- 1Zhengzhou Research Base, State Key Laboratory of Cotton Bio-breeding and Integrated Utilization, School of Agricultural Sciences, Zhengzhou University, Zhengzhou, China
- 2Henan Key Laboratory of Ion-Beam Green Agriculture Bioengineering, Zhengzhou University, Zhengzhou, China
The denitrification process is known to contribute to soil nitrogen (N) loss, which is strongly affected by fertilization strategies; however, the effects of distinct straw retention modes on soil denitrification activity have rarely been discriminated and the underlying mechanisms remain unclear. This study coupled field and incubation experiments to explore the characteristics of soil denitrification activity, soil and standing water physicochemical properties, and the abundance, community diversity, and co-occurrence network of nosZ denitrifiers, based on a paddy field implementing 10-year straw retention under a rice–wheat rotation system. Four straw retention treatments with equivalent chemical fertilizers were applied, namely no straw (NS), wheat straw only (WS), rice straw only (RS), and wheat and rice straw (WRS). Results indicated a significant increase (by 41.93–45.80% when compared to that with NS) in the soil denitrification activity with RS and WRS. Correspondingly, treatments with rice straw retention resulted in the development of a similar community composition (P < 0.05), structure (P = 0.001), and more positively interconnected network, as well as similar specific keystone taxa of nosZ denitrifiers, relative to those in non-rice straw mode. Under long-term rice straw retention conditions, the core nosZ-denitrifying phylogroups shifted (r = 0.83, P < 0.001), with the recruitment of keystone taxa from the phyla Bacteroidetes and Euryarchaeota playing a key role in enhancing denitrification activity and stimulating N loss. Accordingly, in a rice–wheat rotation field, the practice of wheat straw retention in a single season is recommended because it will not markedly sacrifice soil N availability impaired by the denitrification process.
1 Introduction
Owing to nutrient-rich and aerobic–anaerobic interfaces (Du et al., 2024; Nie et al., 2019), the flooded paddy field serves as a hotspot for denitrification processes, which reduce nitrate (NO3−) to atmospheric nitric oxide (NO), nitrous oxide (N2O), or dinitrogen (N2) in a stepwise manner (Seitzinger et al., 2006). The end-products of heterotrophic denitrification under anoxic or microaerophilic conditions are dominated by N2, followed by N2O and NO (Jahangir et al., 2012), jeopardizing N use efficiency in agricultural ecosystems and contributing to global warming (Philippot et al., 2007). Approximately 36% of N fertilizer was estimated escape from paddy fields through denitrification pathways (Aulakh et al., 2001; Ju et al., 2009).
The practice of straw retention has long been recommended to improve soil N availability, as well as organic carbon (C) sequestration (Thangarajan et al., 2013; Xu et al., 2024); moreover, organic C sources comprise one of the crucial drivers of the denitrification process by providing it with energy and electron donors (Wang et al., 2023). However, the effects of crop residue retention on denitrification activity have varied considerably, with 2.62% to 460.0% of denitrification activity enhanced by straw amendment (Wang et al., 2021, 2022, 2024), among previous studies. Those differences among distinct straw retention modes have rarely been categorized, limiting our knowledge of how soil denitrification pathways are affected by crop residues. It warrants a thorough investigation of denitrification activity by incorporating different modes of straw retention in a two-crop rotation field, especially under the same fertilization conditions, which is critical to explore and guide field management strategies with respect to crop residues.
Microorganisms are the key factors driving soil nitrogen (N) transformation. The nitrous oxide reductase, encoded by the nosZ gene, is increasingly assessed to explore the abundance and structure of denitrifiers and determine the denitrification activity in the soil environment (Duan et al., 2018; Qin et al., 2021; Tao et al., 2022; Tang et al., 2024). Recent studies have highlighted the significant diversity of nosZ-denitrifying communities, which vary under different agricultural management practices, including straw retention (Chen et al., 2025; Xie et al., 2024; Zhou et al., 2022). Furthermore, it has been shown that the nosZ gene exists in two distinct clades (nosZ-I and nosZ-II), each contributing differently to the denitrification process under varying soil conditions (Bano et al., 2024). Despite extensive efforts regarding fertilization regimes, much less is available about the effects of different straw retention modes on nosZ-denitrifiers, which directly regulate the organic C sources that might regulate the ability of denitrifiers to compete for soil nitrate (NO3−) or nitrite (N2O) (Giles et al., 2012; Taylor and Townsend, 2010). Moreover, accumulating studies have reported insignificant relationships between denitrification activity and the abundance or composition of denitrifiers (Attard et al., 2011; Kou et al., 2019), suggesting the necessity of exploring key underlying taxa of the denitrifiers, which dominate the specific ecosystem processes in different habitats (Liang et al., 2021; Kong et al., 2023). Therefore, we hypothesized that specific keystone taxa of denitrifiers are likely stimulated under certain straw retention modes, thereby increasing the soil denitrification activity. This hypothesis was verified based on a field experiment employed with a 10-year program of straw retention. The objectives of this study were (1) to discriminate the soil denitrification activity responses across different straw retention modes, (2) to characterize the abundance, diversity, and co-occurrence network of denitrifiers, and (3) to identify the key soil and standing water factors that drive denitrification activity.
2 Materials and methods
2.1 Field experiment and sampling
The field experiment was initiated in 2010 under a summer rice (Oryza sativa L.)–winter wheat (Triticum aestivum L.) rotation at the Agro–Ecological Station of the Chinese Ecosystem Research Network in the Taihu Lake region of China. The experimental soil belongs to the category of Anthrosols and originates from lacustrine sediments. The field experiment adopted a randomized complete block design with three replicates of four treatments as follows: no straw retention (NS), wheat straw retention in the summer season (WS), rice straw retention in the winter season (RS), and wheat and rice straw return in the summer and winter seasons, respectively (WRS). All four treatment groups received the same amount of mineral N, P, and K fertilizers (Supplementary Table S1). Crop residues in the straw retention treatments, including rice and wheat residues, were chopped into pieces measuring 5 to 10 cm in length and incorporated into the soil in a rotary manner approximately one week before the next crop season.
During the rice season in July 2020, three replicate soil (0–15 cm) and standing water samples were randomly collected from each plot and stored in sterile plastic bags and bottles, respectively. The water samples were filtered through 0.45 μm membranes and processed for chemical analysis. The soil samples were homogenized by passing them through a 2 mm sieve, divided into three parts, and then processed for chemical analysis (stored at 4°C), soil incubation experiments (stored at 4°C), and molecular assays (stored at −80°C).
2.2 Determination of chemical properties
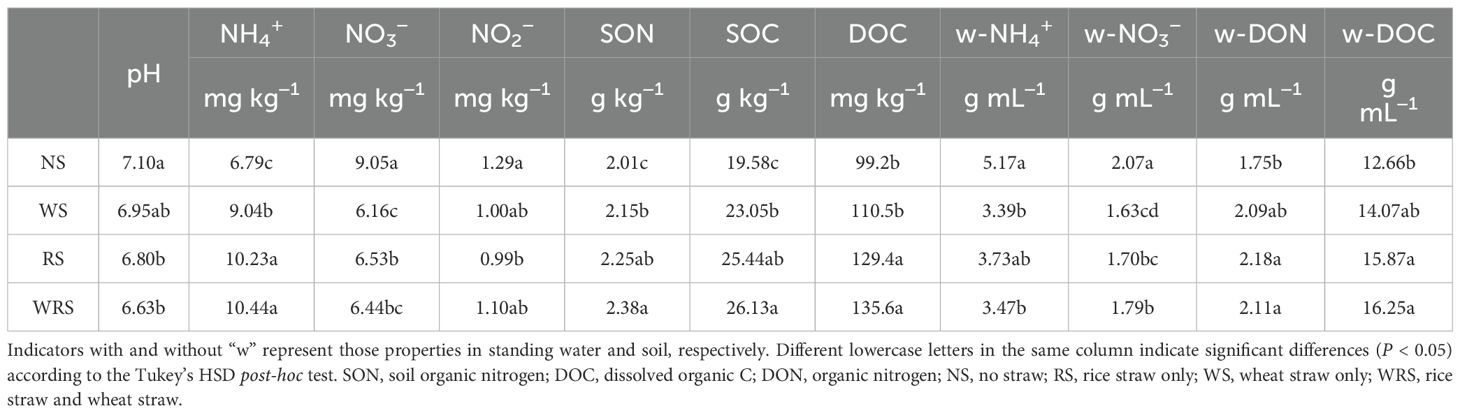
The ammonium nitrogen and nitrite nitrogen in the filtered standing water samples were measured using an ultraviolet spectrophotometer (UV-1280, Shimadzu, Japan) according to Committee of Analytical Method of Water and Wastewater (2002). The concentration of water organic nitrogen was analyzed using a Multi N/C 2100S analyzer (Analytikjena GmbH, Germany) (Ghani et al., 2003). Soil pH was measured using a portable meter (Mettler Toledo, Switzerland) at a soil:water ratio of 1:2.5. Soil electrical conductivity (EC) was detected based on a 1:2.5 soil-to-water ratio using a conductivity salinity meter (Y SI-30, Yellow Springs, USA). Soil organic carbon (SOC) and dissolved organic C (DOC) were measured using TOC-VCPH equipment (Shimadzu, Japan) based on Lu (2000). Soil NH4+-N, NO3−-N, and nitrite nitrogen (NO2−-N) were desorbed with 2 mol L−1 KCl (1:5 soil:solution) and analyzed with an Auto-Analyzer (Skalar, The Netherlands). Soil total nitrogen was measured using the dry combustion method (Lu, 2000). Soil organic nitrogen (SON) was calculated as the difference between soil total nitrogen and the combined soil NH4+, NO3−, and NO2−. Soil available potassium (AK) and phosphorus (AP) were desorbed with 1 mol L−1 NaHCO3 and measured by atomic absorption spectrophotometry and the molybdenum-blue method, respectively. The physicochemical properties of soil and standing water are presented in Table 1.
2.3 Determination of denitrification enzyme activities
The denitrification enzyme activity (DEA) was measured in accordance with the acetylene (C2H2) inhibition method modified from Barton et al. (2000). Briefly, three replicates (10 g) of each fresh soil sample were added to 120 mL glass flasks. Each was amended with 20 mL of solution containing glucose (300 μg C−1 soil) and potassium nitrate (50 μg N g−1 soil). Each flask was sealed with rubber septa and an aluminum crimp cap, and the head space was evacuated and purged with helium gas for 1 min thereafter. Then, 10% of the headspace was replaced with C2H2 (12 mL) through injection. Finally, the replicates were incubated in the dark on a shaker (180 rpm, 25 °C) for 30, 60, 90, and 120 min. Approximately 5 mL of gas samples was transferred into pre-evacuated glass vials and determined using a gas chromatograph (GC-8A, Shimadzu, Japan).
2.4 DNA extraction, real-time PCR assay, and high-throughput sequencing
Genomic DNA was extracted from the triplicate subsamples (0.33 g fresh soil) with Power Soil DNA Isolation Kits (QIAGEN, USA). The quantity and quality of the extracted DNA were measured with a NanoDrop spectrophotometer (Thermo Fisher Scientific, Wilmington, USA). The extracted DNA was divided into two parts, with one fraction for real-time PCR assays and the other fraction for high-throughput sequencing.
The denitrification-associated nosZ gene was amplified using the primer pairs nosZF (5′-CGCRACGGCAASAAGGTSMSSGT-3′) and nosZ (5′-CAKRTGCAKSGCRTGGCAGAA-3′). The reaction mixtures (10 μL) consisted of 5 μL of 2× SYBR green mix II (TaKaRa Biotechnology Co. Ltd., Dalian, China), 0.2 μL of 50× Rox Reference Dye (TaKaRa Biotechnology Co. Ltd., Dalian, China), 0.2 μL (10 μM) of forward and reverse primers, 5 ng of DNA template, and deionized water. The amplification systems consisted of 5 μL of SYBR Master Mix, 0.2 μL (10 μM) of forward and reverse primers, 1 μL of DNA template, and 3.6 μL of double-distilled water (ddH2O). qPCR assay was performed at 95°C for 5 min (denaturation), followed by 40 cycles at 95°C for 30 s, 60°C for 30 s, and 72°C for 60 s (Henry et al., 2006). The qPCR amplification efficiencies and R2 value of the triplicates were 91.4%–95.6% and > 0.99, respectively.
High-throughput sequencing of the nosZ gene was performed using the same primers (nosZF/nosZR) as those for qPCR. The conditions of PCR amplification were as follows: denaturation at 95°C for 3 min; followed by 35 cycles at 94°C for 30 s, 52°C for 45 s, and 72°C for 20 s; and a final elongation at 72°C for 5 min (Delorme et al., 2003). The PCR products were then purified, quantified, and sent for paired-end sequencing (2 × 300 bp) on an Illumina MiSeq platform (Illumina, San Diego, CA, USA).
2.5 Statistical analysis
Raw sequence data were assembled using FLASH2 (Magoč and Salzberg, 2011) and quality filtered using VSEARCH v2.15 (Rognes et al., 2016) and unoise3 in usearch (Edgar, 2016). In total, 769136 high-quality sequences were obtained from 16 samples. The high-quality sequences were clustered into operational taxonomic units (OTUs) based on a 97% nucleotide similarity cutoff (Edgar, 2010), and 2691 OTUs were finally generated. The taxonomic annotations of the OTUs were in accordance with the RDP Functional Gene Repository (FunGene, http://fungene.cme.msu.edu/) using a confidence threshold of 80%. The sequence data have been deposited in the NCBI Sequence Read Archive under the accession number PRJNA668401.
All data were tested for homogeneity of variance with Levene’s tests before analysis. One-way analysis of variance (ANOVA), permutation analysis of variance (PERMANOVA) tests, and the Kruskal-Wallis H-test were performed with SPSS 22.0, R 3.2.1 (http://www.rproject.org/), and STAMP, respectively, to test the significant differences in specific variables among treatment groups. Pearson correlation analysis was used to reveal the relationship between the activity and abundance of denitrifying bacteria using SPSS (version 20.0). Principal coordinate analysis (PCoA, vegan in R) was conducted to visualize assemblage conditions of the denitrifying bacterial community. Random forest analysis (RANDOMFOREST package in R) was used to evaluate the importance of environmental predictors for denitrifying bacteria. The co-occurrence networks were constructed based on the Molecular Ecological Network Analyses Pipeline (MENA) and visualized with Gephi 0.9.2 to detect the interactions among denitrifying bacteria. Structural equation modeling (SEM) was performed to investigate the direct and indirect effects of parameters on DEAs using Amos 21.
3 Results
3.1 Physiochemical characteristics of standing water and soil
The physiochemical characteristics in the paddy field differed among distinct straw retention treatment groups (Table 1). Inorganic N, especially NH4+, comprised the largest proportion of N in the standing water. Compared to those with NS treatment, concentrations of NH4+ and NO3− in the standing water significantly decreased, whereas those of DON and DOC increased after straw retention (P < 0.05). The conditions in the soil were much different. First, organic N (i.e., SON) took the dominant portion of soil N, followed by NH4+, NO3−, and NO2-. More importantly, compared to those with NS treatment, soil pH and soil NO3− and NO2− contents decreased, whereas those of NH4+, SON, SOC, and DOC increased, after the 7-year implementation of straw retention (P < 0.05). Furthermore, whether in the standing water or soil, treatment groups with rice straw amendment (RS or WRS) showed more analogous trends. These results revealed that both in the standing water and soil of the paddy field, the NO3− value was reduced, and the organic C and N values were enhanced by the practice of straw retention, and particularly by rice straw amendment.
3.2 Denitrification enzyme activity and the abundance of nosZ denitrifiers
DEA was used to characterize the potential soil denitrification activity. In the current experimental field, soil DEA was observed to vary from 14.48 to 20.55 mg N2O-N kg−1 h−1 (Figure 1). Although the DEA value increased as an effect of straw retention, only treatments with rice straw amendment (RS and WRS) resulted in significant differences, of which the increments were estimated to be 41.93% and 45.80%, respectively. These results indicate that the employment of rice straw retention might amplify denitrifying N loss in the rice–wheat rotation system.
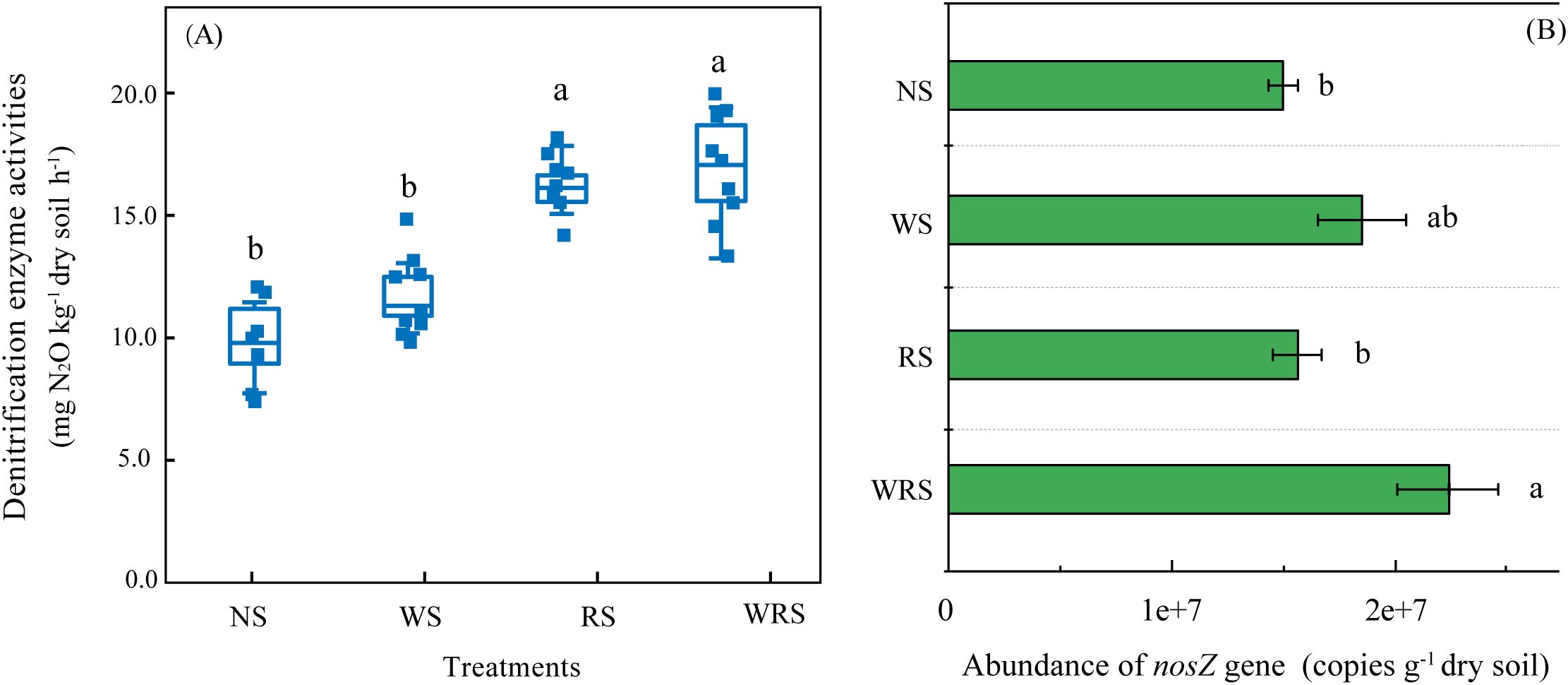
Figure 1. Denitrification enzyme activities (A) and the abundance of the nosZ gene (B) under the four fertilization regimes. Error bars denote the standard errors (n = 3) and are accompanied by different lowercase letters indicating significant differences (P < 0.05) among the four treatment groups according to Tukey’s HSD post-hoc test. NS, no straw; RS, rice straw only; WS, wheat straw only; WRS, rice straw and wheat straw.
Analogous with DEA results, the abundance of the denitrifier-nosZ gene, which ranged from 1.52 to 2.27 × 107 copies g−1 dry soil, was enhanced with straw retention treatments. However, relative to that with NS, only the WRS treatment group showed significantly higher levels of nosZ gene abundance, which also did not correlate with the soil DEA (P > 0.05). The investigation into different phylogroups, but not the copy numbers of denitrifiers, thus appears to be particularly important to account for soil DEA changes as affected by crop residues.
3.3 Beta-diversity and co-occurrence networks of nosZ-denitrifying communities
The characteristics of nosZ-denitrifying communities showed clear variations in response to different straw retention patterns. In the current study, 2691 clustered OTUs were phylogenetically grouped as 69.9% bacteria and 30.1% archaea (Figure 2A), which were predominated by Planctomycetes (19.0%–22.3%), Euryarchaeota (15.1%–22.3%), and Proteobacteria (10.8%–12.7%) at the phylum level (Figure 2B). The ANOVA and STAMP results demonstrated that under treatments with rice straw retention (RS and WRS treatments), whether at the family or genera level, nosZ denitrifiers affiliated with the phyla Euryarchaeota (Haloferacaceae, Archaeoglobaceae, Halonotius) and Bacteroidetes (Cyclobacteriaceae, Lunatimonas) increased in relative abundance (P < 0.05), whereas those affiliated with the phylum Planctomycetes (Phycisphaeraceae, Planctomicrobium, Telmatocola) decreased (P < 0.05) (Figures 3A, B; Supplementary Table S3). PCoA and PERMANOVA results further identified the community distribution characteristics of nosZ denitrifiers, revealing that RS and WRS groups were significantly differentiated relative to the NS and WS groups (Figure 3C).
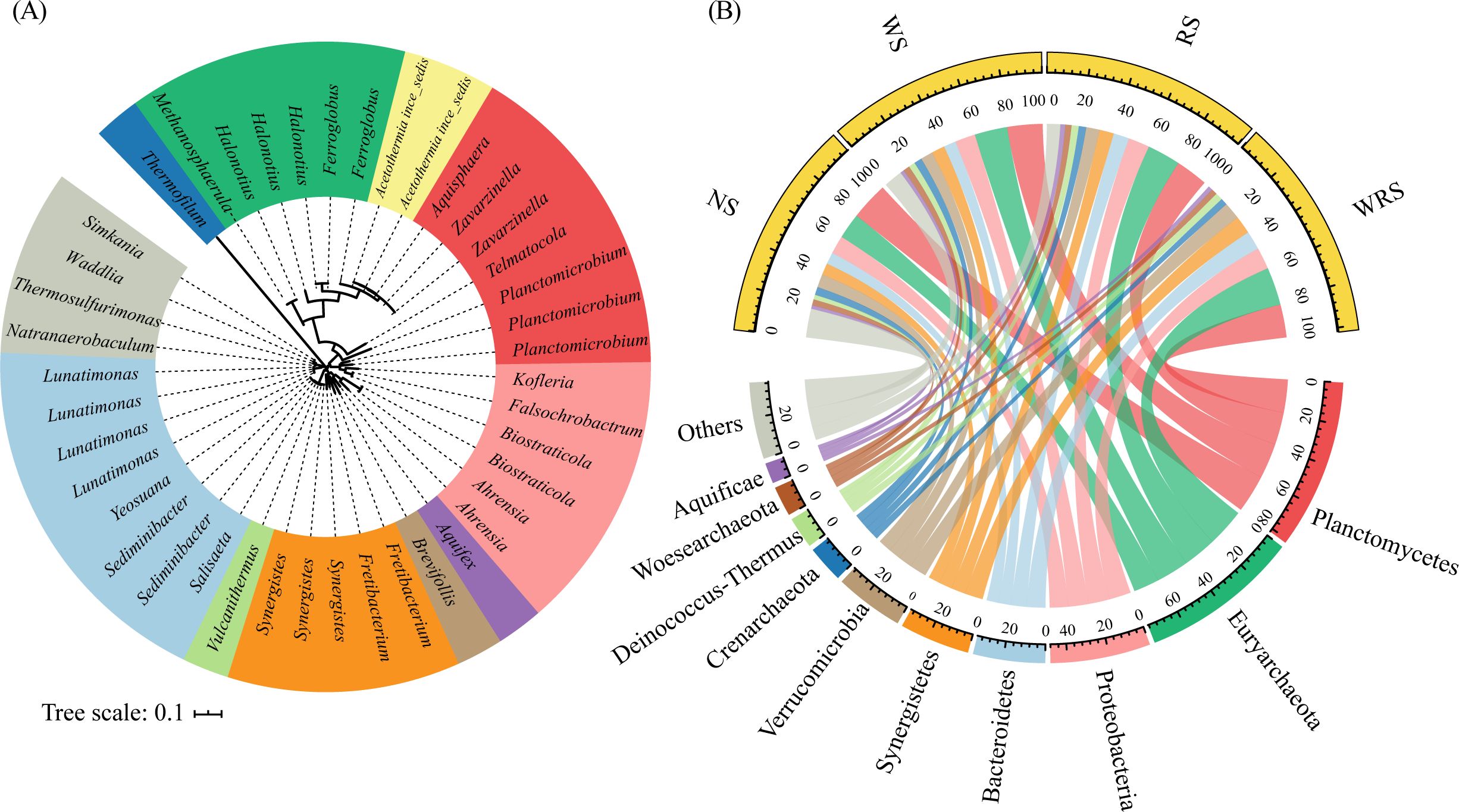
Figure 2. Circular maximum likelihood phylogenetic tree (A) and the community compositions (B), presented based on genus and phylum levels, respectively, of the denitrifying bacteria among treatment groups. The tree is based on the nosZ gene sequences of the most abundant operational taxonomic units. Genera in (A) are color-coded by phylum in (B). NS, no straw; RS, rice straw only; WS, wheat straw only; WRS, rice straw and wheat straw.
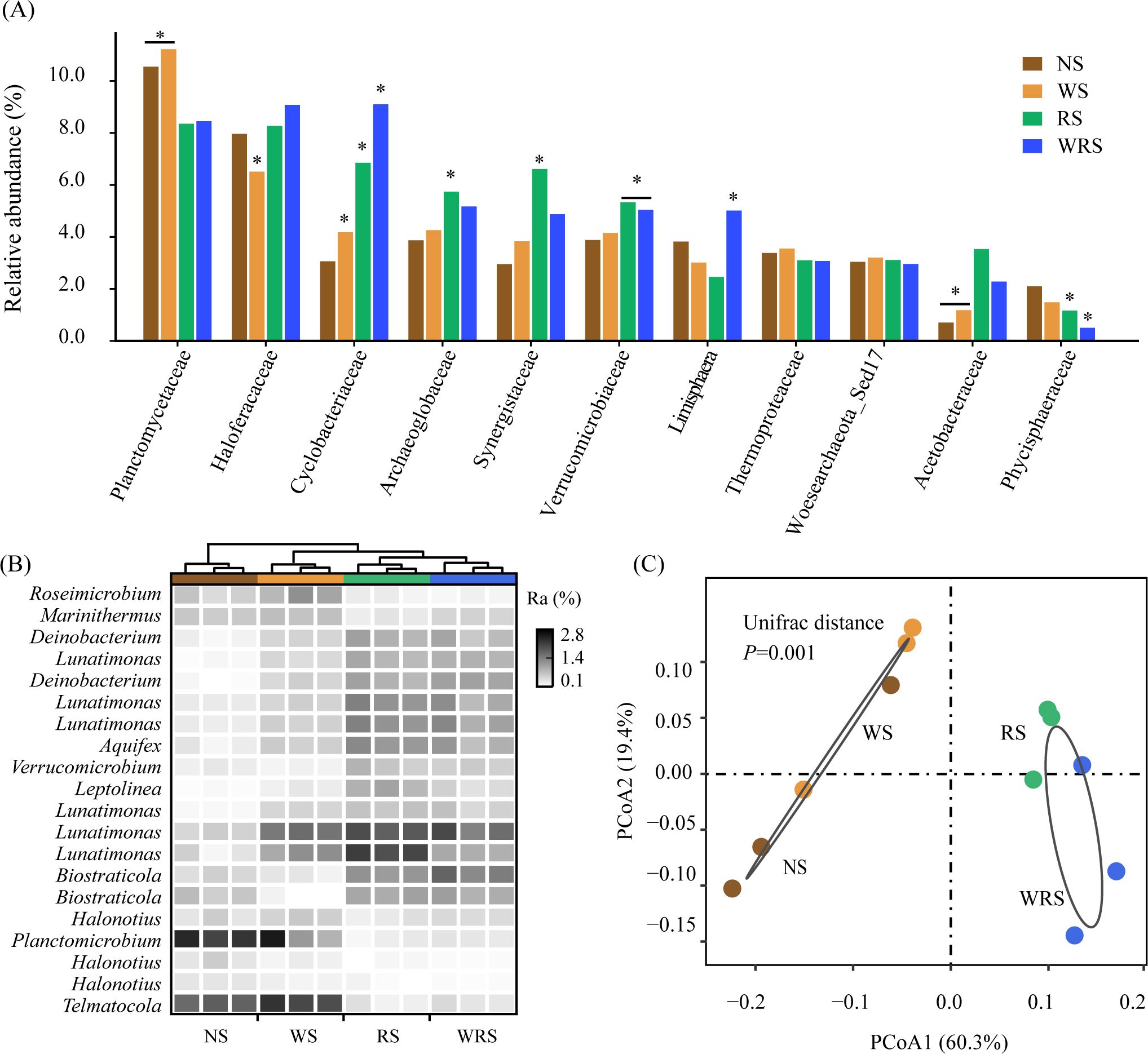
Figure 3. Differences in the community compositions of denitrifying bacteria at family (A) and genus (B) levels and in the bacterial community structures (C) among treatment groups. The statistical differences were calculated using Tukey’s HSD post-hoc test (marked with asterisks, P < 0.05, n = 3), Kruskal-Wallis H-test (P < 0.05, n = 3), and PERMANOVA test (P = 0.001, n = 6) in (A–C), respectively. NS, no straw; RS, rice straw only; WS, wheat straw only; WRS, rice straw and wheat straw.
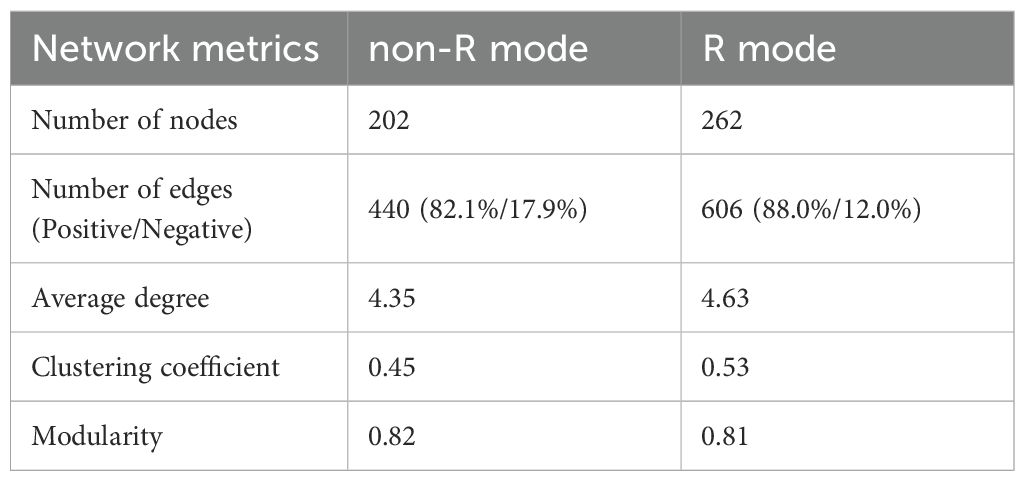
These findings suggested that treatments with and without rice straw resulted in pairwise similarity concerning both the community composition and structure of nosZ denitrifiers. Correspondingly, R mode and non-rice straw (non-R) mode co-occurrence networks were separately constructed to identify the key phylogroups and interactions among the nosZ denitrifiers (Figure 4). Generally, non-R and R mode networks consisted of 202 and 262 nodes and 440 and 606 edges, respectively, the latter possessing a higher ratio of positive associations (88.0% > 82.1%), as well as higher average degree and clustering coefficient values, as shown from the topological features (Figure 4A; Table 2). Correspondingly, the Zi-Pi plot showed four (two module hubs and two connectors) and eight (three module hubs and five connectors) hub nodes (i.e., core OTUs), which are representative of keystone taxa of denitrifiers, within the non-R and R networks, respectively (Figure 4B; Supplementary Table S3). The keystone taxa include OTUs prominently from the phylum Planctomycetes (OTU2, OTU195) in the W network, whereas OTUs were prominently from the phyla Euryarchaeota (OTU585, OTU93, OTU414) and Bacteroidetes (OTU100, OTU90) in the R+S network. These findings revealed that the nosZ denitrifiers in the R mode were more positively interconnected and recruited more and specific keystone taxa, as compared with those in non-R mode.
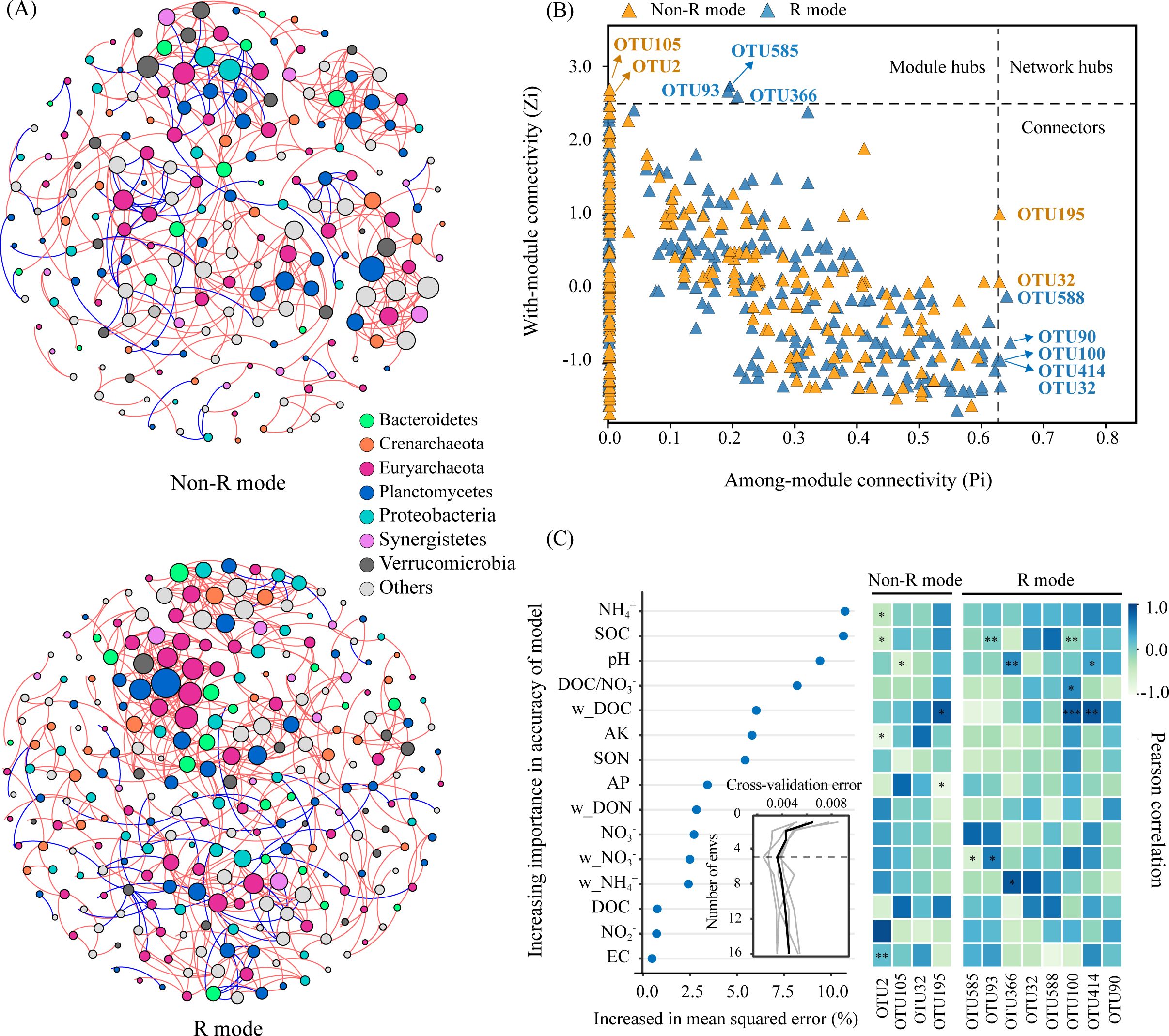
Figure 4. Co-occurrence networks (A) and the keystone taxa (B) of denitrifying bacterial operational taxonomic units (OTUs) based on their topological roles, and the relationships of keystone taxa with soil environmental predictors calculated based on the random forest model (C). Non-R and R modes integrate treatments without (NS+WS) and with (RS+WRS) rice straw retention, respectively. Nodes in (A) are colored based on the phylum level, and their sizes are proportional to the number of degrees; the red and blue edges represent positive and negative correlations between two nodes, respectively, the thicknesses of which are proportional to the value of Spearman’s correlation coefficients. Keystone taxa of co-occurrence networks represented by hubs and connectors in (B) are categorized by threshold lines of Zi = 2.5 and Pi = 0.625, respectively. P-values < 0.05, < 0.01 and < 0.001 are indicated using *, **, and *** in (C), respectively. NS, no straw; RS, rice straw only; WS, wheat straw only; WRS, rice straw and wheat straw.
3.4 Associations between denitrification activity and abiotic or biotic environmental factors
Random forest analyses and Pearson correlation results demonstrated that the nosZ denitrifiers and associated keystone taxa were predominantly adjusted by soil NH4+, SOC, pH, DOC/NO3−, and DOC (in order of significance) (Figure 4C). On this basis, the direct and indirect effects of environmental factors on soil DEA, under the R and non-R modes, respectively, were assessed with SEM (Figure 5; Table 3). Generally, more than 90% of soil DEA variance was explained by these two models. Soil DOC/NO3− (rnon-R = 0.85; rR = 0.03), NH4+ (rnon-R = −0.32; rR = −0.61), and pH (rnon-R = −0.04; rR = −0.11) were all key factors in both models, and these directly and indirectly regulated the soil DEA by modulating biotic factors including nosZ bacterial diversity (rnon-R = 0.05; rR = 0.08) and keystone taxa (rnon-R = 0; rR = 0.83). Interestingly, soil DEA was stimulated by a higher level of soil DOC/NO3−, particularly in the non-R model, whereas it was the nosZ keystone taxa (r = 0.83, P < 0.001) that generated the largest positive effect on soil DEA exclusively in the R model. This underlines the fact that nosZ keystone taxa might play an extremely important role in stimulating soil DEA under rice straw retention conditions.

Figure 5. Structural equation models describing the direct effects of physicochemical and bacterial community characteristics of paddy soil on denitrification enzyme activities under non-R (A) and R (B) modes, respectively.
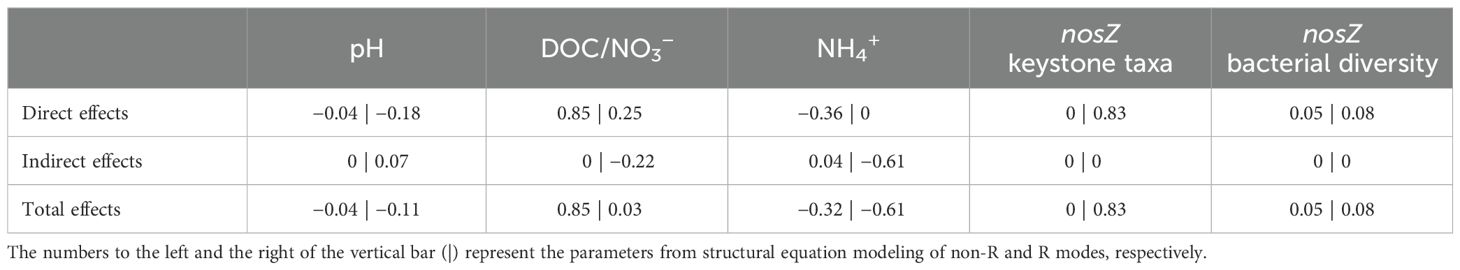
Table 3. Standardized regression weights of direct, indirect, and total effects of the key factors on denitrification enzyme activities.
4 Discussion
4.1 Variations in soil–standing water properties that respond to straw retention
The availability of N is a key factor affecting crop development and the effect of the environment in an agricultural ecosystem (Ju et al., 2009; Zhang et al., 2015). Both in standing water and soil in the present study, the NO3− value was reduced with straw retention treatments (P < 0.05) compared to that with NS treatment. This is beneficial for N loss load reductions, particularly for areas at a high risk of runoff, and was consistent with the results of most studies focused on flooded paddy fields (Chen et al., 2023; Bhattacharyya et al., 2012; Leon and Kohyama, 2017). Distinctively, the NH4+ content in the soil significantly increased after long-term straw retention, particularly for treatments with rice straw amendment (RS and WRS treatments). This was primarily attributed to the characteristics of inorganic N release and microbial immobilization from crop residue decomposition (Kahlon et al., 2013; Xu et al., 2024). More importantly, the available N level, which could be enhanced by stimulation with organic N mineralization following crop residue input, was amplified more with residues having a relatively lower C/N compared to that with residues with a higher C/N (Chen et al., 2021b; Tatti et al., 2017), respectively, corresponding to the rice straw (C/N = 62/1) and wheat straw (C/N = 110/1) in the current study. Nevertheless, the low levels of NO3− with straw retention treatments might imply higher activities of NO3− microbial reduction processes (such as denitrification, anammox, or dissimilatory nitrate reduction to ammonium) rather than its accumulation (Liu et al., 2020; Wang et al., 2021, 2024; Zhang et al., 2021c). Moreover, it is understandable that organic C and N, either in soil or standing water, were enhanced after long-term straw retention (P < 0.05), given the periodic supply of exogenous organic material in conjunction with the amplified microbial activity, which would support the accumulation of organic nutrients (Malhi et al., 2011; Wang et al., 2018; Xu et al., 2024).
4.2 Variations in nosZ denitrifiers in response to straw retention
The responses of the nitrous oxide reductase gene nosZ to fertilization in soil ecosystems have attracted considerable attention (Duan et al., 2018; Li et al., 2022; Philippot et al., 2011; Qin et al., 2021; Tao et al., 2022). In paddy fields, compositions of the dominant nosZ-denitrifying community were found to vary widely against divergent backgrounds (Cucu et al., 2017; Wang et al., 2020). In this experimental field, the nosZ denitrifiers were predominated by Planctomycetes (19.0%–22.3%), Euryarchaeota (15.1%–22.3%), and Proteobacteria (10.8%–12.7%). Most phylogroups of the latter two phyla derived from the nosZ I clade type, which can perform the complete denitrification process (Frostegård et al., 2022; Lin et al., 2023; Zhang et al., 2021a). In contrast, Planctomycetes-affiliated organisms were recently found to belong to the nosZ II clade type based on metagenomics-based analysis (Zhuang et al., 2020). The higher positive association ratio (88%) in the R mode network highlights stronger cooperative interactions among nosZ denitrifiers under rice straw retention, suggesting a more stable and synergistic community structure that facilitates nitrogen cycling (Tao et al., 2022; Zheng et al., 2019). The increased clustering coefficient further indicates the presence of tightly connected microbial clusters, potentially acting as hotspots for efficient nitrogen transformations. These findings underline the diverse ecological roles of nosZ denitrifiers in response to long-term straw retention and the importance of microbial interactions in driving soil nitrogen processes (Hallin et al., 2018; Wei et al., 2015).
Treatments with (R mode) and without (non-R mode) rice straw retention resulted in significant differences, in terms of either the community composition, structure, or keystone taxa of nosZ denitrifiers. Generally, the R mode resulted in the recruitment of more groups from Bacteroidetes and Euryarchaeota phyla as dominant bacteria or keystone taxa. For example, the genera Lunatimonas (phyla Bacteroidetes) and Halonotius (phyla Euryarchaeota), which originate from sea and lake sediments, are typical NO3−-reducing salinophilic bacteria and archaea (Durán-Viseras et al., 2023; Song et al., 2019; Vavourakis et al., 2016). This indicates that the recruitment of these keystone taxa under rice straw retention likely contributes to the observed improvement in denitrification efficiency, aligning with findings that enriched organic carbon conditions stimulate the activity of keystone denitrifiers (Bano et al., 2024; Chen et al., 2025). In contrast, the predominant taxa Planctomicrobium and Telmatocola, which belong to the Planctomycetes phylum in non-R mode, include parthenogenetic anaerobic denitrifying bacteria (Cao et al., 2021) and serve as the dominant bacteria in the anammox process (Chen et al., 2021a; Zheng et al., 2019). This further implies that the specific community of nosZ denitrifiers in the R mode might thereby increase the competitiveness of the denitrification process with respect to nitrate–nitrogen allotropic reduction by increasing the abundance of uncoupled Lunatimonas and Halonotius colonies and their positive interaction ratios, which is partially supported by findings from the co-occurrence networks.
4.2 Roles of abiotic and biotic environmental factors in soil DEA responses to straw retention
In the present paddy field, treatments with rice straw retention (RS and WRS treatments) rather than wheat straw amendment only (WS treatment) significantly increased the soil DEA (by ~ 41.93–45.80%). This corroborates 3-year observation data showing that the application of rice straw results in significant higher seasonal N2O emissions than wheat straw only, under the equivalent input of inorganic fertilizers (Wang et al., 2019). However, neither differences in nosZ gene abundance among distinct straw retention modes nor correlation coefficients for DEA and nosZ gene abundance reach significance. This represents a prevailing controversy based on many studies (Attard et al., 2011; Kou et al., 2019; Kong et al., 2023; Yin et al., 2015), and perhaps in the future, the relationship between gene expression abundance and reactivity can be determined at the mRNA level.
Without considering the abundance of the nosZ gene, the associated microbial community characteristics, as well as the biotic environmental factors, were found to regulate the soil DEA in the experimental paddy field. Specifically, soil DOC and nosZ denitrifier diversity produced a positive effect on the soil DEA, and similar phenomena have been found in many ecosystems (Gao et al., 2019; Jiang et al., 2020; Surey et al., 2020; Yeerken et al., 2024). DOC provides electron donors for denitrification processes and improves microbial competition for soil NO3−-N (Bai et al., 2015; Tao et al., 2022). In contrast, the soil pH, which ranged from 6.63 to 7.10, was partially deviated from the optimum neutral to slightly alkaline range (pH 7–8) (Liu et al., 2010) and had direct negative effects on soil DEA in this study. An interesting finding from this long-term field experiment is that it was rice rather than wheat straw amendment that significantly stimulated soil DEA through the contributions from the keystone taxa of denitrifiers—specifically, a few nitrite-reducing salinophilic bacteria and archaea. This implied that long-term rice straw amendment offers favorable conditions of electron or nitrite competition for these denitrifiers (Wang et al., 2024; Zhang et al., 2021b). The significant role of a few core phylogroups of denitrifiers has been increasingly observed by researchers (Qin et al., 2021; Tao et al., 2022). However, more evidence in terms of microbial cultivation is warranted to directly validate the underlying mechanisms.
5 Conclusions
Results of the present study provide evidence of how and why soil denitrification activity vary among different straw retention modes in a paddy field. Under the equivalent input of chemical fertilizers, soil denitrification activity can be significantly improved by long-term rice straw retention (in single or double crop seasons). Key environmental factors including soil DOC/NO3-, NH4+, and pH, along with keystone taxa and the diversity of nosZ denitrifiers, showed distinct variations between rice and wheat straw retention modes. Particularly, specific nosZ keystone taxa play a prominent role in driving denitrification activity exclusively under treatment with rice straw retention. By contrast, the practice of wheat straw retention in a single season would be recommended to minimize soil N loss from the denitrification process in a rice–wheat rotation field. Given the complexities of denitrification activity determinants, activity denitrification rates cannot be extrapolated to obtain areal fluxes for a paddy field, and intact core or new in situ methods need to be employed and validated in future research.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary Material. Further inquiries can be directed to the corresponding author.
Author contributions
SZ: Data curation, Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. MH: Investigation, Methodology, Writing – original draft. BL: Methodology, Software, Validation, Writing – review & editing. PG: Supervision, Validation, Writing – review & editing. QC: Supervision, Validation, Writing – review & editing. HS: Resources, Validation, Writing – review & editing. HX: Data curation, Validation, Writing – review & editing. DC: Software, Supervision, Writing – review & editing. YZ: Conceptualization, Methodology, Validation, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Natural Science Foundation of China (Grant No. 32301962), China Postdoctoral Science Foundation (Grant No. 2024M752947), Postdoctoral Fellowship Program of CPSF (Grant No. GZC20232437).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2025.1541202/full#supplementary-material
References
Attard, E., Recous, S., Chabbi, A., De Berranger, C., Guillaumaud, N., Labreuche, J., et al. (2011). Soil environmental conditions rather than denitrifier abundance and diversity drive potential denitrification after changes in land uses. Glob. Change Biol. 17, 1975–1989. doi: 10.1111/j.1365-2486.2010.02340.x
Aulakh, M. S., Khera, T. S., Doran, J. W., Bronson, K. F. (2001). Denitrification, N2O and CO2 fluxes in rice-wheat cropping system as affected by crop residues, fertilizer N and legume green manure. Biol. Fertil. Soils 34, 375–389. doi: 10.1007/s003740100420
Bai, R., Xi, D., He, J., Hu, H., Fang, Y., Zhang, L. (2015). Activity, abundance and community structure of anammox bacteria along depth profiles in three different paddy soils. Soil Biol. Biochem. 91, 212–221. doi: 10.1016/j.soilbio.2015.08.040
Bano, S., Wu, Q., Yu, S., Wang, X., Zhang, X. (2024). Soil properties drive nitrous oxide accumulation patterns by shaping denitrifying bacteriomes. Environ. Microbiome 19, 94. doi: 10.1186/s40793-024-00643-9
Barton, L., Schipper, L., Smith, C., McLay, C. (2000). Denitrification enzyme activity is limited by soil aeration in a wastewater-irrigated forest soil. Biol. Fertil. Soils 32, 385–389. doi: 10.1007/s003740000267
Bhattacharyya, P., Roy, K. S., Neogi, S., Adhya, T. K., Rao, K. S., Manna, M. C. (2012). Effects of rice straw and nitrogen fertilization on greenhouse gas emissions and carbon storage in tropical flooded soil planted with rice. Soil Tillage Res. 124, 119–130. doi: 10.1016/j.still.2012.05.015
Cao, X., Zhou, X., Xue, M., Chen, J., Li, S. (2021). Evaluation of nitrogen removal and N2O emission in a novel anammox coupled with sulfite-driven autotrophic denitrification system: Influence of pH. J. Clean. Prod. 321, 128984. doi: 10.1016/j.jclepro.2021.128984
Chen, H., Rosinger, C., Blagodatsky, S., Reichel, R., Li, B., Kumar, A., et al. (2023). Straw amendment and nitrification inhibitor controlling N losses and immobilization in a soil cooling-warming experiment. Sci. Total Environ. 870, 162007. doi: 10.1016/j.scitotenv.2023.162007
Chen, J., Zhou, X., Cao, X., Li, S. (2021a). Optimizing anammox capacity for weak wastewater in an AnSBBR using aerobic activated sludge as inoculation. J. Environ. Manage. 280, 111649. doi: 10.1016/j.jenvman.2020.111649
Chen, P., Cheng, Y., Wang, N., Yu, J. G., Zhao, Y., Xue, L. H. (2025). Roles of straw return in shaping denitrifying bacteria in rice rhizosphere soils through effects on root exudates and soil metabolites. Soil Ecol. Lett. 7, 240261. doi: 10.1007/s42832-024-0261-3
Chen, Z., Tu, X., Meng, H., Chen, C., Chen, Y., Elrys, A. S., et al. (2021b). Microbial process-oriented understanding of stimulation of soil N2O emission following the input of organic materials. Environ. pollut. 284, 117176. doi: 10.1016/j.envpol.2021.117176
Committee of Analytical Method of Water and Wastewater (2002). Analytical Method of Water and Wastewater (Beijing: China Environment Science Press).
Cucu, M. A., Marhan, S., Said-Pullicino, D., Celi, L., Kandeler, E., Rasche, F. (2017). Resource driven community dynamics of NH4+ assimilating and N2O reducing archaea in a temperate paddy soil. Pedobiologia 62, 16–27. doi: 10.1016/j.pedobi.2017.02.001
Delorme, S., Philippot, L., Edel-Hermann, V., Deulvot, C., Mougel, C., Lemanceau, P. (2003). Comparative genetic diversity of the narG, nosZ, and 16S rRNA genes in fluorescent pseudomonads. Appl. Environ. Microbiol. 69, 1004–1012. doi: 10.1128/AEM.69.2.1004-1012.2003
Du, F., Yin, Y., Zhai, L., Zhang, F., Wang, S., Liu, Y., et al. (2024). Increased anaerobic conditions promote the denitrifying nitrogen removal potential and limit anammox substrate acquisition within paddy irrigation and drainage units. Sci. Total Environ. 951, 175616. doi: 10.1016/j.scitotenv.2024.175616
Duan, R., Long, X., Tang, Y., Wen, J., Su, S., Bai, L., et al. (2018). Effects of different fertilizer application methods on the community of nitrifiers and denitrifiers in a paddy soil. J. Soils Sediments 18, 24–38. doi: 10.1007/s11368-017-1738-9
Durán-Viseras, A., Sánchez-Porro, C., Viver, T., Konstantinidis, K. T., Ventosa, A. (2023). Discovery of the streamlined haloarchaeon Halorutilus salinus, comprising a new order widespread in hypersaline environments across the world. Msystems 8, e01198–e01122. doi: 10.1128/msystems.01198-22
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Edgar, R. C. (2016). UNOISE2: improved error-correction for Illumina 16S and ITS amplicon sequencing. BioRxiv, 081257. doi: 10.1101/081257
Frostegård, Å., Vick, S. H., Lim, N. Y., Bakken, L. R., Shapleigh, J. P. (2022). Linking meta-omics to the kinetics of denitrification intermediates reveals pH-dependent causes of N2O emissions and nitrite accumulation in soil. ISME J. 16, 26–37. doi: 10.1038/s41396-021-01045-2
Gao, D., Liu, M., Hou, L., Derrick, Y. L., Wang, W., Li, X., et al. (2019). Effects of shrimp-aquaculture reclamation on sediment nitrate dissimilatory reduction processes in a coastal wetland of southeastern China. Environ. pollut. 255, 113219. doi: 10.1016/j.envpol.2019.113219
Ghani, A., Dexter, M., Perrott, K. (2003). Hot-water extractable carbon in soils: a sensitive measurement for determining impacts of fertilisation, grazing and cultivation. Soil Biol. Biochem. 35, 1231–1243. doi: 10.1016/S0038-0717(03)00186-X
Giles, M., Morley, N., Baggs, E. M., Daniell, T. J. (2012). Soil nitrate reducing processes–drivers, mechanisms for spatial variation, and significance for nitrous oxide production. Front. Microbiol. 3. doi: 10.3389/fmicb.2012.00407
Hallin, S., Philippot, L., Löffler, F. E., Sanford, R. A., Jones, C. M. (2018). Genomics and ecology of novel N2O-reducing microorganisms. Trends Microbiol. 26, 43–55. doi: 10.1016/j.tim.2017.07.003
Henry, S., Bru, D., Stres, B., Hallet, S., Philippot, L. (2006). Quantitative detection of the nosZ gene, encoding nitrous oxide reductase, and comparison of the abundances of 16S rRNA, narG, nirK, and nosZ genes in soils. Appl. Environ. Microbiol. 72, 5181–5189. doi: 10.1128/AEM.00231-06
Jahangir, M. M., Khalil, M. I., Johnston, P., Cardenas, L. M., Hatch, D. J., Butler, M., et al. (2012). Denitrification potential in subsoils: A mechanism to reduce nitrate leaching to groundwater. Agric. Ecosyst. Environ. 147, 13–23. doi: 10.1016/j.agee.2011.04.015
Jiang, X., Gao, G., Zhang, L., Tang, X., Shao, K., Hu, Y. (2020). Denitrification and dissimilatory nitrate reduction to ammonium in freshwater lakes of the Eastern Plain, China: Influences of organic carbon and algal bloom. Sci. Total Environ. 710, 136303. doi: 10.1016/j.scitotenv.2019.136303
Ju, X., Xing, G., Chen, X., Zhang, S., Zhang, L., Liu, X., et al. (2009). Reducing environmental risk by improving N management in intensive Chinese agricultural systems. Proc. Natl. Acad. Sci. U.S.A. 106, 3041–3046. doi: 10.1073/pnas.0813417106
Kahlon, M. S., Lal, R., Ann-Varughese, M. (2013). Twenty two years of tillage and mulching impacts on soil physical characteristics and carbon sequestration in Central Ohio. Soil Tillage Res. 126, 151–158. doi: 10.1016/j.still.2012.08.001
Kong, Y., Zhang, H., Tian, L., Yuan, J., Chen, Y., Li, Y., et al. (2023). Relationships between denitrification rates and functional gene abundance in a wetland: The roles of single-and multiple-species plant communities. Sci. Total Environ. 863, 160913. doi: 10.1016/j.scitotenv.2022.160913
Kou, Y., Li, C., Li, J., Tu, B., Wang, Y., Li, X. (2019). Climate and soil parameters are more important than denitrifier abundances in controlling potential denitrification rates in Chinese grassland soils. Sci. Total Environ. 669, 62–69. doi: 10.1016/j.scitotenv.2019.03.093
Leon, A., Kohyama, K. (2017). Estimating nitrogen and phosphorus losses from lowland paddy rice fields during cropping seasons and its application for life cycle assessment. J. Clean. Prod. 164, 963–979. doi: 10.1016/j.jclepro.2017.06.116
Li, L., Yang, M., Li, J., Roland, B., Du, Z., Wu, D. (2022). Potential denitrification activity response to long-term nitrogen fertilization-A global meta-analysis. J. Clean. Prod. 336, 130451. doi: 10.1016/j.jclepro.2022.130451
Liang, Y., Wu, C., Wei, X., Liu, Y., Chen, X., Qin, H., et al. (2021). Characterization of nirS-and nirK-containing communities and potential denitrification activity in paddy soil from eastern China. Agric. Ecosyst. Environ. 319, 107561. doi: 10.1016/j.agee.2021.107561
Lin, Y., Hu, H. W., Deng, M., Yang, P., Ye, G. (2023). Microorganisms carrying nosZ I and nosZ II share similar ecological niches in a subtropical coastal wetland. Sci. Total Environ. 870, 162008. doi: 10.1016/j.scitotenv.2023.162008
Liu, B., Mørkved, P. T., Frostegård, Å., Bakken, L. R. (2010). Denitrification gene pools, transcription and kinetics of NO, N2O and N2 production as affected by soil pH. FEMS Microbiol. Ecol. 72, 407–417. doi: 10.1111/j.1574-6941.2010.00856.x
Liu, M., Zhang, W., Wang, X., Wang, F., Dong, W., Hu, C., et al. (2020). Nitrogen leaching greatly impacts bacterial community and denitrifiers abundance in subsoil under long-term fertilization. Agric. Ecosyst. Environ. 294, 106885. doi: 10.1016/j.agee.2020.106885
Lu, R. (2000). Methods of soil and agro-chemical analysis (Beijing: China Agricultural Science and Technology Press).
Magoč, T., Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Malhi, S. S., Nyborg, M., Solberg, E. D., Dyck, M. F., Puurveen, D. (2011). Improving crop yield and N uptake with long-term straw retention in two contrasting soil types. Field Crops Res. 124, 378–391. doi: 10.1016/j.fcr.2011.07.009
Nie, S., Zhu, G., Singh, B., Zhu, Y. (2019). Anaerobic ammonium oxidation in agricultural soils-synthesis and prospective. Environ. pollut. 244, 127–134. doi: 10.1016/j.envpol.2018.10.050
Philippot, L., Andert, J., Jones, C. M., Bru, D., Hallin, S. (2011). Importance of denitrifiers lacking the genes encoding the nitrous oxide reductase for N2O emissions from soil. Glob. Change Biol. 17, 1497–1504. doi: 10.1111/j.1365-2486.2010.02334.x
Philippot, L., Hallin, S., Schloter, M. (2007). Ecology of denitrifying prokaryotes in agricultural soil. Adv. Agron. 96, 249–305. doi: 10.1016/S0065-2113(07)96003-4
Qin, H., Wang, D., Xing, X., Tang, Y., Wei, X., Chen, X., et al. (2021). A few key nirK- and nosZ-denitrifier taxa play a dominant role in moisture-enhanced N2O emissions in acidic paddy soil. Geoderma 385, 114917. doi: 10.1016/j.geoderma.2020.114917
Rognes, T., Flouri, T., Nichols, B., Quince, C., Mahé, F. (2016). VSEARCH: a versatile open source tool for metagenomics. PeerJ 4, e2584. doi: 10.7717/peerj.2584
Seitzinger, S., Harrison, J. A., Böhlke, J., Bouwman, A., Lowrance, R., Peterson, B., et al. (2006). Denitrification across landscapes and waterscapes: a synthesis. Ecol. Appl. 16, 2064–2090. doi: 10.1890/1051-0761(2006)016[2064:DALAWA]2.0.CO;2
Song, L., Liu, H., Huang, Y., Dai, X., Zhou, Y. (2019). Pleomorphovibrio marinus gen. nov., sp. nov., isolated from deep-sea sediment. Int. J. Syst. Evol. Microbiol. 69, 3723–3727. doi: 10.1099/ijsem.0.003633
Surey, R., Lippold, E., Heilek, S., Sauheitl, L., Henjes, S., Horn, M. A., et al. (2020). Differences in labile soil organic matter explain potential denitrification and denitrifying communities in a long-term fertilization experiment. Appl. Soil Ecol. 153, 103630. doi: 10.1016/j.apsoil.2020.103630
Tang, Q., Moeskjær, S., Cotton, A., Dai, W., Wang, X., Yan, X., et al. (2024). Organic fertilization reduces nitrous oxide emission by altering nitrogen cycling microbial guilds favouring complete denitrification at soil aggregate scale. Sci. Total Environ. 946, 174178. doi: 10.1016/j.scitotenv.2024.174178
Tao, R., Zhao, X., Zhang, H., Hu, B., Li, J., Chu, G. (2022). Response and recovery of nosZ abundant and rare subcommunities to organic amendment and nitrification inhibitor disturbances, with implications for N2O emissions. Appl. Soil Ecol. 173, 104386. doi: 10.1016/j.apsoil.2022.104386
Tatti, E., Goyer, C., Zebarth, B. J., Wertz, S., Burton, D. L., Chantigny, M., et al. (2017). Over-winter dynamics of soil bacterial denitrifiers and nitrite ammonifiers influenced by crop residues with different carbon to nitrogen ratios. Appl. Soil Ecol. 110, 53–64. doi: 10.1016/j.apsoil.2016.10.014
Taylor, P. G., Townsend, A. R. (2010). Stoichiometric control of organic carbon–nitrate relationships from soils to the sea. Nature 464, 1178–1181. doi: 10.1038/nature08985
Thangarajan, R., Bolan, N. S., Tian, G., Naidu, R., Kunhikrishnan, A. (2013). Role of organic amendment application on greenhouse gas emission from soil. Sci. Total Environ. 465, 72–96. doi: 10.1016/j.scitotenv.2013.01.031
Vavourakis, C. D., Ghai, R., Rodriguez-Valera, F., Sorokin, D. Y., Tringe, S. G., Hugenholtz, P., et al. (2016). Metagenomic insights into the uncultured diversity and physiology of microbes in four hypersaline soda lake brines. Front. Microbiol. 7. doi: 10.3389/fmicb.2016.00211
Wang, H., Shen, M., Hui, D., Chen, J., Sun, G., Wang, X., et al. (2019). Straw incorporation influences soil organic carbon sequestration, greenhouse gas emission, and crop yields in a Chinese rice (Oryza sativa L.)–wheat (Triticum aestivum L.) cropping system. Soil Tillage Res. 195, 104377. doi: 10.1016/j.still.2019.104377
Wang, M., Pendall, E., Fang, C., Li, B. (2018). A global perspective on agroecosystem nitrogen cycles after returning crop residue. Agric. Ecosyst. Environ. 266, 49–54. doi: 10.1016/j.agee.2018.07.019
Wang, N., Luo, J., Juhasz, A. L., Li, H., Yu, J. (2020). Straw decreased N2O emissions from flooded paddy soils via altering denitrifying bacterial community compositions and soil organic carbon fractions. FEMS Microbiol. Ecol. 96, fiaa046. doi: 10.1093/femsec/fiaa046
Wang, N., Zhao, Y., Yu, J., Xue, L., Li, H., Yang, L. (2021). Roles of bulk and rhizosphere denitrifying bacteria in denitrification from paddy soils under straw return condition. J. Soils Sediments 21, 2179–2191. doi: 10.1007/s11368-021-02942-x
Wang, T., Ji, C., Zhou, W., Chen, H., Chen, Y., Liu, Q., et al. (2024). Dry season residual straw reduces nitrous oxide emissions during rice season in upland-paddy rotation systems by inhibiting soil denitrification. Biol. Fertil. Soils 1–14. doi: 10.1007/s00374-024-01842-9
Wang, W., Wang, X., Qin, S., Yuan, J., Luo, J., Bai, Z., et al. (2024). Modifying functional groups of straw-based hydrogel provide soil N2O mitigation potential by complete denitrification. Chem. Eng. J., 157442. doi: 10.1016/j.cej.2024.157442
Wang, X., Li, Y., Ciampitti, I. A., He, P., Xu, X., Qiu, S., et al. (2022). Response of soil denitrification potential and community composition of denitrifying bacterial to different rates of straw return in north-central China. Appl. Soil Ecol. 170, 104312. doi: 10.1016/j.apsoil.2021.104312
Wang, Y., Ren, S., Wang, P., Wang, B., Hu, K., Li, J., et al. (2023). Autotrophic denitrification using Fe (II) as an electron donor: a novel prospective denitrification process. Sci. Total Environ. 858, 159721. doi: 10.1016/j.scitotenv.2022.159721
Wei, W., Isobe, K., Nishizawa, T., Zhu, L., Shiratori, Y., Ohte, N., et al. (2015). Higher diversity and abundance of denitrifying microorganisms in environments than considered previously. ISME J. 9, 1954–1965. doi: 10.1038/ismej.2015.9
Xie, L., Li, L., Xie, J., Wang, J., Mumtaz, M. Z., Effah, Z., et al. (2024). Optimal substitution of inorganic fertilizer with organic amendment sustains rainfed maize production and decreases soil N2O emissions by modifying denitrifying bacterial communities in Northern China. Eur. J. Agron. 160, 127287. doi: 10.1016/j.eja.2024.127287
Xu, L., Chen, H., Zhou, Y., Zhang, J., Nadeem, M. Y., Miao, C., et al. (2024). Long-term straw returning improved soil nitrogen sequestration by accelerating the accumulation of amino acid nitrogen. Agric. Ecosyst. Environ. 362, 108846. doi: 10.1016/j.agee.2023.108846
Yeerken, S., Deng, M., Li, L., Kinh, C. T., Wang, Z., Huang, Y., et al. (2024). Evaluating the role of high N2O affinity complete denitrifiers and non-denitrifying N2O reducing bacteria in reducing N2O emissions in river. J. Hazard. Mater. 479, 135602. doi: 10.1016/j.jhazmat.2024.135602
Yin, C., Fan, F., Song, A., Cui, P., Li, T., Liang, Y. (2015). Denitrification potential under different fertilization regimes is closely coupled with changes in the denitrifying community in a black soil. Appl. Microbiol. Biotechnol. 99, 5719–5729. doi: 10.1007/s00253-015-6461-0
Zhang, B., Penton, C. R., Yu, Z., Xue, C., Chen, Q., Chen, Z., et al. (2021a). A new primer set for Clade I nosZ that recovers genes from a broader range of taxa. Biol. Fertil. Soils 57, 523–531. doi: 10.1007/s00374-021-01544-6
Zhang, S., Zhang, G., Wang, D., Liu, Q. (2021b). Abiotic and biotic effects of long-term straw retention on reactive nitrogen runoff losses in a rice–wheat cropping system in the Yangtze Delta region. Agric. Ecosyst. Environ. 305, 107162. doi: 10.1016/j.agee.2020.107162
Zhang, S., Zhang, G., Wu, M., Wang, D., Liu, Q. (2021c). Straw return and low N addition modify the partitioning of dissimilatory nitrate reduction by increasing conversion to ammonium in paddy fields. Soil Biol. Biochem. 162, 108425. doi: 10.1016/j.soilbio.2021.108425
Zhang, X., Davidson, E. A., Mauzerall, D. L., Searchinger, T. D., Dumas, P., Shen, Y. (2015). Managing nitrogen for sustainable development. Nature 528, 51–59. doi: 10.1038/nature15743
Zheng, M., Han, Y., Han, H., Xu, C., Zhang, Z., Ma, W. (2019). Synergistic degradation on phenolic compounds of coal pyrolysis wastewater (CPW) by lignite activated coke-active sludge (LAC-AS) process: insights into succession of microbial community under selective pressure. Bioresour. Technol. 281, 126–134. doi: 10.1016/j.biortech.2019.02.074
Zhou, L., Li, J., Pokhrel, G. R., Zhao, Y., Zhang, C., Chu, W., et al. (2022). Effects of monoculture regime on the soil nirK- and nosZ-denitrifying bacterial communities of Casuarina equisetifolia. Appl. Soil Ecol. 171, 104326. doi: 10.1016/j.apsoil.2021.104326
Keywords: straw retention, denitrification activity, nosZ gene, denitrifiers, paddy field
Citation: Zhang S, Hou M, Li B, Guan P, Chi Q, Sun H, Xu H, Cui D and Zhu Y (2025) Roles of core nosZ denitrifiers in enhancing denitrification activity under long-term rice straw retention. Front. Plant Sci. 16:1541202. doi: 10.3389/fpls.2025.1541202
Received: 07 December 2024; Accepted: 13 January 2025;
Published: 07 February 2025.
Edited by:
Libin Zhou, Chinese Academy of Sciences (CAS), ChinaReviewed by:
Wenjing Liu, Shandong University, ChinaZhiqiang Cao, Chinese Academy of Sciences (CAS), China
Copyright © 2025 Zhang, Hou, Li, Guan, Chi, Sun, Xu, Cui and Zhu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yupan Zhu, emh1eXVwYW5Aenp1LmVkdS5jbg==
 Shijie Zhang
Shijie Zhang Mengyao Hou1,2
Mengyao Hou1,2 Bing Li
Bing Li Qing Chi
Qing Chi Hao Sun
Hao Sun Dongjie Cui
Dongjie Cui