Explore article hub
- 1NovoCrops Center, Department of Plant and Environmental Sciences, University of Copenhagen, Copenhagen, Denmark
- 2School of Biological Science, University of Western Australia, Perth, WA, Australia
- 3International Research Centre for Environmental Membrane Biology, Foshan University, Foshan, China
Abstract
It is often stated that agricultural outputs need to increase substantially to meet the demands for more food posed by a growing population. However, when accounting for climate change, we argue that current projected increases are unrealistic and a more realistic goal would be to maintain yields per area of food production. This will require breeding for crops with increased tolerance to abiotic stresses such as drought, salinity, waterlogging, and high temperatures. This goal can be accomplished in one of two ways: by introducing stress tolerance genes into present high-yielding crops or by increasing the yields of already tolerant orphan crops and/or wild plants. We argue that the first strategy will require easing the restrictions on the use of gene editing technologies and making substantial improvements to cell-based phenotyping to identify the stress tolerance genes available in the gene pool of a crop and its wild relatives. The success of the second strategy will depend on the number of domestication genes that need to be selected for in order to obtain yields comparable to present-day cultivars. It is still too early to conclude which of the two strategies, rewilding (bringing genes lost from wild ancestors back to domesticated crops) or de novo domestication (domesticating resilient wild plants or underutilized crops directly), will be most effective for future sustainable agriculture. However, given the importance of the issue, some rapid action needs to be taken.
Key points
- Agricultural production is hampered by climate extremes such as heat, drought, waterlogging, and salinity, affecting soil health, water availability, and crop performance.
- Using wild crops to regain the abiotic stress tolerance that was lost or weakened during domestication is fundamental to achieving global food security and sustainable agricultural production in the face of climate change.
- Progress in the field could be achieved by introducing genes from wild relatives, but the lack of understanding of the operation of specific genes in planta remains a major hurdle in breeding programs, calling for broader use of cell-based phenotyping platforms.
- An alternative solution is to engage in direct domestication of wild relatives that already have the genes required for abiotic stress tolerance.
- The success of both strategies will be critically dependent on legislative issues and associated public perception of the technologies used for their implementation.
Introduction
Agriculture is vulnerable to climate change and the implications of global warming on the profitability and sustainability of agricultural production systems are substantial. Climate change is predicted to increase the impact of both biotic and abiotic stresses on crops. The ability to control biotic stresses such as diseases and pests is challenged by the pressure to reduce the use of chemical control agents and thus their environmental footprint. Abiotic stresses negatively affect the fertility of soils and impose massive limitations on crop performance. Many of them (e.g., drought, extreme temperatures, and soil salinization) are tightly interlinked and should be considered in the same context.
Global warming leads to major changes in rainfall patterns, thus increasing both the frequency and severity of extreme weather events (1, 2), with the risk of a 1-in-100 years extreme weather event likely to increase to 1 in 30 by 2040 (3). At present, 40% of the entire land area is currently classified as dryland zones; 70 countries in the world are regularly affected by drought (4), costing agriculture over US$80 billion p.a. in lost opportunities (5). On average, 1 week of drought stress causes a 3–8% yield loss for major staple crops (6), and future predictions are that the drought-related yield reductions will increase by more than 50% by 2050 for the major crops (5). Equally concerning is soil flooding. Large rainfall events can lead to flash flooding, river surges, and rising groundwater tables (7). It is estimated that more than 17 million km² of land surface is affected by waterlogging (which occurs when water fills the root zone of a plant and in this way decreases the oxygen available for respiration of roots), resulting in an annual economic loss of US$74 billion (8). This is hardly surprising: apart from rice, no staple crop can cope with hypoxic conditions in the rhizosphere caused by soil flooding. The 18-fold drop in mitochondrial adenosine triphosphate (ATP) production under hypoxia comes with critical implications for a crop’s ability to acquire nutrients and assimilate carbon, with consequences for growth and development. As a result, waterlogging-induced yield losses in some species may be as high as 70% (9).
High temperature per se also limits crop productivity. Of special concern are increasing nighttime temperatures. It has been reported that nighttime temperatures are rising at a rate that is 1.4 times that of daytime temperatures (10), and every 1°C increase in nighttime temperature causes a 3–6% drop in crop yield (11). While the physiological mechanisms underlying yield decreases in response to elevated nighttime temperatures are complex, the key component is an increase in nighttime respiration, which reduces the amount of photoassimilates available for plant growth and yield. Hotter nights can also negatively affect grain quality (12).
Global warming and changes in soil moisture content also have a significant effect on the soil microbiome (13), thus affecting the biological availability of many essential nutrients. Also, since the Green Revolution (GR), fertilizers have become a cornerstone of modern agriculture (14). As a result, agricultural systems established during the GR have evolved toward an excessive use of intensive production practices. For example, the doubled food production of the past 35 years has come from a 6.9-fold increase in nitrogen (N) fertilization and a 3.5-fold increase in phosphorus (P) fertilization (15). However, these practices are no longer considered to be sustainable under current climate scenarios (16) due to reduced nutrient use efficiency in crops grown under stress conditions (17). This results in massive eutrophication of aquatic ecosystems, where N and P cause algal blooms and result in a loss of biodiversity (18).
Climate change and the challenges it poses to agriculture
What are the future climate scenarios? Will the problem go away? Judging from the current trends (Figure 1), this is highly unlikely. There has been a continuous upward trend in the frequency and intensity of severe drought events since the 1970s (Figure 1A), and models predict this is expected to continue for at least a few decades (Figure 1B). The same trend applies to several extreme heat events (illustrated by the case of Australia in Figure 1C).
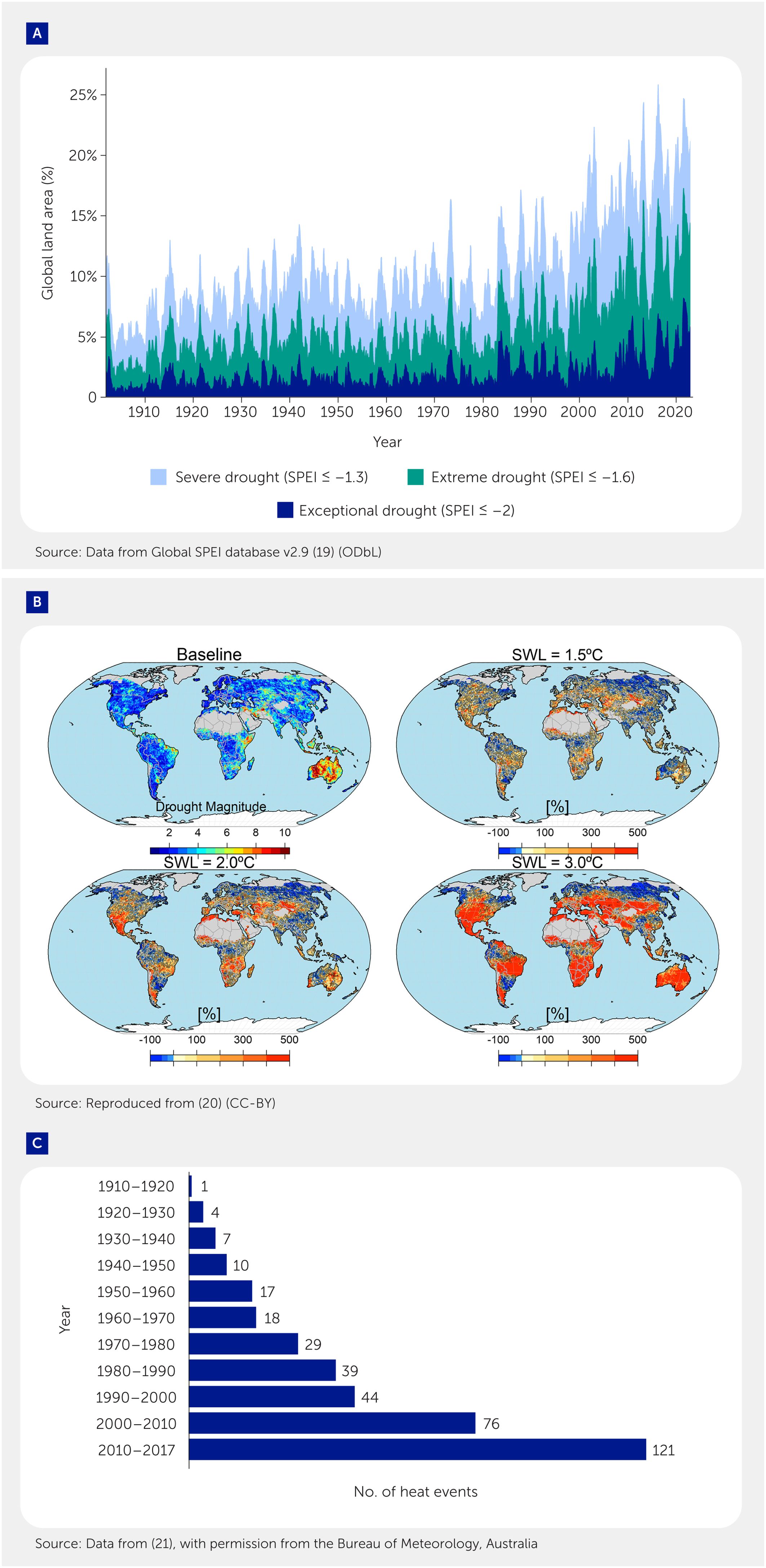
Figure 1. Current trends in occurrences of extreme weather events (heat and drought as examples). (A) Global area (%) affected by drought events on a monthly basis over the last 100 years [based on the Global Standard Precipitation-Evapotranspiration Index (SPEI) database (19)]. (B) Drought magnitude and relative changes (%) in drought magnitude concerning the baseline of three specific warming levels (1.5, 2.0, and 3.0°C). Reproduced from (20), (C) The reported trends in the frequency of extreme heat events over the last 100 years in Australia. Reported numbers are the number of days each year where the averaged daily mean temperature of the Australian area was extreme (e.g., above the 99th percentile of each month). Data from (21), used with permission from the Bureau of Meteorology, Australia. The authors' interpretation of the data shall not be attributed to the Bureau of Meteorology, Australia.
With an increasing number of hot days, soil evaporation is intensified, creating more arid and semi-arid areas (Figure 1B). As a result, agricultural food production will rely heavily on irrigation. Global freshwater consumption has increased progressively over the last century. (22) (Figure 2A); this is partially due to population growth but mostly to an ever-increasing proportion of water being used for irrigation purposes. As such, in the last 100 years, agricultural water consumption has increased almost 6-fold from ∼320 billion m3 in 1900 to over 1900 billion m3 in 2000 (23) (Figure 2B). This is hardly surprising, as in many parts of the world irrigation-based productive systems are an order of magnitude more effective than rainfed systems. In Australia, less than 1% of irrigated agricultural area is responsible for a quarter of the total agricultural value (27). The same is true for many other regions across the globe. In Pakistan (the fifth most populous country in the world) and India (the most populous country in the world), cultivated land that uses irrigation accounts for over 70% and 42%, respectively, of all cultivated land (26).

Figure 2. Global trends in agricultural water withdrawal and increasing salinization of agricultural land. (A) Trends in freshwater consumption over the last century for agricultural, industrial, and domestic use. Adapted from (22). (B) Global trends in water withdrawal for agricultural purposes. Adapted from (23) with inclusion of data from (24), under the “Agricultural water withdrawal” parameter. (C) Essentiality of leaf water evaporation for transpirational cooling. The blue line represents daily changes in the ambient air temperature; the green line reports the actual leaf temperature (using lime tree—Tilia cordata—as an example). Adapted from (25). (D) A heat map showing the trend in changes in electrical conductivity of irrigation water in the five most populated salt-affected countries and Australia. The color scale indicates the range of electrical conductivity (µS cm-1). Adapted from (26).
The increasing reliance on irrigation is unsurprising not only in the context of minimizing the impact of soil drought on plant performance but also as a means to deal with increasing ambient temperatures. The optimal temperature for C3 species is around 23°C, while for C4 species it is around 32°C. Except for maize, all staple crops that provide the bulk of caloric intake for humans (wheat, rice, potatoes, and soybeans) are C3 plants and, therefore, perform better in cooler climates. However, this optimal temperature refers to leaf (not ambient) temperature, and transpiration cooling can bring leaf temperature well below that of ambient air during the hottest time of the day (Figure 2C). This difference may be rather substantial. For example, Gupta et al. (28) conducted assays of the transpiration cooling capacity of five tree species in India, reporting an average leaf-to-air difference between 2.8 and 3.9°C. In extreme cases, this value could be as high as 8–10°C (29). Thus, increased use of irrigation in agricultural production systems may become unavoidable under conditions of global warming if plants are to stay within the optimal “thermal window”.
Agriculture in the 21st century will become saline
The increased reliance on irrigation inevitably comes with the issue of soil salinization. Fresh water is a scarce resource, as most of the water on the planet is saline, and only 0.76% of the biologically available groundwater is considered suitable for irrigation (30). As this is hardly enough to meet the growing demand, agriculture is increasingly relying on the use of poor-quality brackish water (26). Analysis of the electrical conductivity of irrigation water in the five most populous countries affected by salinity shows a trend of increasing electrical conductivity of available irrigation water (Figure 2D). This results in 3–6 tons of salt added per hectare of irrigated land every year (26). As most modern crops are salt excluders [a result of targeted breeding to reduce Na+ accumulation in the shoot (31)], the salinity buildup in the rhizosphere results in land that is not suitable for production. Reversing this trend is a challenging task: it will require either taking the saline land out of production for a few decades or using halophytic species for phytoremediation (desalination).
While the second option has been broadly advocated for in the literature (32, 33), very few conventional staple crops possess sufficient halophytism to perform this role. A notable exception is quinoa (Chenopodium quinoa)—a species that is considered to be nutritionally important by the Food and Agricultural Organization of the United Nations (FAO) (34) and that possesses a remarkable salinity tolerance as well as the ability to accumulate significant amounts of salt in its aboveground parts (35). However, quinoa is cultivated on a much smaller scale (less than 1%) compared to traditional cereal crops such as wheat, rice, and maize, making it an unlikely candidate to fulfill this role. As for taking salt-affected land out of production, this is equally unlikely: the amount of arable land per capita is progressively declining as a result of land degradation and an increased urbanization rate (Figures 3A, B).
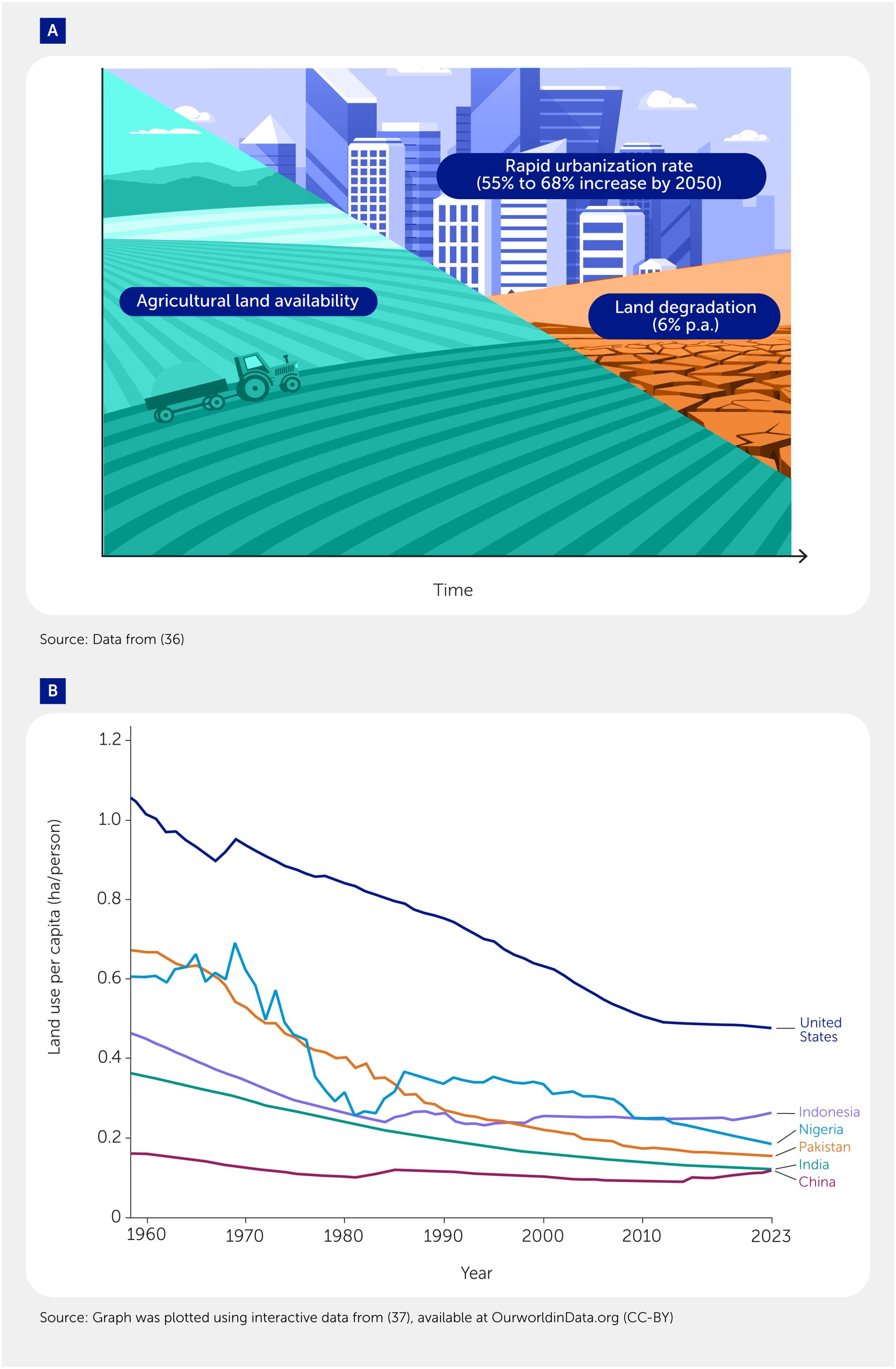
Figure 3. Trends in agricultural land availability. (A) Land urbanization and degradation rate worldwide. Data from (36). (B) Changes in land use per capita in the six most populous countries in the world. The graph was plotted using interactive data from (37).
An urgent need to breed for abiotic stress tolerance
It is a current dogma that the increasing demand for food posed by a growing population urges us to increase food production. Taking into consideration that overall wealth is also increasing, the FAO (38) estimated that an increase in world population by 2050 by 30% will require food production to increase by 70%. As dietary choices change with wealth and in general tend towards increased consumption of meat, the production of which requires more feeds, a 100–110% increase in demand for agricultural products by 2050 has also been forecasted (15). Even these numbers might be underestimates, as an increasing food supply could result in lower prices that may generate increasing demand, creating a vicious circle (39). For example, with wealth, human diets tend to become richer in fats and vegetable oils, and the ever-increasing supply of cheap palm oil [e.g., in Indonesia its production increased dramatically between 1970 and 2020 (40)] has led to ever-increasing demands. It can therefore be argued that an increase in food production may result in a positive feedback loop that only satisfies short-term needs but creates even more demand in the long run (41).
However, even if we believe that a substantial increase in food supply may solve the problem, a question arises: will we be able to meet the projected demand at all? We should test every possibility to increase yields, but increased food production can only be obtained in one of two ways: either by increasing yield per production area or increasing the area of production. Since 1961, food production has doubled twice, mainly due to increased productivity per unit of land, but yearly yield increases have become slower, suggesting that we are approaching a yield ceiling for some of our major crops (42). Will yield increase be able to follow the pace of increasing demand? Taking cereals as an example, this is not the case (43).
The other “solution”, increasing the area of production, is inevitably connected with the loss of nature (44). For example, the Amazonian rainforest is already being cut down to clear land for soybean production and feed for cattle, which threatens the biodiversity that is vital for the survival of humankind. Is this what we want?
The dream of letting supply keep pace with the demand for food could be turned into a nightmare by climate change. With increasing temperatures, drought, extreme weather, and further salinization of soils, yields per unit of land are likely to decrease despite all attempts to realize the opposite, and more land will become marginal and, hence, not suitable for most crops. Considering climate change, maintaining yields per production area therefore emerges as a more realistic but still very ambitious breeding goal for the future.
Breeding for abiotic stress tolerance is a scientific problem, and it will be discussed in the following sections. Predicted challenges from populations that become more urbanized and wealthier, such as changes in dietary choices, food waste, and uneven distribution of food, are not scientific problems but have to be dealt with politically.
Crop domestication and the loss of tolerance to abiotic stress
Domestication of plants has always been a process that favored yield, with early achievements being increases in grain size and loss of seed shattering (45), and continued strong selection for increase in yield has with time diminished genetic diversity in modern crops (46). As abiotic stress resistance was rarely selected for previously (47), this has diminished the adaptability of crops to survive in unfavorable environments (48–50).
In the ancestral tomato species, for example, salt tolerance is facilitated by the gene SlHAK20, which encodes a Na+ and K+ transporter that regulates the balance between Na+ and K+ during salt stress, but in modern sensitive tomato cultivars, this gene is mutated and non-functional (51). Similarly, TmHKT1;5-A, which encodes a high-affinity Na+ transporter at the plasma membrane, contributes to the salt tolerance of Einkorn (Triticum monococcum), a diploid ancestor of tetraploid durum wheat (T. turgidum ssp. durum) and hexaploid bread wheat (T. aestivum), but it is no longer present in durum and bread wheat (52). Regulators of these targets may also have been lost during domestication.
Strategies for regaining lost abiotic stress traits
Rewilding of crops
Reintroducing genes from wild relatives
“Rewilding” has been suggested as a strategy for our modern crops to regain strength (48, 53). According to this approach, genes important for survival in challenging environments—lost during the domestication process—should be identified in wild relatives and reintroduced into modern crops (48). A good example is tomato. Introduction by transgenesis of the functional SlHAK20 allele from the wild ancestor into the modern large fruit tomato improved its salt tolerance (51). Thus, tapping the genetic resources of wild relatives appears to be a viable strategy to improve abiotic stress resistance in crops. However, the potential for introgression of resistance genes from wild relatives into elite cultivars simply by crossing (see next section) remains relatively underexplored, one reason being that even after multiple backcrosses the resulting plants may still have unwanted wild-type traits (54, 55). The latter is hardly surprising, given linkage drag and the multigenic nature of tolerance traits.
Abiotic stress resistance is multigenic
Resistance to specific biotic stresses can be a monogenic trait in which case the responsible gene can be transferred to elite cultivars by transgenesis or crossing and introgression (56). In contrast, resistance towards abiotic stresses as a rule is more complex. It can be monogenic, as is the case for submersion tolerance in rice (57, 58), boron toxicity tolerance in barley (59, 60), and, in the case of constitutive activation of a gene, for aluminum tolerance in barley (61). Typically, however, tolerance to most abiotic stresses involves many elements used in sensing, signaling, and regulation of gene expression and protein function (62). Ways for breeders to circumvent this challenge have been to develop lines that escape the stress, for example, by selecting wheat for early flowering and rapid development so that it completes its life cycle before terminal drought sets (63), or selecting for phenology that helps crops to withstand drought and heat stress conditions, e.g., leaf angle and rolling (64, 65).
Still, there was for a long time hope that “silver bullet” genes could be found and used to confer abiotic stress tolerance when overexpressed (5, 47). Candidate genes that have been identified in the model plant Arabidopsis thaliana, which has neither been domesticated nor adapted to a cropping environment, have often failed to translate to valid targets in crops (66). One previous candidate was the vacuolar Na+/H+ exchanger NHX1, but despite encouraging initial attempts that showed increases in salt tolerance following its overexpression (67, 68), other studies could not reproduce such an effect. This was probably due to a lack of co-expressed transport systems to energize this antiporter (69) as well as a need to ensure efficient Na+ retention in the vacuole, which requires tight control of the operation of Na+-permeable tonoplast fast (FV) and slow (SV) vacuolar channels. Additionally, the energization of NHX1 operation in salt-grown plants is conferred mostly by H+-translocating inorganic pyrophosphatase (H+-PPase) (70), and the latter is K+-dependent and requires significant amounts of K+ in the cytosol for its operation. At the same time, salinity stress results in a massive cytosolic K+ loss (71, 72), thus hampering H+-PPase operation.
Another example is related to the salt overly sensitive (SOS) signaling pathway. A genetic screening of Arabidopsis identified three genes (SOS1, SOS2, and SOS3) with a sos phenotype (73–75). SOS1 turned out to be a Na+/H+ exchanger regulated by SOS2, a protein kinase, which in roots is regulated by SOS3, a Ca2+-dependent protein (76). In shoots, SOS1 is activated by CIPK8, a protein kinase related to SOS2, which is regulated by the Ca2+-dependent protein CBL10 (77). The activated SOS1 is energized by the electrochemical H+ gradient generated by the plasma membrane proton-pumping ATPase (H+-ATPase) (78), which is regulated by multiple regulatory proteins, some of which are activated in salt stress conditions (79).
But do the genes listed above represent the full package? Most likely not. Transgenic Arabidopsis that overexpresses either SOS1 or SOS3 alone has somewhat increased salt tolerance, but no further increase was observed when SOS1, SOS2, and SOS3 were expressed together (80). There can be several reasons for this. First, the SOS1 exchanger is expressed not only in the root epidermis, where it functions by expelling Na+ back into the rhizosphere, but also at the xylem/parenchyma interface (81) (Figure 4A; red circles). Thus, overexpressing SOS1 in the root epidermis to prevent Na+ uptake comes with the danger of a concurrent increase in Na+ loading into the xylem and its delivery to the shoot. Salt tolerance is typically regulated at the cellular or tissue level (85), and “one size will not fit all”. At the very least, either two different isoforms of SOS1 transporters will be required to uncouple Na+ exclusion from uptake from its delivery to the shoot, or the modes of their post-translational regulations should be different. Additionally, SOS1 expression is confined to a small region in the root apex, as shown both by GUS staining and electrophysiological data (Figure 4B). Given that the surface of the mature root zone (where the bulk of Na+ uptake takes place) is several orders of magnitude bigger than that of the root apex, can we expect plants to efficiently expel 98% of all Na+—as per current estimates (86)—through SOS1 activity in this region?

Figure 4. Targeting SOS1 and HKT1 transporters involved in Na+ exclusion from uptake is unlikely to deliver salt-tolerant cultivars that would be performing on par with halophytes. (A) Key membrane transporters mediating plant Na+ and K+ homeostasis that confer salinity stress tolerance in plants. Adapted from (82). The two most targeted genes, namely SOS1 and HKT1, are circled. (B) SOS1 Na+/H+ exchanger is predominantly expressed in the root apex but not the mature zone. The panel on the left show GUS expression in a root tip in a SOS1::GUS Arabidopsis plant. Adapted from (81), with permission from Oxford University Press. The right panel reports the difference in net Na+ efflux (in nmol m-2 s-1) measured from rice roots in elongation (EZ) and mature (MZ) zones by non-invasive microelectrode ion flux estimation technique—based on (83)—that may be used as a proxy for SOS1 operation, as shown by (84). (C) Overexpression of the TmHKT1;5-A gene in the Nax2 locus in wheat that encodes a Na+-selective transporter located on the plasma membrane of root cells surrounding xylem vessels results in a 25% increase in plant yield. However, this beneficial effect was observed in only one (of three) tested field sides (with the highest salinity). Even so, crops overexpressing TmHKT1;5-A showed 50% yield penalties compared with the control. The graph is based on partial data from (52).
The HKT1 gene is another good example. This transporter belongs to the high-affinity family of transporters and plays a critical role in K+ and Na+ homeostasis in plants. There are two different types of HKT transporters, Types I and II, and they differ in their selectivity. Type I transporters are specific to Na+, with a difference conferred by the presence of a serine residue in the first “selectivity filter” (87). In cereals, HKT1;5 transporters are believed to “exclude” Na+ from the xylem into the xylem parenchyma in the root, thus reducing Na+ delivery to the shoot (88–90).
As preventing Na+ delivery to the shoot has been hypothesized to confer better salt tolerance, numerous attempts have been undertaken to genetically manipulate HKT1;5 expression to improve salinity stress tolerance in various species. One of the best examples of this is a work by Munns and co-authors where the authors crossed wheat diploid ancestor Einkorn (T. monococcum) with durum wheat and selected a progeny harboring TmHKT1;5-A in a new backcross (52). The authors did report a 25% increase in plant yield in a salinity-affected field but only in one of three tested sites (Figure 4C).
It was also suggested that OsHKT1;5, first named OsSKC1 (88), was present at a quantitative trait locus (QTL) on chromosome 1 in rice (Oryza sativa). This QTL was termed Saltol and shown to contribute to salt tolerance of rice at a seedling stage. Identified by crossing the salt-tolerant indica landrace Pokkali and the salt-sensitive indica cultivar IR29 followed by several backcrosses to obtain recombinant inbred lines (91, 92), Saltol was suggested to operate in Na+ exclusion from the shoot. This hypothesis was further substantiated by the finding that OsHKT1;5 localizes to cells adjacent to the xylem in roots, and, when rice is salt-stressed, its loss of function causes substantial accumulation of Na+ in shoots (93). However, direct electrophysiological measurements of Na+ fluxes across the plasma membrane of xylem parenchyma cells showed that wild type plants were more efficient in reabsorbing Na+ from the xylem as compared to SKC1 plants (with overexpressed OsHKT1;5) putting into question the functional role of HKT1;5 as a transporter for direct Na+ removal from the xylem (94). Instead, changes in the expression level of HKT1;5 altered the activity of membrane transporters involved in K+ and Ca2+ acquisition and homeostasis in the rice epidermis and stele, explaining the observed phenotype. Thus, the role of HKT1;5 in plant salinity tolerance cannot be attributed to merely reducing Na+ concentration in the xylem sap but triggers a complex feedback regulation of activity of other transporters involved in the maintenance of plant ionic homeostasis and signaling under stress conditions.
It should be also noted that OsHKT1;5 is not the only gene that may be linked to salt tolerance on the Saltol QTL. Approximately 783 protein-coding genes have been mapped within Saltol with about 25% of the genes coding for proteins of unknown function (95). Among these, the intermediate filament (OsIF) stabilizes photosynthesis during salt stress (96); OsSalT, which encodes a mannose-binding lectin protein, is induced by salt mainly in roots (97), shows high diversity between salt-tolerant and sensitive lines (98), and increases salt tolerance when expressed in tobacco (99). Additionally, at least two transcription factors within Saltol add to salt tolerance in rice when overexpressed: the bZIP transcription factor OsHBP1b (100) and OsGATA8, a GATA transcription factor (a family named after the consensus DNA-binding motif) (101).
Consistent with this, the bread wheat gene TaSPL6-D, which encodes a protein with an SBP-type zinc finger required for binding to DNA (102), was recently identified as a transcriptional suppressor of TaHKT1;5-D (103). The corresponding gene in rice is located on chromosome 3 and was found to regulate the transcription of OsHKT1;5 as well (104). Altogether, these findings demonstrate that HKT1;5 is not sufficient for salt tolerance, and upstream regulators of HKT1;5 regulation also have to be considered.
The above findings are not surprising for several reasons. First, excluding Na+ from the shoot may prevent its potential cytotoxicity in leaf mesophyll but comes with a need for plants to rely on de novo synthesis of compatible solutes for osmotic adjustment and turgor maintenance. This is an energetically costly option (104) that comes at the expense of growth/yield. Second, under physiological conditions, Na+ loading into the xylem is a thermodynamically active process (105). Why should plants spend energy loading it first and then trying to retrieve it? Finally, once Na+ is taken from the xylem, where can it go? The storage capacity of the root parenchyma is very limited, and, except for the root tip where the impermeable Casparian strip is absent, the Casparian strip does not allow Na+ to be put back into the cortex and then excluded into the rhizosphere. Also, more recent data from Arabidopsis and crop species show that AtHKT1;1/HKT1;5 alleles also have a strong genetic association with “shoot sodium accumulation” and concomitant salt tolerance (87). Thus, it appears that the effects of AtHKT1;1/HKT1;5 operations are strongly influenced by soil salinity levels, with both hyper-functional or weak AtHKT1;1/HKT1;5 alleles/haplotypes being present.
The above arguments can be made also for other abiotic stresses (drought, waterlogging, and temperature extremes), and the reality of the situation is that the “full package” of any abiotic stress tolerance may remain unknown: it likely includes not only transporters and enzymes but also an elaborate network of regulatory proteins such as receptors, signal transduction components, and transcription factors that are yet to be discovered. Therefore, building such a trait from scratch, i.e., within a species the ancestors of which never had this trait, could prove impossible. The multigenic nature of abiotic stress resistance is the likely reason why efforts to improve the tolerance of crops to such stresses have had limited success so far (47, 106). And—in practical terms—even if we can get a comprehensive understanding of all details of signaling networks, how many genes can breeders practically handle from the required “package”? Five? Ten? Will this be good enough?
Pangenome: the genetic repertoire of a species
If tolerance to abiotic stress is present in a close relative but was lost during domestication, the situation may be less challenging (though not necessarily simple!). In this case, we may hypothesize that many of the genes in the tolerance “package” are still present and intact and only a few elements (in principle, a single element) need to be “repaired”. Thus, if abiotic stress tolerance is a trait involving multiple genes acting in concert, it also follows that the trait could be lost if just a single or a few elements in this assembly are missing. If our present-day crops have retained most of the required elements but are just missing a few, we may hopefully be able to identify the absent pieces. But how do we find them?
Pangenomes have emerged as the haystack in which we might find the lost needle (107). Within a species, many genes (core genes) are common for all accessions, but some individuals or races may have genes (variable genes or cloud genes) that others do not. The pangenome includes both core genes and variable genes and thus covers the genomic diversity of a species. An important finding from studying pangenomes is that the proportion of variable genes in a species can be substantial (107). For example, whereas the reference genome of the tomato (Solanum lycopersicon cult. Heinz 1706) contains 35,496 protein-encoding genes (108), sequencing of 725 tomato accessions in the Lycopersicon clade resulted in a combined gene catalog for the species of 40,369 protein-coding genes (108). This implies that the pangenome of tomato includes as many as 4,873 genes that are not accounted for in the reference genome.
Another important finding has been that genes annotated as being involved in biotic and abiotic stress tolerance are enriched in the group of variable genes, with biotic resistance genes being predominant (102, 107–113). An exception is Amborella trichopoda, which is basal in evolution to all other living flowering plants and is endemic to New Caledonia (114). A total of 11 Amborella accessions were genome-sequenced to capture major presence/absence variation in this species. Among the variable genes, most stress-related genes were linked to abiotic stress, and among these, 152 genes were predicted to be involved in the salt response, 100 in the cold response, and 76 in the drought response (114). Among the dispensable genes for salt tolerance are UWAAmboGene2710 and UWAAmboGene2237, which are homologs of Arabidopsis NHX1 and NHX2, respectively. The total number of genes related to salt stress (core genes plus variable genes) was found to be 480. This represents a large number of candidates for genes that may have gone missing during plant evolution.
Pitfalls of whole-plant phenotyping
Finding the pieces in the complete repertoire of genes of a species that sensitive cultivars are missing appears as a formidable task. For this purpose, forward genetics based on phenotyping remains a possibility.
It is not a surprise that tolerance to various abiotic stresses is multigenic. Most traits require the combined action of multiple genes. A common method to identify the multiple genetic elements that control a trait is QTL analysis (115–117). This method studies the segregation of marker genotypes in combination with the phenotypes of individual plants or plant lines. In this way, their location on chromosomes and the quantitative contribution of the genetic element can be determined. As an example, a resistant and a sensitive line are crossed, and the progeny is allowed to self-fertilize. In the next progeny, the plants are tested for their degree of tolerance and for whether the phenotype is associated with specific genetic markers. A genetic marker is a DNA sequence that varies between genotypes and has a known location on a chromosome. Thus, using specific primers, a polymerase chain reaction (PCR) reaction may amplify a marker in one of the parental lines but result in no product in the other line. If a given marker is found to be associated with a given trait, this reveals information about the chromosomal location and the quantitative contribution of the responsible genetic element.
However, since a QTL analysis typically takes place at the whole plant level, it suffers from two main drawbacks. First, the trait that is scored may be controlled not only by multiple genes but multiple mechanisms that may contribute to the trait. Thus, the analysis does not directly address the mechanism(s) of tolerance but rather overall fitness, which can be conferred by unrelated mechanisms. For example, Zhao et al. (118) carried out a QTL analysis to identify genes contributing to heat tolerance in rice by crossing a heat-tolerant line (Habataki; indica spp.) with a heat-susceptible line (Sasanishiki; japonica ssp.). The F2 generation was subjected to high-temperature stress, and three traits that were influenced by this stress, namely daily flowering time, spikelet fertility, and pollen shedding level, were subsequently scored. Four QTLs were detected for daily flowering time on chromosomes 3, 8, 10, and 11 and seven QTLs were identified for spikelet fertility on chromosomes 1, 2, 4, 5, 7, and 10. While five of the seven QTLs for spikelet fertility overlapped with pollen shedding level (suggesting related mechanisms), none of them overlapped with daily flowering time (suggesting an unrelated contributing mechanism).
A second drawback is that the trait searched for may be misleading. One mechanism of salt tolerance may be salt exclusion, and therefore low Na+ concentration in the leaf blade has been assessed in search for salt tolerance genes (119). A salt-sensitive durum wheat cultivar (T. turgidum ssp. durum cult. Tamaroi) was crossed with a landrace of durum with low Na+ concentration in the leaf blade (Line 149 derived from T. monococcum), and 100 individuals in the F2 generation were scored for the trait. This led to the successful identification of a leaf Na+ “exclusion” locus on the long arm of chromosome 2A, which was designated Nax1 [Na+ exclusion 1 (119)] and was subsequently found to be associated with a Na+ transporter gene HKT7-A2 (of type HKT1;4) (120). However, what is to be done if a mechanism of salt tolerance does not influence overall Na+ in the leaf blade but rather compartmentalization of Na+, e.g., in leaf cell vacuoles? If salt sequestration is the mechanism of tolerance, a salt-tolerant plant and a salt-sensitive plant could well have identical total Na+ content in the shoots (121).
The latter point is further illustrated by analysis of shoot Na+ content in some selected halophyte and glycophyte species (Figure 5A). In this graph, shoot Na+ context (expressed on a water basis) is plotted vs salinity threshold (the level of salt after which plant yield starts to decline). As one can see, the reported strong (R2 = 0.75) correlation is negative: species with higher salinity thresholds have higher overall shoot Na+ content. Thus, the above approach for selecting genotypes with lower shoot Na+ content—used for decades in crop breeding—appears to be fundamentally flawed as it fails to account for plant ability for intracellular Na+ compartmentation. The latter can be done by employing confocal scanning laser microscopy (CSLM) and Na+-specific fluorescent dyes such as CoroNa Green (Figures 5B–D). Analyzing leaf Na+ content by means of inductively coupled plasma (ICP) spectroscopy or flame photometry is much cheaper and quicker than CSLM and, thus, these have been the methods of choice for high-throughput phenotyping. However, as illustrated above, whole-plant ionomic analysis can be misleading as, in the case of salinity stress tolerance, it is Na exclusion from the cytosol—not the shoot—that matters.

Figure 5. The issues related to whole-plant phenotyping using shoot Na+ content as a physiological marker and a need for shifting to cell-based phenotyping. (A) Shoot Na+ content (measured on a water base; in mM) in selected glycophyte (blue dots) and halophyte (green dots) species plotted vs salinity tolerance threshold. Data from (122–130), images of crops from (131–140). Salinity threshold is determined as the salinity level (in dS m−1) above which the plant yield starts to decline. (B, C) Intracellular Na+ distribution measured between vacuolar and cytosolic compartments using CoroNa Green dye in two wheat cultivars contrasting in the salinity tolerance (B: Westonia, tolerant; C: Belgrade 3, sensitive). Adapted from (141). (D) Average CoroNA Green signal (arbitrary units) from two intracellular compartments, the vacuole and cytosol. The third column represents the average signal from the entire cell, calculated from both compartments (B, C). Adapted from (141).
Targeting key mechanisms through cell-based phenotyping
So, where are we now? The reality is that tolerance to many abiotic stresses is conferred by multiple mechanisms. Each of them operates in a specific tissue/cell type and is regulated by multiple genes. In this context, the (empirical) whole-plant phenotyping (regardless of whether it is hyperspectral imaging, or economics, or any other whole-plant-based trait) will always be critical for the final validation of genetic material (e.g., in field trials) but is unlikely to reveal the role of a specific mechanism/gene.
To be more effective, breeding targets can be directed towards specific mechanisms. For example, among multiple other traits (e.g., root architecture, trichome density, leaf rolling, and leaf angle) drought tolerance is consistently linked to the low density of the stomata, the pores through which plants absorb carbon dioxide CO2 but also lose water (142). Transgenic approaches have been employed to lower stomatal abundance, thereby reducing transpiration rates and water loss. For example, overexpressing the peroxidase gene TaPrx109-B1 or the transcription factor PpnMYC2 in wheat and poplar, respectively, resulted in lower stomatal density and increased water use efficiency (143, 144). Transpiration rates can hardly be measured in the field, but another approach is to screen for carbon isotope discrimination. When CO2 is incorporated into plant biomass, the 12C isotope is favored compared to the heavier 13C isotope, and this discrimination between carbon isotopes typically causes the overall abundance of 13C relative to 12C in plant tissue to be less than in atmospheric CO2 (145). Carbon isotope discrimination (Δ13C) is strongly linked to stomatal density, and therefore becomes negatively correlated with transpiration efficiency (146). Carbon discrimination can be measured in dry leaves by mass spectrometry and may be used as a breeding target for improving the efficiency of water usage and yield under drought stress [e.g., Kunz et al. (147)]. A safer way to target a specific mechanism would be using cell-based phenotyping platforms that will account for the cell- and tissue-specificity of the operation of a specific gene(s). For example, one mechanism that confers salt tolerance in plants is their ability to maintain a highly negative membrane potential generated by the plasma membrane H+-ATPase, which allows efficient K+ uptake and retention (148, 149) as well as provides energy for both NHX and SOS1 transporters involved in Na+ exclusion and sequestration (150). Under salt stress, the plasma membrane is depolarized due to a substantial influx of Na+. This is also the case for waterlogging, when the oxygen-limiting conditions create an insufficient ATP supply, leading to membrane depolarization. The outward-rectifying K+ channel GORK is voltage-gated and opens in response to stress-induced membrane depolarization. As a result, a substantial amount of K+ is lost from cells. This K+ efflux can be measured with very high spatial (a few microns) and temporal (5 sec) resolutions by non-invasive microelectrode techniques that can be used as a “physiological marker” for salinity tolerance traits that would account for tissue- and cell-specific operation of transporters involved (151).
Membrane potential can also be measured directly by impaling plant cells with microelectrodes. To test whether a QTL for membrane potential stability could be identified at the cellular level, Gill et al. (121) crossed the Chinese barley landrace TX9425, which is tolerant to waterlogging and salinity, with a Japanese malting cultivar Naso Nijo, which is sensitive to both. In 150 lines of the F2 generation, plasma membrane potential was measured from single epidermal root cells and correlated with the abiotic stress tolerance of the lines. This led to the identification of a major QTL on chromosome 2H that shows a strong linkage with salt tolerance and a weaker but significant linkage with waterlogging tolerance.
Another example of cell-based phenotyping has been carried out using root cell viability markers (152). Salinity stress in plant roots triggers the production and accumulation of reactive oxygen species (ROS), which causes oxidative stress and cell death. Accordingly, following ROS exposure to barley roots, the viability of root cells was performed using a double staining method that included fluorescein diacetate and propidium iodide. Image-based phenotyping of single root cells demonstrated that the viability of salt-tolerant barley was less affected by long-term ROS exposure than salt-sensitive barley varieties. Crosses between tolerant and sensitive lines could help identify QTL contributing to ROS tolerance in root cells during salt stress. Finally, Wu et al. (141) have successfully used CSLM-based CoroNa Green imaging to screen wheat cultivars for their ability to efficiently sequester Na+ in root vacuoles, demonstrating the power of this approach for selecting donors for breeding programs.
Introgression of genes into sensitive cultivars
When the desired variability within a modern crop is no longer present, it may remain in a related cultivar, species, or genera. Assuming that we can identify an ancestral abiotic stress-related gene that has been lost or that lost its function in an elite cultivar, the rewilding task is now to bring it back again. There are three ways for the modern crop to obtain missing genes from a relative.
The first strategy for refurbishment of the trait occurs through the crossing of the two genotypes and subsequent introgression of the gene into the elite cultivar. This can be done by interspecific (within a species) or intergeneric (within a genus) hybridization (Figure 6A). Both strategies combine genomes from different plants for the plant of interest to obtain a trait. If the plants are distantly related, the ability to cross may be low and the hybrids may be sterile and show resistance to chromosome doubling, which can result in the elimination of the chromosomes of one of the parents (153). In both types of hybridization, the breeder will worry about dilution of the elite germplasm, for which reason time-consuming backcrosses are required; this can be complicated if the desired gene is closely linked to a gene that confers an undesirable trait (153). New techniques such as speed breeding (154) and rapid-cycle recurrent genomic selection (155) allow for rapid cycling through generations. However, the risk of dilution of the elite germplasm increases exponentially if multiple genes are to be simultaneously introgressed into the germplasm.

Figure 6. Two strategies for obtaining crops that tolerate abiotic stresses. Current susceptible crops became high yielding due to mutations in the domestication genes of their ancestors. Excessive inbreeding and selection for yield only have resulted in loss of genes that function in gene packages that control stresses such as drought and salinity. (A) The lost tolerance genes may be regained by crossing the modern crop with its wild ancestor. This will dilute the elite crop, but, after several backcrosses, a rewilded tolerant elite crop is the result. (B) In this case, the wild tolerant ancestor is de novo domesticated by directed mutagenesis of domestication genes.
The second strategy is genetic engineering. Here, the donor gene of interest is cloned and inserted into the genome of the elite cultivar by genetic transformation. If the donor and recipient are naturally crossable and produce fertile offspring, the procedure will result in cis-genic plants (as opposed to transgenic plants). Using this method, multiple genes can be inserted at the same time. An inherent problem with such genetic engineering is that the genetic elements that control gene expression must be carefully considered. The natural expression of genes is dependent on cis-regulatory DNA sequences, and these can be placed upstream of the coding sequence, within introns, or downstream of the coding sequence. Even if the complete genomic DNA is used for transformation, its subsequent expression level may be low if the gene happens to be inserted into a chromosomal location with densely packed DNA.
The third strategy can only be employed if the “missing” gene is still there but has become non-functional due to a mutation. Single nucleotide polymorphisms (SNPs) that result in the loss of a trait can be identified by genome-wide association studies (156, 157). A plant with an SNP that causes loss of gene function is in principle carrying a genetic disease, which may be “cured” by gene therapy. This has become possible with the advent of novel gene-editing methods (158). While “traditional” clustered regularly interspaced short palindromic repeats (CRISPR)–CRISPR-associated 9 (Cas9) breaks DNA at defined positions and can create mutations, it most often causes insertions or deletions (indels) of DNA to take place at the breakage site. However, methods that allow editing of single bases and do not involve double-strand cleavage are now available. These are the results of synthetic biology where new abilities have been built into the CRISPR–Cas9 system. Prime editing is the most recent procedure, and it is rapidly being optimized for various purposes (159). A limitation of gene editing may be that the methods require the recipient plant to be amenable to transformation and regeneration, but new methods are being developed that in some cases have overcome these hurdles (160).
De novo domestication of already tolerant wild species
In summary, furnishing a sensitive plant with the complete gene package for any abiotic stress tolerance is not a trivial task and may be impossible if no relative has the desired trait. If a crossable relation is in possession of the trait in question, the task may be possible but is complicated, if not again impossible, depending on the multigenic nature of the trait and whether missing elements can be identified.
As an alternative way to obtain stress-tolerant crops, accelerated domestication of an already resistant relative was proposed (161). Domestication is a method of human-directed evolution where genetic elements that are important for survival in the wild but reduce yield have been lost. An example is the loss of the ability of the plant to shatter its seeds. In barley, this has resulted from loss-of-function mutations in either one of the genes Btr1 or Btr2 (162). The mutated plants were selected for by early farmers during domestication of this crop as they allowed for harvest without grains falling to the ground. In general, domestication of plants does not depend on new genes but results from changes in gene function, changes that in most cases involve loss of gene function.
Wild plants and orphan crops that tolerate abiotic stress already have all the genes required for the trait (163, 164). So, for the domestication of such plants, no new genes are required. Loss of function and/or changes in the strength of gene expression can be obtained relatively easily by new methods of gene editing provided that the editing tools can be introduced into the plant (Figure 6B). This is what makes accelerated domestication of wild plants an attractive possibility to obtain hardy crops.
Domestication genes are the genes that are present in an ancestral plant in an unmodified form and serve a natural purpose but when mutated result in a domestication phenotype (e.g., higher yield). Knowledge of the nature of domestication genes has been steadily increasing, and two main conclusions can be drawn. First, the low-hanging fruits (i.e., the genes that when mutated result in a substantial increase in yield or other important domestication traits) are quite few, maybe fewer than 10; and second, many domestication genes are conserved between species (161). For example, close homologs of barley Btr1 and Btr2 are present in other grass species, which implies that the ability of these grasses to shatter seeds would most likely be lost if any of these homologs were made dysfunctional (165). The barley genus, for example, abounds with species that tolerate extreme salt and drought stress (166, 167), and mutating Btr1 or Btr2 homologs in these species could be a first step to obtaining a domesticated salt-and/or drought-tolerant barley. Examples of resilient wild plants and underutilized crops that could be candidates for accelerated domestication are given in Table 1. Of note is that polyploid and perennial plants may have an increased ability to adapt to changing (221) and challenging environments (163, 222–224), respectively.
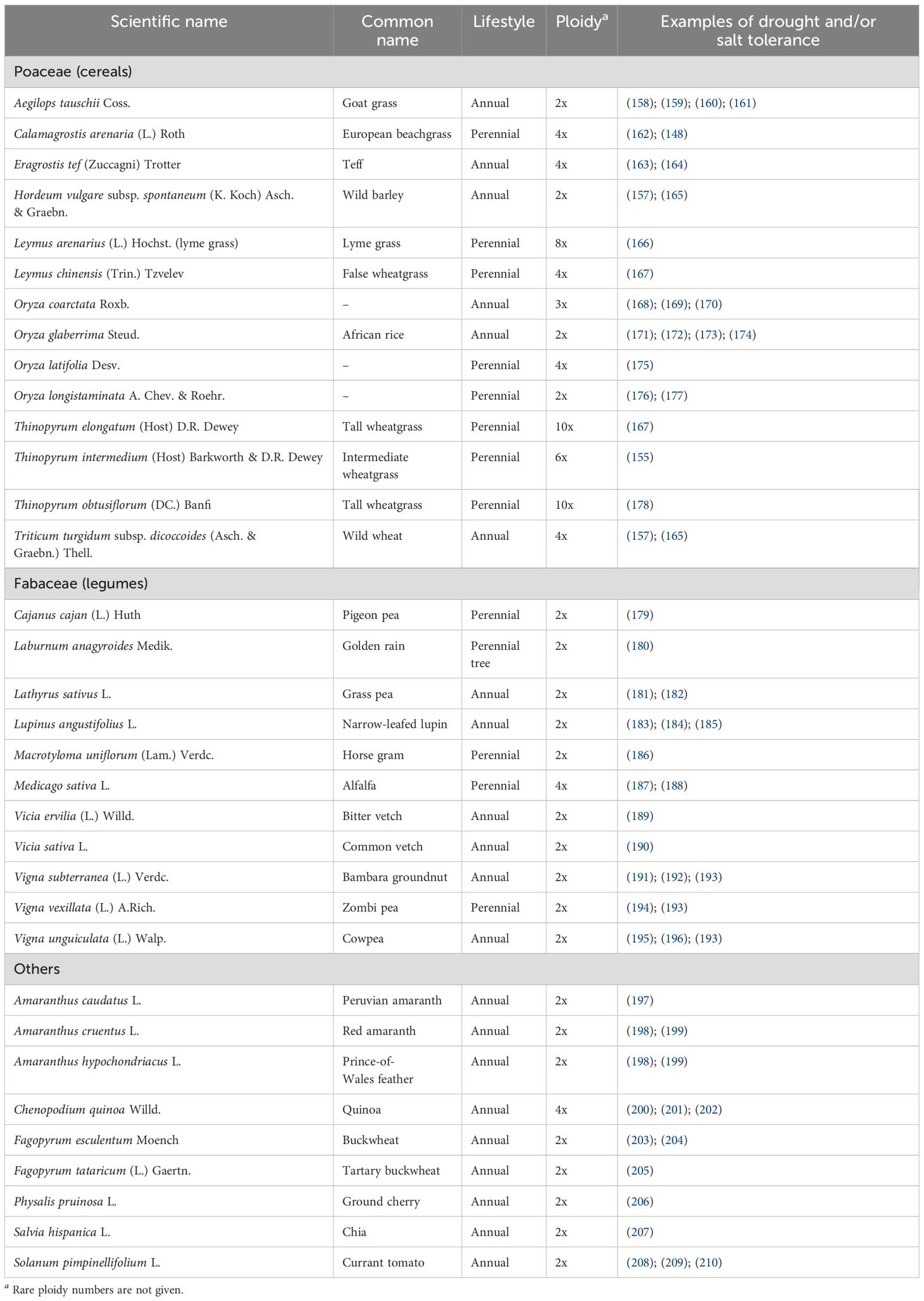
Table 1. Selected resilient wild plants and underutilized crops that are promising candidates for accelerated domestication.
The feasibility of accelerated domestication has been demonstrated by de novo domestication of wild tomato (218, 219) and accelerated domestication of wild groundcherry (Physalis pruinosa) (216), allotetraploid rice O. alta (185), and sheepgrass (Leymus chinensis) (177). It may also be a strategy for the domestication of salt-tolerant crops (106, 218).
Although these results are promising, the question remains whether harvesting the low-hanging fruit will be sufficient to obtain yields that are comparable to our present-day high-yielding crops. If yields are lower, we will need more land to grow plants to maintain food production at the same levels as today, which is unacceptable from a sustainability point of view (225). Many yield-related traits are multigenic and depend on QTLs that each contribute only little to the trait. As yield ultimately is a multigenic trait, a question arises: how many domestication genes need to be modified to transform a wild plant into a high-yielding crop? And how many of those have not been identified so far?
Where do we go from here?
Crop yields are already declining due to increased soil degradation, extreme weather, and salinization of soils. Considering predicted climate change, this problem is likely to be aggravated in the future. The only way forward on to increase crop yields is to bring new land under the plow. A more sustainable but still very demanding breeding goal will be to maintain yields on the land area already producing food. This will require crops with increased tolerance to abiotic stresses such as salinity, drought, and hypoxia. Improving abiotic stress tolerance is therefore a must under the current climate scenario. As tolerance to most abiotic stresses is a highly complex trait (both physiologically and genetically), the search for a “silver bullet” solution (e.g., an attempt to find and introduce a “key gene”) can be counterproductive. Thus, we must accept that either the entire “package” of genes needs to be re-introduced into modern crops or we should follow the path of de novo domestication (for higher yield) of already tolerant orphan crops and/or wild plants (Table 1). It is still too early to conclude which of the two strategies, rewilding or de novo domestication, will be most effective for future sustainable agriculture; both strategies are extremely commanding, and their realization remains a major task for plant science. What is clear, however, is that, given the importance of the issue, rapid action is needed in both spaces. This is echoed by the recent State of Food Security and Nutrition in the World 2021 report, which states that we are not on track to meet our commitments to end world hunger and malnutrition in all its forms by 2030, as was the initial aim (226). To achieve these goals, two major hurdles need to be overcome. The first hurdle is the slowness of acceptance of the need to use cell-based phenotyping and continued reliance on usage of whole-plant physiological indexes for breeding crops for abiotic stress tolerance. The preference for low-cost whole-plant-based high-throughput methods comes with the cost of losing critical information and may be counterproductive and even misleading in the long term.
The second hurdle is related to public acceptance of modern technologies and legislative issues. Worldwide, consumers are showing limited understanding, misconceptions, and unfamiliarity with plant gene technologies in agriculture (227). Such consumer behavior is influenced by psychological, social, cultural, personal, and economic factors, including scientific knowledge, lifestyle, personal welfare, income, religion, and beliefs (226). However, without embracing such new technologies, the issue of abiotic stress tolerance in crops and, hence, global food security, will be a monumental challenge.
Statements
Author contributions
MP: Writing – review & editing, Writing – original draft, Data curation, Conceptualization. SS: Writing – review & editing, Writing – original draft, Data curation, Conceptualization.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This project was supported by Australian Research Council and National Natural Science Foundation of China grants to SS and Novo Nordisk Foundation (NovoCrops), the Carlsberg Foundation (RaisingQuinoa), and Innovationsfonden (DEEPROOTS and PERENNIAL) grants to MP. The funders were not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Frontiers Business Intelligence team assisted in the data analysis and design of Figure 1A in this article using the SPEI12 data from the Global Standardised Precipitation-Evapotranspiration Index SPEI Database, SPEIbase v2.9 Consejo Superior de Investigaciones Cientificas. The data may be accessed at https://spei.csic.es/database.html.
The authors MP and SS declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Generative AI statement
The authors declared that no generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Wheeler T, von Braun J. Climate change impacts on global food security. Science (2013) 341(6145):508–13. doi: 10.1126/science.1239402
2. Seneviratne SI, Zhang X, Adnan M, Badi W, Dereczynski C, Di Luca A, et al. Weather and climate extreme events in a changing climate. In: Masson-Delmotte V, Zhai P, Pirani A, Connors SL, Péan C, Berger S, et al., editors. Climate change: the physical science basis. Contribution of Working Group I to the sixth assessment report of the Intergovernmental Panel on Climate Change. Cambridge: Cambridge University Press (2021). 1513–766. doi: 10.1017/9781009157896.013
3. Bailey R, Benton T, Challinor A, Elliott J, Gustafson D, Hiller B, et al. Extreme weather and resilience of the global food system. Final project report from the UK-US Taskforce on Extreme Weather and Global Food System Resilience. The Global Food Security programme (2015). Available at: https://www.foodsecurity.ac.uk/publications/extreme-weather-resilience-global-food-system.pdf
4. United Nations Convention to Combat Desertification. FACTSHEET: Drought and its socio-economic impacts [online] (2020). Available at: https://www.unccd.int/sites/default/files/2020-03/IWGDrought-Factsheets_EN-final.pdf
5. Razzaq A, Wani SH, Saleem F, Yu M, Zhou M, Shabala S. Rewilding crops for climate resilience: economic analysis and de novo domestication strategies. J Exp Bot (2021) 72(18):6123–39. doi: 10.1093/jxb/erab276
6. Kuwayama Y. The economic impacts of drought on US agriculture. Resources. (2019). Available at: https://www.resources.org/archives/economic-impacts-drought-us-agriculture/
7. Liu K, Harrison MT, Shabala S, Meinke H, Ahmed I, Zhang Y, et al. The state of the art in modelling waterlogging impacts on plants: what do we know and what do we need to know. Earths Future (2020) 8(12):e2020EF001801. doi: 10.1029/2020EF001801
8. Kaur G, Singh G, Motavalli PP, Nelson KA, Orlowski JM, Golden BR. Impacts and management strategies for crop production in waterlogged or flooded soils: a review. Agron J (2020) 112(3):1475–501. doi: 10.1002/agj2.20093
9. Liu K, Harrison MT, Ibrahim A, Manik SMN, Johnson P, Tian XH, et al. Genetic factors increasing barley grain yields under soil waterlogging. Food Energy Sec (2020) 9(4):e238. doi: 10.1002/fes3.238
10. Sadok W, Jagadish SVK. The hidden costs of nighttime warming on yields. Trends Plant Sci (2020) 25(7):644–51. doi: 10.1016/j.tplants.2020.02.003
11. Zhao C, Liu B, Piao SL, Wang XH, Lobell DB, Huang Y, et al. Temperature increase reduces global yields of major crops in four independent estimates. Proc Natl Acad Sci USA (2017) 114(35):9326–31. doi: 10.1073/pnas.1701762114
12. Impa SM, Raju B, Hein NT, Sandhu J, Prasad PVV, Walia H, et al. High night temperature effects on wheat and rice: current status and way forward. Plant Cell Environ (2021) 44(7):2049–65. doi: 10.1111/pce.14028
13. Baldrian P, López-Mondéjar R, Kohout P. Forest microbiome and global change. Nat Rev Microbiol (2023) 21(8):487–501. doi: 10.1038/s41579-023-00876-4
14. Liu Q, Wu K, Song W, Zhong N, Wu Y, Fu X. Improving crop nitrogen use efficiency toward sustainable green revolution. Annu Rev Plant Biol (2022) 73:523–51. doi: 10.1146/annurev-arplant-070121-015752
15. Tilman D, Balzer C, Hill J, Befort BL. Global food demand and the sustainable intensification of agriculture. Proc Natl Acad Sci USA (2011) 108(50):20260–4. doi: 10.1073/pnas.1116437108
16. Pingali PL. Green revolution: impacts, limits, and the path ahead. Proc Natl Acad Sci USA (2012) 109(31):12302–8. doi: 10.1073/pnas.0912953109
17. Prieto I, León-Sánchez L, Nicolás E, Nortes P, Querejeta JI. Warming reduces both photosynthetic nutrient use efficiency and water use efficiency in Mediterranean shrubsWarming reduces nutrient use efficiency. Environ Exp Bot (2023) 210. doi: 10.1016/j.envexpbot.2023.105331
18. Balasuriya BTG, Ghose A, Gheewala SH, Prapaspongsa T. Assessment of eutrophication potential from fertiliser application in agricultural systems in Thailand. Sci Total Environ (2022) 833:154993. doi: 10.1016/j.scitotenv.2022.154993
19. Standardised Precipitation-Evapotranspiration Index. Global Standard Precipitation-Evapotranspiration Index (SPEI) database (2022). Available at: https://spei.csic.es/database.html
20. Naumann G, Alfieri L, Wyser K, Mentaschi L, Betts RA, Carrao H, et al. Global changes in drought conditions under different levels of warming. Geophys Res Lett (2018) 45(7):3285–96. doi: 10.1002/2017GL076521
21. Australian Bureau of Meteorology, Commonwealth Scientific and Industrial Research Organisation. Australia's changing climate. In: State of the Climate 2018. CSIRO (2021). Available at: https://www.csiro.au/en/research/environmental-impacts/climate-change/State-of-the-Climate/Previous/State-of-the-Climate-2018/Australias-changing-climate
22. Ritchie H, Roser M. Water use and stress [online] (2024). Available at: https://ourworldindata.org/water-use-stress
23. Scheierling SM, Treguer DO. Investing in adaptation: the challenge of responding to water scarcity in irrigated agriculture. Econ Rev (2016) 75–100. Available at: https://www.kansascityfed.org/documents/772/Investing_in_Adaptation_The_Challenge_of_Responding_to_Water_Scarcity_in_Irrigated_Agr.pdf
24. Food and Agriculture Organization of the United Nations. AQUASTAT (2020). Available at: https://www.fao.org/aquastat/en/
25. Tötzke C, Cermak J, Nadezhdina N, Tributsch H. Electrochemical in-situ studies of solar mediated oxygen transport and turnover dynamics in a tree trunk of Tilia cordata. iForest Biogeosci For (2017) 10(2):355–61. doi: 10.3832/ifor1681-010
26. Liu MM, Pan T, AllakhyerdievAllakhverdiev SI, Yu M, Shabala S. Crop halophytism: an environmentally sustainable solution for global food security. Trends Plant Sci (2020) 25(7):630–6344. doi: 10.1016/j.tplants.2020.04.008
27. Australian Bureau of Statistics. Gross value of irrigated agricultural production, Australia, year ended 30 June 2017 [online] (2018). Available at: https://www.abs.gov.au/AUSSTATS/abs@.nsf/Lookup/4610.0.55.008Main+Features12016-17?OpenDocument=
28. Gupta SK, Ram J, Singh H. Comparative study of transpiration in cooling effect of tree species in the atmosphere. J Geosci Environ Prot (2018) 06(8):151–66. doi: 10.4236/gep.2018.68011
29. Bahuguna RN, Jagadish KSV, Coast O, Wassmann R. Plant abiotic stress: temperature extremes. In: Van Alfen NK, editor. Encyclopedia of agriculture and food systems. Amsterdam: Academic Press (2014). 330–4. doi: 10.1016/B978-0-444-52512-3.00172-8
30. Gleick PH. Water and conflict: fresh-water resources and international security. Int Sec (1993) 18(1):79–112. doi: 10.2307/2539033
31. van Zelm E, Zhang Y, Testerink C. Salt tolerance mechanisms of plants. Annu Rev Plant Biol (2020) 71:403–33. doi: 10.1146/annurev-arplant-050718-100005
32. Hasanuzzaman M, Nahar K, Alam MM, Bhowmik PC, Hossain MA, Rahman MM, et al. Potential use of halophytes to remediate saline soils. BioMed Res Int (2014) 2014:589341. doi: 10.1155/2014/589341
33. Garcia-Caparros P, Al-Azzawi MJ, Flowers TJ. Economic uses of salt-tolerant plants. Plants (Basel) (2023) 12(14):2669. doi: 10.3390/plants12142669
34. Food and Agriculture Organization of the United Nations. Quinoa: an ancient crop to contribute to world food security. Santiago: FAO Regional Office for Latin America and the Caribbean (2011). Available at: https://www.fao.org/4/aq287e/aq287e.pdf
35. Panta S, Flowers T, Lane P, Doyle R, Haros G, Shabala S. Halophyte agriculture: success stories. Environ Exp Bot (2014) 107:71–83. doi: 10.1016/j.envexpbot.2014.05.006
36. Ritchie H, Samborska V, Roser M. Urbanization [online]. (2024). Available at: https://ourworldindata.org/urbanization
37. Ritchie H, Roser M. Land use [online]. (2024). Available at: https://ourworldindata.org/land-use
38. Food and Agriculture Organization of the United Nations. How to feed the world in 2050: High-level Expert Forum, Rome 12–13 Oct 2009. FAO (2009), 1–35. Available at: https://www.fao.org/fileadmin/templates/wsfs/docs/expert_paper/How_to_Feed_the_World_in_2050.pdf
39. Benton TG, Bailey R. The paradox of productivity: agricultural productivity promotes food system inefficiency. Glob Sustain (2019) 2:1–8 doi: 10.1017/sus.2019.3
40. Food and Agriculture Organization of the United Nations. FAOSTAT statistical database (2023). Available at: http://faostat.fao.org/site/567/default.aspxancor
41. Rull V. Food security: green revolution drawbacks. Science (2010) 328(5975):169. doi: 10.1126/science.328.5975.169-c
42. Hasegawa T, Wilson LT. Raising wheat yield ceiling. Nat Food (2022) 3(7):493–4. doi: 10.1038/s43016-022-00550-7
43. Tester M, Langridge P. Breeding technologies to increase crop production in a changing world. Science (2010) 327(5967):818–22. doi: 10.1126/science.1183700
44. Beckmann M, Gerstner K, Akin-Fajiye M, Ceauşu S, Kambach S, Kinlock NL, et al. Conventional land-use intensification reduces species richness and increases production: a global meta-analysis. Glob Change Biol (2019) 25(6):1941–56. doi: 10.1111/gcb.14606
45. Purugganan MD, Fuller DQ. The nature of selection during plant domestication. Nature (2009) 457(7231):843–8. doi: 10.1038/nature07895
46. Doebley JF, Gaut BS, Smith BD. The molecular genetics of crop domestication. Cell (2006) 127(7):1309–21. doi: 10.1016/j.cell.2006.12.006
47. Gilliham M, Able JA, Roy SJ. Translating knowledge about abiotic stress tolerance to breeding programmes. Plant J (2017) 90(5):898–917. doi: 10.1111/tpj.13456
48. Palmgren MG, Edenbrandt AK, Vedel SE, Andersen MM, Landes X, Østerberg JT, et al. Are we ready for back-to-nature crop breeding? Trends Plant Sci (2015) 20(3):155–64. doi: 10.1016/j.tplants.2014.11.003
49. Menguer PK, Sperotto RA, Ricachenevsky FK. A walk on the wild side: Oryza species as source for rice abiotic stress tolerance. Genet Mol Biol (2017) 40(1):238–52. doi: 10.1590/1678-4685-GMB-2016-0093
50. Yolcu S, Alavilli H, Lee BH. Natural genetic resources from diverse plants to improve abiotic stress tolerance in plants. Int J Mol Sci (2020) 21(22):8567. doi: 10.3390/ijms21228567
51. Wang Z, Hong Y, Zhu G, Li Y, Niu Q, Yao J, et al. Loss of salt tolerance during tomato domestication conferred by variation in a Na+/K+ transporter. EMBO J (2020) 39(10):e103256. doi: 10.15252/embj.2019103256
52. Munns R, James RA, Xu B, Athman A, Conn SJ, Jordans C, et al. Wheat grain yield on saline soils is improved by an ancestral Na+ transporter gene. Nat Biotechnol (2012) 30(4):360–4. doi: 10.1038/nbt.2120
53. Rawat N, Wungrampha S, Singla-Pareek SL, Yu M, Shabala S, Pareek A. Rewilding staple crops for the lost halophytism: toward sustainability and profitability of agricultural production systems. Mol Plant (2022) 15(1):45–64. doi: 10.1016/j.molp.2021.12.003
54. Smýkal P, Nelson MN, Berger JD, Von Wettberg EJB. The impact of genetic changes during crop domestication. Agronomy (2018) 8(7):119. doi: 10.3390/agronomy8070119
55. Burgarella C, Barnaud A, Kane NA, Jankowski F, Scarcelli N, Billot C, et al. Adaptive introgression: an untapped evolutionary mechanism for crop adaptation. Front Plant Sci (2019) 10:4. doi: 10.3389/fpls.2019.00004
56. Kushalappa AC, Gunnaiah R. Metabolo-proteomics to discover plant biotic stress resistance genes. Trends Plant Sci (2013) 18(9):522–31. doi: 10.1016/j.tplants.2013.05.002
57. Xu K, Xu X, Fukao T, Canlas P, Maghirang-Rodriguez R, Heuer S, et al. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature (2006) 442(7103):705–8. doi: 10.1038/nature04920
58. Septiningsih EM, Pamplona AM, Sanchez DL, Neeraja CN, Vergara GV, Heuer S, et al. Development of submergence-tolerant rice cultivars: the Sub1 locus and beyond. Ann Bot (2009) 103(2):151–60. doi: 10.1093/aob/mcn206
59. Sutton T, Baumann U, Hayes J, Collins NC, Shi BJ, Schnurbusch T, et al. Boron-toxicity tolerance in barley arising from efflux transporter amplification. Science (2007) 318(5855):1446–9. doi: 10.1126/science.1146853
60. Schnurbusch T, Hayes J, Hrmova M, Baumann U, Ramesh SA, Tyerman SD, et al. Boron toxicity tolerance in barley through reduced expression of the multifunctional aquaporin HvNIP2;1. Plant Physiol (2010) 153(4):1706–15. doi: 10.1104/pp.110.158832
61. Fujii M, Yokosho K, Yamaji N, Saisho D, Yamane M, Takahashi H, et al. Acquisition of aluminium tolerance by modification of a single gene in barley. Nat Commun (2012) 3:713. doi: 10.1038/ncomms1726
62. Vinocur B, Altman A. Recent advances in engineering plant tolerance to abiotic stress: achievements and limitations. Curr Opin Biotechnol (2005) 16(2):123–32. doi: 10.1016/j.copbio.2005.02.001
63. Shavrukov Y, Kurishbayev A, Jatayev S, Shvidchenko V, Zotova L, Koekemoer F, et al. Early flowering as a drought escape mechanism in plants: how can it aid wheat production? Front Plant Sci (2017) 8:1950. doi: 10.3389/fpls.2017.01950
64. Ahmed K, Shabbir G, Ahmed M, Shah KN. Phenotyping for drought resistance in bread wheat using physiological and biochemical traits. Sci Total Environ (2020) 729:139082. doi: 10.1016/j.scitotenv.2020.139082
65. Merrium S, Ali Z, Habib-Ur-Rahman M, Hakeem S, Khalid MA. Leaf rolling and leaf angle improve fog capturing and transport in wheat; adaptation for drought stress in an arid climate. Bot Stud (2022) 63(1):13. doi: 10.1186/s40529-022-00343-y
66. Inzé D, Nelissen H. The translatability of genetic networks from model to crop species: lessons from the past and perspectives for the future. New Phytol (2022) 236(1):43–8. doi: 10.1111/nph.18364
67. Apse MP, Aharon GS, Snedden WA, Blumwald E. Salt tolerance conferred by overexpression of a vacuolar Na+/H+ antiport in Arabidopsis. Science (1999) 285(5431):1256–8. doi: 10.1126/science.285.5431.1256
68. Zhang HX, Blumwald E. Transgenic salt-tolerant tomato plants accumulate salt in foliage but not in fruit. Nat Biotechnol (2001) 19(8):765–8. doi: 10.1038/90824
69. Nguyen NT, Vu HT, Nguyen TT, Nguyen L-AT, Nguyen M-CD, Hoang KL, et al. Co-expression of Arabidopsis AtAVP1 and AtNHX1 to improve salt tolerance in soybean. Crop Sci (2019) 59(3):1133–43. doi: 10.2135/cropsci2018.10.0640
70. Maeshima M. Vacuolar H+-pyrophosphatase. Biochim Biophys Acta Rev Biomembr (2000) 1465(1–2):37–51. doi: 10.1016/S0005-2736(00)00130-9
71. Shabala S. Signalling by potassium: another second messenger to add to the list? J Exp Bot (2017) 68(15):4003–7. doi: 10.1093/jxb/erx238
72. Wu HH, Zhang XC, Giraldo JP, Shabala S. It is not all about sodium: revealing tissue specificity and signalling roles of potassium in plant responses to salt stress. Plant Soil (2018) 431(1–2):1–17. doi: 10.1007/s11104-018-3770-y
73. Wu SJ, Ding L, Zhu JK. SOS1, a genetic locus essential for salt tolerance and potassium acquisition. Plant Cell (1996) 8(4):617–27. doi: 10.1105/tpc.8.4.617
74. Liu J, Zhu JK. An Arabidopsis mutant that requires increased calcium for potassium nutrition and salt tolerance. Proc Natl Acad Sci USA (1997) 94(26):14960–4. doi: 10.1073/pnas.94.26.14960
75. Zhu JK, Liu J, Xiong L. Genetic analysis of salt tolerance in Arabidopsis. evidence for a critical role of potassium nutrition. Plant Cell (1998) 10(7):1181–91. doi: 10.1105/tpc.10.7.1181
76. Qiu QS, Guo Y, Dietrich MA, Schumaker KS, Zhu JK. Regulation of SOS1, a plasma membrane Na+/H+ exchanger in Arabidopsis thaliana, by SOS2 and SOS3. Proc Natl Acad Sci USA (2002) 99(12):8436–41. doi: 10.1073/pnas.122224699
77. Yin X, Xia Y, Xie Q, Cao Y, Wang Z, Hao G, et al. The protein kinase complex CBL10-CIPK8-SOS1 functions in Arabidopsis to regulate salt tolerance. J Exp Bot (2020) 71(6):1801–14. doi: 10.1093/jxb/erz549
78. Zhou Y, Yin X, Duan R, Hao G, Guo J, Jiang X. SpAHA1 and SpSOS1 coordinate in transgenic yeast to improve salt tolerance. PloS One (2015) 10(9):e0137447. doi: 10.1371/journal.pone.0137447
79. Fuglsang AT, Palmgren M. Proton and calcium pumping P-type ATPases and their regulation of plant responses to the environment. Plant Physiol (2021) 187(4):1856–75. doi: 10.1093/plphys/kiab330
80. Yang Q, Chen ZZ, Zhou XF, Yin HB, Li X, Xin XF, et al. Overexpression of SOS (Salt Overly Sensitive) genes increases salt tolerance in transgenic Arabidopsis. Mol Plant (2009) 2(1):22–31. doi: 10.1093/mp/ssn058
81. Shi HZ, Quintero FJ, Pardo JM, Zhu JK. The putative plasma membrane Na+/H+ antiporter SOS1 controls long-distance Na+ transport in plants. Plant Cell (2002) 14(2):465–77. doi: 10.1105/tpc.010371
82. Zhao C, Zhang H, Song C, Zhu J-K, Shabala S. Mechanisms of plant responses and adaptation to soil salinity. Innovation (Camb) (2020) 1(1):100017. doi: 10.1016/j.xinn.2020.100017
83. Shahzad B, Shabala L, Zhou M, Venkataraman G, CA S, Page D, et al. Comparing essentiality of SOS1-mediated Na+ exclusion in salinity tolerance between cultivated and wild rice species. Int J Mol Sci (2022) 23(17):9900. doi: 10.3390/ijms23179900
84. Wu HH, Shabala L, Zhou MX, Su NN, Wu Q, Ul-Haq T, et al. Root vacuolar Na+ sequestration but not exclusion from uptake correlates with barley salt tolerance. Plant J (2019) 100(1):55–67. doi: 10.1111/tpj.14424
85. Dinneny JR. Analysis of the salt-stress response at cell-type resolution. Plant Cell Environ (2010) 33(4):543–51. doi: 10.1111/j.1365-3040.2009.02055.x
86. Munns R. Genes and salt tolerance: bringing them together. New Phytol (2005) 167(3):645–63. doi: 10.1111/j.1469-8137.2005.01487.x
87. Venkataraman G, Shabala S, Véry AA, Hariharan GN, Somasundaram S, Pulipati S, et al. To exclude or to accumulate? Revealing the role of the sodium HKT1;5 transporter in plant adaptive responses to varying soil salinity. Plant Physiol Biochem (2021) 169:333–42. doi: 10.1016/j.plaphy.2021.11.030
88. Ren ZH, Gao JP, Li LG, Cai XL, Huang W, Chao DY, et al. A rice quantitative trait locus for salt tolerance encodes a sodium transporter. Nat Genet (2005) 37(10):1141–6. doi: 10.1038/ng1643
89. Byrt CS, Xu B, Krishnan M, Lightfoot DJ, Athman A, Jacobs AK, et al. The Na+ transporter, TaHKT1;5-D, limits shoot Na+ accumulation in bread wheat. Plant J (2014) 80(3):516–26. doi: 10.1111/tpj.12651
90. Jiang ZL, Song GS, Shan XH, Wei ZY, Liu YZ, Jiang C, et al. Association analysis and identification of ZmHKT1;5 variation with salt-stress tolerance. Front Plant Sci (2018) 9:1485. doi: 10.3389/fpls.2018.01485
91. Bonilla P, Dvorak J, Mackill D, Deal K, Gregorio G. RFLP and SSLP mapping of salinity tolerance genes in chromosome 1 of rice (Oryza sativa L.) using recombinant inbred lines. Philipp J Agric Sci (2002) 85:68–76
92. Thomson MJ, de Ocampo M, Egdane J, Rahman MA, Sajise AG, Adorada DL, et al. Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice (2010) 3(2–3):148–60. doi: 10.1007/s12284-010-9053-8
93. Kobayashi NI, Yamaji N, Yamamoto H, Okubo K, Ueno H, Costa A, et al. 5 mediates Na+ exclusion in the vasculature to protect leaf blades and reproductive tissues from salt toxicity in rice. Plant J (2017) 91(4):657–70. doi: 10.1111/tpj.13595
94. Al Nayef M, Solis C, Shabala L, Ogura T, Chen ZH, Bose J, et al. Changes in expression level of OsHKT1;5 alters activity of membrane transporters involved in K+ and Ca2+ acquisition and homeostasis in salinized rice roots. Int J Mol Sci (2020) 21(14). doi: 10.3390/ijms21144882
95. Soda N, Kushwaha HR, Soni P, Singla-Pareek SL, Pareek A. A suite of new genes defining salinity stress tolerance in seedlings of contrasting rice genotypes. Funct Integr Genomics (2013) 13(3):351–65. doi: 10.1007/s10142-013-0328-1
96. Soda N, Gupta BK, Anwar K, Sharan A, Govindjee SPSL, Singla-Pareek SL, et al. Rice intermediate filament, OsIF, stabilizes photosynthetic machinery and yield under salinity and heat stress. Sci Rep (2018) 8(1):4072. doi: 10.1038/s41598-018-22131-0
97. Claes B, Dekeyser R, Villarroel R, Van den Bulcke M, Bauw G, Van Montagu M, et al. Characterization of a rice gene showing organ-specific expression in response to salt stress and drought. Plant Cell (1990) 2(1):19–27. doi: 10.1104/pp.108.118117
98. Ganie SA, Karmakar J, Roychowdhury R, Mondal TK, Dey N. Assessment of genetic diversity in salt-tolerant rice and its wild relatives for ten SSR loci and one allele mining primer of salT gene located on 1st chromosome. Plant Syst Evol (2014) 300(7):1741–7. doi: 10.1007/s00606-014-0999-7
99. Kaur N, Prashanth KH, Bhatti MS, Pati PK. OsSalT gene cloned from rice provides evidence of its role in salinity and drought stress tolerance. Plant Sci (2022) 320:111306. doi: 10.1016/j.plantsci.2022.111306
100. Das P, Lakra N, Nutan KK, Singla-Pareek SL, Pareek A. A unique bZIP transcription factor imparting multiple stress tolerance in Rice. Rice (NY) (2019) 12(1):58. doi: 10.1186/s12284-019-0316-8
101. Nutan KK, Singla-Pareek SL, Pareek A. The Saltol QTL-localized transcription factor OsGATA8 plays an important role in stress tolerance and seed development in Arabidopsis and rice. J Exp Bot (2020) 71(2):684–98. doi: 10.1093/jxb/erz368
102. Wang QL, Sun AZ, Chen ST, Chen LS, Guo FQ. SPL6 represses signalling outputs of ER stress in control of panicle cell death in rice. Nat Plants (2018) 4(5):280–8. doi: 10.1038/s41477-018-0131-z
103. Wang M, Cheng J, Wu J, Chen J, Liu D, Wang C, et al. Variation in TaSPL6-D confers salinity tolerance in bread wheat by activating TaHKT1;5-D while preserving yield-related traits. Nat Genet (2024) 56(6):1257–69. doi: 10.1038/s41588-024-01762-2
104. Munns R, Day DA, Fricke W, Watt M, Arsova B, Barkla BJ, et al. Energy costs of salt tolerance in crop plants. New Phytol (2020) 225(3):1072–90. doi: 10.1111/nph.15864
105. Shabala S. Learning from halophytes: physiological basis and strategies to improve abiotic stress tolerance in crops. Ann Bot (2013) 112(7):1209–21. doi: 10.1093/aob/mct205
106. Razzaq A, Saleem F, Wani SH, Abdelmohsen SAM, Alyousef HA, Abdelbacki AMM, et al. De-novo domestication for improving salt tolerance in crops. Front Plant Sci (2021) 12:681367. doi: 10.3389/fpls.2021.681367
107. Bayer PE, Golicz AA, Scheben A, Batley J, Edwards D. Plant pan-genomes are the new reference. Nat Plants (2020) 6(8):914–20. doi: 10.1038/s41477-020-0733-0
108. Gao L, Gonda I, Sun H, Ma Q, Bao K, Tieman DM, et al. The tomato pan-genome uncovers new genes and a rare allele regulating fruit flavor. Nat Genet (2019) 51(6):1044–51. doi: 10.1038/s41588-019-0410-2
109. Yao W, Li G, Zhao H, Wang G, Lian X, Xie W. Exploring the rice dispensable genome using a metagenome-like assembly strategy. Genome Biol (2015) 16:187. doi: 10.1186/s13059-015-0757-3
110. Golicz AA, Bayer PE, Barker GC, Edger PP, Kim H, Martinez PA, et al. The pangenome of an agronomically important crop plant Brassica oleracea. Nat Commun (2016) 7:13390. doi: 10.1038/ncomms13390
111. Jin M, Liu H, He C, Fu J, Xiao Y, Wang Y, et al. Maize pan-transcriptome provides novel insights into genome complexity and quantitative trait variation. Sci Rep (2016) 6:18936. doi: 10.1038/srep18936
112. Montenegro JD, Golicz AA, Bayer PE, Hurgobin B, Lee H, Chan CKK, et al. The pangenome of hexaploid bread wheat. Plant J (2017) 90(5):1007–13. doi: 10.1111/tpj.13515
113. Zhao Q, Feng Q, Lu H, Li Y, Wang A, Tian Q, et al. Pan-genome analysis highlights the extent of genomic variation in cultivated and wild rice. Nat Genet (2018) 50(2):278–84. doi: 10.1038/s41588-018-0041-z
114. Hu H, Scheben A, Verpaalen B, Tirnaz S, Bayer PE, Hodel RGJ, et al. Amborella gene presence/absence variation is associated with abiotic stress responses that may contribute to environmental adaptation. New Phytol (2022) 233(4):1548–55. doi: 10.1111/nph.17658
115. Asins MJ, Bernet GP, Villalta I, Carbonell EA. QTL analysis in plant breeding. In: Jain S, Brar D, editors. Molecular techniques in crop improvement. Dordrecht: Springer (2009) 3–21. doi: 10.1007/978-90-481-2967-6_1
116. Andrade ACB, Viana JMS, Pereira HD, Fonseca e Silva F. Efficiency of Bayesian quantitative trait loci mapping with full-sib progeny. Agron J (2020) 112(4):2759–67. doi: 10.1002/agj2.20297
117. Raj SRG, Nadarajah K. QTL and candidate genes: techniques and advancement in abiotic stress resistance breeding of major cereals. Int J Mol Sci (2022) 24(1):6. doi: 10.3390/ijms24010006
118. Zhao L, Lei J, Huang Y, Zhu S, Chen H, Huang R, et al. Mapping quantitative trait loci for heat tolerance at anthesis in rice using chromosomal segment substitution lines. Breed Sci (2016) 66(3):358–66. doi: 10.1270/jsbbs.15084
119. Lindsay MP, Lagudah ES, Hare RA, Munns R. A locus for sodium exclusion (Nax1), a trait for salt tolerance, mapped in durum wheat. Funct Plant Biol (2004) 31(11):1105–14. doi: 10.1071/FP04111
120. Huang S, Spielmeyer W, Lagudah ES, James RA, Platten JD, Dennis ES, et al. A sodium transporter (HKT7) is a candidate for Nax1, a gene for salt tolerance in durum wheat. Plant Physiol (2006) 142(4):1718–27. doi: 10.1104/pp.106.088864
121. Gill MB, Zeng F, Shabala L, Zhang G, Fan Y, Shabala S, et al. Cell-based phenotyping reveals qtl for membrane potential maintenance associated with hypoxia and salinity stress tolerance in barley. Front Plant Sci (2017) 8:1941. doi: 10.3389/fpls.2017.01941
122. Panta S, Flowers T, Doyle R, Lane P, Haros G, Shabala S. Growth responses of Atriplex lentiformis and Medicago arborea in three soil types treated with saline water irrigation. Environ Exp Bot (2016) 128:39–50. doi: 10.1016/j.envexpbot.2016.04.002
123. Puniran-Hartley N, Hartley J, Shabala L, Shabala S. Salinity-induced accumulation of organic osmolytes in barley and wheat leaves correlates with increased oxidative stress tolerance: in planta evidence for cross-tolerance. Plant Physiol Biochem (2014) 83:32–9. doi: 10.1016/j.plaphy.2014.07.005
124. Seemann JR, Critchley C. Effects of salt stress on the growth, ion content, stomatal behaviour and photosynthetic capacity of a salt-sensitive species, Phaseolus vulgaris L. Planta (1985) 164(2):151–62. doi: 10.1007/BF00396077
125. Shahzad B, Yun P, Rasouli F, Shabala L, Zhou MX, Venkataraman G, et al. Root K+ homeostasis and signalling as a determinant of salinity stress tolerance in cultivated and wild rice species. Environ Exp Bot (2022) 201:104944. doi: 10.1016/j.envexpbot.2022.104944
126. Oi T, Clode PL, Taniguchi M, Colmer TD, Kotula L. Salt tolerance in relation to elemental concentrations in leaf cell vacuoles and chloroplasts of a C4 monocotyledonous halophyte. Plant Cell Environ (2022) 45(5):1490–506. doi: 10.1111/pce.14279
127. Daba AW, Qureshi AS, Nisaren BN. Evaluation of some Rhodes grass (Chloris gayana) genotypes for their salt tolerance, biomass yield and nutrient composition. Appl Sci Basel (2019) 9(1):143. doi: 10.3390/app9010143
128. Chakraborty K, Bose J, Shabala L, Eyles A, Shabala S. Evaluating relative contribution of osmotolerance and tissue tolerance mechanisms toward salinity stress tolerance in three Brassica species. Physiol Plant (2016) 158(2):135–51. doi: 10.1111/ppl.12447
129. Hedrich R, Shabala S. Stomata in a saline world. Curr Opin Plant Biol (2018) 46:87–95. doi: 10.1016/j.pbi.2018.07.015
130. Maas EV, Hoffman GJ. Crop salt tolerance—current assessment. J Irrig Drain Div (1977) 103(2):115–34. doi: 10.1061/JRCEA4.0001137
131. Starr F, Starr K. Phaseolus vulgaris (bush bean, garden bean, pole bean). Flickr (2016). Available at: https://www.flickr.com/photos/starr-environmental/24859099901/
132. NL. Brassica oleracea var. italica (broccoli). Flickr (2008). Available at: https://www.flickr.com/photos/22748341@N00/2638500179
133. Perkins DL. Oryza sativa (rice). Wikimedia Commons (2014). Available at: https://commons.wikimedia.org/wiki/File:Rice_plant.jpg
134. Hobern D. Medicago arborea (moon trefoil). Flickr (2014). Available at: https://www.flickr.com/photos/dhobern/13584169544/
135. Starr F, Starr K. Triticum aestivum (wheat). Flickr (2016). Available at: https://www.flickr.com/photos/starr-environmental/27300584466/
136. Nekonomania Y. Hordeum vulgare (barley). Flickr (2010). Available at: https://www.flickr.com/photos/nekonomania/4674067457/
137. Starr F, Starr K. Chloris gayana (Rhodes grass). Flickr (2016). Available at: https://www.flickr.com/photos/starr-environmental/24522023032/
138. Cyr L. Chenopodium quinoa (quinoa). Flickr (2009). Available at: https://www.flickr.com/photos/borderlys/3831965675/
139. Starr F, Starr K. Atriplex lentiformis (saltbush). Flickr (2016). Available at: https://www.flickr.com/photos/starr-environmental/24135971743/
140. Pisanty G. Suaeda fruticosa (shrubby seablite). Wikimedia Commons (2013). Available at: https://commons.wikimedia.org/wiki/File:Suaeda_fruticosa_1.jpg
141. Wu HH, Shabala L, Liu XH, Azzarello E, Zhou M, Pandolfi C, et al. Linking salinity stress tolerance tissue-specific Na+ sequestration in wheat roots. Front Plant Sci (2015) 6:71. doi: 10.3389/fpls.2015.00071
142. Peláez-Vico MÁ, Zandalinas SI, Devireddy AR, Sinha R, Mittler R. Systemic stomatal responses in plants: coordinating development, stress, and pathogen defense under a changing climate. Plant Cell Environ (2024) 47(4):1171–84. doi: 10.1111/pce.14797
143. Jiao Y, Lv W, Teng W, Li L, Lan H, Bai L, et al. Peroxidase gene TaPrx109-B1 enhances wheat tolerance to water deficit via modulating stomatal density. Plant Cell Environ (2024) 47(8):2954–70. doi: 10.1111/pce.14918
144. Xia Y, Jiang S, Wu W, Du K, Kang X. MYC2 regulates stomatal density and water use efficiency via targeting EPF2/EPFL4/EPFL9 in poplar. New Phytol (2024) 241(6):2506–22. doi: 10.1111/nph.19531
145. Farquhar GD, Ehleringer JR, Hubick KT. Carbon isotope discrimination and photosynthesis. Annu Rev Plant Physiol Plant Mol Biol (1989) 40(1):503–37. doi: 10.1146/annurev.pp.40.060189.002443
146. Buckley TN, Mott KA, Farquhar GD. A hydromechanical and biochemical model of stomatal conductance. Plant Cell Environ (2003) 26(10):1767–85. doi: 10.1046/j.1365-3040.2003.01094.x
147. Kunz K, Hu Y, Schmidhalter U. Carbon isotope discrimination as a key physiological trait to phenotype drought/heat resistance of future climate-resilient German winter wheat compared with relative leaf water content and canopy temperature. Front Plant Sci (2022) 13:1043458. doi: 10.3389/fpls.2022.1043458
148. Chen Z, Pottosin II, Cuin TA, Fuglsang AT, Tester M, Jha D, et al. Root plasma membrane transporters controlling K+/Na+ homeostasis in salt-stressed barley. Plant Physiol (2007) 145(4):1714–25. doi: 10.1104/pp.107.110262
149. Bose J, Rodrigo-Moreno A, Lai DW, Xie YJ, Shen WB, Shabala S. Rapid regulation of the plasma membrane H+-ATPase activity is essential to salinity tolerance in two halophyte species, Atriplex lentiformis and Chenopodium quinoa. Ann Bot (2015) 115(3):481–94. doi: 10.1093/aob/mcu219
150. Shabala S, Bose J, Fuglsang AT, Pottosin I. On a quest for stress tolerance genes: membrane transporters in sensing and adapting to hostile soils. J Exp Bot (2016) 67(4):1015–31. doi: 10.1093/jxb/erv465
151. Shabala L, Zhang JY, Pottosin I, Bose J, Zhu M, Fuglsang AT, et al. Cell-type-specific H+-ATPase activity in root tissues enables K+ retention and mediates acclimation of barley (Hordeum vulgare) to salinity stress. Plant Physiol (2016) 172(4):2445–58. doi: 10.1104/pp.16.01347
152. Wang H, Shabala L, Zhou M, Shabala S. Developing a high-throughput phenotyping method for oxidative stress tolerance in barley roots. Plant Methods (2019) 15:12. doi: 10.1186/s13007-019-0397-9
153. Fedak G. Wide hybridization for cereal improvement [abstract 5]. In: Maluszynski M, editor. Current options for cereal improvement. Doubled haploids, mutants and heterosis. Proceedings of the first FAO/IAEA research co-ordination meeting on “use of induced mutations in connection with haploids and heterosis in cereals”, 8–12 December 1986, Guelph, Canada. Dordrecht: Kluwer Academic Publishers (1989). 39–48.
154. Alahmad S, Dinglasan E, Leung KM, Riaz A, Derbal N, Voss-Fels KP, et al. Speed breeding for multiple quantitative traits in durum wheat. Plant Methods (2018) 14:36. doi: 10.1186/s13007-018-0302-y
155. Zhang X, Pérez-Rodríguez P, Burgueño J, Olsen M, Buckler E, Atlin G, et al. Rapid cycling genomic selection in a multiparental tropical maize population. G3 (Bethesda) (2017) 7(7):2315–26. doi: 10.1534/g3.117.043141
156. Li B. Identification of genes conferring plant salt tolerance using GWAS: current success and perspectives. Plant Cell Physiol (2020) 61(8):1419–26. doi: 10.1093/pcp/pcaa073
157. Alseekh S, Kostova D, Bulut M, Fernie AR. Genome-wide association studies: assessing trait characteristics in model and crop plants. Cell Mol Life Sci (2021) 78(15):5743–54. doi: 10.1007/s00018-021-03868-w
158. Zhang Y, Pribil M, Palmgren M, Gao C. A CRISPR way for accelerating improvement of food crops. Nat Food (2020) 1(4):200–5. doi: 10.1038/s43016-020-0051-8
159. Vu TV, Nguyen NT, Kim J, Hong JC, Kim JY. Prime editing: mechanism insight and recent applications in plants. Plant Biotechnol J (2024) 22(1):19–36. doi: 10.1111/pbi.14188
160. Anjanappa RB, Gruissem W. Current progress and challenges in crop genetic transformation. J Plant Physiol (2021) 261:153411. doi: 10.1016/j.jplph.2021.153411
161. Østerberg JT, Xiang W, Olsen LI, Edenbrandt AK, Vedel SE, Christiansen A, et al. Accelerating the domestication of new crops: feasibility and approaches. Trends Plant Sci (2017) 22(5):373–84. doi: 10.1016/j.tplants.2017.01.004
162. Pourkheirandish M, Hensel G, Kilian B, Senthil N, Chen G, Sameri M, et al. Evolution of the grain dispersal system in barley. Cell (2015) 162(3):527–39. doi: 10.1016/j.cell.2015.07.002
163. Chapman EA, Thomsen HC, Tulloch S, Correia PMP, Luo G, Najafi J, et al. Perennials as future grain crops: opportunities and challenges. Front Plant Sci (2022) 13:898769. doi: 10.3389/fpls.2022.898769
164. Talabi AO, Vikram P, Thushar S, Rahman H, Ahmadzai H, Nhamo N, et al. Orphan crops: a best fit for dietary enrichment and diversification in highly deteriorated marginal environments. Front Plant Sci (2022) 13:839704. doi: 10.3389/fpls.2022.839704
165. DeHaan L, Larson S, López-Marqués RL, Wenkel S, Gao C, Palmgren M. Roadmap for accelerated domestication of an emerging perennial grain crop. Trends Plant Sci (2020) 25(6):525–37. doi: 10.1016/j.tplants.2020.02.004
166. Garthwaite AJ, von Bothmer R, Colmer TD. Salt tolerance in wild Hordeum species is associated with restricted entry of Na+ and Cl− into the shoots. J Exp Bot (2005) 56(419):2365–78. doi: 10.1093/jxb/eri229
167. Nevo E, Chen G. Drought and salt tolerances in wild relatives for wheat and barley improvement. Plant Cell Environ (2010) 33(4):670–85. doi: 10.1111/j.1365-3040.2009.02107.x
168. Farooq S, Niazi MLK, Iqbal N, Shah TM. Salt tolerance potential of wild resources of the tribe Triticeae: II. Screening of species of the genus Aegilops. Plant Soil (1989) 119(2):255–60. doi: 10.1007/BF02370417
169. Gorham J. Salt tolerance in the Triticeae: K+/Na+ discrimination in Aegilops species. J Exp Bot (1990) 41(5):615–21. doi: 10.1093/jxb/41.5.615
170. Schachtman DP, Munns R, Whitecross MI. Variation in sodium exclusion and salt tolerance in Triticum tauschii. Crop Sci (1991) 31(4):992–7. doi: 10.2135/cropsci1991.0011183X003100040030x
171. Abbas A, Hameed R, Saeed M, Shahani AAA, Huang P, Du D, et al. Investigating the dynamic responses of Aegilops tauschii Coss. to salinity, drought, and nitrogen stress: a comprehensive study of competitive growth and biochemical and molecular pathways. Front Plant Sci (2023) 14:1238704. doi: 10.3389/fpls.2023.1238704
172. Huiskes AHL. Ammophila arenaria (L.) Link (Psamma arenaria (L.) Roem. et Schult.; Calamgrostis arenaria (L.) Roth). J Ecol (1979) 67(1):363–82. doi: 10.2307/2259356
173. Cheng A, Mayes S, Dalle G, Demissew S, Massawe F. Diversifying crops for food and nutrition security - a case of teff. Biol Rev Camb Philos Soc (2017) 92(1):188–98. doi: 10.1111/brv.12225
174. Bekele-Alemu A, Ligaba-Osena A. Comprehensive in silico analysis of the underutilized crop tef (Eragrostis tef (Zucc.) Trotter) genome reveals drought tolerance signatures. BMC Plant Biol (2023) 23(1):506. doi: 10.1186/s12870-023-04515-1
175. Wang Y, Chen G, Zeng F, Han Z, Qiu CW, Zeng M, et al. Molecular evidence for adaptive evolution of drought tolerance in wild cereals. New Phytol (2023) 237(2):497–514. doi: 10.1111/nph.18560
176. Greipsson S, Davy AJ. Sand accretion and salinity as constraints on the establishment of Leymus arenarius for land reclamation in Iceland. Ann Bot (1996) 78(5):611–8. doi: 10.1006/anbo.1996.0168
177. Li T, Tang S, Li W, Zhang S, Wang J, Pan D, et al. Genome evolution and initial breeding of the Triticeae grass Leymus chinensis dominating the Eurasian Steppe. Proc Natl Acad Sci USA (2023) 120(44):e2308984120. doi: 10.1073/pnas.2308984120
178. Bal AR, Dutt SK. Mechanism of salt tolerance in wild rice (Oryza coarctata Roxb). Plant Soil (1986) 92(3):399–404. doi: 10.1007/BF02372487
179. Mondal TK, Rawal HC, Gaikwad K, Sharma TR, Singh NK. First de novo draft genome sequence of Oryza coarctata, the only halophytic species in the genus Oryza. F1000Res (2017) 6:1750. doi: 10.12688/f1000research.12414.2
180. Chowrasia S, Nishad J, Pandey R, Mondal TK. Oryza coarctata is a triploid plant with initial events of C4 photosynthesis evolution. Plant Sci (2021) 308:110878. doi: 10.1016/j.plantsci.2021.110878
181. Linares OF. African rice (Oryza glaberrima): history and future potential. Proc Natl Acad Sci USA (2002) 99(25):16360–5. doi: 10.1073/pnas.252604599
182. Sarla N, Swamy BPM. Oryza glaberrima: a source for the improvement of Oryza Sativa. Curr Sci (2005) 89(6):955–63. Available at: https://www.currentscience.ac.in/Volumes/89/06/0955.pdf
183. Cubry P, Pidon H, Ta KN, Tranchant-Dubreuil C, Thuillet AC, Holzinger M, et al. Genome wide association study pinpoints key agronomic QTLs in African rice Oryza Glaberrima. Rice (NY) (2020) 13(1):66. doi: 10.1186/s12284-020-00424-1
184. Lacchini E, Kiegle E, Castellani M, Adam H, Jouannic S, Gregis V, et al. CRISPR-mediated accelerated domestication of African rice landraces. PloS One (2020) 15(3):e0229782. doi: 10.1371/journal.pone.0229782
185. Yu H, Lin T, Meng X, Du H, Zhang J, Liu G, et al. A route to de novo domestication of wild allotetraploid rice. Cell (2021) 184(5):1156–1170.e14. doi: 10.1016/j.cell.2021.01.013
186. Zhang Y, Zhang S, Liu H, Fu B, Li L, Xie M, et al. Genome and comparative transcriptomics of African wild rice Oryza longistaminata provide insights into molecular mechanism of rhizomatousness and self-incompatibility. Mol Plant (2015) 8(11):1683–6. doi: 10.1016/j.molp.2015.08.006
187. Tong S, Ashikari M, Nagai K, Pedersen O. Can the wild perennial, rhizomatous rice species Oryza longistaminata be a candidate for de novo domestication? Rice (NY) (2023) 16(1):13. doi: 10.1186/s12284-023-00630-7
188. Borrajo CI, Sánchez-Moreiras AM, Reigosa MJ. Ecophysiological responses of tall wheatgrass germplasm to drought and salinity. Plants (Basel) (2022) 11(12):1548. doi: 10.3390/plants11121548
189. Varshney RK, Chen W, Li Y, Bharti AK, Saxena RK, Schlueter JA, et al. Draft genome sequence of pigeonpea (Cajanus cajan), an orphan legume crop of resource-poor farmers. Nat Biotechnol (2011) 30(1):83–9. doi: 10.1038/nbt.2022
190. Percival GC. Identification of foliar salt tolerance of woody perennials using chlorophyll fluorescence. HortSci (2005) 40(6):1892–7. doi: 10.21273/HORTSCI.40.6.1892
191. Aloui K, Choukri H, El Haddad N, Gupta P, El Bouhmadi K, Emmrich PMF, et al. Impact of heat and drought stress on grasspea and its wild relatives. Plants (Basel) (2023) 12(19):3501. doi: 10.3390/plants12193501
192. Rajarammohan S, Kaur L, Verma A, Singh D, Mantri S, Roy JK, et al. Genome sequencing and assembly of Lathyrus sativus - a nutrient-rich hardy legume crop. Sci Data (2023) 10(1):32. doi: 10.1038/s41597-022-01903-4
193. Turner NC, Stern WR, Evans P. Water relations and osmotic adjustment of leaves and roots of lupins in response to water deficits. Crop Sci (1987) 27(5):977–83. doi: 10.2135/cropsci1987.0011183X002700050032x
194. French RJ, Buirchell BJ. Lupin: the largest grain legume crop in Western Australia, its adaptation and improvement through plant breeding. Aust J Agric Res (2005) 56(11):1169–80. doi: 10.1071/AR05088
195. Wang P, Zhou G, Jian J, Yang H, Renshaw D, Aubert MK, et al. Whole-genome assembly and resequencing reveal genomic imprint and key genes of rapid domestication in narrow-leafed lupin. Plant J (2021) 105(5):1192–210. doi: 10.1111/tpj.15100
196. Aditya JP, Bhartiya A, Chahota RK, Joshi D, Chandra N, Kant L, et al. Ancient orphan legume horse gram: a potential food and forage crop of future. Planta (2019) 250(3):891–909. doi: 10.1007/s00425-019-03184-5
197. Quan W, Liu X, Wang H, Chan Z. Physiological and transcriptional responses of contrasting alfalfa (Medicago sativa L.) varieties to salt stress. Plant Cell Tiss Organ Cult (2016) 126(1):105–15. doi: 10.1007/s11240-016-0981-x
198. Chiurazzi MJ, Nørrevang AF, García P, Cerdán PD, Palmgren M, Wenkel S. Controlling flowering of Medicago sativa (alfalfa) by inducing dominant mutations. J Integr Plant Biol (2022) 64(2):205–14. doi: 10.1111/jipb.13186
199. Abbo S, Lev-Yadun S, Heun M, Gopher A. On the ‘lost’ crops of the Neolithic near East. J Exp Bot (2013) 64(4):815–22. doi: 10.1093/jxb/ers373
200. Nguyen V, Riley S, Nagel S, Fisk I, Searle IR. Common vetch: a drought tolerant, high protein neglected leguminous crop with potential as a sustainable food source. Front Plant Sci (2020) 11:818. doi: 10.3389/fpls.2020.00818
201. Jørgensen ST, Liu F, Ouédraogo M, Ntundu WH, Sarrazin J, Christiansen JL. Drought responses of two Bambara groundnut (Vigna subterranea L. Verdc.) landraces collected from a dry and a humid area of Africa. J Agron Crop Sci (2010) 196(6):412–22. doi: 10.1111/j.1439-037X.2010.00435.x
202. Mayes S, Ho WK, Chai HH, Gao X, Kundy AC, Mateva KI, et al. Bambara groundnut: an exemplar underutilised legume for resilience under climate change. Planta (2019) 250(3):803–20. doi: 10.1007/s00425-019-03191-6
203. Panzeri D, Guidi Nissim W, Labra M, Grassi F. Revisiting the domestication process of African Vigna species (Fabaceae): background, perspectives and challenges. Plants (Basel) (2022) 11(4):532. doi: 10.3390/plants11040532
204. Iseki K, Takahashi Y, Muto C, Naito K, Tomooka N. Diversity of drought tolerance in the genus Vigna. Front Plant Sci (2018) 9:729. doi: 10.3389/fpls.2018.00729
205. Muñoz-Amatriaín M, Mirebrahim H, Xu P, Wanamaker SI, Luo M, Alhakami H, et al. Genome resources for climate-resilient cowpea, an essential crop for food security. Plant J (2017) 89(5):1042–54. doi: 10.1111/tpj.13404
206. Jayawardhane J, Goyali JC, Zafari S, Igamberdiev AU. The response of cowpea (Vigna unguiculata) plants to three abiotic stresses applied with increasing intensity: hypoxia, salinity, and water deficit. Metabolites (2022) 12(1):38. doi: 10.3390/metabo12010038
207. Stetter MG, Müller T, Schmid KJ. Genomic and phenotypic evidence for an incomplete domestication of South American grain amaranth (Amaranthus caudatus). Mol Ecol (2017) 26(3):871–86. doi: 10.1111/mec.13974
208. Thapa R, Edwards M, Blair MW. Relationship of cultivated grain amaranth species and wild relative accessions. Genes (Basel) (2021) 12(12):1849. doi: 10.3390/genes12121849
209. Blair MW, Londoño JM, Buitrago-Bitar MA, Wu X, Brenner DM. Differentiation of Andean and Mesoamerican accessions in a proposed core collection of grain amaranths. Front Plant Sci (2023) 14:1144681. doi: 10.3389/fpls.2023.1144681
210. Jarvis DE, Ho YS, Lightfoot DJ, Schmöckel SM, Li B, Borm TJA, et al. The genome of Chenopodium quinoa. Nature (2017) 542(7641):307–12. doi: 10.1038/nature21370
211. Zou C, Chen A, Xiao L, Muller HM, Ache P, Haberer G, et al. A high-quality genome assembly of quinoa provides insights into the molecular basis of salt bladder-based salinity tolerance and the exceptional nutritional value. Cell Res (2017) 27(11):1327–40. doi: 10.1038/cr.2017.124
212. López-Marqués RL, Nørrevang AF, Ache P, Moog M, Visintainer D, Wendt T, et al. Prospects for the accelerated improvement of the resilient crop quinoa. J Exp Bot (2020) 71(18):5333–47. doi: 10.1093/jxb/eraa285
213. Zamaratskaia G, Gerhardt K, Knicky M, Wendin K. Buckwheat: an underutilized crop with attractive sensory qualities and health benefits. Crit Rev Food Sci Nutr (2023) 28:1–16. doi: 10.1080/10408398.2023.2249112
214. Fawcett JA, Takeshima R, Kikuchi S, Yazaki E, Katsube-Tanaka T, Dong Y, et al. Genome sequencing reveals the genetic architecture of heterostyly and domestication history of common buckwheat. Nat Plants (2023) 9(8):1236–51. doi: 10.1038/s41477-023-01474-1
215. Zhao H, He Y, Zhang K, Li S, Chen Y, He M, et al. Rewiring of the seed metabolome during Tartary buckwheat domestication. Plant Biotechnol J (2023) 21(1):150–64. doi: 10.1111/pbi.13932
216. Lemmon ZH, Reem NT, Dalrymple J, Soyk S, Swartwood KE, Rodriguez-Leal D, et al. Rapid improvement of domestication traits in an orphan crop by genome editing. Nat Plants (2018) 4(10):766–70. doi: 10.1038/s41477-018-0259-x
217. Gupta P, Geniza M, Elser J, Al-Bader N, Baschieri R, Phillips JL, et al. Reference genome of the nutrition-rich orphan crop chia (Salvia hispanica) and its implications for future breeding. Front Plant Sci (2023) 14:1272966. doi: 10.3389/fpls.2023.1272966
218. Li T, Yang X, Yu Y, Si X, Zhai X, Zhang H, et al. Domestication of wild tomato is accelerated by genome editing. Nat Biotechnol (2018) 36:1160–3. doi: 10.1038/nbt.4273
219. Zsögön A, Čermák T, Naves ER, Notini MM, Edel KH, Weinl S, et al. De novo domestication of wild tomato using genome editing. Nat Biotechnol (2018) 36:1211–6. doi: 10.1038/nbt.4272
220. Xie Y, Zhang T, Huang X, Xu C. A two-in-one breeding strategy boosts rapid utilization of wild species and elite cultivars. Plant Biotechnol J (2022) 20(5):800–2. doi: 10.1111/pbi.13788
221. te Beest M, Le Roux JJ, Richardson DM, Brysting AK, Suda J, Kubesová M, et al. The more the better? The role of polyploidy in facilitating plant invasions. Ann Bot (2012) 109(1):19–45. doi: 10.1093/aob/mcr277
222. Glover JD, Reganold JP, Bell LW, Borevitz J, Brummer EC, Buckler ES, et al. Agriculture. Increased food and ecosystem security via perennial grains. Science (2010) 328(5986):1638–9. doi: 10.1126/science.1188761
223. DeHaan LR, Anderson JA, Bajgain P, Basche A, Cattani DJ, Crain J, et al. Discussion: prioritize perennial grain development for sustainable food production and environmental benefits. Sci Total Environ (2023) 895:164975. doi: 10.1016/j.scitotenv.2023.164975
224. Kapazoglou A, Gerakari M, Lazaridi E, Kleftogianni K, Sarri E, Tani E, et al. Crop wild relatives: A valuable source of tolerance to various abiotic stresses. Plants (Basel) (2023) 12(2):328. doi: 10.3390/plants12020328
225. Luo G, Najafi J, Correia PMP, Trinh MDL, Chapman EA, Østerberg JT, et al. Accelerated domestication of new crops: yield is key. Plant Cell Physiol (2022) 63(11):1624–40. doi: 10.1093/pcp/pcac065
226. Ewa WG, Agata T, Milica P, Anna B, Dennis E, Nick V, et al. Public perception of plant gene technologies worldwide in the light of food security. GM Crops Food (2022) 13(1):218–41. doi: 10.1080/21645698.2022.2111946
Keywords: abiotic stress, adaptation, drought, salinity, flooding, crop domestication
Citation: Palmgren M and Shabala S. Adapting crops for climate change: regaining lost abiotic stress tolerance in crops. Front Sci (2024) 2:1416023. doi: 10.3389/fsci.2024.1416023
Received: 11 April 2024; Accepted: 23 October 2024;
Published: 05 December 2024.
Edited by:
Rosa M. Rivero, Segura Centre for Soil Science and Applied Biology (CEBAS), Spanish National Research Council (CSIC), SpainReviewed by:
Peter Langridge, University of Adelaide, AustraliaFrancesco Loreto, University of Naples Federico II, Italy
Giulia Atzori, Institute for Sustainable Plant Protection (IPSP), National Research Council (CNR), Italy, in collaboration with reviewer FL
Copyright © 2024 Palmgren and Shabala. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sergey Shabala, c2VyZ2V5LnNoYWJhbGFAdXdhLmVkdS5hdQ==; Michael Palmgren, cGFsbWdyZW5AcGxlbi5rdS5kaw==
 Michael Palmgren
Michael Palmgren Sergey Shabala
Sergey Shabala