
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci., 07 March 2025
Sec. Technical Advances in Plant Science
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1535384
This article is part of the Research TopicEmerging Sustainable and Green Technologies for Improving Agricultural ProductionView all 20 articles
 Cecilia Diaz1
Cecilia Diaz1 Steve U. Ayobahan2
Steve U. Ayobahan2 Samson Simon3
Samson Simon3 Luise Zühl3
Luise Zühl3 Andreas Schiermeyer4
Andreas Schiermeyer4 Elke Eilebrecht1
Elke Eilebrecht1 Sebastian Eilebrecht2*
Sebastian Eilebrecht2*RNA interference (RNAi) is a biotechnological tool used for gene silencing in plants, with both endogenous and exogenous applications. Endogenous approaches, such as host-induced gene silencing (HIGS), involve genetically modified (GM) plants, while exogenous methods include spray-induced gene silencing (SIGS). The RNAi mechanism hinges on the introduction of double-stranded RNA (dsRNA), which is processed into short interfering RNAs (siRNAs) that degrade specific messenger RNAs (mRNAs). However, unintended effects on non-target organisms and GM plants are a concern due to sequence homologies or siRNA-induced epigenetic changes. Regulatory bodies such as the EPA and EFSA emphasize the need for comprehensive risk assessments. Detecting unintended effects is complex, often relying on bioinformatic tools and untargeted analyses like transcriptomics and metabolomics, though these methods require extensive genomic data. This review aims to classify mechanisms of RNAi effects induced by short interfering RNA from different sources in plants and to identify technologies that can be used to detect these effects. In addition, practical case studies are summarized and discussed in which previously unintended RNAi effects in genetically modified plants have been investigated. Current literature is limited but suggests RNAi is relatively specific, with few unintended effects observed in GM crops. However, further studies are needed to fully understand and mitigate potential risks, particularly those related to transcriptional gene silencing (TGS) mechanisms, which are less predictable than post-transcriptional gene silencing (PTGS). Particularly the application of untargeted approaches such as small RNA sequencing and transcriptomics is recommended for thorough and comprehensive risk assessments.
RNA interference (RNAi) represents a cutting-edge approach in biotechnology for gene expression silencing, applied e.g. in plant protection, leveraging molecular principles to control gene expression. This innovative strategy encompasses both endogenous and exogenous applications, each with distinct methodologies and implications. Genetically modified (GM) plants harness RNAi to target plant endogenous transcripts e.g. to regulate the gibberellin pathway (maize event MON 94804) or to alter the fatty acid profile (soy event MON 87705). Endogenous applications also involve GM plants in a process known as host-induced gene silencing (HIGS) (Nowara et al., 2010) for pesticidal applications (e.g. maize event MON 87411 containing dsSnf7 against Diabrotica), (see https://euginius.eu). Conversely, exogenous applications, such as spray-induced gene silencing (SIGS), or root soaking of RNAi involve the direct application of RNA molecules to plants (Liu et al., 2020; Werner et al., 2020).
The core mechanism of RNAi in biotechnology application, such as plant protection, lies in its ability to selectively reduce the expression of specific genes within the target organism (Koeppe et al., 2023). In the majority of cases, this is achieved through the introduction of double-stranded RNA (dsRNA), which is subsequently processed by the RNase III Dicer or related enzymes to short interfering (si)RNA, whose base pairing with the complementary sequence of the target messenger (m)RNA leads to its degradation (Guo et al., 2016; Hung and Slotkin, 2021). While this sequence-based mechanism is advantageous for targeting pests and pathogens, there is a potential for unintended effects on non-target organisms (NTOs) and the GM plant itself (Christiaens et al., 2018). These effects may arise due to sequence homologies between the dsRNA and non-target mRNAs or through mechanisms such as siRNA-induced epigenetic changes and disruption of the organism’s endogenous RNAi pathways (Kloc et al., 2008; Zaratiegui and Martienssen, 2012; Swevers et al., 2013).
Recognizing the novel challenges posed by RNAi-based plant protection, regulatory bodies such as the US Environmental Protection Agency (EPA) and the European Food Safety Authority (EFSA) have acknowledged the need for comprehensive risk assessments (Christiaens et al., 2018; Papadopoulou et al., 2020; Christiaens et al., 2022). The Chemicals Committee and the Working Party on Chemicals, Pesticides and Biotechnology of the Organisation for Economic Co-operation and Development (OECD) have compiled considerations to integrate the latest scientific understanding into the environmental risk assessment of RNAi applications (Organisation for Economic Co-operation and Development (OECD) 2020).
One significant concern is the potential for unintended effects on GM plants themselves. Detecting these effects is complex due to several factors. Current prediction methods primarily rely on bioinformatic searches for complementary sequences to the siRNA within the GM plant’s transcriptome (Good et al., 2016; Lück et al., 2019; Farooq et al., 2021). However, these analyses are often hampered by the lack of a complete and accurate reference genome for the GM plant. When available, reference genomes of closely related cultivars may be used, but these can lead to inaccuracies due to sequence polymorphisms, resulting in false positives or negatives in off-target effect predictions.
In this review, we summarize the mechanisms by which RNAi applications could induce unintended effects in plants and evaluate the technologies and approaches available to detect these effects. By assessing the relevance of RNAi-mediated cellular mechanisms to GM plants based on existing literature, we provide a comprehensive overview and aim to rank these mechanisms according to their significance. This detailed examination will contribute to a better understanding of RNAi applications and the development of more accurate risk assessment methodologies.
The principle of RNAi in plant protection relies on reducing or silencing the expression of specific essential genes in the target organism or the GM plant itself. These target genes typically belong to vital metabolic or developmental pathways, leading to a loss-of-function phenotype (Werner et al., 2020; Hernández-Soto and Chacón-Cerdas, 2021). RNAi-based pest control strategies primarily utilize two types of RNA precursors: short hairpin RNAs (shRNA), which consist of two complementary strands forming a stem-loop structure, and complementary dsRNA. The enzyme Dicer, found in nearly all eukaryotes with various isotypes (Zapletal et al., 2023), processes these precursor molecules into short, mostly 21-24 nucleotide (nt) RNA duplexes in the cytoplasm (Figure 1). In plants, Dicer-like (DCL) proteins play an important role in processing dsRNA into siRNAs of different length (Henderson et al., 2006; Mukherjee et al., 2013). The RNA duplexes include a guide strand and a passenger strand [reviewed in (Kim et al., 2009; Borges and Martienssen, 2015)]. While the passenger strand is degraded during further processing, the guide strand, which is complementary to the target gene sequence, is crucial for the silencing of the gene. In the following, we will first focus on the biogenesis of small RNAs in plants and then discuss the mechanisms of RNAi-based silencing before we discuss the implications of these mechanisms for possible off-target effects in GM plants.

Figure 1. siRNA biogenesis in plants. (A) Synthesis of natural antisense transcripts by RNA polymerase II (RNA Pol II) followed by Dicer-like protein (DCL)-mediated cleavage. (B) RNA Pol II-mediated transcription of short hairpin (sh) RNAs, followed by DCL processing. (C) RNA Pol II-mediated synthesis of long non-coding (lnc) RNAs, followed by RNase digestion. (D) miRNA processing of RNA Pol II-transcribed miRNA precursors. (E) Trans acting (ta) siRNA pathway followed by siRNA synthesis by DCL. (F) Virus-derived siRNA synthesis from RNA or DNA viruses via replication/transcription followed by DCL processing. (G) RNA Pol IV-mediated transcription of double-stranded (ds) RNA as precursors for heterochromatic (hc) siRNA, followed by processing via DCL. Generated by the use of Biorender.com.
To investigate the effects of genetic modifications on the RNAi pathway in GM plants, it is essential to consider the natural mechanisms by which RNAi can affect gene expression in plants. To this end, the cellular pathways by which siRNA molecules can be produced in plants are first described here (Figure 1; Table 1). Precursors of siRNA are almost without exception double-stranded RNA molecules, which are either synthesized by endogenous RNA polymerases (RNA Pol) or introduced exogenously (Vazquez et al., 2004; Allen et al., 2005; Axtell et al., 2006). Endogenous precursors include natural antisense transcripts (NAT) synthesized by RNA Pol II, which base-pair with the sense mRNA of the coding gene and thus form the double-stranded substrate for corresponding RNases (Figure 1A) (Borsani et al., 2005; Jen et al., 2005; Zhang et al., 2012). RNA Pol II also synthesizes shRNAs encoded in the genome, which can then be processed by Dicer into siRNA (Figure 1B) (Wesley et al., 2001; Helliwell and Waterhouse, 2003; Senthil-Kumar and Mysore, 2011) or long non-coding (lnc) RNAs (Kim and Sung, 2012; Liu et al., 2012; Wu et al., 2012), whose secondary structures can have hairpins and can thus also be converted into siRNA by corresponding RNases (Figure 1C). Endogenously encoded micro (mi)RNAs are synthesized by Dicer or DCL1 in plants (Kurihara and Watanabe, 2004) from shRNAs, the miRNA precursors (Figure 1D), and either directly regulate the expression of target genes (by miRNA) or base-pair with the precursors of so-called trans-acting (ta)siRNAs, which are then generated by DCL from a double-stranded template (Figure 1E). Exogenously introduced precursors of siRNA are molecules introduced into a cell from an external source. A natural example are viral RNAs, which are either immediately present after infection and replication (RNA viruses, in plant viruses often single-stranded (ss) RNA genome) or are generated by transcription of the viral genome (DNA viruses) and are then templates for DCLs, which produce siRNA from them (Figure 1F) (Ruiz et al., 1998; Lu et al., 2003; Burch-Smith et al., 2004). RNA Pol IV or V can also be involved in siRNA synthesis, for example in the case of the synthesis of precursors of heterochromatic (hc)siRNAs, which are then converted to siRNA by DCL3 (Figure 1G) (Law and Jacobsen, 2010; Zhang and Zhu, 2011; Matzke and Mosher, 2014). In plants, the proteins DCL2, DCL3 and DCL4 generate siRNAs of different lengths mostly with 22 nt, 24 nt and 21 nt, respectively, which in turn trigger different mechanisms of silencing (Henderson et al., 2006; Mukherjee et al., 2013). Of note, DCL2-derived 22 nt siRNAs in plants are involved in a transitive and systemic spread of siRNA especially for antiviral defense, called secondary RNAi (Bouché et al., 2006; Chen et al., 2010; Garcia-Ruiz et al., 2010; Qin et al., 2017). This spread of RNAi involves the amplification and expansion of silencing signals that are mediated by RNA-dependent RNA polymerases (RdRp) (Sanan-Mishra et al., 2021). In this process, siRNAs act on longer RNAs (such as mRNA) as primers for RdRp, whereby a new, long dsRNA is synthesized, which is then eventually processed again by the RNAi machinery into siRNA triggering secondary RNAi.
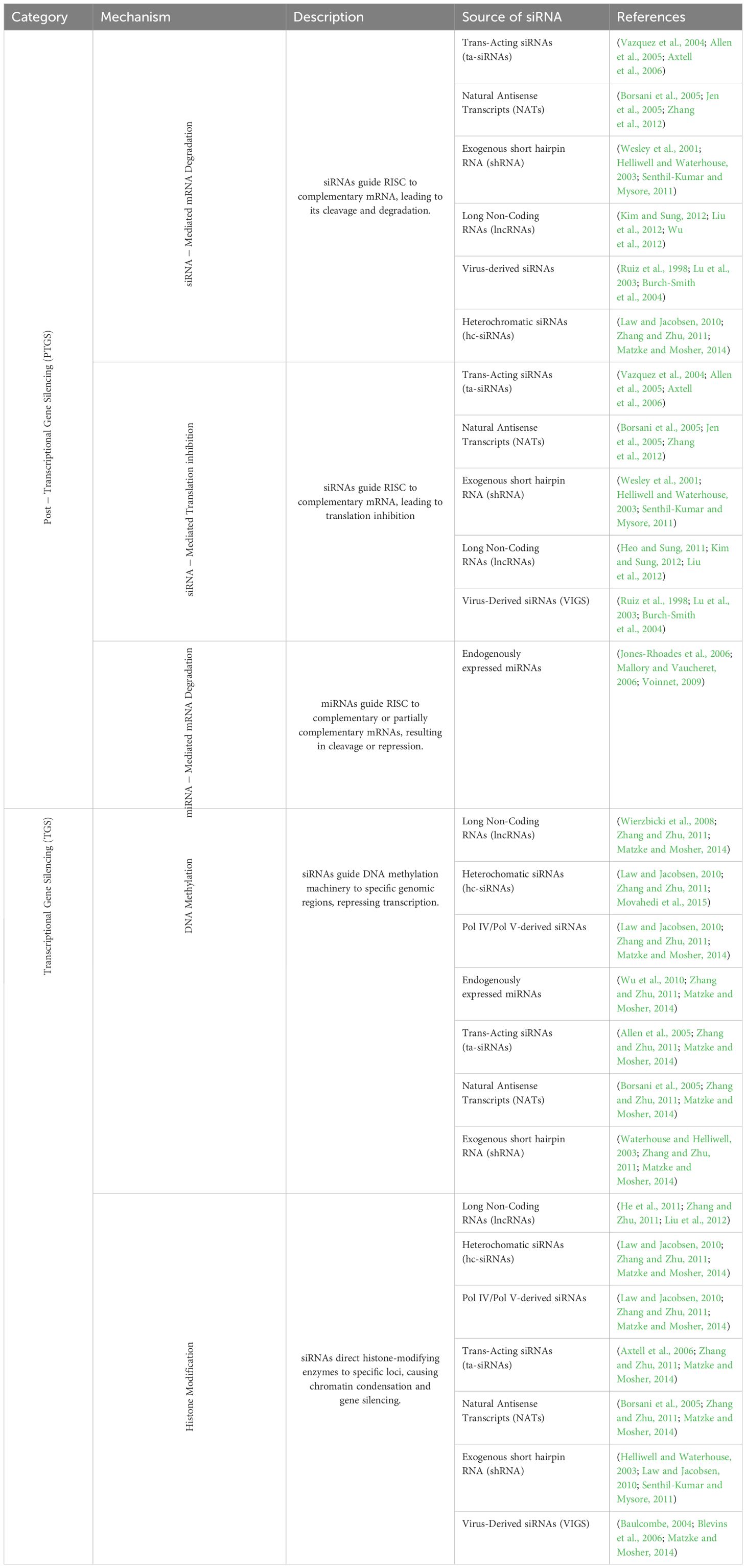
Table 1. Mechanisms by which RNA interference induces gene expression changes in plants, categorized by their general mode of action, including the mechanism, the source of siRNA and corresponding references.
Silencing mechanisms can occur in the GM plant harboring the RNAi construct, at the transcriptional level in the cell nucleus or the translational/post-transcriptional level in the cytoplasm (Figure 2). In the nucleus, siRNA can pair with the nascent mRNA of the target gene, recruiting factors to the transcription machinery that inhibit the transcription elongation by RNA polymerase (Figure 2A, left) (Guang et al., 2010). Similarly, siRNA can recruit enzymes that induce epigenetic silencing of the target gene through DNA methylation (Law and Jacobsen, 2010; Wu et al., 2010; Zhang and Zhu, 2011; Wu et al., 2012; Matzke and Mosher, 2014; Movahedi et al., 2015) or histone modification (Baulcombe, 2004; He et al., 2011; Liu et al., 2012) (Figure 2A, right) (Verdel et al., 2009). In plants, epigenetic silencing via DNA methylation is triggered by DCL3-generated ~24 nt siRNA involving a RISC complex containing the protein Argonaute (Ago)4 (Zilberman et al., 2003; Henderson et al., 2006; Qi et al., 2006; Zheng et al., 2007; Wierzbicki et al., 2009; Havecker et al., 2010; Olmedo-Monfil et al., 2010; Sarkies and Miska, 2014; Lewsey et al., 2016). The most well-studied RNAi silencing mechanism involves the degradation of the target gene’s mRNA (Figure 2B, left). In this process, the protein Ago recruits siRNA to the complementary mRNA sequence to form the RNA-induced silencing complex (RISC) (Wu et al., 2012). In plants, this is triggered by DCL4-generated ~21 nt siRNAs involving a RISC complex containing Ago1 (Xie et al., 2005; Qu et al., 2008; Chen et al., 2010; Wang et al., 2011). If there is perfect complementarity between siRNA and the target gene, the mRNA is degraded, leading to down-regulation of the target protein’s production (Valencia-Sanchez et al., 2006). With incomplete base pairing between siRNA and target mRNA, the RNA is not degraded; instead, ribosome-mediated translation is inhibited, resulting in reduced expression of the target gene (Figure 2B, right) (Brodersen et al., 2008).
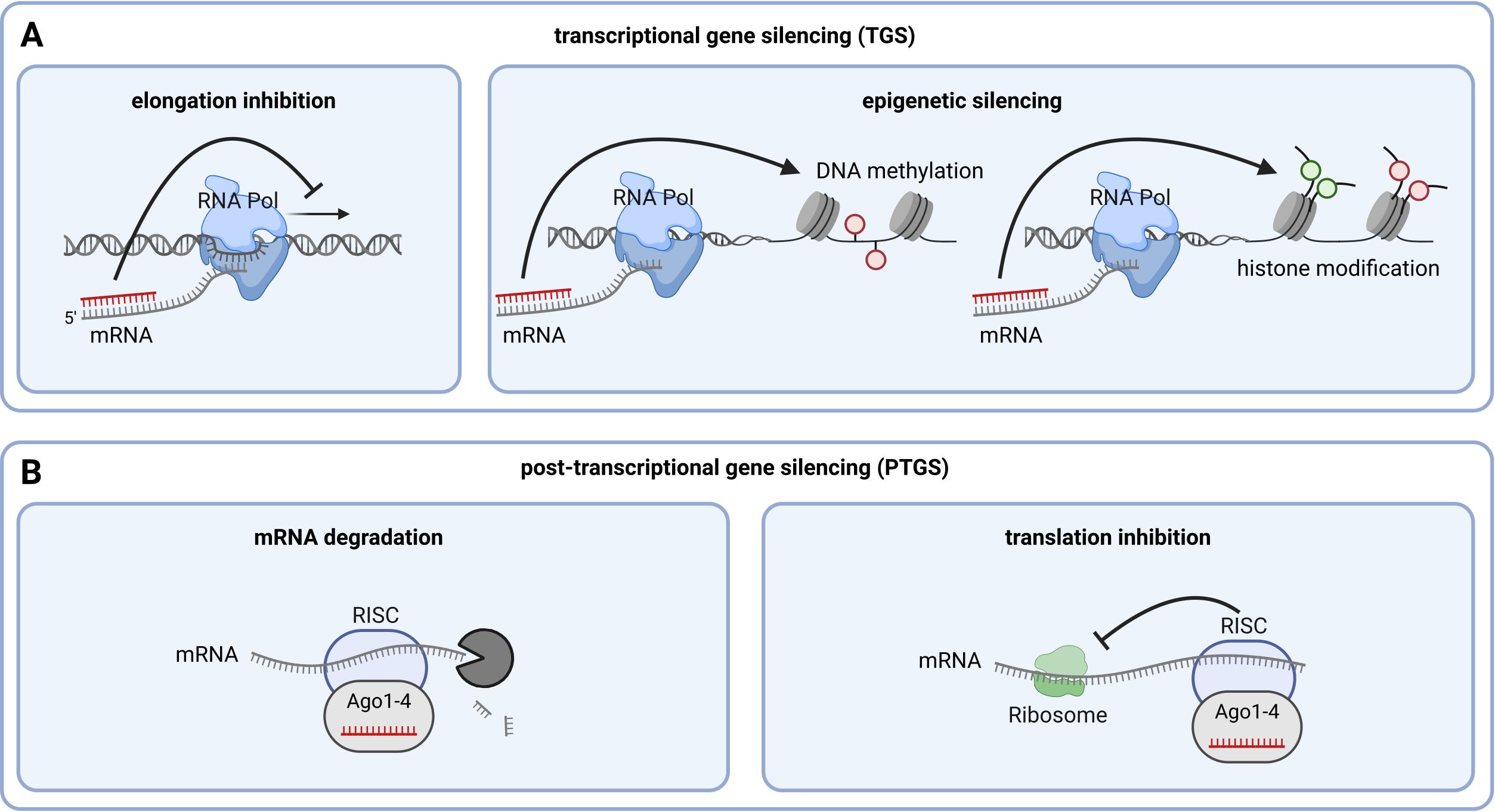
Figure 2. Mechanisms of RNAi-mediated silencing. (A) Mechanisms of transcriptional gene silencing (TGS). (B) Mechanisms of post-transcriptional gene silencing (PTGS). Generated by the use of Biorender.com.
The current literature suggests that siRNA molecules produced via different biogenesis pathways can differ in terms of their length, triggering different types of mechanisms of gene expression regulation described. While DCL4-generated 21 nt siRNA predominantly triggers PTGS via mRNA degradation, DCL3-generated 24 nt siRNA triggers TGS via epigenetic silencing and DCL2-generated 22 nt siRNA induces secondary siRNA. However, all DCL may act on long dsRNA molecules introduced into the plant. Therefore, both TGS and PTGS need to be considered when analyzing RNAi-induced effects in GM plants (Table 1).
With regard to the knowledge about mechanisms by which RNAi can potentially induce unintended effects in the GM plant, the existing literature shows clear bias towards PTGS. For example, a PubMed search with the search term “RNAi AND PTGS NOT TGS” in title and abstract returned 118 hits, whereas the search term “RNAi AND TGS NOT PTGS” only returned 25 hits (as of 18.10.2024). Hence, most published studies are concerned with the investigation of effects resulting from the inhibition of translation or degradation of mRNA (possibly resulting from incomplete complementarity). Relatively fewer studies deal with TGS, possibly because here effects, for example via epigenetic silencing, could also arise upstream or downstream of the gene with sequence complementarity and these cannot be clearly determined on the basis of the pure small RNA sequence by analyzing complementary sequences in the genome.
RNAi can induce different types of off-target effects in the plant, which can be identified and studied using different techniques. Here we provide a brief overview of the different techniques that can be used to study the changes induced by RNAi and RNAi off-target effects. The methods employed to study RNAi effects can be divided into two main approaches: targeted and untargeted analysis (Table 2). Targeted screening of RNAi effects focuses on analyzing the intended silencing effects on specific target genes and includes, for example, validation of gene knockdown, functional assays, validation of phenotypic effects, assessment of specificity and long-term effects (e.g. stability of gene silencing). Targeted screening can also be used to analyze effects on predicted off-target genes. The corresponding techniques include molecular techniques such as RT qPCR (Chi et al., 2008; Sun and Rossi, 2009; Holmes et al., 2010; Varkonyi-Gasic and Hellens, 2011; Augustine et al., 2013; Kitzmann et al., 2013; Liu et al., 2014; Czarnecki et al., 2016; Keykha et al., 2016; Manske et al., 2017; Betti et al., 2021; Sarkar and Roy-Barman, 2021; Xu et al., 2021; Zhou et al., 2021; López-Márquez et al., 2023; Kyslík et al., 2024), northern blotting (Chi et al., 2008; Fukuhara et al., 2011; Augustine et al., 2013; Manske et al., 2017; Sarkar and Roy-Barman, 2021), western blotting (Kumar et al., 2003; Sahin et al., 2007; Sun and Rossi, 2009; Holmes et al., 2010; Liang et al., 2013; Han, 2018; Vidarsdottir et al., 2019; Kyslík et al., 2024), genetic techniques such as reporter gene assays (Kumar et al., 2003; Smart et al., 2005; Rinaldi et al., 2008; Sun and Rossi, 2009; Manske et al., 2017; López-Márquez et al., 2023) or genetic mutations (Chan et al., 2006; Czarnecki et al., 2016; Krzyszton and Kufel, 2022), phenotypic assays (Chi et al., 2008; Liu et al., 2014; Xu et al., 2021; Zhou et al., 2021; Tao et al., 2023), enzyme activity assays (Chi et al., 2008; Betti et al., 2021; Sarkar and Roy-Barman, 2021) or advanced techniques such as genome editing using CRISPR/Cas9 (Moore, 2015; Kanchiswamy et al., 2016; Peretz et al., 2018; Kleter, 2020; Mujtaba et al., 2021; Bock et al., 2022).
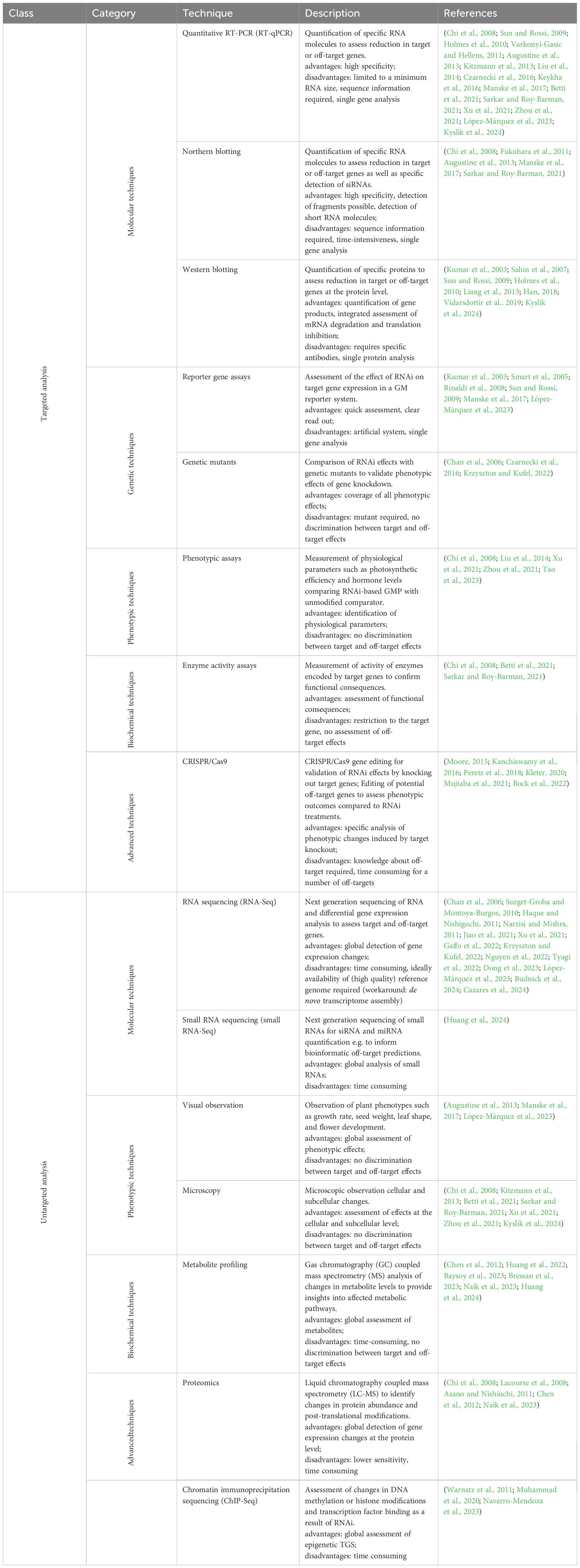
Table 2. Techniques for detecting RNAi off target effects in plants categorized by class and field. Short descriptions of each technique as well as the corresponding references are given.
Untargeted screening of RNAi effects involves comprehensive analyses mainly aimed at identifying unintended consequences and potential unpredicted off-target effects of RNAi treatments. These techniques include analyzing changes in transcriptomic profiles (Chan et al., 2006; Surget-Groba and Montoya-Burgos, 2010; Haque and Nishiguchi, 2011; Narzisi and Mishra, 2011; Jiao et al., 2021; Xu et al., 2021; Gaffo et al., 2022; Krzyszton and Kufel, 2022; Nguyen et al., 2022; Tyagi et al., 2022; Dong et al., 2023; López-Márquez et al., 2023; Budnick et al., 2024; Cazares et al., 2024), changes in protein expression (Chi et al., 2008; Lacourse et al., 2008; Asano and Nishiuchi, 2011; Chen et al., 2012; Naik et al., 2023) and modifications, metabolites (Chen et al., 2012; Huang et al., 2022; Baysoy et al., 2023; Bressan et al., 2023; Naik et al., 2023; Huang et al., 2024) and epigenetic changes (Warnatz et al., 2011; Muhammad et al., 2020; Navarro-Mendoza et al., 2023) to understand the downstream effects of RNAi on cellular processes. In addition, the distribution and potential off-target interactions of RNAi (small RNAs) with unintended mRNA targets can be determined. Furthermore, there are also bioinformatic tools that utilize computational algorithms to predict potential off-target sites based on sequence complementarity and thermodynamic stability (Good et al., 2016; Lück et al., 2019). However, such bioinformatic prediction tools require extensive knowledge, for example of the plant’s genome or its RNAi machinery, in order to apply them effectively.
When studying off-target effects of RNAi, both targeted and untargeted analyses offer unique advantages and disadvantages. Targeted analysis as focuses on predefined genes or pathways, provide specific and efficient validation of RNAi-induced gene silencing. It ensures detailed understanding of intended effects but has a limited scope, potentially missing broader biological impacts and introducing bias by overlooking unexpected interactions. These techniques require fewer technical resources and their costs are reduced, making targeted analysis well suited as validation techniques. In contrast, untargeted analysis provides a comprehensive, genome/proteome/transcriptome-wide assessment, enabling the discovery of both known and unknown off-target interactions. However, this approach depends on high-quality, well-annotated genomes for precise mapping of RNAi-induced changes and understanding the broader implications of gene silencing in plants. While this unbiased method generates extensive datasets that provide deeper insights into RNAi effects, it is resource-intensive and complex, demanding substantial time, computational power, and expertise for analysis and interpretation. Additionally, the large datasets can introduce noise, probably requiring further validation to identify meaningful effects. Despite potential challenges, combining both approaches can offer a balanced perspective, profiting the specificity of targeted analysis and the breadth of untargeted analysis to achieve thorough insights into RNAi effects.
Unintended effects of RNAi applications in GM plants themselves are a critical focus in the safety assessment of food and feed. Consequently, the Food and Agriculture Organization of the United Nations, for example, has issued guidelines for conducting food safety assessments of food derived from recombinant DNA plants (Food and Agriculture Organization of the United Nations, 2003). Also the OECD publishes science-based consensus documents offering information for the regulatory assessments of specific food and feed products, including those derived from transgenic organisms (Organisation for Economic Co-operation and Development, 2021). These documents gather data on the product’s nutrients, anti-nutrients and toxicants, its use as food or feed, and other factors relevant to food and feed safety. Here and in various review articles on the topic of risk assessment of RNAi-based GM crops, primarily untargeted methods for analyzing gene products and their metabolites, such as proteomics and metabolomics, are proposed to investigate RNAi-induced effects in the GM crop itself (Senthil-Kumar and Mysore, 2011; Kleter, 2020; Papadopoulou et al., 2020; Chaudhary et al., 2024).
The mechanisms by which the RNAi pathway can trigger specific gene expression changes in plants include both transcriptional and post-transcriptional regulation. These processes rely on specific base pairing, either with the nascent transcript (TGS) or with the mature target mRNA or a sequence-like mRNA (PTGS). While 21 nt siRNAs are predominantly involved in PTGS, 24 nt siRNAs often trigger TGS via epigenetic changes. In PTGS the target gene is directly known based on the sequence, whereas TGS can also affect genes located in close or distant proximity to the gene with sequence homology, making sequence-based prediction of TGS induced effects more difficult. PTGS is by far the most investigated mechanism in scientific studies to date, while the literature on RNAi-induced TGS is relatively limited. Therefore, the sheer number of scientific studies and the focus on PTGS to date does not necessarily reflect the actual relevance of the respective mechanisms in the plant, making it difficult to rank them according to their potential for causing unintended effects in plants.
Scientific literature on case studies investigating unintended effects in RNAi-based GM crops is currently scarce. However, bioinformatic tools are being dynamically developed to predict intended target genes and potential unintended effects on off-target genes in the GM crop or NTOs in case of HIGS, leveraging sequence homology to enhance the accuracy and scope of these predictions (Chen et al., 2019). While these tools often reach their limits in NTOs due to the lack or deficient annotated-genomes, high quality annotations are available for model plants or major crops, enabling such tools to predict PTGS effects on plant off-target genes with a higher probability. However, there are also mechanisms (such as TGS) that are not based on direct sequence homology to the target and whose unintended effects cannot be easily predicted bioinformatically. In most cases, it can be assumed that off-target effects manifest themselves at the transcriptome level and can be measured using sufficiently sensitive methods.
To detect unintended RNAi-induced effects in GM plants for risk assessment, knowledge about the siRNAs processed in the GM plant, such as size and sequence, compared to the wild type is necessary. Since both intended and possible secondary siRNAs (such as tasiRNA) can play a role, untargeted analyses, such as small RNA sequencing, should be used to identify the sequences of all siRNAs. With this knowledge, bioinformatic tools can be used to predict both intended and unintended effects mediated by sequence homology, primarily through PTGS, and these predictions can be validated using targeted methods such as RT-qPCR. However, a comprehensive bioinformatic search for homologies requires access to the plant´s complete genome, whereas RT-qPCR analyses can also be managed with knowledge of shorter sequence segments. Unintended effects mediated by TGS, on the other hand, are not directly linked to the actual sequence of the siRNA and therefore cannot be adequately detected with targeted methods, but only with untargeted methods. RNA sequencing, for example, can be used for the direct, untargeted investigation of gene expression changes, changes in histone modifications can be detected using ChIP-Seq or altered DNA methylation patterns can be detected using bisulphite sequencing. However, all these methods require the availability of the plant´s genome for accurate analysis. Additionally, there are currently no studies that specifically address the importance of selecting appropriate plant material such as tissue type, developmental stage, and sampling time points or the sensitivities required for untargeted analyses to effectively capture RNAi-induced changes (e.g. alterations in gene expression). Most published studies have focused on using plant tissues, like leaves, without a detailed exploration on how these factors might influence the detection and interpretation of RNAi-induced effects. Likewise, unintended off-target genes may be expressed, for example, in certain tissue types and not in others. These gaps highlight the need for more comprehensive research to optimize experimental designs in RNAi studies aiming to identify unintended effects.
Among the few studies assessing unintended effects of RNAi in GM plants, some have employed untargeted omics methods to analyze changes in gene expression and metabolite profiles. For example, Huang et al. (2022) compared the leaves of three transgenic maize RNAi lines resistant to Apolygus lucorum with those of three conventionally bred maize lines. Using untargeted omics methods at the levels of small RNAs, the transcriptome and the metabolome, the authors observed that the number of differentially expressed genes (DEGs) and differentially accumulated metabolites (DAMs) were greater in RNAi lines than in conventional lines. Additionally, Zörb et al. (2013) using GC-MS-based metabolite profiling showed that RNAi-mediated silencing of the sulfur-rich alpha-gliadin storage protein family in wheat grains did not induce changes in any of the 109 metabolites analyzed. Similarly, Zhang et al. (2020) investigated transcriptomic and metabolomic changes in RNAi-based GM maize resistant to Monolepta hieroglyphica compared to its unmodified variant. This study only identified a single DEG at the transcriptome level and 8 out of 5787 metabolites as DAMs, leading the authors to conclude that the RNAi variant exhibited negligible changes compared to the wild type.
Building on the insights gained from studies exploring off-target effects in RNAi-based GM plants, these findings have helped to inform regulatory approaches, including the one of the first authorization-relevant risk assessments for an RNAi-based genetically modified crop was carried out by the US Environmental Protection Agency (US EPA) for SmartStax Pro (MON 87411/Unique ID: MON-87411-9) (EPA Reg. Number: 62719-707). As part of the product characterization and human risk assessment, in 2016 the US EPA recommended a number of methods to rule out unintended side effects. These include transcriptome analyses using microarray or RNA sequencing, proteome analyses, GC-MS-based metabolomics, and the global detection of changes in DNA methylation patterns. It should be noted that certain recommended methods, such as microarray analyses for transcriptome studies or 2D gel electrophoresis coupled with MS for transcriptome analysis, are no longer state-of-the-art and should be replaced by more up-to-date methods such as RNA sequencing and LC-coupled MS, respectively. The US EPA advised that these analyses should be carried out comparatively between the GM plant containing all modification events (SmartStax Pro), the GM plant lacking the dsRNA cassette (SmartStax) as well as non-genetically modified lines across several generations. Furthermore, they recommended using a combination of different omics methods and to combine them with more sensitive methods such as RT-qPCR, to thoroughly exclude unintended effects.
In summary, the challenges in detecting unintended RNAi effects in GM plants lie in the diversity of siRNAs that can be formed from corresponding precursor molecules and in the fact that TGS (especially via epigenetic mechanisms) can also affect the expression of nearby genes without sequence homology, indicating that targeted/biased bioinformatic methods alone are not sufficient for excluding unintended effects. The few available studies indicate that the RNAi method appears to be relatively specific with minimal unintended effects expected (Zörb et al., 2013; Zhang et al., 2020; Huang et al., 2024).
Untargeted approaches, such as RNA sequencing for transcriptome analysis, LC-MS-based proteomics or GC-MS-based metabolome profiling, offer a promising and increasingly sensitive means of investigating these effects. The current state of well-annotated plant genomes varies significantly across species, with high-quality annotations available for some model plants and major crops, while others remain underrepresented. This variability poses challenges for accurately mapping RNAi-induced changes, as comprehensive and well-annotated reference genomes are crucial for identifying both target and off-target effects, as well as for understanding the broader biological impact of RNAi in diverse plant species. One way around this problem is to perform a de novo assembly of the transcriptome of unannotated plants (Surget-Groba and Montoya-Burgos, 2010; Narzisi and Mishra, 2011). However, this depends on the quality and depth of the sequencing. In combination, bioinformatic approaches with untargeted methods, such as various omics, offer the possibility to detect specific off-target effects in GM plants.
Future research on detecting RNAi-induced effects in GM plants should focus on improving sensitivity and specificity with advanced sequencing technologies, better off-target detection through CRISPR, and more accurate quantification using methods like RT-qPCR and proteomics. Environmental impact studies, long-term monitoring, and standardizing protocols will be key for regulatory safety assessments.
CD: Data curation, Formal Analysis, Investigation, Writing – original draft. SA: Data curation, Investigation, Writing – review & editing. SS: Conceptualization, Writing – review & editing. LZ: Conceptualization, Writing – review & editing. AS: Data curation, Investigation, Writing – review & editing. EE: Data curation, Investigation, Writing – review & editing. SE: Data curation, Formal Analysis, Funding acquisition, Investigation, Supervision, Writing – original draft.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The project was funded by the German Federal Ministry for the Environment, Nature Conservation, Nuclear Safety and Consumer Protection and commissioned by the German Federal Agency for Nature Conservation (BfN); project number 3522 84 2100.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Allen, E., Xie, Z., Gustafson, A. M., Carrington, J. C. (2005). microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell 121, 207–221. doi: 10.1016/j.cell.2005.04.004
Asano, T., Nishiuchi, T. (2011). Comparative analysis of phosphoprotein expression using 2D-DIGE. 1940-6029 744, 225–233. doi: 10.1007/978-1-61779-123-9_16
Augustine, R., Mukhopadhyay, A., Bisht, N. C. (2013). Targeted silencing of BjMYB28 transcription factor gene directs development of low glucosinolate lines in oilseed Brassica juncea. Plant Biotechnol. J. 11, 855–866. doi: 10.1111/pbi.12078
Axtell, M. J., Jan, C., Rajagopalan, R., Bartel, D. P. (2006). A two-hit trigger for siRNA biogenesis in plants. Cell 127, 565–577. doi: 10.1016/j.cell.2006.09.032
Baysoy, A., Bai, Z., Satija, R., Fan, R. (2023). The technological landscape and applications of single-cell multi-omics. Nat. Rev. Mol. Cell Biol. 24, 695–713. doi: 10.1038/s41580-023-00615-w
Betti, F., Ladera-Carmona, M. J., Weits, D. A., Ferri, G., Iacopino, S., Novi, G., et al. (2021). Exogenous miRNAs induce post-transcriptional gene silencing in plants. Nat. Plants 7, 1379–1388. doi: 10.1038/s41477-021-01005-w
Blevins, T., Rajeswaran, R., Shivaprasad, P. V., Beknazariants, D., Si-Ammour, A., Park, H.-S., et al. (2006). Four plant Dicers mediate viral small RNA biogenesis and DNA virus induced silencing. Nucleic Acids Res. 34, 6233–6246. doi: 10.1093/nar/gkl886
Bock, C., Datlinger, P., Chardon, F., Coelho, M. A., Dong, M. B., Lawson, K. A., et al. (2022). High-content CRISPR screening. Nat. Rev. Methods Primers 2, 1–23. doi: 10.1038/s43586-021-00093-4
Borges, F., Martienssen, R. A. (2015). The expanding world of small RNAs in plants. Nat. Rev. Mol. Cell Biol. 16, 727–741. doi: 10.1038/nrm4085
Borsani, O., Zhu, J., Verslues, P. E., Sunkar, R., Zhu, J.-K. (2005). Endogenous siRNAs derived from a pair of natural cis-antisense transcripts regulate salt tolerance in Arabidopsis. Cell 123, 1279–1291. doi: 10.1016/j.cell.2005.11.035
Bouché, N., Lauressergues, D., Gasciolli, V., Vaucheret, H. (2006). An antagonistic function for Arabidopsis DCL2 in development and a new function for DCL4 in generating viral siRNAs. EMBO J. 25, 3347–3356. doi: 10.1038/sj.emboj.7601217
Bressan, D., Battistoni, G., Hannon, G. J. (2023). The dawn of spatial omics. Science 381, eabq4964. doi: 10.1126/science.abq4964
Brodersen, P., Sakvarelidze-Achard, L., Bruun-Rasmussen, M., Dunoyer, P., Yamamoto, Y. Y., Sieburth, L., et al. (2008). Widespread translational inhibition by plant miRNAs and siRNAs. Science 320, 1185–1190. doi: 10.1126/science.1159151
Budnick, A., Franklin, M. J., Utley, D., Edwards, B., Charles, M., Hornstein, E. D., et al. (2024). Long- and short-read sequencing methods discover distinct circular RNA pools in Lotus japonicus. Plant Genome 17, e20429. doi: 10.1002/tpg2.20429
Burch-Smith, T. M., Anderson, J. C., Martin, G. B., Dinesh-Kumar, S. P. (2004). Applications and advantages of virus-induced gene silencing for gene function studies in plants. Plant J. 39, 734–746. doi: 10.1111/j.1365-313X.2004.02158.x
Cazares, T., Higgs, R. E., Wang, J., Ozer, H. G. (2024). SeedMatchR: identify off-target effects mediated by siRNA seed regions in RNA-seq experiments. Bioinformatics 40, 1–4. doi: 10.1093/bioinformatics/btae011
Chan, S. W.-L., Henderson, I. R., Zhang, X., Shah, G., Chien, J. S.-C., Jacobsen, S. E. (2006). RNAi, DRD1, and histone methylation actively target developmentally important non-CG DNA methylation in arabidopsis. PloS Genet. 2, e83. doi: 10.1371/journal.pgen.0020083
Chaudhary, D., Jeena, A. S., Rohit, Gaur, S., Raj, R., Mishra, S., et al. (2024). Advances in RNA interference for plant functional genomics: unveiling traits, mechanisms, and future directions. Appl. Biochem. Biotechnol. 196, 5681–5710. doi: 10.1007/s12010-023-04850-x
Chen, H.-M., Chen, L.-T., Patel, K., Li, Y.-H., Baulcombe, D. C., Wu, S.-H. (2010). 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. U.S.A. 107, 15269–15274. doi: 10.1073/pnas.1001738107
Chen, L., Heikkinen, L., Wang, C., Yang, Y., Sun, H., Wong, G. (2019). Trends in the development of miRNA bioinformatics tools. Brief Bioinform. 20, 1836–1852. doi: 10.1093/bib/bby054
Chen, Y., Pang, Q.-Y., He, Y., Zhu, N., Branstrom, I., Yan, X.-F., et al. (2012). Proteomics and metabolomics of Arabidopsis responses to perturbation of glucosinolate biosynthesis. Mol. Plant 5, 1138–1150. doi: 10.1093/mp/sss034
Chi, Y. H., Moon, J. C., Park, J. H., Kim, H.-S., Zulfugarov, I. S., Fanata, W. I., et al. (2008). Abnormal chloroplast development and growth inhibition in rice thioredoxin m knock-down plants. Plant Physiol. 148, 808–817. doi: 10.1104/pp.108.123547
Christiaens, O., Dzhambazova, T., Kostov, K., Arpaia, S., Joga, M. R., Urru, I., et al. (2018). Literature review of baseline information on RNAi to support the environmental risk assessment of RNAi-based GM plants. EFS3 15, 1–173. doi: 10.2903/sp.efsa.2018.EN-1424
Christiaens, O., Sweet, J., Dzhambazova, T., Urru, I., Smagghe, G., Kostov, K., et al. (2022). Implementation of RNAi-based arthropod pest control: environmental risks, potential for resistance and regulatory considerations. J. Of Pest Sci. 95, 1–15. doi: 10.1007/s10340-021-01439-3
Czarnecki, O., Bryan, A. C., Jawdy, S. S., Yang, X., Cheng, Z.-M., Chen, J.-G., et al. (2016). Simultaneous knockdown of six non-family genes using a single synthetic RNAi fragment in Arabidopsis thaliana. Plant Methods 12, 16. doi: 10.1186/s13007-016-0116-8
Dong, Y., Gao, Q., Chen, Y., Zhang, Z., Du, Y., Liu, Y., et al. (2023). Identification of CircRNA signature associated with tumor immune infiltration to predict therapeutic efficacy of immunotherapy. Nat. Commun. 14, 2540. doi: 10.1038/s41467-023-38232-y
Farooq, R., Hussain, K., Bashir, A., Rashid, K., Ashraf, M. (2021). Databases and bioinformatics tools for genome engineering in plants using RNA interference Nanobiotechnology for Plant Protection. 773–786. doi: 10.1016/B978-0-12-821910-2.00023-0
Food and Agriculture Organization of the United Nations (2003). GUIDELINE FOR THE CONDUCT OF FOOD SAFETY ASSESSMENT OF FOODS DERIVED FROM RECOMBINANT-DNA PLANTS. Available online at: https://www.fao.org/fileadmin/user_upload/gmfp/docs/CAC.GL_45_2003.pdf (Accessed December 9, 2024).
Fukuhara, T., Urayama, S., Okada, R., Kiyota, E., Moriyama, H. (2011). Detection of long and short double-stranded RNAs. Springer Protocols 744, 129–144. doi: 10.1007/978-1-61779-123-9_9
Gaffo, E., Buratin, A., Dal Molin, A., Bortoluzzi, S. (2022). Sensitive, reliable and robust circRNA detection from RNA-seq with CirComPara2. Brief Bioinform. 23, 1–12. doi: 10.1093/bib/bbab418
Garcia-Ruiz, H., Takeda, A., Chapman, E. J., Sullivan, C. M., Fahlgren, N., Brempelis, K. J., et al. (2010). Arabidopsis RNA-dependent RNA polymerases and dicer-like proteins in antiviral defense and small interfering RNA biogenesis during Turnip Mosaic Virus infection. Plant Cell 22, 481–496. doi: 10.1105/tpc.109.073056
Good, R. T., Varghese, T., Golz, J. F., Russell, D. A., Papanicolaou, A., Edwards, O., et al. (2016). OfftargetFinder: a web tool for species-specific RNAi design. Bioinformatics 32, 1232–1234. doi: 10.1093/bioinformatics/btv747
Guang, S., Bochner, A. F., Burkhart, K. B., Burton, N., Pavelec, D. M., Kennedy, S. (2010). Small regulatory RNAs inhibit RNA polymerase II during the elongation phase of transcription. Nature 465, 1097–1101. doi: 10.1038/nature09095
Guo, Q., Liu, Q., Smith, N. A., Liang, G., Wang, M.-B. (2016). RNA silencing in plants: mechanisms, technologies and applications in horticultural crops. Curr. Genomics 17, 476–489. doi: 10.2174/1389202917666160520103117
Han, H. (2018). “RNA interference to knock down gene expression,” in Disease gene identification: methods and protocols. Ed. DiStefano, J. K. (Springer, New York, NY), 293–302.
Haque, N., Nishiguchi, M. (2011). Bisulfite sequencing for cytosine-methylation analysis in plants. 1940-6029 744, 187–197. doi: 10.1007/978-1-61779-123-9_13
Havecker, E. R., Wallbridge, L. M., Hardcastle, T. J., Bush, M. S., Kelly, K. A., Dunn, R. M., et al. (2010). The arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 22, 321–334. doi: 10.1105/tpc.109.072199
He, G., Elling, A. A., Deng, X. W. (2011). The epigenome and plant development. Annu. Rev. Plant Biol. 62, 411–435. doi: 10.1146/annurev-arplant-042110-103806
Helliwell, C., Waterhouse, P. (2003). Constructs and methods for high-throughput gene silencing in plants. Methods 30, 289–295. doi: 10.1016/s1046-2023(03)00036-7
Henderson, I. R., Zhang, X., Lu, C., Johnson, L., Meyers, B. C., Green, P. J., et al. (2006). Dissecting Arabidopsis thaliana DICER function in small RNA processing, gene silencing and DNA methylation patterning. Nat. Genet. 38, 721–725. doi: 10.1038/ng1804
Heo, J. B., Sung, S. (2011). Vernalization-mediated epigenetic silencing by a long intronic noncoding RNA. Science 331, 76–79. doi: 10.1126/science.1197349
Hernández-Soto, A., Chacón-Cerdas, R. (2021). RNAi crop protection advances. Int. J. Mol. Sci. 22, 1–15. doi: 10.3390/ijms222212148
Holmes, K., Williams, C. M., Chapman, E. A., Cross, M. J. (2010). Detection of siRNA induced mRNA silencing by RT-qPCR: considerations for experimental design. BMC Res. Notes 3, 53. doi: 10.1186/1756-0500-3-53
Huang, C., Wang, Z., Zhu, P., Wang, C., Wang, C., Xu, W., et al. (2022). RNA interference-based genetic engineering maize resistant to apolygus lucorum does not manifest unpredictable unintended effects relative to conventional breeding: short interfering RNA, transcriptome, and metabolome analysis. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.745708
Huang, C., Wang, Z., Zhu, P., Xu, W., Du, Z., Qian, Y., et al. (2024). RNAi-based genetically engineered rice resistant to black-streaked dwarf virus does not show adverse genetic effects: A multi-omics analysis. Plants People Planet 6, 622–639. doi: 10.1002/ppp3.10477
Hung, Y.-H., Slotkin, R. K. (2021). The initiation of RNA interference (RNAi) in plants. Curr. Opin. Plant Biol. 61, 102014. doi: 10.1016/j.pbi.2021.102014
Jen, C.-H., Michalopoulos, I., Westhead, D. R., Meyer, P. (2005). Natural antisense transcripts with coding capacity in Arabidopsismay have a regulatory role that is not linked to double-stranded RNA degradation. Genome Biol. 6, R51. doi: 10.1186/gb-2005-6-6-r51
Jiao, S., Wu, S., Huang, S., Liu, M., Gao, B. (2021). Advances in the identification of circular RNAs and research into circRNAs in human diseases. Front. In Genet. 12, 665233. doi: 10.3389/fgene.2021.665233
Jones-Rhoades, M. W., Bartel, D. P., Bartel, B. (2006). MicroRNAS and their regulatory roles in plants. Annu. Rev. Plant Biol. 57, 19–53. doi: 10.1146/annurev.arplant.57.032905.105218
Kanchiswamy, C. N., Maffei, M., Malnoy, M., Velasco, R., Kim, J.-S. (2016). Fine-tuning next-generation genome editing tools. Trends Biotechnol. 34, 562–574. doi: 10.1016/j.tibtech.2016.03.007
Keykha, F., Bagheri, A., Moshtaghi, N., Bahrami, A. R., Sharifi, A. (2016). RNAi-induced silencing in floral tissues of Petunia hybrida by agroinfiltration: a rapid assay for chalcone isomerase gene function analysis. Cell Mol. Biol. (Noisy-le-grand) 62, 26–31. doi: 10.14715/cmb/2016.62.10.4
Kim, V. N., Han, J., Siomi, M. C. (2009). Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10, 126–139. doi: 10.1038/nrm2632
Kim, E.-D., Sung, S. (2012). Long noncoding RNA: unveiling hidden layer of gene regulatory networks. Trends Plant Sci. 17, 16–21. doi: 10.1016/j.tplants.2011.10.008
Kitzmann, P., Schwirz, J., Schmitt-Engel, C., Bucher, G. (2013). RNAi phenotypes are influenced by the genetic background of the injected strain. BMC Genomics 14, 5. doi: 10.1186/1471-2164-14-5
Kleter, G. A. (2020). Food safety assessment of crops engineered withRNAinterference and other methods to modulate expression of endogenous and plant pest genes. Pest Manag Sci. 76, 3333–3339. doi: 10.1002/ps.5957
Kloc, A., Zaratiegui, M., Nora, E., Martienssen, R. (2008). RNA interference guides histone modification during the S phase of chromosomal replication. Curr. Biol. 18, 490–495. doi: 10.1016/j.cub.2008.03.016
Koeppe, S., Kawchuk, L., Kalischuk, M. (2023). RNA interference past and future applications in plants. Int. J. Mol. Sci. 24, 1–18. doi: 10.3390/ijms24119755
Krzyszton, M., Kufel, J. (2022). Analysis of mRNA-derived siRNAs in mutants of mRNA maturation and surveillance pathways in Arabidopsis thaliana. Sci. Rep. 12, 1474. doi: 10.1038/s41598-022-05574-4
Kumar, R., Conklin, D. S., Mittal, V. (2003). High-throughput selection of effective RNAi probes for gene silencing. Genome Res. 13, 2333–2340. doi: 10.1101/gr.1575003
Kurihara, Y., Watanabe, Y. (2004). Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl. Acad. Sci. U.S.A. 101, 12753–12758. doi: 10.1073/pnas.0403115101
Kyslík, J., Born-Torrijos, A., Holzer, A. S., Kosakyan, A. (2024). RNAi-directed knockdown in the cnidarian fish blood parasite Sphaerospora molnari. Sci. Rep. 14, 3545. doi: 10.1038/s41598-024-54171-0
Lacourse, E. J., Perally, S., Hernandez-Viadel, M., Wright, H. A., Brophy, P. M. (2008). A proteomics approach to quantify protein levels following RNA interference: case study with glutathione transferase superfamily from the model metazoan Caenorhabditis elegans. J. Proteome Res. 7, 3314–3318. doi: 10.1021/pr8001035
Law, J. A., Jacobsen, S. E. (2010). Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat. Rev. Genet. 11, 204–220. doi: 10.1038/nrg2719
Lewsey, M. G., Hardcastle, T. J., Melnyk, C. W., Molnar, A., Valli, A., Urich, M. A., et al. (2016). Mobile small RNAs regulate genome-wide DNA methylation. Proc. Natl. Acad. Sci. U.S.A. 113, E801–E810. doi: 10.1073/pnas.1515072113
Liang, W., Mason, A. J., Lam, J. K. W. (2013). “Western Blot Evaluation of siRNA Delivery by pH-Responsive Peptides,” in Target identification and validation in drug discovery: methods and protocols. Ed. Moll (Humana Press Incorporated, Totowa, NJ), 73–87.
Liu, J.-X., Chiou, C.-Y., Shen, C.-H., Chen, P.-J., Liu, Y.-C., Jian, C.-D., et al. (2014). RNA interference-based gene silencing of phytoene synthase impairs growth, carotenoids, and plastid phenotype in Oncidium hybrid orchid. Springerplus 3, 478. doi: 10.1186/2193-1801-3-478
Liu, S., Jaouannet, M., Dempsey, D. A., Imani, J., Coustau, C., Kogel, K.-H. (2020). RNA-based technologies for insect control in plant production. Biotechnol. Adv. 39, 107463. doi: 10.1016/j.bioteChadv.2019.107463
Liu, J., Jung, C., Xu, J., Wang, H., Deng, S., Bernad, L., et al. (2012). Genome-wide analysis uncovers regulation of long intergenic noncoding RNAs in Arabidopsis. Plant Cell 24, 4333–4345. doi: 10.1105/tpc.112.102855
López-Márquez, D., Del-Espino, Á., Ruiz-Albert, J., Bejarano, E. R., Brodersen, P., Beuzón, C. R. (2023). Regulation of plant immunity via small RNA-mediated control of NLR expression. J. Exp. Bot. 74, 6052–6068. doi: 10.1093/jxb/erad268
Lu, R., Martin-Hernandez, A. M., Peart, J. R., Malcuit, I., Baulcombe, D. C. (2003). Virus-induced gene silencing in plants. Methods 30, 296–303. doi: 10.1016/s1046-2023(03)00037-9
Lück, S., Kreszies, T., Strickert, M., Schweizer, P., Kuhlmann, M., Douchkov, D. (2019). siRNA-finder (si-fi) software for RNAi-target design and off-target prediction. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.01023
Mallory, A. C., Vaucheret, H. (2006). Functions of microRNAs and related small RNAs in plants. Nat. Genet. 38 Suppl, S31–S36. doi: 10.1038/ng1791
Manske, U., Landsmann, J., Dietz-Pfeilstetter, A. (2017). Comparison of different methods for the establishment of RNA silencing in plants. Plant Biotechnol. Rep. 11, 115–125. doi: 10.1007/s11816-017-0436-9
Matzke, M. A., Mosher, R. A. (2014). RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat. Rev. Genet. 15, 394–408. doi: 10.1038/nrg3683
Moore, J. D. (2015). The impact of CRISPR-Cas9 on target identification and validation. Drug Discovery Today 20, 450–457. doi: 10.1016/j.drudis.2014.12.016
Movahedi, A., Sun, W., Zhang, J., Wu, X., Mousavi, M., Mohammadi, K., et al. (2015). RNA-directed DNA methylation in plants. Plant Cell Rep. 34, 1857–1862. doi: 10.1007/s00299-015-1839-0
Muhammad, I. I., Kong, S. L., Akmar Abdullah, S. N., Munusamy, U. (2020). RNA-seq and chIP-seq as complementary approaches for comprehension of plant transcriptional regulatory mechanism. IJMS 21, 167. doi: 10.3390/ijms21010167
Mujtaba, M., Wang, D., Carvalho, L. B., Oliveira, J. L., Espirito Santo Pereira, A., Sharif, R., et al. (2021). Nanocarrier-mediated delivery of miRNA, RNAi, and CRISPR-cas for plant protection: current trends and future directions. ACS Agric. Sci. Technol. 1, 417–435. doi: 10.1021/acsagscitech.1c00146
Mukherjee, K., Campos, H., Kolaczkowski, B. (2013). Evolution of animal and plant dicers: early parallel duplications and recurrent adaptation of antiviral RNA binding in plants. Mol. Biol. Evol. 30, 627–641. doi: 10.1093/molbev/mss263
Naik, B., Kumar, V., Rizwanuddin, S., Chauhan, M., Choudhary, M., Gupta, A. K., et al. (2023). Genomics, proteomics, and metabolomics approaches to improve abiotic stress tolerance in tomato plant. Int. J. Mol. Sci. 24. doi: 10.3390/ijms24033025
Narzisi, G., Mishra, B. (2011). Comparing de novo genome assembly: the long and short of it. PloS One 6, e19175. doi: 10.1371/journal.pone.0019175
Navarro-Mendoza, M. I., Pérez-Arques, C., Heitman, J. (2023). Heterochromatin and RNAi act independently to ensure genome stability in Mucorales human fungal pathogens. Proc. Natl. Acad. Sci. U.S.A. 120, 1–12. doi: 10.1073/pnas.2220475120
Nguyen, M. H., Nguyen, H.-N., Vu, T. N. (2022). Evaluation of methods to detect circular RNAs from single-end RNA-sequencing data. BMC Genomics 23, 106. doi: 10.1186/s12864-022-08329-7
Nowara, D., Gay, A., Lacomme, C., Shaw, J., Ridout, C., Douchkov, D., et al. (2010). HIGS: host-induced gene silencing in the obligate biotrophic fungal pathogen Blumeria graminis. Plant Cell 22, 3130–3141. doi: 10.1105/tpc.110.077040
Olmedo-Monfil, V., Durán-Figueroa, N., Arteaga-Vázquez, M., Demesa-Arévalo, E., Autran, D., Grimanelli, D., et al. (2010). Control of female gamete formation by a small RNA pathway in Arabidopsis. Nature 464, 628–632. doi: 10.1038/nature08828
Organisation for Economic Co-operation and Development (2021). REVISED CONSENSUS DOCUMENT ON COMPOSITIONAL CONSIDERATIONS FOR NEW VARIETIES OF POTATO (Solanum tuberosum): key food and feed nutrients, toxicants, allergens, anti-nutrients and other plant metabolite: ENV/JM/MONO(2020). Paris: OECD.
Organisation for Economic Co-operation and Development (OECD). (2020). Considerations for the environmental risk assessment of the application of sprayed or externally applied ds-RNA-based pesticides (OECD).
Papadopoulou, N., Devos, Y., Alvarez-Alfageme, F., Lanzoni, A., Waigmann, E. (2020). Risk assessment considerations for genetically modified RNAi plants: EFSA’s activities and perspective. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00445
Peretz, L., Besser, E., Hajbi, R., Casden, N., Ziv, D., Kronenberg, N., et al. (2018). Combined shRNA over CRISPR/cas9 as a methodology to detect off-target effects and a potential compensatory mechanism. Sci. Rep. 8, 93. doi: 10.1038/s41598-017-18551-z
Qi, Y., He, X., Wang, X.-J., Kohany, O., Jurka, J., Hannon, G. J. (2006). Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 443, 1008–1012. doi: 10.1038/nature05198
Qin, C., Li, B., Fan, Y., Zhang, X., Yu, Z., Ryabov, E., et al. (2017). Roles of dicer-like proteins 2 and 4 in intra- and intercellular antiviral silencing. Plant Physiol. 174, 1067–1081. doi: 10.1104/pp.17.00475
Qu, F., Ye, X., Morris, T. J. (2008). Arabidopsis DRB4, AGO1, AGO7, and RDR6 participate in a DCL4-initiated antiviral RNA silencing pathway negatively regulated by DCL1. Proc. Natl. Acad. Sci. U.S.A. 105, 14732–14737. doi: 10.1073/pnas.0805760105
Rinaldi, G., Morales, M. E., Cancela, M., Castillo, E., Brindley, P. J., Tort, J. F. (2008). Development of functional genomic tools in trematodes: RNA interference and luciferase reporter gene activity in Fasciola hepatica. PloS Negl. Trop. Dis. 2, e260. doi: 10.1371/journal.pntd.0000260
Ruiz, M. T., Voinnet, O., Baulcombe, D. C. (1998). Initiation and maintenance of virus-induced gene silencing. Plant Cell 10, 937–946. doi: 10.1105/tpc.10.6.937
Sahin, O., Löbke, C., Korf, U., Appelhans, H., Sültmann, H., Poustka, A., et al. (2007). Combinatorial RNAi for quantitative protein network analysis. Proc. Natl. Acad. Sci. U.S.A. 104, 6579–6584. doi: 10.1073/pnas.0606827104
Sanan-Mishra, N., Abdul Kader Jailani, A., Mandal, B., Mukherjee, S. K. (2021). Secondary siRNAs in plants: biosynthesis, various functions, and applications in virology. Front. Plant Sci. 12, 610283. doi: 10.1126/science.1079695
Sarkar, A., Roy-Barman, S. (2021). Spray-induced silencing of pathogenicity gene moDES1 via exogenous double-stranded RNA can confer partial resistance against fungal blast in rice. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.733129
Sarkies, P., Miska, E. A. (2014). Small RNAs break out: the molecular cell biology of mobile small RNAs. Nat. Rev. Mol. Cell Biol. 15, 525–535. doi: 10.1038/nrm3840
Senthil-Kumar, M., Mysore, K. S. (2011). Caveat of RNAi in plants: the off-target effect. 1940-6029 744, 13–25. doi: 10.1007/978-1-61779-123-9_2
Smart, N., Scambler, P. J., Riley, P. R. (2005). A rapid and sensitive assay for quantification of siRNA efficiency and specificity. Biol. Procedures Online 7, 1–7. doi: 10.1251/bpo99
Sun, G., Rossi, J. J. (2009). Problems associated with reporter assays in RNAi studies. RNA Biol. 6, 406–411. doi: 10.4161/rna.6.4.9218
Surget-Groba, Y., Montoya-Burgos, J. I. (2010). Optimization of de novo transcriptome assembly from next-generation sequencing data. Genome Res. 20, 1432–1440. doi: 10.1101/gr.103846.109
Swevers, L., Vanden Broeck, J., Smagghe, G. (2013). The possible impact of persistent virus infection on the function of the RNAi machinery in insects: a hypothesis. Front. Physiol. 4. doi: 10.3389/fphys.2013.00319
Tao, Y., Chiu, L.-W., Hoyle, J. W., Dewhirst, R. A., Richey, C., Rasmussen, K., et al. (2023). Enhanced photosynthetic efficiency for increased carbon assimilation and woody biomass production in engineered hybrid poplar. Forests 14, 827. doi: 10.3390/f14040827
Tyagi, P., Singh, D., Mathur, S., Singh, A., Ranjan, R. (2022). Upcoming progress of transcriptomics studies on plants: An overview. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.1030890
Valencia-Sanchez, M. A., Liu, J., Hannon, G. J., Parker, R. (2006). Control of translation and mRNA degradation by miRNAs and siRNAs. Genes Dev. 20, 515–524. doi: 10.1101/gad.1399806
Varkonyi-Gasic, E., Hellens, R. P. (2011). Quantitative stem-loop RT-PCR for detection of microRNAs. Springer Protocols 744, 145–157. doi: 10.1007/978-1-61779-123-9_10
Vazquez, F., Vaucheret, H., Rajagopalan, R., Lepers, C., Gasciolli, V., Mallory, A. C., et al. (2004). Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell 16, 69–79. doi: 10.1016/j.molcel.2004.09.028
Verdel, A., Vavasseur, A., Le Gorrec, M., Touat-Todeschini, L. (2009). Common themes in siRNA-mediated epigenetic silencing pathways. Int. J. Dev. Biol. 53, 245–257. doi: 10.1387/ijdb.082691av
Vidarsdottir, L., Goroshchuk, O., Kolosenko, I., Palm-Apergi, C. (2019). Designing siRNA and Evaluating Its Effect on RNA Targets Using qPCR and Western Blot. Springer Protocols 2036, 53–72. doi: 10.1007/978-1-4939-9670-4_3
Voinnet, O. (2009). Origin, biogenesis, and activity of plant microRNAs. Cell 136, 669–687. doi: 10.1016/j.cell.2009.01.046
Wang, X.-B., Jovel, J., Udomporn, P., Wang, Y., Wu, Q., Li, W.-X., et al. (2011). The 21-nucleotide, but not 22-nucleotide, viral secondary small interfering RNAs direct potent antiviral defense by two cooperative argonautes in Arabidopsis thaliana. Plant Cell 23, 1625–1638. doi: 10.1105/tpc.110.082305
Warnatz, H.-J., Schmidt, D., Manke, T., Piccini, I., Sultan, M., Borodina, T., et al. (2011). The BTB and CNC homology 1 (BACH1) target genes are involved in the oxidative stress response and in control of the cell cycle. J. Of Biol. Chem. 286, 23521–23532. doi: 10.1074/jbc.M111.220178
Waterhouse, P. M., Helliwell, C. A. (2003). Exploring plant genomes by RNA-induced gene silencing. Nat. Rev. Genet. 4, 29–38. doi: 10.1038/nrg982
Werner, B. T., Gaffar, F. Y., Schuemann, J., Biedenkopf, D., Koch, A. M. (2020). RNA-spray-mediated silencing of fusarium graminearum AGO and DCL genes improve barley disease resistance. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00476
Wesley, S. V., Helliwell, C. A., Smith, N. A., Wang, M. B., Rouse, D. T., Liu, Q., et al. (2001). Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 27, 581–590. doi: 10.1046/j.1365-313x.2001.01105.x
Wierzbicki, A. T., Haag, J. R., Pikaard, C. S. (2008). Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135, 635–648. doi: 10.1016/j.cell.2008.09.035
Wierzbicki, A. T., Ream, T. S., Haag, J. R., Pikaard, C. S. (2009). RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat. Genet. 41, 630–634. doi: 10.1038/ng.365
Wu, L., Mao, L., Qi, Y. (2012). Roles of dicer-like and argonaute proteins in TAS-derived small interfering RNA-triggered DNA methylation. Plant Physiol. 160, 990–999. doi: 10.1104/pp.112.200279
Wu, L., Zhou, H., Zhang, Q., Zhang, J., Ni, F., Liu, C., et al. (2010). DNA methylation mediated by a microRNA pathway. Mol. Cell 38, 465–475. doi: 10.1016/j.molcel.2010.03.008
Xie, Z., Allen, E., Wilken, A., Carrington, J. C. (2005). DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl. Acad. Sci. U.S.A. 102, 12984–12989. doi: 10.1073/pnas.0506426102
Xu, K., Zhang, X.-M., Chen, H., Zhang, C., Zhu, J., Cheng, Z., et al. (2021). Fine-tuning florigen increases field yield through improving photosynthesis in soybean. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.710754
Zapletal, D., Kubicek, K., Svoboda, P., Stefl, R. (2023). Dicer structure and function: conserved and evolving features. EMBO Rep. 24, e57215. doi: 10.15252/embr.202357215
Zaratiegui, M., Martienssen, R. A. (2012). SnapShot: small RNA-mediated epigenetic modifications. Cell 151, 456–456.e1. doi: 10.1016/j.cell.2012.10.001
Zhang, X., Xia, J., Lii, Y. E., Barrera-Figueroa, B. E., Zhou, X., Gao, S., et al. (2012). Genome-wide analysis of plant nat-siRNAs reveals insights into their distribution, biogenesis and function. Genome Biol. 13, R20. doi: 10.1186/gb-2012-13-3-r20
Zhang, X. L., Zhang, R. Y., Li, L., Yang, Y., Ding, Y. J., Guan, H. T., et al. (2020). Negligible transcriptome and metabolome alterations in RNAi insecticidal maize againstMonolepta hieroglyphica. Plant Cell Rep. 39, 1539–1547. doi: 10.1007/s00299-020-02582-4
Zhang, H., Zhu, J.-K. (2011). RNA-directed DNA methylation. Curr. Opin. Plant Biol. 14, 142–147. doi: 10.1016/j.pbi.2011.02.003
Zheng, X., Zhu, J., Kapoor, A., Zhu, J.-K. (2007). Role of Arabidopsis AGO6 in siRNA accumulation, DNA methylation and transcriptional gene silencing. EMBO J. 26, 1691–1701. doi: 10.1038/sj.emboj.7601603
Zhou, H., Yang, M., Zhao, L., Zhu, Z., Liu, F., Sun, H., et al. (2021). HIGH-TILLERING AND DWARF 12 modulates photosynthesis and plant architecture by affecting carotenoid biosynthesis in rice. J. Exp. Bot. 72, 1212–1224. doi: 10.1093/jxb/eraa497
Zilberman, D., Cao, X., Jacobsen, S. E. (2003). ARGONAUTE4 control of locus-specific siRNA accumulation and DNA and histone methylation. Science 299, 716–719. doi: 10.1126/science.1079695
Keywords: RNAi GM plants, detection techniques, RNAi mechanism, off-target effects, RNAi pest control
Citation: Diaz C, Ayobahan SU, Simon S, Zühl L, Schiermeyer A, Eilebrecht E and Eilebrecht S (2025) Classification of and detection techniques for RNAi-induced effects in GM plants. Front. Plant Sci. 16:1535384. doi: 10.3389/fpls.2025.1535384
Received: 27 November 2024; Accepted: 08 February 2025;
Published: 07 March 2025.
Edited by:
Ruonan Ma, Zhengzhou University, ChinaReviewed by:
Ming Wang, Chinese Academy of Sciences (CAS), ChinaCopyright © 2025 Diaz, Ayobahan, Simon, Zühl, Schiermeyer, Eilebrecht and Eilebrecht. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sebastian Eilebrecht, c2ViYXN0aWFuLmVpbGVicmVjaHRAaW1lLmZyYXVuaG9mZXIuZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.