- Department of Neuropsychiatry, Oita University Faculty of Medicine, Yufu, Oita, Japan
Bipolar disorder is a chronic disorder characterized by fluctuations in mood state and energy and recurrent episodes of mania/hypomania and depression. Bipolar disorder may be regarded as a neuro-progressive disorder in which repeated mood episodes may lead to cognitive decline and dementia development. In the current review, we employed genome-wide association studies to comprehensively investigate the genetic variants associated with bipolar disorder and dementia. Thirty-nine published manuscripts were identified: 20 on bipolar disorder and 19 on dementia. The results showed that the genes CACNA1C, GABBR2, SCN2A, CTSH, MSRA, and SH3PXD2A were overlapping between patients with bipolar disorder and dementia. In conclusion, the genes CACNA1C, GABBR2, SCN2A, CTSH, MSRA, and SH3PXD2A may be associated with the neuro-progression of bipolar disorder to dementia. Further genetic studies are needed to comprehensively clarify the role of genes in cognitive decline and the development of dementia in patients with bipolar disorder.
Introduction
Bipolar disorder is a chronic disorder characterized by fluctuations in mood state and energy, in addition to recurrent episodes of mania/hypomania and depression (1). Cognitive impairment has been documented in a variety of neuropsychological domains during the mood disturbances associated with the acute episodes of bipolar disorder (2, 3). However, patients with bipolar disorder suffer from cognitive impairment not only during the acute phase but also during the remission phase (4, 5). Bipolar disorder in the first episode is associated with widespread cognitive dysfunction, especially in psychomotor speed, attention, working memory, and cognitive flexibility, suggesting that a broad range of cognitive deficits is already present at this early stage (6). Cognitive decline occurs with repeated manic episodes, hospitalizations, and length of illness in patients with bipolar disorder, suggesting that the recurrence of mania may have a long-term neuropsychological impact (7, 8). Studies on bipolar disorder have shown that neuropsychological deficits are detectable in euthymia, and they contribute to poor outcomes (8, 9). Patients with bipolar disorder have the greatest risk of dementia, followed by those with unipolar depression, schizophrenia, and neurosis (10). A meta-analysis revealed that a history of bipolar disorder significantly increased the risk of dementia (odds ratio: 2.36; 95% CI: 1.36–4.09) (11). Another meta-analysis showed that bipolar disorder increases the risk of dementia (odds ratio: 2.96; 95% CI: 2.09–4.18), and the risk of progression to dementia is higher in bipolar disorder than that in major depressive disorder (12). Thus, bipolar disorder may be seen as a neuro-progressive disorder in which repeated mood episodes may lead to cognitive decline and, finally, the development of dementia.
Genome-wide association studies (GWAS) aim to identify single nucleotide polymorphisms (SNPs) in which allele frequencies vary systematically as a function of phenotypic trait values (13). The identification of trait-associated SNPs distributed throughout the genome may provide new insights into the biological mechanisms underlying psychiatric disorders (13). To date, GWAS have successfully identified SNPs associated with the risk of bipolar disorder and dementia (14, 15). The clinical question arises as to which genetic factors are associated with the association between bipolar disorder and dementia. From a clinical perspective, we previously hypothesized that there is a specific group of patients whose diagnoses longitudinally change from depression to bipolar disorder and finally to dementia, and the glycogen synthase kinase 3β gene may be a common etiological factor in these diseases and diagnostic conversions (16). In the current review, using a completely different perspective, we employed the results of GWAS to comprehensively investigate genetic variants associated with bipolar disorder and dementia (including Alzheimer’s disease, Lewy body dementia, frontotemporal dementia, and vascular dementia).
Methods
This review was qualitative and not systematic in nature. This study was conducted in January, 2024. Using the PubMed database, we conducted searches with keywords “bipolar disorder” and “GWAS”, “dementia” and “GWAS”. In this review, only GWAS that examined the relationship between the diagnosis of bipolar disorder or dementia and genes and SNPs were included. Articles that were not GWAS or were written in languages other than English were excluded. We simply examined the presence of overlapping genes and SNPs reported in patients with bipolar disorder and dementia.
Results
Thirty-nine published manuscripts were identified: 20 on bipolar disorder (14, 17–35) and 19 on dementia (15, 36–53) (see Supplementary Figure 1 for the literature screening flow chart). Table 1 summarizes the characteristics of the included studies. The lists of significant SNPs and genes associated with bipolar disorder and dementia reported by GWAS are provided in Supplementary Tables 1, 2, respectively.
A review revealed that among the genes reported to be associated with the diagnosis of bipolar disorder or dementia in previous GWAS studies, the overlapping gene was calcium voltage-gated channel subunit Alpha1 C (CACNA1C), gamma-aminobutyric acid B receptor 2 (GABBR2), sodium voltage-gated channel Alpha subunit 2 (SCN2A), cathepsin H (CTSH), methionine sulfoxide reductase A (MSRA), and SH3 and PX domains 2A (SH3PXD2A) (Table 2; Supplementary Figure 2). With respect to the type of dementia, rs11062164 of CACNA1C, rs16916777 of GABBR2, and rs17738042 of SH3PXD2A are associated with vascular dementia; rs10184275 and rs2119067 of SCN2A are associated with late-onset Alzheimer’s disease; and, rs12592898 of CTSH and rs4607615 of MSRA are associated with Alzheimer’s disease. No SNPs were found to be reported as associated with both the diagnosis of bipolar disorder and dementia.
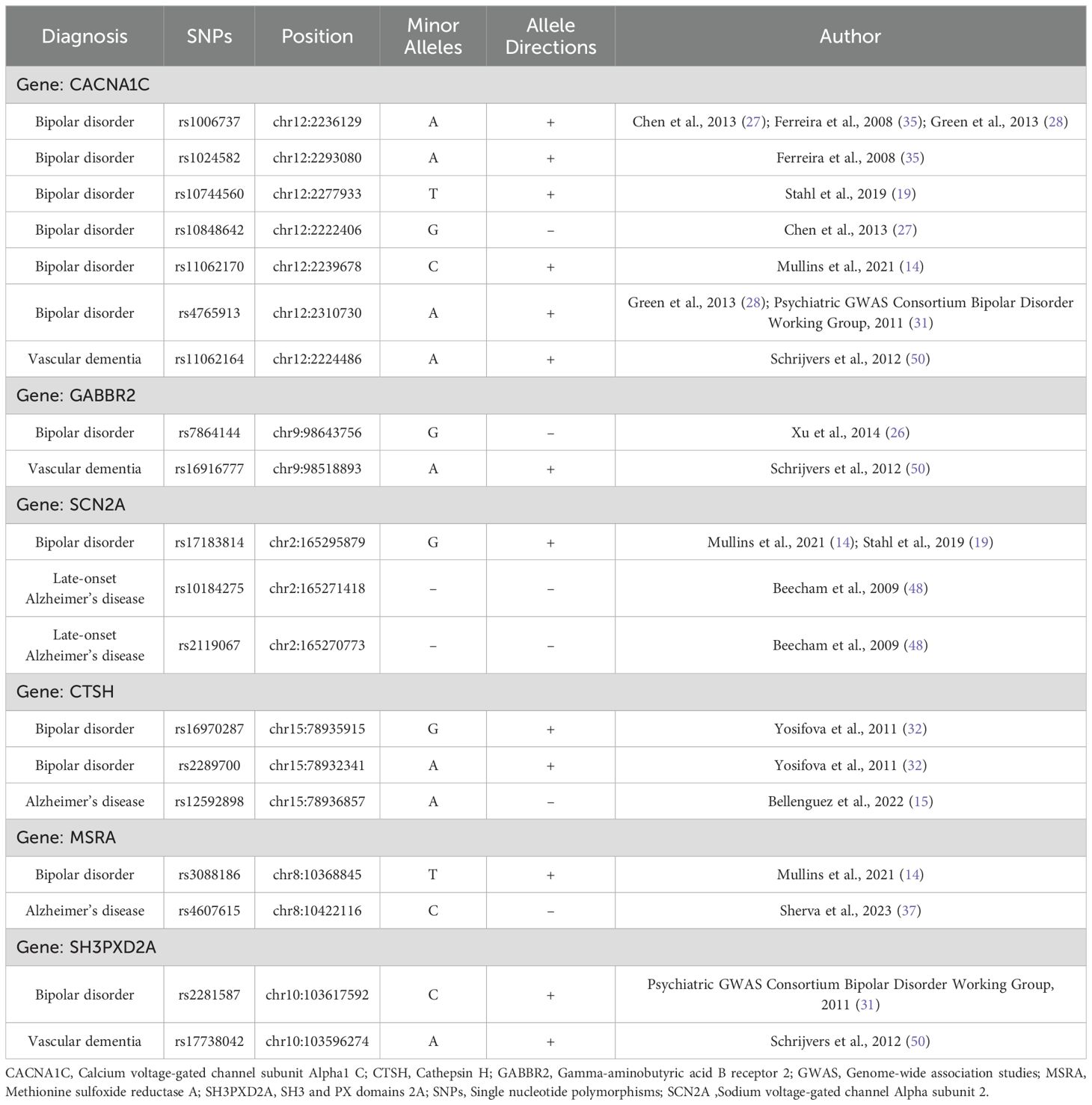
Table 2. The overlapping genes and single nucleotide polymorphisms with bipolar disorder and dementia.
Discussion
According to GWAS, the CACNA1C, GABBR2, SCN2A, CTSH, MSRA, and SH3PXD2A genes were common between bipolar disorder and dementia.
CACNA1C encodes the alpha-1 subunit of the voltage-dependent L-type calcium channel expressed in the human brain (54), which regulates cellular calcium influx and is essential for normal brain development and plasticity (55). Variants of CACNA1C have been associated with bipolar disorder and several neuropsychiatric disorders, such as schizophrenia, major depressive disorder, autism spectrum disorder, attention deficit hyperactivity disorder, and substance-use disorders (56). The CACNA1C gene (especially the rs1006737 A allele) is robustly associated with bipolar disorder and might be crucial in molecular biological research on the set of interacting proteins involved in the calcium channel activity in bipolar disorder (57). The CACNA1C gene may not only be associated with the onset of bipolar disorder but also potentially affects the course of cognitive function and brain imaging. The rs1006737 variant (minor allele: A) of the CACNA1C gene is associated with cognitive impairment in patients with bipolar disorder and schizophrenia spectrum (58). Furthermore, a 2-year longitudinal study on bipolar disorder revealed that patients with the AA genotype of rs1006737 showed poorer cognitive performance, particularly in terms of processing speed (59). The rs10466907 variant of CACNA1C is associated with cognitive recovery after a major depressive episode in bipolar disorder (60). CACNA1C is expressed throughout the mouse brain, including key limbic regions relevant for emotion and cognition, such as the prefrontal cortex, hippocampus, and amygdala (61). Moreover, embryonic deletion of CACNA1C in glutamatergic neurons in the forebrain promotes the manifestation of endophenotypes related to psychiatric disorders, including cognitive decline, impaired synaptic plasticity, reduced sociability, hyperactivity, and increased anxiety (61). In a human brain study on bipolar disorder, the rs1006737 A allele of the CACNA1C gene was associated with gray matter volume, functional connectivity within the corticolimbic frontotemporal neural system, and mean thickness of cortical brain areas (62, 63). Patients with bipolar disorder carrying the rs1006737 A allele showed age-related cortical thinning of the left caudal anterior cingulate cortex (64). In contrast, among non-A carriers, age did not affect cortical thinning in the left caudal anterior cingulate cortex, suggesting an underlying relationship with aging-associated cognitive decline (64). Furthermore, tau phosphorylation is known to increase in the cerebrospinal fluid (CSF) in patients with Alzheimer’s disease (65), and the rs1006737 variant of CACNA1C is significantly associated with hyperphosphorylated tau/total tau ratio in the CSF of patients with bipolar disorder (66). A study on late-onset Alzheimer’s disease focusing on protein–protein network interactions revealed eight genes with strong associations: APOE, SORL1, APOC1, CD33, CLU, TOMM40, CNTNAP2, and CACNA1C (67). Interestingly, CACNA1C (rs10848683 variant) is also associated with ischemic stroke (68). Similarly, CACNA1C (rs11062164 variant) was significantly associated with vascular dementia (50). Thus, CACNA1C is associated with dementia. Furthermore, the rs7297582 T allele of CACNA1C is associated with a risk of bipolar disorder and poor cognitive performance (69). Although no studies have reported an association between rs7297582 and dementia, future studies should consider SNPs of interest in relation to both bipolar disorder and dementia. In the present review, the SNPs of CACNA1C did not match across studies. CACNA1C, especially rs1006737 and rs7297582, may be strongly associated with the onset of bipolar disorder, cognitive decline in bipolar disorder, and dementia.
Human gamma-aminobutyric acid type B receptor (GABABR) is a G protein-coupled receptor central to inhibitory neurotransmission in the brain (70). GABABR is assembled by the heterodimeric interaction of the intracellular C-terminal tails of the two subunits encoded by GABBR1 and GABBR2 (70). In patients with bipolar disorder, GABAergic hypofunction has been observed in the cerebellum (71). Additionally, a post-mortem study revealed reduced protein expressions of GABBR1 and GABBR2 in the cerebellum of patients with bipolar disorder (72). Dysfunction of the GABAergic system may cause cognitive impairment in humans (73). The reduction in GABAergic system components in the brain and lower GABA levels in the CSF of patients with Alzheimer’s disease suggest that the GABAergic system is vulnerable to Alzheimer’s disease pathology and should be considered a potential target for developing pharmacological strategies and novel Alzheimer’s disease biomarkers (74, 75). GABBR2 is also associated with bipolar disorder and dementia.
SCN2A encodes the voltage-gated sodium channel Nav1.2, a major central nervous system sodium channel that plays a role in the initiation and conduction of action potentials (76). SCN2A variants are associated with a range of disorders including autism spectrum disorder, developmental delay, seizures, and epileptic encephalopathy (77). Furthermore, SCN2A is associated with psychiatric disorders such as bipolar disorder and schizophrenia (14, 78). SCN2A contributes to excitability that facilitates synapse formation and development (79). A study on Alzheimer’s disease focusing on protein-protein network interactions revealed six hub genes: SCN2A, SNAP25, GRIN2A, GRIN2B, DLG2, and ATP2B2 (80). Thus, SCN2A is associated with bipolar disorder and dementia.
CTSH is a cysteine cathepsin that primarily acts as an aminopeptidase (81). The main function of cathepsins is to degrade proteins via proteolysis in lysosomes (81). CTSH has been implicated in the cis-regulated mRNA association with Alzheimer’s disease (82). CTSH expression is significantly lower in the brain tissue of healthy controls than in patients with Alzheimer’s disease (83). Moreover, CTSH knockout affected genes related to endocytosis and significantly increased Aβ42 phagocytosis in microglial cells (83). The CTSH gene was significantly associated with Alzheimer’s disease (83). The mechanism by which CTSH is associated with bipolar disorder is unknown; however, a GWAS conducted by Yosifova et al. in a Bulgarian population identified a significant association between bipolar disorder and CTSH (32). Therefore, CTSH is a gene of interest related to bipolar disorder and dementia.
MSRA has been postulated to act as a catalytic antioxidant system that protects against oxidative stress-induced cell injury and is highly expressed in the brain (84, 85). The methionine sulfoxide reductase system may contribute to the development of aging-associated diseases, including neurodegenerative diseases (86). MRSA knockout mice exhibit enhanced neurodegeneration in the brain hippocampus compared to their wild-type counterparts (86). There are hypotheses suggesting that oxidative stress is associated with bipolar disorder and Alzheimer’s disease (87, 88). The rs4840463 polymorphism in MRSA is associated with an increased risk of bipolar I and executive function defects (87). In this review, the rs3088186 polymorphism is associated with bipolar disorder, while rs4607615 is linked to Alzheimer’s disease. Thus, MRSA is implicated in both bipolar disorder and dementia.
The SH3PXD2A gene encodes TKS5, an isoform essential for proper mammalian development (89). Additionally, SH3PXD2A directly interacts with the ADAM metallopeptidase domain 15 gene, which is involved in neurodegeneration and inflammatory processes (90). SH3PXD2A is associated with brain white matter lesions and stroke (90–92). It is noteworthy that SH3PXD2A (rs17738042) is significantly associated with vascular dementia (50). Interestingly, rs3740473 of SH3PXD2A is associated with Alzheimer’s disease (93). The mechanism by which SH3PXD2A is linked to bipolar disorder remains unknown; however, a GWAS revealed a significant association between bipolar disorder and SH3PXD2A (31). Therefore, SH3PXD2A is a gene of interest in relation to bipolar disorder and dementia.
In addition, we previously reviewed that rs334558 of the GSK-3β gene was associated with depression, bipolar disorder, and dementia (16, 94–96). We hypothesized the existence of a mental GSK-3 disease, which comprises a specific group of patients associated with the GSK-3β variant, whose diagnoses longitudinally transition from depression to bipolar disorder and finally to dementia (16). Therefore, although we could not find a significant association between the GSK-3β gene and bipolar disorder and dementia in this GWAS review, rs334558 of the GSK-3β gene is associated with bipolar disorder and dementia.
This review has several limitations. First, it relies on GWAS, which are cross-sectional in nature, and examines bipolar disorder and dementia at each study time point. Therefore, genetic studies involving the longitudinal transitions from bipolar disorder to dementia or cognitive function decline in patients with bipolar disorder are needed. Hence, future longitudinal studies are needed to explore the genetic factors associated with cognitive decline in bipolar disorder and the onset of dementia. Second, bipolar disorder and dementia are believed to be associated with various genes, and it is difficult to explain them based solely on a single gene or SNP. Therefore, it is necessary to consider factors from multiple genes, such as polygenic scores, which can summarize global genomic risk rather than focusing on specific variants. Third, this review only examined whether the identified genes or SNPs were relevant by extracting them from individual studies and did not involve a combined GWAS analysis of the cases in each study. Finally, this review is qualitative, not quantitative, and weighs the effects of candidate genes.
Conclusion
In conclusion, CACNA1C, GABBR2, SCN2A, CTSH, MSRA, and SH3PXD2A may be associated with the neuro-progression of bipolar disorder to dementia. Further genetic studies are needed to comprehensively clarify the role of genes in cognitive decline and dementia development in patients with bipolar disorder.
Author contributions
HH: Writing – original draft, Writing – review & editing. TT: Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The present work was supported by the Japanese Society for the Promotion of Science as Grant-in-Aid for Scientific Research (21K07502).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision
The reviewer HH is currently organizing a Research Topic with the authors.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpsyt.2024.1414776/full#supplementary-material
References
1. Grande I, Berk M, Birmaher B, Vieta E. Bipolar disorder. Lancet. (2016) 387:1561–72. doi: 10.1016/S0140-6736(15)00241-X
2. Martínez-Arán A, Vieta E, Reinares M, Colom F, Torrent C, Sánchez-Moreno J, et al. Cognitive function across manic or hypomanic, depressed, and euthymic states in bipolar disorder. Am J Psychiatry. (2004) 161:262–70. doi: 10.1176/appi.ajp.161.2.262
3. Lin X, Lu D, Huang Z, Chen W, Luo X, Zhu Y. The associations between subjective and objective cognitive functioning across manic or hypomanic, depressed, and euthymic states in Chinese bipolar patients. J Affect Disord. (2019) 249:73–81. doi: 10.1016/j.jad.2019.02.025
4. Solé B, Jiménez E, Torrent C, Reinares M, Bonnin CDM, Torres I, et al. Cognitive impairment in bipolar disorder: treatment and prevention strategies. Int J Neuropsychopharmacol. (2017) 20:670–80. doi: 10.1093/ijnp/pyx032
5. Huang Y, Zhang Z, Lin S, Zhou H, Xu G. Cognitive impairment mechanism in patients with bipolar disorder. Neuropsychiatr Dis Treat. (2023) 19:361–6. doi: 10.2147/NDT.S396424
6. Lee RS, Hermens DF, Scott J, Redoblado-Hodge MA, Naismith SL, Lagopoulos J, et al. A meta-analysis of neuropsychological functioning in first-episode bipolar disorders. J Psychiatr Res. (2014) 57:1–11. doi: 10.1016/j.jpsychires.2014.06.019
7. López-Jaramillo C, Lopera-Vásquez J, Gallo A, Ospina-Duque J, Bell V, Torrent C, et al. Effects of recurrence on the cognitive performance of patients with bipolar I disorder: implications for relapse prevention and treatment adherence. Bipolar Disord. (2010) 12:557–67. doi: 10.1111/j.1399-5618.2010.00835.x
8. Robinson LJ, Ferrier IN. Evolution of cognitive impairment in bipolar disorder: a systematic review of cross-sectional evidence. Bipolar Disord. (2006) 8:103–16. doi: 10.1111/j.1399-5618.2006.00277.x
9. Depp CA, Mausbach BT, Harmell AL, Savla GN, Bowie CR, Harvey PD, et al. Meta-analysis of the association between cognitive abilities and everyday functioning in bipolar disorder. Bipolar Disord. (2012) 14:217–26. doi: 10.1111/j.1399-5618.2012.01011.x
10. Kessing LV, Olsen EW, Mortensen PB, Andersen PK. Dementia in affective disorder: a case-register study. Acta Psychiatr Scand. (1999) 100:176–85. doi: 10.1111/j.1600-0447.1999.tb10843.x
11. Diniz BS, Teixeira AL, Cao F, Gildengers A, Soares JC, Butters MA, et al. History of bipolar disorder and the risk of dementia: A systematic review and meta-analysis. Am J Geriatr Psychiatry. (2017) 25:357–62. doi: 10.1016/j.jagp.2016.11.014
12. Velosa J, Delgado A, Finger E, Berk M, Kapczinski F, de Azevedo Cardoso T. Risk of dementia in bipolar disorder and the interplay of lithium: a systematic review and meta-analyses. Acta Psychiatr Scand. (2020) 141:510–21. doi: 10.1111/acps.13153
13. Marees AT, de Kluiver H, Stringer S, Vorspan F, Curis E, Marie-Claire C, et al. A tutorial on conducting genome-wide association studies: Quality control and statistical analysis. Int J Methods Psychiatr Res. (2018) 27:e1608. doi: 10.1002/mpr.1608
14. Mullins N, Forstner AJ, O'Connell KS, Coombes B, Coleman JRI, Qiao Z, et al. Genome-wide association study of more than 40,000 bipolar disorder cases provides new insights into the underlying biology. Nat Genet. (2021) 53:817–29. doi: 10.1038/s41588-021-00857-4
15. Bellenguez C, Küçükali F, Jansen IE, Kleineidam L, Moreno-Grau S, Amin N, et al. New insights into the genetic etiology of Alzheimer's disease and related dementias. Nat Genet. (2022) 54:412–36. doi: 10.1038/s41588-022-01024-z
16. Terao T, Ishii N, Hirakawa H. A specific group of patients with diagnostic conversion from depression to bipolar disorder and finally to dementia as a mental GSK-3 disease: A hypothesis. Bipolar Disord. (2020) 22:356–9. doi: 10.1111/bdi.12875
17. Li HJ, Zhang C, Hui L, Zhou DS, Li Y, Zhang CY, et al. Novel risk loci associated with genetic risk for bipolar disorder among Han Chinese individuals: A genome-wide association study and meta-analysis. JAMA Psychiatry. (2021) 78:320–30. doi: 10.1001/jamapsychiatry.2020.3738
18. Budde M, Friedrichs S, Alliey-Rodriguez N, Ament S, Badner JA, Berrettini WH, et al. Efficient region-based test strategy uncovers genetic risk factors for functional outcome in bipolar disorder. Eur Neuropsychopharmacol. (2019) 29:156–70. doi: 10.1016/j.euroneuro.2018.10.005
19. Stahl EA, Breen G, Forstner AJ, McQuillin A, Ripke S, Trubetskoy V, et al. Genome-wide association study identifies 30 loci associated with bipolar disorder. Nat Genet. (2019) 51:793–803. doi: 10.1038/s41588-019-0397-8
20. Ikeda M, Takahashi A, Kamatani Y, Okahisa Y, Kunugi H, Mori N, et al. A genome-wide association study identifies two novel susceptibility loci and trans population polygenicity associated with bipolar disorder. Mol Psychiatry. (2018) 23:639–47. doi: 10.1038/mp.2016.259
21. Acikel C, Aydin Son Y, Celik C, Gul H. Evaluation of potential novel variations and their interactions related to bipolar disorders: analysis of genome-wide association study data. Neuropsychiatr Dis Treat. (2016) 12:2997–3004. doi: 10.2147/NDT
22. Hou L, Bergen SE, Akula N, Song J, Hultman CM, Landén M, et al. Genome-wide association study of 40,000 individuals identifies two novel loci associated with bipolar disorder. Hum Mol Genet. (2016) 25:3383–94. doi: 10.1093/hmg/ddw181
23. Li M, Luo XJ, Landén M, Bergen SE, Hultman CM, Li X, et al. Impact of a cis-associated gene expression SNP on chromosome 20q11.22 on bipolar disorder susceptibility, hippocampal structure and cognitive performance. Br J Psychiatry. (2016) 208:128–37. doi: 10.1192/bjp.bp.114.156976
24. Kuo PH, Chuang LC, Liu JR, Liu CM, Huang MC, Lin SK, et al. Identification of novel loci for bipolar I disorder in a multi-stage genome-wide association study. Prog Neuropsychopharmacol Biol Psychiatry. (2014) 51:58–64. doi: 10.1016/j.pnpbp.2014.01.003
25. Mühleisen TW, Leber M, Schulze TG, Strohmaier J, Degenhardt F, Treutlein J, et al. Genome-wide association study reveals two new risk loci for bipolar disorder. Nat Commun. (2014) 5:3339. doi: 10.1038/ncomms4339
26. Xu W, Cohen-Woods S, Chen Q, Noor A, Knight J, Hosang G, et al. Genome-wide association study of bipolar disorder in Canadian and UK populations corroborates disease loci including SYNE1 and CSMD1. BMC Med Genet. (2014) 15:2. doi: 10.1186/1471-2350-15-2
27. Chen DT, Jiang X, Akula N, Shugart YY, Wendland JR, Steele CJ, et al. Genome-wide association study meta-analysis of European and Asian-ancestry samples identifies three novel loci associated with bipolar disorder. Mol Psychiatry. (2013) 18:195–205. doi: 10.1038/mp.2011.157
28. Green EK, Hamshere M, Forty L, Gordon-Smith K, Fraser C, Russell E, et al. Replication of bipolar disorder susceptibility alleles and identification of two novel genome-wide significant associations in a new bipolar disorder case-control sample. Mol Psychiatry. (2013) 18:1302–7. doi: 10.1038/mp.2012.142
29. Cichon S, Mühleisen TW, Degenhardt FA, Mattheisen M, Miró X, Strohmaier J, et al. Genome-wide association study identifies genetic variation in neurocan as a susceptibility factor for bipolar disorder. Am J Hum Genet. (2011) 88:372–81. doi: 10.1016/j.ajhg.2011.01.017
30. Lee MT, Chen CH, Lee CS, Chen CC, Chong MY, Ouyang WC, et al. Genome-wide association study of bipolar I disorder in the Han Chinese population. Mol Psychiatry. (2011) 16:548–56. doi: 10.1038/mp.2010.43
31. Psychiatric GWAS Consortium Bipolar Disorder Working Group. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. (2011) 43:977–83. doi: 10.1038/ng.943
32. Yosifova A, Mushiroda T, Kubo M, Takahashi A, Kamatani Y, Kamatani N, et al. Genome-wide association study on bipolar disorder in the Bulgarian population. Genes Brain Behav. (2011) 10:789–97. doi: 10.1111/j.1601-183X.2011.00721.x
33. Djurovic S, Gustafsson O, Mattingsdal M, Athanasiu L, Bjella T, Tesli M, et al. A genome-wide association study of bipolar disorder in Norwegian individuals, followed by replication in Icelandic sample. J Affect Disord. (2010) 126:312–6. doi: 10.1016/j.jad.2010.04.007
34. Scott LJ, Muglia P, Kong XQ, Guan W, Flickinger M, Upmanyu R, et al. Genome-wide association and meta-analysis of bipolar disorder in individuals of European ancestry. Proc Natl Acad Sci U S A. (2009) 106:7501–6. doi: 10.1073/pnas.0813386106
35. Ferreira MA, O'Donovan MC, Meng YA, Jones IR, Ruderfer DM, Jones L, et al. Collaborative genome-wide association analysis supports a role for ANK3 and CACNA1C in bipolar disorder. Nat Genet. (2008) 40:1056–8. doi: 10.1038/ng.209
36. Dalmasso MC, de Rojas I, Olivar N, Muchnik C, Angel B, Gloger S, et al. The first genome-wide association study in the Argentinian and Chilean populations identifies shared genetics with Europeans in Alzheimer's disease. Alzheimers Dement. (2024) 20:1298–308. doi: 10.1002/alz.13522
37. Sherva R, Zhang R, Sahelijo N, Jun G, Anglin T, Chanfreau C, et al. African ancestry GWAS of dementia in a large military cohort identifies significant risk loci. Mol Psychiatry. (2023) 28:1293–302. doi: 10.1038/s41380-022-01890-3
38. Harper JD, Fan KH, Aslam MM, Snitz BE, DeKosky ST, Lopez OL, et al. Genome-wide association study of incident dementia in a community-based sample of older subjects. J Alzheimers Dis. (2022) 88:787–98. doi: 10.3233/JAD-220293
39. Wightman DP, Jansen IE, Savage JE, Shadrin AA, Bahrami S, Holland D, et al. A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer's disease. Nat Genet. (2021) 53:1276–82. doi: 10.1038/s41588-021-00921-z
40. Jansen IE, Savage JE, Watanabe K, Bryois J, Williams DM, Steinberg S, et al. Genome-wide meta-analysis identifies new loci and functional pathways influencing Alzheimer's disease risk. Nat Genet. (2019) 51:404–13. doi: 10.1038/s41588-018-0311-9
41. Kunkle BW, Grenier-Boley B, Sims R, Bis JC, Damotte V, Naj AC, et al. Genetic meta-analysis of diagnosed Alzheimer's disease identifies new risk loci and implicates Abeta, tau, immunity and lipid processing. Nat Genet. (2019) 51:414–30. doi: 10.1038/s41588-019-0358-2
42. Moreno-Grau S, de Rojas I, Hernández I, Quintela I, Montrreal L, Alegret M, et al. Genome-wide association analysis of dementia and its clinical endophenotypes reveal novel loci associated with Alzheimer's disease and three causality networks: The GR@ACE project. Alzheimers Dement. (2019) 15:1333–47. doi: 10.1016/j.jalz.2019.06.4950
43. Witoelar A, Rongve A, Almdahl IS, Ulstein ID, Engvig A, White LR, et al. Meta-analysis of Alzheimer's disease on 9,751 samples from Norway and IGAP study identifies four risk loci. Sci Rep. (2018) 8:18088. doi: 10.1038/s41598-018-36429-6
44. Jun G, Ibrahim-Verbaas CA, Vronskaya M, Lambert JC, Chung J, Naj AC, et al. A novel Alzheimer disease locus located near the gene encoding tau protein. Mol Psychiatry. (2016) 21:108–17. doi: 10.1038/mp.2015.23
45. Lambert JC, Ibrahim-Verbaas CA, Harold D, Naj AC, Sims R, Bellenguez C, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. (2013) 45:1452–8. doi: 10.1038/ng.2802
46. Seshadri S, Fitzpatrick AL, Ikram MA, DeStefano AL, Gudnason V, Boada M, et al. Genome-wide analysis of genetic loci associated with Alzheimer disease. JAMA. (2010) 303:1832–40. doi: 10.1001/jama.2010.574
47. Mukherjee S, Mez J, Trittschuh EH, Saykin AJ, Gibbons LE, Fardo DW, et al. Genetic data and cognitively defined late-onset Alzheimer's disease subgroups. Mol Psychiatry. (2020) 25:2942–51. doi: 10.1038/s41380-018-0298-8
48. Beecham GW, Martin ER, Li YJ, Slifer MA, Gilbert JR, Haines JL, et al. Genome-wide association study implicates a chromosome 12 risk locus for late-onset Alzheimer disease. Am J Hum Genet. (2009) 84:35–43. doi: 10.1016/j.ajhg.2008.12.008
49. Chia R, Sabir MS, Bandres-Ciga S, Saez-Atienzar S, Reynolds RH, Gustavsson E, et al. Genome sequencing analysis identifies new loci associated with Lewy body dementia and provides insights into its genetic architecture. Nat Genet. (2021) 53:294–303. doi: 10.1038/s41588-021-00785-3
50. Schrijvers EM, Schürmann B, Koudstaal PJ, van den Bussche H, Van Duijn CM, Hentschel F, et al. Genome-wide association study of vascular dementia. Stroke. (2012) 43:315–9. doi: 10.1161/STROKEAHA.111.628768
51. Reus LM, Jansen IE, Mol MO, van Ruissen F, van Rooij J, van Schoor NM, et al. Genome-wide association study of frontotemporal dementia identifies a C9ORF72 haplotype with a median of 12-G4C2 repeats that predisposes to pathological repeat expansions. Transl Psychiatry. (2021) 11:451. doi: 10.1038/s41398-021-01577-3
52. Ferrari R, Grassi M, Salvi E, Borroni B, Palluzzi F, Pepe D, et al. A genome-wide screening and SNPs-to-genes approach to identify novel genetic risk factors associated with frontotemporal dementia. Neurobiol Aging. (2015) 36:2904.e13–26. doi: 10.1016/j.neurobiolaging.2015.06.005
53. Ferrari R, Hernandez DG, Nalls MA, Rohrer JD, Ramasamy A, Kwok JB, et al. Frontotemporal dementia and its subtypes: a genome-wide association study. Lancet Neurol. (2014) 13:686–99. doi: 10.1016/S1474-4422(14)70065-1
54. Rodan LH, Spillmann RC, Kurata HT, Lamothe SM, Maghera J, Jamra RA, et al. Phenotypic expansion of CACNA1C-associated disorders to include isolated neurological manifestations. Genet Med. (2021) 23:1922–32. doi: 10.1038/s41436-021-01232-8
55. Oraki Kohshour M, Papiol S, Ching CRK, Schulze TG. Genomic and neuroimaging approaches to bipolar disorder. BJPsych Open. (2022) 8:e36. doi: 10.1192/bjo.2021.1082
56. Wang Z, Lin X, Luo X, Xiao J, Zhang Y, Xu J, et al. Pleiotropic association of CACNA1C variants with neuropsychiatric disorders. Schizophr Bull. (2023) 49:1174–84. doi: 10.1093/schbul/sbad073
57. Craddock N, Sklar P. Genetics of bipolar disorder. Lancet. (2013) 381:1654–62. doi: 10.1016/S0140-6736(13)60855-7
58. Novaes de Oliveira Roldan AC, Fernandes Júnior LCC, de Oliveira CEC, Nunes SOV. Impact of ZNF804A rs1344706 or CACNA1C rs1006737 polymorphisms on cognition in patients with severe mental disorders: A systematic review and meta-analysis. World J Biol Psychiatry. (2023) 24:195–208. doi: 10.1080/15622975.2022.2097308
59. Arts B, Simons CJ, Os JV. Evidence for the impact of the CACNA1C risk allele rs1006737 on 2-year cognitive functioning in bipolar disorder. Psychiatr Genet. (2013) 23:41–2. doi: 10.1097/YPG.0b013e328358641c
60. Lin K, Xu G, Shi L, Lu W, Guan L, Ouyang H, et al. CACNA1C polymorphisms impact cognitive recovery in patients with bipolar disorder in a six-week open-label trial. Sci Rep. (2017) 7:7022. doi: 10.1038/s41598-017-07368-5
61. Dedic N, Pöhlmann ML, Richter JS, Mehta D, Czamara D, Metzger MW, et al. Cross-disorder risk gene CACNA1C differentially modulates susceptibility to psychiatric disorders during development and adulthood. Mol Psychiatry. (2018) 23:533–43. doi: 10.1038/mp.2017.133
62. Wang F, McIntosh AM, He Y, Gelernter J, Blumberg HP. The association of genetic variation in CACNA1C with structure and function of a frontotemporal system. Bipolar Disord. (2011) 13:696–700. doi: 10.1111/j.1399-5618.2011.00963.x
63. Smedler E, Abé C, Pålsson E, Ingvar M, Landén M. CACNA1C polymorphism and brain cortical structure in bipolar disorder. J Psychiatry Neurosci. (2019) 45:182–7. doi: 10.1503/jpn.190029
64. Soeiro-de-Souza MG, Lafer B, Moreno RA, Nery FG, Chile T, Chaim K, et al. The CACNA1C risk allele rs1006737 is associated with age-related prefrontal cortical thinning in bipolar I disorder. Transl Psychiatry. (2017) 7:e1086. doi: 10.1038/tp.2017.57
65. Blennow K, Hampel H, Weiner M, Zetterberg H. Cerebrospinal fluid and plasma biomarkers in Alzheimer disease. Nat Rev Neurol. (2010) 6:131–44. doi: 10.1038/nrneurol.2010.4
66. Jakobsson J, Pålsson E, Sellgren C, Rydberg F, Ekman A, Zetterberg H, et al. CACNA1C polymorphism and altered phosphorylation of tau in bipolar disorder. Br J Psychiatry. (2016) 208:195–6. doi: 10.1192/bjp.bp.114.159806
67. Bocharova A, Vagaitseva K, Marusin A, Zhukova N, Zhukova I, Minaycheva L, et al. Association and gene-gene interactions study of late-onset alzheimer's disease in the Russian population. Genes (Basel). (2021) 12:1647. doi: 10.3390/genes12101647
68. Peng C, Ding Y, Yi X, Dong Z, Cao L, Li Q, et al. Association study of CACNA1C polymorphisms with large artery atherosclerotic stroke in Chinese Han population. Neurol Res. (2018) 40:677–82. doi: 10.1080/01616412.2018.1461307
69. Smeland OB, Bahrami S, Frei O, Shadrin A, O'Connell K, Savage J, et al. Genome-wide analysis reveals extensive genetic overlap between schizophrenia, bipolar disorder, and intelligence. Mol Psychiatry. (2020) 25:844–53. doi: 10.1038/s41380-018-0332-x
70. Geng Y, Bush M, Mosyak L, Wang F, Fan QR. Structural mechanism of ligand activation in human GABA(B) receptor. Nature. (2013) 504:254–9. doi: 10.1038/nature12725
71. Fatemi SH, Stary JM, Earle JA, Araghi-Niknam M, Eagan E. GABAergic dysfunction in schizophrenia and mood disorders as reflected by decreased levels of glutamic acid decarboxylase 65 and 67 kDa and Reelin proteins in cerebellum. Schizophr Res. (2005) 72:109–22. doi: 10.1016/j.schres.2004.02.017
72. Fatemi SH, Folsom TD, Thuras PD. Deficits in GABA(B) receptor system in schizophrenia and mood disorders: a postmortem study. Schizophr Res. (2011) 128:37–43. doi: 10.1016/j.schres.2010.12.025
73. Solas M, Puerta E, Ramirez MJ. Treatment options in alzheimer´s disease: the GABA story. Curr Pharm Des. (2015) 21:4960–71. doi: 10.2174/1381612821666150914121149
74. Calvo-Flores Guzmán B, Vinnakota C, Govindpani K, Waldvogel HJ, Faull RLM, Kwakowsky A. The GABAergic system as a therapeutic target for Alzheimer's disease. J Neurochem. (2018) 146:649–69. doi: 10.1111/jnc.14345
75. Carello-Collar G, Bellaver B, Ferreira PCL, Ferrari-Souza JP, Ramos VG, Therriault J, et al. The GABAergic system in Alzheimer's disease: a systematic review with meta-analysis. Mol Psychiatry. (2023) 28:5025–5036. doi: 10.1038/s41380-023-02140-w
76. Wolff M, Johannesen KM, Hedrich UBS, Masnada S, Rubboli G, Gardella E, et al. Genetic and phenotypic heterogeneity suggest therapeutic implications in SCN2A-related disorders. Brain. (2017) 140:1316–36. doi: 10.1093/brain/awx054
77. Sanders SJ, Campbell AJ, Cottrell JR, Moller RS, Wagner FF, Auldridge AL, et al. Progress in understanding and treating SCN2A-mediated disorders. Trends Neurosci. (2018) 41:442–56. doi: 10.1016/j.tins.2018.03.011
78. Dickinson D, Straub RE, Trampush JW, Gao Y, Feng N, Xie B, et al. Differential effects of common variants in SCN2A on general cognitive ability, brain physiology, and messenger RNA expression in schizophrenia cases and control individuals. JAMA Psychiatry. (2014) 71:647–56. doi: 10.1001/jamapsychiatry.2014.157
79. Kruth KA, Grisolano TM, Ahern CA, Williams AJ. SCN2A channelopathies in the autism spectrum of neuropsychiatric disorders: a role for pluripotent stem cells? Mol Autism. (2020) 11:23. doi: 10.1186/s13229-020-00330-9
80. Ceylan H. Integrated bioinformatics analysis to identify alternative therapeutic targets for alzheimer's disease: insights from a synaptic machinery perspective. J Mol Neurosci. (2022) 72:273–86. doi: 10.1007/s12031-021-01893-9
81. Rojnik M, Jevnikar ZR, Doljak B, Turk S, Zidar N, Kos J. The influence of differential processing of procathepsin H on its aminopeptidase activity, secretion and subcellular localization in human cell lines. Eur J Cell Biol. (2012) 91:757–64. doi: 10.1016/j.ejcb.2012.05.004
82. Wingo AP, Liu Y, Gerasimov ES, Gockley J, Logsdon BA, Duong DM, et al. Integrating human brain proteomes with genome-wide association data implicates new proteins in Alzheimer's disease pathogenesis. Nat Genet. (2021) 53:143–6. doi: 10.1038/s41588-020-00773-z
83. Li Y, Xu M, Xiang BL, Li X, Zhang DF, Zhao H, et al. Functional genomics identify causal variant underlying the protective CTSH locus for Alzheimer's disease. Neuropsychopharmacology. (2023) 48:1555–66. doi: 10.1038/s41386-023-01542-2
84. Clementi ME, Sampaolese B, Lazzarino G, Giardina B. Effect of punicalagin and resveratrol on methionine sulfoxide reductase: A possible protective contribution against alzheimer's disease. J Prev Alzheimers Dis. (2015) 2:33–7. doi: 10.14283/jpad.2015.40
85. Moskovitz J, Walss-Bass C, Cruz DA, Thompson PM, Bortolato M. Methionine sulfoxide reductase regulates brain catechol-O-methyl transferase activity. Int J Neuropsychopharmacol. (2014) 17:1707–13. doi: 10.1017/S1461145714000467
86. Pal R, Oien DB, Ersen FY, Moskovitz J. Elevated levels of brain-pathologies associated with neurodegenerative diseases in the methionine sulfoxide reductase A knockout mouse. Exp Brain Res. (2007) 180:765–74. doi: 10.1007/s00221-007-0903-6
87. Ni P, Ma X, Lin Y, Lao G, Hao X, Guan L, et al. Methionine sulfoxide reductase A (MsrA) associated with bipolar I disorder and executive functions in A Han Chinese population. J Affect Disord. (2015) 184:235–8. doi: 10.1016/j.jad.2015.06.004
88. Jallow AW, Nguyen DPQ, Sanotra MR, Hsu CH, Lin YF, Lin YF. A comprehensive bibliometric analysis of global research on the role of acrolein in Alzheimer's disease pathogenesis: involvement of amyloid-beta. Front Aging Neurosci. (2024) 16:1378260. doi: 10.3389/fnagi.2024.1378260
89. Cejudo-Martin P, Yuen A, Vlahovich N, Lock P, Courtneidge SA, Díaz B. Genetic disruption of the sh3pxd2a gene reveals an essential role in mouse development and the existence of a novel isoform of tks5. PloS One. (2014) 9:e107674. doi: 10.1371/journal.pone.0107674
90. Van der Auwera S, Ameling S, Wittfeld K, Frenzel S, Bülow R, Nauck M, et al. Circulating microRNA miR-425-5p Associated with Brain White Matter Lesions and Inflammatory Processes. Int J Mol Sci. (2024) 25:887. doi: 10.3390/ijms25020887
91. Armstrong NJ, Mather KA, Sargurupremraj M, Knol MJ, Malik R, Satizabal CL, et al. Common genetic variation indicates separate causes for periventricular and deep white matter hyperintensities. Stroke. (2020) 51:2111–21. doi: 10.1161/STROKEAHA.119.027544
92. Islam T, Rahman MR, Khan A, Moni MA. Integration of Mendelian randomisation and systems biology models to identify novel blood-based biomarkers for stroke. J BioMed Inform. (2023) 141:104345. doi: 10.1016/j.jbi.2023.104345
93. Laumet G, Petitprez V, Sillaire A, Ayral AM, Hansmannel F, Chapuis J, et al. A study of the association between the ADAM12 and SH3PXD2A (SH3MD1) genes and Alzheimer's disease. Neurosci Lett. (2010) 468:1–2. doi: 10.1016/j.neulet.2009.10.040
94. Liu S, Wang L, Sun N, Yang C, Liu Z, Li X, et al. The gender-specific association of rs334558 in GSK3β with major depressive disorder. Med (Baltimore). (2017) 96:e5928. doi: 10.1097/MD.0000000000005928
95. Lin YF, Huang MC, Liu HC. Glycogen synthase kinase 3β gene polymorphisms maybe associated with bipolar I disorder and the therapeutic response to lithium. J Affect Disord. (2013) 147:401–6. doi: 10.1016/j.jad.2012.08.025
Keywords: bipolar disorder, dementia, gene, single nucleotide polymorphisms, genome-wide association study
Citation: Hirakawa H and Terao T (2024) The genetic association between bipolar disorder and dementia: a qualitative review. Front. Psychiatry 15:1414776. doi: 10.3389/fpsyt.2024.1414776
Received: 09 April 2024; Accepted: 05 August 2024;
Published: 20 August 2024.
Edited by:
Tasuku Hashimoto, International University of Health and Welfare (IUHW), JapanCopyright © 2024 Hirakawa and Terao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hirofumi Hirakawa, aGlyYS1oaXJvQG9pdGEtdS5hYy5qcA==
 Hirofumi Hirakawa
Hirofumi Hirakawa Takeshi Terao
Takeshi Terao