- 1Key Laboratory of Bio-Resources and Eco-Environment of Ministry of Education, College of Life Sciences, Sichuan University, Chengdu, China
- 2College of Resources Environment and Chemistry, Chuxiong Normal University, Chuxiong, China
Background: The Peucedanum s.l. genus, the backbone member of subfamily Apioideae, includes many medically and economically important plants. Although previous studies have proved that the genus was not a natural taxonomic unit and taxonomists also conducted several taxonomic revisions for taxa of this genus, classifications of numerous taxa of the genus still have not been satisfactorily resolved, especially for those endemic to China. Therefore, we conducted a comprehensive taxonomic revision of taxa within the polyphyletic Peucedanum s.l. genus in this study.
Methods: We used two molecular datasets (103 plastomes and 43 nrDNA sequences) generated by genome skimming to reconstructed a reliable phylogenetic framework with high support and resolution. In addition, we also investigated the divergence time of core clade of endemic taxa.
Results and Discussion: Both analyses failed to recover Peucedanum s.l. as a monophyletic group and robustly supported that P. morisonii, the representative of Peucedanum s.s., was distantly related to other Peucedanum s.l. members, which implied that these Peucedanum s.l. taxa were not “truly Peucedanum plants”. Among these Peucedanum s.l. members, plastid-based phylogenies recognized two monophyletic clades, clade A (four species) and clade B (10 taxa). Meanwhile, obvious recognized features for morphology, plastome, and chromosome number for each clade were detected: dorsally compressed and glabrous mericarps with filiform dorsal ribs, winged lateral ribs, numerous vittae in commissure and each furrow, IRa/LSC border falling into rpl23 gene, an overall plastome size of 152,288-154,686 bp, and chromosome numbers of 2n=20 were found in clade A; whereas dorsally compressed and pubescent mericarps with filiform dorsal ribs, winged lateral ribs, numerous vittae in commissure and each furrow, IRa/LSC border falling into the ycf2 gene, an overall plastome size of 146,718-147,592 bp, and chromosome numbers of 2n=22 were discovered in clade B. Therefore, we established two new genera (Shanopeucedanum gen. nov. and Sinopeucedanum gen. nov.) to respectively accommodate the taxa of clades A and B. Furthermore, molecular dating analysis showed that the diversification of clades A and B occurred in the early Pleistocene and late Pliocene, respectively, which may have been driven by the complex geological and climate shifts of these periods. In summary, our study impelled a revision of Peucedanum s.l. members and improved the taxonomic system of the Apiaceae family.
1 Introduction
With the advancements of molecular phylogenetics, taxonomists conducted extensively taxonomic revisions in angiosperms and obtained the relatively stable system for orders and families, (Angiosperm Phylogeny Group, 2003, 2009, 2016; Bremer et al., 1998), as well as for those large genera with economic, medicinal and horticultural value (Miller et al., 2014; Li et al., 2012; Razafimandimbison et al., 2014; Drew et al., 2017; Xu and Hong, 2021; Fu et al., 2022; Scatigna et al., 2022). Therefore, molecular phylogenetics provides strong evidence for taxa that are difficult to morphologically identify.
Peucedanum s.l., the backbone member of the subfamily Apioideae, includes 100-120 species. Of these, forty species are distributed in China with 33 of them endemic (Sheh and Watson, 2005). The genus is widely distributed across Europe, Asia, and South Africa, with Europe and East Asia as the distribution centers (Wu et al., 2003; Spalik et al., 2004; Sheh and Watson, 2005; Zhang, 2011). Peucedanum s.l. can be identified by dorsally compressed mericarps with slightly prominent dorsal ribs, narrowly winged lateral ribs, and a broad commissure (Spalik et al., 2004; Sheh and Watson, 2005). However, members of this genus are heterogenous and always exhibit great variability in life-forms, leaf and fruit, and chemical constituents (Shneyer et al., 2003; Zhou et al., 2014; Wang et al., 2015; Ostroumova, 2018). For example, during our field botanical surveys, we found that four species (P. dissolutum, P. guangxiense, P. mashanense and P. medicum) have solitary or numerous stems that are solid, glabrous, and basally usually clothed with fibrous remnant sheaths, branched above; the leaves are basal and cauline, ternate or pinnate, and terminal leaflet diverse; the basal leaves are petiolate with petiole sheathing; the cauline leaves are reduced upwards with the leaf sheath expanded; inflorescences compound umbels, terminal and latera; the bracts are few or absent; the bracteoles are numerous and rarely few; the rays are numerous, unequal, and pubescent; the calyx teeth are conspicuous; and the petals are white and inflexed. Ten species (P. ampliatum, P. formosanum, P. harry-smithii, P. harry-smithii var. grande, P. harry-smithii var. subglabrum, P. huangshanense, P. japonicum, P. praeruptorum, P. turgeniifolium and P. wawrae) possessed stout rootstock, the crown of which was usually clothed with fibrous remnant sheaths; the stems were solitary and branched above; leaves were basal and cauline, ternate or pinnate, and the terminal leaflet was diverse; the basal leaves petiolate with petiole sheathing; the cauline leaves were reduced upwards and the leaf sheath expanded; inflorescences compound umbels, terminal and latera; the bracts were few or absent; the bracteoles were numerous, rarely few; the rays were unequal and pubescent; the calyx teeth were short or obsolete; and the petals were usually white, occasionally purple, and inflexed. Therefore, it noticed that the genus is a taxonomically confused group (Downie et al., 2010).
In addition, all molecular phylogenetic studies on Peucedanum s.l. have failed to recover this genus as a monophyletic group, of which only a few European species, with one species (P. morisonii Besser ex Schult.) extended to East Asia, were consistently clustered with P. officinale L., the type species of this genus (Downie et al., 2000; Spalik et al., 2004; Valiejo-Roman et al., 2006; Feng et al., 2009; Zhou et al., 2009, 2020; Lei et al., 2022; Liu et al., 2022). Therefore, Peucedanum s.s. was adopted (Kadereit and Bittrich, 2018) and several genera were restored or established to accommodate other European taxa, such as Cervaria Wolf, Imperatoria L., Oreoselinum Mill., Pteroselinum Rchb., Thysselinum Adans., Xanthoselinum Schur, Holandrea Reduron, Charpin & Pimenov, and Taeniopetalum Vis (Reduron et al., 1997; Ostroumova et al., 2016). Winter et al. (2008) also revised the African taxa of Peucedanum s.l. by establishing three new genera (Afrosciadium P.J.D. Winter, Nanobubon Magee, and Notobubon B.-E. van Wyk) and transferring 24 species into the Afroligusticum C. Norman, Cynorhiza Eckl. & Zeyh., and Lefebvrea A. Rich. genera based on morphological characteristics and molecular evidence. For Asian taxa previously residing in the Peucedanum s.l., generaKitagawia Pimenov (Pimenov, 1986), Haloselinum Pimenov (Pimenov and Ostroumova, 2012), and Sillaphyton Pimenov (Pimenov et al., 2016) were separated and several transfers have been performed (Pimenov, 2017; Xiao et al., 2017; Gou et al., 2020; Liu et al., 2023; Song et al., 2023). However, classifications of numerous taxa have still not been satisfactorily resolved, especially for those endemic to China, such as P. dissolutum, P. guangxiense, P. mashanense, P. medicum, P. ampliatum, P. formosanum, P. harry-smithii, P. harry-smithii var. grande, P. harry-smithii var. subglabrum, P. huangshanense, P. japonicum, P. praeruptorum, P. turgeniifolium, and P. wawrae. Therefore, it is necessary and urgent to obtain a robust phylogenetic framework to investigate their classifications.
Currently, genomic-level data have begun to replace one or a few fragments for the phylogenetic analyses of plants (Ji et al., 2022). Genome skimming is an effective approach to obtain complete plastome and nrDNA sequences (Straub et al., 2012) and has been extensively and successfully used to solve the phylogeny of plants, especially for the taxonomically difficult and controversial taxa (Ruhsam et al., 2015; Ji et al., 2019, 2022; Li et al., 2019; Liu et al., 2021; Schneider et al., 2021; Choi et al., 2022; Sielemann et al., 2022; Cao et al., 2023; Cui et al., 2023; Lima et al., 2023; Peng et al., 2023; Song et al., 2023; Song et al., 2024a, 2024b). In previously published study (Liu et al., 2022), we also employed the genome skimming approach to generate plastome data to investigate the phylogenetic positions of Peucedanum s.l. members, which significantly improved the taxonomic understanding of this group. However, sampling of this genus was too small to completely aid the taxonomic revision of Peucedanum s.l.
Here, we newly generated nine high-quality plastomes and 43 nrDNA sequences of Peucedanum s.l. using the genome skimming approach. Together with molecular data previously reported by us, 103 plastomes and 43 nrDNA sequences were used in the phylogenetic analyses, including 21 out of the 33 Peucedanum s.l taxa endemic to China. Based on the phylogeny of large data sets, we aim to (1) test whether the previous taxonomic revisions of Peucedanum s.l species were reasonably given; (2) uncover the phylogenetic positions of taxa endemic to China, and combine evidence from plastome features, chromosome number, and morphological features to conduct taxonomic revisions; (3) explore historical diversification for core clades of endemic taxa.
2 Materials and methods
2.1 Taxa sampling, DNA extracting, and genome skimming
In this study, we collected nine Peucedanum s.l. taxa in the wild, including eight taxa endemic to China and one representative of Peucedanum s.s., P. morisonii. Then, the fresh young basal leaves were immediately stored in silica gel and dried for further DNA extraction. Vouchers were deposited in the herbarium of Sichuan University (Chengdu, China) (Supplementary Table S1). In addition, we also obtained root tips from the living plants to count chromosome number, except for P. guangxiense R.H. Shan & M.L. Sheh, P. morisonii, and Semenovia malcolmii (Hemsl. & H. Pearson) Pimenov (P. torilifolium H. Boissieu). Furthermore, the root tips of P. mashanense R.H. Shan & M.L. Sheh and P. medicum Dunn were obtained from our previous collection (Liu et al., 2022). Finally, we collected raw data from genome skimming within the Apiaceae subfamily Apioideae to recover 34 nrDNA sequences (Supplementary Table S2).
The total DNA of the newly collected samples was isolated from the silica-gel-dried leaves using the modified CTAB method (Doyle and Doyle, 1987). Subsequently, pair-end libraries with an average insert size of 300-400 bp were constructed according to the manufacturer’s protocol (Illumina, San Diego, CA, USA). Prepared libraries were sequenced on the Illumina NovaSeq platform at Personalbio (Shanghai, China). About 10 Gb of raw data for each sample were generated by genome skimming and then the raw data were filtered by software fastP v0.15.0 (-n 10 and -q 15) (Chen et al., 2018) to obtain high-quality reads.
2.2 Assembly and annotation of plastome and nrDNA
Based on the high-quality reads, we used GetOrganelle pipeline v1.7.5.0 (Jin et al., 2020) to assemble the complete plastomes with the default parameters and the rbcL sequence of P. japonicum (JF943288) as the seed. Furthermore, GetOrganelle pipeline v1.7.5.0 (Jin et al., 2020) was also employed to recover nrDNA sequences, setting the published nrDNA sequence of P. japonicum (KX757777) as reference. The assembled plastomes were initially annotated using the web server CPGAVAS2 (http://www.herbalgenomics.org/cpgavas2) (Shi et al., 2019). Then, the start and stop codons and intron positions were manually corrected in Geneious v9.0.2 (Kearse et al., 2012). For the annotation of the nrDNA, newly recovered sequences were compared with the reference in Geneious v9.0.2 (Kearse et al., 2012) to determine the borders of the ribosomal RNA genes (18S, 5.8S, and 26S ribosomal RNA genes).
2.3 Phylogenetic analyses
A total of 103 plastomes (40 Peucedanum species are distributed in China with 33 sampled in this study, the coverage of the current samplings accounting for 75%) and 43 nrDNA sequences of the Apiaceae subfamily Apioideae were used to reconstruct the phylogenetic trees (Supplementary Table S3). Of these sequences, nine plastomes were newly sequenced and 94 were downloaded from the National Center for Biotechnology Information (NCBI) that were previously reported by our team, and all 43 nrDNA sequences were newly generated. Among them, Chamaesium mallaeanum Farille & S.B. Malla and Chamaesium viridiflorum (Franch.) H. Wolff ex R.H. Shan were chosen as the outgroup according to the phylogenetic result of Wen et al. (2021). The 43 nrDNA sequences were aligned using MAFFT v7.221 (Katoh and Standley, 2013) to generate the matrix. For the plastome data, 79 commonly shared plastid protein-coding genes (PCGs) were manually extracted in Geneious v9.0.2 (Kearse et al., 2012) and respectively aligned using MAFFT v7.221 (Katoh and Standley, 2013), trimmed by trimAI (Capella-Gutierrez et al., 2009) and then the alignments were concatenated into a super matrix using PhyloSuite v1.2.2 (Zhang et al., 2020).
Two matrixes were subjected to maximum likelihood (ML) and Bayesian inference (BI) analyses. The ML analyses were performed using RAxML v8.2.8 (Stamatakis, 2014) with the GTRGAMMA model, and 1000 bootstrap replicates were used to estimate the support value (BS) for each node. For the BI analyses, phylogenetic trees were generated with MrBayes v3.2.7 (Ronquist et al., 2012). The best-fit substitution model (GTR+I+G) for the matrixes of the concatenated plastid protein-coding genes and nrDNA was determined using Modeltest v3.7 (Posada and Crandall, 1998). Two simultaneous and independent Markov chain Monte Carlo (MCMC) runs with 10 million generations were performed, sampling every 1000 generations. The runs were finished when the average standard deviation of the split frequencies was below 0.01. The first 25% of the trees were abandoned as burn-in and the remainders were used to generate the consensus tree and calculate posterior probabilities (PP). Finally, the phylogenetic trees were edited using FigTree v1.4.2 (Rambaut and Drummond, 2015).
2.4 Molecular dating
According to two pollen fossils within Apiaceae and the strategy adopted by Banasiak et al. (2013) and Wen et al. (2020, 2021), two calibration points constrained to a lognormal distribution were used to constrain the phylogenetic treeL (i) The stem node of Bupleureae was constrained with the lower bound of 33.90 Mya and the upper bound of 58.71 Mya; (ii) whereas the stem node of Pleurospermeae was calibrated with the lower bound of 33.90 Mya and the upper bound of 55.80 Mya. The standard deviation of the lognormal distribution was set to 0.5 for both calibration points. Based on the concatenated data set of 79 plastid protein-coding genes, the divergence times were estimated using BEAST v1.10.4 (Suchard et al., 2018) under the uncorrelated lognormal relaxed clock model, with a Yule speciation process and the best-fit nucleotide substitution model (GTR+I+G) detected using Modeltest v3.7 (Posada and Crandall, 1998). The ML tree inferred from the matrix of concatenated plastid protein-coding genes was used to fix topology. Two MCMC runs of 200,000,000 generations were performed, sampling every 20,000 generations. The convergence of running was checked using Tracer v1.7 (Rambaut et al., 2018), with an effective sample size (ESS) of all parameters not less than 200. The trees generated by the two runs were combined using LogCombiner v1.10.4 (Suchard et al., 2018) after discarding the first 20% of trees as burn-in, and then used to produce the maximum clade credibility tree with median ages and 95% highest posterior density (HPD) intervals with TreeAnotator v2.1.2 (Suchard et al., 2018). The result was exhibited in FigTree v1.4.2 (Rambaut and Drummond, 2015).
2.5 Chromosome counting
The chromosome number varies greatly between genera and species in the Apiaceae family, which is often used as one of the main evidences for exploring the intergeneric relationships, interspecific relationships, and species evolution of Apiaceae plants (Pan and Qin, 1981; Solov’eva et al., 1985; Ci, 1994; Jiang et al., 2002; Marhold, 2006; Pu et al., 2006; Zhang et al., 2006; Marhold et al., 2009; Kumar and Singhal, 2011; Sun et al., 2020; Liu et al., 2023). For example, previous studies reported that the basic chromosome number of the genus Bupleurum L. in China was 4, 6, 7, and 8, indicating that Bupleurum L. is a multi-base genus (Pan and Qin, 1981; Jiang et al., 2002; Liang et al., 2013). Researchers have shown that the chromosome number was different between species in the genus Pleurospermum Hoffm. in the Hengduan Mountains (HDM), with basic chromosome numbers of 9 and 11, which provided vital evidence for interspecific classification within the genus (Pan and Qin, 1981; Marhold, 2006; Pu et al., 2006; Kumar and Singhal, 2011; Sun et al., 2020). Zhang et al. (2006) analyzed the chromosome numbers and karyotypes of three populations of Hansenia forbesii (H. Boissieu) Pimenov & Kljuykov and found significant differences in the karyotype composition of the three populations, indicating that the polymorphism of karyotype composition among the H. forbesii populations was quite obvious, which was likely the result of adaptation to different environments. Ci (1994) found that the karyotype of Saposhnikovia divaricata was 2n=16, which can easily distinguish it from its related genera. Therefore, we collected the root tips of eight Peucedanum s.l. taxa in the current study. All the root tips were pre-treated with a solution of 2mM 8-hydroxyquinoline - 0.2% colchicine (1:1) for 4 h, fixed in Carnoy I (glacial acetic acid–absolute ethanol = 1:3) for 2 h, macerated in 1 N HCl at 60°C for 10 min, and squashed in carbol fuchsin. Finally, we counted the chromosome number under a light microscope (Olympus-BX51).
3 Results
3.1 Features of the plastomes and nrDNA
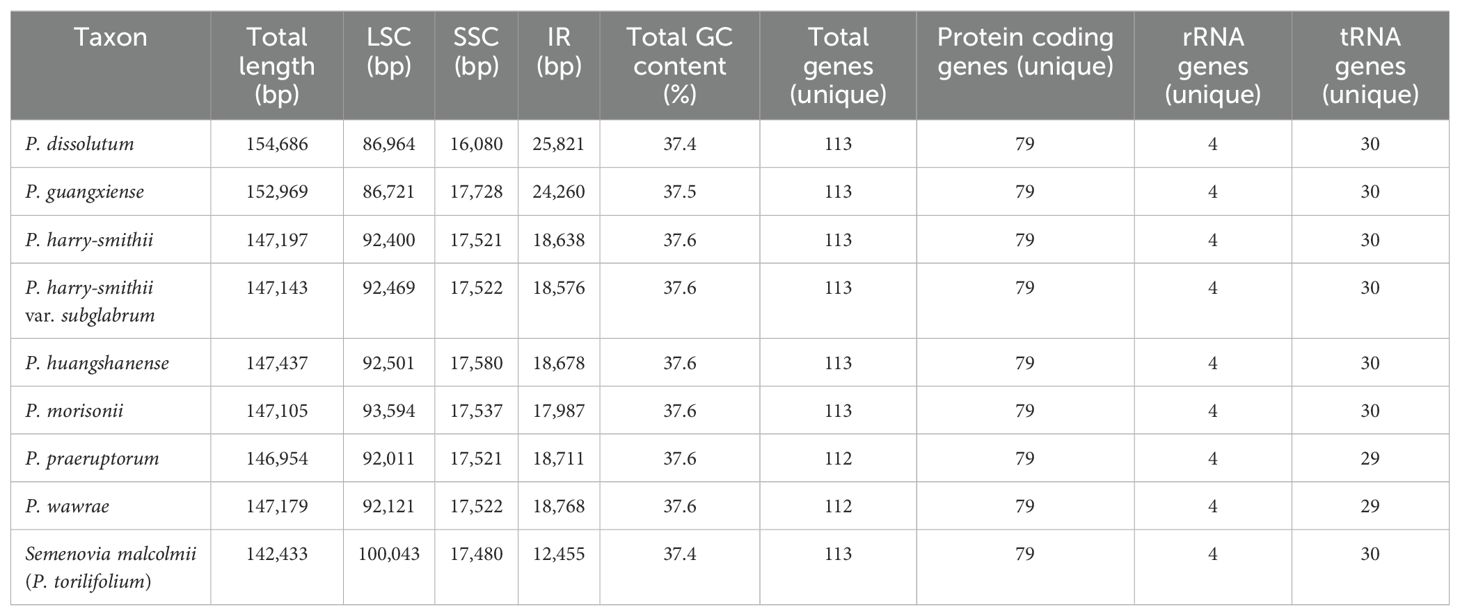
A total of nine high-quality plastomes were newly assembled in the current study. All of them exhibited a typical quadripartite structure, including a pair of inverted repeat regions (IRs: 12,455-25,821 bp) separated by a large single copy region (LSC: 86,721-100,043 bp) and a small single copy region (SSC: 16,080-17,728 bp). The overall sizes of the nine plastomes varied from 142,433 bp to 154,686 bp and the total GC content of which was 37.4%-37.6%. These plastomes encoded 112-113 unique genes, including 79 protein-coding genes, 29-30 tRNA genes, and 4 rRNA genes (Table 1). The trnT-GGU gene was lost in P. praeruptorum Dunn and P. wawrae (H. Wolff) S.W. Su ex M.L. Sheh. In addition, we also recovered 43 nrDNA sequences, including 18S, ITS1, 5.8S, ITS2, and 26S regions, with sequence length varied from 5,195 bp to 5,822 bp.
3.2 Phylogenetic analyses and divergence time estimation
The phylogenetic topologies from the ML and BI analyses based on concatenated plastid protein-coding genes were identical. All the tribes or clades involved in the current study with more than one sample were recovered as a well-supported monophyletic group. However, the monophyly of Peucedanum s.l. failed to recover and members of this genus scattered in four tribes (or clades): (1) Sillaphyton podagraria (H. Boissieu) Pimenov (P. insolens Kitag.) was located in the Arcuatopterus clade (PP = 1.00, BS = 100). (2) Meeboldia delavayi (Franch.) W. Gou & X.J. He (P. delavayi Franch.) was nested in the Meeboldia H. Wolff genus belonging to the Acronema clade (PP = 1.00, BS = 100). (3) Semenovia malcolmii (Hemsl. & H Pearson) Pimenov (P. torilifolium H. Boissieu) was placed in Semenovia Regel & Herder, while Tetrataenium bivittatum (H. Boissieu) Manden. (P. angelicoides H. Wolff ex Kretschmer) was sister to Tetrataenium yunnanense (Franch.) Manden. ex Q.Y. Xiao & X.J. He, belonging to Tordylieae (PP = 1.00, BS = 100). (4) The remainders were included in Selineae (PP = 1.00, BS = 100). Although most of the members of Peucedanum s.l. fell into the Selineae tribe, these taxa have not been grouped in a unit instead of five clades: (i) P. morisonii Besser ex Schult., the representative of Peucedanum s.s., solely represented a clade and was sister to the clade formed by Cortiella hookeri (C.B. Clarke) C. Norman and Ligusticopsis Leute genus (PP = 1.00, BS = 100); (ii) five species, Ligusticopsis acaulis (R.H. Shan & M.L. Sheh) Pimenov (P. acaule R.H. Shan & M.L. Sheh), Ligusticopsis franchetii (C.Y. Wu & F.T. Pu) B.N. Song, C.K. Liu & X.J. He (P. franchetii C.Y. Wu & F.T. Pu), Ligusticopsis nana (R.H. Shan & M.L. Sheh) C.K. Liu & X.J. He (P. nanum R.H. Shan & M.L. Sheh), Ligusticopsis pubescens (Hand.-Mazz.) J.J. Deng, C.K. Liu & X.J. He (P. pubescens Hand.-Mazz.), and Ligusticopsis violacea (R.H. Shan and M.L. Sheh) C.K. Liu & X.J. He (P. violaceum R.H. Shan & M.L. Sheh) were nested in the Ligusticopsis genus with high support (PP = 1.00, BS = 100); (iii) Kitagawia baicalensis (Redowsky ex Willd.) Pimenov (P. baicalense (Redowsky ex Willd.) W.D.J. Koch), Kitagawia komarovii Pimenov (P. elegans Kom.), Kitagawia stepposa (Y.H. Huang) Pimenov (P. stepposum Y.H. Huang), Kitagawia terebinthacea (Fisch. ex Trevir.) Pimenov (P. terebinthaceum (Fisch. ex Trevir.) Turcz.), P. chujaense K. Kim, S.H. Oh, Chan S. Kim & C.W. Park, and P. hakuunense Nakai were clustered into a clade (PP = 1.00, BS = 100); (iv) four species [P. dissolutum (Diels) H. Wolff, P. guangxiense R.H. Shan & M.L. Sheh, P. mashanense R.H. Shan & M.L. Sheh, and P. medicum Dunn] formed clade A (PP = 1.00, BS = 100); (v) the remaining ten taxa (P. ampliatum, P. formosanum, P. harry-smithii, P. harry-smithii var. grande, P. harry-smithii var. subglabrum, P. huangshanense, P. japonicum, P. praeruptorum, P. turgeniifolium, and P. wawrae) constituted clade B with strong support (PP = 1.00, BS = 100) (Figure 1). These five clades were also recovered in the nrDNA tree, whereas P. huangshanense Lu Q.Huang, H.S. Peng & S.S. Chu was not located in clade B but was sister to Pachypleurum alpinum Ledeb. and then nested into Kitagawia Pimenov (Supplementary Figure S1).
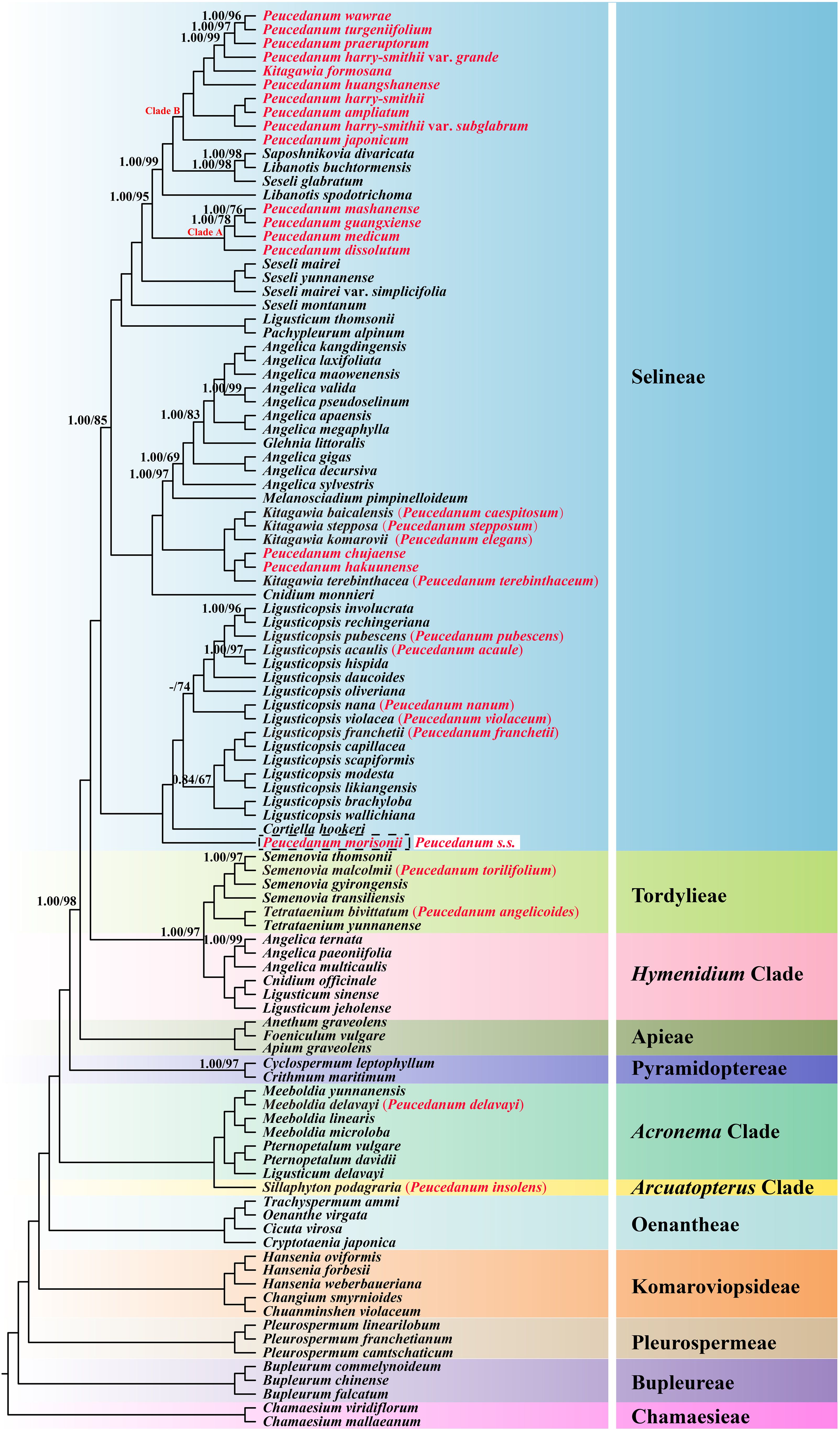
Figure 1. Phylogenetic topologies based on concatenated plastid protein-coding genes inferred by the Bayesian inference (BI) and maximum likelihood (ML) methods. The numbers represent Bayesian posterior probabilities (PP) and maximum likelihood bootstrap values (BS). Nodes with PP=1.00/BS=100 were not displayed. – denotes values < 0.50/50.
In addition, molecular dating analysis based on the ML tree of concatenated plastid protein-coding genes was performed (Figure 2). The molecular dating results indicated that the crown age of the Selineae tribe was estimated to be 9.75 Mya (95% HPD: 12.07-7.58 Mya). Within this tribe, the diversification of clades A and B occurred at 2.30 Mya (95% HPD: 3.47-1.38 Mya) in the early Pleistocene period and 2.73 Mya (95% HPD: 3.76-1.92 Mya) in the late Pliocene period, respectively.
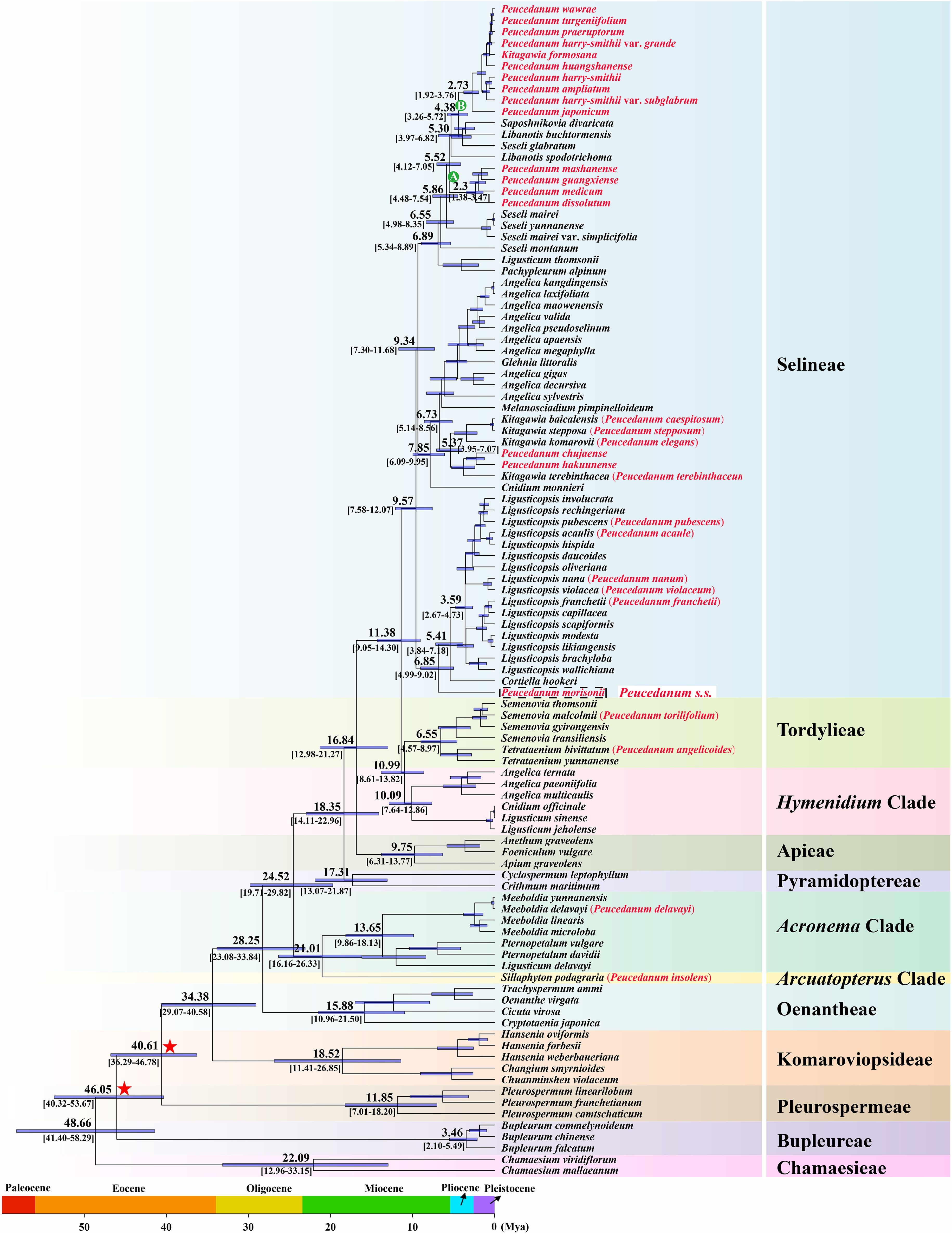
Figure 2. Divergence time estimation based on concatenated plastid protein-coding genes. Numbers above/under branches represent mean divergent age and 95% highest posterior density interval. Red stars indicate the calibration points for the molecular dating.
3.3 Chromosome numbers
The chromosome numbers of eight Peucedanum s.l. taxa were counted. Among them, the chromosome numbers of three species, P. dissolutum (Diels) H. Wolff, P. mashanense R.H. Shan & M.L. Sheh, and P. medicum Dunn, were 2n=20, while the chromosome numbers of the remainders were 2n=22 (Supplementary Figure S2).
4 Discussion
4.1 Phylogenetic framework and taxonomic implications
The phenomenon of incongruences between plastome-based and nrDNA-based phylogenies is common in Apiaceae (Xiao et al., 2017; Lei et al., 2022; Song et al., 2024a, b), and our study was no exception. In our study, the topologies generated from the PCGs and nrDNA were largely similar, except for the discordance regarding the position of P. huangshanense, which may be caused by incomplete lineage sorting due to the young evolutionary history (Wen et al., 2020, 2021). Same as all previous phylogenetic studies (Downie et al., 2000; Spalik et al., 2004; Valiejo-Roman et al., 2006; Feng et al., 2009; Zhou et al., 2009, 2020; Lei et al., 2022; Liu et al., 2022), phylogenetic analyses respectively based on concatenated plastid protein-coding genes and nrDNA sequences in the current study failed to recover Peucedanum s.l. as a monophyletic group. Specifically, both analyses robustly supported that P. morisonii, the representative of Peucedanum s.s., was distantly related to other Peucedanum s.l. members. The distant relationship was further justified by the morphological features: ternate leaves, linear leaflets, yellow petals, and dorsally compressed and glabrous mericarps with 1 vittae in each furrow and 2 on commissure, which were the recognized features of Peucedanum s.s (Kadereit and Bittrich, 2018), and significantly distinguished P. morisonii from other Peucedanum s.l. members. Hence, all these Peucedanum s.l. taxa were not “truly Peucedanum plants” and the taxonomic positions of them need to be re-explored.
In addition, the PCG-based phylogenetic analyses showed that 14 Peucedanum s.l. taxa were divided into two monophyletic clades (clade A and clade B) by Saposhnikovia divaricata (Turcz.) Schischk., Seseli glabratum Willd. ex Schult., Libanotis buchtormensis (Fisch.) DC., and L. spodotrichoma K.T. Fu with robust supports. Furthermore, obvious morphological differences between 14 Peucedanum s.l. taxa and these species were also observed: mericarps with filiform dorsal ribs, winged lateral ribs, and numerous vittae in each furrow and commissure, easily distinguished the 14 Peucedanum s.l. taxa from the mericarps with fake vittae in each rib, and 1 vittae in each furrow and 2 on commissure of Saposhnikovia divaricata, from mericarps with equally filiform or keeled ribs, and 1 vittae in each furrow and 2 on commissure of S. glabratum, L. buchtormensis and L. spodotrichoma (Sheh, 1992; Sheh and Watson, 2005). Thus, it was clearly inappropriate to regard all these taxa as a unit, whether based on morphological features or phylogenetic analyses. Furthermore, we also detected that clades A and B had significant morphological differences, but the morphological characteristics within each clade were unified. In detail, four members of clade A were perennial herbs; stem solitary or numerous, solid, glabrous, basal usually clothed with fibrous remnant sheaths, branched above; leaves basal and cauline, ternate or pinnate, terminal leaflet diverse; basal leaves petiolate, petioles sheathing; cauline leaves reduced upwards, leaf sheath expand; inflorescences compound umbels, terminal and latera; bracts few or absent; bracteoles numerous, rarely few; rays numerous, unequal, pubescent; calyx teeth conspicuous; petals white, inflexed; mericarp dorsally compressed, glabrous; dorsal ribs filiform, lateral ribs narrowly winged; numerous vittae in commissure and each furrow. Ten members of clade B were perennial herbs; rootstock stout, crown usually clothed with fibrous remnant sheaths; stem solitary, branched above; leaves basal and cauline, ternate or pinnate, terminal leaflet diverse; basal leaves petiolate, petioles sheathing; cauline leaves reduced upwards, leaf sheath expand; inflorescences compound umbels, terminal and latera; bracts few or absent; bracteoles numerous, rarely few; rays unequal, pubescent; calyx teeth short or obsolete; petals usually white, occasionally purple, inflexed; mericarp dorsally compressed, pubescent; dorsal ribs filiform, lateral ribs narrowly winged; numerous vittae in commissure and each furrow. These unique features of clades A and B can also easily distinguish them from other related genera. Meanwhile, the plastome structure and chromosome numbers of clades A and B were also different, such as the IRa/LSC border falling into the rpl23 gene, an overall plastome size of 152,288-154,686 bp, and chromosome numbers of 2n=20 in clade A, and the IRa/LSC border falling into the ycf2 gene, an overall plastome size of 146,718-147,592 bp, and chromosome numbers of 2n=22 in clade B (Figure 3). Therefore, we established two new genera (Shanopeucedanum gen. nov. and Sinopeucedanum gen. nov.) to respectively accommodate the taxa of clades A and B and the treatment was accepted based on morphology, plastomes, chromosome number, and phylogeny.
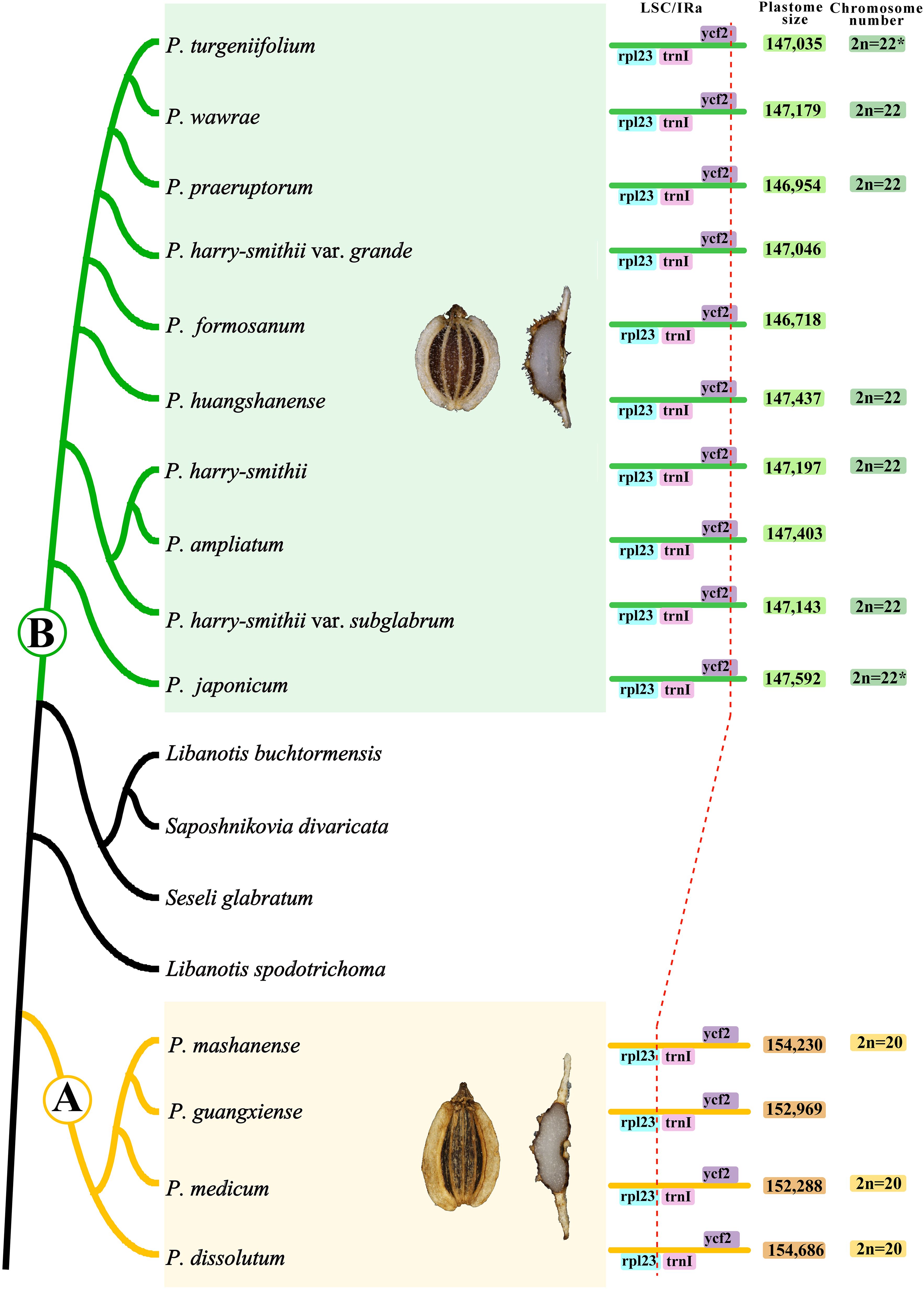
Figure 3. Comparisons of mericarps, plastomes, and chromosome number between clade A and clade B. The chromosome numbers for P. turgeniifolium and P. japonicum marked with a * were obtained from the results of Zhang et al. (2006) and Sun et al. (1996), respectively.
Our PCG-based phylogenies also resolved the positions of the remaining Peucedanum s.l. species with high support. Six species formed a monophyletic clade, in which four species (P. baicalense, P. elegans, P. stepposum, and P. terebinthaceum) have been transferred into the Kitagawia genus (Pimenov, 1986, 2017) and we suggested that the remaining two species (P. chujaense and P. hakuunense) also should be transferred into the Kitagawia genus. Five species (P. acaule, P. franchetii, P. nanum, P. pubescens, and P. violaceum) were nested in Ligusticopsis and have been merged into this genus (Pimenov, 2017; Deng et al., 2022; Liu et al., 2023; Song et al., 2023). P. insolens formed a separate clade and a monotypic genus, Sillaphyton, was established by Pimenov to accommodate this species (Pimenov et al., 2016). P. delavayii and P. torilifolium respectively nested in Meeboldia and Semenovia, while P. angelicoides was sister to Tetrataenium yunnanense, and they have been transferred into Meeboldia, Semenovia, and Tetrataenium, respectively (Xiao, 2017; Xiao et al., 2017; Gou et al., 2020). Hence, with high support and extended samples, our results strongly supported the previous taxonomic revisions for the above species and provide new additional evidence to accept these treatments.
4.2 Estimation divergence time
The diversification of tribes within the Apiaceae family has been inferred based on robust phylogenetic frameworks obtained from transcriptome and plastome datasets by Wen et al. (2020, 2021). However, limited samples for each tribe were employed in both studies, such as only five species of the Selineae tribe in transcriptome datasets and fourteen species of the Selineae tribe in plastome datasets, which may have led to inaccurate results. Due to the majority of the Peucedanum s.l. taxa falling into Selineae, we extended the samples of this tribe to explore the molecular dating analysis in the current study. Our result showed that the diversification of the Selineae tribe occurred at 9.75 Mya (95% HPD: 12.07-7.58 Mya), which was slightly earlier than 5.32 Mya (95% HPD: 7.25-3.60 Mya) and 7.70 Mya (95% HPD: 10.09-5.74 Mya) inferred by Wen et al. (2020, 2021). With more samples, and therefore, more accurate results, we justified that the Selineae tribe was a young lineage and the members of this tribe were the results of recent diversification.
Within the Selineae tribe, the diversification of clades A and B occurred at 2.30 Mya (95% HPD: 3.47-1.38 Mya) (around the early Pleistocene) and 2.73 Mya (95% HPD: 3.76-1.92 Mya) (around the late Pliocene), respectively. Previous studies have proven that the dramatic uplift of Qinhai-Tibet Plateau (QTP) occurred from the late Miocene to the early Pliocene (Harrison et al., 2001; Royden et al., 2008), which significantly modified the global climate (An et al., 2001; Shi et al., 2005), and at least four major glaciations took place in East Asia during the Pleistocene (Shi, 1998). These complex geological and climate shifts drove the expansion/contraction of the species range and isolation in East Asia and thereby facilitated species radiation (Axelrod et al., 1998; Li and Fang, 1999; Qiu et al., 2011), which is thought to have facilitated the diversification of wide spectrum of East Asia plants (Qian and Ricklefs, 2000; Wu et al., 2007; Wen et al., 2014; Favre et al., 2015; Ji et al., 2019) and thus could also have caused isolation and drove species diversification in clades A and B. Thus, we speculate that the diversification in clades A and B may have been driven by complex geological and climate shifts.
4.3 Taxonomic treatments
Shanopeucedanum B.N.Song, C.K. Liu & X.J. He, gen. nov.
Type: Shanopeucedanum medicum (Dunn) C.K. Liu & X.J. He.
Diagnosis: The genus can be easily distinguished from Peucedanum s.s. and related genera by white petals, dorsally compressed and glabrous mericarps with filiform dorsal ribs, winged lateral ribs, and numerous vittae in commissure and each furrow.
Description: Perennial herbs. Stem solitary or numerous, solid, glabrous, basal usually clothed with fibrous remnant sheaths, branched above. Leaves basal and cauline, ternate or pinnate, terminal leaflet diverse; basal leaves petiolate, petioles sheathing; cauline leaves reduced upwards, leaf sheath expanded. Inflorescences compound umbels, terminal and latera; bracts few or absent; bracteoles numerous, rarely few; rays numerous, unequal, pubescent; calyx teeth conspicuous; petals white, inflexed. Mericarp dorsally compressed, glabrous; dorsal ribs filiform, lateral ribs narrowly winged; numerous vittae in commissure and each furrow. 2n=20.
Diversity: Four species.
Distribution: Endemic to China.
Key to species of Shanopeucedanum.
1. Stem solitary; leaves ternate, rarely pinnate……………………
……………………………………..Shanopeucedanum medicum.
1. Stem numerous, leaves pinnate.
2. Terminal leaflet rhombic.......…………….…………………
……………………….…….Shanopeucedanum mashanense.
2. Terminal leaflet ovate.
3. Mericarp large; leaves 3-pinnate.…………………….…
…………………………….Shanopeucedanum dissolutum.
3. Mericarp small; leaves 2-pinnate……………….….……
……………………………Shanopeucedanum guangxiense.
Shanopeucedanum dissolutum (Diels) C.K. Liu & X.J. He, comb. nov.
≡Peucedanum dissolutum (Diels) H. Wolff, Repert. Spec. Nov. Regni Veg. 21: 247. 1925.
≡Angelica dissoluta Diels, Bot. Jahrb. Syst. 29: 499. 1900.
Type: CHINA. Sichuan, Nanchuan, Kên ao p’ing, Abhänge, von Rosthorn 659 (syntype: O 2014068).
Shanopeucedanum guangxiense (R.H. Shan & M.L. Sheh) C.K. Liu & X.J. He, comb. nov.
≡Peucedanum guangxiense R.H. Shan & M.L. Sheh, Acta Phytotax. Sin. 24: 308. 1986.
Type: CHINA. Guangxi, Jingxi, 300 m alt., 8 November 1961, Wu Xinfang 23493 (holotpye: NAS 00082770).
Shanopeucedanum mashanense (R.H. Shan & M.L. Sheh) C.K. Liu & X.J. He, comb. nov.
≡Peucedanum mashanense R.H. Shan & M.L. Sheh, Acta Phytotax. Sin. 24: 304. 1986.
Type: CHINA. Guangxi, Mashan, 300 m alt., 7 September 1958, Zhong Shuquan 301547 (holotype: IBSC; isotype: KUN 0566764).
Shanopeucedanum medicum (Dunn) C.K. Liu & X.J. He, comb. nov.
≡Peucedanum medicum Dunn, J. Linn. Soc. Bot. 35: 496.1903.
Type: CHINA. Hupeh, Ichang and intermediate neighborhood, February 1887, Henry 15462006 (lectotpye, designated by Pimenov (2017): K 000685413, 000685414; isolectotype: BM, E, K 000685416, NY 00406177, P 00752971); Hupeh, Nam-To, 1887, Henry 1906 (syntype: K 000685415); Hupeh, Fang, Henry 5868A (syntypes: K 000685417, LE, NY 00406176, P 00752968); Szechuen, South Wushan, Henry 7473 (syntypes: LE, P 02272028).
Sinopeucedanum B.N.Song, C.K. Liu & X.J. He, gen. nov.
Type: Sinopeucedanum harry-smithii (Fedde ex H. Wolff) C.K. Liu & X.J. He.
Diagnosis: The new genus can be easily recognized from Peucedanum s.s. and related genera by white or purple petals, dorsally compressed and pubescent mericarps with filiform dorsal ribs, winged lateral ribs, and numerous vittae in commissure and each furrow.
Description: Perennial herbs. Rootstock stout, crown usually clothed with fibrous remnant sheaths. Stem solitary, branched above. Leaves basal and cauline, ternate or pinnate, terminal leaflet diverse; basal leaves petiolate, petioles sheathing; cauline leaves reduced upwards, leaf sheath expanded. Inflorescences compound umbels, terminal and latera; bracts few or absent; bracteoles numerous, rarely few; rays unequal, pubescent; calyx teeth short or obsolete; petals usually white, occasionally purple, inflexed. Mericarp dorsally compressed, pubescent; dorsal ribs filiform, lateral ribs narrowly winged; numerous vittae in commissure and each furrow. 2n=22.
Diversity: Ten species.
Distribution: China, Japan, Korea, and Philippines.
Key to species of Sinopeucedanum.
1. Leaves ternate.
2. Calyx teeth inconspicuous.
3. Umbels small; bracteoles linear…………………….……
.………………………………….Sinopeucedanum wawrae.
3. Umbels large; bracteoles pinnate and linear coexisted.…
………………………………Sinopeucedanum ampliatum.
2. Calyx teeth conspicuous.
4. Anthers purple...…………………………………………
……………………….….Sinopeucedanum huangshanense.
4. Anthers white.
5. Stem glabrous…………………………….……………
…………………………….Sinopeucedanum japonicum.
5. Stem tomentose in upper parts.
6. Bracteoles longer than flowers; fruit densely hispid
………………………..Sinopeucedanum formosanum.
6. Bracteoles shorter than flowers; fruit sparsely
puberulent……..Sinopeucedanum praeruptorum.
1. Leaves pinnate.
7. Calyx teeth inconspicuous……………….…………………Sinopeucedanum turgeniifolium.
7. Calyx teeth conspicuous.
8. Stem and leaves densely pubescent………………………..
……………………………....Sinopeucedanum harry-smithii.
8. Stem and leaves glabrous or sparsely pubescent.
9. Umbels small; mericarp suborbicular….……………….
……………………………...Sinopeucedanum subglabrum.
9. Umbels large; mericarp obovate..………….……………
…………………………………...Sinopeucedanum grande.
Sinopeucedanum ampliatum (K.T. Fu) C.K. Liu & X.J. He, comb. nov.
≡Peucedanum ampliatum K.T. Fu, Fl. Tsinling. 1(3): 462. 1981.
Type: CHINA. Shaanxi, Shanyang, Tianzhushan, 1660 m alt., 3 July 1964, Liang Yiming, Yang Jinxiang 3126 (holotype: WUK 228591).
Sinopeucedanum formosanum (Hayata) C.K. Liu & X.J. He, comb. nov.
≡Peucedanum formosanum Hayata, Icon. Pl. Formosan. 10: 22. 1921.
≡Kitagawia formosana (Hayata) Pimenov, Turczaninowia 20(2): 167. 2017.
Type: CHINA. Taiwan, Mt. Niitaka, ad 10000 ped. alt. [Nanto], 19 October 1906, Kawakami, Mori 2052 (holotype: TAIF).
Sinopeucedanum grande (K.T. Fu) C.K. Liu & X.J. He, comb. et stat. nov.
≡Peucedanum harry-smithii Fedde ex H. Wolff var. grande (K.T. Fu) R.H. Shan & M.L. Sheh, Fl. Reipubl. Popularis Sin. 55(3): 164. 1992.
≡Peucedanum praeruptorum Dunn var. grande K.T. Fu, Fl. Tsinling. 1(3): 428. 1981.
Type: CHINA. Shensi, Hwa-in Hsien, Hwa-shan, 1580 m alt., 5 August 1973, Fu Kuntsun 16884 (holotype: WUK).
Sinopeucedanum harry-smithii (Fedde ex H. Wolff) C.K. Liu & X.J. He, comb. nov.
≡Peucedanum harry-smithii Fedde ex H. Wolff, Repert. Spec. Nov. Regni Veg. 33: 247. 1933.
Type: CHINA. Shansi, Chieh-hsiuh-Distr. Sung-lin-miao, Cho mei shan, in prato apricot, ca. 1000 m alt., 3 October 1924, H. Smith 7609 (lectotype, designated by Pimenov (2017): UPS; isolectotypes: MO 150814, PE 00935536).
Sinopeucedanum huangshanense (Lu Q.Huang, H.S. Peng & S.S. Chu) C.K. Liu & X.J. He, comb. nov.
≡Peucedanum huangshanense Lu Q. Huang, H.S. Peng & S.S. Chu, Phytotaxa 430: 21. 2020.
Type: CHINA. Anhui, Mount Huangshan, on the forest margins and cliffs, ca. 1600 m alt., 20 August 2018, Peng 082011 (holotype: ACM; isotype: ACM).
Sinopeucedanum japonicum (Thunb.) C.K. Liu & X.J. He, comb. nov.
≡Peucedanum japonicum Thunb., Nova Acta Regiae Soc. Sci. Upsal. 4: 38. 1783.
≡Anethum japonicum (Thunb.) Koso-Pol., Bull. Soc. Imp. Nat. Mosc. n. s. 29: 117. 1916.
Type: JAPAN. San Bofu, vulgo Fama Bofu feu Iamma Bofu, i. e. Bofu littoralis Kaempf. Am. ex. Fafc. V. p. 825 (lectotype, designated by Thunb. (1784): UPS).
=Peucedanum japonicum Thunb. f. album Q.H. Yang & Q. Tian, Acta Bot. Boreal. Ocid. Sin. 28(2): 399. 2008.
Type: CHINA. Zhejiang, Zhoushan, 22 June 2007, Tian Qi, Yang Qinhua, Zhou Xiangyu 07-0331 (holotype: CSH 0067982).
Sinopeucedanum praeruptorum (Dunn) C.K. Liu & X.J. He, comb. nov.
≡Peucedanum praeruptorum Dunn, J. Linn. Soc. Bot. 35: 497. 1903.
≡Kitagawia praeruptora (Dunn) Pimenov, Turczaninowia 20(2): 167. 2017.
Type: CHINA. Hupeh, Ichang, 1887, Henry 2911 (syntypes: P 02272036, 02272037); Changyang, Henry 7505 (syntype: K); Szechuan, North Wushan, July 1888, Henry 7475 (lectotypes, designated by Pimenov (2017): E 00002618, 00002619; isolectotypes: BM 000885390, G, K 000685408, 000685409, 000685411, 000685412, P 022722038, 022722039).
Sinopeucedanum subglabrum (R.H. Shan & M.L. Sheh) C.K. Liu & X.J. He, comb. et stat. nov.
≡Peucedanum harry-smithii Fedde ex H. Wolff var. subglabrum (R.H. Shan & M.L. Sheh) R.H. Shan & M.L. Sheh, Fl. Reipubl. Popularis Sin. 55(3): 164. 1992.
≡Peucedanum hirsutiusculum (Y.C. Ma) R.H. Shan & M.L. Sheh var. subglabrum R.H. Shan & M.L. Sheh, Acta Phytotax. Sin. 24(4): 310. 1986.
Type: CHINA. Henan, Song Xian, 1000 m alt., 15 September 1965, Wu Peigeng, Wang Wanli 651161 (holotype: NAS 00026834; isotype: NAS 00042627).
Sinopeucedanum turgeniifolium (H. Wolff) C.K. Liu & X.J. He, comb. nov.
≡Peucedanum turgeniifolium H. Wolff, Act. Hort. Gothob. 2: 323. 1926.
Type: CHINA. Sichaun, Ch’osodjo, Ö om älven. Torr buskäng, 18 October 1922, H. Smith 4826 (lectotype, designated by Pimenov (2017): UPS; isolectotype: GB).
Sinopeucedanum wawrae (H. Wolff) C.K. Liu & X.J. He, comb. nov.
≡Peucedanum wawrae (H. Wolff) S.W. Su ex M.L. Sheh, Fl. Reipubl. Popularis Sin. 55(3): 149. 1992.
≡Seseli wawrae H. Wolff, Repert. Spec. Nov. Regni Veg. 27: 315. 1930.
Type: CHINA. Shandong, Chefoo, ravines in hills, 7 August 1920, Cowdry 757 (lectotype, designated by Pimenov (2017): K 000697461; isolectotype: K 000697460).
5 Conclusions
Based on the raw data generated from genome skimming, nine high-quality plastomes and 43 nrDNA sequences were presented. We performed phylogenetic analyses respectively based on concatenated plastid protein-coding genes and nrDNA sequences, including 30 members of Peucedanum s.l. with 21 endemic to China. Both analyses failed to recover Peucedanum s.l. as a monophyletic group and robustly supported that P. morisonii, the representative of Peucedanum s.s., was distantly related to other Peucedanum s.l. members, which implied that all these Peucedanum s.l. taxa were not “truly Peucedanum plants” and the distant relationship was also further supported by morphological evidence. Plastid-based phylogenies recognized two monophyletic clades, clade A (four species) and clade B (ten taxa). Furthermore, obvious recognized features from the morphology, plastomes, and chromosome number for each clade were detected, such as clade A possessing white petals, dorsally compressed and glabrous mericarps with filiform dorsal ribs, winged lateral ribs, and numerous vittae in commissure and each furrow; IRa/LSC border falling into the rpl23 gene; an overall plastome size of 152,288-154,686 bp; and chromosome numbers of 2n=20, while clade B possessed white or purple petals, dorsally compressed and pubescent mericarps with filiform dorsal ribs, winged lateral ribs, and numerous vittae in commissure and each furrow; IRa/LSC border falling into the ycf2 gene; an overall plastome size of 146,718-147,592 bp; and chromosome numbers of 2n=22. Therefore, we established two new genera (Shanopeucedanum and Sinopeucedanum) to accommodate the taxa of clades A and B, respectively. With high support and extended samples, our results also strongly support the previous taxonomic revisions for Peucedanum s.l. taxa. Furthermore, molecular dating analysis showed that the diversification of clades A and B occurred in the early Pleistocene and late Pliocene, respectively, which may have been driven by the complex geological and climate shifts in these periods.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Author contributions
B-NS: Methodology, Software, Writing – original draft, Formal Analysis, Investigation. C-KL: Investigation, Methodology, Software, Writing – original draft, Writing – review & editing, Formal Analysis. J-JD: Formal Analysis, Investigation, Methodology, Software, Writing – review & editing. W-YT: Investigation, Software, Writing – review & editing. S-DZ: Resources, Supervision, Visualization, Writing – review & editing. X-JH: Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by the National Natural Science Foundation of China (Grant Nos. 32170209, 32070221, 32470216).
Acknowledgments
We are grateful to Zhen-Long Liang for his help in the chromosome experiment.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1518418/full#supplementary-material
References
An, Z., Kutzbach, J. E., Prell, W. L., Porter, S. C. (2001). Evolution of Asian monsoons and phased uplift of the Himalaya–Tibetan plateau since Late Miocene times. Nature 411, 62–66.
Angiosperm Phylogeny Group (2003). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG II. Bot. J. Linn. Soc 141, 399–436. doi: 10.1046/j.1095-8339.2003.t01-1-00158.x
Angiosperm Phylogeny Group (2009). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG III. Bot. J. Linn. Soc 161, 105–121. doi: 10.1111/j.1095-8339.2009.00996.x
Angiosperm Phylogeny Group (2016). An update of the Angiosperm Phylogeny Group classification for the orders and families of flowering plants: APG IV. Bot. J. Linn. Soc 181, 1–20. doi: 10.1111/boj.12385
Axelrod, D. I., Al-Shehbaz, I., Raven, P. H. (1998). “History of the modern flora of China,” in Floristic characteristics and diversity of East Asian plants (China Higher Education Press, Beijing), 43–55.
Banasiak, Ł., Piwczy´nski, M., Uli´nski, T., Downie, S. R., Watson, M. F., Shakya, B., et al. (2013). Dispersal patterns in space and time: a case study of Apiaceae subfamily Apioideae. J. Biogeogr. 40, 1324–1335. doi: 10.1111/jbi.12071
Bremer, K., Chase, M., Stevens, P., Thulin, M. (1998). An ordinal classification for the families of flowering plants. Ann. Mo. Bot. Gard. 85, 531–553. doi: 10.2307/2992015
Cao, D. L., Zhang, X. J., Qu, X. J., Fan, S. (2023). Phylogenomics, divergence time estimation, and adaptive evolution in the Polygonoideae (Polygonaceae). J. Syst. Evol. 00, 1–16. doi: 10.1111/jse.12946
Capella-Gutierrez, S., Silla-Martınez, J. M., Gabaldon, T. (2009). trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973. doi: 10.1093/bioinformatics/btp348
Chen, S. F., Zhou, Y. Q., Chen, Y. R., Gu, J. (2018). Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890. doi: 10.1093/bioinformatics/bty560
Choi, I. S., Cardoso, D., Queiroz, L. P., Lima, H. C., Lee, C., Ruhlman, T. A., et al. (2022). Highly resolved papilionoid legume phylogeny based on plastid phylogenomics. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.823190
Ci, Z. L. (1994). Karyotype analysis of Saposchnikovia divaricate. J. Inner. Mongolia. Forestry. College. 16, 29–31.
Cui, Y. F., Zhou, P., Xiang, K. L., Zhang, Q., Yan, H., Zhang, L. G., et al. (2023). Plastome evolution and phylogenomics of Trichosporeae (Gesneriaceae) with its morphological characters appraisal. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1160535
Deng, J. J., Liu, C. K., Zhou, S. D., He, X. J. (2022). Morphological and molecular evidence gives insight into the taxonomic position of Peucedanumpubescens (Apiaceae, Selineae). PhytoKeys 213, 19–33. doi: 10.3897/phytokeys.213.89784
Downie, S. R., Spalik, K., Katz-Downie, D. S., Reduron, J. P. (2010). Major clades within Apiaceae subfamily Apioideae as inferred by phylogenetic analysis of nrDNA ITS sequences. Plant Ecol. Divers. 128, 111–136. doi: 10.1127/1869-6155/2010/0128-0005
Downie, S. R., Watson, M. F., Spalik, K., Katz-Downie, D. S. (2000). Molecular systematics of Old World Apioideae (Apiaceae): relationships among some members of tribe Peucedaneae sensu lato, the placement of several island-endemic species, and resolution within the apioid superclade. Can. J. Bot. 78, 506–528. doi: 10.1139/b00-029
Doyle, J. J., Doyle, J. L. (1987). A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem. Bull. 19, 11–15. doi: 10.1016/0031-9422(80)85004-7
Drew, B. T., González-Gallegos, J. G., Xiang, C., Ricardo, K., Chloe, P. D., Jay, B. W., et al. (2017). Salvia united: The greatest good for the greatest number. Taxon 66, 133–145. doi: 10.12705/661.7
Favre, A., Päckert, M., Pauls, S. U., Jähnig, S. C., Uh, D., Michala, I., et al. (2015). The role of the uplift of the Qinghai-Tibetan plateau for the evolution of Tibetan biotas. Biol. Rev. 90, 236–253. doi: 10.1111/brv.2015.90.issue-1
Feng, T., Downie, S. R., Yu, Y., Zhang, X. M., Chen, W. W., He, X. J., et al. (2009). Molecular systematics of Angelica and allied genera (Apiaceae) from the Hengduan Mountains of China based on nrDNA ITS sequences: phylogenetic affinities and biogeographic implications. J. Plant Res. 122, 403–414. doi: 10.1007/s10265-009-0238-4
Fu, X. G., Liu, S. Y., van Velzen, R., Stull, G. W., Tian, Q., Li, Y. X., et al. (2022). Phylogenomic analysis of the hemp family (Cannabaceae) reveals deep cyto-nuclear discordance and provides new insights into generic relationships. J. Syst. Evol. 61, 806–826. doi: 10.1111/jse.12920
Gou, W., Jia, S. B., Price, M., Guo, X. L., Zhou, S. D., He, X. J. (2020). Complete plastid genome sequencing of eight species from Hansenia, Haplosphaera and Sinodielsia (Apiaceae): comparative analyses and phylogenetic implications. Plants 9, 1523. doi: 10.3390/plants9111523
Harrison, S. P., Yu, G., Takahara, H., Prentice, I. C. (2001). Paleovegetation: diversity of temperate plants in East Asia. Nature 413, 129–130. doi: 10.1038/35093166
Ji, Y. H., Yang, L. F., Chase, M. W., Liu, C. K., Yang, Z. Y., Yang, J., et al. (2019). Plastome phylogenomics, biogeography, and clade diversification of Paris (Melanthiaceae). BMC Plant Biol. 19, 543. doi: 10.1186/s12870-019-2147-6
Ji, Y. H., Yang, J., Landis, J. B., Wang, S. Y., Jin, L., Xie, P. X., et al. (2022). Genome skimming contributes to clarifying species limits in Paris section Axiparis (Melanthiaceae). Front. Plant Sci. 13. doi: 10.3389/fpls.2022.832034
Jiang, C. M., Lu, F., Peng, Y. L. (2002). Chromosome number of Bupleurum L. genus in Northeast China. J. Heilongjiang. Nongken. Normal. College. 04), 59–60.
Jin, J. J., Yu, W. B., Yang, J. B., Song, Y., dePamphilis, C. W., Yi, T. S., et al. (2020). GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 21, 241. doi: 10.1186/s13059-020-02154-5
Kadereit, J. W., Bittrich, V. (2018). “Flowering plants,” in Eudicots: Apiales, Gentianales (except Rubiaceae), 1st edn (Springer, Berlin).
Katoh, K., Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kearse, M., Moir, R., Wilson, A., Stones-Hava, S., Cheung, M., Sturrock, S., et al. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics 28, 1647–1649. doi: 10.1093/bioinformatics/bts199
Kumar, P., Singhal, V. K. (2011). Chromosome number, male meiosis and pollen fertility in selected angiosperms of the cold deserts of Lahaul-Spiti and adjoining areas (Himachal Pradesh, India). Plant Syst. Evol. 297, 271–297. doi: 10.1007/s00606-011-0516-1
Lei, J. Q., Liu, C. K., Cai, J., Price, M., Zhou, S. D., He, X. J. (2022). Evidence from phylogenomics and morphology provide insights into the phylogeny, plastome evolution, and taxonomy of kitagawia. Plants 11, 3275. doi: 10.3390/plants11233275
Li, J. J., Fang, X. M. (1999). Uplift of the Tibetan plateau and environmental changes. China Sci. Bull. 44, 2117–2124. doi: 10.1360/csb1998-43-15-1569
Li, W. P., Yang, F. S., Jivkova, T., Yi, G. S. (2012). Phylogenetic relationships and generic delimitation of Eurasian Aster (Asteraceae: Astereae) inferred from ITS, ETS and trnL-F sequence data. Ann. Bot. 109, 1341–1357. doi: 10.1093/aob/mcs054
Li, H. T., Yi, T. S., Gao, L. M., Ma, P. F., Zhang, T., Yang, J. B., et al. (2019). Origin of angiosperms and the puzzle of the Jurassic gap. Nat. Plants 5, 461–470. doi: 10.1038/s41477-019-0421-0
Liang, Q. L., Wang, C. B., Ma, X. G., Zhao, C., He, X. J. (2013). Chromosomal study on chinese bupleurum L.(Apiaceae). Plant Sci. 31, 11–22. doi: 10.3724/SP.J.1142.2013.10011
Lima, L. V., Salino, A., Kessler, M., Rouhan, G., Testo, W. L., Argolo, C. S., et al. (2023). Phylogenomic evolutionary insights in the fern family Gleicheniaceae. Mol. Phylogenet. Evol. 184, 107782. doi: 10.1016/j.ympev.2023.107782
Liu, C. K., Deng, J. J., Zhou, R. X., Song, B. N., Zhou, S. D., He, X. J. (2023). Plastid phylogenomics provide evidence to accept two new members of ligusticopsis (Apiaceae, angiosperms). Int. J. Mol. Sci. 24, 382. doi: 10.3390/ijms24010382
Liu, C. K., Lei, J. Q., Jiang, Q. P., Zhou, S. D., He, X. J. (2022). The complete plastomes of seven Peucedanum plants: comparative and phylogenetic analyses for the Peucedanum genus. BMC Plant Biol. 22, 101. doi: 10.1186/s12870-022-03488-x
Liu, C. K., Yang, J., Jin, L., Wang, S. Y., Yang, Z. Y., Ji, Y. H. (2021). Plastome phylogenomics of the East Asian endemic genus Dobinea. Plant Divers. 43, 35–42. doi: 10.1016/j.pld.2020.05.002
Marhold, K., Chepinoga, V. V., Gnutikov, A. A., Enushchenko, I. V., Rosbakh, S. A. (2009). IAPT/IOPB chromosome data 8. Taxon 58, 1281–1289. doi: 10.1002/tax.2009.58.issue-1
Miller, J. T., Seigler, D., Mishler, B. D. (2014). A phylogenetic solution to the Acacia problem. Taxon 63, 653–658. doi: 10.12705/633.2
Ostroumova, T. A. (2018). Fruit micromorphology in the umbelliferae of the Russian far east. Bot. Garden-Inst. 7, 41–49. doi: 10.17581/bp.2018.07107
Ostroumova, T. A., Pimenov, M. G., Degtjareva, G. V., Samigullin, T. H. (2016). Taeniopetalum Vis. (Apiaceae), a neglected segregate of Peucedanum L., supported as a remarkable genus by morphological and molecular data. Skvortsovia 3, 20–44.
Pan, Z. H., Qin, H. Z. (1981). A report on the chromosome numbers of Chinese Umbelliferae. J. Univ. Chin. Acad. Sci. 19, 447–450.
Peng, C., Guo, X. L., Zhou, S. D., He, X. J. (2023). Backbone phylogeny and adaptive evolution of Pleurospermum sl: New insights from phylogenomic analyses of complete plastome data. Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1148303
Pimenov, M. G. (1986). Kitagawia—A new asiatic genus of the family umbelliferae. Bot. Zhurn. 71, 942–949.
Pimenov, M. G., Ostroumova, T. A. (2012). Umbelliferae of Russia (Moscow, Russia: Association of Scientifific Publications), ISBN: ISBN 978-5-87317-813-1.
Pimenov, M. G., Ostroumova, T. A., Degtjareva, G. V., Samigullin, T. H. (2016). Sillaphyton, a new genus of the umbelliferae, endemic to the korean peninsula. Bot. Garden-Inst. 5, 31–41. doi: 10.17581/bp.2016.05204
Pimenov, M. G. (2017). Updated checklist of Chinese Umbelliferae: nomenclature, synonymy, typifcation, distribution. Turczaninowia 20, 106–239. doi: 10.14258/turczaninowia.21.1.10
Posada, D., Crandall, K. A. (1998). Modeltest: testing the model of DNA substitution. Bioinformatics 14, 817–818. doi: 10.1093/bioinformatics/14.9.817
Pu, J. X., He, X. J., Zhang, X. M., Chen, W. W. (2006). Karyotypes of seven populations belonging to four species of umbelliferae in hengduan mountains. Acta Bot. Boreali-Occident. Sin. 26, 1 989–11995.
Qian, H., Ricklefs, R. E. (2000). Large-scale processes and the Asian bias in species diversity of temperate plants. Nature 407, 180–182. doi: 10.1038/35025052
Qiu, Y. X., Fu, C. X., Comes, H. P. (2011). Plant molecular phylogeography in China and adjacent regions: tracing the genetic imprints of quaternary climate and environmental change in the world’s most diverse temperate flora. Mol. Phylogenet. Evol. 59, 225–244. doi: 10.1016/j.ympev.2011.01.012
Rambaut, A., Drummond, A. (2015). FigTree, version 1.4.2. Available online at: http://tree.bio.ed.ac.uk/software/fgtree/ (Accessed Jul 1, 2024).
Rambaut, A., Drummond, A. J., Xie, D., Baele, G., Suchard, M. A. (2018). Posterior summarization in bayesian phylogenetics using Tracer 1.7. Syst. Biol. 67, 901–904. doi: 10.1093/sysbio/syy032
Razafimandimbison, S. G., Taylor, C. M., Wikström, N. (2014). Phylogeny and generic limits in the sister tribes Psychotrieae and Palicoureeae (Rubiaceae): Evolution of schizocarps in Psychotria and origins of bacterial leaf nodules of the Malagasy species. Am. J. Bot. 101, 1102–1126. doi: 10.3732/ajb.1400076
Reduron, J. P., Charpin, A., Pimenov, M. G. (1997). Contribution à la nomenclature générique des Apiaceae (Ombellifères). J. Bot. Soc Bot. France. 1, 91–104. doi: 10.3406/jobot.1997.1657
Ronquist, F., Teslenko, M., Mark, P. H. D., Ayres, D. L., Darling, A., Hhna, S., et al. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 61, 539–542. doi: 10.1093/sysbio/sys029
Royden, L. H., Burchfiel, B. C., Van, D. H. R. D. (2008). The geological evolution of the Tibetan plateau. Science 321, 1054–1058. doi: 10.1126/science.1155371
Ruhsam, M., Rai, H. S., Mathews, S., Ross, T. G., Hollingsworth, P. M. (2015). Does complete plastid genome sequencing improve species discrimination and phylogenetic resolution in Araucaria? Mol. Ecol. Resour. 15, 1067–1078. doi: 10.1111/1755-0998.12375
Scatigna, A. V., Souza, V. C., Sosa, M. M., Colletta, D. G., MaChado, R. M., Simões, A. O. (2022). Phylogenetics of Gratioleae (Plantaginaceae): paraphyly of Stemodia and its implications for generic circumscriptions, with insights from floral evolution. Bot. J. Linn. Soc 200, 194–217. doi: 10.1093/botlinnean/boac013
Schneider, J. V., Paule, J., Jungcurt, T., Cardoso, D., Zizka, G. (2021). Resolving recalcitrant clades in the pantropical Ochnaceae: insights from comparative phylogenomics of plastome and nuclear genomic data derived from targeted sequencing. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.638650
Sheh, M. L. (1992). “Peucedanum,” in Flora Reipublicae Popularis Sinicae, vol. 55. (Science Press, Beijing, China), 127–158.
Sheh, M. L., Watson, M. F. (2005). “Peucedanum linnaeus,” in Flora of China, vol. 14. (Science Press & Missouri Botanical Garden Press, Beijing, China; St. Louis, MO, USA), 182–192.
Shi, Y. F. (1998). Evolution of the cryosphere in the Tibetan plateau, China, and its relationship with the global change in the mid-Quaternary. J. Glaciol. Geocryol. 20, 197–208. doi: 10.1088/0256-307X/16/9/027
Shi, L. C., Chen, H. M., Jiang, M., Wang, L. Q., Wu, X., Huang, L. F., et al. (2019). CPGAVAS2, an integrated plastome sequence annotator and analyzer. Nucleic Acids Res. 47, W65–W73. doi: 10.1093/nar/gkz345
Shi, Y. F., Cui, Z. J., Su, Z. (2005). The quaternary glaciations and environmental variations in China (Shijiangzhuang: Hebei Science and Technology Publishing House).
Shneyer, V. S., Kutyavina, N. G., Pimenov, M. G. (2003). Systematic relationships within and between Peucedanum and Angelica (Umbelliferae–Peucedaneae) inferred from immunological studies of seed proteins. Plant Syst. Evol. 236, 175–194. doi: 10.1007/s00606-002-0239-4
Sielemann, K., Pucker, B., Schmidt, N., Viehver, P., Holtgrwe, D. (2022). Complete pan-plastome sequences enable high resolution phylogenetic classification of sugar beet and closely related crop wild relatives. BMC Genome 23, 113. doi: 10.1186/s12864-022-08336-8
Solov’eva, N. M., Pimenov, M. G., Vasil’eva, M. G., Žigareva, N. N., Turkov, V. D. (1985). Karyotaxonomic study of some species of Peucedanum (Umbelliferae). Pl. Syst. Evol. 151, 89–101. doi: 10.1007/BF02418022
Song, B. N., Liu, C. K., Ren, T., Xiao, Y. L., Chen, L., Xie, D. F., et al. (2024b). Plastid phylogenomics contributes to the taxonomic revision of taxa within the genus Sanicula L. and acceptance of two new members of the genus. Front. Plant Sci. 15. doi: 10.3389/fpls.2024.1351023
Song, B. N., Liu, C. K., Xie, D. F., Xiao, Y. L., Tian, R. M., Li, Z. X., et al. (2023). Plastid phylogenomic analyses reveal the taxonomic position of peucedanum franchetii. Plants 12, 97. doi: 10.3390/plants12010097
Song, B. N., Liu, C. K., Zhao, A. Q., Tian, R. M., Xie, D. F., Xiao, Y. L., et al. (2024a). Phylogeny and diversification of genus Sanicula L. (Apiaceae): novel insights from plastid phylogenomic analyses. BMC Plant Biol. 24, 70. doi: 10.1186/s12870-024-04750-0
Spalik, K., Reduron, J. P., Downie, S. R. (2004). The phylogenetic position of Peucedanum sensu lato and allied genera and their placement in tribe Selineae (Apiaceae, subfamily Apioideae). Plant Syst. Evol. 243, 189–210. doi: 10.1007/s00606-003-0066-2
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Straub, S. C., Parks, M., Weitemier, K., Fishbein, M., Cronn, R. C., Liston, A. (2012). Navigating the tip of the genomic iceberg: next-generation sequencing for plant systematics. Am. J. Bot. 99, 349–364. doi: 10.3732/ajb.1100335
Suchard, M. A., Lemey, P., Baele, G., Ayres, D. L., Drummond, A. J., Rambaut, A. (2018). Bayesian phylogenetic and phylodynamic data integration using BEAST 1.10. Virus Evol. 4, vey016. doi: 10.1093/ve/vey016
Sun, B. Y., Park, J. H., Kwak, M. J., Kim, C. H., Kim, K. S. (1996). Chromosome counts from the flora of Korea with emphasis on Apiaceae. J. Plant Biol. 39, 15–22.
Sun, W. Y., Sun, W. G., Li, Z. M. (2020). Karyomorphological study of two endemic plants (Apiaceae: pleurospermum) from the alpine subnival belt in hengduan mountains. J. Yunnan. Normal. Univ. 40, 59–64.
Valiejo-Roman, C. M., Terentieva, E. I., Samigullin, T. H., Pimenov, M. G., Ghahremani-Nejad, F., Mozaffarian, V. (2006). Molecular data (nrITS-sequencing) reveal relationships among Iranian endemic taxa of the Umbelliferae. Feddes. Repert. 117, 367–388. doi: 10.1002/fedr.200611106
Wang, M. Y., Liu, M., Cheng, X. Y., Wang, Y. T. (2015). The phylogenetic significance of fruit structures in Chinese Peucedanum of Apiaceae. Acta Phys. Sin. 24, 68–76. doi: 10.11686/cyxb2014512
Wen, J., Xie, D. F., Price, M., Ren, T., Deng, Y. Q., Gui, L. J., et al. (2021). Backbone phylogeny and evolution of Apioideae (Apiaceae): new insights from phylogenomic analyses of plastome data. Mol. Phylogenet. Evol. 161, 107183. doi: 10.1016/j.ympev.2021.107183
Wen, J., Yu, Y., Xie, D. F., Peng, C., Liu, Q., Zhou, S. D., et al. (2020). A transcriptome based study on the phylogeny and evolution of the taxonomically controversial subfamily Apioideae (Apiaceae). Ann. Bot. 125, 937–953. doi: 10.1093/aob/mcaa011
Wen, J., Zhang, J. Q., Nie, Z. L., Zhong, Y., Sun, H. (2014). Evolutionary diversifications of plants on the Qinghai-Tibetan plateau. Front. Genet. 5, 4. doi: 10.3389/fgene.2014.00004
Winter, P. J. D., Magee, A. R., Phephu, N., Tilney, P. M., Downie, S. R., Ben-Erik (2008). A new generic classification for African peucedanoid species (Apiaceae). Taxon 57, 347–364. doi: 10.2307/25066009
Wu, Z. Y., Lu, A. M., Tang, Y. C., Chen, Z. D., Li, D. Z. (2003). The families and genera of Angiosperms in China: a comprehensive analysis (Beijing: Science Press).
Wu, Z. Y., Sun, H., Zhou, Z. K., Peng, H., Li, D. Z. (2007). Origin and differentiation of endemism in the flora of China. Front. Biol. 2, 125–143. doi: 10.1007/s11515-007-0020-8
Xiao, Q. (2017). Phylogeny of subtribe Tordyliinae (Apiaceae tribe Tordylieae) in China (Chengdu: Sichuan University).
Xiao, Q. Y., Hu, H. Y., Tong, F., Li, M. J., He, X. J. (2017). Semenovia torilifolia is conspecific with S. Malcolmii (Apiaceae) based on morphology and molecular data. Phytotaxa 321, 225. doi: 10.11646/phytotaxa.321.3.1
Xu, C., Hong, D. Y. (2021). Phylogenetic analyses confirm polyphyly of the genus Campanula (Campanulaceae s. str.), leading to a proposal for generic reappraisal. J. Syst. Evol. 59, 475–489. doi: 10.1111/jse.12586
Zhang, X. (2011). Studies on geographic distribution of Peucedanum L. @ in southwest of China. Chin. Agric. Sci. Bull. 27, 177–180.
Zhang, D., Gao, F. L., Jakovlić, I., Zou, H., Zhang, J., Li, W. X., et al. (2020). PhyloSuite: an integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 20, 348–355. doi: 10.1111/1755-0998.13096
Zhang, X. M., He, X. J., Pi, J. X., Chen, W. W. (2006). Studies on karyotypes of five populations of three species in umbelliferae from China. Plant Sci. 24, 408–412.
Zhou, J., Gao, Y. Z., Wei, J., Liu, Z. W., Downie, S. R. (2020). Molecular phylogenetics of Ligusticum (Apiaceae) based on nrDNA ITS sequences: rampant polyphyly, placement of the Chinese endemic species, and a much-reduced circumscription of the genus. Int. J. Plant Sci. 181, 306–323. doi: 10.1086/706851
Zhou, J., Gong, X., Downie, S. R., Peng, H. (2009). Towards a more robust molecular phylogeny of Chinese Apiaceae subfamily Apioideae: additional evidence from nrDNA ITS and cpDNA intron (rpl16 and rps16) sequences. Mol. Phylogenet. Evol. 53, 56–68. doi: 10.1016/j.ympev.2009.05.029
Keywords: phylogenomics, divergence time, plastome, Shanopeucedanum, Sinopeucedanum
Citation: Song B-N, Liu C-K, Deng J-J, Tan W-Y, Zhou S-D and He X-J (2025) Genome skimming provides evidence to accept two new genera (Apiaceae) separated from the Peucedanum s.l.. Front. Plant Sci. 15:1518418. doi: 10.3389/fpls.2024.1518418
Received: 28 October 2024; Accepted: 19 December 2024;
Published: 20 January 2025.
Edited by:
Robert Philipp Wagensommer, Free University of Bozen-Bolzano, ItalyReviewed by:
Hengchang Wang, Chinese Academy of Sciences (CAS), ChinaEnrico Vito Perrino, University of Foggia, Italy
Copyright © 2025 Song, Liu, Deng, Tan, Zhou and He. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Song-Dong Zhou, enNkQHNjdS5lZHUuY24=
†These authors have contributed equally to this work
 Bo-Ni Song
Bo-Ni Song Chang-Kun Liu
Chang-Kun Liu Jiao-Jiao Deng1
Jiao-Jiao Deng1 Song-Dong Zhou
Song-Dong Zhou Xing-Jin He
Xing-Jin He