- 1School of Animal Technology and Innovation, Institute of Agricultural Technology, Suranaree University of Technology, Nakhon Ratchasima, Thailand
- 2Postharvest Technology and Innovation in Animal Unit, Institute of Agricultural Technology, Suranaree University of Technology, Nakhon Ratchasima, Thailand
- 3Research and Development Institute Suranaree University of Technology, Nakhon Ratchasima, Thailand
The maintenance of host health and immune function is heavily dependent on the gut microbiota. However, the precise contribution of individual microbial taxa to regulating the overall functionality of the gut microbiome remains inadequately investigated. Chickens are commonly used as models for studying poultry gut microbiota, with high-throughput 16S rRNA sequencing has emerged as a valuable tool for assessing both its composition and functionality. The interactions between the gut’s microbial community and its host significantly influence health outcomes, disease susceptibility, and various mechanisms affecting gastrointestinal function. Despite substantial research efforts, the dynamic nature of this microbial ecosystem has led to inconsistencies in findings related to chicken gut microbiota, which is largely attributed to variations in rearing conditions. Consequently, the interaction between the chickens’ gut microflora and its host remains inadequately explored. This review highlights recent advances in understanding these relationships, with a specific focus on microbial composition, diversity, functional mechanisms, and their potential implications for improving poultry production.
1 Introduction
The gut microbiota refers to the diverse collection of microorganisms, including bacteria, archaea, fungi, and viruses, residing in the digestive tracts of animals (1, 2). The gastrointestinal metagenome encompasses the collective genomes of these gut microorganisms (3, 4). These microbiota exert wide-ranging effects, influencing factors such as colonization resistance against pathogens, maintenance of the intestinal epithelium, metabolism of dietary and regulation of immune function, and even modulation of host behavior through the gut-brain axis (4). The microbial composition of the gut varies across different regions of the digestive tract (5), the gastrointestinal tract (GIT), which is densely populated with microorganisms that interact with both the host and ingested feed (6). In birds, the GIT contains a complex microbiota that plays a crucial role in nutrient absorption and pathogen defense (7). Specifically, in terms of pathogen defense, the gut microbiota establishes a protective barrier that limits the colonization of pathogenic bacteria (8). Additionally, dietary changes and treatment interventions have been shown to enhance poultry growth while reducing the risk of enteric infections (9). The intricate relationship between the intestinal microbiome, microorganisms, host, and diet has profound effects on both the nutrition and health of poultry (10). These interactions involve nutrient exchange, changes in gut morphology, as well as host physiology and immunity (6).
Interestingly, various feed additives, including probiotics, prebiotics, enzymes, amino acids, and phytobiotics, have been demonstrated to enhance gut health (10–14). By optimizing the gut environment, these interventions lead to better feed conversion and overall health (Figure 1). Following the hatching process, the gastrointestinal tract of the chick is rapidly exposed to a diverse array of external microorganisms, fostering an optimal environment for the proliferation of anaerobic species. As these microorganisms colonize the gut, they interact with the host’s digestive processes. Over time, microbial communities diversify, and by the time chickens reach maturity, a relatively stable microbial balance is established (15). Notably, the chicken gut microbiota differs from those of other animals due to the chicken’s smaller and faster digestive tracts, resulting in a unique microbial composition (16, 17). As chickens develop, the interaction between gut microbes and host performance becomes increasingly complex (6, 18). The enzymatic degradation of indigestible dietary polysaccharides produces fermentable sugars and short-chain fatty acids (SCFAs) (19, 20). SCFAs promote the proliferation of epithelial cells in the gastrointestinal tract, thereby enhancing the surface area available for nutrient absorption (21). The bacteriostatic effects of SCFAs contribute to the elimination of foodborne pathogens, such as Salmonella spp. (22). Moreover, the microbiota facilitates nitrogen metabolism, incorporating nitrogen into bacterial cellular proteins. This process enables these bacteria to provide essential amino acids, proteins, and vitamins to the host (23). This review investigates the factors that influence the development of the poultry gut microbiome, with a particular emphasis on the interactions between the host and its microbiome, as well as the repercussions of microbiome disturbances on poultry health, disease susceptibility, and productivity.
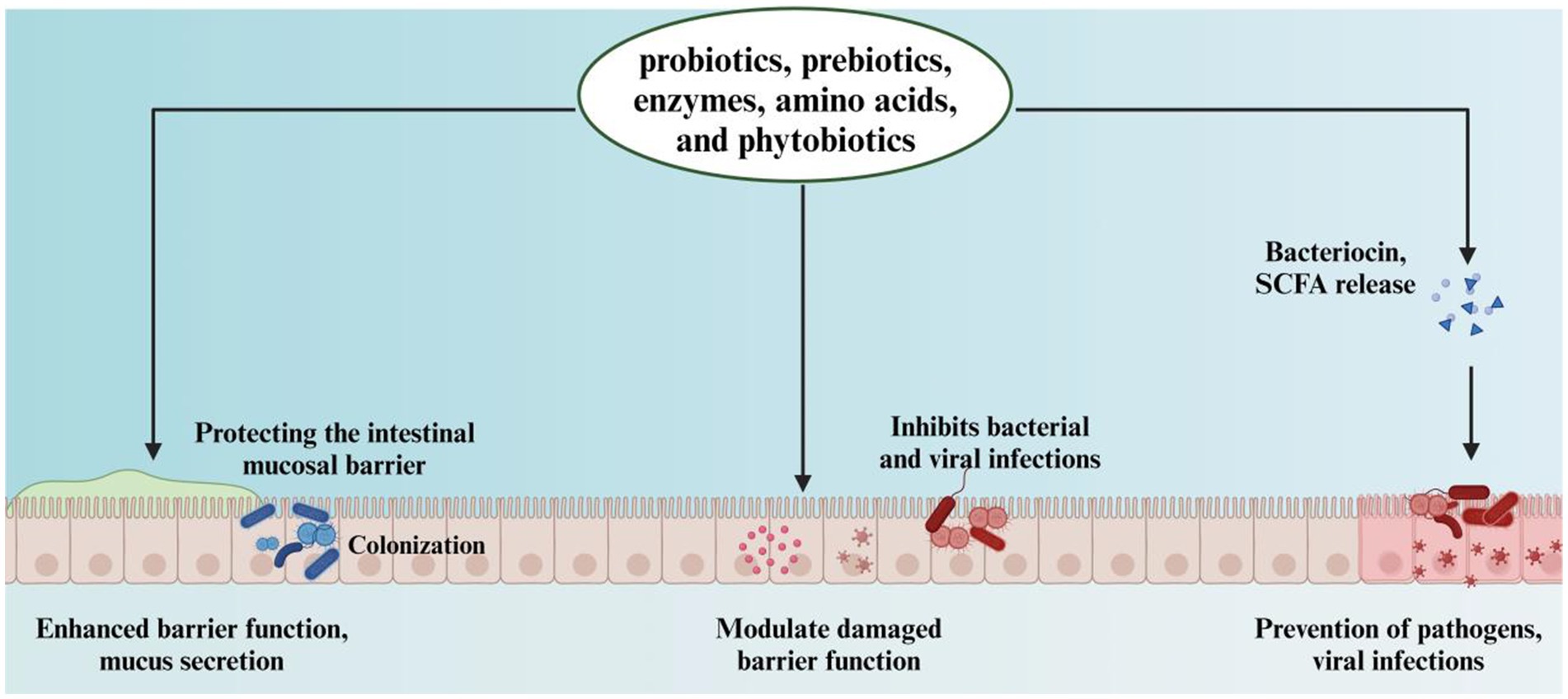
Figure 1. The role of probiotics, prebiotics, enzymes, amino acids, and phytobiotics in enhancing intestinal barrier and preventing infections.
2 Limitations and potential of 16S rRNA sequencing in microbial identification
The 16S ribosomal RNA is widely recognized as a fundamental genetic marker for the classification and identification of bacterial species. Its ubiquitous presence across diverse bacterial taxa, conserved function in critical cellular processes, and adequate sequence length facilitate precise differentiation among various bacterial species (24). The methodology for sequencing the 16S rRNA gene targets a specific segment of microbial DNA, providing valuable insights into microbial community diversity and identification. Encoded within prokaryotic cells encompassing bacteria and archaea, the 16S rRNA gene specifies the RNA component of the 30S ribosomal subunit. Recognized as a “molecular clock” owing to its ubiquitous presence in bacteria and archaea, this gene plays an essential role in delineating phylogenetic relationships and species divergence. Leveraging the structure and functionality of the 16S rRNA gene enables researchers to trace evolutionary lineages while gaining deeper comprehension of genetic interrelationships among diverse microbial species (25). The structure of 16S rRNA comprises highly conserved segments interspersed with nine hypervariable regions facilitating concurrent sequencing of multiple species using universal primers while offering potential discrimination based solely on variable regions (26). Numerous studies have shown that 16S rRNA gene sequencing can achieve genus-level identification in over 90% of cases, but its ability to identify species-level ranges from approximately 65 to 83% (27). The Ribosomal Database Project (RDP) is a comprehensive, publicly accessible resource dedicated to the acquisition, analysis, and dissemination of ribosomal RNA (rRNA) gene sequences. It serves as an essential tool for researchers investigating microbial diversity, taxonomy, and phylogenetics. The Ribosomal Database Project (28) and SILVA databases encompass comprehensive rRNA sequence data spanning bacteria, eukarya, and archaea (29). However, 16S rRNA sequencing has several limitations (Figure 2). It does not offer insights into the metabolic potential or activity of microbial communities. The 16S rRNA gene, as a conserved housekeeping gene present in all prokaryotes, primarily reflects phylogenetic relationships rather than functional attributes such as metabolic capabilities, virulence, or antibiotic resistance. Moreover, the sequenced region of the 16S rRNA gene may lack sufficient variability to distinguish between closely related species or strains, for instance, differentiating between pathogenic and commensal strains of Escherichia coli.
3 Composition and function of the gut microbiota in chickens
3.1 Composition of the gut microbiota in chickens
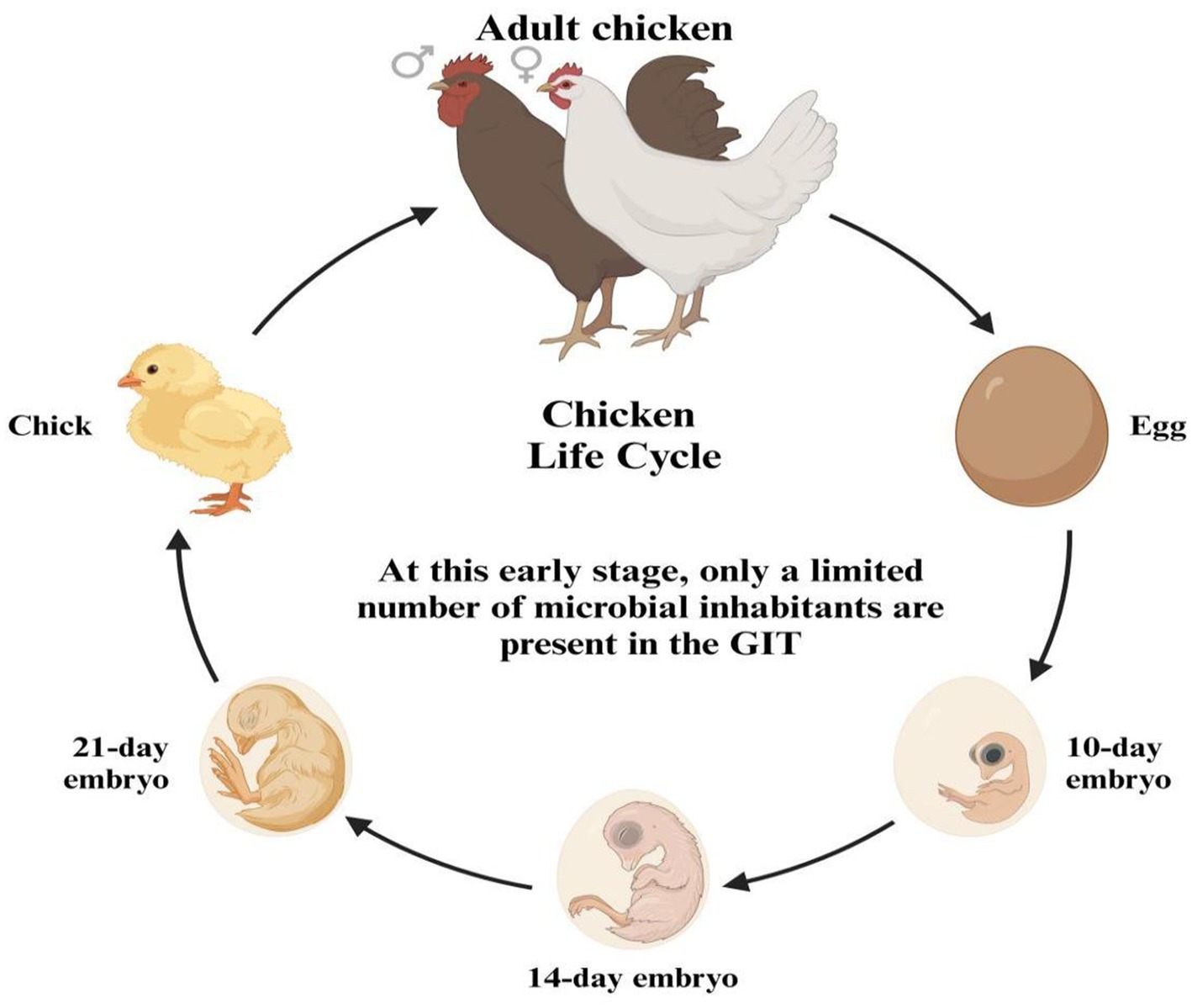
The GIT of a newly hatched chick is nearly devoid of microorganisms, indicating a state close to sterility. In this early stage, only a limited number of microbial inhabitants are present in the GIT (Figure 3). These initial microbes are not randomly acquired from the environment but are instead vertically transmitted from the mother hen to her chicks, either through the oviduct or via the eggshell pores. This early microbial colonization plays a crucial role in the chick’s development and health by establishing a foundation for a more complex and diverse gut microbiome as the chick matures (30, 31). Additionally, microbiota can be transferred to the gastrointestinal tract during hatchery handling and transportation (32), and microbes have been detected in the chick’s GIT while it is still inside the shell (33). The early stage of the post-hatch microbial contamination affects the immune system and intestinal microbiota (34). The natural intestinal microflora develops after hatching and rapidly increases (35), from the 1st to the 19th day of life (36). The microbial colonization continuously grows until the GIT population reaches its balance (37). The fungi are more inhabited in the upper GIT site than the lower parts, while the bacterial inhabitance follows an opposite pattern (38). The microbiota of young chickens exhibits high variability (39–41) and defining the core chicken gut microbiota necessitates a focus on the microbiota of adult chickens, specifically those aged at least 20 weeks. This is due to the high variability and incomplete establishment of microbial communities in younger chickens. By studying adult chickens, researchers can discern the stable and consistent microbial populations that typify a well-developed gut microbiota. These matured adult chickens have undergone significant maturation, leading to a more stable and representative state of their gut microbiota, thereby offering a clearer understanding of the essential core microbial species for maintaining gut health and function (42–45). The microbial diversity within the gastrointestinal tract of chickens exhibits regional variations (Figure 4). Lactobacilli dominate the proximal parts of the digestive tract in adult chickens, although other species are also present (46, 47). Composition and complexity of the microbiota increase significantly in the distal segments of the intestinal tract, such as the cecum and colon. However, variations in colonic microbiota can be attributed to the chicken’s intestinal physiology and may exhibit similarities to either ileal or cecal microbiota.
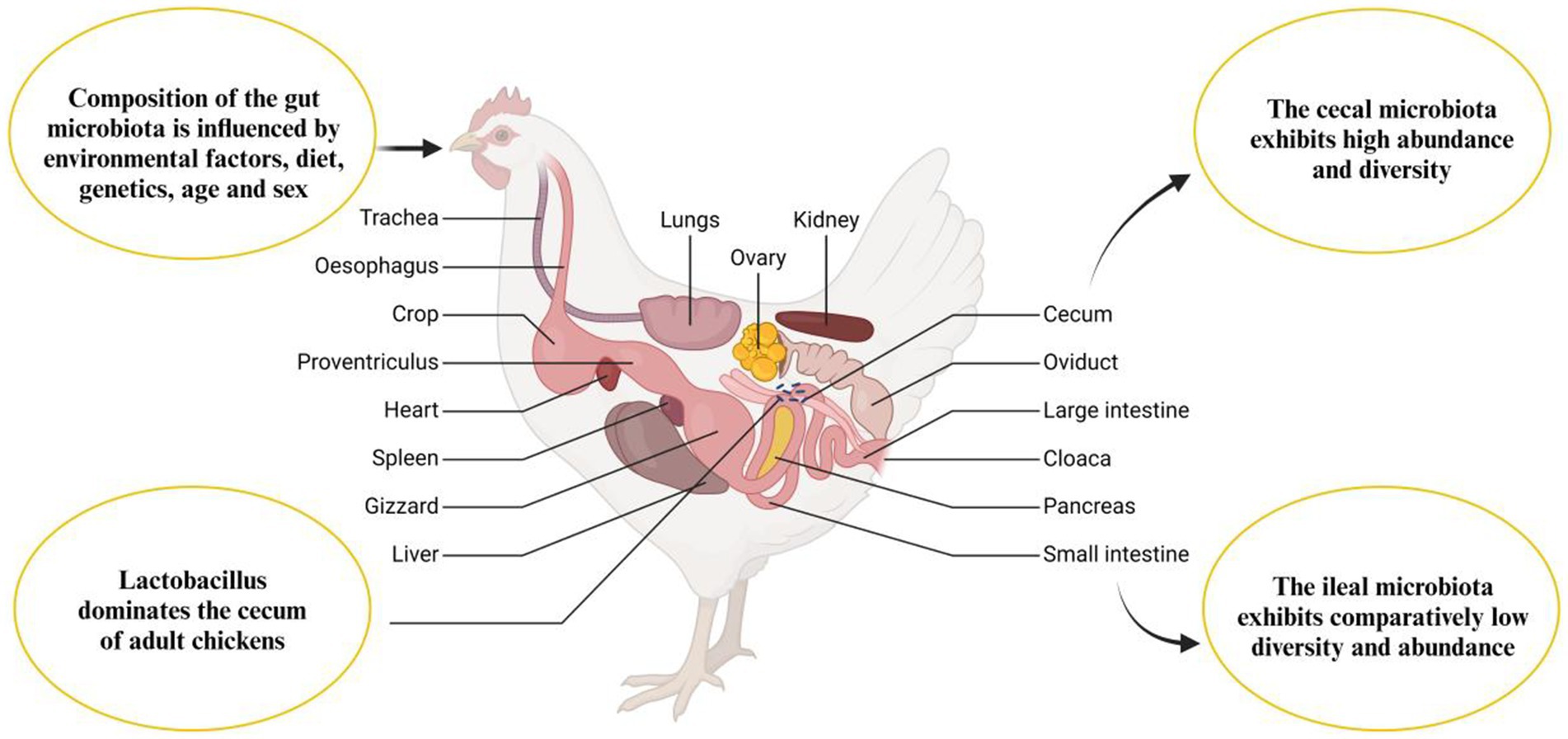
Figure 4. Anatomy of the chicken gastrointestinal tract and factors influencing gut microbiota composition.
3.2 Microbiota composition in the cecum
In the composition of the gut microbiota in chickens, bacteria represent the predominant group, constituting over 90% of the total microbial population. The most common bacterial groups include Lactobacillus, Bacteroides, Bifidobacterium, Clostridium, and Bacillus cereus, as well as Escherichia coli, yeast, and other fungi (48). The complexity and absolute numbers of gut microbiota significantly increase with age and environmental exposure. Most of these microbial species are classified into two major phyla, including the Gram-positive Firmicutes and the Gram-negative Bacteroidetes. The diversity and abundance of these groups highlight their essential role in digestion and maintaining gut health. Several studies emphasize the importance of a balanced microbial community within the cecum for optimal gut ecosystem function (49, 50).
In addition to Firmicutes and Bacteroidetes, two less dominant phyla, Actinobacteria (Gram-positive) and Proteobacteria (Gram-negative), are also present. In healthy adult hens, Firmicutes and Bacteroidetes typically represent about 45% each of the total microbiota, while Actinobacteria and Proteobacteria usually range from 2 to 3%. It is important to note that studies using 16S rRNA sequencing may slightly underestimate the abundance of Actinobacteria due to the low copy number of the 16S rRNA genes in species like Olsenella, Collinsella, and Bifidobacterium (51). While there is broad agreement on the overall composition of the cecal microbiota, significant individual variation persists. For instance, the proportion of Bacteroidetes can range from 10 to 90% without any signs of abnormality, and cases have been documented where chicks exhibit over 10% Actinobacteria and Proteobacteria. Despite variations in abundance, representatives from all four phyla are consistently found in the ceca of nearly all adult chickens.
3.3 Composition of small intestine microbiota
The small intestinal microbiome of chickens is a sophisticated and dynamic system, characterized by intricate interactions among numerous microorganisms. The bacterial composition in the small intestine is primarily dominated by four major phyla: Firmicutes, Bacteroidetes, Proteobacteria, and Actinobacteria. Among these, Firmicutes, represented by genera such as Lactobacillus and Clostridium, typically dominate. These bacteria can act either as symbionts or pathogens, depending on the strain type (52–54). The composition of the microbiota in the different sections of the small intestine, such as the duodenum, jejunum, and ileum, shows a high degree of similarity, as indicated by previous studies (45, 55). However, further comprehensive research is necessary to precisely identify the bacterial species uniquely adapted to each specific compartment of the small intestine. Occasionally, the microbiota of the ileum can intermingle with microorganisms originating from the caecum (55).
Firmicutes remain the most prevalent phylum in the small intestine, with genera such as Lactobacillus, Enterococcus, Turicibacter, Clostridium sensu stricto, as well as isolates from the Clostridium cluster within the family Peptostreptococcaceae, and the genus Romboutsia. In addition to Firmicutes, bacteria from the phylum Proteobacteria, including Escherichia coli and Helicobacter, can also be found in the small intestine. Notably, the presence of Helicobacter has been associated with reduced performance in chickens (56).
Moreover, despite the diversity of microbial species, the small intestine’s microbiota demonstrates limited diversity, with one to five genera accounting for around 50% of the entire ileal microbiota. This suggests a simpler microbial structure compared to other regions of the gastrointestinal tract (56). The ileum, a crucial component of the chicken’s digestive system, plays an essential role in nutrient absorption and immune regulation. It harbors a diverse microbial community that contributes significantly to maintaining intestinal health, enhancing nutrient absorption, and protecting against pathogens. These microorganisms break down complex carbohydrates, proteins, and lipids to produce short-chain fatty acids such as acetic acid, propionic acid, and butyric acid. These fatty acids not only provide energy for intestinal epithelial cells but also improves feed efficiency by promoting sodium and water absorption (40, 57, 58).
3.4 Fecal microbial composition
Many scientific studies utilize fecal samples to characterize the microbiota of chickens (46, 55, 59, 60). When experiments require repeated sampling from the same bird, researchers must often rely on fecal material. However, several considerations must be addressed during sample collection. First, it is difficult to prompt each chicken to defecate on command. Consequently, researchers frequently collect fecal material directly from the floor, making it impossible to control how long the droppings were exposed to air, whether for 10 min or 5 h. This is important because many gut colonizers are strict anaerobes, and exposure to air may reduce their viability or alter their community structure, potentially influencing the study’s final results.
Apart from environmental exposure, chicken digestion physiology significantly impacts the composition of colonic and fecal microbiota, chickens have a remarkably short digestive transit time, with digesta passing from ingestion to excretion in as little as 2 h (61, 62). Unlike mammals such as pigs or humans, adult chickens have a relatively short colon, measuring only about 10 centimeters, which limits the retention time of digesta. After stomach processing, most digesta quickly moves from the small intestine to the colon and is excreted approximately every 2 h (63). In contrast, only a small portion of the digesta moves from the ileum to the caecum, where it undergoes fermentation, for 8 to 12 h (64, 65). This rapid transit and short retention time shape the microbial composition found in the colonic and fecal matter, which must be considered when interpreting microbiota data. Additionally, the periodic voiding of caecal contents into the colon, which typically occurs twice daily, adds another layer of variability to sample composition (66, 67). If samples are collected immediately after caecal voiding, the colonic or fecal microbiota may closely resemble the caecal microbiota. Conversely, if samples are collected before the next caecal voiding, the microbiota may more closely resemble the ileal community. When digesta from the small intestine passes through the colon after caecal excretion, the microbiota could represent a mixture of both caecal and ileal communities. Thus, variations in the timing of sample collection related to periodic caecal voiding and intestinal transit contribute to substantial variability in colonic or fecal microbial composition (55, 59, 68, 69), and researchers must account for this in their experimental designs.
3.5 Major bacterial taxa colonising chicken intestinal tract
3.5.1 Classification of intestinal bacteria at the major phyla
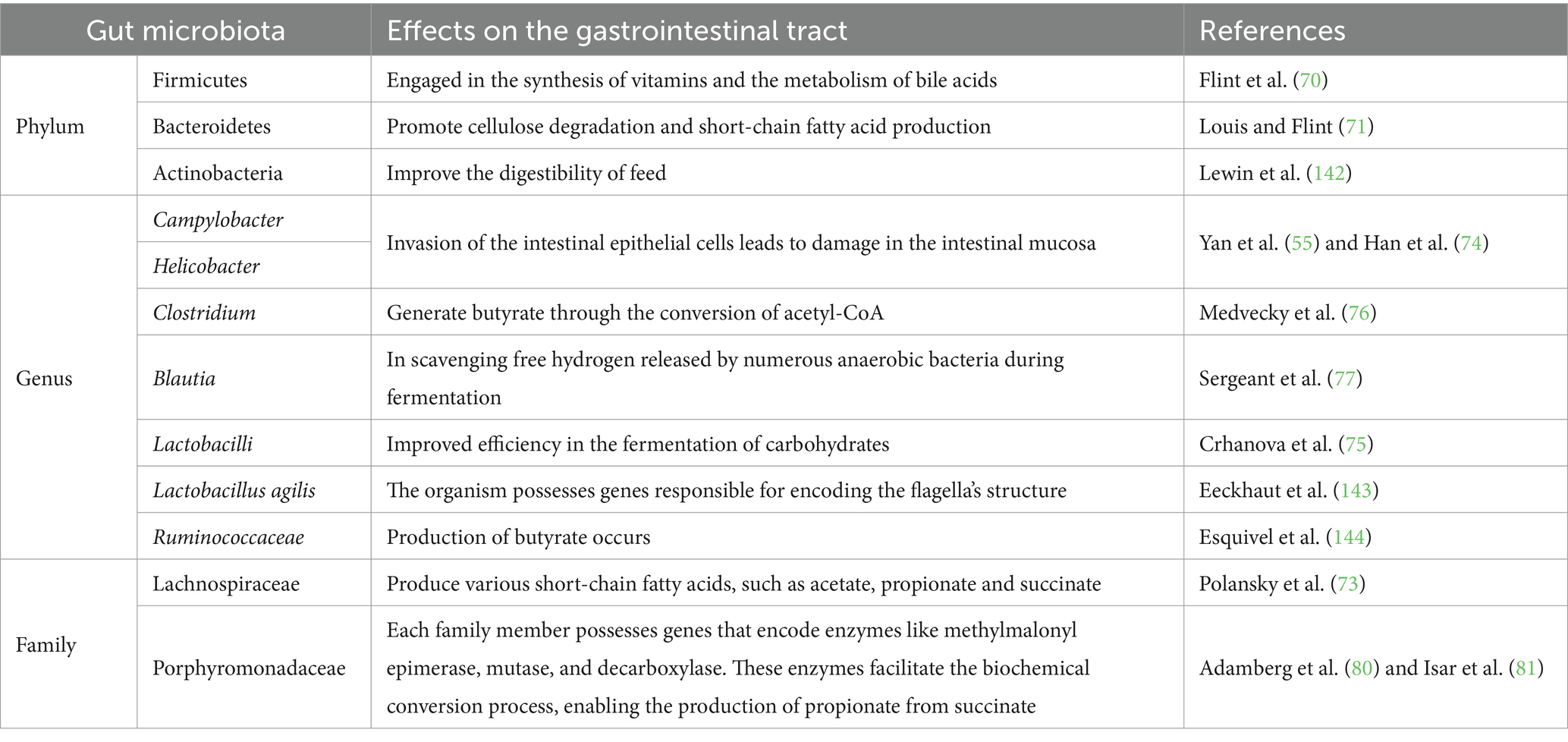
The gut microbiome consists of bacteria classified across various taxonomic ranks, including phylum, class, order, family, genus, and species. These bacteria exhibit distinct distribution patterns and perform a range of functions in different segments of the intestinal tract. Among the major bacterial phyla, Bacteroides predominate in the chicken cecum (Table 1). These bacteria are well-known for their ability to degrade complex polysaccharides and plant cellulose, engaging in fermentation processes that produce short-chain fatty acids such as acetic acid, propionic acid, and butyric acid. These fatty acids play a crucial role in maintaining intestinal acid–base balance and inhibiting pathogen proliferation (69). It should be noted that Bacteroides are involved in vitamin synthesis and bile acid metabolism, both of which are essential for nutrient absorption and host metabolism (70). Additionally, Firmicutes represent a significant group of anaerobic bacteria in the chicken gut, including a substantial number of Clostridium species. While some Clostridium species can produce potentially harmful metabolites, they also contribute to cellulose degradation and short-chain fatty acid production (71). Especially, certain Firmicutes bacteria possess probiotic properties, helping to modulate the intestinal microecological balance and enhance the host’s immune response (72).
3.5.2 Classification of intestinal bacteria at the major generic
Actinobacteria are a group of Gram-positive bacteria with high GC content (around 65%), playing a key role in the breakdown of organic matter and are known for their production of secondary metabolites such as antibiotics, enzymes, and other bioactive compounds. In the chicken cecum, the predominant genera of colonizing bacteria include Coriobacteriaceae, represented by Olsenella and Collinsella from the Bifidobacteraceae family, which also includes Bifidobacterium. Proteobacteria, which are non-spore-forming, Gram-negative bacteria, are also present. Common colonizing bacteria in the chicken cecum include facultative organisms like E. coli, as well as Desulfovibrio, Sutterella, Parasutterella, Anaerobiospirillum, and Succinatomonas. Additionally, Actinomycetes contribute to poultry health by producing nutrients and metabolizing short-chain fatty acids, enhancing digestive efficiency and improving feed conversion rates (73).
Helicobacter and Campylobacter are also prevalent components of the chicken microbiota. In highly infected chickens (55, 74), E. coli and Salmonella constitute approximately 0.1% of the total microbiota, while Campylobacter and Helicobacter can account for over 10%. This highlights differing colonization patterns between E. coli, Salmonella, and bacteria like Helicobacter and Campylobacter. Other bacterial families in the cecum include Lactobacillaceae, Veillonellaceae, and Erysipelotrichaceae. Trichiidae, which consist of strictly anaerobic spore-forming bacteria with a genomic GC content of about 45%, produce butyrate from acetyl-CoA. Notable species include Bacillus coli, Clostridium lactofermens, and Clostridium saccharolytica, which are significant butyrate producers (75, 76).
Cyanobacteria contribute to microbial metabolism by encoding enzymes like 5-methyltetrahydrofolate: cobalamin methyltransferase and acetyl-CoA synthetase (73). These enzymes enable Cyanobacteria to use CO2 and H2 to generate acetic acid through reductive acetogenesis (77). Similarly, Blautia plays a key role in scavenging free hydrogen released by anaerobic bacteria during fermentation.
Lactobacilli efficiently ferment carbohydrates (78), producing lactic acid and lowering the environmental pH. This acidic condition inhibits the growth of other bacterial species, giving Lactobacilli a competitive advantage in diverse environments, including the gastrointestinal tract and fermented foods. Certain strains, like Lactobacillus ruminis and Lactobacillus agilis, possess flagella, enhancing their motility and ability to colonize and acquire nutrients in complex environments, such as the gut (78, 79).
Ruminococcaceae are primary butyrate producers. Through carbohydrate fermentation, most Ruminococcaceae convert two acetyl-CoA molecules to crotonyl-CoA, resulting in butyrate production (73). Flavonoids and pseudoflavonoids can also produce butyrate through lysine fermentation or succinic acid reduction. Anaerotruncus represents a potentially mobile intestinal colonizer. Due to their high sensitivity to oxygen, Ruminococcaceae and Lachnospiraceae are often among the first bacterial families to diminish during inflammatory diseases, as reactive oxygen species from macrophages and granulocytes damage the gut microbiome. Therefore, the reduction of Ruminococcaceae and Lachnospiraceae is typically a consequence, rather than a causative factor (78, 79).
3.5.3 Taxonomic classification of predominant bacterial families in the intestines
The family characteristics frequently observed in the chicken cecum include members of the Bacteroidaceae, as well as Gram-positive families, like Lachnospiraceae and Ruminococcaceae. The genus Bacteroides comprises numerous species that show host-specific adaptations. For instance, human-adapted species such as B. dorei, B. uniformis, and B. clarus contrast with chicken-associated species like B. salanitronis, B. caecigallinarum, and B. coprocola. Interestingly, although not yet fully understood, certain chicken-associated Bacteroides strains have acquired KUP genes encoding potassium ion transporters. In adult chickens, Bacteroidaceae genomes contain a high number of genes responsible for the degradation of complex polysaccharides. These bacteria produce short-chain fatty acids (SCFAs) such as acetate, propionate, and succinate. Notably, in vitro, Bacteroides species show nearly the same acidification capability as Lactobacilli, which are well-known for their acid production (Table 1). As a result, the extensive fermentation by Bacteroides species significantly contributes to the acidic environment in the caecum and plays a critical role in digestion (73, 76).
The phylum Bacteroidetes includes several distinct families, such as Rikenellaceae, Bacteroidaceae, Prevotellaceae, and Porphyromonadaceae. Bacteroidetes genomes are relatively large, ranging from approximately 3 to over 6 megabase pairs (Mbp). Members of this phylum possess genes encoding enzymes like methylmalonyl-CoA epimerase, mutase, and decarboxylase, which are crucial for the biochemical conversion of succinate to propionate (80–82).
The Rikenellaceae and Alistipes stand out with a GC content of 58–60%, distinguishing them from other members of Bacteroideae. Remarkably, Alistipes is one of the earliest colonizers of the chick cecum within the Bacteroidetes group (83, 84). The Porphyromonadaceae family consists of genera such as Barnesiella, Odoribacter, Butyricimonas, and Parabacteroides. Both Bacillus putitidis and Bacillus butyricomonas can produce butyrate through lysine fermentation and succinic acid reduction pathways. Additionally, butyrate can be synthesized from acetyl-CoA (76).
Prevotellaceae is primarily responsible for the degradation of cellulose and other complex carbohydrates. Members of this family are renowned for their diverse and intricate metabolic capabilities (83, 85). The characteristics of chicken isolates from the Prevotellaceae family remain largely unexplored. The isolates obtained in pure culture show only distant relations to characterized Prevotella species, with 16S rRNA sequences displaying about 90% similarity to the closest GenBank entries. This suggests that the Prevotella family in chickens may represent new genera distinct from those in humans, mice, or pigs. Omics studies have shown that chicken Prevotellaceae specialize in digesting complex polysaccharides (75), paralleling the presence of Prevotellaceae in the rural African gut microbiota (84, 86).
4 Gut microbiota and host interactions
4.1 Pathogen colonization interacts with the gastrointestinal tract
Bacteria pathogenic to poultry include species such as Staphylococcus, Escherichia coli, Clostridium, Campylobacter, and Salmonella spp. (49). One of the most notable pathogens is Salmonella, a bacterial threat in poultry that presents significant risks for human health due to its prevalence and potential for contamination (87, 88). Salmonella can easily colonize the intestines of poultry. However, research is underway to explore the use of targeted bacteriophages to eliminate Salmonella from the microbiome of broiler chickens (89). Among the various strains, Salmonella typhimurium is particularly concerning for humans, as it can colonize the poultry digestive system without causing harm to the birds themselves (90). Fortunately, current research focuses on microbial interventions, such as probiotics and metabolites, to prevent Salmonella colonization and promote the restoration of a healthy gut microbiota after infection (90–92).
Another pathogen that affects the poultry intestines is Clostridium perfringens (93), particularly types A and C, which can lead to necrotic enteritis (NE), characterized by decaying and inflamed intestinal tissue. This condition generally arises under specific dietary conditions, often in conjunction with the presence of the parasite coccidia (93, 94). In addition to intestinal pathogens, respiratory infections (RI) in poultry are also a concern. A 2021 study on the bacterial composition of the turkey respiratory tract revealed that the presence of Ornithobacterium and Mycoplasma increases the risk of RI in turkeys (95). Moreover, domesticated turkeys were found to have less microbial diversity and a higher prevalence of pathogenic and antibiotic-resistant bacterial strains compared to their wild counterparts (96).
4.2 Nutritional role of the chicken gut microbiome and host
4.2.1 Fermentation and the production of short-chain fatty acids
A key function of the chicken gut microbiome is the fermentation of dietary fibers and other complex carbohydrates (Figure 5). Since chickens lack the necessary enzymes to break down certain fibers found in plant-based feeds, their gut microbiota compensates by fermenting these substrates in the cecum and colon. This fermentation process produces short-chain fatty acids (SCFAs), such as acetate, propionate, and butyrate (97), which are crucial energy sources for chickens. For example, acetate is absorbed into the bloodstream and serves as an energy substrate for various tissues, including muscles, and the liver (98). In contrast, propionate and butyrate are mainly utilized by the gut epithelium, with butyrate playing a particularly critical role in maintaining intestinal cell health, enhancing gut barrier function, and reducing inflammation (99). Besides providing energy, these SCFAs also contribute to the structural integrity and immune function of the gut, which are vital for overall health and efficient nutrient absorption.
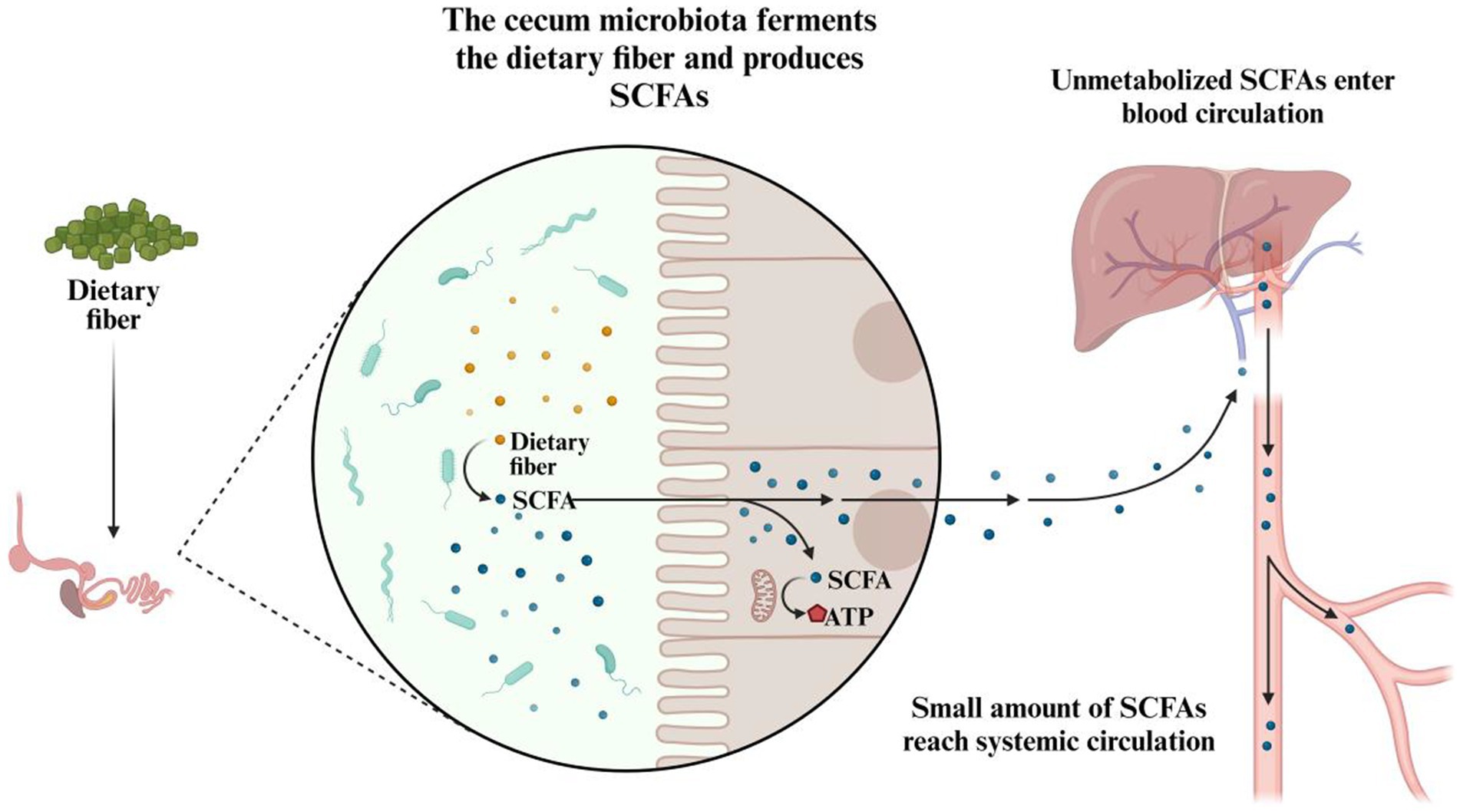
Figure 5. The role of cecal microbiota in the fermentation of dietary fiber and production of SCFAs.
It is important to emphasize that dietary carbohydrates are broken down and absorbed primarily within the proximal gastrointestinal tract, while undigested carbohydrates, along with residual digestible ones, proceed to microbial fermentation in the distal gastrointestinal tract (100). The gut microbiota can hydrolyze non-digestible disaccharides, oligosaccharides, and polysaccharides into their monosaccharide components through fermentation, producing SCFAs, which are then used by the host for energy generation and as a carbon source (101, 102). This fermentation activity predominantly occurs in the ceca, where the microbial density is highest (103), and increases as the chicken matures. While absent in the ceca of one-day-old chicks, optimal SCFA concentrations are typically achieved by day 15 post-hatch and remain stable afterward (104). Through passive diffusion across the cecal epithelium, SCFAs enter various metabolic pathways. Research has also highlighted the regulatory roles of SCFAs, which include modulating colonic blood flow, stimulating enterocyte development and proliferation, enhancing mucin production, and regulating the intestinal immune response (105, 106).
4.2.2 Vitamin interaction
The gut microbiota of chickens plays a crucial role in the complex biosynthesis of various B vitamins, such as biotin, folate, and riboflavin. Research has demonstrated that specific bacterial species within the chicken gut can produce these vitamins through fermentation processes (107). For instance, Lactobacilli and Bifidobacteria are capable of producing folate and biotin, which are essential for cellular functions and metabolism (107, 108). However, certain gut microbes can degrade vitamins, potentially leading to deficiencies. For example, some strains of Clostridium spp. have been shown to degrade vitamin B12, reducing its availability to the host (109). This degradation may result in vitamin deficiencies and subsequent health issues in chickens. Additionally, the gut microbiota influences the absorption of fat-soluble vitamins, such as vitamins A, D, E, and K. Specific microbial populations can either enhance or inhibit the absorption of these vitamins. For example, a study found that a diverse gut microbiota improves the absorption of vitamins A and E by altering bile acid metabolism and strengthening intestinal barrier function (110). Deficiencies or imbalances in vitamins due to microbial activity can lead to poor growth rates, immune dysfunction, and increased susceptibility to diseases (109).
4.2.3 Protein metabolism
Metabolism of proteins in chickens is a crucial component of their nutritional needs and growth requirements. The gut microbiota play a significant role in this process by influencing protein digestion, amino acid absorption, and the generation of bioactive peptides. Although the digestion of dietary proteins begins in the stomach and small intestine, a substantial portion of protein digestion and fermentation occurs in the cecum and colon, where the gut microbiota are abundant. These microbial communities assist in breaking down proteins that the host’s enzymes cannot fully digest.
Proteolytic bacteria in the gut ferment dietary proteins into various peptides and amino acids (107). For instance, bacteria such as Clostridium and Lactobacillus produce enzymes that degrade proteins into shorter peptides and free amino acids. This microbial proteolysis provides an essential secondary mechanism to break down proteins that escape digestion in the upper gastrointestinal tract (111). The amino acids released through microbial protein degradation are absorbed by the host and utilized in various metabolic processes (Figure 6). Gut microbes affect the availability of these amino acids by influencing their absorption and conversion. Some microbial enzymes can release amino acids bound in dietary proteins or peptide bonds that resist degradation by host enzymes. For example, Bacteroides and Bifidobacteria contribute to protein hydrolysis and amino acid release, which the host’s intestinal cells subsequently absorb (112).
Figure 6. The role of gut microbiota in host interactions: metabolism, immunity, and the gut-brain axis.
In addition to aiding digestion, gut microbes produce bioactive peptides that exert physiological effects on the host. These peptides can influence immune responses, gut health, and growth. Some of these microbial-derived peptides possess antimicrobial properties or modulate immune functions. For instance, certain peptides produced by gut bacteria have been shown to enhance the host’s immune system and protect against pathogenic infections (109). Furthermore, efficient microbial protein digestion and amino acid absorption promote improved growth rates and better feed conversion ratios. Research indicates that maintaining a balanced gut microbiota optimizes protein utilization, enhancing feed efficiency. For example, beneficial microbes such as Lactobacillus improve protein digestibility while reducing the production of nitrogenous waste, leading to better growth outcomes and improved feed conversion (112).
4.2.4 Mineral absorption
Composition of gut microbiota impacts the absorption of essential minerals, including calcium, phosphorus and magnesium, by modulating the intestinal microenvironment. Beneficial bacteria, in particular, can produce short-chain fatty acids (SCFAs) through the fermentation of dietary fibers, which enhances the solubility and uptake of these minerals. Additionally, certain gut bacteria can biosynthesize vitamins and other compounds that further facilitate mineral assimilation (113). The presence of specific microbial species is linked to increased mineral bioavailability. Research shows that probiotic strains positively impact the availability of calcium and phosphorus in the gastrointestinal tract, leading to improved mineral absorption and utilization (114). In contrast, dysbiosis, or an imbalance in gut microbiota, can impair mineral absorption and cause deficiencies. Diets high in fiber and prebiotics promote a diverse and beneficial microbiota, which in turn optimizes mineral absorption. For example, dietary fibers encourage the growth of beneficial bacteria that produce SCFAs, thereby aiding in mineral absorption (115).
4.3 Modulation of the chicken immune system by gut microbiota
The development and function of gut-associated lymphoid tissue (GALT) in chickens are strongly influenced by the gut microbiome. A well-balanced microbiome strengthens the body’s defense mechanisms against threats and promotes a robust immune response (116). Additionally, this balance helps prevent excessive immune reactions to harmless antigens, such as food or commensal bacteria, thus protecting against unnecessary immune activation. Conversely, when microbial balance is disrupted, a condition known as dysbiosis (117), it can trigger inflammatory responses, increasing the risk of autoimmune disorders and making chickens more vulnerable to infections.
The colonization of beneficial bacteria promotes immune tolerance, effectively training the immune system to differentiate between pathogenic and non-pathogenic stimuli. This symbiotic relationship enhances the body’s capacity to mount efficient responses to pathogens while mitigating unnecessary immune reactions (118). Moreover, the gut microbiota influences the production of immune cells, such as macrophages, dendritic cells, and T cells, by releasing microbial metabolites (Figure 6). These metabolites act as signaling molecules that modulate immune responses both locally in the gut and systemically throughout the body. Research suggests that gut microbiota can enhance the production of secretory immunoglobulin A (sIgA) in the gut mucosa (119). sIgA is a key component of the immune system that serves as the first line of defense by neutralizing pathogens and toxins before they cross the epithelial barrier. Continuous production of sIgA is essential for protecting chickens from gastrointestinal infections, especially during periods of stress or exposure to pathogens. Dysbiosis not only compromises the gut’s physical barrier but also impairs the immune system’s ability to defend against infections. This disruption increases the risk of diseases such as necrotic enteritis, coccidiosis, and other gastrointestinal disorders that are common in poultry (119).
4.4 Gut microbiota mediated protection against viral diseases
Recent research highlights the essential role of gut microbiota in shaping immune responses to viral infections in chickens. Commensal bacteria contribute to pathogen defense through direct competition, antibody production, and the activation of cytokines, which modulate both innate and adaptive immune responses (120). However, several significant viral diseases can disrupt the diversity of the intestinal microbiota, leading to dysbiosis, a state associated with various pathological conditions that facilitate acute viral infections in chickens.
The gastrointestinal tract’s highly dynamic environment presents opportunities for pathogens to upset the balance between the host and microflora, resulting in dysbiosis and subsequent mucosal infections (121). Bacterial dysbiosis has been linked to inflammation and alterations in immune functions (122), particularly affecting type I interferons (IFNs) and inflammatory responses (123, 124). Diseases that destabilize intestinal microflora (125–127) further increase chickens’ vulnerability to bacterial infections during dysbiosis (128). Moreover, research has established links between gut microbiota and distant organs, forming key pathways such as the gut-lung, gut-brain, gut-skin, and gut-liver axes (129). In particular, species such as Bifidobacterium, Firmicutes, Faecalibacterium, Blautia, and Clostridium are essential for preventing and managing viral diseases. They outcompete pathogens and colonize the gastrointestinal mucosal surface, thereby maintaining microbial balance.
These beneficial microbiota also support digestion and produce short-chain fatty acids (SCFAs), which serve as an energy source and modulate antiviral immune responses. SCFAs stimulate the production of interferons (IFN-α and IFN-β) and enhance the function of T regulatory cells. This process promotes the secretion of anti-inflammatory cytokines such as IL-22 and strengthens humoral immune responses through the production of IgA and IgG antibodies. Together, these mechanisms contribute to controlling the severity of viral infections in chickens (130–132).
4.5 Interaction between gut microbiota and the gut-brain axis
The gut-brain axis (GBA) serves as a bidirectional communication network that connects the gut microbiota with the central nervous system (CNS). The gut microbiota can influence the production of neurotransmitters, which act as chemical messengers transmitting signals between the gut and the brain (Figure 6). Specific gut bacteria, for example, have the ability to produce neurotransmitters like serotonin and gamma-aminobutyric acid (GABA), which play crucial roles in mood regulation and behavior. Studies conducted on chickens have demonstrated that changes in the composition of gut microbiota can affect levels of these neurotransmitters, thereby influencing their overall well-being and stress responses (133). The modulation of behavioral responses in chickens is closely linked to the functioning of the gut-brain axis. Changes in gut microbiota can impact stress levels, anxiety, and behavior, suggesting that the gut microbiota, through its connection with the gut-brain axis, may modulate behavioral traits, potentially affecting productivity and welfare in poultry (134). Additionally, alterations in immune function due to changes in short-chain fatty acids (SCFAs) produced by gut bacteria can affect brain health by interacting with inflammatory cytokines and influencing neuroinflammation. In chickens, dysregulated microbial communities may lead to increased intestinal permeability and systemic inflammation, potentially exerting negative effects on brain function and behavior (135). Notably, stress responses are influenced by the gut microbiota, and stress itself can alter the microbial composition, creating an imbalanced state that worsens the stress response and affects overall health. Variations in microbial communities have been associated with changes in stress hormone levels and intestinal integrity, all of which contribute to how chickens respond to stress and influence their general health condition (136). Unfortunately, the relationship between the gut microbiota and serotonergic activity, as well as related psychological health, has not been thoroughly investigated.
The gut microbiota can also affect levels of neurotransmitters such as dopamine and serotonin. Certain gut bacteria participate in the production of these neurotransmitters or their precursors. For example, the microbiota can influence the synthesis of tryptophan and tyrosine, the amino acids that are precursors for serotonin and dopamine, respectively (137). Alterations in gut microbiota composition can lead to changes in neurotransmitter levels, which may, in turn, affect behavior and stress responses in chickens. Imbalances in the gut microbiota can result in decreased serotonin levels, potentially impacting mood and increasing stress (138). Changes in dopamine levels may also affect motivational and reward-related behaviors. Research indicates that dietary changes influencing the gut microbiota can modulate these neurotransmitter levels, thereby altering behavioral responses in chickens (139). Moreover, the gut microbiota can affect the function of the blood-brain barrier and modulate levels of neurotransmitters like dopamine and serotonin, impacting cognitive functions and emotional states (140). It should be noted that imbalances in gut microbiota affecting neurotransmitter levels can contribute to stress, anxiety, and other health issues (141). Understanding these interactions can help in formulating dietary and management strategies to maintain a healthy gut microbiota and promote an optimal neurotransmitter balance, ultimately enhancing the well-being of chickens.
5 Conclusion
Although advances in biotechnology have improved our understanding of the poultry microbiome and its role in poultry health and disease, further exploration of the GIT ecosystem, including the factors that influence it, and the interactions between the host and microbiome in chickens, is necessary to fully grasp the dynamics at play. This review aims to enhance our understanding of intestinal ecosystems while simultaneously establishing a robust foundation for the development of more effective strategies in the management and treatment of poultry diseases. A central focus of future research should be on investigating how changes in the composition of the poultry microbiome correspond to environmental factors, growth conditions and specific feed formulations. Additionally, it is essential to identify advantageous microbial strains that play a pivotal role in sustaining poultry health through the application of diverse methodologies. The potential of nanotechnology to enhance the nutritional value of dietary factors presents an exciting opportunity for improving poultry health. Lastly, integrating metagenomics and metabolomics approaches will advance the study of GIT diseases in poultry, offering a more holistic understanding of the relationship between the gut microbiome and overall health.
Author contributions
YY: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Software, Supervision, Visualization, Writing – original draft, Writing – review & editing. PLu: Supervision, Validation, Writing – review & editing. JL: Investigation, Supervision, Validation, Writing – review & editing. PLa: Software, Supervision, Writing – review & editing. PS: Formal analysis, Funding acquisition, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the SUT Research and Development Fund, Thailand Science Research and Innovation (TSRI), and the National Science, Research, and Innovation Fund (NSRF) with a project codes: FF3-303-68-12.
Acknowledgments
The authors gratefully recognizes the Suranaree University of Technology scholarship for External Grants and Scholarships for Graduate Students (SUT-OROG scholarship).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Moszak, M, Szulińska, M, and Bogdański, P. You are what you eat-the relationship between diet, microbiota, and metabolic disorders—a review. Nutrients. (2020) 12:1096. doi: 10.3390/nu12041096
2. Engel, P, and Moran, NA. The gut microbiota of insects—diversity in structure and function. FEMS Microbiol Rev. (2013) 37:699–735. doi: 10.1111/1574-6976.12025
3. Segata, N, Boernigen, D, Tickle, TL, Morgan, XC, Garrett, WS, and Huttenhower, C. Computational meta’omics for microbial community studies. Mol Syst Biol. (2013) 9:666. doi: 10.1038/msb.2013.22
4. Saxena, R, and Sharma, VK. A metagenomic insight into the human microbiome In: Medical and health genomics. Amsterdam: Elsevier (2016). 107–19.
5. Liu, W, Yuan, Y, Sun, C, Balasubramanian, B, Zhao, Z, and An, L. Effects of dietary betaine on growth performance, digestive function, carcass traits, and meat quality in indigenous yellow-feathered broilers under long-term heat stress. Animals. (2019) 9:506. doi: 10.3390/ani9080506
6. Pan, D, and Yu, Z. Intestinal microbiome of poultry and its interaction with host and diet. Gut Microbes. (2014) 5:108–19. doi: 10.4161/gmic.26945
7. O’Hara, AM, and Shanahan, F. The gut flora as a forgotten organ. EMBO Rep. (2006) 7:688–93. doi: 10.1038/sj.embor.7400731
8. Yegani, M, and Korver, DR. DR factors affecting intestinal health in poultry. Poult Sci. (2008) 87:2052–63. doi: 10.3382/ps.2008-00091
9. Kohl, KD. Diversity and function of the avian gut microbiota. J Comp Physiol. (2012) 182:591–602. doi: 10.1007/s00360-012-0645-z
10. Daniel, N, Lécuyer, E, and Chassaing, B. Host/microbiota interactions in health and diseases—time for mucosal microbiology. Mucosal Immunol. (2021) 14:1006–16. doi: 10.1038/s41385-021-00383-w
11. Murate, LS, Paião, FG, de Almeida, AM, Berchieri, A Jr, and Shimokomaki, M. Efficacy of prebiotics, probiotics, and synbiotics on laying hens and broilers challenged with Salmonella enteritidis. J Poult Sci. (2015) 52:52–6. doi: 10.2141/jpsa.0130211
12. Llamas-Moya, S, Girdler, CP, Shalash, SM, Atta, AM, Gharib, HB, Morsy, EA, et al. Effect of a multicarbohydrase containing α-galactosidase enzyme on the performance, carcass yield, and humoral immunity of broilers fed corn-soybean meal–based diets of varying energy density. J Appl Poult Res. (2020) 29:142–51. doi: 10.1016/j.japr.2019.10.001
13. Adil, S, Banday, T, Bhat, GA, Mir, MS, and Rehman, M. Effect of dietary supplementation of organic acids on performance, intestinal histomorphology, and serum biochemistry of broiler chicken. Veterinary medicine international, (2010) 2010:479485. doi: 10.4061/2010/479485
14. Abd El-Hack, ME, Alqhtani, AH, Swelum, AA, El-Saadony, MT, Salem, HM, Babalghith, AO, et al. Pharmacological, nutritional and antimicrobial uses of Moringa oleifera Lam. leaves in poultry nutrition: an updated knowledge. Poult Sci. (2022) 101:102031. doi: 10.1016/j.psj.2022.102031
15. Yang, X, Liang, S, Guo, F, Ren, Z, Yang, X, and Long, F. Gut microbiota mediates the protective role of Lactobacillus plantarum in ameliorating deoxynivalenol-induced apoptosis and intestinal inflammation of broiler chickens. Poult Sci. (2020) 99:2395–406. doi: 10.1016/j.psj.2019.10.034
16. Herrero-Encinas, J, Blanch, M, Pastor, JJ, Mereu, A, Ipharraguerre, IR, and Menoyo, D. Effects of a bioactive olive pomace extract from Olea europaea on growth performance, gut function, and intestinal microbiota in broiler chickens. Poult Sci. (2020) 99:2–10. doi: 10.3382/ps/pez467
17. Zheng, M, Mao, P, Tian, X, Guo, Q, and Meng, L. Effects of dietary supplementation of alfalfa meal on growth performance, carcass characteristics, meat and egg quality, and intestinal microbiota in Beijing-you chicken. Poult Sci. (2019) 98:2250–9. doi: 10.3382/ps/pey550
18. Zhou, C, Xu, P, Huang, C, Liu, G, Chen, S, Hu, G, et al. Effects of subchronic exposure of mercuric chloride on intestinal histology and microbiota in the cecum of chicken. Ecotoxicol Environ Saf. (2020) 188:109920. doi: 10.1016/j.ecoenv.2019.109920
19. Dunkley, KD, Dunkley, CS, Njongmeta, NL, Callaway, TR, Hume, ME, Kubena, LF, et al. Comparison of in vitro fermentation and molecular microbial profiles of high-fiber feed substrates incubated with chicken cecal inocula. Poult Sci. (2007) 86:801–10. doi: 10.1093/ps/86.5.801
20. Hai, Y, Zhou, T, Gong, J, Young, C, Su, X, Li, X, et al. Isolation of deoxynivalenol-transforming bacteria from the chicken intestines using the approach of PCR-DGGE guided microbial selection. BMC Microbiol. (2010) 10:182. doi: 10.1186/1471-2180-10-182
21. Dibner, J, and Richards, D. Antibiotic growth promoters in agriculture: history and mode of action. Poult Sci. (2005) 84:634–43. doi: 10.1093/ps/84.4.634
22. Ricke, S. Perspectives on the use of organic acids and short chain fatty acids as antimicrobials. Poult Sci. (2003) 82:632–9. doi: 10.1093/ps/82.4.632
23. LeBlanc, J, Milani, C, De Giori, G, Sesma, F, van Sinderen, D, and Ventura, M. Bacteria as vitamin suppliers to their host: a gut microbiota perspective. Curr Opin Biotechnol. (2013) 24:160–8. doi: 10.1016/j.copbio.2012.08.005
24. Janda, JM, and Abbott, SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J Clin Microbiol. (2007) 45:2761–4. doi: 10.1128/jcm.01228-07
25. Duchêne, S, Holt, KE, Weill, FX, Le Hello, S, Hawkey, J, Edwards, DJ, et al. Genome-scale rates of evolutionary change in bacteria. Microb Genom. (2016) 2:e000094. doi: 10.1099/mgen.0.000094
26. Chakravorty, S, Helb, D, Burday, M, Connell, N, and Alland, D. A detailed analysis of 16S ribosomal RNA gene segments for the diagnosis of pathogenic bacteria. J Microbiol Methods. (2007) 69:330–9. doi: 10.1016/j.mimet.2007.02.005
27. Drancourt, M, Bollet, C, Carlioz, A, Martelin, R, Gayral, JP, and Raoult, D. 16S ribosomal DNA sequence analysis of a large collection of environmental and clinical unidentifiable bacterial isolates. J Clin Microbiol. (2000) 38:3623–30. doi: 10.1128/jcm.38.10.3623-3630.2000
28. Cole, JR, Wang, Q, Fish, JA, Chai, B, McGarrell, DM, Sun, Y, et al. Ribosomal database project: data and tools for high throughput rRNA analysis. Nucleic Acids Res. (2014) 42:D633–42. doi: 10.1093/nar/gkt1244
29. Quast, C, Pruesse, E, Yilmaz, P, Gerken, J, Schweer, T, Yarza, P, et al. The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. (2013) 41:D590–6. doi: 10.1093/nar/gks1219
30. Gantois, I, Ducatelle, R, Pasmans, F, Haesebrouck, F, Gast, R, Humphrey, TJ, et al. Mechanisms of egg contamination by Salmonella enteritidis. FEMS Microbiol Rev. (2009) 33:718–38. doi: 10.1111/j.1574-6976.2008.00161.x
31. Roto, SM, Kwon, YM, and Ricke, SC. Applications of in ovo technique for the optimal development of the gastrointestinal tract and the potential influence on the establishment of its microbiome in poultry. Front Vet Sci. (2016) 3:63. doi: 10.3389/fvets.2016.00063
32. Pedroso, AA, Menten, JFM, and Lambais, MR. The structure of bacterial community in the intestines of newly hatched chicks. J Appl Poult Res. (2005) 14:232–7. doi: 10.1093/japr/14.2.232
33. Lumpkins, BS, Batal, AB, and Lee, MD. Evaluation of the bacterial community and intestinal development of different genetic lines of chickens. Poult Sci. (2010) 89:1614–21. doi: 10.3382/ps.2010-00747
34. Apajalahti, J, Kettunen, A, and Graham, H. Characteristics of the gastrointestinal microbial communities, with special reference to the chicken. Worlds Poult Sci J. (2004) 60:223–32. doi: 10.1079/WPS200415
35. Lan, Y, Verstegen, MWA, Tamminga, S, and Williams, BA. The role of the commensal gut microbial community in broiler chickens. Worlds Poult Sci J. (2005) 61:95–104. doi: 10.1079/WPS200445
36. Crhanova, M, Hradecka, H, Faldynova, M, Matulova, M, Havlickova, H, Sisak, F, et al. Immune response of chicken gut to natural colonization by gut microflora and to Salmonella enterica serovar enteritidis infection. Infect Immun. (2011) 79:2755–63. doi: 10.1128/iai.01375-10
37. Borda-Molina, D, Seifert, J, and Camarinha-Silva, A. Current perspectives of the chicken gastrointestinal tract and its microbiome. Comput Struct Biotechnol J. (2018) 16:131–9. doi: 10.1016/j.csbj.2018.03.002
38. Robinson, K, Xiao, Y, Johnson, TJ, Chen, B, Yang, Q, Lyu, W, et al. Chicken intestinal mycobiome: initial characterization and its response to bacitracin methylene disalicylate. Appl Environ Microbiol. (2020) 86:86. doi: 10.1128/aem.00304-20
39. Stanley, D, Geier, MS, Chen, H, Hughes, RJ, and Moore, RJ. Comparison of fecal and cecal microbiotas reveals qualitative similarities but quantitative differences. BMC Microbiol. (2015) 15:51. doi: 10.1186/s12866-015-0388-6
40. Stanley, D, Hughes, RJ, Geier, MS, and Moore, RJ. Bacteria within the gastrointestinal tract microbiota correlated with improved growth and feed conversion: challenges presented for the identification of performance enhancing probiotic bacteria. Front Microbiol. (2016) 7:187. doi: 10.3389/fmicb.2016.00187
41. Willson, NL, Nattrass, GS, Hughes, RJ, Moore, RJ, Stanley, D, Hynd, PI, et al. Correlations between intestinal innate immune genes and cecal microbiota highlight potential for probiotic development for immune modulation in poultry. Appl Microbiol Biotechnol. (2018) 102:9317–29. doi: 10.1007/s00253-018-9281-1
42. Faldynova, M, Prikrylova, H, Sebkova, A, Volf, J, Karasova, D, Crhanova, M, et al. Contact with adult hens affects the composition of skin and respiratory tract microbiota in newly hatched chicks. Poult Sci. (2024) 103:103302. doi: 10.1016/j.psj.2023.103302
43. Lee, JY, Kang, SK, Heo, YJ, Shin, DW, Park, TE, Han, GG, et al. Influence of flaxseed oil on fecal microbiota, egg quality and fatty acid composition of egg yolks in laying hens. Curr Microbiol. (2016) 72:259–66. doi: 10.1007/s00284-015-0946-z
44. Wongkuna, S, Ambat, A, Ghimire, S, Mattiello, SP, Maji, A, Kumar, R, et al. Identification of a microbial sub-community from the feral chicken gut that reduces Salmonella colonization and improves gut health in a gnotobiotic chicken model. Microbiol Spectr. (2024) 12:e01621–3. doi: 10.1128/spectrum.01621-23
45. Long, C, Wang, J, Zhang, HJ, Wu, SG, and Qi, GH. Effects of dietary rapeseed meal supplementation on cecal microbiota in laying hens with different flavin-containing monooxygenase 3 genotypes. Poult Sci. (2017) 96:1748–58. doi: 10.3382/ps/pew449
46. Videnska, P, Faldynova, M, Juricova, H, Babak, V, Sisak, F, Havlickova, H, et al. Chicken faecal microbiota and disturbances induced by single or repeated therapy with tetracycline and streptomycin. BMC Vet Res. (2013) 9:30. doi: 10.1186/1746-6148-9-30
47. Wang, J, Fan, H, Han, Y, Wei, J, Zhao, J, and Zhou, Z. Pyrosequencing of the broiler chicken gastrointestinal tract reveals the regional similarity and dissimilarity of microbial community. Can J Anim Sci. (2016) 97:302–13. doi: 10.1139/cjas-2015-0120
48. Mancabelli, L, Milani, C, Lugli, GA, Turroni, F, Mangifesta, M, Viappiani, A, et al. Unveiling the gut microbiota composition and functionality associated with constipation through metagenomic analyses. Sci Rep. (2017) 7:9879. doi: 10.1038/s41598-017-10663-w
49. Aruwa, CE, Pillay, C, Nyaga, MM, and Sabiu, S. Poultry gut health–microbiome functions, environmental impacts, microbiome engineering and advancements in characterization technologies. J. Anim. Sci. Biotechnol, (2021). 12:1–15. doi: 10.1186/s40104-021-00640-9
50. Nordentoft, S, Mølbak, L, Bjerrum, L, De Vylder, J, Van Immerseel, F, and Pedersen, K. The influence of the cage system and colonisation of Salmonella enteritidis on the microbial gut flora of laying hens studied by T-RFLP and 454 pyrosequencing. BMC Microbiol. (2011) 11:187. doi: 10.1186/1471-2180-11-187
51. Candela, M, Vitali, B, Matteuzzi, D, and Brigidi, P. Evaluation of the rrn operon copy number in Bifidobacterium using real-time PCR. Lett Appl Microbiol. (2004) 38:229–32. doi: 10.1111/j.1472-765x.2003.01475.x
52. Kers, JG, Velkers, FC, Fischer, EAJ, Hermes, GDA, Stegeman, JA, and Smidt, H. Host and environmental factors affecting the intestinal microbiota in chickens. Front Microbiol. (2018) 9:235. doi: 10.3389/fmicb.2018.00235
53. Duggett, N. High-throughput sequencing of the chicken gut microbiome In: PhD dissertation. Birmingham: University of Birmingham (2016)
54. Wickramasuriya, SS, Park, I, Lee, K, Lee, Y, Kim, WH, Nam, H, et al. Role of physiology, immunity, microbiota, and infectious diseases in the gut health of poultry. Vaccines. (2022) 10:172. doi: 10.3390/vaccines10020172
55. Yan, W, Sun, C, Zheng, J, Wen, C, Ji, C, Zhang, D, et al. Efficacy of fecal sampling as a gut proxy in the study of chicken gut microbiota. Front Microbiol. (2019) 10:2126. doi: 10.3389/fmicb.2019.02126
56. Kollarcikova, M, Kubasova, T, Karasova, D, Crhanova, M, Cejkova, D, Sisak, F, et al. Use of 16S rRNA gene sequencing for prediction of new opportunistic pathogens in chicken ileal and cecal microbiota. Poult Sci. (2019) 98:2347–53. doi: 10.3382/ps/pey594
57. Oakley, BB, Morales, CA, Line, J, Berrang, ME, Meinersmann, RJ, Tillman, GE, et al. The poultry-associated microbiome: network analysis and farm-to-fork characterizations. PLoS One. (2013) 8:e57190. doi: 10.1371/journal.pone.0057190
58. Clavijo, V, and Flórez, MJV. The gastrointestinal microbiome and its association with the control of pathogens in broiler chicken production: a review. Poult Sci. (2018) 97:1006–21. doi: 10.3382/ps/pex359
59. Rothrock, MJ Jr, Locatelli, A, Feye, KM, Caudill, AJ, Guard, J, Hiett, K, et al. A microbiomic analysis of a pasture-raised broiler flock elucidates foodborne pathogen ecology along the farm-to-fork continuum. Front Vet Sci. (2019) 6:260. doi: 10.3389/fvets.2019.00260
60. Gao, P, Ma, C, Sun, Z, Wang, L, Huang, S, Su, X, et al. Feed-additive probiotics accelerate yet antibiotics delay intestinal microbiota maturation in broiler chicken. Microbiome. (2017) 5:91. doi: 10.1186/s40168-017-0315-1
61. Svihus, B, Hetland, H, Choct, M, and Sundby, F. Passage rate through the anterior digestive tract of broiler chickens fed on diets with ground and whole wheat. Br Poult Sci. (2002) 43:662–8. doi: 10.1080/0007166021000025037
62. Hughes, RJ. Relationship between digesta transit time and apparent metabolisable energy value of wheat in chickens. Br Poult Sci. (2008) 49:716–20. doi: 10.1080/00071660802449145
63. Warriss, PD, Wilkins, LJ, Brown, SN, Phillips, AJ, and Allen, V. Defaecation and weight of the gastrointestinal tract contents after feed and water withdrawal in broilers. Br Poult Sci. (2004) 45:61–6. doi: 10.1080/0007166041668879
64. Svihus, B, Choct, M, and Classen, HL. Function and nutritional roles of the avian caeca: a review. World Poult Sci J. (2013) 69:249–64. doi: 10.1017/S0043933913000287
65. Hinton, A Jr, Buhr, RJ, and Ingram, KD. Physical, chemical, and microbiological changes in the ceca of broiler chickens subjected to incremental feed withdrawal. Poult Sci. (2000) 79:483–8. doi: 10.1093/ps/79.4.483
66. Duke, GE. Relationship of cecal and colonic motility to diet, habitat, and cecal anatomy in several avian species. J Exp Zool Suppl. (1989) 252:38–47. doi: 10.1002/jez.1402520507
67. Duke, GE, Eccleston, E, Kirkwood, S, Louis, CF, and Bedbury, HP. Cellulose digestion by domestic turkeys fed low or high fiber diets. J Nutr. (1984) 114:95–102. doi: 10.1093/jn/114.1.95
68. Varmuzova, K, Kubasova, T, Davidova-Gerzova, L, Sisak, F, Havlickova, H, Sebkova, A, et al. Composition of gut microbiota influences resistance of newly hatched chickens to Salmonella enteritidis infection. Front Microbiol. (2016) 7:957. doi: 10.3389/fmicb.2016.00957
69. Stanley, D, Denman, SE, Hughes, RJ, Geier, MS, Crowley, TM, Chen, H, et al. Intestinal microbiota associated with differential feed conversion efficiency in chickens. Appl Microbiol Biotechnol. (2012) 96:1361–9. doi: 10.1007/s00253-011-3847-5
70. Flint, HJ, Bayer, EA, Rincon, MT, Lamed, R, and White, BA. Polysaccharide utilization by gut bacteria: potential for new insights from genomic analysis. Nat Rev Microbiol. (2008) 6:121–31. doi: 10.1038/nrmicro1817
71. Louis, P, and Flint, HJ. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett. (2009) 294:1–8. doi: 10.1111/j.1574-6968.2009.01514.x
72. Chica Cardenas, LA, Clavijo, V, Vives, M, and Reyes, A. Bacterial meta-analysis of chicken cecal microbiota. PeerJ. (2021) 9:e10571. doi: 10.7717/peerj.10571
73. Polansky, O, Sekelova, Z, Faldynova, M, Sebkova, A, Sisak, F, and Rychlik, I. Important metabolic pathways and biological processes expressed by chicken cecal microbiota. Appl Environ Microbiol. (2016) 82:1569–76. doi: 10.1128/AEM.03473-15
74. Han, Z, Willer, T, Pielsticker, C, Gerzova, L, Rychlik, I, and Rautenschlein, S. Differences in host breed and diet influence colonization by Campylobacter jejuni and induction of local immune responses in chicken. Gut Pathog. (2016) 8:56. doi: 10.1186/s13099-016-0133-1
75. Crhanova, M, Karasova, D, Juricova, H, Matiasovicova, J, Jahodarova, E, Kubasova, T, et al. Systematic culturomics shows that half of chicken caecal microbiota members can be grown in vitro except for two lineages of Clostridiales and a single lineage of Bacteroidetes. Microorganisms. (2019) 7:496. doi: 10.3390/microorganisms7110496
76. Medvecky, M, Cejkova, D, Polansky, O, Karasova, D, Kubasova, T, Cizek, A, et al. Whole genome sequencing and function prediction of 133 gut anaerobes isolated from chicken caecum in pure cultures. BMC Genomics. (2018) 19:561. doi: 10.1186/s12864-018-4959-4
77. Sergeant, MJ, Constantinidou, C, Cogan, TA, Bedford, MR, Penn, CW, and Pallen, MJ. Extensive microbial and functional diversity within the chicken cecal microbiome. PLoS One. (2014) 9:e91941. doi: 10.1371/journal.pone.0091941
78. Thiennimitr, P, Winter, SE, Winter, MG, Xavier, MN, Tolstikov, V, Huseby, DL, et al. Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci USA. (2011) 108:17480–5. doi: 10.1073/pnas.1107857108
79. Winter, SE, Thiennimitr, P, Winter, MG, Butler, BP, Huseby, DL, Crawford, RW, et al. Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature. (2010) 467:426–9. doi: 10.1038/nature09415
80. Adamberg, S, Tomson, K, Vija, H, Puurand, M, Kabanova, N, Visnapuu, T, et al. Degradation of fructans and production of propionic acid by Bacteroides thetaiotaomicron are enhanced by the shortage of amino acids. Front Nutr. (2014) 1:21. doi: 10.3389/fnut.2014.00021
81. Isar, J, Agarwal, L, Saran, S, and Saxena, RK. Succinic acid production from Bacteroides fragilis: process optimization and scale up in a bioreactor. Anaerobe. (2006) 12:231–7. doi: 10.1016/j.anaerobe.2006.07.001
82. Strobel, HJ. Vitamin B12-dependent propionate production by the ruminal bacterium Prevotella ruminicola 23. Appl Environ Microbiol. (1992) 58:2331–3. doi: 10.1128/aem.58.7.2331-2333.1992
83. Vital, M, Howe, AC, and Tiedje, JM. Revealing the bacterial butyrate synthesis pathways by analyzing (meta)genomic data. mBio. (2014) 5:e00889. doi: 10.1128/mBio.00889-14
84. Schnorr, SL, Candela, M, Rampelli, S, Centanni, M, Consolandi, C, Basaglia, G, et al. Gut microbiome of the Hadza hunter-gatherers. Nat Commun. (2014) 5:3654. doi: 10.1038/ncomms4654
85. Biddle, A, Stewart, L, Blanchard, J, and Leschine, S. Untangling the genetic basis of fibrolytic specialization by Lachnospiraceae and Ruminococcaceae in diverse gut communities. Diversity. (2013) 5:627–40. doi: 10.3390/d5030627
86. Gorvitovskaia, A, Holmes, SP, and Huse, SM. Interpreting Prevotella and Bacteroides as biomarkers of diet and lifestyle. Microbiome. (2016) 4:15. doi: 10.1186/s40168-016-0160-7
87. Chambers, JR, and Gong, J. The intestinal microbiota and its modulation for Salmonella control in chickens. Food Res Int. (2011) 44:3149–59. doi: 10.1016/j.foodres.2011.08.017
88. Rama, EN. Characterization of broiler gut microbiomes and pathogen prevalence in conventional and no antibiotics ever poultry production systems In: Doctoral dissertation. Athens: University of Georgia (2021)
89. Lorenzo-Rebenaque, L, Casto-Rebollo, C, Diretto, G, Frusciante, S, Rodríguez, JC, Ventero, MP, et al. Examining the effects of Salmonella phage on the caecal microbiota and metabolome features in Salmonella-free broilers. Front Genet. (2022) 13:1060713. doi: 10.3389/fgene.2022.1060713
90. Robinson, K, Assumpcao, ALFV, Arsi, K, Erf, GF, Donoghue, A, and Jesudhasan, PRR. Effect of Salmonella typhimurium colonization on microbiota maturation and blood leukocyte populations in broiler chickens. Animals. (2022) 12:2867. doi: 10.3390/ani12202867
91. Khan, S, and Chousalkar, KK. Salmonella typhimurium infection disrupts but continuous feeding of Bacillus based probiotic restores gut microbiota in infected hens. J Anim Sci Biotechnol. (2020) 11:29. doi: 10.1186/s40104-020-0433-7
92. Jacobson, A, Lam, L, Rajendram, M, Tamburini, F, Honeycutt, J, Pham, T, et al. A gut commensal-produced metabolite mediates colonization resistance to Salmonella infection. Cell Host Microbe. (2018) 24:296–307.e7. doi: 10.1016/j.chom.2018.07.002
93. Van Immerseel, F, De Buck, J, Pasmans, F, Huyghebaert, G, Haesebrouck, F, and Ducatelle, R. Clostridium perfringens in poultry: an emerging threat for animal and public health. Avian Pathol. (2004) 33:537–49. doi: 10.1080/03079450400013162
94. Prescott, JF, Parreira, VR, Mehdizadeh Gohari, I, Lepp, D, and Gong, J. The pathogenesis of necrotic enteritis in chickens: what we know and what we need to know: a review. Avian Pathol. (2016) 45:288–94. doi: 10.1080/03079457.2016.1139688
95. Kursa, O, Tomczyk, G, Sawicka-Durkalec, A, Giza, A, and Słomiany-Szwarc, M. Bacterial communities of the upper respiratory tract of turkeys. Sci Rep. (2021) 11:2544. doi: 10.1038/s41598-021-81984-0
96. Craft, J, Eddington, H, Christman, ND, Pryor, W, Chaston, JM, Erickson, DL, et al. Increased microbial diversity and decreased prevalence of common pathogens in the gut microbiomes of wild turkeys compared to domestic turkeys. Appl Environ Microbiol. (2022) 88:e0142321. doi: 10.1128/aem.01423-21
97. Morrison, DJ, and Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes. (2016) 7:189–200. doi: 10.1080/19490976.2015.1134082
98. Kim, CH, Park, J, and Kim, M. Gut microbiota-derived short-chain fatty acids, T cells, and inflammation. Immune Netw. (2014) 14:277–88. doi: 10.4110/in.2014.14.6.277
99. Hosseini, E, Grootaert, C, Verstraete, W, and Van de Wiele, T. Propionate as a health-promoting microbial metabolite in the human gut. Nutr Rev. (2011) 69:245–58. doi: 10.1111/j.1753-4887.2011.00388.x
100. Sullivan, DJ, Azlin-Hasim, S, Cruz-Romero, M, Cummins, E, Kerry, JP, and Morris, MA. Antimicrobial effect of benzoic and sorbic acid salts and nano-solubilisates against Staphylococcus aureus, Pseudomonas fluorescens and chicken microbiota biofilms. Food Control. (2020) 107:106786. doi: 10.1016/j.foodcont.2019.106786
101. Tellez, G, Higgins, SE, Donoghue, AM, and Hargis, BM. Digestive physiology and the role of microorganisms. J Appl Poult Res. (2006) 15:136–44. doi: 10.1093/japr/15.1.136
102. Xia, Y, Kong, J, Zhang, G, Zhang, X, Seviour, R, and Kong, Y. Effects of dietary inulin supplementation on the composition and dynamics of cecal microbiota and growth-related parameters in broiler chickens. Poult Sci. (2019) 98:6942–53. doi: 10.3382/ps/pez483
103. Rehman, HU, Vahjen, W, Awad, WA, and Zentek, J. Indigenous bacteria and bacterial metabolic products in the gastrointestinal tract of broiler chickens. Arch Anim Nutr. (2007) 61:319–35. doi: 10.1080/17450390701556817
104. van der Wielen, PW, Biesterveld, S, Notermans, S, Hofstra, H, Urlings, BA, and van Knapen, F. Role of volatile fatty acids in development of the cecal microflora in broiler chickens during growth. Appl Environ Microbiol. (2000) 66:2536–40. doi: 10.1128/aem.66.6.2536-2540.2000
105. Guan, L, Hu, A, Ma, S, Liu, J, Yao, X, Ye, T, et al. Lactiplantibacillus plantarum postbiotic protects against Salmonella infection in broilers via modulating NLRP3 inflammasome and gut microbiota. Poult Sci. (2024) 103:103483. doi: 10.1016/j.psj.2024.103483
106. Wei, S, Morrison, M, and Yu, Z. Bacterial census of poultry intestinal microbiome. Poult Sci. (2013) 92:671–83. doi: 10.3382/ps.2012-02822
107. Luo, YH, Peng, HW, Wright, AD, Bai, SP, Ding, XM, Zeng, QF, et al. Broilers fed dietary vitamins harbor higher diversity of cecal bacteria and higher ratio of Clostridium, Faecalibacterium, and Lactobacillus than broilers with no dietary vitamins revealed by 16S rRNA gene clone libraries. Poult Sci. (2013) 92:2358–66. doi: 10.3382/ps.2012-02935
108. Kc, M, Ngunjiri, JM, Lee, J, Ahn, J, Elaish, M, Ghorbani, A, et al. Avian Toll-like receptor 3 isoforms and evaluation of toll-like receptor 3-mediated immune responses using knockout quail fibroblast cells. Poult Sci. (2020) 99:6513–24. doi: 10.1016/j.psj.2020.09.029
109. Anwar, MN, Ravindran, V, Morel, PCH, Ravindran, G, and Cowieson, AJ. Measurement of the true ileal calcium digestibility of some feed ingredients for broiler chickens. Anim Feed Sci Technol. (2018) 237:118–28. doi: 10.1016/j.anifeedsci.2018.01.010
110. Polycarpo, GV, Burbarelli, MF, CarÃo, AC, Merseguel, CE, Dadalt, JC, Maganha, SR, et al. Effects of lipid sources, lysophospholipids and organic acids in maize-based broiler diets on nutrient balance, liver concentration of fat-soluble vitamins, jejunal microbiota and performance. Br Poult Sci. (2016) 57:788–98. doi: 10.1080/00071668.2016.1219019
111. Costa, MM, Pio, LB, Bule, P, Cardoso, VA, Duarte, M, Alfaia, CM, et al. Recalcitrant cell wall of Ulva lactuca seaweed is degraded by a single ulvan lyase from family 25 of polysaccharide lyases. Anim Nutr. (2022) 9:184–92. doi: 10.1016/j.aninu.2022.01.004
112. Liu, L, Li, Q, Yang, Y, and Guo, A. Biological function of short-chain fatty acids and its regulation on intestinal health of poultry. Front Vet Sci. (2021) 8:736739. doi: 10.3389/fvets.2021.736739
113. Watanabe, M, Kojima, H, Umezawa, K, and Fukui, M. Genomic characteristics of Desulfonema ishimotonii Tokyo 01T implying horizontal gene transfer among phylogenetically dispersed filamentous gliding bacteria. Front Microbiol. (2019) 10:227. doi: 10.3389/fmicb.2019.00227
114. Lu, P-y, Wang, J, Wu, S-g, Gao, J, Dong, Y, Zhang, H-j, et al. Standardized ileal digestible amino acid and metabolizable energy content of wheat from different origins and the effect of exogenous xylanase on their determination in broilers. Poult Sci. (2020) 99:992–1000. doi: 10.1016/j.psj.2019.10.013
115. Melo, TP, Fortes, MRS, Bresolin, T, Mota, LFM, Albuquerque, LG, and Carvalheiro, R. Multitrait meta-analysis identified genomic regions associated with sexual precocity in tropical beef cattle. J Anim Sci. (2018) 96:4087–99. doi: 10.1093/jas/sky289
116. Kogut, M, and Arsenault, R. Immunometabolic phenotype alterations associated with the induction of disease tolerance and persistent asymptomatic infection of Salmonella in the chicken intestine. Front Immunol. (2017) 8:372. doi: 10.3389/fimmu.2017.00372
117. Kogut, MH. The effect of microbiome modulation on the intestinal health of poultry. Anim Feed Sci Technol. (2019) 250:32–40. doi: 10.1016/j.anifeedsci.2018.10.008
118. Pirr, S, and Viemann, D. Host factors of favorable intestinal microbial colonization. Front Immunol. (2020) 11:584288. doi: 10.3389/fimmu.2020.584288
119. León, E, and Francino, M. Roles of secretory immunoglobulin A in host-microbiota interactions in the gut ecosystem. Front Microbiol. (2022) 13:880484. doi: 10.3389/fmicb.2022.880484
120. Abaidullah, M, Peng, S, Kamran, M, Song, X, and Yin, Z. Current findings on gut microbiota mediated immune modulation against viral diseases in chicken. Viruses. (2019) 11:681. doi: 10.3390/v11080681
121. Spor, A, Koren, O, and Ley, R. Unravelling the effects of the environment and host genotype on the gut microbiome. Nat Rev Microbiol. (2011) 9:279–90. doi: 10.1038/nrmicro2540
122. Chang, C, and Lin, H. Dysbiosis in gastrointestinal disorders. Best Pract Res Clin Gastroenterol. (2016) 30:3–15. doi: 10.1016/j.bpg.2016.02.001
123. Deriu, E, Boxx, GM, He, X, Pan, C, Benavidez, SD, Cen, L, et al. Influenza virus affects intestinal microbiota and secondary Salmonella infection in the gut through type I interferons. PLoS Pathog. (2016) 12:e1005572. doi: 10.1371/journal.ppat.1005572
124. Winter, SE, Winter, MG, Xavier, MN, Thiennimitr, P, Poon, V, Keestra, AM, et al. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science. (2013) 339:708–11. doi: 10.1126/science.1232467
125. Barnes, EM. The intestinal microflora of poultry and game birds during life and after storage. Address of the president of the Society for Applied Bacteriology delivered at a meeting of the society on 10 January 1979. J Appl Bacteriol. (1979) 46:407–19. doi: 10.1111/j.1365-2672.1979.tb00838.x
126. Gong, J, Si, W, Forster, RJ, Huang, R, Yu, H, Yin, Y, et al. 16S rRNA gene-based analysis of mucosa-associated bacterial community and phylogeny in the chicken gastrointestinal tracts: from crops to ceca. FEMS Microbiol Ecol. (2007) 59:147–57. doi: 10.1111/j.1574-6941.2006.00193.x
127. Ma, X, Wang, Q, Li, H, Xu, C, Cui, N, and Zhao, X. 16S rRNA genes Illumina sequencing revealed differential cecal microbiome in specific pathogen free chickens infected with different subgroup of avian leukosis viruses. Vet Microbiol. (2017) 207:195–204. doi: 10.1016/j.vetmic.2017.05.016
128. Stanley, D, Wu, SB, Rodgers, N, Swick, RA, and Moore, RJ. Differential responses of cecal microbiota to fishmeal, Eimeria and Clostridium perfringens in a necrotic enteritis challenge model in chickens. PLoS One. (2014) 9:e104739. doi: 10.1371/journal.pone.0104739
129. Chen, CJ, Wu, GH, Kuo, RL, and Shih, SR. Role of the intestinal microbiota in the immunomodulation of influenza virus infection. Microbes Infect. (2017) 19:570–9. doi: 10.1016/j.micinf.2017.09.002
130. Clarke, TB. Early innate immunity to bacterial infection in the lung is regulated systemically by the commensal microbiota via nod-like receptor ligands. Infect Immun. (2014) 82:4596–606. doi: 10.1128/iai.02212-14
131. Fagundes, CT, Amaral, FA, Vieira, AT, Soares, AC, Pinho, V, Nicoli, JR, et al. Transient TLR activation restores inflammatory response and ability to control pulmonary bacterial infection in germfree mice. J Immunol. (2012) 188:1411–20. doi: 10.4049/jimmunol.1101682
132. Schuijt, TJ, Lankelma, JM, Scicluna, BP, de Sousa e Melo, F, Roelofs, JJTH, de Boer, JD, et al. The gut microbiota plays a protective role in the host defence against pneumococcal pneumonia. Gut. (2016) 65:575–83. doi: 10.1136/gutjnl-2015-309728
133. Althammer, F, and Grinevich, V. Diversity of oxytocin neurones: beyond magno- and parvocellular cell types? J Neuroendocrinol. (2018):30. doi: 10.1111/jne.12549
134. Ziółkowski, H, Jasiecka-Mikołajczyk, A, Madej-Śmiechowska, H, Janiuk, J, Zygmuntowicz, A, and Dąbrowski, M. Comparative pharmacokinetics of chlortetracycline, tetracycline, minocycline, and tigecycline in broiler chickens. Poult Sci. (2020) 99:4750–7. doi: 10.1016/j.psj.2020.06.038
135. Cao, C, Chowdhury, VS, Cline, MA, and Gilbert, ER. The microbiota-gut-brain axis during heat stress in chickens: a review. Front Physiol. (2021) 12:752265. doi: 10.3389/fphys.2021.752265
136. Seo, KW, and Lee, YJ. Prevalence and characterization of plasmid mediated quinolone resistance genes and class 1 integrons among multidrug-resistant Escherichia coli isolates from chicken meat. J Appl Poult Res. (2019) 28:761–70. doi: 10.3382/japr/pfz016
137. Alves, MN, Neto, SE, Alves, AS, Fonseca, BM, Carrêlo, A, Pacheco, I, et al. Characterization of the periplasmic redox network that sustains the versatile anaerobic metabolism of Shewanella oneidensis MR-1. Front Microbiol. (2015) 6:665. doi: 10.3389/fmicb.2015.00665
138. Cryan, JF, and Dinan, TG. Mind-altering microorganisms: the impact of the gut microbiota on brain and behaviour. Nat Rev Neurosci. (2012) 13:701–12. doi: 10.1038/nrn3346
139. Uyanga, VA, Onagbesan, OM, Oke, OE, Abiona, JA, and Egbeyale, LT. Influence of age of broiler breeders and storage duration on egg quality and blastoderm of Marshall broiler breeders. J Appl Poult Res. (2020) 29:535–44. doi: 10.1016/j.japr.2020.03.001
140. Wu, M, Tian, T, Mao, Q, Zou, T, Zhou, CJ, Xie, J, et al. Associations between disordered gut microbiota and changes of neurotransmitters and short-chain fatty acids in depressed mice. Transl Psychiatry. (2020) 10:350. doi: 10.1038/s41398-020-01038-3
141. Jadhav, V, Han, J, Fasina, Y, and Harrison, S. Connecting gut microbiomes and short chain fatty acids with the serotonergic system and behavior in Gallus gallus and other avian species. Front Physiol. (2022) 13:1035538. doi: 10.3389/fphys.2022.1035538
142. Lewin, GR, Carlos, C, Chevrette, MG, Horn, HA, McDonald, BR, et al., Stankey, RJ. Evolution and ecology of Actinobacteria and their bioenergy applications. Annu. Rev. Microbiol. (2016) 70:235–254. doi: 10.1146/annurev-micro-102215-095748
143. Eeckhaut, V, Wang, J, Van Parys, A, Haesebrouck, F, Joossens, M, Falony, G, et al. The Probiotic Butyricicoccus pullicaecorum Reduces Feed Conversion and Protects from Potentially Harmful Intestinal Microorganisms and Necrotic Enteritis in Broilers. Front Microbiol. (2016) 7:1416. doi: 10.3389/fmicb.2016.01416
Keywords: gut microbiota, immune function, microbial composition, intestinal health, diseases
Citation: Yue Y, Luasiri P, Li J, Laosam P and Sangsawad P (2024) Research advancements on the diversity and host interaction of gut microbiota in chickens. Front. Vet. Sci. 11:1492545. doi: 10.3389/fvets.2024.1492545
Edited by:
Jianzhu Liu, Shandong Agricultural University, ChinaReviewed by:
Daniel Hernandez-Patlan, National Autonomous University of Mexico, MexicoRajesh Durairaj, Institut de Recherche en Sémiochimie et Ethologie Appliquée (IRSEA), France
Copyright © 2024 Yue, Luasiri, Li, Laosam and Sangsawad. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Papungkorn Sangsawad, cGFwdW5na29ybkBzdXQuYWMudGg=
†ORCID: Yong Yue, orcid.org/0009-0004-0904-8390
Pichitpon Luasiri, orcid.org/0009-0007-2332-324X
Phanthipha Laosam, orcid.org/0000-0003-1684-338X
Papungkorn Sangsawad, orcid.org/0000-0003-2420-8634
 Yong Yue
Yong Yue Pichitpon Luasiri1,2†
Pichitpon Luasiri1,2† Papungkorn Sangsawad
Papungkorn Sangsawad