- 1Bioinformatics Infrastructure Facility Center, University of Kalyani, Kalyani, West Bengal, India
- 2Department of Biochemistry and Biophysics, University of Kalyani, Kalyani, West Bengal, India
Circular RNAs (circRNAs) have gained prominence as important players in various biological processes such as gastric cancer (GC). Identification of several dysregulated circRNAs may serve as biomarkers for early diagnosis or as novel therapeutic targets. Predictive models can suggest potential new interactions and regulatory roles of circRNAs in GCs. Experimental validations of key interactions are being performed using in vitro models, confirming the significance of identified circRNA networks. The aim of this review is to highlight the important circRNAs associated with GC. On top of that an overview of the mechanistic details of the biogenesis and functionalities of the circRNAs are also presented. Furthermore, the potentialities of the circRNAs in the field of new drug discovery are deciphered.
1 Introduction
The circular RNAs (circRNAs), a member of the group of RNAs apparently having no-coding potentials, have a profound influence on regulatory pathways and are associated with different types of malignancies, notably gastric cancers (GCs) (Li X. W. et al, 2020). An important feature of such RNAs is their unusual stability due to their covalently closed structure, which allows them to withstand the attacks of the exonucleases (Liu et al., 2022a). The prime activities of the circRNAs include.
• interactions with RNA-binding proteins (RBPs),
• functioning as micro RNA (miRNA) sponges,
and
• altering gene expression profiles (Zang et al., 2020).
Identifying circRNAs is a bit challenging compared to linear RNAs due to their unique structure. The specific characteristics of circRNAs which set them apart from their linear counterparts are the absence of a capping at the 5′ end and polyadenylated tail at the 3′ end. Instead of traditional markers, specific methods and computational strategies are used to identify and verify circRNAs. Key techniques and markers for identifying circRNAs are as follows:
• Back-splicing junction (Zhang et al., 2016)
• RNaseR resistance (Hansen, 2018)
• Divergent primer PCR (Zhong et al., 2018)
• Northern blot (Schneider et al., 2018)
• Fluorescence in situ hybridization (FISH) (Zirkel and Papantonis, 2018)
• Ribosomal RNA depletion (López-Jiménez et al., 2018)
CircRNAs have their abilities to modulate gene expressions and establish their roles in several diseases, like cancer, neurodegenerations, cardiovascular disorders etc. The localization of circRNAs plays a crucial role in their function, and their invasive potential, particularly in the context of cancer. CircRNAs that are localized in the cytoplasm are often associated with invasive properties, especially in cancer. In the cytoplasm, they commonly act as miRNA sponges, which is a mechanism of competitive inhibition (Ma et al., 2018). CircRNAs are also found in exosomes or other extracellular vesicles, where they can be secreted from cells and taken up by neighboring cells or distant tissues. Exosomal circRNAs can also contribute to invasiveness by facilitating long-range communication between cells and thereby promoting metastasis (Di Bella and Taverna, 2024). While circRNAs in the nucleus are typically involved in regulating transcriptional activity or splicing, they are less often associated with invasiveness (Huang et al., 2017). Their unusual stability and tissue-specific expressions make them an ideal choice in personalized medicine (Matsuoka and Yashiro, 2023). As mentioned earlier, the circRNAs have become the focal point of attraction in GC onsets. In GC patients, the circRNAs influence the expressions of specific genes thereby exerting control over tumor growth and metastasis (Li et al., 2019). GC refers to the unwanted growth of malignant cells in the stomach lining or gut tissues. There are several types of GCs, and the most significant among them are adenocarcinoma, gastric lymphoma, gastrointestinal stromal, and carcinoid tumors. This review aims to explain the roles of circRNAs in GC, focusing on tumorigenesis, progression, diagnosis, and therapeutic resistance. The authors further investigate circRNAs’ potential as therapeutic agents and prognostic indicators. The review distinguishes itself by delivering new insights into this field of study by addressing the biotechnological therapeutic potential of circRNAs, analyzing how circRNAs are controlled by drugs or microbes in GC, and providing an updated literature mining on the associated molecular processes.
2 Latest trends in circRNA research
Recent researches on circRNAs highlight their significant potential in health maintenance, disease detection, and therapeutic advancements (Fischer and Leung, 2017; Zhou et al., 2022; Haque and Harries, 2017; Verduci et al., 2021).
CircRNAs are also emerging as powerful biomarkers due to their stabilities and specific expression patterns to detect various diseases (Zaiou, 2019). These characteristics make them ideal for non-invasive disease detection methods like liquid biopsies (Wen et al., 2021; Roy et al., 2022). In therapeutic contexts, circRNA-based approaches, including circRNA vaccines, are showing considerable promise. These vaccines may offer enhanced stability and long-lasting effects compared to traditional mRNA vaccines, potentially revolutionizing immunization with fewer doses (Hu et al., 2024; Ren et al., 2024; Wang X. et al, 2022; Li and Xing, 2024). Furthermore, circRNAs have interacting potentials with miRNAs, a process referred to as miRNA sponging, modulating gene expressions (Zang et al., 2020), and influencing therapeutic strategies that may connect to gene therapy research to target specific pathways (TGF-β, β-catenin, MAPK, PI3K/Akt/mTOR signalling pathways) involved in various diseases including GC.
3 Biogenesis of circRNAs
3.1 Biogenesis of circRNA through intron-pairing-driven circularization forming ecircRNAs
This biogenesis process starts with the transcription of pre-mRNA containing exons and introns. Complementary sequences within flanking introns pair together (Zhao et al., 2022) bringing splice sites close enough to facilitate back-splicing. The event finally leads to the joining of 3′hydroxyl group to a 5′phosphate group present downstream and upstream respectively at the splice site junctions to form a lasso intermediate. This results in exons being joined in a circular form, excising the intervening intronic sequences. The resulting circRNA is covalently closed and does not have a capping at the 5′end and a poly(A) tail at the 3′end forming the exonic circRNAs (ecRNAs/ecircRNAs). This circularization is regulated by RBPs and specialized RNA secondary structures are formed to enhance or inhibit the process (Zhao et al., 2022; Hwang and Kim, 2024; Ebbesen et al., 2017).
3.2 Biogenesis of circRNA through RBP-mediated circularization forming exonic-intronic circRNA (EIciRNAs)
Another process of the biogenesis of circRNAs is through RBP-mediated circularization where RBPs bind to specific motifs within the pre-mRNA, often within flanking introns, to stabilize the RNA structure and facilitate the back-splicing process (Hwang and Kim, 2024). These proteins act as scaffolds, promoting the proximity of splice sites, which enhance circularization. Proteins like Quaking can bind intronic sequences to promote circRNA formation by looping out intervening exons and facilitating back-splicing to form EIciRNAs. The resulting circRNA has a covalent linkage between its 5′ and 3′ termini due to absence of the capping and polyadenylation at 5′ and 3′ ends respectively (Conn et al., 2015; Hwang and Kim, 2024; Zang et al., 2020; Chaudhary and Banerjee, 2024).
3.3 Biogenesis of circRNA through lariat-driven circularization forming ciRNAs
This mechanism involves the formation of a lariat intermediate during the splicing of pre-mRNA. When classical GU/AG splicing takes place in the pre-mRNAs, exons are skipped, and a lariat-like intermediate containing intron-exon sequences can form (Brown and Simpson, 1998; Zhao et al., 2022). The 5′splice site of an exon joins a branch point within an intron, forming a lariat structure. Back-splicing occurs within this lariat, resulting in a circRNA molecule (Pisignano et al., 2023) named as intronic circRNAs. The excised lariat intron is debranched by enzymes, leaving behind the circRNA. The efficiency of this process is influenced by sequence elements within the introns and exons, as well as cellular factors that regulate splicing (Wilusz, 2018).
4 Functions of circRNAs associated with GC
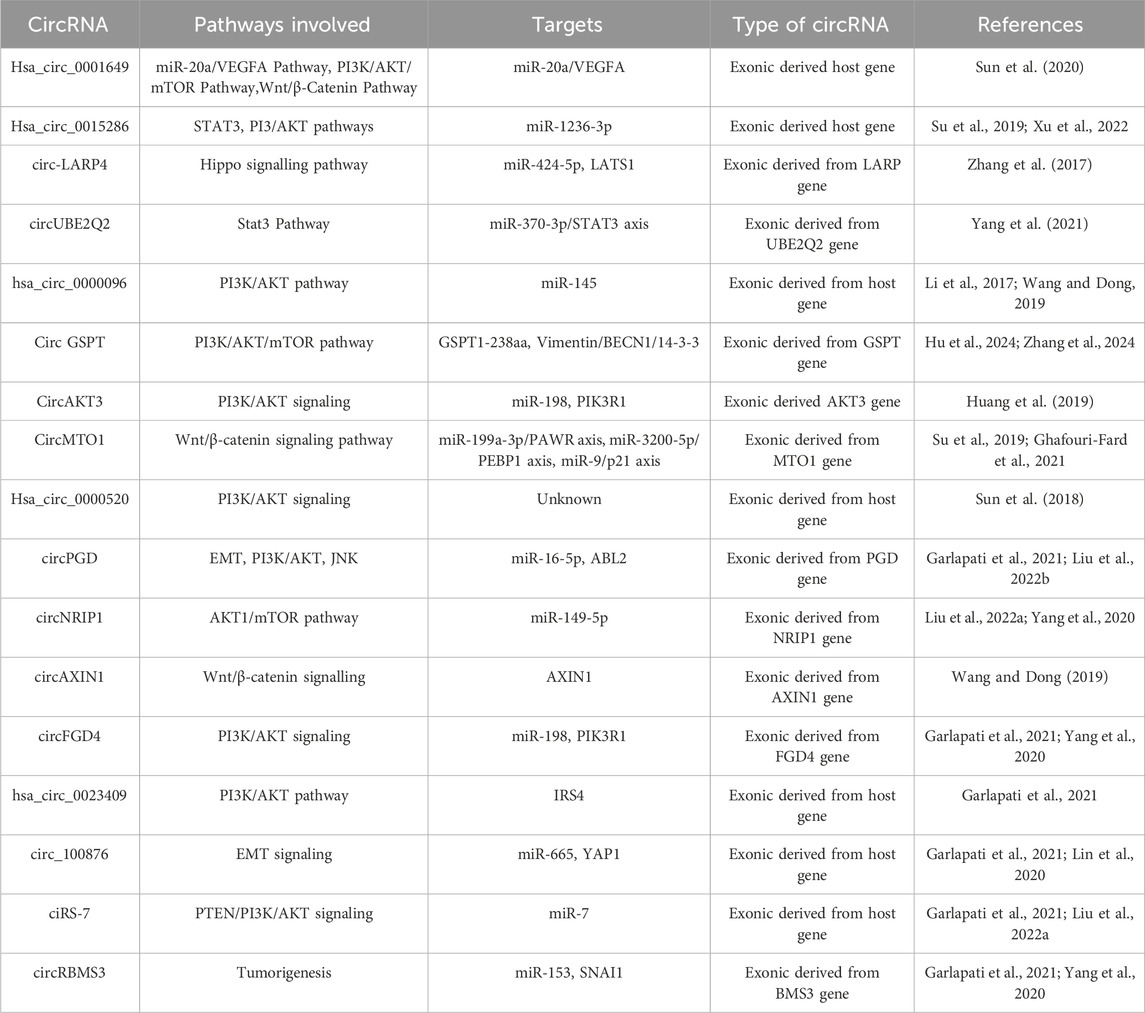
CircRNAs of various kinds have been implicated in the initiation and advancement of GC through their regulation of distinct signalling pathways. These circRNAs typically function through a variety of mechanisms, including the regulation of particular signalling pathways (TGF-β, β-catenin, MAPK, PI3K/Akt/mTOR, Hippo signalling pathways etc.), activation and deactivation of the phosphorylation sites of those proteins, binding with particular proteins that can activate or inhibit those pathways (shown in Table 1), acting as translated proteins, interacting with RBPs, miRNA to affect gene expressions.
4.1 miRNA sponge
CircRNAs, either competitively or non-competitively, bind to miRNAs, a process referred to as miRNA sponging, and thereby regulating their availability and activity within cells. In contrast to their linear counterparts, circRNAs are resistant to degradation due to their closed-loop structure, making them stable miRNA decoys. In this way the circRNAs prevent them from getting tagged to their target mRNA, leading to the de-repression of miRNA-targeted mRNAs. This regulatory mechanism is crucial in various biological processes (Zang et al., 2020), including development, cellular differentiation and homeostasis (Di Timoteo et al., 2020). Dysregulation of circRNA-mediated miRNA sponging is linked to diseases such as cancer, cardiovascular disorders and neurological disorders, thus highlighting their roles in pathophysiology (Min et al., 2021; van Zonneveld et al., 2021; Shi et al., 2022). Moreover, circRNA-mediated miRNA sponging is one of the prime events that enhances or represses different physiological activities like cell cycle regulation, proliferation, and metastasis (Khanipouyani et al., 2021; Wang Y. et al, 2022). For example, circAKT3 (Huang et al., 2019) interacts with miR-198 to influence the PI3K/AKT pathway in GC. Similarly, circRNA hsa_circ_0023409 sponges miR-542-3p, promoting GC progression through the PI3K/AKT axis. Several other circRNAs, like CDR1as and circ_100876, also regulate the migrations of the GC cells and their corresponding invasion by exerting control over the miRNAs (Lin et al., 2020). YAP1 activation via epithelial-mesenchymal transition (EMT)-related transcriptional pathways are modulated by circRPL15, which sequesters miR-502-3p from OLFM4 mRNA and activates the STAT3 pathway, leading to enhanced GC tissue growth, increased cell motility, invasion and reduced apoptosis (Li Y. et al, 2020). CircRanGAP1 promotes VEGFA expression, angiogenesis, and GC metastasis by inhibiting miR-877-3p (Lu et al., 2020). Upregulation of circRNAs in GC drives proliferation and reduces apoptosis, as shown by various in vitro and in vivo studies. Repressor circRNAs like circYAP1 inhibit GC progression by exerting control over miR-367-5p thereby modulating the p27 Kip1 expression (Liu et al., 2018). CircOXCT1 interacts with miR-136 to alleviate SMAD4 repression, limiting GC EMT and metastasis (Liu et al., 2020). CircFGD4 expression, reduced in tissues of the GC patients, is connected to a low level of metastasis of the lymph nodes and better prognosis by regulating the miR-532-3p/APC/β-catenin interaction pathway (Dai et al., 2020). Similarly, circREPS2 impedes the metastasis of GC tumours by repressing the RUNX3/β-catenin axis and removing miR-558 (Guo et al., 2020).
4.2 Transcriptional regulation by interactions with RBPs
CircRNAs can regulate transcription through interactions with RBPs and chromatin-modifying complexes. CircRNAs may act as scaffolds or decoys for RBPs, altering their availability and, consequently, the transcriptional activity of specific genes. By sequestering these RBPs away from their chromatin targets, circRNAs can alter the accessibility of gene promoters or enhancers, thereby modulating transcriptional activity (Hwang and Kim, 2024). Additionally, circRNAs can interact with RNA polymerase II and transcription factors, affecting their bindings to gene promoters and influencing the initiation or elongation phases of transcription. This interaction can either promote or inhibit the transcription of specific genes, depending on the sequence and structure of the circRNA and its binding partners (Bose and Ain, 2018). Moreover, some circRNAs have been found to localize in the nucleus (Li et al., 2017), where they can directly interact with transcriptional machinery or chromatin to exert regulatory effects. RBP IGF2BP3 (insulin-like growth factor 2 binding protein 3, also called IMP3) performs several post-transcriptional functions. In GC, IGF2BP3 (Yu et al., 2021) acts as an oncogene that stimulates cancer cell proliferation and invasion. One of the most important EMT-related transcription factors SNAIL helps in GC progression. Yu et al. described that circTNPO3 is significantly downregulated in GC. It was revealed from in vivo as well in vitro analyses that circTNPO3 inhibits the growth and metastasis of GC. In terms of mechanism, circTNPO3 competitively interacts with IGF2BP3 (Yu et al., 2021), which leads to the destruction of the MYC mRNA. This inhibition of MYC and SNAIL which is the target of MYC, is the chief and crucial player in the inducing EMT. CircFNDC3B is another up-regulatory circRNA, which can be seen in GC in cell lines. IGF2BP3, an RBP linked to several malignant tumours, can bind to circFNDC3B. Additionally, IGF2BPs have the potential to stabilize CD44 mRNA by targeting it (Hong et al., 2019). Upregulated circFNDC3B helps in the process of migrating and subsequent invading of GC cells when a tri-complex of circFNDC3B-IGF2BP3-CD44 mRNA is formed (Hong et al., 2019) and the regulation of E-cadherin in GC. Furthermore, the expression of E-cadherin is controlled by the presence of circFNDC3B. This was achieved through a novel mechanism. These encouraging findings offer fresh perspectives on treatment and GC prognosis forecasts.
4.3 Translation and translocation potential
CircRNAs can influence protein translation through internal ribosome entry sites (IRES), enabling them to produce proteins independently of the canonical mRNA cap structure (Shi et al., 2020). CircRNAs influence protein translocation by interacting with RBPs like HuR and IGF2BP3, which regulate protein localization and control diseases (Granados-Riveron and Aquino-Jarquin, 2016; Shi et al., 2020; Okholm et al., 2020). Such bindings allow circRNAs to guide or sequester RBPs, potentially impacting subcellular protein localization. CircRNAs may also indirectly influence protein translocation by modulating the expression of miRNAs and RBPs involved in protein trafficking (Wan and Hopper, 2018). In GC, circRNAs like CircMAPK1, which is a derivative of MAPK1, can bind to MEK1 competitively and prevent the phosphorylation event that controls the MAPK signalling pathway.
This circMAPK1 can downregulate GC progression by inhibiting MAPK1 protein synthesis, demonstrating their role in translation regulation (Wang Y. et al, 2022). CircRNAs can produce novel peptides with oncogenic or tumor-suppressive functions, adding complexity to their role. Subcellular localization is crucial for circRNA function, with many circRNAs found in the cytoplasm, influencing post-transcriptional gene regulation, while others are nuclear, participating in transcriptional regulation (Zang et al., 2022). For instance, hsa_circ_0000467 derived from exosomes promotes GC progression by facilitating nuclear translocation of transcription factors or stabilizing mRNA targets (Jiang et al., 2024). CircAGO2, derived from the AGO2 gene, regulates AGO2-miRNA complexes and cancer progression by interacting with HuR protein and thereby reducing the gene silencing pathways mediated by AGO2/miRNA network (Chen et al., 2019). Another circRNA, circTHBS1, associated with poor prognosis in GC, promotes malignant behaviours and EMT through the INHBA/TGF-β pathway by sponging miR-204-5p and stabilizing HuR-mediated INHBA mRNA (Qiu et al., 2022). In terms of mechanism, circTHBS1 sponges the miR-204-5p to increase INHBA expression. The circularRNA helps in stabilizing HuR-mediated INHBA mRNA, which activates the TGF-β pathway and can promote GC.
4.4 Protein sponge and direct interaction
CircRNAs can directly interact with proteins, influencing their stability, localization, and activity. They may serve as sponges for multiple miRNAs or RBPs, forming complex regulatory networks. CircUBE2Q2, an upregulated regulatory circRNA in GC, promotes tumor metastasis by inhibiting the STAT3 pathway through the circUBE2Q2-miR-370-3p-STAT3 axis and exosomal communication (Yang et al., 2021). Similarly, CircURI1, derived from exons 3 and 4 of URI1, is highly expressed in GC in comparison to the wild type tissues (paraGC). CircURI1 inhibits movements and corresponding metastasis of the GC cells both in vitro and in vivo by interacting with hnRNPM, thereby regulating alternative splicing of migration-related genes and suppressing metastasis (Wang et al., 2020; Wang et al., 2021). Additionally, circMRPS35 impedes the growth and invasion of gastrointestinal cancer cells in vivo and in vitro. Mechanistically, circMRPS35 acts as a scaffold, recruiting histone acetyltransferase KAT7 to the promoters of FOXO1 and FOXO3a, leading to H4K5 acetylation. This enhances the transcription of FOXO1/3a and their downstream targets, including p21, p27, Twist1, and E-cadherin, thereby inhibiting cell proliferation and invasion. Moreover, a positive correlation exists between circMRPS35 and FOXO1/3a expression in GC tissues (Jie et al., 2020). All these observations demonstrate that circRNAs have pivotal roles in regulating biological processes associated with GC metastasis. The biogenesis and functional mechanism of circRNAs are presented in Figure 1.
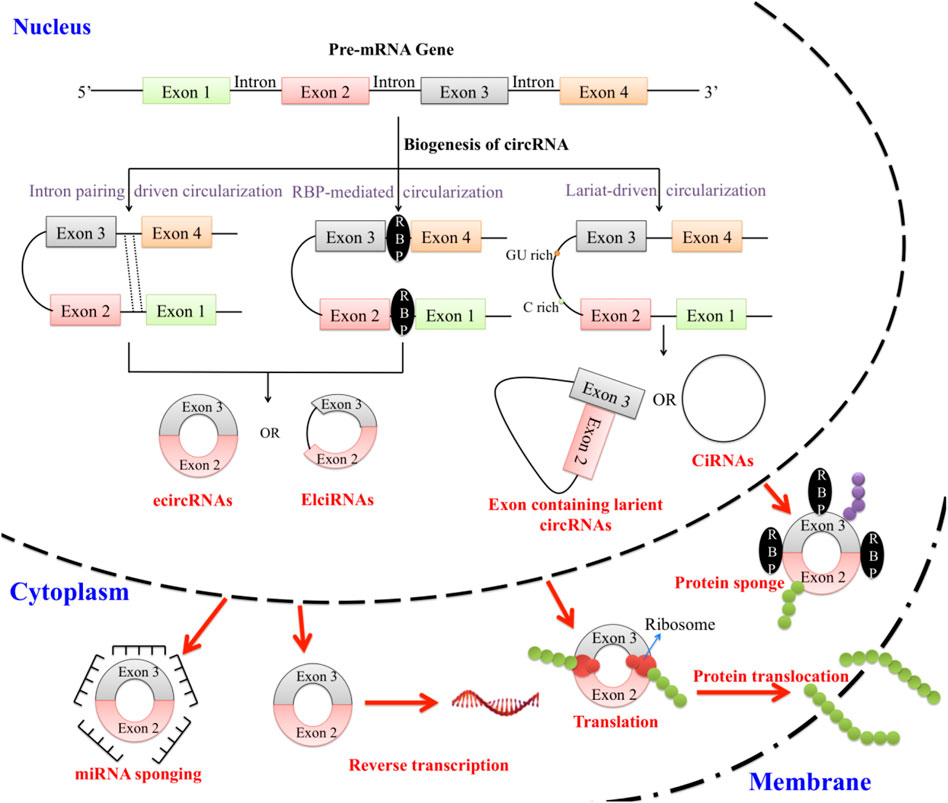
Figure 1. Pre-mRNAs are the parent molecule from which circRNAs are created through reverse splicing. Reverse splicing along with the regular splicing mechanism can exclude the intronic portion to form exonic circRNAs (ecRNAs/ecircRNAs). Few exonic circRNAs have their introns retained and are known as exon-intron circRNAs (EIciRNAs). CircRNAs can also form through intron lariat formation or whole intronic circularization forming intronic circRNAs (ciRNAs). All the pre-mentioned circRNAs are localized inside the nucleus except ecircRNA, which localizes in the cytoplasm. CircRNAs can sponge miRNAs and RBPs. They are also involved in reverse transcription, translation, and protein translocation.
5 CircRNAs as potential biomarkers for GC
5.1 As indicators for diagnosis
Currently, the best biomarker for diagnosing GC early is CA72-4; however, its sensitivity and specificity are not very high. On the other hand, when compared to healthy individuals, circRNAs exhibit unique expression patterns in GC patients, which makes them potentially useful biomarkers for diagnostic purposes of GC while conducting tissue biopsies. Since gastritis is associated with a higher risk of GC, Xie et al. (2018) observed that hsa_circ_0074362 was downregulated in both GC and gastritis tissues, suggesting it could be an early indication of GC (Xie et al., 2018). The levels of hsa_circ_0074362, despite having a poor sensitivity, can be correlated with lymph node metastases and the serum tumor marker CA19-9, suggesting that it may be useful in combination with other markers. A reliable GC biomarker, hsa_circ_0001649 is another circRNA that is downregulated in GC tissues and exhibits great diagnostic accuracy with a sensitivity of 71.1%, and specificity of 81.6% (Li et al., 2017). Moreover, exosomal hsa_circ_0015286 levels in GC patients decrease dramatically after surgery, indicating that it may very well be considered as a tool for the detection and diagnosis of GC in a non-invasive manner exemplifying its capability to act as a potential biomarker (Zheng et al., 2022). Recently, Roy et al. (2022) have discovered a set of eight circRNAs as possible non-invasive indicators for early GC diagnosis.
5.2 As biomarkers in prognosis
Patients with GC may have a better prognosis if secondary prevention measures are taken, such as early detection, diagnosis, and treatment. It is now known that circRNAs may be predictive biomarkers. For instance, CircLARP4, which is primarily present in the cytoplasm, sponging miR-424, affects the behaviour of GC cells. According to Zhang et al. (2017) decreased circLARP4 expression in GC tissues is a reliable indicator of GC patients’ overall survival. In vivo research revealed that circUBE2Q2 knockdown in conjunction with a STAT3 inhibitor synergistically suppresses the growth of GC, indicating that circUBE2Q2 targeting may improve the efficacy of targeted therapies (Yang et al., 2021). A different serum hsa_circ_0007507 expression levels across GC patients with gastritis, intestinal metaplasia, post-surgery, and recurrence cases suggest that it may be a novel biomarker for GC monitoring and diagnosis (Zhang et al., 2021). Furthermore, a substantial downregulation of hsa_circ_0000096 in GC tissues was reported by Li et al. (2017) have strong diagnostic accuracy.
5.3 As therapeutic targets
CircRNAs have been implicated in the development of GC and are important; these findings raise the possibility that circRNAs could be targets for therapeutic intervention. For instance, in GC patients being treated by cisplatin (CDDP), the overexpression of circAKT3 hampers the chances of disease-free survival (DFS) and is independently linked to aggressive tumor characteristics. The expression profile of circAKT3 is found to be higher in the tissues and cells which are resistant to CDDP as compared to their CDDP sensitive counterparts. According to Sun et al. (2018) there is a positive correlation between TNM stage and the expression of CEA in GC plasma, but a negative correlation exists between the concentrations of hsa_circ_0000520 in GC tissues and TNM stage. Hu et al. (2022) found that the novel protein GSPT1-238aa, encoded by circGSPT1, suppresses the growth of GC tumors, suggesting, it may be a special therapeutic target. Circ-MTO1, another circRNA, has been linked to enhanced chemotherapeutic sensitivity, longer DFS, and decreased lymph node metastases in GC (Chang et al., 2022).
Various diagnostic methods have been developed to accurately detect and quantify circRNA expression levels. RNA sequencing (Wu et al., 2022; Szabo and Salzman, 2016) and thereafter different computational tools (Wu et al., 2022; Nguyen et al., 2022; Singh et al., 2022) are needed to detect circRNAs for different purposes.
6 Conclusion and future perspectives
Since the last decade, circRNAs have emerged as significant players in the regulation of gene expression, with growing evidence suggesting their critical roles in GC. One of the most cost-effective ways to detect the interplay between circRNAs is through the bioinformatics approach, which is also found to be coming very fast to catalyze this area of research (Figure 2). These stable, covalently closed RNA molecules, circRNAs exhibit unique characteristics, such as high resistance to exonucleases and the ability to act as miRNA sponges, regulators of transcription, and protein scaffolds. In GC, circRNAs have been implicated in key pathological processes, including protein aggregation, and healthy cell death. Researches indicate that circRNAs can influence disease progression by modulating gene expression networks and interacting with disease-related proteins. Future perspectives on circRNAs in GC involve several promising directions. Advanced high-throughput sequencing and bioinformatics tools will facilitate the comprehensive identification and characterization of circRNAs in diseased versus healthy tissues. Functional studies using in vitro and in vivo models may pave the pathways for the elucidations of the precise functionalities of circRNAs in GC cells and numerous other diseases. Moreover, the development of circRNA-based therapeutics, such as synthetic circRNAs or circRNA inhibitors, holds potential for innovative treatments. Overall, continued research into circRNAs will enhance our understanding of GC and pave the way for novel diagnostic and therapeutic strategies.
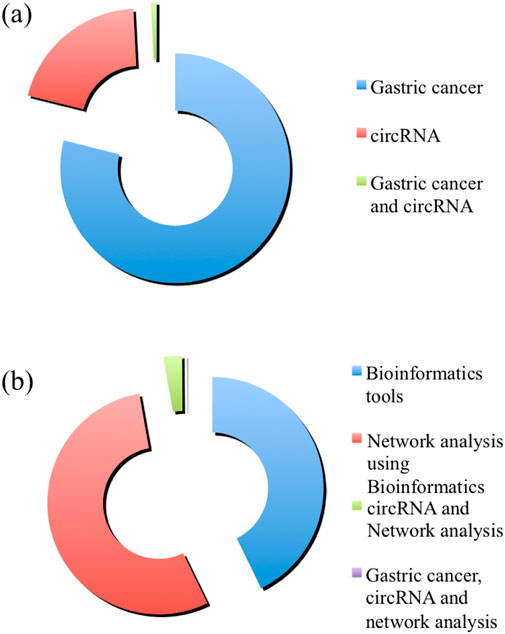
Figure 2. The number of publications in “PubMed” is shown in the chart. Panel (A) shows the amount of publications with the keywords “Gastric cancer” (in blue), “circRNA” (in red), and “Gastric cancer and circRNA” (in green). Panel (B) shows the amount of publications with the keywords “Bioinformatics tools” (in blue), “Network analysis using Bioinformatics” (in red), “circRNA and Network analysis” (in green), and “Gastric cancer, circRNA and network analysis” (in purple).
Author contributions
DG: Writing–original draft. JM: Writing–original draft. AN: Writing–original draft. SB: Writing–original draft. AB: Conceptualization, Formal Analysis, Funding acquisition, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The financial support from the Department of Biotechnology (DBT), Government of India is duly acknowledged. The infrastructural support from the DBT-funded Bioinformatics Infrastructure Facility Centre (Project Sanction no: BT/PR40162/BTIS/137/48/2022, dated 31.10.2022) and National Network Project (Project Sanction no: BT/PR40192/BTIS/137/69/2023, dated 19.12.2023), sanctioned to Angshuman Bagchi, were utilized in this work. Jit Mondal (Student ID: 211610197059) receives funding from UGC.
Acknowledgments
The authors acknowledge the financial and infrastructural support from the Department of Biotechnology (DBT), the Government of India, and the University of Kalyani. The machinery support from the DBT-funded Bioinformatics Infrastructure Facility Centre is duly acknowledged. Dipanjan Guha receives salary from the DBT-funded Bioinformatics Infrastructure Facility Centre. Sima Biswas and Anirban Nandy receives fellowship from DBT funded NNP, Govt. of India.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Bose, R., and Ain, R. (2018). Regulation of transcription by circular RNAs. Adv. Exp. Med. Biol. 1087, 81–94. doi:10.1007/978-981-13-1426-1_7
Brown, J. W., and Simpson, C. G. (1998). Splice site selection in plant pre-mRNA splicing. Annu. Rev. Plant Physiology Plant Mol. Biol. 49, 77–95. doi:10.1146/annurev.arplant.49.1.77
Chang, C., Zheng, A., Wang, P., and Teng, X. (2022). Circular RNA mitochondrial translation optimization 1 correlates with less lymph node metastasis, longer disease free survival, and higher chemotherapy sensitivity in gastric cancer. J. Clin. Laboratory Analysis 36 (6), e23918. doi:10.1002/jcla.23918
Chaudhary, U., and Banerjee, S. (2024). Decoding the non-coding: tools and databases unveiling the hidden world of “junk” RNAs for innovative therapeutic exploration. ACS Pharmacol. and Transl. Sci. 7 (7), 1901–1915. doi:10.1021/acsptsci.3c00388
Chen, Y., Yang, F., Fang, E., Xiao, W., Mei, H., Li, H., et al. (2019). Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death and Differ. 26 (7), 1346–1364. doi:10.1038/s41418-018-0220-6
Conn, S. J., Pillman, K. A., Toubia, J., Conn, V. M., Salmanidis, M., Phillips, C. A., et al. (2015). The RNA binding protein quaking regulates formation of circRNAs. Cell 160 (6), 1125–1134. doi:10.1016/j.cell.2015.02.014
Dai, X., Liu, J., Guo, X., Cheng, A., Deng, X., Guo, L., et al. (2020). Circular RNA circFGD4 suppresses gastric cancer progression via modulating miR-532-3p/APC/β-catenin signalling pathway. Clin. Sci. 134 (13), 1821–1839. doi:10.1042/CS20191043
Di Bella, M. A., and Taverna, S. (2024). Extracellular vesicles: diagnostic and therapeutic applications in cancer. Biology 13 (9), 716. doi:10.3390/biology13090716
Di Timoteo, G., Rossi, F., and Bozzoni, I. (2020). Circular RNAs in cell differentiation and development. Dev. Camb. Engl. 147 (16), dev182725. doi:10.1242/dev.182725
Ebbesen, K. K., Hansen, T. B., and Kjems, J. (2017). Insights into circular RNA biology. RNA Biol. 14 (8), 1035–1045. doi:10.1080/15476286.2016.1271524
Fischer, J. W., and Leung, A. K. (2017). CircRNAs: a regulator of cellular stress. Crit. Rev. Biochem. Mol. Biol. 52 (2), 220–233. doi:10.1080/10409238.2016.1276882
Garlapati, P., Ling, J., Chiao, P. J., and Fu, J. (2021). Circular RNAs regulate cancer-related signaling pathways and serve as potential diagnostic biomarkers for human cancers. Cancer Cell Int. 21 (1), 317. doi:10.1186/s12935-021-02017-4
Ghafouri-Fard, S., Khoshbakht, T., Bahranian, A., Taheri, M., and Hallajnejad, M. (2021). CircMTO1: a circular RNA with roles in the carcinogenesis. Biomed. and Pharmacother. 142, 112025. doi:10.1016/j.biopha.2021.112025
Granados-Riveron, J. T., and Aquino-Jarquin, G. (2016). The complexity of the translation ability of circRNAs. Biochimica Biophysica Acta 1859 (10), 1245–1251. doi:10.1016/j.bbagrm.2016.07.009
Guo, X., Dai, X., Liu, J., Cheng, A., Qin, C., and Wang, Z. (2020). Circular RNA circREPS2 acts as a sponge of miR-558 to suppress gastric cancer progression by regulating RUNX3/β-catenin signaling. Mol. Therapy-Nucleic Acids 21, 577–591. doi:10.1016/j.omtn.2020.06.026
Hansen, T. B. (2018). Improved circRNA identification by combining prediction algorithms. Front. cell Dev. Biol. 6, 20. doi:10.3389/fcell.2018.00020
Haque, S., and Harries, L. W. (2017). Circular RNAs (circRNAs) in health and disease. Genes 8 (12), 353. doi:10.3390/genes8120353
Hong, Y., Qin, H., Li, Y., Zhang, Y., Zhuang, X., Liu, L., et al. (2019). FNDC3B circular RNA promotes the migration and invasion of gastric cancer cells via the regulation of E-cadherin and CD44 expression. J. Cell. physiology 234 (11), 19895–19910. doi:10.1002/jcp.28588
Hu, C., Bai, Y., Liu, J., Wang, Y., He, Q., Zhang, X., et al. (2024). Research progress on the quality control of mRNA vaccines. Expert Rev. vaccines 23 (1), 570–583. doi:10.1080/14760584.2024.2354251
Hu, F., Peng, Y., Chang, S., Luo, X., Yuan, Y., Zhu, X., et al. (2022). Vimentin binds to a novel tumor suppressor protein, GSPT1-238aa, encoded by circGSPT1 with a selective encoding priority to halt autophagy in gastric carcinoma. Cancer Lett. 545, 215826. doi:10.1016/j.canlet.2022.215826
Huang, S., Yang, B., Chen, B. J., Bliim, N., Ueberham, U., Arendt, T., et al. (2017). The emerging role of circular RNAs in transcriptome regulation. Genomics 109 (5-6), 401–407. doi:10.1016/j.ygeno.2017.06.005
Huang, X., Li, Z., Zhang, Q., Wang, W., Li, B., Wang, L., et al. (2019). Circular RNA AKT3 upregulates PIK3R1 to enhance cisplatin resistance in gastric cancer via miR-198 suppression. Mol. Cancer 18 (1), 71. doi:10.1186/s12943-019-0969-3
Hwang, H. J., and Kim, Y. K. (2024). Molecular mechanisms of circular RNA translation. Exp. and Mol. Med. 56 (6), 1272–1280. doi:10.1038/s12276-024-01220-3
Jiang, T., Xu, L., Qu, X., Li, R., Cheng, Y., and He, H. (2024). Hsa_circ_0014606 derived from exosomes promotes gastric carcinoma tumorigenesis and proliferation by sponging miR-514b-3p to upregulate HNRNPC. Dig. Dis. Sci. 69 (3), 811–820. doi:10.1007/s10620-023-08254-z
Jie, M., Wu, Y., Gao, M., Li, X., Liu, C., Ouyang, Q., et al. (2020). CircMRPS35 suppresses gastric cancer progression via recruiting KAT7 to govern histone modification. Mol. cancer 19, 56–16. doi:10.1186/s12943-020-01160-2
Khanipouyani, F., Akrami, H., and Fattahi, M. R. (2021). Circular RNAs as important players in human gastric cancer. Clin. Transl. Oncol. 23 (1), 10–21. doi:10.1007/s12094-020-02419-2
Li, P., Chen, H., Chen, S., Mo, X., Li, T., Xiao, B., et al. (2017). Circular RNA 0000096 affects cell growth and migration in gastric cancer. Br. J. cancer 116 (5), 626–633. doi:10.1038/bjc.2016.451
Li, R., Jiang, J., Shi, H., Qian, H., Zhang, X., and Xu, W. (2019). CircRNA: a rising star in gastric cancer. Cell. Mol. Life Sci. 77 (9), 1661–1680. doi:10.1007/s00018-019-03345-5
Li, X. W., Yang, W. H., and Xu, J. (2020a). Circular RNA in gastric cancer. Chin. Med. J. 133 (15), 1868–1877. doi:10.1097/CM9.0000000000000908
Li, Y., Gong, Y., Ma, J., and Gong, X. (2020b). Overexpressed circ-RPL15 predicts poor survival and promotes the progression of gastric cancer via regulating miR-502-3p/OLFM4/STAT3 pathway. Biomed. and Pharmacother. 127, 110219. doi:10.1016/j.biopha.2020.110219
Li, Z., and Xing, J. (2024). Potential therapeutic applications of circular RNA in acute kidney injury. Biomed. and Pharmacother. 174, 116502. doi:10.1016/j.biopha.2024.116502
Lin, X., Huang, C., Chen, Z., Wang, H., and Zeng, Y. (2020). CircRNA_100876 is upregulated in gastric cancer (GC) and promotes the GC cells’ growth, migration and invasion via miR-665/YAP1 signaling. Front. Genet. 11, 546275. doi:10.3389/fgene.2020.546275
Liu, H., Liu, Y., Bian, Z., Zhang, J., Zhang, R., Chen, X., et al. (2018). Circular RNA YAP1 inhibits the proliferation and invasion of gastric cancer cells by regulating the miR-367-5p/p27 Kip1 axis. Mol. Cancer 17 (1), 151. doi:10.1186/s12943-018-0902-1
Liu, J., Dai, X., Guo, X., Cheng, A., Mac, S. M., and Wang, Z. (2020). Circ-OXCT1 suppresses gastric cancer EMT and metastasis by attenuating TGF-β pathway through the circ-OXCT1/miR-136/SMAD4 Axis. OncoTargets Ther. 13, 3987–3998. doi:10.2147/OTT.S239789
Liu, Y., Cao, J., Zhu, L., Zhao, W., Zhou, Y., Shao, C., et al. (2022b). Circular RNA circPGD contributes to gastric cancer progression via the sponging miR-16-5p/ABL2 axis and encodes a novel PGD-219aa protein. Cell death Discov. 8 (1), 384. doi:10.1038/s41420-022-01177-0
Liu, Y., Cheng, X., Li, H., Hui, S., Zhang, Z., Xiao, Y., et al. (2022a). Non-coding RNAs as novel regulators of neuroinflammation in Alzheimer's disease. Front. Immunol. 13, 908076. doi:10.3389/fimmu.2022.908076
López-Jiménez, E., Rojas, A. M., and Andrés-León, E. (2018). RNA sequencing and prediction tools for circular RNAs analysis. Circular RNAs Biogenesis Funct. 1087, 17–33. doi:10.1007/978-981-13-1426-1_2
Lu, J., Wang, Y. H., Yoon, C., Huang, X. Y., Xu, Y., Xie, J. W., et al. (2020). Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877–3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 471, 38–48. doi:10.1016/j.canlet.2019.11.038
Ma, H. B., Yao, Y. N., Yu, J. J., Chen, X. X., and Li, H. F. (2018). Extensive profiling of circular RNAs and the potential regulatory role of circRNA-000284 in cell proliferation and invasion of cervical cancer via sponging miR-506. Am. J. Transl. Res. 10 (2), 592–604.
Matsuoka, T., and Yashiro, M. (2023). Novel biomarkers for early detection of gastric cancer. World J. Gastroenterology 29 (17), 2515–2533. doi:10.3748/wjg.v29.i17.2515
Min, X., Liu, D. L., and Xiong, X. D. (2021). Circular RNAs as competing endogenous RNAs in cardiovascular and cerebrovascular diseases: molecular mechanisms and clinical implications. Front. Cardiovasc. Med. 8, 682357. doi:10.3389/fcvm.2021.682357
Nguyen, M. H., Nguyen, H. N., and Vu, T. N. (2022). Evaluation of methods to detect circular RNAs from single-end RNA-sequencing data. BMC genomics 23 (1), 106. doi:10.1186/s12864-022-08329-7
Okholm, T. L. H., Sathe, S., Park, S. S., Kamstrup, A. B., Rasmussen, A. M., Shankar, A., et al. (2020). Transcriptome-wide profiles of circular RNA and RNA-binding protein interactions reveal effects on circular RNA biogenesis and cancer pathway expression. Genome Med. 12 (1), 112. doi:10.1186/s13073-020-00812-8
Pisignano, G., Michael, D. C., Visal, T. H., Pirlog, R., Ladomery, M., and Calin, G. A. (2023). Going circular: history, present, and future of circRNAs in cancer. Oncogene 42 (38), 2783–2800. doi:10.1038/s41388-023-02780-w
Qiu, S., Li, B., Xia, Y., Xuan, Z., Li, Z., Xie, L., et al. (2022). CircTHBS1 drives gastric cancer progression by increasing INHBA mRNA expression and stability in a ceRNA- and RBP-dependent manner. Cell Death and Dis. 13 (3), 266. doi:10.1038/s41419-022-04720-0
Ren, Y., Manoharan, T., Liu, B., Cheng, C. Z. M., En Siew, B., Cheong, W. K., et al. (2024). Circular RNA as a source of neoantigens for cancer vaccines. J. Immunother. Cancer 12 (3), e008402. doi:10.1136/jitc-2023-008402
Roy, S., Kanda, M., Nomura, S., Zhu, Z., Toiyama, Y., Taketomi, A., et al. (2022). Diagnostic efficacy of circular RNAs as noninvasive, liquid biopsy biomarkers for early detection of gastric cancer. Mol. Cancer 21 (1), 42. doi:10.1186/s12943-022-01527-7
Schneider, T., Schreiner, S., Preußer, C., Bindereif, A., and Rossbach, O. (2018). Northern blot analysis of circular RNAs. Circular RNAs Methods Protoc. 1724, 119–133. doi:10.1007/978-1-4939-7562-4_10
Shi, H., Zhou, Y., Jia, E., Liu, Z., Pan, M., Bai, Y., et al. (2022). Comparative analysis of circular RNA enrichment methods. RNA Biol. 19 (1), 55–67. doi:10.1080/15476286.2021.2012632
Shi, Y., Jia, X., and Xu, J. (2020). The new function of circRNA: translation. Clin. and Transl. Oncol. 22 (12), 2162–2169. doi:10.1007/s12094-020-02371-1
Singh, D., Kesharwani, P., Alhakamy, N. A., and Siddique, H. R. (2022). Accentuating CircRNA-miRNA-transcription factors Axis: a conundrum in cancer research. Front. Pharmacol. 12, 784801. doi:10.3389/fphar.2021.784801
Su, M., Xiao, Y., Ma, J., Tang, Y., Tian, B., Zhang, Y., et al. (2019). Circular RNAs in Cancer: emerging functions in hallmarks, stemness, resistance and roles as potential biomarkers. Mol. cancer 18 (1), 90. doi:10.1186/s12943-019-1002-6
Sun, H., Tang, W., Rong, D., Jin, H., Fu, K., Zhang, W., et al. (2018). Hsa_circ_0000520, a potential new circular RNA biomarker, is involved in gastric carcinoma. Cancer Biomarkers 21 (2), 299–306. doi:10.3233/CBM-170379
Sun, H., Wang, Q., Yuan, G., Quan, J., Dong, D., Lun, Y., et al. (2020). Hsa_circ_0001649 restrains gastric carcinoma growth and metastasis by downregulation of miR-20a. J. Clin. Laboratory Analysis 34 (6), e23235. doi:10.1002/jcla.23235
Szabo, L., and Salzman, J. (2016). Detecting circular RNAs: bioinformatic and experimental challenges. Nat. Rev. Genet. 17 (11), 679–692. doi:10.1038/nrg.2016.114
van Zonneveld, A. J., Kölling, M., Bijkerk, R., and Lorenzen, J. M. (2021). Circular RNAs in kidney disease and cancer. Nat. Rev. Nephrol. 17 (12), 814–826. doi:10.1038/s41581-021-00465-9
Verduci, L., Tarcitano, E., Strano, S., Yarden, Y., and Blandino, G. (2021). CircRNAs: role in human diseases and potential use as biomarkers. Cell Death and Dis. 12 (5), 468. doi:10.1038/s41419-021-03743-3
Wan, Y., and Hopper, A. K. (2018). Size matters: conserved proteins function in length-dependent nuclear export of circular RNAs. Genes and Dev. 32 (9-10), 600–601. doi:10.1101/gad.316216.118
Wang, K. W., and Dong, M. (2019). Role of circular RNAs in gastric cancer: recent advances and prospects. World J. Gastrointest. Oncol. 11 (6), 459–469. doi:10.4251/wjgo.v11.i6.459
Wang, X., Li, J., Bian, X., Wu, C., Hua, J., Chang, S., et al. (2021). CircURI1 interacts with hnRNPM to inhibit metastasis by modulating alternative splicing in gastric cancer. Proc. Natl. Acad. Sci. 118 (33), e2012881118. doi:10.1073/pnas.2012881118
Wang, X., Zhang, J., Cao, G., Hua, J., Shan, G., and Lin, W. (2022a). Emerging roles of circular RNAs in gastric cancer metastasis and drug resistance. J. Exp. and Clin. cancer Res. CR 41 (1), 218. doi:10.1186/s13046-022-02432-z
Wang, Y., Li, Z., Xu, S., and Guo, J. (2020). Novel potential tumor biomarkers: circular RNAs and exosomal circular RNAs in gastrointestinal malignancies. J. Clin. Laboratory Analysis 34 (7), e23359. doi:10.1002/jcla.23359
Wang, Y., Wu, C., Du, Y., Li, Z., Li, M., Hou, P., et al. (2022b). Expanding uncapped translation and emerging function of circular RNA in carcinomas and noncarcinomas. Mol. Cancer 21 (1), 13. doi:10.1186/s12943-021-01484-7
Wen, G., Zhou, T., and Gu, W. (2021). The potential of using blood circular RNA as liquid biopsy biomarker for human diseases. Protein and Cell 12 (12), 911–946. doi:10.1007/s13238-020-00799-3
Wilusz, J. E. (2018). A 360° view of circular RNAs: from biogenesis to functions. Wiley Interdiscip. Rev. RNA 9 (4), e1478. doi:10.1002/wrna.1478
Wu, W., Zhang, J., Cao, X., Cai, Z., and Zhao, F. (2022). Exploring the cellular landscape of circular RNAs using full-length single-cell RNA sequencing. Nat. Commun. 13 (1), 3242. doi:10.1038/s41467-022-30963-8
Xie, Y. I., Shao, Y., Sun, W., Ye, G., Zhang, X., Xiao, B., et al. (2018). Downregulated expression of hsa_circ_0074362 in gastric cancer and its potential diagnostic values. Biomarkers Med. 12 (1), 11–20. doi:10.2217/bmm-2017-0114
Xu, C. Y., Zeng, X. X., Xu, L. F., Liu, M., and Zhang, F. (2022). Circular RNAs as diagnostic biomarkers for gastric cancer: a comprehensive update from emerging functions to clinical significances. Front. Genet. 13, 1037120. doi:10.3389/fgene.2022.1037120
Yang, C. M., Qiao, G. L., Song, L. N., Bao, S., and Ma, L. J. (2020). Circular RNAs in gastric cancer: biomarkers for early diagnosis. Oncol. Lett. 20 (1), 465–473. doi:10.3892/ol.2020.11623
Yang, J., Zhang, X., Cao, J., Xu, P., Chen, Z., Wang, S., et al. (2021). Circular RNA UBE2Q2 promotes malignant progression of gastric cancer by regulating signal transducer and activator of transcription 3-mediated autophagy and glycolysis. Cell death and Dis. 12 (10), 910. doi:10.1038/s41419-021-04216-3
Yu, T., Ran, L., Zhao, H., Yin, P., Li, W., Lin, J., et al. (2021). Circular RNA circ-TNPO3 suppresses metastasis of GC by acting as a protein decoy for IGF2BP3 to regulate the expression of MYC and SNAIL. Mol. Therapy-Nucleic Acids 26, 649–664. doi:10.1016/j.omtn.2021.08.029
Zaiou, M. (2019). Circular RNAs as potential biomarkers and therapeutic targets for metabolic diseases. Adv. Exp. Med. Biol. 1134, 177–191. doi:10.1007/978-3-030-12668-1_10
Zang, J., Lu, D., and Xu, A. (2020). The interaction of circRNAs and RNA binding proteins: an important part of circRNA maintenance and function. J. Neurosci. Res. 98 (1), 87–97. doi:10.1002/jnr.24356
Zang, X., Jiang, J., Gu, J., Chen, Y., Wang, M., Zhang, Y., et al. (2022). Circular RNA EIF4G3 suppresses gastric cancer progression through inhibition of β-catenin by promoting δ-catenin ubiquitin degradation and upregulating SIK1. Mol. Cancer 21 (1), 141. doi:10.1186/s12943-022-01606-9
Zhang, B., Li, Z., Ye, G., and Hu, K. (2024). Biologic activity and treatment resistance to gastrointestinal cancer: the role of circular RNA in autophagy regulation. Front. Oncology, 14. 1393670, doi:10.3389/fonc.2024.1393670
Zhang, J., Liu, H., Hou, L., Wang, G., Zhang, R., Huang, Y., et al. (2017). Circular RNA_LARP4 inhibits cell proliferation and invasion of gastric cancer by sponging miR-424-5p and regulating LATS1 expression. Mol. Cancer 16 (1), 151. doi:10.1186/s12943-017-0719-3
Zhang, W., Zheng, M., Kong, S., Li, X., Meng, S., Wang, X., et al. (2021). Circular RNA hsa_circ_0007507 may serve as a biomarker for the diagnosis and prognosis of gastric cancer. Front. Oncol. 11, 699625. doi:10.3389/fonc.2021.699625
Zhang, X. O., Dong, R., Zhang, Y., Zhang, J. L., Luo, Z., Zhang, J., et al. (2016). Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res. 26 (9), 1277–1287. doi:10.1101/gr.202895.115
Zhao, X., Zhong, Y., Wang, X., Shen, J., and An, W. (2022). Advances in circular RNA and its applications. Int. J. Med. Sci. 19 (6), 975–985. doi:10.7150/ijms.71840
Zheng, P., Gao, H., Xie, X., and Lu, P. (2022). Plasma exosomal hsa_circ_0015286 as a potential diagnostic and prognostic biomarker for gastric cancer. Pathology Oncol. Res. 28, 1610446. doi:10.3389/pore.2022.1610446
Zhong, S., Wang, J., Zhang, Q., Xu, H., and Feng, J. (2018). CircPrimer: a software for annotating circRNAs and determining the specificity of circRNA primers. BMC Bioinforma. 19, 292–295. doi:10.1186/s12859-018-2304-1
Zhou, S., Chen, R., She, Y., Liu, X., Zhao, H., Li, C., et al. (2022). A new perspective on depression and neuroinflammation: non-coding RNA. J. Psychiatric Res. 148, 293–306. doi:10.1016/j.jpsychires.2022.02.007
Keywords: circRNA, biogenesis, gastric cancer, early diagnosis, therapeutics
Citation: Guha D, Mondal J, Nandy A, Biswas S and Bagchi A (2024) Interplay of circular RNAs in gastric cancer - a systematic review. Front. Syst. Biol. 4:1497510. doi: 10.3389/fsysb.2024.1497510
Received: 17 September 2024; Accepted: 31 October 2024;
Published: 13 November 2024.
Edited by:
Sema Misir, Cumhuriyet University, TürkiyeReviewed by:
Osman Akıdan, Mengücek Gazi Education and Research Hospital, TürkiyeImran Ince Akca, Tokat Gaziosmanpaşa University, Türkiye
Copyright © 2024 Guha, Mondal, Nandy, Biswas and Bagchi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Angshuman Bagchi, YW5nc2h1bWFuYkBnbWFpbC5jb20=, YW5nc2h1QGtseXVuaXYuYWMuaW4=
†These authors have contributed equally to this work and share first authorship
 Dipanjan Guha
Dipanjan Guha Jit Mondal
Jit Mondal Anirban Nandy
Anirban Nandy Sima Biswas
Sima Biswas Angshuman Bagchi
Angshuman Bagchi