- 1State Key Laboratory of Agricultural Microbiology, The Cooperative Innovation Center for Sustainable Pig Production, College of Animal Science and Veterinary Medicine, Huazhong Agricultural University, Wuhan, China
- 2Hubei Key Laboratory of Agricultural Bioinformatics, College of Informatics, Huazhong Agricultural University, Wuhan, China
- 3Key Laboratory of Prevention and Control Agents for Animal Bacteriosis (Ministry of Agriculture and Rural Affairs), Animal Husbandry and Veterinary Institute, Hubei Academy of Agricultural Sciences, Wuhan, China
- 4Department of Microbiology, School of Molecular and Cellular Biology, University of Illinois at Urbana-Champaign, Urbana, IL, United States
Pasteurella multocida is a versatile zoonotic pathogen. Multiple systems have been applied to type P. multocida from different diseases in different hosts. Recently, we found that assigning P. multocida strains by combining their capsular, lipopolysaccharide, and MLST genotypes (marked as capsular: lipopolysaccharide: MLST genotype) could help address the biological characteristics of P. multocida circulation in different hosts. However, there is still lack of a rapid and efficient tool to diagnose P. multocida according to this system. Here, we developed an intelligent genotyping platform PmGT for P. multocida strains according to their whole genome sequences using the web 2.0 technologies. By using PmGT, we determined capsular genotypes, LPS genotypes, and MLST genotypes as well as the main virulence factor genes (VFGs) of P. multocida isolates from different host species based on their whole genome sequences published on NCBI. The results revealed a closer association between the genotypes and pasteurellosis rather than between genotypes and host species. With the advent of high-quality, inexpensive DNA sequencing, PmGT represents a more efficient tool for P. multocida diagnosis in both epidemiological studies and clinical settings.
Introduction
Rapid and accurate diagnosis of sources of infections is critical for both medical and veterinary activities, and it is important for improved understanding of disease mechanisms and measures to control the illness (1). Microbial typing is an important link for the diagnosis of pathogens associated with diseases. The most widely used typing methods consist of serological typing systems and PCR-based molecular typing methods (2, 3). The establishment of discriminatory typing systems help in the understanding and control of pathogens, especially those with multiple serovars and/or genotypes from different environmental or host sources. Whole genome sequencing combined with the high-end computational technology is such an emerging approach for microbial diagnosis (4). Using the whole genome sequencing technologies, it is possible to determine the causative agent of infectious diseases rapidly and accurately, including newly emerged ones (5, 6). However, interpretation of the sequencing results to formulate a definitive diagnosis still requires technical experts with computational and bioinformatics skills. Therefore, a practical, automated platform that combines whole genome sequencing with computational technologies to provide diagnostic outcomes would be beneficial in advancing the field.
Pasteurella multocida is an important zoonotic pathogen and it can colonize and cause infections in a wide range of domestic and wild animals including food producing animals (e.g., poultry, pigs, beef, sheep) and companion animals (e.g., cats and dogs) as well as in humans (7–9). Animal diseases associated with P. multocida such as fowl cholera in poultry and other birds, progressive atrophic rhinitis and pneumonic pasteurellosis in pigs, haemorrhagic septicaemia and respiratory diseases in cattle and buffalos, leporine atrophic rhinitis and pneumonic pasteurellosis, are of great economic significance in agriculture (9). In humans, opportunistic infections of soft tissue, including wound dermonecrosis, respiratory disease with chronic pulmonary, urinary tract infection and bacteremic meningitis have also been reported (9). Most of these infections are associated with animal biting, scratching, kissing, and/or licking (10–12). In this regard, P. multocida represents a risk to public health. P. multocida strains from different hosts are serologically classified into five serogroups (A, B, D, E, F) (13–15) and/or 16 serovars (serovars 1 to 16) (16), according to their capsular and lipopolysaccharide (LPS) antigens, respectively. However, these two traditional serological typing methods require high-quantity antisera that are challenging to prepare, particularly for clinical use, such those methods are no longer widely used for large-scale epidemiological studies (7, 17).
In 2001, a multiplex PCR-based method was established to type the five serogroups into five capsular genotypes (A, B, D, E, F) (18), and in 2015, another multiplex PCR-based method was also developed to classified the 16 serovars into eight LPS genotypes (L1~L8) (19). In 2004 and 2010, two multilocus sequencing typing systems were also developed to genotype P. multocida strains (https://pubmlst.org/pmultocida/) from multiple mammalian hosts and birds, respectively (20, 21). In 2017, a virulence genotyping system based on the detection of different virulence factor gene (VFG) profiles was also reported for distinguishing P. multocida strains from different hosts (22). Compared to the traditional serological typing methods, these molecular DNA-based typing systems are indeed highly effective and accurate, and they are now widely used to determine the epidemiological and genetic characteristics of clinical isolates (23–27).
Despite of more than 135 years of research, differences on the molecular biological characteristics of P. multocida prevalence in different host species remain to be addressed. For example, P. multocida type A strains have been recovered from avian species, pigs, bovine species, and many other host species (8, 9), but little is known about differences on those type A isolates from different hosts. Recently, we developed a system to assign P. multocida strains from different host species by combining their capsular, LPS, and MLST genotypes (marked as capsular genotype: LPS genotype: MLST genotype), as well as determine the VFG profiles, which contributes to address the molecular biological characteristics of P. multocida prevalence in different host species (7, 23, 27). However, this strategy requires bioinformatics experts for data analysis and interpretation. Here, we report the development of an automated platform to type P. multocida strains from multiple hosts that combines the use of whole genome sequencing.
Materials and Methods
Bacterial Strains and Nucleotide Sequences
P. multocida strains used in this study include one isolate of bovine origin (strain HB01), one isolate of avian origin (strain HB02), and 50 isolates of porcine origin (strains HB03, HN04, HN05, HN06, HN07, HNA01~HNA22, HND01~HND21, HNF01 and HNF02) (Supplementary Table S1). All of these strains are from our laboratory collection, for which we have previously sequenced their whole genome sequences (27–30).
Nucleotide sequences specific for the determination of P. multocida strains (KMT1, 460 bp), and their the five capsular genotypes (A, 1044 bp; B, 760 bp; D, 657 bp; E, 511 bp; F, 851 bp); as well as their eight LPS genotypes (L1, 1307 bp; L2, 810 bp; L3, 474 bp; L4, 550 bp; L5, 1175 bp; L6, 668 bp; L7, 931 bp; L8, 255 bp) were extracted from the genome sequences of the different P. multocida strains according to the positions documented in previous publications (18, 19) and were deposited in GenBank under accession numbers MT570166, MN938443~MN938455 (Supplementary Text 1).
The nucleotide sequences of 23 types of virulence genes commonly detected in P. multocida epidemiological studies, including those encoding fimbriae and other adhesins (ptfA, fimA, hsf-1, hsf-2, pfhA, and tadD), toxin (toxA), iron acquisition proteins (exbB, exbD, tonB, hgbA, hgbB, fur, and tbpA), sialidases (nanB and nanH), hyaluronidase (pmHAS), outer membrane proteins (OMPs) (ompA, ompH, oma87, and plpB), and superoxide dismutase (sodA and sodC), were amplified from the genomic DNA of P. multocida HN06 and HB01 by PCR assays using the protocols documented elsewhere (23, 31). These nucleotide sequences were deposited in GenBank under accession numbers MT570167~ MT570189 (Supplementary Text 1).
The publicly available whole genome sequences of 262 P. multocida strains from bovine species [n = 106; including those recovered bovine haemorrhagic septicaemia cases (32)], avian species (n = 39), porcine species (n = 66), leporine species (n = 20), ovine species (n = 6), humans (n = 13), canines (n = 3), murine species (n = 2), horses (n = 2), cats (n = 2), alpacas (n = 2) and 1 synthetic DNA sequence in NCBI genome database were downloaded for use (Supplementary Table S1).
System Implementation
The PmGT platform was integrated on a CentOS server, mainly providing two kinds of online services: genotyping tool, and data query and display. To establish the genotyping online service, we first used Apache (https://www.apache.org) as the web container. Then, we downloaded the BLAST package (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/LATEST/) from NCBI, which was thereafter installed and configured on the web container. PHP was used as the server-side language and the browser-side script used jQuery, which is a fast, small, and feature-rich JavaScript library. The view pages were constructed with Hypertext Markup Language (HTML) and Cascading Style Sheets (CSS). For the target strain, the format of the sequence was first verified by the web user interface and then the sequence data was uploaded to the server through the PHP program which subsequently called the localized BLAST to align the uploaded sequence with the reference database. The nucleotide sequences specific for the determination of P. multocida strains, capsular genotypes, LPS genotypes, and the 23 types of virulence factor genes (VFGs) were packaged and used as the reference database for sequence alignment. Finally, the result was returned and displayed in the web page. In addition, if the user selected the option of “MLST genotyping,” the http request function “curl_setopt” in PHP was used to request PubMLST's RESTful interface (http://rest.pubmlst.org/db/pubmlst_Pmultocida_seqdef/sequence) and the function “curl_exec” was used to catch the response which thereafter was parsed to the result and displayed in the genotyping page.
PCR Detection of Capsular Genotypes, LPS Genotypes, MLST Genotypes, and Virulence Genes of P. multocida Strains From Pigs
Capsular genotypes and LPS genotypes of P. multocida strains from our laboratory collection were determined using multiplex PCR-based assays, as documented elsewhere (18, 19). Profiles of 23 types of virulence genes mentioned above were determined by PCR assays, as described previously (23). Sequence types (STs) were determined according to the protocols described in Pasteurella multocida MLST database (https://pubmlst.org/organisms/pasteurella-multocida/multi-host).
Data Availability
Nucleotide sequences specific for P. multocida and its capsular genotypes, LPS genotypes, as well as VFGs were publicly available in GenBank under accession numbers MN938443-MN938455 and MT570167~MT570189. The typing system developed in the present study is available at: http://vetinfo.hzau.edu.cn/PmGT.
Results
Development and Implementation of PmGT
The general process for genotyping is summarized as: when a query sequence is submitted via the web user interface, this sequence will be then submitted to the CentOS server via HTTP protocol. Thereafter, the sequence is evaluated by the PHP program, and the passed sequence will be BLASTed against the genotype database to yield a result, which will be returned to the webpage through the PHP program (Figures 1A,B). Through the above procedures, the genotyping module of PmGT (http://vetinfo.hzau.edu.cn/PmGT) was developed (Figure 1).
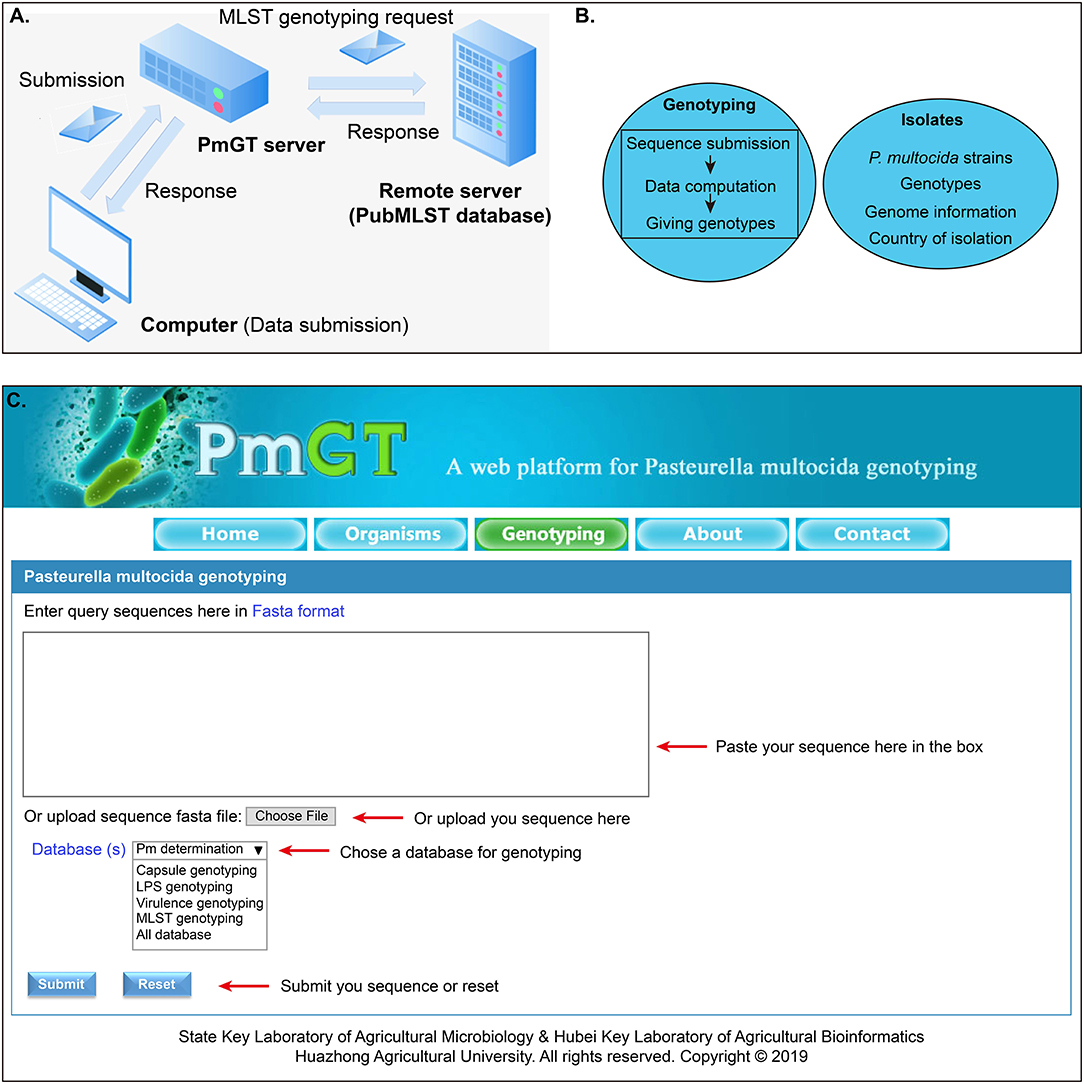
Figure 1. Development of the P. multocida genotyping and host prediction platform. (A) Flowchart showing the system design; (B) Main functions of the web platform; (C) Overview of the genotyping system of P. multocida.
Currently, PmGT provides the above services includes five menus: (1) the “Home” page gives a brief introduction of P. multocida etiological characteristics to help the users understand the bacterium; (2) the “Isolates” page displays the genotypes of P. multocida strains based on their whole genome sequences that are publicly available in NCBI; this page also provides the link for the users to download the genomes of these P. multocida strains from NCBI; (3) the “Genotyping” page enables the users to determine whether a putative isolate is a P. multocida and genotype P. multocida strains by using the whole genome sequence assembled from the sequencing reads (Figure 1C); (4) the “About” page summarizes the guidelines for the use of this web tool; (5) the “Contact” page provides the contact information of the developers.
PmGT Shows the Same Accuracy With PCR Methods in Genotyping P. multocida Strains
To test the accuracy of PmGT, we used two methods to type 52 P. multocida isolates (HB01, HB02, HB03, HN04, HN05, HN06, HN07, HNA01~HNA22, HND01~HND21, HNF01, and HNF02) from our laboratory collection (27). First, we submitted their whole genome sequences to PmGT for genotyping. As a comparison, we also determined the capsular genotypes, LPS genotypes, sequence types, as well as the profile of the abovementioned 23-kinds of virulence genes by using PCR assays. All these 52 strains were genotyped by PmGT and through this online genotyping platform (Table 1). Genotyping by PCR assays confirmed these capsular, LPS, and MLST genotypes. PCR results of capsular and LPS genotypes are provided in Supplementary Figures S1, S2.
Determination of the 23 types of virulence genes for each of the 52 strains by using this online system revealed that several genes (ptfA, fimA, oma87, and sodC) were broadly presented in the genome sequences genotyped (Figure 2). However, several genes (hsf-1, hsf-2, pfhA, and tadD) were heterogeneously distributed, and in particularly, none of the 52 sequences genotyped carried the toxA or tbpA genes (Figure 2). These results were also confirmed by PCR assays (Supplementary Table S1).
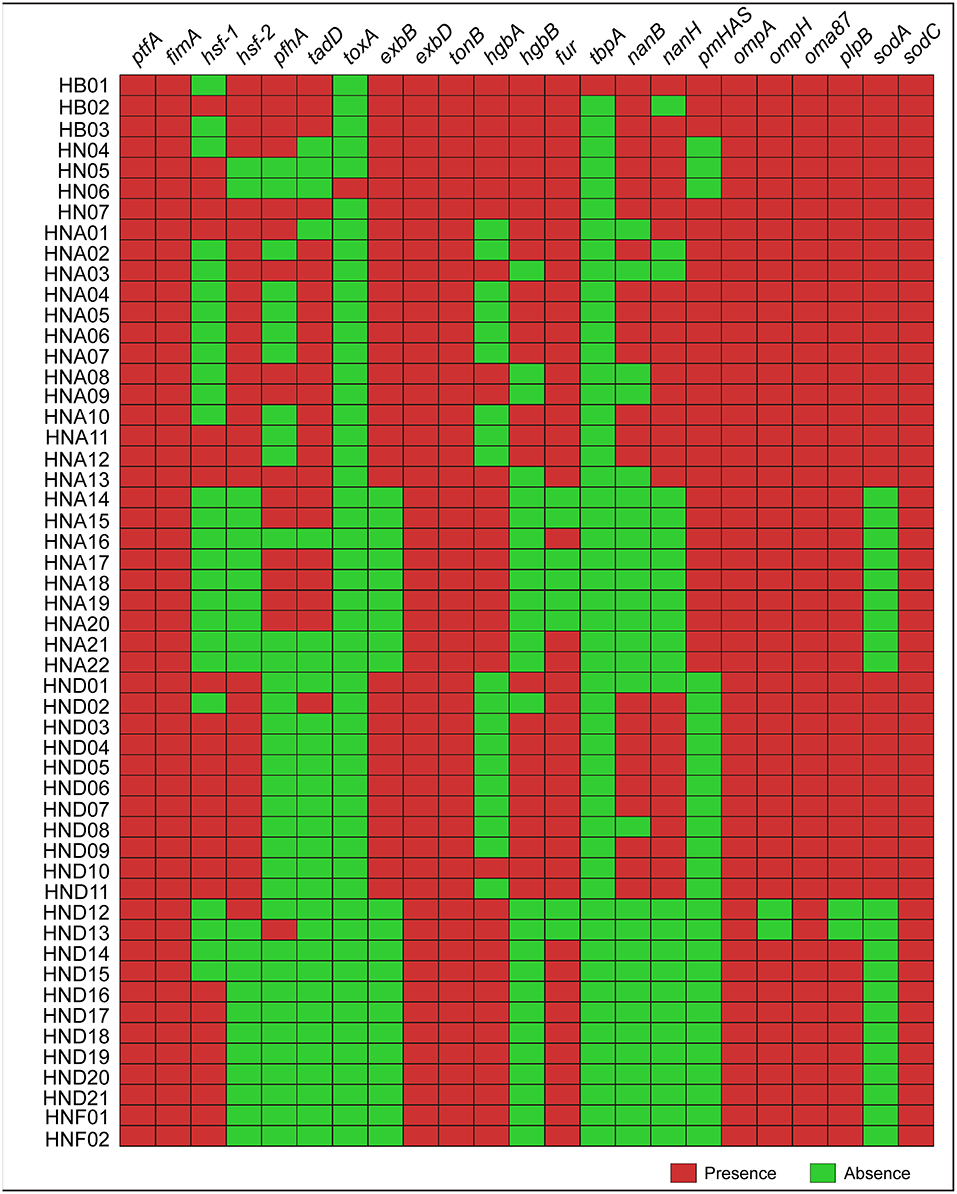
Figure 2. Heatmap showing the distribution of the 23 types of virulence genes (VFGs) among the 52 P. multocida strains from pigs. Boxes in red indicate a VFG is presence in the strain while boxes in green represent a VFG is missing in the strain.
Genotypes of P. multocida From Different Hosts
To understand the genotypes of P. multocida strains circulation in different host species, the 262 whole genome sequences of P. multocida strains were genotyped by PmGT. The results revealed that P. multocida isolates from different hosts displayed a certain preference for “capsular/LPS/MLST genotypes” (Figure 3). For example, most of the porcine strains were determined as capsular genotypes A (52%) and D (39%), LPS genotypes L3 (36%) and L6 (61%), sequence types ST3 (29%), ST11 (22%), and ST10 (34%), respectively; while most of the genotyped bovine strains were determined as capsular genotypes A (72%) and B (28%), LPS genotypes L3 (67%) and L2 (27%), and sequence types ST1 (59%) and ST44 (25%), respectively (Figure 3). When combining the capsular genotypes and the LPS genotypes, it revealed that most of the genotyped avian P. multocida were typed as A:L1 and A:L3, while most of the genotyped bovine P. multocida were typed as A:L3 and B:L2; the genotyped porcine P. multocida mainly belonged to D:L6, A:L3, and A:L6; while the genotyped leporine P. multocida mainly belonged to A:L3; most of the genotyped human P. multocida were typed as A:L3 and A:L1 (Figure 4A). If the capsular genotypes, LPS genotypes, and MLST genotypes were combined, most of the genotyped avian P. multocida were typed as A:L1:ST128, while most of the genotyped bovine P. multocida were typed as A:L3:ST1 and B:L2:ST44; the genotyped porcine P. multocida mainly belonged to D:L6:ST11, A:L3:ST3, and A:L6:ST10; while the genotyped leporine P. multocida mainly belonged to A:L3:ST12 (Figure 4).
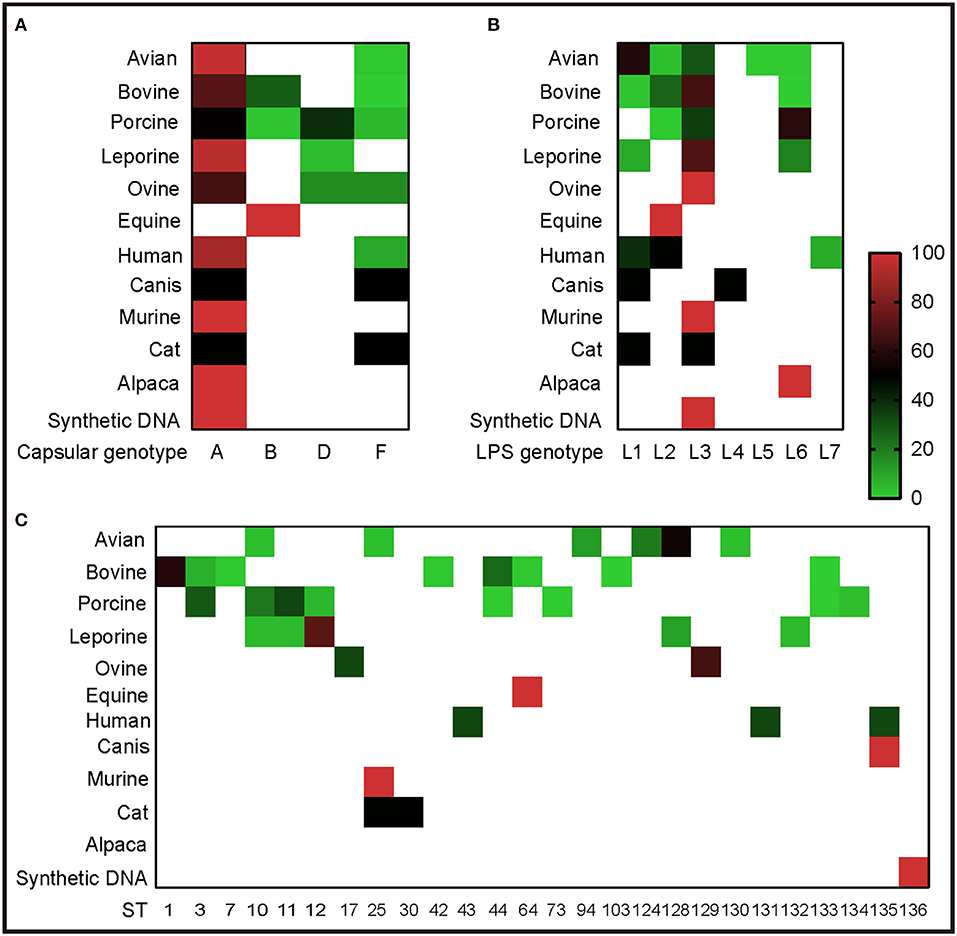
Figure 3. Heatmap revealing the association between capsular/LPS/MLST genotypes and P. multocida strains from different host species determined by PmGT. (A) Heatmap revealing the association between capsular genotypes and P. multocida strains from different host species; (B) Heatmap revealing the association between LPS genotypes and P. multocida strains from different host species; (C) Heatmap revealing the association between MLST genotypes and P. multocida strains from different host species. Percentages of sequences typed are shown with different colors displayed at right corner.
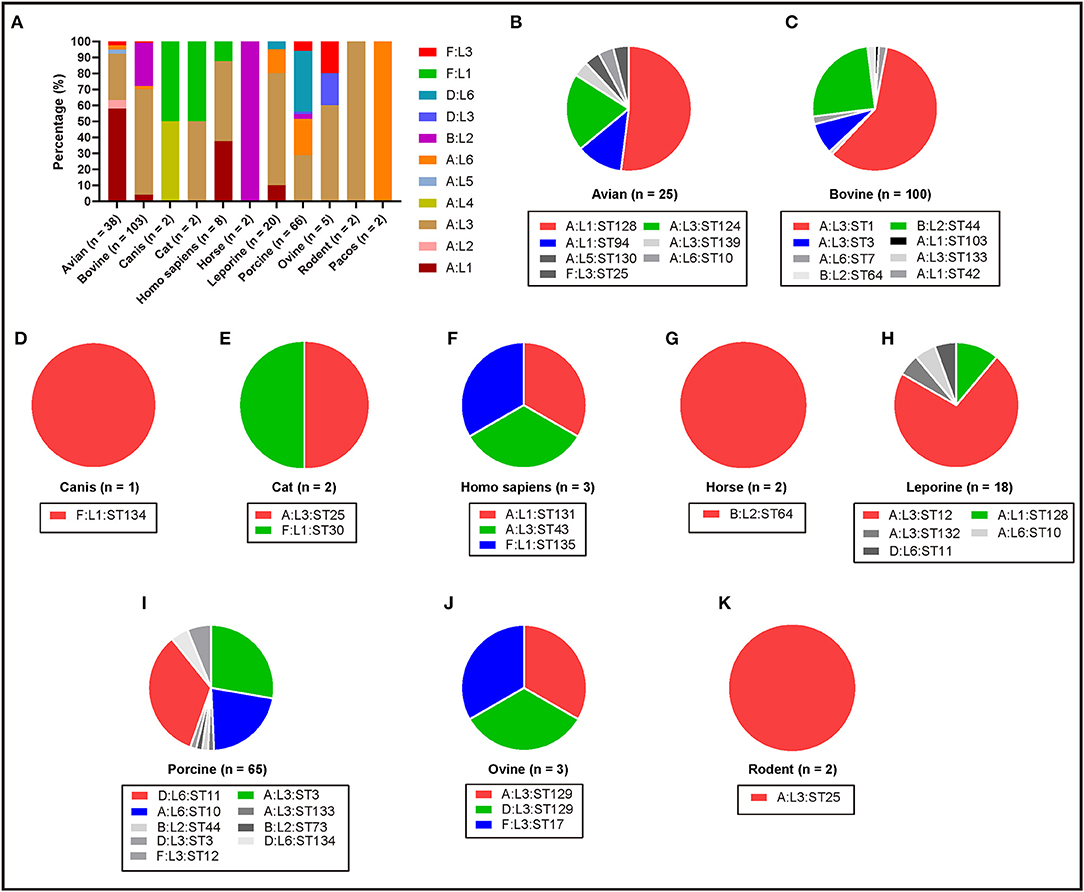
Figure 4. Column and pie charts showing the distribution of capsular: LPS genotypes and/or the capsular: LPS: MLST genotypes of P. multocida strains from different host species determined by PmGT by using the whole genome sequences. (A) Column chart showing the distribution of capsular: LPS genotypes of P. multocida strains from different host species; (B–K) Pie charts showing the distribution of capsular: LPS: MLST genotypes of P. multocida strains from avian species, bovine species, canis, cats, humans, horses, leporine species, pigs, ovine species, and rodents, respectively.
Virulence genotyping using the system developed herein revealed that the presence of multiple VFGs, including ptfA, fimA, hsf-2, exbB, exbD, tonB, hgbA, hgbB, fur, nanB, nanH, ompA, ompH, oma87, plpB, sodA, and sodC, was a broad characteristic of P. multocida strains from multiple host species (Figure 5). However, several VFGs were only determined in the genome sequences of P. multocida from certain hosts. For example, toxA, a gene encoding a dermonecrotic toxin, was found only in strains from pig, sheep, and alpacas, while tbpA, a transferrin binding protein coding gene, was found only in strains from cattle, sheep, and alpacas (Figure 5).
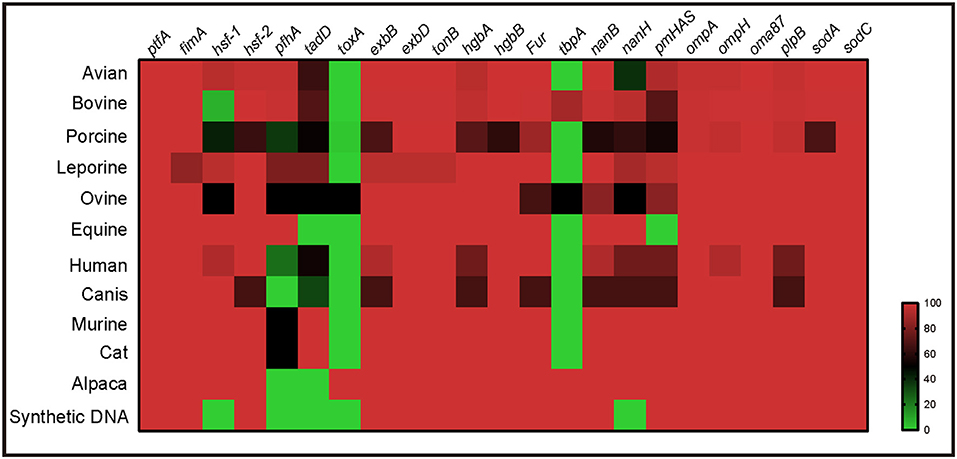
Figure 5. Heatmap revealing the association between virulence genes and P. multocida strains from different host species.
Discussion
P. multocida is the causative agent of multiple diseases with a wide spectrum of host species, including humans and other primates (7–9). In addition, P. multocida isolates recovered from different hosts with different diseases can be classified in many different serovars/genotypes according to different typing systems (7, 9). Relying on only one or two typing systems is difficult to address the characteristics of P. multocida isolates from different host species and/or their association with different diseases. For example, P. multocida isolates from different host species might have the same capsular genotypes but possess different LPS genotypes and/or MLST genotypes; even those from different host species that share the same capsular, LPS, and MLST genotypes might carry different VFGs (27, 33). Therefore, we have proposed a combined “capsular: LPS: MLST” genotyping system that includes virulence genotyping to discriminate P. multocida isolates from different host species and/or those associated with different diseases (7). However, this combined genotyping system is multiplex PCR-based and is laborious and time-consuming.
Advances in bioinformatics and bioinformatical tools enable the application of whole genome sequence data for inclusion of various demographic information for bacterial characterization, such as capsular and LPS genotyping; the presence of adhesins, toxins, or other virulence factors (34). In the present study, we reported the development of a genotyping platform for distinguishing P. multocida isolates according to the bacterial whole genome sequences. Validation of the PmGT platform was performed on a collection of P. multocida isolates from our laboratory. Results revealed that this genotyping system provides consistent results of determining the capsular-, LPS-, MLST genotypes, and VFGs, as compared with that obtained using multiplex PCR-based typing systems. Compared to the multiplex PCR-based typing systems (18, 19, 21, 22) and traditional serological typing systems (13, 16), this genotyping system takes less time to yield results and does not require high-quality antisera, which represents a more efficient and cost-saving tool for characterizing P. multocida isolates in both epidemiological studies and clinical settings.
By using PmGT, the capsular-, LPS-, MLST genotypes, and VFGs of P. multocida strains from different hosts were determined according to the whole genome sequences. These results agree with those of the epidemiological studies (23, 24, 26, 35). For example, P. multocida serovars B: 2 and A: 3 strains are frequently associated with bovine haemorrhagic septicaemia and respiratory diseases, respectively (36, 37). It is known that P. multocida serogroups A and B are assigned to capsular genotypes A and B by multiplex PCR, respectively (18); while P. multocida Heddleston serovars 2 and 3 are assigned to LPS genotypes L2 and L3 by multiplex PCR, respectively (19). That is why the capsular: LPS genotypes of most of the bovine strains were determined as A: L3 and B: L2, respectively. In addition, P. multocida strains isolated from bovine haemorrhagic septicaemia are commonly determined as ST122 (38), this sequence type can be reassigned to ST44 by using the multihost MLST database (27). These findings could explain why P. multocida strains associated with bovine haemorrhagic septicaemia were typed as capsular: LPS: MLST genotype B: L2: ST44. Similar findings were also observed in P. multocida strains from the other host species. In particularly, most of the P. multocida strains from pigs were determined as capsular: LPS: MLST genotypes D: L6: ST11, A: L3: ST3, and A: L6: ST10. These results are also in agreement with the results of our previously epidemiological study (23), suggesting that these three genotypes, particularly genotype D/L6/ST11, are likely to be strongly associated with swine respiratory diseases. However, during our test we also found the capsular-, LPS-, and/or MLST-genotypes of several strains could not be determined by PmGT according to the whole genome sequences. After check the data we put forward several reasons to explain this result: (1) most of these non-typeable genomes are sequenced and assembled using the second-generation sequencing technologies and the quality of these genomes are not high, some of the genes used for capsular/LPS/MLST genotyping fell within the gaps between genome contigs in the assemblies (7); (2) the genome sequences might be those of the capsular nontypeable strains reported (23, 39); (3) several strains belong to novel sequence types and the current Pasteurella multocida MLST database do not include these sequence types.
In conclusion, we developed an online platform for P. multocida genotyping (PmGT platform), which combines whole genome sequence analysis tools with web 2.0 technologies. By using this system, we determined the genotypes of P. multocida isolates from different host species. Overall, this system represents a more convenient tool for P. multocida diagnosis in both epidemiological studies and clinical settings. More importantly, our study provides an example to develop rapid and efficient tools for bacterial diagnosis by using their whole genome sequences in the coming age of artificial intelligence.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/genbank/, MN938443-MN938455 and MT570166~MT570166.
Author Contributions
ZP, BWi, JW, and BWu contributed to conception and design of the study. ZP, JL, WL, FW, LW, XW, and LH performed the experiments. ZP, JL, LW, and JW performed the statistical analysis. ZP wrote the first draft of the manuscript. ZP, HC, BWi, JW, and BWu revised the manuscript. All authors contributed to manuscript revision, read, and approved the submitted version.
Funding
This work was supported in part by China Postdoctoral Science Foundation (grant number 2020T130232), the Fundamental Research Funds for the Central Universities of China (grant number 2662018JC034), Guangdong Provincial Key Laboratory of Livestock Disease Prevention (grant number YDWS1901), and Hubei Provincial Key Research & Development Program (grant number 2021BBA085).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We sincerely acknowledge Huazhong Agricultural University College of Informatics for providing the CentOS server and the PubMLST database for the RESTful port for connection.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fvets.2021.771157/full#supplementary-material
Supplementary Text 1. Nucleotide sequences and their GenBank accession numbers for the construction of a comparative database for P. multocida genotyping.
Supplementary Figure S1. PCR results of capsular genotypes of tested P. multocida strains in the present study.
Supplementary Figure S2. PCR results of LPS genotypes of tested P. multocida strains in the present study.
Supplementary Table S1. Pasteurella multocida genome sequences used in the present study.
References
1. Kessel M. Why microbial diagnostics need more than money. Nat Biotechnol. (2015) 33:898–900. doi: 10.1038/nbt.3328
2. Peng Z, Ling L, Stratton CW, Li C, Polage CR, Wu B, et al. Advances in the diagnosis and treatment of Clostridium difficile infections. Emerg Microbes Infect. (2018) 7:15. doi: 10.1038/s41426-017-0019-4
3. Schmitz JE, Tang YW. The GenMark ePlex((R)): another weapon in the syndromic arsenal for infection diagnosis. Future Microbiol. (2018) 13:1697–708. doi: 10.2217/fmb-2018-0258
4. Lecuit M, Eloit M. The diagnosis of infectious diseases by whole genome next generation sequencing: a new era is opening. Front Cell Infect Microbiol. (2014) 4:25. doi: 10.3389/fcimb.2014.00025
5. Zhang YZ, Holmes EC. A genomic perspective on the origin and emergence of SARS-CoV-2. Cell. (2020) 181:223–7. doi: 10.1016/j.cell.2020.03.035
6. Török ME, Peacock SJ. Rapid whole-genome sequencing of bacterial pathogens in the clinical microbiology laboratory–pipe dream or reality? J Antimicrob Chemother. (2012) 67:2307–8. doi: 10.1093/jac/dks247
7. Peng Z, Wang X, Zhou R, Chen H, Wilson BA, Wu B. Pasteurella multocida: genotypes and genomics. Microbiol Mol Biol Rev. (2019) 83:e00014–9. doi: 10.1128/MMBR.00014-19
8. Wilkie IW, Harper M, Boyce JD, Adler B. Pasteurella multocida: diseases and pathogenesis. Curr Top Microbiol Immunol. (2012) 361:1–22. doi: 10.1007/82_2012_216
9. Wilson BA, Ho M. Pasteurella multocida: from zoonosis to cellular microbiology. Clin Microbiol Rev. (2013) 26:631–55. doi: 10.1128/CMR.00024-13
10. Ryan JM, Feder HM Jr. Dog licks baby Baby gets Pasteurella multocida meningitis. Lancet. (2019) 393:e41. doi: 10.1016/S0140-6736(19)30953-5
11. Dryden MS, Dalgliesh D. Pasteurella multocida from a dog causing Ludwig's angina. Lancet. (1996) 347:123. doi: 10.1016/S0140-6736(96)90250-0
12. Godey B, Morandi X, Bourdinière J, Heurtin C. Beware of dogs licking ears. Lancet. (1999) 354:1267–8. doi: 10.1016/S0140-6736(99)04197-5
13. Carter GR. Studies on Pasteurella multocida. I A hemagglutination test for the identification of serological types. Am J Vet Res. (1955) 16:481–4.
14. Carter GR. A new serological type of Pasteurella multocida from Central Africa. Veterinary Record. (1961) 73:1052.
15. Rimler RB, Rhoades KR. Serogroup F. A new capsule serogroup of Pasteurella multocida. J Clin Microbiol. (1987) 25:615–8. doi: 10.1128/jcm.25.4.615-618.1987
16. Heddleston KL, Gallagher JE, Rebers PA. Fowl cholera: gel diffusion precipitin test for serotyping Pasteruella multocida from avian species. Avian Dis. (1972) 16:925–36. doi: 10.2307/1588773
17. Peng Z, Liang W, Wu B. Molecular typing methods for Pasteurella multocida-a review. Wei Sheng Wu Xue Bao. (2016) 56:1521–9. doi: 10.13343/j.cnki.wsxb.20160002
18. Townsend KM, Boyce JD, Chung JY, Frost AJ, Adler B. Genetic organization of Pasteurella multocida cap loci and development of a multiplex capsular PCR typing system. J Clin Microbiol. (2001) 39:924–9. doi: 10.1128/JCM.39.3.924-929.2001
19. Harper M, John M, Turni C, Edmunds M, St Michael F, Adler B, et al. Development of a rapid multiplex PCR assay to genotype Pasteurella multocida strains by use of the lipopolysaccharide outer core biosynthesis locus. J Clin Microbiol. (2015) 53:477–85. doi: 10.1128/JCM.02824-14
20. Davies RL, MacCorquodale R, Reilly S. Characterisation of bovine strains of Pasteurella multocida and comparison with isolates of avian, ovine and porcine origin. Vet Microbiol. (2004) 99:145–58. doi: 10.1016/j.vetmic.2003.11.013
21. Subaaharan S, Blackall LL, Blackall PJ. Development of a multi-locus sequence typing scheme for avian isolates of Pasteurella multocida. Vet Microbiol. (2010) 141:354–61. doi: 10.1016/j.vetmic.2010.01.017
22. Garcia-Alvarez A, Vela AI, San Martin E, Chaves F, Fernandez-Garayzabal JF, Lucas D, et al. Characterization of Pasteurella multocida associated with ovine pneumonia using multi-locus sequence typing (MLST) and virulence-associated gene profile analysis and comparison with porcine isolates. Vet Microbiol. (2017) 204:180–7. doi: 10.1016/j.vetmic.2017.04.015
23. Peng Z, Wang H, Liang W, Chen Y, Tang X, Chen H, et al. A capsule/lipopolysaccharide/MLST genotype D/L6/ST11 of Pasteurella multocida is likely to be strongly associated with swine respiratory disease in China. Arch Microbiol. (2018) 200:107–18. doi: 10.1007/s00203-017-1421-y
24. Li Z, Cheng F, Lan S, Guo J, Liu W, Li X, et al. Investigation of genetic diversity and epidemiological characteristics of Pasteurella multocida isolates from poultry in southwest China by population structure, multi-locus sequence typing and virulence-associated gene profile analysis. J Vet Med Sci. (2018) 80:921–9. doi: 10.1292/jvms.18-0049
25. Devi LB, Bora DP, Das SK, Sharma RK, Mukherjee S, Hazarika RA. Virulence gene profiling of porcine Pasteurella multocida isolates of Assam. Vet World. (2018) 11:348–54. doi: 10.14202/vetworld.2018.348-354
26. Massacci FR, Magistrali CF, Cucco L, Curcio L, Bano L, Mangili P, et al. Characterization of Pasteurella multocida involved in rabbit infections. Vet Microbiol. (2018) 213:66–72. doi: 10.1016/j.vetmic.2017.11.023
27. Peng Z, Liang W, Wang F, Xu Z, Xie Z, Lian Z, et al. Genetic and phylogenetic characteristics of Pasteurella multocida isolates from different host species. Front Microbiol. (2018) 9:1408. doi: 10.3389/fmicb.2018.01408
28. Peng Z, Liang W, Liu W, Wu B, Tang B, Tan C, et al. Genomic characterization of Pasteurella multocida HB01, a serotype A bovine isolate from China. Gene. (2016) 581:85–93. doi: 10.1016/j.gene.2016.01.041
29. Liu W, Yang M, Xu Z, Zheng H, Liang W, Zhou R, et al. Complete genome sequence of Pasteurella multocida HN06, a toxigenic strain of serogroup D. J Bacteriol. (2012) 194:3292–3. doi: 10.1128/JB.00215-12
30. Peng Z, Liang W, Wang Y, Liu W, Zhang H, Yu T, et al. Experimental pathogenicity and complete genome characterization of a pig origin Pasteurella multocida serogroup F isolate HN07. Vet Microbiol. (2017) 198:23–33. doi: 10.1016/j.vetmic.2016.11.028
31. Khamesipour F, Momtaz H, Azhdary Mamoreh M. Occurrence of virulence factors and antimicrobial resistance in Pasteurella multocida strains isolated from slaughter cattle in Iran. Front Microbiol. (2014) 5:536. doi: 10.3389/fmicb.2014.00536
32. Moustafa AM, Seemann T, Gladman S, Adler B, Harper M, Boyce JD, et al. Comparative genomic analysis of Asian Haemorrhagic Septicaemia-associated strains of Pasteurella multocida identifies more than 90 Haemorrhagic Septicaemia-specific genes. PLoS One. (2015) 10:e0130296. doi: 10.1371/journal.pone.0130296
33. Ujvári B, Makrai L, Magyar T. Virulence gene profiling and ompA sequence analysis of Pasteurella multocida and their correlation with host species. Vet Microbiol. (2019) 233:190–5. doi: 10.1016/j.vetmic.2019.05.005
34. Stoesser N, Sheppard AE, Pankhurst L, De Maio N, Moore CE, Sebra R, et al. Evolutionary history of the global emergence of the escherichia coli epidemic clone ST131. MBio. (2016) 7:e02162. doi: 10.1128/mBio.02162-15
35. Ewers C, Lübke-Becker A, Bethe A, Kiebling S, Filter M, Wieler LH. Virulence genotype of Pasteurella multocida strains isolated from different hosts with various disease status. Vet Microbiol. (2006) 114:304–17. doi: 10.1016/j.vetmic.2005.12.012
36. Shivachandra SB, Viswas KN, Kumar AA. A review of hemorrhagic septicemia in cattle and buffalo. Anim Health Res Rev. (2011) 12:67–82. doi: 10.1017/S146625231100003X
37. Welsh RD, Dye LB, Payton ME, Confer AW. Isolation and antimicrobial susceptibilities of bacterial pathogens from bovine pneumonia: 1994–2002. J Vet Diagn Invest. (2004) 16:426–31. doi: 10.1177/104063870401600510
38. Hotchkiss EJ, Hodgson JC, Lainson FA, Zadoks RN. Multilocus sequence typing of a global collection of Pasteurella multocida isolates from cattle and other host species demonstrates niche association. BMC Microbiol. (2011) 11:115. doi: 10.1186/1471-2180-11-115
Keywords: Pasteurella multocida, genotyping, whole genome sequence, PmGT, genotypes
Citation: Peng Z, Liu J, Liang W, Wang F, Wang L, Wang X, Hua L, Chen H, Wilson BA, Wang J and Wu B (2021) Development of an Online Tool for Pasteurella multocida Genotyping and Genotypes of Pasteurella multocida From Different Hosts. Front. Vet. Sci. 8:771157. doi: 10.3389/fvets.2021.771157
Received: 06 September 2021; Accepted: 30 November 2021;
Published: 17 December 2021.
Edited by:
Doreene Hyatt, Colorado State University, United StatesReviewed by:
Patrick Blackall, The University of Queensland, AustraliaKeyvan Tadayon, Razi Vaccine and Serum Research Institute, Iran
A. Jesse, Putra Malaysia University, Malaysia
Andreas Petersen, Statens Serum Institut (SSI), Denmark
Copyright © 2021 Peng, Liu, Liang, Wang, Wang, Wang, Hua, Chen, Wilson, Wang and Wu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brenda A. Wilson, d2lsc29uN0BpbGxpbm9pcy5lZHU=; Jia Wang, d2FuZy5qaWFAbWFpbC5oemF1LmVkdS5jbg==; Bin Wu, d3ViQG1haWwuaHp1YS5lZHUuY24=
†ORCID: Zhong Peng orcid.org/0000-0001-5249-328X
Bin Wu orcid.org/0000-0001-9078-386X
 Zhong Peng
Zhong Peng Junyang Liu2
Junyang Liu2 Wan Liang
Wan Liang Fei Wang
Fei Wang Huanchun Chen
Huanchun Chen Brenda A. Wilson
Brenda A. Wilson Jia Wang
Jia Wang Bin Wu
Bin Wu