- Keshan Branch of Heilongjiang Academy of Agricultural Sciences, Qiqihar, China
Plant architecture determines canopy coverage, photosynthetic efficiency, and ultimately productivity in soybean (Glycine max). Optimizing plant architecture is a major goal of breeders to develop high yield soybean varieties. Over the past few decades, the yield per unit area of soybean has not changed significantly; however, rice and wheat breeders have succeeded in achieving high yields by generating semi‐dwarf varieties. Semi-dwarf crops have the potential to ensure yield stability in high-density planting environments because they can significantly improve responses to fertilizer input, lodging resistance, and enhance resistance to various abiotic and biotic stresses. Soybean has a unique plant architecture, with leaves, inflorescences, and pods growing at each node; internode number greatly affects the final yield. Therefore, producing high-yielding soybean plants with an ideal architecture requires the coordination of effective node formation, effective internode formation, and branching. Dozens of quantitative trait loci (QTLs) controlling plant architecture have been identified in soybean, but only a few genes that control this trait have been cloned and characterized. Here, we review recent progress in understanding the genetic basis of soybean plant architecture. We provide our views and perspectives on how to breed new high-yielding soybean varieties.
Introduction
Soybean (Glycine max [L.] Merr.) is an economically important crop, and provides approximately one-quarter of the world’s plant protein for food and animal feed (Graham and Vance, 2003; Carter et al., 2004; Wilson, 2008; Hartman et al., 2011). Cultivated soybean was domesticated from wild soybean (G. soja Sieb. & Zucc.) approximately 5000 years ago in China, and subsequently spread worldwide (Carter et al., 2004; Wilson, 2008). Soybean yield is ultimately determined by the number of seeds per unit area and seed mass, both of which are affected by number of internodes, branches, pods per plant, seeds per pod, seed size, and plant height (Pedersen and Lauer, 2004; Liu et al., 2020). In addition, soybean yield also is affected by the angle of petiole and length of petiole, both of which are associated with canopy structure and photosynthetic efficiency (Gao et al., 2017; Liu et al., 2020; Zhang et al., 2022). Soybean yield component traits are significantly correlated with both phenotype and genotype (Zhang et al., 2015).
Plant architecture is an essential target trait for developing high-yielding soybean cultivars. This trait can be altered by modulating genes that control stem growth habit, node number, internode length, branch number, leaf size and shape, and leaf angle (Hartung et al., 1981; Bao et al., 2019; Sun et al., 2019; Chen et al., 2021). In the past decade, many quantitative trait loci (QTLs) controlling important agronomic traits have been identified in soybean, some of which have been integrated into the soybase database (https://www.soybase.org/). However, only a small number of the responsible genes for these QTLs have been cloned and functionally characterized. Here, we focus on the genes that have been functionally validated (Table 1).
Genetic basis of stem growth habit
Stem growth habit is a major agronomic trait affecting soybean seed yield because it is related to plant height, flowering time, maturity, abiotic stress tolerance, root architecture, node production (Bernard, 1972; Specht et al., 2001; Heatherly and Smith, 2004; Zhang et al., 2019). Semi-dwarf soybean plant is one of the most important target traits for enhancing lodging resistance and improving yield. Over the past few decades, great efforts have been done to improve soybean yields by stem growth habit-based selection for a semi-dwarf soybean plant (Liu et al., 2010; Tian et al., 2010; Ping et al., 2014). It has been demonstrated that stem growth habit is controlled by two classical genetic loci Dt1 and Dt2 in soybean (Bernard, 1972; Ping et al., 2014). Dt1Dt2 genotypes produce semi-determinate phenotypes, Dt1dt2 genotypes produce indeterminate phenotypes, dt1Dt2 and dt1dt2 genotypes produce determinate, indicating that the dt1 allele has an epistatic effect on the Dt2/dt2 locus.
Dt1 encodes a TERMINAL FLOWER 1 (TFL1) protein in soybean (Liu et al., 2010; Tian et al., 2010). It has been showed that the transition from indeterminate to determinate stem growth habit was caused by independent human selection of four distinct single-nucleotide substitutions in the coding sequence of Dt1 gene during soybean domestication, each of which led to a single amino acid change that resulted in a recessive dt1 allele specifying determinate stem growth (Tian et al., 2010). Dt2 encodes a gain-of-function MADS-domain transcription factor belonging to the APETALA (AP1)/SQUAMOSA subfamily in soybean (Bowman et al., 1993; Gu et al., 1998; Ferrandiz et al., 2000; Ping et al., 2014). Dt2 interacts with SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1 (SOC1) in the shoot apical meristem, where they directly bind to the promoter of Dt1 to repress its transcription and modulate the semi-determinate growth habit in soybean (Liu et al., 2016). Recently, a third locus Dt3 that controlling soybean stem growth habit was discovered, and confirmed that recessive allele dt3 was responsible for semi-determinate stem growth habit in soybean (Clark et al., 2023).
Genes responsible for internode length
Plant height is a key plant architecture trait that directly affects lodging resistance and soybean yield (Chapman et al., 2003; Liu et al., 2020). Internode length and main stem node number determine plant height in soybean (Liu et al., 2013; Chang et al., 2018). Reduced plant height due to shortened stems is beneficial for improving crop yield potential, increasing resilience to abiotic stress, and the use of agronomic and management practices for rapid crop production (Peng et al., 1999; Hedden, 2003; Liu et al., 2020; Lee et al., 2022). A shorter stem due to shortened internodes is typically observed in plants deficient in endogenous gibberellin (GA) biosynthesis or defective in the perception of GA (Yamaguchi, 2008; Binenbaum et al., 2018).
In soybean, DWARF MUTANT 1 (DW1) encodes an ent-kaurene synthase, a key enzyme in the GA biosynthetic pathway that plays a crucial role in GA-regulated cell elongation in stem internodes (Li et al., 2018). The dw1 mutant shows reduced bioactive GA contents, resulting in a dwarf phenotype (Li et al., 2018). Overexpressing the cryptochrome genes CRY1s increased the abundance of STF1 and STF2 proteins, which directly activated the expression of GA2ox genes to deactivate GA1 and repress stem elongation (Lyu et al., 2021). Meanwhile, overexpressing gibberellin 2-oxidase 8 genes (GA2ox8A and GA2ox8B) reduced bioactive GA contents to decrease internode and suppress trailing growth (Wang et al., 2021). Meanwhile, there is a strong artificial selection in cultivated soybean in the genomic region of GA2ox8A and GA2ox8B (Wang et al., 2021).
A quadruple mutant of soybean LATE ELONGATED HYPOCOTYL (LHY) genes exhibited reduced expression of GA pathway genes, reduced plant height, and shortened internodes (Cheng et al., 2019; Dong et al., 2021). In addition, multiple genes involved in regulating plant height by shortening internode length have been reported. For example, overexpression of GmMYB14 transgenic soybean plants shows reduced plant height, internode length, leaf area, and leaf petiole length and angle as well as improved soybean yield when grown in the field under high-density conditions (Chen et al., 2021). Recent research shows that two homologous SUPPRESSOR OF PHYA (SPA) genes Plant Height 13 (PH13) and reduced internode 1 (rin1) play an important role in regulating internode length in soybean. Loss-of-function of RIN1 and PH13 significantly reduced internode length and enhanced grain yield under high-density planting conditions in field trials (Li et al., 2023; Qin et al., 2023).
Molecular basis of branch number
Shoot architecture plays a pivotal role in determining high-yielding crops, and shoot branching is a major component of shoot architecture (Mathan et al., 2016; Barbier et al., 2017). Meanwhile, shoot branching also plays an important role in controlling soybean yield (Liang et al., 2022), and modulating branch number is crucial for high-yield soybean breeding (Liu et al., 2020). Shoot branching is an agronomically important and complex developmental trait controlled by a group of genes and influenced by environment and genotype × environment interactions. Genome-wide analysis using homology searches identified 406 genes that might be associated with branching in soybean, 57 of which colocalize with QTLs for soybean branching (Tan et al., 2013). However, to date, few genes associated with soybean branching have been described.
Overexpressing miR156b in soybean significantly increased the number of long branches and the 100-seed weight, resulting in a 46%–63% increase in yield per plant (Sun et al., 2019). GmmiR156b regulated plant architecture by directly cleaving the SQUAMOSA PROMOTER BINDING PROTEIN-LIKE9d (SPL9d) transcript. SPL9d physically interacted with the homeobox protein WUSCHELa/b (WUSa/b) to regulate axillary bud formation and shoot branching (Sun et al., 2019). The soybean genome contains four SPL9 homologs (SPL9a, SPL9b, SPL9c, and SPL9d), all of which are negatively regulated by GmmiR156b (Cao et al., 2015; Sun et al., 2019). The spl9abcd homozygous quadruple mutant of Williams 82 has more branches and nodes than the wild type (Bao et al., 2019). Dt2 interacted with Agl22 and SOC1a to bind the promoters of Ap1a and Ap1d to activate their transcription, resulting in reduced branching (Liang et al., 2022). In addition, Overexpression of GmMYB181 could increase the branch number in Arabidopsis (Yang et al., 2018).
Critical genes for leaf architecture
Leaf architecture affects photosynthetic efficiency, canopy coverage, and ultimately plant productivity in many legume crops (Gao et al., 2017). Leaf growth direction is controlled by the curvature of the petiole, which is defined as the angle between the leaf petiole and the main stem (Rodrigues and MaChado, 2008; Gao et al., 2017). A few genes that control leaf shape and leaf petiole angle in soybean have been identified.
Leaves and flowers develop continuously at the flanks of the shoot apical meristem in flowering plants. A single mutation often causes pleiotropic phenotypes during leaf and flower development (Tsukaya, 2006), suggesting that a common regulatory circuit is involved in the production of leaves and flowers. One major Ln locus that contributes to the variation in leaflet and seed number per pod (Domingo, 1945; Tischner et al., 2003). Broad leaflets are usually associated with non-4-seeded pods, and narrow leaflets are linked to 4-seeded pods. Broad leaflets and non-4-seeded pods are thought to be dominant over narrow leaflets and 4-seeded pods (Domingo, 1945; Jeong et al., 2011). It has been demonstrated that Ln encodes JAGGED1 (JAG1) protein, which regulates lateral organ development; variants of JAG1 have pleiotropic effects on fruit patterning (Dinneny et al., 2004; Ohno et al., 2004; Jeong et al., 2012; Fang et al., 2013). The transition from broad (Ln) to narrow leaflets (ln) is associated with an amino acid substitution in the EAR motif of JAG1 (Jeong et al., 2012; Fang et al., 2013).
Leaf petiole angle is particularly important for determining plant architecture in soybean and many other legumes (Rodrigues and MaChado, 2008; Zhou et al., 2012). A soybean mutant Increased Leaf Petiole Angle1 (ilpa1) with increased leaf petiole angle is a gamma ray-induced mutant derived from Chinese soybean cultivar Hedou 12 (Song et al., 2015). The ILPA1 locus encodes an APC8-like protein that functions as a subunit of the anaphase-promoting complex/cyclosome. Loss-of- function alleles ILPA1 lead to leaf development defects and alter petiole angle by promoting cell proliferation (Gao et al., 2017).
The auxin signaling regulators Auxin/Indole‐3‐Acetic Acid (Aux/IAA) and Auxin Response Factor (ARF), the auxin co‐receptor Transport Inhibitor Response1/Auxin‐related F‐box Protein (TIR1/AFB), and the auxin‐conjugating enzyme Gretchen Hagen 3 (GH3) all influence the establishment of petiole angle in monocots (Song et al., 2009; Bian et al., 2012; Zhao et al., 2013; Chen et al., 2018; Liu et al., 2018). The auxin efflux transporter genes PINFORMED1a (PIN1a) and PIN1c determined polar auxin transport and controlled plant architecture and petiole angle in soybean. The pin1abc triple mutant shows a semidwarf stature and a small leaf petiole angle (Zhang et al., 2022). Meanwhile, (Iso)flavonoids inhibit the transcript of PIN1a/c to regulate petiole angle in soybean (Zhang et al., 2022).
Conclusions and perspectives
Plant architecture plant critical role in affecting crop production (Huyghe, 1998; Jiao et al., 2010; Miura et al., 2010; Tian et al., 2019). The application of semi dwarf varieties has significantly improved crop yield by increasing the planting density and the lodging resistance. The gains in grain productivity during the Green Revolution were a direct consequence of optimal plant height. Mutant alleles of the Green Revolution genes Semidwarf1 (Sd1) and Reduced height (Rht) are utilized to improve crop yields by decreasing overall plant (Peng et al., 1999; Sasaki et al., 2002). In addition, maize cultivars with more upright leaf angles can produce more grains per unit land area when grown in the field under high-density conditions (Lu et al., 2007; Tian et al., 2019). Soybeans exhibit a unique plant architecture, as each node generates leaves, inflorescences, and pods; internode number greatly affects final soybean yields (Sun et al., 2019; Liu et al., 2020). It is currently difficult to achieve high yields by decreasing the number of nodes to reduce plant height and increasing the planting density in soybean. Several studies have shown that introducing the brachytic stem trait (shortened internodes with a zigzag arrangement of the main stem) into elite modern soybean varieties altered plant architecture to facilitate high‐density planting, reduce lodging, and increase yields (Adams and Weaver, 1998; Cui et al., 2007). Therefore, instead of changing the number of nodes, reducing plant height by shortening internodes to increase planting density may be an effective strategy for increasing soybean yields. To achieve this goal, we propose that soybean varieties with ideal plant architecture should have shorter internodes, more internodes, lodging tolerance, narrow leaflets, a higher proportion of four-seeded pods, smaller petiole angle, shorter petioles, and few or no short branches, allowing them to tolerate high‐density planting (Figure 1).
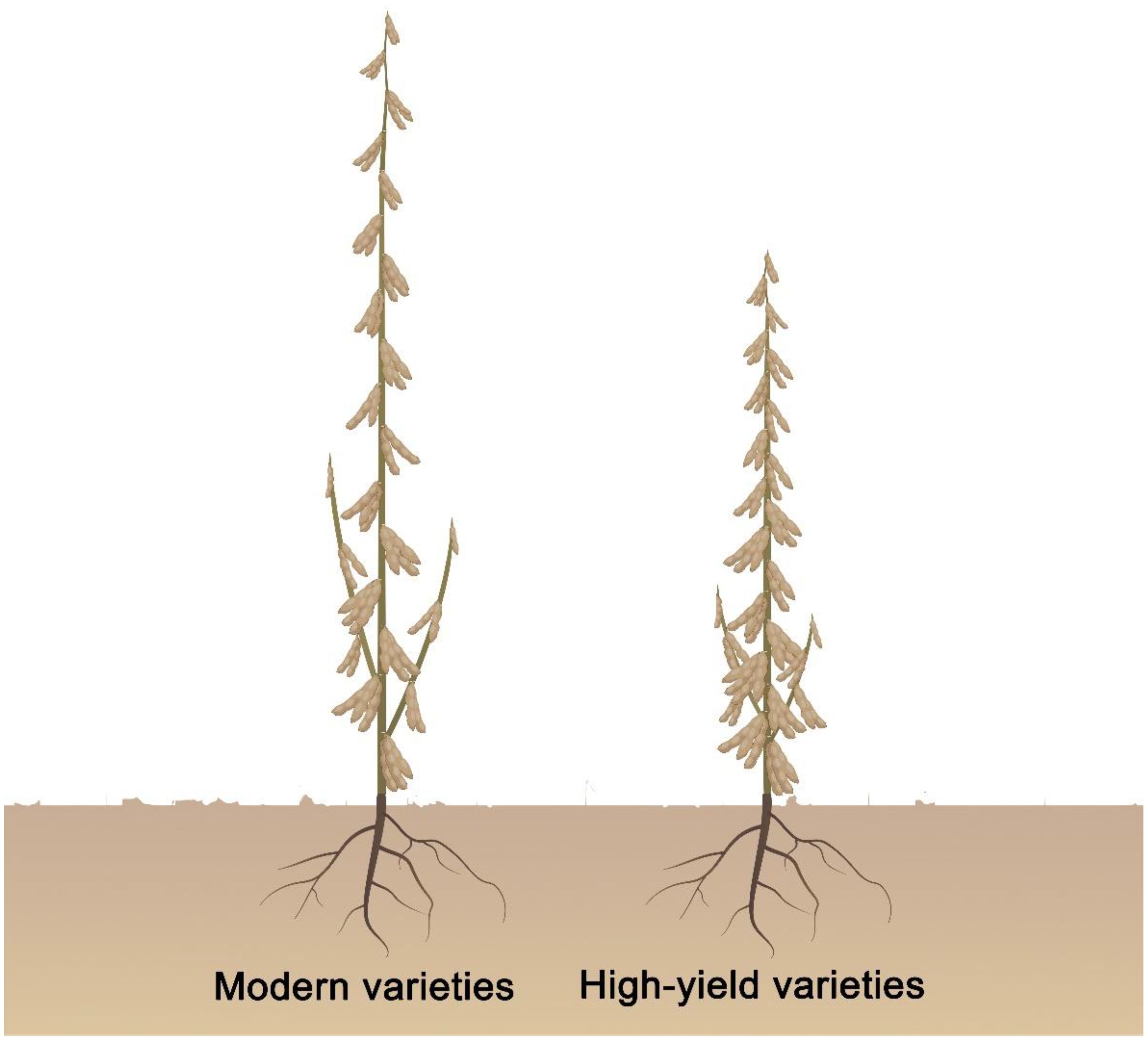
Figure 1. Proposed high yield and ideal plant architecture in soybean. High yield soybean varieties should have shorter internode length, more internodes, lodging tolerance, narrow leaflet and higher the ratio of four seed per pod, smaller petiole angle, and shorter petiole, few or no short branches, tolerate high‐density planting.
Modern crops have much lower genetic diversity than their wild relatives because artificial selection and population/genetic bottlenecks (Hyten et al., 2006; Lam et al., 2010; Qiu et al., 2017; Fernie and Yan, 2019). Wild species are rich sources of natural variation, which is important for improving the yield and quality of crops (Tian et al., 2019; Liu et al., 2021; Goettel et al., 2022; Huang et al., 2022). To understand the genetic architecture and networks underlying agronomic traits, it is crucial to isolate and characterize the genes responsible for plant architecture in soybean has been an important research topic for decades, but only a few genes controlling this trait have been characterized. Wild soybean represents an excellent germplasm resource for identifying key genes or alleles that could be used to develop high-yielding soybean varieties that tolerate dense planting via molecular breeding and gene editing.
Author contributions
WL: Writing – original draft, Writing – review & editing. LW: Writing – review & editing, Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization. HX: Resources, Writing – review & editing. MZ: Writing – review & editing. HS: Writing – review & editing. MQ: Writing – review & editing. QD: Writing – review & editing, Writing – original draft.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the National Natural Science Foundation Cultivation Project of Heilongjiang Academy of Agricultural Sciences (2020FJZX013); Agricultural science technology innovation leapfrog and major demand science technology innovation project of Heilongjiang Province (CX23ZD03); Applied Research and Development Project of Heilongjiang Academy of Agricultural Sciences (2021YYYF035).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adams, P. A., Weaver, D. B. (1998). Brachytic stem trait, row spacing and population effects on soybean yield. Crop Sci. 38, 750–755. doi: 10.2135/cropsci1998.0011183X003800030022x
Bao, A., Chen, H., Chen, L., Chen, S., Hao, Q., Guo, W., et al. (2019). CRISPR/Cas9-mediated targeted mutagenesis of GmSPL9 genes alters plant architecture in soybean. BMC Plant Biol. 19, 131. doi: 10.1186/s12870-019-1746-6
Barbier, F. F., Dun, E. A., Beveridge, C. A. (2017). Apical dominance. Curr. Biol 27, R864–R865. doi: 10.1016/j.cub.2017.05.024
Bernard, R. L. (1972). Two genes affecting stem termination in soybeans. Crop Sci. 12, 235–239. doi: 10.2135/cropsci1972.0011183X001200020028x
Bian, H., Xie., Y., Guo., F., Han., N., Ma, S., Zeng, Z., et al. (2012). Distinctive expression patterns and roles of the miRNA393/TIR1 homolog module in regulating flag leaf inclination and primary and crown root growth in rice (Oryza sativa). New Phytol. 196, 149–161. doi: 10.1111/j.1469-8137.2012.04248.x
Binenbaum, J., Weinstain., R., Shani., E. (2018). Gibberellin localization and transport in plants. Trends Plant Sci. 23, 410–421. doi: 10.1016/j.tplants.2018.02.005
Bowman, J. L., Alvarez, J., Weigel, D., Meyerowitz, E. M., Smyth, D. R. (1993). Control of flower development in Arabidopsis thaliana by Apetala1 and interacting Genes. Development 119, 721–743. doi: 10.1242/dev.119.3.721
Cao, D., Li, Y., Wang, J. L., Nan, H. Y., Wang, Y. N., Lu, S. J., et al. (2015). GmmiR156b overexpression delays flowering time in soybean. Plant Mol. Biol. 89, 353–363. doi: 10.1007/s11103-015-0371-5
Carter, T. E., Nelson, R. L., Sneller, C. H., Cui, Z. (2004). In Soybeans: Improvement, Production and Uses, Third, H.R. Boerma and J.E. Specht, eds. (American Society of Agronomy-Crop Science Society of America-Soil Science Society of America). 303–416.
Chang, F., Guo, C., Sun, F., Zhang, J., Wang, Z., Kong, J., et al. (2018). Genome-wide association studies for dynamic plant height and number of nodes on the Main stem in summer sowing soybeans. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01184
Chapman, A., Pantalone, V. R., Ustun, A., Allen, F. L., Landau-Ellis, D., Trigiano, R. N., et al. (2003). Quantitative trait loci for agronomic and seed quality traits in an F2 and F4:6 soybean population. Euphytica 129, 387–393. doi: 10.1023/A:1022282726117
Chen, L., Yang, H., Fang, Y., Guo, W., Chen, H., Zhang, X., et al. (2021). Overexpression of GmMYB14 improves high-density yield and drought tolerance of soybean through regulating plant architecture mediated by the brassinosteroid pathway. Plant Biotechnol. J. 19, 702–716. doi: 10.1111/pbi.13496
Chen, S. H., Zhou, L. J., Xu, P., Xue, H. W. (2018). SPOC domain containing protein leaf inclination3 interacts with LIP1 to regulate rice leaf inclination through auxin signaling. PloS Genet. 14, e1007829. doi: 10.1371/journal.pgen.1007829
Cheng, Q., Dong, L., Su, T., Li, T., Gan, Z., Nan, H., et al. (2019). CRISPR/Cas9-mediated targeted mutagenesis of GmLHY genes alters plant height and internode length in soybean. BMC Plant Biol. 19, 562. doi: 10.21203/rs.2.14148/v2
Clark, C. B., Zhang, D. J., Wang, W. D., Ma, J. X. (2023). Identification and mapping of a recessive allele, dt3, specifying semideterminate stem growth habit in soybean. Theor. Appl. Genet. 136, 258. doi: 10.1007/s00122-023-04493-w
Cui, S., Meng, Q., Gai, J., Yu, D. (2007). Gene mapping of brachytic stem and its effects on yield-related traits in soybean. Aust. J. Agrl. Res. 58, 774–779. doi: 10.1071/AR06358
Dinneny, J. R., Yadegari, R., Fischer, R. L., Yanofsky, M. F., Weigel, D. (2004). The role of JAGGED in shaping lateral organs. Development 131, 1101–1110. doi: 10.1242/dev.00949
Domingo, W. E. (1945). Inheritance of number of seeds per pod and leaflet shape in the soybean. J. Agric. Res. 70, 251–268. doi: 10.1093/jhered/93.5.346
Dong, L., Fang, C., Cheng, Q., Su, T., Kou, K., Kong, L., et al. (2021). Genetic basis and adaptation trajectory of soybean from its temperate origin to tropics. Nat. Commun. 12, 5445. doi: 10.1038/s41467-021-25800-3
Fang, C., Li, W., Li, G., Wang, Z., Zhou, Z., Ma, Y., et al. (2013). Cloning of Ln gene through combined approach of map-based cloning and association study in soybean. J. Genet. Genomics 40, 93e96. doi: 10.1016/j.jgg.2013.01.002
Fernie, A. R., Yan, J. (2019). De Novo Domestication: an alternative route toward new crops for the future. Mol. Plant 12, 615–631. doi: 10.1016/j.molp.2019.03.016
Ferrandiz, C., Gu, Q., Martienssen, R., Yanofsky, M. F. (2000). Redundant regulation of meristem identity and plant architecture by FRUITFULL, APETALA1 and CAULIFLOWER. Development 127, 725–734. doi: 10.1007/PL00008234
Gao, J., Yang., S., Cheng., W., Fu., Y., Leng., J., Yuan., X., et al. (2017). GmILPA1, encoding an APC8-like protein, controls leaf petiole angle in soybean. Plant Physiol. 174, 1167–1176. doi: 10.1104/pp.16.00074
Goettel, W., Zhang, H., Li, Y., Qiao, Z., Jiang, H., Hou, D., et al. (2022). POWR1 is a domestication gene pleiotropically regulating seed quality and yield in soybean. Nat. Commun. 13, 3051. doi: 10.1038/s41467-022-30314-7
Graham, P. H., Vance, C. P. (2003). Legumes: importance and constraints to greater use. Plant Physiol. 131, 872–877. doi: 10.1104/pp.017004
Gu, Q., Ferrandiz, C., Yanofsky, M. F., Martienssen, R. (1998). The FRUITFULL MADS-box gene mediates cell differentiation during Arabidopsis fruit development. Development 125, 1509–1517. doi: 10.1242/dev.125.8.1509
Hartman, G. L., West, E. D., Herman, T. K. (2011). Crops that feed the World 2. Soybean-worldwide production, use, and constraints caused by pathogens and pests. Food Secur. 3, 5–17. doi: 10.1007/s12571-010-0108-x
Hartung, R. C., Specht, J. E., Williams, J. H. (1981). Modification of soybean plant architecture by genes for stem growth habit and maturity. Crop Sci. 21, 51–56. doi: 10.2135/cropsci1981.0011183X002100010015x
Heatherly, L. G., Smith, J. R. (2004). Effect of soybean stem growth habit on height and node number after beginning bloom in the midsouthern USA. Crop Sci. 44, 1855–1858. doi: 10.2135/cropsci2004.1855
Hedden, P. (2003). The genes of the green revolution. Trends Genet. 19, 5–9. doi: 10.1016/s0168-9525(02)00009-4
Huang, Y., Wang, H., Zhu, Y., Huang, X., Li, S., Wu, X., et al. (2022). THP9 enhances seed protein content and nitrogen-use efficiency in maize. Nature 612, 292–300. doi: 10.1038/s41586-022-05441-2
Huyghe, C. (1998). Genetics and genetic modifications of plant architecture in grain legumes: a review. Agronomie 118, 383–411. doi: 10.1051/agro:19980505
Hyten, D. L., Song, Q., Zhu, Y., Choi, I. Y., Nelson, R. L., Costa, J. M., et al. (2006). Impacts of genetic bottlenecks on soybean genome diversity. Proc. Natl. Acad. Sci. U.S.A. 103, 16666–16671. doi: 10.1073/pnas.0604379103
Jeong, N., Moon, J. K., Kim, H. S., Kim, C. G., Jeong, S. C. (2011). Fine genetic mapping of the genomic region controlling leaflet shape and number of seeds per pod in the soybean. Theor. Appl. Genet. 122, 865–874. doi: 10.1007/s00122-010-1492-5
Jeong, N., Suh, S. J., Kim, M. H., Lee, S., Moon, J. K., Kim, H. S., et al. (2012). Ln is a key regulator of leaflet shape and number of seeds per pod in soybean. Plant Cell 24, 4807–4818. doi: 10.1105/tpc.112.104968
Jiao, Y., Wang, Y., Xue, D., Wang, J., Yan, M., Liu, G., et al. (2010). Regulation of OsSPL14 by OsmiR156 defines ideal plant architecture in rice. Nat. Genet. 42, 541–544. doi: 10.1038/ng.591
Kou, K., Yang, H., Li, H., Fang, C., Chen, L., Yue, L., et al. (2022). A functionally divergent SOC1 homolog improves soybean yield and latitudinal adaptation. Curr. Biol. 32, 1728–1742. doi: 10.1016/j.cub.2022.02.046
Lam, H. M., Xu, X., Liu, X., Chen, W., Yang, G., Wong, F. L., et al. (2010). Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat. Genet. 42, 1053–1059. doi: 10.1038/ng0411-387
Lee, M. B., Shekasteband, R., Hutton, S. F., Lee, T. G. (2022). A mutant allele of the flowering promoting factor 1 gene at the tomato BRACHYTIC locus reduces plant height with high quality fruit. Plant Direct 6, e422. doi: 10.1002/pld3.422
Li, S. C., Sun, Z. H., Sang, Q., Qin, C., Kong, L. P., Huang, X., et al. (2023). Soybean reduced internode 1 determines internode length and improves grain yield at dense planting. Nat. Commun. 14, 7939. doi: 10.1038/s41467-023-42991-z
Li, Z. F., Guo, Y., Ou, L., Hong, H., Wang, J., Liu, Z. X., et al. (2018). Identification of the dwarf gene GmDW1 in soybean (Glycine max L.) by combining mapping-by-sequencing and linkage analysis. Theor. Appl. Genet. 131, 1001–1016. doi: 10.1007/s00122-017-3044-8
Liang, Q., Chen, L., Yang, X., Yang, H., Liu, S., Kou, K., et al. (2022). Natural variation of Dt2 determines branching in soybean. Nat. Commun 12, 6429. doi: 10.1038/s41467-022-34153-4
Liu, Y., Li, Y., Reif, J. C., Mette, M. F., Liu, Z., Liu, B., et al. (2013). Identification of quantitative trait loci underlying plant height and seed weight in soybean. Plant Genome 6, 841–856. doi: 10.3835/plantgenome2013.03.0006
Liu, Y., Wang, H., Jiang, Z., Wang, W., Xu, R., Wang, Q., et al. (2021). Genomic basis of geographical adaptation to soil nitrogen in rice. Nature 590, 600–605. doi: 10.1038/s41586-020-03091-w
Liu, B., Watanabe, S., Uchiyama, T., Kong, F., Kanazawa, A., Xia, Z., et al. (2010). The soybean stem growth habit gene Dt1 is an ortholog of Arabidopsis TERMINAL FLOWER1. Plant Physiol. 153, 198–210. doi: 10.1104/pp.109.150607
Liu, X., Yang, C. Y., Miao, R., Zhou, C. L., Cao, P. H., Lan, J., et al. (2018). DS1/OsEMF1 interacts with OsARF11 to control rice architecture by regulation of brassinosteroid signaling. Rice 11, 46. doi: 10.1186/s12284-018-0239-9
Liu, S., Zhang, M., Feng, F., Tian, Z. (2020). Toward a “Green revolution” for soybean. Mol. Plant 13, 688–697. doi: 10.1016/j.molp.2020.03.002
Liu, Y., Zhang, D., Ping, J., Li, S., Chen, Z., Ma, J. (2016). Innovation of a regulatory mechanism modulating semi-determinate stem growth through artificial selection in soybean. PloS Genet. 12, e1005818. doi: 10.1371/journal.pgen.1005818
Lu, M., Zhou, F., Xie, C. X., Li, M. S., Xu, Y. B., Marilyn, W., et al. (2007). Construction of a SSR linkage map and mapping of quantitative trait loci (QTL) for leaf angle and leaf orientation with an elite maize hybrid. Yi Chuan 29, 1131–1138. doi: 10.1360/yc-007-1131
Lyu, X., Cheng, Q., Qin, C., Li, Y., Xu, X., Ji, R., et al. (2021). GmCRY1s modulate gibberellin metabolism to regulate soybean shade avoidance in response to reduced blue light. Mol. Plant 14, 298–314. doi: 10.1016/j.molp.2020.11.016
Mathan, J., Bhattacharya, J., Ranjan, A. (2016). Enhancing crop yield by optimizing plant developmental features. Development 143, 3283–3294. doi: 10.1242/dev.134072
Miura, K., Ikeda, M., Matsubara, A., Song, X. J., Ito, M., Asano, K., et al. (2010). OsSPL14 promotes panicle branching and higher grain productivity in rice. Nat. Genet. 42, 545–549. doi: 10.1038/ng.592
Ohno, C. K., Reddy, G. V., Heisler, M. G., Meyerowitz, E. M. (2004). The Arabidopsis JAGGED gene encodes a zinc finger protein that promotes leaf tissue development. Development 131, 1111–1122. doi: 10.1242/dev.00991
Pedersen, P., Lauer, J. G. (2004). Response of soybean yield components to management system and planting date. Agron. J. 96, 1372–1381. doi: 10.1051/agro:2004034
Peng, J., Richards, D. E., Hartley, N. M., Murphy, G. P., Devos, K. M., Flintham, J. E., et al. (1999). [amp]]lsquo;Green revolution’ genes encode mutant gibberellin response modulators. Nature 400, 256–261. doi: 10.1038/22307
Ping, J. Q., Liu, Y. F., Sun, L. J., Zhao, M. X., Li, Y. H., She, M. Y., et al. (2014). Dt2 is a Gain-of-function MADS-domain factor gene that specifies semideterminacy in soybean. Plant Cell 26, 2831–2842. doi: 10.1105/tpc.114.126938
Qin, C., Li, Y. H., Li, D. L., Zhang, X. R., Kong, L. P., Zhou, Y. G., et al. (2023). PH13 improves soybean shade traits and enhances yield for high-density planting at high latitudes. Nat. Commun. 14, 6813. doi: 10.1038/s41467-023-42608-5
Qiu, J., Zhou, Y., Mao, L., Ye, C., Zhang, W. J., Yu, Y., et al. (2017). Genomic variation associated with local adaptation of weedy rice during de-domestication. Nat. Commun. 8, 15323. doi: 10.1038/ncomms15323
Rodrigues, T. M., MaChado, S. R. (2008). Pulvinus functional traits in relation to leaf movements: a light and transmission electron microscopy study of the vascular system. Micron 39, 7–16. doi: 10.1016/j.micron.2007.09.001
Sasaki, A., Ashikari, M., Ueguchi-Tanaka, M., Itoh, H., Nishimura, A., Swapan, D., et al. (2002). A mutant gibberellin-synthesis gene in rice. Nature 416, 701–702. doi: 10.1038/416701a
Song, X., Wei, H., Cheng, W., Yang, S., Zhao, Y., Li, X., et al. (2015). Development of INDEL markers for genetic mapping based on whole-genome re-sequencing in soybean. G3 (Bethesda) 5, 2793–2799. doi: 10.1534/g3.115.022780
Song, Y., You, J., Xiong, L. (2009). Characterization of OsIAA1 gene, a member of rice Aux/IAA family involved in auxin and brassinosteroid hormone responses and plant morphogenesis. Plant Mol. Biol. 70, 297–309. doi: 10.1007/s11103-009-9474-1
Specht, J. E., Chase, K., Macrander, M., Graef, G. L., Chung, J., Markwell, J. P., et al. (2001). Soybean response to water: A QTL analysis of drought tolerance. Crop Sci. 41, 493–509. doi: 10.2135/cropsci2001.412493x
Sun, Z., Su, C., Yun, J., Jiang, Q., Wang, L., Wang, Y., et al. (2019). Genetic improvement of the shoot architecture and yield in soya bean plants via the manipulation of GmmiR156b. Plant Biotechnol. J. 17, 50–62. doi: 10.1111/pbi.12946
Tan, B., Guo, Y., Qiu, L. J. (2013). Whole genome discovery of genes related to branching and co-localization with QTLs in soybean. Hereditas (Beijing) 35, 793–804. doi: 10.3724/SP.J.1005.2013.00793
Tian, Z., Wang, X., Lee, R., Li, Y., Specht, J. E., Nelson, R. L., et al. (2010). Artificial selection for determinate growth habit in soybean. Proc. Natl. Acad. Sci. U.S.A. 107, 8563–8568. doi: 10.1073/pnas.1000088107
Tian, J., Wang, C., Xia, J., Wu, L., Xu, G., Wu, W., et al. (2019). Teosinte ligule allele narrows plant architecture and enhances high-density maize yields. Science 365, 658–664. doi: 10.1126/science.aax5482
Tischner, T., Allphin, L., Chase, K., Orf, J. H., Lark, K. G. (2003). Genetics of seed abortion and reproductive traits in soybean. Crop Sci. 43, 464–473. doi: 10.2135/cropsci2003.0464
Tsukaya, H. (2006). Mechanism of leaf-shape determination. Annu. Rev. Plant Biol. 57, 477–496. doi: 10.1146/annurev.arplant.57.032905.105320
Wang, X., Li, M. W., Wong, F. L., Luk, C. Y., Chung, C. Y. L., Yung, W. S., et al. (2021). Increased copy number of gibberellin 2-oxidase 8 genes reduced trailing growth and shoot length during soybean domestication. Plant J. 107, 1739–1755. doi: 10.1111/tpj.15414
Wilson, R. F. (2008). Soybean: Market Driven Research Needs in Genetics and Genomics of Soybean (New York, NY: Springer Science Press).
Yamaguchi, S. (2008). Gibberellin metabolism and its regulation. Annu. Rev. Plant Biol. 59, 225–251. doi: 10.1007/s003440010039
Yang, H., Xue, Q., Zhang, Z. Z., Du, J. Y., Yu, D. Y., Huang, F. (2018). GmMYB181, a soybean R2R3-MYB protein, increases branch number in transgenic Arabidopsis. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.01027
Zhang, Z., Gao, L., Ke, M., Gao, Z., Tu, T., Huang, L., et al. (2022). GmPIN1-mediated auxin asymmetry regulates leaf petiole angle and plant architecture in soybean. J. Integr. Plant Biol. 64, 1325–1338. doi: 10.1111/jipb.13269
Zhang, H., Hao, D., Sitoe, H. M., Yin, Z., Hu, Z., Zhang, G., et al. (2015). Genetic dissection of the relationship between plant architecture and yield component traits in soybean (Glycine max) by association analysis across multiple environments. Plant Breed. 134, 564–572. doi: 10.1038/srep21625
Zhang, D., Wang, X., Li, S., Wang, C., Gosney, M. J., Mickelbart, M. V., et al. (2019). A post-domestication mutation, Dt2, triggers systemic modification of divergent and convergent pathways modulating multiple agronomic traits in soybean. Mol. Plant 12, 1366–1382. doi: 10.1016/j.molp.2019.05.010
Zhao, S. Q., Xiang, J. J., Xue, H. W. (2013). Studies on the rice LEAF INCLINATION1 (LC1), an IAA-amido synthetase, reveal the effects of auxin in leaf inclination control. Mol. Plant 6, 174–187. doi: 10.1093/mp/sss064
Keywords: soybean, plant architecture, stem growth habit, internode length, branch, leaf architecture
Citation: Li W, Wang L, Xue H, Zhang M, Song H, Qin M and Dong Q (2024) Molecular and genetic basis of plant architecture in soybean. Front. Plant Sci. 15:1477616. doi: 10.3389/fpls.2024.1477616
Received: 08 August 2024; Accepted: 20 September 2024;
Published: 07 October 2024.
Edited by:
Huatao Chen, Jiangsu Academy of Agricultural Sciences (JAAS), ChinaReviewed by:
Wei Zhang, Jiangsu Academy of Agricultural Sciences (JAAS), ChinaQun Cheng, Guangzhou University, China
Copyright © 2024 Li, Wang, Xue, Zhang, Song, Qin and Dong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Quanzhong Dong, a3NkcXpkcXpAMTI2LmNvbQ==
†These authors have contributed equally to this work
 Weiwei Li†
Weiwei Li† Huan Song
Huan Song Quanzhong Dong
Quanzhong Dong