
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Synth. Biol., 07 March 2025
Sec. Genetic Systems and Circuit Design
Volume 3 - 2025 | https://doi.org/10.3389/fsybi.2025.1548572
This article is part of the Research TopicEngineering the Future: Advances in Synthetic Gene Regulatory NetworksView all articles
Devices sensing inputs and generating outputs are fundamental regulatory units, and as such are the basis of more complex regulatory networks. We provide an overview of regulatory devices used as fundamental regulatory building blocks in synthetic biology, and how complex genetic circuitry is being constructed from them. We first comprehensively explore devices operating at different levels of gene regulation, with action modes on the DNA sequence, to transcriptional, translational and post-translational control. We then discuss design principles of constructing genetic circuits from basic regulatory units, addressing challenges such as orthogonality, context-dependence, noise, and complexity. We present examples of genetic circuitry, including bistable switches, logic gates, signal amplification, memory devices and circuitry for biocomputation. How artificial genetic circuitry can be useful in real-life applications is illustrated with examples from bioproduction, living therapeutics, and biosafety. Our aim is to provide a comprehensive overview of the toolbox of regulatory devices and a profound understanding of their potential for constructing diverse genetic circuits and their applications.
Sensing and reacting to external and internal stimuli is a fundamental property of all living systems. This capability is enabled by molecular regulatory systems that can sense a specific signal (“sensor”) and create an output in response to that signal (“effector” or “actuator”). Typically, several of such regulatory systems can interface with one another to e.g., integrate, amplify, or remember signals, forming regulatory networks.
In synthetic biology, a discipline dedicated to engineering life, engineering goals frequently focus on rational programming of cellular behavior in response to defined input signals. For this purpose, regulatory systems have frequently been lifted from nature and “re-wired”, meaning put into different regulatory contexts. Increasingly, entirely new synthetic regulatory systems are being developed. By now, the synthetic biologist’s toolbox boasts a staggering selection of regulatory devices to choose from, with a variety of modes of action.
The ability to engineer cellular behavior through synthetic regulatory systems has enabled numerous applications across biotechnology and medicine, from sustainable bioproduction to therapeutic applications. As the field matures, increasing emphasis is being placed on creating robust and predictable systems through careful characterization of parts, adherence to engineering principles, and computational approaches for automated design of genetic parts and circuits.
This review aims to provide a comprehensive overview of the current state-of-the-art toolkit of regulatory parts for synthetic circuit design and illustrates their implementation into more sophisticated devices and systems through selected examples. First, we present a thorough survey of regulatory devices, from DNA-based controls to post-translational regulation. We then explore fundamental design principles and considerations for constructing artificial genetic circuitry and illustrate these with selected examples of typical circuit architectures and functions. Finally, we highlight some applications that demonstrate the implementation of sophisticated artificial regulatory systems in real-world contexts.
Molecular devices that sense inputs and generate outputs are the fundamental units of gene regulatory networks, both natural and synthetic ones. Regulatory devices have been used and further developed based on a diverse array of molecular mechanisms, enabling control at multiple levels of gene expression. In this section, we present a comprehensive overview of these devices, organized by their mode of action, from affecting DNA sequence, target gene transcription, translation, or post-translational effects on the target protein (Figure 1).
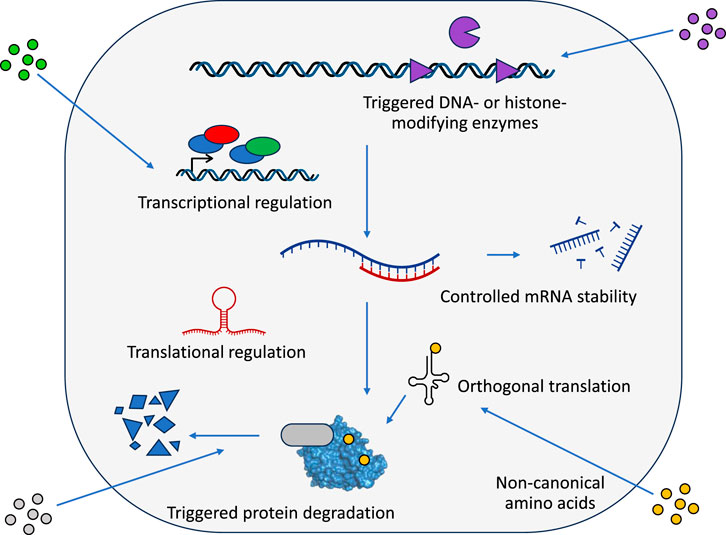
Figure 1. Overview of regulatory devices acting on different levels of gene expression. Schematic representation of various control points for gene regulation and the corresponding regulatory devices employed in synthetic biology. DNA-level regulation (top) includes site-specific recombinases that can invert or excise DNA segments, CRISPR-based systems for targeted DNA modifications, and epigenetic regulators that can modify DNA or histones to control gene accessibility. Transcriptional control (middle) encompasses prokaryotic and eukaryotic transcription factors, synthetic transcription factors based on programmable DNA-binding domains, orthogonal RNA polymerases and sigma factors, and RNA-based regulation through riboswitches. Translational regulation (bottom) includes RNA structure-based controllers such as riboswitches and toehold switches, as well as RNA interference mechanisms. Post-translational control can be achieved through conditional protein degradation, protein localization, or protein activity modulation. Regulatory devices can be made responsive to various inputs including small molecules, light, temperature, and macromolecules.
For modulating the activity of a target gene, its presence and integrity on the DNA level is a first point of possible interference along the flow of genetic information. Being permanent and inheritable, conditional alterations on the DNA sequence are particularly well-suited to implement devices intended to have stable states, such as bistable switches or higher-order memory devices, which are being discussed in more detail in subsequent sections.
Commonly used effectors for genetic circuit control belong to the family of tyrosine recombinases (e.g., lambda, Cre, Flp, FimB/FimE) and serine integrases (e.g., Bxb1, PhiC31). Gene expression regulation is commonly achieved by inversion of DNA segments, thus controlling whether or not a promoter is aligned with the target gene, resulting in a distinct stable ON or OFF state on that DNA molecule. This approach had initially been used to create inducible expression systems in bacteria leveraging tyrosine recombinases of different origins for construction of unidirectional switches, e.g., the integrase of the lambda bacteriophage (Podhajska et al., 1985; Sektas et al., 2001), flippase (Flp) recombinase from the 2 micron plasmid of Saccharomyces cerevisiae (Sektas and Szybalski, 1998), and FimE recombinase from Escherichia coli (Ham et al., 2006). Moving beyond sole control of heterologous gene expression, the latter was used to switch between chemotactic systems, thus regulating cell behavior (Moon et al., 2011). The FimB/FimE system in its endogenous context controls the presence or absence of type 1 fimbriae much in the same way, but works as a bidirectional switch (Abraham et al., 1985). Designed bidirectional switchability can be achieved using a pair of unidirectionally active recombinases catalyzing the opposite recombination reaction (Fernandez-Rodriguez et al., 2015), or using a serine integrase together with a cognate excisionase controlling the directionality of the integrase reaction (Bonnet et al., 2012). Interleaving recombined rafts can create inheritable states that scale exponentially with the number of used recombinases (Ham et al., 2008; Roquet et al., 2016). Using suitable topologies, recombinase-driven inversions have been employed to implement counting circuitry (Friedland et al., 2009) and numerous Boolean logic gates (Bonnet et al., 2013; Siuti et al., 2013). Irreversible deletions have also been used to regulate gene activity in such circuitry, for instance by transcriptional terminators removable through recombinase activity (Weinberg et al., 2017). These fundamental recombinase-based regulatory units serve as building blocks for more complex genetic circuits, which will be discussed in detail in Section 3 (Design principles and examples of genetic circuits).
Regulation of recombinase activity is usually achieved through controlling their expression in response to external stimuli, typically through transcriptional regulation systems. However, it is also possible to interfere with their local activity at specific sites using switchable transcription factors (discussed in Section 2.3) for circuit control (Short et al., 2023). In eukaryotes, global activity can be made conditional by fusing the recombinase to the ligand binding domain of the estrogen receptor. This has been done for instance for Cre recombinase (Metzger et al., 1995) and flippase recombinase (Hunter et al., 2005), making their activity dependent on estrogen receptor agonists. This kind of inducible recombinase technology quickly after its inception was adopted for inducible gene knockouts in whole animals (Feil et al., 1996).
Recombinase activity has also been made light-dependent, allowing optogenetic device control. One possibility to control its activity is by splitting the recombinase, and reconstituting it through a light-inducible dimerization system (Kawano et al., 2016; Morikawa et al., 2020; Jung et al., 2019). Another way uses the plant-derived light receptor domain LOV2, which unfolds a C-terminal helix from the protein core upon blue-light illumination. Through a suitable chimeric fusion to LOV2, Cre recombinase activity has been made dependent on blue-light illumination (Duplus-Bottin et al., 2021).
Aside from tyrosine recombinases and serine integrases, CRISPR-Cas-derived devices can also be used as effectors acting on the DNA sequence. The advantage of Cas nuclease-based devices is that sequence specificity is determined through the sequence of guide RNA. This RNA programmability of Cas nucleases has been the basis for engineering synthetic gene editing devices that do not introduce double-strand breaks. Base editors, consisting of a Cas9 nickase with a cytidine (Komor et al., 2016) or adenosine deaminase (Gaudelli et al., 2017), allow targeted single nucleotide changes. So-called prime editors, consisting of a Cas9 nickase and reverse transcriptase, allow more complex site-directed edits (Anzalone et al., 2019). Further, Cas1-Cas2 integrase has been used for sequential insertions of arbitrary DNA sequences (Shipman et al., 2016). In the context of synthetic biology devices, the mentioned Cas-based effectors have a prominent role in memory devices for ‘recording’ of internal or external stimuli (see 3.3.5 Synthetic Memory Circuit).
Beyond direct DNA sequence modifications, synthetic regulatory systems have been developed that enable programmable epigenetic control through modifications of DNA bases and histones. Park et al. established an orthogonal epigenetic regulatory system using N6-methyladenine (m6A) DNA modifications. They engineered a synthetic initiator module combining E. coli DNA adenine methyltransferase (Dam) as a writer domain with a zinc finger protein for sequence-specific targeting. The reader module was constructed by fusing the m6A-binding domain of DpnI with various transcriptional effector domains, allowing m6A marks to be translated into defined transcriptional outputs. This combination of engineered writers and readers created circuits capable of establishing and propagating stable transcriptional states (Park et al., 2019).
While this system relies on direct DNA base modification, epigenetic regulation can also be achieved through modifications of histone proteins that affect chromatin state and accessibility. The CRISPRoff/CRISPRon system demonstrates this broader approach by combining dead Cas9 (dCas9) with either a DNA methyltransferase (DNMT3A/3L) and a transcriptional repressor (KRAS) for programmable epigenetic silencing (CRISPRoff) or by combining dCas9 with a demethylase (TET, ten-eleven translocation family enzyme) to remove the methylation mark (CRISPRon). This combination creates stable and heritable gene silencing that can be reversed when desired (Nuñez et al., 2021).
Transcriptional regulation in the lac operon was the first gene regulation mechanism to be understood on a molecular level thanks to Jacob and Monod’s seminal work on regulation of lactose metabolism in E. coli (Jacob and Monod, 1961). In nature, transcriptional gene regulation systems consist of two core elements: binding sites on the DNA (the ‘cis’ acting element) within or near a promoter whose activity is regulated, and a transcription factor (the ‘trans’ acting element) whose binding to its cognate binding site alters that promoter’s activity. The transcription factor itself can possess signal-sensing capability, or have its expression or activity regulated by another system. Such transcriptional regulation devices have seen extensive adoption and are likely the most commonly used mode to control target gene expression in recombinant systems. This section covers both transcription factor-based systems and alternative approaches using orthogonal polymerases and RNA-based regulatory motifs.
Owing to their comparative simplicity with few necessary components, prokaryotic devices have been used extensively. Prototypical examples are allosteric transcription factors and their binding sites from bacterial operons like the lactose operon (Lewis, 2005), the arabinose operon (Schleif, 2000), and the tetracycline operon (Bertram and Hillen, 2008). Generally, transcription factor binding may be inhibitory on transcription (negative regulation), like in the case of the lactose and tetracycline repressors (LacI, TetR), or activate transcription (positive regulation), which is the case for the majority of eukaryotic transcription factors. AraC, the regulatory protein of the arabinose operon does both: it acts as an inhibitor in the absence of its inducer arabinose, and as an activator in its presence (Schleif, 2000).
Regulatory devices found in nature have frequently been engineered, for instance to improve, shift or invert the dynamic range of the response, or to alter the stimulus eliciting a response. Both cis and trans elements have been subject to engineering efforts. Particularly for the paradigmatic lacO/LacI system there is a wealth of respective studies engineering its properties [reviewed in (Hersey et al., 2023)]. In this system, an allosteric transcription factor, the lac repressor (LacI), silences promoter activity by binding to the lac operator in the absence of its inducer. In its natural context, a degree of leaky expression is required (Jobe and Bourgeois, 1972), whereas that characteristic is typically undesirable in designed circuitry. One approach to make the system more stringent is through engineering the operator for tighter binding of the transcription factor (Sadler et al., 1983; Milk et al., 2010). However, there is a much larger body of research on engineering of the transcription factor, from making control more stringent, improving inducibility (Satya Lakshmi and Rao, 2009), to profound functional changes such as inverting the response to its inducer (Poelwijk et al., 2011; Hoffmann et al., 2016; Richards et al., 2017; Groseclose et al., 2020), or altering its inducer (Taylor et al., 2016) or operator specificity (Lehming et al., 1987; Milk et al., 2010).
The LacI/GalR transcription factor family with >500 known members is extremely well characterized and functionally understood (Sousa et al., 2016). Swapping domains within this protein family has allowed creating functional chimeric transcription factors (Tungtur et al., 2007; Shis et al., 2014; Dimas et al., 2019; Jiang et al., 2021), a strategy also working within other prokaryotic transcription factor families [reviewed in (Chan et al., 2024)]. Apart from engineering the regulatory elements themselves, the system’s response may also be tuned by changing how import of the inducer is regulated. For instance, the arabinose inducible system has been converted from an autocatalytic regulation with an all-or-nothing response to a system with titratable induction by decoupling expression of the arabinose importer (Khlebnikov et al., 2000).
Regulatory devices of prokaryotic origins are also widely being used in eukaryotic systems. For instance, the lac repressor with carefully placed lac operators can be used to negatively regulate gene expression in mammalian cells from viral promoters (Chan et al., 2024; Khlebnikov et al., 2000; Brown et al., 1987), and even endogenous promoters (Hannan et al., 1993). However, it is more common to fuse prokaryotic transcription factors to transactivating domains for use in eukaryotic systems. There are numerous examples of genetic switches engineered for eukaryotic systems in which a prokaryotic allosteric transcription factor acts as DNA binding and ligand sensing part, and a fused transcriptional effector domain as the actuator. This has been done for the lac repressor (Labow et al., 1990), but more commonly used are systems based on the tetracycline repressor (TetR), typically employing doxycycline as effector [reviewed in (Das et al., 2016)]. Those TetR based devices have been developed for diverse eukaryotes, from yeast (Garí et al., 1997), filamentous fungi (Wanka et al., 2016), to mammalian cells (Gossen and Bujard, 1992) and plants (Weinmann et al., 1994). While fusions with the original TetR result in gene de-activation upon effector addition (so called ‘Tet-off’ systems), using a logic-inverted TetR variant has allowed doxycycline-inducible transcription activation (“Tet-on”) (Gossen et al., 1995). The system was later improved through directed evolution to create Tet-On3G, featuring enhanced doxycycline sensitivity, lower background expression and an optimized promoter (Zhou et al., 2006).
There are also notable widely used transcription regulation systems lifted from eukaryotes. A paradigmatic example originates from yeast’s GAL regulon, which responds to the availability of galactose. The principal regulatory elements in this regulon are the transcriptional activator Gal4, its inhibitor Gal80, the signal transducer Gal3 and the cognate cis elements, which Gal4 is binding to, called upstream activating sequences (UAS). In the presence of galactose, Gal3 sequesters Gal80, allowing Gal4 to specifically bind to its UAS and activate transcription (Rajeshkannan et al., 2022). This system has been widely used in yeast, employing Gal4-activated promoters for galactose-inducible expression of transgenes (Ro et al., 2006). However, as Gal4 regulates endogenous genes in yeast and induction involves a change of the supplied carbon source, use of this system comes with pleiotropic effects. To avoid perturbations by the change of carbon source, chimeric Gal4-based transcription factors have been made that bind to the GAL UAS in reaction to estradiol. This has been accomplished by fusing the Gal4 DNA binding domain and a strong viral activation domain with the hormone-binding domain of the estradiol receptor, a type of nuclear receptor (Louvion et al., 1993; McIsaac et al., 2011). In the absence of its inducer, it is sequestered in the cytoplasm, rendering the chimeric transcription factor inactive. Upon addition of estradiol, it is translocated to the nucleus and thereby activated. However, estradiol induction of Gal4 still leads to activation of GAL responsive genes. In higher eukaryotes, Gal4 is orthogonal and Gal4-mediated transcription activation of UAS associated promoters has been used in cell culture (Webster et al., 1988; Kakidani and Ptashne, 1988), as well as in whole animals, both in invertebrates (Brand and Perrimon, 1993), and vertebrates (Hartley et al., 2002; Köster and Fraser, 2001; Rowitch et al., 1999). In fact, Gal4/UAS systems have become a foundational tool for genetics studies in Drosophila (Duffy, 2002).
Another distinct class of eukaryotic transcription factors widely used in engineered genetic systems are nuclear receptors, to which the estradiol receptor mentioned above belongs to. These proteins found in animals typically respond to lipophilic effectors, such as steroids or retinoids acting as hormones or vitamins (Sladek, 2011). When bound to their cognate response elements on the DNA, nuclear receptors in their unliganded state are either inactive or actively silencing their target genes through recruitment of co-repressors. Upon ligand binding, they switch to recruiting co-activators and thereby activate transcription of their target genes. In mammalian cells, the glucocorticoid receptor has been adopted to regulate transgene expression (Ko et al., 1989; James et al., 2000), typically with dexamethasone as inducer. To avoid crosstalk with endogenous regulation, host-orthogonal systems have been used, such as an insect ecdysone receptor in mammalian cells (Christopherson et al., 1992) or a mammalian steroid receptor in plant cells (Schena et al., 1991).
In eukaryotes, another important class of molecular receptors are the membrane-bound G-protein coupled receptors (GPCRs). They are the most versatile class in terms of recognized ligands, ranging from various small molecules to entire proteins. Ligand specificity has been successfully engineered using both structure-based rational design (Gao et al., 2006) and directed evolution (Di Roberto et al., 2017). This has allowed the creation of receptors recognizing non-natural ligands, which is useful to create receptor-ligand pairs that are orthogonal to endogenous ones (Jacobson et al., 2007).
GPCRs typically act on transcription of multiple genes by G-protein mediated signal transduction through a variety of routes, such as cAMP, phospholipase C, and MAPK/ERK signaling pathways (Jiang et al., 2022), often involving numerous second messenger molecules. In higher eukaryotes, due to the large number of GPCRs, their signaling typically has considerable crosstalk. This regulatory complexity poses challenges for using GPCRs in artificial genetic circuitry. However, S. cerevisiae has only two GPCR signaling pathways. One of them, the mating pheromone pathway, has been engineered to allow swapping the GPCR for heterologous receptors (King et al., 1990), and has also been converted into a tunable regulatory system (Shaw et al., 2019). In each case, numerous genomic edits have been necessary for the desired refactoring.
A plethora of new-to-nature transcriptional regulatory devices for eukaryotes have been developed by combining DNA binding domains of various sources with transcriptional regulatory domains. In order to achieve orthogonal regulatory devices, DNA binding domains and cognate cis-acting elements can be lifted from phyla distant to the host organism, e.g., from plants (Naseri et al., 2017) or mammals (McIsaac et al., 2014) to yeast.
However, when taking the DNA-binding domain from a given naturally occurring transcription factor, specificity for the cognate cis-acting element is typically largely fixed. This limitation has been overcome by using programmable DNA-binding proteins like zinc fingers (Beerli et al., 2000; Khalil et al., 2012), transcription activator-like effectors (TALEs) (Machens et al., 2017) and Cas proteins (Bikard et al., 2013). This approach allows targeting designed, synthetic cis-acting elements, but also native endogenous promoters (Park et al., 2003). Due to their ease of programming target specificity through their guide RNA, Cas proteins have seen particularly widespread use in synthetic transcription factors, both with activating or repressing effector domains (Du et al., 2021). CRISPR/Cas systems are widely being used to inhibit transcription initiation or elongation for negative expression control (Qi et al., 2013), an approach called CRISPR interference (CRISPRi). This suppression can be enhanced by fusing a transcriptional repressor domain (Gilbert et al., 2013).
Activity of synthetic transcription factors is often regulated through transcriptional control of their expression using ‘conventional’ inducible systems. To impart sensing to modular synthetic transcription factors themselves, different ways have been explored. The approach of controlling transcriptional activity through the ligand-binding domain of a nuclear receptor (Webster et al., 1988; McIsaac et al., 2014) has already been mentioned. Another, widely used strategy is using ligand (Tak et al., 2017) or light-dependent (Shimizu-Sato et al., 2002; Polstein and Gersbach, 2012; Nihongaki et al., 2015) dimerization systems to control conditional interaction between the DNA binding and the effector domain. Light-dependent DNA binding has also been achieved using light-responsive allosteric proteins like LOV domains (Strickland et al., 2008; 2010) or photoactive yellow protein (PYP) (Morgan et al., 2010; Fan et al., 2011), to control accessibility of the DNA binding domain. However, these systems are more difficult to engineer, typically requiring a combination of structure-guided design and directed evolution to achieve the desired photoresponse (Mazumder et al., 2015).
Beyond creating new synthetic transcription factors, they have also been engineered for improved performance. De Carluccio et al. combined the CRISPR-Cas endoribonuclease CasRx with the Tet-On system to create an inducible gene expression platform with minimal leakiness called CASwitch. The system employs two tetracycline-responsive promoters working in opposite directions: one activates target gene expression in presence of doxycycline, while the other, controlling CasRx expression, is repressed. CasRx recognizes and cleaves specific sequences in the target transcript, preventing leaky expression in the absence of doxycycline. This dual control achieves minimal leakiness while maintaining high maximal expression levels, demonstrating over 3000-fold induction (De Carluccio et al., 2024).
Instead of using transcription factors to direct the endogenous transcriptional machinery, orthogonal transcription systems have been employed to control transcriptional activity of genes. A paradigmatic system is the RNA polymerase and its promoters of the T7 bacteriophage. Being highly active and selective for its cognate promoter sequence, T7 RNA polymerase has been leveraged to drive high-level expression of transgenes in bacterial (Tabor and Richardson, 1985; Studier and Moffatt, 1986) and eukaryotic hosts (Fuerst et al., 1986; Chen et al., 1994; Nguyen et al., 2004). The system taken from T7 has been engineered to create multiple mutually orthogonal transcription systems (Temme et al., 2012).
To convey sensing function to the system itself, rather than relying on expression control of the polymerase through another system, T7 RNA polymerase has been made inducible using different strategies. One is based on incorporation of a non-canonical amino acid in a carefully chosen position, suppressing polymerase activity. Upon cleavage of the amino acid by UV light irradiation, the T7 RNA polymerase is activated (Chou et al., 2010). However, more common are strategies based on a split-protein approach, in which two parts of the T7 RNA polymerase are each fused to a partner of an inducible dimerization system, which allows bringing the two-halves together. Through the choice of the dimerization system, polymerase activity has been made inducible by small molecules (Pu et al., 2017), light (Baumschlager et al., 2017), and even macromolecules (Komatsu et al., 2023).
In prokaryotes, the endogenous RNA polymerase can also be leveraged for orthogonal transcription by using orthogonal sigma factors. These proteins help initiate transcription by interacting with the core RNA polymerase and targeting it to their cognate promoters. By introducing sigma factors whose recognition sequences are sufficiently different from the ones of endogenous sigma factors, orthogonal transcription can be achieved from recognized promoters (Rhodius et al., 2013). Sigma factors from Bacillus subtilis have been well established for orthogonal transcription in E. coli (Bervoets et al., 2018), along with predictive design of cognate promoters of desired expression strength (Van Brempt et al., 2020). Moreover, many sigma factors possess anti-sigma factors, inhibiting their sigma factors in response to stimuli. Leveraging combinations of orthogonal sigma and anti-sigma factors supports construction of sophisticated genetic circuitry such as bistable switches (Chen and Arkin, 2012).
Transcriptional regulation systems covered so far rely on trans-acting proteins, modulating whether transcription takes place from a given promoter. However, regulation can also occur through functional, ligand-sensing elements in the 5′-untranslated region of mRNA itself. Such regulator elements in the mRNA are called riboswitches, and a subset of naturally occurring riboswitches control transcription of their mRNA. They work by switching between a terminator hairpin configuration and an antiterminator configuration, depending on ligand availability, with adoption of the terminator fold leading to premature transcription termination (Figure 2A) (Mironov et al., 2002). These transcriptional switches generally have a ligand binding domain, the aptamer, and the effector domain adopting the termination hairpin. By replacing the aptamer in natural riboswitches with other aptamers, including artificially generated ones, synthetic transcriptional riboswitches can be created that react to other small molecules (Ceres et al., 2013a).
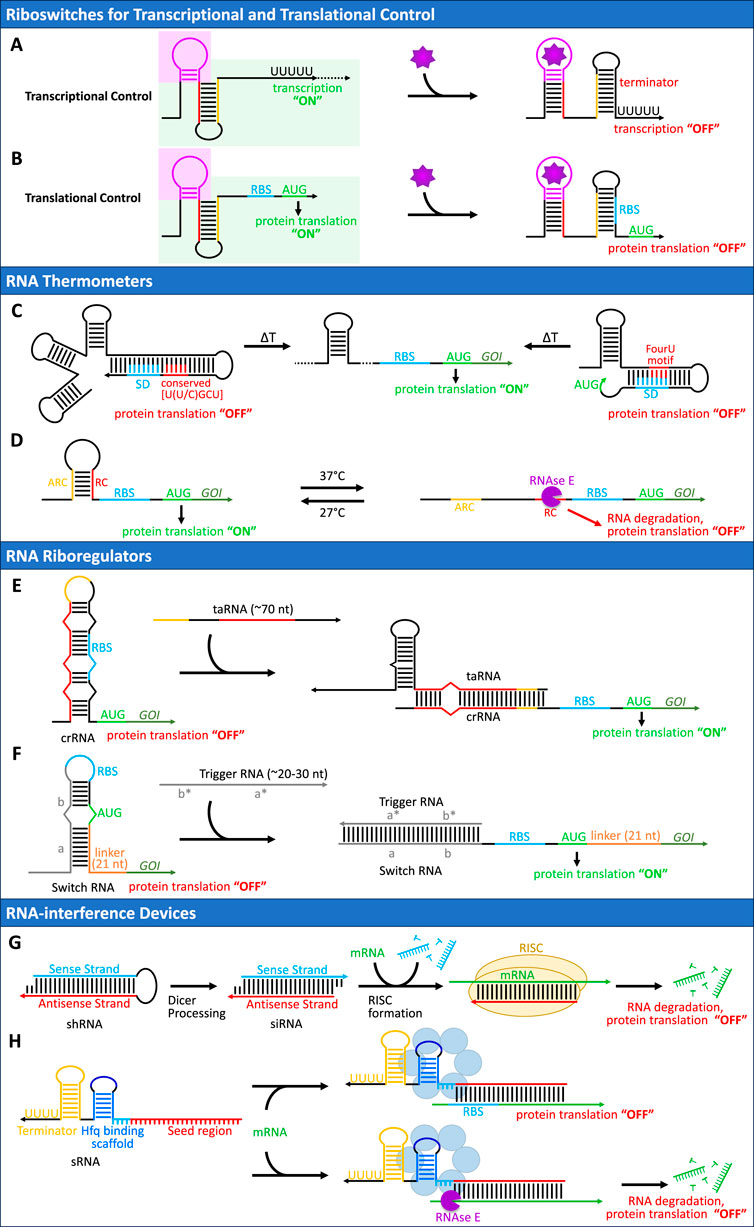
Figure 2. Overview of RNA-based regulatory mechanisms for gene expression control. Arrowheads indicate 3′ ends throughout all (A, B) Riboswitches for Transcriptional and Translational Control. Riboswitches regulate gene expression through ligand-induced conformational changes, consisting of an aptamer domain (purple) that specifically binds its cognate ligand (purple star) and a regulatory domain (green shading) (A) In transcriptional control, the switching sequence (yellow) base-pairs with an anti-terminator sequence (red) in the ligand-free state, enabling transcription (“ON”). Ligand binding induces a conformational change that releases these sequences, allowing the formation of a terminator stem-loop structure (yellow/black) that halts transcription (“OFF”) (B) Translational riboswitches operate through a similar mechanism but control access to the ribosome binding site (RBS, blue). In the absence of the ligand, the RBS remains exposed, allowing translation initiation (“ON”). Upon ligand binding, structural rearrangement leads to RBS sequestration, preventing ribosome access and inhibiting translation (“OFF”) (C, D) RNA Thermometers. (C) The ROSE-type RNA thermometer (left) uses a conserved U(U/C)GCU motif and multiple hairpins that sequester the SD sequence (blue) at low temperature, while the FourU RNA thermometer (right) employs four consecutive uridines. Temperature elevation disrupts these structures, enabling translation (D) A synthetic heat-repressible RNA thermometer employs an RNase E cleavage site (RC, red) protected by base-pairing with an anti-RNase E cleavage sequence (ARC, yellow) at low temperatures. At 37°C, the protective structure unfolds, exposing the RC site to RNase E (purple)-mediated degradation, preventing protein expression (Hoynes-O’Connor et al., 2015) (E, F) RNA Riboregulators and toehold switches. (E) Conventional riboregulators use a cis-repressed RNA (crRNA) that sequesters the RBS (blue) in a stem structure, resulting in translational repression (“OFF” state). Activation occurs through a trans-activating RNA (taRNA, ∼70 nt) that initiates interaction via loop-mediated base-pairing, leading to structural reorganization to expose the RBS (blue) and enable translation (“ON” state). This design requires specific loop sequences, constraining the programmability of the system (F) Toehold switches represent an advanced riboregulatory design where the switch RNA sequesters the region around the start codon instead of directly binding to the RBS or start codon. The switch RNA consists of a single-stranded toehold domain (gray, a, 12 nt) that initiates binding with the trigger RNA, followed by a stem structure (gray, b, 18 nt). Importantly, both the RBS (blue) and start codon (green) remain unpaired within an 11-nt loop and 3-nt bulge respectively, imposing less design constraints. A 21-nt linker sequence coding for low-molecular-weight amino acids (orange) follows the stem to connect to the regulated gene. Translation is activated when a trigger RNA binds to the toehold through complementary sequences (a*, b*) and displaces the stem through linear-linear interactions. Variable sequences are shown in gray, whereas conserved or constrained sequences are represented in different colors (G, H) RNA-interference Devices. (G) Design of short hairpin RNA (shRNA) where the antisense strand (red), complementary to the target mRNA, forms a stem-loop structure (∼21–23 nt) with the sense strand (blue). Dicer processing removes the loop, generating siRNA with 2-nt 3′overhangs. The siRNA is loaded into the RISC complex (yellow), which uses the antisense strand to target complementary mRNA sequences, leading to mRNA degradation and translational repression. (H) Design of bacterial sRNA consisting of three major components: (i) a 5′ seed region (red, 12–24 nt) complementary to the target mRNA, (ii) an Hfq binding scaffold that comprises an AU-rich region (light blue, 4 nt), a stem (blue, 4–6 nt) and a loop (dark blue, ∼6 nt), and (iii) a terminator (yellow) consisting of a stem loop structure followed by four U at the 3′ end. Alternative architectures exist where the seed region is positioned between the Hfq binding scaffold and the terminator. Upon binding to the target mRNA, the Hfq protein complex (blue) facilitates either translational repression through RBS sequestration (top) or recruitment of RNase E (purple), leading to mRNA degradation (bottom).
Naturally occurring transcriptional riboswitches typically terminate transcription in the presence of their ligand (Figure 2A). However, devices which activate or de-repress gene expression upon ligand addition can be more versatile for use in synthetic circuity. Different rational designs have enabled the creation of transcriptional riboswitches with such ON-switching behavior (Wachsmuth et al., 2013; Ceres et al., 2013b). Beyond transcriptional control, riboswitches can also affect translation or stability of their mRNA; these are discussed below under devices for translational regulation.
Translational regulation is a fundamental mechanism in gene expression control, allowing cells to adjust protein synthesis rates rapidly in response to internal and external stimuli. This regulation occurs through different mechanisms, including mRNA stability, translation initiation, and elongation. Understanding and utilizing these systems provides another level of control for the construction of sophisticated genetic circuits.
As mentioned in the previous section, riboswitches are natural regulatory elements that typically reside in the 5′ untranslated regions (UTRs) of bacterial mRNAs and are composed of two interacting domains, the ligand sensing domain (aptamer) and the expression platform (device). Ligand binding leads to switching between two mutually exclusive conformations of the expression platform, resulting in gene expression (ON switch) or repression (OFF switch) (Figure 2B). Riboswitches can function as transcriptional control elements by forming terminator structures in response to ligand binding modulating RNA polymerase activity and transcription termination (outlined above, Figure 2A). Additionally, riboswitches can function as translational control elements through direct mechanisms by changing the mRNA conformation to expose or sequester the RBS (Figure 2B), or through indirect mechanisms by alternative splicing of mRNA or modulating mRNA stability (Breaker, 2018). One example of a riboswitch sequestering the RBS is the riboswitch for adenosylcobalamin (AdoCbl), which has been investigated at atomic resolution by x-ray crystallography (Johnson Jr et al., 2012).
Riboswitches have been widely used as regulatory tools in synthetic biology because of their modular nature, and they have been engineered in multiple ways to alter and fine-tune input as well as output behavior (Etzel and Mörl, 2017). One of the best studied systems is the theophylline riboswitch, which has been engineered to function as transcriptional or translational control element and as ON or OFF switch, respectively (Wang et al., 2023).
Next to ligand-mediated control of RNA structure, temperature can likewise influence RNA structure and thereby control protein translation. This is found in RNA thermometers (RNAT), which are located within the 5′ UTR and, at low temperature, form stable secondary structures that block ribosome access (Figure 2C). Elevating the temperature leads to a zipper-like gradual shift from the closed to the open conformation, thereby exposing the RBS and enabling the translation process (Kortmann and Narberhaus, 2012; Sharma et al., 2022). The ROSE (Repression Of heat Shock gene Expression) family is the most common class of RNAT, controlling the expression of small heat shock genes in many alphaproteobacteria and gammaproteobacteria (Narberhaus et al., 2006). They usually contain two to four hairpins, with the 3′-most hairpin sequestering the Shine-Dalgarno sequence at low temperatures, while the other hairpin structures most likely aid in proper folding of the temperature-sensitive hairpin (Figure 2C left).
Another class of thermosensors are the “FourU thermometers”, named after their characteristic sequence of four consecutive uridines that form a zipper-like RNA structure occluding the Shine-Dalgarno sequence at low temperatures (Figure 2C right) (Waldminghaus et al., 2007). Found in bacterial virulence genes and heat shock proteins, FourU thermometers are typically shorter but melt at higher temperatures compared to the widespread ROSE elements (Tong et al., 2023).
Synthetic RNATs have been designed to create temperature-responsive genetic circuits, sometimes referred to as “thermogenetics”. Furthermore, synthetic RNATs can be simplified and modularized compared to their natural counterparts. For instance, Neupert et al. developed a modular approach separating promoter, start codon, SD sequence and a complementary anti-SD (ASD) sequence with four restriction sites in such a way that at low temperature the ASD sequesters the SD preventing protein translation (Neupert and Bock, 2009). Using a fluorescent reporter, SD and anti-SD as well as the spacing between SD sequence and initiation codon could be optimized by easily exchanging modules between the restriction sites (Neupert and Bock, 2009).
Based on two natural RNATs with only one base difference but significantly different temperature response, Sen et al. constructed a library of RNATs guided by thermodynamic computations and evaluated their temperature dependence in a cell-free assay (Sen et al., 2017). While computational predictions showed only weak correlation with the experimental data, this systematic approach nevertheless yielded a toolbox of RNA thermometers with varying temperature sensitivities and thresholds (Sen et al., 2017).
Natural RNA thermosensors typically activate gene expression at elevated temperatures by giving access to the SD sequence. In contrast, Hoynes-O'Connor et al. engineered a heat-repressible RNA thermosensor using RNase E, an endogenous endoribonuclease in E. coli that preferentially cleaves single-stranded RNA (Figure 2D) (Hoynes-O’Connor et al., 2015). They incorporated an RNase E cleavage site (RC) that gets sequestered by a complementary anti-RNase E cleavage sequence (ARC) in a stem-loop structure. At low temperatures, this structure remains stable, sequestering the RC site and protecting the RNA from degradation, thereby allowing translation. At higher temperature (37°C), the stem-loop unfolds, exposing the RC site to RNase E, which leads to RNA degradation and subsequent translation inhibition.
A significant advancement in RNA-based gene regulation was the development of engineered riboregulators that enable post-transcriptional control of gene expression. The first generation of synthetic RNA regulators consisted of two parts: a cis-repressed mRNA (crRNA) that forms a stem-loop structure sequestering the ribosome binding site (RBS), and a trans-activating RNA (taRNA) that can base-pair with the crRNA to expose the RBS and enable translation (Figure 2E). Isaacs et al. demonstrated this concept by engineering a series of riboregulators in E. coli, achieving up to 19-fold activation of gene expression (Isaacs et al., 2004).
Toehold switches represent an important advancement over riboregulators, by significantly facilitating the design of synthetic regulators. Unlike conventional riboregulators, which repress translation by directly base-pairing to the RBS, toehold switches achieve translational control through base-pairing interactions that sequester the region around the start codon within an RNA stem, while leaving the RBS unpaired within a loop region (Figure 2F) (Green et al., 2014). This design strategy frees the RBS and start codon regions from sequence constraints that limit conventional riboregulators. Another key innovation is the use of linear-linear RNA interactions through a single-stranded toehold domain rather than the loop-mediated interactions employed by conventional riboregulators. The trans-activating RNA, called trigger RNA, is designed to be perfectly complementary to the toehold domain and stem region, enabling efficient strand displacement upon binding.
Both experimental approaches and computational tools have advanced the development of synthetic toehold switches. These include deep learning approaches that apply techniques from computer vision and natural language processing (Valeri et al., 2020), automated design software incorporating experimental constraints (Cisneros et al., 2023), as well a systematic evaluation of mutation effects on truncated switches, reducing the length from 30 nt (Green et al., 2014) down to 18–23 nt (McSweeney et al., 2023).
Modulation of gene expression through mRNA stabilizing and destabilizing effects are ubiquitous in all organisms. RNA interference (RNAi) is an evolutionarily conserved post-transcriptional gene regulation mechanism that responds to double-stranded RNA (dsRNA) in eukaryotic cells, and plays a key role in gene silencing (Fire et al., 1998). Common types of effector RNAs are microRNAs (miRNA), which are encoded in the genome, and small interfering RNAs (siRNA), which are often from exogenous sources (Ahmadzada et al., 2018). Both RNA types are processed by Dicer, a member of the RNAse III family, to produce duplexes of approximately 21–23 nucleotides that are integrated into the RNA-induced silencing complex (RISC) (Elbashir et al., 2001). The Argonaute two component cleaves the duplex, degrading the passenger (sense) strand and retaining the guide (antisense) strand to direct RISC to complementary mRNA sequences (Matranga et al., 2005; Alshaer et al., 2021).
Based on target location and complementarity, RNAi can induce different outcomes: miRNAs typically show partial complementarity in the 3′ UTR, resulting in protein recruitment and translational repression, while siRNAs usually display full complementarity within the coding sequence (CDS), triggering endonucleolytic cleavage and mRNA degradation (Hutvágner and Zamore, 2002). Following cleavage, the RISC complex is released and can bind additional mRNA targets (Hutvágner and Zamore, 2002; Haley and Zamore, 2004).
Efficient and specific RNAi relies on a set of critical criteria during the siRNA design process, which is further detailed in a review from (Fakhr et al., 2016). For therapeutic purposes, siRNA can be delivered through different approaches: either as chemically synthesized short oligonucleotides, often packaged in various vesicles (Hu et al., 2020), or through in vivo transcription methods (Fu et al., 2021). Alternatively, short hairpin RNAs (shRNAs), first created in 2002 (Brummelkamp et al., 2002; Paddison et al., 2002), can be used, which after Dicer processing provide siRNA-like oligos (Figure 2G) (Sheng et al., 2020). These shRNAs can be encoded in DNA constructs, consisting of a 5′ overhang, targeting sequence, loop, reverse-complement targeting sequence, transcriptional terminator sequence, and 3′ overhang, and can be integrated into the genome to be used, e.g., for inducible gene knockdown (Moore et al., 2010; Frank et al., 2017). Various methods have been developed to use RNAi for Boolean logic gates in mammalian cells (see also Section 3.3.3 and Figure 6) (Rinaudo et al., 2007; Xie et al., 2011; Groves et al., 2016; Matsuura et al., 2018).
Prokaryotic organisms use small RNAs (sRNAs) for post-transcriptional gene regulation in a variety of physiological processes (Modi et al., 2011). These sRNAs, typically 50–300 nt in length, target mRNAs at or near the ribosomal binding site in a cis- or trans-acting manner. Most sRNAs contain three functional domains: a seed region complementary to the target mRNA, a scaffold region aiding in Hfq chaperone and DNAse E recruitment, and a Rho-independent transcription terminator, consisting of a pyrimidine-rich palindromic sequence followed by a stretch of U nucleotides (Figure 2H) (Noh et al., 2019; Bandyra et al., 2012). Transcriptional repression usually occurs through base pairing, often mediated by the Hfq chaperon, leading to translational blocking and RNAse E mediated mRNA decay (De Lay et al., 2013).
Synthetic small RNAs (sRNAs) have been developed as an extension to transcription factors, offering an easier method for targeting specific mRNA sequences. Unlike transcription factors, which typically exhibit sigmoidal response curves, sRNAs display a linear response, enabling more gradual control of gene expression (Hussein and Lim, 2012). This characteristic makes synthetic sRNAs particularly suitable for fine-tuning gene regulation in diverse applications such as metabolic engineering and high-throughput screening (Lin et al., 2019; Bhatnagar et al., 2019; Na et al., 2013). Recent work has further analyzed the underlying mechanism and expanded their application across a wide range of bacterial species (Brück et al., 2024). For example, novel sRNA constructs were designed, achieving over 50% knockdown efficiency in 12 bacterial species, including Gram-positive bacteria (Cho et al., 2023). Additionally, systematic analysis of seed region length has revealed design rules that enhance the efficiency and specificity of synthetic sRNAs (Brück et al., 2024).
In prokaryotes, translation can be controlled using engineered, orthogonal ribosomes that exclusively translate specific transcripts. Altering the anti-Shine-Dalgarno sequence on the 16S rRNA results in an altered specificity of the ribosome to Shine-Dalgarno sequences, the ribosome-binding sites on mRNA transcripts. This has long been used to direct translation to specific heterologous mRNA species (Hui and De Boer, 1987) by creating a population of ribosomes orthogonal to the native ribosome pool. Multiple orthogonal ribosome/mRNA pairs have been developed and shown to allow implementation of logic circuits (Rackham and Chin, 2005). Orthogonal ribosomes have been further engineered to support genetic code expansion by efficient decoding of amber stop codons (Wang et al., 2007) and quadruplet codons (Neumann et al., 2010).
Translation elongation control through genetic code expansion presents another angle of regulating translation that works both in prokaryotes and eukaryotes. Here, gene-specific control can be achieved by leveraging orthogonal translation systems, consisting of an aminoacyl tRNA synthetase and a cognate suppressor tRNA, able to decode stop codons in the presence of non-canonical amino acids. These pairs are taken from organisms phylogenetically very distant to the target host to be orthogonal to its endogenous translation machinery. The synthetase has to specifically incorporate non-canonical amino acids whilst being orthogonal to canonical ones (Vargas-Rodriguez et al., 2018). Expression control of a target gene is achieved by placing suppressible stop codons within its coding sequence. Thus, full-length protein translation is made dependent on the availability of a suitable non-canonical amino acid necessary to suppress termination at these positions. This principle has been used, for example, to control expression of a recombinase (Zhang et al., 2022), or essential proteins to create strains with synthetic auxotrophies dependent on the availability of non-canonical amino acids (Mandell et al., 2015; Rovner et al., 2015; Chang et al., 2023).
Control of target genes is also possible on the protein level by engineering target proteins such that their stability, location or function can be modulated in response to stimuli. A general advantage of control on the protein level is that responses are typically faster than those of control systems operating on preceding processes of gene regulation.
One approach is the control of protein half-life, which allows rapid changes in protein levels through regulated degradation. For bacteria, different degradation systems have been engineered, primarily based on the native proteases ClpXP, ClpAP, or Lon. A widely used approach employs the SspB adaptor protein, which delivers tagged proteins to the ClpXP protease complex. Inducible degradation can be achieved by controlling SspB expression or by engineering conditional exposure of the degradation tag. This has been successfully demonstrated for metabolic control, where the DAS+4 degradation tag system was used to create metabolic switches responding to phosphate levels (Ye et al., 2021). The Lon protease system has also been employed in synthetic circuits, for instance in the design of kill switches where it provides an additional layer of control through targeted protein degradation (Chan et al., 2016).
In eukaryotes, regulated protein degradation is typically achieved through the proteasomal system. Regulatable degradation tags fused to proteins of interest can trigger proteasomal degradation under specific conditions, typically the presence or absence of a small-molecule ligand. There are a number of different approaches to create these conditional degrons. Destabilizing domain (DD) degrons are based on a ligand-binding protein engineered to be in an unstable conformation, and thus be directed for degradation, without their ligand. Binding the ligand stabilizes the conformation and increases the protein’s half-life. DD degrons have been based on different protein scaffolds, such as FKBP12 (Banaszynski et al., 2006), dihydrofolate reductase (Iwamoto et al., 2010), UnaG (Navarro et al., 2016), and the human estrogen receptor (Miyazaki et al., 2012), each reacting to different ligands. A DD class degron has been leveraged to create stringent dependence on the availability of beta-estrogen in yeast, by fusing the degron to suitable essential genes (Hoffmann and Cai, 2024).
The auxin-inducible degron has been lifted from plants (Nishimura et al., 2009). Here, an E3 ubiquitin ligase complex polyubiquitinates the degron in the presence of auxin [or an auxin derivative in an improved version of the system (Yesbolatova et al., 2020)] and targets it for degradation. The SMASh tag is a degron that cleaves itself off the fused protein, unless its specific protease activity is inhibited by an hepatitis C virus (HCV) protease inhibitor, leading to degradation of the tagged protein (Chung et al., 2015). There are also systems in which the bound ligand itself directs degradation. Proteolysis targeting chimera (PROTAC) ligands do so through recruitment of an endogenous E3 ubiquitin ligase. In this case, the degrons being fused to the target protein are protein domains for which efficient PROTACs are available, such as the HaloTag (Buckley et al., 2015) or the dTAG (Nabet et al., 2018). Further, ligands with a hydrophobic moiety have been used to direct HaloTag fusion proteins for degradation through the cell’s quality control (Neklesa et al., 2011).
There are also light-responsive post-translational control systems. Degrons have been made conditional on light illumination instead of small ligands, using a photoresponsive LOV domain and chimerizing its C-terminal helix with a degron (Renicke et al., 2013; Bonger et al., 2014). Thus, blue light illumination exposes the degron and leads to its degradation along with the protein it is fused to. Light-inducible systems have also been used for fine-grained spatiotemporal control of the location of target proteins. Such control over Rho-family GTPases has allowed targeted remodeling of the cytoskeleton (Levskaya et al., 2009).
Apart from controlling protein half-life through conditional degradation, intein-based protein splicing presents another option of post-translational control. Inteins catalyze their own excision from a protein. Their action can be made conditional, e.g., through a split-protein approach, or by inserting a sensory domain in the intein (Topilina and Mills, 2014; Sarmiento and Camarero, 2019). Either way, intein splicing activity leads to creation of a protein producing an output (Jillette et al., 2019; Anastassov et al., 2023). Such intein-based approaches have been used to create biosensory systems for a variety of stimuli. For example, split intein systems lend themselves to sensing protein-protein interactions by reconstituting functional reporter proteins upon interaction of the split parts, as demonstrated in studies using split luciferase or fluorescent proteins (Ozawa et al., 2001; Paulmurugan et al., 2002). Conversely, single protein reporters have been used for instance for small molecule sensing (Buskirk et al., 2004).
Generally, systems may also obtain expression control through post-translational modalities such as phosphorylation status cellular localization, dimerization, or allosteric state. However, examples for such systems often act on transcription factors or signalling cascades (Spencer et al., 1993; Wu et al., 2009; Yang et al., 2025), and as such may be hard to delineate from transcriptional control.
Using regulatory parts detailed in Section 2 (Regulatory devices), genetic circuits can be composed, in which multiple parts work together to achieve more complex cellular behaviors. This section outlines basic design principles, highlights considerations for the combination of several regulatory elements and provides selected examples of fundamental circuit architectures.
The design of genetic circuits relies on several key principles that form the foundation of synthetic biology as an engineering discipline. These principles guide the creation of robust, tunable, and predictable biological systems. Similar to other engineering disciplines, having abstractable and reliable components facilitates the creation of systems of higher complexity. Engineering biology faces particular challenges inherent with biological systems, such as a high degree of complexity and interconnectedness. This section presents the key principles of modularization and standardization, orthogonality, and robustness and tunability to tackle these hurdles and enable the development of increasingly complex and sophisticated synthetic genetic networks.
Modularization, a fundamental engineering concept, involves creating independent, interchangeable parts that can be combined to build complex systems. Such parts include promoters, ribosome binding sites (RBS), coding sequences, regulatory devices, and terminators (Figure 3A). Closely related to modularity is composability, which ensures that individual parts can be combined in predictable ways to create functional systems. Such modularity facilitates systematic design, testing, optimization and adaptation of genetic circuits. The concept of modularity has been central to the development of standardized biological parts, aiming to create a common “language” for describing genetic parts and their interactions (Müller and Arndt, 2012). This facilitates knowledge sharing and enables the development of tools for automated circuit design. The BioBrick standard, for instance, uses specific restriction enzyme sites to allow for easy, automatable, assembly of genetic parts (Endy, 2005; Müller and Arndt, 2012).
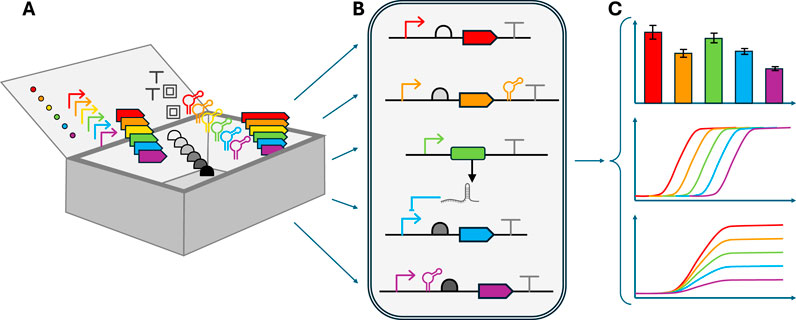
Figure 3. Key design principles for construction of synthetic genetic circuits (A) Modularization and standardization are achieved through libraries of well-characterized genetic parts that can be combined in predictable ways. Individual parts like promoters, ribosome binding sites, coding sequences, terminators, and regulatory devices (represented in different colors) are designed to be functionally independent to enable their combination in various arrangements (B) Orthogonality between different regulatory elements is crucial for predictable circuit behavior. Independent genetic modules showing minimal cross-talk between host cell machinery and other circuit components can be assembled into more complex circuits. (C) Robustness and tunability of genetic circuits can be assessed through characterization of individual parts and complete circuits under different conditions. Circuits should ideally show robust behavior under different conditions with little variation (top) as measured by consistent output levels across varying environmental conditions, and should be tunable in terms of input threshold and dynamic range (middle) and output signal strength (bottom panel).
The Synthetic Biology Open Language (SBOL) represents another major standardization effort, providing a machine-readable format for representing genetic circuits and their components (Galdzicki et al., 2014; Buecherl et al., 2023). SBOL enables researchers to describe DNA components and their interactions, exchange genetic designs between different software tools, and facilitate the reproducibility of synthetic biology experiments. Later versions have been expanded to also represent multicellular systems (Brown et al., 2020). Complementing the SBOL data standard, SBOL Visual provides standardized graphical notations for genetic circuit diagrams, further enhancing communication and design in the field (Beal et al., 2019). While standardization has greatly advanced the field, it is important to note that biological systems often exhibit context-dependent behavior, which poses challenges to the ideal of fully modular, standardized parts.
Orthogonality refers to the ability of genetic circuit components to function independently without interfering with each other or the host cell’s native processes. Achieving orthogonality is crucial for predictable circuit behavior but remains a significant challenge, since–unlike in, e.g., electrical engineering–there is little to no spatial separation in biological systems (Figure 3B). Multiple strategies have been developed to improve orthogonality, including the use of heterologous components from different organisms, engineering of existing components to reduce cross-talk, and the development of entirely new synthetic parts (Rao, 2012; Brödel et al., 2016; Naseri et al., 2017). The importance of orthogonality for scaling up circuit complexity was demonstrated by Nielsen et al., who leveraged a large set of orthogonal repressor-operator pairs to construct sophisticated logic gates in E. coli (Nielsen et al., 2016).
Genetic circuits should function reliably across various conditions and be easily adjustable (Figure 3C). This involves considering factors such as gene expression noise, metabolic burden on the host cell, and environmental fluctuations. Strategies to enhance robustness include incorporating autoregulatory negative feedback loops, redundancy and degeneracy (Becskei and Serrano, 2000; Macia and Solé, 2009; Randall et al., 2011). Tunability of circuit behavior is another important aspect, which can be achieved through various mechanisms such as titratable promoters engineered ribosome binding sites or riboswitches that respond to external stimuli (Ang et al., 2013).
A crucial factor affecting robustness and tunability is the context dependence of genetic parts, posing challenges for the design and implementation of reliable genetic circuits. Köbbing et al. conducted a comprehensive study on the effects of genetic context on synthetic promoters in Pseudomonas putida by systematically characterizing how the performance of stacked (concatenated) promoters behave depending on their context (Köbbing et al., 2020). Their findings revealed that adjacent genetic elements can significantly alter promoter activity, highlighting the importance of considering genetic context in circuit design.
Another important aspect to consider, especially for larger circuits with increasing numbers of components, is the concept of “load balancing”, which addresses the challenge of maintaining circuit function as it scales up in complexity. This is achieved by considering the metabolic burden imposed by synthetic circuits on the host cell, aiming to optimize circuit design for better overall performance (Ceroni et al., 2015; Borkowski et al., 2016). A significant challenge in this context is resource competition between circuit modules or between the circuit and the host cell. Zhang et al. demonstrated how growth feedback can interfere with memory maintenance in a topology-dependent manner (Zhang et al., 2020), while resource competition can lead to “winner-takes-all” behavior in cascading bistable circuits, disrupting expected dynamics (Zhang et al., 2021). They proposed a microbial consortia strategy to mitigate these effects by decoupling resource pools. Further strategies addressing context dependence have been reviewed elsewhere (Stone et al., 2024).
In general, these principles of robustness and expression load are closely related to the concept of “evolutionary stability” in synthetic biology. Circuits that are more robust and have a low expressional load, which has also been termed fitness threshold, are often more likely to maintain their function over multiple generations, even in the face of evolutionary pressures (Sleight and Sauro, 2013). This is particularly important for applications where long-term stability of the synthetic circuit is crucial, such as in therapeutic applications or environmental biosensors.
The engineering principles described above provide a degree of abstraction from the complexity inherent to biological systems, enabling forward design of systems involving multiple regulatory elements with acceptable predictability. These design principles, combined with our increased ability to model biological systems, have enabled computer-aided tools for design of complex synthetic circuitry. A pivotal development in this area was the introduction of Cello (Cellular Logic), a design automation platform for genetic circuits (Nielsen et al., 2016). Cello allows users to describe desired circuit function using Verilog, a hardware description language, which is then translated into a DNA sequence encoding the specified logic. Its successor, Cello 2.0, further expanded these capabilities with support for a wider range of logic gates and improved optimization algorithms (Jones et al., 2022).
Recent work has focused on various approaches to automate robust genetic circuit design. These include adapting machine-learning algorithms to optimize gene circuit designs (Hiscock, 2019), as well as methods that account for structural variants and parameter uncertainty, combining evolutionary algorithms with stochastic simulations (Schladt et al., 2021).
While these computational tools have greatly advanced the field, it is important to note their limitations. Current modeling approaches often struggle to fully capture the complexity of biological systems, particularly when predicting the behavior of circuits in different cellular contexts or over long time scales.
The following section presents seminal examples of genetic circuits. These examples demonstrate how the principles discussed above have been applied to create functional biological systems with predictable behaviors from regulatory ‘building blocks’ outlined in Section 2 (Regulatory devices).
Genetic circuits have progressed in complexity from single-node to two-node and three-node designs, showcasing principles like noise reduction, bistability, and oscillatory dynamics. Foundational circuits such as the autoregulatory circuit (Becskei and Serrano, 2000), toggle switch (Gardner et al., 2000), and repressilator (Elowitz and Leibler, 2000) exemplify these advances.
Bistable switches are genetic circuits that can exist in one of two stable states and switch between them in response to specific inputs. A classic example is the toggle switch, consisting of two repressor proteins, each inhibiting the expression of the other. External stimuli can flip the switch between these states (Figure 4A) (Gardner et al., 2000).
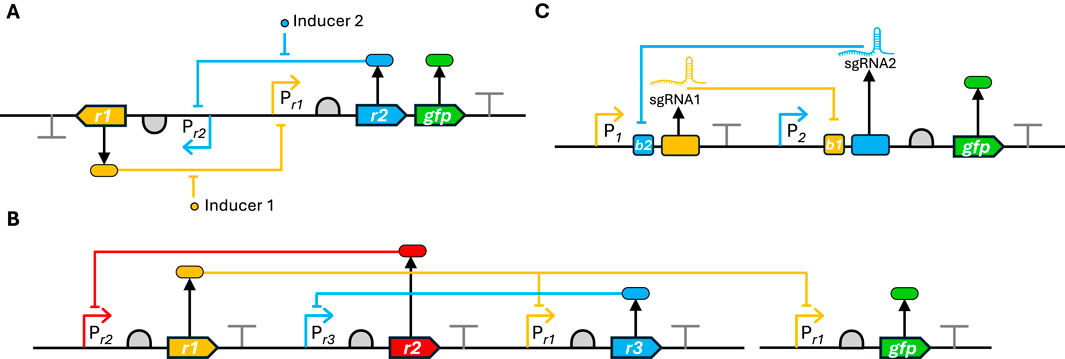
Figure 4. Illustration of bistable and oscillating circuit designs (A) Bistable switch based on mutual repression, in which repressor 1 (yellow) inhibits expression of repressor 2 (blue) and vice versa, creating two stable states. Each state is maintained through the dominant repressor blocking expression of the other repressor. Addition of inducer 1 (yellow) or inducer 2 (blue), respectively, inactivates the corresponding repressor, allowing transition to the opposite state. GFP (green) expression in one state allows monitoring of the switch (Gardner et al., 2000) (B) Repressilator with coupled orthogonal promoter-repressor-inducer sets showing oscillatory behavior between three distinct states, monitored by oscillating GFP levels. The system achieves oscillations through cyclic repression where three repressors sequentially inhibit each other’s expression (Elowitz and Leibler, 2000) (C) Re-design of the bistable switch using CRISPRi, where sgRNA1 (yellow) binds b1 sites, blocking transcription of sgRNA2 (blue) and vice versa. Not shown is constitutively expressed dCas9 (Santos-Moreno et al., 2020).
Oscillatory circuits generate periodic changes in gene expression. The repressilator consists of three transcriptional repressors arranged in a ‘cycle’; each repressor inhibits the expression of its successor, resulting in oscillatory behavior (Figure 4B) (Elowitz and Leibler, 2000). Subsequent work has improved the robustness and tunability of synthetic oscillators. By reducing circuit complexity and incorporating elements that reduce gene expression noise, a robust bacterial oscillator was built that maintained persistent oscillations for hundreds of generations (Potvin-Trottier et al., 2016).
More recently, the versatility of CRISPR-based circuits was demonstrated by constructing both a bistable toggle switch and repressilator using CRISPRi instead of repressor proteins (Figure 4C) (Santos-Moreno et al., 2020). This work illustrates how modern CRISPR technology can be applied to recreate and potentially improve upon these foundational synthetic biology designs.
Biological systems require mechanisms to maintain stable function despite fluctuations in cellular resources and environmental conditions, making robustness likewise a key objective in the design of synthetic gene circuits. Building on early demonstrations of noise reduction through simple negative feedback loops, where autoregulation dampened fluctuations in protein expression (Becskei and Serrano, 2000), several more sophisticated control mechanisms have been developed to achieve robust performance and adaptation to different kinds of perturbations.
Incoherent feedforward loops represent one strategy for achieving robustness. Bleris et al. demonstrated that synthetic incoherent feedforward circuits can adapt to changes in the genetic template abundance, providing a system for gene dosage compensation (Bleris et al., 2011). The field advanced further with the development of antithetic integral feedback controllers for robust perfect adaptation (Briat et al., 2016; Aoki et al., 2019). These controllers use molecular sensors and actuators that mutually annihilate each other, enabling dynamic response to deviations to ensure stable performance even in noisy environments (Figure 5). Frei et al. adapted the concept of annihilation of the controllers to mammalian cells using sense and antisense RNA to design a mammalian genetic proportional-integral controller that achieved precise and robust gene regulation, demonstrating the potential for applying such control systems in complex biological contexts (Frei et al., 2022).
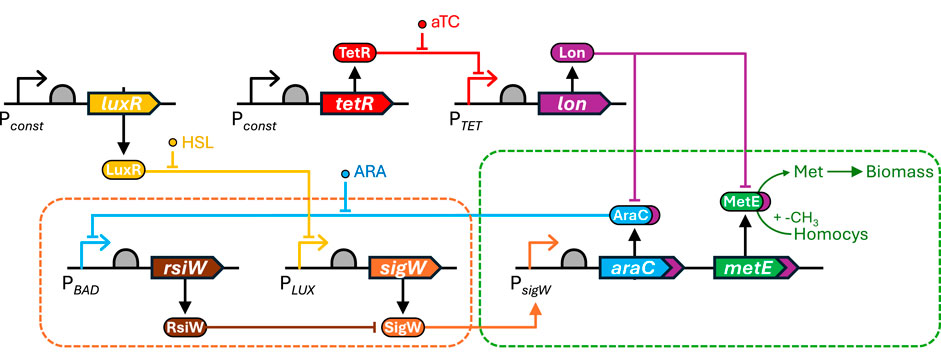
Figure 5. Antithetic integral feedback control circuit. The circuit consists of an antithetic controller (orange dashed box) and a controlled output module (green dashed box) (Aoki et al., 2019). The antithetic controller module is based on the σ factor SigW (orange) and anti-σ factor RsiW (brown) from Bacillus subtilis that annihilate each other upon interaction. SigW expression is regulated by LuxR (yellow) in response to external homoserine lactone (HSL), while rsiW expression is controlled via a negative feedback loop mediated by AraC (blue) from the controlled output module and regulated via the external inducer arabinose (ARA). This interplay creates an antithetic integral feedback mechanism that ensures dynamic regulation and robustness to perturbations. The controlled output module translates the regulatory dynamics into a measurable output, which can be a reporter molecule or biomass production as in this example. Non-annihilated SigW directly regulates both araC expression (enabling feedback control via RsiW) and the expression of the gene of interest. The system was validated using either GFP as reporter or metE (green) encoding methionine synthase, which catalyzes the conversion of homocysteine to methionine supporting biomass production. Additionally, orthogonal perturbation can be applied using anhydrotetracycline (aTc)-inducible Lon protease (purple), which can degrade both AraC and the output protein via a Lon-specific degradation tag (purple).
Huang et al. demonstrated how quasi-integral control through a synthetic small RNA-based feedback controller can enable adaptation of genetic modules to variable ribosome demand, providing another strategy to adapt to cellular resource fluctuations (Huang et al., 2018). Combining metabolic regulation with genetic control mechanisms offers another approach to enhance robustness. Lv et al. demonstrated how coupling metabolic addiction with negative autoregulation could both stabilize strain performance and improve pathway yield in metabolic engineering applications (Lv et al., 2020).
A recent approach by Glass et al. implemented a biphasic fitness strategy in a synthetic differentiation circuit in E. coli to generate robustness against environmental changes and mutant takeover (Glass et al., 2024). Their Biphasically Differentiating E. coli (BDEC) system contains a synthetic differentiation circuit system to mimic stem, progenitor and differentiated cells in E. coli. This was achieved using an integrase to irreversibly remove a plasmid-encoded antibiotic resistance gene while simultaneously restoring an essential metabolic pathway, creating a biphasic control mechanism. Consequently, the authors defined a stem cell with no cut plasmid and a fully differentiated cell with all plasmids cut, while a progenitor cell contains a mixture of uncut and cut plasmids. Due to the biphasic control, this design selected for a specific differentiation rate and showed remarkable robustness to environmental changes and resistance to mutant takeover in long-term evolution experiments.
Logic gates in synthetic biology perform Boolean operations on biological inputs to produce specific outputs. Such gates form the basis for more complex genetic circuits and allow cells to process multiple inputs and make decisions. The most common types of logic gates implemented in biological systems include.
1. AND gate: Requires all inputs to be present to produce an output.
2. OR gate: Produces an output when at least one of the inputs is present.
3. NOT gate: Inverts the input signal, producing an output when the input is absent.
4. NAND and NOR gates: Universal gates that can be used to construct any other logic function. NAND produces an output unless all inputs are present, while NOR produces an output only when no inputs are present.
These basic gates can be combined to create more complex logical operations such as XOR (exclusive OR) and XNOR (exclusive NOR). The implementation of logic gates in biological systems has been achieved through various molecular mechanisms.
Genetic AND gates, for example, were constructed using a modified T7 RNA polymerase with internal amber stop codons combined with an amber suppressor tRNA as second input signal (Anderson et al., 2007) or with a split T7 RNA polymerase (Shis and Bennett, 2013). A simple example of a NOT gate is the use of repressor proteins that inhibit gene expression, as used in the repressilator circuit (Elowitz and Leibler, 2000). Building on this, Tamsir et al. implemented multiple gate types (AND, OR, and NOR) in E. coli using a library of simple regulatory circuits and linking them through diffusible chemical signals to construct more complex logical operations (Tamsir et al., 2011). Siuti et al. built logic gates using two different inputs (AHL and aTc) to activate two recombinases through orthogonal riboregulated systems. Specific arrangement of the modules allowed them to create all 16 two-input Boolean logic functions without coupling multiple gates (Siuti et al., 2013). Likewise, recombinases were used to construct all Boolean logic functions in mammalian cells (Weinberg et al., 2017).
RNAi-based circuits offer another powerful approach for implementing Boolean logic in living cells (Rinaudo et al., 2007). Applying this RNAi-based regulation, Xie et al. constructed a classifier circuit that integrated sensing of six endogenous miRNAs through a combination of AND and AND NOT logic operations to selectively trigger apoptosis in HeLa cancer cells while sparing other cell types based on their miRNA expression profiles (Figure 6) (Xie et al., 2011). This demonstrated how RNAi-based logic could be used to sense complex cellular states to execute specific responses. In a similar approach, Matsuura et al. developed synthetic mRNA-delivered circuits that could implement multiple types of logic gates (AND, OR, NAND, NOR and XOR) in mammalian cells by combining miRNA sensing with RNA-binding proteins as regulators (Matsuura et al., 2018).

Figure 6. RNAi-based logic circuit for cell-type selective induction of apoptosis. The circuit integrates sensing of multiple microRNAs (miR-A to miR-F) through Boolean logic to control apoptosis (Xie et al., 2011). The top level shows constitutive expression of reverse tetracyclin-controlled transactivator (rtTA, blue) from a CMV promoter, negatively regulated by miR-A (yellow) or miR-B (orange) and miR-C (brown), respectively. On the second level, rtTA activates LacI (green) expression from a tetracycline-responsive promoter (PTRE), which again is negatively regulated by miR-A, B and C to prevent any leakage. As an additional safety measure, LacI is linked to anti-apoptotic Bcl2 (dark red) through a 2A peptide (white) that enables production of separate LacI and Bcl2 proteins. LacI represses the expression of the pro-apoptotic protein Bax (red), which can be inhibited by Bcl2 on the protein level and whose expression is additionally negatively regulated by three further miRNAs (miR-D, E and F; purple colors). This multi-layered regulation ensures tight control of the apoptotic response, which is triggered only by the specific input of miR-A ∧ miR-B ∧ miR-C ∧ ¬(miR-D) ∧ ¬(miR-E) ∧ ¬(miR-F).
CRISPR-Cas9-based systems have been increasingly used for implementing logic gates. An AND gate was constructed in yeast where dCas9 and MCP-VP64 expression was controlled by galactose and β-estradiol, respectively, and the scaffold RNA (scRNA) was used to connect both parts and direct them to the promoter site for VP64-mediated gene expression (Hofmann et al., 2019). Similarly, a NOR gate was engineered based on the CRISPR-dCas9 system, which used gRNAs as input signals to a specific target sequence on the NOR gate promoter. The generated output is also a gRNA that matches the target sequence on other NOR gate promoters, allowing for interconnected logic circuits (Gander et al., 2017). In a third example, a NOT gate was constructed by combining two RNA regulation systems, CRISPR and antisense RNA (asRNA). The CRISPR system represses the target gene, which can be derepressed by the expression of an antisense RNA, enabling an ON/OFF switching behavior (Lee et al., 2016).
Biological logic gates have also been used in medical applications. For example, a CRISPR-dCas9-based AND gate with two cancer-specific promoters was designed for detection and control of bladder cancer cell growth in vitro (Liu et al., 2014). Similarly, Courbet et al. implemented Boolean logic when engineering an E. coli-based biosensor cell to detect biomarkers such as nitrogen oxides and glucose in human blood and urine samples (Courbet et al., 2015). They created a sensor module that enables multiple detection of biomarkers and coupled the output signal to Boolean integrase logic gate modules, enabling signal digitization and amplification.
While most logic gates in synthetic biology are implemented through genetic circuits, Vishweshwaraiah et al. developed a system where logical operations were achieved at the protein level (Vishweshwaraiah et al., 2021). They engineered a single protein to function as a two-input OR gate by rationally incorporating both a rapamycin-inducible domain and a light-sensitive LOV2 domain into focal adhesion kinase (FAK), achieving orthogonal control through chemical and optical inputs.
As of now, an astonishing breadth of sophisticated biosensors are available, and for instance can allow cost-effective environmental surveillance. Here, it is often desirable to detect specific environmental pollutants or pathogens at very low levels. In such cases, high sensitivity is needed, which can require circuitry for signal amplification.
One such area is the detection of heavy metals. Several groups have designed whole-cell biosensors for the detection of arsenate and arsenite contamination in water, making use of the natural arsenic resistance operon in E. coli adding a reporter gene controlled by this operon (Stocker et al., 2003; Siegfried et al., 2012; Jia et al., 2019; Wan et al., 2019; Chen et al., 2022). It has been shown possible to improve signal output by optimization of individual components within a given circuit topology, for instance improving dynamic range through the choice of promoters (Chen et al., 2022). However, circuits can be specifically constructed for increasing sensitivity, for instance by implementing an additional positive feedback loop (Jia et al., 2019) or by cascading signal amplification. Wan et al. added up to three orthogonal amplifier modules, in which the original output of the arsenic resistance operon serves as input for the first amplifier, whose output then drives the second amplifier, and similarly the second amplifier’s output feeds into the third amplifier module (Figure 7) (Wan et al., 2019). Likewise, a plethora of other heavy metal biosensors have been designed, making use of different sensor (input) and reporter (output) modules, implementing amplifier circuits, as well as features such as logic gates and feedback loops [reviewed in (Kim et al., 2018; Liu et al., 2022)].

Figure 7. Cascaded amplifying circuit design for ultrasensitive heavy metal detection. The genetic circuit consists of an input module sensing arsenic (As3+) and three sequential amplifier modules driving GFP expression as output (Wan et al., 2019). The input module uses the natural ArsR-based arsenic sensing system, which is expressed under a weak promoter and represses the promoter of the first amplifier A (ParsR) in absence of arsenic. A similar system can be constructed for mercury detection by replacing the input module with MerR controlling the PmerT promoter. Upon heavy metal binding, ArsR releases from ParsR, allowing expression of the first amplifier module (hrpR/hrpS). HrpR and HrpS proteins form a complex that activates the hrpL promoter controlling ECF (extracytoplasmic function sigma factor) expression. ECF in turn activates Pe11 to drive expression of RinA, which binds to the rinA promoter on a high-copy plasmid to trigger GFP expression. The sequential amplification through three stages (HrpRS, ECF, and RinA) enables ultrasensitive detection of arsenic. Elements are organized on two plasmids: a low-copy plasmid carrying the sensing and amplification modules, and a high-copy plasmid carrying the output module. Arrows indicate activation, T-bars indicate repression.
Memory circuits allow cells to “remember” past events and maintain a specific state over time, even after the initial stimulus has been removed. In yeast, a synthetic memory circuit has been constructed using a transcriptional positive feedback loop. Once activated by a transient stimulus, the circuit maintains its active state through self-sustaining feedback (Ajo-Franklin et al., 2007).
In contrast to states being maintained by transcriptional control, devices changing the DNA sequence do not require sustained metabolic expenditure to upkeep memory. The SCRIBE system uses inducible DNA recombinases in combination with a retron, an inducible bacterial reverse transcriptase system, to produce ssDNA which introduces specific mutations in the genome based on sequence homology (Farzadfard and Lu, 2014), allowing long-term storage of cellular memories across multiple generations. Another approach used recombinases to irreversibly flip DNA sequences, creating a permanent record of transient signals in bacteria as well as mammalian cells (Figure 8A) (Siuti et al., 2013; Weinberg et al., 2017). A synthetic cellular memory device capable of storing more than 1 byte of information in the DNA of living cells has been constructed from a memory array of 11 orthogonal integrases (Yang et al., 2014).

Figure 8. DNA-based memory systems employing recombinases and prime editing (A) An input signal triggers the recombinase-based genetic memory system. The input signal (red) drives expression of a recombinase (Rec, purple) from a signal-responsive promoter (Psignal, red). The recombinase recognizes specific sites (purple triangles) and catalyzes either deletion or inversion of the intervening DNA part (yellow), creating permanent genetic modifications, which, depending on the part can trigger a specific output, by, e.g., flipping promoter or coding sequences for activation or deactivation or deleting a terminator (Siuti et al., 2013; Weinberg et al., 2017) (B) The peCHYRON (prime editing CHYRON) system allows sequential DNA recording of different signals (Loveless et al., 2024). A prime editor (PE, blue), consisting of a nicking Cas9 and a reverse transcriptase, is guided to the target sequence by prime editing guide RNAs (pegRNAs). Each pegRNA contains a spacer sequence (spA, orange or spB, purple), a constant sgRNA scaffold (c, dark blue), and a reverse transcription template (RTT). The RTT consists of an overhang region (RTo, dark blue), propagation sequence (PropA, orange or PropB, purple), containing a 3-nucleotide signature (yellow symbols), and the primer binding sites (PBS A, orange or PBS B, purple). Sequential recording begins when PE binds and nicks the target DNA strand, allowing the RTT to hybridize via its primer binding site. This initiates reverse transcription to insert the propagation sequence containing one of several possible 3-nucleotide signatures (yellow symbols) that encode up to 6 bits of information. Sets of alternating A→B and B→A pegRNAs enable sequential recording, as each inserted propagation sequence serves as the target site for the next recording round. This self-propagating architecture enables theoretically unlimited rounds of sequential recording.
More recently, CRISPR-based systems have been employed for genomic recording, leading to several advanced memory systems. Sheth et al. introduced TRACE (Temporal Recording in Arrays by CRISPR Expansion), which enabled the recording of multiple cellular events over time (Sheth et al., 2017). This system makes use of the bacterial CRISPR-Cas adaptation process, which can integrate exogenous nucleic acid into the genomic CRISPR arrays as spacer. To convert the input signal into such trigger DNA, the authors linked the input signal to the expression of phage P1 lytic replication protein RepL, which in turn initiates the replication of a trigger plasmid (pTrig). The authors could show that the system responds to the amount of input signal and can also be adapted for different input signals.
CAMERA (CRISPR-mediated Analog Multi-event Recording Apparatus) is capable of recording multiple cellular events in both bacterial and mammalian cells (Tang and Liu, 2018). Beyond simple presence/absence detection, this approach enables analog recording that captures amplitude or signal duration. This system consists of a writer plasmid expressing either Cas9 with sgRNA or a base editing Cas system under inducible promoters, and recorder plasmids that serve as writing substrate. Upon activation, the system modifies the recorder plasmids through either Cas9-mediated cutting or base editing, and this recording can then be detected by measuring the ratio between modified and unmodified recorder plasmids.
Building on the SCRIBE system, the DOMINO (DNA-based Ordered Memory and Iteration Network Operator) system utilizes CRISPR-based base editing to introduce memory in the form of single-nucleotide mutations as a response to external stimuli (Farzadfard et al., 2019). The used base-editing fusion protein consists of a nickase Cas9 (nCas9), a cytidine deaminase (CDA), and a uracil glycosylase inhibitor (ugi), guided by a single guide RNA (sgRNA) to specific genomic loci, where it performs targeted C-to-T mutations. By controlling the expression of the fusion protein and one or more sgRNAs with inducible promoters, the system can operate in both “analog” mode, where the mutation frequency correlates with the intensity and duration of the input signals, and “digital” mode, where transition of fully converted states are considered.
Two recent approaches enable ordered recording in mammalian cells through sequential genome editing. CHYRON (Cell History Recording by Ordered Insertion) combines CRISPR-Cas9 with terminal deoxynucleotidyl transferase to achieve ordered insertions at a synthetic recording array (Loveless et al., 2021). Building on this, two prime editing-based DNA recorders were developed: DNA Typewriter records through sequential insertions at predefined target arrays (Choi et al., 2022), being able to maintain sequential edits across at least 20 generations and 25 days. PeCHYRON (prime editing CHYRON) iteratively adds 3-nucleotide signatures (theoretically encoding up to 6 bits each) alongside sequences that create new target sites (Loveless et al., 2024). By generating its own target sites rather than requiring a synthetic array, this self-propagating architecture allows theoretically unlimited rounds of recording (Figure 8B). These approaches overcome earlier limitations by achieving ordered accumulation of information-rich edits while avoiding double-strand breaks.
The progression from simple feedback loops to sophisticated CRISPR-based systems has opened up new possibilities for recording and storing intracellular and extracellular events over time in living cells. Such DNA-based memory systems have potential applications in tracking and recording biologically relevant information, e.g., in difficult to assess areas such as bioreactors, host-associated microbiomes or environments (Sheth and Wang, 2018).
Building on the principles of basic genetic circuits and memory systems, researchers have developed increasingly sophisticated genetic circuits capable of complex computations and multi-state operations.
Friedland et al. created a riboregulated transcriptional cascade counter (RTC) in E. coli capable of counting up to three induction events (Friedland et al., 2009). The circuit has three nodes in the cascade: T7 RNAP drives transcription of T3 RNAP, which drives transcription of a GFP reporter. Both transcripts are cis-repressed by riboregulators, which form a stem-loop structure with the RBS preventing translation. Repression can be relieved by a trans-activating RNA, which is driven by an arabinose-inducible promoter. Using short arabinose pulses, the authors could show that three pulses are necessary to produce T7 RNAP, T3 RNAP and finally, GFP.
Green et al. have constructed multiple toehold switches forming AND, OR and NOT gates (Figure 9) and constructed from these larger ribocomputing devices by combining up to five AND, five OR and 2 NOT gates, demonstrating how computational tasks can be implemented in living cells using RNA control mechanisms (Green et al., 2017).
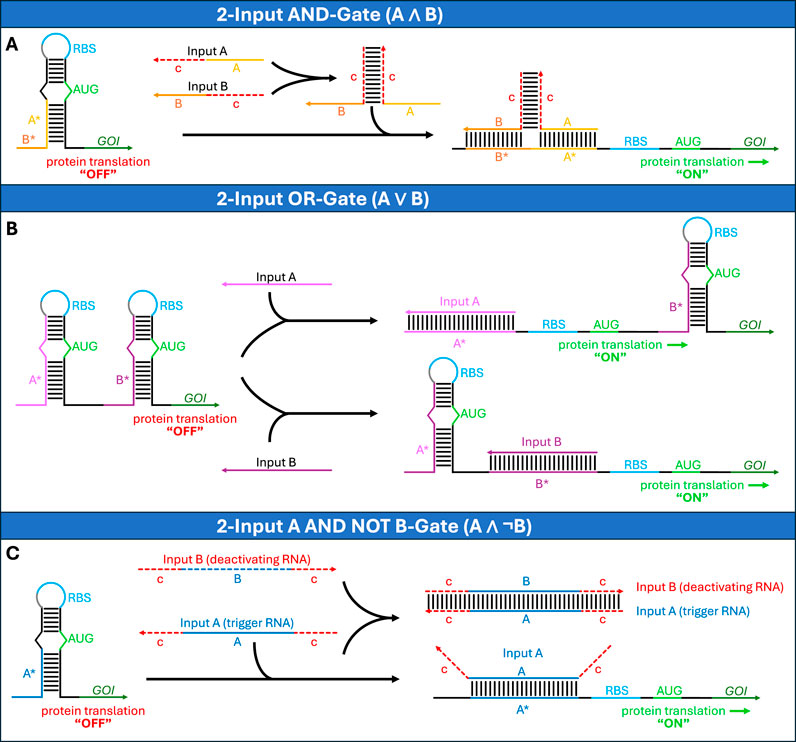
Figure 9. Fundamental RNA-based logic gate designs for cellular computation (A) The two-input AND gate consists of a toehold switch that regulates gene expression through sequestration of the ribosome binding site (RBS, blue) and start codon (AUG, green) within a stem-loop structure. Two input RNAs (A ∧ B) are required, each containing half of the trigger sequence (A, yellow or B, orange) and complementary regions (c, red) that enable their hybridization. Only when both inputs are present, they form a complete trigger RNA complex that can bind to the gate RNA (via A* and B* regions), unfolding the stem-loop and enabling protein translation (B) The two-input OR gate contains two toehold switch modules in series, each with its own RBS and start codon. Either input RNA (A ∨ B) can independently bind to its corresponding region (A* or B*) in the gate RNA to activate translation. Translation from different start codons results in the same functional protein with different N-terminal extensions (C) The A AND NOT B gate (A ∧ ¬B) employs a toehold switch activated by input A (trigger RNA). Input B (deactivating RNA) contains sequences complementary to input A (regions c for initial binding and region B complementary to A) and can either prevent trigger RNA binding to the gate RNA or displace already bound trigger RNA through strand displacement, thereby maintaining translational repression.
CRISPR-based systems have also been employed for cellular computing. The DOMINO (DNA-based Ordered Memory and Iteration Network Operator) system described in the previous section has been used for order-independent, sequential and temporal logic operations (Farzadfard et al., 2019).
In mammalian cells, Weinberg et al. developed a strategy called BLADE (Boolean logic and arithmetic through DNA excision) to implement biocomputing operations (Weinberg et al., 2017). Using a set of 12 recombinases, they constructed 113 circuits, from which 96.5% worked as predicted without further optimization. This allowed them to construct not only all possible Boolean logic functions but also arithmetic logic circuits such as a three-input-2-output full adder or a half adder-subtractor, which can add or subtract two inputs depending on the presence of a third select input.
The previous sections introduced 1) a variety of fundamental approaches that genetic devices for controlling gene function have been based on, and 2) basic principles and considerations to build higher-order genetic circuitry from these basic regulatory parts. Leveraging the ever-growing toolbox of regulatory parts through engineering principles has fueled the creation of increasingly complex biological systems. This foundation has enabled novel bio-based solutions and step-changing advances in a variety of domains. Here, we want to highlight some relevant advances and use cases in sustainable bioproduction, novel therapeutics, and survival control over engineered microbes.
Industrial biotechnology has historically relied on the existing physiological capabilities of microorganisms for bioproduction. Initially, metabolic flux was directed towards the desired product through strategic control of fermentation parameters such as medium composition, oxygenation and pH. A prime example of such a process is the acetone–butanol–ethanol (ABE) fermentation using Clostridium acetobutylicum. However, the natural metabolic capabilities of microorganisms severely limit the range of compounds that can be (economically) produced.
Recombinant technology dramatically changed the field by enabling the transfer of biosynthetic genes from different organisms to the production host. A prominent example is the production of artemisinic acid, a precursor to the antimalarial drug artemisinin, in yeast. Paddon et al. engineered S. cerevisiae to produce high titers of artemisinic acid from inexpensive carbon substrates by introducing the biosynthetic pathway from Artemisia annua and fine-tuning expression levels of key enzymes, balancing metabolic flux and mitigating toxicity issues (Paddon et al., 2013).
Advances in gene regulatory networks have introduced sophisticated regulatory circuits that enable dynamic control over gene expression and metabolic flux. These circuits can optimize yields by reducing toxicity and metabolic burden, making production processes more robust. For example, in fatty acid biosynthesis, which provides precursors for diverse high-value products from pharmaceuticals to cosmetics, simple overexpression of the rate-limiting enzyme acetyl-CoA carboxylase (ACC) is generally toxic to cells. This challenge was addressed in E. coli by implementing a sensor-actuator system with dynamic feedback control using the malonyl-CoA-sensitive transcriptional repressor FapR from Bacillus subtilis to link ACC expression to the amount of malonyl-CoA (Figure 10A) (Liu et al., 2015).
Figure 10. Sensor-Actuator System and metabolic switches for dynamic pathway control (A) The malonyl-CoA sensor-actuator system enables dynamic feedback regulation of fatty acid precursor biosynthesis. The rate-limiting step catalyzed by acetyl-CoA carboxylase (ACC, red) converts acetyl-CoA to malonyl-CoA. To enhance malonyl-CoA production while preventing toxic ACC accumulation, a negative feedback loop was implemented using the malonyl-CoA-sensitive transcriptional repressor FapR (blue) and a synthetic promoter (PFR1). In the absence of malonyl-CoA, FapR binds to PFR1, preventing LacI (orange) expression, which results in ACC expression from PT7. Increased ACC expression raises malonyl-CoA levels, which trigger FapR/promoter dissociation, thereby increasing LacI expression and reducing ACC levels until a new steady state is reached (Liu et al., 2015) (B) CRISPR-based metabolic switch for phosphate-dependent gene silencing. The system consists of the native Escherichia coli CRISPR/Cascade machinery (brown) (with cas3 deleted) under constitutive expression (Pc), and a phosphate-sensitive promoter (PugpBp) controlling gRNA (yellow) expression. Under low phosphate conditions, expressed gRNAs direct the CASCADE complex to the promoter region of the target gene (goi, gene of interest; green), reducing its expression (Ye et al., 2021) (C) Proteolysis-based metabolic switch utilizing the SspB/ClpXP system. The target enzyme (EOI, enzyme of interest; green) is chromosomally tagged with a C-terminal DAS+4 degron tag (red) and constitutively expressed (Pc). Under low phosphate conditions, the PugpBp promoter drives expression of the SspB chaperone (blue), which binds to the DAS+4 tag and targets the enzyme for ClpXP-mediated proteolysis. These switches can be used to control expression of key metabolic enzymes such as citrate synthase (GltA), glucose-6-phosphate dehydrogenase (Zwf), enoyl-ACP reductase (FabI), or soluble transhydrogenase (UdhA) to enable transition from growth to production phase through phosphate-dependent control (Ye et al., 2021).
Beyond mitigating toxicity, dynamic metabolic flux control strategies have emerged to optimize feedstock utilization in bioproduction. A notable approach is to limit biomass production by decoupling growth from bioproduction, for instance in response to an external trigger. Harder et al. used temperature to control the production of itaconic acid (Harder et al., 2018). By replacing the promoter of isocitrate dehydrogenase with a promoter controlled by a temperature-sensitive repressor, metabolism proceeds normally with biomass production at higher temperatures, while lowering the temperature activates the repressor, thereby blocking the TCA cycle and redirecting the flux to itaconic acid. This temperature-controlled metabolic switch exemplifies how external physical stimuli can be strategically used to dynamically regulate metabolic pathways.
Gupta et al. constructed a pathway-independent system for autonomous dynamic regulatory circuit control in E. coli that can respond to metabolic states and adjust pathway flux under a variety of process parameters (Gupta et al., 2017). Leveraging parts of a quorum-sensing system from Pantoea stewartii, they constructed a system that can autonomously regulate gene expression without the need for external inducers or human intervention. Pandit et al. defined the concept of pathway orthogonality, postulating optimal target production when the production pathway and natural metabolism were largely independent (Pandit et al., 2017). They validated this concept by introducing a metabolic valve—a control reaction that directs the desired flux towards either biomass production or product formation.
To improve process robustness across different production scales, a two-stage dynamic regulation strategy was developed (Ye et al., 2021). In this system, engineered synthetic metabolic valves respond to low phosphate levels, triggering both gene silencing through CRISPRi and proteolysis through engineered degron tags to downregulate key metabolic enzymes (Figures 10B,C). Shifting the cellular state from growth to stationary phase, metabolic flux is redirected towards product synthesis while simultaneously suppressing inhibitory feedback loops. The concept of metabolic valves has also been applied in P. putida, a promising chassis for industrial biotechnology (Batianis et al., 2023). In this study, pyruvate was used as a control node as it constitutes a key metabolic molecule for the TCA cycle and fatty acid biosynthesis, but also serves as a precursor for several industrially relevant products such as branched-chain alcohols, diol, and isoprenoids.
A large variety of novel therapeutic approaches have been enabled by synthetic biology tools and concepts. Here we highlight two examples of therapeutic areas: one based on engineered bacteria and the other on engineered mammalian cells, demonstrating how designed regulatory circuitry helps address specific challenges.
Engineering probiotics for targeted drug delivery represents a promising approach, particularly for treating metabolic gastrointestinal disorders. Early efforts in this field focused on simple expression systems, which laid the groundwork for more sophisticated genetic circuits that provide better control of production and release of therapeutic molecules in response to specific environmental cues.
One example of engineered probiotics for metabolic disorders is an approach targeting phenylketonuria (PKU), a genetic disorder with impaired activity of phenylalanine hydroxylase (PAH) resulting in hyperphenylalaninemia (HPA) and intellectual disability. Sarkissian et al. demonstrated the potential of using recombinant phenylalanine ammonia lyase (PAL) for PKU treatment, although their approach involved direct enzyme administration rather than probiotic delivery (Sarkissian et al., 1999). Building on this concept, a probiotic strain of Lactobacillus reuteri was later engineered to express PAL for PKU treatment from a constitutive promoter. While the system resulted in decreased plasma Phe levels, it failed to reduce them to physiologic ranges in a PKU mouse model (Durrer et al., 2017).
This issue was addressed with an engineered E. coli Nissle 1917 strain, SYNB1618, with a more advanced genetic circuit for PKU treatment (Isabella et al., 2018). SYNB1618 incorporates two complementary phenylalanine-metabolizing pathways functioning at different oxygen levels while also reducing the metabolic load on the engineered strain and ensuring genetic stability (Figure 11). L-amino acid deaminase (LAAD), a highly active, but oxygen-dependent enzyme, was expressed under Arabinose control, whereas the expression of PAL as well as the Phe transporter PheP, which ensures intracellular transport of Phe for PAL-mediated degradation, was controlled by an anaerobic-inducible promoter to be triggered by increasingly anoxic conditions in the gastrointestinal tract. Additionally, a separate circuit produced PAL in response to IPTG to ensure high enzyme levels during strain production. Genetic stability was further achieved by genomic integration of several copies of the respective genes at different positions such that sequences between insertion loci contained essential genes to avoid any recombination events. Furthermore, SYNB1618 included an additional safety switch based on 4-hydroxy-tetrahydropicolinate synthase gene (dapA) deletion, making the strain dependent on exogenous diaminopimelate (DAP) for cell wall biosynthesis and growth. This added an extra layer of control and safety to the system, addressing concerns about the application of genetically-modified organisms.

Figure 11. Circuit design for probiotic Escherichia coli Nissle strain as treatment strategy for Phenylketonuria. The strain contains two inducible Phe degradation modules (blue and green dashed box) (Isabella et al., 2018). LAAD (blue), which is expressed under arabinose control, leads to oxygen-dependent extracellular Phe degradation to Phenylpyruvate (PP). The second module (green dashed box) works in anoxic environment, where low oxygen triggers expression of both PAL (green) and the Phe-transporter PheP (orange) to enable cytosolic Phe degradation to trans-cinnamate (TCA). To ensure PAL expression also during aerobic strain production, additional gene copies under the Ptac promoter allowed IPTG-induced PAL expression. Both Phe degradation modules were inserted genomically at different numbers (as indicated by the numbers for each circuit) at sites previously identified as suitable integration sites. The biocontainment module (violet) consists of a deletion of the dapA gene, rendering the cells dependent on DAP for cell wall biosynthesis and growth.
The strain was further refined by improving PAL activity through a biosensor-based selection process using an allosteric transcription factor that triggers GFP expression in the presence of trans-cinnamate (TCA), the Phe degradation product of PAL (Adolfsen et al., 2021). The resulting strain SYNB1934 performed successfully in a phase 2 clinical trial (SynPheny-1; NCT04534842) in adults with PKU (Vockley et al., 2023), however a proceeding study (SynPheny-3, NCT05764239) was terminated as it was deemed unlikely to meet the primary endpoint. Importantly, the termination was not due to any safety or tolerability concerns illustrating the potential for safe use of such probiotics.
Engineered probiotics can also be used for diagnostics, as explored in the work from Riglar et al., who developed a commensal murine E. coli strain to sense inflammation signals in the gastrointestinal tract of mice and retain this “memory” for later analysis by fecal testing (Riglar et al., 2017). The system combines the TtrR/TtrS/PttrBCA system from Saccharomyces typhimurium and the Cro-inducible CI/Cro transcriptional switch from phage lambda in two circuits. In the trigger circuit, tetrathionate, a transient product of reactive oxygen species (ROS) characteristic for inflammation, leads to phosphorylation of TtrS and TtrR and subsequent activation of the PttrBCA, driving Cro expression. The memory circuit is initially in the OFF-state, dominated by the CI repressor. When Cro is expressed from the trigger circuit, it acts on the memory circuit, switching it from the CI-dominated OFF-state to the Cro-dominated ON state. This switch leads to further Cro expression in the memory circuit to maintain the memory state, as well as expression of lacZ as a reporter element. The authors demonstrated that their engineered bacteria could successfully colonize the mouse gut and maintain their responsiveness to inflammation signals in vivo for over 6 months. This long-term functionality suggests good genetic stability and low metabolic burden imposed by the synthetic pathway, addressing key concerns in the development of engineered probiotics for clinical applications.
To facilitate circuit design not only in well-characterized lab stains but also in clinically relevant strains such as E. coli Nissle 1917 (EcN), Lebovich et al. developed a scalable computational approach for designing sequential logic and gene circuits to allow multiplex sensing and signal recording (Lebovich et al., 2023). They characterized a library of 16 transcriptional NOT gates and nine biochemical sensors specifically in EcN, demonstrating that these components could be used to create circuits capable of sensing multiple inputs and recording memory through sequential logic. The study highlighted the differences in signal processing between EcN and laboratory strains, which necessitated the establishment of strain-specific response functions for accurate predictive design. Using this strain-specific characterization data, they implemented a computational design approach for creating robust sequential logic circuits. The authors successfully constructed various genetic circuits, including combinational logic circuits, memory circuits (set-reset (SR) latches), and a more complex concentration-recording circuit by combining SR latches with different sensitivities for input signals. This latter circuit can detect, record, and report three distinct concentration ranges of a biochemical signal using sequential logic, showcasing the potential for more sophisticated sensing and reporting of gut conditions. Their work provides both a valuable toolkit of characterized components and design algorithms, as well as proof-of-concept circuits that could serve as a foundation for future engineering of probiotics as living diagnostic or even therapeutic devices for applications in gut health monitoring or treatment.
Chimeric Antigen Receptor (CAR) T-cell therapy is an innovative immune therapy approach where patient T cells are modified ex vivo to express a chimeric antigen receptor targeting a tumor-specific antigen. This approach has been particularly successful in treating hematological malignancies (Lu et al., 2024), with the first FDA approvals in 2017 for tisagenlecleucel (KYMRIAH) and axicabtagene ciloleucel (YESCERTA) (FDA, 2017a; 2017b). While CAR-T cell therapy has shown remarkable outcomes, a major challenge remains controlling potential adverse effects, such as on-target off-tumor (OTOT) toxicity, which occurs when CAR-T cells target healthy tissues expressing low levels of the targeted antigen (Sterner and Sterner, 2021). To improve the safety and efficacy of CAR-T cell therapy, synthetic gene circuits have been integrated into CAR designs to provide better control over T-cell activity.
One fundamental approach to addressing these safety concerns involves implementing safety switches. Gargett and Brown designed an inducible caspase-9 (iCasp9) suicide switch by fusing a modified caspase-9 protein to a drug-binding domain (Gargett and Brown, 2014). Administration of a small molecule dimerizer triggers caspase-9 dimerization, activating downstream caspase-3 and inducing rapid apoptosis in the CAR-T cells. The system has already demonstrated efficacy in a phase 1 clinical trial (Gargett et al., 2024).
While safety switches provide crucial emergency control, more sophisticated genetic circuits have been developed to prevent adverse effects by enhancing the precision of CAR-T cell activation. Roybal et al. introduced a combinatorial antigen-sensing circuit design to enhance the precision of CAR-T cell therapies and reduce off-target activation (Roybal et al., 2016). They designed a synthetic Notch (synNotch) receptor to detect the first antigen, which then induces the expression of a CAR binding the second antigen (Figure 12A). Building on this two-step “AND-gate” approach, similar systems were developed for better discrimination between tumor and healthy tissue (Srivastava et al., 2019; Choe et al., 2021).
Figure 12. Advanced genetic circuits for enhanced control of CAR-T cell activity. Schematic representation of four strategies to improve the specificity and safety of CAR-T cell therapy (A) The synthetic Notch (synNotch) receptor system (green) enables sequential antigen detection. While the CAR (blue) targets tumor cell antigen 1 (Ag 1, pink), cell activation only occurs when synNotch recognizes antigen 2 (Ag 2, purple), releasing a transcription factor (TF, green) that drives CAR expression. This creates an AND-gate logic requiring both antigens for full T cell activation (Roybal et al., 2016) (B) The oxygen-sensing system restricts CAR activity to hypoxic environments as found in solid tumors. Fusion of CAR (blue) to an oxygen-dependent degradation domain (ODD, gray) ensures CAR degradation under normoxic conditions, while the hypoxic tumor microenvironment stabilizes CAR expression. To prevent leakiness, CAR expression is additionally controlled by a hypoxia-responsive promoter (HRE) (Kosti et al., 2021) (C) The conditional payload expression system enables antigen-dependent enhancement of T cell function. CAR-mediated antigen recognition triggers expression of a payload gene (e.g., IL2, orange) from an NR4A-based promoter (PNR4A). The secreted factors support T cell proliferation and function through autocrine signaling, particularly under suboptimal stimulation conditions. The system provides a feedback mechanism where payload expression is coupled to the level of T cell activation (Guo et al., 2022) (D) The drug-dependent control system provides temporal regulation of T cell activity. A small molecule (brown) activates a synthetic transcription factor (synTF, orange) to control CAR expression, enabling precise timing of therapeutic activity (Li et al., 2022). Pc: constitutive promoter; HRE: hypoxia response element.
Environmental cues can also be leveraged to enhance targeting specificity. Utilizing the hypoxic tumor environment, a dual-input genetic circuit was developed responding to both antigen recognition and low oxygen levels (Figure 12B) (Juillerat et al., 2017; Kosti et al., 2021).
Beyond controlling activation, genetic circuits can enhance CAR-T cell function in challenging environments. Allen et al. engineered a synthetic cytokine circuit where tumor-antigen detection through a synthetic Notch receptor triggers IL-2 production independently of T cell receptor or CAR activation (Allen et al., 2022). This autocrine IL-2 release enables T cells to proliferate and infiltrate immune-excluded tumors while minimizing systemic toxicity. In a similar approach, Guo et al. coupled expression of a payload directly to CAR activation (Figure 12C) (Guo et al., 2022). This system provides tight control over payload expression with minimal background activity and could be particularly valuable for delivering immune-enhancing molecules when CAR-T cell activation is suboptimal.
Temporal control over CAR-T cell activity represents another important aspect of precision therapy. Li et al. developed a system where small molecules regulate synthetic transcription factors based on zinc finger proteins to control CAR expression (Figure 12D). By using FDA-approved drugs as triggers, this strategy enables precise timing of therapeutic activity while prioritizing clinical compatibility (Li et al., 2022).
While this review focuses on genetic control circuits, many advances in CAR-T cell engineering have been achieved through protein engineering, including programmable receptor designs with various logic gates (AND, OR) as well as internal or external control mechanisms for improved specificity and safety, as reviewed recently (Lee et al., 2022; Young et al., 2022; Neeser et al., 2023).
The deployment of engineered organisms outside laboratory settings necessitates robust safety measures to prevent environmental release or unauthorized proliferation. Various genetic safeguard strategies have been developed to address these concerns, primarily focusing on conditional survival mechanisms and contained genetic material [reviewed in (Simon and Ellington, 2016; Lee et al., 2018; Hoffmann et al., 2023)]. Kill switches represent a major category of biosafety mechanisms, where typically toxic proteins are activated or de-repressed under specific conditions such as the presence or absence of particular signals. Stringent control of toxin activity is crucial to ensure activation only when desired and to then induce reliable cell death.
Chan et al. demonstrated two sophisticated circuitry approaches for bacterial containment (Chan et al., 2016). The “deadman” switch, based on a monostable toggle design with reciprocal repression between LacI and TetR, requires continuous presence of an input signal (aTc, anhydrotetracyclin) for cell survival (Figure 13A). Upon loss of the input signal, the circuit triggers expression of both a toxin and a heterologous protease, which degrades the LacI repressor as well as essential cellular proteins tagged for degradation, thereby ensuring efficient cell death through multiple mechanisms (Chan et al., 2016).

Figure 13. Genetic circuits for bacterial containment through engineered kill switches (A) The “deadman” switch circuit is based on a monostable toggle with reciprocal repression between LacI and TetR transcription factors (Chan et al., 2016). Continuous presence of anhydrotetracycline (aTc, blue) maintains LacI (green) expression, resulting in cell survival by blocking toxin expression. Without aTc, the circuit triggers expression of a toxin (red) and a heterologous protease (mf-Lon, yellow). The protease accelerates switching dynamics by degrading LacI tagged with the mf-Lon degradation sequence (yellow), and provides an additional safety layer through degradation of the essential protein MurC (purple), involved in cell wall biosynthesis, which was similarly tagged with the mf-Lon recognition sequence. The dark shaded RBS symbol (semicircle) indicates a strong RBS, while the dashed line represents weak repression. (B) The “passcode” switch links cell survival to a specific combination of input signals (Chan et al., 2016). The circuit with two sequential AND gates was constructed with designed hybrid transcription factors, in which the DNA recognition modules (DRMs, green and dark red) and the environmental sensing modules (ESMs, pink, orange, cyan) from different transcription factors were combined. The first AND gate requires two different input molecules (input A and input B) for activation. The output of this gate then feeds into a second AND gate, which also takes the inverse of a third input signal (NOT C). This arrangement ensures cell survival only when both A AND B are present, AND C is absent, creating a specific molecular “passcode” for survival. (Pc, constitutive promoter; Ptet, aTc-dependent promoter; Plac, LacI-dependent promoter; Pscr, ScrR-dependent promoter from Klebsiella pneumoniae; A, B, C, ESM from transcriptions factors responsive to input molecules A, B and C; L, DRM from LacI; S, DRM from ScrR).
The “passcode” switch provides tight control through a defined combination of input signals (Figure 13B) (Chan et al., 2016). Using designed hybrid transcription factors, it implements Boolean logic through sequential AND gates, requiring both the presence of two inputs (A AND B) and absence of a third input (NOT C) for cell survival (survival = (A ∧ B) ∧ ¬C). Incorrect input combinations lead to toxin expression and cell death (death = (¬A ∨ ¬B) ∨ C), creating a specific molecular “passcode”. Both systems achieved robust control of cell survival. However, rigorous testing produced some escape mutants. Analysis revealed that these bypassed the kill switch mainly through inactivating mutations in the toxin genes, highlighting the need for additional containment strategies. While this review has focused on synthetic circuit-based containment strategies, it is important to note that other strategies such as metabolic auxotrophies have been developed as well, and combining multiple containment mechanisms will provide a more comprehensive approach to ensuring the safe deployment of engineered organisms.
The engineering of gene regulatory networks draws from decades of cumulative research into gene regulation. From aiming to understand regulation in natural systems, it has branched to using natural parts in different contexts and altering them, to developing new-to-nature regulatory devices. As laid out here, there now exists a staggering variety of molecular parts that allow regulatory interference of gene expression and activity at virtually any point along the flow of genetic information. This ever-growing toolbox has underpinned the construction of artificial genetic circuitry, consisting of several or multiple fundamental regulatory units, performing increasingly complex functions.
The application of engineering principles in pursuit of predictable forward design has enabled systematic approaches for circuit development and largely turned the field into an engineering discipline. While traditional engineering approaches continue to be refined, machine learning and artificial intelligence tools are increasingly being employed in synthetic biology (Goshisht, 2024). A persistent bottleneck remains the availability of sufficient experimental data connecting genotypes to phenotypes. To address this limitation, high-throughput approaches are required for DNA assembly and delivery, and assaying of resulting system behavior, either leveraging lab automation platforms, or employing pooled approaches typically underpinned by genetic barcoding and deep sequencing.
However, considering not only the vast number of parts, but also the extremely high dimensionality and context sensitivity of biological systems—for instance pertaining to the particular host strain, culture conditions, physiological state, sequence context, combination of regulatory systems—it is safe to assume that experimental data will continue to be a limiting factor in the development of a ‘general’ forward design tool. The intrinsic complexity of biological systems inherently limits the degree of possible abstraction, affecting predictability of the behavior of derived higher-order systems across a wide parameter range. Still, we expect that computer-aided tools for the design of synthetic circuitry will increasingly be integrated with comprehensive whole-cell models describing cellular processes at different scales to make forward design more robust (Marucci et al., 2020).
This integration of multiple modeling approaches, combined with experimental and computational advances in creating and transplanting artificial genomes (James et al., 2024), is expected to pave the way towards the bottom-up creation of designed microorganisms tailored for particular tasks. These advancements hold the promise to unlock transformative applications, addressing global challenges in health, environmental protection, and sustainable production.
MM: Visualization, Writing–original draft, Writing–review and editing. KA: Conceptualization, Visualization, Writing–original draft, Writing–review and editing. SH: Conceptualization, Writing–original draft, Writing–review and editing.
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abraham, J. M., Freitag, C. S., Clements, J. R., and Eisenstein, B. I. (1985). An invertible element of DNA controls phase variation of type 1 fimbriae of Escherichia coli. Proc. Natl. Acad. Sci. 82, 5724–5727. doi:10.1073/pnas.82.17.5724
Adolfsen, K. J., Callihan, I., Monahan, C. E., Greisen, P., Spoonamore, J., Momin, M., et al. (2021). Improvement of a synthetic live bacterial therapeutic for phenylketonuria with biosensor-enabled enzyme engineering. Nat. Commun. 12, 6215. doi:10.1038/s41467-021-26524-0
Ahmadzada, T., Reid, G., and McKenzie, D. R. (2018). Fundamentals of siRNA and miRNA therapeutics and a review of targeted nanoparticle delivery systems in breast cancer. Biophys. Rev. 10, 69–86. doi:10.1007/s12551-017-0392-1
Ajo-Franklin, C. M., Drubin, D. A., Eskin, J. A., Gee, E. P. S., Landgraf, D., Phillips, I., et al. (2007). Rational design of memory in eukaryotic cells. Genes Dev. 21, 2271–2276. doi:10.1101/gad.1586107
Allen, G. M., Frankel, N. W., Reddy, N. R., Bhargava, H. K., Yoshida, M. A., Stark, S. R., et al. (2022). Synthetic cytokine circuits that drive T cells into immune-excluded tumors. Science 378, eaba1624. doi:10.1126/science.aba1624
Alshaer, W., Zureigat, H., Al Karaki, A., Al-Kadash, A., Gharaibeh, L., Hatmal, M. M., et al. (2021). siRNA: mechanism of action, challenges, and therapeutic approaches. Eur. J. Pharmacol. 905, 174178. doi:10.1016/j.ejphar.2021.174178
Anastassov, S., Filo, M., Chang, C.-H., and Khammash, M. (2023). A cybergenetic framework for engineering intein-mediated integral feedback control systems. Nat. Commun. 14, 1337. doi:10.1038/s41467-023-36863-9
Anderson, J. C., Voigt, C. A., and Arkin, A. P. (2007). Environmental signal integration by a modular AND gate. Mol. Syst. Biol. 3, 133. doi:10.1038/msb4100173
Ang, J., Harris, E., Hussey, B. J., Kil, R., and McMillen, D. R. (2013). Tuning response curves for synthetic biology. ACS Synth. Biol. 2, 547–567. doi:10.1021/sb4000564
Anzalone, A. V., Randolph, P. B., Davis, J. R., Sousa, A. A., Koblan, L. W., Levy, J. M., et al. (2019). Search-and-replace genome editing without double-strand breaks or donor DNA. Nature 576, 149–157. doi:10.1038/s41586-019-1711-4
Aoki, S. K., Lillacci, G., Gupta, A., Baumschlager, A., Schweingruber, D., and Khammash, M. (2019). A universal biomolecular integral feedback controller for robust perfect adaptation. Nature 570, 533–537. doi:10.1038/s41586-019-1321-1
Banaszynski, L. A., Chen, L., Maynard-Smith, L. A., Ooi, A. G. L., and Wandless, T. J. (2006). A rapid, reversible, and tunable method to regulate protein function in living cells using synthetic small molecules. Cell 126, 995–1004. doi:10.1016/j.cell.2006.07.025
Bandyra, K. J., Said, N., Pfeiffer, V., Górna, M. W., Vogel, J., and Luisi, B. F. (2012). The seed region of a small RNA drives the controlled destruction of the target mRNA by the endoribonuclease RNase E. Mol. Cell 47, 943–953. doi:10.1016/j.molcel.2012.07.015
Batianis, C., Van Rosmalen, R. P., Major, M., Van Ee, C., Kasiotakis, A., Weusthuis, R. A., et al. (2023). A tunable metabolic valve for precise growth control and increased product formation in Pseudomonas putida. Metab. Eng. 75, 47–57. doi:10.1016/j.ymben.2022.10.002
Baumschlager, A., Aoki, S. K., and Khammash, M. (2017). Dynamic blue light-inducible T7 RNA polymerases (Opto-T7RNAPs) for precise spatiotemporal gene expression control. ACS Synth. Biol. 6, 2157–2167. doi:10.1021/acssynbio.7b00169
Beal, J., Nguyen, T., Gorochowski, T. E., Goñi-Moreno, A., Scott-Brown, J., McLaughlin, J. A., et al. (2019). Communicating structure and function in synthetic biology diagrams. ACS Synth. Biol. 8, 1818–1825. doi:10.1021/acssynbio.9b00139
Becskei, A., and Serrano, L. (2000). Engineering stability in gene networks by autoregulation. Nature 405, 590–593. doi:10.1038/35014651
Beerli, R. R., Dreier, B., and Barbas, C. F. (2000). Positive and negative regulation of endogenous genes by designed transcription factors. Proc. Natl. Acad. Sci. 97, 1495–1500. doi:10.1073/pnas.040552697
Bertram, R., and Hillen, W. (2008). The application of Tet repressor in prokaryotic gene regulation and expression. Microb. Biotechnol. 1, 2–16. doi:10.1111/j.1751-7915.2007.00001.x
Bervoets, I., Van Brempt, M., Van Nerom, K., Van Hove, B., Maertens, J., De Mey, M., et al. (2018). A sigma factor toolbox for orthogonal gene expression in Escherichia coli. Nucleic Acids Res. 46, 2133–2144. doi:10.1093/nar/gky010
Bhatnagar, K., Hinz, A., Kohlman, M., and Wong, A. (2019). An sRNA screen for reversal of quinolone resistance in Escherichia coli. G3 GenesGenomesGenetics 10, 79–88. doi:10.1534/g3.119.400199
Bikard, D., Jiang, W., Samai, P., Hochschild, A., Zhang, F., and Marraffini, L. A. (2013). Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic Acids Res. 41, 7429–7437. doi:10.1093/nar/gkt520
Bleris, L., Xie, Z., Glass, D., Adadey, A., Sontag, E., and Benenson, Y. (2011). Synthetic incoherent feedforward circuits show adaptation to the amount of their genetic template. Mol. Syst. Biol. 7, 519. doi:10.1038/msb.2011.49
Bonger, K. M., Rakhit, R., Payumo, A. Y., Chen, J. K., and Wandless, T. J. (2014). General method for regulating protein stability with light. ACS Chem. Biol. 9, 111–115. doi:10.1021/cb400755b
Bonnet, J., Subsoontorn, P., and Endy, D. (2012). Rewritable digital data storage in live cells via engineered control of recombination directionality. Proc. Natl. Acad. Sci. 109, 8884–8889. doi:10.1073/pnas.1202344109
Bonnet, J., Yin, P., Ortiz, M. E., Subsoontorn, P., and Endy, D. (2013). Amplifying genetic logic gates. Science 340, 599–603. doi:10.1126/science.1232758
Borkowski, O., Ceroni, F., Stan, G.-B., and Ellis, T. (2016). Overloaded and stressed: whole-cell considerations for bacterial synthetic biology. Curr. Opin. Microbiol. 33, 123–130. doi:10.1016/j.mib.2016.07.009
Brand, A. H., and Perrimon, N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401–415. doi:10.1242/dev.118.2.401
Breaker, R. R. (2018). Riboswitches and translation control. Cold Spring Harb. Perspect. Biol. 10, a032797. doi:10.1101/cshperspect.a032797
Briat, C., Gupta, A., and Khammash, M. (2016). Antithetic integral feedback ensures robust perfect adaptation in noisy biomolecular networks. Cell Syst. 2, 15–26. doi:10.1016/j.cels.2016.01.004
Brödel, A. K., Jaramillo, A., and Isalan, M. (2016). Engineering orthogonal dual transcription factors for multi-input synthetic promoters. Nat. Commun. 7, 13858. doi:10.1038/ncomms13858
Brown, B., Bartley, B., Beal, J., Bird, J. E., Goñi-Moreno, Á., McLaughlin, J. A., et al. (2020). Capturing multicellular system designs using synthetic biology Open Language (SBOL). ACS Synth. Biol. 9, 2410–2417. doi:10.1021/acssynbio.0c00176
Brown, M., Figge, J., Hansen, U., Wright, C., Jeang, K.-T., Khoury, G., et al. (1987). Lac repressor can regulate expression from a hybrid SV40 early promoter containing a lac operator in animal cells. Cell 49, 603–612. doi:10.1016/0092-8674(87)90536-8
Brück, M., Köbel, T. S., Dittmar, S., Ramírez Rojas, A. A., Georg, J., Berghoff, B. A., et al. (2024). A library-based approach allows systematic and rapid evaluation of seed region length and reveals design rules for synthetic bacterial small RNAs. iScience 27, 110774. doi:10.1016/j.isci.2024.110774
Brummelkamp, T. R., Bernards, R., and Agami, R. (2002). A system for stable expression of short interfering RNAs in mammalian cells. Science 296, 550–553. doi:10.1126/science.1068999
Buckley, D. L., Raina, K., Darricarrere, N., Hines, J., Gustafson, J. L., Smith, I. E., et al. (2015). HaloPROTACS: use of small molecule PROTACs to induce degradation of HaloTag fusion proteins. ACS Chem. Biol. 10, 1831–1837. doi:10.1021/acschembio.5b00442
Buecherl, L., Mitchell, T., Scott-Brown, J., Vaidyanathan, P., Vidal, G., Baig, H., et al. (2023). Synthetic biology open language (SBOL) version 3.1.0. J. Integr. Bioinforma. 20, 20220058. doi:10.1515/jib-2022-0058
Buskirk, A. R., Ong, Y.-C., Gartner, Z. J., and Liu, D. R. (2004). Directed evolution of ligand dependence: small-molecule-activated protein splicing. Proc. Natl. Acad. Sci. 101, 10505–10510. doi:10.1073/pnas.0402762101
Ceres, P., Garst, A. D., Marcano-Velázquez, J. G., and Batey, R. T. (2013a). Modularity of select riboswitch expression platforms enables facile engineering of novel genetic regulatory devices. ACS Synth. Biol. 2, 463–472. doi:10.1021/sb4000096
Ceres, P., Trausch, J. J., and Batey, R. T. (2013b). Engineering modular ‘ON’ RNA switches using biological components. Nucleic Acids Res. 41, 10449–10461. doi:10.1093/nar/gkt787
Ceroni, F., Algar, R., Stan, G.-B., and Ellis, T. (2015). Quantifying cellular capacity identifies gene expression designs with reduced burden. Nat. Methods 12, 415–418. doi:10.1038/nmeth.3339
Chan, C. T. Y., Kennedy, V., and Kinshuk, S. (2024). A domain swapping strategy to create modular transcriptional regulators for novel topology in genetic network. Biotechnol. Adv. 72, 108345. doi:10.1016/j.biotechadv.2024.108345
Chan, C. T. Y., Lee, J. W., Cameron, D. E., Bashor, C. J., and Collins, J. J. (2016). “Deadman” and “Passcode” microbial kill switches for bacterial containment. Nat. Chem. Biol. 12, 82–86. doi:10.1038/nchembio.1979
Chang, T., Ding, W., Yan, S., Wang, Y., Zhang, H., Zhang, Y., et al. (2023). A robust yeast biocontainment system with two-layered regulation switch dependent on unnatural amino acid. Nat. Commun. 14, 6487. doi:10.1038/s41467-023-42358-4
Chen, D., and Arkin, A. P. (2012). Sequestration-based bistability enables tuning of the switching boundaries and design of a latch. Mol. Syst. Biol. 8, 620. doi:10.1038/msb.2012.52
Chen, S.-Y., Zhang, Y., Li, R., Wang, B., and Ye, B.-C. (2022). De novo design of the ArsR regulated P ars promoter enables a highly sensitive whole-cell biosensor for arsenic contamination. Anal. Chem. 94, 7210–7218. doi:10.1021/acs.analchem.2c00055
Chen, X., Li, Y., Xiong, K., and Wagner, T. E. (1994). A self-initiating eukaryotic transient gene expression system based on contransfection of bacteriophage T7 RNA polymerase and DNA vectors containing a T7 autogene. Nucleic Acids Res. 22, 2114–2120. doi:10.1093/nar/22.11.2114
Cho, J. S., Yang, D., Prabowo, C. P. S., Ghiffary, M. R., Han, T., Choi, K. R., et al. (2023). Targeted and high-throughput gene knockdown in diverse bacteria using synthetic sRNAs. Nat. Commun. 14, 2359. doi:10.1038/s41467-023-38119-y
Choe, J. H., Watchmaker, P. B., Simic, M. S., Gilbert, R. D., Li, A. W., Krasnow, N. A., et al. (2021). SynNotch-CAR T cells overcome challenges of specificity, heterogeneity, and persistence in treating glioblastoma. Sci. Transl. Med. 13, eabe7378. doi:10.1126/scitranslmed.abe7378
Choi, J., Chen, W., Minkina, A., Chardon, F. M., Suiter, C. C., Regalado, S. G., et al. (2022). A time-resolved, multi-symbol molecular recorder via sequential genome editing. Nature 608, 98–107. doi:10.1038/s41586-022-04922-8
Chou, C., Young, D. D., and Deiters, A. (2010). Photocaged T7 RNA polymerase for the light activation of transcription and gene function in pro- and eukaryotic cells. ChemBioChem 11, 972–977. doi:10.1002/cbic.201000041
Christopherson, K. S., Mark, M. R., Bajaj, V., and Godowski, P. J. (1992). Ecdysteroid-dependent regulation of genes in mammalian cells by a Drosophila ecdysone receptor and chimeric transactivators. Proc. Natl. Acad. Sci. 89, 6314–6318. doi:10.1073/pnas.89.14.6314
Chung, H. K., Jacobs, C. L., Huo, Y., Yang, J., Krumm, S. A., Plemper, R. K., et al. (2015). Tunable and reversible drug control of protein production via a self-excising degron. Nat. Chem. Biol. 11, 713–720. doi:10.1038/nchembio.1869
Cisneros, A. F., Rouleau, F. D., Bautista, C., Lemieux, P., and Dumont-Leblond, N. (2023). Toeholder: a software for automated design and in silico validation of toehold riboswitches. PeerJ Phys. Chem. 5, e28. doi:10.7717/peerj-pchem.28
Courbet, A., Endy, D., Renard, E., Molina, F., and Bonnet, J. (2015). Detection of pathological biomarkers in human clinical samples via amplifying genetic switches and logic gates. Sci. Transl. Med. 7, 289ra83. doi:10.1126/scitranslmed.aaa3601
Das, A. T., Tenenbaum, L., and Berkhout, B. (2016). Tet-on systems for doxycycline-inducible gene expression. Curr. Gene Ther. 16, 156–167. doi:10.2174/1566523216666160524144041
De Carluccio, G., Fusco, V., and di Bernardo, D. (2024). Engineering a synthetic gene circuit for high-performance inducible expression in mammalian systems. Nat. Commun. 15, 3311. doi:10.1038/s41467-024-47592-y
De Lay, N., Schu, D. J., and Gottesman, S. (2013). Bacterial small RNA-based negative regulation: Hfq and its accomplices. J. Biol. Chem. 288, 7996–8003. doi:10.1074/jbc.R112.441386
Dimas, R. P., Jiang, X.-L., Alberto de la Paz, J., Morcos, F., and Chan, C. T. Y. (2019). Engineering repressors with coevolutionary cues facilitates toggle switches with a master reset. Nucleic Acids Res. 47, 5449–5463. doi:10.1093/nar/gkz280
Di Roberto, R. B., Chang, B., and Peisajovich, S. G. (2017). The directed evolution of ligand specificity in a GPCR and the unequal contributions of efficacy and affinity. Sci. Rep. 7, 16012. doi:10.1038/s41598-017-16332-2
Du, P., Lou, C., Zhao, X., Wang, Q., Ji, X., and Wei, W. (2021). CRISPR-based genetic switches and other complex circuits: research and application. Life 11, 1255. doi:10.3390/life11111255
Duffy, J. B. (2002). GAL4 system in drosophila: a fly geneticist’s swiss army knife. genesis 34, 1–15. doi:10.1002/gene.10150
Duplus-Bottin, H., Spichty, M., Triqueneaux, G., Place, C., Mangeot, P. E., Ohlmann, T., et al. (2021). A single-chain and fast-responding light-inducible Cre recombinase as a novel optogenetic switch. eLife 10, e61268. doi:10.7554/eLife.61268
Durrer, K. E., Allen, M. S., and Hunt Von Herbing, I. (2017). Genetically engineered probiotic for the treatment of phenylketonuria (PKU); assessment of a novel treatment in vitro and in the PAHenu2 mouse model of PKU. PLOS ONE 12, e0176286. doi:10.1371/journal.pone.0176286
Elbashir, S. M., Harborth, J., Lendeckel, W., Yalcin, A., Weber, K., and Tuschl, T. (2001). Duplexes of 21-nucleotide RNAs mediate RNA interference in cultured mammalian cells. Nature 411, 494–498. doi:10.1038/35078107
Elowitz, M. B., and Leibler, S. (2000). A synthetic oscillatory network of transcriptional regulators. Nature 403, 335–338. doi:10.1038/35002125
Etzel, M., and Mörl, M. (2017). Synthetic riboswitches: from plug and pray toward plug and play. Biochemistry 56, 1181–1198. doi:10.1021/acs.biochem.6b01218
Fakhr, E., Zare, F., and Teimoori-Toolabi, L. (2016). Precise and efficient siRNA design: a key point in competent gene silencing. Cancer Gene Ther. 23, 73–82. doi:10.1038/cgt.2016.4
Fan, H. Y., Morgan, S.-A., Brechun, K. E., Chen, Y.-Y., Jaikaran, A. S. I., and Woolley, G. A. (2011). Improving a designed photocontrolled DNA-binding protein. Biochemistry 50, 1226–1237. doi:10.1021/bi101432p
Farzadfard, F., Gharaei, N., Higashikuni, Y., Jung, G., Cao, J., and Lu, T. K. (2019). Single-nucleotide-resolution computing and memory in living cells. Mol. Cell 75, 769–780. doi:10.1016/j.molcel.2019.07.011
Farzadfard, F., and Lu, T. K. (2014). Synthetic biology. Genomically encoded analog memory with precise in vivo DNA writing in living cell populations. Science 346, 1256272. doi:10.1126/science.1256272
FDA (2017a). FDA approval letter - KYMRIAH. Available at: https://www.fda.gov/vaccines-blood-biologics/cellular-gene-therapy-products/kymriah (Accessed October 7, 2024).
FDA (2017b). FDA approval letter - YESCARTA. Available at: https://www.fda.gov/vaccines-blood-biologics/cellular-gene-therapy-products/yescarta (Accessed October 7, 2024).
Feil, R., Brocard, J., Mascrez, B., LeMeur, M., Metzger, D., and Chambon, P. (1996). Ligand-activated site-specific recombination in mice. Proc. Natl. Acad. Sci. 93, 10887–10890. doi:10.1073/pnas.93.20.10887
Fernandez-Rodriguez, J., Yang, L., Gorochowski, T. E., Gordon, D. B., and Voigt, C. A. (2015). Memory and combinatorial logic based on DNA inversions: dynamics and evolutionary stability. ACS Synth. Biol. 4, 1361–1372. doi:10.1021/acssynbio.5b00170
Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. doi:10.1038/35888
Frank, S. B., Schulz, V. V., and Miranti, C. K. (2017). A streamlined method for the design and cloning of shRNAs into an optimized Dox-inducible lentiviral vector. BMC Biotechnol. 17, 24. doi:10.1186/s12896-017-0341-x
Frei, T., Chang, C.-H., Filo, M., Arampatzis, A., and Khammash, M. (2022). A genetic mammalian proportional–integral feedback control circuit for robust and precise gene regulation. Proc. Natl. Acad. Sci. 119, e2122132119. doi:10.1073/pnas.2122132119
Friedland, A. E., Lu, T. K., Wang, X., Shi, D., Church, G., and Collins, J. J. (2009). Synthetic gene networks that count. Science 324, 1199–1202. doi:10.1126/science.1172005
Fu, Z., Zhang, X., Zhou, X., Ur-Rehman, U., Yu, M., Liang, H., et al. (2021). In vivo self-assembled small RNAs as a new generation of RNAi therapeutics. Cell Res. 31, 631–648. doi:10.1038/s41422-021-00491-z
Fuerst, T. R., Niles, E. G., Studier, F. W., and Moss, B. (1986). Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. 83, 8122–8126. doi:10.1073/pnas.83.21.8122
Galdzicki, M., Clancy, K. P., Oberortner, E., Pocock, M., Quinn, J. Y., Rodriguez, C. A., et al. (2014). The Synthetic Biology Open Language (SBOL) provides a community standard for communicating designs in synthetic biology. Nat. Biotechnol. 32, 545–550. doi:10.1038/nbt.2891
Gander, M. W., Vrana, J. D., Voje, W. E., Carothers, J. M., and Klavins, E. (2017). Digital logic circuits in yeast with CRISPR-dCas9 NOR gates. Nat. Commun. 8, 15459. doi:10.1038/ncomms15459
Gao, Z.-G., Duong, H. T., Sonina, T., Kim, S.-K., Van Rompaey, P., Van Calenbergh, S., et al. (2006). Orthogonal activation of the reengineered A 3 adenosine receptor (neoceptor) using tailored nucleoside agonists. J. Med. Chem. 49, 2689–2702. doi:10.1021/jm050968b
Gardner, T. S., Cantor, C. R., and Collins, J. J. (2000). Construction of a genetic toggle switch in Escherichia coli. Nature 403, 339–342. doi:10.1038/35002131
Gargett, T., and Brown, M. P. (2014). The inducible caspase-9 suicide gene system as a “safety switch” to limit on-target, off-tumor toxicities of chimeric antigen receptor T cells. Front. Pharmacol. 5, 235. doi:10.3389/fphar.2014.00235
Gargett, T., Truong, N. T. H., Gardam, B., Yu, W., Ebert, L. M., Johnson, A., et al. (2024). Safety and biological outcomes following a phase 1 trial of GD2-specific CAR-T cells in patients with GD2-positive metastatic melanoma and other solid cancers. J. Immunother. Cancer 12, e008659. doi:10.1136/jitc-2023-008659
Garí, E., Piedrafita, L., Aldea, M., and Herrero, E. (1997). A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast 13, 837–848. doi:10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T
Gaudelli, N. M., Komor, A. C., Rees, H. A., Packer, M. S., Badran, A. H., Bryson, D. I., et al. (2017). Programmable base editing of A⋅T to G⋅C in genomic DNA without DNA cleavage. Nature 551, 464–471. doi:10.1038/nature24644
Gilbert, L. A., Larson, M. H., Morsut, L., Liu, Z., Brar, G. A., Torres, S. E., et al. (2013). CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell 154, 442–451. doi:10.1016/j.cell.2013.06.044
Glass, D. S., Bren, A., Vaisbourd, E., Mayo, A., and Alon, U. (2024). A synthetic differentiation circuit in Escherichia coli for suppressing mutant takeover. Cell 187, 931–944.e12. doi:10.1016/j.cell.2024.01.024
Goshisht, M. K. (2024). Machine learning and deep learning in synthetic biology: key architectures, applications, and challenges. ACS Omega 9, 9921–9945. doi:10.1021/acsomega.3c05913
Gossen, M., and Bujard, H. (1992). Tight control of gene expression in mammalian cells by tetracycline-responsive promoters. Proc. Natl. Acad. Sci. 89, 5547–5551. doi:10.1073/pnas.89.12.5547
Gossen, M., Freundlieb, S., Bender, G., Müller, G., Hillen, W., and Bujard, H. (1995). Transcriptional activation by tetracyclines in mammalian cells. Science 268, 1766–1769. doi:10.1126/science.7792603
Green, A. A., Kim, J., Ma, D., Silver, P. A., Collins, J. J., and Yin, P. (2017). Complex cellular logic computation using ribocomputing devices. Nature 548, 117–121. doi:10.1038/nature23271
Green, A. A., Silver, P. A., Collins, J. J., and Yin, P. (2014). Toehold switches: de-novo-designed regulators of gene expression. Cell 159, 925–939. doi:10.1016/j.cell.2014.10.002
Groseclose, T. M., Rondon, R. E., Herde, Z. D., Aldrete, C. A., and Wilson, C. J. (2020). Engineered systems of inducible anti-repressors for the next generation of biological programming. Nat. Commun. 11, 4440. doi:10.1038/s41467-020-18302-1
Groves, B., Chen, Y.-J., Zurla, C., Pochekailov, S., Kirschman, J. L., Santangelo, P. J., et al. (2016). Computing in mammalian cells with nucleic acid strand exchange. Nat. Nanotechnol. 11, 287–294. doi:10.1038/nnano.2015.278
Guo, T., Ma, D., and Lu, T. K. (2022). Sense-and-Respond payload delivery using a novel antigen-inducible promoter improves suboptimal CAR-T activation. ACS Synth. Biol. 11, 1440–1453. doi:10.1021/acssynbio.1c00236
Gupta, A., Reizman, I. M. B., Reisch, C. R., and Prather, K. L. J. (2017). Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat. Biotechnol. 35, 273–279. doi:10.1038/nbt.3796
Haley, B., and Zamore, P. D. (2004). Kinetic analysis of the RNAi enzyme complex. Nat. Struct. Mol. Biol. 11, 599–606. doi:10.1038/nsmb780
Ham, T. S., Lee, S. K., Keasling, J. D., and Arkin, A. P. (2006). A tightly regulated inducible expression system utilizing the fim inversion recombination switch. Biotechnol. Bioeng. 94, 1–4. doi:10.1002/bit.20916
Ham, T. S., Lee, S. K., Keasling, J. D., and Arkin, A. P. (2008). Design and construction of a double inversion recombination switch for heritable sequential genetic memory. PLoS ONE 3, e2815. doi:10.1371/journal.pone.0002815
Hannan, G. N., Lehnert, S. A., MacAvoy, E. S., Jennings, P. A., and Molloy, P. L. (1993). An engineered PGK promoter and lac operator-repressor system for the regulation of gene expression in mammalian cells. Gene 130, 233–239. doi:10.1016/0378-1119(93)90424-2
Harder, B., Bettenbrock, K., and Klamt, S. (2018). Temperature-dependent dynamic control of the TCA cycle increases volumetric productivity of itaconic acid production by Escherichia coli. Biotechnol. Bioeng. 115, 156–164. doi:10.1002/bit.26446
Hartley, K. O., Nutt, S. L., and Amaya, E. (2002). Targeted gene expression in transgenic Xenopus using the binary Gal4-UAS system. Proc. Natl. Acad. Sci. 99, 1377–1382. doi:10.1073/pnas.022646899
Hersey, A. N., Kay, V. E., Lee, S., Realff, M. J., and Wilson, C. J. (2023). Engineering allosteric transcription factors guided by the LacI topology. Cell Syst. 14, 645–655. doi:10.1016/j.cels.2023.04.008
Hiscock, T. W. (2019). Adapting machine-learning algorithms to design gene circuits. BMC Bioinforma. 20, 214. doi:10.1186/s12859-019-2788-3
Hoffmann, S. A., and Cai, Y. (2024). Engineering stringent genetic biocontainment of yeast with a protein stability switch. Nat. Commun. 15, 1060. doi:10.1038/s41467-024-44988-8
Hoffmann, S. A., Diggans, J., Densmore, D., Dai, J., Knight, T., Leproust, E., et al. (2023). Safety by design: biosafety and biosecurity in the age of synthetic genomics. iScience 26, 106165. doi:10.1016/j.isci.2023.106165
Hoffmann, S. A., Kruse, S. M., and Arndt, K. M. (2016). Long-range transcriptional interference in E. coli used to construct a dual positive selection system for genetic switches. Nucleic Acids Res. 44, e95. doi:10.1093/nar/gkw125
Hofmann, A., Falk, J., Prangemeier, T., Happel, D., Köber, A., Christmann, A., et al. (2019). A tightly regulated and adjustable CRISPR-dCas9 based AND gate in yeast. Nucleic Acids Res. 47, 509–520. doi:10.1093/nar/gky1191
Hoynes-O’Connor, A., Hinman, K., Kirchner, L., and Moon, T. S. (2015). De novo design of heat-repressible RNA thermosensors in E. coli. Nucleic Acids Res. 43, 6166–6179. doi:10.1093/nar/gkv499
Hu, B., Zhong, L., Weng, Y., Peng, L., Huang, Y., Zhao, Y., et al. (2020). Therapeutic siRNA: state of the art. Signal Transduct. Target. Ther. 5, 101–125. doi:10.1038/s41392-020-0207-x
Huang, H.-H., Qian, Y., and Del Vecchio, D. (2018). A quasi-integral controller for adaptation of genetic modules to variable ribosome demand. Nat. Commun. 9, 5415. doi:10.1038/s41467-018-07899-z
Hui, A., and De Boer, H. A. (1987). Specialized ribosome system: preferential translation of a single mRNA species by a subpopulation of mutated ribosomes in Escherichia coli. Proc. Natl. Acad. Sci. 84, 4762–4766. doi:10.1073/pnas.84.14.4762
Hunter, N. L., Awatramani, R. B., Farley, F. W., and Dymecki, S. M. (2005). Ligand-activated Flpe for temporally regulated gene modifications. genesis 41, 99–109. doi:10.1002/gene.20101
Hussein, R., and Lim, H. N. (2012). Direct comparison of small RNA and transcription factor signaling. Nucleic Acids Res. 40, 7269–7279. doi:10.1093/nar/gks439
Hutvágner, G., and Zamore, P. D. (2002). A microRNA in a multiple-turnover RNAi enzyme complex. Science 297, 2056–2060. doi:10.1126/science.1073827
Isaacs, F. J., Dwyer, D. J., Ding, C., Pervouchine, D. D., Cantor, C. R., and Collins, J. J. (2004). Engineered riboregulators enable post-transcriptional control of gene expression. Nat. Biotechnol. 22, 841–847. doi:10.1038/nbt986
Isabella, V. M., Ha, B. N., Castillo, M. J., Lubkowicz, D. J., Rowe, S. E., Millet, Y. A., et al. (2018). Development of a synthetic live bacterial therapeutic for the human metabolic disease phenylketonuria. Nat. Biotechnol. 36, 857–864. doi:10.1038/nbt.4222
Iwamoto, M., Björklund, T., Lundberg, C., Kirik, D., and Wandless, T. J. (2010). A general chemical method to regulate protein stability in the mammalian central nervous system. Chem. Biol. 17, 981–988. doi:10.1016/j.chembiol.2010.07.009
Jacob, F., and Monod, J. (1961). Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 3, 318–356. doi:10.1016/S0022-2836(61)80072-7
Jacobson, K. A., Gao, Z.-G., and Liang, B. T. (2007). Neoceptors: reengineering GPCRs to recognize tailored ligands. Trends Pharmacol. Sci. 28, 111–116. doi:10.1016/j.tips.2007.01.006
James, J. S., Dai, J., Chew, W. L., and Cai, Y. (2024). The design and engineering of synthetic genomes. Nat. Rev. Genet. doi:10.1038/s41576-024-00786-y
James, R. I., Elton, J. P., Todd, P., and Kompala, D. S. (2000). Engineering CHO cells to overexpress a secreted reporter protein upon induction from mouse mammary tumor virus promoter. Biotechnol. Bioeng. 67, 134–140. doi:10.1002/(SICI)1097-0290(20000120)67:2<134::AID-BIT2>3.0.CO;2-V
Jia, X., Bu, R., Zhao, T., and Wu, K. (2019). Sensitive and Specific Whole-Cell Biosensor for Arsenic Detection. Appl. Environ. Microbiol. 85, e00694-19. doi:10.1128/AEM.00694-19
Jiang, H., Galtes, D., Wang, J., and Rockman, H. A. (2022). G protein-coupled receptor signaling: transducers and effectors. Am. J. Physiol.-Cell Physiol. 323, C731–C748. doi:10.1152/ajpcell.00210.2022
Jiang, X.-L., Dimas, R. P., Chan, C. T. Y., and Morcos, F. (2021). Coevolutionary methods enable robust design of modular repressors by reestablishing intra-protein interactions. Nat. Commun. 12, 5592. doi:10.1038/s41467-021-25851-6
Jillette, N., Du, M., Zhu, J. J., Cardoz, P., and Cheng, A. W. (2019). Split selectable markers. Nat. Commun. 10, 4968. doi:10.1038/s41467-019-12891-2
Jobe, A., and Bourgeois, S. (1972). Lac Repressor-operator interaction. VI. The natural inducer of the lac operon. J. Mol. Biol. 69, 397–408. doi:10.1016/0022-2836(72)90253-7
Johnson Jr, J. E., Reyes, F. E., Polaski, J. T., and Batey, R. T. (2012). B12 cofactors directly stabilize an mRNA regulatory switch. Nature 492, 133–137. doi:10.1038/nature11607
Jones, T. S., Oliveira, S. M. D., Myers, C. J., Voigt, C. A., and Densmore, D. (2022). Genetic circuit design automation with Cello 2.0. Nat. Protoc. 17, 1097–1113. doi:10.1038/s41596-021-00675-2
Juillerat, A., Marechal, A., Filhol, J. M., Valogne, Y., Valton, J., Duclert, A., et al. (2017). An oxygen sensitive self-decision making engineered CAR T-cell. Sci. Rep. 7, 39833. doi:10.1038/srep39833
Jung, H., Kim, S.-W., Kim, M., Hong, J., Yu, D., Kim, J. H., et al. (2019). Noninvasive optical activation of Flp recombinase for genetic manipulation in deep mouse brain regions. Nat. Commun. 10, 314. doi:10.1038/s41467-018-08282-8
Kakidani, H., and Ptashne, M. (1988). GAL4 activates gene expression in mammalian cells. Cell 52, 161–167. doi:10.1016/0092-8674(88)90504-1
Kawano, F., Okazaki, R., Yazawa, M., and Sato, M. (2016). A photoactivatable Cre–loxP recombination system for optogenetic genome engineering. Nat. Chem. Biol. 12, 1059–1064. doi:10.1038/nchembio.2205
Khalil, A. S., Lu, T. K., Bashor, C. J., Ramirez, C. L., Pyenson, N. C., Joung, J. K., et al. (2012). A synthetic biology framework for programming eukaryotic transcription functions. Cell 150, 647–658. doi:10.1016/j.cell.2012.05.045
Khlebnikov, A., Risa, Ø., Skaug, T., Carrier, T. A., and Keasling, J. D. (2000). Regulatable arabinose-inducible gene expression system with consistent control in all cells of a culture. J. Bacteriol. 182, 7029–7034. doi:10.1128/JB.182.24.7029-7034.2000
Kim, H. J., Jeong, H., and Lee, S. J. (2018). Synthetic biology for microbial heavy metal biosensors. Anal. Bioanal. Chem. 410, 1191–1203. doi:10.1007/s00216-017-0751-6
King, K., Dohlman, H. G., Thorner, J., Caron, M. G., and Lefkowitz, R. J. (1990). Control of yeast mating signal transduction by a mammalian beta 2-adrenergic receptor and Gs alpha subunit. Science 250, 121–123. doi:10.1126/science.2171146
Ko, M. S. H., Naomi, T., Norifumi, S., and Toshiya, T. (1989). An auto-inducible vector conferring high glucocorticoid inducibility upon stable transformant cells. Gene 84, 383–389. doi:10.1016/0378-1119(89)90512-X
Köbbing, S., Blank, L. M., and Wierckx, N. (2020). Characterization of context-dependent effects on synthetic promoters. Front. Bioeng. Biotechnol. 8, 551. doi:10.3389/fbioe.2020.00551
Komatsu, S., Ohno, H., and Saito, H. (2023). Target-dependent RNA polymerase as universal platform for gene expression control in response to intracellular molecules. Nat. Commun. 14, 7256. doi:10.1038/s41467-023-42802-5
Komor, A. C., Kim, Y. B., Packer, M. S., Zuris, J. A., and Liu, D. R. (2016). Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature 533, 420–424. doi:10.1038/nature17946
Kortmann, J., and Narberhaus, F. (2012). Bacterial RNA thermometers: molecular zippers and switches. Nat. Rev. Microbiol. 10, 255–265. doi:10.1038/nrmicro2730
Köster, R. W., and Fraser, S. E. (2001). Tracing transgene expression in living zebrafish embryos. Dev. Biol. 233, 329–346. doi:10.1006/dbio.2001.0242
Kosti, P., Opzoomer, J. W., Larios-Martinez, K. I., Henley-Smith, R., Scudamore, C. L., Okesola, M., et al. (2021). Hypoxia-sensing CAR T cells provide safety and efficacy in treating solid tumors. Cell Rep. Med. 2, 100227. doi:10.1016/j.xcrm.2021.100227
Labow, M. A., Baim, S. B., Shenk, T., and Levine, A. J. (1990). Conversion of the lac repressor into an allosterically regulated transcriptional activator for mammalian cells. Mol. Cell. Biol. 10, 3343–3356. doi:10.1128/mcb.10.7.3343
Lebovich, M., Zeng, M., and Andrews, L. B. (2023). Algorithmic programming of sequential logic and genetic circuits for recording biochemical concentration in a probiotic bacterium. ACS Synth. Biol. 12, 2632–2649. doi:10.1021/acssynbio.3c00232
Lee, J. W., Chan, C. T. Y., Slomovic, S., and Collins, J. J. (2018). Next-generation biocontainment systems for engineered organisms. Nat. Chem. Biol. 14, 530–537. doi:10.1038/s41589-018-0056-x
Lee, S., Khalil, A. S., and Wong, W. W. (2022). Recent progress of gene circuit designs in immune cell therapies. Cell Syst. 13, 864–873. doi:10.1016/j.cels.2022.09.006
Lee, Y. J., Hoynes-O’Connor, A., Leong, M. C., and Moon, T. S. (2016). Programmable control of bacterial gene expression with the combined CRISPR and antisense RNA system. Nucleic Acids Res. 44, 2462–2473. doi:10.1093/nar/gkw056
Lehming, N., Sartorius, J., Niemöller, M., Genenger, G., V Wilcken-Bergmann, B., and Müller-Hill, B. (1987). The interaction of the recognition helix of lac repressor with lac operator. EMBO J. 6, 3145–3153. doi:10.1002/j.1460-2075.1987.tb02625.x
Levskaya, A., Weiner, O. D., Lim, W. A., and Voigt, C. A. (2009). Spatiotemporal control of cell signalling using a light-switchable protein interaction. Nature 461, 997–1001. doi:10.1038/nature08446
Li, H.-S., Israni, D. V., Gagnon, K. A., Gan, K. A., Raymond, M. H., Sander, J. D., et al. (2022). Multidimensional control of therapeutic human cell function with synthetic gene circuits. Science 378, 1227–1234. doi:10.1126/science.ade0156
Lin, P., Pu, Q., Wu, Q., Zhou, C., Wang, B., Schettler, J., et al. (2019). High-throughput screen reveals sRNAs regulating crRNA biogenesis by targeting CRISPR leader to repress Rho termination. Nat. Commun. 10, 3728. doi:10.1038/s41467-019-11695-8
Liu, C., Yu, H., Zhang, B., Liu, S., Liu, C., Li, F., et al. (2022). Engineering whole-cell microbial biosensors: design principles and applications in monitoring and treatment of heavy metals and organic pollutants. Biotechnol. Adv. 60, 108019. doi:10.1016/j.biotechadv.2022.108019
Liu, D., Xiao, Y., Evans, B. S., and Zhang, F. (2015). Negative feedback regulation of fatty acid production based on a malonyl-CoA sensor–actuator. ACS Synth. Biol. 4, 132–140. doi:10.1021/sb400158w
Liu, Y., Zeng, Y., Liu, L., Zhuang, C., Fu, X., Huang, W., et al. (2014). Synthesizing AND gate genetic circuits based on CRISPR-Cas9 for identification of bladder cancer cells. Nat. Commun. 5, 5393. doi:10.1038/ncomms6393
Louvion, J.-F., Havaux-Copf, B., and Picard, D. (1993). Fusion of GAL4-VP16 to a steroid-binding domain provides a tool for gratuitous induction of galactose-responsive genes in yeast. Gene 131, 129–134. doi:10.1016/0378-1119(93)90681-R
Loveless, T. B., Carlson, C. K., Dentzel Helmy, C. A., Hu, V. J., Ross, S. K., Demelo, M. C., et al. (2024). Open-ended molecular recording of sequential cellular events into DNA. Nat. Chem. Biol. doi:10.1038/s41589-024-01764-5
Loveless, T. B., Grotts, J. H., Schechter, M. W., Forouzmand, E., Carlson, C. K., Agahi, B. S., et al. (2021). Lineage tracing and analog recording in mammalian cells by single-site DNA writing. Nat. Chem. Biol. 17, 739–747. doi:10.1038/s41589-021-00769-8
Lu, L., Xie, M., Yang, B., Zhao, W., and Cao, J. (2024). Enhancing the safety of CAR-T cell therapy: synthetic genetic switch for spatiotemporal control. Sci. Adv. 10, eadj6251. doi:10.1126/sciadv.adj6251
Lv, Y., Gu, Y., Xu, J., Zhou, J., and Xu, P. (2020). Coupling metabolic addiction with negative autoregulation to improve strain stability and pathway yield. Metab. Eng. 61, 79–88. doi:10.1016/j.ymben.2020.05.005
Machens, F., Balazadeh, S., Mueller-Roeber, B., and Messerschmidt, K. (2017). Synthetic promoters and transcription factors for heterologous protein expression in Saccharomyces cerevisiae. Front. Bioeng. Biotechnol. 5, 63. doi:10.3389/fbioe.2017.00063
Macia, J., and Solé, R. V. (2009). Distributed robustness in cellular networks: insights from synthetic evolved circuits. J. R. Soc. Interface 6, 393–400. doi:10.1098/rsif.2008.0236
Mandell, D. J., Lajoie, M. J., Mee, M. T., Takeuchi, R., Kuznetsov, G., Norville, J. E., et al. (2015). Biocontainment of genetically modified organisms by synthetic protein design. Nature 518, 55–60. doi:10.1038/nature14121
Marucci, L., Barberis, M., Karr, J., Ray, O., Race, P. R., De Souza Andrade, M., et al. (2020). Computer-aided whole-cell design: taking a holistic approach by integrating synthetic with systems biology. Front. Bioeng. Biotechnol. 8, 942. doi:10.3389/fbioe.2020.00942
Matranga, C., Tomari, Y., Shin, C., Bartel, D. P., and Zamore, P. D. (2005). Passenger-strand cleavage facilitates assembly of siRNA into ago2-containing RNAi enzyme complexes. Cell 123, 607–620. doi:10.1016/j.cell.2005.08.044
Matsuura, S., Ono, H., Kawasaki, S., Kuang, Y., Fujita, Y., and Saito, H. (2018). Synthetic RNA-based logic computation in mammalian cells. Nat. Commun. 9, 4847. doi:10.1038/s41467-018-07181-2
Mazumder, M., Brechun, K. E., Kim, Y. B., Hoffmann, S. A., Chen, Y. Y., Keiski, C.-L., et al. (2015). An Escherichia coli system for evolving improved light-controlled DNA-binding proteins. Protein Eng. Des. Sel. 28, 293–302. doi:10.1093/protein/gzv033
McIsaac, R. S., Gibney, P. A., Chandran, S. S., Benjamin, K. R., and Botstein, D. (2014). Synthetic biology tools for programming gene expression without nutritional perturbations in Saccharomyces cerevisiae. Nucleic Acids Res. 42, e48. doi:10.1093/nar/gkt1402
McIsaac, R. S., Silverman, S. J., McClean, M. N., Gibney, P. A., Macinskas, J., Hickman, M. J., et al. (2011). Fast-acting and nearly gratuitous induction of gene expression and protein depletion in Saccharomyces cerevisiae. Mol. Biol. Cell 22, 4447–4459. doi:10.1091/mbc.e11-05-0466
McSweeney, M. A., Zhang, Y., and Styczynski, M. P. (2023). Short activators and repressors of RNA toehold switches. ACS Synth. Biol. 12, 681–688. doi:10.1021/acssynbio.2c00641
Metzger, D., Clifford, J., Chiba, H., and Chambon, P. (1995). Conditional site-specific recombination in mammalian cells using a ligand-dependent chimeric Cre recombinase. Proc. Natl. Acad. Sci. 92, 6991–6995. doi:10.1073/pnas.92.15.6991
Milk, L., Daber, R., and Lewis, M. (2010). Functional rules for lac repressor–operator associations and implications for protein–DNA interactions. Protein Sci. 19, 1162–1172. doi:10.1002/pro.389
Mironov, A. S., Gusarov, I., Rafikov, R., Lopez, L. E., Shatalin, K., Kreneva, R. A., et al. (2002). Sensing small molecules by nascent RNA: a mechanism to control transcription in bacteria. Cell 111, 747–756. doi:10.1016/S0092-8674(02)01134-0
Miyazaki, Y., Imoto, H., Chen, L., and Wandless, T. J. (2012). Destabilizing domains derived from the human estrogen receptor. J. Am. Chem. Soc. 134, 3942–3945. doi:10.1021/ja209933r
Modi, S. R., Camacho, D. M., Kohanski, M. A., Walker, G. C., and Collins, J. J. (2011). Functional characterization of bacterial sRNAs using a network biology approach. Proc. Natl. Acad. Sci. 108, 15522–15527. doi:10.1073/pnas.1104318108
Moon, T. S., Clarke, E. J., Groban, E. S., Tamsir, A., Clark, R. M., Eames, M., et al. (2011). Construction of a genetic multiplexer to toggle between chemosensory pathways in Escherichia coli. J. Mol. Biol. 406, 215–227. doi:10.1016/j.jmb.2010.12.019
Moore, C. B., Guthrie, E. H., Huang, M. T.-H., and Taxman, D. J. (2010). Short hairpin RNA (shRNA): design, delivery, and assessment of gene knockdown. Methods Mol. Biol. Clifton N. J. 629, 141–158. doi:10.1007/978-1-60761-657-3_10
Morgan, S.-A., Al-Abdul-Wahid, S., and Woolley, G. A. (2010). Structure-based design of a photocontrolled DNA binding protein. J. Mol. Biol. 399, 94–112. doi:10.1016/j.jmb.2010.03.053
Morikawa, K., Furuhashi, K., De Sena-Tomas, C., Garcia-Garcia, A. L., Bekdash, R., Klein, A. D., et al. (2020). Photoactivatable Cre recombinase 3.0 for in vivo mouse applications. Nat. Commun. 11, 2141. doi:10.1038/s41467-020-16030-0
Müller, K. M., and Arndt, K. M. (2012). “Standardization in synthetic biology,” in Synthetic gene networks. Editors W. Weber, and M. Fussenegger (Totowa, NJ: Humana Press), 23–43. doi:10.1007/978-1-61779-412-4_2
Na, D., Yoo, S. M., Chung, H., Park, H., Park, J. H., and Lee, S. Y. (2013). Metabolic engineering of Escherichia coli using synthetic small regulatory RNAs. Nat. Biotechnol. 31, 170–174. doi:10.1038/nbt.2461
Nabet, B., Roberts, J. M., Buckley, D. L., Paulk, J., Dastjerdi, S., Yang, A., et al. (2018). The dTAG system for immediate and target-specific protein degradation. Nat. Chem. Biol. 14, 431–441. doi:10.1038/s41589-018-0021-8
Narberhaus, F., Waldminghaus, T., and Chowdhury, S. (2006). RNA thermometers. FEMS Microbiol. Rev. 30, 3–16. doi:10.1111/j.1574-6976.2005.004.x
Naseri, G., Balazadeh, S., Machens, F., Kamranfar, I., Messerschmidt, K., and Mueller-Roeber, B. (2017). Plant-derived transcription factors for orthologous regulation of gene expression in the yeast Saccharomyces cerevisiae. ACS Synth. Biol. 6, 1742–1756. doi:10.1021/acssynbio.7b00094
Navarro, R., Chen, L., Rakhit, R., and Wandless, T. J. (2016). A novel destabilizing domain based on a small-molecule dependent fluorophore. ACS Chem. Biol. 11, 2101–2104. doi:10.1021/acschembio.6b00234
Neeser, A., Ramasubramanian, R., Wang, C., and Ma, L. (2023). Engineering enhanced chimeric antigen receptor-T cell therapy for solid tumors. Immuno-Oncol. Technol. 19, 100385. doi:10.1016/j.iotech.2023.100385
Neklesa, T. K., Tae, H. S., Schneekloth, A. R., Stulberg, M. J., Corson, T. W., Sundberg, T. B., et al. (2011). Small-molecule hydrophobic tagging–induced degradation of HaloTag fusion proteins. Nat. Chem. Biol. 7, 538–543. doi:10.1038/nchembio.597
Neumann, H., Wang, K., Davis, L., Garcia-Alai, M., and Chin, J. W. (2010). Encoding multiple unnatural amino acids via evolution of a quadruplet-decoding ribosome. Nature 464, 441–444. doi:10.1038/nature08817
Neupert, J., and Bock, R. (2009). Designing and using synthetic RNA thermometers for temperature-controlled gene expression in bacteria. Nat. Protoc. 4, 1262–1273. doi:10.1038/nprot.2009.112
Nguyen, H. T., Leelavathi, S., and Reddy, V. S. (2004). Bacteriophage T7 RNA polymerase-directed, inducible and tissue-specific over-expression of foreign genes in transgenic plants. Plant Biotechnol. J. 2, 301–310. doi:10.1111/j.1467-7652.2004.00071.x
Nielsen, A. A. K., Der, B. S., Shin, J., Vaidyanathan, P., Paralanov, V., Strychalski, E. A., et al. (2016). Genetic circuit design automation. Science 352, aac7341. doi:10.1126/science.aac7341
Nihongaki, Y., Yamamoto, S., Kawano, F., Suzuki, H., and Sato, M. (2015). CRISPR-Cas9-based photoactivatable transcription system. Chem. Biol. 22, 169–174. doi:10.1016/j.chembiol.2014.12.011
Nishimura, K., Fukagawa, T., Takisawa, H., Kakimoto, T., and Kanemaki, M. (2009). An auxin-based degron system for the rapid depletion of proteins in nonplant cells. Nat. Methods 6, 917–922. doi:10.1038/nmeth.1401
Noh, M., Yoo, S. M., Yang, D., and Lee, S. Y. (2019). Broad-spectrum gene repression using scaffold engineering of synthetic sRNAs. ACS Synth. Biol. 8, 1452–1461. doi:10.1021/acssynbio.9b00165
Nuñez, J. K., Chen, J., Pommier, G. C., Cogan, J. Z., Replogle, J. M., Adriaens, C., et al. (2021). Genome-wide programmable transcriptional memory by CRISPR-based epigenome editing. Cell 184, 2503–2519.e17. doi:10.1016/j.cell.2021.03.025
Ozawa, T., Kaihara, A., Sato, M., Tachihara, K., and Umezawa, Y. (2001). Split luciferase as an optical probe for detecting Protein−Protein interactions in mammalian cells based on protein splicing. Anal. Chem. 73, 2516–2521. doi:10.1021/ac0013296
Paddison, P. J., Caudy, A. A., Bernstein, E., Hannon, G. J., and Conklin, D. S. (2002). Short hairpin RNAs (shRNAs) induce sequence-specific silencing in mammalian cells. Genes Dev. 16, 948–958. doi:10.1101/gad.981002
Paddon, C. J., Westfall, P. J., Pitera, D. J., Benjamin, K., Fisher, K., McPhee, D., et al. (2013). High-level semi-synthetic production of the potent antimalarial artemisinin. Nature 496, 528–532. doi:10.1038/nature12051
Pandit, A. V., Srinivasan, S., and Mahadevan, R. (2017). Redesigning metabolism based on orthogonality principles. Nat. Commun. 8, 15188. doi:10.1038/ncomms15188
Park, K.-S., Lee, D., Lee, H., Lee, Y., Jang, Y.-S., Kim, Y. H., et al. (2003). Phenotypic alteration of eukaryotic cells using randomized libraries of artificial transcription factors. Nat. Biotechnol. 21, 1208–1214. doi:10.1038/nbt868
Park, M., Patel, N., Keung, A. J., and Khalil, A. S. (2019). Engineering epigenetic regulation using synthetic read-write modules. Cell 176, 227–238. doi:10.1016/j.cell.2018.11.002
Paulmurugan, R., Umezawa, Y., and Gambhir, S. S. (2002). Noninvasive imaging of protein–protein interactions in living subjects by using reporter protein complementation and reconstitution strategies. Proc. Natl. Acad. Sci. 99, 15608–15613. doi:10.1073/pnas.242594299
Podhajska, A. J., Hasan, N., and Szybalski, W. (1985). Control of cloned gene expression by promoter inversion in vivo: construction of the heat-pulse-activated att-nutL-p-att-N module. Gene 40, 163–168. doi:10.1016/0378-1119(85)90038-1
Poelwijk, F. J., de Vos, M. G. J., and Tans, S. J. (2011). Tradeoffs and optimality in the evolution of gene regulation. Cell 146, 462–470. doi:10.1016/j.cell.2011.06.035
Polstein, L. R., and Gersbach, C. A. (2012). Light-inducible spatiotemporal control of gene activation by customizable zinc finger transcription factors. J. Am. Chem. Soc. 134, 16480–16483. doi:10.1021/ja3065667
Potvin-Trottier, L., Lord, N. D., Vinnicombe, G., and Paulsson, J. (2016). Synchronous long-term oscillations in a synthetic gene circuit. Nature 538, 514–517. doi:10.1038/nature19841
Pu, J., Zinkus-Boltz, J., and Dickinson, B. C. (2017). Evolution of a split RNA polymerase as a versatile biosensor platform. Nat. Chem. Biol. 13, 432–438. doi:10.1038/nchembio.2299
Qi, L. S., Larson, M. H., Gilbert, L. A., Doudna, J. A., Weissman, J. S., Arkin, A. P., et al. (2013). Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell 152, 1173–1183. doi:10.1016/j.cell.2013.02.022
Rackham, O., and Chin, J. W. (2005). A network of orthogonal ribosome x mRNA pairs. Nat. Chem. Biol. 1, 159–166. doi:10.1038/nchembio719
Rajeshkannan, E., Mahilkar, A., and Saini, S. (2022). GAL regulon in the yeast S. cerevisiae is highly evolvable via acquisition in the coding regions of the regulatory elements of the network. Front. Mol. Biosci. 9, 801011. doi:10.3389/fmolb.2022.801011
Randall, A., Guye, P., Gupta, S., Duportet, X., and Weiss, R. (2011). “Design and connection of robust genetic circuits,” in Methods in enzymology (Elsevier), 159–186. doi:10.1016/B978-0-12-385075-1.00007-X
Rao, C. V. (2012). Expanding the synthetic biology toolbox: engineering orthogonal regulators of gene expression. Curr. Opin. Biotechnol. 23, 689–694. doi:10.1016/j.copbio.2011.12.015
Renicke, C., Schuster, D., Usherenko, S., Essen, L.-O., and Taxis, C. (2013). A LOV2 domain-based optogenetic tool to control protein degradation and cellular function. Chem. Biol. 20, 619–626. doi:10.1016/j.chembiol.2013.03.005
Rhodius, V. A., Segall-Shapiro, T. H., Sharon, B. D., Ghodasara, A., Orlova, E., Tabakh, H., et al. (2013). Design of orthogonal genetic switches based on a crosstalk map of σs, anti-σs, and promoters. Mol. Syst. Biol. 9, 702. doi:10.1038/msb.2013.58
Richards, D. H., Meyer, S., and Wilson, C. J. (2017). Fourteen ways to reroute cooperative communication in the lactose repressor: engineering regulatory proteins with alternate repressive functions. ACS Synth. Biol. 6, 6–12. doi:10.1021/acssynbio.6b00048
Riglar, D. T., Giessen, T. W., Baym, M., Kerns, S. J., Niederhuber, M. J., Bronson, R. T., et al. (2017). Engineered bacteria can function in the mammalian gut long-term as live diagnostics of inflammation. Nat. Biotechnol. 35, 653–658. doi:10.1038/nbt.3879
Rinaudo, K., Bleris, L., Maddamsetti, R., Subramanian, S., Weiss, R., and Benenson, Y. (2007). A universal RNAi-based logic evaluator that operates in mammalian cells. Nat. Biotechnol. 25, 795–801. doi:10.1038/nbt1307
Ro, D.-K., Paradise, E. M., Ouellet, M., Fisher, K. J., Newman, K. L., Ndungu, J. M., et al. (2006). Production of the antimalarial drug precursor artemisinic acid in engineered yeast. Nature 440, 940–943. doi:10.1038/nature04640
Roquet, N., Soleimany, A. P., Ferris, A. C., Aaronson, S., and Lu, T. K. (2016). Synthetic recombinase-based state machines in living cells. Science 353, aad8559. doi:10.1126/science.aad8559
Rovner, A. J., Haimovich, A. D., Katz, S. R., Li, Z., Grome, M. W., Gassaway, B. M., et al. (2015). Recoded organisms engineered to depend on synthetic amino acids. Nature 518, 89–93. doi:10.1038/nature14095
Rowitch, D. H., St.-Jacques, B., Lee, S. M. K., Flax, J. D., Snyder, E. Y., and McMahon, A. P. (1999). Sonic hedgehog regulates proliferation and inhibits differentiation of CNS precursor cells. J. Neurosci. 19, 8954–8965. doi:10.1523/JNEUROSCI.19-20-08954.1999
Roybal, K. T., Rupp, L. J., Morsut, L., Walker, W. J., McNally, K. A., Park, J. S., et al. (2016). Precision tumor recognition by T cells with combinatorial antigen-sensing circuits. Cell 164, 770–779. doi:10.1016/j.cell.2016.01.011
Sadler, J. R., Sasmor, H., and Betz, J. L. (1983). A perfectly symmetric lac operator binds the lac repressor very tightly. Proc. Natl. Acad. Sci. 80, 6785–6789. doi:10.1073/pnas.80.22.6785
Santos-Moreno, J., Tasiudi, E., Stelling, J., and Schaerli, Y. (2020). Multistable and dynamic CRISPRi-based synthetic circuits. Nat. Commun. 11, 2746. doi:10.1038/s41467-020-16574-1
Sarkissian, C. N., Shao, Z., Blain, F., Peevers, R., Su, H., Heft, R., et al. (1999). A different approach to treatment of phenylketonuria: phenylalanine degradation with recombinant phenylalanine ammonia lyase. Proc. Natl. Acad. Sci. 96, 2339–2344. doi:10.1073/pnas.96.5.2339
Sarmiento, C., and Camarero, J. A. (2019). Biotechnological applications of protein splicing. Curr. Protein Pept. Sci. 20, 408–424. doi:10.2174/1389203720666190208110416
Satya Lakshmi, O., and Rao, N. M. (2009). Evolving Lac repressor for enhanced inducibility. Protein Eng. Des. Sel. 22, 53–58. doi:10.1093/protein/gzn069
Schena, M., Lloyd, A. M., and Davis, R. W. (1991). A steroid-inducible gene expression system for plant cells. Proc. Natl. Acad. Sci. 88, 10421–10425. doi:10.1073/pnas.88.23.10421
Schladt, T., Engelmann, N., Kubaczka, E., Hochberger, C., and Koeppl, H. (2021). Automated design of robust genetic circuits: structural variants and parameter uncertainty. ACS Synth. Biol. 10, 3316–3329. doi:10.1021/acssynbio.1c00193
Schleif, R. (2000). Regulation of the l-arabinose operon of Escherichia coli. Trends Genet. 16, 559–565. doi:10.1016/S0168-9525(00)02153-3
Sektas, M., Hasan, N., and Szybalski, W. (2001). Expression plasmid with a very tight two-step control: int/att-mediated gene inversion with respect to the stationary promoter. Gene 267, 213–220. doi:10.1016/S0378-1119(01)00395-X
Sektas, M., and Szybalski, W. (1998). Tightly controlled two-stage expression vectors employing the Flp/FRT-mediated inversion of cloned genes. Mol. Biotechnol. 9, 17–24. doi:10.1007/BF02752694
Sen, S., Apurva, D., Satija, R., Siegal, D., and Murray, R. M. (2017). Design of a toolbox of RNA thermometers. ACS Synth. Biol. 6, 1461–1470. doi:10.1021/acssynbio.6b00301
Sharma, P., Mondal, K., Kumar, S., Tamang, S., Najar, I. N., Das, S., et al. (2022). RNA thermometers in bacteria: role in thermoregulation. Biochim. Biophys. Acta BBA - Gene Regul. Mech. 1865, 194871. doi:10.1016/j.bbagrm.2022.194871
Shaw, W. M., Yamauchi, H., Mead, J., Gowers, G.-O. F., Bell, D. J., Öling, D., et al. (2019). Engineering a model cell for rational tuning of GPCR signaling. Cell 177, 782–796. doi:10.1016/j.cell.2019.02.023
Sheng, P., Flood, K. A., and Xie, M. (2020). Short hairpin RNAs for strand-specific small interfering RNA production. Front. Bioeng. Biotechnol. 8, 940. doi:10.3389/fbioe.2020.00940
Sheth, R. U., and Wang, H. H. (2018). DNA-based memory devices for recording cellular events. Nat. Rev. Genet. 19, 718–732. doi:10.1038/s41576-018-0052-8
Sheth, R. U., Yim, S. S., Wu, F. L., and Wang, H. H. (2017). Multiplex recording of cellular events over time on CRISPR biological tape. Science 358, 1457–1461. doi:10.1126/science.aao0958
Shimizu-Sato, S., Huq, E., Tepperman, J. M., and Quail, P. H. (2002). A light-switchable gene promoter system. Nat. Biotechnol. 20, 1041–1044. doi:10.1038/nbt734
Shipman, S. L., Nivala, J., Macklis, J. D., and Church, G. M. (2016). Molecular recordings by directed CRISPR spacer acquisition. Science 353, aaf1175. doi:10.1126/science.aaf1175
Shis, D. L., and Bennett, M. R. (2013). Library of synthetic transcriptional AND gates built with split T7 RNA polymerase mutants. Proc. Natl. Acad. Sci. 110, 5028–5033. doi:10.1073/pnas.1220157110
Shis, D. L., Hussain, F., Meinhardt, S., Swint-Kruse, L., and Bennett, M. R. (2014). Modular, multi-input transcriptional logic gating with orthogonal LacI/GalR family chimeras. ACS Synth. Biol. 3, 645–651. doi:10.1021/sb500262f
Short, A. E., Kim, D., Milner, P. T., and Wilson, C. J. (2023). Next generation synthetic memory via intercepting recombinase function. Nat. Commun. 14, 5255. doi:10.1038/s41467-023-41043-w
Siegfried, K., Endes, C., Bhuiyan, A. F.Md. K., Kuppardt, A., Mattusch, J., Van Der Meer, J. R., et al. (2012). Field testing of arsenic in groundwater samples of Bangladesh using a test kit based on lyophilized bioreporter bacteria. Environ. Sci. Technol. 46, 3281–3287. doi:10.1021/es203511k
Simon, A. J., and Ellington, A. D. (2016). Recent advances in synthetic biosafety. F1000Research 5, F1000 Faculty Rev-2118. doi:10.12688/f1000research.8365.1
Siuti, P., Yazbek, J., and Lu, T. K. (2013). Synthetic circuits integrating logic and memory in living cells. Nat. Biotechnol. 31, 448–452. doi:10.1038/nbt.2510
Sladek, F. M. (2011). What are nuclear receptor ligands? Mol. Cell. Endocrinol. 334, 3–13. doi:10.1016/j.mce.2010.06.018
Sleight, S. C., and Sauro, H. M. (2013). Visualization of evolutionary stability dynamics and competitive fitness of Escherichia coli engineered with randomized multigene circuits. ACS Synth. Biol. 2, 519–528. doi:10.1021/sb400055h
Sousa, F. L., Parente, D. J., Shis, D. L., Hessman, J. A., Chazelle, A., Bennett, M. R., et al. (2016). AlloRep: a repository of sequence, structural and mutagenesis data for the LacI/GalR transcription regulators. J. Mol. Biol. 428, 671–678. doi:10.1016/j.jmb.2015.09.015
Spencer, D. M., Wandless, T. J., Schreiber, S. L., and Crabtree, G. R. (1993). Controlling signal transduction with synthetic ligands. Science 262, 1019–1024. doi:10.1126/science.7694365
Srivastava, S., Salter, A. I., Liggitt, D., Yechan-Gunja, S., Sarvothama, M., Cooper, K., et al. (2019). Logic-gated ROR1 chimeric antigen receptor expression rescues T cell-mediated toxicity to normal tissues and enables selective tumor targeting. Cancer Cell 35, 489–503. doi:10.1016/j.ccell.2019.02.003
Sterner, R. C., and Sterner, R. M. (2021). CAR-T cell therapy: current limitations and potential strategies. Blood Cancer J. 11, 69–11. doi:10.1038/s41408-021-00459-7
Stocker, J., Balluch, D., Gsell, M., Harms, H., Feliciano, J., Daunert, S., et al. (2003). Development of a set of simple bacterial biosensors for quantitative and rapid measurements of arsenite and arsenate in potable water. Environ. Sci. Technol. 37, 4743–4750. doi:10.1021/es034258b
Stone, A., Youssef, A., Rijal, S., Zhang, R., and Tian, X.-J. (2024). Context-dependent redesign of robust synthetic gene circuits. Trends Biotechnol. 42, 895–909. doi:10.1016/j.tibtech.2024.01.003
Strickland, D., Moffat, K., and Sosnick, T. R. (2008). Light-activated DNA binding in a designed allosteric protein. Proc. Natl. Acad. Sci. 105, 10709–10714. doi:10.1073/pnas.0709610105
Strickland, D., Yao, X., Gawlak, G., Rosen, M. K., Gardner, K. H., and Sosnick, T. R. (2010). Rationally improving LOV domain–based photoswitches. Nat. Methods 7, 623–626. doi:10.1038/nmeth.1473
Studier, F. W., and Moffatt, B. A. (1986). Use of bacteriophage T7 RNA polymerase to direct selective high-level expression of cloned genes. J. Mol. Biol. 189, 113–130. doi:10.1016/0022-2836(86)90385-2
Tabor, S., and Richardson, C. C. (1985). A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc. Natl. Acad. Sci. 82, 1074–1078. doi:10.1073/pnas.82.4.1074
Tak, Y. E., Kleinstiver, B. P., Nuñez, J. K., Hsu, J. Y., Horng, J. E., Gong, J., et al. (2017). Inducible and multiplex gene regulation using CRISPR–Cpf1-based transcription factors. Nat. Methods 14, 1163–1166. doi:10.1038/nmeth.4483
Tamsir, A., Tabor, J. J., and Voigt, C. A. (2011). Robust multicellular computing using genetically encoded NOR gates and chemical ‘wires. Nature 469, 212–215. doi:10.1038/nature09565
Tang, W., and Liu, D. R. (2018). Rewritable multi-event analog recording in bacterial and mammalian cells. Science 360, eaap8992. doi:10.1126/science.aap8992
Taylor, N. D., Garruss, A. S., Moretti, R., Chan, S., Arbing, M. A., Cascio, D., et al. (2016). Engineering an allosteric transcription factor to respond to new ligands. Nat. Methods 13, 177–183. doi:10.1038/nmeth.3696
Temme, K., Hill, R., Segall-Shapiro, T. H., Moser, F., and Voigt, C. A. (2012). Modular control of multiple pathways using engineered orthogonal T7 polymerases. Nucleic Acids Res. 40, 8773–8781. doi:10.1093/nar/gks597
Tong, A. Y., Caudill, E. E., Jones, A. R., Passalacqua, L., and Abdelsayed, M. M. (2023). Characterization of a FourU RNA thermometer in the 5′ untranslated region of autolysin gene blyA in the Bacillus subtilis 168 prophage SPβ. Biochemistry 62, 2902–2907. doi:10.1021/acs.biochem.3c00368
Topilina, N. I., and Mills, K. V. (2014). Recent advances in in vivo applications of intein-mediated protein splicing. Mob. DNA 5, 5. doi:10.1186/1759-8753-5-5
Tungtur, S., Egan, S. M., and Swint-Kruse, L. (2007). Functional consequences of exchanging domains between LacI and PurR are mediated by the intervening linker sequence. Proteins Struct. Funct. Bioinforma. 68, 375–388. doi:10.1002/prot.21412
Valeri, J. A., Collins, K. M., Ramesh, P., Alcantar, M. A., Lepe, B. A., Lu, T. K., et al. (2020). Sequence-to-function deep learning frameworks for engineered riboregulators. Nat. Commun. 11, 5058. doi:10.1038/s41467-020-18676-2
Van Brempt, M., Clauwaert, J., Mey, F., Stock, M., Maertens, J., Waegeman, W., et al. (2020). Predictive design of sigma factor-specific promoters. Nat. Commun. 11, 5822. doi:10.1038/s41467-020-19446-w
Vargas-Rodriguez, O., Sevostyanova, A., Söll, D., and Crnković, A. (2018). Upgrading aminoacyl-tRNA synthetases for genetic code expansion. Curr. Opin. Chem. Biol. 46, 115–122. doi:10.1016/j.cbpa.2018.07.014
Vishweshwaraiah, Y. L., Chen, J., Chirasani, V. R., Tabdanov, E. D., and Dokholyan, N. V. (2021). Two-input protein logic gate for computation in living cells. Nat. Commun. 12, 6615. doi:10.1038/s41467-021-26937-x
Vockley, J., Sondheimer, N., Puurunen, M., Diaz, G. A., Ginevic, I., Grange, D. K., et al. (2023). Efficacy and safety of a synthetic biotic for treatment of phenylketonuria: a phase 2 clinical trial. Nat. Metab. 5, 1685–1690. doi:10.1038/s42255-023-00897-6
Wachsmuth, M., Findeiss, S., Weissheimer, N., Stadler, P. F., and Morl, M. (2013). De novo design of a synthetic riboswitch that regulates transcription termination. Nucleic Acids Res. 41, 2541–2551. doi:10.1093/nar/gks1330
Waldminghaus, T., Heidrich, N., Brantl, S., and Narberhaus, F. (2007). FourU: a novel type of RNA thermometer in Salmonella. Mol. Microbiol. 65, 413–424. doi:10.1111/j.1365-2958.2007.05794.x
Wan, X., Volpetti, F., Petrova, E., French, C., Maerkl, S. J., and Wang, B. (2019). Cascaded amplifying circuits enable ultrasensitive cellular sensors for toxic metals. Nat. Chem. Biol. 15, 540–548. doi:10.1038/s41589-019-0244-3
Wang, K., Neumann, H., Peak-Chew, S. Y., and Chin, J. W. (2007). Evolved orthogonal ribosomes enhance the efficiency of synthetic genetic code expansion. Nat. Biotechnol. 25, 770–777. doi:10.1038/nbt1314
Wang, X., Fang, C., Wang, Y., Shi, X., Yu, F., Xiong, J., et al. (2023). Systematic comparison and rational design of theophylline riboswitches for effective gene repression. Microbiol. Spectr. 11, e0275222. doi:10.1128/spectrum.02752-22
Wanka, F., Cairns, T., Boecker, S., Berens, C., Happel, A., Zheng, X., et al. (2016). Tet-on, or Tet-off, that is the question: advanced conditional gene expression in Aspergillus. Fungal Genet. Biol. 89, 72–83. doi:10.1016/j.fgb.2015.11.003
Webster, N., Jin, J. R., Green, S., Hollis, M., and Chambon, P. (1988). The yeast UASG is a transcriptional enhancer in human hela cells in the presence of the GAL4 trans-activator. Cell 52, 169–178. doi:10.1016/0092-8674(88)90505-3
Weinberg, B. H., Pham, N. T. H., Caraballo, L. D., Lozanoski, T., Engel, A., Bhatia, S., et al. (2017). Large-scale design of robust genetic circuits with multiple inputs and outputs for mammalian cells. Nat. Biotechnol. 35, 453–462. doi:10.1038/nbt.3805
Weinmann, P., Gossen, M., Hillen, W., Bujard, H., and Gatz, C. (1994). A chimeric transactivator allows tetracycline-responsive gene expression in whole plants. Plant J. 5, 559–569. doi:10.1046/j.1365-313X.1994.5040559.x
Wu, Y. I., Frey, D., Lungu, O. I., Jaehrig, A., Schlichting, I., Kuhlman, B., et al. (2009). A genetically encoded photoactivatable Rac controls the motility of living cells. Nature 461, 104–108. doi:10.1038/nature08241
Xie, Z., Wroblewska, L., Prochazka, L., Weiss, R., and Benenson, Y. (2011). Multi-input RNAi-based logic circuit for identification of specific cancer cells. Science 333, 1307–1311. doi:10.1126/science.1205527
Yang, L., Nielsen, A. A. K., Fernandez-Rodriguez, J., McClune, C. J., Laub, M. T., Lu, T. K., et al. (2014). Permanent genetic memory with >1-byte capacity. Nat. Methods 11, 1261–1266. doi:10.1038/nmeth.3147
Yang, X., Rocks, J. W., Jiang, K., Walters, A. J., Rai, K., Liu, J., et al. (2025). Engineering synthetic phosphorylation signaling networks in human cells. Science 387, 74–81. doi:10.1126/science.adm8485
Ye, Z., Li, S., Hennigan, J. N., Lebeau, J., Moreb, E. A., Wolf, J., et al. (2021). Two-stage dynamic deregulation of metabolism improves process robustness and scalability in engineered E. coli. Metab. Eng. 68, 106–118. doi:10.1016/j.ymben.2021.09.009
Yesbolatova, A., Saito, Y., Kitamoto, N., Makino-Itou, H., Ajima, R., Nakano, R., et al. (2020). The auxin-inducible degron 2 technology provides sharp degradation control in yeast, mammalian cells, and mice. Nat. Commun. 11, 5701. doi:10.1038/s41467-020-19532-z
Young, R. M., Engel, N. W., Uslu, U., Wellhausen, N., and June, C. H. (2022). Next-generation CAR T-cell therapies. Cancer Discov. 12, 1625–1633. doi:10.1158/2159-8290.CD-21-1683
Zhang, H., Fu, X., Gong, X., Wang, Y., Zhang, H., Zhao, Y., et al. (2022). Systematic dissection of key factors governing recombination outcomes by GCE-SCRaMbLE. Nat. Commun. 13, 5836. doi:10.1038/s41467-022-33606-0
Zhang, R., Goetz, H., Melendez-Alvarez, J., Li, J., Ding, T., Wang, X., et al. (2021). Winner-takes-all resource competition redirects cascading cell fate transitions. Nat. Commun. 12, 853. doi:10.1038/s41467-021-21125-3
Zhang, R., Li, J., Melendez-Alvarez, J., Chen, X., Sochor, P., Goetz, H., et al. (2020). Topology-dependent interference of synthetic gene circuit function by growth feedback. Nat. Chem. Biol. 16, 695–701. doi:10.1038/s41589-020-0509-x
Keywords: genetic circuit design, gene expression control, synthetic parts, regulatory parts, synthetic gene networks, transcriptional and translational control
Citation: Müller MM, Arndt KM and Hoffmann SA (2025) Genetic circuits in synthetic biology: broadening the toolbox of regulatory devices. Front. Synth. Biol. 3:1548572. doi: 10.3389/fsybi.2025.1548572
Received: 19 December 2024; Accepted: 10 February 2025;
Published: 07 March 2025.
Edited by:
Corentin Briat, University of Applied Sciences and Arts Northwestern Switzerland, SwitzerlandReviewed by:
Maurice Filo, ETH Zürich, SwitzerlandCopyright © 2025 Müller, Arndt and Hoffmann. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Katja M. Arndt, a2F0amEuYXJuZHRAdW5pLXBvdHNkYW0uZGU=; Stefan A. Hoffmann, c3RlZmFuLmhvZmZtYW5uQHd1ci5ubA==
†ORCID: Marik M. Müller, orcid.org/0000-0003-2173-5631; Katja M. Arndt, orcid.org/0000-0001-9220-0920; Stefan A. Hoffmann, orcid.org/0000-0002-3184-8640
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.