- Laboratory of Applied Biological Control, School of Agroecology, Mongolian University of Life Sciences, Ulaanbaatar, Mongolia
Mentha arvensis L. (M. arvensis) is an aromatic plant of the Mentha genus, renowned for its medicinal and economic importance. The primary components of its essential oils (EOs) are monoterpenoids, synthesized and stored in peltate glandular trichomes (PGTs). In general, the EO content in M. arvensis is relatively low. Methyl jasmonate (MJ) has been reported as an effective elicitor of terpenoid biosynthesis in medicinal plants, but the specific mechanisms underlying MJ’s influence on M. arvensis remain unclear. In this study, exogenous application of MJ significantly increased the EO content, yield, and PGT density in a dose-dependent manner. At a 5 mM dose, the EO content and PGT density peaked, with increases of 71.20% and 53.69%, respectively. Gas chromatography-mass spectrometry (GC-MS) analysis indicated that, in general, MJ treatment did not significantly alter the types or relative proportions of EO components of M. arvensis. However, L-menthol content decreased slightly by 7.90% under 5 mM MJ treatment. Transcriptome analysis identified 4,659 differentially expressed genes (DEGs) in MJ-treated leaves. KEGG enrichment analysis revealed that “Monoterpenoid biosynthesis” was among the most significantly enriched metabolic pathways. Key genes involved in jasmonic acid (JA) signaling (JAZs and MYCs) and monoterpenoid biosynthesis (GPPSs, LSs, L3Hs, and IPRs) were significantly up-regulated. Co-expression analysis, promoter binding element analysis and weighted gene co-expression network analysis (WGCNA) indicated that transcription factors (TFs) such as AP2/ERF, WRKY, MYB, and bHLH play crucial roles in regulating MJ-mediated monoterpenoid biosynthesis. Several key candidate TFs potentially involved in regulating monoterpenoid biosynthesis in M. arvensis were identified. These findings provide valuable insights into the molecular mechanisms regulating monoterpenoid accumulation in the Mentha genus.
1 Introduction
Mentha arvensis L., commonly known as Japanese mint, is widely distributed across various regions of the world. Its geographical range mainly includes Asia, Europe, and North America (Kumar et al., 2012; Vining et al., 2020). Cultivated primarily for the production of essential oil (EO), this plant thrives in moist, temperate climates and is often found in wetlands, riverbanks, and other water-abundant areas (Bussmann et al., 2020). The EO of Mentha species is widely utilized in medicine, cosmetics, food and other industries. It is primarily composed of monoterpenoids such as L-menthol, neomenthol, isomenthol, and carvone, which are synthesized and stored in peltate glandular trichomes (PGTs) of above-ground plant tissues (Ahkami et al., 2015; Lange, 2015). In M. arvensis, L-menthol is the predominant monoterpenoid, accounting for more than 70% of the total EO content (Batool et al., 2020). This compound is highly sought after in global markets for its cooling and fragrance properties, making it a key natural product in pharmaceutical and cosmetic applications (Khaliq and Mushtaq, 2023). However, the EO content in Mentha plants is relatively low, and the production does not meet the growing demand in pharmaceutical and other industries (Riaz et al., 2021).
The biosynthesis of terpenoids involves a series of biochemical processes, including precursor formation, intermediate conversion, end product generation, and post-modification (Mahmoud and Croteau, 2002). In mint, monoterpenoid biosynthesis starts with the methylerythritol phosphate (MEP) pathway, where pyruvate and 3-phosphoglyceraldehyde are converted into isopentenyl diphosphate (IPP) via six enzymatic steps. Key enzymes in this process include 1-deoxy-D-xylulose-5-phosphate synthase (DXS) and 1-deoxy-D-xylulose-5-phosphate reductoisomerase (DXR). IPP is then partially converted to dimethylallyl diphosphate (DMAPP) by isopentenyl diphosphate isomerase (IPPI), and these isomers combine through geranyl diphosphate synthase (GPPS) to form geranyl diphosphate (GPP) (Ahkami et al., 2015; Mahmoud and Croteau, 2002). Subsequently, GPP is converted to limonene by limonene synthase (LS), then catalyzed by limonene-3-hydroxylase (L3H), isopiperitenol dehydrogenase (IPD), isopiperitenone reductase (IPR), pulegone reductase (PR), and L-menthol reductase (MD), etc., resulting in the formation of valuable secondary metabolites such as menthone, isomenthone, and L-menthol (Mahmoud and Croteau, 2002; Croteau et al., 2005). Earlier studies on the Mentha genus primarily focused on identifying active compounds and conducting bioactivity assays (Zeljkovi et al., 2020; Mamadalieva et al., 2020). Recent research highlights the roles of structural genes in terpenoid biosynthesis. For instance, in Mentha piperita, overexpression of DXS and DXR and IPPI enhances its EO content by about 15%, 60% and 26%, respectively (Lange et al., 2011; Mahmoud and Croteau, 2001, 2002; Lange et al., 2011). While overexpression of GPPS enhanced Mentha piperita EO by 18% and increased tobacco limonene content approximately 35-fold (Yin et al., 2017; Lange et al., 2011).
Strategies to enhance terpenoid production now focus on modulating biosynthesis pathways and increasing the density or size of PGTs (Pandey et al., 2021; Pribat et al., 2013; Singh et al., 2024). Jasmonic acid (JA), a key phytohormone, plays a crucial role in plant defense and stress responses (Wang et al., 2020), while also regulating secondary metabolite synthesis and secretory structure development by reprogramming gene expression (Maes et al., 2011; Wasternack and Hause, 2013). For example, JA and its derivatives methyl jasmonates (MJ) promote terpenoid synthesis and PGT development in medicinal plants like Artemisia annua (A. annua), Isodon rubescens, and Chrysanthemum indicum var. aromaticum (Maes et al., 2011; Li, 2015; Zhang, 2021). High-throughput RNA sequencing (RNA-seq) has been extensively utilized to elucidate the molecular mechanisms of MJ in regulating terpenoid biosynthesis in plants. In Pogostemon Cablin, MJ treatment significantly increased the content of sesquiterpene patchouli alcohol, through upregulation of MEP pathway genes (Chen et al., 2019). In Sindora glabra, MJ increased the content of the sesquiterpenes α-copaene and β-caryophyllene, leading to the upregulation of most mevalonate (MVA) pathway genes, while the majority of MEP pathway genes were significantly down-regulated (Yu et al., 2021a). Although MJ treatment led to an increase in terpenoid content both in Sindora glabra and Pogostemon Cablin, the response patterns of structural genes in the MVA and MEP pathways differed between the two species. These findings highlight the species-specific mechanisms in JA-regulated terpenoid biosynthesis.
Transcription factors (TFs) induced by MJ are pivotal in regulating terpenoid biosynthesis and PGT development, such as AP2/ERF (apetala2/ethylene-responsive factor), bHLH (basic/helix-loop-helix), MYB (myeloblastosis DNA-binding protein), WRKY (WRKY-type DNA binding protein), and NAC (NAM, ATAF, and CUC domain protein) family genes. For example, the AP2/ERF family TF CrORCA3 in Catharanthus roseus and AaERF1 in A. annua positively regulate terpenoid synthesis by activating structural gene promoters (van der Fits and Memelink, 2000; Tan et al., 2015; Yu et al., 2012). Similarly, the bHLH TF MYC2 is a central regulator of JA signaling, coordinating transcriptional networks across multiple pathways. CrMYC2, induced by JA, promotes the biosynthesis of vinblastine by activating the transcription of CrORCA3, thereby actively regulating vinblastine synthesis (Pan et al., 2012; Sui et al., 2018). In A. annua, AaMYC2 can directly activate the expression of artemisinin synthesis structural genes, or actively regulate artemisinin synthesis through the MYC2-GSW1 (WRKY)-ORA (AP2/ERF) transcription cascade regulatory module (Chen et al., 2017; Shen et al., 2016). Negative regulators like JAZ proteins repress terpenoid biosynthesis by inhibiting TFs or structural genes. In A. annua, 9 AaJAZs bind to AabHLH1 and inhibit its activation of the artemisinin biosynthesis structural genes AaADS and AaCYP71AV1 (Li et al., 2019). Overexpression of AaJAZ8 significantly reduces the PGT density, AaJAZ8 negatively regulates the initiation of PGT by inhibiting the expression of AaSEP1 and AaHD1, thereby reducing artemisinin accumulation. When JAs and light are sufficient, AaJAZ8 is degraded by the 26S proteasome system, releasing AaSEP1, and promoting the activation of the AaSEP1-AaMYB16-AaHD1-AaGSW2 transcriptional cascade regulatory module (Chen et al., 2023; Xie et al., 2021a). Despite these advances, the regulatory mechanisms underlying monoterpenoid biosynthesis and PGT formation remain poorly understood.
In this study, MJ was used as an exogenous hormone to investigate its effects on the EO content, yield, and the PGT density in M. arvensis. Gas chromatography-mass spectrometry (GC-MS) was used to analyze chemical components in the EO. High-throughput RNA sequencing was performed to identify potential molecular regulatory mechanisms of MJ in regulating monoterpenoid biosynthesis of mint. This research provides new insights into molecular basis of monoterpenoid biosynthesis and lays the groundwork for further studies in Mentha species.
2 Materials and methods
2.1 Plant cultivation and long-term plant hormone treatment for essential oil and peltate glandular trichome analysis
A commercial variety of M. arvensis was propagated by planting root segments under natural conditions. Each root segment contained one growth point and was planted in a substrate composed of peat soil and perlite in a 3:1 ratio. Six plants were planted per pot, with pot dimensions of 49 × 21 × 15 cm. The experiment included 3 independent replicates. Approximately 10 weeks after planting, once the plants had developed 5 leaves and entered the vigorous growth phase, the plants were treated with varying concentrations of MJ or SA in order to screen for effective hormone. The MJ concentrations used were 10 μM, 100 μM, 1 mM, 5 mM, and 10 mM, the solution (Aladdin, Shanghai, China) was prepared with 0.2% Tween 20 (Sangon, Shanghai, China). The SA concentrations used were 100 μM and 1 mM, and the solution (Aladdin, Shanghai, China) was prepared with 0.2% Tween 20 (Sangon, Shanghai, China). The control group was treated with a 0.2% Tween 20 aqueous solution. Hormone treatments were applied weekly and sprayed on leaves until there was runoff for a total of 8 treatments. Twenty-four hours after the final treatment, the aboveground parts of the MJ-treated plants were harvested, and measurements were taken for fresh weight, PGT density, EO content, and yield. Similarly, twenty-four hours after the final treatment, the aboveground parts of the SA-treated plants were harvested, and measurements were taken for EO content.
2.2 MJ treatment and transcriptomic sampling strategy
M. arvensis plants were cultured in a temperature-controlled tissue culture room maintained at 25 °C with 70% humidity, under a 16-hour light/8-hour dark cycle with a light density of 150 μmol·m-2·s-1. Once the plants developed the first 5 leaves, they were transitioned to continuous light conditions and pre-cultured for one week. The plants were then sprayed with 1 mM MJ solution until runoff. The MJ solution (Aladdin, Shanghai, China) was prepared with 0.2% Tween 20 (Sangon, Shanghai, China). The control group was treated with a 0.2% Tween 20 aqueous solution. The third pair of leaf samples were collected at 0 hours (CK), 4 hours (H4), 8 hours (H8), and 24 hours (H24) after MJ treatment. Three biological replicates were taken for each time point. All samples were immediately frozen in liquid nitrogen and stored at -80°C for further analysis.
2.3 Essential oil extraction and its component analysis
The EO of M. arvensis was extracted via hydro-distillation using a Clevenger apparatus. A sample of 200 g of above-ground fresh plants was subjected to hydro-distillation for 50 minutes. EO content was expressed as a percentage of fresh weight (w/w). EO yield was quantified in grams per plant. The composition of the EO was analyzed using GC-MS. An Agilent 7890B gas chromatograph coupled with a 5977A mass spectrometer (Agilent Technologies, Santa Clara, USA) was employed for the GC-MS analysis. The chromatographic separation was performed on a DB-WAX column (30 m × 0.25 mm, 0.25 µm film thickness; Agilent Technologies, Santa Clara, USA). Helium was used as the carrier gas at a flow rate of 1 mL/min. The quadrupole temperature was maintained at 150°C, and the ionization mode was electron ionization (EI+) (Mahmoud and Croteau, 2001).
2.4 Peltate glandular trichome density analysis
From our previous preliminary experiments, we observed that the PGT density was the highest in young leaves and the lowest in old leaves. However, despite their higher PGT density, young leaves often contain immature PGTs that have not yet synthesized or stored EO. Consequently, leaves may contain PGTs at various developmental stages and sizes, complicating accurate observation and statistical analysis. The third and fourth pairs of leaves typically have mature and rounded PGTs filled with EO. Therefore, we selected the third pair of leaves of uniform size as representative samples for PGT density analysis. Fluorescence microscopy (Olympus, Tokyo, Japan) was used to capture images of the abaxial side of each leaf, focusing on the top, middle, and bottom sections. A 10x objective lens was used for imaging, and fluorescence was induced using UV light. The quantification of PGT was performed using ImageJ software (version 1.54k, National Institutes of Health, Bethesda, USA) with appropriate image processing and analysis protocols (Yan et al., 2017). This approach ensured precise quantification of trichome density across different leaf sections.
2.5 Total RNA extraction, cDNA library construction, and sequencing
Total RNA from plant leaves was extracted using the Plant RNA Extraction Kit (Takara, Dalian, China) following the manufacturer’s protocol. The purity and concentration of the extracted RNA were assessed with a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, USA). RNA samples with an A260/A280 ratio between 1.8 and 2.0, and an A260/A230 ratio above 2.0 were selected for further analysis. The RNA sequencing was performed by Biomarker Technologies (Qingdao, China). Sequencing libraries were prepared using the NEBNext® Ultra™ RNA Library Prep Kit for Illumina® (New England Biolabs, Ipswich, USA) following the manufacturer’s protocol, with index codes added for sample identification. mRNA was purified from total RNA using poly-T magnetic beads. First-strand and second-strand cDNA synthesis were carried out using M-MuLV Reverse Transcriptase, DNA Polymerase I, and RNase H. After adenylation of 3’ ends, NEBNext Adaptors were ligated. cDNA fragments (~240 bp) were purified using the AMPure XP system (Beckman Coulter, Beverly, USA). The USER enzyme was applied to the adaptor-ligated cDNA before PCR amplification using Phusion High-Fidelity DNA polymerase and specific primers. Finally, PCR products were purified and the library quality was assessed on an Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, USA). Clean reads were obtained by processing raw fastq data with in-house scripts to remove adapters, poly-N sequences, and low-quality reads, while calculating Q20, Q30, GC content, and sequence duplication levels for quality control. Clean reads were assembled de novo using Trinity software (version 2.8.4) with default parameters. Functional annotation of the assembled sequences was carried out using several databases, including the NR (NCBI non-redundant protein sequences), Pfam (Protein family), KOG/COG/eggNOG (Clusters of Orthologous Groups of proteins), Swiss-Prot (A manually annotated and reviewed protein sequence database), KEGG (Kyoto Encyclopedia of Genes and Genomes), and GO (Gene Ontology), to provide comprehensive insights into the biological functions of the identified genes (Li et al., 2023).
2.6 Differentially expressed genes identification and pathway enrichment analysis
Differentially expressed genes (DEGs) were identified using DESeq2 software with the threshold false discovery rate (FDR) < 0.05 and |log2 Fold Change| ≥ 1 (Love et al., 2014). The GO classification analysis and KEGG pathway enrichment analysis were performed using the GO database (https://github.com/tanghaibao/Goatools) and KOBAS program (http://kobas.cbi.pku.edu.cn/) with p < 0.05. ClusterProfiler (version 4.12.6, Yulab, Guangzhou, China) was used to visualize the enrichment results using bar graphs (Klopfenstein et al., 2018). Volcano plots were constructed and analyzed using TBtools software (version 2.119, Guangzhou, China), allowing for the identification of significant DEGs by plotting the log2 fold change against the -log10 p-value (Chen et al., 2020). Veen diagram analysis was performed using the VENNY platform (https://bioinfogp.cnb.csic.es/tools/venny/index). Heatmaps were constructed from log2(TPM) values using TBtools (Chen et al., 2020).
2.7 Transcription factor identify, gene co-expression analysis, and promoter binding element analysis
TFs were predicted using the Biomarker Cloud platform (http://www.biocloud.net/) (Du et al., 2021). Gene co-expression analysis was conducted via the SRplot platform (http://www.bioinformatics.com.cn) (Tang et al., 2023). DEGs involved in monoterpenoid biosynthesis of M. arvensis, along with key enzyme genes previously reported in monoterpenoid biosynthesis pathways (Lange and Croteau, 1999b; Lange et al., 2011), and differentially expressed transcription factors (DETFs), were used to construct a co-expression trend network diagram (Supplementary Table S1). Eight monoterpenoid biosynthesis enzyme genes (Supplementary Table S2) were mapped to the transcriptome database with a reference genome by local Blast, and the promoter sequence was searched. Promoter binding elements analysis was then performed using the PlantRegMap platform (https://plantregmap.gao-lab.org/) (Du et al., 2021).
2.8 Weighted gene co-expression network analysis
DEGs involved in monoterpenoid biosynthesis and DETFs were used to perform weighted gene co-expression network analysis (WGCNA). The WGCNA was conducted using the Biomarker Cloud platform (http://www.biocloud.net/) (Du et al., 2021). Five co-expressed modules were identified by WGCNA. Genes in module 5 were selected to predict and analyze protein–protein interaction (PPI) using the STRING database (https://cn.string-db.org). The interactions were filtered based on a confidence score threshold of 0.7 to ensure high reliability. The resulting PPI network was then imported into Cytoscape (version 3.10.2, Cytoscape Consortium, San Diego, USA). To identify hub genes, the CytoHubba plugin was employed, and the top 15 hub genes were selected based on their MCC scores, indicating their potential key roles in monoterpenoid biosynthesis biological processes under study (Li et al., 2024).
2.9 Quantitative real-time PCR analysis
The total RNA of the leaf samples was extracted using the RNAprep Pure Plant Kit (Tiangen, Beijing, China). The quality and concentration of the RNA were assessed by agarose gel electrophoresis (Major Science, Saratoga, USA) and Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific, Waltham, USA). The cDNA was synthesized using the HiScript II Reverse Transcriptase kit (Vazyme, Nanjing, China). The gene-specific primers were designed with Primer Premier software (version 6, PREMIER Biosoft International, Palo Alto, USA). The specific primers used are shown in Supplementary Table S3. Quantitative real-time PCR (qRT-PCR) was performed using AceQ qPCR SYBR Green Master Mix kit (Vazyme, Nanjing, China) and a Bio-Rad MiniOpticon Real-Time PCR machine (Bio-Rad, Hercules, USA). The PCR reaction conditions were: pre-denaturation at 95°C for 1 min, followed by denaturation at 95°C for 10 seconds, and annealing at 60°C for 30 seconds, a total of 40 cycles were performed. All the data were normalized using Actin gene as reference, and the gene expression level was calculated using 2−ΔΔCT (Li et al., 2023).
2.10 Data analysis
The values are presented as the mean ± standard deviation (SD) of at least 3 replicates. One-way analysis of variance (ANOVA) was performed using GraphPad Prism (version 8.0, GraphPad Software, San Diego, USA), and statistical significance was set at *P < 0.05, **P < 0.01, ***P < 0.001, and ****P< 0.0001 (Park et al., 2024).
3 Results
3.1 Effect of methyl jasmonate on the production of essential oil and peltate glandular trichome density in Mentha arvensis
The biosynthesis of metabolites in many medicinal plants is closely linked to defense mechanisms (Bednarek and Osbourn, 2009). MJ and salicylic acid (SA), two key defense-related phytohormones, were applied to mint leaves to evaluate their effects. MJ significantly increased the EO content, while SA showed minimal to no effect (Supplementary Figure 1). Further experiments demonstrated that MJ promoted M. arvensis EO synthesis in a concentration-dependent manner (Figures 1A–C). As the MJ concentration increased from 10 μM to 5 mM, a corresponding increase in EO content was observed. At the 5 mM dose of MJ, the EO content peaked, increasing by 71.20% compared to the control group. While high concentrations of MJ slightly reduced plant growth (e.g., a 9.51% decrease in fresh weight under 10 mM treatment), this did not hinder EO production. Notably, under 1 mM MJ treatment, the EO production increased by 58.50% compared to the control group, although the highest EO content was achieved under 5 mM MJ treatment.
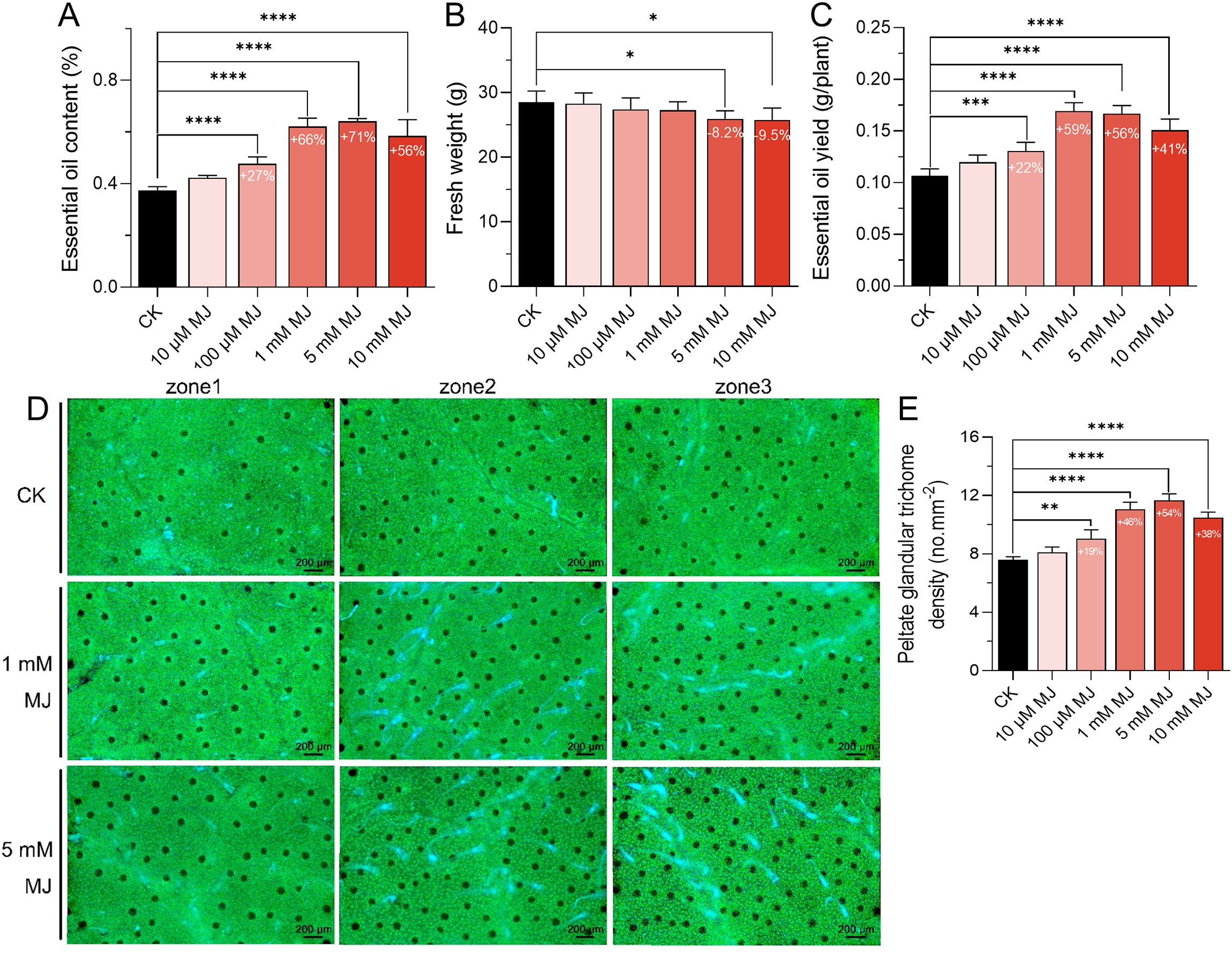
Figure 1. Effects of different concentrations of MJ on essential oil (EO) production and peltate glandular trichome (PGT) density. (A) EO content. (B) Plant growth. (C) EO yield. (D) PGTs were observed and recorded using a fluorescence microscope. The green background represents the autofluorescence of chlorophyll, the black dots indicate the autofluorescence of PGTs. (E) Histogram showing PGT density statistics. *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.001.
PGTs are where mint EO is synthesized and stored (Croteau et al., 2005). The density of mint PGT was assessed using fluorescence microscopy. The results showed that the trends in PGT density were closely aligned with the changes in EO content (Figures 1D, E). Specifically, the PGT density peaked under 5 mM MJ treatment, with an increase of 53.69% compared to the control group.
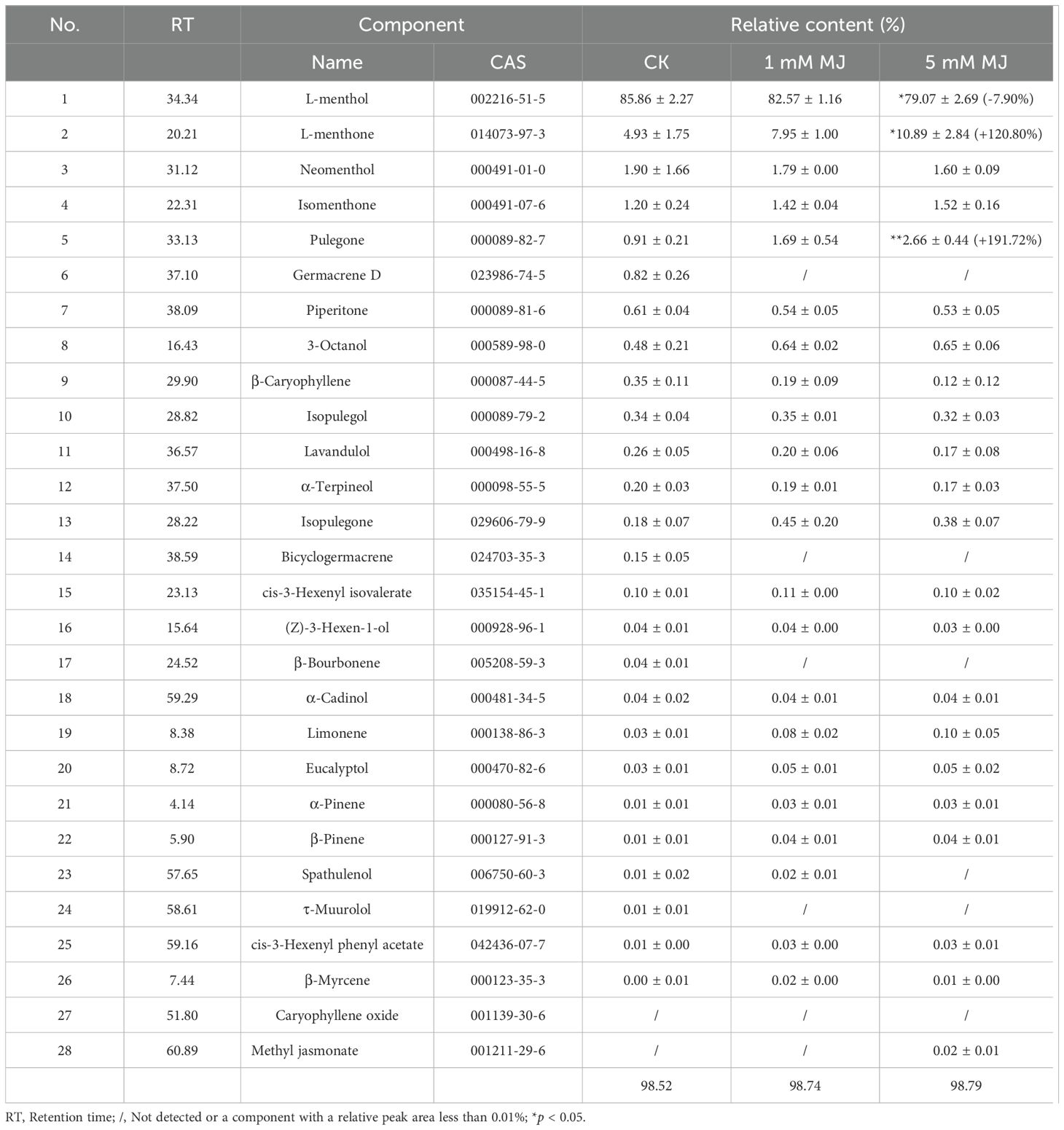
3.2 Effects of methyl jasmonate on the essential oil composition of Mentha arvensis
Across all treatments, 28 chemical components were identified in M. arvensis EOs, representing over 98.52% of the total composition (Table 1). In general, the types and relative proportions of the compounds identified remained consistently similar across all treatments. In each group, monoterpenoids were the predominant components. The top 5 compounds were L-menthol (79.07%~85.86%), L-menthone (4.93%~10.89%), neomenthol (1.60%~1.90%), isomenthone (1.20%~1.52%) and pulegone (0.91%~2.66%). These compounds represented over 94.80% of the total volatiles in each treatment. However, subtle variations were observed between different groups, particularly under the 5 mM MJ treatment. L-menthol, the predominant component of M. arvensis EO, serves as a key quality indicator (Adlard, 2010; Wei et al., 2023). In all treatments, L-menthol content exceeded 79.07%. The control group exhibited the highest L-menthol content (85.86%). As MJ concentration increased, L-menthol content decreased slightly. Under 1 mM MJ treatment, L-menthol content was 82.57%, while at a 5 mM MJ dose, it decreased to 79.07%, a significant reduction of 7.90% compared to the control group (p < 0.05). In contrast, under 5 mM MJ treatment, menthone and pulegone content increased significantly by 120.80% (p < 0.05) and 191.72% (p < 0.01), respectively, compared to the control.
3.3 RNA sequencing data and quality assessment
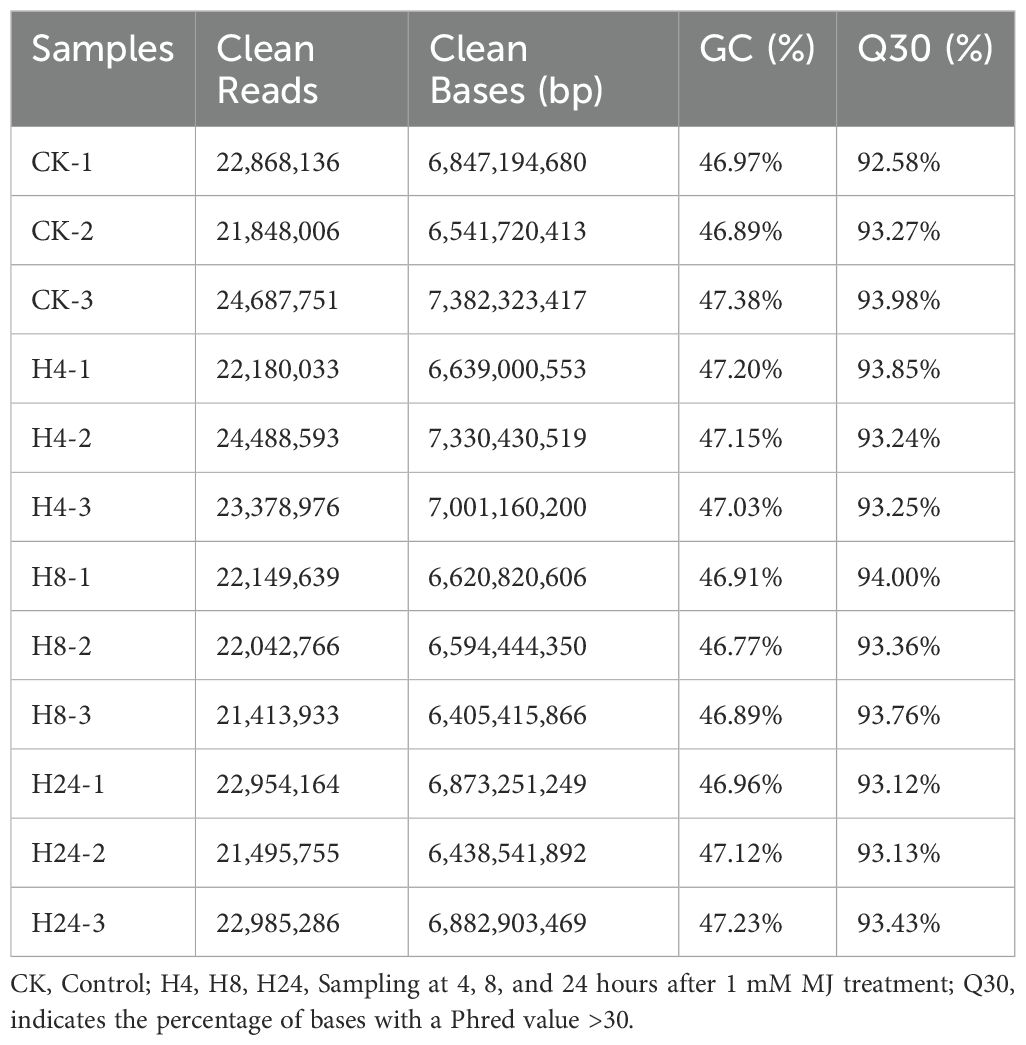
Transcriptome sequencing was performed to explore how MJ regulates EO biosynthesis and PGT development in M. arvensis at the transcriptional level. Total RNA was extracted from M. arvensis leaves at 0, 4, 8, and 24 hours after MJ treatment. Twelve cDNA libraries (3 replicates per treatment) were sequenced, yielding 81.56 Gb of clean data. Each library produced no less than 6.41 Gb of clean data. GC content ranged from 46.91% to 47.38% across the 12 transcriptome samples. The percentage of Q30 bases was at least 92.58% (Table 2). Sample replicate correlation analysis (Supplementary Figure S2A) showed that the Pearson’s correlation coefficients within the same treatment group were not less than 0.899. Furthermore, the principal component analysis (Supplementary Figure S2B) revealed that the values of PC1 and PC2 were 17.14% and 13.23%, respectively. These results indicated that the dataset is robust, with high reproducibility between samples, making it suitable for further analysis.
3.4 Functional annotation of unigenes and identification of differentially expressed genes
A total of 55,154 unigenes were annotated using public protein databases (NR, Swiss-Prot, GO, COG, KOG, KEGG, etc.) with a BLAST E-value cutoff of 1.0 × 10-5 (Supplementary Table S4). To evaluate the effect of MJ on M. arvensis gene expression, DEG analysis was performed. A total of 7,428 DEGs were identified across six comparative groups (Figure 2). In the CK vs H4 comparison, 2,765 DEGs were identified, including 1,317 up-regulated and 1,448 down-regulated genes. In the CK vs H8 comparison, 1,674 DEGs were identified, including 745 up-regulated and 929 down-regulated genes. In the CK vs H24 comparison, 2,443 DEGs were identified, including 1,828 up-regulated and 615 down-regulated genes. In the H4 vs H8 comparison, 2,069 DEGs were identified, including 929 up-regulated and 1,140 down-regulated genes. In the H4 vs H24 comparison, 4,511 DEGs were identified, including 2,971 up-regulated and 1,540 down-regulated genes. In the H8 vs H24 comparison, 3,885 DEGs were identified, including 2,731 up-regulated and 1,154 down-regulated genes (Figures 2A, B; Supplementary Figure S3). In addition, a total of 4,659 DEGs were identified across the CK vs H4, CK vs H8, and CK vs H24 comparisons. The analysis revealed that 96 genes were consistently up-regulated, while 124 genes consistently down-regulated after MJ treatment (Figure 2C). Over time, different genes showed varied expression patterns in response to MJ (Figure 2D). Overall, MJ significantly affected gene transcription in M. arvensis.
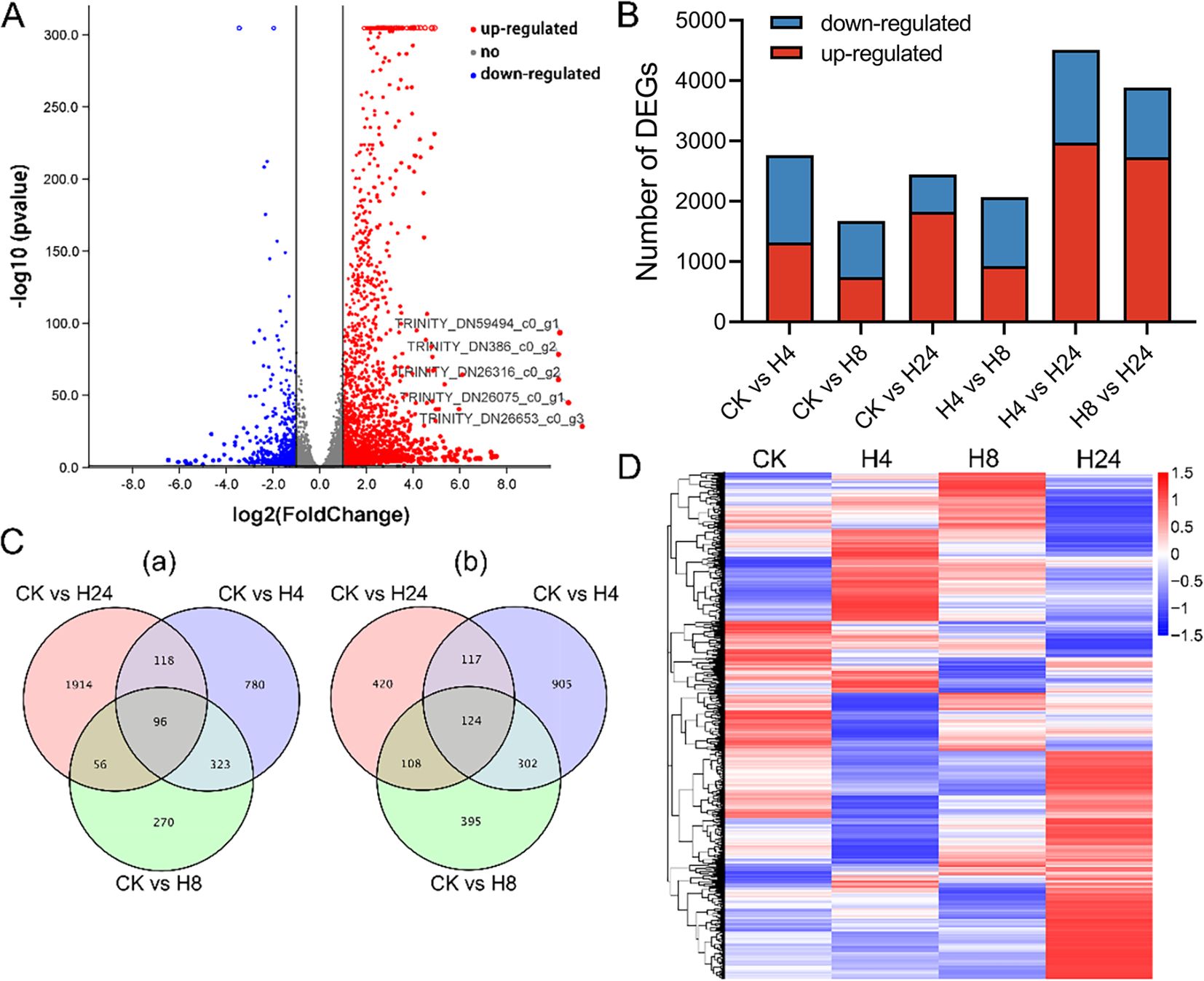
Figure 2. Characteristics of DEGs under MJ treatment. (A) Volcano plot of MJ treatment for 24 hours. (B) Histogram showing DEG expression changes under MJ treatment for 4, 8, and 24 hours. (C) Venn diagrams of up-regulated and down-regulated genes: (a) Up-regulated genes; (b) Down-regulated genes. (D) Heat map of DEGs.
3.5 GO enrichment and KEGG pathway analysis of differentially expressed genes
GO enrichment analysis was performed to explore the potential functions of these DEGs from the CK vs H4, CK vs H8, and CK vs H24 comparisons (Figure 3), categorizing them into 3 major groups: biological process (BP), cellular component (CC), and molecular function (MF). Within BP, the top 4 subcategories were “metabolic process”, “cellular process”, “single-organism process” and “biological regulation”. In CC, the top 3 subcategories were “membrane”, “cell” and “cell part”. In MF, most DEGs clustered in “binding”, followed by “catalytic activity” (Figure 3A). Notably, 1,569 DEGs were annotated in the “metabolic process” subcategory, making it the most abundant category in the GO enrichment analysis. Additionally, 409, 203, 45, 41, and 20 genes were annotated in the subcategories of “response to stimulus”, “detoxification”, “immune system process”, “signal transducer activity”, and “antioxidant activity”, respectively (Figure 3A; Supplementary Table S5). These subcategories have been shown to play key roles in plant resistance to stress.
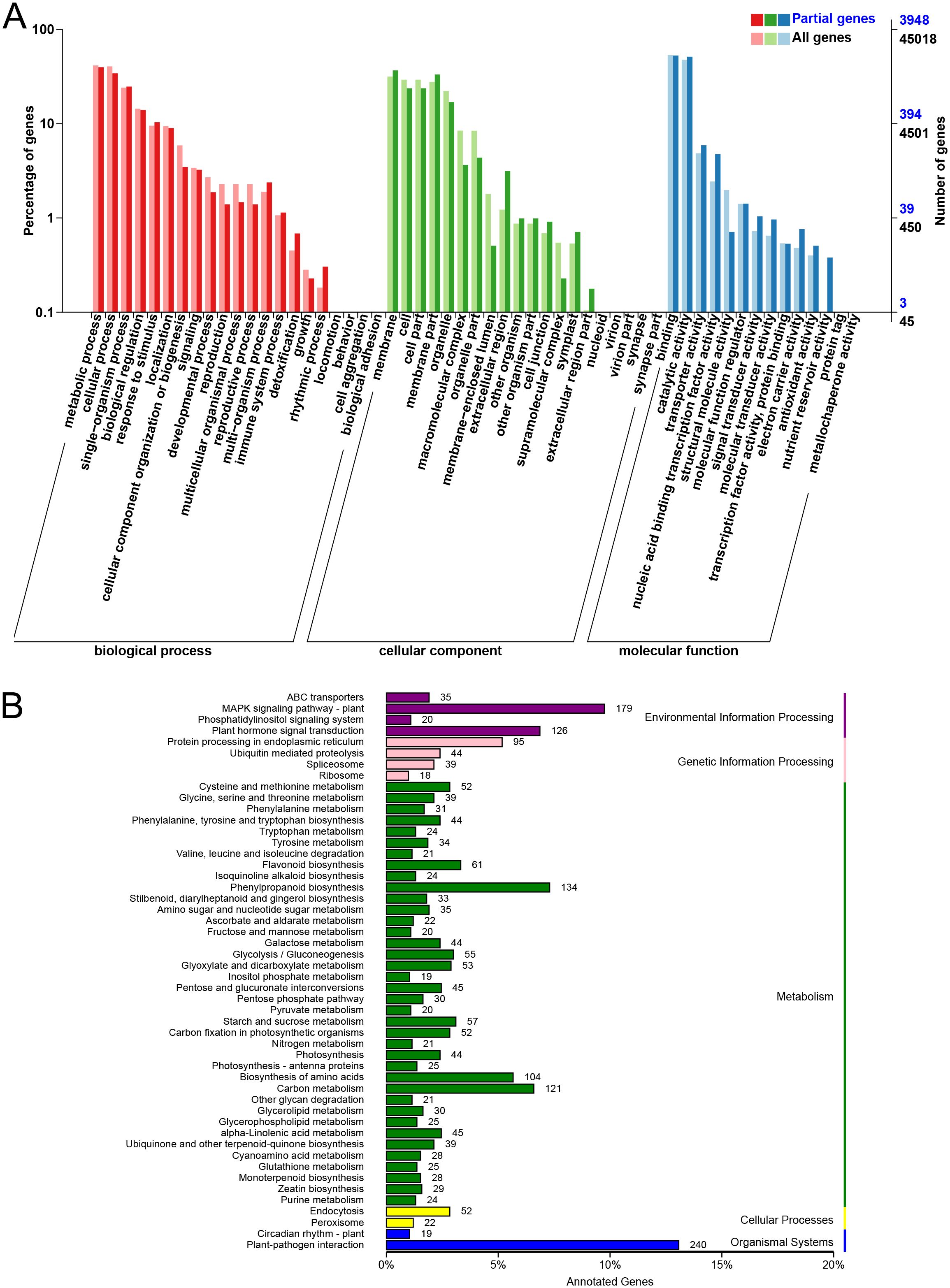
Figure 3. GO enrichment and KEGG pathway characterization of DEGs after MJ treatment. (A) GO enrichment analysis. (B) KEGG pathway analysis.
KEGG pathway analysis showed that 130 pathways were significantly enriched under MJ treatment. DEGs were predominantly enriched in the plant-pathogen interaction (240 genes), MAPK signaling (179 genes), phenylpropanoid biosynthesis (134 genes), and plant hormone signal transduction pathways (126 genes). Notably, 28 genes were enriched in the monoterpenoid biosynthesis pathway (Figure 3B; Supplementary Table S6). Additionally, 12 genes (1.17%), 9 genes (1.53%), and 19 genes (2.13%) were enriched in this pathway in the CK vs H4, CK vs H8, and CK vs H24 comparisons, respectively (Supplementary Figure S4), indicating that H24 induced a higher number of genes involved in monoterpenoid biosynthesis. For example, in the CK vs H24 comparison, 3 genes encoding cytochrome P450 enzyme (CYP450) (TRINITY_DN14738_c1_g1, TRINITY_DN32509_c0_g1, and TRINITY_DN46431_c0_g1), 2 genes encoding IPR (TRINITY_DN60151_c0_g1 and TRINITY_DN90669_c0_g1), 1 gene encoding LS (TRINITY_DN5134_c0_g1), and 1 gene encoding MD (TRINITY_DN28821_c1_g1 encoded) were identified.
3.6 Gene expression profiles related to JA signaling and monoterpenoid biosynthesis
In Arabidopsis thaliana, the F-box protein coronatine insensitive 1 (COI1) acts as the receptor for JA. Perception of JA-Ile by the SCFCOI1 complex triggers the degradation of JAZ proteins through the 26S proteasome. This process activates downstream TFs involved in JA responses, such as Octadecanoid responsive 3 (ORA3), a member of the APETALA2/ethylene responsive factor (AP2/ERF) family (Montiel et al., 2011; Yu et al., 2021b). In this study, 10 DEGs were identified as key factors of JA signaling in M. arvensis (Figure 4A). Among them, 1 JAR6 gene (TRINITY_DN3071_c0_g1) and 4 JAZ9 genes (TRINITY_DN16183_c0_g1, TRINITY_DN19929_c0_g1, TRINITY_DN11251_c0_g1 and TRINITY_DN834_c0_g2) were significantly up-regulated at 4 hours after MJ treatment. Two MYC2 genes (TRINITY_DN4107_c0_g2 and TRINITY_DN16914_c0_g1) were significantly up-regulated at 24 hours after MJ treatment (Figure 4A; Supplementary Table S1).
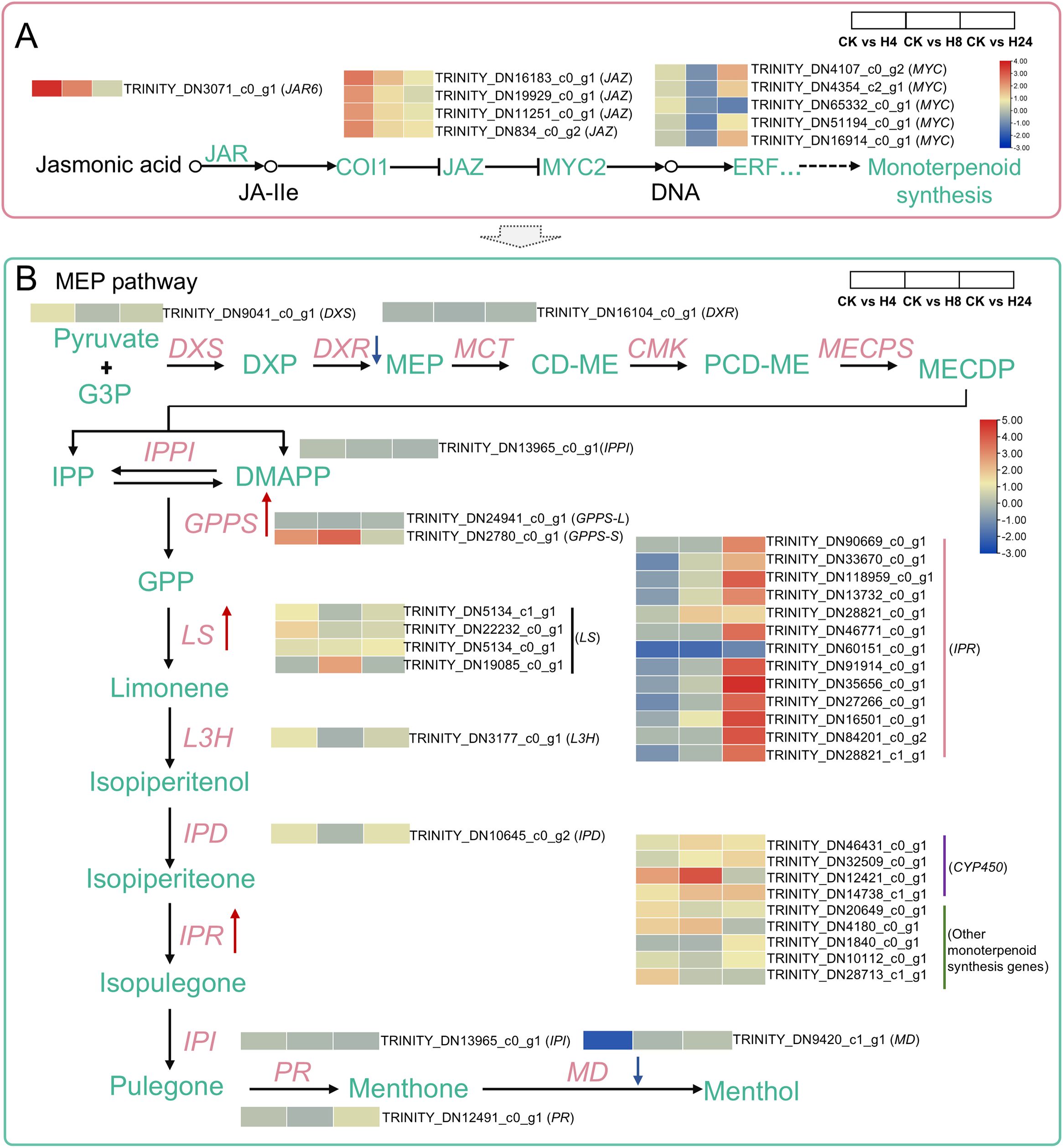
Figure 4. Heatmaps of the DEGs in the JA signaling and monoterpenoid biosynthesis pathways under MJ treatment. (A) JA signaling pathway: JAR6, Jasmonoyl-L-amino acid synthetase; COI1, Coronatine insensitive 1; JAZ, Jasmonate ZIM domain protein; MYC2, bHLH transcription factor MYC2. (B) Monoterpenoid biosynthesis pathway: DXS, 1-deoxy-D-xylulose-5-phosphate synthase; DXR, 1-deoxy-D-xylulose-5-phosphate reductoisomerase; MCT, 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase; CMK, 4-diphosphocytidyl-2-C-methyl-D-erythritol kinase; MECPS, 2-C-methylerythritol-2,4-cyclodiphosphate synthase; IPPI, Isopentenyl diphosphate isomerase; GPPS, Geranyl diphosphate synthase; LS, Limonene synthase; L3H, Limonene-3-hydroxylase; IPD, trans-Isopiperitenol dehydrogenase; IPR, Isopiperitenone reductase; IPI, cis-Isopulegone isomerase; PR, Pulegone reductase; MD, L-menthol dehydrogenase.
In M. arvensis, monoterpenoids are the primary components of its EOs. KEGG enrichment results indicated that “Monoterpenoid biosynthesis” was one of the most significantly enriched pathways under MJ treatment. Twenty-eight DEGs associated with monoterpenoid biosynthesis were identified in response to MJ treatment. Notably, GPPSs were significantly up-regulated at 4 and 8 hours after MJ treatment, 4 LS genes were up-regulated both at 4, 8 and 24 hours, and 12 IPR genes were significantly up-regulated at 24 hours after MJ treatment. CYP450, catalyze the decoration of terpenoid basic skeletons and thereby contribute significantly to their structural diversity (Weitzel and Simonsen, 2015). In this study, 4 CYP450 genes were identified as significantly expressed (Figure 4B; Supplementary Table S1). L-menthol is the predominant component of M. arvensis EO, which is synthesized through a series of enzymatic reactions. The enzymatic catalysis mechanism of L-menthol synthesis has been extensively studied in 3 Mentha varieties: Mentha piperita, Mentha spicata, and Mentha haplocalyx (Ahkami et al., 2015). Using reference genes from these varieties as queries, 11 orthologous genes were identified in M. arvensis (Supplementary Table S1), including the previously reported LS and MD genes (Akhtar et al., 2017; Wang et al., 2013).
3.7 Identification of DETFs and co-expression analysis with monoterpenoid biosynthesis genes
TFs regulate plant growth, development, and secondary metabolite synthesis (Yang et al., 2012). In this study, 260 DETFs belonging to 22 families were identified after MJ treatment for 4, 8, and 24 hours (Figure 5A; Supplementary Table S1). The top 8 families with the highest number of DEGs were AP2/ERF (42 genes), MYB (including MYB-related, 38 genes), WRKY (33 genes), NAC (22 genes), HSF (heat shock transcription factors, 16 genes), bHLH (14 genes), C2H2 (C2H2 zinc-finger protein, 14 genes), and GRAS (GAI-RGA-and-SCR, 14 genes). These TFs responded positively to MJ treatment. To further explore the regulatory relationship between TFs and monoterpenoid biosynthesis, a co-expression pattern analysis of DETFs and monoterpenoid biosynthetic genes was performed (Figure 5B). The analysis identified 7 gene clusters. Each cluster exhibited a unique expression pattern, indicating a close association between TFs and monoterpenoid biosynthetic enzyme genes. Most of the highly expressed monoterpenoid biosynthesis genes (Log2FC > 3) were grouped into cluster 6. Therefore, further analysis focused on this cluster. In cluster 6, the top 3 TF families with the highest number of DEGs were AP2/ERF (17 genes), WRKY (16 genes), and MYB (10 genes) (Figure 5B; Supplementary Table S2). In this study, 8 monoterpenoid biosynthesis genes, including DXS, DXR, GPPS-L, LS, IPR, PR, and MD, which are either highly expressed genes (Log2FC > 3) or key genes in this process as previously reported (Lange and Croteau, 1999a; Lange et al., 2011; Mahmoud et al., 2004) were selected for promoter analysis. These genes contained the highest number of AP2/ERF binding sites (2,763), followed by bHLH (266), WRKY (251), and MYB (197) (Figure 5C; Supplementary Table S2).
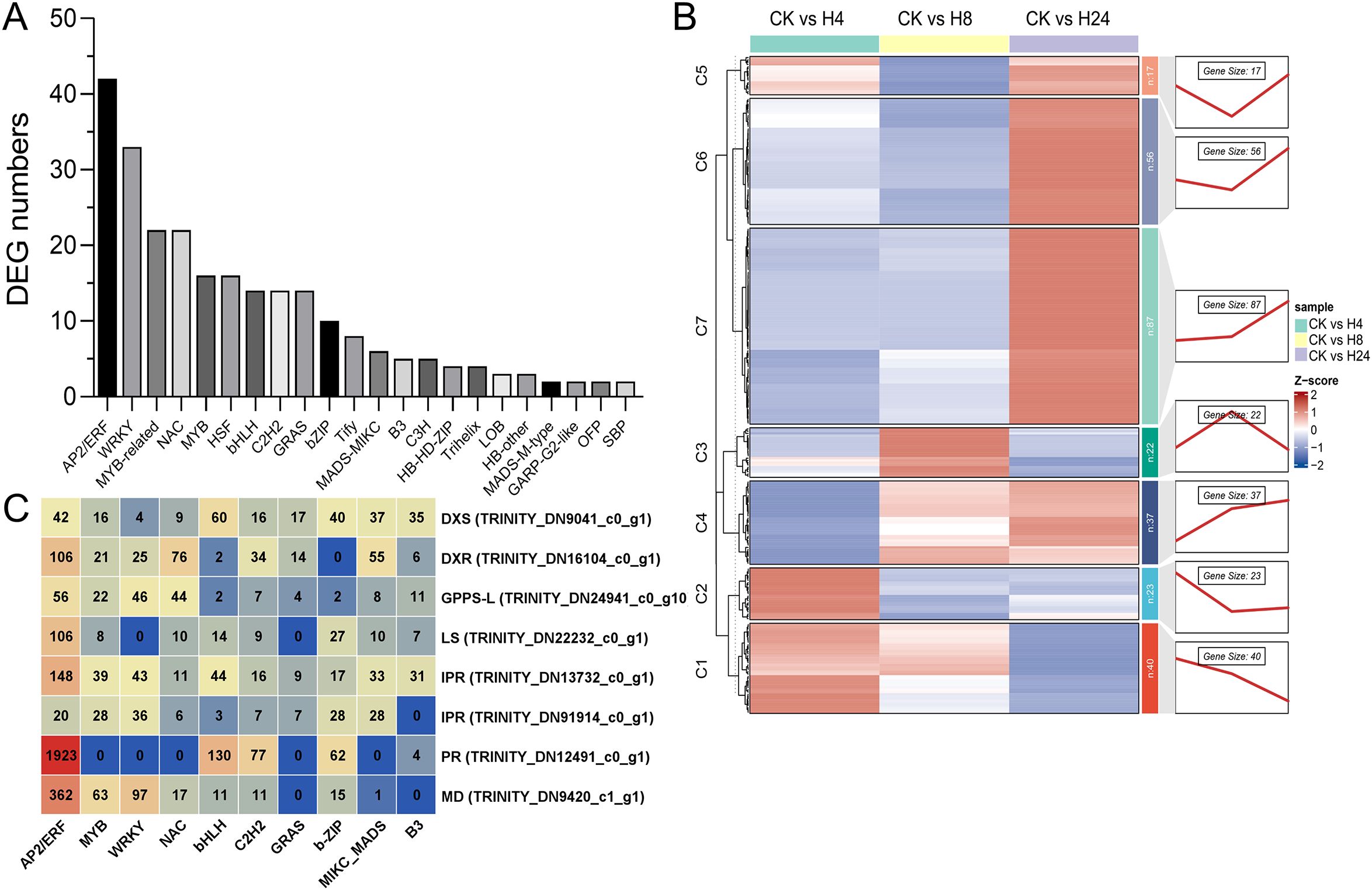
Figure 5. Analysis of the co-expression patterns of monoterpenoid biosynthesis genes and differentially expressed transcription factors (DETFs). (A) Identification of DETFs. (B) Gene clusters classified based on co-expression patterns. (C) Statistics of promoter binding elements for key enzyme genes.
3.8 WGCNA of differentially expressed monoterpenoid biosynthesis genes and transcription factors
WGCNA was employed to construct a gene co-expression network, aiming to identify DETFs involved in MJ-induced monoterpenoid biosynthesis. Hierarchical clustering identified 5 co-expressed modules (Figure 6A). Most of the high expressed monoterpenoid biosynthesis genes (10 out of 11, Log2FC > 3) peaked at 24 hours after MJ treatment and were classified in module 5 (Figure 6B; Supplementary Table S7). KEGG analysis was performed for this module. Among these, 18 unigenes were annotated in the plant hormone signal transduction pathway (35.29%), 16 unigenes in the plant pathogen interaction pathway (31.37%), and 11 unigenes in the monoterpenoid biosynthesis pathway (21.57%) (Supplementary Figure S4). Additionally, module 5 was highly associated with the H24 group (R = 0.99, p = 0.006). These results closely mirrored the analysis of the monoterpenoid biosynthesis pathway in Figure 4, prompting a focus on module 5 (Figure 6B; Supplementary Table S7). Furthermore, all genes of module 5 were used to conduct PPI analysis, the results were visualized using Cytoscape, and hub genes were identified using CytoHubba (Figure 6C). A venn diagram analysis was used to identify common genes between the top 15 hub genes from the CytoHabba analysis and cluster 6 genes from the co-expression analysis (Log2FC > 1, FPKM > 10) (Figure 5B), which were considered as candidate genes. The results revealed 5 common genes, including 4 AP2/EFR genes—TRINITY_DN8517_c0_g1 (ERF108), TRINITY_DN4459_c0_g1 (ERF1B), TRINITY_DN11107_c0_g1 (DREB1D), TRINITY_DN688_c0_g1 (DREB1C), and 1 WRKY gene TRINITY_DN1548_c0_g1 (WRKY33), which were significantly up-regulated (Log2FC > 2) at 24 hours after MJ treatment (Figures 6D, E).
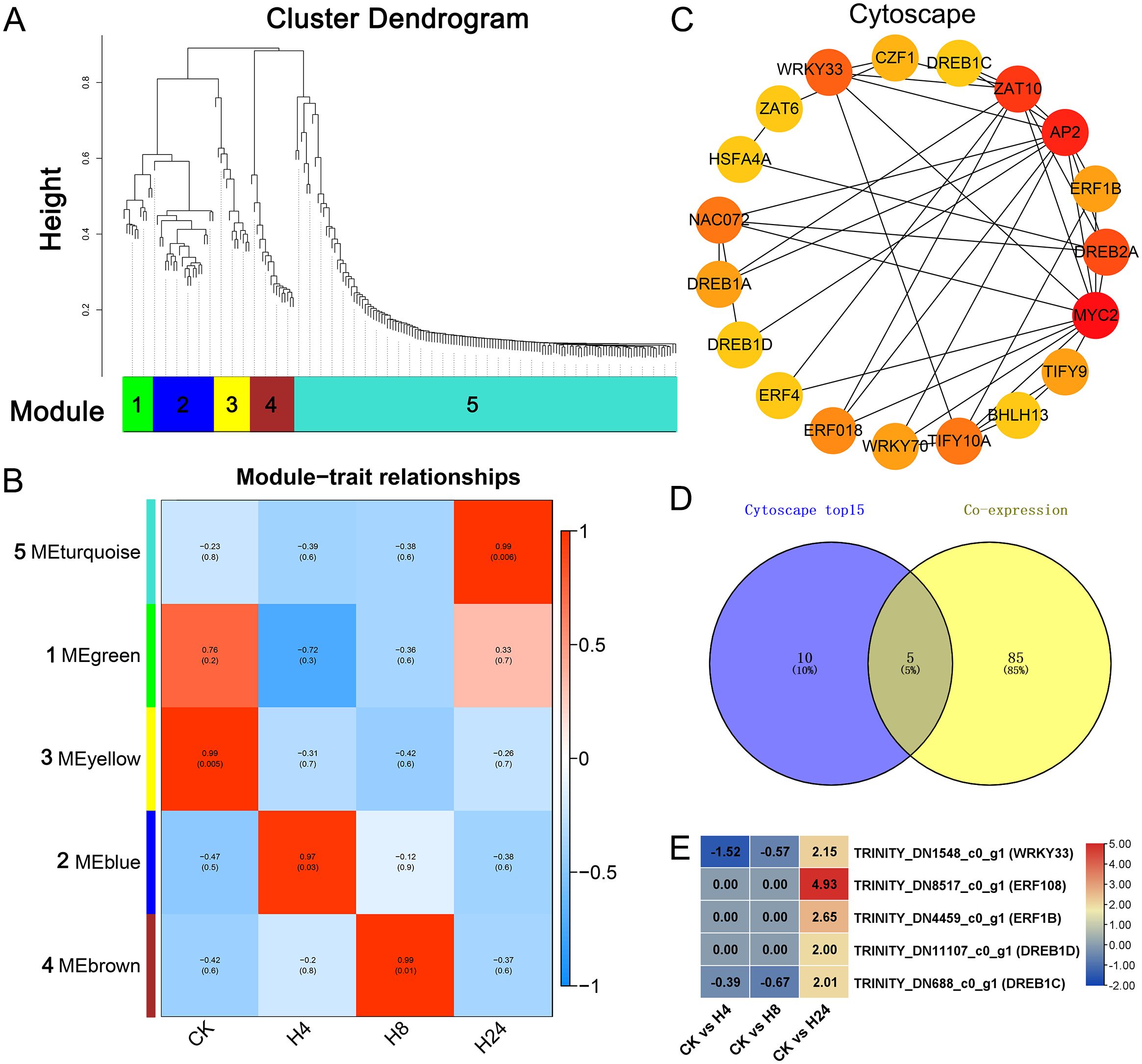
Figure 6. WGCNA of differentially expressed monoterpenoid biosynthesis genes and transcription factors. (A) Hierarchical clustering of differentially expressed gene modules. (B) Module-trait relationships. (C) Cytoscape visualization of the top 15 connectivity pairs in module 5. (D) Venn diagram analysis of common genes between module 5 from WGCNA and cluster 6 from co-expression analysis. (E) Gene expression analysis of common genes identified in the Venn diagram analysis.
3.9 Validation of RNA-seq data
A qRT-PCR assay with independent samples from the control and MJ treatment group (H24) was conducted to verify the expression changes of several key genes involved in monoterpenoid biosynthesis. In total, 10 genes, including 6 from the monoterpenoid biosynthesis pathway, and 4 TFs were selected to confirm the RNA-seq data. The expression levels of these selected genes, as determined by qRT-PCR, were generally consistent with the RNA-seq results (Figure 7).
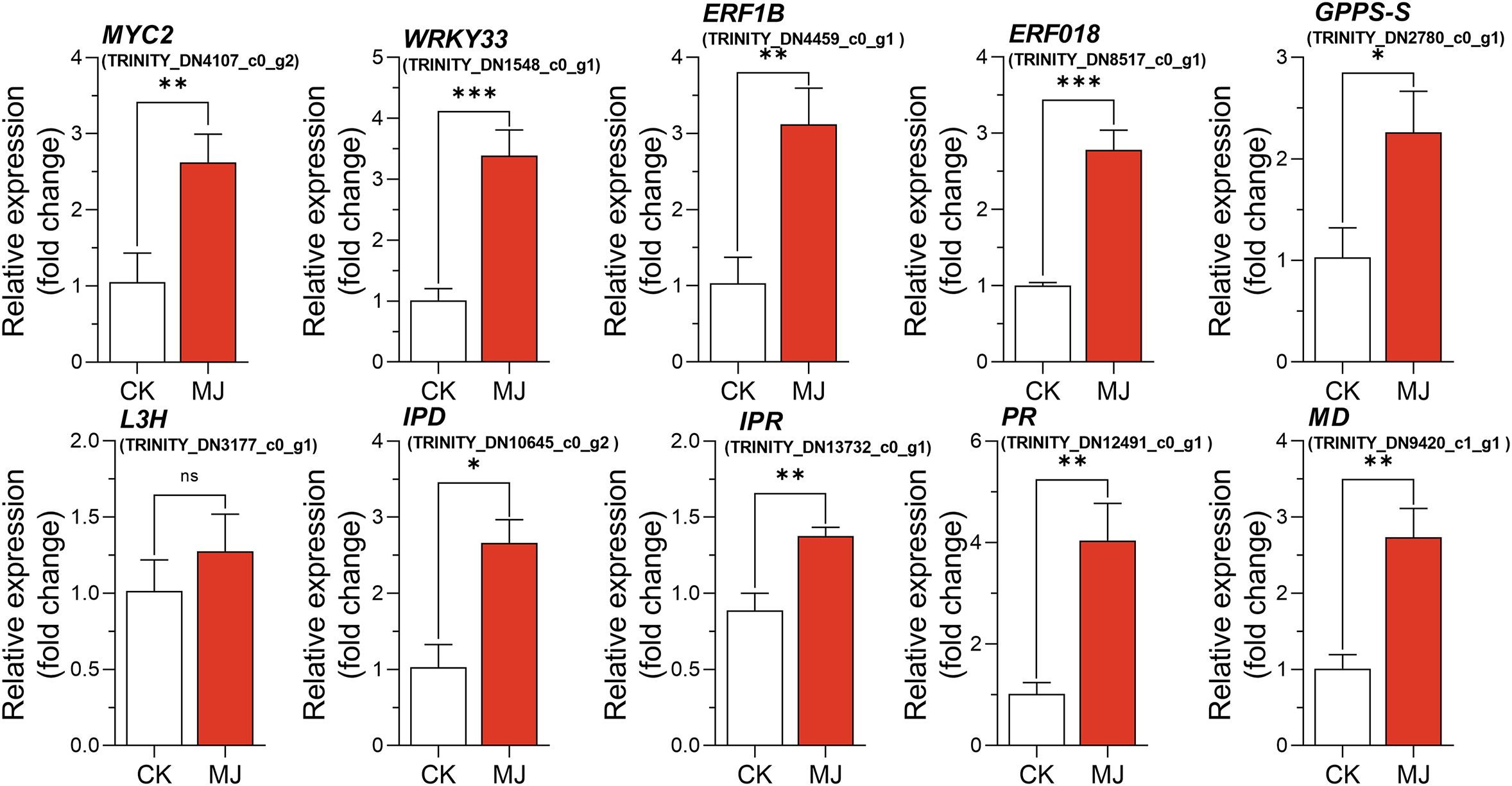
Figure 7. Quantitative real-time PCR verification of the RNA-seq data of 10 selected genes in the MJ treatment group (H24) and the control group (CK). *P < 0.05, **P < 0.01, ***P < 0.001.
4 Discussion
The EO of M. arvensis, rich in monoterpenoids like L-menthol and menthone, is highly valued for its medicinal properties and is widely used in industries such as medicine, cosmetics, and food (Kumar et al., 2012; Souza et al., 2020). Previous studies have shown that exogenous defense hormones such as MJ and SA can promote the biosynthesis of plant secondary metabolites including terpenoids, anthocyanins and alkaloids (Kumari et al., 2018; Li et al., 2024; Xiang et al., 2015). In some cases, these hormones also increase the density of PGT, as observed in Artemisia annua (Kumari et al., 2018). In our study, MJ treatment significantly enhanced EO content in M. arvensis, whereas SA showed minimal to no effect (Supplementary Figure S1). Although SA is known to antagonize JA signaling pathways and can reduce JA levels, such as in Arabidopsis thaliana (Barbara and David, 2002), its lack of impact on EO content in M. arvensis may explained by several factors. First, plant genotype plays a critical role. For example, in Pyrus pyrifolia (pear), exogenous SA treatment had minimal effect on the total amount of cuticle wax in leaves (mainly composed of terpenoids), suggesting insensitivity of terpenoid biosynthesis to SA in this species (Wu et al., 2018). Second, EO synthesis in M. arvensis may involve mechanisms beyond trichome development, which could make it less responsive to SA-induced interference (Han et al., 2022). Third, the dose, frequency, and application method of SA are important factors. Studies have shown that applying SA once or twice can promote the accumulation of terpenoid lactones, while increasing the frequency to three or four applications suppresses their synthesis (Wang et al., 2021a). Lastly, it remains necessary to confirm the changes in endogenous levels of these hormones following JA and SA treatments in this study to fully understand their interactions.
JA plays a critical role in increasing secondary metabolite production in aromatic and medicinal plants (Patt et al., 2018; Wasternack and Strnad, 2019). For example, in Picea abies, JA treatment induced the accumulation of monoterpenoid and diterpene in resin ducts (Schmidt et al., 2011). Similarly, MJ treatment increased the artemisinin content by about 300% in A. annua (Kumari et al., 2018; Maes et al., 2011) and the EO content by 36.80% in Ocimum basilicum (Zlotek et al., 2016). Furthermore, MJ promotes trichome development, not only in both non-glandular trichome-bearing species like Arabidopsis thaliana and Medicago truncatula (Maes and Goossens, 2010; Traw and Bergelson, 2003), but also in glandular trichome-producing species such as Solanum lycopersicum and A. annua (Maes and Goossens, 2010; Maes et al., 2011). In this study, PGT density was increased by 53.69% under 5 mM MJ treatment in M. arvensis. These results suggest that JA is a potent inducer of EO production through stimulating both monoterpenoid biosynthesis and PGT development in M. arvensis. Although MJ significantly increased the EO content, it also inhibited mint growth in a concentration-dependent manner. Treatment with 10 mM MJ reduced the fresh weight of M. arvensis by 9.51% (Figure 1B), consistent with previous studies indicating that excessive JA accumulation can suppress gibberellin biosynthesis, thereby delaying plant growth (Heinrich et al., 2013). In this study, while high concentrations of MJ slightly reduced L-menthol content, the 1 mM MJ treatment achieved the highest EO yield without significantly affecting L-menthol levels. These findings highlight the importance of optimizing MJ doses to maximize EO production (Table 1).
To further elucidate the molecular mechanisms underlying MJ’s effects on EO biosynthesis, RNA-seq analysis was conducted. A total of 7,428 DEGs were identified across six comparison groups (Figure 2). Interestingly, the observed increase in DEGs between H4 vs H24 compared to the CK vs H24, suggests a dynamic transcriptional response to MJ (Figure 2B). This pattern aligns with the known rapid and transient nature of JA-induced gene expression. Previous studies have shown that JA signaling triggers immediate transcriptional changes, peaking within a few hours of treatment, followed by a decline as the response stabilizes (Hickman et al., 2017). In our study, the H4 sample likely captures this peak transcriptional activity, encompassing genes involved in defense responses, secondary metabolite biosynthesis, and stress signaling pathways. By 24 hours (H24), many of these responses may have been downregulated due to feedback mechanisms or metabolic adjustments, resulting in fewer DEGs when compared to CK. For example, some core factors of JA signaling were highly expressed in H4, while some were decreased in H24 (Figure 4). GO enrichment analysis revealed that MJ treatment significantly influenced metabolic processes, with DEGs predominantly involved in the “response to stimulus”, “detoxification”, “immune system process”, and “signal transducer activity” categories (Figure 3A; Supplementary Table S2), which are crucial for plant responses to biotic and abiotic stress (Chi et al., 2018; Li et al., 2013; Nie et al., 2020). KEGG analysis revealed that DEGs were enriched in key pathways such as plant-pathogen interaction, MAPK signaling, phenylpropanoid biosynthesis, plant hormone signal transduction, and monoterpenoid biosynthesis (Figure 3B; Supplementary Table S6). These results indicate that JA may enhance terpenoid production in plants to effectively respond to environmental challenges (Unsicker et al., 2009).
The core elements of JA signaling were significantly differentially expressed in this study, including 1 JAR6, 4 JAZ9, and 2 MYC2 genes (Figure 4A; Supplementary Table S1). The result is consistent with previous research in Chrysanthemum indicum var. aromaticum (Gao et al., 2020), indicating that MJ mediates JA signaling pathways, thereby regulating a range of downstream genes in M. arvensis. Monoterpenoids, the primary constituents of M. arvensis EO, are crucial for plant defense against pathogens and herbivores (Unsicker et al., 2009). KEGG analysis indicated that MJ treatment triggered significant differential expression of 28 genes involved in monoterpenoid biosynthesis and 12 genes related to terpenoid backbone biosynthesis, including genes encoding GPPS, LS, IPR, MD, and CYP450 enzymes (Figure 4B; Supplementary Tables S1, S6). Notably, early-stage genes such as DXS showed peak expression at H4, intermediate-stage genes like GPPS and LS peaked at H8, and late-stage genes like IPR and MD reached their highest expression at H24 (Figure 4B; Supplementary Figure S1). These findings align with previous studies on peppermint—in the later stages of monoterpenoid biosynthesis, L-menthol becomes the predominant component. Correspondingly, structural genes involved in the later reactions, such as MD, exhibit a delayed developmental timeline compared to the earlier enzymes in the monoterpenoid biosynthesis pathway (Turner et al., 2000).
The production and the accumulation of terpenoids, meticulously controlled in plants in a spatiotemporal manner, are orchestrated by TFs (Singh et al., 2024). In this study, 260 DETFs belonged to 22 families, including AP2/ERF, WRKY, MYB, and bHLH etc., were identified in M. arvensis after MJ treatment (Figure 5A, Supplementary Table S1). Many TFs have been reported to regulate the terpenoid biosynthesis, including bHLH, MYB, WRKY, AP2/ERF, TCP, bZIP, and NAC in previous studies (Su et al., 2019; Zhang et al., 2022). These TFs enhance terpenoid accumulation by directly or indirectly promoting the transcriptional activation of both TF and structural genes. For instance, AtMYB21 in Arabidopsis thaliana and FhMYB21 in Freesia hybrida promote terpenoid accumulation by binding to TPS promoters (Yang et al., 2020). In A. annua, bHLH1 and bHLH112 significantly promote the accumulation of sesquiterpene artemisinin by binding to the promoters of the artemisinin synthesis genes and the transcription factor ERF1, respectively (Ji et al., 2014; Xiang et al., 2019). In Phalaenopsis orchids, bHLH4 increases the monoterpenoid content approximately 950-fold by activating the GPPS promoter (Chuang et al., 2018). In Dendrobium officinale, bHLH4 and in Arabidopsis thaliana, MYC2 promote terpenoid synthesis by binding to the TPS gene (Hong et al., 2012; Yu et al., 2021c). In Catharanthus roseus and A. annua, MYC enhances terpenoid synthesis by regulating AP2/ERF and WRKY transcription factors (Chen et al., 2017; Paul et al., 2017). In A. annua, under JA induction, WRKY9 activates the transcription of GSW1, thereby positively regulating artemisinin synthesis (Fu et al., 2021; Jiang et al., 2016). AP2/ERF transcription factors AaERF1, AaERF2, AaTAR1 and AaORA are induced by JA and positively regulate artemisinin synthesis by binding to the promoter of artemisinin synthesis genes (Tan et al., 2015; Yu et al., 2012). NACs in A. annua and Actinidia arguta can both promote monoterpenoid synthesis by activating TPS expression (Nieuwenhuizen et al., 2015). Despite significant progress in understanding the molecular mechanisms of plant terpenoid biosynthesis, certain key mechanisms remain poorly understood. Additionally, different TFs have specific roles in regulating terpenoid biosynthesis in various plants, and these regulatory mechanisms may vary between species (Singh et al., 2024). In this study, co-expression analysis revealed that AP2/ERF, WRKY, and MYB families were strongly associated with monoterpenoid biosynthesis genes (Figure 5B; Supplementary Table S2). Promoter analysis indicated that key monoterpenoid biosynthesis genes contained binding sites for these TFs (Figure 5C), suggesting that AP2/ERF, WRKY, and MYB may play important roles in regulating MJ-induced monoterpenoid biosynthesis in M. arvensis. Furthermore, WGCNA, PPI, and CytoHubba analysis revealed that MYC2 is positioned at the center of the regulatory network and is involved the biosynthesis of various secondary metabolites, such as anthocyanins and terpenoids (Luo et al., 2023). A venn analysis identified 5 common genes between cluster 6 of the co-expression analysis and module 5 of WGCNA, including 4 AP2/ERF genes and 1 WRKY gene (Figure 6). Among them, ERF1B is homologous to CrORCA3 from Catharanthus roseus (Montiel et al., 2011). The WRKY33 homolog from Arabidopsis thaliana has been reported to participate in defense responses and positively regulate the synthesis of the secondary metabolite camalexin (Zhou et al., 2020). These findings suggest that AP2/ERF TFs play an important role in regulating mint monoterpenoid biosynthesis. However, the functions of these candidate genes require further verification.
Trichomes are specialized structures formed by the highly differentiated epidermal cells of plant’s aboveground organs. Depending on their secretory function, trichomes can be divided into glandular and non-glandular types (Dai et al., 2010). The mint EO is synthesized and stored exclusively in the peltate glandular trichome (Croteau et al., 2005). Trichome development is regulated by complex molecular networks (Han et al., 2022). Currently, the molecular regulatory mechanisms governing non-glandular trichome development in plants are relatively well-understood (Han et al., 2022; Wang et al., 2021b). In plants like Arabidopsis thaliana, key regulatory pathways for non-glandular trichome development have been identified, involving TFs such as GLABRA1 (GL1), GLABRA3 (GL3), and TRANSPARENT TESTA GLABRA1 (TTG1). These TFs form the MYB-bHLH-WD40 (MBW) complex (Pesch and Hulskamp, 2009; Zhao et al., 2008), which activates downstream genes GLABRA2 (GL2) and TRANSPARENT TESTA GLABRA2 (TTG2), thereby initiating trichome differentiation (Han et al., 2022; Wang and Chen, 2008). In contrast, negative regulators such as R3-MYB transcription factors TRICHOMELESS (TCL), TRIPTYCHON (TRY), CAPRICE (CPC), and ENHANCER OF TRY AND CPC (ETC), competitively bind to TTG1-GL3/EGL3 complex, forming an inactive complex that inhibits the expression of downstream genes GL2 and TTG2 (Esch et al., 2003; Zhao et al., 2008). However, research on the molecular regulatory networks involved in glandular trichome development remains limited (Chalvin et al., 2020). Some progress has been made in species like A. annua and Solanum lycopersicum, where key regulatory pathways have been identified (Chalvin et al., 2020). In A. annua, AaMIXTA1 (MYB) interacts with AaHD8 (HD-ZIP) forming a complex that activates the downstream AaHD1 (HD-ZIP) and AaGSW2 (WRKY), then promoting glandular trichome initiation (Xie et al., 2021b; Yan et al., 2017, 2018). However, attempts to overexpress MIXTA from Antirrhinum majus in Arabidopsis thaliana failed to rescue the trichome loss phenotype, and overexpression of non-glandular trichome regulators gene AtGL1 from Arabidopsis thaliana in Solanaceae species did not induce glandular trichome initiation, suggesting distinct regulatory pathways for glandular and non-glandular trichomes (Payne et al., 1999).
However, little is known about the molecular mechanisms underlying PGT development in the Mentha genus (Tissier, 2012). Recent studies have reported that in Mentha canadensis, the HD-ZIP transcription factor McHD-ZIP3 interacts with McMIXTA to form a complex that promotes PGT development (Qi et al., 2022). In this study, we identified homologous genes involved in non-glandular trichome development from Arabidopsis thaliana in M. arvensis transcriptome, including positive regulators like GL3, GL2, and TTG2, as well as negative regulators like TRY, CPC, and ETC3. These genes were significantly differentially expressed after MJ treatment (Supplementary Figure 6, Supplementary Table S9). Unfortunately, through local BLAST, we did not find homologous genes for the positive regulators AaMIXTA, AaHD8, and AaHD1 from A. annua. However, homologs of McMIXTA and HD-ZIP3 from Mentha canadensis were identified but did not show significant differential expression compared to the control, possibly due to the differences in the leaf age of the samples. Studies have reported that genes related to PGT initiation in mint, like McMIXTA, are highly expressed in young leaves, while the leaves collected in this experiment were mature (Qi et al., 2022). Further investigation is needed to validate the role of these candidate genes in PGT formation in mint.
5 Conclusion
Exogenous application of MJ enhanced the EO accumulation and increased the PGT density in M. arvensis. RNA-seq analysis indicated that numerous unigenes were differentially expressed, particularly those involved in JA signal transduction, monoterpenoid biosynthesis, and TFs. MJ treatment up-regulated genes associated with JA signal transduction, including JAZs and MYCs, as well as key structural genes in the monoterpenoid biosynthesis pathway, such as GPPSs, LSs, L3Hs, and IPRs. Additionally, TFs such as AP2/ERF, WRKY, MYB, and bHLH were identified as potential regulators of monoterpenoid biosynthesis or PGT development. Several key candidate genes which regulate monoterpenoid biosynthesis and PGT development were identified, even though their specific functions require further validation. These findings provide valuable insights into the molecular mechanisms governing monoterpenoid biosynthesis and PGT development, and offer potential strategies for enhancing EO production in M. arvensis.
Data availability statement
The data presented in the study are deposited in the NCBI repository, accession number PRJNA1202940, for more information regarding our data policies, refer to our guidelines.
Author contributions
TH: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Project administration, Software, Visualization, Writing – original draft, Writing – review & editing. WM: Funding acquisition, Methodology, Writing – review & editing. AM: Methodology, Writing – review & editing. UD: Supervision, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors would like to thank the reviewers for their valuable comments on this manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1517851/full#supplementary-material
References
Adlard, E. (2010). Handbook of essential oils: science, technology and applications. Chromatographia 72, 1021–1021. doi: 10.1365/s10337-010-1680-0
Ahkami, A., Johnson, S., Srividya, N., Lange, B. (2015). Multiple levels of regulation determine monoterpenoid essential oil compositional variation in the mint family. Mol. Plant 8, 188–191. doi: 10.1016/j.molp.2014.11.009
Akhtar, M., Qamar, N., Yadav, P., Kulkarni, P., Kumar, A., Shasany, A. (2017). Comparative glandular trichome transcriptome-based gene characterization reveals reasons for differential (-)-L-menthol biosynthesis in Mentha species. Physiol. Plant 160, 128–141. doi: 10.1111/ppl.12550
Barbara, N., David, M. (2002). Cross talk between signaling pathways in pathogen defense. Curr. Opin. Plant Biol. 5, 325–331. doi: 10.1016/S1369-5266(02)00275-3
Batool, I., Nisar, S., Hamrouni, L., Jilani, M. (2020). Extraction, production and analysis techniques for L-menthol: A review. Int. J. Chem. Biochem. Sci. 14, 71–76. Available at: https://www.iscientific.org/wp-content/uploads/2020/05/8-IJCBS-18-14-8.pdf (Accessed November 20, 2024).
Bednarek, P., Osbourn, A. (2009). Plant-microbe interactions: chemical diversity in plant defense. Science 324, 746–748. doi: 10.1126/science.1171661
Bussmann, R., Batsatsashvili, K., Kikvidze, Z., Kadirova, I., Ghorbani, A., de Boer, H. (2020). “Mentha arvensis L. Mentha longifolia (L.) L. Mentha spicata L. Menta × piperita L. Lamiaceae,” in Ethnobotany of mountain regions (Springer, Cham), 1–9. doi: 10.1007/978-3-319-77087-1_89-1
Chalvin, C., Drevensek, S., Dron, M., Bendahmane, A., Boualem, A. (2020). Genetic control of glandular trichome development. Trends Plant Sci. 25, 477–487. doi: 10.1016/j.tplants.2019.12.025
Chen, C., Chen, H., Zhang, Y., Thomas, H., Frank, M., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Chen, X., Li, J., Wang, X., Zhong, L., Chen, L. (2019). Full-length transcriptome sequencing and methyl jasmonate-induced expression profile analysis of genes related to patchoulol biosynthesis and regulation in Pogostemon cablin. BMC Plant Biol. 19, 1–18. doi: 10.1186/s12870-019-1884-x
Chen, T., Liu, H., Li, Y., Yao, X., Qin, W., Yan, X., et al. (2023). AaSEPALLATA1 integrates JA and light-regulated glandular secretory trichome initiation in Artemisia annua. Plant Physiol. 192, 1483–1497. doi: 10.1093/plphys/kiad113
Chen, M., Yan, T., Shen, Q., Lu, X., Pan, Q., Huang, Y., et al. (2017). GLANDULAR TRICHOME-SPECIFIC WRKY 1 promotes artemisinin biosynthesis in Artemisia annua. New Phytol. 214, 304–316. doi: 10.1111/nph.14373
Chi, C., Giri, S., Jun, J., Kim, S., Kim, H., Kang, J., et al. (2018). Detoxification-and immune-related transcriptomic analysis of gills from bay scallops (Argopectenirradians) in response to algal toxin okadaic acid. Toxins 10, 308. doi: 10.3390/toxins10080308
Chuang, Y., Hung, Y., Tsai, W., Chen, W., Chen, H. (2018). PbbHLH4 regulates floral monoterpene biosynthesis in Phalaenopsis orchids. J. Exp. Bot. 69, 4363–4377. doi: 10.1093/jxb/ery246
Croteau, R., Davis, E., Ringer, K., Wildung, M. (2005). (-)-L-menthol biosynthesis and molecular genetics. Naturwissenschaften 92, 562–577. doi: 10.1007/s00114-005-0055-0
Dai, X., Wang, G., Yang, D., Tang, Y., Broun, P., Marks, M., et al. (2010). Trichome: a comparative omics database for plant trichomes. Plant Physiol. 152, 44–54. doi: 10.1104/pp.109.145813
Du, T., Fan, Y., Cao, H., Song, Z., Dong, B., Liu, T., et al. (2021). Transcriptome analysis revealed key genes involved in flavonoid metabolism in response to jasmonic acid in pigeon pea (Cajanus cajan (L.) Millsp.). Plant Physiol. Biochem. 168, 410–422. doi: 10.1016/j.plaphy.2021.10.022
Esch, J., Chen, M., Sanders, M., Hillestad, M., Ndkium, S., Idelkope, B., et al. (2003). A contradictory GLABRA3 allele helps define gene interactions controlling trichome development in Arabidopsis. Development 130, 5885–5894. doi: 10.1242/dev.00812
Fu, X., Peng, B., Hassani, D., Xie, L., Liu, H., Li, Y., et al. (2021). AaWRKY9 contributes to light- and jasmonate-mediated to regulate the biosynthesis of artemisinin in Artemisia annua. New Phytol. 231, 1858–1874. doi: 10.1111/nph.17453
Gao, W., Meng, Q., Luo, H., Chen, F., Zhou, Y., He, M. (2020). Transcriptional responses for biosynthesis of flavor volatiles in methyl jasmonate-treated Chrysanthemum indicum var. aromaticum leaves. Ind. Crop Prod. 147, 112254. doi: 10.1016/j.indcrop.2020.112254
Han, G., Li, Y., Yang, Z., Wang, C., Zhang, Y., Wang, B. (2022). Molecular mechanisms of plant trichome development. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.910228
Heinrich, M., Hettenhausen, C., Lange, T., Wunsche, H., Fang, J., Baldwin, I., et al. (2013). High levels of jasmonic acid antagonize the biosynthesis of gibberellins and inhibit the growth of Nicotiana attenuata stems. Plant J. 73, 591–606. doi: 10.1111/tpj.12058
Hickman, R., Van Verk, M., Van Dijken, A., Pereira Mendes, M., Vroegop-Vos, I., Caarls, L., et al. (2017). Architecture and dynamics of the jasmonic acid gene regulatory network. Plant Cell. 29, 2086. doi: 10.1105/tpc.16.00958
Hong, G., Xue, X., Mao, Y., Wang, L., Chen, X. (2012). Arabidopsis MYC2 interacts with DELLA proteins in regulating sesquiterpene synthase gene expression. Plant Cell. 24, 2635–2648. doi: 10.1105/tpc.112.098749
Ji, Y., Xiao, J., Shen, Y., Ma, D., Li, Z., Pu, G., et al. (2014). Cloning and characterization of AabHLH1, a bHLH transcription factor that positively regulates artemisinin biosynthesis in Artemisia annua. Plant Cell Physiol. 55, 1592–1604. doi: 10.1093/pcp/pcu090
Jiang, W., Fu, X., Pan, Q., Tang, Y., Shen, Q., Lv, Z., et al. (2016). Overexpression of AaWRKY1 leads to an enhanced content of artemisinin in Artemisia annua. Biomed. Res. Int. 2016, 7314971. doi: 10.1155/2016/7314971
Khaliq, F., Mushtaq, A. (2023). The five most traded compounds worldwide: importance, opportunities, and risks. Int. J. Chem. Biochem. Sci. 23, 185–194. Available at: https://www.iscientific.org/wp-content/uploads/2023/05/21-IJCBS-23-23-25.pdf (Accessed November 23, 2024).
Klopfenstein, D., Zhang, L., Pedersen, B., Ramírez, F., Warwick Vesztrocy, A., Naldi, A., et al. (2018). GOATOOLS: A python library for gene ontology analyses. Sci. Rep. 8, 1–17. doi: 10.1038/s41598-018-28948-z
Kumar, A., Khajuria, V., Aggarwal, S. (2012). Secondary metabolites of Mentha arvensis and their biological activities. Anal. Chem. Lett. 2, 373–400. doi: 10.1080/22297928.2012.10662623
Kumari, A., Pandey, N., Pandey-Rai, S. (2018). Exogenous salicylic acid-mediated modulation of arsenic stress tolerance with enhanced accumulation of secondary metabolites and improved size of glandular trichomes in Artemisia annua L. Protoplasma 255, 139–152. doi: 10.1007/s00709-017-1136-6
Lange, B. (2015). Biosynthesis and biotechnology of high-value p-menthane monoterpenes, including L-menthol, carvone, and limonene. Biotechnol. Isoprenoids. 148, 319–353. doi: 10.1007/10_2014_289
Lange, B., Croteau, R. (1999a). Isopentenyl diphosphate biosynthesis via a mevalonate-independent pathway: Isopentenyl monophosphate kinase catalyzes the terminal enzymatic step. PNAS 96, 13714–13719. doi: 10.1073/pnas.96.24.13714
Lange, B., Croteau, R. (1999b). Isoprenoid biosynthesis via a mevalonate-independent pathway in plants: cloning and heterologous expression of 1-deoxy-D-xylulose-5-phosphate reductoisomerase from peppermint. Arch. Biochem. Biophys. 365, 170–174. doi: 10.1006/abbi.1999.1168
Lange, B., Mahmoud, S., Wildung, M., Turner, G., Davis, E., Lange, I., et al. (2011). Improving peppermint essential oil yield and composition by metabolic engineering. PNAS 108, 16944–16949. doi: 10.1073/pnas.1111558108
Li, H. (2015). Effects of Melthyl jasmonate and light intensity on the density of glandular trichomes and glandular trichome secretions of Dendranthema indicum var.aromaticum (Harbin (IL: Northeast Forestry University).
Li, S., Dong, Y., Li, D., Shi, S., Zhao, N., Liao, J., et al. (2024). Eggplant transcription factor SmMYB5 integrates jasmonate and light signaling during anthocyanin biosynthesis. Plant Physiol. 194, 1139–1165. doi: 10.1093/plphys/kiad531
Li, C., Gao, Z., Hu, W., Zhu, X., Li, Y., Li, N., et al. (2023). Integration of comparative transcriptomics and WGCNA characterizes the regulation of anthocyanin biosynthesis in mung bean (Vigna radiata L.). Front. Plant Sci. 14. doi: 10.3389/fpls.2023.1251464
Li, L., Hao, X., Liu, H., Wang, W., Fu, X., Ma, Y., et al. (2019). Jasmonic acid-responsive AabHLH1 positively regulates artemisinin biosynthesis in Artemisia annua. Biotechnol. Appl. Biochem. 66, 369–375. doi: 10.1002/bab.1733
Li, S., Liu, Q., Wang, Y., Gu, Y., Liu, D., Wang, C., et al. (2013). Differential gene expression profiling and biological process analysis in proximal nerve segments after sciatic nerve transection. PloS One 8, e57000. doi: 10.1371/journal.pone.0057000
Love, M., Huber, W., Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Luo, L., Wang, Y., Qiu, L., Han, X., Zhu, Y., Liu, L., et al. (2023). MYC2: a master switch for plant physiological processes and specialized metabolite synthesis. Int. J. Mol. Sci. 24, 3511. doi: 10.3390/ijms24043511
Maes, L., Goossens, A. (2010). Hormone-mediated promotion of trichome initiation in plants is conserved but utilizes species and trichome-specific regulatory mechanisms. Plant Signal. Behav. 5, 205–207. doi: 10.4161/psb.5.2.11214
Maes, L., Van Nieuwerburgh, F., Zhang, Y., Reed, D., Pollier, J., Vande Casteele, S., et al. (2011). Dissection of the phytohormonal regulation of trichome formation and biosynthesis of the antimalarial compound artemisinin in Artemisia annua plants. New Phytol. 189, 176–189. doi: 10.1111/j.1469-8137.2010.03466.x
Mahmoud, S., Croteau, R. (2001). Metabolic engineering of essential oil yield and composition in mint by altering expression of deoxyxylulose phosphate reductoisomerase and menthofuran synthase. PNAS 98, 8915–8920. doi: 10.1073/pnas.141237298
Mahmoud, S., Croteau, R. (2002). Strategies for transgenic manipulation of monoterpene biosynthesis in plants. Trends Plant Sci. 7, 366–373. doi: 10.1016/s1360-1385(02)02303-8
Mahmoud, S., Williams, M., Croteau, R. (2004). Cosuppression of limonene-3-hydroxylase in peppermint promotes accumulation of limonene in the essential oil. Phytochemistry 65, 547–554. doi: 10.1016/j.phytochem.2004.01.005
Mamadalieva, N., Hussain, H., Xiao, J. (2020). Recent advances in genus Mentha: phytochemistry, antimicrobial effects, and food applications. Food Frontiers. 1, 435–458. doi: 10.1002/fft2.53
Montiel, G., Zarei, A., Korbes, A., Memelink, J. (2011). The jasmonate-responsive element from the ORCA3 promoter from Catharanthus roseus is active in Arabidopsis and is controlled by the transcription factor AtMYC2. Plant Cell Physiol. 52, 578–587. doi: 10.1093/pcp/pcr016
Nie, H., Wang, H., Jiang, K., Yan, X. (2020). Transcriptome analysis reveals differential immune related genes expression in Ruditapes philippinarum under hypoxia stress: potential HIF and NF-kappaB crosstalk in immune responses in clam. BMC Genom. 21, 318. doi: 10.1186/s12864-020-6734-6
Nieuwenhuizen, N., Chen, X., Wang, M., Matich, A., Perez, R., Allan, A., et al. (2015). Natural variation in monoterpene synthesis in kiwifruit: transcriptional regulation of terpene synthases by NAC and ETHYLENE-INSENSITIVE3-like transcription factors. Plant Physiol. 167, 1243–1258. doi: 10.1104/pp.114.254367
Pan, Q., Wang, Q., Yuan, F., Xing, S., Zhao, J., Choi, Y., et al. (2012). Overexpression of ORCA3 and G10H in Catharanthus roseus plants regulated alkaloid biosynthesis and metabolism revealed by NMR-metabolomics. PloS One 7, e43038. doi: 10.1371/journal.pone.0043038
Pandey, N., Tiwari, A., Rai, S., Pandey-Rai, S. (2021). “Accumulation of secondary metabolites and improved size of glandular trichomes in Artemisia annua” in. Fundamentals Applications., 99–116. doi: 10.1007/978-3-030-30185-9_31
Park, K., Kim, J., Seo, N., Baek, J., Lee, Y., Yun, J., et al. (2024). Study on method validation of assay for detecting undifferentiated cells in cell therapy using digital droplets pcr (ddpcr). Cytotherapy 26, S119–S119. doi: 10.1016/j.jcyt.2024.03.226
Patt, J. M., Robbins, P., Niedz, R., McCollum, G., Alessandro, R. (2018). Exogenous application of the plant signalers methyl jasmonate and salicylic acid induces changes in volatile emissions from citrus foliage and influences the aggregation behavior of Asian citrus psyllid (Diaphorina citri), vector of Huanglongbing. PLoS One 13, e0193724. doi: 10.1371/journal.pone.0193724
Paul, P., Singh, S., Patra, B., Sui, X., Pattanaik, S., Yuan, L. (2017). A differentially regulated AP2/ERF transcription factor gene cluster acts downstream of a MAP kinase cascade to modulate terpenoid indole alkaloid biosynthesis in Catharanthus roseus. New Phytol. 213, 1107–1123. doi: 10.1111/nph.14252
Payne, T., Clement, J., Arnold, D., Lloyd, A. (1999). Heterologous myb genes distinct from GL1 enhance trichome production when overexpressed in Nicotiana tabacum. Development 126, 671–682. doi: 10.1023/B:FISH.0000030638.56031.ed
Pesch, M., Hulskamp, M. (2009). One, two, three … models for trichome patterning in Arabidopsis? Curr. Opin. Plant Biol. 12, 587–592. doi: 10.1016/j.pbi.2009.07.015
Pribat, A., Boureau, L., Mortain-Bertrand, A., Bert, L., Gallusci, P. (2013). “Metabolic engineering of isoprenoid biosynthesis” in. Phenolics Terpenes., 2813–2851. doi: 10.1007/978-3-642-22144-6_126
Qi, X., Chen, Z., Yu, X., Li, L., Bai, Y., Fang, H., et al. (2022). Characterisation of the Mentha canadensis R2R3-MYB transcription factor gene McMIXTA and its involvement in peltate glandular trichome development. BMC Plant Biol. 22, 219. doi: 10.1186/s12870-022-03614-9
Riaz, U., Iqbal, S., Sohail, M., Samreen, T., Ashraf, M., Akmal, F., et al. (2021). A comprehensive review on emerging importance and economical potential of medicinal and aromatic plants (MAPs) in current scenario. Pak. J. Agric. 34, 381–392. doi: 10.17582/journal.pjar/2021/34.2.381.392
Schmidt, A., Nagel, R., Krekling, T., Christiansen, E., Gershenzon, J., Krokene, P. (2011). Induction of isoprenyl diphosphate synthases, plant hormones and defense signalling genes correlates with traumatic resin duct formation in Norway spruce (Picea abies). Plant Mol. Biol. 77, 577–590. doi: 10.1007/s11103-011-9832-7
Shen, Q., Lu, X., Yan, T., Fu, X., Lv, Z., Zhang, F., et al. (2016). The jasmonate-responsive AaMYC2 transcription factor positively regulates artemisinin biosynthesis in Artemisia annua. New Phytol. 210, 1269–1281. doi: 10.1111/nph.13874
Singh, S., Chhatwal, H., Pandey, A. (2024). Deciphering the complexity of terpenoid biosynthesis and its multi-level regulatory mechanism in plants. J. Plant Growth Regul. 43, 3320–3336. doi: 10.1007/s00344-024-11347-2
Souza, M., Braga, R., Santos, A., Rocha, J., Castro, R., Fernandes, M., et al. (2020). Productive, metabolic and anatomical parameters of L-menthol mint are influenced by light intensity. An. Acad. Bras. Cienc. 92, e20180321. doi: 10.1590/0001-3765202020180321
Su, W., Jiang, Y., Bai, Y., Gan, X., Liu, Y., Lin, S. (2019). Advances in transcription factors regulation on plant terpene biosynthesi. J. Agric. Biotechnol. 27, 919–926. doi: 10.3969/j.issn.1674-7968.2019.05.015
Sui, X., Singh, S., Patra, B., Schluttenhofer, C., Guo, W., Pattanaik, S., et al. (2018). Cross-family transcription factor interaction between MYC2 and GBFs modulates terpenoid indole alkaloid biosynthesis. J. Exp. Bot. 69, 4267–4281. doi: 10.1093/jxb/ery229
Tan, H., Xiao, L., Gao, S., Li, Q., Chen, J., Xiao, Y., et al. (2015). TRICHOME AND ARTEMISININ REGULATOR 1 is required for trichome development and artemisinin biosynthesis in Artemisia annua. Mol. Plant 8, 1396–1411. doi: 10.1016/j.molp.2015.04.002
Tang, D., Chen, M., Huang, X., Zhang, G., Zeng, L., Zhang, G., et al. (2023). SRplot: A free online platform for data visualization and graphing. PloS One 18, e0294236. doi: 10.1371/journal.pone.0294236
Tissier, A. (2012). Glandular trichomes: what comes after expressed sequence tags? Plant J. 70, 51–68. doi: 10.1111/j.1365-313X.2012.04913.x
Traw, M., Bergelson, J. (2003). Interactive effects of jasmonic acid, salicylic acid, and gibberellin on induction of trichomes in Arabidopsis. Plant Physiol. 133, 1367–1375. doi: 10.1104/pp.103.027086
Turner, G., Gershenzon, J., Croteau, R. (2000). Distribution of peltate glandular trichomes on developing leaves of peppermint. Plant Physiol. 124, 655–664. doi: 10.1104/pp.124.2.655
Unsicker, S., Kunert, G., Gershenzon, J. (2009). Protective perfumes: the role of vegetative volatiles in plant defense against herbivores. Curr. Opin. Plant Biol. 12, 479–485. doi: 10.1016/j.pbi.2009.04.001
van der Fits, L., Memelink, J. (2000). ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 289, 295–297. doi: 10.1126/science.289.5477.295
Vining, K., Hummer, K., Bassil, N., Lange, B., Khoury, C., Carver, D. (2020). Crop wild relatives as germplasm resource for cultivar improvement in mint (Mentha L.). Front. Plant Sci. 11. doi: 10.3389/fpls.2020.01217
Wang, S., Chen, J. G. (2008). Arabidopsis transient expression analysis reveals that activation of GLABRA2 may require concurrent binding of GLABRA1 and GLABRA3 to the promoter of GLABRA2. Plant Cell Physiol. 49, 1792–1804. doi: 10.1093/pcp/pcn159
Wang, G., Guo, Y., Feng, Y. (2021a). Method, application and process for inducing accumulation of flavonoids and terpene lactone compounds in plant leaves using salicylic acid. China. Patent No CN109953026B (Nanjing, China: China National Intellectual Property Administration (CNIPA)).
Wang, X., Shen, C., Meng, P., Tan, G., Lv, L. (2021b). Analysis and review of trichomes in plants. BMC Plant Biol. 21, 1–11. doi: 10.1186/s12870-021-02840-x
Wang, J., Song, L., Gong, X., Xu, J., Li, M. (2020). Functions of jasmonic acid in plant regulation and response to abiotic stress. Int. J. Mol. Sci. 21, 1446. doi: 10.3390/ijms21041446
Wang, H., Yu, X., Liu, Y., Liang, C., Li, W. (2013). Analysis of genetic variability and relationships among Mentha L. using the limonene synthase gene, LS. Gene 524, 246–252. doi: 10.1016/j.gene.2013.04.012
Wasternack, C., Hause, B. (2013). Jasmonates: biosynthesis, perception, signal transduction and action in plant stress response, growth and development. An update to the 2007 review in Annals of Botany. Ann. botany. 111, 1021–1058. doi: 10.1016/j.pbi.2009.04.001
Wasternack, C., Strnad, M. (2019). Jasmonates are signals in the biosynthesis of secondary metabolites-Pathways, transcription factors and applied aspects-A brief review. N Biotechnol. 48, 1–11. doi: 10.1016/j.nbt.2017.09.007
Wei, H., Kong, S., Jayaraman, V., Selvaraj, D., Soundararajan, P., Manivannan, A. (2023). Mentha arvensis and Mentha × piperita-vital herbs with myriads of pharmaceutical benefits. Horticulturae 9, 224. doi: 10.3390/horticulturae9020224
Weitzel, C., Simonsen, H. (2015). Cytochrome P450-enzymes involved in the biosynthesis of mono-and sesquiterpenes. Phytochem. Rev. 14, 7–24. doi: 10.1007/s11101-013-9280-x
Wu, X., Chen, Y., Shi, X., Qi, K., Cao, P., Yin, H., et al. (2018). Effects of spraying exogenous hormones on cuticular wax composition, structure and permeability of the leaves in ‘Yuluxiang’ pear. J. Nanjing Agric. Univ. 41, 647–654. doi: 10.7685/jnau.201710007
Xiang, L., Jian, D., Zhang, F., Yang, C., Bai, G., Lan, X., et al. (2019). The cold-induced transcription factor bHLH112 promotes artemisinin biosynthesis indirectly via ERF1 in Artemisia annua. J. Exp. Bot. 70, 4835–4848. doi: 10.1093/jxb/erz220
Xiang, L., Zhu, S., Zhao, T., Zhang, M., Liu, W., Chen, M., et al. (2015). Enhancement of artemisinin content and relative expression of genes of artemisinin biosynthesis in Artemisia annua by exogenous MeJA treatment. Plant Growth Regul. 75, 435–441. doi: 10.1007/s10725-014-0004-z
Xie, L., Yan, T., Li, L., Chen, M., Hassani, D., Li, Y., et al. (2021a). An HD-ZIP-MYB complex regulates glandular secretory trichome initiation in Artemisia annua. New Phytol. 231, 2050–2064. doi: 10.1111/nph.17514
Xie, L., Yan, T., Li, L., Chen, M., Ma, Y., Hao, X., et al. (2021b). The WRKY transcription factor AaGSW2 promotes glandular trichome initiation in Artemisia annua. J. Exp. Bot. 72, 1691–1701. doi: 10.1093/jxb/eraa523
Yan, T., Chen, M., Shen, Q., Li, L., Fu, X., Pan, Q., et al. (2017). HOMEODOMAIN PROTEIN 1 is required for jasmonate-mediated glandular trichome initiation in Artemisia annua. New Phytol. 213, 1145–1155. doi: 10.1111/nph.14205
Yan, T., Li, L., Xie, L., Chen, M., Shen, Q., Pan, Q., et al. (2018). A novel HD-ZIP IV/MIXTA complex promotes glandular trichome initiation and cuticle development in Artemisia annua. New Phytol. 218, 567–578. doi: 10.1111/nph.15005
Yang, C., Fang, X., Wu, X., Mao, Y., Wang, L., Chen, X. (2012). Transcriptional regulation of plant secondary metabolism. J. Integr. Plant Biol. 54, 703–712. doi: 10.1111/j.1744-7909.2012.01161.x
Yang, Z., Li, Y., Gao, F., Jin, W., Li, S., Kimani, S., et al. (2020). MYB21 interacts with MYC2 to control the expression of terpene synthase genes in flowers of Freesia hybrida and Arabidopsis thaliana. J. Exp. Bot. 71, 4140–4158. doi: 10.1093/jxb/eraa184
Yin, J., Wong, W., Jang, I., Chua, N. (2017). Co-expression of peppermint geranyl diphosphate synthase small subunit enhances monoterpene production in transgenic tobacco plants. New Phytol. 213, 1133–1144. doi: 10.1111/nph.14280
Yu, N., Chen, Z., Yang, J., Li, R., Zou, W. (2021a). Integrated transcriptomic and metabolomic analyses reveal regulation of terpene biosynthesis in the stems of Sindora glabra. Tree Physiol. 41, 1087–1102. doi: 10.1093/treephys/tpaa168
Yu, Z., Li, J., Yang, C., Hu, W., Wang, L., Chen, X. (2012). The jasmonate-responsive AP2/ERF transcription factors AaERF1 and AaERF2 positively regulate artemisinin biosynthesis in Artemisia annua L. Mol. Plant 5, 353–365. doi: 10.1093/mp/ssr087
Yu, X., Qi, X., Li, S., Fang, H., Bai, Y., Li, L., et al. (2021b). Transcriptome analysis of light-regulated monoterpenes biosynthesis in leaves of Mentha canadensis L. Plants 10, (5). doi: 10.3390/plants10050930
Yu, Z., Zhang, G., Teixeira da Silva, J., Zhao, C., Duan, J. (2021c). The methyl jasmonate-responsive transcription factor DobHLH4 promotes DoTPS10, which is involved in linalool biosynthesis in Dendrobium officinale during floral development. Plant Sci. 309, 110952. doi: 10.1016/j.plantsci.2021.110952
Zeljkovi, S., Jana iková, K., Diego, N., Tarkowski, P. (2020). Phenolic compounds and biological activity of selected Mentha species. Plants 10, 550. doi: 10.3390/plants10030550
Zhang, J. (2021). Regulation mechanism of Methyl jasmonate on secondary metabolism of isodon rubescens (Zhengzhou(IL: Zhengzhou University).
Zhang, C., Wu, Y., Yu, J., Yang, D., Yao, G., Yang, H., et al. (2022). Molecular mechanism of terpenoids synthesis intermediated by light and jasmonates signals. Biotechnol. Bull. 38, 32–40. doi: 10.13560/j.cnki.biotech.bull.1985.2021-1267
Zhao, M., Morohashi, K., Hatlestad, G., Grotewold, E., Lloyd, A. (2008). The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 135, 1991–1999. doi: 10.1242/dev.016873
Zhou, J., Wang, X., He, Y., Sang, T., Wang, P., Dai, S., et al. (2020). Differential phosphorylation of the transcription factor WRKY33 by the protein kinases CPK5/CPK6 and MPK3/MPK6 cooperatively regulates camalexin biosynthesis in Arabidopsis. Plant Cell. 32, 2621–2638. doi: 10.1105/tpc.19.00971
Keywords: Mentha arvensis (mint), essential oil, jasmonic acid, monoterpenoid, glandular trichome, transcriptome
Citation: Huang T, Men W, Myanganbayar A and Davaasambuu U (2025) Transcriptome analysis reveals regulatory mechanism of methyl jasmonate-induced monoterpenoid biosynthesis in Mentha arvensis L.. Front. Plant Sci. 15:1517851. doi: 10.3389/fpls.2024.1517851
Received: 27 October 2024; Accepted: 16 December 2024;
Published: 15 January 2025.
Edited by:
Shaojun Dai, Shanghai Normal University, ChinaReviewed by:
Anastasia L. Lagopodi, Aristotle University of Thessaloniki, GreeceBayarmaa Gun-aajav, National University of Mongolia, Mongolia
Copyright © 2025 Huang, Men, Myanganbayar and Davaasambuu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Undarmaa Davaasambuu, dW5kYXJtYWFAbXVscy5lZHUubW4=
 Tingting Huang
Tingting Huang Wenjin Men
Wenjin Men Undarmaa Davaasambuu
Undarmaa Davaasambuu