- 1Plant Genetic Engineering Laboratory, National Center for Biotechnology, Astana, Kazakhstan
- 2Plant Genomics and Bioinformatics Laboratory, National Center for Biotechnology, Astana, Kazakhstan
- 3Applied Genetics Laboratory, National Center for Biotechnology, Astana, Kazakhstan
- 4General Biology and Genomics Department, Faculty of Natural Sciences, L.N. Gumilyov Eurasian National University, Astana, Kazakhstan
Species of Tulipa are important ornamental plants used for horticultural purposes in various countries, across Asia, Europe, and North Africa. The present study is the first report on typical features of the complete chloroplast genome sequence of four local and endangered species including T. alberti, T. kaufmanniana, T. greigii, and T. dubia from Kazakhstan using Illumina sequencing technology. The comparative analyses revealed that the complete genomes of four species were highly conserved in terms of total genome size (152. 006 bp - 152. 382 bp), including a pair of inverted repeat regions (26. 330 bp - 26. 371 bp), separated by a large single copy region (82.169 bp - 82,378 bp) and a small copy region (17.172 bp -17.260 bp). Total GC content (36.58-36.62 %), gene number (131), and intron length (540 bp - 2620 bp) of 28 genes. The complete genomes of four species showed nucleotide diversity (π =0,003257). The total number of SSR loci was 159 in T. alberti, 158 in T. kaufmanniana, 174 in T. greigii, and 163 in T. dubia. The result indicated that ten CDS genes, namely rpoC2, cemA, rbcL, rpl36, psbH, rps3, rpl22, ndhF, ycf1, and matK, with effective polymorphic simple sequence repeats (SSRs), high sequence variability (SV) ranging from 2.581 to 6.102, and high nucleotide diversity (Pi) of these loci ranging from 0,004 to 0,010. For all intergenic regions longer than 150 bp, twenty one most variable regions were found with high sequence variability (SV) ranging from 4,848 to 11,862 and high nucleotide diversity (Pi) ranging from 0,01599 to 0,01839. Relative synonymous codon usage (RSCU) analysis was used to identify overrepresented and underrepresented codons for each amino acid. Based on the phylogenic analysis, the sequences clustered into four major groups, reflecting distinct evolutionary lineages corresponding to the subgenera Eriostemons, Tulipa, and Orithyia. Notably, T. greigii was distinctively grouped with species from Orithyia and Eriostemons rather than with other Tulipa species, suggesting a unique evolutionary history potentially shaped by geographical isolation or specific ecological pressures. The complete chloroplast genome of the four Tulipa species provides fundamental information for future research studies, even for designing the high number of available molecular markers.
1 Introduction
The Liliaceae family includes approximately 250 genera and 3500 species distributed worldwide (Ju et al., 2021). Tulipa. L. belongs to the subfamily Lilioideae and tribe Lilieae, and includes four subgenera Clusianae, Tulipa, Eriostemones, and Orythia (Christenhusz et al., 2013). The center of diversity of the genus is in the Pamir and Hindu Kush mountains and the steppes of Kazakhstan. Approximately 150 species of the genus Tulipa grow and develop well in the geographical regions of Asia, Europe, and North Africa (Marasek-Ciolakowska et al., 2018). Tulipa is a perennial plant that produces flowers in the spring during March to May. Tulips come in a wide range of colors, including red, pink, yellow, and white, with warm colors being the most common. Detailed accession-related passport information on 1257 Tulipa accessions is available on the online platform of the Genesys database and major Tulipa germplasm banks. The majority of accessions in the Genesys database are from Poland (636 accessions), followed by the Czech Republic (340 accessions), Ukraine (121 accessions), United Kingdom (72 accessions), Israel (64 accessions), United States (9 accessions), Russia (7 accessions), and Armenia (2 accessions) (https://www.genesys-pgr.org/). The Gardenia and Missouri Botanical Garden websites provide a comprehensive platform for exploring the botanical characteristics of various tulip species (https://www.gardenia.net, https://www.missouribotanicalgarden.org/).
Tulips are economically critical ornamental plants used for horticultural purposes in many countries (Marasek-Ciolakowska et al., 2018). In addition to their horticultural use, tulips have great value in the culinary world. Many varieties of edible flowers have been popular since ancient Greek and Roman times for enhancing the flavor of sweet and savory dishes (Rop et al., 2012). Tulip flowers are eaten to gain strength and as a classic dish for special occasions and have a wide range of medical and health benefits. For example, tulip flowers are the best remedy for coughs and colds, reduce the risk of cancer, used for sinus pain, hay fever and headaches; tulip flower extracts are an excellent poultice for insect bites, bee stings, burns, and rashes on the skin, as it gave quick relief with a soothing effect; tulip extracts have cosmetic uses in creams, hand lotions and in essential oils and perfumes. So, focusing on the nutritional and beneficial health benefits is essential. It is worth noting that not much research has been done on the medicinal use of Tulipa species.
The chemical composition of flowers from five T. gesneriana cultivars with different flower colors was analyzed for phenolic compounds (phenolic acids and flavonoids) and organic acids by Krzymińska et al. (Krzymińska et al., 2020). According to the results of this study, the total phenolic content in tulip petals is not affected by the place of cultivation, but the accumulation of organic acids in the petals is strongly correlated with the cultivars used, the duration of storage and the field or greenhouse conditions.
Tulipa species are believed to originate from Central Asia and is represented by 63 species in Central Asia (Vvedensky and Kovalevskaja, 1971). Scientists have confirmed that southern Kazakhstan and adjacent areas of Central Asia were the centers of origin of wild tulips (Rouhi et al., 2010). In the wonderful book «Tulips and other bulbous plants of Kazakhstan», published by Ivaschenko in 2005, a total of 34 species belonging to the three subgenera Tulipa, Eriostemones, and Orythia were described. These species are widely distributed throughout Kazakhstan (Ivaschenko, 2005). Eighteen species are listed in the Red Book and are protected by the state (https://astana.citypass.kz/en/2021/03/10/v-kazakhstane-35-vidov-dikih-tyulpanov/).This study discussed the typical characteristics of four local and endangered species of Tulipa, including T. alberti, T. kaufmanniana, and T. greigii. These species are listed in the Red Book and are protected at the state level.
T. alberti is native to Kazakhstan, Kyrgyzstan and Uzbekistan, and its distribution spans several regions of Kazakhstan, including the Karatau Mountains in the Syrdaria area, Chu Ili, the south of the Betpakdala desert, southwestern foothills of the Zhungar Alatau (in southern Kazakhstan). Its historical importance dates back to 1877, when Edward Regel described the flower of T. alberti based on samples collected by his son, Albert E. Regel, in the Karatau Mountains. The Karatau Mountains (are mainly located in the Zhambyl region of Kazakhstan, and the reference samples are now housed in the herbarium of the St. Petersburg Botany Research Institute. Morphologically, T. alberti is characterized by ovoid bulbs up to 3-4 cm in diameter, adorned with coriaceous, dark, fulvous, elongated scales. Its stems are robust, typically reaching 20 cm in height, while each bulb produces 3-4 linear, glaucous, wavy, broadly lanceolate, bluish-green leaves without spots (Zonneveld, 2009). The flower shows wide range of colors, from pure yellow and orange to mixed shades of crimson and incarnadine vermilion. Its fruits, up to 6 cm long and 2.5 cm wide, are produced from early April to the first decade of May, with fruiting occurring in May-June (Ivaschenko, 2005).
T. kaufmanniana, commonly known as the water lily tulip, is a species native to Central Asia, initially discovered in Turkestan region and described by Eduard August von Regel in the botanical journal “Gartenflora” in 1877 (POWO, 2024). This tulip species is well suited for rock gardens, beds, and borders. The bulbs of T. kaufmanniana vary in shape, from oblong to egg-shaped and turnip-like, up to 4 cm thick, covered with black-fulvous or golden-brown coriaceous scales. Their stems range from 10 to 50 cm in height, are often anthocyanin-colored, and typically bear two bluish grey leaves, occasionally three or four. Flower shapes vary greatly, from cup or wineglass to flat radial, with pointed or blunt tips of the perianth leaves. The colors are diverse, ranging from white, cream, gold, bright yellow, and orange to light red and burgundy. T. kaufmanniana blooms from late March to early July (Ivaschenko, 2005).
T. greigii was originally discovered in Turkestan region and widely distributed throughout Kazakhstan, from the northern deserts around Kyzylorda to the mountains and gentle foothills of Karatau. It is also found in the mountain ranges of West Tien-Shan, Kyrgyz, and Chu-lli, up to the Kordai Pass in the Ile Alatau Mountains, covering areas in Zhambyl, South Kazakhstan regions, and the eastern part of Kyzylorda region. It grows in loamy soils in valleys, foothills, and rocky slopes up to 2400 m altitude. The first sample of T. greigii was brought from the Karatau Mountains by Eduard August von Regel, who published its description in Gartenflora Vol. 22, on page 290 in 1873. The species was named after Samuel A. Greig (1827-1887), and the reference specimen was kept in the herbarium of the Botanical Research Institute of St. Petersburg. The bulb of T. greigii has ovoid bulb of up to 2-4 cm in diameter, 10-50 cm long fuzzy stem. The plant typically bears four leaves, although three or five leaves have also been observed, with leaf size decreasing towards the top. The lower leaf is oval-elliptic or broadly elliptic, while the upper leaf is spear-shaped. The leaves are bluish-grey with dark purple or violet spots of varying intensity. The flowers of T. greigii are usually single, quite large, and wineglass-shaped, reaching a height of 10-12 cm and a width of 10 cm. Their colors range from predominantly red with shades of orange, bright yellow, and light cream, with the undersides of red flowers being either black or yellow. The fruit can be up to 8 cm long and 25 cm wide. Propagation is primarily by seed, with cloning extremely rare (Ivaschenko, 2005). T. greigii flowers from early April to early June and bears fruit in June and July. The bulbs of this species can be eaten fresh or baked, and in Uzbek and Kazakh folk medicine, the petals are used to relieve headaches, while the seeds are used to treat boils.
T. dubia is endemic to the western Tian-Shan region and is widespread in Kazakhstan, Kyrgyzstan, and Uzbekistan. It grows mainly on the rubbly-melkosem slopes of the Tallas and Ugam ridges (Ivaschenko, 2005). It is distributed on dry, stony slopes and screes and typically inhabits the middle and upper mountain zones at altitudes from 1,500 to 3,300 meters above sea level. The T. dubia populations occur at higher altitudes compared to T. kaufmanniana. However, in the Aksay Valley, all three species discussed in this study grow at almost the exact altitudes but occupy different habitats. The plant is characterized by a short, perennial bulb that grows up to 20 cm high. Its broad green leaves are adorned with red stripes. The flower opens as a wide star with perigone segments that are 2-4 cm long, equal, yellow, red, or variegated yellow-red with a small indistinct yellow spot in red form or an orange spot in yellow form (Tojibaev et al., 2022).
It’s worth noting T. alberti, T. kaufmanniana, and T. dubia are listed as Critically Endangered or near-threatened on the IUCN Red List of Threatened Species, underscoring the importance of conservation efforts for these species. On the other hand, T. greigii was listed as Least Concern in the 2022 assessment, indicating a relatively stable population status for this species (https://www.iucnredlist.org/).
Chloroplast is an essential double membrane-bound organelle responsible for photosynthesis, primarily found in plant and algal cells. The complete chloroplast genome exists in both circular and linear configurations. It typically ranges in length from 120,000 and 170,000 base pairs (bp) and consists of two copies of inverted repeat regions (IRA and IRB) each spanning 20-28 kb, a large single copy (LSC) region of 80-90 kb, and a small single copy (SSC) region of 16-27 kb (Li and Zheng, 2018). The chloroplast genome comprises 120-130 genes, primarily involved in photosynthesis, transcription, and translation (Wicke et al., 2011; Turudić et al., 2023). Advances in high-throughput sequencing technologies have revolutionized the sequencing of chloroplast genomes due to their time-saving and cost-effective advantages. The complete chloroplast genome sequences of Nicotiana tabacum (tobacco) (Shinozaki et al., 1986) and Marchantia polymorpha (liverwort) (Ohyama et al., 1986) were first determined by Japanese scientists in 1986. Since then, numerous studies and reviews have discussed the structure and composition of chloroplast genomes from various land plant species (Wicke et al., 2011; Higgs, 2009; She et al., 2020; Lu et al., 2017; Daniell et al., 2016b; Ruang-Areerate et al., 2021; Dobrogojski et al., 2020; Tonti-Filippini et al., 2017). Chloroplast genomes have several favorable characteristics compared to nuclear genomes: their high abundance in the cell, small genome size, and haploid nature (Lu et al., 2017; Song et al., 2017). In addition, chloroplast genomes are predominantly maternally inherited (Park et al., 2021; Raspé, 2001; Villanueva-Corrales et al., 2021) although rare exceptions have been observed (Mccauley et al., 2007; Korpelainen, 2004). For example, Park et al. showed that the cucumber (Cucumis sativus var. sativus) chloroplast genome is maternally inherited in F1 hybrids, consistent with observations in other plant species (Park et al., 2021). However, investigations by McCauley et al. revealed rare non-maternal inheritance of chloroplast DNA in Silene vulgaris (Mccauley et al., 2007). Furthermore, chloroplast genomes exhibit highly conserved genome structure and gene order (Song et al., 2017; Lu et al., 2017; Chang et al., 2021; Ruang-Areerate et al., 2021). Despite their high conservation, chloroplast genomes exhibit significant microstructural variation at the boundaries of the four regions. As a result of these features, they serve as valuable sources for exploring neutral DNA markers for intraspecific and interspecific identification, evolutionary studies and phylogenetic relationships (Park et al., 2019a, Park et al., 2019b; Li et al., 2021a, Li et al., 2021b; Lu et al., 2017; Song et al., 2017). For example, Song et al. (2017); Lu et al. (2017), and Li et al. (2021c) studied the complete chloroplast genomes of three Cardiocrinum, three Paris, and five Tulipa species. They reported that while length, gene content, and gene order were nearly identical, these genomes exhibited nucleotide variability (Pi) in simple sequence repeats (SSRs, 1-10 bp), single nucleotide polymorphisms (SNPs), and long repeat sequences (greater than 30 bp) such as forward, palindromic, and complement repeats. Comprehensive studies have shown that microstructural variations are higher in non-coding regions than in coding regions (Li and Zheng, 2018). In addition to microstructural variation, major structural rearrangements occur during chloroplast genome evolution, including pseudogenization (Abdullah et al., 2021; Scobeyeva et al., 2021; Esfeld et al., 2018), gene deletions, and intron or exon losses (Frailey et al., 2018). Events such as the complete loss of one of the inverted repeats (Sabir et al., 2014), and large inversion of LSC and SSC regions as well as switching between these regions (Choi et al., 2021; Liang et al., 2022) have also been documented. These evolutionary processes contribute to the dynamic nature of chloroplast genomes and provide valuable insights into plant evolution and adaptation.
Whole chloroplast genome sequence analyses of species from different taxa are crucial for understanding chloroplast structures, gene organizations, diversity, genetic changes, recurrent adaptive evolution, and relationships among different groups of plant species (Ju et al., 2020a; Wicke et al., 2011; Ruang-Areerate et al., 2021; Fan et al., 2018; Cui et al., 2006). In addition, they can address challenging problems such the characterization of plastid-to-nucleus signaling mutants (Leister, 2003), plastome transformation (Olejniczak et al., 2016; Daniell et al., 2016b), developing chloroplast-derived vaccines against human diseases (Lössl and Waheed, 2011; Daniell et al., 2016a), understanding biogeographic history (Zhai et al., 2023), phylogeographic structure (Zhou et al., 2017) and obtaining information from plant fossils (Middleton et al., 2014).
The genus Tulipa contains about 150 species (Ju et al., 2021), however, based on the NCBI database, whole chloroplast genome sequences of several Tulipa species are available, including T. altaica, T. thianschanica, T. iliensis, T. patens, T. sinkiangensis, T. schrenkii, T. gesneriana, T. buhseana, T. sylvestris, T. brachystemon, T. kolpakowskiana, T. fosteriana, T. zenaidae, T. alberti, and T. lemmersii (Ju et al., 2021; Yuan et al., 2022; Ju et al., 2020b; Do et al., 2020; Zhou et al., 2019; Ju et al., 2020a; Li et al., 2021c). The present study is the first report on the complete chloroplast genome sequence of T. alberti, T. kaufmanniana, T. greigii, and T. dubia from Kazakhstan. The objectives of this study were (1) to determine and characterize the organization of the complete chloroplast genome sequence of T. greigii, T. kaufmanniana, T. alberti and T. dubia from Kazakhstan (2) to gain insight into the overall polymorphism and evolutionary dynamics of Tulipa chloroplast genomes; (3) to provide genes with effective SSRs and high nucleotide diversities for species identification in the genus Tulipa, and (4) to calibrate the phylogenetic position of T. alberti, T. kaufmanniana, T. greigii and T. dubia based on phylogenomic analysis by comparing with published complete chloroplast sequences of Tulipa species from the NCBI database. These results provide a more comprehensive understanding of the phylogeny of Tulipa and contribute primary genetic information for the phylogenetic relationship analysis of the genus and other relevant research.
2 Materials and methods
2.1 Materials
In this study, all plant materials of T. alberti (43о38’16’’ N, 68о37’46’’ E), T. kaufmanniana (42о20’51’’ N, 70о28’9’’ E), T. greigii (42о20’39’’ N, 70°25’29’’ E), and T. dubia (42о24’7’’ N, 70о35’43’’ E) were gathered at the collection sites in Karatau State Nature Reserve and Aksu-Zhabagly State Nature Reserve with the guidance of state reserve botanists in May of 2021 and 2022. Permission to collect samples of endangered species was obtained from the Forestry and Wildlife Committee of the Ministry of Ecology, Geology, and Natural Resources of the Republic of Kazakhstan. The detailed source information of species is described in Table 1.
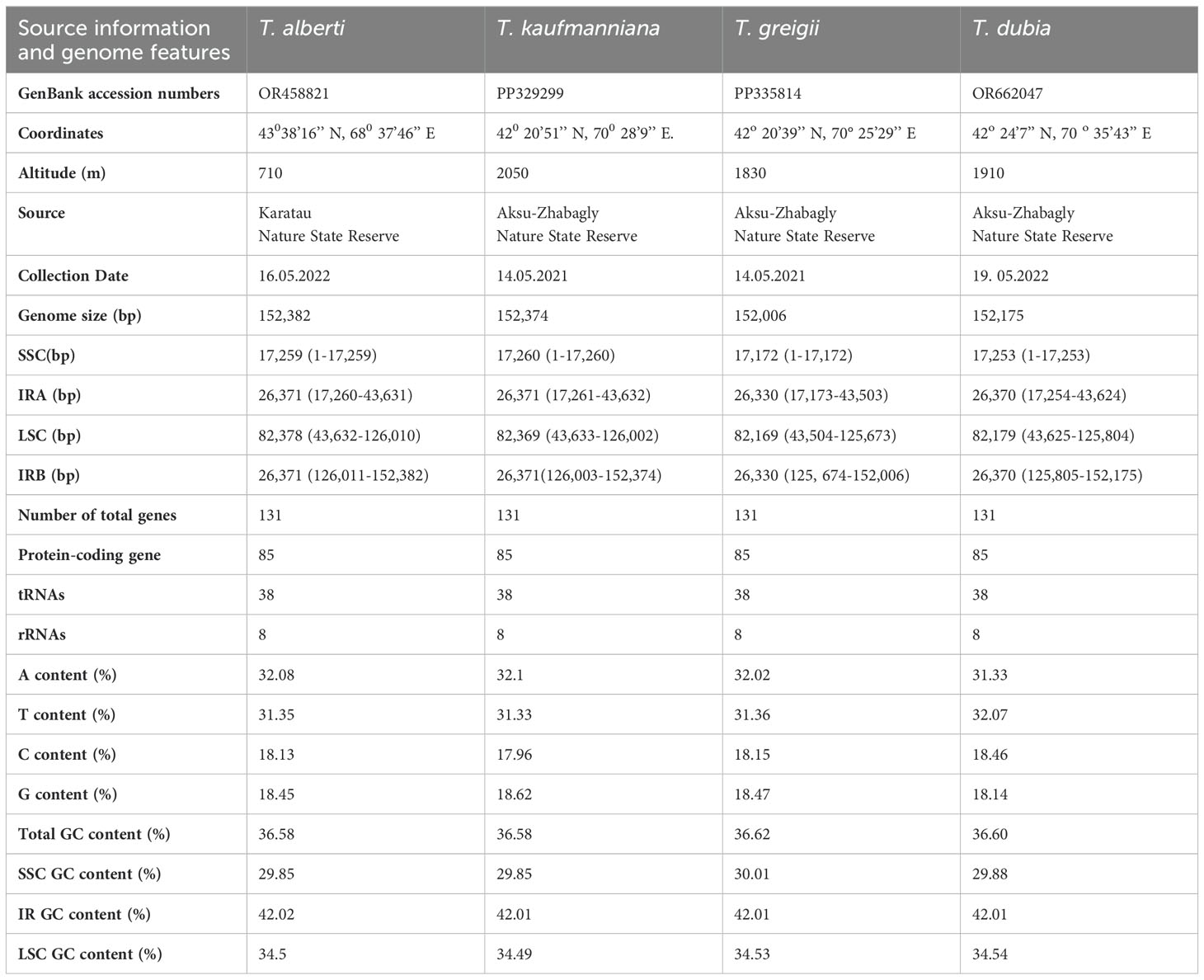
Table 1. The detailed Source information and genome features of four species of Tulipa L. in this study.
2.2 Plant material, DNA extraction and sequencing
Fresh leaves of Tulipa species were stored at -80°C until DNA was extracted. DNA was extracted following the protocol as described by Shi et al. (2012). The extracted DNA was checked for its intactness, homogeneity, and purity by 1% agarose gel electrophoresis and run at 120V for 30 min. The concentration and quantitative analysis were performed by NanoDrop™ 2000/2000c spectrophotometers (Thermo Fisher Scientific). To prepare the DNA libraries, Illumina® DNA Prep, (M) Tagmentation (96 samples) (Illumina, 20018705) was used. Libraries were sequenced on the MiSeq sequencer using MiSeq Reagent Kit v3 (600-cycle) (MS-102-3003). All reagents and protocols were used according to the manufacturer’s instructions.
2.3 Chloroplast genome assembly and annotation
Raw data assembly was performed using Geneious Prime 2024.0.4 software (https://www.geneious.com). Genome annotation was performed using the MPI-MP CHLOROBOX GeSeq (Tillich et al., 2017) software tool based on the reference chloroplast genome of T. altaica (NC_044780.1), manually checked for errors and corrected. In addition, circular maps of the complete chloroplast genomes were illustrated by Chloe (Zhong, 2020) and were edited in Inkscape. Finally, the complete chloroplast genomes of four tulips were submitted to the NCBI Nucleotide database.
2.4 Analysis of chloroplast genome features
The alignment of four complete chloroplast genome sequences of Tulipa species was performed using BioEdit 7.7 (Alzohairy, 2011) and then subsequently used for further analyses. MEGA 11 (Tamura et al., 2021) software was used to calculate codon usage frequencies and relative synonymous codon usage (RSCU) values. RSCU values greater than 1.6 indicated overrepresented codons and RSCU values less than 0.6 indicated underrepresented codons (Noor et al., 2023). Next, boundary regions were visualized using IRscope (Amiryousefi et al., 2018) online software. All gene structures and varying relative positions near junctions were defined. Furthermore, Krait V1.5.1 (Du et al., 2017) was also used for the simple sequence repeat (SSR) identification and localization. The SSR sequences detected from the genome were divided into ten groups by length, 10 bp apart, namely 10-20, 21-30, 31-40, 41-50, 51-60, 61-70, 71-80, 81-90, >91 (Zhao et al., 2023). Genes with effective SSR-rich regions within the CDS and introns were selected and analyzed to identify divergent hotspots across Tulipa genomes). The value of nucleotide diversity (Pi) was calculated using DnaSP v5.10 (Librado and Rozas, 2009).
2.5 Characterization of sequence variability hotspot in CDS genes and intergenic regions
Sequences of each CDS and intergenic regions of more than 150 bp were blasted by the GenBank nucleotide BLAST of NCBI (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Blast parameters were as follows: percent identity = 99-100, query coverage=99-100, and organism optional was Tulipa (taxid:13305). DnaSP v5.10.1 was used to sequence variability (SV) in each protein-coding gene (CDS) and intergenic regions. The number of mutations, indel events, conserved sites and nucleotide diversity (Pi) for each sequence were obtained by DnaSP (Librado and Rozas, 2009). SV was calculated according to the method of (Shaw et al., 2014; Smidt et al., 2020): SV = (number of nucleotide mutations + the number of indel events)/(the number of conserved sites + the number of nucleotide mutations + the number of indel events) × 100%.
2.6 Phylogenetic analysis based on SNPs
The whole chloroplast genome sequences of four Tulipa species in this study, together with previously published whole chloroplast genomes from NCBI were used to infer the phylogenetic relationships. SNPs were identified by comparing the mapped reads to the reference genome T. altaica (NC_044780) using BioNumerics. The tree was conducted using the Maximum Likelihood method and Kimura 2-parameter model in MEGA 11 with a bootstrap value of 500. The whole chloroplast genome of Smilax China (accession number: HM_536959) was used as an outgroup.
3 Results
3.1 Structure and content characteristics of the four Tulipa chloroplast genomes
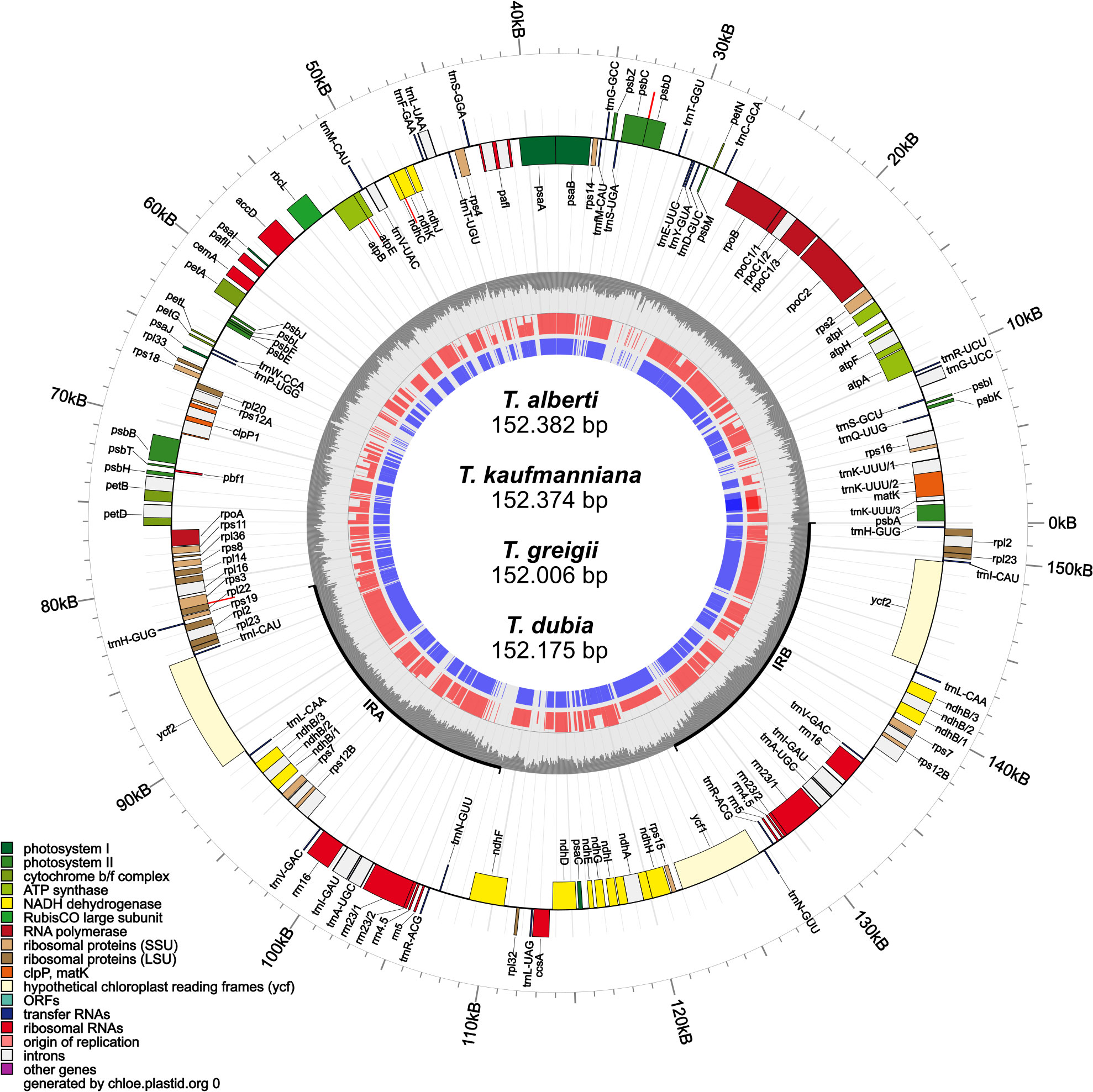
The complete chloroplast genomes of T. alberti (152,382 bp), T. kaufmanniana (152,374bp), T. greigii (152,006 bp), and T. dubia (152,175 bp) were analyzed. The structure of these Tulipa chloroplast genomes consisted of a pair of IRA and IRB regions with lengths ranging from 26,330 to 26,371 bp, an SSC region with lengths ranging from 17,172 to 17,260 bp, and an LSC region with lengths ranging from 82,169 to 82,378 bp. The total GC content of T. alberti and T. kaufmanniana species were 36.58%, and that of T. greigii and T. dubia were 36.62% and 36.60%, respectively, in the whole genome. The GC contents of the SSC (29.85-30.01%) and LSC (34.49-34.54%) regions were almost identical among the species, while the IR regions showed higher GC contents, up to 42.02% (Figure 1; Table 1). A total of 151. 478 nucleotide pairs were analyzed for nucleotide pair frequency analysis, which revealed 99,64% identical pairs, 0,19% transitional pairs, and 0,17% transversional pairs in the four chloroplast genomes of Tulipa species (Supplementary Figure S1). Based on the results from Tajima's Neutrality Test, the nucleotide diversity ((π) was 0,003257. The annotated genome sequences of T. alberti (accession number OR458821.1), T. kaufmanniana (PP329299), T. greigii (PP335814) and T. dubia (OR662047.1) are available in the NCBI database Further details on the genome characteristics are given in Table 1.
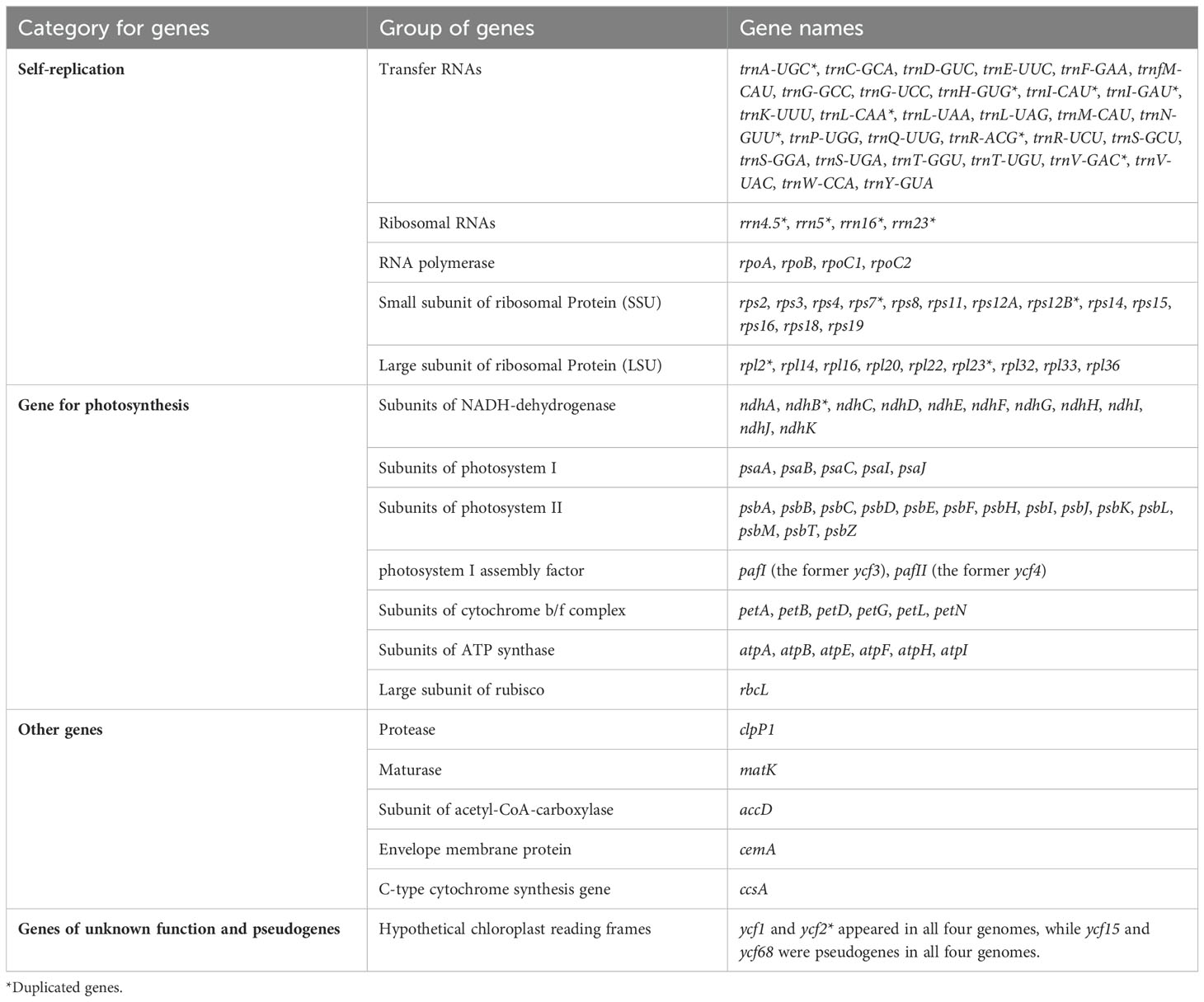
A total of 131 functional genes, including 85 protein-coding genes, were annotated in T. Alberti and T. greigi, while 130 genes, including 84 protein-coding genes, were annotated in T. kaufmanniana and T. dubia (Table 1). All these chloroplast genomes had 38 tRNA and 8 rRNA genes. In addition, all four genomes had 18 duplicated genes located in IR regions, including six coding genes (ndhB, rpl2, rpl23, rps7, rps12B and ycf2), four RNA genes (rrn4.5, rrn5, rrn16, rrn23), and eight tRNA genes (trnA-UGC, trnH-GUG, trnI-CAU, trnI-GAU, trnL-CAA, trnN-GUU, trnR-ACG, and trnV-GAC). ycf1 and ycf2 were found in the genomes of T. alberti, T. greigii, T. kaufmanniana, and T. dubia, while ycf15 and ycf68 were pseudogenes in all four genomes (Table 2). The intron lengths (540 bp- 2620 bp) of 28 genes were well conserved across the four genomes (Supplementary Table S1). The matK gene was located in the intron of trnK gene in all four genomes.
3.2 Comparison of LSC, IR, and SSC junction position among the four Tulipa species
Analysis of the IR/SC boundary areas for the four Tulipa chloroplast genomes was conducted by comparing them to closely related species within the common genus: T. sinkiangensis and T. uniflora from subgenus Orithyia, T. sylvestris from subgenus Eriostemones, T. altaica and T. schrenkii from subgenus Tulipa (Figure 2). In general, plastid genome regions’ lengths and gene number as well as order among 9 Tulipa species were conserved.
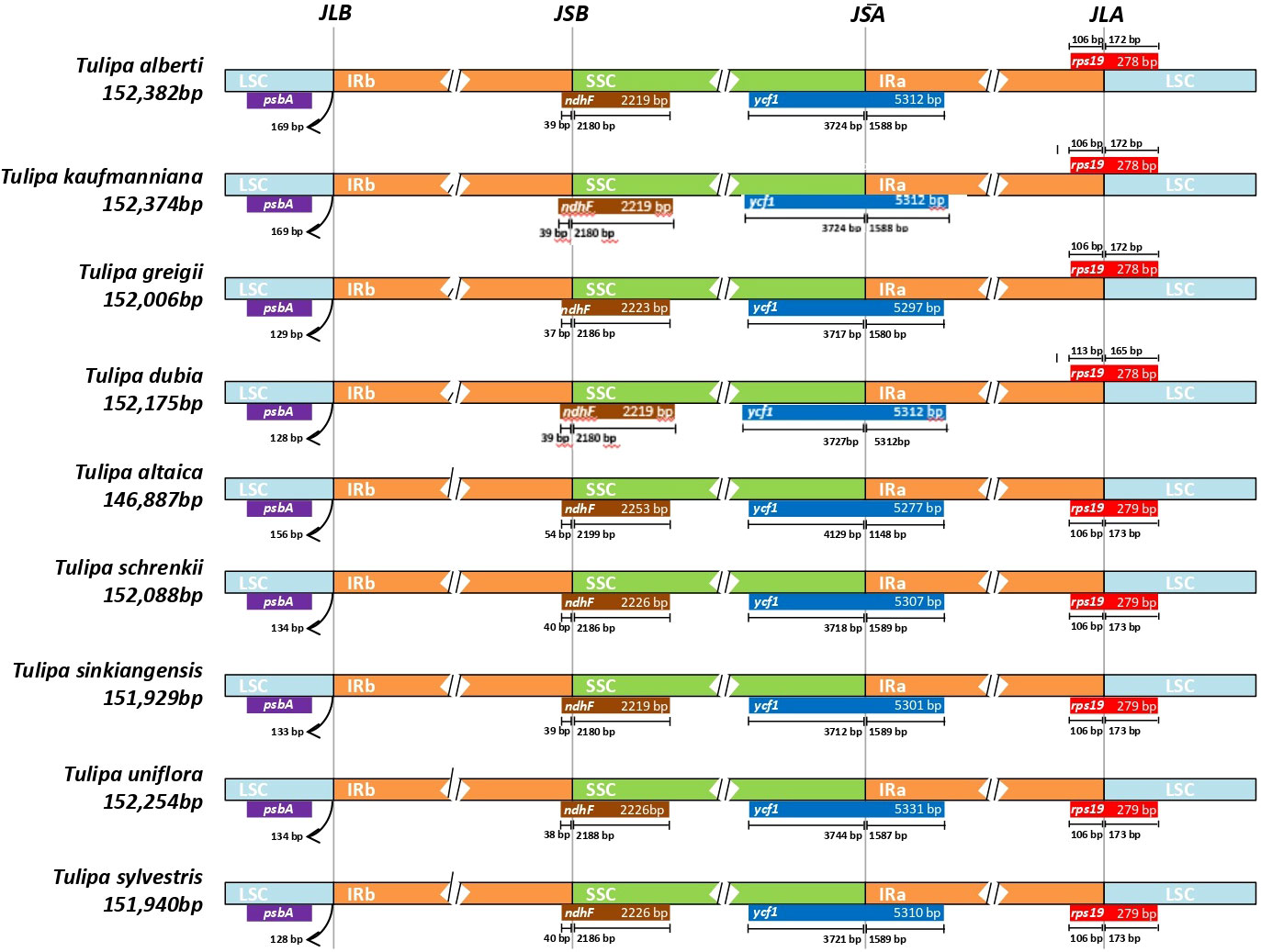
Figure 2. Comparison of SSC, IR, and LSC junction position among four Tulipa species. JSA, junction of SSC and IRA; JLA, junction of LSC and IRA; JLB, junction of LSC and IRB; JSB, junction of SSC and IRB.
An identical haplotype was observed in within the SSC regions of T. alberti, T. kaufmanniana, T. greigii, and T. dubia, resulting in the maintenance of approximately equal amounts of base pairs, number of genes, and orientations. The ycf1 gene was located in the boundary area of the IRA and SSC regions (JSA), while the ndhF gene was found in the boundary area of the SSC and IRB (JSB) regions of these species (Figures 1, 2).
In the LSC regions of all four species, the same haplotype was identified, with approximately equal amounts of base pairs, number of genes, and orientations. The rps19 gene was located in the boundary area of the LSC and IRA regions (JLA), ranging from 43,398 bp to 43,805 bp in the genome of these species. psbA gene was located in the boundary area of the IRB and LSC regions (JLB), ranging from 124,484 bp to 125,842 bp in the genome of these species (Figures 1, 2).
3.3 Simple sequence repeat (SSR) pattern and variation analysis
The total number of SSR loci was 159 in T. alberti, 158 in T. kaufmanniana, 174 in T. greigii, and 163 in T. dubia. SSRs are repeated DNA sequences consisting of tandem repeats of 10-20 in length per unit distributed throughout the genomes of all four Tulipa species, followed by 21-30 bp and 31-40 bp in length. Through comparative genomic analysis, abundant SSRs were detected in the LSC region ranging from 63.19% to 67.2%, followed by the SSC region ranging from 18.39% to19.63% and the IR region ranging from 14.37% to 17.72%. There were 59 types and six categories of SSRs (mono-, di-, tri-, tetra-, penta-, and hexanucleotide repeats) in the chloroplast genomes of studied Tulipa species. The distribution of motif types consisted of 57-70 mono, 37-39 di, 51-53 tri, 7-10 tetra, 1-3 penta, and 2-3 hexa SSRs. The frequency of occurrence was 35.85-40.80% for mono, 22.41-23.93% for di, 29.31-33.54% for tri, 5.03-5.75% for tetra, 0.63-1.72% for penta, and 1.26-1.84% for hexa SSRs (Figure 3; Supplementary Table S2).
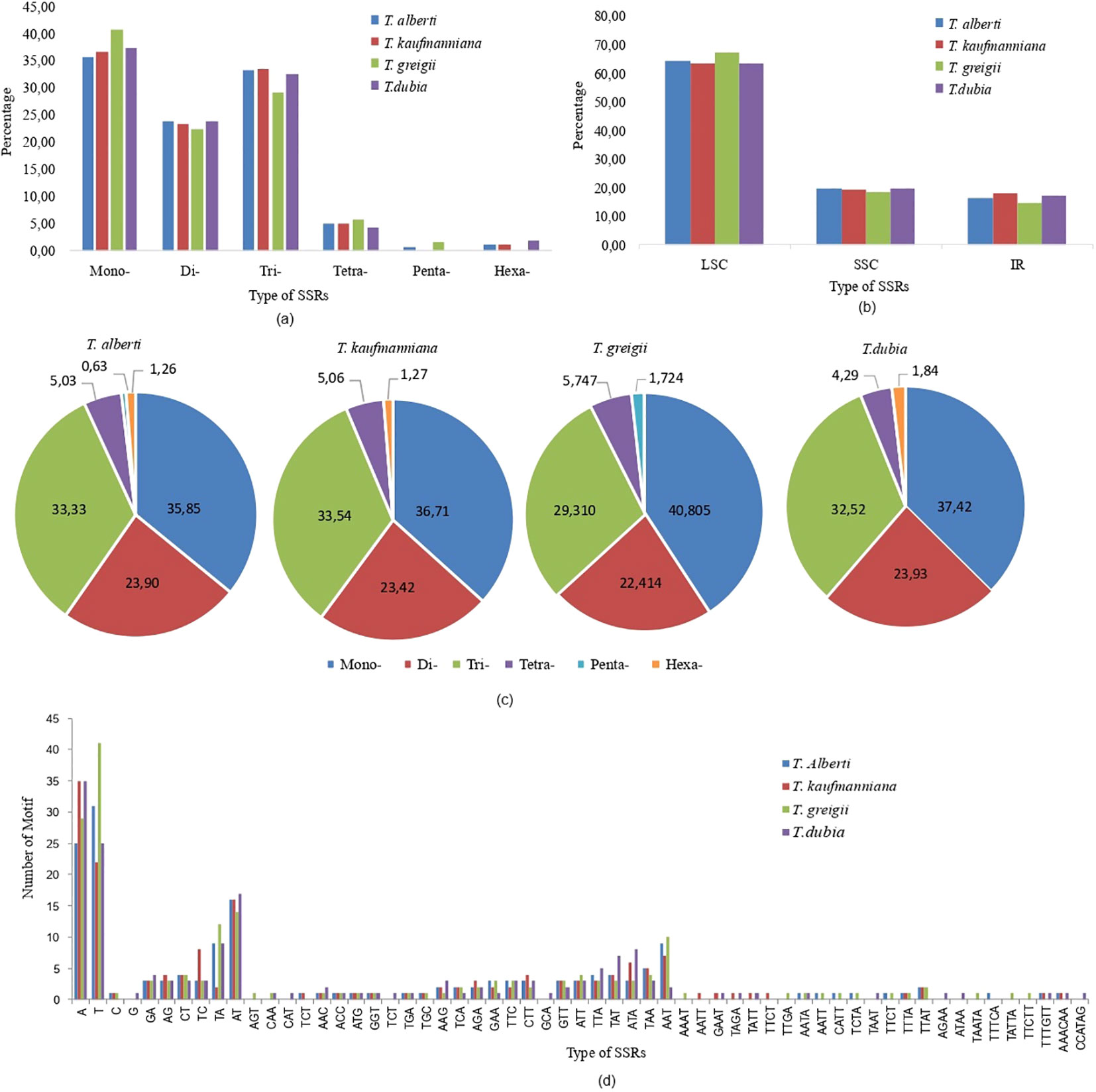
Figure 3. Type and number of identified SSR motifs (mono, di, tri, tetra, penta and hexa) in the whole chloroplast genome of four Tulipa species. (A) frequency of different SSR motif types in the whole chloroplast genome; (B) frequency of Identified SSR motifs in the SSC, IRA, LSC, and IRB regions; (C) percentage of mono, di, tri, tetra, and penta SSR motifs in each chloroplast genomes; (D) type and number of identified SSR motifs (mono, di, tri, tetra, penta, and compound).
3.3.1 Common types of SSRs in the four genomes of Tulipa species
Twenty types of SSRs, including two types of monomers, six types of dinucleotides, and eighteen trinucleotides, were found in all chloroplast genomes of Tulipa species. T and A nucleotides were reported as the most abundant motifs, according to 35.22%, 36.08%, 40.23%, and 36.81% of all SSR motifs in the chloroplast genomes of T. alberti, T. kaufmanniana, T. greigii, and T. dubia, respectively. The major types of dinucleotide SSRs were AT and TA, followed by TC, CT, GA, and AG. The most abundant trimotifs were AAT, TAA, TTA, TAT, TTC, GTT, GAA, CTT, ATT, ATA, TCA, AGA, AAG, TGA, GGT, ATG, ACC and AAC.
3.3.2 Common types of SSRs in the three genomes of Tulipa species
Monomers of type C, trimotifs of type TGC, tetranucleotide repeats of type TTAT and TTTA were common for the genomes of T. alberti, T. kaufmanniana, and T. greigii.
3.3.3 Common types of SSRs in the two genomes of Tulipa species
The TCT type of tri-motif was found in the genomes of T. albert and T. kaufmanniana, while the CAA of tri-motif was found in the genomes of T. greigii and T. dubia. The tetranucleotide repeats, including AATA, AATT, CATT, TCTA, and TTCT, were distributed in the genomes of T. alberti and T. greigii, and GAAT, TAGA, and TATT were distributed in T. kaufmanniana, and T. dubia. Only three types of hexanucleotide repeats were found, among which TTTGTT and AAACAA were distributed in the genomes of T. alberti and T. kaufmanniana. No hexanucleotide repeats were found in the genome of T. greigii.
3.3.4 Unique SSRs only found in one genome of Tulipa species
The AGT type of tri-motif was found in T. greigii. G type of monomer, CAT, TCT, and GCA type of tri-motifs, AGAA, ATAA, and TAAT of tetranucleotide repeats and CCATAG type of hexanucleotide repeats were found in only T. dubia.
3.3.5 SSRs in the intergenic, protein coding gene sequences (CDS), intronic, tRNA regions
The intergenic regions of the four genomes of Tulipa species had the highest SSR density, ranging from 84 to 102, followed by CDS (38-64), intron (16-22), and tRNA (1-3). T. dubia had the most intergenic and tRNA SSRs, T. greigii had the most CDS SSRs, and T. alberti had the most intronic SSRs (Supplementary Figure S2).
3.3.6 Characterization of sequence variability hotspot in CDS genes and intergenic regions
We found a total of 10 CDS genes that have effective polymorphic SSRs, with high sequence variability (SV) ranging from 2.581 to 6.102 and high nucleotide diversity (Pi) of these loci ranging from 0,004 to 0,010. They are rpoC2, cemA, rbcL, rpl36, psbH, rps3, rpl22, ndhF, ycf1 and matK (Supplementary Table S3) (Figure 4).

Figure 4. Sequence variability (SV) and nucleotide diversity (Pi) within each protein-coding sequences (CDS) and intergenic regions.
For all intergenic regions longer than 150 bp, twenty one most variable regions were found with high sequence variability (SV) ranging from 4,848 to 11,862 and high nucleotide diversity (Pi) ranging from 0,01599 to 0,01839. They are ndhF_rpl32, trnR-UCU_trnG-UCC, trnY-GUA_trnD-GUC, trnT-GGU_trnE-UUC, rpoA_petD, rps15_ycf1, psbI_psbK, trnF-GAA_trnL-UAA, psbB_clpP1, trnfM-CAU_trnG-GCC, ndhE-ndhG, psaJ_trnP-UGG, rps8_rpl36, pafII_psaI, rpl14_rps8, ndhG-ndhI, trnQ-UUG_rps16, cemA_pafII, petL_psbE, rpl32_trnl-UAG, and rpl33_psaJ (Figure 4).
3.4 Codon usage patterns
The CDS of four Tulipa species were used for codon usage analysis. The total number of codons in the genomes of the analyzed species ranged from 22,457 (T. dubia) to 25,695 (T. greigii) (Supplementary Figure S3). The third most abundant codons were T (36.065%), followed by A (31.065%), G (17.02%), and C (15.85%) in the genomes T. alberti, T. kaufmanianna and T. greigii, whereas in the genome of T. dubia, A (29.68%) was the most abundant codons followed by T (26.65%), G (24.6%), and C (19%). Identified twenty-one high frequency (HF) codons (RSCU>1) were general for all Tulip species (Figure 5). Two HF codons (RSCU>1), GCT and CTT, were identified in the genomes of T. alberti, T. kaufmanianna and T. greigii. ACT (RSCU>1) was found in T. kaufmanianna, T. greigii, and T. dubia. GGG and AGG (RSCU>1) were identified in the genomes of T. alberti and T. dubia. Five HF codons (RSCU>1), including TTA, CGA, CGT, GGT, and AGT were found in T. kaufmanianna and T. greigii. Four unique codons (RSCU>1), namely GTT, TCG, TGT, and ATA, were found only in the genome of T. dubia. Seven codons (TTA, AGA, GCT, TCT, TAT, ACT, and GAT) were considered overrepresented codons (RSCU>1.6) in the genomes of T. greigii and T. kaufmanianna. In contrast, only one (AGA) overrepresented codon (RSCU>1.6) was found in the genomes of T. alberti and T. dubia. Twenty-four underrepresented (RSCU<0.6) codons were found in the genomes of T. greigii and T. kaufmanianna. They are AAC, AAG, ACG, AGC, AGG, ATC, CAC, CAG, CCG, CGC, CGG, CTC, CTG, GAC, GAG, GCG, GGC, GTC, GTG, TAC, TCG, and TGC. Five underrepresented (RSCU<0.6) codons, such as AGC, CGC, CGT, CTC and CTG were found in the genomes of T. alberti and T. dubia. According to the codon usage analysis, at amino acid levels, Leucine was the most abundant amino acid in the genomes of Tulipa species, with an average of 11.51%. Cysteine was the least abundant amino acid in the genomes of T. greigii, T. kaufmanianna, and T. dubia, with an average of 1.38%, and methionine was the least frequent amino acid in the genome of T. alberti. The results of the total RSCU values are shown in Supplementary Table S4.
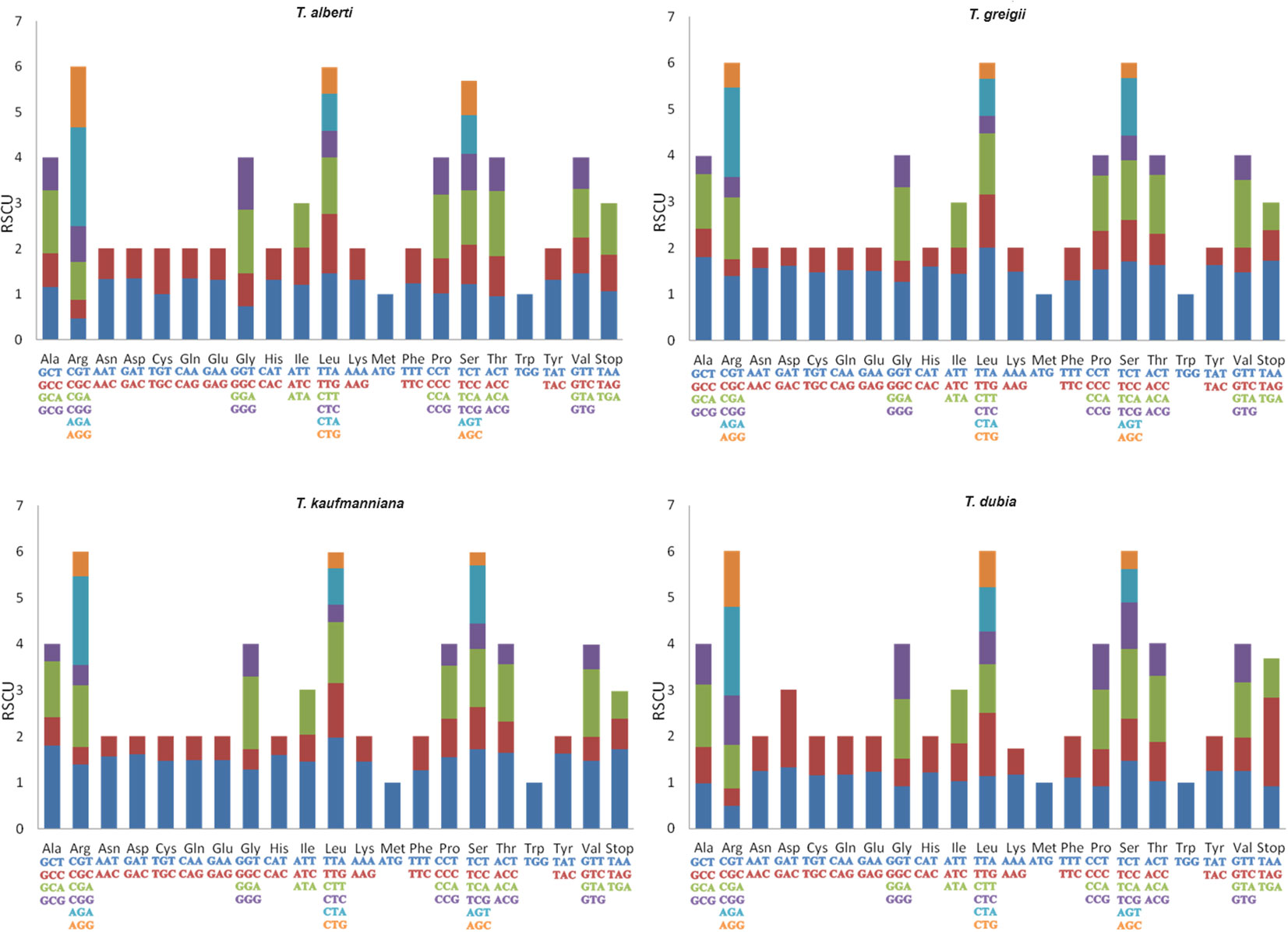
Figure 5. Codon content of 20 amino acids and stop codons in the CDS of four Tulipa species. The color of the histogram corresponds to the color of the codons.
3.5 Phylogenetic analysis
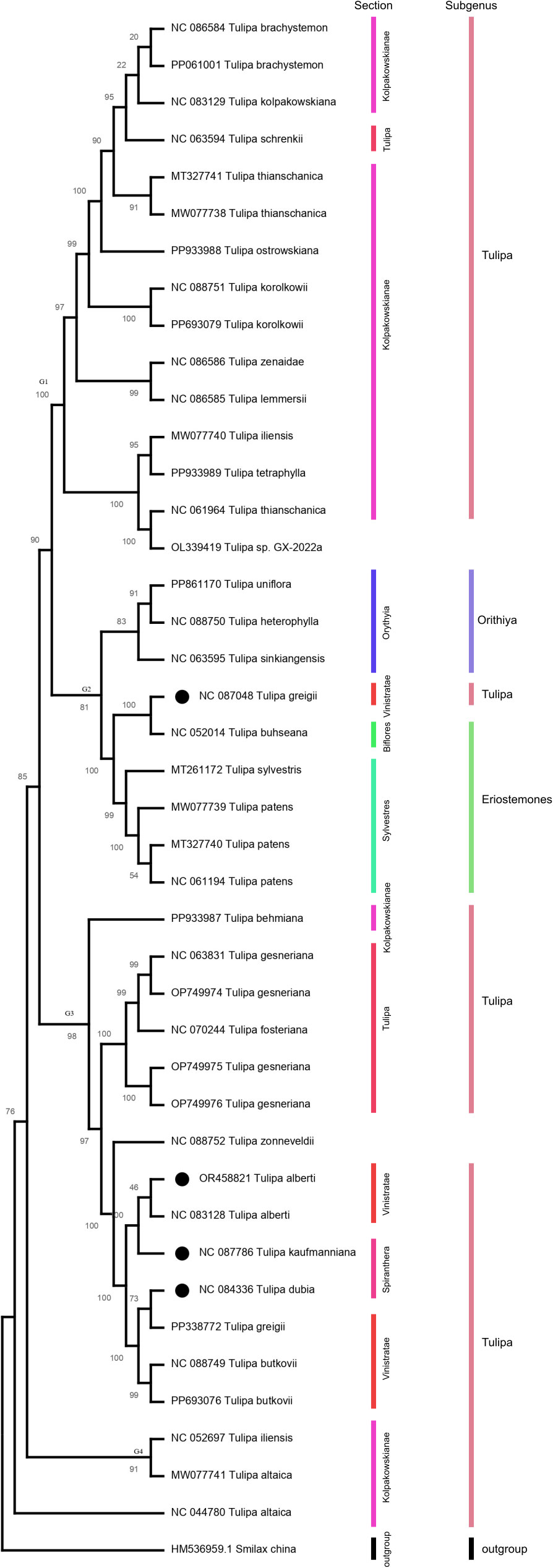
According to SNP-based on the phylogenic analysis, all 40 sequences across 25 species were clustered into four main groups (Figure 6). Group 1, group 3, and group 4 contained species of subgenus Tulipa, while group 2 contained species of subgenus Orithyia and Eriostemons.
Three Tulipa species including T. alberti, T. kaufmanniana, and T. dubia from Kazakhstan were formed one group with species from subgenus Tulipa and the sections Tulipa and Vinistriatae, however, T.greigii in formed a separate with species from subgenus Orithyia and Eriostemons.
4 Discussion
4.1 Structure and content characteristics of the four Tulipa chloroplast genomes
The results presented in this study provide a comprehensive analysis of the chloroplast genomes of four Tulipa species, namely T. alberti, T. kaufmanniana, T. greigii, and T. dubia. These genomes shared typical structural features, including a pair of IR, SSC, and LSC regions, with slight vitiations in the lengths ranging from 152,006 bp to 152,382 bp. Consistency in chloroplast genome sizes across all species in the Liliacae family has been observed previously in other studies (Li et al., 2021c; Lu et al., 2017; Do et al., 2020). All Tulipa species that we analyzed from NCBI database for phylogenetic relationship had whole genome sizes between 151,691 bp and 152,382 bp, except for T. altaica (146,887 bp, NC_044780) and T. fosteriana (153,308 bp, NC_070244). The GC content of the chloroplast genomes was consistent across species, with minor variations (36.58-36.62%) observed. Notably, the GC content was higher in the IR regions compared to the SSC and LSC regions. This phenomenon is consistent with findings in other plant species (Li et al., 2021d; Jiang et al., 2023; Li et al., 2022b) and is thought to be related to the IR regions’ functional significance and structural constraints. Nucleotide pair frequency analysis reveals the distribution of identical, transition, and transversion pairs, and the nucleotide diversity of the four chloroplast genomes showed differences, providing insight into the genetic diversity within these genomes.
In this study, based on the relative order and the orientation of the genes, identical haplotypes were identified in the SSC and LSC regions. There were no detected inversion events in the regions of all four species. However, inversion events in the SSC regions were first reported in the chloroplast genomes of Gossypium barbadense (Ibrahim et al., 2006), Phoenix dactylifera (Yang et al., 2010), Artemisia frigida (Liu et al., 2013), Lasthenia burkei (Walker et al., 2014), and various Asteraceae species (Zhang et al., 2014). Despite being overlooked in several studies, Walker et al. discussed the sources of inversion variation in the SSC region of chloroplast genomes in the American journal of botany in 2015 (Walker et al., 2015). Three structural haplotypes were reported in 61 land plant species from 19 orders of angiosperms, gymnosperms, and pteridophytes. Wild variation in the orientation of the SSC region was found in the Malvaceae family (Cheng et al., 2020). Using the relative order of genes, Liang et al. (2022) revealed SSC region switching in the chloroplast genome of wild rice (Liang et al., 2022).
Inversion events in the LSC regions have also been reported in many plant studies. In addition, the occurrence of flip-flop recombination on short inverted repeat sequences has been demonstrated, generating different isoforms of the transformed plastid genome that differ in the orientation of a 70 kb segment in the LSC region of tobacco (Rogalski et al., 2006). A structural inversion occurred in the LSC region of wild rice, resulting in the reverse orientation of 36 genes (Liang et al., 2022). The LSC region between the accD and rps16 genes contains a 54 kb inversion, the most prominent difference in the chloroplast genome of Oenothera elata (Hupfer et al., 2000). Three inversions (Inversion I, rps4; Inversion II, trnH-GUG/rps16; and Inversion III, trnS-GCU/trnS-GGA) occurred in Anemoclema, Anemone, and Pulsatilla (Choi et al., 2021). A total of 35 gene inversions spanning 44.8 kb, including 21 protein-coding genes and 14 tRNAs from trnT-UGU to rps16 genes, were detected in the chloroplast genome of Adonis mongolica (Nyamgerel et al., 2023).
Ibrahim et al. (2006) suggested that the faster divergence of SSC and LSC regions than IR regions may be caused by the differences between their introns and intergenic regions. Liang et al. (2022) explained it by comparing it with the analogous to the mating-type (MAT) region of a yeast genome in the study of Haber (2012) and suggested that SSC switching, MAT switching, or even more transposon-like elements may share common mechanisms. All research studies attempted to explain this phenomenon, generally attributing it to the results of intramolecular recombination event (Ibrahim et al., 2006; Liu et al., 2013; Walker et al., 2015).
4.2 CDS Genes and intergenic regions could be used as novel chloroplast markers for species identification in the genus Tulipa
The results of previous studies have shown that comparative analyses of chloroplast genome sequences of species in a family help to identify sequence variability (SV) and nucleotide variability and subsequently to determine highly variable genes and intergenic regions as genetic markers used for genetic or phylogenetic studies (Hong et al., 2020; Li et al., 2021c; Cheng et al., 2020; Fan and Ma, 2022). In this study, the comparative analysis of four Tulipa species and species from NCBI showed that ten genes, and twenty-one intergenic regions with high nucleotide diversity (Pi) and sequence variability (SV) could be used as molecular markers for future phylogenetic analysis and species identification studies in the Liliaceae family.
These genes, including rpoC2, rbcL, psbH, rpl22, ndhF, ycf1, and matK have been proposed as high divergence and novel chloroplast markers for species identification, discrimination, and in Tulipa (Li et al., 2021c; Almerekova et al., 2024; Sutula et al., 2024), Poaceae (Moon et al., 2016), Meconopsis (Li et al., 2019), Orchidaceae (Huyen-Trang et al., 2017), Siraitia Merrill (Huyen-Trang et al., 2017), Paris (Song et al., 2017) species. There was no information about cemA, rpl36 and rps3 used as markers.
ndhF_rpl32, trnR-UCU_trnG-UCC, rpoA_petD, rps15_ycf1, ndhG-ndhI, trnY-GUA_trnD-GUC, ndhE-ndhG, psaJ_trnP-UGG, trnT-GGU_trnE-UUC, trnQ-UUG_rps16, rpl14_rps8, cemA_pafII, rpl33-psaJ, and petL_psbE were most variable loci in Adenophora racemose, Iridaceae species, Broomcorn Millet, Glycyrrhiza eurycarpa, Camellia ‘Xiari Qixin’, Apiaceae species, Primula sinensis (Primulaceae), Hibiscus sinosyriacus, Atropa vs. Nicotiana, Lotus vs. Medicago, and Saccharum vs. Oryza, Medicago sativ (Kim and Cheon, 2021; Xiao et al., 2024; Oyuntsetseg et al., 2024; Nie et al., 2018; Zhang et al., 2022; Elyor et al., 2023; Xu et al., 2023; Gou et al., 2020; Kwon et al., 2023; Shaw et al., 2007; Li et al., 2022a).
psbI-psbK (Baraket et al., 2011), trnL (UAA)-trnF (GAA) (Sen et al., 2020; Abdessamad et al., 2016; Baraket et al., 2008), rps8_rpl36 (Seberg and Petersen, 2009), rpl14-rps8 (Ikinci et al., 2011), were used as robust markers for sequence variations, molecular evolution phylogenetic analysis in Pisum sativum L. Palisota (Commelinaceae), Ficus carica L., Quercus suber L., Crocus L., Scorpiris. To our knowledge, trnfM-CAU_trnG-GCC and pafII_psaI were found as newly explored regions.
4.3 Codon usage patterns
SCUB analysis is a powerful tool for investigating species specificity, evolutionary relationships, mRNA translation, and discovering novel genes, all of which are important for understanding gene function and molecular phylogeny (Song et al., 2022). This study analyzed the CDS of four Tulipa species to investigate codon usage patterns and amino acid frequencies. The total number of codons in the genomes of the four species ranged from 22,457 to 25,695, indicating slight differences in genome size among the species. Interestingly, the distribution of codons showed different patterns among the species. For example, in the genomes of T. alberti, T. greigii, and T. kaufmanianna, the most abundant third codon base was T, followed by A, G and C. In contrast, in the genome of T. dubia, it was A, followed by T, G, and C. This variation suggests potential differences in evolutionary pressures and genome composition among the species. In T. greigii and T. kaufmanianna, significantly overrepresented codons reflect conserved codon preferences within the Tulipa genus. However, only one unique and overrepresented codon was specific to the T. alberti and T. dubia chloroplast genomes, indicating species-specific evolutionary dynamics. Furthermore, the analysis of underrepresented codons revealed species-specific trends. T. greigii and T. kaufmanianna exhibited a larger number of underrepresented codons compared to T. alberti and T. dubia, indicating potential differences in codon bias and evolutionary constraints between the species. Analysis of amino acid frequencies revealed that leucine was the most abundant amino acid in all Tulipa species, which is consistent with a previous study (Li et al., 2021c) on chloroplast genomes. Conversely, cysteine was the least abundant amino acid in T. greigii, T. kaufmanianna, and T. dubia, while methionine was the least frequent in T. alberti. Overall, the codon usage patterns observed in this study suggest a complex interplay between evolutionary forces, functional constraints, and environmental adaptation in shaping the chloroplast genomes of Tulipa species. Further research is needed to elucidate the precise mechanisms underlying these codon usage patterns and their implications for the biology and conservation of these endangered species.
4.4 Phylogenetic analysis
The evolutionary development of Tulipa species has been extensively studied through phylogenetic analysis. Phylogenetic studies use various methods, including molecular data analysis and morphological comparisons, to understand the evolutionary relationships among species within the genus. These studies aim to elucidate the genetic ancestry, divergence patterns, and evolutionary history of Tulipa species, shedding light on their evolutionary trajectories and the factors driving their diversification.
The results of the phylogenetic analysis revealed interesting patterns in the relationships among the 40 sequences from 25 species. The analysis indicates that the sequences were grouped into four major clusters, suggesting a significant degree of genetic variation among the species. These clusters likely represent evolutionary connections and may signify different evolutionary lineages or adaptations. Groups 1, 3, and 4 all contain species from the subgenus Tulipa. This indicates that the species within this subgenus are relatively closely related compared to species from other subgenera. This clustering is consistent with the idea that subgenus Tulipa represents a coherent evolutionary unit. Group 2, which contains species from subgenus Orithyia and Eriostemons, is separate from the groups containing Tulipa species. This separation highlights distinct evolutionary pathways or other three adaptations that have occurred in these subgenera.
The separation of T. greigii from other three Tulipa species and its clustering with species from subgenus Orithyia and Eriostemons is particularly noteworthy. It suggests that T. greigii might have a unique evolutionary history or genetic background that sets it apart from other Tulipa species, even though it belongs to the same genus. This could be due to historical geographic isolation, specific ecological adaptations, or other evolutionary pressures. These findings collectively contribute to our understanding of Tulipa species evolutionary relationships and highlight the impact of geographical and ecological factors on their genetic diversity and divergence. Further studies incorporating larger datasets and refined methodologies will continue to enhance our knowledge of Tulipa evolution and its underlying mechanisms.
5 Conclusion
In conclusion, this study achieved its objectives by determining and characterizing the complete chloroplast genome sequences of T. greigii, T. kaufmanniana, T. alberti, and T. dubia from Kazakhstan. Through this, we gained insights into the overall evolutionary dynamics of Tulipa chloroplast genomes. Our research also aimed to provide novel chloroplast markers for species identification within the genus Tulipa. We identified ten genes with effective polymorphic SSRs and high nucleotide diversity, serving as valuable molecular markers for Tulipa species identification. Furthermore, by comparing our sequences with published data from the NCBI database, we calibrated the phylogenetic position of T. alberti, T. kaufmanniana, T. greigii, and T. dubia, contributing to a more comprehensive understanding of Tulipa phylogeny. The newly identified chloroplast genes with high SSRs offer valuable tools for species identification and population studies, aiding conservation efforts and future Tulipa genetics and evolution research.
Data availability statement
The datasets generated and analyzed for this study can be found in the NCBI database (https://www.ncbi.nlm.nih.gov/) under the accession numbers OR458821, PP329299, PP335814, and OR662047.
Author contributions
DT: Data curation, Formal analysis, Methodology, Software, Visualization, Writing – original draft. VS: Software, Writing – original draft. MR: Software, Writing – original draft, Data curation, Formal analysis, Visualization. AR: Writing – original draft, Methodology. AS: Methodology, Data curation, Software, Writing – review & editing. SM: Data curation, Writing – review & editing, Conceptualization, Formal analysis, Investigation, Resources, Supervision.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded by the program, titled «Study of the current state of the diversity of tubular plants in Kazakhstan using modern methods of botany, molecular genetics, and bioinformatics» (BR18574125).
Acknowledgments
We express our gratitude to the Ministry of Science and Higher Education of the Republic of Kazakhstan for providing financial support. We are grateful to the Karatau and Aksu-Zhabagly State Nature Reserves for providing biological material.We would like to thank Kalysh Berdimuratova for assisting us in DNA extraction, Asylulan Amirgazin for helping us with sample sequencing, also to Samatulla Dzhumanov and Ayan Kakanay for their assistance in collecting experimental materials, and M.A.R. Rashid and A. Bakhsh for proofreading.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2024.1433253/full#supplementary-material
References
Abdessamad, A., Baraket, G., Sakka, H., Ammari, Y., Ksontini, M., Hannachi, A. S. (2016). Chloroplast DNA analysis of Tunisian cork oak populations (Quercus suber L.): sequence variations and molecular evolution of the trnL (UAA)-trnF (GAA) region. Gen. Mol. Res. 15, 1–19. doi: 10.4238/gmr15048749
Abdullah, Mehmood, F., Heidari, P., Rahim, A., Ahmed, I., Poczai, P. (2021). Pseudogenization of the chloroplast threonine (trnT-GGU) gene in the sunflower family (Asteraceae). Sci. Rep. 11, 21122. doi: 10.1038/s41598-021-00510-4
Almerekova, S., Yermagambetova, M., Ivaschenko, A., Turuspekov, Y., Abugalieva, S. (2024). Comparative analysis of plastome sequences of seven tulipa L. (Liliaceae juss.) species from section kolpakowskianae raamsd. Ex zonn and veldk. Int. J. Mol. Sci. 25, 7874. doi: 10.3390/ijms25147874
Alzohairy, A. (2011). BioEdit: An important software for molecular biology. GERF Bull. Biosci. 2, 60–61.
Amiryousefi, A., Hyvönen, J., Poczai, P. (2018). IRscope: an online program to visualize the junction sites of chloroplast genomes. Bioinformatics 34, 3030–3031. doi: 10.1093/bioinformatics/bty220
Baraket, G., Ben Abdelkrim, A., Mars, M., Salhi-Hannachi, A. (2011). Cyto-nuclear discordance in the genetic relationships among Tunisian fig cultivars (Ficus carica L.): Evidence from non coding trnL–trnF and ITS regions of chloroplast and ribosomal DNAs. Scientia Hortic. 130, 203–210. doi: 10.1016/j.scienta.2011.06.038
Baraket, G., Saddoud, O., Chatti, K., Mars, M., Mohamed, M., Mokhtar, T., et al. (2008). Chloroplast DNA analysis in Tunisian fig cultivars (Ficus carica L.): Sequence variations of the trnL-trnF intergenic spacer. Biochem. Systematics Ecol. 36, 828–835. doi: 10.1016/j.bse.2008.09.005
Chang, H., Zhang, L., Xie, H., Liu, J., Xi, Z., Xu, X. (2021). The conservation of chloroplast genome structure and improved resolution of infrafamilial relationships of crassulaceae. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.631884
Cheng, Y., Zhang, L., Qi, J., Zhang, L. (2020). Complete chloroplast genome sequence of hibiscus cannabinus and comparative analysis of the malvaceae family. Front. Genet. 11, 227. doi: 10.3389/fgene.2020.00227
Choi, K. S., Ha, Y.-H., Gil, H.-Y., Choi, K., Kim, D.-K., Oh, S.-H. (2021). Two korean endemic clematis chloroplast genomes: inversion, reposition, expansion of the inverted repeat region, phylogenetic analysis, and nucleotide substitution rates. Plants 10, 397. doi: 10.3390/plants10020397
Christenhusz, M., Govaerts, R., David, J., Hall, T., Borland, K., Roberts, P., et al. (2013). Tiptoe through the tulips – cultural history, molecular phylogenetics and classification of Tulipa (Liliaceae). Botanical J. Linn. Soc. 172, 280–328. doi: 10.1111/boj.12061
Cui, L., Leebens-Mack, J., Wang, L.-S., Tang, J., Rymarquis, L., Stern, D. B., et al. (2006). Adaptive evolution of chloroplast genome structure inferred using a parametric bootstrap approach. BMC Evolutionary Biol. 6, 13. doi: 10.1186/1471-2148-6-13
Daniell, H., Chan, H. T., Pasoreck, E. K. (2016a). Vaccination via chloroplast genetics: affordable protein drugs for the prevention and treatment of inherited or infectious human diseases. Annu. Rev. Genet. 50, 595–618. doi: 10.1146/annurev-genet-120215-035349
Daniell, H., Lin, C.-S., Yu, M., Chang, W.-J. (2016b). Chloroplast genomes: diversity, evolution, and applications in genetic engineering. Genome Biol. 17, 134. doi: 10.1186/s13059-016-1004-2
Do, H. D. K., Kim, C., Chase, M. W., Kim, J. H. (2020). Implications of plastome evolution in the true lilies (monocot order Liliales). Mol. Phylogenet Evol. 148, 106818. doi: 10.1016/j.ympev.2020.106818
Dobrogojski, J., Adamiec, M., Luciński, R. (2020). The chloroplast genome: a review. Acta Physiologiae Plantarum 42, 98. doi: 10.1007/s11738-020-03089-x
Du, L., Zhang, C., Liu, Q., Zhang, X., Yue, B. (2017). Krait: an ultrafast tool for genome-wide survey of microsatellites and primer design. Bioinformatics 34, 681–683. doi: 10.1093/bioinformatics/btx665
Elyor, O., Kurbonalieva, M., Aliyeva, K., Khudoyberdieva, S., Asatulloev, T., Yusupov, Z., et al. (2023). Plastid genomes of four species of Iris from subgenus Scorpiris. Plant Diversity Cent. Asia 2, 102–123. doi: 10.54981/PDCA/vol2_iss2/a4
Esfeld, K., Berardi, A. E., Moser, M., Bossolini, E., Freitas, L., Kuhlemeier, C. (2018). Pseudogenization and resurrection of a speciation gene. Curr. Biol. 28, 3776–3786.e7. doi: 10.1016/j.cub.2018.10.019
Fan, W.-B., Wu, Y., Yang, J., Shahzad, K., Li, Z.-H. (2018). Comparative chloroplast genomics of dipsacales species: insights into sequence variation, adaptive evolution, and phylogenetic relationships. Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00689
Fan, Z.-F., Ma, C.-L. (2022). Comparative chloroplast genome and phylogenetic analyses of Chinese Polyspora. Sci. Rep. 12, 15984. doi: 10.1038/s41598-022-16290-4
Frailey, D. C., Chaluvadi, S. R., Vaughn, J. N., Coatney, C. G., Bennetzen, J. L. (2018). Gene loss and genome rearrangement in the plastids of five Hemiparasites in the family Orobanchaceae. BMC Plant Biol. 18, 30. doi: 10.1186/s12870-018-1249-x
Gou, W., Jia, S.-B., Price, M., Guo, X.-L., Zhou, S.-D., He, X.-J. (2020). Complete plastid genome sequencing of eight species from hansenia, haplosphaera and sinodielsia (Apiaceae): comparative analyses and phylogenetic implications. Plants 9, 1523. doi: 10.3390/plants9111523
Haber, J. E. (2012). Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics 191, 33–64. doi: 10.1534/genetics.111.134577
Higgs, D. C. (2009). “Chapter 24 - the chloroplast genome,” in The Chlamydomonas Sourcebook, 2nd ed. Eds. Harris, E. H., Stern, D. B., Witman, G. B. (Academic Press, London).
Hong, Z., Wu, Z., Zhao, K., Yang, Z., Zhang, N., Guo, J., et al. (2020). Comparative analyses of five complete chloroplast genomes from the genus pterocarpus (Fabacaeae). Int. J. Mol. Sci. 21 (11), 3758. doi: 10.3390/ijms21113758
Hupfer, H., Swiatek, M., Hornung, S., Herrmann, R. G., Maier, R. M., Chiu, W. L., et al. (2000). Complete nucleotide sequence of the Oenothera elata plastid chromosome, representing plastome I of the five distinguishable euoenothera plastomes. Mol. Gen. Genet. 263, 581–585. doi: 10.1007/PL00008686
Huyen-Trang, V., Le, L., Nguyen, T., Duong, T., Tran, H.-D. (2017). Review on molecular markers for identification of Orchids. Vietnam J. Science Technol. Eng. 59, 62–75. doi: 10.31276/VJSTE.vol(number).page
Ibrahim, R. I., Azuma, J., Sakamoto, M. (2006). Complete nucleotide sequence of the cotton (Gossypium barbadense L.) chloroplast genome with a comparative analysis of sequences among 9 dicot plants. Genes Genet. Syst. 81, 311–321. doi: 10.1266/ggs.81.311
Ikinci, N., Hall, T., Lledó, M. D., Clarkson, J. J., Tillie, N., Seisums, A., et al. (2011). Molecular phylogenetics of the juno irises, Iris subgenus Scorpiris (Iridaceae), based on six plastid markers. Botanical J. Linn. Soc. 167, 281–300. doi: 10.1111/j.1095-8339.2011.01176.x
Ivaschenko, A. A. (2005). Tulips and other bulbs plants of Kazakhstan (Almaty: Printing house), 192.
Jiang, D., Cai, X., Gong, M., Xia, M., Xing, H., Dong, S., et al. (2023). Complete chloroplast genomes provide insights into evolution and phylogeny of Zingiber (Zingiberaceae). BMC Genomics 24, 30. doi: 10.1186/s12864-023-09115-9
Ju, X., Shi, G., Chen, S., Dai, W., He, T. (2021). Characterization and phylogenetic analysis of the complete chloroplast genome of Tulipa patens (Liliaceae). Mitochondrial DNA B Resour 6, 2750–2751. doi: 10.1080/23802359.2021.1967799
Ju, X., Shi, G., Hou, Z., Wu, C., Liu, G., Cao, C., et al. (2020a). Characterization of the complete chloroplast genome of Tulipa iliensis (Liliaceae). Mitochondrial DNA B Resour 5, 2362–2363. doi: 10.1080/23802359.2020.1773333
Ju, X., Tang, N., Shi, G., Ye, R., Hou, Z. (2020b). Complete chloroplast genome of Tulipa buhseana (Liliaceae). Mitochondrial DNA B Resour 5, 2360–2361. doi: 10.1080/23802359.2020.1773331
Kim, K.-A., Cheon, K.-S. (2021). Complete chloroplast genome sequence of Adenophora racemosa (Campanulaceae): Comparative analysis with congeneric species. PLoS One 16, e0248788. doi: 10.1371/journal.pone.0248788
Korpelainen, H. (2004). The evolutionary processes of mitochondrial and chloroplast genomes differ from those of nuclear genomes. Naturwissenschaften 91, 505–518. doi: 10.1007/s00114-004-0571-3
Krzymińska, A., Gąsecka, M., Magdziak, Z. (2020). Content of phenolic compounds and organic acids in the flowers of selected tulipa gesneriana cultivars. Molecules 25 (23), 5627. doi: 10.3390/molecules25235627
Kwon, S.-H., Kwon, H.-Y., Choi, Y.-I., Shin, H. (2023). Comprehensive analysis of chloroplast genome of hibiscus sinoSyriacus: evolutionary studies in related species and genera. Forests 14, 2221. doi: 10.3390/f14112221
Leister, D. (2003). Chloroplast research in the genomic age. Trends Genet. 19, 47–56. doi: 10.1016/S0168-9525(02)00003-3
Li, B., Zheng, Y. (2018). Dynamic evolution and phylogenomic analysis of the chloroplast genome in Schisandraceae. Sci. Rep. 8, 9285. doi: 10.1038/s41598-018-27453-7
Li, B., Liu, T., Ali, A., Xiao, Y., Shan, N., Sun, J., et al. (2022a). Complete chloroplast genome sequences of three aroideae species (Araceae): lights into selective pressure, marker development and phylogenetic relationships. BMC Genomics 23, 218. doi: 10.1186/s12864-022-08400-3
Li, C., Cai, C., Tao, Y., Sun, Z., Jiang, M., Chen, L., et al. (2021a). Variation and evolution of the whole chloroplast genomes of fragaria spp. (Rosaceae). Front. Plant Sci. 12. doi: 10.3389/fpls.2021.754209
Li, D., Gan, G., Li, W., Li, W., Jiang, Y., Liang, X., et al. (2021b). Inheritance of Solanum chloroplast genomic DNA in interspecific hybrids. Mitochondrial DNA Part B 6, 351–357. doi: 10.1080/23802359.2020.1866450
Li, J., Price, M., Su, D.-M., Zhang, Z., Yu, Y., Xie, D.-F., et al. (2021c). Phylogeny and comparative analysis for the plastid genomes of five tulipa (Liliaceae). BioMed. Res. Int. 2021, 6648429. doi: 10.1155/2021/6648429
Li, L., Wu, Q., Fang, L., Wu, K., Li, M., Zeng, S. (2022b). Comparative chloroplast genomics and phylogenetic analysis of thuniopsis and closely related genera within coelogyninae (Orchidaceae). Front. Genet. 13, 850201. doi: 10.3389/fgene.2022.850201
Li, X., Tan, W., Sun, J., Du, J., Zheng, C., Tian, X., et al. (2019). Comparison of four complete chloroplast genomes of medicinal and ornamental meconopsis species: genome organization and species discrimination. Sci. Rep. 9, 10567. doi: 10.1038/s41598-019-47008-8
Li, Y., Dong, Y., Liu, Y., Yu, X., Yang, M., Huang, Y. (2021d). Comparative analyses of Euonymus chloroplast genomes: genetic structure, screening for loci with suitable polymorphism, positive selection genes, and phylogenetic relationships within Celastrineae. Front. Plant Sci. 11, 593984. doi: 10.3389/fpls.2020.593984
Liang, J., Chen, R., Zhang, F., Wang, Q., Yang, Y., Lv, M., et al. (2022). Full-length chloroplast genome of Dongxiang wild rice reveals small single-copy region switching. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.929352
Librado, P., Rozas, J. (2009). DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25, 1451–1452. doi: 10.1093/bioinformatics/btp187
Liu, Y., Huo, N., Dong, L., Wang, Y., Zhang, S., Young, H. A., et al. (2013). Complete chloroplast genome sequences of Mongolia medicine Artemisia frigida and phylogenetic relationships with other plants. PLoS One 8, e57533. doi: 10.1371/journal.pone.0057533
Lössl, A. G., Waheed, M. T. (2011). Chloroplast-derived vaccines against human diseases: achievements, challenges and scopes. Plant Biotechnol. J. 9, 527–539. doi: 10.1111/j.1467-7652.2011.00615.x
Lu, R.-S., Li, P., Qiu, Y.-X. (2017). The complete chloroplast genomes of three cardiocrinum (Liliaceae) species: comparative genomic and phylogenetic analyses. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.02054
Marasek-Ciolakowska, A., Nishikawa, T., Shea, D. J., Okazaki, K. (2018). Breeding of lilies and tulips-Interspecific hybridization and genetic background. Breed Sci. 68, 35–52. doi: 10.1270/jsbbs.17097
Mccauley, D. E., Sundby, A. K., Bailey, M. F., Welch, M. E. (2007). Inheritance of chloroplast DNA is not strictly maternal in Silene vulgaris (Caryophyllaceae): evidence from experimental crosses and natural populations. Am. J. Bot. 94, 1333–1337. doi: 10.3732/ajb.94.8.1333
Middleton, C. P., Senerchia, N., Stein, N., Akhunov, E. D., Keller, B., Wicker, T., et al. (2014). Sequencing of chloroplast genomes from wheat, barley, rye and their relatives provides a detailed insight into the evolution of the triticeae tribe. PLoS One 9, e85761. doi: 10.1371/journal.pone.0085761
Moon, J.-C., Kim, J.-H., Jang, C. S. (2016). Development of multiplex PCR for species-specific identification of the Poaceae family based on chloroplast gene, rpoC2. Appl. Biol. Chem. 59, 201–207. doi: 10.1007/s13765-016-0155-x
Nie, X., Zhao, X., Wang, S., Zhang, T., Li, C., Liu, H., et al. (2018). Complete chloroplast genome sequence of broomcorn millet (Panicum miliaceum L.) and comparative analysis with other panicoideae species. Agronomy 8, 159. doi: 10.3390/agronomy8090159
Noor, F., Ashfaq, U. A., Bakar, A., Qasim, M., Masoud, M. S., Alshammari, A., et al. (2023). Identification and characterization of codon usage pattern and influencing factors in HFRS-causing hantaviruses. Front. Immunol. 14. doi: 10.3389/fimmu.2023.1131647
Nyamgerel, N., Baasanmunkh, S., Oyuntsetseg, B., Bayarmaa, G. A., Erst, A., Park, I., et al. (2023). Insight into chloroplast genome structural variation of the Mongolian endemic species Adonis mongolica (Ranunculaceae) in the Adonideae tribe. Sci. Rep. 13, 22014. doi: 10.1038/s41598-023-49381-x
Ohyama, K., Fukuzawa, H., Kohchi, T., Shirai, H., Sano, T., Sano, S., et al. (1986). Chloroplast gene organization deduced from complete sequence of liverwort Marchantia polymorpha chloroplast DNA. Nature 322, 572–574. doi: 10.1038/322572a0
Olejniczak, S. A., Łojewska, E., Kowalczyk, T., Sakowicz, T. (2016). Chloroplasts: state of research and practical applications of plastome sequencing. Planta 244, 517–527. doi: 10.1007/s00425-016-2551-1
Oyuntsetseg, D., Nyamgerel, N., Baasanmunkh, S., Oyuntsetseg, B., Urgamal, M., Yoon, J. W., et al. (2024). The complete chloroplast genome and phylogentic results support the species position of Swertia banzragczii and Swertia marginata (Gentianaceae) in Mongolia. Botanical Stud. 65, 11. doi: 10.1186/s40529-024-00417-z
Park, J., Kim, Y., Kwon, W., Xi, H., Kwon, M. (2019b). The complete chloroplast genome of tulip tree, Liriodendron tulifipera L. (Magnoliaceae): investigation of intra-species chloroplast variations. Mitochondrial DNA B Resour 4, 2523–2524. doi: 10.1080/23802359.2019.1598822
Park, H.-S., Lee, W. K., Lee, S.-C., Lee, H. O., Joh, H. J., Park, J. Y., et al. (2021). Inheritance of chloroplast and mitochondrial genomes in cucumber revealed by four reciprocal F1 hybrid combinations. Sci. Rep. 11, 2506. doi: 10.1038/s41598-021-81988-w
Park, I., Yang, S., Kim, W. J., Song, J. H., Lee, H. S., Lee, H. O., et al. (2019a). Sequencing and comparative analysis of the chloroplast genome of angelica polymorpha and the development of a novel indel marker for species identification. Molecules 24 (6), 1038. doi: 10.3390/molecules24061038
POWO (2024). Plants of the World Online. Facilitated by the Royal Botanic Gardens, Kew. https://powo.science.kew.org/, Retrieved 21 October 2024.
Raspé, O. (2001). Inheritance of the chloroplast genome in sorbus aucuparia L. (Rosaceae). J. Heredity 92, 507–509. doi: 10.1093/jhered/92.6.507
Rogalski, M., Ruf, S., Bock, R. (2006). Tobacco plastid ribosomal protein S18 is essential for cell survival. Nucleic Acids Res. 34, 4537–4545. doi: 10.1093/nar/gkl634
Rop, O., Mlcek, J., Jurikova, T., Neugebauerova, J., Vabkova, J. (2012). Edible flowers—A new promising source of mineral elements in human nutrition. Molecules 17, 6672–6683. doi: 10.3390/molecules17066672
Rouhi, H., Shakarami, K., Afshari, R. T. (2010). Seed treatments to overcome dormancy of waterlily tulip ('Tulipa kaufmanniana'Regel.). Aust. J. Crop Sci. 4, 718–721. doi: 10.14720/aas.2019.113.2
Ruang-Areerate, P., Kongkachana, W., Naktang, C., Sonthirod, C., Narong, N., Jomchai, N., et al. (2021). Complete chloroplast genome sequences of five Bruguiera species (Rhizophoraceae): comparative analysis and phylogenetic relationships. PeerJ 9, e12268. doi: 10.7717/peerj.12268
Sabir, J., Schwarz, E., Ellison, N., Zhang, J., Baeshen, N. A., Mutwakil, M., et al. (2014). Evolutionary and biotechnology implications of plastid genome variation in the inverted-repeat-lacking clade of legumes. Plant Biotechnol. J. 12, 743–754. doi: 10.1111/pbi.2014.12.issue-6
Scobeyeva, V. A., Artyushin, I. V., Krinitsina, A. A., Nikitin, P. A., Antipin, M. I., Kuptsov, S. V., et al. (2021). Gene loss, pseudogenization in plastomes of genus allium (Amaryllidaceae), and putative selection for adaptation to environmental conditions. Front. Genet. 12. doi: 10.3389/fgene.2021.674783
Seberg, O., Petersen, G. (2009). How many loci does it take to DNA barcode a crocus? PLoS One 4, e4598. doi: 10.1371/journal.pone.0004598
Sen, F., Uncu, A. O., Uncu, A. T., Erdeger, S. N. (2020). The trnL (UAA)-trnF (GAA) intergenic spacer is a robust marker of green pea (Pisum sativum L.) adulteration in economically valuable pistachio nuts (Pistacia vera L.). J. Sci. Food Agric. 100, 3056–3061. doi: 10.1002/jsfa.v100.7
Shaw, J., Lickey, E. B., Schilling, E. E., Small, R. L. (2007). Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am. J. Bot. 94, 275–288. doi: 10.3732/ajb.94.3.275
Shaw, J., Shafer, H. L., Leonard, O. R., Kovach, M. J., Schorr, M., Morris, A. B. (2014). Chloroplast DNA sequence utility for the lowest phylogenetic and phylogeographic inferences in angiosperms: the tortoise and the hare IV. Am. J. Bot. 101, 1987–2004. doi: 10.3732/ajb.1400398
She, R., Zhao, P., Zhou, H., Yue, M., Yan, F., Hu, G., et al. (2020). Complete chloroplast genomes of Liliaceae (s.l.) species: comparative genomic and phylogenetic analyses. Nordic J. Bot. 38, e02477. doi: 10.1111/njb.02477
Shi, C., Hu, N., Huang, H., Gao, J., Zhao, Y. J., Gao, L. Z. (2012). An improved chloroplast DNA extraction procedure for whole plastid genome sequencing. PLoS One 7, e31468. doi: 10.1371/journal.pone.0031468
Shinozaki, K., Ohme, M., Tanaka, M., Wakasugi, T., Hayashida, N., Matsubayashi, T., et al. (1986). The complete nucleotide sequence of the tobacco chloroplast genome: its gene organization and expression. EMBO J. 5, 2043–2049. doi: 10.1002/j.1460-2075.1986.tb04464.x
Smidt, E. D. C., Páez, M. Z., Vieira, L. D. N., Viruel, J., De Baura, V. A., Balsanelli, E., et al. (2020). Characterization of sequence variability hotspots in Cranichideae plastomes (Orchidaceae, Orchidoideae). PLoS One 15, e0227991. doi: 10.1371/journal.pone.0227991
Song, Y., Wang, S., Ding, Y., Xu, J., Li, M. F., Zhu, S., et al. (2017). Chloroplast genomic resource of paris for species discrimination. Sci. Rep. 7, 3427. doi: 10.1038/s41598-017-02083-7
Song, Y.-F., Yang, Q.-H., Yi, X.-G., Zhu, Z.-Q., Wang, X.-R., Li, M. (2022). Comparative analysis of codon usage patterns in chloroplast genomes of cherries. Forests 13, 1891. doi: 10.3390/f13111891
Sutula, M., Kakanay, A., Tussipkan, D., Dzhumanov, S., Manabayeva, S. (2024). Phylogenetic Analysis of Rare and Endangered Tulipa Species (Liliaceae) of Kazakhstan Based on Universal Barcoding Markers. Biology 13 (6), 365. doi: 10.3390/biology13060365
Tamura, K., Stecher, G., Kumar, S. (2021). MEGA11: molecular evolutionary genetics analysis version 11. Mol. Biol. Evol. 38, 3022–3027. doi: 10.1093/molbev/msab120
Tillich, M., Lehwark, P., Pellizzer, T., Ulbricht-Jones, E. S., Fischer, A., Bock, R., et al. (2017). GeSeq - versatile and accurate annotation of organelle genomes. Nucleic Acids Res. 45, W6–w11. doi: 10.1093/nar/gkx391
Tojibaev, K., Dekhkonov, D., Ergashov, I., Sun, H., Deng, T., Yusupov, Z. (2022). The synopsis of the genus Tulipa (Liliaceae) in Uzbekistan. Phytotaxa 573, 163–214. doi: 10.11646/phytotaxa.573.2.2
Tonti-Filippini, J., Nevill, P. G., Dixon, K., Small, I. (2017). What can we do with 1000 plastid genomes? Plant J. 90, 808–818. doi: 10.1111/tpj.13491
Turudić, A., Liber, Z., Grdiša, M., Jakše, J., Varga, F., Šatović, Z. (2023). Variation in chloroplast genome size: biological phenomena and technological artifacts. Plants (Basel) 12 (2), 254. doi: 10.3390/plants12020254
Villanueva-Corrales, S., García-Botero, C., Garcés-Cardona, F., Ramírez-Ríos, V., Villanueva-Mejía, D. F., Álvarez, J. C. (2021). The complete chloroplast genome of plukenetia volubilis provides insights into the organelle inheritance. Front. Plant Sci. 12. doi: 10.3389/fpls.2021.667060
Vvedensky, A. I., Kovalevskaja, S. S. (1971). Tulipa L. In: vvedensky, A.I., ed., conspectus florae asiae mediae. Acad. Sci. Press Tashkent 2, 94–109. doi: 10.1111/njb.00374
Walker, J. F., Jansen, R. K., Zanis, M. J., Emery, N. C. (2015). Sources of inversion variation in the small single copy (SSC) region of chloroplast genomes. Am. J. Bot. 102, 1751–1752. doi: 10.3732/ajb.1500299
Walker, J. F., Zanis, M. J., Emery, N. C. (2014). Comparative analysis of complete chloroplast genome sequence and inversion variation in Lasthenia burkei (Madieae, Asteraceae). Am. J. Bot. 101, 722–729. doi: 10.3732/ajb.1400049
Wicke, S., Schneeweiss, G. M., Depamphilis, C. W., Müller, K. F., Quandt, D. (2011). The evolution of the plastid chromosome in land plants: gene content, gene order, gene function. Plant Mol. Biol. 76, 273–297. doi: 10.1007/s11103-011-9762-4
Xiao, F., Zhao, Y., Wang, X., Jian, X. (2024). Characterization of the chloroplast genome of Gleditsia species and comparative analysis. Sci. Rep. 14, 4262. doi: 10.1038/s41598-024-54608-6
Xu, Y., Liu, Y., Yu, Z., Jia, X. (2023). Complete chloroplast genome sequence of the long blooming cultivar camellia 'Xiari qixin': genome features, comparative and phylogenetic analysis. Genes (Basel) 14. doi: 10.3390/genes14020460
Yang, M., Zhang, X., Liu, G., Yin, Y., Chen, K., Yun, Q., et al. (2010). The complete chloroplast genome sequence of date palm (Phoenix dactylifera L.). PLoS One 5, e12762. doi: 10.1371/journal.pone.0012762
Yuan, L., Yan, X., Chen, X., Zhu, X. (2022). The complete chloroplast genome of Tulipa gesneriana (Liliaceae) and its phylogenetic analysis. Mitochondrial DNA B Resour 7, 1255–1256. doi: 10.1080/23802359.2022.2093676
Zhai, Y., Zhang, T., Guo, Y., Gao, C., Zhou, L., Feng, L., et al. (2023). Phylogenomics, phylogeography and germplasms authentication of the Rheum palmatum complex based on complete chloroplast genomes. J. Plant Res. 136, 291–304. doi: 10.1007/s10265-023-01440-0
Zhang, Y., Li, L., Yan, T. L., Liu, Q. (2014). Complete chloroplast genome sequences of Praxelis (Eupatorium catarium Veldkamp), an important invasive species. Gene 549, 58–69. doi: 10.1016/j.gene.2014.07.041
Zhang, J., Lu, J. H., Wang, Q. Q., Liu, M. N., Xu, K. (2022). Characteristics of the chloroplast genome of Glycyrrhiza eurycarpa P.C.Li from Xinjiang with comparison and phylogenetic analysis of the chloroplast genomes of the medicinal plants of Glycyrrhiza. Yaoxue Xuebao 57, 1516–1525. doi: 10.16438/j.0513-4870.2021-1661
Zhao, M., Shu, G., Hu, Y., Cao, G., Wang, Y. (2023). Pattern and variation in simple sequence repeat (SSR) at different genomic regions and its implications to maize evolution and breeding. BMC Genomics 24, 136. doi: 10.1186/s12864-023-09156-0
Zhong, X. (2020). Assembly, annotation and analysis of chloroplast genomes [Doctoral Thesis, The University of Western Australia]. doi: 10.26182/5f333d9ac2bee
Zhou, W., Barrett, S. C. H., Li, H.-D., Wu, Z.-K., Wang, X.-J., Wang, H., et al. (2017). Phylogeographic insights on the evolutionary breakdown of heterostyly. New Phytol. 214, 1368–1380. doi: 10.1111/nph.2017.214.issue-3
Zhou, J.-T., Yin, P.-P., Chen, Y., Zhao, Y.-P. (2019). The complete chloroplast genome of Tulipa altaica (Liliaceae), a wild relative of tulip. Mitochondrial DNA Part B 4, 2017–2018. doi: 10.1080/23802359.2019.1618221
Keywords: Tulipa L., chloroplast genome, comparative analysis, simple sequence repeat (SSR), codon usage patterns, phylogenetic analysis
Citation: Tussipkan D, Shevtsov V, Ramazanova M, Rakhimzhanova A, Shevtsov A and Manabayeva S (2024) Kazakhstan tulips: comparative analysis of complete chloroplast genomes of four local and endangered species of the genus Tulipa L.. Front. Plant Sci. 15:1433253. doi: 10.3389/fpls.2024.1433253
Received: 15 May 2024; Accepted: 14 October 2024;
Published: 12 November 2024.
Edited by:
Niaz Ahmad, National Institute for Biotechnology and Genetic Engineering, PakistanReviewed by:
Ibrar Ahmed, Alpha Genomics Private Limited, PakistanXiaoyu Ding, Nanjing Normal University, China
Copyright © 2024 Tussipkan, Shevtsov, Ramazanova, Rakhimzhanova, Shevtsov and Manabayeva. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shuga Manabayeva, bWFuYWJheWV2YUBiaW9jZW50ZXIua3o=
 Dilnur Tussipkan
Dilnur Tussipkan Vladislav Shevtsov2
Vladislav Shevtsov2 Shuga Manabayeva
Shuga Manabayeva