developed, offering flexibility in testing locations without the need for specialized skills. These tests rely on the detection of viral protein(s) in patient specimens through antibody interactions. Despite their advantages, antigen tests have been reported to exhibit lower sensitivity compared to nucleic acid-based tests (6, 7). To enhance the sensitivity of antigen tests, monoclonal antibodies with high affinity binding to target proteins may provide a better strategy and has already been shown to have potential for success (10, 57–60).
Optimizing antigen targets for effective detection of SARS-CoV-2
Rapid antigen tests (RATs) have become pivotal in the timely detection of SARS-CoV-2 infection. Currently, available RATs target the N protein, while in some cases, both N and S proteins are targeted. The N protein, with its high abundance in SARS-CoV-2 viral particles, is an ideal target for detecting the virus (43, 46, 61–63). Additionally, the conserved nature of the N protein within the coronavirus family suggests its potential utility in detecting newly emerging variants. Test utilizing a combination of monoclonal and polyclonal antibodies as capturing and detection reagents showed a higher sensitivity than tests using only monoclonal antibodies. In contrast, the S protein as a target antigen for virus detection may present disadvantages due to its lower abundance in viral particles as well as the high rate of genetic variations in the S protein, which can compromise its interaction with diagnostic antibodies used for detection. A strategy involving protein-protein interactions between the S protein and ACE2 has been considered for developing robust diagnostic tests (9). While current vaccines such as mRNA vaccines (BNT162b2 and mRNA-1273), and adenoviral vector vaccines (ChAdOx1 nCoV-19), are designed for spike protein expression in cells of vaccinees, the low-level expression for a limited-time allows for S protein to remain a viable antigen for virus detection (23).
Exploring the potential of M protein as an antigen target for enhanced detection of SARS-CoV-2
The M protein exhibits the highest number of molecules in the envelope of virus particles, which may make it a promising candidate for detection assays along with N protein (27). Hence, M protein may serve as an additional antigen target for SARS-CoV-2 detection. Given that the antigen assays are unable to rely on signal amplification as occurs with nucleic acid-based detection, targeting high expression antigens within virions may be necessary for improved sensitivity. Therefore, a combination of N and M proteins as targets may offer improved sensitivity due to their abundance in virus particles, as summarized in Table 2. As noted earlier, M protein consists of multiple membrane spanning domains. This feature may pose problems in the expression and purification of protein for antigen assays. Alternatively, the amino acids representing the loop region between the transmembrane domains, which are exposed outside the particle, could be tested for their potential as antigen in the assays. The utilization of a RAT based on combined N and M proteins could improve the accuracy and efficiency of viral infection detection, contributing to better management and control of diseases in addition to proving valuable in the detection of emerging coronaviruses in the future.
Insights into platform technologies and antibody interactions for COVID-19 immunotherapy and vaccine development
The development of COVID-19 vaccines has relied on various platform technologies to combat the Covid pandemic. These include inactivated SARS-CoV-2 vaccines, nucleic acid vaccines (DNA and mRNA) (64–67), vectored virus vaccines engineered to express the spike protein, and virus-like particle vaccines displaying the S protein on their outer surface. Inactivated virus vaccines have been widely used in the development of vaccines for viral diseases such as flu, yellow fever, and rabies, employing inactivation agents like formaldehyde or β-propiolactone (68–72). Figure 3 outlines the various vaccine technologies for SARS-CoV-2 virus.
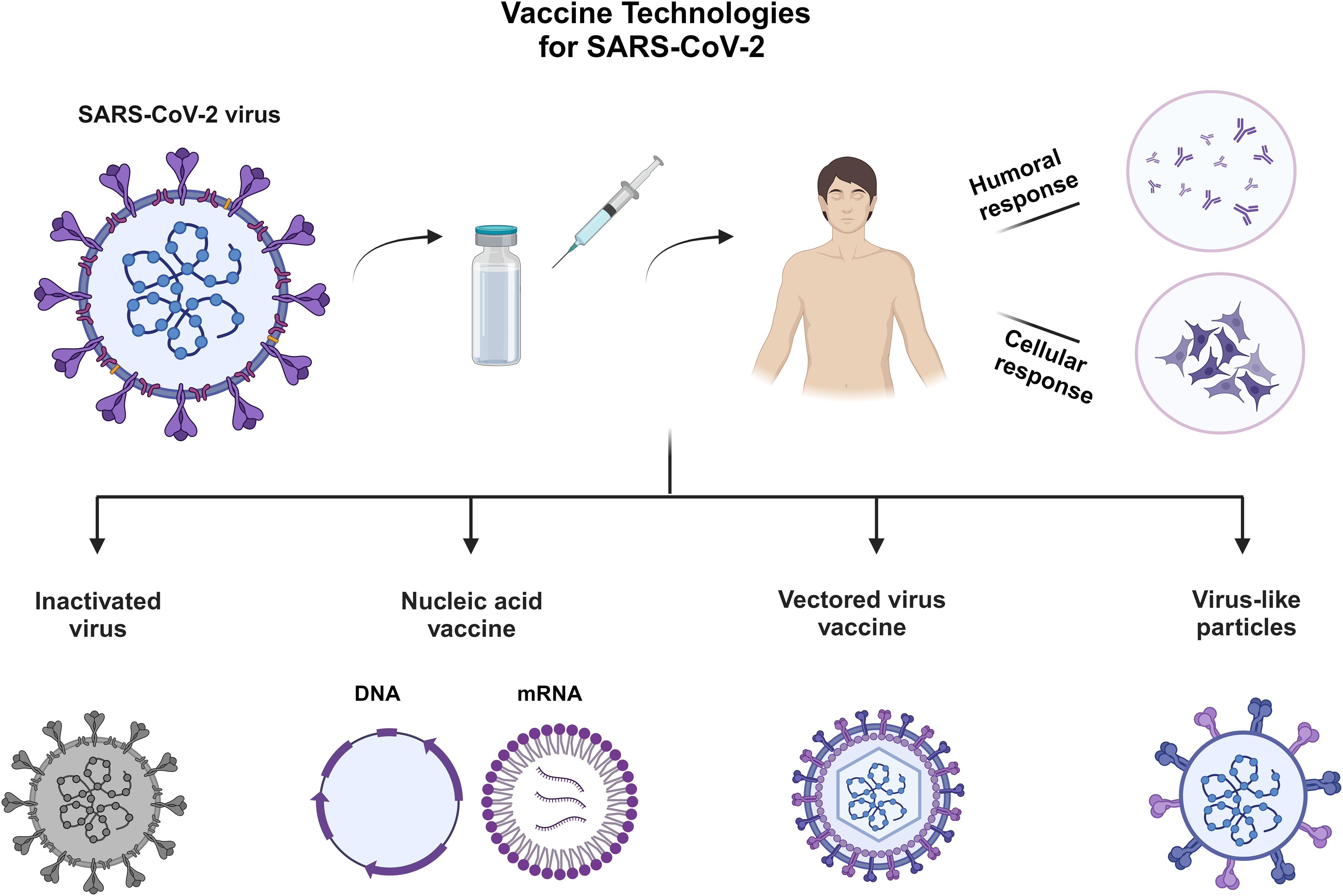
Figure 3. Vaccine technologies that have been applied for the development of vaccines for SARS-CoV-2. Created with BioRender.com.
Antibody neutralization challenges with SARS-CoV-2 variants in vaccinated individuals: insights from a study on Pfizer and AstraZeneca vaccines
The emergence of the Delta variant of COVID-19 has presented new challenges in terms of antibody neutralization, particularly for individuals who have received COVID-19 vaccinations. A study conducted on a group of 59 vaccinated individuals aimed to assess the neutralizing antibody levels against the Delta variant at different timepoints (73). The study participants were divided into two cohorts based on the vaccine they received: BNT162b2 (Pfizer), a nucleoside-modified mRNA expressing a prefusion-stabilized spike protein (16 individuals) and AZD1222 (AstraZeneca) a non-replicating ChAdOx1 Vector Vaccine expressing spike protein (43 individuals) (24). The neutralizing antibody levels were measured after a single dose and multiple time intervals following the second dose. The results showed that after a single dose, a relatively small proportion of the BNT162b2- cohort (13%) and AZD1222 cohort (9%) were able to neutralize the Delta variant (74, 75). However, five weeks after the administration of the second dose, both groups exhibited high levels of neutralization. Specifically, the BNT162b2 group demonstrated 94% neutralization, while the AZD1222 group showed 95% neutralization. It is worth noting that only the BNT162b2 group was monitored up to 13 weeks after the second dose, and their neutralization efficacy against the Delta variant decreased to 85%. This finding suggests a potential decline in neutralizing antibody levels over time, emphasizing the importance of ongoing monitoring and potential need for booster doses.
Furthermore, the study also examined the neutralization efficacy against the Beta variant. After the first dose, both the BNT162b2 and AZD1222 groups displayed low levels of neutralization against the Beta variant, with only 4% and 6% efficacy, respectively. Even after the second dose, the neutralization efficacy against the Beta variant remained significantly lower compared to the Delta variant. The BNT162b2 group showed 46% efficacy against the Beta variant 13 weeks after the second dose (73). These findings underscore the importance of considering the impact of emerging variants on vaccine efficacy. The study highlights the need for two doses to achieve high levels of neutralization against the Delta variant for both BNT162b2r and AZD1222 vaccines. Additionally, it suggests that the new variants may pose challenges in terms of neutralization, even after the administration of two or three doses. The study emphasizes the importance of considering the impact of emerging variants on vaccine efficacy and highlights the need for two or three doses to achieve high levels of neutralization against the variant for both BNT162b2 and AZD1222 vaccines. The findings suggest potential challenges in neutralizing the Beta variant, indicating the need for continued research and surveillance to address the neutralization of emerging variants. As the COVID-19 landscape continues to evolve, ongoing research and surveillance will be essential to monitor the effectiveness of existing vaccines and develop strategies, such as booster doses, to address emerging variants and maintain optimal protection against the virus.
Status of viral structural proteins in inactivated viruses
It has been reported that inactivated virus vaccines efficacy rates are different with different viruses. In the case of SARS-CoV-2, the efficacy of inactivated virus vaccines is reported to be at a reduced level in comparison to mRNA vaccines. Inactivated virus vaccines, such as Sinovac and Sinopharm, have reported efficacy rates of 51% and 79%, respectively. These lower rates may be attributed to alterations in viral proteins resulting from purification and inactivation methods. Liu et al. (11) analyzed inactivated viruses upon treatment with β-propiolactone and the virus particles were found to be in spherical to pleomorphic shape (11). Furthermore, it was shown that majority of spike protein existed in the post-fusion state, indicating the release of S1 from S2. The prefusion conformation state of the spike protein is characterized by a club-like structure with RBD on the top and sequestering of S2. RBD in the open conformation binds to ACE2 and the club shaped structure changes to a thin and long nail-like structure. The post-fusion conformation of the spike protein is achieved only once it has engaged with the ACE2 receptor on the host cell and interacted with host cell derived proteases. Both cryo-EM and tomography studies of inactivated SARS-CoV-2 vaccine revealed that around 74% of spike proteins were in a post-fusion state on virus particles (11). Based on imaging of intact SARS-CoV-2 viral particles, Ke et al. (76) noted predominant prefusion conformation and a minority of spike protein in a post-fusion conformation (76). The changes in spike protein on the surface of virus particles, likely the result of the inactivation and/or virus purification process, are of concern because the vaccine generated immune responses may not confer protection against infection.
Developments in antibody therapeutics
In the ongoing pursuit of effective therapeutic strategies against SARS-CoV-2, numerous monoclonal antibodies and nanobodies have been developed to target the spike protein. These antibodies were meticulously characterized for their interactions with the spike protein, specifically aiming to disrupt the binding between the RBD and ACE2, thereby impeding virus entry into host cells (77, 78). A recent study conducted by Tragni et al. (79) employed a computational approach to assess the interaction energies of 14 antibodies and 5 nanobodies (79). Among the antibodies, five were derived from SARS-CoV-2 convalescent patients, and a humanized antibody, ADG20, currently undergoing global clinical trials, was also included (79). Structural alignment was performed with 3D models of the spike RBD from seven variants, including the recently crystallized structures of Omicron B.1.1.529 and BA.4/5, using the Wuhan spike RBD as a reference. The modeling procedure facilitated the calculation of free energy of interaction between Spike RBD and antibodies or nanobodies. Four conformations of the trimeric spike protein were considered (79). The findings revealed that none of the investigated antibodies could simultaneously bind to all three RBDs of a spike trimer. Fab antibody portions exhibited a preference for interacting with an RBD in the up conformation. Contrary to expectations, only two out of the five investigated nanobodies demonstrated the ability to bind all three RBDs simultaneously, specifically when the RBDs were in either all-up or all-down conformation. These observations provide insights into the current limitations of antibodies targeting the spike protein and emphasize the importance of nanobodies in investigational therapeutics, particularly concerning structural orientation and spike flexibility (79).
Engineered protein and small molecule as therapeutics for SARS-CoV-2
Other novel approaches involve the use of soluble or engineered ACE2, or ACE2 “decoys”, to exploit the high affinity interaction between viral S protein and ACE2 receptors to inhibit viral entry and treat infection (25, 80, 81). Monteil et al. (80) was able to show enhanced binding affinity of SARS-CoV-2 variants Alpha, Gamma, Delta, and Omicron to recombinant soluble human ACE2 (80). The significance of their findings was augmented by the demonstrated neutralization of SARS-CoV-2 infection by soluble ACE2, against multiple variants of concern in multiple cell lines (80). They found that the clinical grade soluble ACE2 was highly effective in reducing viral load, and that the degree of reduction corresponded with the Spike RBD/ACE2 binding affinities revealed earlier through surface plasmon resonance.
Another study by Torchia et al. (25) describes the effectiveness of engineered ACE2-Fc decoys in neutralization of SARS-CoV-2 viral infection in vivo (25). Their findings suggest that neutralization is not only mediated through competitive inhibition but also through the structural shift in the viral S protein upon binding to ACE2 decoys – specifically a permanent transition from the “prefusion” state whereby S1 and S2 subunits are noncovalently bonded to a “post-fusion” separation of the S1 and S2 subunits and subsequent “refolding” of the S2 subunit. Such a change renders the virus particle incapable of rebinding to ACE2 on host cells and therefore unable to enter and infect cells (25). It was found that efficacy of viral clearance by the ACE2-Fc compounds evaluated in this study corresponded with affinity of each compound for the S protein trimer. Other studies have demonstrated that further modifications, such as trimerizing the engineered ACE2, can enhance binding affinity of the decoys for spike protein (39). The findings in these papers are highly relevant to the development of therapeutics against SARS-CoV-2 infection as a primary concern is identifying and developing therapeutic approaches which will remain effective against future strains of the virus. Furthermore, ACE2 decoys as “universal” agents in blocking the Spike/ACE2 interaction are advantageous as their use is not limited to SARS-CoV-2 infection; they are effective against any novel coronaviruses and variants utilizing the highly conserved Spike/ACE2 interaction for viral entry (80).
Compared to protein-based therapeutics, there are several advantages to the small molecule inhibitors against SARS-CoV-2 including ease of production and entry into cells. In addition, combination therapy approach against multiple targets is also possible with this system. Based on the number of molecules present in the virus particles, efforts should be directed using proteins such as S, N and M as targets for the identification of small molecule inhibitors.
COVID-19 testing: engineering solutions and perspectives
Identifying the structure and sequence of the SARS-CoV-2 virus at an early stage of the pandemic allowed for a quick response against the disease. A significant part of this response was the ability to accurately diagnose the disease in patients through rapid testing programs (82). Furthermore, the review by (78, 83) provides a comprehensive overview of diagnostic strategies and testing methodologies for COVID-19. The focus lies on their role in disease transmission prevention, understanding epidemiology, case management, and transmission suppression (83). Two primary approaches dominate COVID-19 testing: nucleic acid-based assays (RT-PCR) and protein-based antigen assays. This review serves as a guiding framework for navigating diagnostic pathways amidst the pandemic’s evolving landscape. While these methods are well established, several engineering solutions have been developed to support existing diagnostic procedures. These novel approaches aim to provide alternative testing platforms that are user-friendly, fast, accurate, accessible, and most importantly, cost-effective. This section will highlight some of the methods used for COVID-19 testing as outlined in Figure 4.

Figure 4. Diagnostic techniques for detection of SARS-CoV-2 infection. Created with BioRender.com.
The lateral flow technique
In this technique, capillary forces laterally transport the fluid test sample through a membrane toward locations where the target genes, antibodies, or antigens are present (10, 84). In the case of SARS-CoV-2, S and N proteins are the most valuable biomarkers for diagnosis of COVID-19 (85). Typically, the targets are conjugated with visual reporters (nanoparticles or fluorescent reporters) (10, 84). The test line captures sample bound to targets while unbound targets are captured on the control line. The advantages of this technique are the low cost, high accessibility, and short duration (15-30 minutes) needed to complete the test (10). However, commercial test kits developed during the pandemic (home tests or at the point of care) targeting viral proteins had low sensitivity and specificity compared to RT-PCR (10).
Current prototypes are designed to target genes and antibodies. Several new devices developed have shown superior sensitivity and specificity, reaching up to 100% (60, 86–88). These new devices include the detection of ORF1ab, N, and E genes, as well as SARS-CoV-2 specific anti-spike IgM and IgG. Common to all prototypes is the ability to detect multiple biomarkers simultaneously alongside optimized visual reporters. In addition, some other prototypes are also employing pre-amplification, filtration, and concentration of the test samples to enhance detection accuracy (84).
Microfluidic devices
Microfluidic devices have gained significant attraction in recent decades as they are easy to manufacture and are used in various applications such as diagnosis, cell culture, and drug delivery as presented in Figure 4. Due to the channels’ microscale, the flow is characterized by a low ratio of inertial to viscous forces (Reynolds number), where the capillary forces are dominant and responsible for driving the fluid mechanically through the channels (89). This results in laminar flow, which is highly predictable and can therefore be readily controlled and modeled. Microfluidic device prototypes have been developed for COVID testing (82).
Using microfluidic devices as biosensors has demonstrated potential in COVID testing. Several prototypes used microfluidic devices to detect antibodies and proteins with electrical/electrochemical, optical, and fluorescence-based sensors (90). A significant limitation of such devices is the need for secondary equipment to electrically or visually amplify the signal (90). In contrast to the microfluidic devices themselves, these secondary devices are expensive, limiting the use of microfluidic devices to POC sites. To tackle such a problem, Li and Lillehaug proposed a smartphone-based diagnostic tool to detect and analyze data based on their microfluidic device, which showed high sensitivity and specificity (91). This approach has the potential to reduce the cost of secondary equipment needed alongside microfluidic devices while allowing for rapid and accurate test results. It can also enable microfluidic devices to be used as home-testing tools in addition to POC sites.
Conclusion: a blueprint to tackle future pandemics
The rapid emergence of the SARS-CoV-2 pandemic has underscored the urgent need for global preparedness against infectious diseases. Extensive research efforts after the pandemic started quickly led to an understanding of virus biology that informed the rapid development of vaccines and other therapeutics to slow the spread of SARS-CoV-2 and/or reduce the severity of COVID-19. These efforts saved millions of lives, reopened businesses and countries, and now provide a comprehensive blueprint for effectively combating future outbreaks. Key to this blueprint are two crucial areas: early detection of infected individuals and the development of therapeutics and vaccines. By leveraging advanced technologies such as scanning electron microscopy, cryo-electron microscopy, mass spectrometry, and sequence analysis, we can gain valuable insights into the composition and behavior of a virus or other pathogen. These insights can then pave the way for the development of assays for rapid detection of viral antigens and nucleic acids, enabling early identification of infected individuals. Furthermore, these analyses of viral structural proteins provide useful information to aid the development of more effective vaccines and therapeutics.
In particular, understanding the dynamic conformational changes in proteins such as the spike protein of SARS-CoV-2 has informed the design of mRNA vaccines and highlighted strategies to enhance the immunogenicity and efficacy of traditional inactivated virus vaccines. Improved purification and inactivation methods are paramount to maintaining the consistency and reliability of vaccine formulations. Moving forward, further research is warranted to optimize antigen capture strategies, identify protein modifications to enhance vaccine efficacy, and innovate novel therapeutic approaches. By building upon the knowledge gained from the study of SARS-CoV-2, we can fortify our defenses against future pandemics, whether caused by coronaviruses or other infectious agents. This summary underscores the importance of ongoing research and collaboration in safeguarding global health security.
Author contributions
CC: Resources, Writing – review & editing, Formal analysis. PI: Writing – review & editing, Resources, Data curation, Validation. EL: Data curation, Resources, Writing – review & editing. ZX: Resources, Writing – review & editing. YP: Resources, Writing – review & editing. LM: Resources, Writing – review & editing. SK: Resources, Writing – review & editing. AS: Resources, Writing – review & editing. KM: Resources, Writing – review & editing, Project administration, Writing – original draft.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
All figures were created with “BioRender.com” and templates used can be retrieved from “https://app.biorender.com/biorender-templates” as follows: Figure 1 was adapted from “Human Coronavirus Structure” (2023), Figure 2 was adapted from “COVID-19 Drug Mechanism of Action (Layout)” (2023), Figure 3 was adapted from “Approaches to Viral Vaccine Development” (2023), and Figure 4 was adapted from “COVID-19 Serologic Diagnostic Test through Antibody Detection” and “COVID-19 Fact Sheet” (2023).
Conflict of interest
Authors CC, PI, EL, YP, SK, KM were employed by the company GeneOne Life Science Inc. Author AS was employed by the company NanoBio Diagnostics.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Cevik M, Kuppalli K, Kindrachuk J, Peiris M. Virology, transmission, and pathogenesis of SARS-CoV-2. BMJ. (2020) 371:m3862. doi: 10.1136/bmj.m3862
2. Rossi G, Salmanton-García J, Cattaneo C, Marchesi F, Dávila-Valls J, Martín-Pérez S, et al. Age, successive waves, immunization, and mortality in elderly COVID-19 hematological patients: EPICOVIDEHA findings. Int J Infect Dis. (2023) 137:98–110. doi: 10.1016/j.ijid.2023.10.013
3. Chan JF, Yuan S, Kok KH, To KK, Chu H, Yang J, et al. A familial cluster of pneumonia associated with the 2019 novel coronavirus indicating person-to-person transmission: a study of a family cluster. Lancet. (2020) 395:514–23. doi: 10.1016/S0140-6736(20)30154-9
4. Ghinai I, Mcpherson TD, Hunter JC, Kirking HL, Christiansen D, Joshi K, et al. First known person-to-person transmission of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) in the USA. Lancet. (2020) 395:1137–44. doi: 10.1016/S0140-6736(20)30607-3
5. Huang C, Wang Y, Li X, Ren L, Zhao J, Hu Y, et al. Clinical features of patients infected with 2019 novel coronavirus in Wuhan, China. Lancet. (2020) 395:497–506. doi: 10.1016/S0140-6736(20)30183-5
6. Mak GC, Cheng PK, Lau SS, Wong KK, Lau CS, Lam ET, et al. Evaluation of rapid antigen test for detection of SARS-CoV-2 virus. J Clin Virol. (2020) 129:104500. doi: 10.1016/j.jcv.2020.104500
7. Yamayoshi S, Sakai-Tagawa Y, Koga M, Akasaka O, Nakachi I, Koh H, et al. Comparison of rapid antigen tests for COVID-19. Viruses. (2020) 12(12):1420. doi: 10.3390/v12121420
8. Li D, Li J. Immunologic testing for SARS-CoV-2 infection from the antigen perspective. J Clin Microbiol. (2021) 59(5):e02160-20. doi: 10.1128/JCM.02160-20
9. Mahmoudinobar F, Britton D, Montclare JK. Protein-based lateral flow assays for COVID-19 detection. Protein Eng Des Sel. (2021) 34:gzab010. doi: 10.1093/protein/gzab010
10. Budd J, Miller BS, Weckman NE, Cherkaoui D, Huang D, Decruz AT, et al. Lateral flow test engineering and lessons learned from COVID-19. Nat Rev Bioeng. (2023) 1:13–31. doi: 10.1038/s44222-022-00007-3
11. Liu C, Mendonca L, Yang Y, Gao Y, Shen C, Liu J, et al. The architecture of inactivated SARS-CoV-2 with postfusion spikes revealed by cryo-EM and cryo-ET. Structure. (2020) 28:1218–1224.e1214. doi: 10.1016/j.str.2020.10.001
12. Yao H, Song Y, Chen Y, Wu N, Xu J, Sun C, et al. Molecular architecture of the SARS-CoV-2 virus. Cell. (2020) 183:730–738.e713. doi: 10.1016/j.cell.2020.09.018
13. Calder LJ, Calcraft T, Hussain S, Harvey R, Rosenthal PB. Electron cryotomography of SARS-CoV-2 virions reveals cylinder-shaped particles with a double layer RNP assembly. Commun Biol. (2022) 5:1210. doi: 10.1038/s42003-022-04183-1
14. Jackson CB, Farzan M, Chen B, Choe H. Mechanisms of SARS-CoV-2 entry into cells. Nat Rev Mol Cell Biol. (2022) 23:3–20. doi: 10.1038/s41580-021-00418-x
15. Desai D, Khan AR, Soneja M, Mittal A, Naik S, Kodan P, et al. Effectiveness of an inactivated virus-based SARS-CoV-2 vaccine, BBV152, in India: a test-negative, case-control study. Lancet Infect Dis. (2022) 22:349–56. doi: 10.1016/S1473-3099(21)00674-5
16. Law M, Ho SSH, Tsang GKC, Ho CMY, Kwan CM, Yan VKC, et al. Efficacy and effectiveness of inactivated vaccines against symptomatic COVID-19, severe COVID-19, and COVID-19 clinical outcomes in the general population: a systematic review and meta-analysis. Lancet Reg Health West Pac. (2023) 37:100788. doi: 10.1016/j.lanwpc.2023.100788
17. Liu DX, Fung TS, Chong KK, Shukla A, Hilgenfeld R. Accessory proteins of SARS-CoV and other coronaviruses. Antiviral Res. (2014) 109:97–109. doi: 10.1016/j.antiviral.2014.06.013
18. Bar-On YM, Flamholz A, Phillips R, Milo R. SARS-coV-2 (COVID-19) by the numbers. Elife. (2020) 9:e57309. doi: 10.7554/eLife.57309
19. Kim D, Lee JY, Yang JS, Kim JW, Kim VN, Chang H. The architecture of SARS-CoV-2 transcriptome. Cell. (2020) 181:914–921 e910. doi: 10.1016/j.cell.2020.04.011
20. Redondo N, Zaldivar-Lopez S, Garrido JJ, Montoya M. SARS-CoV-2 accessory proteins in viral pathogenesis: knowns and unknowns. Front Immunol. (2021) 12:708264. doi: 10.3389/fimmu.2021.708264
21. Zhang J, Ejikemeuwa A, Gerzanich V, Nasr M, Tang Q, Simard JM, et al. Understanding the role of SARS-CoV-2 ORF3a in viral pathogenesis and COVID-19. Front Microbiol. (2022) 13:854567. doi: 10.3389/fmicb.2022.854567
22. Koonin EV, Starokadomskyy P. Are viruses alive? The replicator paradigm sheds decisive light on an old but misguided question. Stud Hist Philos Biol BioMed Sci. (2016) 59:125–34. doi: 10.1016/j.shpsc.2016.02.016
23. Andrews N, Stowe J, Kirsebom F, Toffa S, Rickeard T, Gallagher E, et al. Covid-19 vaccine effectiveness against the omicron (B.1.1.529) variant. N Engl J Med. (2022) 386:1532–46. doi: 10.1056/NEJMoa2119451
24. Belik M, Jalkanen P, Lundberg R, Reinholm A, Laine L, Vaisanen E, et al. Comparative analysis of COVID-19 vaccine responses and third booster dose-induced neutralizing antibodies against Delta and Omicron variants. Nat Commun. (2022) 13:2476. doi: 10.1038/s41467-022-30162-5
25. Torchia JA, Tavares AH, Carstensen LS, Chen D-Y, Huang J, Xiao T, et al. Optimized ACE2 decoys neutralize antibody-resistant SARS-CoV-2 variants through functional receptor mimicry and treat infection in vivo. Sci Adv. (2022) 8:eabq6527. doi: 10.1126/sciadv.abq6527
26. Verbeke R, Hogan MJ, Lore K, Pardi N. Innate immune mechanisms of mRNA vaccines. Immunity. (2022) 55:1993–2005. doi: 10.1016/j.immuni.2022.10.014
27. Zhang Z, Nomura N, Muramoto Y, Ekimoto T, Uemura T, Liu K, et al. Structure of SARS-CoV-2 membrane protein essential for virus assembly. Nat Commun. (2022) 13:4399. doi: 10.1038/s41467-022-32019-3
28. Jahirul Islam M, Nawal Islam N, Siddik Alom M, Kabir M, Halim MA. A review on structural, non-structural, and accessory proteins of SARS-CoV-2: Highlighting drug target sites. Immunobiology. (2023) 228:152302. doi: 10.1016/j.imbio.2022.152302
29. Markov PV, Ghafari M, Beer M, Lythgoe K, Simmonds P, Stilianakis NI, et al. The evolution of SARS-CoV-2. Nat Rev Microbiol. (2023) 21:361–79. doi: 10.1038/s41579-023-00878-2
30. Giannessi F, Aiello A, Franchi F, Percario ZA, Affabris E. The role of extracellular vesicles as allies of HIV, HCV and SARS viruses. Viruses. (2020) 12(5):571. doi: 10.3390/v12050571
31. Cao C, Cai Z, Xiao X, Rao J, Chen J, Hu N, et al. The architecture of the SARS-CoV-2 RNA genome inside virion. Nat Commun. (2021) 12:3917. doi: 10.1038/s41467-021-22785-x
32. Bojkova D, Klann K, Koch B, Widera M, Krause D, Ciesek S, et al. Proteomics of SARS-CoV-2-infected host cells reveals therapy targets. Nature. (2020) 583:469–72. doi: 10.1038/s41586-020-2332-7
33. Cubuk J, Alston JJ, Incicco JJ, Singh S, Stuchell-Brereton MD, Ward MD, et al. The SARS-CoV-2 nucleocapsid protein is dynamic, disordered, and phase separates with RNA. Nat Commun. (2021) 12:1936. doi: 10.1038/s41467-021-21953-3
34. Nelson CA, Pekosz A, Lee CA, Diamond MS, Fremont DH. Structure and intracellular targeting of the SARS-coronavirus Orf7a accessory protein. Structure. (2005) 13:75–85. doi: 10.1016/j.str.2004.10.010
35. Huang Y, Yang C, Xu X-F, Xu W, Liu S-W. Structural and functional properties of SARS-CoV-2 spike protein: potential antivirus drug development for COVID-19. Acta Pharmacologica Sin. (2020) 41:1141–9. doi: 10.1038/s41401-020-0485-4
36. Korber B, Fischer WM, Gnanakaran S, Yoon H, Theiler J, Abfalterer W, et al. Tracking changes in SARS-CoV-2 spike: evidence that D614G increases infectivity of the COVID-19 virus. Cell. (2020) 182:812–827.e819. doi: 10.1016/j.cell.2020.06.043
37. Dokainish HM, Sugita Y. Structural effects of spike protein D614G mutation in SARS-CoV-2. Biophys J. (2023) 122:2910–20. doi: 10.1016/j.bpj.2022.11.025
38. Pierri CL. SARS-CoV-2 spike protein: flexibility as a new target for fighting infection. Signal Transduction Targeted Ther. (2020) 5:254. doi: 10.1038/s41392-020-00369-3
39. Guo L, Bi W, Wang X, Xu W, Yan R, Zhang Y, et al. Engineered trimeric ACE2 binds viral spike protein and locks it in “Three-up” conformation to potently inhibit SARS-CoV-2 infection. Cell Res. (2021) 31:98–100. doi: 10.1038/s41422-020-00438-w
40. Mercurio I, Tragni V, Busto F, De Grassi A, Pierri CL. Protein structure analysis of the interactions between SARS-CoV-2 spike protein and the human ACE2 receptor: from conformational changes to novel neutralizing antibodies. Cell Mol Life Sci. (2021) 78:1501–22. doi: 10.1007/s00018-020-03580-1
41. Dokainish HM, Re S, Mori T, Kobayashi C, Jung J, Sugita Y. The inherent flexibility of receptor binding domains in SARS-CoV-2 spike protein. Elife. (2022) 11:e75720. doi: 10.7554/eLife.75720
42. Malik YA. Covid-19 variants: Impact on transmissibility and virulence. Malays J Pathol. (2022) 44:387–96.
43. Tragni V, Preziusi F, Laera L, Onofrio A, Mercurio I, Todisco S, et al. Modeling SARS-CoV-2 spike/ACE2 protein-protein interactions for predicting the binding affinity of new spike variants for ACE2, and novel ACE2 structurally related human protein targets, for COVID-19 handling in the 3PM context. EPMA J. (2022) 13:149–75. doi: 10.1007/s13167-021-00267-w
44. Mittal A, Khattri A, Verma V. Structural and antigenic variations in the spike protein of emerging SARS-CoV-2 variants. PloS Pathog. (2022) 18:e1010260. doi: 10.1371/journal.ppat.1010260
45. Rahman MS, Islam MR, Alam ASMRU, Islam I, Hoque MN, Akter S, et al. Evolutionary dynamics of SARS-CoV-2 nucleocapsid protein and its consequences. J Med Virol. (2021) 93:2177–95. doi: 10.1002/jmv.26626
46. Carlson CR, Asfaha JB, Ghent CM, Howard CJ, Hartooni N, Safari M, et al. Phosphoregulation of phase separation by the SARS-CoV-2 N protein suggests a biophysical basis for its dual functions. Mol Cell. (2020) 80:1092–1103.e1094. doi: 10.1016/j.molcel.2020.11.025
47. Klein S, Cortese M, Winter SL, Wachsmuth-Melm M, Neufeldt CJ, Cerikan B, et al. SARS-CoV-2 structure and replication characterized by in situ cryo-electron tomography. Nat Commun. (2020) 11:5885. doi: 10.1038/s41467-020-19619-7
48. Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, et al. Structural basis of receptor recognition by SARS-CoV-2. Nature. (2020) 581:221–4. doi: 10.1038/s41586-020-2179-y
49. Fehr AR, Perlman S. Coronaviruses: an overview of their replication and pathogenesis. Methods Mol Biol. (2015) 1282:1–23. doi: 10.1007/978-1-4939-2438-7_1
50. Siu YL, Teoh KT, Lo J, Chan CM, Kien F, Escriou N, et al. The M, E, and N structural proteins of the severe acute respiratory syndrome coronavirus are required for efficient assembly, trafficking, and release of virus-like particles. J Virol. (2008) 82:11318–30. doi: 10.1128/JVI.01052-08
51. Zhou Z, Huang C, Zhou Z, Huang Z, Su L, Kang S, et al. Structural insight reveals SARS-CoV-2 ORF7a as an immunomodulating factor for human CD14(+) monocytes. iScience. (2021) 24:102187. doi: 10.1016/j.isci.2021.102187
52. Laue M, Kauter A, Hoffmann T, Moller L, Michel J, Nitsche A. Morphometry of SARS-CoV and SARS-CoV-2 particles in ultrathin plastic sections of infected Vero cell cultures. Sci Rep. (2021) 11:3515. doi: 10.1038/s41598-021-82852-7
53. Ogando NS, Dalebout TJ, Zevenhoven-Dobbe JC, Limpens R, van der Meer Y, Caly L, et al. SARS-coronavirus-2 replication in Vero E6 cells: replication kinetics, rapid adaptation and cytopathology. J Gen Virol. (2020) 101:925–40. doi: 10.1099/jgv.0.001453
54. Bezstarosti K, Lamers MM, Doff W, Wever PC, Thai KTD, Van Kampen JJA, et al. Targeted proteomics as a tool to detect SARS-CoV-2 proteins in clinical specimens. PloS One. (2021) 16:e0259165. doi: 10.1371/journal.pone.0259165
55. Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. (2003) 361:1319–25. doi: 10.1016/S0140-6736(03)13077-2
56. Poon LL, Chan KH, Wong OK, Yam WC, Yuen KY, Guan Y, et al. Early diagnosis of SARS coronavirus infection by real time RT-PCR. J Clin Virol. (2003) 28:233–8. doi: 10.1016/j.jcv.2003.08.004
57. Axiaq A, Almohtadi A, Massias SA, Ngemoh D, Harky A. The role of computed tomography scan in the diagnosis of COVID-19 pneumonia. Curr Opin Pulm Med. (2021) 27:163–8. doi: 10.1097/MCP.0000000000000765
58. Corti D, Purcell LA, Snell G, Veesler D. Tackling COVID-19 with neutralizing monoclonal antibodies. Cell. (2021) 184:3086–108. doi: 10.1016/j.cell.2021.05.005
59. Kassania SH, Kassanib PH, Wesolowskic MJ, Schneidera KA, Detersa RJB. Automatic detection of coronavirus disease (COVID-19) in X-ray and CT images: a machine learning based approach. Biocybern Biomed Eng. (2021) 41:867–79. Engineering, B. doi: 10.1016/j.bbe.2021.05.013
60. Liu H, Dai E, Xiao R, Zhou Z, Zhang M, Bai Z, et al. Development of a SERS-based lateral flow immunoassay for rapid and ultra-sensitive detection of anti-SARS-CoV-2 IgM/IgG in clinical samples. Sens Actuators B Chem. (2021) 329:129196. doi: 10.1016/j.snb.2020.129196
62. López-Muñoz AD, Kosik I, Holly J, Yewdell JW. Cell surface SARS-CoV-2 nucleocapsid protein modulates innate and adaptive immunity. Sci Adv. (2022) 8:eabp9770. doi: 10.1126/sciadv.abp9770
63. Song W, Fang Z, Ma F, Li J, Huang Z, Zhang Y, et al. The role of SARS-CoV-2 N protein in diagnosis and vaccination in the context of emerging variants: present status and prospects. Front Microbiol. (2023) 14:1217567. doi: 10.3389/fmicb.2023.1217567
64. Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, et al. Safety and efficacy of the BNT162b2 mRNA covid-19 vaccine. N Engl J Med. (2020) 383:2603–15. doi: 10.1056/NEJMoa2034577
65. Smith TRF, Patel A, Ramos S, Elwood D, Zhu X, Yan J, et al. Immunogenicity of a DNA vaccine candidate for COVID-19. Nat Commun. (2020) 11:2601. doi: 10.1038/s41467-020-16505-0
66. Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, et al. Efficacy and safety of the mRNA-1273 SARS-coV-2 vaccine. N Engl J Med. (2021) 384:403–16. doi: 10.1056/NEJMoa2035389
67. Kim WJ, Roberts CC, Song JY, Yoon JG, Seong H, Hyun HJ, et al. Safety and immunogenicity of the bi-cistronic GLS-5310 COVID-19 DNA vaccine delivered with the GeneDerm suction device. Int J Infect Dis. (2023) 128:112–20. doi: 10.1016/j.ijid.2022.12.037
68. Gao Q, Bao L, Mao H, Wang L, Xu K, Yang M, et al. Development of an inactivated vaccine candidate for SARS-CoV-2. Science. (2020) 369:77–81. doi: 10.1126/science.abc1932
69. Xia S, Duan K, Zhang Y, Zhao D, Zhang H, Xie Z, et al. Effect of an inactivated vaccine against SARS-CoV-2 on safety and immunogenicity outcomes: interim analysis of 2 randomized clinical trials. Jama. (2020) 324:951–60. doi: 10.1001/jama.2020.15543
70. Chen F, Seong Seo H, Ji HJ, Yang E, Choi JA, Yang JS, et al. Characterization of humoral and cellular immune features of gamma-irradiated influenza vaccine. Hum Vaccin Immunother. (2021) 17:485–96. doi: 10.1080/21645515.2020.1780091
71. Cajaraville A, Gomes MPB, Azamor T, Pereira RC, Neves P, De Luca PM, et al. Evaluation of two adjuvant formulations for an inactivated yellow fever 17DD vaccine candidate in mice. Vaccines (Basel). (2022) 11(1):73. doi: 10.3390/vaccines11010073
72. Zhao H, Li P, Bian L, Zhang W, Jiang C, Chen Y, et al. Immune response of inactivated rabies vaccine inoculated via intraperitoneal, intramuscular, subcutaneous and needle-free injection technology-based intradermal routes in mice. Int J Mol Sci. (2023) 24(17):13587. doi: 10.3390/ijms241713587
73. Lopez Bernal J, Andrews N, Gower C, Gallagher E, Simmons R, Thelwall S, et al. Effectiveness of covid-19 vaccines against the B.1.617.2 (Delta) variant. N Engl J Med. (2021) 385:585–94. doi: 10.1056/NEJMoa2108891
74. Meo SA, Bukhari IA, Akram J, Meo AS, Klonoff DC. COVID-19 vaccines: comparison of biological, pharmacological characteristics and adverse effects of Pfizer/BioNTech and Moderna Vaccines. Eur Rev Med Pharmacol Sci. (2021) 25:1663–9. doi: 10.26355/eurrev_202102_24877
75. Darweesh O, Khatab N, Kheder R, Mohammed T, Faraj T, Ali S, et al. Assessment of COVID-19 vaccination among healthcare workers in Iraq; adverse effects and hesitancy. PloS One. (2022) 17:e0274526. doi: 10.1371/journal.pone.0274526
76. Ke Z, Oton J, Qu K, Cortese M, Zila V, Mckeane L, et al. Structures and distributions of SARS-CoV-2 spike proteins on intact virions. Nature. (2020) 588:498–502. doi: 10.1038/s41586-020-2665-2
77. Brouwer PJM, Caniels TG, van der Straten K, Snitselaar JL, Aldon Y, Bangaru S, et al. Potent neutralizing antibodies from COVID-19 patients define multiple targets of vulnerability. Science. (2020) 369:643–50. doi: 10.1126/science.abc5902
78. Kevadiya BD, Machhi J, Herskovitz J, Oleynikov MD, Blomberg WR, Bajwa N, et al. Pharmacotherapeutics of SARS-CoV-2 infections. J Neuroimmune Pharmacol. (2021a) 16:12–37. doi: 10.1007/s11481-020-09968-x
79. Tragni V, Mercurio I, Paoletti DP, Onofrio A, Laera L, Cafferati Beltrame L, et al. Deconstructing SARS-CoV-2 neutralization: A modular molecular framework for computational design and comparison of antibodies and nanobodies targeting the spike RBD. J Med Virol. (2023) 95:e28875. doi: 10.1002/jmv.28875
80. Monteil V, Eaton B, Postnikova E, Murphy M, Braunsfeld B, Crozier I, et al. Clinical grade ACE2 as a universal agent to block SARS-CoV-2 variants. EMBO Mol Med. (2022) 14:e15230. doi: 10.15252/emmm.202115230
81. Kegler A, Drewitz L, Arndt C, Daglar C, Rodrigues Loureiro L, Mitwasi N, et al. A novel ACE2 decoy for both neutralization of SARS-CoV-2 variants and killing of infected cells. Front Immunol. (2023) 14. doi: 10.3389/fimmu.2023.1204543
82. Moulahoum H, Ghorbanizamani F, Zihnioglu F, Turhan K, Timur S. How should diagnostic kits development adapt quickly in COVID 19-like pandemic models? Pros and cons of sensory platforms used in COVID-19 sensing. Talanta. (2021) 222:121534. doi: 10.1016/j.talanta.2020.121534
83. Kevadiya BD, Machhi J, Herskovitz J, Oleynikov MD, Blomberg WR, Bajwa N, et al. Diagnostics for SARS-coV-2 infections. Nat Mater. (2021b) 20:593–605. doi: 10.1038/s41563-020-00906-z
84. Hsiao WW-W, Le T-N, Pham DM, Ko H-H, Chang H-C, Lee C-C, et al. Recent advances in novel lateral flow technologies for detection of COVID-19. Biosensors (Basel). (2021) 11:295. doi: 10.3390/bios11090295
85. Zhou Y, Wu Y, Ding L, Huang X, Xiong Y. Point-of-care COVID-19 diagnostics powered by lateral flow assay. Trends Analyt Chem. (2021) 145:116452. doi: 10.1016/j.trac.2021.116452
86. Wang C, Yang X, Gu B, Liu H, Zhou Z, Shi L, et al. Sensitive and simultaneous detection of SARS-CoV-2-specific IgM/IgG using lateral flow immunoassay based on dual-mode quantum dot nanobeads. Anal Chem. (2020) 92:15542–9. doi: 10.1021/acs.analchem.0c03484
87. Wang D, He S, Wang X, Yan Y, Liu J, Wu S, et al. Rapid lateral flow immunoassay for the fluorescence detection of SARS-CoV-2 RNA. Nat Biomed Eng. (2020) 4:1150–8. doi: 10.1038/s41551-020-00655-z
88. Zhu X, Wang X, Han L, Chen T, Wang L, Li H, et al. Multiplex reverse transcription loop-mediated isothermal amplification combined with nanoparticle-based lateral flow biosensor for the diagnosis of COVID-19. Biosens Bioelectron. (2020) 166:112437. doi: 10.1016/j.bios.2020.112437
89. Convery N, Gadegaard NJM. 30 years of microfluidics. Micro and Nano Engineering. (2019) 2:76–91. Engineering, N. doi: 10.1016/j.mne.2019.01.003
90. Tayyab M, Sami MA, Raji H, Mushnoori S, Javanmard M. Potential microfluidic devices for COVID-19 antibody detection at point-of-care (POC): A review. IEEE Sens J. (2020) 21:4007–17. doi: 10.1109/JSEN.2020.3034892
Keywords: SARS-CoV-2 viral structural proteins, infections and variants, clinical diagnostics, therapeutics, vaccine development
Citation: Chung C, Irudayaraj P, Lallow E, Xu Z, Park YK, Kudchodkar SB, Montaner LJ, Srinivasan A and Muthumani K (2024) An overview of SARS-CoV-2 viral proteins with relevance to improved diagnostic and therapeutic platforms. Front. Virol. 4:1399993. doi: 10.3389/fviro.2024.1399993
Received: 12 March 2024; Accepted: 09 September 2024;
Published: 03 October 2024.
Edited by:
Andreu Comas-Garcia, Autonomous University of San Luis Potosí, MexicoReviewed by:
George William Carnell, University of Cambridge, United KingdomAhmed M. Senan, Süleyman Demirel University, Türkiye
Copyright © 2024 Chung, Irudayaraj, Lallow, Xu, Park, Kudchodkar, Montaner, Srinivasan and Muthumani. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kar Muthumani, a211dGh1bWFuaUBnbWFpbC5jb20=
 Christopher Chung
Christopher Chung Pratiba Irudayaraj
Pratiba Irudayaraj Emran Lallow
Emran Lallow Ziyang Xu2
Ziyang Xu2 Sagar B. Kudchodkar
Sagar B. Kudchodkar Luis J. Montaner
Luis J. Montaner Alagarsamy Srinivasan
Alagarsamy Srinivasan Kar Muthumani
Kar Muthumani