- 1Transplant Research Program, Boston Children’s Hospital, Boston, MA, United States
- 2Division of Nephrology, Boston Children’s Hospital, Boston, MA, United States
- 3The Department of Pediatrics, Boston Children’s Hospital, Boston, MA, United States
- 4The Department of Pediatrics, Harvard Medical School, Boston, MA, United States
- 5Computational Health Informatics Program, Boston Children’s Hospital, Boston, MA, United States
It is increasingly appreciated that the expression of immunoregulatory molecules within tumors have potential to shape a microenvironment that promotes local immunoevasion and immunoregulation. However, little is known about tissue-intrinsic immunomodulatory mechanisms following transplantation. We propose that differences in the phenotype of microvascular endothelial cells impact the alloantigenicity of the graft and its potential to promote immunoregulation following transplantation. We focus this review on the concept that graft-dependent immunoregulation may evolve post-transplantation, and that it is dependent on the phenotype of select subsets of intragraft endothelial cells. We also discuss evidence that long-term graft survival is critically dependent on adaptive interactions among immune cells and endothelial cells within the transplanted tissue microenvironment.
Introduction
The development of rejection involves a marked inflammatory reaction characterized by effector T cell and B cell activation, an intense cellular and humoral response and an associated trafficking of alloreactive leukocytes into the allograft (1–3). Rejection is initiated by the recipient's immunological response to donor antigen, which is coordinated by CD4+ T cells that actively undergo expansion and differentiation into effectors and/or memory T cells (1, 4, 5). However, multiple pathways operate concurrently in order to control and regulate effector alloimmunity, and it is proposed that this process of immunoregulation can be a more potent component of the overall response (4–9). Indeed, the regulation of effector alloimmunity is complex, and classically involves several immune cell types (5, 9–15) including the expansion and function of CD4+Foxp3+ Tregulatory (Treg) cells (15–17). But importantly, it is also dependent on critical adaptive responses that occur within the graft itself (18–26). In this review, we focus our discussion on how obligate interactions among alloresponsive immune cells and multiple subsets of intragraft microvascular endothelial cells (EC) dictate the outcome of the rejection response. We postulate that the process of rejection is shaped by the immunogenic phenotype of select subsets of EC within a graft. Also, we suggest that it is possible to regulate the initiation of rejection through a process we have called graft-dependent immunoregulation (Figure 1).
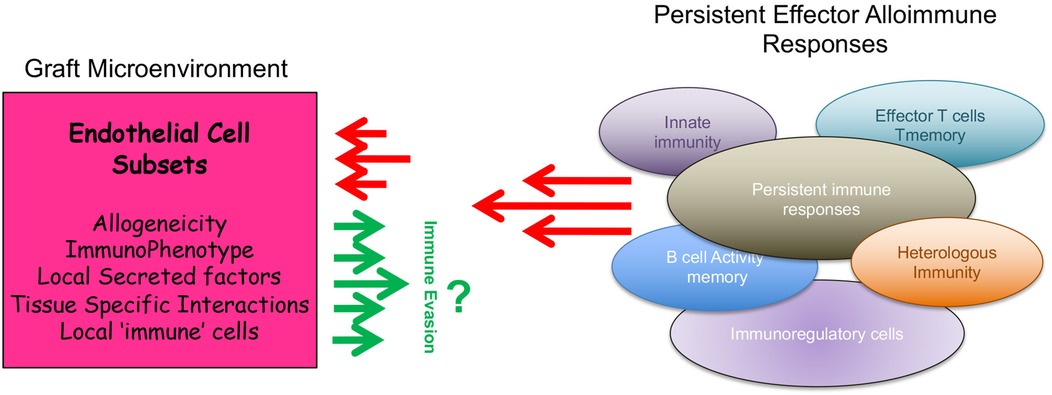
Figure 1. Cartoon illustration of how the phenotype of intragraft endothelial cell subsets regulate local alloimmunity to promote graft-dependent immunoregulation.
The phenotype of intragraft endothelial cells (EC) and graft-dependent immunoregulation
Multiple observations have established a functional role for graft vascular EC in the development of acute and chronic rejection (19, 22–24, 27–31). The expression of adhesion molecules and chemokines promote the recruitment of leukocytes into the graft, and the expression of MHC class I and II molecules, costimulatory molecules and cytokines promotes allogeneicity (25–27, 29, 32). These features enable EC to serve as semi-professional antigen presenting cells (APCs) and promote the activation of subsets of T cells (23, 27). Furthermore, the unique location of EC ensures obligate interactions with recipient immune cells. Since there are greater numbers of microvascular EC within a graft vs. professional APCs, there is a high likelihood that their interactions with infiltrating effector T cells dictate the nature of the reactivation response (23, 25, 33–36). Although controversial (37), direct interactions between CD4+ or CD8+ T cells and intragraft EC is sufficient to initiate and sustain allogeneic T cell activation as well as the rejection response (20, 25, 26, 38). Thus, the immunogenic phenotype of the EC has great relevance for the outcome of the local intragraft T cell immune response. Consistent with this interpretation, molecular profiling data and computational analysis in humans has confirmed that the state of activation, immunophenotype and allogeneicity of the graft microvasculature is associated with a microenvironment that is predictive of sustained rejection (39, 40). To date however bioinformatic approaches have not yet addressed the process of graft-dependent immunoregulation (24, 36).
Nevertheless, recent studies indicate that EC may express immunoregulatory molecules, including PD-L1, PD-L2, Tim-3, B7-H3, IDO and others, that are well established to modulate cell-mediated and alloimmune inflammatory reactions (24, 41–45). While little is known about EC-dependent immunoregulation following transplantation, several studies support a working model whereby the expression of select coinhibitory molecules on intragraft EC is both necessary and sufficient to support graft-dependent immunoregulation (18, 46–48).
Chalasani et al. (18) used a model in which fully MHC mismatched cardiac allografts were transplanted into splenectomized alymphoplastic (aly/aly) mice which lack secondary lymphoid tissues and accept allografts indefinitely. They found that the adoptive transfer of alloactivated T cells on day 2 post-transplantation resulted in graft failure, whereas transfer of the identical T cells on day 70 post-transplantation failed to precipitate rejection, all grafts surviving for >100 days. They also transferred alloactivated T cells into fully MHC mismatched C57BL/6 recipients of Balb/c cardiac allografts at similar time points following multidose treatment of recipients with CTLA4Ig and anti-CD154. Again, after a period of conditioning with costimulatory blockade (day 50 post-transplantation), adoptive transfer of alloreactive T cells failed to initiate acute rejection and all grafts survived long-term. Although these findings allowed for the interpretation that the graft itself has potential to determine the outcome of rejection, they did not identify the mechanism of graft-dependent immunoregulation in these studies. It is however most intriguing to consider that immunosuppressive agents and/or conditioning may induce select intragraft immunoevasive and/or immunomodulatory factors that shape these outcomes.
Riella et al (46). used a similar fully MHC mismatched C57BL/6 into Balb/c cardiac transplantation model and multi-dose CTLA4Ig treatment to prolong graft survival. These authors found that PD-L1 knockout grafts are rejected at an accelerated pace suggesting that local tissue expression is both necessary and sufficient to elicit graft-dependent immunoregulation. Consistent with this interpretation and the possibility that PD-L1 is functional on intragraft EC subsets, they also found accelerated rejection of allografts from bone-marrow chimeric mice in which PD-L1 is deficient in non-hematopoietic cells.
In another study, Koga et al. (48) used the minor MHC mismatched C57BL6 into B6.C-H2bm12 cardiac transplant model, which is known to result in a chronic insidious rejection process for >45 days (49, 50). They found that treatment with anti-PD-L1 resulted in accelerated rejection, characterized by marked inflammatory infiltrates, intragraft cytokine production and accelerated graft vascular arteriosclerosis vs. controls. In the same minor MHC mismatched B6.C-H2bm12 transplant model, Yang et al. used PD-L1 and PD-L2 knockout mice as donors and found that intragraft PD-L1, but not PD-L2, was functional to prolong graft survival (47). Furthermore, they also demonstrated that neither PD-L1 nor PD-L2 is functional in the regulation of the peripheral alloimmune response, conclusively discovering that intragraft PD-L1 is sufficient to elicit graft-dependent immunoregulation. Interestingly, it was also found that intragraft PD-L1 is functional to support early graft survival in a murine model of kidney transplantation, but these authors did not evaluate its expression on EC following immunosuppressive conditioning at later times post transplantation (51).
In vitro studies have demonstrated that select EC phenotypes suppress local alloimmune Teffector responses and/or augment the local activity of Tregs (24, 41, 42, 52). Thus, it is possible that select immunosuppressive therapeutics have potential to alter the phenotype of distinct subsets of intragraft EC to promote graft-dependent immunoregulation. Indeed, consistent with this hypothesis, pilot studies in our laboratory using a model of graft-dependent immunoregulation have revealed that EC within these grafts have a unique phenotype that includes regulation of the mTOR intracellular signaling pathways and the expression of multiple costimulatory, coinhibitory and immunoevasive molecules (53). Although mTOR inhibition can regulate coinhibitory gene expression by EC in vitro (42), understanding the mechanisms underlying immunoregulatory and immunoevasive gene expression in vivo will likely have significant implications for long-term transplant outcomes.
Overall, while little is known about EC- and graft-dependent immunoregulation following transplantation, the mechanism of tissue-dependent immunoevasion is an area of intense research in the tumor immunology field (24, 54–56). This process is functionally associated with coinhibitory molecule expression on EC. But it is not yet known if intrinsic heterogeneity within EC subsets, or differences in organ-specific production of immunoevasive and immunomodulatory genes impact the potential for graft-dependent immunoregulation.
Heterogeneity in microvascular endothelial cell (EC) phenotypes both within and across different organs
Over the past 5–10 years, high-throughput single-cell RNA sequencing (scRNAseq) and spatial imaging technologies have brought new insights into the diversity and broad functions of microvascular EC subsets within tissues (36, 57). Several studies have determined that there is significant heterogeneity within microvascular EC subpopulations, and there are notable differences in EC phenotypes within different organs (36, 58–65). Although the significance of these differences has not yet been explored following transplantation, it appears that specialized EC subsets within different organs (notably, heart, lung and kidney) express unique gene signatures (36, 57, 60, 61, 65). Also, transcriptomic and epigenomic studies have demonstrated EC subset-specific differences in activation responses to pro-inflammatory stimuli (36, 59, 61, 63, 64, 66–71). This insight has brought new concepts to the transplantation field, for example that EC subsets within different organs respond with unique intracellular signals and gene expression signatures in the course of rejection and/or that EC subsets from different microvascular beds have potential to express immunoregulatory/immunoevasive gene signatures and thus resist Teffector mediated injury (36, 59, 63, 64, 66–74). Although previous studies indicated some heterogeneity in activation responses in human transplant biopsies by immunohistochemistry [for example (29, 75–77)], these new findings are suggestive of a paradigm whereby EC activation responses are not uniformal across subsets, but are rather unique to distinct subsets within each microvascular bed and/or across organs. The Valenzuela group reported that cultured human EC from heart, lung, liver, kidney and skin exhibited distinct inflammatory phenotypes at the mRNA level as well as in response to the pro-inflammatory cytokines TNFα and IL-1β (59). They speculated that diversity in activation phenotypes may contribute to differences in the injury response following transplantation. Moreover, tissue staining, microarray analysis and several other published scRNAseq studies of murine tissues indicate that there are at least 7 major EC subtypes within each organ microenvironment and that capillary EC are a most heterogeneous cell type with phenotypic differences both within and across different tissues (57, 61, 64, 65, 78).
To highlight microvascular heterogeneity that evolves in established in vivo models of transplantation, we evaluated EC subset phenotypes and gene expression patterns in the initial post transplantation period by evaluating a recently published murine heart transplant scRNAseq dataset (79). As illustrated in Figure 2, we identify 12 distinct capillary EC subsets with marked phenotypic heterogeneity including distinct patterns of expression of pro-inflammatory [ICAM-1, VCAM-1 (29)], immunoregulatory [PD-L1 (46, 47)] and immunoevasive [Sema3F (24)] genes. We also compared gene expression patterns in C57BL/6 heart isografts and allografts harvested on day 5 post-transplantation (from C57BL/6 and Balb/c recipients respectively). As expected, the phenotype of EC subsets, expression levels and the distribution of well-established activation and immunoregulatory molecules differ within isografts and allografts (Figure 2). Furthermore, pseudotime trajectory analysis of pro-inflammatory chemokines (CXCL9, CXCL10), activation (VCAM-1), immunoregulatory (PD-L1) and immunoevasive (Sema3F) genes within this dataset identified distinct EC subset-specific patterns of expression of each gene within the microvasculature (Figure 3). Expression levels of each selected gene varied across the microvascular bed, with notable increases and decreases in expression in selected EC subsets (Figures 2, 3). These collective findings are consistent with the concept that distinct EC subsets within the transplanted tissue have potential to contribute to either pro-inflammation, immunoregulation and/or immunoevasion. Indeed, immunomodulation by select EC subsets can be influenced by the expression of coinhibitory molecules (23, 24, 36, 47, 55), cytokine-induced responses (24, 72, 73, 85), intracellular signaling responsiveness (24, 62, 77, 86) and the capacity to induce apoptosis in immune cells [for example, via FAS ligand (68)].
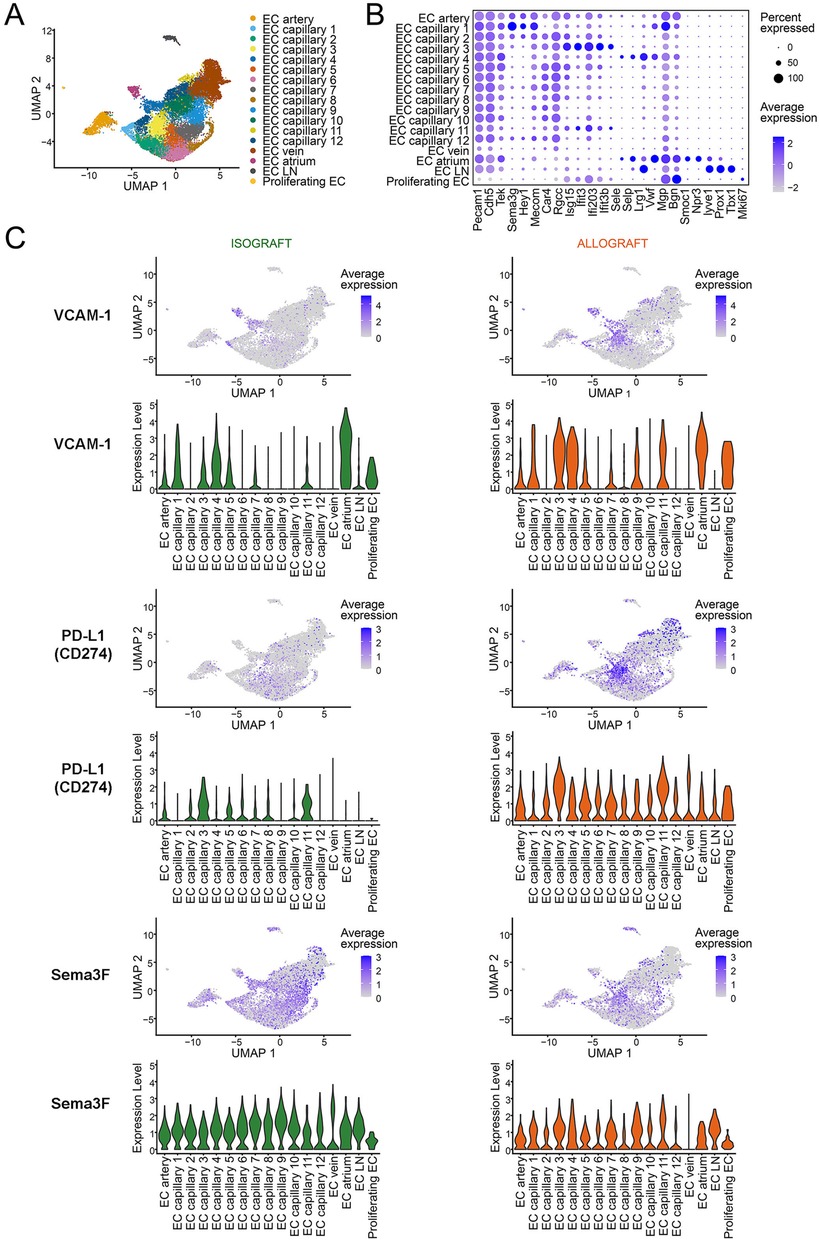
Figure 2. Intragraft microvascular endothelial cell heterogeneity from murine cardiac allo- and iso grafts. Single cell RNA-sequencing data from post-transplant day 5 murine cardiac isografts (Balb/c → Balb/c, n = 2) and allografts (Balb/c → C57BL6, n = 2) were downloaded from the NCBI GEO (accession number: GSE151048) (79). Seurat objects were generated from each sample, integrated using Harmony, and cluster resolution determined using the Clustree method (80–83). EC were identified using Pecam1 and Cdh5 expression and were clustered to identify subsets using selected EC annotation transcripts (57, 62, 80, 81). (A) UMAP scatter plot, color-coded for EC subclusters. EC were clustered based on established arterial, venous and capillary gene expression; a total of 12 capillary subsets are color coded. (B) Dot plot illustrating the transcripts used for EC subset annotation. The percent and level of expression of each transcript is illustrated by the size and color (blue) of each dot. (C) Feature and Violin plots of intragraft EC subsets isolated from isografts (left panels) or allografts (right panels) depicting select transcript expression of pro-inflammatory (VCAM-1), immunoregulatory (PD-L1) and immunoevasive (Sema3F) molecules. The color (blue dot) illustrates the level of expression of each gene in each Feature plot. Violin plots illustrate the relative level of expression of each gene in each EC subset.
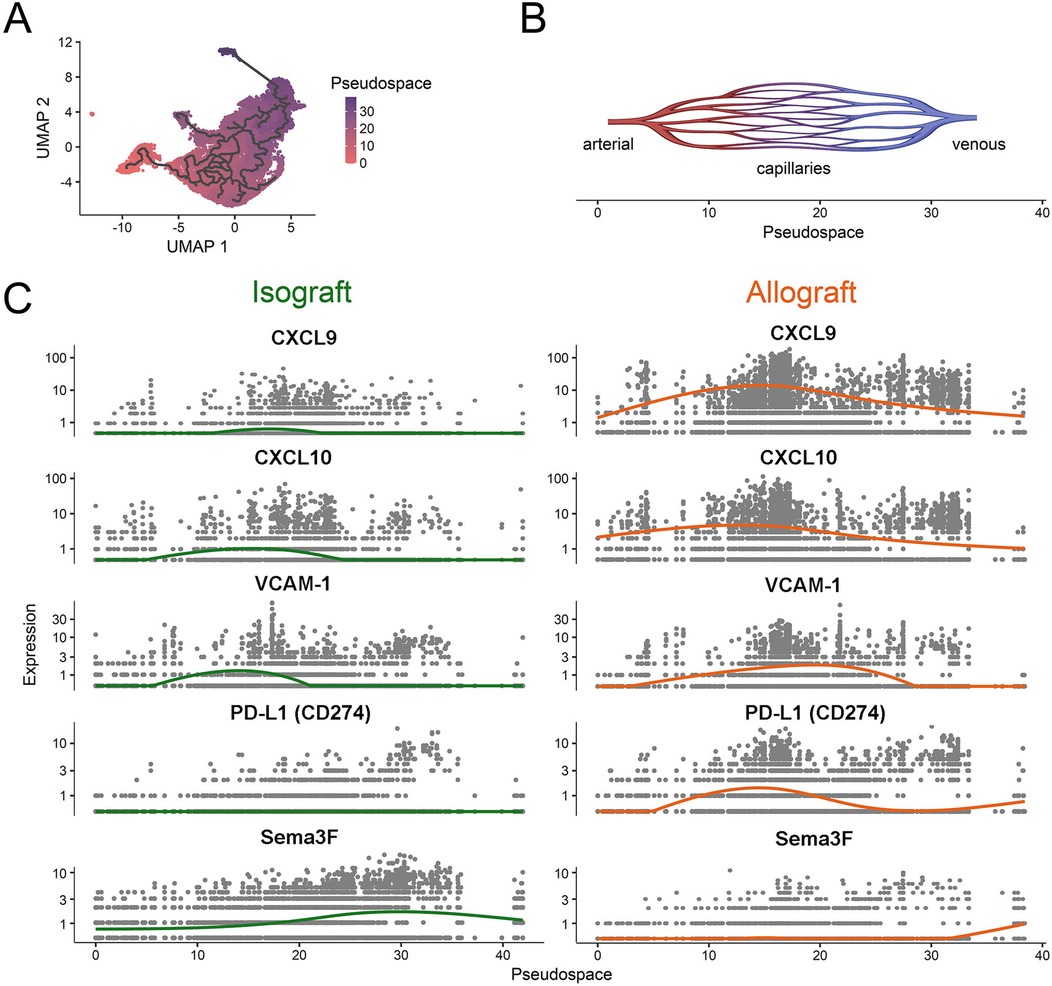
Figure 3. Pseudospatial expression of proinflammatory, immunoregulatory and immunoevasive transcripts in intragraft endothelial cells. A pseudotime estimation method was used to generate a pseudospatial resolution of single cell RNA-sequencing data from Figure 2 within the microvascular bed (84). The spatial trajectory starts in arterial microvasculature (as shown in Figure 2A), passes through capillaries and ends in venous EC subsets. (A) UMAP scatter plot color coded for the pseudospace. The trajectory is highlighted. (B) Schematic representation of the pseudospace within the microvasculature. (C) Scatter plots of isografts (left; green) and allografts (right; orange) depicting patterns of select proinflammatory (CXCL9, CXCL-10 and VCAM-1) or immunoregulatory/immunoevasive (PD-L1, Semaphorin3F) transcript expression over the pseudospace within each EC subset. Each line (green, isograft vs. orange, allograft) represents the average transcript expression within EC subsets along the pseudospace.
To add to the complexity of this biology, it has also been reported that EC within different organs express unique profiles of inflammatory or regulatory genes. For example, EC within the lung express gene signatures associated with immune activation, consistent with barrier organ biology, whereas EC derived from non-barrier organs such as the kidney and liver express genes associated with tissue-specific immune regulation (36). Jambusaria et al. (61) also found that EC within the brain, lung, and heart adapt to signatures from the surrounding tissue parenchyma; EC from the brain express genes associated with neuronal function and EC from the heart express genes associated with cardiac muscle development. Dumas et al. (60) identified 24 distinct EC subsets within the kidney, each with a unique transcriptional profile, for example with selective metabolic, IFN-responsive or antigen presentation phenotypes, and heterogeneity in EC phenotypes has been confirmed within transplanted kidneys (87). Thus, EC subsets within allografts may adopt profiles/phenotypes based on the cellular composition and/or immune events within the local intragraft microenvironment to support either chronic inflammation or immune regulation (88, 89).
Mechanisms of graft-dependent immunoregulation
These new insights into the heterogeneity of EC within organs and tissues suggest that existing pro-inflammatory paradigms based on studies of single populations of EC are incomplete. For example, proinflammatory responses are not uniform across different EC subsets [artery, vein, capillary, see Figure 2 and (36)] or between EC from different organs (59). Furthermore, current paradigms suggest that pathological immune events within the intragraft microenvironment are shaped by local EC responsiveness to inflammation including local tissue hypoxia (19, 22, 90), cytokine production by infiltrating effector T cells (91) or resident immune cells (92), and tissue expression of local growth factors including VEGF-A (19, 31). Effector cytokines produced locally are well established to promote the activation of EC and support ongoing immune cell infiltration (19, 23, 24, 27). In contrast, during inflammation resolution multiple mediators produced by EC including pro-resolution lipids (93–95), anti-inflammatory cytokines (96) and/or neuronal guidance molecules (24, 97) promote immunoevasion to regulate leukocyte subset trafficking into tissues.
Physiological post-inflammatory mechanisms that resolve cell-mediated immune inflammation and sustain immune homeostasis are associated with the production and secretion of multiple families of molecules by EC, including neuronal guidance Netrins, Semaphorins and Slit family molecules that inhibit leukocyte trafficking into the tissue (24, 73, 93, 98–104). These immunoevasive proteins were originally described in the formation of the nervous system (105, 106), but they are expressed by multiple non-neuronal cell types, including EC, and their receptors are expressed on leukocyte subsets (97, 101, 103, 105). In this manner, neuronal guidance cues interact with immune cells and the response(s) elicit either chemoattractive or chemorepulsive signals (96). Thus, in the context of transplantation, intragraft expression of Netrins, Semaphorins and/or Slits have potential to influence the local phenotype of rejection response.
The Netrins are a family of secreted molecules that are structurally related to laminins and bind to uncoordinated 5 (UNC5) A-D, deleted in colorectal cancer (DCC), Neogenin, and the Down Syndrome cell adhesion molecule (DSCAM) receptors (98, 107–110). Immunoevasion elicited by Netrin-1 has been studied in immunity, and is dominantly attributed to interactions with the UNC5 family of receptors (98, 99, 111). The chemorepellent receptor UNC5B is expressed by peripheral blood mononuclear cells including neutrophils, T cells and monocytes, where it acts as an inhibitor of migration towards chemotactic stimuli (98, 109, 111) including inflammation that is associated with ischemia-reperfusion injury (99). In contrast, interactions between Netrin-1 and its neogenin receptor that is induced on activated CD4+ T cell subsets result in chemoattraction (98, 110). Thus, chemorepulsive UNC5 receptors or promigratory neogenin that are differentially expressed on CD4+ T cells may determine the immune response to local Netrin-1 expression within a tissue. Of note, neogenin can also bind to additional ligands involved in the regulation of T cell activation (112), indicating that its expression on leukocytes may dictate the relative immunomodulatory function of local EC-derived Netrin-1 in the course of cell-mediated immune inflammation and/or rejection.
The semaphorins (Semas) are immunomodulatory proteins that were also discovered as neuronal guidance cues (97, 113–117). Semas consist of eight families, most of which are membrane bound, and vertebrate members (Sema families 3–7) are reported to function in the immune response (114–116, 118). The Sema3 family members (Sema3A-G) are soluble secreted proteins, and some (for example Sema3F and 3G) are expressed at high levels by EC (65). These proteins bind to neuropilin (NRP) -1 and NRP-2 that are expressed by T cell subsets (73, 118–121). NRP-1 is a marker of activated CD4+ Foxp3+ Treg cells (118, 122, 123), but recent studies have also identified its expression on antigen-activated and exhausted CD8+ T cells (119, 120). We (73) and others (118, 119) have observed that the interaction between Sema3 and Sema4 proteins with NRP1/2 results in an inhibition of PI-3K/Akt/mTOR signalling as well as cytoskeletal collapse and reduced migration in multiple cells types including lymphocytes. Knockout of NRP-1 on lymphocytes is associated with enhanced migration and effector function of CD4+ and CD8+ T cells (119). In contrast, the stimulation of NRP-1 on Tregs enhances stability and function (118). Thus, EC expression of Sema3 family proteins (see Figures 2, 3) is likely to have potent implications for both the migration and activation of NRP-expressing effector and regulatory cells within allografts.
The Slit family of proteins are also immunoevasive neuronal guidance cues that are expressed by EC at lower levels (103, 104), but little is known about their tissue expression or biology in the resolution of cell-mediated immunity. Nevertheless, Slit-2 has been shown to inhibit the migration of leukocytes in response to chemokines via interactions with the Roundabout (Robo) family of receptors that are expressed on leukocytes (104, 124). Expression is also reported to protect and inhibit neutrophil-induced chemotaxis (96, 104) as well as ischemia-reperfusion injury (102), but to our knowledge their biology has not yet been explored following transplantation.
Since little is known about the biology of immunoevasion, in previous studies, we developed an in vitro platform to evaluate attraction and inhibition of leukocyte migration simultaneously. In this manner, it was possible to evaluate the effects of migratory guidance cues on bidirectional leukocyte trafficking patterns (96). We discovered that migration and leukocyte trafficking is more complex than previously described (125–129), as the migratory response (or lack of) does not simply relate to chemoattraction. Rather, migratory responses occur in at least four distinct patterns, called chemoattraction, chemorepulsion, chemoinhibition, and chemokinesis (96), as illustrated in Figure 4.
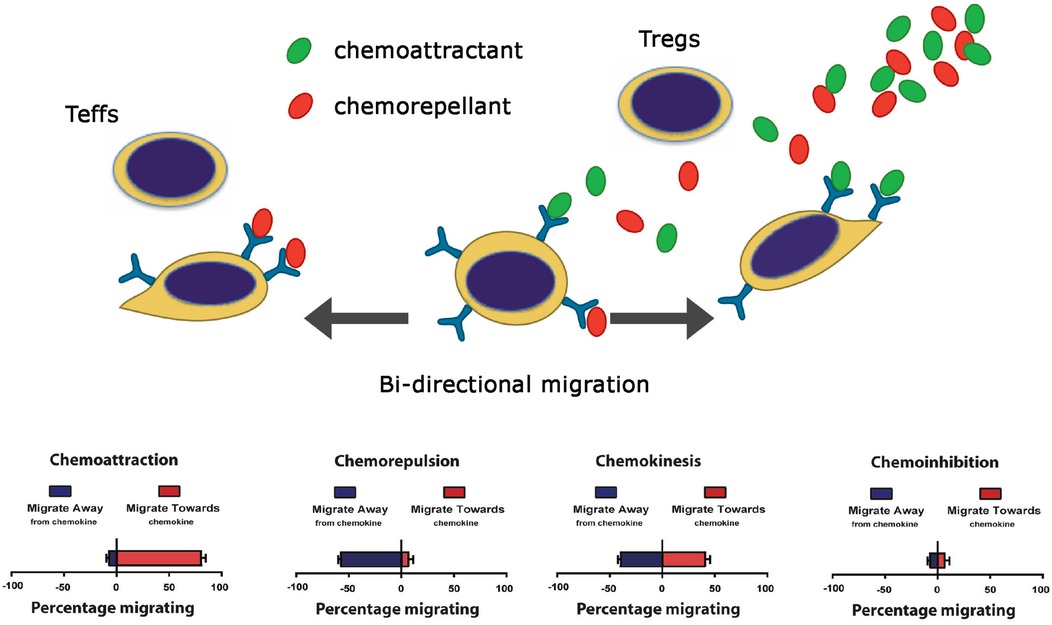
Figure 4. Cartoon illustration of the four patterns of leukocyte migration (96).
Tissue-dependent immunoevasion may be elicited in part through the process of chemorepulsion that results in migration in the opposite direction to chemoattraction (96). Furthermore, a chemoinhibitory stimulus [such as a response to Slit family molecules (96, 102, 103)] reduces migration in random directions to a guidance cue. Importantly, some molecules [for example Netrin-1 (98, 100, 111)] promotes a bidirectional migratory response with potential to elicit both chemorepulsion and chemoattraction depending upon the relative expression of its receptor(s) on each leukocyte subset(s). Also, members of the semaphorin family, including endothelial Sema3F [(73) and Figures 2, 3] or SDF-1/CXCL12 (96) elicits chemorepulsive and/or dispersive signals via receptors expressed on distinct subsets of immune cells, including CD4+ T cells (122, 130). Collectively, these studies indicate that combinations of guidance molecules expressed by EC subsets may serve to inhibit leukocyte migration and extravasation into allografts but the process of chemoinhibition and/or chemorepulsion is also dependent on the relative levels of chemotactic receptors expressed on individual infiltrating immune cells.
Another important consideration is whether the characteristics of resident immune cells within the graft alter the EC subset phenotype, or whether the EC subset phenotype regulates the characteristics of the local intragraft immune infiltrate. The Lakkis group demonstrated that initial effector cell infiltration into an allograft requires recognition of alloantigen, likely expressed on locally activated and MHC-expressing EC subsets (26). Furthermore, they found that the differentiation of effectors into pathological T resident memory cells (TRM) requires antigen presentation as well as cytokine-induced activation (88, 92). Abou-Daya et al. (88) found that recipient graft infiltrating effector T cells acquire a TRM phenotype and that these cells sustain their localization within an allograft where they produce effector proinflammatory cytokines (88). Although Tieu et al. later demonstrated that the maintenance of TRM within the graft was dependent on antigen presentation by intragraft dendritic cells (92), the role of interacting EC subsets in the persistence of TRM localization was not evaluated.
In addition, the function of EC subsets in the recruitment of Tregs into an allograft is poorly understood, but it is also likely to involve the recognition of antigen as well as local activation responses (131, 132). In select transplant models and/or following immunosuppressive conditioning, perivascular aggregates of Tregs are recruited into the graft where they localize into Treg-rich Organized Lymphoid Structures (TOLS) that are reported to promote immunoregulation and support long-term graft survival (89, 133–135). TOLS-containing allografts elicit an immunoregulatory response following retransplantation into fully MHC mismatched recipients (133) and early depletion of Foxp3+ Tregs within TOLS results in allograft rejection (134, 136). These findings confirm a role for TOLS in graft-dependent immunoregulation. Nevertheless, it is not known if the presence of intragraft TOLS is associated with changes in the phenotype of local EC within a graft, or whether local EC subsets adapt and express immunomodulatory genes (e.g., Sema proteins) that support Treg localization and thus graft-dependent immunoregulation. Furthermore, as discussed above, EC- and graft-dependent mechanisms of immunoregulation may occur in the absence of CD4+ T regulatory cell recruitment or TOLS development, for example following a period of immunosuppression after transplantation in recipients treated with costimulatory blockade (18, 46). Thus, the development of EC phenotypes that support graft-dependent immunoregulation likely involves an independent cell-intrinsic mechanism and/or a modulatory signaling response(s) within the local tissue microenvironment (24, 36, 42).
Therapeutic implications
Cell-intrinsic mTOR signaling in EC is well established to play a central role in EC activation responses (86, 137–141), and its biology in EC is implicated in a large number of human inflammatory diseases (86, 142, 143). Targeting mTOR in EC with pharmacological mTOR inhibitors, even at low concentrations (144), inhibits EC activation (30, 86, 138) and has marked effects on the augmentation of coinhibitory PD-L1 and PD-L2 expression (42). This response has been reported to be associated with graft-dependent immunoregulation (as discussed above) and to enhance local immunoregulation in part via the augmentation of Treg function (42, 52, 145). Thus, treatment with mTOR inhibitors [event at low doses (144)] may target the graft EC to promote immunomodulation independent of its effects on the peripheral alloimmune response (86).
DEPTOR is a potent cell-intrinsic regulator of mTOR (146) that is expressed at variable levels within EC subsets in vitro and in vivo (77). It was originally identified to modulate mTOR signaling activity via its dominant ability to bind and inhibit mTORC1 complex assembly (146–148), but it also regulates the MAPK and STAT signaling pathways in EC (77, 146). Interestingly, rapamycin augments DEPTOR expression (147), suggesting another mechanism whereby it may be therapeutic to target EC activation. siRNA knockdown of DEPTOR in EC has a striking effect on the induction of activation gene expression signatures with up to a 1,000-fold increase in the expression of select chemokines (77). In addition, a recently published study indicated that knockout of EC DEPTOR has similar biological consequences in vivo (149). Since DEPTOR is a potent regulator of mTOR, its biology in EC is thus directly linked to intragraft inflammation and immunoregulation. Overall, these findings suggest that cell intrinsic modulation of mTOR signaling is both necessary and sufficient for EC-dependent immunoregulation. They also suggest that therapeutics that inhibit mTOR activity and/or sustain the expression of cell-intrinsic DEPTOR will be of great significance to support the development of graft-dependent immunoregulation.
Summary and future outlook
The understanding of tissue-dependent immunoregulation is driven by studies in the tumor literature, and little is known about underlying mechanisms within allografts. In this review, we discuss the literature demonstrating that it is possible to augment graft-dependent immunoregulation following a period of immune conditioning. We also review literature showing that the inhibition of mTOR signaling and/or cell intrinsic modulators of proinflammatory signals in EC have potential to induce an immunomodulatory phenotype. However, there is a need to evaluate and study the heterogeneity in phenotypes within the allograft microvasculature, differences in EC phenotypes and responses across different organs and changes that occur following transplantation. In this manner, it may be possible to uncover unique signals that drive EC phenotypes that are associated within immunomodulation and long-term graft survival. Future research studies may also identify mechanisms whereby EC adopt microenvironmental cues to promote either pro-inflammation or immunoregulation. Deciphering fundamental mechanisms underlying how different EC subsets within different organs adapt in order to regulate and modulate the local immune response will have significant clinical implications in the field. We predict a future whereby different graft-targeted therapeutics will be used following organ transplantation to sustain the induction of local genes that promote immunomodulation. Another potential future outlook relates to the monitoring of grafts for immunoregulatory gene expression as a determinant of long-term outcome.
Author contributions
SB: Data curation, Formal Analysis, Writing – review & editing. VD: Writing – review & editing, Data curation, Formal Analysis. CT: Writing – review & editing, Data curation, Formal Analysis. SJ: Writing – review & editing, Data curation. NS: Writing – review & editing, Data curation. AK: Writing – review & editing, Formal Analysis. MK: Data curation, Writing – review & editing. LB: Writing – review & editing, Data curation, Formal Analysis. SK: Formal Analysis, Writing – review & editing. JW: Data curation, Formal Analysis, Methodology, Supervision, Writing – original draft, Writing – review & editing. DB: Conceptualization, Data curation, Formal Analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The work cited in this review was supported by National Institutes of Health grants R01AI136503 and R01AI148539, the Isabella Julian Forrest Foundation and the Casey Lee Ball Foundation (to DMB). CT was supported by the Swedish Vetenskapsrådet and the Swedish Hjärt-Lungfonden. JW is supported by a Basic Science Career Transition Grant from the American Society of Transplantation.
Acknowledgments
The authors wish to thank past members of the laboratory (post docs and technicians) for their support of the studies referenced in this review. Some illustrations were created with BioRender.com.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Nankivell BJ, Alexander SI. Rejection of the kidney allograft. N Engl J Med. (2010) 363(15):1451–62. doi: 10.1056/NEJMra0902927
2. Ingulli E, Alexander SI, Briscoe DM. Immunology of pediatric renal transplantation. In: Avner ED, Harmon WE, Niaudet P, Yoshikawa N, Emma F, Goldstein SL, editors. Pediatric Nephrology. New York, NY: Springer (2016). p. 2457–500.
3. Zhang X, Reed EF. Effect of antibodies on endothelium. Am J Transplant. (2009) 9(11):2459–65. doi: 10.1111/j.1600-6143.2009.02819.x
4. Rothstein DM, Sayegh MH. T-cell costimulatory pathways in allograft rejection and tolerance. Immunol Rev. (2003) 196:85–108. doi: 10.1046/j.1600-065X.2003.00088.x
5. Li XC, Turka LA. An update on regulatory T cells in transplant tolerance and rejection. Nat Rev Nephrol. (2010) 6(10):577–83. doi: 10.1038/nrneph.2010.101
6. Brusko TM, Putnam AL, Bluestone JA. Human regulatory T cells: role in autoimmune disease and therapeutic opportunities. Immunol Rev. (2008) 223:371–90. doi: 10.1111/j.1600-065X.2008.00637.x
7. Wing K, Sakaguchi S. Regulatory T cells exert checks and balances on self tolerance and autoimmunity. Nat Immunol. (2010) 11(1):7–13. doi: 10.1038/ni.1818
8. Mueller DL. Mechanisms maintaining peripheral tolerance. Nat Immunol. (2010) 11(1):21–7. doi: 10.1038/ni.1817
9. Tang Q, Bluestone JA. The Foxp3+ regulatory T cell: a jack of all trades, master of regulation. Nat Immunol. (2008) 9(3):239–44. doi: 10.1038/ni1572
10. Mohib K, Cherukuri A, Rothstein DM. Regulatory B cells and transplantation: almost prime time? Curr Opin Organ Transplant. (2018) 23(5):524–32. doi: 10.1097/MOT.0000000000000559
11. Kim JI, Rothstein DM, Markmann JF. Role of B cells in tolerance induction. Curr Opin Organ Transplant. (2015) 20(4):369–75. doi: 10.1097/MOT.0000000000000204
12. Newell KA, Asare A, Kirk AD, Gisler TD, Bourcier K, Suthanthiran M, et al. Identification of a B cell signature associated with renal transplant tolerance in humans. J Clin Invest. (2010) 120(6):1836–47. doi: 10.1172/JCI39933
13. Newell KA, Turka LA. Tolerance signatures in transplant recipients. Curr Opin Organ Transplant. (2015) 20(4):400–5. doi: 10.1097/MOT.0000000000000207
14. Yeung MY, Ding Q, Brooks CR, Xiao S, Workman CJ, Vignali DA, et al. Tim-1 signaling is required for maintenance and induction of regulatory B cells. Am J Transplant. (2015) 15(4):942–53. doi: 10.1111/ajt.13087
15. Shevach EM. Biological functions of regulatory T cells. Adv Immunol. (2011) 112:137–76. doi: 10.1016/B978-0-12-387827-4.00004-8
16. Huynh A, Zhang R, Turka LA. Signals and pathways controlling regulatory T cells. Immunol Rev. (2014) 258(1):117–31. doi: 10.1111/imr.12148
17. Bettencourt IA, Powell JD. Targeting metabolism as a novel therapeutic approach to autoimmunity, inflammation, and transplantation. J Immunol. (2017) 198(3):999–1005. doi: 10.4049/jimmunol.1601318
18. Chalasani G, Li Q, Konieczny BT, Smith-Diggs L, Wrobel B, Dai Z, et al. The allograft defines the type of rejection (acute versus chronic) in the face of an established effector immune response. J Immunol. (2004) 172(12):7813–20. doi: 10.4049/jimmunol.172.12.7813
19. Bruneau S, Woda CB, Daly KP, Boneschansker L, Jain NG, Kochupurakkal N, et al. Key features of the intragraft microenvironment that determine long-term survival following transplantation. Front Immunol. (2012) 3:54. doi: 10.3389/fimmu.2012.00054
20. Kreisel D, Krupnick AS, Gelman AE, Engels FH, Popma SH, Krasinskas AM, et al. Non-Hematopoietic allograft cells directly activate Cd8+ T cells and trigger acute rejection: an alternative mechanism of allorecognition. Nat Med. (2002) 8(3):233–9. doi: 10.1038/nm0302-233
21. Al-Lamki RS, Bradley JR, Pober JS. Endothelial cells in allograft rejection. Transplantation. (2008) 86(10):1340–8. doi: 10.1097/TP.0b013e3181891d8b
22. Jiang X, Sung YK, Tian W, Qian J, Semenza GL, Nicolls MR. Graft microvascular disease in solid organ transplantation. J Mol Med. (2014) 92(8):797–810. doi: 10.1007/s00109-014-1173-y
23. Pober JS, Sessa WC. Evolving functions of endothelial cells in inflammation. Nat Rev Immunol. (2007) 7(10):803–15. doi: 10.1038/nri2171
24. Wedel J, Nakayama H, Kochupurakkal NM, Koch J, Klagsbrun M, Bielenberg DR, et al. The intragraft microenvironment as a central determinant of chronic rejection or local immunoregulation/tolerance. Curr Opin Organ Transplant. (2017) 22(1):55–63. doi: 10.1097/MOT.0000000000000373
25. Valujskikh A, Lantz O, Celli S, Matzinger P, Heeger PS. Cross-primed Cd8(+) T cells mediate graft rejection via a distinct effector pathway. Nat Immunol. (2002) 3(9):844–51. doi: 10.1038/ni831
26. Walch JM, Zeng Q, Li Q, Oberbarnscheidt MH, Hoffman RA, Williams AL, et al. Cognate antigen directs Cd8+ T cell migration to vascularized transplants. J Clin Invest. (2013) 123(6):2663–71. doi: 10.1172/JCI66722
27. Abrahimi P, Liu R, Pober JS. Blood vessels in allotransplantation. Am J Transplant. (2015) 15(7):1748–54. doi: 10.1111/ajt.13242
28. Pober JS, Jane-wit D, Qin L, Tellides G. Interacting mechanisms in the pathogenesis of cardiac allograft vasculopathy. Arterioscler Thromb Vasc Biol. (2014) 34(8):1609–14. doi: 10.1161/ATVBAHA.114.302818
29. Briscoe DM, Yeung AC, Schoen FJ, Allred EN, Stavrakis G, Ganz P, et al. Predictive value of inducible endothelial cell adhesion molecule expression for acute rejection of human cardiac allografts. Transplantation. (1995) 59:204–11. doi: 10.1097/00007890-199501000-00009
30. Dormond O, Dufour M, Seto T, Bruneau S, Briscoe DM. Targeting the intragraft microenvironment and the development of chronic allograft rejection. Hum Immunol. (2012) 73(12):1261–1268. doi: 10.1016/j.humimm.2012.07.334
31. Reinders ME, Rabelink TJ, Briscoe DM. Angiogenesis and endothelial cell repair in renal disease and allograft rejection. J Am Soc Nephrol. (2006) 17(4):932–42. doi: 10.1681/ASN.2005121250
32. Melter M, Exeni A, Reinders ME, Fang JC, McMahon G, Ganz P, et al. Expression of the chemokine receptor Cxcr3 and its ligand Ip-10 during human cardiac allograft rejection. Circulation. (2001) 104(21):2558–64. doi: 10.1161/hc4601.098010
33. Pober JS, Gimbrone MA Jr., Collins T, Cotran RS, Ault KA, Fiers W, et al. Interactions of T lymphocytes with human vascular endothelial cells: role of endothelial cells surface antigens. Immunobiology. (1984) 168(3-5):483–94. doi: 10.1016/s0171-2985(84)80132-1
34. Epperson DE, Pober JS. Antigen-presenting function of human endothelial cells. Direct activation of resting Cd8T cells. J Immunol. (1994) 153(12):5402–12. doi: 10.4049/jimmunol.153.12.5402
35. Briscoe DM, DesRoches LE, Kiely JM, Lederer JA, Lichtman AH. Antigen-dependent activation of T helper cell subsets by endothelium. Transplantation. (1995) 59(11):1638–41.7778184
36. Amersfoort J, Eelen G, Carmeliet P. Immunomodulation by endothelial cells - partnering up with the immune system? Nat Rev Immunol. (2022) 22(9):576–88. doi: 10.1038/s41577-022-00694-4
37. Lakkis FG. Where is the alloimmune response initiated? Am J Transplant. (2003) 3(3):241–2. doi: 10.1034/j.1600-6143.2003.00054.x
38. Briscoe DM, Sayegh MH. A rendezvous before rejection: where do T cells meet transplant antigens? Nat Med. (2002) 8(3):220–2. doi: 10.1038/nm0302-220
39. Madill-Thomsen KS, Halloran PF. Precision diagnostics in transplanted organs using microarray-assessed gene expression: concepts and technical methods of the molecular microscope(R) diagnostic system (Mmdx). Clin Sci. (2024) 138(11):663–85. doi: 10.1042/CS20220530
40. Halloran PF, Madill-Thomsen KS, Reeve J. The molecular phenotype of kidney transplants: insights from the Mmdx project. Transplantation. (2024) 108(1):45–71. doi: 10.1097/tp.0000000000004624
41. Rodig N, Ryan T, Allen JA, Pang H, Grabie N, Chernova T, et al. Endothelial expression of Pd-L1 and Pd-L2 down-regulates Cd8+ T cell activation and cytolysis. Eur J Immunol. (2003) 33(11):3117–26. doi: 10.1002/eji.200324270
42. Wang C, Yi T, Qin L, Maldonado RA, von Andrian UH, Kulkarni S, et al. Rapamycin-treated human endothelial cells preferentially activate allogeneic regulatory T cells. J Clin Invest. (2013) 123(4):1677–93. doi: 10.1172/JCI66204
43. Freeman GJ, Long AJ, Iwai Y, Bourque K, Chernova T, Nishimura H, et al. Engagement of the Pd-1 immunoinhibitory receptor by a novel B7 family member leads to negative regulation of lymphocyte activation. J Exp Med. (2000) 192(7):1027–34. doi: 10.1084/jem.192.7.1027
44. Han G, Chen G, Shen B, Li Y. Tim-3: an activation marker and activation limiter of innate immune cells. Front Immunol. (2013) 4:449. doi: 10.3389/fimmu.2013.00449
45. Cong Y, Wang X, Wang S, Qiao G, Li Y, Cao J, et al. Tim-3 promotes tube formation and decreases tight junction formation in vascular endothelial cells. Biosci Rep. (2020) 40(10):BSR20202130. doi: 10.1042/BSR20202130
46. Riella LV, Watanabe T, Sage PT, Yang J, Yeung M, Azzi J, et al. Essential role of Pdl1 expression on nonhematopoietic donor cells in acquired tolerance to vascularized cardiac allografts. Am J Transplant. (2011) 11(4):832–40. doi: 10.1111/j.1600-6143.2011.03451.x
47. Yang J, Popoola J, Khandwala S, Vadivel N, Vanguri V, Yuan X, et al. Critical role of donor tissue expression of programmed death ligand-1 in regulating cardiac allograft rejection and vasculopathy. Circulation. (2008) 117(5):660–9. doi: 10.1161/CIRCULATIONAHA.107.741025
48. Koga N, Suzuki J, Kosuge H, Haraguchi G, Onai Y, Futamatsu H, et al. Blockade of the interaction between Pd-1 and Pd-L1 accelerates graft arterial disease in cardiac allografts. Arterioscler Thromb Vasc Biol. (2004) 24(11):2057–62. doi: 10.1161/01.ATV.0000145015.23656.e4
49. Schenk S, Kish DD, He C, El-Sawy T, Chiffoleau E, Chen C, et al. Alloreactive T cell responses and acute rejection of single class Ii Mhc-disparate heart allografts are under strict regulation by Cd4+ Cd25+ T cells. J Immunol. (2005) 174(6):3741–8. doi: 10.4049/jimmunol.174.6.3741
50. Nagano H, Mitchell RN, Taylor MK, Hasegawa S, Tilney NL, Libby P. Interferon-gamma deficiency prevents coronary arteriosclerosis but not myocardial rejection in transplanted mouse hearts. J Clin Invest. (1997) 100:550–7. doi: 10.1172/JCI119564
51. Shim YJ, Khedraki R, Dhar J, Fan R, Dvorina N, Valujskikh A, et al. Early T cell infiltration is modulated by programed cell death-1 protein and its ligand (Pd-1/Pd-L1) interactions in murine kidney transplants. Kidney Int. (2020) 98(4):897–905. doi: 10.1016/j.kint.2020.03.037
52. Krupnick AS, Gelman AE, Barchet W, Richardson S, Kreisel FH, Turka LA, et al. Murine vascular endothelium activates and induces the generation of allogeneic Cd4 + 25 + Foxp3+ regulatory T cells. J Immunol. (2005) 175(10):6265–70. doi: 10.4049/jimmunol.175.10.6265
53. Wedel J, Liu K, Seto T, Kong S, Briscoe D. Novel phenotype of intragraft endothelial cells that modulates Cd4+ T effector-mediated rejection. Am J Transplant. (2022) 22:371. doi: 10.1111/ajt.16878
54. Alnaqbi H, Becker LM, Mousa M, Alshamsi F, Azzam SK, Emini Veseli B, et al. Immunomodulation by endothelial cells: prospects for cancer therapy. Trends Cancer. (2024) 10:1072–1091. doi: 10.1016/j.trecan.2024.08.002
55. Lanitis E, Irving M, Coukos G. Targeting the tumor vasculature to enhance T cell activity. Curr Opin Immunol. (2015) 33:55–63. doi: 10.1016/j.coi.2015.01.011
56. Kean LS, Turka LA, Blazar BR. Advances in targeting co-inhibitory and co-stimulatory pathways in transplantation settings: the yin to the yang of cancer immunotherapy. Immunol Rev. (2017) 276(1):192–212. doi: 10.1111/imr.12523
57. Becker LM, Chen SH, Rodor J, de Rooij L, Baker AH, Carmeliet P. Deciphering endothelial heterogeneity in health and disease at single-cell resolution: progress and perspectives. Cardiovasc Res. (2023) 119(1):6–27. doi: 10.1093/cvr/cvac018
58. He L, Vanlandewijck M, Mae MA, Andrae J, Ando K, Del Gaudio F, et al. Single-Cell Rna sequencing of mouse brain and lung vascular and vessel-associated cell types. Sci Data. (2018) 5:180160. doi: 10.1038/sdata.2018.160
59. Gunawardana H, Romero T, Yao N, Heidt S, Mulder A, Elashoff DA, et al. Tissue-specific endothelial cell heterogeneity contributes to unequal inflammatory responses. Sci Rep. (2021) 11(1):1949. doi: 10.1038/s41598-020-80102-w
60. Dumas SJ, Meta E, Borri M, Goveia J, Rohlenova K, Conchinha NV, et al. Single-cell Rna sequencing reveals renal endothelium heterogeneity and metabolic adaptation to water deprivation. J Am Soc Nephrol. (2020) 31(1):118–38. doi: 10.1681/ASN.2019080832
61. Jambusaria A, Hong Z, Zhang L, Srivastava S, Jana A, Toth PT, et al. Endothelial heterogeneity across distinct vascular beds during homeostasis and inflammation. Elife. (2020) 9:e51413. doi: 10.7554/eLife.51413
62. Geldhof V, de Rooij L, Sokol L, Amersfoort J, De Schepper M, Rohlenova K, et al. Single cell atlas identifies lipid-processing and immunomodulatory endothelial cells in healthy and malignant breast. Nat Commun. (2022) 13(1):5511. doi: 10.1038/s41467-022-33052-y
63. Krausgruber T, Fortelny N, Fife-Gernedl V, Senekowitsch M, Schuster LC, Lercher A, et al. Structural cells are key regulators of organ-specific immune responses. Nature. (2020) 583(7815):296–302. doi: 10.1038/s41586-020-2424-4
64. Nolan DJ, Ginsberg M, Israely E, Palikuqi B, Poulos MG, James D, et al. Molecular signatures of tissue-specific microvascular endothelial cell heterogeneity in organ maintenance and regeneration. Dev Cell. (2013) 26(2):204–19. doi: 10.1016/j.devcel.2013.06.017
65. Kalucka J, de Rooij L, Goveia J, Rohlenova K, Dumas SJ, Meta E, et al. Single-cell transcriptome atlas of murine endothelial cells. Cell. (2020) 180(4):764–79.e20. doi: 10.1016/j.cell.2020.01.015
66. Butler JM, Nolan DJ, Vertes EL, Varnum-Finney B, Kobayashi H, Hooper AT, et al. Endothelial cells are essential for the self-renewal and repopulation of notch-dependent hematopoietic stem cells. Cell Stem Cell. (2010) 6(3):251–64. doi: 10.1016/j.stem.2010.02.001
67. Ding BS, Nolan DJ, Butler JM, James D, Babazadeh AO, Rosenwaks Z, et al. Inductive angiocrine signals from sinusoidal endothelium are required for liver regeneration. Nature. (2010) 468(7321):310–5. doi: 10.1038/nature09493
68. Motz GT, Santoro SP, Wang LP, Garrabrant T, Lastra RR, Hagemann IS, et al. Tumor endothelium fasl establishes a selective immune barrier promoting tolerance in tumors. Nat Med. (2014) 20(6):607–15. doi: 10.1038/nm.3541
69. Georganaki M, van Hooren L, Dimberg A. Vascular targeting to increase the efficiency of immune checkpoint blockade in cancer. Front Immunol. (2018) 9:3081. doi: 10.3389/fimmu.2018.03081
70. Shetty S, Lalor PF, Adams DH. Liver sinusoidal endothelial cells—gatekeepers of hepatic immunity. Nat Rev Gastroenterol Hepatol. (2018) 15(9):555–67. doi: 10.1038/s41575-018-0020-y
71. Sonar SA, Lal G. Blood-brain barrier and its function during inflammation and autoimmunity. J Leukoc Biol. (2018) 103(5):839–53. doi: 10.1002/JLB.1RU1117-428R
72. Sha X, Meng S, Li X, Xi H, Maddaloni M, Pascual DW, et al. Interleukin-35 inhibits endothelial cell activation by suppressing Mapk-Ap-1 pathway. J Biol Chem. (2015) 290(31):19307–18. doi: 10.1074/jbc.M115.663286
73. Nakayama H, Bruneau S, Kochupurakkal N, Coma S, Briscoe DM, Klagsbrun M. Regulation of Mtor signaling by semaphorin 3f-neuropilin 2 interactions in vitro and in vivo. Sci Rep. (2015) 5:11789. doi: 10.1038/srep11789
74. Wedel J, Bruneau S, Liu K, Kong SW, Sage PT, Sabatini DM, et al. Deptor modulates activation responses in Cd4(+) T cells and enhances immunoregulation following transplantation. Am J Transplant. (2019) 19(1):77–88. doi: 10.1111/ajt.14995
75. Denton MD, Davis SF, Baum MA, Melter M, Reinders ME, Exeni A, et al. The role of the graft endothelium in transplant rejection: evidence that endothelial activation may serve as a clinical marker for the development of chronic rejection. Pediatr Transplant. (2000) 4(4):252–60. doi: 10.1034/j.1399-3046.2000.00031.x
76. Hill PA, Main IW, Atkins RC. Icam-1 and Vcam-1 in human renal allograft rejection. Kidney Int. (1995) 47(5):1383–91. doi: 10.1038/ki.1995.194
77. Bruneau S, Nakayama H, Woda CB, Flynn EA, Briscoe DM. Deptor regulates vascular endothelial cell activation and proinflammatory and angiogenic responses. Blood. (2013) 122(10):1833–42. doi: 10.1182/blood-2013-03-488486
78. Augustin HG, Koh GY. Organotypic vasculature: from descriptive heterogeneity to functional pathophysiology. Science. (2017) 357(6353):eaal2379. doi: 10.1126/science.aal2379
79. Tang Y, Wang J, Zhang Y, Li J, Chen M, Gao Y, et al. Single-cell Rna sequencing identifies intra-graft population heterogeneity in acute heart allograft rejection in mouse. Front Immunol. (2022) 13:832573. doi: 10.3389/fimmu.2022.832573
80. Zheng GX, Terry JM, Belgrader P, Ryvkin P, Bent ZW, Wilson R, et al. Massively parallel digital transcriptional profiling of single cells. Nat Commun. (2017) 8:14049. doi: 10.1038/ncomms14049
81. Hao Y, Stuart T, Kowalski MH, Choudhary S, Hoffman P, Hartman A, et al. Dictionary learning for integrative, multimodal and scalable single-cell analysis. Nat Biotechnol. (2024) 42(2):293–304. doi: 10.1038/s41587-023-01767-y
82. Korsunsky I, Millard N, Fan J, Slowikowski K, Zhang F, Wei K, et al. Fast, sensitive and accurate integration of single-cell data with harmony. Nat Methods. (2019) 16(12):1289–96. doi: 10.1038/s41592-019-0619-0
83. Zappia L, Oshlack A. Clustering trees: a visualization for evaluating clusterings at multiple resolutions. Gigascience. (2018) 7(7):giy083. doi: 10.1093/gigascience/giy083
84. Cao J, Spielmann M, Qiu X, Huang X, Ibrahim DM, Hill AJ, et al. The single-cell transcriptional landscape of mammalian organogenesis. Nature. (2019) 566(7745):496–502. doi: 10.1038/s41586-019-0969-x
85. Mai J, Nanayakkara G, Lopez-Pastrana J, Li X, Li YF, Wang X, et al. Interleukin-17a promotes aortic endothelial cell activation via transcriptionally and post-translationally activating P38 mitogen-activated protein kinase (Mapk) pathway. J Biol Chem. (2016) 291(10):4939–54. doi: 10.1074/jbc.M115.690081
86. Contreras AG, Dormond O, Edelbauer M, Calzadilla K, Hoerning A, Pal S, et al. Mtor-understanding the clinical effects. Transplant Proc. (2008) 40(10 Suppl):S9–S12. doi: 10.1016/j.transproceed.2008.10.011
87. Wu H, Malone AF, Donnelly EL, Kirita Y, Uchimura K, Ramakrishnan SM, et al. Single-cell transcriptomics of a human kidney allograft biopsy specimen defines a diverse inflammatory response. J Am Soc Nephrol. (2018) 29(8):2069–80. doi: 10.1681/ASN.2018020125
88. Abou-Daya KI, Tieu R, Zhao D, Rammal R, Sacirbegovic F, Williams AL, et al. Resident memory T cells form during persistent antigen exposure leading to allograft rejection. Sci Immunol. (2021) 6:eabc8122. doi: 10.1126/sciimmunol.abc8122
89. Rosales IA, Yang C, Farkash EA, Ashry T, Ge J, Aljabban I, et al. Novel intragraft regulatory lymphoid structures in kidney allograft tolerance. Am J Transplant. (2022) 22(3):705–16. doi: 10.1111/ajt.16880
90. Babu AN, Murakawa T, Thurman JM, Miller EJ, Henson PM, Zamora MR, et al. Microvascular destruction identifies murine allografts that cannot be rescued from airway fibrosis. J Clin Invest. (2007) 117(12):3774–85. doi: 10.1172/JCI32311
91. Zhao D, Abou-Daya KI, Dai H, Oberbarnscheidt MH, Li XC, Lakkis FG. Innate allorecognition and memory in transplantation. Front Immunol. (2020) 11:918. doi: 10.3389/fimmu.2020.00918
92. Tieu R, Zeng Q, Zhao D, Zhang G, Feizi N, Manandhar P, et al. Tissue-resident memory T cell maintenance during antigen persistence requires both cognate antigen and interleukin-15. Sci Immunol. (2023) 8(82):eadd8454. doi: 10.1126/sciimmunol.add8454
93. Serhan CN, Chiang N, Dalli J. The resolution code of acute inflammation: novel pro-resolving lipid mediators in resolution. Semin Immunol. (2015) 27(3):200–15. doi: 10.1016/j.smim.2015.03.004
94. Serhan CN, Levy BD. Resolvins in inflammation: emergence of the pro-resolving superfamily of mediators. J Clin Invest. (2018) 128(7):2657–69. doi: 10.1172/JCI97943
95. Li W, Shepherd HM, Terada Y, Shay AE, Bery AI, Gelman AE, et al. Resolvin D1 prevents injurious neutrophil swarming in transplanted lungs. Proc Natl Acad Sci. (2023) 120(31):e2302938120. doi: 10.1073/pnas.2302938120
96. Boneschansker L, Yan J, Wong E, Briscoe DM, Irimia D. Microfluidic platform for the quantitative analysis of leukocyte migration signatures. Nat Commun. (2014) 5:4787. doi: 10.1038/ncomms5787
97. Mirakaj V, Rosenberger P. Immunomodulatory functions of neuronal guidance proteins. Trends Immunol. (2017) 38(6):444–56. doi: 10.1016/j.it.2017.03.007
98. Boneschansker L, Nakayama H, Eisenga M, Wedel J, Klagsbrun M, Irimia D, et al. Netrin-1 augments chemokinesis in Cd4+ T cells in vitro and elicits a proinflammatory response in vivo. J Immunol. (2016) 197(4):1389–98. doi: 10.4049/jimmunol.1502432
99. Tadagavadi RK, Wang W, Ramesh G. Netrin-1 regulates Th1/Th2/Th17 cytokine production and inflammation through Unc5b receptor and protects kidney against ischemia-reperfusion injury. J Immunol. (2010) 185(6):3750–8. doi: 10.4049/jimmunol.1000435
100. Ranganathan PV, Jayakumar C, Mohamed R, Dong Z, Ramesh G. Netrin-1 regulates the inflammatory response of neutrophils and macrophages, and suppresses ischemic acute kidney injury by inhibiting Cox-2-mediated Pge2 production. Kidney Int. (2013) 83(6):1087–98. doi: 10.1038/ki.2012.423
101. Mizui M, Kumanogoh A, Kikutani H. Immune semaphorins: novel features of neural guidance molecules. J Clin Immunol. (2009) 29:1–11. doi: 10.1007/s10875-008-9263-7
102. Chaturvedi S, Yuen DA, Bajwa A, Huang YW, Sokollik C, Huang L, et al. Slit2 prevents neutrophil recruitment and renal ischemia-reperfusion injury. J Am Soc Nephrol. (2013) 24(8):1274–87. doi: 10.1681/ASN.2012090890
103. Mehlen P, Delloye-Bourgeois C, Chedotal A. Novel roles for slits and netrins: axon guidance cues as anticancer targets? Nat Rev Cancer. (2011) 11(3):188–97. doi: 10.1038/nrc3005
104. Wu JY, Feng L, Park HT, Havlioglu N, Wen L, Tang H, et al. The neuronal repellent slit inhibits leukocyte chemotaxis induced by chemotactic factors. Nature. (2001) 410(6831):948–52. doi: 10.1038/35073616
105. Tessier-Lavigne M, Goodman CS. The molecular biology of axon guidance. Science. (1996) 274(5290):1123–33. doi: 10.1126/science.274.5290.1123
106. Antipenko A, Himanen JP, van Leyen K, Nardi-Dei V, Lesniak J, Barton WA, et al. Structure of the semaphorin-3a receptor binding module. Neuron. (2003) 39(4):589–98. doi: 10.1016/S0896-6273(03)00502-6
107. Sun KL, Correia JP, Kennedy TE. Netrins: versatile extracellular cues with diverse functions. Development. (2011) 138(11):2153–69. doi: 10.1242/dev.044529
108. Zhang H, Vreeken D, Leuning DG, Bruikman CS, Junaid A, Stam W, et al. Netrin-4 expression by human endothelial cells inhibits endothelial inflammation and senescence. Int J Biochem Cell Biol. (2021) 134:105960. doi: 10.1016/j.biocel.2021.105960
109. van Gils JM, Derby MC, Fernandes LR, Ramkhelawon B, Ray TD, Rayner KJ, et al. The neuroimmune guidance cue netrin-1 promotes atherosclerosis by inhibiting the emigration of macrophages from plaques. Nat Immunol. (2012) 13(2):136–43. doi: 10.1038/ni.2205
110. Konig K, Gatidou D, Granja T, Meier J, Rosenberger P, Mirakaj V. The axonal guidance receptor neogenin promotes acute inflammation. PLoS One. (2012) 7(3):e32145. doi: 10.1371/journal.pone.0032145
111. Ly NP, Komatsuzaki K, Fraser IP, Tseng AA, Prodhan P, Moore KJ, et al. Netrin-1 inhibits leukocyte migration in vitro and in vivo. Proc Natl Acad Sci. (2005) 102(41):14729–34. doi: 10.1073/pnas.0506233102
112. Xiao Y, Yu S, Zhu B, Bedoret D, Bu X, Francisco LM, et al. Rgmb is a novel binding partner for Pd-L2 and its engagement with Pd-L2 promotes respiratory tolerance. J Exp Med. (2014) 211(5):943–59. doi: 10.1084/jem.20130790
113. Moretti S, Procopio A, Boemi M, Catalano A. Neuronal semaphorins regulate a primary immune response. Curr Neurovasc Res. (2006) 3(4):295–305. doi: 10.2174/156720206778792939
114. Capparuccia L, Tamagnone L. Semaphorin signaling in cancer cells and in cells of the tumor microenvironment–two sides of a coin. J Cell Sci. (2009) 122(Pt 11):1723–36. doi: 10.1242/jcs.030197
115. Kumanogoh A, Kikutani H. Immunological functions of the neuropilins and plexins as receptors for semaphorins. Nat Rev Immunol. (2013) 13(11):802–14. doi: 10.1038/nri3545
116. Takamatsu H, Kumanogoh A. Diverse roles for semaphorin-plexin signaling in the immune system. Trends Immunol. (2012) 33(3):127–35. doi: 10.1016/j.it.2012.01.008
117. Goodman CS, Kolodkin AL, Luo Y, Püschel AW, Raper JA. Unified nomenclature for the semaphorins/collapsins. Semaphorin nomenclature committee. Cell. (1999) 97(5):551–2. doi: 10.1016/S0092-8674(00)80766-7
118. Delgoffe GM, Woo SR, Turnis ME, Gravano DM, Guy C, Overacre AE, et al. Stability and function of regulatory T cells is maintained by a neuropilin-1-semaphorin-4a axis. Nature. (2013) 501(7466):252–6. doi: 10.1038/nature12428
119. Barnkob MB, Michaels YS, Andre V, Macklin PS, Gileadi U, Valvo S, et al. Semaphorin 3A causes immune suppression by inducing cytoskeletal paralysis in tumour-specific Cd8(+) T cells. Nat Commun. (2024) 15(1):3173. doi: 10.1038/s41467-024-47424-z
120. Liu C, Somasundaram A, Manne S, Gocher AM, Szymczak-Workman AL, Vignali KM, et al. Neuropilin-1 is a T cell memory checkpoint limiting long-term antitumor immunity. Nat Immunol. (2020) 21(9):1010–21. doi: 10.1038/s41590-020-0733-2
121. Mendes-da-Cruz DA, Brignier AC, Asnafi V, Baleydier F, Messias CV, Lepelletier Y, et al. Semaphorin 3f and neuropilin-2 control the migration of human T-cell precursors. PLoS One. (2014) 9(7):e103405. doi: 10.1371/journal.pone.0103405
122. Yadav M, Louvet C, Davini D, Gardner JM, Martinez-Llordella M, Bailey-Bucktrout S, et al. Neuropilin-1 distinguishes natural and inducible regulatory T cells among regulatory T cell subsets in vivo. J Exp Med. (2012) 209(10):1713–22. doi: 10.1084/jem.20120822
123. Hansen W, Hutzler M, Abel S, Alter C, Stockmann C, Kliche S, et al. Neuropilin 1 deficiency on Cd4+ Foxp3+ regulatory T cells impairs mouse melanoma growth. J Exp Med. (2012) 209(11):2001–16. doi: 10.1084/jem.20111497
124. Feng L, Shu HP, Sun LL, Tu YC, Liao QQ, Yao LJ. Role of the slit-robo signaling pathway in renal pathophysiology and various renal diseases. Front Physiol. (2023) 14:1226341. doi: 10.3389/fphys.2023.1226341
125. Moser B, Loetscher P. Lymphocyte traffic control by chemokines. Nat Immunol. (2001) 2(2):123–8. doi: 10.1038/84219
126. Melter M, Exeni A, Briscoe DM. Chemokines and their receptors in human clinical solid organ transplantation. Curr Opin Organ Transplantation. (2002) 7:77–84. doi: 10.1097/00075200-200203000-00016
127. Hancock WW. Chemokines and transplant immunobiology. J Am Soc Nephrol. (2002) 13(3):821–4. doi: 10.1681/ASN.V133821
128. Schenk AD, Rosenblum JM, Fairchild RL. Chemokine-directed strategies to attenuate allograft rejection. Clin Lab Med. (2008) 28(3):441–54. doi: 10.1016/j.cll.2008.07.004
129. el-Sawy T, Fahmy NM, Fairchild RL. Chemokines: directing leukocyte infiltration into allografts. Curr Opin Immunol. (2002) 14(5):562–8. doi: 10.1016/s0952-7915(02)00382-5
130. Curreli S, Arany Z, Gerardy-Schahn R, Mann D, Stamatos NM. Polysialylated neuropilin-2 is expressed on the surface of human dendritic cells and modulates dendritic cell-T lymphocyte interactions. J Biol Chem. (2007) 282(42):30346–56. doi: 10.1074/jbc.M702965200
131. Hoerning A, Koss K, Datta D, Boneschansker L, Jones CN, Wong IY, et al. Subsets of human Cd4(+) regulatory T cells express the peripheral homing receptor Cxcr3. Eur J Immunol. (2011) 41(8):2291–302. doi: 10.1002/eji.201041095
132. Dai H, Pena A, Bauer L, Williams A, Watkins SC, Camirand G. Treg suppression of immunity within inflamed allogeneic grafts. JCI Insight. (2022) 7(16):e160579. doi: 10.1172/jci.insight.160579
133. Li W, Bribriesco AC, Nava RG, Brescia AA, Ibricevic A, Spahn JH, et al. Lung transplant acceptance is facilitated by early events in the graft and is associated with lymphoid neogenesis. Mucosal Immunol. (2012) 5(5):544–54. doi: 10.1038/mi.2012.30
134. Miyajima M, Chase CM, Alessandrini A, Farkash EA, Della Pelle P, Benichou G, et al. Early acceptance of renal allografts in mice is dependent on Foxp3(+) cells. Am J Pathol. (2011) 178(4):1635–45. doi: 10.1016/j.ajpath.2010.12.024
135. Li W, Gauthier JM, Higashikubo R, Hsiao HM, Tanaka S, Vuong L, et al. Bronchus-associated lymphoid tissue-resident Foxp3+ T lymphocytes prevent antibody-mediated lung rejection. J Clin Invest. (2019) 129(2):556–68. doi: 10.1172/JCI122083
136. Hu M, Wang C, Zhang GY, Saito M, Wang YM, Fernandez MA, et al. Infiltrating Foxp3(+) regulatory T cells from spontaneously tolerant kidney allografts demonstrate donor-specific tolerance. Am J Transplant. (2013) 13(11):2819–30. doi: 10.1111/ajt.12445
137. Guba M, von Breitenbuch P, Steinbauer M, Koehl G, Flegel S, Hornung M, et al. Rapamycin inhibits primary and metastatic tumor growth by antiangiogenesis: involvement of vascular endothelial growth factor. Nat Med. (2002) 8(2):128–35. doi: 10.1038/nm0202-128
138. Dormond O, Madsen JC, Briscoe DM. The effects of Mtor-Akt interactions on anti-apoptotic signaling in vascular endothelial cells. J Biol Chem. (2007) 282(32):23679–86. doi: 10.1074/jbc.M700563200
139. Phung TL, Ziv K, Dabydeen D, Eyiah-Mensah G, Riveros M, Perruzzi C, et al. Pathological angiogenesis is induced by sustained Akt signaling and inhibited by rapamycin. Cancer Cell. (2006) 10(2):159–70. doi: 10.1016/j.ccr.2006.07.003
140. Sun JF, Phung T, Shiojima I, Felske T, Upalakalin JN, Feng D, et al. Microvascular patterning is controlled by fine-tuning the Akt signal. Proc Natl Acad Sci. (2005) 102(1):128–33. doi: 10.1073/pnas.0403198102
141. Wang C, Qin L, Manes TD, Kirkiles-Smith NC, Tellides G, Pober JS. Rapamycin antagonizes Tnf induction of Vcam-1 on endothelial cells by inhibiting Mtorc2. J Exp Med. (2014) 211(3):395–404. doi: 10.1084/jem.20131125
142. Laplante M, Sabatini DM. Mtor signaling in growth control and disease. Cell. (2012) 149(2):274–93. doi: 10.1016/j.cell.2012.03.017
143. Saxton RA, Sabatini DM. Mtor signaling in growth, metabolism, and disease. Cell. (2017) 168(6):960–76. doi: 10.1016/j.cell.2017.02.004
144. Phung TL, Eyiah-Mensah G, O'Donnell RK, Bieniek R, Shechter S, Walsh K, et al. Endothelial Akt signaling is rate-limiting for rapamycin inhibition of mouse mammary tumor progression. Cancer Res. (2007) 67(11):5070–5. doi: 10.1158/0008-5472.CAN-06-3341
145. Couzin-Frankel J. Breakthrough of the year 2013. Cancer immunotherapy. Science. (2013) 342(6165):1432–3. doi: 10.1126/science.342.6165.1432
146. Caron A, Briscoe DM, Richard D, Laplante M. Deptor at the nexus of cancer, metabolism, and immunity. Physiol Rev. (2018) 98(3):1765–803. doi: 10.1152/physrev.00064.2017
147. Peterson TR, Laplante M, Thoreen CC, Sancak Y, Kang SA, Kuehl WM, et al. Deptor is an Mtor inhibitor frequently overexpressed in multiple myeloma cells and required for their survival. Cell. (2009) 137(5):873–86. doi: 10.1016/j.cell.2009.03.046
148. Proud CG. Dynamic balancing: deptor tips the scales. J Mol Cell Biol. (2009) 1(2):61–3. doi: 10.1093/jmcb/mjp012
Keywords: endothelial cell, allograft (ALLO), immunoregulation, transplantation, graft survival
Citation: Bose S, Do V, Testini C, Jadhav SS, Sailliet N, Kho AT, Komatsu M, Boneschansker L, Kong SW, Wedel J and Briscoe DM (2025) Immunomodulation by allograft endothelial cells. Front. Transplant. 4:1518772. doi: 10.3389/frtra.2025.1518772
Received: 28 October 2024; Accepted: 15 January 2025;
Published: 4 February 2025.
Edited by:
Xian C Li, Houston Methodist Hospital, United StatesReviewed by:
Lisha Mou, Shenzhen Second People's Hospital, ChinaMaria Coronel, University of Michigan, United States
Copyright: © 2025 Bose, Do, Testini, Jadhav, Sailliet, Kho, Komatsu, Boneschansker, Kong, Wedel and Briscoe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Johannes Wedel, am9oYW5uZXMud2VkZWxAY2hpbGRyZW5zLmhhcnZhcmQuZWR1; David M. Briscoe, ZGF2aWQuYnJpc2NvZUBjaGlsZHJlbnMuaGFydmFyZC5lZHU=
 Sayantan Bose
Sayantan Bose Vicki Do
Vicki Do Chiara Testini
Chiara Testini Suchita S. Jadhav1,2,3,4
Suchita S. Jadhav1,2,3,4 Nicolas Sailliet
Nicolas Sailliet Sek Won Kong
Sek Won Kong Johannes Wedel
Johannes Wedel David M. Briscoe
David M. Briscoe