
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 20 February 2025
Sec. Plant Pathogen Interactions
Volume 16 - 2025 | https://doi.org/10.3389/fpls.2025.1527944
This article is part of the Research Topic Innovative Molecular Strategies for Enhancing Plant Defense Against Biotic Stresses View all 9 articles
The RNA-based spray-induced gene silencing (SIGS) technology represents an ecologically sustainable approach to crop protection and pathogen management. Following the recent approval of Ledprona as the first sprayable double-stranded RNA (dsRNA) biopesticide by the EPA at the end of 2023, SIGS has emerged as a focal point in both academic and industrial sectors. This review analyzes recent advances and emerging trends in SIGS. The application of SIGS for crop protection, including the control of insects, fungal pathogens, and viruses, is briefly summarized. Distinguishing this review from others, we delve into practical aspects of the technology, such as the selection and screening of target genes, large-scale production methods, and delivery systems, highlighting major advancements in these areas and also addressing the remaining questions and issues, particularly concerning safety concerns and controlling harmful weeds. Finally, this review emphasizes the emerging trends in SIGS technology, particularly its integration with nanotechnology and other methodologies. Collectively, the rapid progress in SIGS studies is poised to accelerate the maturation and application of this technology.
It was demonstrated that some pathogens can deliver small RNAs (sRNAs) into host cells to suppress host immunity. Conversely, hosts also transfer sRNAs into pathogens and pests to inhibit their virulence (Wang et al., 2016, 2017). Travel of sRNAs between interacting organisms can induce gene silencing in the counter party, a mechanism termed cross -kingdom RNA interference (RNAi) or trans-kingdom RNAi (Cai et al., 2018a). As an example, trans-kingdom RNAi has been recently proposed for potential management of Fusarium wilt disease (Huang et al., 2025). In fact, trans-kingdom RNAi can be achieved either by host-induced gene silencing (HIGS) or spray-induced gene silencing (SIGS). The HIGS technology has become an important disease-control method by transgenic expression of pathogen gene-targeting double-stranded RNA (dsRNA) in plants (Cai et al., 2018b; Koch and Wassenegger, 2021). However, transgenic approaches have limitations during development and promotion. To circumvent the release of genetic modified plants, spray-induced gene silencing (SIGS), a phenomenon also referred as environmental RNAi, has been explored by direct application of dsRNAs or small interfering RNA (siRNA) onto host plants or post-harvest products, which leads to silencing of the target microbe/pest gene and confers efficient disease control (Lucena-Leandro et al., 2022). Environmental RNAi is powerful, environment-friend, and can be easily adapted to control multiple diseases simultaneously. As a novel, eco-friendly approach for managing plant pests and diseases, SIGS does not alter the host genome, therefore is widely accepted as an alternative to HIGS that needs genetic modification (Nityagovsky et al., 2022; Parise et al., 2024). Rooted in the natural RNAi process, where RNA molecules inhibit gene expression by neutralizing target mRNA, SIGS provides precise, targeted gene regulation (Niehl et al., 2018; Vetukuri et al., 2021). Early studies highlighted its potential to significantly reduce disease severity in plants, particularly through the suppression of fungal pathogen genes (Koch et al., 2016, 2019; Wu et al., 2024). SIGS has since expanded into pest and disease control strategies, offering high specificity, minimal off-target effects, and high environmental safety (Zotti and Smagghe, 2015; Das and Sherif, 2020). Unlike traditional pesticides, which persist in the environment and harm non-target species, SIGS degrades naturally, reducing ecological and health risks (Aktar et al., 2009; Imfeld and Vuilleumier, 2012). The specificity of dsRNA in SIGS targets pathogen genomes without affecting beneficial organisms such as insects, animals, or soil microbiota, making it a sustainable tool for integrated pest management (IPM) (Zotti and Smagghe, 2015; Christiaens et al., 2020). Studies show that dsRNA degrades before crops reach consumers, addressing concerns about pesticide residues in food and complying with stringent food safety standards (Christiaens et al., 2020). Moreover, SIGS supports soil health, as RNA biodegradation prevents disruption of microbial diversity and soil fertility, contrasting with the long-term detrimental effects of chemical pesticides (Imfeld and Vuilleumier, 2012; Dubrovina and Kiselev, 2019). This aligns SIGS with global initiatives like the European Commission’s Go-Green plan to reduce pesticide use by 2030 (Schebesta et al., 2020; Stojanovic, 2021). Research advances in SIGS include optimizing dsRNA delivery, improving stability, and exploring nanomaterials for enhanced uptake, ensuring its viability for large-scale applications. Despite challenges in foliar absorption due to leaf surface properties and environmental factors like UV exposure, innovations such as clay nanosheets and nanovesicles have extended dsRNA protection in plants (Taning et al., 2020; Hoang et al., 2022). As costs for dsRNA production decrease, products like Ledprona® demonstrate promising commercial potential, positioning SIGS as a key component of sustainable agriculture.
The dsRNA-based SIGS technology involves applying dsRNA to plant surfaces, allowing pathogens to absorb it and silence target genes without requiring transgenic plants, making it a faster and more adaptable solution (Beernink et al., 2024). By applying dsRNA to foliage, plants absorb it, triggering RNAi that silences specific genes in pests and pathogens by disrupting their life cycles, boosts disease resistance. Once applied, dsRNA is processed into siRNAs by Dicer, incorporated into the RISC complex, and binds to complementary mRNA, leading to its degradation and enhanced disease resistance in plants (Figure 1).
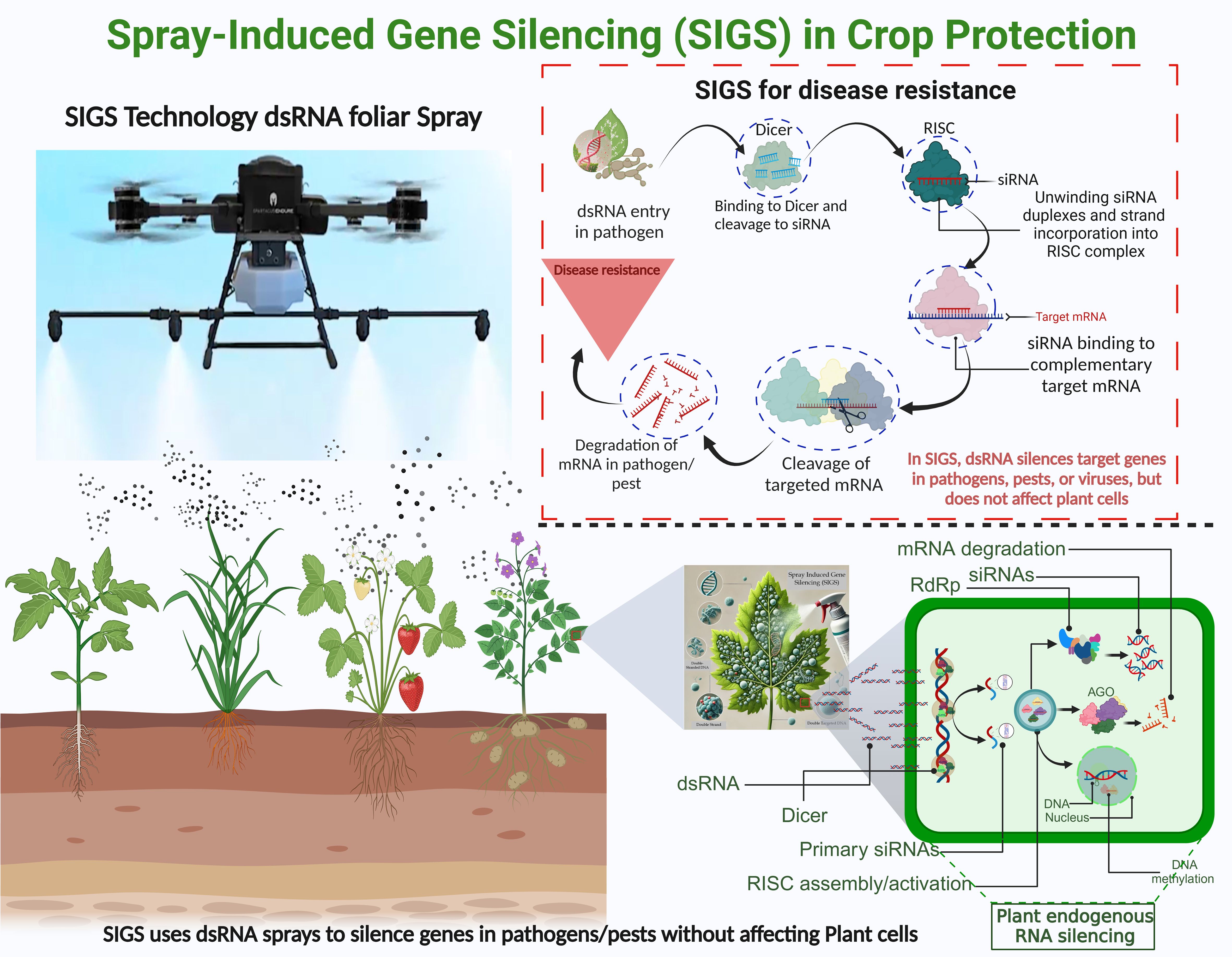
Figure 1. Spray-Induced Gene Silencing (SIGS) technology uses foliar dsRNA sprays to protect crops by silencing target genes. dsRNA is processed into siRNAs by Dicer, incorporated into the RISC complex, and binds to complementary mRNA, leading to its degradation and enhanced disease resistance in plants. The lower panel highlights the molecular mechanism of dsRNA processing and gene silencing within pathogen cells.
Importantly, SIGS offers a non-transgenic alternative with more immediate applicability, although its effectiveness depends on successful dsRNA uptake (Sang and Kim, 2020). To achieve the desired RNAi response, sprayed dsRNA must ultimately be delivered into responsive cells in the target organism at a sufficient level. The absorption of dsRNA might be relied on endocytosis. On one side, exogenous dsRNA can be directly absorbed by some pathogens or pests. For example, it was demonstrated that Botrytis cinerea (B. cinerea) can take up external dsRNAs, and applying dsRNAs on the surface of fruits, vegetables and flowers significantly inhibits grey mould disease (Wang et al., 2016). However, the ability to uptake dsRNA varies among different pathogens, e.g., it was observed efficient uptake in fungal pathogens, like B. cinerea, Sclerotinia sclerotiorum, Rhizoctonia solani, Aspergillus niger and Verticillium dahliae, but no uptake in Colletotrichum gloeosporioides, and weak uptake in a beneficial fungus, Trichoderma virens (Qiao et al., 2021). Effective uptake of dsRNAs in both Fusarium oxysporum and tomato tissues has been presented by fluorescence tracing (Ouyang et al., 2023). In contrast, we found weak uptake of dsRNA in Fusarium graminearum, a prevalent pathogen causing fusarium head blight. It was showed that uptake of dsRNA in Sclerotinia sclerotiorum through clathrin-mediated endocytosis (Wytinck et al., 2020b). For insects, the dsRNA molecules are absorbed through intestinal uptake following feeding, allowing for systemic spread. This systemic RNAi in nematodes is mediated by multiple SID proteins that are dsRNA specific membrane channels (Whangbo et al., 2017; Wytinck et al., 2020a). On the other side, sprayed dsRNA can be absorbed by plant through different organs and then secreted exosome-like extracellular vesicles to deliver sRNAs into fungal pathogen (Cai et al., 2018b). It was reported that sRNAs can be efficiently taken up and systemically transported to Malus domestica, Vitis vinifera, and Nicotiana benthamiana based on trunk injection and/or petiole absorption, and systemically transportation was strictly restricted to the xylem and apoplast (Dalakouras et al., 2018). Anyhow, for the indirect absorption after plant processing, dsRNA must travel from the surface of a leaf through the waxy cuticle, and then traverse the apoplast, cell wall, and plasma membrane to gain access to the plant cell’s RNAi machinery (Bennett et al., 2020). Once inside the plant cell, the applied dsRNA can move to adjacent cells through plasmodesmata and subsequently to distal cells through the phloem vasculature. Out of cell, trace amounts of dsRNA were detected in plant root excretions and in small brown planthoppers honeydew, and hoppers could transfer dsRNA via vomit (Zhang et al., 2022). Furthermore, ingestion of consumer hoppers could also result in localized RNAi in the midguts of the predator spiders, suggesting transmission along food chain.
Since the discovery of RNAi, advances have enabled its application in crop protection through precise, sequence-specific gene targeting via complementary dsRNA molecules. This specificity surpasses conventional pesticides, allowing for targeted pest management with minimal off-target effects. While synthetic pesticides, first widely used in the 1930s, have improved crop yields and quality, they have also led to growing pesticide resistance, particularly in insects (Mota-Sanchez and Wise, 2024). The advent of RNAi, first elucidated in 1998, marked a significant advancement in reverse genetics and pest management (Fire et al., 1998). As a post-transcriptional gene silencing (PTGS) mechanism, RNAi serves as a natural defense system in eukaryotic cells, targeting and degrading specific mRNA to inhibit gene expression (Fire et al., 1998; Baum and Roberts, 2014). This continued exploration of RNAi-based methods highlights its potential as a sustainable and precise alternative to traditional chemical solutions in modern agriculture. In fact, RNAi technology is increasingly recognized for its potential to manage various agricultural challenges, including insect pests, fungal pathogens, and viruses.
The application of dsRNA for insect control typically involves spraying it onto plant surfaces, allowing it to be directly ingested or absorbed by plants and later ingested by insects during feeding from plants (Pitino et al., 2011). The dsRNA passes through digestive system and enters insect cells, where it is processed by the Dicer complex into small interfering RNAs (siRNAs). These siRNAs are then incorporated into the RNA-induced silencing complex (RISC), which targets and cleaves complementary mRNA sequences, preventing the production of essential proteins for insect survival (Fire et al., 1998; Lundgren and Duan, 2013). This disruption affects vital processes such as growth, reproduction, and metabolism, ultimately leading to insect death (Bachman et al., 2013). As summarized, some studies have demonstrated the effectiveness of dsRNA in managing certain insect pests (Table 1). For example, a study by Pinheiro et al. (2020) have reported that after three days of injecting dsRNAs on Sri Lankan weevils for target genes, their transcript levels were significantly reduced (up to 91.4%) whereas, feeding of weevils with targeted dsRNAs showed significant decreases in gene transcript levels and significant mortality of insects treated with Prosα2 and Snf7 dsRNAs (78.6 to 92.7%). In another study, microinjection of dsRNA into the larvae of Frankliniella occidentalis thrips effectively silenced key genes (V-ATPase-B, CYP3653A2, and ApoLp-II/I), confirmed at 48 and 72 hours post-injection during the first and second instar stages, improving tissue health and survival whereas silencing CYP3653A2 or ApoLp-II/I increased larval mortality, proving their essential role in vitality (Han and Rotenberg, 2024).However, sensitivity to dsRNA varies across insect orders, with Coleopterans (beetles) being highly responsive, requiring only small amounts of dsRNA to achieve significant mortality by silencing key genes (Nitnavare et al., 2021). In contrast, Hemiptera (sucking insects) and Lepidoptera (moths and butterflies) display more variable responses, depending on their feeding habits and RNAi pathway efficiency (Niehl et al., 2016; Christiaens et al., 2020). For example, foliar application of dsRNA targeting Leptinotarsa decemlineata (Colorado potato beetle) genes such as Actin and V-ATPase resulted in reduced population growth, while plant-mediated RNAi targeting the CYP6AE14 gene in Helicoverpa armigera (cotton bollworm) led to impaired growth and reduced survival (Mao et al., 2007; San Miguel and Scott, 2016). Additionally, dsRNA targeting V-ATPase or tubulin in the western corn rootworm agar diet effectively increased Diabrotica virgifera larval mortality and/or development stunting (Baum et al., 2007). Similar dsRNA-based approaches have shown promising results in controlling pests like Myzus persicae (green peach aphid), Tuta absoluta (tomato leaf miner), and Plutella xylostella (diamondback moth), reducing their reproduction, survival, and feeding behavior (Pitino et al., 2011; Camargo et al., 2016). Various delivery methods, including foliar sprays, artificial diet feeding, and bacterial expression of dsRNA, have been employed to enhance dsRNA efficacy (Zha et al., 2011; Sang and Kim, 2020; Beernink et al., 2024). Delivering dsRNA via bacteria offers advantages over in vitro synthesis, with studies showing significant mortality in Leptinotarsa decemlineata larvae fed on E. coli expressing dsRNA targeting multiple genes (Zhu et al., 2011). Similar RNAi effects have been observed in pests like Spodoptera exigua, Tuta absoluta, and Chilo infuscatellus following ingestion of bacteria-expressed dsRNA (Tian et al., 2009; Vatanparast and Kim, 2017; Bento et al., 2020). Anyhow, achieving effective gene silencing under field conditions often requires larger quantities of dsRNA.
Fungal plant diseases are a major threat to global food security, leading to annual crop yield losses of up to 20% and post-harvest losses of about 10% worldwide (Delgado-Baquerizo et al., 2020). Traditional methods for managing fungal pathogens rely heavily on fungicides, which have contributed to the rise of fungicide-resistant in fungal pathogens (Fisher et al., 2018; Imran et al., 2021; Wang et al., 2023a). To address this, eco-friendly alternatives such as RNAi technologies have gained attention and recent discoveries demonstrated that many fungal pathogens can absorb environmental RNAs, which can then trigger gene silencing of fungal targets with complementary sequences (Koch et al., 2016; Werner et al., 2020; Qiao et al., 2021). This has enabled the development of SIGS where dsRNAs or sRNAs targeting fungal virulence genes are applied to plants to combat infections. Foliar application of dsRNA has been effective in controlling several fungal pathogens by targeting specific genes (Qiao et al., 2021). The uptake of dsRNA by fungi can inhibit growth, reduce pathogenicity by targeting the specific genes and cause mortality (Table 2), presenting a promising alternative to conventional pesticides. However, the efficiency of dsRNA uptake varies across fungal species, with some fungi showing negligible uptake, limiting RNAi effectiveness (Zhang et al., 2016). For instance, dsRNA targeting Botrytis cinerea genes DC-L1 and DC-L2 significantly reduced its growth and virulence in grapes, decreasing lesion size by over 80% (Nerva et al., 2020). In another study, application of sRNA or dsRNA target DCL1 and DCL2 genes in botrytis and demonstrated significant reduction in gray mold disease (Wang et al., 2016). Similarly, targeting ergosterol biosynthesis genes in Fusarium graminearum (F. graminearum) reduced fungal transcript levels by 58%(CYP51A), 50% (CYP51B), 48% (CYP51C) in leaves sprayed with CYP3-dsRNA and decreased pathogen DNA in barley (Koch et al., 2016). Targeting the ergosterol biosynthesis pathway in F. graminearum not only restricted fungal growth but also disrupted respiration, highlighting the link between ergosterol production and fungal metabolism (Koch et al., 2016). Moreover, dsRNA applied to combat Austropuccinia psidii (myrtle rust) showed both preventive and curative effects, reducing disease severity (Degnan et al., 2023). Studies also show the potential of dsRNA to target key respiratory genes in fungi, leading to impaired mitochondrial respiration, reduced ATP production, and overall growth inhibition (Table 2). For example, targeting mitochondrial TIM44 genes in B. cinerea resulted in decreased ATP synthesis, affecting fungal energy metabolism (Koch et al., 2016). Similar results were seen in Sclerotinia sclerotium, where dsRNA reduced respiratory gene expression by 50%, correlating with lower ATP levels and impaired growth (Wytinck et al., 2020b). Fungal pathogens primarily take up dsRNA through clathrin-mediated endocytosis (CME), with studies confirming that inhibiting clathrin-related genes reduces dsRNA uptake and RNAi efficacy (Wytinck et al., 2020b). While CME is the dominant mechanism, other less-understood endocytic pathways may also contribute to dsRNA absorption. Understanding these mechanisms is critical for optimizing RNAi-based strategies to improve fungal disease management in agriculture.
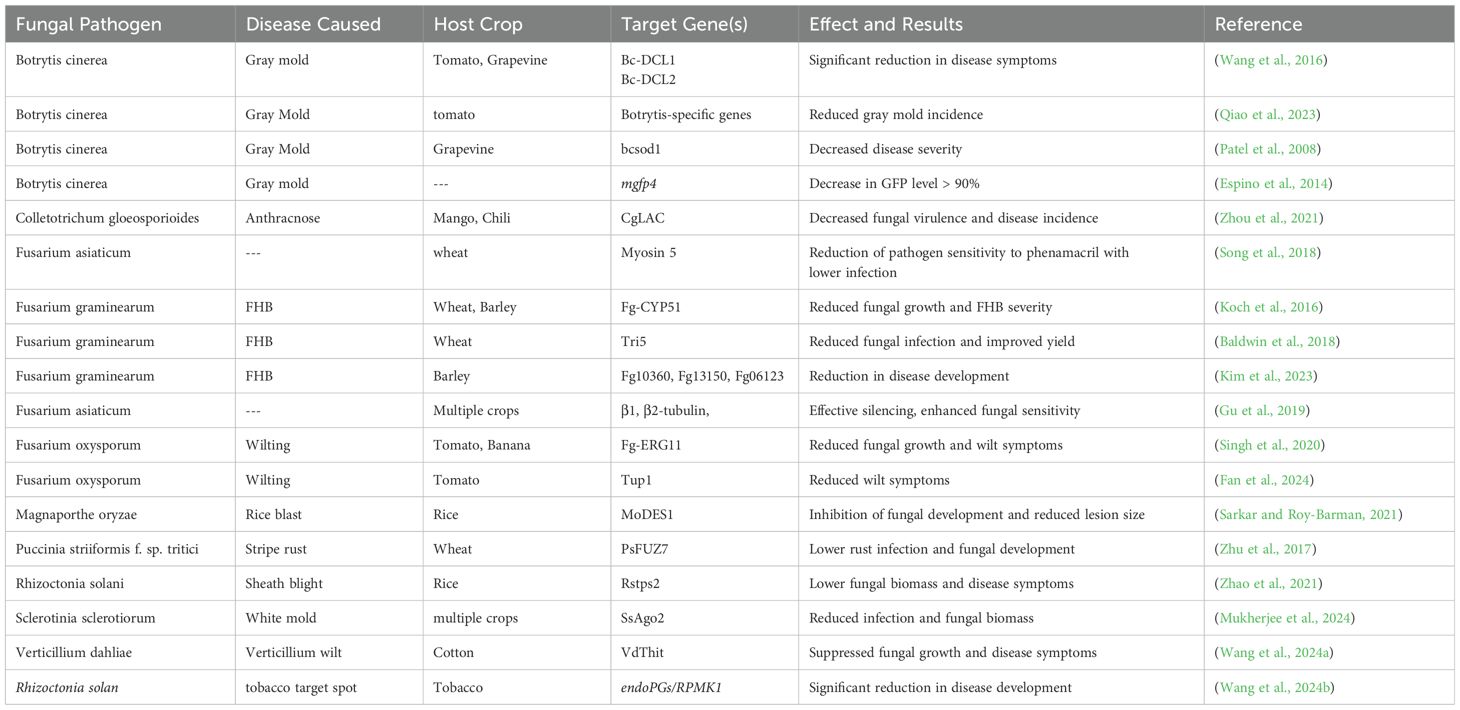
Table 2. Some reports of dsRNA-based SIGS for the control of fungal plant pathogens by foliar spray.
Plant viruses are one of the most significant threats and emerging challenge in agriculture, leading to severe crop losses, from stunted growth to total crop failure, impacting both yield and quality (Tatineni and Hein, 2023; Hamim et al., 2023). Conventional control methods often fail due to the high mutation rates of plant viruses, whereas RNAi-based approaches, particularly dsRNA technologies, offer a promising, sustainable alternative for managing viral diseases, and various studies have documented the use of dsRNA for the control of plant viruses (Table 3). When plants are infected, viral small RNAs (vsRNAs) are produced by RNAi machinery of plants, which processes viral dsRNAs or hairpin RNAs (hpRNAs). These vsRNAs degrade complementary viral single-stranded RNAs, acting as a natural antiviral defense (Agius et al., 2012). Building on this, dsRNA-based silencing technologies targeting viral genes have been developed, including engineered dsRNAs or hpRNAs (Marjanac et al., 2009; Guo et al., 2016; Hameed et al., 2017). The key advantage of SIGS technology in the control of viral diseases is its ability to provide targeted control of plant viruses with minimal off-target effects on non-target microorganisms (Baulcombe, 2015; Mitter et al., 2017b; Rêgo-MaChado et al., 2023). Once dsRNA corresponding to viral genes is introduced into the plant, the RNAi pathway is activated, leading to viral RNA degradation and suppression of viral replication, without harming beneficial organisms in the ecosystem (Delgado-Martín et al., 2022). This systemic effect can extend protection beyond the treated areas, enhancing overall plant health and resilience (Mitter et al., 2017b). The uptake of dsRNA by plant cells is facilitated through natural pathways like endocytosis or via viral assistance (Mitter et al., 2017a). Inside the plant cells, Dicer-like (DCL) enzymes cleave dsRNA into small interfering RNAs (siRNAs), which are then incorporated into the RNA-Induced Silencing Complex (RISC). This complex cleaves complementary viral RNA, preventing viral replication (Zotti et al., 2018). The RNAi response can spread throughout the plant via the vascular system, amplifying the silencing signal and enhancing viral defense (Singh et al., 2019). The ability of dsRNA to target essential viral genes makes it a versatile tool for managing various viral threats in agriculture (Mitter et al., 2017b; Cagliari et al., 2019). For example, it was demonstrated that SIGS significantly delayed symptoms of Tomato spotted wilt virus (TSWV) (Tabein et al., 2020), and dsRNA stability was enhanced through delivery methods like nanoparticle encapsulation and biopolymer incorporation (Mitter et al., 2017a). Additionally, dsRNA applied to crops like papaya, zucchini, and cucumber significantly reduced viral symptoms and virus accumulation (Delgado-Martín et al., 2022; Rêgo-MaChado et al., 2023). Studies have also shown that dsRNA targeting specific viral genes (e.g., RP gene in pepper against mild mottle virus, HC gene in tobacco against tobacco etch virus, RNA3 gene in alfalfa against alfalfa mosaic virus) can lower viral loads and delay systemic symptoms (Tenllado and Díaz-Ruíz, 2001; Tenllado et al., 2003). Similarly, hpRNA targeting Sugarcane mosaic virus (SCMV) in maize reduced viral load, and dsRNA targeting the Nib gene in potato enhanced resistance to potato virus Y (PVY) (Gan et al., 2010; Sun et al., 2010). Tobacco mosaic virus was also controlled by targeting multiple viral genes (MP, CP, RP, RNA) with dsRNA and hpRNA, significantly reducing viral loads and infection rates (Konakalla et al., 2016). Thus, SIGS-dsRNA technology shows great potential for managing viral pathogens. Advances in dsRNA stabilization techniques, such as UV protection and temperature resilience, will be key to improving the practicality and commercial viability of SIGS in agriculture. Additionally, the immune-priming effects of dsRNA may improve plant resilience to other pathogens and stressors, promoting better nutrient uptake and overall plant vigor.
Recent developments in the use of dsRNAs for plant protection have focused on target selection because optimized targets allow the precise gene silencing of specific pests or diseases, minimizing collateral damage to beneficial microbiota and microorganisms, and ultimately promoting ecosystem health. The integration of biomarkers into the target selection process enables researchers to predict biological responses with greater accuracy, thereby refining SIGS application to enhance the effectiveness while reducing non-target impacts. Simultaneously, advancements in the production methods aim to scale up the availability of these siRNAs, making them more accessible for sustainable agricultural practices. Improved microbial and enzyme-based synthesis techniques, alongside innovations in nanotechnology, significantly enhanced the scalability and practicality of dsRNA applications in agriculture. Furthermore, optimizing dsRNA design, considering factors such as length and nucleotide composition, enhances the silencing efficiency by improving cellular uptake and initiating RNAi more effectively. These advancements not only lower production costs but also improve the stability and delivery efficacy of dsRNA under various environmental conditions, protecting it from rapid degradation. Consequently, these innovations ensure the prolonged effectiveness of SIGS applications, expanding their utility across diverse crops and agricultural settings and opening new horizons in sustainable agricultural production.
The success of SIGS relies heavily on the precise selection and screening of target genes (Zhao et al., 2024). Identifying the genes that are crucial for survival, virulence, or reproduction of insect pests and plant pathogens is essential for effective control strategies. Advancements in bioinformatics and multi-omics technologies have significantly improved the efficiency of this gene-targeting process (Sigoillot et al., 2012; Ahmed et al., 2015). Among these technologies, high-throughput sequencing and gene expression profiling have enabled the researchers to identify differentially expressed genes during the infection or infestation, making the genes as a prime candidate for silencing. In contrast, comparative genomics have facilitated the identification of conserved genes across multiple pest species, ensuring that SIGS technology maintains a broad spectrum of effectiveness. An illustrative example is the Functional Representation of Gene Signatures (FRoGS) technology, which employs machine learning to enhance target prediction by integrating the gene functions within a comparative analysis framework (Namba et al., 2022) and this approach has demonstrated greater accuracy in identifying the potential therapeutic targets compared to traditional identity-based methods (Chen et al., 2024). Moreover, combining transcriptomic data with genetic perturbation signatures allow the researchers to differentiate between inhibitory and active targets, thereby refining the selection process for SIGS applications in crop protection. Advanced bioinformatics approaches streamline the target identification through in silico analyses, which significantly reduce the time and resources required for experimental validation and this optimization is critical for developing effective and precise SIGS strategies.
In SIGS technology, the production methods have evolved to meet the growing demand for effective gene silencing agents in agricultural applications. Traditional methods, such as chemical synthesis and in vitro transcription, are now replaced by advanced techniques such as recombinant fermentation and cell-free synthesis, which facilitate the large-scale production of RNA molecules essential for SIGS and ensuring a consistent supply of high-quality silencing agents.
Recombinant fermentation utilizes genetically modified organisms (GMOs) for producing specific RNA sequences, offering several advantages for dsRNA production. Utilizing microorganisms like Escherichia coli or yeasts such as Pichia pastoris allow the rapid growth and high-density cultures, making this approach more cost-effective and efficient (Vieira Gomes et al., 2018). Additionally, recombinant fermentation provides the precise control over production process, allowing the optimization of conditions like temperature and nutrient supply to enhance the yield and quality (Ross et al., 2024; da Rosa et al., 2024). Thus, recombinant fermentation emerges as a reliable and scalable solution for producing high quality silencing agents in agricultural biotechnology.
Cell-free synthesis (CFS) facilitates the direct assembly of RNA molecules without the need for living cells, presenting lower contamination risks and faster production and this makes the CFS particularly suitable for rapid response scenarios in managing insect pests. Further, CFS allow the quick adjustments of target sequences by simply altering the DNA template which enables the simultaneous production of multiple dsRNA targets within a single reaction vessel (Hough et al., 2022). The scalability of CFS significantly enhances its cost-effectiveness, allowing the rapid production and flexibility in manufacturing processes. Unlike traditional cell-based systems that often require lengthy culture periods and complex scaling procedures, CFS can reduce production time from weeks to hours, facilitating the rapid market response thereby decreasing the labour costs associated with cell maintenance and culture management. Furthermore, CFS systems can be easily scaled up or down without compromising the yield or quality, accommodating the specific production needs without substantial additional costs and this adaptability is beneficial particularly for producing the small batches of specialized products. However, a major bottleneck of CFS is the stability and cost of transcriptase.
Ongoing research on optimizing these production methods to enhance the yield and reduce the costs should be prioritized. Innovations in bioprocessing technologies can improve SIGS production efficiency and promote wider adoption in crop protection strategies against insects, fungi and viruses. These advancements in production methods are fundamental for enhanced efficacy and applicability of SIGS technology in agriculture and ultimately pave the way for sustainable crop protection and promote green agriculture. Once upon a time, cost is one primary factor constraining the large-scale application of SIGS. However, this is not a problem now. In fact, the cost for dsRNA production is quickly decreased along with the industrialization. For example, RNAGri had the ability to produce tons of dsRNA at a cost of 1$/g (Guan et al., 2021), while Greenlight Biosciences further reduced the cost of dsRNA synthesis by combination of microbial fermentation and CFS technologies (https://www.greenlightbiosciences.com/how-do-we-make-rna).
Efficient delivery of dsRNA remains a core challenge in achieving effective outcomes in SIGS, particularly for protecting the crops from a variety of insect pests and pathogens. The delivery vehicle should safeguard the RNA molecules from environmental degradation, and may also facilitate their uptake into plant tissues because successful delivery requires that dsRNA reaches the target organism in sufficient quantities to elicit a silencing response. As summarized in Figure 2, the current delivery approaches include microinjection, controlled release, feeding of dsRNA-containing diet, foliar spray, and trunk inoculation (Wuriyanghan et al., 2011; Pinheiro et al., 2020). The choice of carriers significantly influences the delivery efficiency. Many recent studies have focused on enhancing RNA stability and controlled release through diverse nanocarrier systems and encapsulation technologies (Pal et al., 2024). Thus, various carriers including nanoparticles (NPs), have been explored to improve the stability and uptake by target organisms (Koch et al., 2019).
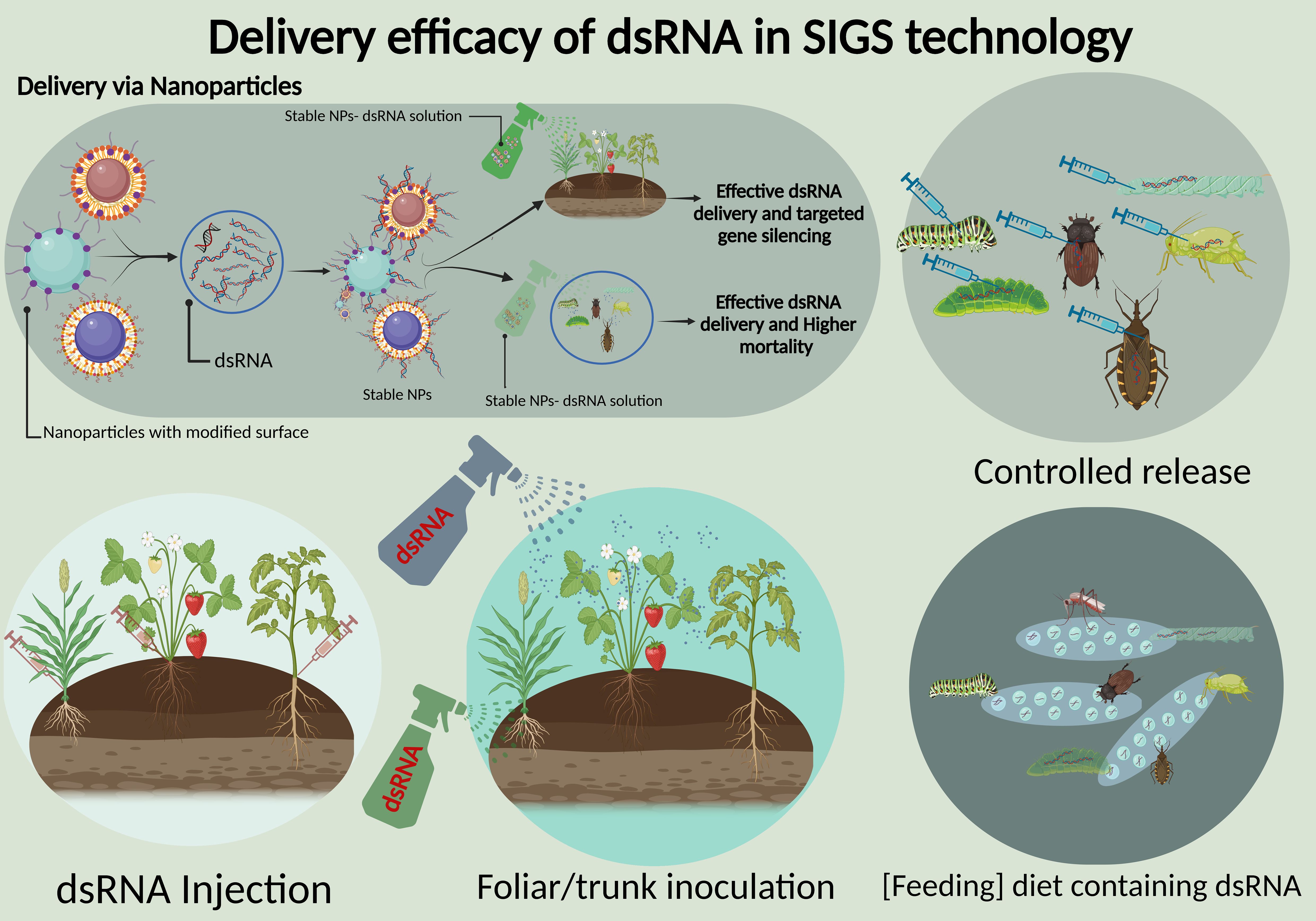
Figure 2. Delivery efficacy of dsRNA in SIGS technology. The figure highlights multiple dsRNA delivery methods: nanoparticle-based delivery for effective gene silencing and insect mortality, controlled release mechanisms, dsRNA injection into plant tissues, foliar/trunk inoculation via sprays, and feeding diets containing dsRNA for targeted pest control. These approaches improve stability, delivery, and efficiency of dsRNA in crop protection.
To optimize these conditions, various NPs, such as chitosan, polyethyleneimine, and layered double hydroxide (LDH) clay nanosheets, have been explored. They can not only protect the RNA from environmental degradation but also demonstrate facilitated absorption by target pathogens thereby significantly increased the gene silencing efficacy. Studies have demonstrated that chitosan and LDH considerably improved the dsRNA stability, and prolonged the protective effects against pathogens for up to 20 days, making them highly suitable for agricultural applications where durability is the most crucial factor (Koch et al., 2019). A study has highlighted the effectiveness of NPs in delivering dsRNA for targeted protection against Rhizoctonia solani, the pathogen responsible for rice sheath blight, where RsAGO1 and RsAGO2 were identified as the effective targets for dsRNA interference (Wang et al., 2023b). NPs of protamine, carbon quantum dots and graphene quantum dots demonstrated the ability to form stable nanoparticle-dsRNA structures and effectively silenced MGV1 and RAS1 genes of F. graminearum (Gyawali et al., 2024). Notably, carbon quantum dots and chitosan/SPc complexes enhanced the dsRNA loading capacity by maintaining the functionality with only a 7% reduction in fluorescence intensity post-nuclease treatment (Wang et al., 2023b). Some other carriers, like liposomes and carbon nanotubes, have also been utilized as NPs offer superior stability and delivery efficiency, which makes them the optimal for SIGS applications because NP carriers reduce the frequency of applications and minimize the chemical runoff into ecosystems (Mittal et al., 2020). This targeted delivery effectively silences the specific genes in pests and pathogens without affecting non-target organisms by reducing the reliance on broad-spectrum pesticides. Additionally, these biodegradable carriers, derived from renewable resources, also mitigate the environmental toxicity associated with traditional chemical treatments.
Despite the promising potential of SIGS technology for crop protection, several critical challenges must be addressed to optimize its efficacy and safety. One of the primary concerns is the stability of the RNA molecules utilized in gene silencing, as various environmental factors and enzymatic activities in plants can degrade these RNA molecules. Although researchers have documented that the structural modifications to RNA can enhance its stability, these modifications may also influence silencing efficiency (Dubrovina and Kiselev, 2019). Achieving consistent delivery and expression levels in targeted plants remains a significant challenge, as variations in plant responses to SIGS treatments can lead to inconsistent outcomes. This variability highlights the necessity for further optimization of application protocols and formulations. To tackle these issues, researchers have investigated various encapsulation methods, and embedding dsRNA in protective nanocarriers considerably increased the stability and persistence on plant surfaces (Guo et al., 2022; Pal et al., 2024). For example, the delivery of dsRNA to soybean aphid Aphis glycines demonstrated rapid penetration into the body wall within 4 hours with the help of nanocarrier. Through the topical application effectively silenced the target gene expression with the knockdown effects ranging from 86.86 to 58.87% and demonstrated higher mortality up to 81.67% (Yan et al., 2020). Among these nanocarriers, clay nanosheets have demonstrated considerable potential by effectively shielding the dsRNA from degradation due to UV exposure and heat while facilitating a gradual release. This sustained release allows for a prolonged presence of RNA on the plant, thereby maintaining the bioactivity of the molecules and dwindling the gap for gene silencing effects (Mitter et al., 2017a).
Another significant challenge associated with SIGS is the risk of off-target effects. While SIGS is designed to specifically target certain genes, the inherent specificity of RNAi relies heavily on the sequence complementarity between the dsRNA/siRNA and the intended target gene. Unintended interactions with non-target genes can lead to adverse effects on plant health and development, as evidenced by studies documenting off-target silencing due to sequence homology (Chen et al., 2021). To mitigate these risks, bioinformatics tools, including homology-based sequence screening and predictive modelling, are increasingly utilized to refine target selection and minimize off-target effects. Nevertheless, extensive in vivo testing across diverse plant species remains crucial for further assessing and mitigating these risks, underscoring the need for robust methods to predict and evaluate off-target activity (Dubrovina and Kiselev, 2019; Vetukuri et al., 2021). As dsRNA might persist in soil and enter the food chain, posing potential risks to biodiversity and human health (Papadopoulou et al., 2020). Thus, addressing these challenges requires developing the biosafety frameworks and advanced predictive models to assess dsRNA persistence and its pathways in non-target organisms (San Miguel and Scott, 2016). The integration of SIGS with traditional chemical controls and advanced nanotechnology offers promising potential for sustainable IPM strategies. This combination may significantly enhance the effectiveness of agricultural practices in managing plant pathogens and pests.
In addition, weed species significantly reduce agricultural productivity by competing for essential resources, such as water, light, and nutrients (Horvath et al., 2023). Their rapid reproduction is aided by traits like deep root systems and allelopathic substance release, which inhibit crop growth and promote pathogens, ultimately increasing cultivation costs (Trognitz et al., 2016). Studies across Europe have identified numerous weed species in various crops, with notable examples including Fallopia convolvulus and Amaranthus retroflexus (Gerhards et al., 2017; Hofmeijer et al., 2021). A patent has explored SIGS to combat herbicide resistance, such as restoring glyphosate efficacy in Amaranthus palmeri (Sammons et al., 2011). An interesting study designed three types of RNAi-based herbicides that specifically silenced endogenous target genes and controlled the growth of Mikania micrantha Kunth (Mai et al., 2021). In particular, the study shown that weed leaves turned yellow and eventually wilted after spray of dsRNA targeting Chlorophyll a/b-binding protein. However, genetic similarity between crops and weeds complicates the design of dsRNAs that risking off-target effects in related crops. Addressing the challenges of delivering stable siRNA formulations and enhancing genomic knowledge of weeds is crucial for making SIGS a viable alternative to conventional herbicides (Zabala-Pardo et al., 2022). Despite advancements in RNAi for insect pests and viruses, its application for weed management remains limited, and to the best of our knowledge and based on available literature.
Generally, SIGS technology offers targeted pest control, rapid response, and reduced pesticide reliance, enhancing crop resilience. However, in almost all of these studies pathogens were inoculated simultaneously with or shortly after dsRNA treatment, which is different from the real situation. Under natural conditions, pathogens often have already existed in the plant tissue, therefore limiting the value in application. Meanwhile, most of the studies were conducted using disease models and little field studies were conducted. From a practical point, SIGS also faces challenges like variable efficacy, regulatory hurdles, and high costs, including risks like off-target effects, resistance development, and concerns about dsRNA stability and public perception of genetic manipulation (Figure 3). Thus, to enhance the application of SIGS technology, several key scientific questions need exploration, which includes identifying the specific RNA modifications that improve stability while preserving silencing efficiency, refining predictive models for off-target effects to increase accuracy across diverse plant species, and assessing the ecological impacts of dsRNA persistence in soil on non-target organisms. Additionally, understanding how SIGS integration with other pest management strategies can influence the resistance development in target pathogens is vital, and establishing the regulatory frameworks for the safe application of SIGS under field conditions is essential. Thus, addressing these questions will be crucial for optimizing the efficacy and safety of SIGS in sustainable agricultural practices.
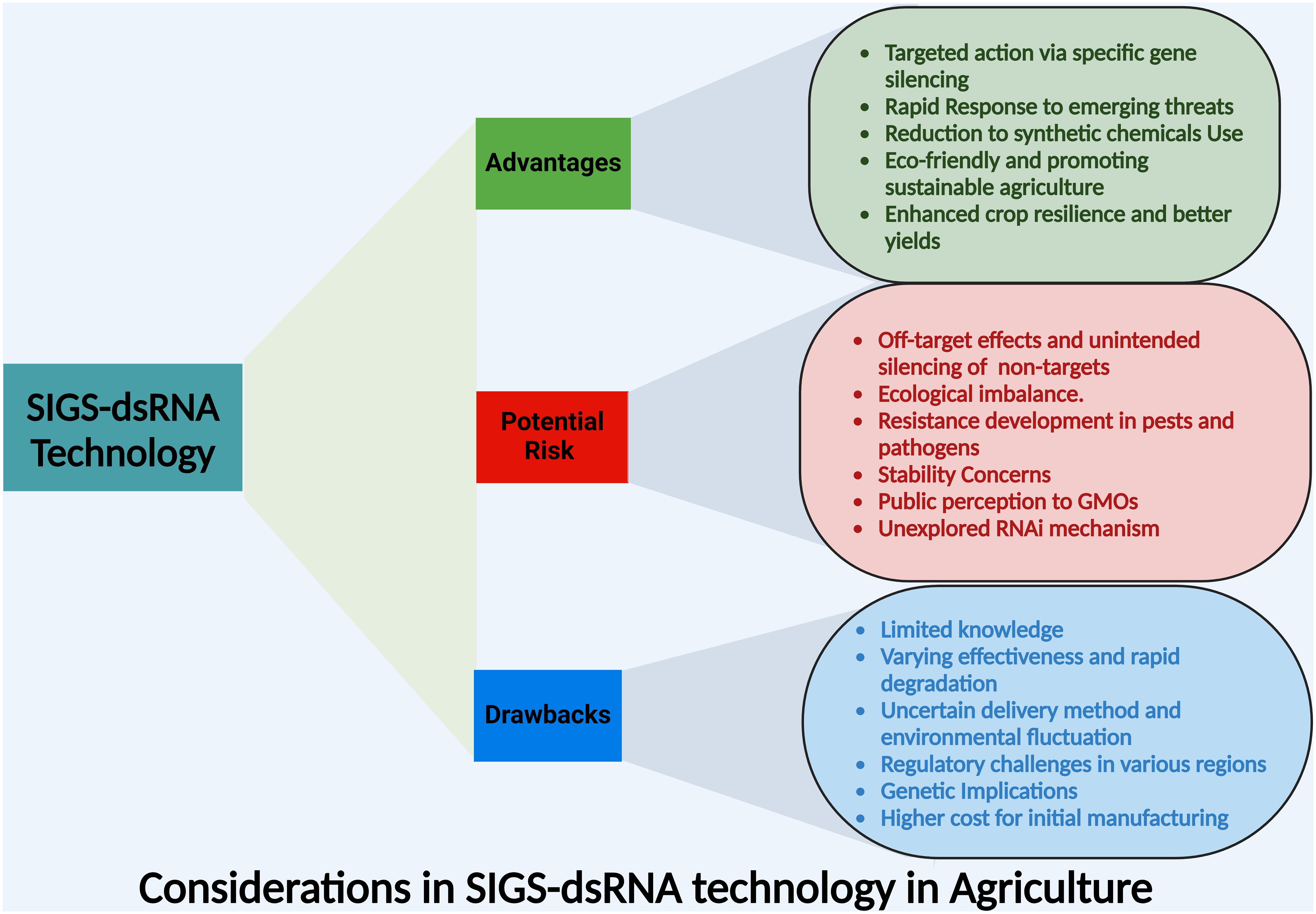
Figure 3. An overview of the core advantages, potential risks and drawbacks of dsRNA application in agriculture associated with SIGS technology for crop protection.
In green agriculture, fungal and bacterial communities are emerging as effective biocontrol agents in sustainable agriculture due to their eco-friendly performance (Aldayel et al., 2024; Imran et al., 2024). These biocontrol agents produced various volatile metabolites and hydrolytic enzymes that can enhance the efficacy of dsRNA applications for pest and pathogen management because these compounds serve as signalling molecules, triggering plant defence mechanisms and complementing the effects of dsRNA, which targets specific genes in pests and pathogens to inhibit their growth (Imran et al., 2022). Further, these species respond to the volatile compounds emitted from pathogens and upregulate biocontrol-related genes, thereby producing antifungal metabolites (Giorgio et al., 2015; Li et al., 2018). This interaction not only amplifies the inhibitory effects of dsRNA but also bolsters plant resilience against biotic stresses, suggesting that combining volatile compounds with dsRNA strategies could yield synergistic benefits. High-throughput screening methods for identifying effective bacterial biocontrol agents can further enhance the application of dsRNA technologies, allowing simultaneous screening of thousands of candidates and improving overall biocontrol strategies (Hough et al., 2022; Kjeldgaard et al., 2022). Emerging trends in SIGS technology highlight its potential to revolutionize crop protection through innovative applications and methodologies as it enables targeted gene silencing to control pests and pathogens by delivering dsRNA molecules to crops, positioning it as a promising alternative to traditional pesticides. Further, it offers a more sustainable, targeted, and environmentally friendly solution. Current trends in SIGS emphasize overcoming these challenges by integrating new technologies, particularly focusing on nanotechnology and the production of dsRNA by biocontrol agents to enhance the efficacy of RNA-based applications.
The integration of nanotechnology into SIGS represents a significant advancement in enhancing the stability, delivery efficiency, and targeted action of dsRNA molecules. Encapsulating dsRNA within NPs protects it from environmental factors that influence its effectiveness under open field conditions, thereby extending its functional longevity. This encapsulation reduces the frequency of applications needed for gene silencing and minimizes the costs. Various NP carriers, including liposomes, biopolymer nanoparticles, and virus-like particles (VLPs), have been explored (Pal et al., 2024) for their potential to deliver dsRNA effectively. For example, liposomes encapsulate dsRNA in lipid bilayers that mimic cellular membranes, facilitating the absorption by plant cells, while biopolymer NPs derived from natural or synthetic polymers offer biocompatibility and controlled release properties, making them more suitable for agricultural applications (Ghosh et al., 2023). Contrary to VLPs, which resemble viruses but lack infectious genetic material, also deliver dsRNA by evading the host defense that typically degrades foreign genetic material and enhances the stability and precision (Sarkar and Roy-Barman, 2021; Ghosh et al., 2023). One notable advantage of NP use is the improved stability of dsRNA in alkaline environments, such as insect guts, where it would otherwise degrade rapidly, as studies have shown that specific NP formulations can protect the dsRNA from high pH and enzymatic changes, prolonging its activity and enhancing its insecticidal efficacy (Qiao et al., 2023). This is particularly relevant for controlling pests with alkaline gut environments, where traditional dsRNA applications may fail without NP protection (Yang et al., 2022). Furthermore, nanotechnology enables controlled release mechanisms that enhance the persistence of RNAi agents on plant surfaces by minimizing their application frequency required for effective pest management (Dhandapani et al., 2022). Thus, the combination of SIGS with nanotechnology not only addresses RNA stability challenges but also paves the way for more targeted and efficient crop protection strategies.
As major drivers for controlling host populations and evolution, viruses are potential biological control agents (Wagemans et al., 2022). Mycoviruses are viruses that infect fungi, and some mycoviruses may alter fungal host pathogenicity resulting in hypervirulence or hypovirulence and therefore could be used for plant protection. Hypovirulence-inducing mycoviruses represent a powerful means to defeat fungal epidemics on crop plants. Infections of fungi by mycoviruses are sometimes fatal, as they perturb sporulation, growth, and, if applicable, virulence of the fungal host. A dsRNA chrysovirus-like mycovirus (FgV-ch9) debilitates Fusarium graminearum, the causal agent of fusarium head blight (Bormann et al., 2018). The chrysovirus FodV1 also induces hypovirulence of its host F. oxysporum, which causes vascular wilt in carnation by decreasing the colonizing efficiency of its fungal host (Torres-Trenas et al., 2019). A novel mycovirus, designated as PtCV1, from the fungus Pestalotiopsis theae (P. theae), a pathogen of tea, has four dsRNAs as its genome. PtCV1 can significantly reduce the growth rates of its host fungus in vitro and abolish its virulence in planta, converting its host fungus to a non-pathogenic endophyte on tea leaves, while PtCV1-free isolates were highly virulent (Zhou et al., 2021). Moreover, the presence of PtCV1 conferred high resistance to the host plants against the virulent P. theae strains. In fact, it was found that a large proportion of Fusarium isolates (46%) were infected with mycoviruses and five mycoviruses were shared between F. graminearum and F. culmorum (Buivydaitė et al., 2024). Therefore, the presence of these hypovirulence-inducing mycoviruses may reduce the fitness of a fungal pathogen and enhance the effectiveness of RNAi-based control. For example, it was found a fungal host’s RNAi machinery is upregulated in the presence of mycovirus that lacks a virus-encoded suppressor (VSR), compared to one that has an active VSR (Zhang et al., 2008).
On the other side, the presence of mycoviruses may disrupt the fungal RNAi and derail the efficacy of SIGS control strategy, which requires functional RNAi in the target cell. This is because some mycoviruses produce VSRs, that disrupt their host’s RNAi (Aulia et al., 2021; Ko et al., 2021). These VSRs either suppress the transcription of key enzymes (like DCL2 and AGL2) or reduce the accumulation of siRNA to repress fungal RNAi (Rodriguez et al., 2022; Myers and James, 2022). For example, Aspergillus virus 1816 was capable of suppressing RNAi and this resulted in reduced siRNA in A. nidulans (Hammond et al., 2008). Similarly, the Rosellinia necatrix mycoreovirus 3 showed VSR activity and suppressed RNAi in Nicotiana benthamiana, possibly by less accumulation of siRNA (Yaegashi et al., 2013). Therefore, these VSRs might be novel add-on targets for dsRNA-based SIGS, as silencing them can maintain or even enhance the RNAi machinery of the fungal host. Meanwhile, co-application of mycovirus that lacks VSRs and dsRNA will probably enhance the stability of dsRNA by counteracting degradation, achieving prolonged and effective gene silencing. Moreover, mycovirus can be engineered by inserting the target transcript in both sense and antisense orientations, which can convert pathogenic fungi into hypovirulent strains by silencing the target gene. As reported, a mycovirus, FgGMTV1, has been successfully engineered and efficiently triggered gene silencing in F. graminearum (Zhang et al., 2023). This is highly promising, as the strategy combined the infectious property of mycovirus and the RNAi mechanism, also simplified the requirements for production and storage.
Plants utilize dsRNA for producing siRNA, driving gene silencing and enhancing resistance to pathogens, and this RNAi mechanism allows crops to downregulate critical genes, effectively preventing their further development while minimizing unintended effects through precise, targeted action. However, enhancing target specificity through multi-omics gene selection approaches can minimize off-target effects by preserving the biodiversity, but for this, long-term field studies are required that will inform the regulatory guidelines. Besides, integrating SIGS with IPM and precision agriculture can reduce crop losses and support soil health and beneficial microbiota, as topical application of dsRNA can trigger a protective response, providing reliable control without the adverse impacts associated with traditional chemicals.
Further research on optimal dsRNA concentration, stability, and delivery systems is crucial for realizing the potential of SIGS in sustainable crop disease management. Efficient carriers, such as nanoparticles and bio-based polymers, can enhance dsRNA uptake, while cost-effective production methods like microbial fermentation and cell-free synthesis make SIGS accessible to resource-limited farmers in the form of powders and gels by simplifying transportation and application. To some extent, recent advancements have addressed the production costs, stability, and off-target effects, positioning RNAi as a promising eco-friendly alternative to synthetic pesticides. In SIGS, pathogen-specific genes are employed to inhibit the growth and pathogenicity, which offers a viable alternative and minimizes the adverse effects to non-target species. However, in-depth addressing of these challenges can effectively scale this technology in a broad spectrum. With ongoing advancements, SIGS is poised to play a pivotal role in meeting global food demands, aligning with the “Third Green Revolution” to ensure effective crop protection and food security.
CC: Funding acquisition, Investigation, Methodology, Writing – original draft, Writing – review & editing. IM: Writing – original draft, Writing – review & editing. XF: Data curation, Formal analysis, Writing – review & editing. XS: Conceptualization, Project administration, Supervision, Writing – review & editing. ZS: Conceptualization, Funding acquisition, Supervision, Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. Supported by the Open Project Program of State Key Laboratory for Crop Stress Resistance and High-Efficiency Production (CSBAAKF2021005); Training Plan of Young Backbone Teachers in Colleges and Universities of Henan Province, China (2021GGJS139); the Key Scientific and Technological Project of Henan Province, China (242102110214).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Agius, C., Eamens, A. L., Millar, A. A., Watson, J. M., Wang, M. B. (2012). RNA silencing and antiviral defense in plants. Methods Mol. Biol. 894, 17–38. doi: 10.1007/978-1-61779-882-5_2
Ahmed, F., Dai, X., Zhao, P. X. (2015). Bioinformatics tools for achieving better gene silencing in plants. Methods Mol. Biol. (Clifton N.J.) 1287, 43–60. doi: 10.1007/978-1-4939-2453-0_3
Aktar, W., Sengupta, D., Chowdhury, A. (2009). Impact of pesticides use in agriculture: their benefits and hazards. Interdiscip. Toxicol. 2, 1–12. doi: 10.2478/v10102-009-0001-7
Aldayel, M. F., Alrajeh, H. S., Sallam, N. M. A., Imran, M. (2024). Bacillus amyloliquefaciens IKMM and zinc nanoparticles as biocontrol candidate induce the systemic resistance by producing antioxidants in tomato plants challenged with early blight pathogen. J. Crop Health 76, 87–103. doi: 10.1007/s10343-023-00942-0
Aulia, A., Hyodo, K., Hisano, S., Kondo, H., Hillman, B. I., Suzuki, N. (2021). Identification of an RNA silencing suppressor encoded by a symptomless fungal hypovirus, cryphonectria hypovirus 4. Biology 10 (2), 100. doi: 10.3390/biology10020100
Bachman, P. M., Bolognesi, R., Moar, W. J., Mueller, G. M., Paradise, M. S., Ramaseshadri, P., et al. (2013). Characterization of the spectrum of insecticidal activity of a double-stranded RNA with targeted activity against Western Corn Rootworm (Diabrotica virgifera virgifera LeConte). Transgenic Res. 22, 1207–1222. doi: 10.1007/s11248-013-9744-1
Baldwin, T., Islamovic, E., Klos, K., Schwartz, P., Gillespie, J., Hunter, S., et al. (2018). Silencing efficiency of dsRNA fragments targeting Fusarium graminearum TRI6 and patterns of small interfering RNA associated with reduced virulence and mycotoxin production. PloS One 13, e0202798. doi: 10.1371/journal.pone.0202798
Baulcombe, D. C. (2015). VIGS, HIGS and FIGS: small RNA silencing in the interactions of viruses or filamentous organisms with their plant hosts. Curr. Opin. Plant Biol. 26, 141–146. doi: 10.1016/j.pbi.2015.06.007
Baum, J. A., Bogaert, T., Clinton, W., Heck, G. R., Feldmann, P., Ilagan, O., et al. (2007). Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25, 1322–1326. doi: 10.1038/nbt1359
Baum, J. A., Roberts, J. K. (2014). “Advances in insect physiology,” in Progress Towards RNAi-Mediated Insect Pest Management. Eds. Dhadialla, T. S., Gill, S. S. (Elsevier, London, UK), 249–295. doi: 10.1016/B978-0-12-800197-4.00005-1
Beernink, B. M., Amanat, N., Li, V. H., Manchur, C. L., Whyard, S., Belmonte, M. F. (2024). SIGS vs. HIGS: opportunities and challenges of RNAi pest and pathogen control strategies. Can. J. Plant Pathol. 46, 1–15. doi: 10.1080/07060661.2024.2392610
Bennett, M., Deikman, J., Hendrix, B., Iandolino, A. (2020). Barriers to Efficient Foliar Uptake of dsRNA and Molecular Barriers to dsRNA Activity in Plant Cells. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00816
Bento, F. M., Marques, R. N., Campana, F. B., Demétrio, C. G., Leandro, R. A., Parra, J. R. P., et al. (2020). Gene silencing by RNAi via oral delivery of DsRNA by bacteria in the South American tomato pinworm, tuta absoluta. Pest Manage. Sci. 76, 287–295. doi: 10.1002/ps.5513
Bormann, J., Heinze, C., Blum, C., Mentges, M., Brockmann, A., Alder, A., et al. (2018). Expression of a structural protein of the mycovirus FgV-ch9 negatively affects the transcript level of a novel symptom alleviation factor and causes virus infection-like symptoms in Fusarium graminearum. J. Virol. 92, e00326–e00318. doi: 10.1128/JVI.00326-18
Buivydaitė, Ž., Winding, A., Jørgensen, L. N., Zervas, A., Sapkota, R. (2024). New insights into RNA mycoviruses of fungal pathogens causing Fusarium head blight. Virus Res. 349, 199462. doi: 10.1016/j.virusres.2024.199462
Cagliari, D., Dias, N. P., Galdeano, D. M., Dos Santos, E. Á., Smagghe, G., Zotti, M. J. (2019). Management of pest insects and plant diseases by non-transformative RNAi. Front. Plant Sci. 10, 1319. doi: 10.3389/fpls.2019.01319
Cai, Q., He, B., Kogel, K. H., Jin, H. (2018a). Cross-kingdom RNA trafficking and environmental RNAi-nature’s blueprint for modern crop protection strategies. Curr. Opin. Microbiol. 46, 58–64. doi: 10.1016/j.mib.2018.02.003
Cai, Q., Qiao, L., Wang, M., He, B., Lin, F. M., Palmquist, J., et al. (2018b). Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Sci. (New York N.Y.) 360, 1126–1129. doi: 10.1126/science.aar4142
Camargo, R. A., Barbosa, G. O., Possignolo, I. P., Peres, L. E., Lam, E., Lima, J. E., et al. (2016). RNA interference as a gene silencing tool to control Tuta absoluta in tomato (Solanum lycopersicum). PeerJ 4, e2673. doi: 10.7717/peerj.2673
Chen, H., King, F. J., Zhou, B., Wang, Y., Canedy, C. J., Hayashi, J., et al. (2024). Drug target prediction through deep learning functional representation of gene signatures. Nat. Commun. 15, 853. doi: 10.1038/s41467-024-46089-y
Chen, J., Peng, Y., Zhang, H., Wang, K., Zhao, C., Zhu, G., et al. (2021). Off-target effects of RNAi correlate with the mismatch rate between dsRNA and non-target mRNA. RNA Biol. 18, 1747–1759. doi: 10.1080/15476286.2020.1868680
Chen, X., Li, L., Hu, Q., Zhang, B., Wu, W., Jin, F., et al. (2015). Expression of dsRNA in recombinant Isaria fumosorosea strain targets the TLR7 gene in Bemisia tabaci. BMC Biotechnol. 15, 64. doi: 10.1186/s12896-015-0170-8
Choi, M. Y., Vander Meer, R. K. (2019). Phenotypic effects of PBAN RNAi using oral delivery of DsRNA to corn earworm (Lepidoptera: Noctuidae) and tobacco budworm larvae. J. Econ. Entomol. 112, 434–439. doi: 10.1093/jee/toy356
Christiaens, O., Whyard, S., Vélez, A. M., Smagghe, G. (2020). Double-stranded RNA technology to control insect pests: current status and challenges. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00451
Dalakouras, A., Jarausch, W., Buchholz, G., Bassler, A., Braun, M., Manthey, T., et al. (2018). Delivery of hairpin RNAs and small RNAs into woody and herbaceous plants by trunk injection and petioleaAbsorption. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01253
da Rosa, J., Viana, A. J. C., Ferreira, F. R. A., Koltun, A., Mertz-Henning, L. M., Marin, S. R. R., et al. (2024). Optimizing dsRNA engineering strategies and production in E. coli HT115 (DE3). J. Ind. Microbiol. Biotechnol. 51, kuae028. doi: 10.1093/jimb/kuae028
Das, P. R., Sherif, S. M. (2020). Application of exogenous dsRNAs-induced RNAi in agriculture: challenges and triumphs. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00946
Degnan, R. M., Shuey, L. S., Radford-Smith, J., Gardiner, D. M., Carroll, B. J., Mitter, N., et al. (2023). Double-stranded RNA prevents and cures infection by rust fungi. Commun. Biol. 6, 1234. doi: 10.1038/s42003-023-05618-z
Delgado-Baquerizo, M., Guerra, C. A., Cano-Dıaz, C., Egidi, E., Wang, J. T., Eisenhauer, N., et al. (2020). The proportion of soil-borne pathogens increases with warming at the global scale. Nat. Clim. Change 10, 550–554. doi: 10.1038/s41558-020-0759-3
Delgado-Martín, J., Ruiz, L., Janssen, D., Velasco, L. (2022). Exogenous application of dsRNA for the control of viruses in cucurbits. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.895953
Dhandapani, R. K., Gurusamy, D., Palli, S. R. (2022). Protamine-lipid-dsRNA nanoparticles improve RNAi efficiency in the fall armyworm, Spodoptera frugiperda. J. Agric. Food Chem. 70, 6634–6643. doi: 10.1021/acs.jafc.2c00901
Dubrovina, A. S., Kiselev, K. V. (2019). Exogenous RNAs for gene regulation and plant resistance. Int. J. Mol. Sci. 20, 2282. doi: 10.3390/ijms20092282
Espino, J., González, M., González, C., Brito, N. (2014). Efficiency of different strategies for gene silencing in Botrytis cinerea. Appl. Microbiol. Biotechnol. 98, 9413–9424. doi: 10.1007/s00253-014-6087-7
Fan, S., Zhou, Y., Zhu, N., Meng, Q., Zhao, Y., Xu, J., et al. (2024). Exogenous Application of dsRNA-Inducing Silencing of the Fusarium oxysporum Tup1 Gene and Reducing Its Virulence. Int. J. Mol. Sci. 25, 10286. doi: 10.3390/ijms251910286
Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., Mello, C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. doi: 10.1038/35888
Fisher, M. C., Hawkins, N. J., Sanglard, D., Gurr, S. J. (2018). Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 360, 739–742. doi: 10.1126/science.aap7999
Gan, D., Zhang, J., Jiang, H., Jiang, T., Zhu, S., Cheng, B. (2010). Bacterially expressed dsRNA protects maize against SCMV infection. Plant Cell Rep. 29, 1261–1268. doi: 10.1007/s00299-010-0911-z
Ganbaatar, O., Cao, B., Zhang, Y., Bao, D., Bao, W., Wuriyanghan, H. (2017). Knockdown of Mythimna separata chitinase genes via bacterial expression and oral delivery of RNAi effectors. BMC Biotechnol. 17, 1–11. doi: 10.1186/s12896-017-0328-7
Gerhards, R., Bezhin, K., Santel, H. J. (2017). Sugar beet yield loss predicted by relative weed cover, weed biomass and weed density. Plant Protect. Sci. 53, 118–125. doi: 10.17221/57/2016-PPS
Ghosh, S., Patra, S., Ray, S. (2023). A combinatorial nanobased spray-induced gene silencing technique for crop protection and improvement. ACS omega 8, 22345–22351. doi: 10.1021/acsomega.3c01968
Giorgio, A., De Stradis, A., Lo Cantore, P., Iacobellis, N. S. (2015). Biocide effects of volatile organic compounds produced by potential biocontrol rhizobacteria on Sclerotinia sclerotiorum. Front. Microbiol. 6 1056. doi: 10.3389/fmicb.2015.01056
Gu, K. X., Song, X. S., Xiao, X. M., Duan, X. X., Wang, J. X., Duan, Y. B., et al. (2019). A β 2-tubulin dsRNA derived from Fusarium asiaticum confers plant resistance to multiple phytopathogens and reduces fungicide resistance. Pestic. Biochem. Physiol. 153, 36–46. doi: 10.1016/j.pestbp.2018.10.005
Guan, R., Chu, D., Han, X., Miao, X., Li, H. (2021). Advances in the development of microbial double-stranded RNA production systems for application of RNA interference in agricultural pest control. Front. Bioeng. Biotechnol. 9. doi: 10.3389/fbioe.2021.753790
Guo, Q., Liu, Q., Smith, N. A., Liang, G. L., Wang, M. B. (2016). RNA silencing in plants: mechanisms, technologies and applications in horticultural crops. Curr. Genom. 17, 476–489. doi: 10.2174/1389202917666160520103117
Guo, Y., Fan, Y., Teng, Z., Wang, L., Tan, X., Wan, F., et al. (2022). Efficacy of RNA interference using nanocarrier-based transdermal dsRNA delivery system in the woolly apple aphid, Eriosoma lanigerum. Arch. Insect Biochem. Physiol. 110, e21888. doi: 10.1002/arch.21888
Gyawali, B., Rahimi, R., Alizadeh, H., Mohammadi, M. (2024). Graphene quantum dots (GQD)-mediated dsRNA delivery for the control of fusarium head blight disease in wheat. ACS Appl. Bio Mater 7, 1526–1535. doi: 10.1021/acsabm.3c00972
Hameed, A., Tahir, M., Asad, S., Bilal, R., Eck, J., Jander, G., et al. (2017). RNAi-mediated simultaneous resistance against three RNA viruses in potato. Mol. Biotechnol. 59, 73–83. doi: 10.1007/s12033-017-9995-9
Hamim, I., Komatsu, K., Prager, S. M., Wu, B. (2023). Viruses in agricultural systems: Interactions with plants, insect pollinators and fungi. Front. Microbiol. 14. doi: 10.3389/fmicb.2023.1170402
Hammond, T. M., Andrewski, M. D., Roossinck, M. J., Keller, N. P. (2008). Aspergillus mycoviruses are targets and suppressors of RNA silencing. Eukaryotic Cell 7 (2), 350–357. doi: 10.1128/EC.00356-07
Han, J., Rotenberg, D. (2024). Microinjection-enabled gene silencing in first instar larvae of western flower thrips, Frankliniella occidentalis, reveals vital genes for larval survival. Ins Sci. 0, 1–13. doi: 10.1111/1744-7917.13478
Hoang, B. T. L., Fletcher, S. J., Brosnan, C. A., Ghodke, A. B., Manzie, N., Mitter, N. (2022). RNAi as a foliar spray: efficiency and challenges to field applications. Int. J. Mol. Sci. 23, 6639. doi: 10.3390/ijms23126639
Hofmeijer, M. A., Melander, B., Salonen, J., Lundkvist, A., Zarina, L., Gerowitt, B. (2021). Crop diversification affects weed communities and densities in organic spring cereal fields in northern Europe. Agric. Ecosyst. Environ. 308, 107251. doi: 10.1016/j.agee.2020.107251
Horvath, D. P., Clay, S. A., Swanton, C. J., Anderson, J. V., Chao, W. S. (2023). Weed-induced crop yield loss: a new paradigm and new challenges. Trends Plant Sci. 28, 567–582. doi: 10.1016/j.tplants.2022.12.014
Hough, J., Howard, J. D., Brown, S., Portwood, D. E., Kilby, P. M., Dickman, M. J. (2022). Strategies for the production of dsRNA biocontrols as alternatives to chemical pesticides. Front. Bioeng. Biotechnol. 10. doi: 10.3389/fbioe.2022.980592
Huang, X. N., Wang, Y., Li, Y. T., Xiao, Y., Ouyang, S. Q. (2025). Trans-kingdom sRNA silencing in the prevention and control of crop fusarium wilt disease. Phytopathol. Res. 7, 18. doi: 10.1186/s42483-024-00298-x
Imfeld, G., Vuilleumier, S. (2012). Measuring the effects of pesticides on bacterial communities in soil: a critical review. Eur. J. Soil Biol. 49, 22–30. doi: 10.1016/j.ejsobi.2011.11.010
Imran, M., Abo-Elyousr, K. A., Mousa, M. A., Saad, M. M. (2022). Screening and biocontrol evaluation of indigenous native Trichoderma spp. against early blight disease and their field assessment to alleviate natural infection. Egyptian J. Biol. Pest Control 32, p.40. doi: 10.1186/s41938-022-00544-4
Imran, M., Ali, E. F., Hassan, S., Abo-Elyousr, K. A., Sallam, N. M., Khan, M. M. M., et al. (2021). Characterization and sensitivity of Botrytis cinerea to benzimidazole and succinate dehydrogenase inhibitors fungicides, and illustration of the resistance profile. Australas. Plant Pathol. 50, 589–601. doi: 10.1007/s13313-021-00803-2
Imran, M., Sun, Z., Abo-Elyousr, K. A., Ali, H., Aldayel, M. F., Li, C. (2024). One stone two birds: Endophytes alleviating trace elements accumulation and suppressing soilborne pathogen by stimulating plant growth, photosynthetic potential and defense related gene expression. J. Hazard. Mater. 476, 135084. doi: 10.1016/j.jhazmat.2024.135084
Jin, H., Abouzaid, M., Lin, Y., Hull, J. J., Ma, W. (2021). Cloning and RNAi-mediated three lethal genes that can be potentially used for Chilo suppressalis (Lepidoptera: Crambidae) management. Pestic. Biochem. Physiol. 174, 104828. doi: 10.1016/j.pestbp.2021.104828
Kim, S., Lee, R., Jeon, H., Lee, N., Park, J., Moon, H., et al. (2023). Identification of essential genes for the establishment of sprayinduced gene silencing-based disease control in Fusarium graminearum. J. Agric. Food Chem. 71, 19302–19311. doi: 10.1021/acs.jafc.3c04557
Kjeldgaard, B., Neves, A. R., Fonseca, C., Kovács, Á.T., Domínguez-Cuevas, P. (2022). Quantitative high-throughput screening methods designed for identification of bacterial biocontrol strains with antifungal properties. Microbiol. Spectr. 10, e0143321. doi: 10.1128/spectrum.01433-21
Ko, Y. H., So, K. K., Chun, J., Kim, D. H. (2021). Distinct roles of two DNA methyltransferases from cryphonectria parasitica in fungal virulence, responses to hypovirus infection, and viral clearance. MBio 12 (1), e02890–e02820. doi: 10.1128/mBio.02890-20
Koch, A., Biedenkopf, D., Furch, A., Weber, L., Rossbach, O., Abdellatef, E., et al. (2016). An RNAi-Based Control of Fusarium graminearum Infections Through Spraying of Long dsRNAs Involves a Plant Passage and Is Controlled by the Fungal Silencing Machinery. PLoS Pathog. 12, e1005901. doi: 10.1371/journal.ppat.1005901
Koch, A., Höfle, L., Werner, B. T., Imani, J., Schmidt, A., Jelonek, L., et al. (2019). SIGS vs HIGS: a study on the efficacy of two dsRNA delivery strategies to silence Fusarium FgCYP51 genes in infected host and non-host plants. Mol. Plant Pathol. 20, 1636–1644. doi: 10.1111/mpp.12866
Koch, A., Wassenegger, M. (2021). Host-induced gene silencing - mechanisms and applications. New Phytol. 231, 54–59. doi: 10.1111/nph.17364
Konakalla, N. C., Kaldis, A., Berbati, M., Masarapu, H., Voloudakis, A. E. (2016). Exogenous application of double-stranded RNA molecules from TMV p126 and CP genes confers resistance against TMV in tobacco. Planta 244, 961–969. doi: 10.1007/s00425-016-2567-6
Kottaipalayam-Somasundaram, S. R., Jacob, J. P., Aiyar, B., Merzendorfer, H., Nambiar-Veetil, M. (2022). Chitin metabolism as a potential target for RNAi-based control of the forestry pest Hyblaea Puera Cramer (Lepidoptera: Hyblaeidae). Pest Manage. Sci. 78, 296–303. doi: 10.1002/ps.6634
Kumar, P., Pandit, S. S., Baldwin, I. T. (2012). Tobacco rattle virus vector: A rapid and transient means of silencing Manduca sexta genes by plant mediated RNA interference. PLoS One 7, e31347. doi: 10.1371/journal.pone.0031347
Li, N., Alfiky, A., Wang, W., Islam, M., Nourollahi, K., Liu, X., et al. (2018). Volatile compound-mediated recognition and inhibition between Trichoderma biocontrol agents and Fusarium oxysporum. Front. Microbiol. 9. doi: 10.3389/fmicb.2018.02614
Lucena-Leandro, V. S., Abreu, E. F. A., Vidal, L. A., Torres, C. R., Junqueira, C. I. C. V. F., Dantas, J., et al. (2022). Current scenario of exogenously induced RNAi for lepidopteran agricultural pest control: from dsRNA design to topical application. Int. J. Mol. Sci. 23, 15836. doi: 10.3390/ijms232415836
Lundgren, J. G., Duan, J. J. (2013). RNAi-based insecticidal crops: potential effects on nontarget species. Bioscience 63, 657–665. doi: 10.1525/bio.2013.63.8.8
Mai, J., Liao, L., Ling, R., Guo, X., Lin, J., Mo, B., et al. (2021). Study on RNAi-based herbicide for Mikania micrantha. Synth Syst. Biotechnol. 6, 437–445. doi: 10.1016/j.synbio.2021.11.005
Mao, Y. B., Cai, W. J., Wang, J. W., Hong, G. J., Tao, X. Y., Wang, L. J., et al. (2007). Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 25, 1307–1313. doi: 10.1038/nbt1352
Marjanac, G., Karimi, M., Naudts, M., Beeckman, T., Depicker, A., De Buck, S. (2009). Gene silencing induced by hairpin or inverted repeated sense transgenes varies among promoters and cell types. New Phytol. 184, 851–864. doi: 10.1111/j.1469-8137.2009.03011.x
Mittal, D., Kaur, G., Singh, P., Yadav, K., Ali, S. A. (2020). Nanoparticle-based sustainable agriculture and food science: Recent advances and future outlook. Front. Nanotechnol. 2. doi: 10.3389/fnano.2020.579954
Mitter, N., Worrall, E. A., Robinson, K. E., Li, P., Jain, R. G., Taochy, C., et al. (2017a). Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants. 3, 16207. doi: 10.1038/nplants.2016.207
Mitter, N., Worrall, E. A., Robinson, K. E., Xu, Z. P., Carroll, B. J. (2017b). Induction of virus resistance by exogenous application of double-stranded RNA. Curr. Opin. Virol. 26, 49–55. doi: 10.1016/j.coviro.2017.07.009
Mota-Sanchez, D., Wise, J. C. (2024). The Arthropod Pesticide Resistance Database (East Lansing, MI, USA: Michigan State University). Available at: http://www.pesticideresistance.org (Accessed November 8, 2024).
Mukherjee, S., Beligala, G., Feng, C., Marzano, S. Y. (2024). Double-stranded RNA targeting white mold Sclerotinia sclerotiorum argonaute 2 for disease control via spray-induced gene silencing. Phytopathol 114, 1253–1262. doi: 10.1094/PHYTO-11-23-0431-R
Myers, J. M., James, T. Y. (2022). Mycoviruses. Curr. Biol. 32 (4), R150–R155. doi: 10.1016/j.cub.2022.01.049
Namba, S., Iwata, M., Yamanishi, Y. (2022). From drug repositioning to target repositioning: prediction of therapeutic targets using genetically perturbed transcriptomic signatures. Bioinformatics 38, i68i76. doi: 10.1093/bioinformatics/btac240
Namgial, T., Kaldis, A., Chakraborty, S., Voloudakis, A. (2019). Topical application of double-stranded RNA molecules containing sequences of Tomato leaf curl virus and Cucumber mosaic virus confers protection against the cognate viruses. Physiol. Mol. Plant Pathol. 108, 101432. doi: 10.1016/j.pmpp.2019.101432
Nerva, L., Sandrini, M., Gambino, G., Chitarra, W. (2020). Double-stranded RNAs (dsRNAs) as a sustainable tool against gray mold (Botrytis cinerea) in grapevine: Effectiveness of different application methods in an open-air environment. Biomolecules 10, 200. doi: 10.3390/biom10020200
Niehl, A., Soininen, M., Poranen, M. M., Heinlein, M. (2018). Synthetic biology approach for plant protection using dsRNA. Plant Biotechnol. J. 16, 1679–1687. doi: 10.1111/pbi.12904
Niehl, A., Wyrsch, I., Boller, T., Heinlein, M. (2016). Double-stranded RNAs induce a pattern-triggered immune signaling pathway in plants. New Phytol. 211, 1008–1019. doi: 10.1111/nph.13944
Nitnavare, R. B., Bhattacharya, J., Singh, S., Kour, A., Hawkesford, M. J., Arora, N. (2021). Next generation dsRNA-based insect control: Success so far and challenges. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.673576
Nityagovsky, N. N., Kiselev, K. V., Suprun, A. R., Dubrovina, A. S. (2022). Exogenous dsRNA induces RNA interference of a chalcone synthase gene in Arabidopsis thaliana. Int. J. Mol. Sci. 23, p.5325. doi: 10.3390/ijms23105325
Ouyang, S. Q., Ji, H. M., Feng, T., Luo, S. J., Cheng, L., Wang, N. (2023). Artificial trans-kingdom RNAi of FolRDR1 is a potential strategy to control tomato wilt disease. PloS Pathog. 19, e1011463. doi: 10.1371/journal.ppat.1011463
Pal, G., Ingole, K. D., Yavvari, P. S., Verma, P., Kumari, A., Chauhan, C., et al. (2024). Exogenous application of nanocarrier-mediated double-stranded RNA manipulates physiological traits and defence response against bacterial diseases. Mol. Plant Pathol. 25, e13417. doi: 10.1111/mpp.13417
Papadopoulou, N., Devos, Y., Álvarez-Alfageme, F., Lanzoni, A., Waigmann, E. (2020). Risk assessment considerations for genetically modified RNAi plants: EFSA’s activities and perspective. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00445
Parise, C., Galetto, L., Abbà, S., Bodino, N., Marzachì, C., Bosco, D. (2024). RNA interference protocols for gene silencing in the spittlebug Philaenus spumarius, vector of Xylella fastidiosa. Sci. Rep. 14, p.25812. doi: 10.1038/s41598-024-73889-5
Patel, R. M., Van Kan, J. A. L., Bailey, A. M., Foster, G. D. (2008). RNA-mediated gene silencing of superoxide dismutase (bcsod1) in Botrytis cinerea. Phytopathology 98, 1334–1339. doi: 10.1094/PHYTO-98-12-1334
Pinheiro, D. H., Taylor, C. E., Wu, K., Siegfried, B. D. (2020). Delivery of gene-specific dsRNA by microinjection and feeding induces RNAi response in Sri Lanka Weevil, Myllocerus Undecimpustulatus Undatus Marshall. Pest Manage. Sci. 76, 936–943. doi: 10.1002/ps.5601
Pitino, M., Coleman, A. D., Maffei, M. E., Ridout, C. J., Hogenhout, S. A. (2011). Silencing of aphid genes by dsRNA feeding from plants. PloS One 6, e25709. doi: 10.1371/journal.pone.0025709
Qiao, L., Lan, C., Capriotti, L., Ah-Fong, A., Nino Sanchez, J., Hamby, R., et al. (2021). Spray-induced gene silencing for disease control is dependent on the efficiency of pathogen RNA uptake. Plant Biotechnol. J. 19, 1756–1768. doi: 10.1111/pbi.13589
Qiao, L., Niño-Sánchez, J., Hamby, R., Capriotti, L., Chen, A., Mezzetti, B., et al. (2023). Artificial nanovesicles for dsRNA delivery in spray-induced gene silencing for crop protection. Plant Biotechnol. J. 21, 854–865. doi: 10.1111/pbi.14001
Ramkumar, G., Asokan, R., Prasannakumar, N. R., Kariyanna, B., Karthi, S., Alwahibi, M. S., et al. (2021). RNA interference suppression of V-ATPase B and juvenile hormone binding protein genes through topically applied dsRNA on tomato leaves: developing biopesticides to control the south American pinworm, Tuta Absoluta (Lepidoptera: Gelechiidae). Front. Physiol. 12. doi: 10.3389/fphys.2021.742871
Rêgo-MaChado, C. M., Inoue-Nagata, A. K., Nakasu, E. Y. T. (2023). Topical application of dsRNA for plant virus control: a review. Trop. Plant Pathol. 48, 11–22. doi: 10.1007/s40858-022-00534-9
Rego-MaChado, C. M., Nakasu, E. Y. T., Silva, J. M. F., Lucinda, N., Nagata, T., Inoue-Nagata, A. K. (2020). siRNA biogenesis and advances in topically applied dsRNA for controlling virus infections in tomato plants. Sci. Rep. 10, 22277. doi: 10.1038/s41598-020-79360-5
Rodriguez, C. L., Plummer, K. M., Khalifa, M. E., MacDiarmid, R. M. (2022). Mycovirus-encoded suppressors of RNA silencing: Possible allies or enemies in the use of RNAi to control fungal disease in crops. Front. Fungal Biol. 3. doi: 10.3389/ffunb.2022.965781
Ross, S. J., Owen, G. R., Hough, J., Philips, A., Maddelein, W., Ray, J., et al. (2024). Optimizing the production of dsRNA biocontrols in microbial systems using multiple transcriptional terminators. Biotechnol. bioeng 121, 3582–3599. doi: 10.1002/bit.28805
Sammons, R., Ivashuta, S., Liu, H., Wang, D., Feng, P., Kouranov, A. Y., et al. (2011). Polynucleotide molecule for gene regulation in plants (St. Louis: Monsanto Technology).
Sang, H., Kim, J. I. (2020). Advanced strategies to control plant pathogenic fungi by host-induced gene silencing (HIGS) and spray-induced gene silencing (SIGS). Plant Biotechnol. Rep. 14, 1–8. doi: 10.1007/s11816-019-00588-3
San Miguel, K., Scott, J. G. (2016). The next generation of insecticides: dsRNA is stable as a foliar-applied insecticide. Pest Manage. Sci. 72, 801–809. doi: 10.1002/ps.4056
Sarkar, A., Roy-Barman, S. (2021). Spray-induced silencing of pathogenicity gene MoDES1 via exogenous double-stranded RNA can confer partial resistance against fungal blast in rice. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.733129
Schebesta, H., Bernaz, N., Macchi, C. (2020). The European union farm to fork strategy. Eur. Food Feed Law Rev. 15, 420–427.
Sharath Chandra, G., Asokan, R., Manamohan, M., Krishna Kumar, N. (2019). Enhancing RNAi by using concatemerized doubleStranded RNA. Pest Manage. Sci. 75, 506–514. doi: 10.1002/ps.5149
Sigoillot, F. D., Lyman, S., Huckins, J. F., Adamson, B., Chung, E., Quattrochi, B., et al. (2012). A bioinformatics method identifies prominent off-targeted transcripts in RNAi screens. Nat. Methods 9, 363–366. doi: 10.1038/nmeth.1898
Singh, K., Dardick, C., Kumar, K. J. (2019). RNAi-mediated resistance against viruses in perennial fruit plants. Plants (Basel Switzerland) 8, 359. doi: 10.3390/plants8100359
Singh, N., Mukherjee, S. K., Rajam, M. V. (2020). Silencing of the ornithine decarboxylase gene of Fusarium oxysporum f. sp. lycopersici by host-induced RNAi confers resistance to Fusarium wilt in tomato. Plant Mol. Biol. Rep. 38, 419–429. doi: 10.1007/s11105-020-01205-2
Song, X. S., Gu, K. X., Duan, X. X., Xiao, X. M., Hou, Y. P., Duan, Y. B., et al. (2018). A myosin5 dsRNA that reduces the fungicide resistance and pathogenicity of Fusarium asiaticum. Pestic. Biochem. Physiol. 150, 1–9. doi: 10.1016/j.pestbp.2018.03.020
Stojanovic, N. (2021). European green deal and” Farm-to-fork” Strategy for a fair, healthy and environmentally-friendly food system. Collection Papers Conf. Org. Occasion Day Fac. L, 130. doi: 10.1126/science.341.6147.730
Sun, Z. N., Song, Y. Z., Yin, G. H., Zhu, C. X., Wen, F. J. (2010). HpRNAs derived from different regions of the NIb gene have different abilities to protect tobacco from infection with potato virus y. J. Phytopathol. 158, 566–568. doi: 10.1111/j.1439-0434.2009.01650.x
Tabein, S., Jansen, M., Noris, E., Vaira, A. M., Marian, D., Behjatnia, S. A. A., et al. (2020). The induction of an effective dsRNA-mediated resistance against tomato spotted wilt virus by exogenous application of double-stranded RNA largely depends on the selection of the viral RNA target region. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.533338
Taning, C. N., Arpaia, S., Christiaens, O., Dietz-Pfeilstetter, A., Jones, H., Mezzetti, B., et al. (2020). RNA-based biocontrol compounds: current status and perspectives to reach the market. Pest Manage. Sci. 76, 841–845. doi: 10.1002/ps.5686
Tatineni, S., Hein, G. L. (2023). Plant viruses of agricultural importance: Current and future perspectives of virus disease management strategies. Phytopathol. 113, pp.117–pp.141. doi: 10.1094/PHYTO-05-22-0167-RVW
Tenllado, F., Díaz-Ruíz, J. R. (2001). Double-stranded RNA-mediated interference with plant virus infection. J. Virol. 75, 12288–12297. doi: 10.1128/JVI.75.24.12288-12297.2001
Tenllado, F., Martínez-García, B., Vargas, M., Díaz-Ruíz, J. R. (2003). Crude extracts of bacterially expressed dsRNA can be used to protect plants against virus infections. BMC Biotechnol. 3, 3. doi: 10.1186/1472-6750-3-3
Tian, H., Peng, H., Yao, Q., Chen, H., Xie, Q., Tang, B., et al. (2009). Developmental control of a lepidopteran pest Spodoptera exigua by ingestion of bacteria expressing DsRNA of a non-midgut gene. PLoS One 4, e6225. doi: 10.1371/journal.pone.0006225
Torres-Trenas, A., Prieto, P., Cañizares, M. C., García-Pedrajas, M. D., Pérez-Artés, E. (2019). Mycovirus Fusarium oxysporum f. sp. dianthi Virus 1 Decreases the Colonizing Efficiency of Its Fungal Host. Front. Cell Infect. Microbiol. 9. doi: 10.3389/fcimb.2019.00051
Trognitz, F., Hackl, E., Widhalm, S., Sessitsch, A. (2016). The role of plant-microbiome interactions in weed establishment and control. FEMS Microbiol. Ecol. 92, fiw138. doi: 10.1093/femsec/fiw138
Vatanparast, M., Kim, Y. (2017). Optimization of recombinant bacteria expressing dsRNA to enhance insecticidal activity against a lepidopteran insect, Spodoptera exigua. PloS One 12, 183054. doi: 10.1371/journal.pone.0183054
Vetukuri, R. R., Dubey, M., Kalyandurg, P. B., Carlsson, A. S., Whisson, S. C., Ortiz, R. (2021). Spray-induced gene silencing: an innovative strategy for plant trait improvement and disease control. Crop Breed. Appl. Biotechnol. 21, 387921S11. doi: 10.1590/1984-70332021v21sa24
Vieira Gomes, A. M., Souza Carmo, T., Silva Carvalho, L., Mendonça Bahia, F., Parachin, N. S. (2018). Comparison of yeasts as hosts for recombinant protein production. Microorganisms 6, 38. doi: 10.3390/microorganisms6020038
Wagemans, J., Holtappels, D., Vainio, E., Rabiey, M., Marzachì, C., Herrero, S., et al. (2022). Going viral: virus-based biological control agents for plant protection. Annu. Rev. Phytopathol. 60, 21–42. doi: 10.1146/annurev-phyto-021621-114208
Wang, X., Aboughanem-Sabanadzovic, N., Sabanadzovic, S., Tomaso-Peterson, M., Wilkerson, T. H., Allen, T. W. (2023a). Defining fungicide resistance mechanisms in the Corynespora cassiicola population from Mississippi soybean. Plant Dis. 107, 2365–2374. doi: 10.1094/PDIS-06-22-1297-RE
Wang, Y., Guo, Y., Guo, S., Qi, L., Li, B., Jiang, L., et al. (2024b). RNA interference-based exogenous double-stranded RNAs confer resistance to Rhizoctonia solani AG-3 on Nicotiana tabacum. Pest Manag Sci. 80, 2170–2178. doi: 10.1002/ps.7962
Wang, Q., Pan, G., Wang, X., Sun, Z., Guo, H., Su, X., et al. (2024a). Host-induced gene silencing of the Verticillium dahliae thiamine transporter protein gene (VdThit) confers resistance to Verticillium wilt in cotton. J. Integr. Agric. 23, 3358–3369. doi: 10.1016/j.jia.2024.03.024
Wang, M., Thomas, N., Jin, H. (2017). Cross-kingdom RNA trafficking and environmental RNAi for powerful innovative pre- and post-harvest plant protection. Curr. Opin. Plant Biol. 38, 133–141. doi: 10.1016/j.pbi.2017.05.003
Wang, M., Weiberg, A., Lin, F. M., Thomma, B. P., Huang, H. D., Jin, H. (2016). Bidirectional cross-kingdom RNAi and fungal uptake of external RNAs confer plant protection. Nat. Plants. 2, 16151. doi: 10.1038/nplants.2016.151
Wang, Y., Yan, Q., Lan, C., Tang, T., Wang, K., Shen, J., et al. (2023b). Nanoparticle carriers enhance RNA stability and uptake efficiency and prolong the protection against Rhizoctonia solani. Phytopathol. Res. 5, 2. doi: 10.1186/s42483-023-00157-1
Werner, B. T., Gaffar, F. Y., Schuemann, J., Biedenkopf, D., Koch, A. M. (2020). RNA-spray-mediated silencing of Fusarium graminearum AGO and DCL genes improve barley disease resistance. Front. Plant Sci. 11. doi: 10.3389/fpls.2020.00476
Whangbo, J. S., Weisman, A. S., Chae, J., Hunter, C. P. (2017). SID-1 Domains Important for dsRNA Import in Caenorhabditis elegans. G3 (Bethesda Md.) 7, 3887–3899. doi: 10.1534/g3.117.300308
Worrall, E. A., Bravo-Cazar, A., Nilon, A. T., Fletcher, S. J., Robinson, K. E., Carr, J. P., et al. (2019). Exogenous application of RNAi-inducing double-stranded RNA inhibits aphid-mediated transmission of a plant virus. Front. Plant Sci. 10. doi: 10.3389/fpls.2019.00265
Wu, F., Yan, L., Zhao, X., Lv, C., Jin, W. (2024). Development of an RNA Nanostructure for Effective Botrytis cinerea Control through Spray-Induced Gene Silencing without an Extra Nanocarrier. J. Fungi 10, 483. doi: 10.3390/jof10070483
Wuriyanghan, H., Rosa, C., Falk, B. W. (2011). Oral delivery of double-stranded RNAs and siRNAs induces RNAi effects in the potato/tomato psyllid, Bactericerca cockerelli. PloS One 6, e27736. doi: 10.1371/journal.pone.0027736
Wytinck, N., Manchur, C. L., Li, V. H., Whyard, S., Belmonte, M. F. (2020a). dsRNA uptake in plant pests and pathogens: insights into RNAi-based insect and fungal control technology. Plants 9, 1780. doi: 10.3390/plants9121780
Wytinck, N., Sullivan, D. S., Biggar, K. T., Crisostomo, L., Pelka, P., Belmonte, M. F., et al. (2020b). Clathrin mediated endocytosis is involved in the uptake of exogenous double-stranded RNA in the white mold phytopathogen Sclerotinia sclerotiorum. Sci. Rep. 10, 12773. doi: 10.1038/s41598-020-69771-9
Yaegashi, H., Nakamura, H., Sawahata, T., Sasaki, A., Iwanami, Y., Ito, T., et al. (2013). Appearance of mycovirus-like double-stranded RNAs in the white root rot fungus, rosellinia necatrix, in an apple orchard. FEMS Microbiol. Ecol. 83 (1), 49–62. doi: 10.1111/j.1574-6941.2012.01454.x
Yan, S., Qian, J., Cai, C., Ma, Z., Li, J., Yin, M., et al. (2020). Spray method application of transdermal dsRNA delivery system for efficient gene silencing and pest control on soybean aphid Aphis glycines. J. Pest Sci. 93, 449–459. doi: 10.1007/s10340-019-01157-x
Yang, W., Wang, B., Lei, G., Chen, G., Liu, D. (2022). Advances in nanocarriers to improve the stability of dsRNA in the environment. Front. Bioeng. Biotechnol. 10. doi: 10.3389/fbioe.2022.974646
Zabala-Pardo, D., Gaines, T., Lamego, F. P., Avila, L. A. (2022). RNAi as a tool for weed management: challenges and opportunities. Adv. Weed Sci. 40, e020220096. doi: 10.51694/AdvWeedSci/2022;40:seventy-five006
Zha, W., Peng, X., Chen, R., Du, B., Zhu, L., He, G. (2011). Knockdown of midgut genes by dsRNA-transgenic plant-mediated RNA interference in the hemipteran insect Nilaparvata lugens. PloS One 6, e20504. doi: 10.1371/journal.pone.0020504
Zhang, H., Chen, J., Gao, J., Zhang, Q., Liu, X., Han, Z. (2022). New insights into transmission pathways and possible off-target effects of insecticidal dsRNA released by treated plants. Pestic Biochem. Physiol. 188, 105281. doi: 10.1016/j.pestbp.2022.105281
Zhang, X., Segers, G. C., Sun, Q., Deng, F., Nuss, D. L. (2008). Characterization of hypovirus-derived small RNAs generated in the chestnut blight fungus by an inducible DCL-2-dependent pathway. J. Virology. 82, 2613–2619. doi: 10.1128/jvi.02324-07
Zhang, L., Wang, S., Ruan, S., Nzabanita, C., Wang, Y., Guo, L. (2023). A mycovirus VIGS vector confers hypovirulence to a plant pathogenic fungus to control wheat FHB. Adv. Sci. (Weinheim Baden-Wurttemberg Germany) 10, e2302606. doi: 10.1002/advs.202302606
Zhang, T., Zhao, Y. L., Zhao, J. H., Wang, S., Jin, Y., Chen, Z. Q., et al. (2016). Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants 2, 1–6. doi: 10.1038/nplants.2016.153
Zhao, J. H., Liu, Q. Y., Xie, Z. M., Guo, H. S. (2024). Exploring the challenges of RNAi-based strategies for crop protection. Adv. Biotech. 2, 23. doi: 10.1007/s44307-024-00031-x
Zhao, M., Wang, C., Wan, J., Li, Z., Liu, D., Yamamoto, N., et al. (2021). Functional validation of pathogenicity genes in rice sheath blight pathogen Rhizoctonia solani by a novel host-induced gene silencing system. Mol. Plant Pathol. 22, 1587–1598. doi: 10.1111/mpp.13130
Zhou, L., Li, X., Kotta-Loizou, I., Dong, K., Li, S., Ni, D., et al. (2021). A mycovirus modulates the endophytic and pathogenic traits of a plant associated fungus. ISME J. 15, 1893–1906. doi: 10.1038/s41396-021-00892-3
Zhu, X., Qi, T., Yang, Q., He, F., Tan, C., Ma, W., et al. (2017). Host-induced gene silencing of the MAPKK gene PsFUZ7 confers stable resistance to wheat stripe rust. Plant Physiol. 175, 1853–1863. doi: 10.1104/pp.17.01223
Zhu, F., Xu, J., Palli, R., Ferguson, J., Palli, S. R. (2011). Ingested RNA interference for managing the populations of the colorado potato beetle, Leptinotarsa decemlineata. Pest Manage. Sci. 67, 175–182. doi: 10.1002/ps.2048
Zotti, M., Dos Santos, E. A., Cagliari, D., Christiaens, O., Taning, C. N. T., Smagghe, G. (2018). RNA interference technology in crop protection against arthropod pests, pathogens and nematodes. Pest Manage. Sci. 74, 1239–1250. doi: 10.1002/ps.4813
Keywords: biopesticides, SIGS, dsRNA, crop protection, nanotechnology, RNAi, sustainable agriculture
Citation: Chen C, Imran M, Feng X, Shen X and Sun Z (2025) Spray-induced gene silencing for crop protection: recent advances and emerging trends. Front. Plant Sci. 16:1527944. doi: 10.3389/fpls.2025.1527944
Received: 14 November 2024; Accepted: 27 January 2025;
Published: 20 February 2025.
Edited by:
Wen-Ming Wang, Sichuan Agricultural University, ChinaReviewed by:
Ming-Bo Wang, Commonwealth Scientific and Industrial Research Organisation (CSIRO), AustraliaCopyright © 2025 Chen, Imran, Feng, Shen and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xihui Shen, eGlodWlzaGVuQG53c3VhZi5lZHUuY24=; Zhongke Sun, c3VuemhAZGFhZC1hbHVtbmkuZGU=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.