- 1Jiangsu Agri-Animal Husbandry Vocational College, Jiangsu, China
- 2State Key Laboratory for Animal Disease Control and Prevention, Harbin Veterinary Research Institute of Chinese Academy of Agricultural Sciences, Harbin, China
- 3Heilongjiang Provincial Research Center for Veterinary Biomedicine, Harbin, China
The porcine reproductive and respiratory syndrome virus (PRRSV) is a highly significant infectious disease that poses a substantial threat to the global pig industry. In recent years, the NADC30-like strain has gradually emerged as prevalent in China, causing a profound impact on the country’s pig farming industry. Therefore, it is important to conduct an in-depth study on the characteristics and gene functions of the NADC30-like strain. An infectious cDNA clone is an indispensable tool for investigating the functions of viral genes. In this current study, we successfully isolated a NADC30-like strain and constructed its full-length infectious cDNA clone. The utilization of this clone will facilitate our investigation into the viral replication, pathogenesis, and immune response associated with the PRRSV NADC30-like strain.
Introduction
Porcine reproductive and respiratory syndrome virus (PRRSV) is a highly contagious viral disease that affects pigs worldwide, posing a significant threat to the global pig industry. The PRRSVs, belonging to the family Arteriviridae, are typically classified into two distinguished species: PRRSV-1 and PRRSV-2 (1–3). China is the largest country for pig production and has the world’s largest market for pork consumption. PRRSV-2, identified as the epidemic strain in China since its outbreak, has caused significant losses for most farms (4–6). The PRRSV-2 can be classified into nine lineages (Lineage 1–Lineage 9), among which Lineage 1, Lineage 3, Lineage 5, and Lineage 8 have emerged in China since 1996 (7, 8). The prevalent PRRSV-2 strains in China are primarily classified into four lineages: Lineage1 (NADC30-like/NADC34-like), Lineage3 (QYYZ-like), Lineage5 (VR2332-like), and Lineage8 (HP-PRRSV-like/CH-1a-like) (9). Currently, Lineage 1 NADC30-like PRRSV and NADC34-like PRRSV have become the main endemic strains in China (9). These new PRRSV variants pose additional challenges as they may exhibit different characteristics compared to previously known strains.
To better understand the characteristics and gene functions of this NADC30-like strain, it is crucial to conduct an in-depth study. One essential tool for investigating viral gene functions is an infectious cDNA clone, which will allow researchers to manipulate specific genes of the virus’s genome, enabling us to study their effects on viral replication, pathogenesis, and immune response (10–12). In current study, we first isolated a NADC30-like PRRSV. Moreover, we constructed a full-length cDNA of this strain. Understanding the characteristics and gene functions of the NADC30-like strain will provide valuable insights into its pathogenicity, aiding in the development of effective control strategies against PRRSV outbreaks in China and globally.
Materials and methods
Cells, virus, reagent, and plasmids
MARC-145 cells were stored in our lab as previous works (10, 11). Immortalized porcine alveolar macrophages (iPAMs) were described as our previous work (13). HeN-L1 strain was isolated in a farm at Henan province. The sequence of HeN-L1 strain was confirmed by amplifying the genome as previously described (14). Briefly, viral RNA was extracted using the QIAamp Viral RNA Mini Kit (QIAGEN) according to the instructions, followed by cDNA synthesis using PrimeScript™ II 1st Strand cDNA Synthesis Kit (TaKaRa). Indicated viral fragments were amplified by Q5 High-Fidelity polymerase (NEB). 5′ and 3′ RACE reactions (Invitrogen) were performed to acquire the terminal untranslated regions. The full-length HeN-L1 genome sequence assembled and subsequently submitted to GenBank (Accession No. PQ062578). Our laboratory stores the pOK12, pcDNA3.1(+), and pUC19 vectors. The pEASY-Blunt3 Cloning Kit, a high-efficiency cloning vector kit, purchased from Beijing Quanshi Jin Biotechnology Co., Ltd. (China). ThermoFisher (United States) provided the restriction endonucleases used in this study. Roche (United States) provides the X-tremeGENE HP DNA Transfection Reagent for transfection. Monoclonal antibodies against PRRSV N protein stored in our lab and FITC-labeled goat anti-mouse IgG antibody purchased from Sigma (United States).
Sequence analysis
Sequence analysis was described as our previous work (13).
Assembly of full-length cDNA
The restriction enzyme sites present in both the pOK12 vector sequence and HeN-L1 full genome sequence were analyzed using SnapGene software. Subsequently, the pOK12 vector was first modified as shown in Table 1. The MARC-145 cells were infected with HeN-L1 virus at a multiplicity of infection (MOI) of 0.1. When cytopathic effects (CPE) occurred, the viral culture underwent three cycles of freezing and thawing at −80°C, followed by extraction of total RNA from the supernatant using an RNA extraction kit. Then, using the reverse-transcribed cDNA as a template, we employed specific primers listed in Table 1 to amplify the complete sequence of HeN-L1. The fragments were cloned into modified pOK12 vectors individually. Subsequently, the full-length cDNA was assembled.
Virus rescue
The MARC-145 cells were seeded into a six-well plate and cultured until reaching a confluent monolayer density of 70–80%. Subsequently, the cells were transfected with pOK-HeN-L1 using X-tremeGENE HP DNA Transfection Reagent, while pOK-MCS empty vector used as a control. Once CPE were observed, the viral particles were collected for subsequent experiments as our recent work (10).
Biomarker detection
The viral RNA of rescued HeN-L1 was extracted, followed by reverse transcription into cDNA. Subsequently, PCR amplification was conducted, and the resulting PCR products were subjected to DNA sequencing.
Immunofluorescence assay and Western blot
The rescued HeN-L1 was inoculated into MARC-145 cells, followed by IFA or Western blot analysis 48 h post-infection as our previous work (15–17). Additionally, a negative control without virus inoculation was included.
Results and discussion
HeN-L1 isolation and sequence analysis
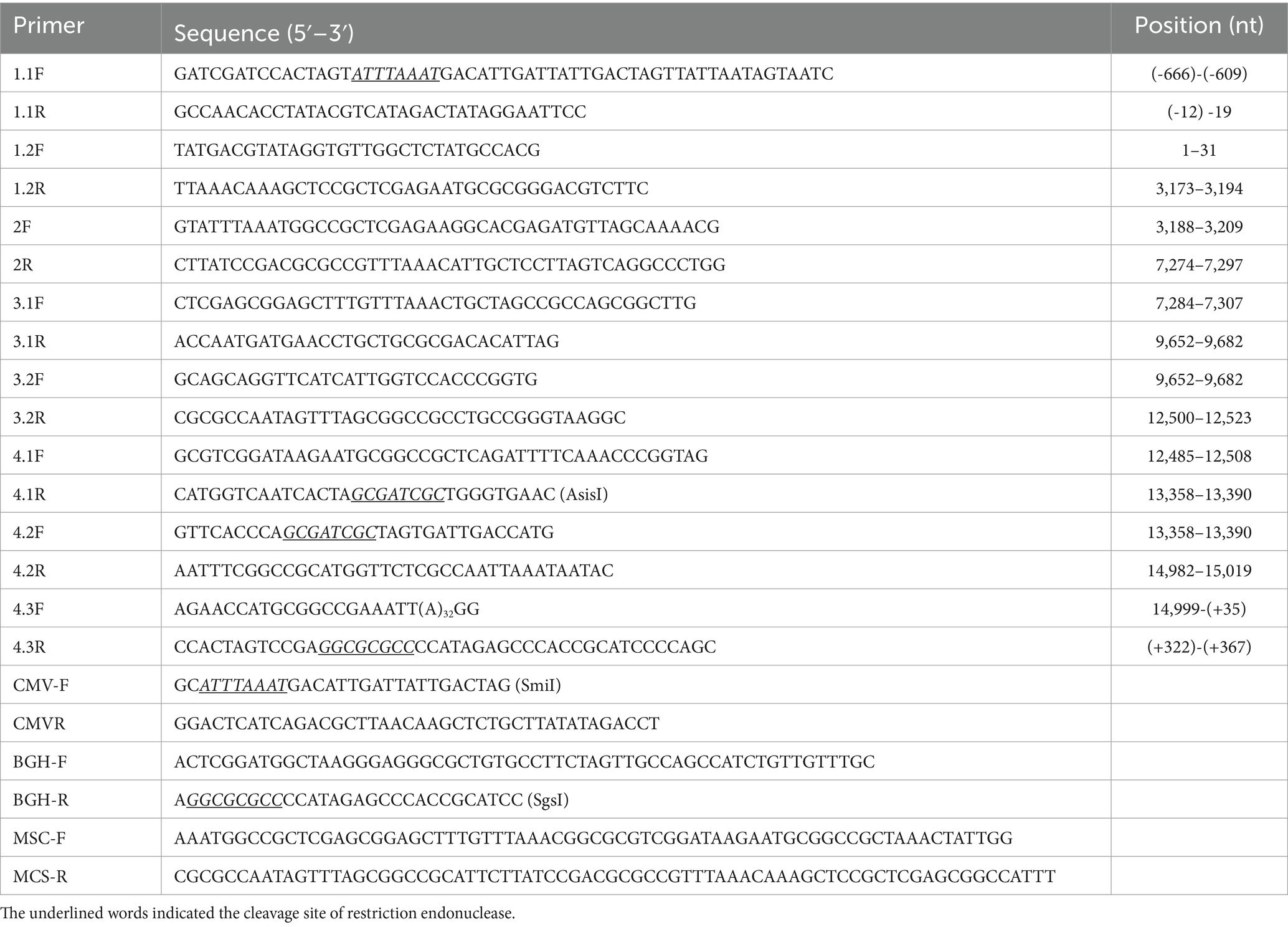
The HeN-L1 strain of NADC30-like PRRSV was isolated from an aborted fetus in Henan province, China. This virus was capable of infecting MARC-145 cells and iPAM-Tang cells (Figure 1A). The full-length genome sequence of HeN-L1 was determined to be 15,017 bp [excluding the poly (A) tail] through DNA sequencing. Phylogenetic analysis revealed that the HeN-L1 strain clustered with other NADC30-like PRRSV isolates such as HNjz15, CHsx1401, and SD-A19 (Figure 1B). To further characterize the HeN-L1 strain, its NSP2 sequence was aligned with reference PRRSV strains. Sequence alignment indicated that HeN-L1 exhibited three discontinuous deletions in NSP2: a 111-amino acid deletion from position 322 to 432, a single amino acid deletion at position 483, and a 19-amino acid deletion from position 504 to 522 (Figure 1C). The recombinant analysis revealed that HeN-L1 is a strain resulting from recombination (Figure 2). These deletions are consistent with those observed in SD-A19 and NADC30 strains. Collectively, these findings preliminarily suggest that HeN-L1 represents one of the circulating strains of NADC30-like PRRSVs in China.
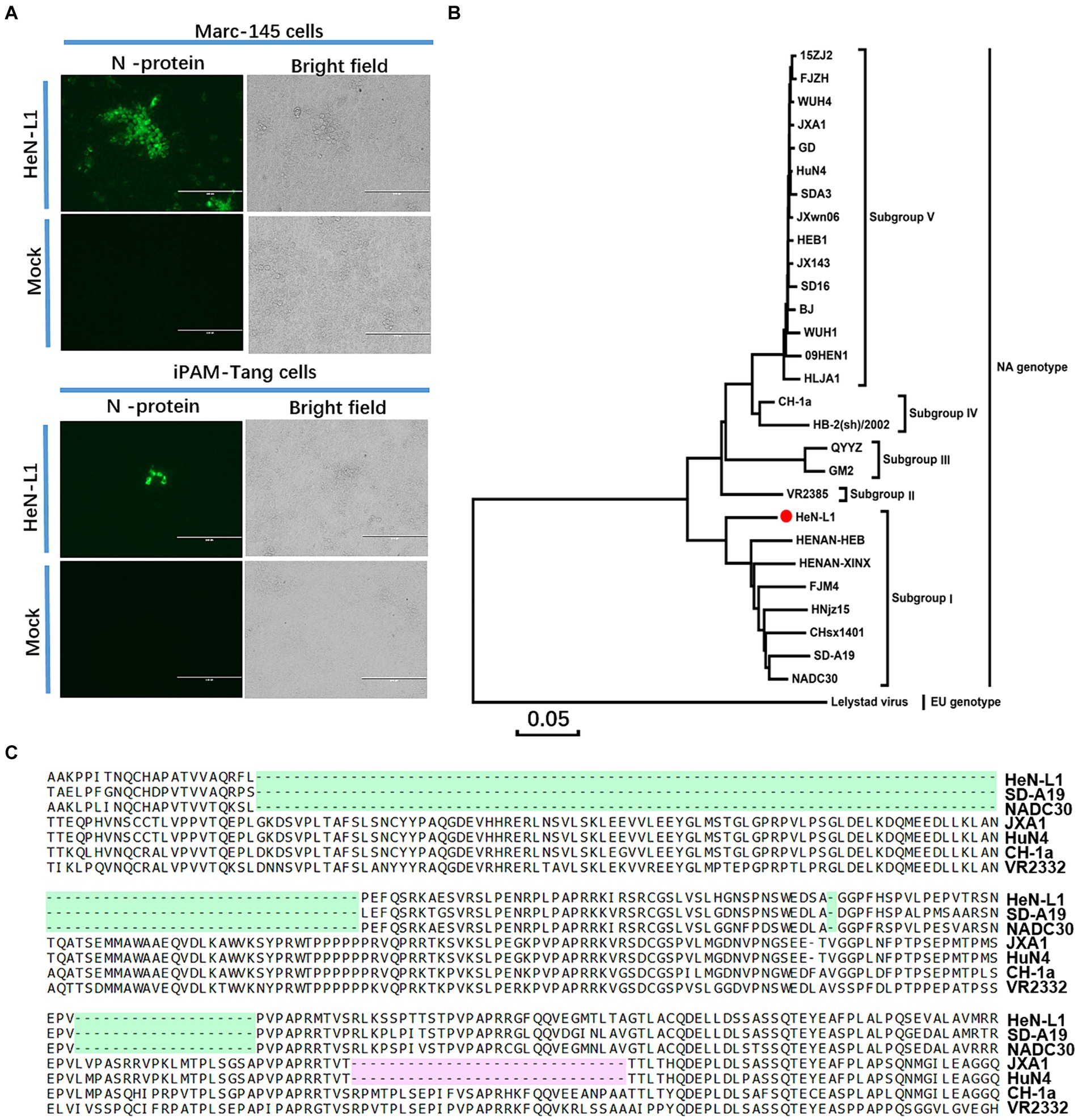
Figure 1. HeN-L1 isolation and sequence analysis. (A) MARC-145 cells and iPAM-Tang cells were infected with HeN-L1 strain, respectively. Mock infection as a control. (B) Phylogenies of HeN-L1 strain. (C) Amino acids alignment for nsp2.
Figure 2. The HeN-L1 strain was subjected to genome recombination analysis using Simplot 3.5.1 software. The complete genome of HeN-L1 was utilized as the query sequence, and the recombination locations are presented at the bottom.
Construction of full-length HeN-L1 cDNA clone
The infectious clone serves as a crucial platform for investigating the functional aspects of specific viruses and plays a pivotal role in the development of novel vaccines (18–20). A DNA-based approach was employed to generate the infectious clone of HeN-L1 (Figure 3A). The infectious cDNA clone of HeN-L1 strain was generated by inserting full-length genomic cDNA into the low-copy-number vector pOK12 under the control of the eukaryotic RNA polymerase II (Pol II) cytomegalovirus (CMV) promoter. To ensure the release of the authentic 5′ end and 3′end of the viral RNA, the hammerhead ribozyme (HamRz) and hepatitis delta ribozyme (HdvRz) were inserted prior to or after the HeN-L1 genome. Bovine growth hormone polyadenylation signal (BGH) sequence were utilized for efficient transcription termination. In order to differentiate parental virus or clone-derived virus, CT to GC was introduced at 13,281–13,282 nt position of the viral genome. To construct a full-length HeN-L1 cDNA clone, we first infected MARC-145 cells with HeN-L1 at an MOI of 0.01. After 24 h post-infection, the viruses were collected for viral RNA extraction and then reverse transcribed using random primers. The assembly strategy was illustrated in Figure 3A, and we amplified the fragments of HeN-L1 using specific primers listed in Table 1, with the reverse-transcribed cDNA serving as a template.
Figure 3. The schematic diagram of the HeN-L1 infectious cDNA clone. (A) S protein consists of S1 and S2 domain, and 652 and 661 near the fusion peptide is illustrated. (B) MARC-145 cells infected with rescued HeN-L1 strain. Mock infection as a control. (C) Sequence analysis of biomarker of rescued HeN-L1 strain. (D) The MARC-145 cells were infected with wild-type and rescued virus at a multiplicity of infection (MOI) of 0.1, followed by cell lysis after 24 h for subsequent Western blot analysis.
After assembling the full-length cDNA, we recovered infectious virus by transfecting the viral cDNA clone into MARC-145 cells. The cytopathic effect (CPE) became visible on day 4 post-transfection (data not shown). The rescued virus was further confirmed by re-infecting MARC-145 cells. An indirect immunofluorescence assay against PRRSV N protein revealed a significant number of cells expressing viral N protein on day 3 post-infection (Figure 3B). To ensure that this virus was not contaminated with wild-type virus, we extracted RNAs and amplified the fragment containing a biomarker of the viral genome using RT-PCR. After DNA sequencing, the genetic marker was identified in the rescued virus (Figure 3C). We further confirmed rescued virus by Western blot (Figure 3D).
Overall, the present study successfully isolated a NADC30-like PRRSV and constructed an infectious cDNA clone of this virus. The utilization of this platform will enhance our future investigations aimed at comprehending the characteristics of NADC30-like PRRSV.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Author contributions
Y-YQ: Data curation, Funding acquisition, Investigation, Writing – original draft. H-MW: Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Resources, Writing – original draft. HL: Project administration, Resources, Software, Writing – original draft. Y-JW: Methodology, Resources, Software, Validation, Writing – original draft. WZ: Methodology, Software, Validation, Visualization, Writing – original draft. HG: Methodology, Resources, Writing – original draft. X-HC: Conceptualization, Resources, Writing – original draft. Q-SX: Conceptualization, Resources, Supervision, Writing – original draft, Writing – review & editing. Z-YC: Conceptualization, Resources, Supervision, Writing – original draft, Writing – review & editing. Y-DT: Conceptualization, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was funded by the grants from Jiangsu Agri-Animal Husbandry Vocational College (NSF2023CB06 and NSF2023CB17).
Acknowledgments
We thank Qian Wang for providing N protein antibody.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Gorbalenya, AE, Enjuanes, L, Ziebuhr, J, and Snijder, EJ. Nidovirales: evolving the largest RNA virus genome. Virus Res. (2006) 117:17–37. doi: 10.1016/j.virusres.2006.01.017
2. Allende, R, Lewis, TL, Lu, Z, Rock, DL, Kutish, GF, Ali, A, et al. North American and European porcine reproductive and respiratory syndrome viruses differ in non-structural protein coding regions. J Gen Virol. (1999) 80:307–15. doi: 10.1099/0022-1317-80-2-307
3. Ruedas-Torres, I, Rodriguez-Gomez, IM, Sanchez-Carvajal, JM, Larenas-Munoz, F, Pallares, FJ, Carrasco, L, et al. The jigsaw of PRRSV virulence. Vet Microbiol. (2021) 260:109168. doi: 10.1016/j.vetmic.2021.109168
4. An, TQ, Tian, ZJ, Xiao, Y, Li, R, Peng, JM, Wei, TC, et al. Origin of highly pathogenic porcine reproductive and respiratory syndrome virus, China. Emerg Infect Dis. (2010) 16:365–7. doi: 10.3201/eid1602.090005
5. Zhou, L, and Yang, H. Porcine reproductive and respiratory syndrome in China. Virus Res. (2010) 154:31–7. doi: 10.1016/j.virusres.2010.07.016
6. Li, C, Zhao, J, Li, W, Xu, H, Gong, B, Sun, Q, et al. Prevalence and genetic evolution of porcine reproductive and respiratory syndrome virus in commercial fattening pig farms in China. Porcine Health Manag. (2024) 10:5. doi: 10.1186/s40813-024-00356-y
7. Shi, M, Lam, TT, Hon, CC, Murtaugh, MP, Davies, PR, Hui, RK, et al. Phylogeny-based evolutionary, demographical, and geographical dissection of north American type 2 porcine reproductive and respiratory syndrome viruses. J Virol. (2010) 84:8700–11. doi: 10.1128/JVI.02551-09
8. Guo, Z, Chen, XX, Li, R, Qiao, S, and Zhang, G. The prevalent status and genetic diversity of porcine reproductive and respiratory syndrome virus in China: a molecular epidemiological perspective. Virol J. (2018) 15:2. doi: 10.1186/s12985-017-0910-6
9. Xu, H, Li, C, Li, W, Zhao, J, Gong, B, Sun, Q, et al. Novel characteristics of Chinese NADC34-like PRRSV during 2020-2021. Transbound Emerg Dis. (2022) 69:e3215–24. doi: 10.1111/tbed.14485
10. Bai, YZ, Sun, Y, Liu, YG, Zhang, HL, An, TQ, Wang, Q, et al. Minor envelope proteins from GP2a to GP4 contribute to the spread pattern and yield of type 2 PRRSV in MARC-145 cells. Front Cell Infect Microbiol. (2024) 14:1376725. doi: 10.3389/fcimb.2024.1376725
11. Wang, TY, Fang, QQ, Cong, F, Liu, YG, Wang, HM, Zhang, HL, et al. The Nsp12-coding region of type 2 PRRSV is required for viral subgenomic mRNA synthesis. Emerg Microbes Infect. (2019) 8:1501–10. doi: 10.1080/22221751.2019.1679010
12. Tang, YD, Fang, QQ, Liu, JT, Wang, TY, Wang, Y, Tao, Y, et al. Open reading frames 1a and 1b of the porcine reproductive and respiratory syndrome virus (PRRSV) collaboratively initiate viral minus-strand RNA synthesis. Biochem Biophys Res Commun. (2016) 477:927–31. doi: 10.1016/j.bbrc.2016.06.161
13. Wang, TY, Liu, YG, Li, L, Wang, G, Wang, HM, Zhang, HL, et al. Porcine alveolar macrophage CD163 abundance is a pivotal switch for porcine reproductive and respiratory syndrome virus infection. Oncotarget. (2018) 9:12174–85. doi: 10.18632/oncotarget.24040
14. Wang, HM, Liu, YG, Tang, YD, Liu, TX, Zheng, LL, Wang, TY, et al. A natural recombinant PRRSV between HP-PRRSV JXA1-like and NADC30-like strains. Transbound Emerg Dis. (2018) 65:1078–86. doi: 10.1111/tbed.12852
15. Zhang, YY, Liang, R, Wang, SJ, Ye, ZW, Wang, TY, Chen, M, et al. SARS-CoV-2 hijacks macropinocytosis to facilitate its entry and promote viral spike-mediated cell-to-cell fusion. J Biol Chem. (2022) 298:102511. doi: 10.1016/j.jbc.2022.102511
16. Yang, YL, Liu, J, Wang, TY, Chen, M, Wang, G, Yang, YB, et al. Aminopeptidase N is an entry co-factor triggering porcine deltacoronavirus entry via an endocytotic pathway. J Virol. (2021) 95:e0094421. doi: 10.1128/JVI.00944-21
17. Yang, YL, Meng, F, Qin, P, Herrler, G, Huang, YW, and Tang, YD. Trypsin promotes porcine deltacoronavirus mediating cell-to-cell fusion in a cell type-dependent manner. Emerg Microbes Infect. (2020) 9:457–68. doi: 10.1080/22221751.2020.1730245
18. Tang, YD, Yu, C, and Cai, XH. Novel technologies are turning a dream into reality: conditionally replicating viruses as vaccines. Trends Microbiol. (2024) 32:292–301. doi: 10.1016/j.tim.2023.09.002
19. Wang, TY, Meng, FD, Sang, GJ, Zhang, HL, Tian, ZJ, Zheng, H, et al. A novel viral vaccine platform based on engineered transfer RNA. Emerg Microbes Infect. (2023) 12:2157339. doi: 10.1080/22221751.2022.2157339
Keywords: NADC30-like, infectious cDNA clone, PRRSV, NADC30, PRRSV-2
Citation: Qiao Y-Y, Wang H-M, Lu H, Wang Y-J, Zhang W, Gu H, Cai X-H, Xu Q-S, Chen Z-Y and Tang Y-D (2024) Generation of an infectious cDNA clone for NADC30-like PRRSV. Front. Vet. Sci. 11:1468981. doi: 10.3389/fvets.2024.1468981
Edited by:
Shixing Yang, Jiangsu University, ChinaReviewed by:
Junhua Huang, Texas A&M University, United StatesZhibang Zhang, Yichun University, China
Copyright © 2024 Qiao, Wang, Lu, Wang, Zhang, Gu, Cai, Xu, Chen and Tang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Qin-Se Xu, MTM4NTI4NDU1OTlAMTYzLmNvbQ==; Zhang-Yan Chen, NjMzNzQ5M0AxMjYuY29t; Yan-Dong Tang, dGFuZ3lhbmRvbmcyMDA4QDE2My5jb20=
†These authors have contributed equally to this work
 Yang-Yang Qiao1†
Yang-Yang Qiao1† Yan-Dong Tang
Yan-Dong Tang