
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Vet. Sci. , 05 November 2020
Sec. Veterinary Infectious Diseases
Volume 7 - 2020 | https://doi.org/10.3389/fvets.2020.586826
Coronaviruses are widespread in nature and infect humans, mammals and poultry. They cause harm to humans and animals. Virus-mediated cell cycle arrest is an essential strategy for viral survival and proliferation in the host cells. A clarification system of the mechanisms of virus-induced cell cycle arrest is highly desirable to promote the development of antiviral therapies. In this review, molecular mechanisms of coronavirus-induced cell cycle arrest were systematically summarized. Moreover, the common features of coronavirus-mediated cell cycle arrest were discussed. This review will provide a theoretical basis for further studies on the infection mechanisms and prevention of coronaviruses.
Coronaviruses (CoVs) are the largest positive-sense, single-stranded RNA viruses. CoVs belong to the Coronaviridae family of the Nidovirales order. CoVs are classified into four genera: Alphacoronavirus (α-CoV), Betacoronavirus (β-CoV), Gammacoronavirus (γ-CoV), and Deltacoronavirus (δ-CoV) (1). CoVs share a common basic genomic organization. The genome size is approximately 25000–30000 nt. CoV contains at least six open reading frames (ORFs) in the order: ORF1a, ORF1b, spike (S), envelope (E), membrane (M), and nucleocapsid (N), and certain CoVs encode a hemagglutinin esterase (HE). Each virus contains one to several accessory genes, the ORF3 of α-CoV porcine epidemic diarrhea virus (PEDV), the ORF3a, ORF3b, ORF6, ORF7a, ORF7b, ORF8a, ORF8b, and ORF9 of severe acute respiratory syndrome coronavirus (SARS-CoV), the ORF3a, ORF3b, ORF5a, ORF5b of γ-CoV infectious bronchitis virus (IBV), NS6 and NS7 of δ-CoV porcine deltacoronavirus (PDCoV). Several large-scale epidemics of human and animal CoVs have occurred over the last few decades, especially the SARS-CoV outbreak in 2003, the Middle East respiratory syndrome coronavirus (MERS-CoV) outbreak in 2012, and the current global pandemic of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which has threatened global public health security. Therefore, CoVs have become a topic of research interest in the field of virology.
The interaction between the virus and the host is a symbiotic relationship. As obligate parasites, viruses entirely depend on the cellular pathways of the host to obtain resources necessary for replication. Eukaryotic cell cycle progression is a highly ordered and tightly regulated process that performs a crucial role in maintaining cell metabolism and homeostasis. When cells are stressed or injured, the host cells initiate repair mechanisms by regulating cell cycle progression, which is a primary response to external stimulation. Several viruses often interfere with the normal cell cycle progression to obtain sufficient resources for viral replication by inducing host cell cycle arrest in an active metabolic state, such as the DNA synthesis phase (2–4). Studies have confirmed that CoVs such as transmissible gastroenteritis virus (TGEV), PEDV, murine hepatitis virus (MHV), SARS-CoV, and IBV induce cell cycle arrest to facilitate viral replication (5–19). Although studies on CoV-regulated cell cycle have had a high research output, there has been no systematic overview of the studies. In this review, we aimed to summarize molecular mechanisms that are used by CoVs to manipulate the cell cycle and to provide insights into the common features of CoV-mediated cell cycle arrest.
A eukaryotic cell cycle comprises four different phases: Gap 1 (G1), Synthesis (S), Gap 2 (G2), and Mitosis (M) (20, 21). During the G1 phase, the cell primarily synthesizes proteins, saccharides and RNA, which are necessary for DNA replication in preparation for entry into the S phase (DNA synthesis phase). After DNA replication is completed, the cell enters the G2 phase and prepares for mitosis, cell division. Cyclin-dependent kinases (CDKs) and cyclins are the major regulators of cell cycle progression (20, 21). In mammalian cells, different cyclin-CDK complexes are involved in the regulation of different cell cycle transitions (20, 21). During the G1 phase, cyclin D/CDK4 (or CDK6) complexes phosphorylate the downstream factor, retinoblastoma (Rb), which in turn releases the transcription factor E2F promoting transcription of cyclin E, cyclin A, and CDK1 genes (22, 23). Transition from G1 to S phase is regulated by the cyclin A-CDK2 and cyclin E-CDK2 complexes (24). During the G2 phase, the cyclin A (or cyclin B)-CDK1 complexes phosphorylate histone H1, which initiates the cells into the M phase (25, 26). In addition, the cell cycle is negatively regulated by CDK inhibitors (CKIs) (20, 21). CKIs such as p21, can either bind to isolated CDKs or to the cyclin-CDK complex to prevent activation of CDKs. Furthermore, the tumor suppressor p53 that indirectly regulates more than 250 cell cycle-associated genes, performs an essential role in the regulation of the cell cycle (27, 28). Activation and induction of p53 in response to DNA damages or other stresses, and upregulation of p21 expression can inhibit CDK activity, thereby inducing cell cycle arrest or apoptosis.
CoVs are pathogens with a wide range of hosts that critically endanger human health and development of animal husbandry. Notably, the prevention and control of emerging and re-emerging CoVs such as SARS-CoV-2 and PEDV, has been a great challenge globally. Therefore, in addition to the development of diagnostic reagents, antiviral drugs, and vaccines, it is imperative to have a deeper understanding of the interaction mechanisms between CoVs and host cells.
CoV-regulated cell cycle has become one of the research hot spots in CoVs research in recent years. Viruses have evolved multiple mechanisms that induce cell cycle arrest to generate resources and cellular conditions favorable for viral replication to enhance replication efficiency. Numerous CoVs have evolved diverse strategies to manipulate host cell cycle for their own replication (Table 1, Figure 1). The various types of CoVs not only exhibit distinct characteristics of cell cycle regulation, but also exhibit potentially common regulatory mechanisms.
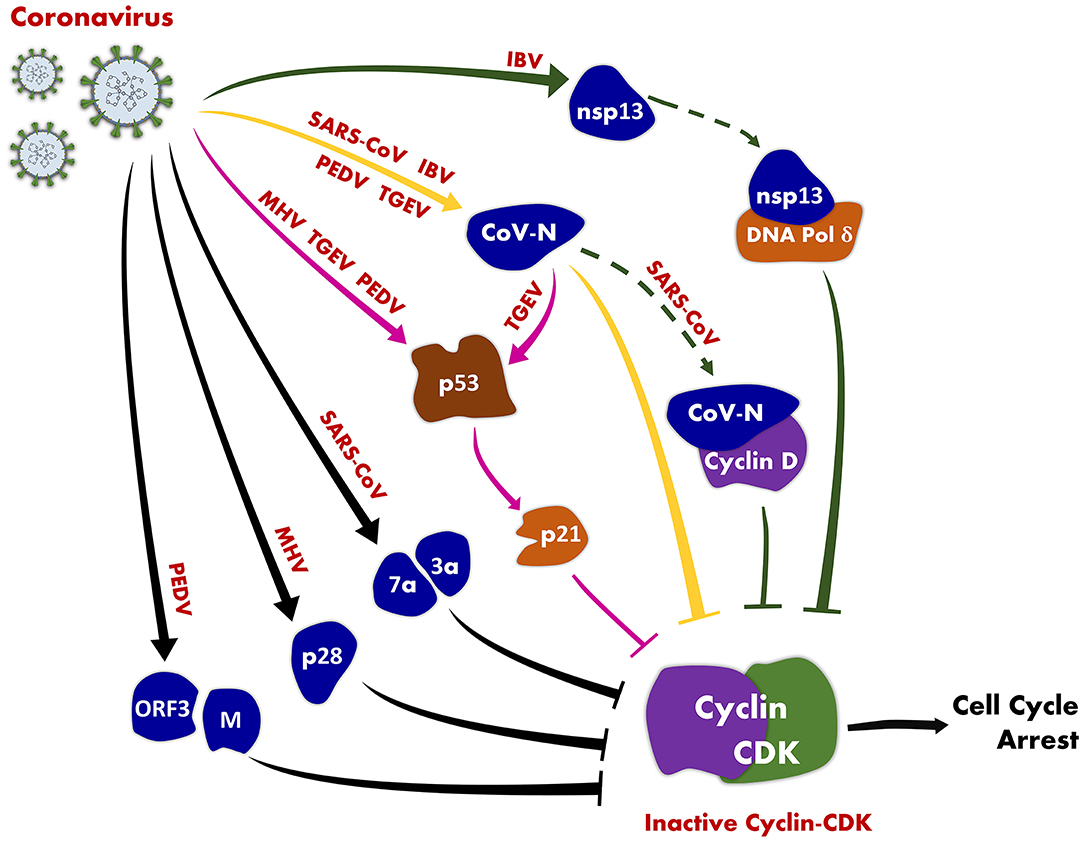
Figure 1. An illustration of coronavirus-induced cell cycle arrest. CoV induces host cell cycle arrest by inhibiting the activity of cyclin-CDK complexes through the following pathways: CoV (MHV, TGEV, and PEDV) inhibits the activity of cyclin-CDK complexes through the p53 signaling pathway (pink solid lines); CoV N protein (SARS-CoV, TGEV, and PEDV) suppresses the activity of cyclin-CDK complexes (yellow solid lines); CoV (SARS-CoV, IBV) inhibits the activity of cyclin-CDK complexes by directly interacting with host cell cycle-associated proteins (solid and dotted green lines).
Cyclin-CDK complexes are the core components of the cell cycle regulatory network. Similar to other viruses, CoVs affect the activity of cyclin-CDK complexes, which is a common strategy of manipulating cell cycle progression. SARS-CoV accessory proteins, 3a and 7b inhibit Rb phosphorylation by limiting the expression of cyclin D3, which in turn blocks the G0/G1 phase of the cell cycle in HEK 239 cells (7, 8). The N protein of SARS-CoV inhibits the activity of cyclin-CDK complex and blocks progression of the S phase in Huh7 cells (9). MHV-infected cells cause a reduction in the amounts of CDKs (CDK4 and CDK6) and cyclins (cyclin D1, D2, D3, and E) in 17Cl-1 cells, and insufficient hyperphosphorylation of pRb, resulting in inhibition of the cell cycle in the G0/G1 phase (10, 11). PEDV infection can induce cell cycle arrest in the G0/G1 phase in Vero cells, by up-regulating p21, cdc2, CDK2, CDKk4, cyclin A, cyclin B1 protein and down-regulating cyclin D1 and cyclin E1 protein (12, 13). However, other studies have revealed that PEDV N and M proteins prolong the S phase in IEC cells by down-regulating cyclin A (14, 15). IBV infection induces cell cycle arrest in the S and G2/M phases in Vero cells, by down-regulating cyclin D1 and cyclin D2, and the redistribution of subcellular location of cyclin D1 was observed (16, 17). TGEV N proteins induce accumulation of PK-15 cells in the S and G2/M phases by inhibiting expression of cyclin B1 and CDK2 (18).
The tumor suppressor, p53 has a key role in modulating response to cellular stress (27, 28). Activated p53 regulates hundreds of genes involved in multiple biological processes, including DNA damage repair, cell cycle arrest, apoptosis and senescence. Furthermore, p53 is involved in the manipulation of virus-host cell cycle. Previous studies have reported that CoV infections induce cell cycle arrest via the p53-dependent pathway (11, 12, 18, 19). MHV, PEDV and TGEV induce p53 activation and upregulate the expression of p21, which binds to cyclin-CDK complexes and inhibits kinase activity resulting in host cell cycle arrest. In addition, a previous study revealed that p53 phosphorylation on Ser-15 was upregulated in both 3a and M proteins of SARS-CoV expressed in HEK 239 cells (7); SARS-CoV and HCoV-NL63 infections affect the degradation pathway of p53 (29, 30); IBV infection redistributes the subcellular location of p53 (31). Such studies indicate that p53 has a crucial role in CoV-mediated cell cycle arrest. However, a previous study indicated that cell cycle arrest induced by IBV infection in H1299 cells was p53-independent (16). Therefore, further studies are required to investigate if p53 is a common regulatory mechanism of the CoV-mediated cell cycle arrest.
The N protein of CoVs is a multifunctional protein involved in multiple steps of viral replication (32). The N protein of CoV has been demonstrated to have a key role in regulating cell cycle progression, such as apoptosis, autophagy, and antagonizing innate immune responses (18, 33, 34). The N protein also modulates host cell cycle. Previous studies have reported that SARS-CoV, PEDV, and TGEV N proteins induce host cell cycle arrest in the S phase (9, 15, 18). SARS-CoV N protein inhibits the activity of cyclin-CDK complex by directly binding to cyclin D or cyclin-CDK2 complex, and by indirectly down-regulating protein levels of CDK2, cyclin E, and cyclin A (9). The nucleolus is a sub-nuclear compartment associated with numerous biological processes, including the cell cycle (35). Nuclear localization of the N protein is a common feature of CoVs, and the N protein interacts with nucleolar proteins such as nucleophosmin (NPM1) and fibrillarin (34, 36). In addition, Wurm et al. (35) reported that nucleolar localization of N proteins correlates with the disruption of host cell division caused by CoVs (37). A study by Cawood et al. (38) revealed that the different stages of the cell cycle can affect dynamic trafficking of N proteins in the nucleus and nucleolus (38). Moreover, Liu et al. (39) demonstrated that PEDV-induced cell cycle arrest in Vero cells depended on the nuclear localization signals (39).
The direct interaction between viral and host proteins is one of the effective strategies used by virus to manipulate the host cell cycle (40, 41). CoVs can subvert the host cell cycle to facilitate viral replication through its interactions with host proteins. SARS-CoV N protein prolongs cell cycle of the S phase by directly interacting with cyclin D (9). Nsp13 proteins of IBV and SARS-CoV interact with the p125 subunit of DNA polymerase δ to induce DNA damage, consequently causing cell cycle arrest in the S phase (42). MHV nsp15 interacts with pRb, increases expression of genes that are normally repressed by pRb, and affects the cell cycle by blocking cell cycle progression at the S phase in NIH 3T3 cells (43). In addition, binding of PEDV N protein with NPM1 prevents proteolytic cleavage of NPM1 and enhances survival of Vero cells (34). Moreover, PEDV M protein interacts with the cell cycle-associated protein cell division cycle 42 (CDC42) (44).
The molecular mechanisms of CoV-mediated regulation of cell cycle have not been systematically investigated, especially the common features of CoV-mediated cell cycle arrest. Further studies should be conducted in the future to explore the mechanisms according to the characteristics of CoVs and regulation of the cell cycle induced by each CoV. The N protein of CoVs and p53 of host proteins that are the key factors of coronavirus-mediated cell cycle regulation in this review form potentially valuable foundations for future studies on the common mechanisms of CoV-mediated cell cycle arrest.
Virus infection is a complex, multistage, and highly dynamic process. Virus dynamic regulates progression of the host cell cycle at various phases of infection based on the replication requirements. For example, certain viruses can induce cell-cycle arrest during the initial stages of a viral infection to utilize a host's resources, subsequently resulting in apoptosis of host cells for virion release (40). A few studies have been conducted on the dynamics of regulation of the host cell cycle process in CoVs, and the studies have focused on the effect of the virus on cell cycle at a specific stage of infection, and individual viral proteins on the cell cycle. This has resulted in the negligence of the impacts of different phases of infection and synergy between viral proteins and host cell cycle progression, resulting in limited understanding of the host cell cycle regulation during a CoV infection.
To date, numerous studies on the regulation of cell cycle by CoVs are based on grounded theory to explore how viruses regulate the cell cycle, and a few are based on applied research. Targeted blocking of key pathways by designing small-molecule drugs based on the pathways of viral regulation of the cell cycle is an effective strategy of inhibiting viral replication. CoVs can convert the host cell cycle into a microenvironment for virus proliferation through direct interaction with host cell cycle-associated proteins. Based on this, the interaction sites may be an ideal target for the small molecule drugs. In addition, based on the viral replication requirements, the microenvironment for viral replication can be enhanced by optimizing the cell culture media for viral vaccines, which can enhance the efficiency of vaccine production. However, this has not been successfully applied in the production of vaccines due to lack of systematic scientific theories.
DS: conceptualization. MS, SQ, DS, and LF: investigation. MS: writing. MS and YC: revision. All authors contributed to the article and approved the submitted version.
This study was supported by the National Natural Science Foundation of China (grant no. 31873011).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
1. Brian DA, Baric RS. Coronavirus genome structure and replication. Curr Top Microbiol Immunol. (2005) 287:1–30. doi: 10.1007/3-540-26765-4_1
2. Bagga S, Bouchard MJ. Cell cycle regulation during viral infection. Methods Mol Biol. (2014) 1170:165–227. doi: 10.1007/978-1-4939-0888-2_10
3. Nascimento R, Costa H, Parkhouse RM. Virus manipulation of cell cycle. Protoplasma. (2012) 249:519–28. doi: 10.1007/s00709-011-0327-9
4. Li LY, Yao K. Cell cycle regulation during viral infection. Int J Virol. (2015) 22:137–40. doi: 10.3760/cma.j.issn.1673-4092.2015.02.019
5. Ye S, Li Z, Chen F, Li W, Guo X, Hu H, et al. Porcine epidemic diarrhea virus ORF3 gene prolongs S-phase, facilitates formation of vesicles and promotes the proliferation of attenuated PEDV. Virus Genes. (2015) 51:385–92. doi: 10.1007/s11262-015-1257-y
6. Yuan X, Shan Y, Zhao Z, Chen J, Cong Y. G0/G1 arrest and apoptosis induced by SARS-CoV 3b protein in transfected cells. Virol J. (2005) 2:66. doi: 10.1186/1743-422X-2-66
7. Yuan X, Yao Z, Wu J, Wu J, Zhou Y, Shan Y, et al. G1 phase cell cycle arrest induced by SARS-CoV 3a protein via the cyclin D3/pRb pathway. Am J Respir Cell Mol Biol. (2007) 37:9–19. doi: 10.1165/rcmb.2005-0345RC
8. Yuan X, Wu J, Shan Y, Yao Z, Dong B, Chen B, et al. SARS coronavirus 7a protein blocks cell cycle progression at G0/G1 phase via the cyclin D3/pRb pathway. Virology. (2006) 346:74–85. doi: 10.1016/j.virol.2005.10.015
9. Surjit M, Liu B, Chow VTK, Lal SK. The nucleocapsid protein of severe acute respiratory syndrome-coronavirus inhibits the activity of cyclin-cyclin-dependent kinase complex and blocks S phase progression in mammalian cells. J Biol Chem. (2006) 281:10669–81. doi: 10.1074/jbc.M509233200
10. Chen CJ, Sugiyama K, Kubo H, Huang C, Makino S. Murine coronavirus nonstructural protein p28 arrests cell cycle in G0/G1 phase. J Virol. (2004) 78:10410–9. doi: 10.1128/JVI.78.19.10410-10419.2004
11. Chen CJ, Makino S. Murine coronavirus replication induces cell cycle arrest in G0/G1 phase. J Virol. (2004) 78:5658–69. doi: 10.1128/JVI.78.11.5658-5669.2004
12. Sun P, Wu H, Huang J, Xu Y, Yang F, Zhang Q, et al. Porcine epidemic diarrhea virus through p53-dependent pathway causes cell cycle arrest in the G0/G1 phase. Virus Res. (2018) 253:1–11. doi: 10.1016/j.virusres.2018.05.019
13. Luo YR, Zhou ST, Yang L, Liu YP, Jiang SY, Dawuli Y, et al. Porcine epidemic diarrhoea virus induces cell-cycle arrest through the dna damage-signalling pathway. J Vet Res. (2020) 64:25–32. doi: 10.2478/jvetres-2020-0024
14. Xu XG, Zhang HL, Zhang Q, Dong J, Huang Y, Tong DW. Porcine epidemic diarrhea virus M protein blocks cell cycle progression at S-phase and its subcellular localization in the porcine intestinal epithelial cells. Acta Virol. (2015) 59:265–75. doi: 10.4149/av_2015_03_265
15. Xu X, Zhang H, Zhang Q, Huang Y, Dong J, Liang Y, et al. Porcine epidemic diarrhea virus N protein prolongs S-phase cell cycle, induces endoplasmic reticulum stress, and up-regulates interleukin-8 expression. Vet Microbiol. (2013) 164:212–21. doi: 10.1016/j.vetmic.2013.01.034
16. Li FQ, Tam JP, Liu DX. Cell cycle arrest and apoptosis induced by the coronavirus infectious bronchitis virus in the absence of p53. Virology. (2007) 365:435–45. doi: 10.1016/j.virol.2007.04.015
17. Harrison SM, Dove BK, Rothwell L, Kaiser P, Tarpey I, Brooks G, et al. Characterisation of cyclin D1 down-regulation in coronavirus infected cells. FEBS Lett. (2007) 581:1275–86. doi: 10.1016/j.febslet.2007.02.039
18. Ding L, Huang Y, Du Q, Dong F, Zhao X, Zhang W, et al. TGEV nucleocapsid protein induces cell cycle arrest and apoptosis through activation of p53 signaling. Biochem Biophys Res Commun. (2014) 445:497–503. doi: 10.1016/j.bbrc.2014.02.039
19. Ding L, Huang Y, Dai M, Zhao X, Du Q, Dong F, et al. Transmissible gastroenteritis virus infection induces cell cycle arrest at S and G2/M phases via p53-dependent pathway. Virus Res. (2013) 178:241–51. doi: 10.1016/j.virusres.2013.09.036
20. Vermeulen K, van Bockstaele DR, Berneman ZN. The cell cycle: a review of regulation, deregulation and therapeutic targets in cancer. Cell Prolif. (2003) 36:131–49. doi: 10.1046/j.1365-2184.2003.00266.x
21. Harper JV, Brooks G. The mammalian cell cycle: an overview. Methods Mol Biol. (2005) 296:113–53. doi: 10.1385/1-59259-857-9:113
23. Nurse P. Ordering S phase and M phase in the cell cycle. Cell. (1994) 79:547–50. doi: 10.1016/0092-8674(94)90539-8
24. Ohtsubo M, Theodoras AM, Schumacher J, Roberts JM, Pagano M. Human cyclin E, a nuclear protein essential for the G1-to-S phase transition. Mol Cell Biol. (1995) 15:2612–24. doi: 10.1128/MCB.15.5.2612
25. Girard F, Strausfeld U, Fernandez A, Lamb NJ. Cyclin A is required for the onset of DNA replication in mammalian fibroblasts. Cell. (1991) 67:1169–79. doi: 10.1016/0092-8674(91)90293-8
26. Walker DH, Maller JL. Role for cyclin A in the dependence of mitosis on completion of DNA replication. Nature. (1991) 354:314–7. doi: 10.1038/354314a0
27. Vogelstein B, Lane D, Levine AJ. Surfing the p53 network. Nature. (2000) 408:307–10. doi: 10.1038/35042675
28. Engeland K. Cell cycle arrest through indirect transcriptional repression by p53: I have a DREAM. Cell Death Differ. (2018) 25:114–32. doi: 10.1038/cdd.2017.172
29. Yuan L, Chen Z, Song S, Wang S, Tian C, Xing G, et al. p53 degradation by a coronavirus papain-like protease suppresses type I interferon signaling. J Biol Chem. (2015) 290:3172–82. doi: 10.1074/jbc.M114.619890
30. Ma-Lauer Y, Carbajo-Lozoya J, Hein MY, Muller MA, Deng W, Lei J, et al. p53 down-regulates SARS coronavirus replication and is targeted by the SARS-unique domain and PLpro via E3 ubiquitin ligase RCHY1. Proc Natl Acad Sci USA. (2016) 113:E5192–201. doi: 10.1073/pnas.1603435113
31. Dove B, Brooks G, Bicknell K, Wurm T, Hiscox JA. Cell cycle perturbations induced by infection with the coronavirus infectious bronchitis virus and their effect on virus replication. J Virol. (2006) 80:4147–56. doi: 10.1128/JVI.80.8.4147-4156.2006
32. McBride R, van Zyl M, Fielding BC. The coronavirus nucleocapsid is a multifunctional protein. Viruses. (2014) 6:2991–3018. doi: 10.3390/v6082991
33. Kong N, Shan T, Wang H, Jiao Y, Zuo Y, Li L, et al. BST2 suppresses porcine epidemic diarrhea virus replication by targeting and degrading virus nucleocapsid protein with selective autophagy. Autophagy. (2019) 16:1737–52. doi: 10.1080/15548627.2019.1707487
34. Shi D, Shi H, Sun D, Chen J, Zhang X, Wang X, et al. nucleocapsid interacts with npm1 and protects it from proteolytic cleavage, enhancing cell survival, and is involved in PEDV growth. Sci Rep. (2017) 7:39700. doi: 10.1038/srep39700
35. Wurm T, Chen H, Hodgson T, Britton P, Brooks G, Hiscox JA. Localization to the nucleolus is a common feature of coronavirus nucleoproteins, and the protein may disrupt host cell division. J Virol. (2001) 75:9345–56. doi: 10.1128/JVI.75.19.9345-9356.2001
36. Carmo-Fonseca M, Mendes-Soares L, Campos I. To be or not to be in the nucleolus. Nat Cell Biol. (2000) 2:E107–12. doi: 10.1038/35014078
37. Chen H, Wurm T, Britton P, Brooks G, Hiscox JA. Interaction of the coronavirus nucleoprotein with nucleolar antigens and the host cell. J Virol. (2002) 76:5233–50. doi: 10.1128/JVI.76.10.5233-5250.2002
38. Cawood R, Harrison SM, Dove BK, Reed ML, Hiscox JA. Cell cycle dependent nucleolar localization of the coronavirus nucleocapsid protein. Cell Cycle. (2007) 6:863–7. doi: 10.4161/cc.6.7.4032
39. Liu SX, Shi D, Chen JF, Zhang X, Shi HY, Liu XZ, et al. Effects on host cell cycle by nucleoli localization signal peptides of porcine epidemic diarrhea virus nucleocapsid protein. Zhong Guo Yu Fang Shou Yi Xue Bao. (2013) 35:173–6. doi: 10.3969/j.issn.1008-0589.2013.03.01
40. Li S, Lu LF, Liu SB, Zhang C, Li ZC, Zhou XY, et al. Spring viraemia of carp virus modulates p53 expression using two distinct mechanisms. PLoS Pathog. (2019) 15:e1007695. doi: 10.1371/journal.ppat.1007695
41. Liu S, Liu H, Kang J, Xu L, Zhang K, Li X, et al. The severe fever with thrombocytopenia syndrome virus nss protein interacts with CDK1 To Induce G2 cell cycle arrest and positively regulate viral replication. J Virol. (2020) 94:e01575–19. doi: 10.1128/JVI.01575-19
42. Xu LH, Huang M, Fang SG, Liu DX. Coronavirus infection induces DNA replication stress partly through interaction of its nonstructural protein 13 with the p125 subunit of DNA polymerase δ. J Biol Chem. (2011) 286:39546–559. doi: 10.1074/jbc.M111.242206
43. Bhardwaj K, Liu P, Leibowitz JL, Kao CC. The coronavirus endoribonuclease Nsp15 interacts with retinoblastoma tumor suppressor protein. J Virol. (2012) 86:4294–304. doi: 10.1128/JVI.07012-11
Keywords: coronavirus, cell cycle arrest, cyclin-CDK complex, p53, nucleocapsid protein
Citation: Su M, Chen Y, Qi S, Shi D, Feng L and Sun D (2020) A Mini-Review on Cell Cycle Regulation of Coronavirus Infection. Front. Vet. Sci. 7:586826. doi: 10.3389/fvets.2020.586826
Received: 24 July 2020; Accepted: 19 October 2020;
Published: 05 November 2020.
Edited by:
Maureen T. Long, University of Florida, United StatesReviewed by:
Rahul K. Nelli, Iowa State University, United StatesCopyright © 2020 Su, Chen, Qi, Shi, Feng and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Dongbo Sun, ZG9uZ2Jvc3VuQDEyNi5jb20=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.