- 1Institute of Biotechnology, Brandenburg University of Technology Cottbus-Senftenberg, Senftenberg, Germany
- 2Institute of Microbiology, University of Agriculture, Faisalabad, Pakistan
- 3Department of Life Sciences, The Milner Centre for Evolution, University of Bath, Bath, United Kingdom
Introduction: The dissemination of antimicrobial resistance (AMR) in critical priority pathogens is a significant threat. Non-clinical reservoirs of AMR, such as agriculture and food production facilities, may contribute to the transmission of clinically relevant pathogens such as multidrug-resistant (MDR) Klebsiella pneumoniae. There is currently very limited knowledge regarding the population structure and genomic diversity of K. pneumoniae in poultry production in Pakistan.
Methods: We explored healthy broilers in a commercial farm from Faisalabad, Pakistan, and identified six K. pneumoniae strains from 100 broiler birds. We characterized the strains, determining clonality, virulence and antimicrobial resistance genes using next generation sequencing.
Results: The evaluation of antimicrobial susceptibility revealed that all the strains were MDR. Genomic analysis showed that 3/6 strains belonged to ST152, harbouring acquired resistance aminoglycosides [aadA2, aph(4′)-Ia], β-lactams (blaSHV-187, blaLAP2), fosfomycin (fosA6), tetracycline (tetA), trimethoprim (dfrA12), quinolone (qnrS1), sulphonamides (sul2) and phenicol (floR). All the strains harboured the efflux pump genes oqxA, oqxB, emrR, kpnG, kpnH, kpnF, baeR, mtdB and mtdC. All six strains encoded identical virulence profiles possessing six genes, i.e., ureA, iutA, entB, allS, fimH and mrkD. Phylogenomic analysis of the dominant sequence type (ST152) present in our dataset with publicly available genomes showed that the isolates clustered to strains mainly from human sources and could pose a potential threat to food safety and public health.
Discussion: The combination of these findings with antimicrobial use data would allow a better understanding of the selective pressures that may be driving the spread of AMR. This is the first report of MDR K. pneumoniae isolated from broiler hens in Pakistan, and the finding suggests that routine surveillance of WHO critical priority pathogens in such settings would be beneficial to the development of effective control strategies to reduce AMR.
1 Introduction
Klebsiella pneumoniae, a member of the Enterobacteriaceae, is commonly carried in healthy humans and other animals (1) but is also an important cause of community- and hospital-acquired infections (2). The rise of antimicrobial resistance (AMR) in K. pneumoniae led to its inclusion as an ESKAPE pathogen, organisms that are increasing threats to public health (3), as well as being on the World Health Organisation (WHO) bacterial priority pathogens list (4). In addition, a hypervirulent phenotype has emerged, in particular in East and Southeast Asia, with increasing reports of convergent strains carrying both AMR and hypervirulence (5). With an open pangenome and large accessory genome, including many genes likely to have been acquired from other bacterial taxa, K. pneumoniae has great genomic plasticity and acquisition of resistance and virulence determinants is common (6); K. pneumoniae is recognised as an important source of AMR globally (7).
The ‘One Health’ approach, which recognises the interconnection between human, animal and environmental health, lays bare the links between use of antibiotics and the spread of AMR within and between the three domains (8). The presence of K. pneumoniae in healthy animals, particularly ones in the human food chain, is one possible source for the spread to humans. K. pneumoniae has been shown to be common in the intestinal flora of poultry grown for human consumption in Norway (9) and Portugal (10). In these countries, where the use of antibiotics is low, isolates of K. pneumoniae were found that were either hypervirulent or multidrug-resistant (MDR), including some in Portugal that were identical to clinical isolates. In contrast, in Pakistan there have been high levels of antibiotic use in poultry production, for treatment, prophylactic, and growth promotion purposes (11, 12). Such overuse of antibiotics can lead to an increase in AMR, with a clear risk to human health (13).
MDR K. pneumoniae and isolates carrying virulence genes have been documented in human infections in Pakistan (14). However, no studies have investigated the presence of AMR in K. pneumoniae in poultry produced for human consumption, which is likely to be an important reservoir for the spread to humans (15). Since the inception of the Pakistan National Action Plan on AMR (11), little effort has been made to investigate this WHO critical priority pathogen in food animals such as the broiler chicken. In this study we report the presence of K. pneumoniae in broiler chickens on a single farm in Faisalabad, Pakistan. Using phenotypic antimicrobial susceptibility and whole genome sequencing to characterise the isolates, we focus on clinically relevant AMR genes, MLST and serotypes.
2 Materials and methods
2.1 Isolation and identification of K. pneumoniae
This pilot observational study was conducted to detect MDR K. pneumoniae and investigate its phenotypic and genomic features in a commercial poultry farm in Faisalabad, Pakistan. During winter 2023, a total of 100 cloacal swabs from broilers birds were screened. In order to selectively isolate and screen K. pneumoniae, cloacal swabs were spread onto Simmons citrate agar (SCAI, Oxoid) with amoxicillin (10 μg/mL) and myo-inositol (10%) and incubated at 37°C for 48 h. Possible K. pneumoniae isolates were streaked on CHROMagar orientation plates (Mast Diagnostica GmbH, Reinfeld, Germany) and blue coloured pure colonies were subjected to MALDI-TOF (microflex, Bruker, Billerica, MA, United States) for confirmation.
2.2 In vitro antimicrobial susceptibility testing
The antimicrobial susceptibility was determined using the VITEK®2 at the Laboratory Diagnostics and Microbiology of Klinikum Niederlausitz GmbH. In case of tigecycline, minimum inhibitory concentration (MIC) of six strains was determined on a range of concentrations (0–32 μg/mL) using broth microdilution method. Three technical and biological repetitions were performed for each strain and E. coli strain ATCC 25922 was used as control. MDR was defined as resistance to three or more antibiotics of different classes (16). Results were interpreted according to the breakpoints of Clinical and Laboratory Standards Institute documents and European Commission on Antimicrobial Susceptibility Testing (EUCAST) documents (17, 18).
2.3 Whole genome sequencing and analysis
Genomic DNA was extracted from an overnight culture (2 mL) of K. pneumoniae strains with QIAcube automated system (Qiagen). DNA concentration was measured using Qubit (ThermoFisher Scientific, United States) and sent for commercial sequencing to Azenta Life Sciences, Leipzig, Germany. The Illumina NovaSeq platform carried out paired-end sequencing, generating 150 bp pair end reads and resulting in a genome coverage of 100x. The initial processing of the raw reads involved quality trimming and removal of Illumina-specific adapters with the Trimmomatic (19). For quality assessment, the sequences were further analysed using FastQC, and all samples exhibited a quality score not less than 30. The genome was assembled using SPAdes (20) and further assessed for assembly quality via Quast (21). The contigs were annotated using Prokka (22) for the identification of virulence genes and factors. Antibiotic resistance genes (ARGs) were ascertained by utilizing the Comprehensive Antibiotic Resistance Database (CARD) (23) and ABRicate1 (24). Plasmids were identified from each of the WGS using Staramr which employs PlasmidFinder database (25, 26). Furthermore, Kleborate v2.2 (5) was used identify Klebsiealla–specific information on pathogenicity and virulence genes and to determine ST. Virulence genes (n = 19) involved in formucoviscosity, LPS synthesis, adhesins, allantoin metabolism, iron acquisition and siderophores, cytotoxicity and urease (27) were analysed using BLAST (28). The phylogenetic analysis was conducted by comparing dominant STs to global types found in published literature using RAxML version 8.2.12 by 100 bootstrap (29). Moreover, pangenome analysis through Roary (30) to identify core genome (4,278 genes) was performed and a maximum-likelihood phylogenetic tree with 100 bootstrap was constructed and visualized using iTOL (31).
3 Results
3.1 Occurrence and antibiotic susceptibility profiles of K. pneumoniae
A total of 6 K. pneumoniae isolates were recovered from 100 chicken cloacal swabs. MICs of 16 antimicrobial agents showed that the strains had different resistance profiles, and all were designated as MDR (Figure 1). Most of the strains were resistant to second generation cephalosporins (cefuroxime and cefuroxime+axetil) and five of them were also resistant to penicillin and β-lactam in combination. Two of the strains (SAMN37674196 and SAMN3764199) showed resistance to third generation cephalosporins (cefotaxime, ceftazidime and ceftriaxone). Notably, tigecycline resistance was observed in all six strains. All the strains were susceptible to carbapenems (imipenem and meropenem) and aminoglycoside (gentamicin).
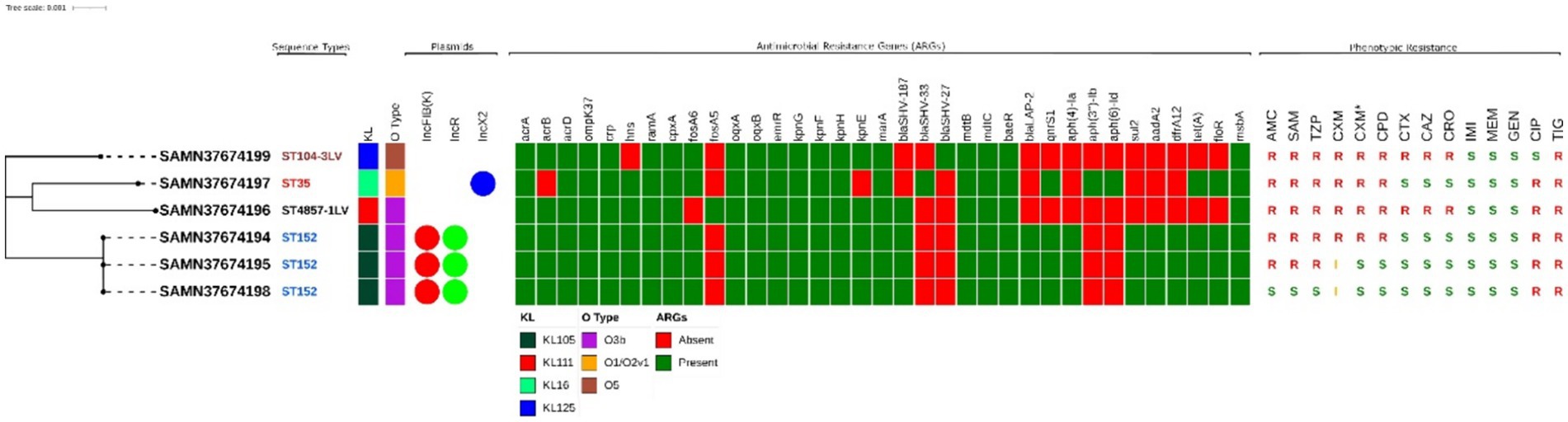
Figure 1. Maximum likelihood tree representing the phylogenetic relationship between 6 K. pneumoniae strains from this study. Associated metadata shown here represents the STs, KL and O-type identified using Kleborate, ARGs are determined through CARD database using Abricate. The phenotypic antimicrobial profiling for all antimicrobials except tigecycline is done by VITEK®2 illustrated as (R = Resistant, I = Intermediate, S = Sensitive). AMC, Amoxicillin + Clavulanic acid; SAM, Ampicillin + sulbactam; TZP, Piperacillin + Tazobactam; CXM, Cefuroxime; CXM*, Cefuroxime + Axetil; CPD, Cefpodoxime; CTX, Cefotaxime; CAZ, Ceftazidime; CRO, Ceftriaxone; IMI, Imipenem; MEM, Meropenem; GEN, Gentamicin; CIP, Ciprofloxacin; TIG, Tigecycline; FOS Fosfomycin; SXT, Trimethoprim + Sulfamethoxazol. All the metadata is illustrated using iTOL.
3.1.1 Genetic divergence of K. pneumoniae strains
The genetic diversity of MDR-K. pneumoniae strains in our study were examined using MLST (32). Four STs were identified (ST104-3LV, ST35, ST4857-1LV and ST152), with ST152 being the predominant type, detected in three strains which also possessed the same plasmid types, i.e., IncFIB(K) and IncR, as shown in Figure 1.
3.2 Drug-resistance genes and virulence genes
Analysis of ARGs showed that the K. pneumoniae strains possessed a variety of genes conferring resistance to multiple antimicrobials (Figure 1). The number of ARGs detected in the strains ranged from 21 to 30. We did not detect any isolate harbouring resistance to carbapenems or colistin. Strains belonging to ST152 were found to encode resistance to aminoglycosides [aadA2, aph(4′)-Ia], β-lactams (blaSHV-187, blaLAP2), fosfomycin (fosA6), tetracycline (tetA), trimethoprim (dfrA12), quinolone (qnrS1), sulphonamides (sul2) and phenicols (floR). All the strains encoded the efflux pump genes, i.e., oqxA, oqxB, emrR, kpnG, kpnH, kpnF, baeR, mtdB and mtdC.
The strains were also analysed for major virulence encoding factors, resulting in identical profile as shown in Figure 2. A total of 19 factors were examined, only six genes were found. Notably, none of the strains possessed formucoviscosity genes (i.e., rmpA, rmpA2, magA) and LPS biosynthesis genes (i.e., uge and wabG). However, all strains encoded allantoin metabolism gene allS and urease encoding ureA gene. Only two of the genes encoding siderophores, which are also important virulence factors, were detected (i.e., iutA and entB). Similarly, only two out of three important adhesins (fimH and mrkD) were found among all six K. pneumoniae strains.
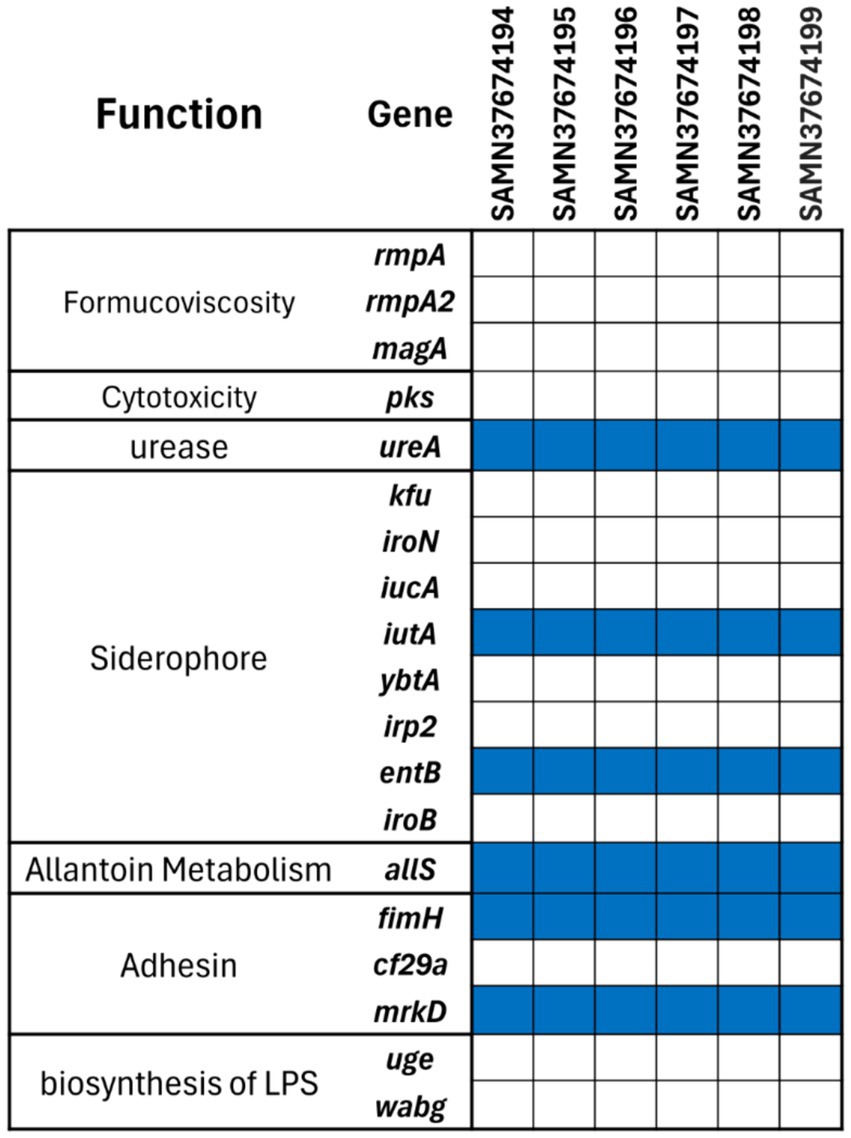
Figure 2. Virulence gene profiles of the six MDR K. pneumoniae strains. The blue tile in the figure shows presence and white tile exhibits the absence of the gene.
3.3 Global genomic epidemiology of ST152
In order to analyse global genomic epidemiology of ST152, we conducted a single nucleotide polymorphism (SNP)-based phylogenetic analysis to compare the genetic similarities and evolutionary relationships among the three ST152 K. pneumoniae isolates in our study and 39 other publicly available ST152 isolates. According to Pathogenwatch database, 196 strains have been reported belonging to ST152 with only one strain isolated from bovine and the rest were isolated from humans (33). We selected the 39 ST152 isolates used based on their location (Pakistan), K type or O type from the Pathogenwatch database. All 39 strains (Supplementary Table S1) were derived from humans, with 29 of them originating in United States (Figure 3). As a result, the strains investigated in our study were primarily classified into four clusters. This is the first report of ST152 being isolated from poultry, specifically chicken. All the isolates included in this analysis had O-type of O3b except one other human isolate reported from Pakistan with O-type of O4. In case of K-types of ST152, most of them were unknown but there were six strains of KL105 (three strains from the present study), four strains of KL46 and only one strain from Senegal typed as KL74. Isolates from this study were all categorized as KL105 (Figure 3).
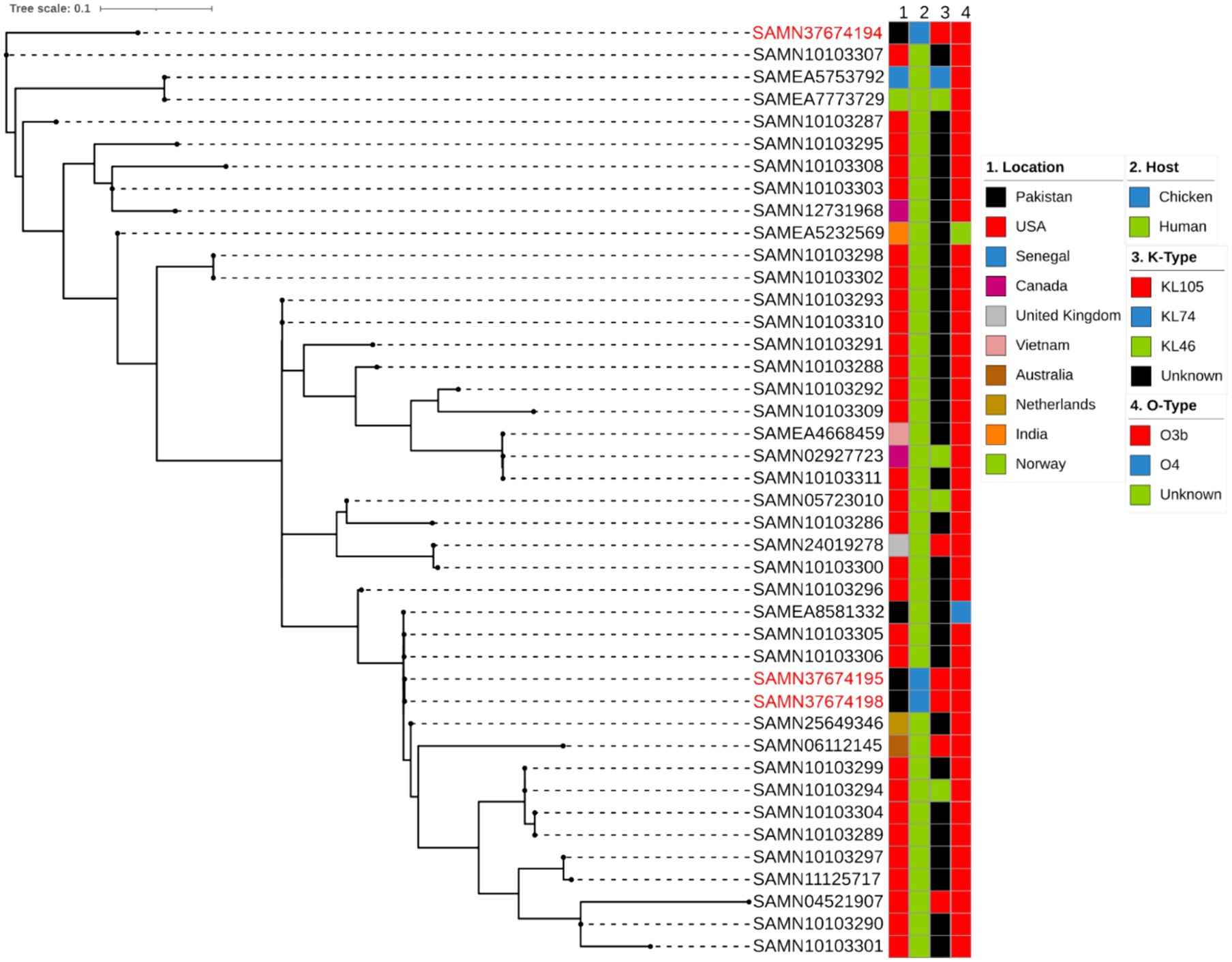
Figure 3. Phylogenetic comparison of the K. pneumoniae ST152 depicting location, host, K and O type.
4 Discussion
This study explores the occurrence, antimicrobial resistance, and genetic diversity of K. pneumoniae in healthy broiler chickens in Faisalabad, Pakistan. We isolated six K. pneumoniae strains from a total of 100 healthy broiler chicken. Two studies at poultry farms in China and Bangladesh reported recovery rates of K. pneumoniae of 4.67 and 43.9%, respectively, (34, 35), but both included environmental samples from the poultry farms and so are not directly comparable. A study of veterinary and agricultural waste in Pakistan found two isolates of MDR K. pneumoniae in sludge and wastewater from poultry farms (36).
All six of the K. pneumoniae isolates in this study are MDR. We observed high rates of resistance to multiple classes of antimicrobials including second generation cephalosporin (C CXM and CXM*) (83.3%), third generation cephalosporin (CTX, CAZ AND CRO) (33.3%) penicillin and β-lactam combination (66.6%), fluoroquinolone (83.3%), sulphonamide (50%) and tetracycline (100%). Resistance to all of these antimicrobials is of concern for human health (7). Resistance to third generation cephalosporins is a global problem; in Europe third-generation cephalosporin-resistant K. pneumoniae has been reported as the third largest cause of infections and attributable deaths (37). Assessing the health burden of infections with antibiotic-resistant bacteria in the EU/EEA (37). Piperacillin/tazobactam (TZP) is prescribed to immune-compromised and critically ill patients (38). Fluoroquinolones are used in clinical settings due to their broad-spectrum of activity, and resistance is increasing rapidly in K. pneumoniae (39). Tigecycline resistant strains of K. pneumoniae were first isolated in hospitals and have been increasingly reported since its introduction (7). Thus all of these resistance phenotypes are potentially a threat to human health.
We employed WGS to identify potential resistance mechanisms of MDR the K. pneumoniae isolates. The six isolates in this study encoded multiple MDR efflux pumps, which can reduce susceptibility in clinically significant pathogenic bacteria (40). One of the most important multidrug efflux systems, detected in five out six K. pneumoniae isolates, was acriflavine resistance B (AcrB) encoded by acrAB. This pump, belonging to resistance nodulation cell diving (RND) superfamily of efflux transporters, allows bacteria to exhibit resistance to various antimicrobial classes of antibiotics, i.e., quinolones, beta-lactams, tetracyclines, macrolides, aminoglycosides, and chloramphenicol (40, 41). kpnEFGH operon, also detected across six strains, with the exception of kpnE in SAMN37674197, encodes for a small multidrug resistant (SMR) type efflux pump (42). The KpnEF efflux pump confers resistance to a variety of antibiotics; antimicrobial susceptibility testing of the kpnEF deletion mutant of different K. pneumoniae serotypes established its role in broad-spectrum AMR (42). Another antibiotic efflux pump encoded by all the six strains was OqxAB. The overexpression of oqxAB confers resistance against not only multiple drugs such as quinoxalines, quinolones, tigecycline, nitrofurantoin and chloramphenicol but also detergents and disinfectants (43). mdtBC, encoding a RND-type drug exporter, stimulated by response regulator BaeR in Escherichia coli (44), were detected in all six K. pneumoniae isolates. MdtBC contains two different transporter proteins, MdtB and MdtC, and appears to function only as a B2C heterotrimer. When overexpressed, it pumps out norfloxacin, novobiocin, cloxacillin, and deoxycholate (45).
The isolates in this study were not hypervirulent, carrying only a small number of virulence determinants. Although all of the isolates had one gene from the locus encoding the siderophore aerobactin, the other genes were absent. The K and O loci in these isolates are not associated with clones known to pose a threat to human health (46).
Against a background of other ST152 isolates from Pakistan and with the same K- and O- loci, the two isolates SAMN37674195 and SAMN37674198 appear to be very similar to each other, and to other isolates from Pakistan and United States. SAMN37674194 is more distinct, being less closely related to the other ST152 isolates selected. It would be interesting to compare this isolate with a wider selection of K. pneumoniae isolates, perhaps including other, related, STs.
The other three STs represented in this study were ST104-3LV, ST35 and ST4857-1LV. For ST35, there have been reports of MDR isolates which are also hypervirulent, have acquired yersiniabactin (47, 48). None of the isolates in this study carry yersiniabactin, but has been identified in agricultural sources in Pakistan in recent years. In a study of dairy cows in three states in Pakistan, our group isolated K pneumoniae with otherwise low virulence profiles but from two sites some isolates did carry yersiniabactin (49).
Overall, we detected a variety of MDR K. pneumoniae strains harbouring important AMR genes in broiler hens in a single commercial farm in Faisalabad, Pakistan. Some of these strains were very similar to strains that have been found to cause disease in humans, highlighting the potential risk of spread of such strains from commercially produced poultry to humans. A useful addition to this investigation would be antimicrobial usage data from the farm, which could allow a better understanding of the drivers of the spread of the resistance genes. In addition, the use of short read sequencing did not allow a complete study of the plasmids found to be present in the strains, particularly the ST152 isolates. As AMR genes and virulence factors are often harboured on plasmids (6) better characterisation of them would be beneficial to our understanding of the risk of their spread. Despite these limitations, this study has provided a useful addition to our knowledge of this important pathogen in the poultry industry in Pakistan, further highlighting the need for a one-health approach to control of the spread of AMR.
Data availability statement
The data presented in the study are deposited in the online repository NCBI under the accession number PRJNA1023867. The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Ethics statement
The animal study was approved by Institutional Biosafety Committee D. No. 170/ORIC, University of Agriculture, Faisalabad. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
MK: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Supervision, Validation, Writing – original draft, Writing – review & editing. MAM: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – review & editing. NA: Data curation, Methodology, Writing – review & editing. FF: Data curation, Methodology, Writing – review & editing. MG: Formal analysis, Writing – review & editing. PS: Conceptualization, Funding acquisition, Project administration, Resources, Validation, Writing – review & editing. MM: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research work was supported by funding from the International Foundation for Science, Sweden (IFS I3-B-5793-2).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2024.1433124/full#supplementary-material
Footnotes
References
1. Bagley, ST. Habitat association of Klebsiella species. Infect Control Hosp Epidemiol. (1985) 6:52–8. doi: 10.1017/S0195941700062603
2. Podschun, R, and Ullmann, U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. (1998) 11:589–603. doi: 10.1128/cmr.11.4.589
3. Miller, WR, and Arias, CA. ESKAPE pathogens: antimicrobial resistance, epidemiology, clinical impact and therapeutics. Nat Rev Microbiol. (2024) 22:1–19. doi: 10.1038/s41579-024-01054-w
4. WHO Bacterial Priority Pathogens List. Bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance (2024). Available at: https://www.who.int/publications/i/item/9789240093461 (Accessed July 14, 2024).
5. Lam, MMC, Wick, RR, Watts, SC, Cerdeira, LT, Wyres, KL, and Holt, KE. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun. (2021) 12:4188. doi: 10.1038/s41467-021-24448-3
6. Holt, KE, Wertheim, H, Zadoks, RN, Baker, S, Whitehouse, CA, Dance, D, et al. Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc Natl Acad Sci. (2015) 112:E3574–81. doi: 10.1073/pnas.1501049112
7. Navon-Venezia, S, Kondratyeva, K, and Carattoli, A. Klebsiella pneumoniae: a major worldwide source and shuttle for antibiotic resistance. FEMS Microbiol Rev. (2017) 41:252–75. doi: 10.1093/femsre/fux013
8. Robinson, TP, Bu, DP, Carrique-Mas, J, Fèvre, EM, Gilbert, M, Grace, D, et al. Antibiotic resistance is the quintessential one health issue. Trans R Soc Trop Med Hyg. (2016) 110:377–80. doi: 10.1093/trstmh/trw048
9. Franklin-Alming, FV, Kaspersen, H, Hetland, MAK, Bakksjø, R-J, Nesse, LL, Leangapichart, T, et al. Exploring Klebsiella pneumoniae in healthy poultry reveals high genetic diversity, good biofilm-forming abilities and higher prevalence in turkeys than broilers. Front Microbiol. (2021) 12:725414. doi: 10.3389/fmicb.2021.725414
10. Mourão, J, Magalhães, M, Ribeiro-Almeida, M, Rebelo, A, Novais, C, Peixe, L, et al. Decoding Klebsiella pneumoniae in poultry chain: unveiling genetic landscape, antibiotic resistance, and biocide tolerance in non-clinical reservoirs. Front Microbiol. (2024) 15:1365011. doi: 10.3389/fmicb.2024.1365011
11. WHO. Pakistan: Antimicrobial resistance national action plan. Available at: https://www.who.int/publications/m/item/pakistan-antimicrobial-resistance-national-action-plan (Accessed July 14, 2024)
12. Mohsin, M, Van Boeckel, TP, Saleemi, MK, Umair, M, Naseem, MN, He, C, et al. Excessive use of medically important antimicrobials in food animals in Pakistan: a five-year surveillance survey. Glob Health Action. (2019) 12:1697541. doi: 10.1080/16549716.2019.1697541
13. Van Boeckel, TP, Pires, J, Silvester, R, Zhao, C, Song, J, Criscuolo, NG, et al. Global trends in antimicrobial resistance in animals in low- and middle-income countries. Science. (2019) 365:eaaw1944. doi: 10.1126/science.aaw1944
14. Fatima, S, Liaqat, F, Akbar, A, Sahfee, M, Samad, A, Anwar, M, et al. Virulent and multidrug-resistant Klebsiella pneumoniae from clinical samples in Balochistan. Int Wound J. (2021) 18:510–8. doi: 10.1111/iwj.13550
15. Gržinić, G, Piotrowicz-Cieślak, A, Klimkowicz-Pawlas, A, Górny, RL, Ławniczek-Wałczyk, A, Piechowicz, L, et al. Intensive poultry farming: a review of the impact on the environment and human health. Sci Total Environ. (2023) 858:160014. doi: 10.1016/j.scitotenv.2022.160014
16. Magiorakos, A-P, Srinivasan, A, Carey, RB, Carmeli, Y, Falagas, ME, Giske, CG, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. (2012) 18:268–81. doi: 10.1111/j.1469-0691.2011.03570.x
17. European Committee on Antimicrobial Susceptibility Testing. Breakpoint tables for interpretation of MICs and zone diameters (2019).
18. Clinical and Laboratory Standards Institute. Performance standards for antimicrobial susceptibility testing. CLSI Supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute (2022).
19. Bolger, AM, Lohse, M, and Usadel, B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. (2014) 30:2114–20. doi: 10.1093/bioinformatics/btu170
20. Seemann, T. Shovill: Faster (SPAdes) assembly of (Illumina) reads (2017). Available at: https://github.com/tseemann/shovill
21. Gurevich, A, Saveliev, V, Vyahhi, N, and Tesler, G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. (2013) 29:1072–5. doi: 10.1093/bioinformatics/btt086
22. Seemann, T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. (2014) 30:2068–9. doi: 10.1093/bioinformatics/btu153
23. Jia, B, Raphenya, AR, Alcock, B, Waglechner, N, Guo, P, Tsang, KK, et al. CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. (2017) 45:D566–73. doi: 10.1093/nar/gkw1004
24. Seemann, T. GitHub-tseemann/abricate: Mass screening of contigs for antimicrobial resistance or virulence genes (2022)
25. Bharat, A, Petkau, A, Avery, BP, Chen, JC, Folster, JP, Carson, CA, et al. Correlation between phenotypic and in silico detection of antimicrobial resistance in Salmonella enterica in Canada using Staramr. Microorganisms. (2022) 10:292. doi: 10.3390/microorganisms10020292
26. Carattoli, A, Zankari, E, García-Fernández, A, Voldby Larsen, M, Lund, O, Villa, L, et al. In silico detection and typing of plasmids using PlasmidFinder and plasmid multilocus sequence typing. Antimicrob Agents Chemother. (2014) 58:3895–903. doi: 10.1128/AAC.02412-14
27. Kim, D, Park, BY, Choi, MH, Yoon, E-J, Lee, H, Lee, KJ, et al. Antimicrobial resistance and virulence factors of Klebsiella pneumoniae affecting 30 day mortality in patients with bloodstream infection. J Antimicrob Chemother. (2019) 74:190–9. doi: 10.1093/jac/dky397
28. Sayers, EW, Bolton, EE, Brister, JR, Canese, K, Chan, J, Comeau, DC, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. (2021) 50:D20–6. doi: 10.1093/nar/gkab1112
29. Stamatakis, A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. (2014) 30:1312–3. doi: 10.1093/bioinformatics/btu033
30. Page, AJ, Cummins, CA, Hunt, M, Wong, VK, Reuter, S, Holden, MTG, et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. (2015) 31:3691–3. doi: 10.1093/bioinformatics/btv421
31. Letunic, I, and Bork, P. Interactive tree of life (iTOL) v4: recent updates and new developments. Nucleic Acids Res. (2019) 47:W256–9. doi: 10.1093/nar/gkz239
32. Urwin, R, and Maiden, MCJ. Multi-locus sequence typing: a tool for global epidemiology. Trends Microbiol. (2003) 11:479–87. doi: 10.1016/j.tim.2003.08.006
33. Argimón, S, David, S, Underwood, A, Abrudan, M, Wheeler, NE, Kekre, M, et al. Rapid genomic characterization and global surveillance of Klebsiella using pathogenwatch. Clin Infect Dis. (2021) 73:S325–35. doi: 10.1093/cid/ciab784
34. Li, Z, Xin, L, Peng, C, Liu, C, Wang, P, Yu, L, et al. Prevalence and antimicrobial susceptibility profiles of ESBL-producing Klebsiella pneumoniae from broiler chicken farms in Shandong Province, China. Poult Sci. (2022) 101:102002. doi: 10.1016/j.psj.2022.102002
35. Saha, O, Basri, R, Hossain, MA, and Sultana, M. Characterization of multidrug and heavy metal resistance of carbapenemases producing Klebsiella pneumoniae from poultry samples in Bangladesh. Vet Med Sci. (2023) 9:1685–701. doi: 10.1002/vms3.1168
36. Chaudhry, TH, Aslam, B, Arshad, MI, Alvi, RF, Muzammil, S, Yasmeen, N, et al. Emergence of blaNDM-1 harboring Klebsiella pneumoniae ST29 and ST11 in veterinary settings and waste of Pakistan. Infect Drug Resist. (2020) 13:3033–43. doi: 10.2147/IDR.S248091
37. European Centre for Disease Prevention and Control. Assessing the health burden of infections with antibiotic-resistant bacteria in the EU/EEA, 2016–2020. (2022) Available at: https://www.ecdc.europa.eu/en/publications-data/health-burden-infections-antibiotic-resistant-bacteria-2016-2020 (Accessed July 14, 2024)
38. Abdelraouf, K, Chavda, KD, Satlin, MJ, Jenkins, SG, Kreiswirth, BN, and Nicolau, DP. Piperacillin-Tazobactam-resistant/third-generation cephalosporin-susceptible Escherichia coli and Klebsiella pneumoniae isolates: resistance mechanisms and in vitro-in vivo discordance. Int J Antimicrob Agents. (2020) 55:105885. doi: 10.1016/j.ijantimicag.2020.105885
39. Geetha, PV, Aishwarya, KVL, Mariappan, S, and Sekar, U. Fluoroquinolone resistance in clinical isolates of Klebsiella pneumoniae. J Lab Physicians. (2020) 12:121–5. doi: 10.1055/s-0040-1716478
40. Piddock, LJV. Multidrug-resistance efflux pumps? Not just for resistance. Nat Rev Microbiol. (2006) 4:629–36. doi: 10.1038/nrmicro1464
41. Zhang, Z, Morgan, CE, Bonomo, RA, and Yu, EW. Cryo-EM structures of the Klebsiella pneumoniae AcrB multidrug efflux pump. MBio. (2023) 14:e0065923. doi: 10.1128/mbio.00659-23
42. Srinivasan, VB, and Rajamohan, G. KpnEF, a new member of the Klebsiella pneumoniae cell envelope stress response regulon, is an SMR-type efflux pump involved in broad-spectrum antimicrobial resistance. Antimicrob Agents Chemother. (2013) 57:4449–62. doi: 10.1128/aac.02284-12
43. Li, J, Zhang, H, Ning, J, Sajid, A, Cheng, G, Yuan, Z, et al. The nature and epidemiology of OqxAB, a multidrug efflux pump. Antimicrob Resist Infect Control. (2019) 8:44. doi: 10.1186/s13756-019-0489-3
44. Nagakubo, S, Nishino, K, Hirata, T, and Yamaguchi, A. The putative response regulator BaeR stimulates multidrug resistance of Escherichia coli via a novel multidrug exporter system, MdtABC. J Bacteriol. (2002) 184:4161–7. doi: 10.1128/JB.184.15.4161-4167.2002
45. Li, X-Z, Plésiat, P, and Nikaido, H. The challenge of efflux-mediated antibiotic resistance in gram-negative bacteria. Clin Microbiol Rev. (2015) 28:337–418. doi: 10.1128/cmr.00117-14
46. Wyres, KL, Lam, MMC, and Holt, KE. Population genomics of Klebsiella pneumoniae. Nat Rev Microbiol. (2020) 18:344–59. doi: 10.1038/s41579-019-0315-1
47. Shen, Z, Gao, Q, Qin, J, Liu, Y, and Li, M. Emergence of an NDM-5-producing Hypervirulent Klebsiella pneumoniae sequence type 35 strain with chromosomal integration of an integrative and conjugative element, ICEKp1. Antimicrob Agents Chemother. (2019) 64:e01675-19. doi: 10.1128/aac.01675-19
48. Dong, N, Yang, X, Chan, EW-C, Zhang, R, and Chen, S. Klebsiella species: taxonomy, hypervirulence and multidrug resistance. EBioMedicine. (2022) 79:103998. doi: 10.1016/j.ebiom.2022.103998
Keywords: antimicrobial resistance, poultry, genomics, K. pneumoniae , sequence type
Citation: Khan MM, Mushtaq MA, Abbas N, Fatima F, Gibbon MJ, Schierack P and Mohsin M (2024) Occurrence, antimicrobial resistance and genomic features of Klebsiella pneumoniae from broiler chicken in Faisalabad, Pakistan. Front. Vet. Sci. 11:1433124. doi: 10.3389/fvets.2024.1433124
Edited by:
Homer Pantua, BioAssets Corporation, PhilippinesReviewed by:
Rachel Amanda Hickman, Independent Researcher, Uppsala, SwedenMihaela Niculae, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, Romania
Copyright © 2024 Khan, Mushtaq, Abbas, Fatima, Gibbon, Schierack and Mohsin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mashkoor Mohsin, bWFzaGtvb3Jtb2hzaW5AZ21haWwuY29t; bWFzaGtvb3Jtb2hzaW5AdWFmLmVkdS5waw==
 Muhammad Moman Khan
Muhammad Moman Khan Muhammad Ahmed Mushtaq
Muhammad Ahmed Mushtaq Nayyar Abbas
Nayyar Abbas Fariha Fatima
Fariha Fatima Marjorie J. Gibbon3
Marjorie J. Gibbon3 Peter Schierack
Peter Schierack Mashkoor Mohsin
Mashkoor Mohsin