
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
MINI REVIEW article
Front. Psychol., 08 March 2022
Sec. Psychology for Clinical Settings
Volume 13 - 2022 | https://doi.org/10.3389/fpsyg.2022.751041
This article is part of the Research TopicPersonalized Treatment in Mental Illness: From Genetics to Clinical SettingView all 4 articles
Attention-deficit/hyperactivity disorder (ADHD) is a neurodevelopmental disorder with a childhood prevalence of 5%. In about two-thirds of the cases, ADHD symptoms persist into adulthood and often cause significant functional impairment. Based on the results of family and twin studies, the estimated heritability of ADHD approximates 80%, suggests a significant genetic component in the etiological background of the disorder; however, the potential genetic effects on disease risk, symptom severity, and persistence are unclear. This article provides a brief review of the genome-wide and candidate gene association studies with a focus on the clinical aspects, summarizing findings of ADHD disease risk, ADHD core symptoms as dimensional traits, and other traits frequently associated with ADHD, which may contribute to the susceptibility to other comorbid psychiatric disorders. Furthermore, neuropsychological impairment and measures from neuroimaging and electrophysiological paradigms, emerging as potential biomarkers, also provide a prominent target for molecular genetic studies, since they lie in the pathway from genes to behavior; therefore, they can contribute to the understanding of the underlying neurobiological mechanisms and the interindividual heterogeneity of clinical symptoms. Beyond the aforementioned aspects, throughout the review, we also give a brief summary of the genetic results, including polygenic risk scores that can potentially predict individual response to different treatment options and may offer a possibility for personalized treatment for the therapy of ADHD in the future.
Due to its high heritability and neurodevelopmental nature, Attention-Deficit Hyperactivity Disorder (ADHD) is a condition receiving significant interest in psychiatric genetics. The childhood prevalence is 5–7% (Polanczyk et al., 2007), and in one-third of patients, symptoms persist into adulthood (Franke et al., 2018), causing functional impairment of everyday life. Due to the dimensional feature of the core symptoms – inattention, hyperactivity, and impulsivity– and somewhat subjective approach to determine the extent of functional impairment, mapping of possible genetic risk factors for ADHD remains important in both clinical and population samples. In this review, we present the selected literature to give a summary of the current knowledge about the genetic perspective of ADHD, which highlights the gaps that emerge from the approaches taken by candidate gene and genome-wide association studies (GWAS). Besides the aforementioned aspects, we also present the relevant results of the polygenic risk score (PRS) approach (Torkamani et al., 2018; Martin et al., 2019; Ronald et al., 2021), which can individually estimate the genetic liability for the disorder and ADHD-related traits, thereby offering further insight into the background of the symptomatic and genetic heterogeneity characterizing ADHD. Despite the many limitations to its application, it might hold the promise of creating a bridge between research and the clinic, from ‘bench to bedside.’
The heritability of ADHD is high compared to most psychiatric disorders and compares to the heritability rates found in autism spectrum disorder (ASD), bipolar disorder, and schizophrenia (Sullivan et al., 2012). However, our knowledge of the underlying genetic architecture of ADHD remains limited. The familial aggregation of the disorder is strong, and current results suggest that the relative risk of ADHD is 5- to 10-fold for first-degree relatives (Biederman et al., 1990; Biederman, 2005; Franke et al., 2012), which is applicable to core symptoms and is gender-independent (Taylor et al., 2016; Martin et al., 2018). However, regarding the exact characterization of heritability, our results are inconsistent. Based on family and twin studies in recent decades, the heritability of ADHD has been estimated at 77–88% (Faraone and Larsson, 2019). Surprisingly, a mega-analysis of the results from existing GWAS found that the proportion of heritability based on single nucleotide polymorphisms (SNPs) is only 22% (Demontis et al., 2019). Thus, the gap between the results originating from family or twin studies and SNP heritability based on GWAS is huge, a phenomenon commonly referred to as ‘hidden or lost heritability.’ There are several potential explanations for the inconsistent results, of which the methodological differences and the currently incomplete knowledge of the genetic architecture underlying ADHD receive the most attention.
One explanation is that the overall effect of the SNPs that impact ADHD is relatively small. Moreover, the allele frequency of rare variants is below the detection threshold, because linkage disequilibrium (LD) is low; therefore, the GWAS do not have enough statistical power to detect them (Hong and Park, 2012; Visscher et al., 2017). Even in the mega-analysis summarizing the currently available GWAS results, there were only 12 significant hits in a sample over 20,000 cases (Demontis et al., 2019). Therefore, rare variants (having an allele frequency lower than 0.05) that are potentially related to ADHD, such as copy number variants (CNVs), which are genomic segments ranging from 1 Kb to several Mb in the DNA occurring as multiple copies or deletions of a certain chromosomal section, and single nucleotide variants (SNVs) have become the focus of research. The role of CNVs in both childhood (Williams et al., 2010; Stergiakouli et al., 2012; Martin et al., 2015) and adult ADHD (Ramos-Quiroga et al., 2014) has been found to be significant in several studies, and a significant overlap with the loci previously identified in ASD and schizophrenia has been demonstrated (Thapar et al., 2016; Gudmundsson et al., 2019). Overall, whereas the high heritability of ADHD can be estimated with confidence, the underlying genetic factors are complex, both common and rare variants play an important role in the susceptibility to the disorder.
Although genetic effects are more pronounced in the etiology of ADHD, 22% of phenotypic variance can be attributed to environmental factors (Faraone et al., 2005; Nikolas and Burt, 2010). In the case of certain genes, the gene expression and thus the phenotypic trait are influenced by environmental and consequential epigenetic effects. Epidemiological studies have identified several environmental risk factors in the background of ADHD, which include maternal substance use, stress, the presence of environmental toxins in the prenatal or perinatal period, and low birth weight. Similarly, ADHD risk was increased in preterm birth, in the presence of psychosocial adversities during early childhood, or in the case of suboptimal nutritional factors (Loche and Ozanne, 2016). These results significantly overlap with the results of epigenetic studies, although the risk factors associated with the disorder are not necessarily causal. Studies to date have shown that maternal smoking, alcohol and substance use, and also suboptimal nutrition can have a significant effect on transcription. In adolescence, increased stress, trauma, and abuse play a role in the epigenetic modifications. Besides the epigenetic effects, which are exerted through chemical modifications of DNA molecules, such as cytosine methylation, histone modifications, or RNA-mediated modifications, there are candidate genes for which the presence of a certain variation in itself is associated with increased environmental vulnerability to ADHD. Examples include interactions between dopamine receptor D4 (DRD4) and maternal smoking (Pluess et al., 2009), dopamine transporter (DAT1) and maternal alcohol consumption during pregnancy (Brookes et al., 2006), or between serotonin transporter (5HTT) and adverse psychosocial events (Müller et al., 2008) (see review Palladino et al., 2019).
The clinical diagnosis of ADHD is based on the lifelong presence of the core symptom domains such as inattention, hyperactivity, and impulsivity, which are associated with functional impairment in daily life, and the onset of these symptoms in childhood. Although the heritability of ADHD dimensional traits is somewhat lower than that of ADHD itself, clinically diagnosed ADHD and population traits of the disorder overlap significantly in terms of genetic risk (Demontis et al., 2019).
Research by Taylor et al. (2019) used a joint categorical/continuous twin method to estimate the genetic correlation between psychiatric diagnoses (ADHD, Tic disorders, obsessive–compulsive disorder, anxiety, major depressive disorder, and schizophrenia), and the corresponding continuous traits of these disorders offered a better understanding of the partial discrepancy. Subsequent to examining the association between PRS for each disorder and associated traits, additional analyses were done after excluding individuals diagnosed with the relevant psychiatric disorder. Authors found that ADHD PRS was associated with ADHD population traits [ß (SE) = 0.27 (0.03)] at the age of 9 years. The phenotypic correlation between ADHD diagnosis and ADHD-related traits was 0.52 (0.50–0.54), and both ADHD dimensions displayed moderate phenotypic (mean estimate: 0.49; range: 0.45–0.53) and genetic (mean estimate: 0.53; range: 0.49–0.57) correlations with ADHD diagnosis. Cross-trait analyses yielded a correlation of 0.47 (0.43–0.51) in monozygotic (MZ) and 0.17 (0.13–0.21) for dizygotic (DZ) samples (Taylor et al., 2019).
Besides the high genetic correlation observed between ADHD case–control status and ADHD-related traits (Bidwell et al., 2017; Demontis et al., 2019), the symptom dimensions of ADHD also show a significant genetic correlation with each other [rg (SE) = 0.73 (0.08)] (Ebejer et al., 2013; Bidwell et al., 2017). Consequently, distinguishing between the clinical presentations of ADHD is of practical importance due to the differences in the lifelong continuation of the symptoms, the nature of the related functional impairment, and also the vulnerability to other psychiatric comorbidities which impact treatment decisions (Lahey et al., 2002). Nevertheless, current genetic results are more in favor of a trait-based, rather than a cutoff (the presence or absence of ADHD diagnosis) approach, whereby the symptoms are considered as quantitaive traits along a dimensional spectrum, with ADHD itself corresponding to extremes of the spectrum in which the symptom is present with sufficient intensity, and continuity over time required for the diagnosis (Larsson et al., 2012). Based on current studies, ADHD PRS values, that is, an individual estimate of overall SNP effects, have been consistently associated with ADHD diagnosis and functions in a dose-dependent manner (Ronald et al., 2021). ADHD PRS was significantly associated with ADHD dimensional symptom severity scores both in clinical and population samples (Albaugh et al., 2019; Burton et al., 2019; Stojanovski et al., 2019; Nigg et al., 2020) in parent-report (Nigg et al., 2018), self-report (Burton et al., 2019), and also teacher-rated scales (de Zeeuw et al., 2020) (see review Ronald et al., 2021). As gene–environment interactions play an important role in the background of ADHD symptom severity, the study of Selzam et al. (2019) using DZ twin samples and comparing between-family and within-family factors is particularly interesting. The between-family ADHD PRS effect, which was estimated independent of the within-family effect, significantly predicted more ADHD traits. The within-family ADHD PRS effect showed that, within pairs, the twin with higher ADHD PRS had more ADHD traits than their cotwins. Another remarkable finding is that the ADHD genome-wide polygenic score within-family prediction was significantly lower than between-family prediction for educational achievement, and the between-family ADHD PRS on educational achievement was significantly reduced when socioeconomic status was controlled for, but remained significant (Selzam et al., 2019).
Based on the results of recent studies, ADHD PRS is also positively associated with ADHD-related traits. In two studies, significant association was found with both symptom dimensions (inattention and hyperactivity/impulsivity) (Burton et al., 2019; Taylor et al., 2019), whereas in two other studies, association only with hyperactivity/impulsivity was reported (Sudre et al., 2020; Vuijk et al., 2020). The heterogeneity of the symptoms, the dynamic changes in the ADHD symptom presentation over the lifetime of the patient, or the presence of subthreshold symptoms in the other symptom domain, which may not cause functional impairment at a given life period, are also consistent with this result (5th ed.; DSM-5; American Psychiatric Association, 2013; Stojanovski et al., 2019).
Several genetic findings, such as the genetic correlation for both common and rare variants, significantly overlap with ASD (Grove et al., 2019; Satterstrom et al., 2019), or the correlation between ADHD and lower IQ underscored the neurodevelopmental nature of ADHD (Frazier et al., 2004; Pinares-Garcia et al., 2018). A meta-analysis of longitudinal studies found that at least 15% of children diagnosed with ADHD continue to meet diagnostic criteria at age of 25, and further 50% of the cases reach only partial remission still causing impairment in their everyday life (Faraone et al., 2006). As the heritability of ADHD is stable across the lifespan (Kuntsi et al., 2005; Kan et al., 2013; Brikell et al., 2015), there is an emerging research interest in capturing persistence at the genetic level. In a well-powered meta-analysis of GWAS results, which included 17,149 cases and 32,411 controls, evolving childhood ADHD and adult ADHD cases were analyzed separately and jointly (Rovira et al., 2020). Nine independent loci were identified that overlapped in the childhood and adult ADHD groups, all of which were common variants playing a role in certain stages of brain development such as neuronal migration, myelination, or diencephalon development. These hits did not overlap with the results of previous candidate gene studies. In this study, the genetic correlation between childhood ADHD and adult persistent ADHD was found to be remarkably high (rg = 0.81, CI: 95% 0.64–0.94). Further analysis was aimed at investigating whether latent persistent cases genetically differed from the non-persistent cases; however, variants specifically associated with ADHD persistence could not be identified (Rovira et al., 2020), and persistence was also independent from gender (Caye et al., 2016). Given the familial aggregation of ADHD persistence demonstrated in earlier studies, the notion arises that rare variants and also gene–environment interactions may also play a role in the genetic background of persistence. Since persistence is a dynamic process over time, longitudinal studies with the potential of examining remitting cases could provide more insight into the potential gene by age interactions (Kuntsi et al., 2005; Chang et al., 2013; Thissen et al., 2015).
The evolutionary perspective of the ADHD symptoms raises the question why these symptoms have not bred out through the processes of natural selection despite that they are related to adverse functional outcomes (Keller and Miller, 2006; Nesse, 2006). One explanation for this – also known as the mismatch theory (Crawford and Salmon, 2002; Durisko et al., 2016) or the anachronism of ADHD (Arcos-Burgos and Acosta, 2007) – could be that changes in human societies have occurred so rapidly that they have outpaced the much slower evolutionary changes required to select for these traits (Jensen et al., 1997). Another potential approach is the natural positive selection theory (Thagaard et al., 2016), which suggests that ADHD traits may have been beneficial in certain circumstances. For example, in a hunter-gatherer environment which is typically characterized by the depletion of resources, and also time-critical and novel-rapidly changing conditions, hyperactivity could have been advantageous in spotting new opportunities or migrating toward better climates, whereas impulsivity as related to the response-readiness and the ability to fight-or-flee, and inattention, as a high-scanning behavior could have likely been adaptive under these environmental conditions (Jensen et al., 1997). Results of two studies (Ding et al., 2002; Wang et al., 2004) support this theory as they found that the increased 7R frequency in DRD4 gene related to ADHD may be the result of positive selection since it is associated with both ADHD and the personality trait of novelty seeking (Benjamin et al., 1996; Gizer et al., 2009). An alternative hypothesis for this positive selection is simply that ADHD traits have persisted because they increased reproductive success, as it is well known that compared with individuals without ADHD, patients are more likely to be younger at first sexual intercourse, to have more sexual partners and to be involved in teenage parenthood (Barkley et al., 2006; Flory et al., 2006; Østergaard et al., 2017; Esteller-Cucala et al., 2020).
The findings in ASD substantiating that the PRS for ASD is positively correlated with general cognitive ability in the general population (Clarke et al., 2016) are also thought-provoking in an evolutionary context, especially given the considerable overlap between the ASD and the ADHD phenotype. However, in their study, Clarke et al. (2016) did not find consistent evidence in support of the association between the polygenic risk for ADHD and cognitive function. Beyond the theoretical hypotheses, behavioral observations are of major importance. Arildskov et al. (2021) applied behavioral tests to mimic ancestral environmental conditions. Studying school age children in a response-readiness laboratory test, continuous ADHD traits (measured by the ADHD-RS-IV) were not found to be related to the test performance. Nevertheless, it is important to emphasize the limitation of such studies, namely that it is difficult to create a situation mimicking all ancestral environmental conditions that can have all potential effects on the performance; moreover, there could be other ADHD-related advantages of these traits, which may not be in the focus of the given study.
It is estimated that around 60–100% of children with ADHD also exhibit one or more comorbid disorders that often continue into adulthood complicating the diagnosis and the treatment (Biederman et al., 1993; Gillberg et al., 2004). When compared with non-ADHD subjects, significantly higher rates of comorbid major depression, dysthymia, bipolar disorder, anxiety disorders, substance use disorders, and personality disorders were consistently reported in adults with ADHD (Jacob et al., 2007; Sobanski et al., 2007; Cumyn et al., 2009; Asherson et al., 2014; Perroud et al., 2014; Bitter et al., 2019). Results of cross-disorder studies show that there is a significant genetic overlap between the genetic backgrounds of mental disorders, the so-called general psychopathological factor accounting for 10–57% of the phenotypic variance (Lahey et al., 2011; Caspi et al., 2014; Pettersson et al., 2016; Waldman et al., 2016; Selzam et al., 2018; Allegrini et al., 2020; Brikell et al., 2020). One possible explanation is that the different clinical phenotypes are the consequence of the highly pleiotropic effects of the genetic variants that contribute to the risk for developing psychiatric disorders. Another hypothesis is that these variants primarily define traits that are present in a subclinical form in many diseases, whereas in the case of other disorders, their expression becomes more dominant (Plomin et al., 2009). For example, in terms of psychiatric vulnerability, ADHD PRS has been shown to be positively associated with sensation-seeking, risk-taking behavior, and irritability (Du Rietz et al., 2018; Riglin et al., 2019; Nigg et al., 2020). PRS differences within the ADHD groups based on the clinical presence of emotional lability or different impulsivity factors suggest additional clinical subtypes with different genetic risks (Grimm et al., 2020; Nigg et al., 2020).
Observations of family and twin studies indicated a significant genetic overlap between ADHD and ASD (Rommelse et al., 2010; Ghirardi et al., 2018), schizophrenia (Larsson et al., 2013), bipolar disorder (Faraone et al., 2012), major depression (Faraone and Biederman, 1997), and antisocial personality disorder (Christiansen et al., 2008). The ADHD PRS results from GWAS are only partially consistent with these findings. ADHD PRS showed a significant positive association with the general psychopathology factor in children (Riglin et al., 2019), the ADHD + bipolar disorder group was characterized by a higher PRS (only compared to the control group), and no association was found with schizophrenia. In depression, anxiety, neuroticism, and also eating disorders, the results were contradictory, in many cases showing age-related effects (Ronald et al., 2021). In a GWAS mega-analysis of the PGC Cross-Disorder Group (Lee et al., 2019), data from 232,964 cases and 494,162 controls were analyzed across eight disorders: ADHD, anorexia nervosa, ASD, schizophrenia, bipolar disorder, major depression, obsessive–compulsive disorder, and Tourette syndrome. They identified 109 pleiotropic loci confirming the significantly shared genetic background of psychiatric disorders. ADHD showed the strongest SNP-based genetic correlation with major depression (rg = 0.44), followed by a positive association with neurodevelopmental disorders (ASD rg = 0.37, Tourette syndrome rg = 0.27) ascertained by LD score regression analysis.
It is important to note that in the five studies which investigated either diagnosed autism or autistic traits in most cases, ADHD PRS did not predict these traits (Jansen et al., 2020; Serdarevic et al., 2020; Torske et al., 2020), only one study (LaBianca et al., 2021) (on autism) reported a significant positive association with the ADHD PRS, and another study (Serdarevic et al., 2020) reported a significant positive association only in male participants. ADHD PRS did not associate with other neurodevelopmental conditions; however, a forward-looking result is that in non-ADHD disorders, ADHD PRS appears to have transdiagnostic utility in characterizing subgroups of individuals with early-onset symptoms. For example, although ADHD PRS did not associate with schizophrenia, within a schizophrenia sample, it is associated with the cognitive trajectory from adolescence into adulthood, showing the strongest association with the subgroup presenting with the earliest preadolescent cognitive impairment (Dickinson et al., 2020).
The correlation between ADHD and internalizing, externalizing, and neurodevelopmental diseases has been targeted in several studies (Du Rietz et al., 2018; Demontis et al., 2019; Lee et al., 2019). In a large longitudinal study (Du Rietz et al., 2021b), ADHD indicated a significant association with all three groups (r = 0.67–0.75); however, after correcting for the general psychopathology factor, only the association with neurodevelopmental disorders remained moderately strong (r = 0.43, 95% CI: 0.42–0.45), which is largely influenced by genetic factors. The association with externalizing disorders was lower (r = 0.25, 95% CI: 0.24–0.27), which is largely influenced by environmental effects, and there was no significant association with internalizing disorders. Overall, current studies suggest that although the genetic overlap between ADHD and other psychiatric disorders is significant, it could likely be explained by general psychopathology factors, and the role of unique genetic effects may be plausible mostly between ADHD and other neurodevelopmental disorders.
The diagnosis of ADHD is hindered by the heterogeneity of the disease, variability of symptom presentation over time, and subjectivity of symptom severity, as potential confounding factors; therefore, interest in predictive biomarkers that could aid the diagnosis, prognosis, and assessment of the response to pharmacological interventions has increased substantially (Faraone et al., 2014; Mehta et al., 2020). Endophenotypes (Gottesman and Gould, 2003), a subtype of biomarkers (Biomarkers Definitions Working Group, 2001), are quantitative indicators of the biological processes underlying the disease rather than of the clinical phenotypes. These measures should be quantifiable, state independent, they are expressed regardless of whether the disorder is manifest, and they are more prevalent in the unaffected relatives of patients than in the general population. Genetic determination is a prerequisite for the definition of endophenotypes, that is, the endophenotype is heritable, the endophenotype and the disorder are associated within the family, it must show association and/or linkage with one or more of the candidate genes, and the endophenotype should mediate the association and/or the linkage between the candidate gene and the disorder. The hope of identifying endophenotypes has reoriented research interest to candidate gene studies.
Of the ADHD candidate genes linked to dopaminergic, noradrenergic, and serotonergic neurotransmitter systems (Gizer et al., 2009), most results are available for DAT1 and DRD4 in terms of various endophenotypes (Faraone et al., 2014). Summary of findings is provided in Table 1.
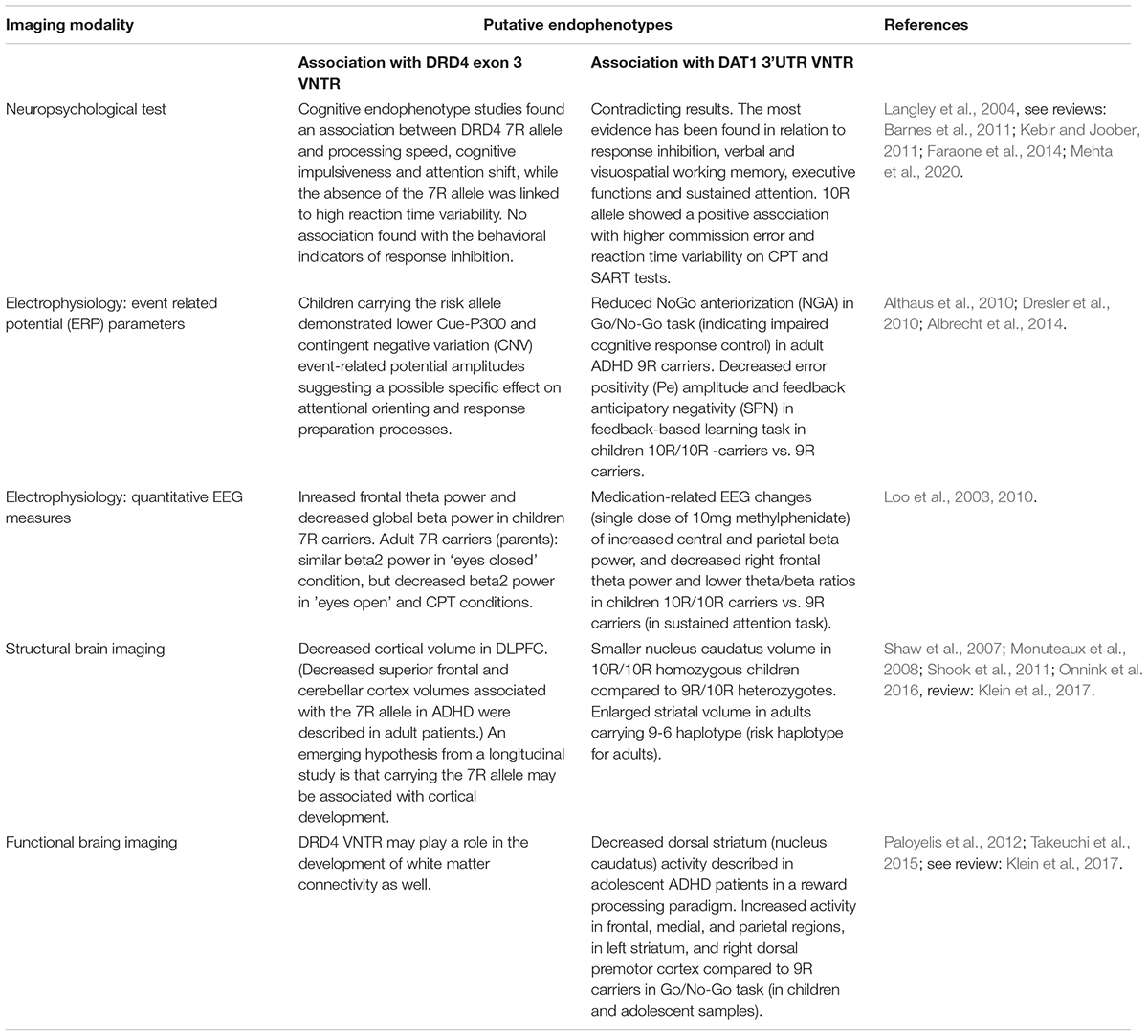
Table 1. Association of VNTR polymorphisms in candidate genes DRD4 and DAT1 with various ADHD-related neuropsychological, eletrophysiological and neuroimaging measures emerging as putative endophenotypes.
The dopamine transporter gene (DAT1) codes for the solute carrier protein responsible for the reuptake of dopamine from the synaptic cleft. Gene expression is most pronounced in the striatum. The most intensively studied variant of DAT1 is variable tandem repeats (VNTRs) of 40 base pairs located at the 3’untranslated region (3′UTR) of which 10-repeat (10R) and 9-repeat (9R) alleles occur most frequently. An additional VNTR polymorphism located in intron 8 containing 5R and 6R alleles, which has also been associated with increased susceptibility to ADHD, is also being studied as a haplotype. Interestingly, based on the previous studies, whereas the 10R/10R genotype and 10/6 haplotypes are likely to be risk factors in children (Brookes et al., 2006; Asherson et al., 2007), the 9R/9R genotypes and 9/6 haplotypes were associated with the disease in adult patients with ADHD (Franke et al., 2010). The DRD4 gene is predominantly expressed in the anterior cingulate cortex (and orbitofrontal cortex), a brain region of major importance for attentional and inhibitory processes. The most commonly studied polymorphism of the DRD4 gene is the 48-bp VNTR in exon3, the 7R allele of which is linked to an increased risk of ADHD in the Caucasian population (Wu et al., 2012).
The heterogeneity of the ADHD symptoms and the complexity of its genetic architecture have put endophenotype studies in a new context in recent years, drawing the attention to the difficulty of defining the genotype and the phenotype, let alone creating a bridge between them, which would in fact be key to a more complete understanding of the genetic or biological determinants and the clinical phenotype. The challenges associated with and the implications of linking genes to structural and functional variations in the brain systems responsible for cognition and emotion are considerable.
A major difficulty is that the methodological aspects of neuroimaging (Shaw et al., 2007; Monuteaux et al., 2008; Shook et al., 2011; Paloyelis et al., 2012; Takeuchi et al., 2015; Onnink et al., 2016; Klein et al., 2017), electrophysiological (Loo et al., 2003, 2010; Althaus et al., 2010; Dresler et al., 2010; Albrecht et al., 2014), and neuropsychological (Langley et al., 2004; Barnes et al., 2011; Kebir and Joober, 2011; Faraone et al., 2014; Mehta et al., 2020) studies, which served as potential endophenotypes, differ from the methodological preferences of genetic studies. For example, as the case–control design by gender, age, and education is of high importance for cognitive performance, the majority of these studies have low sample sizes; moreover, the heterogeneity of methodological parameters of the paradigms applied in the studies resulting in task-dependent changes in cognitive performance constitutes a limitation to the aggregation of certain phenotypic variables.
Chauvin et al. (2021) offer an overarching approach to this issue facilitating the comparison between task paradigms that are shared across multiple cognitive functions, resembling a cognitive core, from those that are task-specific (Konrad and Eickhoff, 2010; Lin et al., 2014). The results of the study (Chauvin et al., 2021) are promising for the endophenotype approach, as ADHD siblings displayed a task connectivity modulation profile that is an intermediate between diagnosed ADHD siblings and control participants; namely, they showed a similar degree of task generic connectivity modulation as controls, but significantly more task-specific connectivity modulation than ADHD probands. In the context of neuroimaging techniques, it is also important to mention that even for the same genetic mechanism, the effect size can vary widely depending on the imaging target measure (structural variation, functional activation, or functional connections) examined (Hariri et al., 2002; Meyer-Lindenberg and Weinberger, 2006).
The difficulty in selecting the optimal psychological test and test indicators is another factor that adds to the complexity of endophenotype studies. Neuropsychological tests that are characterized with higher effect sizes in patients with ADHD (Bálint et al., 2009) – such as the continuous performance tests (Conners et al., 2003) or the standardized neuropsychological measures, for example, Stroop task (Golden, 1975), the Digit Span subtest of WAIS (Wechsler, 1981) or the Wisconsin Card Sorting Test (Heaton et al., 1993) – are also widely used in endophenotype investigations; however, the results are inconsistent (Kebir and Joober, 2011; Faraone et al., 2014). A study by Acosta-López et al. (2021) offers a promising approach to overcome the gap between the genotype and the phenotype, whereby applying a family-based design and using an extensive test battery of neuropsychological tasks (Stroop test, Cross-Out-Squares Test, and Trail Making Test) and also reaction time-based task paradigms (Conners’ Continuous Performance Test and Go/No-Go Tasks (Jiménez-Figueroa et al., 2017; Jimenez-Figueroa et al., 2020); temporal processing is evaluated as a potential endophenotype in ADHD. Estimating the effect sizes for neuropsychologically based variables related to a cognitive mechanism (e.g., temporal processing) in a case–control design and the parallel estimation of the heritability of these variables within one sample group can provide variables which are likely to be relevant to both clinical (neuropsychological) and genetic perspectives.
The current results of electrophysiological studies in terms of endophenotypes are still limited. Based on the recently available twin and family studies, the resting state EEG measures are related to higher heritability rates compared with event-related potentials (Iacono, 2014). Iacono’s publication (Iacono et al., 2017) reviews a variety of electrophysiological measures and classifies them as the biomarkers or putative endophenotypes. For ADHD, moderate evidence was considered for error-related negativity (ERN) amplitude (Albrecht et al., 2008; Anokhin et al., 2008; McLoughlin et al., 2009), and suggestive evidence was considered for very low-frequency EEG activity (Tye et al., 2012), increased theta and delta power (Loo et al., 2010), and decreased beta power (Loo et al., 2010; Rudo-Hutt, 2015). From event-related EEG measures, ITPC (intertrial phase coherence) (McLoughlin et al., 2014), No-Go N2 amplitude observed in the flanker task (Albrecht et al., 2008; McLoughlin et al., 2009), and the extensively investigated amplitude of the P300 component (van Beijsterveldt and van Baal, 2002; Malone et al., 2014) have suggestive evidence in terms of the endophenotype criteria (Iacono et al., 2017). Interestingly, the ratio of theta to beta resting EEG power may constitute only a biomarker for ADHD, as it does not appear to be genetically influenced (Snyder and Hall, 2006; Arns et al., 2013).
Notably, there are additional emerging approaches such as the investigation of oscillation potential change related to a trait, since the hierarchy of brain oscillations has remained remarkably preserved during the course of mammalian evolution (Buzsáki et al., 2013). Examples include the examination of oscillatory activity in psychiatric disorders frequently associated with impaired language skills such as schizophrenia (Murphy and Benítez-Burraco, 2016) or ASD (Benítez-Burraco and Murphy, 2016). Considering that the symptoms of inattention and hyperactivity often coexist with language problems in both clinical and community samples (Hawkins et al., 2016) and the reciprocal presence of the disorder-related traits between ASD and ADHD symptoms (Baixauli-Fortea et al., 2019), examining oscillopathic alterations could be a promising opportunity to construct successful endophenotypes also in patients with ADHD. It is highly important in terms of potential electrophysiological endophenotypes to emphasize that during development, the longitudinal stability of the different EEG measures is different; consequently, the relationship between the gene and the endophenotype may not be constant across the development; therefore, the utility of the endophenotype may be limited to certain developmental periods (Poil et al., 2014; Giertuga et al., 2017).
There are also several methodological issues associated with genotyping. For example, the notion that endophenotypes are genetically less complex than psychiatric disorders is controversial and not necessarily true (Roffman, 2019). Additionally, the potential gene–gene interactions or the effect of environmental factors on gene transcription are often neglected aspects. As genetic studies have moved from the univariate candidate gene risk polymorphism approach to multimarker analysis, it is reasonable to examine the relationship between multiple genetic variables and ADHD phenotypes. To explore the genetic background of endophenotypes in genetic factors associated with psychiatric phenotypes (Iacono et al., 2017; Dick, 2018), the association between ADHD PRS and neuroimaging or neuropsychological indicators has been increasingly investigated in recent years. Of the neuropsychological indicators, the association between working memory and ADHD PRS is the most consistent result (Nigg et al., 2018; Hermosillo et al., 2020; Shen et al., 2020; Sudre et al., 2020; Torske et al., 2020; Vuijk et al., 2020); nevertheless, there are also positive findings for focused attention, delay discounting, and vigilance or arousal, whereas surprisingly, no association was found with indicators of executive functions (such as response inhibition). Due to the significant clinical overlap between ADHD and other psychiatric disorders, there is an increasing effort to define clinical phenotypes that are etiologically related. For example, considerable attention is paid to the investigation of developmental endophenotype with the intention of also elucidating whether these indicators reflect disease- or condition-specific biological pathways or may be more linked to general psychopathological processes (Gui et al., 2020).
In summary, although there have been several studies aimed at identifying endophenotypes, they still represent early attempts yielding contradictory results, and so far, none of the indicators have met all criteria required in the definition. Studies to date have had small sample sizes, differed in study design, and have been inconsistent in the interpretation of the association with the genotype. The lack of evidence substantiating a causal relationship between the genetic risk factors and the phenotype should also be highlighted, given that only few studies conducted a mediation analysis are available. A further limitation of endophenotype studies is the difficulty of capturing gene–environment interactions which are of great importance in ADHD and emphasizes the need for further longitudinal studies.
Several biological mechanisms, including dopaminergic, serotoninergic, and glutamatergic signaling, have been implicated in the etiology of ADHD (Bonvicini et al., 2016). Medications approved by the US Food and Drug Administration (FDA) (Multiple authors, 2018) for the treatment of ADHD are stimulants such as methylphenidate (MPH) and amphetamines, and non-stimulant medications such as the selective noradrenaline reuptake inhibitor atomoxetine (ATX), the alpha-2 agonists guanfacine and clonidine, and the recently approved serotonin noradrenaline-modulating viloxazine. Pharmacogenetic studies are primarily focused on predicting drug responsiveness, and given the heterogeneity of drug responses, they are also aimed at exploring whether genetic determinants determine individual treatment response (Elsayed et al., 2020). This is even more relevant considering that the current pharmacological ADHD treatment is effective only in about 70% of the cases (Jensen et al., 2007), and the effect size is 30–50% decreased in adults compared with children (Cortese et al., 2018a). Significantly, fewer studies target the investigation of the effective dose and the optimal drug dose for undesired side effects, despite them being highly relevant for therapeutic adherence (Storebø et al., 2018; Brown et al., 2019; Bousman et al., 2021).
Although only a small proportion of the vulnerability to ADHD is linked to a single gene, given the monoaminergic attack points of drugs, pharmacogenetic studies initially focused on the potential associations of candidate ADHD genes and drug efficacy. Most data are available for methylphenidate and atomoxetine. Results from a meta-analysis of childhood and adolescent ADHD indicated significant associations between several common variants, namely DAT1, DRD4 VNTRs, and SNPs in the ADRA2A, catechol-O-methyltransferase (COMT), and noradrenaline transporter (SLC6A2) genes, and MPH responsiveness (Myer et al., 2018). In addition, the latrophilin-3 gene (ADGRL3 = LPHN3) carrying the G allele has also been correlated with MPH treatment response (Arcos-Burgos et al., 2010; Labbe et al., 2012; Özaslan et al., 2021). In adult patients with ADHD, Contini et al. (2012, 2013) found no associations with DRD4, COMT, serotonin receptor 1B (HTR1B), tryptophan hydroxylase (TPH2), dopamine β-hydroxylase (DBH), 5HTT, and synaptosomal-associated protein 25 (SNAP25) genes, only with DAT1 (Contini et al., 2012; see review Contini et al., 2013). In contrast, in a pharmacogenetic review and meta-analysis specifically targeting DAT1, no association was detected between the 40-bp VNTR and the MPH response (Bonvicini et al., 2016). An additional interesting finding is the possible correlation between SNARE complex polymorphism and MPH responsiveness in an adult sample (da Silva et al., 2018).
In the case of atomoxetine, significant correlation was found between the response status in childhood ADHD and DBH and SLC6A2 in a Chinese sample, and later in a broader sample (Yang et al., 2013; Fang et al., 2015; Gul et al., 2021). Further pharmacogenetic studies of atomoxetine target the cytochrome P450 polymorphisms to determine the possible genetic factors for poor, extensive, high, and ultrarapid metabolism (Michelson et al., 2007; Trzepacz et al., 2008; Fijal et al., 2015, see review Yu et al., 2016).
A summary of pharmacogenetic studies (Mick et al., 2006; Kooij et al., 2008; Nemoda et al., 2009; Ramoz et al., 2009; Contini et al., 2010, 2011, 2012; Johnson et al., 2013; Yang et al., 2013; Fang et al., 2015; Hegvik et al., 2016; Gomez-Sanchez et al., 2017; Pagerols et al., 2017; Angyal et al., 2018; Huang et al., 2018; Naumova et al., 2019; Bonvicini et al., 2020; Gul et al., 2021; Yuan et al., 2021) assessing the most investigated candidate genes is provided in Table 2.
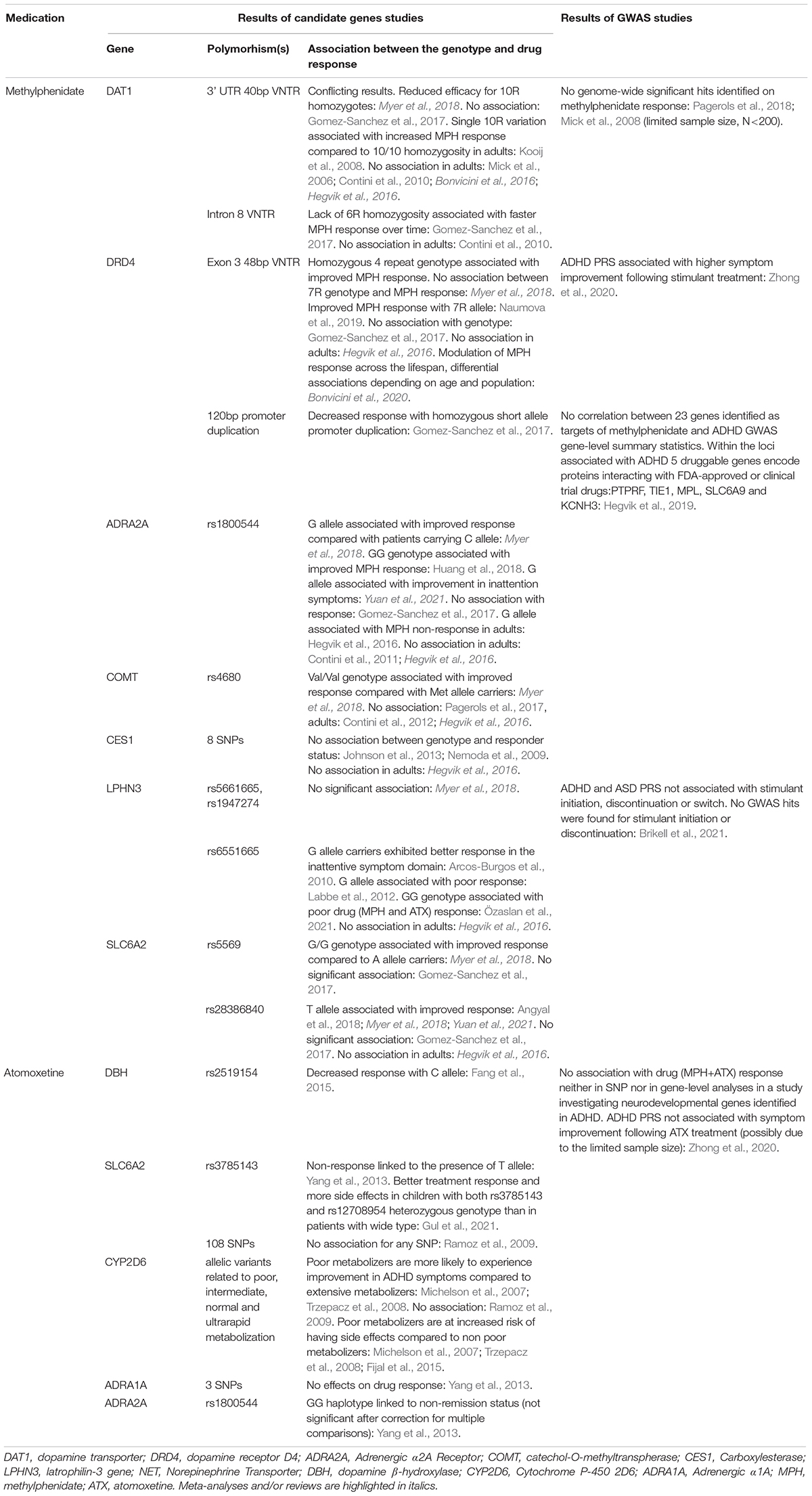
Table 2. Summary of the pharmacogenetic studies assessing the most extensively investigated candidate genes and/or using genome-wide method in ADHD patients on methylphenidate and atomoxetine treatment.
In terms of pharmacogenetic studies, genome-wide association studies, which are not hypothesis-driven, do not overlap with the results of candidate gene studies. In a GWAS study with the highest sample size and statistical power available to date (Hegvik et al., 2019), no association was found between ADHD and the effectiveness of FDA-approved first-line pharmacological agents, suggesting that they may exert their effect through mechanisms different from the ones underlying ADHD. One of the main advantages of GWAS is that they better capture the heterogeneity of ADHD and the cross-disorder nature of the characteristic traits, thereby facilitating the identification of potential new therapeutic options. It opens the possibility of drug repurposing, that is, using other drugs that have been shown to be effective in other psychiatric disorders. The successful utilization of the metabotropic glutamate receptor activating fasoracetam, an agent previously used in vascular dementia, in ADHD adolescents in whom the glutamatergic signaling is affected serves as a good example of drug-repurposing (Elia et al., 2018). The high psychiatric comorbidity of ADHD, especially in adulthood, and also the significant symptomatic and genetic overlap with other psychiatric disorders raise the possibility of another therapeutic approach whereby the medication is selected based on the presence of a characteristic trait. Symptoms that are often associated with ADHD such as emotional or affect lability for which the efficacy of current agents is inadequate can serve as the examples for this approach. Summary of findings using genome-wide method (Mick et al., 2008; Pagerols et al., 2018; Hegvik et al., 2019; Zhong et al., 2020; Brikell et al., 2021) is also provided in Table 2.
Compared with the extensive description of the psychiatric comorbidities, the somatic comorbidities of ADHD have received much less attention in the research literature despite the fact that ADHD has been associated with a broad range of medical health problems, such as obesity (Fuemmeler et al., 2011), asthma (Cortese et al., 2018b), migraine (Hansen et al., 2018), type 2 diabetes mellitus, and hypertension (Instanes et al., 2018) and also other somatic diseases. On the other hand, the significant genetic correlations between ADHD and certain somatic traits or diseases (such as weight and weight-related traits, smoking-related cancer, and reproductive traits) reveal a great degree of overlap between the genetic risk factors (Demontis et al., 2019). At this point, it is important to mention that current results support that non-coding variants, such as intronic indels, have been shown to play an important role (Liu et al., 2021) not only in identifying ADHD risk genes (Al-Mubarak et al., 2020), but also for medical diseases (Tan, 2020), as the non-coding elements can regulate the transcription and translation of protein-coding genes. Consistently, recent recommendations for complex, neurodevelopmental disorders increasingly raise the usefulness of whole genome analysis, since the identification of the underlying genetic etiology between neurodevelopmental and somatic conditions can provide a more precise clinical management impacting patient care. This may include initiation of surveillance for disease-related conditions and referrals for further evaluation of associated medical conditions or the possibility to prevent medical comorbidities that may develop later in life (Schaefer and Mendelsohn, 2013; Srivastava et al., 2020; Vanzo et al., 2020). However, there is a need for studies which help to decide whether the same genetic variants influence genetic vulnerability to multiple (psychiatric and somatic) phenotypes (horizontal/independent pleiotropy), or the genetic variants influence vulnerability to one phenotype, and that phenotype in turn causes the other phenotype (Paaby and Rockman, 2013; Verbanck et al., 2018; Vink et al., 2020; Du Rietz et al., 2021a; Zhu, 2021).
Finally, it is important to emphasize that the drugs currently used in ADHD are not curative and thus only allow symptomatic treatment of the disorder. A clearer understanding of the molecular biological background underlying ADHD would be essential for finding causal therapy, to this end, animal models and pluripotent stem cell studies could provide a promising perspective (Tong et al., 2019).
In our brief review, without claiming to be exhaustive, we intended to give an overview of the genetic results relevant to the clinical practice of ADHD and the emerging issues. The clinical need to be addressed by genetic studies can be summarized as the prediction of disease heritability, the identification of diagnostic aids, and the assessment of responsiveness to medication, thus taking into account individual characteristics in the hope of establishing an individualized treatment approach (Zayats and Neale, 2019; Grimm et al., 2020). Table 3 summarizes the potential approaches between research and the clinic. Current genetic results support the trait-based approach, namely that the symptoms of ADHD (similarly to ASD) represent the extremes of a continuous trait that varies in the population. Although the recent genetic results present some new perspectives (e.g., drug-repurposing), they still have many limitations. For example, due to the relative isolation of child and adult studies, we have little bit of knowledge about the genetic background and lifespan perspective of persistence, which is a key issue due to the neurodevelopmental nature of ADHD. The results of the studies with different approaches (candidate gene studies and GWAS) currently show few overlaps, which calls attention to the currently limited knowledge of the genetic architecture of ADHD. These gaps warrant further research and a deeper understanding of the genetic and neurobiological processes underlying ADHD. Moreover, the use of genetic testing in clinical practice should be approached cautiously to avoid the possibility of severe ethical issues.
The applicability of the current genetic results to the clinical practice is still limited; nevertheless, the focus and the methodology of the studies increasingly accommodate clinical needs. It is also important to highlight that genetic testing seems to be a promising approach for predicting response to treatment and drug tolerability; however, the study results available are still limited. In addition, a more comprehensive determination of the clinical phenotype entailing not only the core symptoms of ADHD but also the related traits and the associated somatic symptoms would prove beneficial for the routine clinical practice. In conclusion, the findings are encouraging for the long-term development of the clinical practice, which is exactly what we need to strive for.
LB drafted the first version of the manuscript. JR and AP participated in writing and critical revision of the manuscript. All authors approved the submitted version.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acosta-López, J. E., Suárez, I., Pineda, D. A., Cervantes-Henríquez, M. L., Martínez-Banfi, M. L., Lozano-Gutiérrez, S. G., et al. (2021). Impulsive and omission errors: potential temporal processing endophenotypes in ADHD. Brain. Sci. 11:1218. doi: 10.3390/brainsci11091218
Albaugh, M. D., Hudziak, J. J., Ing, A., Chaarani, B., Barker, E., Jia, T., et al. (2019). White matter microstructure is associated with hyperactive/inattentive symptomatology and polygenic risk for attention-deficit/hyperactivity disorder in a population-based sample of adolescents. Neuropsychopharmacology 44, 1597–1603. doi: 10.1038/s41386-019-0383-y
Albrecht, B., Brandeis, D., Uebel, H., Heinrich, H., Mueller, U. C., Hasselhorn, M., et al. (2008). Action monitoring in boys with attention-deficit/hyperactivity disorder, their nonaffected siblings, and normal control subjects: evidence for an endophenotype. Biol. Psychiatry 64, 615–625. doi: 10.1016/j.biopsych.2007.12.016
Albrecht, B., Brandeis, D., Uebel-von Sandersleben, H., Valko, L., Heinrich, H., Xu, X., et al. (2014). Genetics of preparation and response control in ADHD: the role of DRD4 and DAT1. J. Child. Psychol. Psychiatry 55, 914–923. doi: 10.1111/jcpp.12212
Allegrini, A. G., Cheesman, R., Rimfeld, K., Selzam, S., Pingault, J.-B., Eley, T. C., et al. (2020). The p factor: genetic analyses support a general dimension of psychopathology in childhood and adolescence. J. Child. Psychol. Psychiatry 61, 30–39. doi: 10.1111/jcpp.13113
Al-Mubarak, B. R., Omar, A., Baz, B., Al-Abdulaziz, B., Magrashi, A. I., Al-Yemni, E., et al. (2020). Whole exome sequencing in ADHD trios from single and multi-incident families implicates new candidate genes and highlights polygenic transmission. Eur. J. Hum. Genet. 28, 1098–1110. doi: 10.1038/s41431-020-0619-7
Althaus, M., Groen, Y., Wijers, A. A., Minderaa, R. B., Kema, I. P., Dijck, J. D., et al. (2010). Variants of the SLC6A3 (DAT1) polymorphism affect performance monitoring-related cortical evoked potentials that are associated with ADHD. Biol. Psychol. 85, 19–32. doi: 10.1016/j.biopsycho.2010.04.007
Angyal, N., Horvath, E. Z., Tarnok, Z., Richman, M. J., Bognar, E., Lakatos, K., et al. (2018). Association analysis of norepinephrine transporter polymorphisms and methylphenidate response in ADHD patients. Prog. Neurpsychopharmacol. Biol. Psychiatry 84, 122–128. doi: 10.1016/j.pnpbp.2018.01.013
Anokhin, A. P., Golosheykin, S., and Heath, A. C. (2008). Heritability of frontal brain function related to action monitoring. Psychophysiology 45, 524–534. doi: 10.1111/j.1469-8986.2008.00664.x
Arcos-Burgos, M., and Acosta, M. T. (2007). Tuning major gene variants conditioning human behavior: the anachronism of ADHD. Curr. Opin. Genet. Dev. 17, 234–238. doi: 10.1016/j.gde.2007.04.011
Arcos-Burgos, M., Jain, M., Acosta, M. T., Shively, S., Stanescu, H., Wallis, D., et al. (2010). A common variant of the latrophilin 3 gene, LPHN3, confers susceptibility to ADHD and predicts effectiveness of stimulant medication. Mol. Psychiatry. 15, 1053–1066. doi: 10.1038/mp.2010.6
Arildskov, T. W., Virring, A., Thomsen, P. H., and Østergaard, S. D. (2021). Testing the evolutionary advantage theory of attention-deficit/hyperactivity disorder traits. Eur. Child. Adolesc. Psychiatry doi: 10.1007/s00787-020-01692-4 Online ahead of print
Arns, M., Conners, C. K., and Kraemer, H. C. (2013). A decade of EEG theta/beta ratio research in ADHD: a meta-analysis. J. Atten. Disord. 17, 374–383. doi: 10.1177/1087054712460087
Asherson, P., Brookes, K., Franke, B., Chen, W., Gill, M., Ebstein, R. P., et al. (2007). Confirmation that a specific haplotype of the dopamine transporter gene is associated with combined-type ADHD. Am. J. Psychiatry 164, 674–677. doi: 10.1176/ajp.2007.164.4.674
Asherson, P., Young, A. H., Eich-Höchli, D., Moran, P., Porsdal, V., and Deberdt, W. (2014). Differential diagnosis, comorbidity, and treatment of attention-deficit/hyperactivity disorder in relation to bipolar disorder or borderline personality disorder in adults. Curr. Med. Res. Opin. 30, 1657–1672. doi: 10.1185/03007995.2014.915800
American Psychiatric Association (2013). Diagnostic and Statistical Manual of Mental Disorders, 5th Edn. Arlington, VA: American Psychiatric Association. doi: 10.1176/appi.books.9780890425596
Baixauli-Fortea, I., Miranda Casas, A., Berenguer-Forner, C., Colomer-Diago, C., and Roselló-Miranda, B. (2019). Pragmatic competence of children with autism spectrum disorder. Impact of theory of mind, verbal working memory, ADHD symptoms, and structural language. Appl. Neuropsychol. Child. 8, 101–112. doi: 10.1080/21622965.2017.1392861
Bálint, S., Czobor, P., Komlósi, S., Mészáros, A., Simon, V., and Bitter, I. (2009). Attention deficit hyperactivity disorder (ADHD): gender- and age-related differences in neurocognition. Psychol. Med. 39, 1337–1345. doi: 10.1017/s0033291708004236
Barkley, R. A., Fischer, M., Smallish, L., and Fletcher, K. (2006). Young adult outcome of hyperactive children: adaptive functioning in major life activities. J. Am. Acad. Child. Adolesc. Psychiatry 45, 192–202. doi: 10.1097/01.chi.0000189134.97436.e2
Barnes, J. J., Dean, A. J., Nandam, L. S., O’Connell, R. G., and Bellgrove, M. A. (2011). The molecular genetics of executive function: role of monoamine system genes. Biol. Psychiatry 69, e127–e143. doi: 10.1016/j.biopsych.2010.12.040
Benítez-Burraco, A., and Murphy, E. (2016). The oscillopathic nature of language deficits in autism: from genes to language evolution. Front. Hum. Neurosci. 10:120. doi: 10.3389/fnhum.2016.00120
Benjamin, J., Li, L., Patterson, C., Greenberg, B. D., Murphy, D. L., and Hamer, D. H. (1996). Population and familial association between the D4 dopamine receptor gene and measures of Novelty Seeking. Nat. Genet. 12, 81–84. doi: 10.1038/ng0196-81
Bidwell, L. C., Gray, J. C., Weafer, J., Palmer, A. A., de Wit, H., and MacKillop, J. (2017). Genetic influences on ADHD symptom dimensions: examination of a priori candidates, gene-based tests, genome-wide variation, and SNP heritability. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 174, 458–466. doi: 10.1002/ajmg.b.32535
Biederman, J. (2005). Attention-deficit/hyperactivity disorder: a selective overview. Biol. Psychiatry 57, 1215–1220. doi: 10.1016/j.biopsych.2004.10.020
Biederman, J., Faraone, S. V., Keenan, K., Knee, D., and Tsuang, M. T. (1990). Family-genetic and psychosocial risk factors in DSM-III attention deficit disorder. J. Am. Acad. Child. Adolesc. Psychiatry 29, 526–533. doi: 10.1097/00004583-199007000-00004
Biederman, J., Faraone, S. V., Spencer, T., Wilens, T., Norman, D., Lapey, K. A., et al. (1993). Patterns of psychiatric comorbidity, cognition, and psychosocial functioning in adults with attention deficit hyperactivity disorder. Am. J. Psychiatry 150, 1792–1798. doi: 10.1176/ajp.150.12.1792
Biomarkers Definitions Working Group (2001). Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin. Pharmacol. Ther. 69, 89–95. doi: 10.1067/mcp.2001.113989
Bitter, I., Mohr, P., Balogh, L., Látalová, K., Kakuszi, B., Stopková, P., et al. (2019). ADHD: a hidden comorbidity in adult psychiatric patients. Atten. Defic. Hyperact. Disord. 11, 83–89. doi: 10.1007/s12402-019-00285-9
Bonvicini, C., Cortese, S., Maj, C., Baune, B. T., Faraone, S. V., and Scassellati, C. (2020). DRD4 48bp multiallelic variants as age-population-specific biomarkers in attention-deficit/hyperactivity disorder. Transl. Psychiatry 10:70. doi: 10.1038/s41398-020-0755-4
Bonvicini, C., Faraone, S. V., and Scassellati, C. (2016). Attention-deficit hyperactivity disorder in adults: a systematic review and meta-analysis of genetic, pharmacogenetic and biochemical studies. Mol. Psychiatry 21:1643. doi: 10.1038/mp.2016.128
Bousman, C. A., Bengesser, S. A., Aitchison, K. J., Amare, A. T., Aschauer, H., Baune, B. T., et al. (2021). Review and consensus on pharmacogenomic testing in psychiatry. Pharmacopsychiatry 54, 5–17. doi: 10.1055/a-1288-1061
Brikell, I., Kuja-Halkola, R., and Larsson, H. (2015). Heritability of attention-deficit hyperactivity disorder in adults. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 168, 406–413. doi: 10.1002/ajmg.b.32335
Brikell, I., Larsson, H., Lu, Y., Pettersson, E., Chen, Q., Kuja-Halkola, R., et al. (2020). The contribution of common genetic risk variants for ADHD to a general factor of childhood psychopathology. Mol. Psychiatry 25, 1809–1821. doi: 10.1038/s41380-018-0109-2
Brikell, I., Wimberley, T., Albiñana, C., Pedersen, E. M., Vilhjálmsson, B. J., Agerbo, E., et al. (2021). Genetic, clinical, and sociodemographic factors associated with stimulant treatment outcomes in ADHD. Am. J. Psychiatry 178, 854–864. doi: 10.1176/appi.ajp.2020.20121686
Brookes, K. J., Mill, J., Guindalini, C., Curran, S., Xu, X., Knight, J., et al. (2006). A common haplotype of the dopamine transporter gene associated with attention-deficit/hyperactivity disorder and interacting with maternal use of alcohol during pregnancy. Arch. Gen. Psychiatry 63, 74–81. doi: 10.1001/archpsyc.63.1.74
Brown, J. T., Bishop, J. R., Sangkuhl, K., Nurmi, E. L., Mueller, D. J., Dinh, J. C., et al. (2019). Clinical Pharmacogenetics implementation consortium guideline for cytochrome P450 (CYP)2D6 genotype and atomoxetine therapy. Clin. Pharmacol. Ther. 106, 94–102. doi: 10.1002/cpt.1409
Burton, C. L., Wright, L., Shan, J., Xiao, B., Dupuis, A., Goodale, T., et al. (2019). SWAN scale for ADHD trait-based genetic research: a validity and polygenic risk study. J. Child. Psychol. Psychiatry 60, 988–997. doi: 10.1111/jcpp.13032
Buzsáki, G., Logothetis, N., and Singer, W. (2013). Scaling brain size, keeping timing: evolutionary preservation of brain rhythms. Neuron 80, 751–764. doi: 10.1016/j.neuron.2013.10.002
Caspi, A., Houts, R. M., Belsky, D. W., Goldman-Mellor, S. J., Harrington, H., Israel, S., et al. (2014). The p factor:one general psychopathology factor in the structure of psychiatric disorders? Clin. Psychol. Sci. 2, 119–137. doi: 10.1177/2167702613497473
Caye, A., Spadini, A. V., Karam, R. G., Grevet, E. H., Rovaris, D. L., Bau, C. H. D., et al. (2016). Predictors of persistence of ADHD into adulthood: a systematic review of the literature and meta-analysis. Eur. Child. Adolesc. Psychiatry 25, 1151–1159. doi: 10.1007/s00787-016-0831-8
Chang, Z., Lichtenstein, P., Asherson, P. J., and Larsson, H. (2013). Developmental twin study of attention problems: high heritabilities throughout development. JAMA Psychiatry 70, 311–318. doi: 10.1001/jamapsychiatry.2013.287
Chauvin, R. J., Buitelaar, J. K., Sprooten, E., Oldehinkel, M., Franke, B., Hartman, C., et al. (2021). Task-generic and task-specific connectivity modulations in the ADHD brain: an integrated analysis across multiple tasks. Transl. Psychiatry 11:159. doi: 10.1038/s41398-021-01284-z
Christiansen, H., Chen, W., Oades, R. D., Asherson, P., Taylor, E. A., Lasky-Su, J., et al. (2008). Co-transmission of conduct problems with attention-deficit/hyperactivity disorder: familial evidence for a distinct disorder. J. Neural. Transm. 115, 163–175. doi: 10.1007/s00702-007-0837-y
Clarke, T. K., Lupton, M. K., Fernandez-Pujals, A. M., Starr, J., Davies, G., Cox, S., et al. (2016). Common polygenic risk for autism spectrum disorder (ASD) is associated with cognitive ability in the general population. Mol. Psychiatry 21, 419–425. doi: 10.1038/mp.2015.12
Conners, C. K., Epstein, J. N., Angold, A., and Klaric, J. (2003). Continuous performance test performance in a normative epidemiological sample. J. Abnorm. Child. Psychol. 31, 555–562. doi: 10.1023/a:1025457300409
Contini, V., Rovaris, D. L., Victor, M. M., Grevet, E. H., Rohde, L. A., and Bau, C. H. (2013). Pharmacogenetics of response to methylphenidate in adult patients with attention-deficit/hyperactivity disorder (ADHD): a systematic review. Eur. Neuropsychopharmacol. 23, 555–560. doi: 10.1016/j.euroneuro.2012.05.006
Contini, V., Victor, M. M., Bertuzzi, G. P., Salgado, C. A., Picon, F. A., Grevet, E. H., et al. (2012). No significant association between genetic variants in 7 candidate genes and response to methylphenidate treatment in adult patients with ADHD. J. Clin. Psychopharmacol. 32, 820–823. doi: 10.1097/JCP.0b013e318270e727
Contini, V., Victor, M. M., Cerqueira, C. C., Polina, E. R., Grevet, E. H., Salgado, C. A., et al. (2011). Adrenergic α2A receptor gene is not associated with methylphenidate response in adults with ADHD. Eur. Arch. Psychiatry. Clin. Neurosci. 261, 205–211. doi: 10.1007/s00406-010-0172-4
Contini, V., Victor, M. M., Marques, F. Z., Bertuzzi, G. P., Salgado, C. A., Silva, K. L., et al. (2010). Response to methylphenidate is not influenced by DAT1 polymorphisms in a sample of Brazilian adult patients with ADHD. J. Neural. Transm. 117, 269–276. doi: 10.1007/s00702-009-0362-2
Cortese, S., Adamo, N., Del Giovane, C., Mohr-Jensen, C., Hayes, A. J., Carucci, S., et al. (2018a). Comparative efficacy and tolerability of medications for attention-deficit hyperactivity disorder in children, adolescents, and adults: a systematic review and network meta-analysis. Lancet Psychiatry 5, 727–738. doi: 10.1016/s2215-0366(18)30269-4
Cortese, S., Sun, S., Zhang, J., Sharma, E., Chang, Z., Kuja-Halkola, R., et al. (2018b). Association between attention deficit hyperactivity disorder and asthma: a systematic review and meta-analysis and a Swedish population-based study. Lancet Psychiatry 5, 717–726. doi: 10.1016/s2215-0366(18)30224-4
Crawford, C., and Salmon, C. (2002). Psychopathology or adaptation? Genetic and evolutionary perspectives on individual differences and psychopathology. Neuro. Endocrinol. Lett. 23(Suppl. 4), 39–45.
Cumyn, L., French, L., and Hechtman, L. (2009). Comorbidity in adults with attention-deficit hyperactivity disorder. Can. J. Psychiatry 54, 673–683. doi: 10.1177/070674370905401004
da Silva, B. S., Cupertino, R. B., Rovaris, D. L., Schuch, J. B., Kappel, D. B., Müller, D., et al. (2018). Exocytosis-related genes and response to methylphenidate treatment in adults with ADHD. Mol. Psychiatry 23, 1446–1452. doi: 10.1038/mp.2017.90
de Zeeuw, E. L., Hottenga, J. J., Ouwens, K. G., Dolan, C. V., Ehli, E. A., Davies, G. E., et al. (2020). Intergenerational transmission of education and ADHD: effects of parental genotypes. Behav. Genet. 50, 221–232. doi: 10.1007/s10519-020-09992-w
Demontis, D., Walters, R. K., Martin, J., Mattheisen, M., Als, T. D., Agerbo, E., et al. (2019). Discovery of the first genome-wide significant risk loci for attention deficit/hyperactivity disorder. Nat. Genet. 51, 63–75. doi: 10.1038/s41588-018-0269-7
Dick, D. M. (2018). Mapping risk from genes to behavior: the enduring and evolving influence of irving gottesman’s endophenotype concept. Twin. Res. Hum. Genet. 21, 306–309. doi: 10.1017/thg.2018.35
Dickinson, D., Zaidman, S. R., Giangrande, E. J., Eisenberg, D. P., Gregory, M. D., and Berman, K. F. (2020). Distinct polygenic score profiles in schizophrenia subgroups with different trajectories of cognitive development. Am. J. Psychiatry 177, 298–307. doi: 10.1176/appi.ajp.2019.19050527
Ding, Y. C., Chi, H. C., Grady, D. L., Morishima, A., Kidd, J. R., Kidd, K. K., et al. (2002). Evidence of positive selection acting at the human dopamine receptor D4 gene locus. Proc. Natl. Acad. Sci. U.S.A. 99, 309–314. doi: 10.1073/pnas.012464099
Dresler, T., Ehlis, A. C., Heinzel, S., Renner, T. J., Reif, A., Baehne, C. G., et al. (2010). Dopamine transporter (SLC6A3) genotype impacts neurophysiological correlates of cognitive response control in an adult sample of patients with ADHD. Neuropsychopharmacology 35, 2193–2202. doi: 10.1038/npp.2010.91
Du Rietz, E., Coleman, J., Glanville, K., Choi, S. W., O’Reilly, P. F., and Kuntsi, J. (2018). Association of polygenic risk for attention-deficit/hyperactivity disorder with co-occurring traits and disorders. Biol. Psychiatry. Cogn. Neurosci. Neuroimaging 3, 635–643. doi: 10.1016/j.bpsc.2017.11.013
Du Rietz, E., Pettersson, E., Brikell, I., Ghirardi, L., Chen, Q., Hartman, C., et al. (2021b). Overlap between attention-deficit hyperactivity disorder and neurodevelopmental, externalising and internalising disorders: separating unique from general psychopathology effects. Br. J. Psychiatry 218, 35–42. doi: 10.1192/bjp.2020.152
Du Rietz, E., Brikell, I., Butwicka, A., Leone, M., Chang, Z., Cortese, S., et al. (2021a). Mapping phenotypic and aetiological associations between ADHD and physical conditions in adulthood in Sweden: a genetically informed register study. Lancet Psychiatry 8, 774–783. doi: 10.1016/s2215-0366(21)00171-1
Durisko, Z., Mulsant, B. H., McKenzie, K., and Andrews, P. W. (2016). Using evolutionary theory to guide mental health research. Can. J. Psychiatry 61, 159–165. doi: 10.1177/0706743716632517
Ebejer, J. L., Duffy, D. L., van der Werf, J., Wright, M. J., Montgomery, G., Gillespie, N. A., et al. (2013). Genome-wide association study of inattention and hyperactivity-impulsivity measured as quantitative traits. Twin. Res. Hum. Genet. 16, 560–574. doi: 10.1017/thg.2013.12
Elia, J., Ungal, G., Kao, C., Ambrosini, A., De Jesus-Rosario, N., Larsen, L., et al. (2018). Fasoracetam in adolescents with ADHD and glutamatergic gene network variants disrupting mGluR neurotransmitter signaling. Nat. Commun. 9:4. doi: 10.1038/s41467-017-02244-2
Elsayed, N. A., Yamamoto, K. M., and Froehlich, T. E. (2020). Genetic influence on efficacy of pharmacotherapy for pediatric attention-deficit/hyperactivity disorder: overview and current status of research. CNS Drugs 34, 389–414. doi: 10.1007/s40263-020-00702-y
Esteller-Cucala, P., Maceda, I., Børglum, A. D., Demontis, D., Faraone, S. V., Cormand, B., et al. (2020). Genomic analysis of the natural history of attention-deficit/hyperactivity disorder using Neanderthal and ancient Homo sapiens samples. Sci. Rep. 10:8622. doi: 10.1038/s41598-020-65322-4
Fang, Y., Ji, N., Cao, Q., Su, Y., Chen, M., Wang, Y., et al. (2015). Variants of dopamine beta hydroxylase gene moderate atomoxetine response in children with attention-deficit/hyperactivity disorder. J. Child. Adolesc. Psychopharmacol. 25, 625–632. doi: 10.1089/cap.2014.0178
Faraone, S. V., and Biederman, J. (1997). Do attention deficit hyperactivity disorder and major depression share familial risk factors? J. Nerv. Ment. Dis. 185, 533–541. doi: 10.1097/00005053-199709000-00001
Faraone, S. V., and Larsson, H. (2019). Genetics of attention deficit hyperactivity disorder. Mol. Psychiatry 24, 562–575. doi: 10.1038/s41380-018-0070-0
Faraone, S. V., Biederman, J., and Mick, E. (2006). The age-dependent decline of attention deficit hyperactivity disorder: a meta-analysis of follow-up studies. Psychol. Med. 36, 159–165. doi: 10.1017/s003329170500471x
Faraone, S. V., Biederman, J., and Wozniak, J. (2012). Examining the comorbidity between attention deficit hyperactivity disorder and bipolar I disorder: a meta-analysis of family genetic studies. Am. J. Psychiatry 169, 1256–1266. doi: 10.1176/appi.ajp.2012.12010087
Faraone, S. V., Bonvicini, C., and Scassellati, C. (2014). Biomarkers in the diagnosis of ADHD–promising directions. Curr. Psychiatry. Rep. 16:497. doi: 10.1007/s11920-014-0497-1
Faraone, S. V., Perlis, R. H., Doyle, A. E., Smoller, J. W., Goralnick, J. J., Holmgren, M. A., et al. (2005). Molecular genetics of attention-deficit/hyperactivity disorder. Biol. Psychiatry 57, 1313–1323. doi: 10.1016/j.biopsych.2004.11.024
Fijal, B. A., Guo, Y., Li, S. G., Ahl, J., Goto, T., Tanaka, Y., et al. (2015). CYP2D6 predicted metabolizer status and safety in adult patients with attention-deficit hyperactivity disorder participating in a large placebo-controlled atomoxetine maintenance of response clinical trial. J. Clin. Pharmacol. 55, 1167–1174. doi: 10.1002/jcph.530
Flory, K., Molina, B. S., Pelham, W. E. Jr., Gnagy, E., and Smith, B. (2006). Childhood ADHD predicts risky sexual behavior in young adulthood. J. Clin. Child. Adolesc. Psychol. 35, 571–577. doi: 10.1207/s15374424jccp3504_8
Franke, B., Faraone, S. V., Asherson, P., Buitelaar, J., Bau, C. H., Ramos-Quiroga, J. A., et al. (2012). The genetics of attention deficit/hyperactivity disorder in adults, a review. Mol. Psychiatry 17, 960–987. doi: 10.1038/mp.2011.138
Franke, B., Michelini, G., Asherson, P., Banaschewski, T., Bilbow, A., Buitelaar, J. K., et al. (2018). Live fast, die young? A review on the developmental trajectories of ADHD across the lifespan. Eur. Neuropsychopharmacol. 28, 1059–1088. doi: 10.1016/j.euroneuro.2018.08.001
Franke, B., Vasquez, A. A., Johansson, S., Hoogman, M., Romanos, J., Boreatti-Hümmer, A., et al. (2010). Multicenter analysis of the SLC6A3/DAT1 VNTR haplotype in persistent ADHD suggests differential involvement of the gene in childhood and persistent ADHD. Neuropsychopharmacology 35, 656–664. doi: 10.1038/npp.2009.170
Frazier, T. W., Demaree, H. A., and Youngstrom, E. A. (2004). Meta-analysis of intellectual and neuropsychological test performance in attention-deficit/hyperactivity disorder. Neuropsychology 18, 543–555. doi: 10.1037/0894-4105.18.3.543
Fuemmeler, B. F., Østbye, T., Yang, C., McClernon, F. J., and Kollins, S. H. (2011). Association between attention-deficit/hyperactivity disorder symptoms and obesity and hypertension in early adulthood: a population-based study. Int. J. Obes. 35, 852–862. doi: 10.1038/ijo.2010.214
Ghirardi, L., Brikell, I., Kuja-Halkola, R., Freitag, C. M., Franke, B., Asherson, P., et al. (2018). The familial co-aggregation of ASD and ADHD: a register-based cohort study. Mol. Psychiatry 23, 257–262. doi: 10.1038/mp.2017.17
Giertuga, K., Zakrzewska, M. Z., Bielecki, M., Racicka-Pawlukiewicz, E., Kossut, M., and Cybulska-Klosowicz, A. (2017). Age-related changes in resting-state EEG activity in attention deficit/hyperactivity disorder: a cross-sectional study. Front. Hum. Neurosci. 11:285. doi: 10.3389/fnhum.2017.00285
Gillberg, C., Gillberg, I. C., Rasmussen, P., Kadesjö, B., Söderström, H., Råstam, M., et al. (2004). Co-existing disorders in ADHD – implications for diagnosis and intervention. Eur. Child. Adolesc. Psychiatry 13(Suppl. 1), I80–I92. doi: 10.1007/s00787-004-1008-4
Gizer, I. R., Ficks, C., and Waldman, I. D. (2009). Candidate gene studies of ADHD: a meta-analytic review. Hum. Genet. 126, 51–90. doi: 10.1007/s00439-009-0694-x
Golden, C. J. (1975). A group version of the stroop color and word test. J. Pers. Assess. 39, 386–388. doi: 10.1207/s15327752jpa3904_10
Gomez-Sanchez, C. I., Carballo, J. J., Riveiro-Alvarez, R., Soto-Insuga, V., Rodrigo, M., Mahillo-Fernandez, I., et al. (2017). Pharmacogenetics of methylphenidate in childhood attention-deficit/hyperactivity disorder: long-term effects. Sci. Rep. 7:10391. doi: 10.1038/s41598-017-10912-y
Gottesman, I. I., and Gould, T. D. (2003). The endophenotype concept in psychiatry: etymology and strategic intentions. Am. J. Psychiatry 160, 636–645. doi: 10.1176/appi.ajp.160.4.636
Grimm, O., Kranz, T. M., and Reif, A. (2020). Genetics of ADHD: what should the clinician know? Curr. Psychiatry. Rep. 22:18. doi: 10.1007/s11920-020-1141-x
Grove, J., Ripke, S., Als, T. D., Mattheisen, M., Walters, R. K., Won, H., et al. (2019). Identification of common genetic risk variants for autism spectrum disorder. Nat. Genet. 51, 431–444. doi: 10.1038/s41588-019-0344-8
Gudmundsson, O. O., Walters, G. B., Ingason, A., Johansson, S., Zayats, T., Athanasiu, L., et al. (2019). Attention-deficit hyperactivity disorder shares copy number variant risk with schizophrenia and autism spectrum disorder. Transl. Psychiatry 9:258. doi: 10.1038/s41398-019-0599-y
Gui, A., Mason, L., Gliga, T., Hendry, A., Begum Ali, J., Pasco, G., et al. (2020). Look duration at the face as a developmental endophenotype: elucidating pathways to autism and ADHD. Dev. Psychopathol. 32, 1303–1322. doi: 10.1017/s0954579420000930
Gul, M. K., Sener, E. F., Onal, M. G., and Demirci, E. (2021). Role of the norepinephrine transporter polymorphisms in atomoxetine treatment: from response to side effects in children with ADHD. J. Psychopharmacol. doi: 10.1177/02698811211015245 Online ahead of print
Hansen, T. F., Hoeffding, L. K., Kogelman, L., Haspang, T. M., Ullum, H., Sørensen, E., et al. (2018). Comorbidity of migraine with ADHD in adults. BMC. Neurol. 18:147. doi: 10.1186/s12883-018-1149-6
Hariri, A. R., Mattay, V. S., Tessitore, A., Kolachana, B., Fera, F., Goldman, D., et al. (2002). Serotonin transporter genetic variation and the response of the human amygdala. Science 297, 400–403. doi: 10.1126/science.1071829
Hawkins, E., Gathercole, S., Astle, D., The Calm, T., and Holmes, J. (2016). Language problems and ADHD symptoms: how specific are the links? Brain. Sci. 6:50. doi: 10.3390/brainsci6040050
Heaton, R., Chelune, G., Talley, J., Kay, G., and Curtiss, G. (1993). Wisconsin Card Sorting Test Manual. Odessa: Psychological Assessment Resources, Inc.
Hegvik, T. A., Jacobsen, K. K., Fredriksen, M., Zayats, T., and Haavik, J. (2016). A candidate gene investigation of methylphenidate response in adult attention-deficit/hyperactivity disorder patients: results from a naturalistic study. J. Neural. Transm. 123, 859–865. doi: 10.1007/s00702-016-1540-7
Hegvik, T. A., Waløen, K., Pandey, S. K., Faraone, S. V., Haavik, J., and Zayats, T. (2019). Druggable genome in attention deficit/hyperactivity disorder and its co-morbid conditions. New avenues for treatment. Mol. Psychiatry 26, 4004–4015. doi: 10.1038/s41380-019-0540-z
Hermosillo, R. J. M., Mooney, M. A., Fezcko, E., Earl, E., Marr, M., Sturgeon, D., et al. (2020). Polygenic risk score-derived subcortical connectivity mediates attention-deficit/hyperactivity disorder diagnosis. Biol. Psychiatry. Cogn. Neurosci. Neuroimaging 5, 330–341. doi: 10.1016/j.bpsc.2019.11.014
Hong, E. P., and Park, J. W. (2012). Sample size and statistical power calculation in genetic association studies. Genomics. Inform. 10, 117–122. doi: 10.5808/gi.2012.10.2.117
Huang, H. C., Wu, L. S., Yu, S. C., Wu, B. J., Lua, A. C., Lee, S. M., et al. (2018). The Alpha-2A adrenergic receptor gene -1291C/G single nucleotide polymorphism is associated with the efficacy of methylphenidate in treating taiwanese children and adolescents with attention-deficit hyperactivity disorder. Psychiatry. Investig. 15, 306–312. doi: 10.30773/pi.2017.07.24
Iacono, W. G. (2014). Genome-wide scans of genetic variants for psychophysiological endophenotypes: introduction to this special issue of Psychophysiology. Psychophysiology 51, 1201–1202. doi: 10.1111/psyp.12340
Iacono, W. G., Malone, S. M., and Vrieze, S. I. (2017). Endophenotype best practices. Int. J. Psychophysiol. 111, 115–144. doi: 10.1016/j.ijpsycho.2016.07.516
Instanes, J. T., Klungsøyr, K., Halmøy, A., Fasmer, O. B., and Haavik, J. (2018). Adult ADHD and comorbid somatic disease: a systematic literature review. J. Atten. Disord. 22, 203–228. doi: 10.1177/1087054716669589
Jacob, C. P., Romanos, J., Dempfle, A., Heine, M., Windemuth-Kieselbach, C., Kruse, A., et al. (2007). Co-morbidity of adult attention-deficit/hyperactivity disorder with focus on personality traits and related disorders in a tertiary referral center. Eur. Arch. Psychiatry. Clin. Neurosci. 257, 309–317. doi: 10.1007/s00406-007-0722-6
Jansen, A. G., Dieleman, G. C., Jansen, P. R., Verhulst, F. C., Posthuma, D., and Polderman, T. J. C. (2020). Psychiatric polygenic risk scores as predictor for attention deficit/hyperactivity disorder and autism spectrum disorder in a clinical child and adolescent sample. Behav. Genet. 50, 203–212. doi: 10.1007/s10519-019-09965-8
Jensen, P. S., Arnold, L. E., Swanson, J. M., Vitiello, B., Abikoff, H. B., Greenhill, L. L., et al. (2007). 3-year follow-up of the NIMH MTA study. J. Am. Acad. Child. Adolesc. Psychiatry 46, 989–1002. doi: 10.1097/CHI.0b013e3180686d48
Jensen, P. S., Mrazek, D., Knapp, P. K., Steinberg, L., Pfeffer, C., Schowalter, J., et al. (1997). Evolution and revolution in child psychiatry: ADHD as a disorder of adaptation. J. Am. Acad. Child. Adolesc. Psychiatry 36, 1672–1679. doi: 10.1097/00004583-199712000-00015
Jiménez-Figueroa, G., Ardila-Duarte, C., Pineda, D. A., Acosta-López, J. E., Cervantes-Henríquez, M. L., Pineda-Alhucema, W., et al. (2017). Prepotent response inhibition and reaction times in children with attention deficit/hyperactivity disorder from a caribbean community. Atten. Defic. Hyperact. Disord. 9, 199–211. doi: 10.1007/s12402-017-0223-z
Jimenez-Figueroa, G., Vidarte Claros, J. A., and Restrepo de Mejía, F. (2020). Interference control in attention deficit and hyperactivity disorder (ADHD). CES Psicol. 13, 104–124.
Johnson, K. A., Barry, E., Lambert, D., Fitzgerald, M., McNicholas, F., Kirley, A., et al. (2013). Methylphenidate side effect profile is influenced by genetic variation in the attention-deficit/hyperactivity disorder-associated CES1 gene. J. Child. Adolesc. Psychopharmacol. 23, 655–664. doi: 10.1089/cap.2013.0032
Kan, K. J., Dolan, C. V., Nivard, M. G., Middeldorp, C. M., van Beijsterveldt, C. E., Willemsen, G., et al. (2013). Genetic and environmental stability in attention problems across the lifespan: evidence from the Netherlands twin register. J. Am. Acad. Child. Adolesc. Psychiatry 52, 12–25. doi: 10.1016/j.jaac.2012.10.009
Kebir, O., and Joober, R. (2011). Neuropsychological endophenotypes in attention-deficit/hyperactivity disorder: a review of genetic association studies. Eur. Arch. Psychiatry. Clin. Neurosci. 261, 583–594. doi: 10.1007/s00406-011-0207-5
Keller, M. C., and Miller, G. (2006). Resolving the paradox of common, harmful, heritable mental disorders: which evolutionary genetic models work best? Behav. Brain. Sci. 29, 385–404. doi: 10.1017/s0140525x06009095
Klein, M., Onnink, M., van Donkelaar, M., Wolfers, T., Harich, B., Shi, Y., et al. (2017). Brain imaging genetics in ADHD and beyond - mapping pathways from gene to disorder at different levels of complexity. Neurosci. Biobehav. Rev. 80, 115–155. doi: 10.1016/j.neubiorev.2017.01.013
Konrad, K., and Eickhoff, S. B. (2010). Is the ADHD brain wired differently? A review on structural and functional connectivity in attention deficit hyperactivity disorder. Hum. Brain. Mapp. 31, 904–916. doi: 10.1002/hbm.21058
Kooij, J. S., Boonstra, A. M., Vermeulen, S. H., Heister, A. G., Burger, H., Buitelaar, J. K., et al. (2008). Response to methylphenidate in adults with ADHD is associated with a polymorphism in SLC6A3 (DAT1). Am. J. Med. Genet. B Neuropsychiatr. Genet. 147b, 201–208. doi: 10.1002/ajmg.b.30586
Kuntsi, J., Rijsdijk, F., Ronald, A., Asherson, P., and Plomin, R. (2005). Genetic influences on the stability of attention-deficit/hyperactivity disorder symptoms from early to middle childhood. Biol. Psychiatry 57, 647–654. doi: 10.1016/j.biopsych.2004.12.032
Labbe, A., Liu, A., Atherton, J., Gizenko, N., Fortier, M., Sengupta, S. M., et al. (2012). Refining psychiatric phenotypes for response to treatment: contribution of LPHN3 in ADHD. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 159b, 776–785. doi: 10.1002/ajmg.b.32083
LaBianca, S., LaBianca, J., Pagsberg, A. K., Jakobsen, K. D., Appadurai, V., Buil, A., et al. (2021). Copy number variants and polygenic risk scores predict need of care in autism and/or ADHD families. J. Autism. Dev. Disord. 51, 276–285. doi: 10.1007/s10803-020-04552-x
Lahey, B. B., Loeber, R., Burke, J., and Rathouz, P. J. (2002). Adolescent outcomes of childhood conduct disorder among clinic-referred boys: predictors of improvement. J. Abnorm. Child. Psychol. 30, 333–348. doi: 10.1023/a:1015761723226
Lahey, B. B., Van Hulle, C. A., Singh, A. L., Waldman, I. D., and Rathouz, P. J. (2011). Higher-order genetic and environmental structure of prevalent forms of child and adolescent psychopathology. Arch. Gen. Psychiatry. 68, 181–189. doi: 10.1001/archgenpsychiatry.2010.192
Langley, K., Marshall, L., van den Bree, M., Thomas, H., Owen, M., O’Donovan, M., et al. (2004). Association of the dopamine D4 receptor gene 7-repeat allele with neuropsychological test performance of children with ADHD. Am. J. Psychiatry 161, 133–138. doi: 10.1176/appi.ajp.161.1.133
Larsson, H., Anckarsater, H., Råstam, M., Chang, Z., and Lichtenstein, P. (2012). Childhood attention-deficit hyperactivity disorder as an extreme of a continuous trait: a quantitative genetic study of 8,500 twin pairs. J. Child. Psychol. Psychiatry 53, 73–80. doi: 10.1111/j.1469-7610.2011.02467.x
Larsson, H., Rydén, E., Boman, M., Långström, N., Lichtenstein, P., and Landén, M. (2013). Risk of bipolar disorder and schizophrenia in relatives of people with attention-deficit hyperactivity disorder. Br. J. Psychiatry 203, 103–106. doi: 10.1192/bjp.bp.112.120808
Lee, P. H., Anttila, V., Won, H., Feng, Y.-C. A., Rosenthal, J., Zhu, Z., et al. (2019). Genomic relationships, novel loci, and pleiotropic mechanisms across eight psychiatric disorders. Cell 179, 1469.e1411–1482.e1411. doi: 10.1016/j.cell.2019.11.020
Lin, P., Sun, J., Yu, G., Wu, Y., Yang, Y., Liang, M., et al. (2014). Global and local brain network reorganization in attention-deficit/hyperactivity disorder. Brain. Imaging. Behav. 8, 558–569. doi: 10.1007/s11682-013-9279-3
Liu, Y., Chang, X., Qu, H. Q., Tian, L., Glessner, J., Qu, J., et al. (2021). Rare recurrent variants in noncoding regions impact attention-deficit hyperactivity disorder (ADHD) gene networks in children of both african american and european american ancestry. Genes. 12:310. doi: 10.3390/genes12020310
Loche, E., and Ozanne, S. E. (2016). Early nutrition, epigenetics, and cardiovascular disease. Curr. Opin. Lipidol. 27, 449–458. doi: 10.1097/mol.0000000000000338
Loo, S. K., Hale, S. T., Hanada, G., Macion, J., Shrestha, A., McGough, J. J., et al. (2010). Familial clustering and DRD4 effects on electroencephalogram measures in multiplex families with attention deficit/hyperactivity disorder. J. Am. Acad. Child. Adolesc. Psychiatry 49, 368–377.
Loo, S. K., Specter, E., Smolen, A., Hopfer, C., Teale, P. D., and Reite, M. L. (2003). Functional effects of the DAT1 polymorphism on EEG measures in ADHD. J. Am. Acad. Child. Adolesc. Psychiatry 42, 986–993. doi: 10.1097/01.Chi.0000046890.27264.88
Malone, S. M., Vaidyanathan, U., Basu, S., Miller, M. B., McGue, M., and Iacono, W. G. (2014). Heritability and molecular-genetic basis of the P3 event-related brain potential: a genome-wide association study. Psychophysiology 51, 1246–1258. doi: 10.1111/psyp.12345
Martin, A. R., Daly, M. J., Robinson, E. B., Hyman, S. E., and Neale, B. M. (2019). Predicting polygenic risk of psychiatric disorders. Biol. Psychiatry 86, 97–109. doi: 10.1016/j.biopsych.2018.12.015
Martin, J., O’Donovan, M. C., Thapar, A., Langley, K., and Williams, N. (2015). The relative contribution of common and rare genetic variants to ADHD. Transl. Psychiatry 5:e506. doi: 10.1038/tp.2015.5
Martin, J., Walters, R. K., Demontis, D., Mattheisen, M., Lee, S. H., Robinson, E., et al. (2018). A genetic investigation of sex bias in the prevalence of attention-deficit/hyperactivity disorder. Biol. Psychiatry 83, 1044–1053. doi: 10.1016/j.biopsych.2017.11.026
McLoughlin, G., Albrecht, B., Banaschewski, T., Rothenberger, A., Brandeis, D., Asherson, P., et al. (2009). Performance monitoring is altered in adult ADHD: a familial event-related potential investigation. Neuropsychologia 47, 3134–3142. doi: 10.1016/j.neuropsychologia.2009.07.013
McLoughlin, G., Palmer, J. A., Rijsdijk, F., and Makeig, S. (2014). Genetic overlap between evoked frontocentral theta-band phase variability, reaction time variability, and attention-deficit/hyperactivity disorder symptoms in a twin study. Biol. Psychiatry 75, 238–247. doi: 10.1016/j.biopsych.2013.07.020
Mehta, T., Mannem, N., Yarasi, N. K., and Bollu, P. C. (2020). Biomarkers for ADHD: the present and future directions. Curr. Dev. Disord. Rep. 7, 85–92. doi: 10.1007/s40474-020-00196-9
Meyer-Lindenberg, A., and Weinberger, D. R. (2006). Intermediate phenotypes and genetic mechanisms of psychiatric disorders. Nat. Rev. Neurosci. 7, 818–827. doi: 10.1038/nrn1993
Michelson, D., Read, H. A., Ruff, D. D., Witcher, J., Zhang, S., and McCracken, J. (2007). CYP2D6 and clinical response to atomoxetine in children and adolescents with ADHD. J. Am. Acad. Child. Adolesc. Psychiatry 46, 242–251. doi: 10.1097/01.chi.0000246056.83791.b6
Mick, E., Biederman, J., Spencer, T., Faraone, S. V., and Sklar, P. (2006). Absence of association with DAT1 polymorphism and response to methylphenidate in a sample of adults with ADHD. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 141B, 890–894. doi: 10.1002/ajmg.b.30376
Mick, E., Neale, B., Middleton, F. A., McGough, J. J., and Faraone, S. V. (2008). Genome-wide association study of response to methylphenidate in 187 children with attention-deficit/hyperactivity disorder. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 147B, 1412–1418. doi: 10.1002/ajmg.b.30865
Monuteaux, M. C., Seidman, L. J., Faraone, S. V., Makris, N., Spencer, T., Valera, E., et al. (2008). A preliminary study of dopamine D4 receptor genotype and structural brain alterations in adults with ADHD. Am. J. Med. Genet. B Neuropsychiatr. Genet. 147B, 1436–1441. doi: 10.1002/ajmg.b.30870
Müller, D. J., Mandelli, L., Serretti, A., DeYoung, C. G., De Luca, V., Sicard, T., et al. (2008). Serotonin transporter gene and adverse life events in adult ADHD. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 147B, 1461–1469. doi: 10.1002/ajmg.b.30706
Murphy, E., and Benítez-Burraco, A. (2016). Bridging the gap between genes and language deficits in schizophrenia: an oscillopathic approach. Front. Hum. Neurosci. 10:422. doi: 10.3389/fnhum.2016.00422
Myer, N. M., Boland, J. R., and Faraone, S. V. (2018). Pharmacogenetics predictors of methylphenidate efficacy in childhood ADHD. Mol. Psychiatry 23, 1929–1936. doi: 10.1038/mp.2017.234
Naumova, D., Grizenko, N., Sengupta, S. M., and Joober, R. (2019). DRD4 exon 3 genotype and ADHD: randomised pharmacodynamic investigation of treatment response to methylphenidate. World J. Biol. Psychiatry 20, 486–495. doi: 10.1080/15622975.2017.1410221
Nemoda, Z., Angyal, N., Tarnok, Z., Gadoros, J., and Sasvari-Szekely, M. (2009). Carboxylesterase 1 gene polymorphism and methylphenidate response in ADHD. Neuropharmacology 57, 731–733. doi: 10.1016/j.neuropharm.2009.08.014
Nigg, J. T., Gustafsson, H. C., Karalunas, S. L., Ryabinin, P., McWeeney, S. K., Faraone, S. V., et al. (2018). Working memory and vigilance as multivariate endophenotypes related to common genetic risk for attention-deficit/hyperactivity disorder. J. Am. Acad. Child. Adolesc. Psychiatry 57, 175–182. doi: 10.1016/j.jaac.2017.12.013
Nigg, J. T., Karalunas, S. L., Gustafsson, H. C., Bhatt, P., Ryabinin, P., Mooney, M. A., et al. (2020). Evaluating chronic emotional dysregulation and irritability in relation to ADHD and depression genetic risk in children with ADHD. J. Child. Psychol. Psychiatry 61, 205–214. doi: 10.1111/jcpp.13132
Nikolas, M. A., and Burt, S. A. (2010). Genetic and environmental influences on ADHD symptom dimensions of inattention and hyperactivity: a meta-analysis. J. Abnorm. Psychol. 119, 1–17. doi: 10.1037/a0018010
Multiple authors. (2018). FDA-Approved Drugs to Treat ADHD. J. Psychosoc. Nurs. Ment. Health. Serv. 56, 13–14. doi: 10.3928/02793695-20180207-79
Onnink, A. M., Franke, B., van Hulzen, K., Zwiers, M. P., Mostert, J. C., Schene, A. H., et al. (2016). Enlarged striatal volume in adults with ADHD carrying the 9-6 haplotype of the dopamine transporter gene DAT1. J. Neural. Transm. 123, 905–915. doi: 10.1007/s00702-016-1521-x
Østergaard, S. D., Dalsgaard, S., Faraone, S. V., Munk-Olsen, T., and Laursen, T. M. (2017). Teenage parenthood and birth rates for individuals with and without attention-deficit/hyperactivity disorder: a nationwide cohort study. J. Am. Acad. Child. Adolesc. Psychiatry 56, 578.e573–584.e573. doi: 10.1016/j.jaac.2017.05.003
Özaslan, A., Güney, E., Ergün, M. A., Okur, İ, and Yapar, D. (2021). CDH13 and LPHN3 gene polymorphisms in attention-deficit/hyperactivity disorder: their relation to clinical characteristics. J. Mol. Neurosci. 71, 394–408. doi: 10.1007/s12031-020-01662-0
Paaby, A. B., and Rockman, M. V. (2013). The many faces of pleiotropy. Trends. Genet. 29, 66–73. doi: 10.1016/j.tig.2012.10.010
Pagerols, M., Richarte, V., Sánchez-Mora, C., Garcia-Martínez, I., Corrales, M., Corominas, M., et al. (2017). Pharmacogenetics of methylphenidate response and tolerability in attention-deficit/hyperactivity disorder. Pharmacogenomics. J. 17, 98–104. doi: 10.1038/tpj.2015.89
Pagerols, M., Richarte, V., Sánchez-Mora, C., Rovira, P., Soler Artigas, M., Garcia-Martínez, I., et al. (2018). Integrative genomic analysis of methylphenidate response in attention-deficit/hyperactivity disorder. Sci. Rep. 8:1881. doi: 10.1038/s41598-018-20194-7
Palladino, V. S., McNeill, R., Reif, A., and Kittel-Schneider, S. (2019). Genetic risk factors and gene-environment interactions in adult and childhood attention-deficit/hyperactivity disorder. Psychiatr. Genet. 29, 63–78. doi: 10.1097/ypg.0000000000000220
Paloyelis, Y., Mehta, M. A., Faraone, S. V., Asherson, P., and Kuntsi, J. (2012). Striatal sensitivity during reward processing in attention-deficit/hyperactivity disorder. J. Am. Acad. Child. Adolesc. Psychiatry 51, 722.e729–732.e729. doi: 10.1016/j.jaac.2012.05.006
Perroud, N., Cordera, P., Zimmermann, J., Michalopoulos, G., Bancila, V., Prada, P., et al. (2014). Comorbidity between attention deficit hyperactivity disorder (ADHD) and bipolar disorder in a specialized mood disorders outpatient clinic. J. Affect. Disord. 168, 161–166. doi: 10.1016/j.jad.2014.06.053
Pettersson, E., Larsson, H., and Lichtenstein, P. (2016). Common psychiatric disorders share the same genetic origin: a multivariate sibling study of the Swedish population. Mol. Psychiatry 21, 717–721. doi: 10.1038/mp.2015.116
Pinares-Garcia, P., Stratikopoulos, M., Zagato, A., Loke, H., and Lee, J. (2018). Sex: a significant risk factor for neurodevelopmental and neurodegenerative disorders. Brain. Sci. 8:154. doi: 10.3390/brainsci8080154
Plomin, R., Haworth, C. M., and Davis, O. S. (2009). Common disorders are quantitative traits. Nat. Rev. Genet. 10, 872–878. doi: 10.1038/nrg2670
Pluess, M., Belsky, J., and Neuman, R. J. (2009). Prenatal smoking and attention-deficit/hyperactivity disorder: DRD4-7R as a plasticity gene. Biol. Psychiatry 66, e5–e6. doi: 10.1016/j.biopsych.2009.04.019
Poil, S. S., Bollmann, S., Ghisleni, C., O’Gorman, R. L., Klaver, P., Ball, J., et al. (2014). Age dependent electroencephalographic changes in attention-deficit/hyperactivity disorder (ADHD). Clin. Neurophysiol. 125, 1626–1638. doi: 10.1016/j.clinph.2013.12.118
Polanczyk, G., de Lima, M. S., Horta, B. L., Biederman, J., and Rohde, L. A. (2007). The worldwide prevalence of ADHD: a systematic review and metaregression analysis. Am. J. Psychiatry 164, 942–948. doi: 10.1176/ajp.2007.164.6.942
Ramos-Quiroga, J. A., Sánchez-Mora, C., Casas, M., Garcia-Martínez, I., Bosch, R., Nogueira, M., et al. (2014). Genome-wide copy number variation analysis in adult attention-deficit and hyperactivity disorder. J. Psychiatr. Res. 49, 60–67. doi: 10.1016/j.jpsychires.2013.10.022
Ramoz, N., Boni, C., Downing, A. M., Close, S. L., Peters, S. L., Prokop, A. M., et al. (2009). A haplotype of the norepinephrine transporter (Net) gene Slc6a2 is associated with clinical response to atomoxetine in attention-deficit hyperactivity disorder (ADHD). Neuropsychopharmacology 34, 2135–2142. doi: 10.1038/npp.2009.39
Riglin, L., Eyre, O., Thapar, A. K., Stringaris, A., Leibenluft, E., Pine, D. S., et al. (2019). Identifying novel types of irritability using a developmental genetic approach. Am. J. Psychiatry 176, 635–642. doi: 10.1176/appi.ajp.2019.18101134
Roffman, J. L. (2019). Endophenotype research in psychiatry-the grasshopper grows up. JAMA Psychiatry 76, 1230–1231. doi: 10.1001/jamapsychiatry.2019.2194
Rommelse, N. N., Franke, B., Geurts, H. M., Hartman, C. A., and Buitelaar, J. K. (2010). Shared heritability of attention-deficit/hyperactivity disorder and autism spectrum disorder. Eur. Child. Adolesc. Psychiatry 19, 281–295. doi: 10.1007/s00787-010-0092-x
Ronald, A., de Bode, N., and Polderman, T. J. C. (2021). Systematic review: how the attention-deficit/hyperactivity disorder polygenic risk score adds to our understanding of ADHD and associated traits. J. Am. Acad. Child. Adolesc. Psychiatry. 60, 1234–1277. doi: 10.1016/j.jaac.2021.01.019
Rovira, P., Demontis, D., Sánchez-Mora, C., Zayats, T., Klein, M., Mota, N. R., et al. (2020). Shared genetic background between children and adults with attention deficit/hyperactivity disorder. Neuropsychopharmacology 45, 1617–1626. doi: 10.1038/s41386-020-0664-5
Rudo-Hutt, A. S. (2015). Electroencephalography and externalizing behavior: a meta-analysis. Biol. Psychol. 105, 1–19. doi: 10.1016/j.biopsycho.2014.12.005
Satterstrom, F. K., Walters, R. K., Singh, T., Wigdor, E. M., Lescai, F., Demontis, D., et al. (2019). Autism spectrum disorder and attention deficit hyperactivity disorder have a similar burden of rare protein-truncating variants. Nat. Neurosci. 22, 1961–1965. doi: 10.1038/s41593-019-0527-8
Schaefer, G. B., and Mendelsohn, N. J. (2013). Clinical genetics evaluation in identifying the etiology of autism spectrum disorders: 2013 guideline revisions. Genet. Med. 15, 399–407. doi: 10.1038/gim.2013.32
Selzam, S., Coleman, J. R. I., Caspi, A., Moffitt, T. E., and Plomin, R. (2018). A polygenic p factor for major psychiatric disorders. Transl. Psychiatry 8, 205. doi: 10.1038/s41398-018-0217-4
Selzam, S., Ritchie, S. J., Pingault, J. B., Reynolds, C. A., O’Reilly, P. F., and Plomin, R. (2019). Comparing within- and between-family polygenic score prediction. Am. J. Hum. Genet. 105, 351–363. doi: 10.1016/j.ajhg.2019.06.006
Serdarevic, F., Tiemeier, H., Jansen, P. R., Alemany, S., Xerxa, Y., Neumann, A., et al. (2020). Polygenic risk scores for developmental disorders, neuromotor functioning during infancy, and autistic traits in childhood. Biol. Psychiatry 87, 132–138. doi: 10.1016/j.biopsych.2019.06.006
Shaw, P., Gornick, M., Lerch, J., Addington, A., Seal, J., Greenstein, D., et al. (2007). Polymorphisms of the dopamine D4 receptor, clinical outcome, and cortical structure in attention-deficit/hyperactivity disorder. Arch. Gen. Psychiatry 64, 921–931. doi: 10.1001/archpsyc.64.8.921
Shen, C., Luo, Q., Jia, T., Zhao, Q., Desrivières, S., Quinlan, E. B., et al. (2020). Neural correlates of the dual-pathway model for ADHD in adolescents. Am. J. Psychiatry 177, 844–854. doi: 10.1176/appi.ajp.2020.19020183
Shook, D., Brady, C., Lee, P. S., Kenealy, L., Murphy, E. R., Gaillard, W. D., et al. (2011). Effect of dopamine transporter genotype on caudate volume in childhood ADHD and controls. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 156b, 28–35. doi: 10.1002/ajmg.b.31132
Snyder, S. M., and Hall, J. R. (2006). A meta-analysis of quantitative EEG power associated with attention-deficit hyperactivity disorder. J. Clin. Neurophysiol. 23, 440–455. doi: 10.1097/01.wnp.0000221363.12503.78
Sobanski, E., Brüggemann, D., Alm, B., Kern, S., Deschner, M., Schubert, T., et al. (2007). Psychiatric comorbidity and functional impairment in a clinically referred sample of adults with attention-deficit/hyperactivity disorder (ADHD). Eur. Arch. Psychiatry. Clin. Neurosci. 257, 371–377. doi: 10.1007/s00406-007-0712-8
Srivastava, S., Love-Nichols, J. A., Dies, K. A., Ledbetter, D. H., Martin, C. L., Chung, W. K., et al. (2020). Correction: meta-analysis and multidisciplinary consensus statement: exome sequencing is a first-tier clinical diagnostic test for individuals with neurodevelopmental disorders. Genet. Med. 22, 1731–1732. doi: 10.1038/s41436-020-0913-3
Stergiakouli, E., Hamshere, M., Holmans, P., Langley, K., Zaharieva, I., Hawi, Z., et al. (2012). Investigating the contribution of common genetic variants to the risk and pathogenesis of ADHD. Am. J. Psychiatry 169, 186–194. doi: 10.1176/appi.ajp.2011.11040551
Stojanovski, S., Felsky, D., Viviano, J. D., Shahab, S., Bangali, R., Burton, C. L., et al. (2019). Polygenic risk and neural substrates of attention-deficit/hyperactivity disorder symptoms in youths with a history of mild traumatic brain injury. Biol. Psychiatry 85, 408–416. doi: 10.1016/j.biopsych.2018.06.024
Storebø, O. J., Pedersen, N., Ramstad, E., Kielsholm, M. L., Nielsen, S. S., Krogh, H. B., et al. (2018). Methylphenidate for attention deficit hyperactivity disorder (ADHD) in children and adolescents - assessment of adverse events in non-randomised studies. Cochrane Database Syst. Rev. 5, Cd012069. doi: 10.1002/14651858.CD012069.pub2
Sudre, G., Frederick, J., Sharp, W., Ishii-Takahashi, A., Mangalmurti, A., Choudhury, S., et al. (2020). Mapping associations between polygenic risks for childhood neuropsychiatric disorders, symptoms of attention deficit hyperactivity disorder, cognition, and the brain. Mol. Psychiatry 25, 2482–2492. doi: 10.1038/s41380-019-0350-3
Sullivan, P. F., Daly, M. J., and O’Donovan, M. (2012). Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat. Rev. Genet. 13, 537–551. doi: 10.1038/nrg3240
Takeuchi, H., Tomita, H., Taki, Y., Kikuchi, Y., Ono, C., Yu, Z., et al. (2015). Cognitive and neural correlates of the 5-repeat allele of the dopamine D4 receptor gene in a population lacking the 7-repeat allele. Neuroimage 110, 124–135. doi: 10.1016/j.neuroimage.2015.01.053
Tan, H. (2020). Somatic mutation in noncoding regions: the sound of silence. EBioMedicine 61:103084. doi: 10.1016/j.ebiom.2020.103084
Taylor, M. J., Lichtenstein, P., Larsson, H., Anckarsäter, H., Greven, C. U., and Ronald, A. (2016). Is there a female protective effect against attention-deficit/hyperactivity disorder? evidence from two representative twin samples. J. Am. Acad. Child. Adolesc. Psychiatry 55, 504.e502–512.e502. doi: 10.1016/j.jaac.2016.04.004
Taylor, M. J., Martin, J., Lu, Y., Brikell, I., Lundström, S., Larsson, H., et al. (2019). Association of genetic risk factors for psychiatric disorders and traits of these disorders in a swedish population twin sample. JAMA Psychiatry 76, 280–289. doi: 10.1001/jamapsychiatry.2018.3652
Thagaard, M. S., Faraone, S. V., Sonuga-Barke, E. J., and Østergaard, S. D. (2016). Empirical tests of natural selection-based evolutionary accounts of ADHD: a systematic review. Acta Neuropsychiatr. 28, 249–256. doi: 10.1017/neu.2016.14
Thapar, A., Martin, J., Mick, E., Arias Vásquez, A., Langley, K., Scherer, S. W., et al. (2016). Psychiatric gene discoveries shape evidence on ADHD’s biology. Mol. Psychiatry 21, 1202–1207. doi: 10.1038/mp.2015.163
Thissen, A. J., Bralten, J., Rommelse, N. N., Arias-Vasquez, A., Greven, C. U., Heslenfeld, D., et al. (2015). The role of age in association analyses of ADHD and related neurocognitive functioning: a proof of concept for dopaminergic and serotonergic genes. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 168, 471–479. doi: 10.1002/ajmg.b.32290
Tong, J., Lee, K. M., Liu, X., Nefzger, C. M., Vijayakumar, P., Hawi, Z., et al. (2019). Generation of four iPSC lines from peripheral blood mononuclear cells (PBMCs) of an attention deficit hyperactivity disorder (ADHD) individual and a healthy sibling in an Australia-Caucasian family. Stem Cell. Res. 34:101353. doi: 10.1016/j.scr.2018.11.014
Torkamani, A., Wineinger, N. E., and Topol, E. J. (2018). The personal and clinical utility of polygenic risk scores. Nat. Rev. Genet. 19, 581–590. doi: 10.1038/s41576-018-0018-x
Torske, T., Naerland, T., Bettella, F., Bjella, T., Malt, E., Høyland, A. L., et al. (2020). Autism spectrum disorder polygenic scores are associated with every day executive function in children admitted for clinical assessment. Autism. Res. 13, 207–220. doi: 10.1002/aur.2207
Trzepacz, P. T., Williams, D. W., Feldman, P. D., Wrishko, R. E., Witcher, J. W., and Buitelaar, J. K. (2008). CYP2D6 metabolizer status and atomoxetine dosing in children and adolescents with ADHD. Eur. Neuropsychopharmacol. 18, 79–86. doi: 10.1016/j.euroneuro.2007.06.002
Tye, C., Rijsdijk, F., Greven, C. U., Kuntsi, J., Asherson, P., and McLoughlin, G. (2012). Shared genetic influences on ADHD symptoms and very low-frequency EEG activity: a twin study. J. Child. Psychol. Psychiatry 53, 706–715. doi: 10.1111/j.1469-7610.2011.02501.x
van Beijsterveldt, C. E., and van Baal, G. C. (2002). Twin and family studies of the human electroencephalogram: a review and a meta-analysis. Biol. Psychol. 61, 111–138. doi: 10.1016/s0301-0511(02)00055-8
Vanzo, R. J., Prasad, A., Staunch, L., Hensel, C. H., Serrano, M. A., Wassman, E. R., et al. (2020). The temple grandin genome: comprehensive analysis in a scientist with high-functioning autism. J. Pers. Med. 11:21. doi: 10.3390/jpm11010021
Verbanck, M., Chen, C. Y., Neale, B., and Do, R. (2018). Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 50, 693–698. doi: 10.1038/s41588-018-0099-7
Vink, J. M., Treur, J. L., Pasman, J. A., and Schellekens, A. (2020). Investigating genetic correlation and causality between nicotine dependence and ADHD in a broader psychiatric context. Am. J. Med. Genet. B. Neuropsychiatr. Genet. 186, 423–429. doi: 10.1002/ajmg.b.32822
Visscher, P. M., Wray, N. R., Zhang, Q., Sklar, P., McCarthy, M. I., Brown, M. A., et al. (2017). 10 years of GWAS discovery: biology, function, and translation. Am. J. Hum. Genet. 101, 5–22. doi: 10.1016/j.ajhg.2017.06.005
Vuijk, P. J., Martin, J., Braaten, E. B., Genovese, G., Capawana, M. R., O’Keefe, S. M., et al. (2020). Translating discoveries in attention-deficit/hyperactivity disorder genomics to an outpatient child and adolescent psychiatric cohort. J. Am. Acad. Child. Adolesc. Psychiatry 59, 964–977. doi: 10.1016/j.jaac.2019.08.004
Waldman, I. D., Poore, H. E., van Hulle, C., Rathouz, P. J., and Lahey, B. B. (2016). External validity of a hierarchical dimensional model of child and adolescent psychopathology: tests using confirmatory factor analyses and multivariate behavior genetic analyses. J. Abnorm. Psychol. 125, 1053–1066. doi: 10.1037/abn0000183
Wang, E., Ding, Y. C., Flodman, P., Kidd, J. R., Kidd, K. K., Grady, D. L., et al. (2004). The genetic architecture of selection at the human dopamine receptor D4 (DRD4) gene locus. Am. J. Hum. Genet. 74, 931–944. doi: 10.1086/420854
Wechsler, D. (1981). Wechsler Adult Intelligence Scale- Revised. New York, NY: The Psychological Corporation.
Williams, N. M., Zaharieva, I., Martin, A., Langley, K., Mantripragada, K., Fossdal, R., et al. (2010). Rare chromosomal deletions and duplications in attention-deficit hyperactivity disorder: a genome-wide analysis. Lancet 376, 1401–1408. doi: 10.1016/s0140-6736(10)61109-9
Wu, J., Xiao, H., Sun, H., Zou, L., and Zhu, L. Q. (2012). Role of dopamine receptors in ADHD: a systematic meta-analysis. Mol. Neurobiol. 45, 605–620. doi: 10.1007/s12035-012-8278-5
Yang, L., Qian, Q., Liu, L., Li, H., Faraone, S. V., and Wang, Y. (2013). Adrenergic neurotransmitter system transporter and receptor genes associated with atomoxetine response in attention-deficit hyperactivity disorder children. J. Neural. Transm. 120, 1127–1133. doi: 10.1007/s00702-012-0955-z
Yu, G., Li, G. F., and Markowitz, J. S. (2016). Atomoxetine: a review of its pharmacokinetics and pharmacogenomics relative to drug disposition. J. Child. Adolesc. Psychopharmacol. 26, 314–326. doi: 10.1089/cap.2015.0137
Yuan, D., Zhang, M., Huang, Y., Wang, X., Jiao, J., and Huang, Y. (2021). Noradrenergic genes polymorphisms and response to methylphenidate in children with ADHD: a systematic review and meta-analysis. Medicine 100:e27858. doi: 10.1097/MD.0000000000027858
Zayats, T., and Neale, B. M. (2019). Recent advances in understanding of attention deficit hyperactivity disorder (ADHD): how genetics are shaping our conceptualization of this disorder. F1000Res 8:F1000FacultyRev-2060. doi: 10.12688/f1000research.18959.2
Zhong, Y., Yang, B., Su, Y., Qian, Y., Cao, Q., Chang, S., et al. (2020). The association with quantitative response to attention-deficit/hyperactivity disorder medication of the previously identified neurodevelopmental network genes. J. Child. Adolesc. Psychopharmacol. 30, 348–354. doi: 10.1089/cap.2018.0164
Keywords: attention deficit hyperactivity disorder (ADHD), genetics, polygenic risk score (PRS), neurodevelopment, endophenotype, comorbidity, psychiatry
Citation: Balogh L, Pulay AJ and Réthelyi JM (2022) Genetics in the ADHD Clinic: How Can Genetic Testing Support the Current Clinical Practice? Front. Psychol. 13:751041. doi: 10.3389/fpsyg.2022.751041
Received: 31 July 2021; Accepted: 03 January 2022;
Published: 08 March 2022.
Edited by:
Dora Koller, Yale University, United StatesReviewed by:
Antonio Benítez-Burraco, Seville University, SpainCopyright © 2022 Balogh, Pulay and Réthelyi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lívia Balogh, YmFsb2doLmxpdmlhQG1lZC5zZW1tZWx3ZWlzLXVuaXYuaHU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.