
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Vet. Sci., 22 September 2023
Sec. Livestock Genomics
Volume 10 - 2023 | https://doi.org/10.3389/fvets.2023.1229369
Transmissible spongiform encephalopathies (TSEs) have been reported in a broad spectrum of hosts. The genetic polymorphisms and characteristics of the prion protein (PRNP) gene have a vital impact on the development of TSEs. Notably, natural TSE infection cases have never been reported in rabbits, and genetic variations of the leporine PRNP gene have not been investigated to date. To identify leporine PRNP gene polymorphism, we performed amplicon sequencing in 203 rabbits. We report a novel single nucleotide polymorphism on the leporine PRNP gene. In addition, we performed a comparative analysis of amino acid sequences of prion protein (PrP) across several hosts using ClustalW2. Furthermore, we evaluated the effect of changes of unique leporine PrP amino acids with those conserved among various species using Swiss-Pdb Viewer. Interestingly, we found seven unique leporine amino acids, and the change of unique leporine amino acids with those conserved among other species, including S175N, Q221K, Q221R, A226Y, A230G, and A230S, was predicted to reduce hydrogen bonds in leporine PrP.
Transmissible spongiform encephalopathies (TSEs), which are infectious and lethal neurodegenerative diseases, manifests as various infections in a wide range of mammalian species, such as scrapie in sheep and goats, chronic wasting disease (CWD) in elk, bovine spongiform encephalopathy (BSE) in cattle, transmissible mink encephalopathy (TME) in mink, feline spongiform encephalopathy (FSE) in cats as well as Creutzfeldt–Jakob disease (CJD), fatal familial insomnia (FFI), Gerstmann-Sträussler Scheinker syndrome (GSS), and kuru in humans (1–9). TSEs are induced by the β-sheet-rich pathogenic isoform of prion protein (PrPSc) originating from a cellular form of prion protein (PrPC). In PrPSc and PrPC, Sc and C stand for scrapie isoform of the prion protein and cellular prion protein, respectively (10, 11). However, no naturally TSE-infected cases have been reported in rabbits, dogs, and horses. Thus, rabbits, dogs, and horses are considered TSE-resistant animals. Structural stability of PrP has been reported in TSE-resistant animals. In addition, unique amino acids of PrPs in TSE-resistant animals, including S175 in rabbits, S167 in horses, and D163 in dogs, play a major role in PrP stability (12–14). In a circular dichroism (CD) study, rabbit, horse, and dog PrPs showed more stable characteristics against harsh pH conditions compared to those of TSE-susceptible animals, including hamsters and mice (15). In addition, recombinant human PrP with the M209 allele showed increased thermodynamic stability and an inhibitory effect on the conformational conversion of PrPC to PrPSc (16). In dog PrP, a dog-unique amino acid (D163) contributes to the stability of protein by expanding helix 1 and introducing a negative surface charge (17, 18). In addition, transgenic mice expressing mouse PrP with the D163 allele showed resistance to diverse prion strains, including RML, 301C, 22L, and ME7 (13, 19). In equine PrP, the horse-unique amino acid S167 stabilizes the β2-α2 loop of equine PrP (18, 20). In leporine PrP, the rabbit-unique amino acid S175 has a stronger interaction with N172 than amino acids conserved among other species. The strong side-chain interaction between S175 and N172 forms a helix-capping domain motif that regulates the formation of the β-sheet and is related to an increase in the global stability of leporine PrP (21). Interestingly, the S175N variation disrupts the long-distance interaction of loop 165–172 (22). In addition, rabbits showed resistance to infection by several prion strains, including kuru, CJD, and sheep scrapie (23, 24).
Similar to the major amino acids of TSE-resistant animals mediating resistance to TSEs, PRNP gene polymorphisms are vulnerable to mediate TSE infections in sheep, goats, cattle, deer, and humans. In humans, M129V heterozygosity in the PRNP gene is rarely observed in variant and sporadic CJD patients (25, 26). In Papua New Guinea highlands, the V127 polymorphism of PRNP has been reported as a resistance factor for kuru (27). The key residues of the ovine PRNP gene at codons 136, 154, and 171 have been reported as major determinants of susceptibility to scrapie, and the V136R154Q171 allele was considered the most susceptible factor to scrapie (28). Although PRNP gene polymorphisms are important factors associated with susceptibility to TSE (12, 29–31) and these polymorphisms have been reported in diverse animals (32–37), PRNP gene polymorphisms in rabbits have not been reported to date.
To identify PRNP gene polymorphism in rabbits, we performed amplicon sequencing using gene-specific primers. We also performed phylogenetic analysis using Molecular Evolutionary Genetics Analysis (MEGA X Ver. 10.2.0) to estimate evolutionary PrP relationship among 10 species, including deer, mink, cats, mice, cattle, sheep, humans, goats, camels, and rabbits (38). In addition, to identify rabbit-unique amino acids, multiple sequence alignments were performed with amino acid sequences of PrPs from the above-mentioned 10 species. Furthermore, to evaluate the impact of rabbit-unique amino acids on leporine PrP, we evaluated the effect of changes of rabbit-unique amino acids with those conserved among other species on leporine PrP using AMYCO, PolyPhen-2, PROVEAN, and Swiss-Pdbviewer (39–42).
Brain samples from 203 crossbreed rabbits (New Zealand white and Flemish Giant [FG] rabbits) were provided from the Nonghyup slaughterhouse (Korea).
Genomic DNA was isolated from 20 mg of brain tissue from 203 rabbits using Labopass Tissue Genomic DNA Isolation Kit Mini (Cosmogenetech, Seoul, Korea) following the manufacturer's instructions.
Polymerase chain reaction (PCR) was performed using the leporine PRNP gene-specific primers: leporine PRNP-F (AGACAGGTCCAGGCTGTGAT) and leporine PRNP-R (GGA CCAAGAGAGAAGCGAGA). PCR was performed using BioFACT™ Taq DNA Polymerase (Biofact, Daejeon, Korea) following the manufacturer's instructions. Amplified PCR products (965 bp) were obtained by electrophoresis on a 1.0% agarose gel and were purified using a Favor Prep Gel/PCR Purification Mini Kit (FAVORGEN, Pingtung County, Taiwan). PCR products were analyzed using an ABI 3730 sequencer (ABI, Foster City, California, USA). Sequencing results were examined by Finch TV software (Geospiza Inc, Seattle, WA, USA).
Phylogenetic analysis according to amino acid sequences of PrPs from 10 species (deer, mink, cats, mice, cattle, sheep, humans, goats, camels, and rabbits) was performed using MEGA X Ver. 10.2.0 (Swiss Institute of Bioinformatics, Geneva, Switzerland) (Supplementary Table S1) (38). The evolutionary tree was constructed by the neighbor-joining method with 3,000 bootstrap tests. The Poisson correction method was used to compute the evolutionary distances, which are reported in units of the number of amino acid changes per site.
Multiple sequence alignments of sequences from 10 species-specific PrPs (deer, mink, cats, mice, cattle, sheep, humans, goats, camels, and rabbits) were conducted using ClustalW2 (43).
The effect on leporine PrP amino acid changes was predicted using three in silico programs, PolyPhen-2, PROVEAN, and AMYCO. PolyPhen-2 evaluated the impact on protein function based on numerous features, including the sequence, structure, and phylogenetic information. The predicted effects ranged from benign to possibly damaging or probably damaging. PROVEAN scores were calculated to predict the impact of protein sequence variations on protein function by clustering BLAST hits based on the homologs obtained from the National Center for Biotechnology Information (NCBI) database. AMYCO evaluated the impact on the aggregation propensity of proteins using pWALTZ and PAPA algorithms. An AMYCO score less than 0.45 indicates low amyloid property.
The nuclear magnetic resonance (NMR) of leporine PrP (protein data bank [PDB] ID: 2FJ3) was visualized using Swiss-PdbViewer 4.1. The structural change resulting from the amino acid changes was analyzed using Swiss-PdbViewer 4.1. Hydrogen bonds were predicted when a hydrogen atom fell within the range of 1.2 to 2.76 Å from a “compatible” donor atom.
The Hardy-Weinberg equilibrium (HWE) test was estimated using Haploview version 4.2 (Broad Institute, Cambridge, MA, USA).
The leporine PRNP gene is composed of three exons on chromosome 4. We performed PCR amplification to target the open reading frame (ORF) of the leporine PRNP gene, which is located in exon 3, using gene-specific primers. The length of the amplicon was 965 bp, and the sequence was identical to that reported in GenBank (Gene ID: 100008658). To investigate genetic variations in the leporine PRNP gene, we performed PCR with genomic DNA from 203 rabbits. Notably, we identified only one novel synonymous SNP (c.234C>T, p.G78G) in the ORF region (Figure 1, Supplementary Figure S1). Of 203 rabbits, 181 (89.2%) were homozygous for the C allele, 1 (0.4%) was homozygous for the T allele, and 21 (10.4%) were heterozygous (10.4%) at codon 78. The allele frequency of c.234C>T was 383 C alleles (94.3%) and 23 T alleles (5.7%) in 203 rabbits. This SNP was in HWE (Table 1).
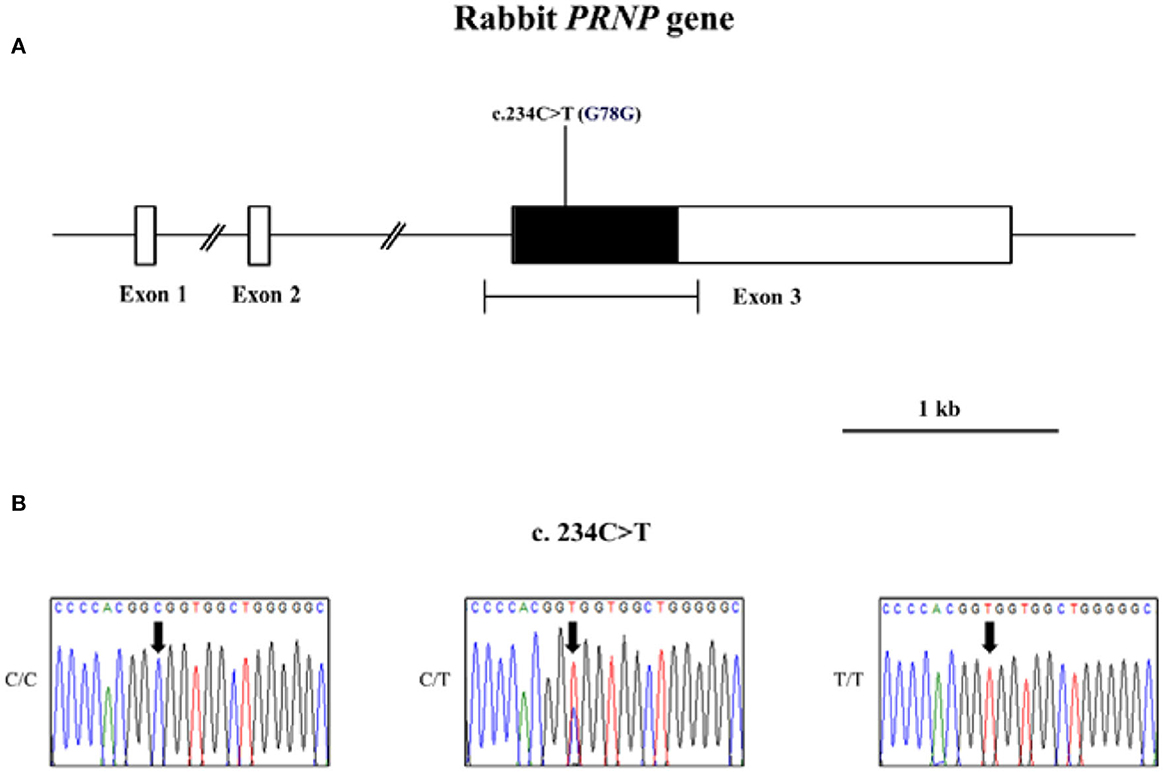
Figure 1. Gene map and electropherograms of a novel single nucleotide polymorphism (SNP) identified in the leporine prion protein (PRNP) gene on chromosome 4. (A) Map and a novel polymorphism identified in the leporine PRNP gene. The open reading frame (ORF) is visually represented by a shaded block, and the 5′ and 3′ untranslated regions (UTRs) are depicted by white blocks. The horizontal bar with edges indicates the regions sequenced. (B) Electropherograms of the novel SNP c.234C>T (p.G78G). Electropherograms showing the three genotypes at codon 78. Left panel: GGC/GGC; middle panel: GGT/GGC; right panel: GGT/GGT. Gene map and novel polymorphism identified in the leporine PRNP gene. The ORF is indicated by a shaded block, and the 5′ and 3′ UTRs are indicated by white blocks. The horizontal bar with edges indicates the regions sequenced. (B) Electropherograms of the novel SNP c.234C>T (p.G78G). Electropherograms showing the three genotypes at codon 78. Left panel: GGC/GGC; middle panel: GGT/GGC; right panel: GGT/GGT.
Phylogenetic analysis was performed to estimate the evolutionary relationship among the amino acid sequences of PrPs in 10 species (deer, mink, cats, mice, cattle, sheep, humans, goats, camels, and rabbits). The rabbit showed a closer evolutionary relationship with mice and human than deer, mink, cats, cattle, sheep, goats, and camels. The subclades of cattle, sheep, goats, camels, and deer, which are known as artiodactyla, were clustered with the farthest evolutionary distances from rabbits (Figure 2A).
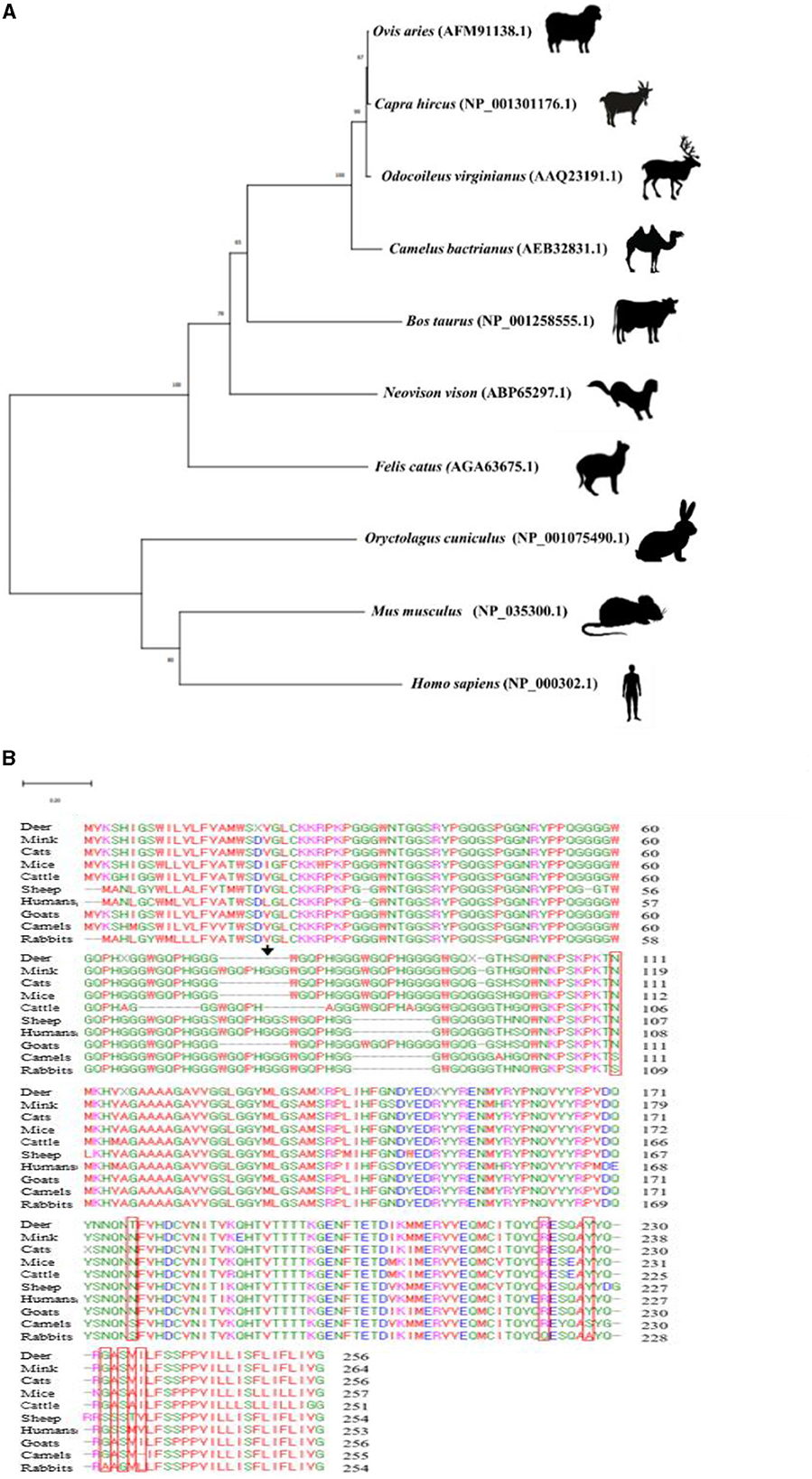
Figure 2. Phylogenetic analysis and multiple sequence alignments of the amino acid sequences of the PrPs in 10 species. (A) Phylogenetic analysis included amino acid sequences of deer (Odocoileus virginianus, AAQ23191.1), cattle (Bos taurus, NP_001258555.1), sheep (Ovis aries, AFM91138.1), mink (Neovison vison, ABP65297.1), cats (Felis catus, AGA63675.1), rabbits (Oryctolagus cuniculus, NP_001075490.1), mice (Mus musculus, NP_035300.1), goats (Capra hircus, NP_001301176.1), camels (Camelus bactrianus, AEB32831.1), and humans (Homo sapiens, NP_000302.1). Evolutionary analysis was carried out using Molecular Evolutionary Genetics Analysis (MEGA) X software based on the neighbor-joining method. The branch lengths indicating evolutionary distances were computed by using the minimum evolution method. The numbers displayed at the branches represent the percentage of bootstrap replicates in which the associated taxa formed a cluster together in the bootstrap test (3,000 replicates). The scale bar represents a length of 0.20. (B) The PrP sequences of deer (Odocoileus virginianus, AAQ23191.1), mink (Neovison vison, ABP65297.1), cats (Felis catus, AGA63675.1), mice (Mus musculus, NP_035300.1), cattle (Bos taurus, NP_001258555.1), sheep (Ovis aries, AFM91138.1), humans (Homo sapiens, NP_000302.1), goats (Capra hircus, NP_001301176.1), camels (Camelus bactrianus, AEB32831.1), and rabbits (Oryctolagus cuniculus, NP_001075490.1) were aligned with ClustalW2. The red box indicates rabbit-unique amino acids. The black arrow indicates a novel SNP found in this study.
To identify rabbit-unique amino acids, we aligned the amino acid sequences of PrPs of mink, deer, cats, mice, cattle, sheep, humans, goats, camels, and rabbits (Supplementary Table S1). The amino acid residue numbering was determined by reference sequence of the leporine PrP. A total of seven rabbit-unique amino acids, including S109, S175, Q221, A226, A230, G232, and L234 were found. In addition, interspecies-specific conserved amino acids were found at 11 residues (N109, N175, K221, R221, Y226, F226, G230, S230, S232, I234, and V234) (Figure 2B).
To investigate the influence on rabbit-unique amino acids, we estimated the effect of amino acid changes from rabbit-unique amino acids into interspecies-specific conserved amino acids (Figure 2B) on leporine PrP using PolyPhen-2, PROVEAN, and AMYCO (Supplementary Table S2). The amyloid propensity of wild-type leporine PrP was predicted to have a score of 0.27 using AMYCO. Notably, except for S175T (0.23), all amino acid changes, including S109N, S175N, Q221K, Q221R, A226Y, A230G, A230S, G232S, L234I, and L234V, were predicted to have a score of 0.27 using AMYCO (Supplementary Table S2). AMTCO scores <0.45 and >0.45 denote low and high aggregation propensity, respectively. In addition, the effects of all amino acid changes on leporine PrP were evaluated to be “Benign” and “Neutral” using PolyPhen-2 and PROVEAN, respectively (Supplementary Table S2).
The effects of changes of rabbit-unique amino acids with interspecies-unique conserved amino acids on the 3D protein structure and hydrogen bonds of leporine PrP were analyzed using the Swiss-Pdb Viewer (Figure 3). S175 was expected to have hydrogen bonds with D179 (2.36 Å) and with S172 (2.18 Å), respectively (Figure 3A). However, N175 expected to form a hydrogen bond (2.36 Å) with D179 (Figure 3B). T175 was also expected to form a hydrogen bond with D179 (2.36 Å) (Figure 3C). Q221 was expected to form a with T217 (1.93 Å) and with A225 (1.69 Å), respectively (Figure 3D). K221 was expected to have one hydrogen bond with Q220 (2.04 Å) and with A225 (1.69 Å), respectively (Figure 3E). R221 was expected to form a hydrogen bond with A225 (1.69 Å) (Figure 3F). A226 was expected to form two hydrogen bonds with E222 and S223 (1.91 and 2.60 Å) (Figure 3G). Y226 was expected to form no hydrogen bonds (Figure 3H). A230 was expected to have single E227 (2.65 Å) (Figure 3I). G230 and S230 were predicted to have no hydrogen bonds (Figures 3J, K).
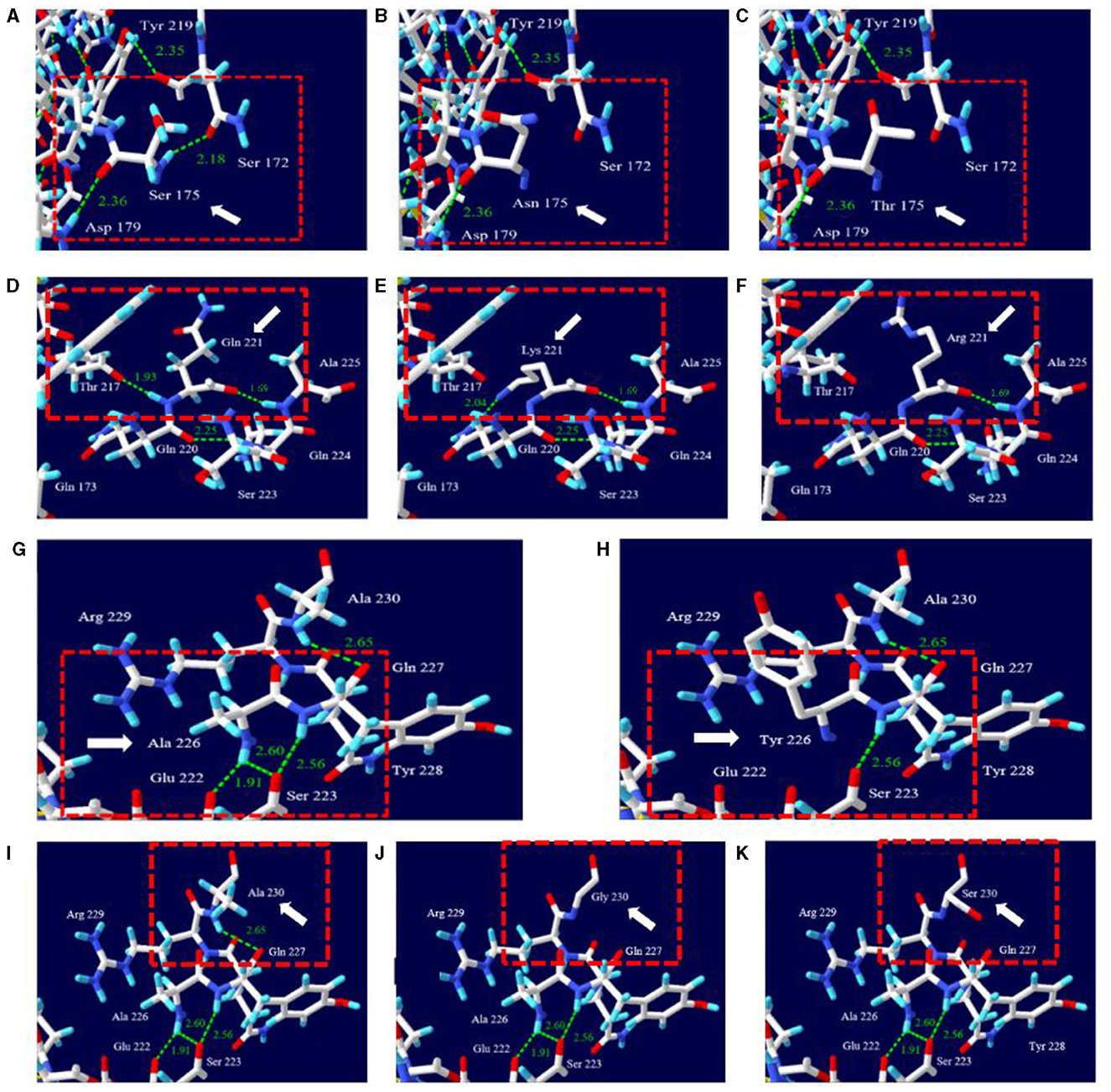
Figure 3. Prediction of the 3D structure and hydrogen bonds of PrP according to amino acid changes of rabbit-unique amino acids with interspecies-specific conserved amino acids. (A) 3D structure of leporine PrP carrying the S175 allele, (B) 3D structure of leporine PrP carrying the N175 allele, (C) 3D structure of leporine PrP carrying the T175 allele, (D) 3D structure of leporine PrP carrying the Q221 allele, (E) 3D structure of leporine PrP carrying the K221 allele, (F) 3D structure of leporine PrP carrying the R221 allele, (G) 3D structure of leporine PrP carrying the A226 allele, (H) 3D structure of leporine PrP carrying the Y226 allele, (I) 3D structure of leporine PrP carrying the A230 allele, (J) 3D structure of leporine PrP carrying the G230 allele, and (K) 3D structure of leporine PrP carrying the S230 allele. The target amino acid residues are indicated by the white arrow. The adjacent amino acid residues are highlighted in the red box indicates adjacent amino acid residues. The presence of hydrogen bonds is represented by green dotted lines. The distance of the hydrogen bonds is indicated by the green numbers.
To identify susceptibility/resistance factors for prion diseases, the structure of PrP was analyzed using NMR, molecular dynamics (MD), and X-ray crystallography in previous studies (18, 44, 45). In rabbits, a TSE-resistant animal, PrP has a helix-capping domain, a highly ordered β2–α2 loop and stable salt bridges and exhibits stable characteristics that may contribute to resistance to TSE infection. However, leporine PRNP polymorphism affecting protein structure has not been reported in previous studies. Interestingly, we found only one novel synonymous SNP located on the tandem repeat region of the leporine PrP that does not affect the amino acid sequences and structure of leporine PrP (Figures 1A, B). To the best of our knowledge, an association between synonymous SNPs and susceptibility to prion diseases has not been reported thus far. Notably, among TSE-resistant animals, SNPs have not been reported in chicken PRNP. In addition, a nonsynonymous SNP has been reported in that of horse PRNP, but this SNP did not affect the stability of the horse-unique PrP structure (12, 46, 47). Compared to TSE-susceptible animals, which have several PRNP gene polymorphisms, TSE-resistant animals have a small number of polymorphisms. Based on these studies, the number of polymorphisms could be associated with resistance to TSEs.
Synonymous SNPs may modulate translation speed and protein stability (48, 49). Since prion diseases are associated with protein stability, further validation of the relationship between the synonymous SNP of the leporine PRNP gene and the structural stability of leporine PrP at the molecular level is highly desirable. In the present study, a polymorphism was identified within the ORF of the leporine PRNP gene in exon 3. However, it is possible that other influencing genetic polymorphisms exist in non-ORF regions. Therefore, further investigation in the non-ORF regions of the leporine PRNP gene is needed in the future.
Some evidence regarding prion resistance in rabbits exists. Firstly, no naturally occurring cases of TSE infection have been reported in rabbits. Secondly, in vitro screening has identified key amino acids related to the misfolding of leporine PrP (50). However, transgenic mice overexpressing leporine PrP and transgenic rabbits carrying ovine PrP showed vulnerability to prion infection (51, 52). However, since the path mechanisms of prion diseases are influenced by various factors, including the PrP sequence, cofactors, and infection route, etc., studies using transgenic animals may not accurately represent the natural prion disease-resistance observed in rabbits. Consequently, there is a strong need for further investigation using a direct infection model using rabbits.
To identify an additional prion resistance factor of leporine PrP, we found seven rabbit-unique amino acids (Figure 2B). Unexpectedly, deleterious effects based on changes of rabbit-unique amino acids with interspecies-specific conserved amino acids have not been estimated by PolyPhen-2, PROVEAN, and AMYCO (Supplementary Table S2). Given that PolyPhen-2, PROVEAN, and AMYCO programs evaluate the alteration of protein function based on homologs collected from databases, stability-related properties may haven't been predicted by the programs (39, 40, 42, 53). Our results showed that PolyPhen-2, PROVEAN, and AMYCO haven't been predicted to have deleterious effects according to the changes of rabbit-unique amino acids. However, since the rabbit-unique amino acids may affect the TSE resistance, we evaluated the amino acid changes effect of rabbit-unique amino acids by 3D structure analysis (Figure 3).
Among seven rabbit-unique amino acids, S175 has been previously reported to be a crucial factor in TSE resistance that confers stability and expansion of helix 2 (21, 22). We also observed that leporine PrP with the N175 allele has fewer hydrogen bonds (Figure 3), which are essential to the secondary structure and stability of proteins (54–56). In addition to S175, rabbit-unique amino acids, including Q221, A226, and A230, were observed, and these amino acids play a crucial role in the formation of hydrogen bonds (Figure 3). Given that one hydrogen bond provides protein stabilization (1.0 kcal/mol), further studies are needed to investigate the impact of rabbit-unique amino acids on leporine PrP. To determine the effect of rabbit-unique amino acids on the stability of leporine PrP, further analysis using CD, NMR, and X-ray crystallography should be performed in the future. Furthermore, TSE infection in transgenic rabbit using leporine PrP carrying interspecies-specific conserved amino acids instead of rabbit-unique amino acids should be performed to evaluate susceptibility of unique amino acids to prion diseases.
In summary, we identified one novel synonymous polymorphism of the leporine PRNP gene. In addition, leporine PrP has seven rabbit-unique amino acids compared with nine species including deer, mink, cats, mice, cattle, sheep, humans, goats, and camels. Hydrogen bonds of leporine PrP were decreased by the changes of rabbit-unique amino-acids into amino acids conserved among species, including S175, Q221, A226, and A230.
The data presented in the study are deposited in the DRYAD repository (https://datadryad.org/stash/share/kG8H4tE-LwkAk6VQcdQcuX3vlfbmem5DlKxqZNMgdWU).
The animal study was approved by Institute of Animal Care and Use Committee of Jeonbuk National University. The study was conducted in accordance with the local legislation and institutional requirements.
D-JK, YC-K, and B-HJ conceived and designed the experiment, analyzed the data, and wrote the paper. D-JK and Y-CK performed the experiments. All authors read and approved the final manuscript.
This research was supported by the Basic Science Research Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education (Grant Nos. 2017R1A6A1A03015876 and 2021R1A6A3A01086488), the National Research Foundation of Korea (NRF) grant funded by the Korean government (MSIT) (Grant Nos. 2021R1A2C1013213 and 2022R1C1C2004792), and Korea Basic Science Institute (National Research Facilities and Equipment Center) grant funded by the Ministry of Education (Grant No. 2021R1A6C101C369). DJ-K was supported by the BK21 Plus Program in the Department of Bioactive Material Sciences.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2023.1229369/full#supplementary-material
1. Gibbs CJ Jr, Gajdusek DC, Asher DM, Alpers MP, Beck E, Daniel PM, et al. Creutzfeldt-Jakob disease (spongiform encephalopathy): transmission to the chimpanzee. Science. (1968) 161:388–9. doi: 10.1126/science.161.3839.388
2. Lantos PL, Bhatia K, Doey LJ, Al-Sarraj S, Doshi R, Beck J, Collinge J. Is the neuropathology of new variant Creutzfeldt-Jakob disease and kuru similar? The Lancet. (1997) 350:187–8. doi: 10.1016/S0140-6736(05)62355-0
3. Benestad SL, Mitchell G, Simmons M, Ytrehus B, Vikøren T. First case of chronic wasting disease in Europe in a Norwegian free-ranging reindeer. Vet Res. (2016) 47:1–7. doi: 10.1186/s13567-016-0375-4
4. Spraker TR, Miller MW, Williams ES, Getzy DM, Adrian WJ, Schoonveld GG, et al. Spongiform encephalopathy in free-ranging mule deer (Odocoileus hemionus), white-tailed deer (Odocoileus virginianus) and Rocky Mountain elk (Cervus elaphus nelsoni) in northcentral Colorado. J Wildl Dis. (1997) 33:1–6. doi: 10.7589/0090-3558-33.1.1
5. Wells GA, Scott AC, Johnson CT, Gunning RF, Hancock RD, Jeffrey M, et al. A novel progressive spongiform encephalopathy in cattle. Vet Rec. (1987) 121:419–20.
6. Marsh RF, Bessen RA, Lehmann S, Hartsough GR. Epidemiological and experimental studies on a new incident of transmissible mink encephalopathy. J Gen Virol. (1991) 72:589–94. doi: 10.1099/0022-1317-72-3-589
7. Wyatt JM, Pearson GR, Smerdon TN, Gruffydd-Jones TJ, Wells GA, Wilesmith JW. Naturally occurring scrapie-like spongiform encephalopathy in five domestic cats. Vet Rec. (1991) 129:233–6. doi: 10.1136/vr.129.11.233
8. Uttley L, Carroll C, Wong R, Hilton DA, Stevenson M. Creutzfeldt-Jakob disease: a systematic review of global incidence, prevalence, infectivity, and incubation. Lancet Infect Dis. (2020) 20:e2–10. doi: 10.1016/S1473-3099(19)30615-2
9. Baiardi S, Rossi M, Capellari S, Parchi P. Recent advances in the histo-molecular pathology of human prion disease. Brain Pathol. (2019) 29:278–300. doi: 10.1111/bpa.12695
10. Donne DG, Viles JH, Groth D, Mehlhorn I, James TL, Cohen FE, et al. Structure of the recombinant full-length hamster prion protein PrP (29–231): the N terminus is highly flexible. Proc Nat Acad Sci. (1997) 94:13452–7. doi: 10.1073/pnas.94.25.13452
11. Pan KM, Baldwin M, Nguyen J, Gasset M, Serban AN, Groth D, et al. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc Nat Acad Sci. (1993) 90:10962–6. doi: 10.1073/pnas.90.23.10962
12. Kim YC, Jeong BH. The first report of polymorphisms and genetic characteristics of the prion protein gene (PRNP) in horses. Prion. (2018) 12:245–52. doi: 10.1080/19336896.2018.1513316
13. Vidal E, Fernández-Borges N, Eraña H, Parra B, Pintado B, Sánchez-Martín MA, et al. Dogs are resistant to prion infection, due to the presence of aspartic or glutamic acid at position 163 of their prion protein. The FASEB Journal. (2020) 34:3969–82. doi: 10.1096/fj.201902646R
14. Fernández-Borges N, Parra B, Vidal E, Eraña H, Sánchez-Martín MA, de Castro J, et al. Unraveling the key to the resistance of canids to prion diseases. PLoS Pathog. (2017) 13:e1006716. doi: 10.1371/journal.ppat.1006716
15. Li-Li QI, Hui ZH, Lin-Lin LI. Progress on low susceptibility mechanisms of transmissible spongiform encephalopathies. Zoological Research. (2014) 35:436. doi: 10.13918/j.issn.2095-8137.2014.5.436
16. Kong Q, Mills JL, Kundu B, Li X, Qing L, Surewicz K, et al. Thermodynamic stabilization of the folded domain of prion protein inhibits prion infection in vivo. Cell Rep. (2013) 4:248–54. doi: 10.1016/j.celrep.2013.06.030
17. Lysek DA, Schorn C, Nivon LG, Esteve-Moya V, Christen B, Calzolai L, et al. Prion protein NMR structures of cats, dogs, pigs, and sheep. Proc Nat Acad Sci. (2005) 102:640–5. doi: 10.1073/pnas.0408937102
18. Sanchez-Garcia J, Fernandez-Funez P. D159 and S167 are protective residues in the prion protein from dog and horse, two prion-resistant animals. Neurobiol Dis. (2018) 119:1–2. doi: 10.1016/j.nbd.2018.07.011
19. Otero A, Bolea R, Hedman C, Fernández-Borges N, Marín B, López-Pérez Ó, et al. An amino acid substitution found in animals with low susceptibility to prion diseases confers a protective dominant-negative effect in prion-infected transgenic mice. Mol Neurobiol. (2018) 55:6182–92. doi: 10.1007/s12035-017-0832-8
20. Pérez DR, Damberger FF, Wüthrich K. Horse prion protein NMR structure and comparisons with related variants of the mouse prion protein. J Mol Biol. (2010) 400:121–8. doi: 10.1016/j.jmb.2010.04.066
21. Khan MQ, Sweeting B, Mulligan VK, Arslan PE, Cashman NR, Pai EF, et al. Prion disease susceptibility is affected by β-structure folding propensity and local side-chain interactions in PrP. Proc Nat Acad Sci. (2010) 107:19808–13. doi: 10.1073/pnas.1005267107
22. Wen Y, Li J, Yao W, Xiong M, Hong J, Peng Y, et al. Unique structural characteristics of the rabbit prion protein. J Biol Chem. (2010) 285:31682–93. doi: 10.1074/jbc.M110.118844
23. Gibbs CJ Jr, Gajdusek DC. Experimental subacute spongiform virus encephalopathies in primates and other laboratory animals. Science. (1973) 182:67–8. doi: 10.1126/science.182.4107.67
24. Barlow RM, Rennie JC. The fate of ME7 scrapie infection in rats, guinea-pigs and rabbits. Res Vet Sci. (1976) 21:110–1. doi: 10.1016/S0034-5288(18)33406-4
25. Salvatore M, Genuardi M, Petraroli R, Masullo C, D'Alessandro M, Pocchiari M. Polymorphisms of the prion protein gene in Italian patients with Creutzfeldt-Jakob disease. Hum Genet. (1994) 94:375–9. doi: 10.1007/BF00201596
26. Jeong BH, Lee KH, Kim NH, Jin JK, Kim JI, Carp RI, et al. Association of sporadic Creutzfeldt–Jakob disease with homozygous genotypes at PRNP codons 129 and 219 in the Korean population. Neurogenetics. (2005) 6:229–32. doi: 10.1007/s10048-005-0016-y
27. Mead S, Whitfield J, Poulter M, Shah P, Uphill J, Campbell T, et al. A novel protective prion protein variant that colocalizes with kuru exposure. N Engl J Med. (2009) 361:2056–65. doi: 10.1056/NEJMoa0809716
28. Curcio L, Sebastiani C, Di Lorenzo P, Lasagna E, Biagetti M. A review on classical and atypical scrapie in caprine: Prion protein gene polymorphisms and their role in the disease. Animal. (2016) 10:1585–93. doi: 10.1017/S1751731116000653
29. Won SY, Kim YC, Do K, Jeong BH. Absence of strong genetic linkage disequilibrium between single nucleotide polymorphisms (SNPs) in the prion protein gene (PRNP) and the prion-like protein gene (PRND) in the horse, a prion-resistant species. Genes. (2020) 11:518. doi: 10.3390/genes11050518
30. Won SY, Kim YC, Kim SK, Jeong BH. The first report of genetic and structural diversities in the SPRN gene in the horse, an animal resistant to prion disease. Genes. (2019) 11:39. doi: 10.3390/genes11010039
31. Kim DJ, Kim YC, Kim AD, Jeong BH. Novel polymorphisms and genetic characteristics of the prion protein gene (PRNP) in dogs—a resistant animal of prion disease. Int J Mol Sci. (2020) 21:4160. doi: 10.3390/ijms21114160
32. Jeong BH, Nam JH, Lee YJ, Lee KH, Jang MK, Carp RI, et al. Polymorphisms of the prion protein gene (PRNP) in a Korean population. J Hum Genet. (2004) 49:319–24. doi: 10.1007/s10038-004-0150-7
33. Kim SK, Kim YC, Won SY, Jeong BH. Potential scrapie-associated polymorphisms of the prion protein gene (PRNP) in Korean native black goats. Sci Rep. (2019) 9:15293. doi: 10.1038/s41598-019-51621-y
34. Kim YC, Won SY, Jeong BH. Absence of single nucleotide polymorphisms (SNPs) in the open reading frame (ORF) of the prion protein gene (PRNP) in a large sampling of various chicken breeds. BMC Genomics. (2019) 20:1–7. doi: 10.1186/s12864-019-6315-8
35. Roh IS, Kim YC, Won SY, Jeong MJ, Park KJ, Park HC, et al. First report of a strong association between genetic polymorphisms of the prion protein gene (PRNP) and susceptibility to chronic wasting disease in sika deer (Cervus nippon). Transbound Emerg Dis. (2022) 69:e2073–83. doi: 10.1111/tbed.14543
36. Kim HH, Kim YC, Kim K, Kim AD, Jeong BH. Novel polymorphisms and genetic features of the Prion Protein Gene (Prnp) in cats, hosts of feline spongiform encephalopathy. Genes. (2020) 12:13. doi: 10.3390/genes12010013
37. Jeong BH, Sohn HJ, Lee JO, Kim NH, Kim JI, Lee SY, et al. Polymorphisms of the prion protein gene (PRNP) in Hanwoo (Bos taurus coreanae) and Holstein cattle. Genes Genet Syst. (2005) 80:303–8. doi: 10.1266/ggs.80.303
38. Kumar S, Stecher G, Li M, Knyaz C, Tamura K. MEGA XL: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol. (2018) 35:1547. doi: 10.1093/molbev/msy096
39. Iglesias V, Conchillo-Sole O, Batlle C, Ventura S. AMYCO: evaluation of mutational impact on prion-like proteins aggregation propensity. BMC Bioinformatics. (2019) 20:1–5. doi: 10.1186/s12859-019-2601-3
40. Adzhubei I, Jordan DM, Sunyaev SR. Predicting functional effect of human missense mutations using PolyPhen-2. Curr Prot Human Gen. (2013) 76:7–20. doi: 10.1002/0471142905.hg0720s76
41. Kaplan W, Littlejohn TG. Swiss-PDB viewer (deep view). Brief Bioinform. (2001) 2:195–7. doi: 10.1093/bib/2.2.195
42. Choi Y, Chan AP. PROVEAN web server: a tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics. (2015) 31:2745–7. doi: 10.1093/bioinformatics/btv195
43. Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, et al. Clustal W and Clustal X version 20. Bioinformatics. (2007) 23:2947–8. doi: 10.1093/bioinformatics/btm404
44. Zhang J, Wang F, Zhang Y. Molecular dynamics studies on the NMR structures of rabbit prion protein wild type and mutants: surface electrostatic charge distributions. J Biomol Struct Dyn. (2015) 33:1326–35. doi: 10.1080/07391102.2014.947325
45. Zhang J. Studies on the structural stability of rabbit prion probed by molecular dynamics simulations of its wild-type and mutants. J Theor Biol. (2010) 264:119–22. doi: 10.1016/j.jtbi.2010.01.024
46. Kim YC, Jeong MJ, Jeong BH. The first report of genetic variations in the chicken prion protein gene. Prion. (2018) 12:197–203. doi: 10.1080/19336896.2018.1471922
47. Kim YC, Won SY, Do K, Jeong BH. Identification of the novel polymorphisms and potential genetic features of the prion protein gene (PRNP) in horses, a prion disease-resistant animal. Sci Rep. (2020) 10:8926. doi: 10.1038/s41598-020-65731-5
48. Fung KL, Pan J, Ohnuma S, Lund PE, Pixley JN, Kimchi-Sarfaty C, et al. MDR1 synonymous polymorphisms alter transporter specificity and protein stability in a stable epithelial monolayer. Cancer Res. (2014) 74:598–608. doi: 10.1158/0008-5472.CAN-13-2064
49. Im EH, Choi SS. Synonymous codon usage controls various molecular aspects. Genomics Inform. (2017) 15:123. doi: 10.5808/GI.2017.15.4.123
50. Erana H, Fernandez-Borges N, Elezgarai SR, Harrathi C, Charco JM, Chianini F, et al. In vitro approach to identify key amino acids in low susceptibility of rabbit prion protein to misfolding. J Virol. (2017) 91:10–128. doi: 10.1128/JVI.01543-17
51. Vidal E, Fernandez-Borges N, Pintado B, Erana H, Ordóñez M, Márquez M, et al. Transgenic mouse bioassay: evidence that rabbits are susceptible to a variety of prion isolates. PLoS Pathog. (2015) 11:e1004977. doi: 10.1371/journal.ppat.1004977
52. Sarradin P, Viglietta C, Limouzin C, Andreoletti O, Daniel-Carlier N, Barc C, et al. Transgenic rabbits expressing ovine PrP are susceptible to scrapie. PLoS Pathog. (2015) 11:e1005077. doi: 10.1371/journal.ppat.1005077
53. Toombs JA, Petri M, Paul KR, Kan GY, Ben-Hur A, Ross ED, et al. De novo design of synthetic prion domains. Proc Nat Acad Sci. (2012) 109:6519–24. doi: 10.1073/pnas.1119366109
54. Myers JK, Pace CN. Hydrogen bonding stabilizes globular proteins. Biophys J. (1996) 71:2033–9. doi: 10.1016/S0006-3495(96)79401-8
55. Alber T, Dao-Pin S, Wilson K, Wozniak JA, Cook SP, Matthews BW. Contributions of hydrogen bonds of Thr 157 to the thermodynamic stability of phage T4 lysozyme. Nature. (1987) 330:41–6. doi: 10.1038/330041a0
Keywords: prion, PRNP, rabbit, polymorphism, SNP
Citation: Kim D-J, Kim Y-C and Jeong B-H (2023) First report of a novel polymorphism and genetic characteristics of the leporine prion protein (PRNP) gene. Front. Vet. Sci. 10:1229369. doi: 10.3389/fvets.2023.1229369
Received: 26 May 2023; Accepted: 04 September 2023;
Published: 22 September 2023.
Edited by:
Peter Dovc, University of Ljubljana, SloveniaReviewed by:
Eva Bagyinszky, Gachon University, Republic of KoreaCopyright © 2023 Kim, Kim and Jeong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Byung-Hoon Jeong, YmhqZW9uZ0BqYm51LmFjLmty
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.