- 1Faculty of Agriculture, Setsunan University, Osaka, Japan
- 2RNA Biology Laboratory, Faculty of Pharmaceutical Sciences, Hokkaido University, Sapporo, Japan
Short Interspersed Elements (SINEs) comprise a significant portion of the genomes of higher eukaryotes, including humans and mice. This review focuses on SINE-derived noncoding RNAs (ncRNAs), particularly BC1, BC200, and 4.5SH RNA, which are expressed abundantly and in a species-specific manner. These ncRNAs seem to have independently evolved their functions during evolutionary processes: BC1 and BC200 have become cytoplasmic translation inhibitors, while 4.5SH RNA has developed into a nuclear ncRNA that regulates splicing. This review delves into the unique roles of these ncRNAs, with a special emphasis on the recently discovered splicing regulation function of 4.5SH RNA. Furthermore, we discuss their evolutionary trajectories and potential implications for understanding the complexities of gene regulation.
Introduction
SINEs (Short Interspersed Elements) (Singer, 1982) are among the most “successful” retroelements within mammalian genomes, judging by copy number, reaching up to 1.9 × 106 and 1.6 × 106 copies in humans and mice, respectively (Smit, 1996). In both primates and rodents, two major classes of SINEs, SINE B1s (or the Alu family in humans) and SINE B2s, have evolved from distinct, abundant, ribosome-associated noncoding RNAs (ncRNAs): 7SL and tRNAs, respectively (Okada, 1991; Deininger, 2011; Kramerov and Vassetzky, 2011). SINEs are thought to amplify themselves non-autonomously, utilizing the reverse transcriptase of autonomous retroelements, known as LINEs (Long Interspersed Elements) (Singer, 1982), via a mechanism called target-primed reverse transcription (Deininger, 2011; Kramerov and Vassetzky, 2011). Retro-transposition of SINEs can be potentially hazardous to host genomes if inserted into coding sequences or regulatory elements. Furthermore, SINE insertions can introduce deleterious exons by providing consensus splice sites inherent to certain SINE groups (Sela et al., 2007; Sorek, 2007). Indeed, SINE insertions, particularly Alu elements, have been implicated in a range of human genetic disorders, including Fukuyama-type muscular dystrophy (Taniguchi-Ikeda et al., 2011), neurofibromatosis type 1 (Wallace et al., 1991), among others. Consequently, the activity of SINEs, along with other transposing elements, must be strictly regulated to maintain genomic integrity. In higher eukaryotes, including humans and mice, small RNAs known as piRNAs, processed from precursor ncRNAs containing retroelements, help keep the SINE expression under control. These piRNAs are loaded onto piwi-family proteins, introducing repressive epigenetic modifications to retroelements, including SINEs, through their association with chromatin modification complexes (Onishi et al., 2021; Wang and Lin, 2021). As such, the expression of SINEs is typically kept at an extremely low level, if any expression occurs at all.
Despite the global repression of SINEs, SINE-related short ncRNAs, distinct from the ancestral 7SL or tRNAs, are abundantly expressed in a species-specific manner (Figure 1A). The small rodent-specific 4.5SH RNA is one of the earliest examples of such species-specific short ncRNA, with its sequence being determined almost simultaneously with the determination of the Alu-element sequence, the firstly discovered SINE (Harada and Kato, 1980; Rubin et al., 1980). The sequence of 4.5SH RNA is highly homologous to a variant of SINE B1 known as pB1d10, suggesting that this could be a direct precursor of 4.5SH RNA (Gogolevskaya et al., 2005). BC1 and BC200, identified as abundant, brain-specific, cytoplasmic RNAs in rats and primates respectively, are other examples of such SINE-related ncRNAs (Sutcliffe et al., 1984; Martignetti and Brosius, 1993; Tiedge et al., 1993). Although they are thought to be functional counterparts, BC1 exhibits homology to Identifier (ID) elements, a tRNA-derived SINE (Sutcliffe et al., 1982), while BC200 originates from a distinct group of SINEs, the Alu family retrotransposon (Figure 1A). These highly expressed SINE-related endogenous ncRNAs are transcribed by RNA polymerase III, similar to ancestral SINEs, but lack an A-rich tail required for retrotranspositions (Kramerov and Vassetzky, 2011). Hence, they are located at a single region in each genome. While BC1 and BC200 are single copy genes (DeChiara and Brosius, 1987; Martignetti and Brosius, 1993), 4.5SH genes are tandemly arrayed at a distinct region in the genome, chromosome 6 in the case of mice (Schoeniger and Jelinek, 1986). The tandem organization of 4.5SH is reminiscent of the tandemly repeated ribosomal RNA (rRNA) genes, which are generated via unequal crossover during meiosis (Eickbush and Eickbush, 2007).
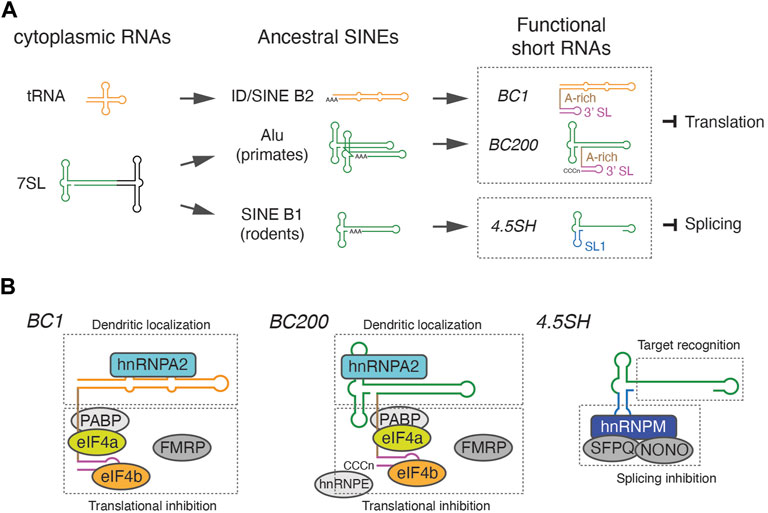
FIGURE 1. Origin of retrotransposon-derived short ncRNAs and their modular structures. (A) Schematic representation of the evolutionary origin of BC1, BC200, and 4.5SH RNA. BC1 and BC200 are derived from distinct SINEs, ID and Alu respectively, but independently acquired an A-rich linker and 3′ stem-loop (SL) structure to become functionally similar translational inhibitors. 4.5SH RNA acquired a unique stem-loop 1 (SL1) that associates with splicing repressors. (B) The modular organization of functional short RNAs. BC1 and BC2 associate with a similar set of proteins that mediate dendritic localization or translational inhibition. 4.5SH RNA consists of an effector binding module and a target recognition module, which bind splicing inhibitor proteins and base pair with target exons, respectively.
Even though these ncRNAs were discovered as early as the 1970-80s, their precise molecular functions and physiological roles have only become clearer in the last decade. In this short review, we will summarize the origins, mechanisms of action, and physiological roles of these abundant SINE-derived ncRNAs.
BC1 RNA: rodent-specific short RNA function as a translational regulator in dendrites
The brain cytoplasmic 1 (BC1) RNA was initially identified through cross-hybridization of multiple cDNA clones with a short, ncRNA specifically expressed in the brain (Sutcliffe et al., 1984). This cross-hybridization was due to the presence of an 82 base ID element within the cDNA clones (Sutcliffe et al., 1982), a member of the SINE B2 family of retrotransposons homologous to tRNAs (Kramerov and Vassetzky, 2011). Although ID elements were initially presumed to be enriched in brain-specific transcripts (Sutcliffe et al., 1982), current genome sequence information and gene annotation do not support this initial assumption. Instead, ID elements are widely distributed throughout the genome, with their numbers ranging from 200 to 120,000 copies in a given rodent genome (Kim et al., 1994). Despite these ID elements typically being silenced and not transcribed into RNAs, BC1, which is highly homologous to ID elements, is abundantly expressed in the brain. BC1 is a single copy gene in rats, while two copy loci have been identified in guinea pigs, possibly due to a DNA-mediated duplication event (Kim et al., 1994). BC1 RNA is transcribed by RNA polymerase III and measures approximately 150 nucleotides in length (DeChiara and Brosius, 1987). BC1 is thought to have originated from a tRNAAla (CGC), however, evolutionary base substitution led to a significant reorganization of the secondary structure. Instead of forming the well-known clover-leaf structure typical of tRNAs, the 5′ region of BC1 homologous to tRNA (or ID) forms a single long stem loop structure (Rozhdestvensky et al., 2001). The 5′ stem loop of BC1 RNA is followed by an A-rich linker region and a 3′ stem loop sequence, both unique to BC1 (Rozhdestvensky et al., 2001) (Figure 1A).
BC1 exhibits a unique distribution pattern, being specifically enriched in the dendrites of various types of neurons (Tiedge et al., 1991), a localization mediated by its 5′ stem loop structure through association with hnRNPA2 (Muslimov et al., 2006; Muslimov et al., 2011). This has led to long-standing speculation that BC1 could control local translation at synaptic terminals. The first concrete evidence that BC1 can inhibit translation came from in vitro studies. BC1 has been shown to inhibit the in vitro translation of mRNAs in reticulocyte lysates in a manner dependent on eIF4A, an ATP-dependent helicase required for translational initiation (Wang et al., 2002). It is postulated that BC1 may repress translation through multiple pathways (Figure 1B). The central A-rich linker of BC1 directly binds to eIF4A, inhibiting its helicase activity (Lin et al., 2008; Eom et al., 2011). This A-rich linker also binds to PABP, which may also contribute to the translation inhibition by BC1 (Wang et al., 2002). The 3′ stem loop of BC1 associates with eIF4B, inhibiting its interaction with the small ribosomal subunit (Eom et al., 2011). Upon neuronal stimulation, eIF4B is dephosphorylated and dissociates from BC1, suggesting that BC1-mediated translational inhibition is controlled by neuronal activities (Eom et al., 2014).
The physiological significance of BC1 has been illuminated through a series of studies utilizing BC1 knockout (KO) mice. While BC1 KO mice do not exhibit gross morphological abnormalities (Skryabin et al., 2003), behavioral studies have identified a decrease in exploratory behavior and an increase in anxiety in these mice (Lewejohann et al., 2004). At the electrophysiological level, dopamine-mediated synaptic regulation responses in striatal neurons and GABA-mediated spontaneous inhibitory activities are both enhanced in BC1 KO mice (Centonze et al., 2007; Centonze et al., 2008). Notably, BC1 KO mice demonstrate a pronounced increase in susceptibility to high-decibel (dB) auditory stimuli, with exposure to 120-dB sound potentially causing seizures or even death (Zhong et al., 2009). In these mice, mGluR-stimulated translation is anomalously upregulated, leading to the overexpression of synaptic proteins, including FMRP and PSD-95, upon mGluR activation. Electrophysiological studies reveal heightened synaptic activities in BC1 KO mice, potentially explaining their extreme sensitivity to sound stimuli. This phenotype can be mitigated by the administration of anisomycin, a translational inhibitor, further supporting the notion that BC1 typically attenuates local protein translation induced by neural stimuli (Zhong et al., 2009). More recent studies suggest that BC1 RNA regulates the structural plasticity of pyramidal neurons by inhibiting the translation of synaptic proteins, which results in enlarged synaptic spines in BC1 KO mice (Briz et al., 2017).
In terms of the molecular mechanisms that inhibit translation at synaptic terminals, it was previously proposed that BC1 binds to FMRP, an RNA-binding protein abundant in dendrites (Richter and Zhao, 2021), and recruits this translational inhibitor to target mRNAs with sequences complementary to BC1 (Zalfa et al., 2003). Indeed, BC1 and FMRP partially colocalize at striatal axon bundles, and GABA transmission is similarly upregulated in both FMRP/BC1 double knockout (dKO) mice and single knockout of either gene (Centonze et al., 2008). However, later studies revealed that FMRP binds various types of mRNAs and other RNAs in a sequence non-specific manner, including BC1 and FMRP-regulated target mRNAs (Iacoangeli et al., 2008). Furthermore, double knockout (dKO) mice lacking both FMRP and BC1 exhibit increased responses to loud sounds compared to single KO mice (Zhong et al., 2010). This genetic interaction suggests that FMRP and BC1 may regulate translation independently, potentially via distinct mechanisms. Recent findings that FMRP forms liquid droplets could reconcile these contradicting interpretations (Tsang et al., 2019). The C-terminal region of FMRP, known to be highly disordered, facilitates multivalent weak interactions necessary for the formation of phase-separated liquid droplets that contain RNAs in vitro (Tsang et al., 2019). Typically, intrinsically disordered regions of certain proteins interact with various RNAs in a sequence-independent manner, leading to the formation of phase-separated granules (Protter et al., 2018). The formation of mRNA-containing granules through FMRP may establish a molecular environment conducive to efficient translational inhibition by multiple mechanisms, including BC1-mediated translational suppression. Consequently, while the interaction between FMRP and BC1 may be non-specific, they appear to be functionally interconnected.
BC200: primate-specific short RNA function as a translational regulator in dendrites
The original identification of BC200 RNA resulted from the cross-hybridization of ID probes (which are tRNA-derived SINEs) to monkey RNA (Watson and Sutcliffe, 1987). These ID probes had previously been used to identify BC1 RNA (Sutcliffe et al., 1982). However, BC200 RNA shares significant sequence homology with Alu (a SINE derived from 7SL RNA), rather than with tRNA-derived SINEs such as ID (Watson and Sutcliffe, 1987) (Figure 1A). This seemingly contradictory outcome can be explained by the fortunate presence of short sequences complementary to Alu in the ID probe (Watson and Sutcliffe, 1987). Despite their differences in origin and primary sequences, BC1 and BC200 share numerous common properties: both are short RNAs transcribed by RNA polymerase III, exclusively expressed in the brain, and localized in the dendrites of specific neurons (Tiedge et al., 1991). Both are composed of three distinct domains: the 5′ element homologous to Alu, the central A-rich domain, and the unique C-rich 3′ stem-loop structure (Martignetti and Brosius, 1993). Similar to BC1, BC200 localizes to dendrite through 5′ stem loop region that associates with hnRNPA2 (Muslimov et al., 2006; Muslimov et al., 2011; Muslimov et al., 2019), directly binds to eIF4a/eIF4b in vitro and inhibits translation (Lin et al., 2008; Eom et al., 2014), associates with PABP via the A-rich central domain (Muddashetty et al., 2002; Kondrashov et al., 2005), and forms a complex with FMRP (Zalfa et al., 2003) (Figure 1B). As a result, BC200 RNA influence mRNA translation in a similar fashion to BC1 RNA. Thus, these two ncRNAs, despite originating from completely different groups of SINEs, may play a convergent role in negatively regulating local translation at the synapse.
In addition to these common protein components, BC200 has been found to associate with a unique set of proteins. The 5′ region of BC200 is highly homologous to 7SL, an RNA component of signal recognition particles (SRP), and binds to SRP9/14 (Kremerskothen et al., 1998). BC200 also binds to the Synaptotagmin Binding Cytoplasmic RNA Interacting Protein (SYNCRIP) via the central A-rich linker domain (Duning et al., 2008), and to HNRNPE1 and HNRNPE2 via the 3′-located C-rich domain (Jang et al., 2017). Since in vitro translation inhibition mediated by BC200 is counteracted by the addition of HNRNPE1/2, these proteins might function as cellular regulators to control local translation in dendrites (Jang et al., 2017).
Although no inherited diseases have been directly associated with BC200, it has been implicated in a variety of disorders such as cancers and neurodegenerative diseases. For instance, BC200 is found in abnormally high levels in Alzheimer’s disease (Mus et al., 2007) and in various cancers (Chen et al., 1997). BC200 is also thought to be involved in the regulation of alternative splicing of apoptosis-related genes, such as Bcl-x, in breast cancer (Singh et al., 2016). Additional requirements for specific article types and further information please refer to “Article types” on every Frontiers journal page.
4.5SH RNA: a novel splicing regulator that function as a molecular antidote for toxic exonization of SINE B1 in small rodents
4.5SH RNA, a 90-nucleotide nuclear ncRNA, was initially identified in the 1970s as an RNA that co-purified with poly-A (+) RNAs (Jelinek and Leinwand, 1978; Harada et al., 1979; Harada and Kato, 1980; Leinwand et al., 1982). This RNA is specifically found in a group of small, short-lived rodents, including mice, hamsters, and rats (Gogolevskaya et al., 2005). 4.5SH RNA shares substantial homology with SINE B1, the most abundant SINE in rodent species, and is believed to have originated from the ancient B1 subfamily pB1d10 (Quentin, 1994; Gogolevskaya et al., 2005) (Figures 1A, 2B). Unlike BC1 or BC200, 4.5SH is a multicopy gene. Contrary to ancestral SINEs that are interspersed throughout the genome, 4.5SH forms a large cluster at a specific genomic region in each species. In the case of the mouse, the transcription unit of 4.5SH is embedded in a 4.2 kb fragment, which is arranged in tandem to form large clusters of ∼200 copies in a ∼900 MB region of chromosome 6 (Schoeniger and Jelinek, 1986; Yoshimoto et al., 2022). Interestingly, while the 4.5SH transcription unit is highly conserved, the other regions of the 4.2 kb repeat unit exhibit less conservation, suggesting the functional importance of 4.5SH. The expression level of 4.5SH RNA is exceptionally high, with over 10,000 molecules produced per cell (Schoeniger and Jelinek, 1986), and these transcripts are localized in nuclear speckles (Ishida et al., 2015) (Figure 2A), nuclear compartments containing poly-A (+) RNAs as well as splicing factors (Spector and Lamond, 2011). 4.5SH RNA has been demonstrated to interact with multiple proteins, including La and Nucleolin (Leinwand et al., 1982; Hirose and Harada, 2008). Despite these molecular descriptions, the biological functions of 4.5SH remained largely unknown for more than 40 years since its early discovery.
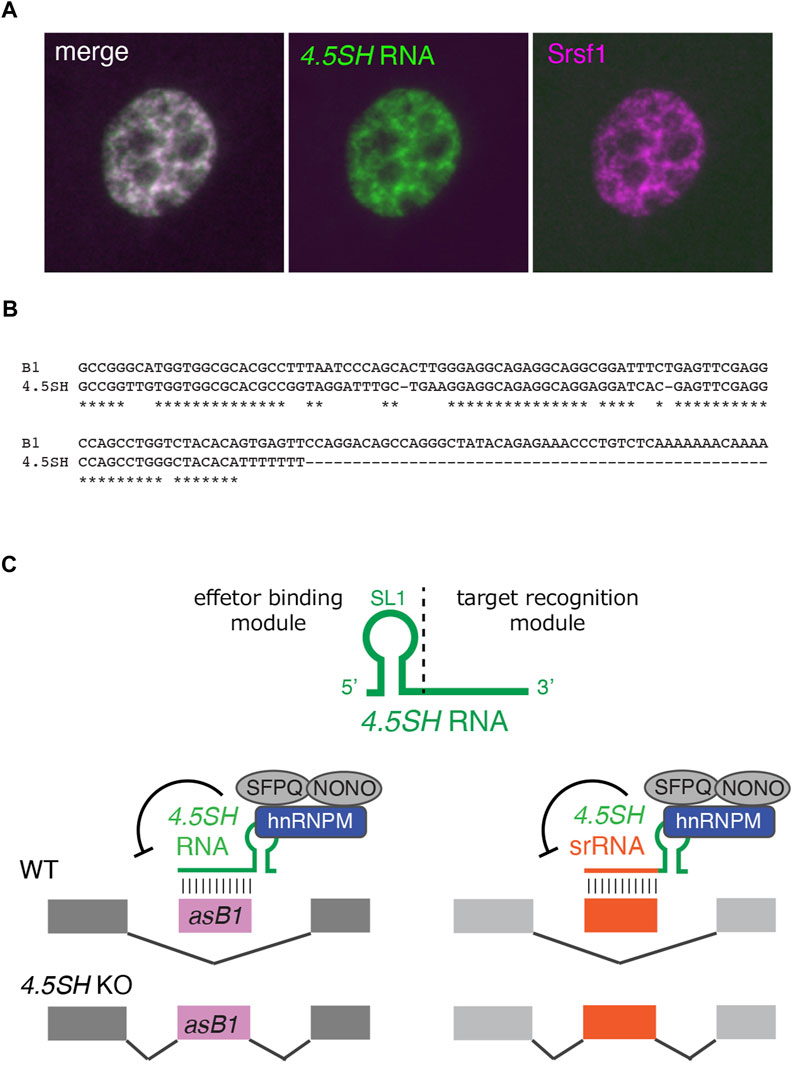
FIGURE 2. 4.5SH induces skipping of deleterious exons. (A) Subcellular localization of 4.5SH RNA in 3T3 cells. 4.5SH RNA (green) is localized to nuclear speckles enriched in splicing regulators such as Srsf1 (magenta). (B) Alignment of 4.5SH RNA and SINE B1. 4.5SH RNA is highly homologous to SINE B1 but lacks 3′ A-rich sequences required for retrotranspositions. (C) Schematics of 4.5SH-mediated exon skipping and the generation of artificial splicing regulators. 4.5SH RNA base pairs with antisense insertions of SINE B1 (asB1) and inhibits their exonizations. In the 4.5SH KO cells, these asB1-containing exons are abnormally included in hundreds of host transcripts, introducing premature stop codons and frame-shift mutations, leading to embryonic lethality. The modular organization of 4.5SH RNA enables the generation of programmable splicing regulatory RNA (srRNA) consisting of the effector binding module of 4.5SH RNA and sequences complementary to target exons of interest.
The first molecular function of 4.5SH was demonstrated through experiments using antisense oligonucleotides (ASO) to deplete this nuclear ncRNA (Ishida et al., 2015). Treating murine cells, such as Neuro2A or 3T3, with ASO led to decreased cellular proliferation, whereas no such effect was observed in human cells that lack the 4.5SH gene. Remarkably, 4.5SH RNA formed an intra-molecular double-stranded RNA structure with reporter mRNAs that contain antisense insertions of SINE B1 (asB1) in the 3′ untranslated region, detectable by A to I base substitution dependent on the adenosine deaminase ADAR that recognizes a long stretch of double-stranded RNA (Ishida et al., 2015). However, the precise molecular mechanisms leading to decreased cellular proliferation remained unclear.
More definitive evidence for the molecular and physiological functions of 4.5SH RNA comes from very recent studies using knockout (KO) mice (Yoshimoto et al., 2022). The mutant mice exhibited early embryonic lethality, suggesting that 4.5SH is an essential gene in mice. Subsequent analyses of RNA-Sequencing data led to the discovery of hundreds of exons that were aberrantly included in 4.5SH KO cells. Notably, 75% of these KO-enriched exons were unannotated in RefSeq and were identified as cryptic exons. A detailed examination of these abnormal exons revealed that their consensus sequences matched asB1. Indeed, asB1 naturally contains near-consensus splicing acceptor and donor sequences, excluding the branch point sequences (Sela et al., 2007). Hence, when asB1 is inserted downstream of branch point consensus sequences in introns, it can potentially introduce deleterious premature stop codons or induce frameshift mutations. 4.5SH counteracts the toxic exonization of asB1, serving as a molecular antidote that protects the transcriptome from the aberrant inclusion of asB1 (Figure 2C).
Mechanistically, 4.5SH RNA consists of two modules: the 5′ effector-binding module and the 3′ target recognition module (Yoshimoto et al., 2022). The 5′ effector-binding module features a characteristic stem-loop structure and associates with splicing inhibitory effector proteins, including Hnrnpm, Sfpq, and Nono. The complexes are recruited to asB1-containing exons via the 3′ target recognition region, which base pairs with the complementary asB1 sequences. Interestingly, the modular organization of 4.5SH RNA offers an opportunity to engineer programmable splicing regulator RNAs (srRNAs) (Figure 2C). By replacing the target recognition module with sequences complementary to exons of interest, one can create a chimeric srRNA that induces skipping of target exons. This finding opens up new avenues in genetic research, with potential applications as a molecular biology tool and as a basis for RNA therapeutics.
Concluding remarks
The field of ncRNAs continues to reveal increasingly complex regulatory networks influencing cellular physiology and pathology (Mattick et al., 2023). This review focused on three retrotransposon-derived short RNAs–BC1, BC200, and 4.5SH RNA–highlighting the multifaceted roles of ncRNAs. Intriguingly, the functions of these short ncRNAs appear to have been acquired independently throughout each species’ evolution, not confined by their ancestral origin: BC1 and BC200, derived from tRNA and 7SL, respectively, seem to have undergone convergent evolution to become translation inhibitors. Conversely, 4.5SH, derived from 7SL, has obtained nuclear localization properties unlike BC200, evolving to become a splicing regulatory ncRNA, possibly through modification of the 5′ stem loop structure that binds strongly to Hnrnpm, a splicing inhibitor (Yoshimoto et al., 2022).
A pertinent question is why these short ncRNAs emerged in a species-specific manner despite their essential roles during development (as seen with 4.5SH RNA) or under certain environmental conditions (as with BC1). They may have co-evolved with the stochastic emergence of deleterious SINE insertions. For instance, the insertion of the exonization-prone asB1 into the intron of an essential gene may give rise to toxic exons detrimental to animal survival. However, if this happens in animals expressing ancestral 4.5SH, the intronic asB1 insertions are likely neutral and don’t substantially affect the transcriptome’s integrity. Hence, SINE B1 can freely propagate and embed its copy without harm in animals expressing 4.5SH RNA, and the copy number of 4.5SH also increases to suppress the rising number of potentially toxic exons. The same scenario might be applicable to the emergence and evolution of BC1 and BC200, especially if the insertions of the complementary retrotransposon into the 3′ UTR of certain neuronal genes could affect harmful effects by enhancing the translation of the host mRNAs.
It is crucial to emphasize that there are other SINE-derived or repeat-derived short ncRNAs whose functions have not been fully elucidated (Matylla-Kulinska et al., 2014; O'Neill, 2020). Future research on these ncRNAs will not only deepen our understanding of gene regulation complexities but also offer potential implications for the diagnosis, prevention, and treatment of human diseases. We anticipate that the insights presented here will spur further studies and potential applications for these fascinating RNAs.
Author contributions
RY: Conceptualization, Funding acquisition, Writing–original draft, Writing–review and editing. SN: Conceptualization, Funding acquisition, Writing–original draft, Writing–review and editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was supported by JSPS KAKENHI Grant Number JP 16H06276 (AdAMS), 21H05274 and 21K19246 granted to SN, 22K05565 granted to RY.
Acknowledgments
We thank intellectual input from Dr. Norihiro Okada and Dr. Fumio Harada.
Conflict of interest
RY and SN declare that a patent application has been submitted based on the paper “Yoshimoto et al., 2022”.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Briz, V., Restivo, L., Pasciuto, E., Juczewski, K., Mercaldo, V., Lo, A. C., et al. (2017). The non-coding RNA BC1 regulates experience-dependent structural plasticity and learning. Nat. Commun. 8, 293. doi:10.1038/s41467-017-00311-2
Centonze, D., Rossi, S., Mercaldo, V., Napoli, I., Ciotti, M. T., De Chiara, V., et al. (2008). Abnormal striatal GABA transmission in the mouse model for the fragile X syndrome. Biol. Psychiatry 63, 963–973. doi:10.1016/j.biopsych.2007.09.008
Centonze, D., Rossi, S., Napoli, I., Mercaldo, V., Lacoux, C., Ferrari, F., et al. (2007). The brain cytoplasmic RNA BC1 regulates dopamine D2 receptor-mediated transmission in the striatum. J. Neurosci. 27, 8885–8892. doi:10.1523/jneurosci.0548-07.2007
Chen, W., Bocker, W., Brosius, J., and Tiedge, H. (1997). Expression of neural BC200 RNA in human tumours. J. Pathol. 183, 345–351. doi:10.1002/(sici)1096-9896(199711)183:3<345:aid-path930>3.0.co;2-8
Dechiara, T. M., and Brosius, J. (1987). Neural BC1 RNA: cDNA clones reveal nonrepetitive sequence content. Proc. Natl. Acad. Sci. U. S. A. 84, 2624–2628. doi:10.1073/pnas.84.9.2624
Deininger, P. (2011). Alu elements: know the SINEs. Genome Biol. 12, 236. doi:10.1186/gb-2011-12-12-236
Duning, K., Buck, F., Barnekow, A., and Kremerskothen, J. (2008). SYNCRIP, a component of dendritically localized mRNPs, binds to the translation regulator BC200 RNA. J. Neurochem. 105, 351–359. doi:10.1111/j.1471-4159.2007.05138.x
Eickbush, T. H., and Eickbush, D. G. (2007). Finely orchestrated movements: evolution of the ribosomal RNA genes. Genetics 175, 477–485. doi:10.1534/genetics.107.071399
Eom, T., Berardi, V., Zhong, J., Risuleo, G., and Tiedge, H. (2011). Dual nature of translational control by regulatory BC RNAs. Mol. Cell. Biol. 31, 4538–4549. doi:10.1128/mcb.05885-11
Eom, T., Muslimov, I. A., Tsokas, P., Berardi, V., Zhong, J., Sacktor, T. C., et al. (2014). Neuronal BC RNAs cooperate with eIF4B to mediate activity-dependent translational control. J. Cell Biol. 207, 237–252. doi:10.1083/jcb.201401005
Gogolevskaya, I. K., Koval, A. P., and Kramerov, D. A. (2005). Evolutionary history of 4.5SH RNA. Mol. Biol. Evol. 22, 1546–1554. doi:10.1093/molbev/msi140
Harada, F., Kato, N., and Hoshino, H. (1979). Series of 4.5S RNAs associated with poly(A)-containing RNAs of rodent cells. Nucleic Acids Res. 7, 909–918. doi:10.1093/nar/7.4.909
Harada, F., and Kato, N. (1980). Nucleotide sequences of 4.5S RNAs associated with poly(A)-containing RNAs of mouse and hamster cells. Nucleic Acids Res. 8, 1273–1286. doi:10.1093/nar/8.6.1273
Hirose, Y., and Harada, F. (2008). Mouse nucleolin binds to 4.5S RNAh, a small noncoding RNA. Biochem. Biophysical Res. Commun. 365, 62–68. doi:10.1016/j.bbrc.2007.10.117
Iacoangeli, A., Rozhdestvensky, T. S., Dolzhanskaya, N., Tournier, B., Schutt, J., Brosius, J., et al. (2008). On BC1 RNA and the fragile X mental retardation protein. Proc. Natl. Acad. Sci. U. S. A. 105, 734–739. doi:10.1073/pnas.0710991105
Ishida, K., Miyauchi, K., Kimura, Y., Mito, M., Okada, S., Suzuki, T., et al. (2015). Regulation of gene expression via retrotransposon insertions and the noncoding RNA 4.5S RNA H. Genes cells. 20, 887–901. doi:10.1111/gtc.12280
Jang, S., Shin, H., Lee, J., Kim, Y., Bak, G., and Lee, Y. (2017). Regulation of BC200 RNA-mediated translation inhibition by hnRNP E1 and E2. FEBS Lett. 591, 393–405. doi:10.1002/1873-3468.12544
Jelinek, W., and Leinwand, L. (1978). Low molecular weight RNAs hydrogen-bonded to nuclear and cytoplasmic poly(A)-terminated RNA from cultured Chinese hamster ovary cells. Cell 15, 205–214. doi:10.1016/0092-8674(78)90095-8
Kim, J., Martignetti, J. A., Shen, M. R., Brosius, J., and Deininger, P. (1994). Rodent BC1 RNA gene as a master gene for ID element amplification. Proc. Natl. Acad. Sci. U. S. A. 91, 3607–3611. doi:10.1073/pnas.91.9.3607
Kondrashov, A. V., Kiefmann, M., Ebnet, K., Khanam, T., Muddashetty, R. S., and Brosius, J. (2005). Inhibitory effect of naked neural BC1 RNA or BC200 RNA on eukaryotic in vitro translation systems is reversed by poly(A)-binding protein (PABP). J. Mol. Biol. 353, 88–103. doi:10.1016/j.jmb.2005.07.049
Kramerov, D. A., and Vassetzky, N. S. (2011). SINEs. Wiley Interdiscip. Rev. RNA 2, 772–786. doi:10.1002/wrna.91
Kremerskothen, J., Zopf, D., Walter, P., Cheng, J. G., Nettermann, M., Niewerth, U., et al. (1998). Heterodimer SRP9/14 is an integral part of the neural BC200 RNP in primate brain. Neurosci. Lett. 245, 123–126. doi:10.1016/s0304-3940(98)00215-8
Leinwand, L. A., Wydro, R. M., and Nadal-Ginard, B. (1982). Small RNA molecules related to the Alu family of repetitive DNA sequences. Mol. Cell Biol. 2, 1320–1330. doi:10.1128/mcb.2.11.1320
Lewejohann, L., Skryabin, B. V., Sachser, N., Prehn, C., Heiduschka, P., Thanos, S., et al. (2004). Role of a neuronal small non-messenger RNA: behavioural alterations in BC1 RNA-deleted mice. Behav. Brain Res. 154, 273–289. doi:10.1016/j.bbr.2004.02.015
Lin, D., Pestova, T. V., Hellen, C. U., and Tiedge, H. (2008). Translational control by a small RNA: dendritic BC1 RNA targets the eukaryotic initiation factor 4A helicase mechanism. Mol. Cell. Biol. 28, 3008–3019. doi:10.1128/mcb.01800-07
Martignetti, J. A., and Brosius, J. (1993). BC200 RNA: a neural RNA polymerase III product encoded by a monomeric Alu element. Proc. Natl. Acad. Sci. U. S. A. 90, 11563–11567. doi:10.1073/pnas.90.24.11563
Mattick, J. S., Amaral, P. P., Carninci, P., Carpenter, S., Chang, H. Y., Chen, L. L., et al. (2023). Long non-coding RNAs: definitions, functions, challenges and recommendations. Nat. Rev. Mol. Cell Biol. 24, 430–447. doi:10.1038/s41580-022-00566-8
Matylla-Kulinska, K., Tafer, H., Weiss, A., and Schroeder, R. (2014). Functional repeat-derived RNAs often originate from retrotransposon-propagated ncRNAs. WIREs RNA 5, 591–600. doi:10.1002/wrna.1243
Muddashetty, R., Khanam, T., Kondrashov, A., Bundman, M., Iacoangeli, A., Kremerskothen, J., et al. (2002). Poly(A)-binding protein is associated with neuronal BC1 and BC200 ribonucleoprotein particles. J. Mol. Biol. 321, 433–445. doi:10.1016/s0022-2836(02)00655-1
Mus, E., Hof, P. R., and Tiedge, H. (2007). Dendritic BC200 RNA in aging and in Alzheimer's disease. Proc. Natl. Acad. Sci. U. S. A. 104, 10679–10684. doi:10.1073/pnas.0701532104
Muslimov, I. A., Iacoangeli, A., Brosius, J., and Tiedge, H. (2006). Spatial codes in dendritic BC1 RNA. J. Cell Biol. 175, 427–439. doi:10.1083/jcb.200607008
Muslimov, I. A., Iacoangeli, A., Eom, T., Ruiz, A., Lee, M., Stephenson, S., et al. (2019). Neuronal BC RNA transport impairments caused by systemic lupus erythematosus autoantibodies. J. Neurosci. 39, 7759–7777. doi:10.1523/jneurosci.1657-18.2019
Muslimov, I. A., Patel, M. V., Rose, A., and Tiedge, H. (2011). Spatial code recognition in neuronal RNA targeting: role of RNA-hnRNP A2 interactions. J. Cell Biol. 194, 441–457. doi:10.1083/jcb.201010027
O'neill, R. J. (2020). Seq'ing identity and function in a repeat-derived noncoding RNA world. Chromosome Res. 28, 111–127. doi:10.1007/s10577-020-09628-z
Onishi, R., Yamanaka, S., and Siomi, M. C. (2021). piRNA- and siRNA-mediated transcriptional repression in Drosophila, mice, and yeast: new insights and biodiversity. EMBO Rep. 22, e53062. doi:10.15252/embr.202153062
Protter, D. S. W., Rao, B. S., Van Treeck, B., Lin, Y., Mizoue, L., Rosen, M. K., et al. (2018). Intrinsically disordered regions can contribute promiscuous interactions to RNP granule assembly. Cell Rep. 22, 1401–1412. doi:10.1016/j.celrep.2018.01.036
Quentin, Y. (1994). Emergence of master sequences in families of retroposons derived from 7sl RNA. Genetica 93, 203–215. doi:10.1007/bf01435252
Richter, J. D., and Zhao, X. (2021). The molecular biology of FMRP: new insights into fragile X syndrome. Nat. Rev. Neurosci. 22, 209–222. doi:10.1038/s41583-021-00432-0
Rozhdestvensky, T. S., Kopylov, A. M., Brosius, J., and Huttenhofer, A. (2001). Neuronal BC1 RNA structure: evolutionary conversion of a tRNA(ala) domain into an extended stem-loop structure. RNA 7, 722–730. doi:10.1017/s1355838201002485
Rubin, C. M., Houck, C. M., Deininger, P. L., Friedmann, T., and Schmid, C. W. (1980). Partial nucleotide sequence of the 300-nucleotide interspersed repeated human DNA sequences. Nature 284, 372–374. doi:10.1038/284372a0
Schoeniger, L. O., and Jelinek, W. R. (1986). 4.5S RNA is encoded by hundreds of tandemly linked genes, has a short half-life, and is hydrogen bonded in vivo to poly(A)-terminated RNAs in the cytoplasm of cultured mouse cells. Mol. Cell. Biol. 6, 1508–1519. doi:10.1128/mcb.6.5.1508-1519.1986
Sela, N., Mersch, B., Gal-Mark, N., Lev-Maor, G., Hotz-Wagenblatt, A., and Ast, G. (2007). Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu's unique role in shaping the human transcriptome. Genome Biol. 8, R127. doi:10.1186/gb-2007-8-6-r127
Singer, M. F. (1982). SINEs and LINEs: highly repeated short and long interspersed sequences in mammalian genomes. Cell 28, 433–434. doi:10.1016/0092-8674(82)90194-5
Singh, R., Gupta, S. C., Peng, W. X., Zhou, N., Pochampally, R., Atfi, A., et al. (2016). Regulation of alternative splicing of Bcl-x by BC200 contributes to breast cancer pathogenesis. Cell Death Dis. 7, e2262. doi:10.1038/cddis.2016.168
Skryabin, B. V., Sukonina, V., Jordan, U., Lewejohann, L., Sachser, N., Muslimov, I., et al. (2003). Neuronal untranslated BC1 RNA: targeted gene elimination in mice. Mol. Cell. Biol. 23, 6435–6441. doi:10.1128/mcb.23.18.6435-6441.2003
Smit, A. F. (1996). The origin of interspersed repeats in the human genome. Curr. Opin. Genet. Dev. 6, 743–748. doi:10.1016/s0959-437x(96)80030-x
Sorek, R. (2007). The birth of new exons: mechanisms and evolutionary consequences. RNA 13, 1603–1608. doi:10.1261/rna.682507
Spector, D. L., and Lamond, A. I. (2011). Nuclear speckles. Cold Spring Harb. Perspect. Biol. 3, a000646. doi:10.1101/cshperspect.a000646
Sutcliffe, J. G., Milner, R. J., Bloom, F. E., and Lerner, R. A. (1982). Common 82-nucleotide sequence unique to brain RNA. Proc. Natl. Acad. Sci. U. S. A. 79, 4942–4946. doi:10.1073/pnas.79.16.4942
Sutcliffe, J. G., Milner, R. J., Gottesfeld, J. M., and Lerner, R. A. (1984). Identifier sequences are transcribed specifically in brain. Nature 308, 237–241. doi:10.1038/308237a0
Taniguchi-Ikeda, M., Kobayashi, K., Kanagawa, M., Yu, C. C., Mori, K., Oda, T., et al. (2011). Pathogenic exon-trapping by SVA retrotransposon and rescue in Fukuyama muscular dystrophy. Nature 478, 127–131. doi:10.1038/nature10456
Tiedge, H., Chen, W., and Brosius, J. (1993). Primary structure, neural-specific expression, and dendritic location of human BC200 RNA. J. Neurosci. 13, 2382–2390. doi:10.1523/jneurosci.13-06-02382.1993
Tiedge, H., Fremeau, R. T., Weinstock, P. H., Arancio, O., and Brosius, J. (1991). Dendritic location of neural BC1 RNA. Proc. Natl. Acad. Sci. U. S. A. 88, 2093–2097. doi:10.1073/pnas.88.6.2093
Tsang, B., Arsenault, J., Vernon, R. M., Lin, H., Sonenberg, N., Wang, L. Y., et al. (2019). Phosphoregulated FMRP phase separation models activity-dependent translation through bidirectional control of mRNA granule formation. Proc. Natl. Acad. Sci. U. S. A. 116, 4218–4227. doi:10.1073/pnas.1814385116
Wallace, M. R., Andersen, L. B., Saulino, A. M., Gregory, P. E., Glover, T. W., and Collins, F. S. (1991). A de novo Alu insertion results in neurofibromatosis type 1. Nature 353, 864–866. doi:10.1038/353864a0
Wang, C., and Lin, H. (2021). Roles of piRNAs in transposon and pseudogene regulation of germline mRNAs and lncRNAs. Genome Biol. 22, 27. doi:10.1186/s13059-020-02221-x
Wang, H., Iacoangeli, A., Popp, S., Muslimov, I. A., Imataka, H., Sonenberg, N., et al. (2002). Dendritic BC1 RNA: functional role in regulation of translation initiation. J. Neurosci. 22, 10232–10241. doi:10.1523/jneurosci.22-23-10232.2002
Watson, J. B., and Sutcliffe, J. G. (1987). Primate brain-specific cytoplasmic transcript of the Alu repeat family. Mol. Cell. Biol. 7, 3324–3327. doi:10.1128/mcb.7.9.3324-3327.1987
Yoshimoto, R., Nakayama, Y., Yamamoto, I., Tanaka, S., Kurihara, M., Suzuki, Y., et al. (2022). 4.5SH RNA counteracts deleterious exonization of SINE B1 in mice. Res. Square. doi:10.21203/rs.21203.rs-1949270/v1949271
Zalfa, F., Giorgi, M., Primerano, B., Moro, A., Di Penta, A., Reis, S., et al. (2003). The fragile X syndrome protein FMRP associates with BC1 RNA and regulates the translation of specific mRNAs at synapses. Cell 112, 317–327. doi:10.1016/s0092-8674(03)00079-5
Zhong, J., Chuang, S. C., Bianchi, R., Zhao, W., Lee, H., Fenton, A. A., et al. (2009). BC1 regulation of metabotropic glutamate receptor-mediated neuronal excitability. J. Neurosci. 29, 9977–9986. doi:10.1523/jneurosci.3893-08.2009
Keywords: SINE, ncRNA, BC1, BC200, 4.5SH RNA, translation, splicing
Citation: Yoshimoto R and Nakagawa S (2023) SINE-derived short noncoding RNAs: their evolutionary origins, molecular mechanisms, and physiological significance. Front. RNA Res. 1:1257775. doi: 10.3389/frnar.2023.1257775
Received: 12 July 2023; Accepted: 31 July 2023;
Published: 10 August 2023.
Edited by:
Chandrasekhar Kanduri, University of Gothenburg, SwedenReviewed by:
Gordon Carmichael, University of Connecticut Health Center, United StatesCopyright © 2023 Yoshimoto and Nakagawa. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shinichi Nakagawa, bmFrYWdhd2FzQHBoYXJtLmhva3VkYWkuYWMuanA=
 Rei Yoshimoto
Rei Yoshimoto Shinichi Nakagawa
Shinichi Nakagawa