
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Plant Sci. , 27 June 2024
Sec. Technical Advances in Plant Science
Volume 15 - 2024 | https://doi.org/10.3389/fpls.2024.1407625
 Yan Jiang1
Yan Jiang1 Yanyun Jin1,2
Yanyun Jin1,2 Yiyi Shan1,2
Yiyi Shan1,2 Quanzhou Zhong1,2
Quanzhou Zhong1,2 Huizhong Wang1,2
Huizhong Wang1,2 Chenjia Shen1,2
Chenjia Shen1,2 Shangguo Feng1,2*
Shangguo Feng1,2*The plants of the genus Physalis L. have been extensively utilized in traditional and indigenous Chinese medicinal practices for treating a variety of ailments, including dermatitis, malaria, asthma, hepatitis, and liver disorders. The present review aims to achieve a comprehensive and up-to-date investigation of the genus Physalis, a new model crop, to understand plant diversity and fruit development. Several chloroplast DNA-, nuclear ribosomal DNA-, and genomic DNA-based markers, such as psbA-trnH, internal-transcribed spacer (ITS), simple sequence repeat (SSR), random amplified microsatellites (RAMS), sequence-characterized amplified region (SCAR), and single nucleotide polymorphism (SNP), were developed for molecular identification, genetic diversity, and phylogenetic studies of Physalis species. A large number of functional genes involved in inflated calyx syndrome development (AP2-L, MPF2, MPF3, and MAGO), organ growth (AG1, AG2, POS1, and CNR1), and active ingredient metabolism (24ISO, DHCRT, P450-CPL, SR, DUF538, TAS14, and 3β-HSB) were identified contributing to the breeding of novel Physalis varieties. Various omic studies revealed and functionally identified a series of reproductive organ development-related factors, environmental stress-responsive genes, and active component biosynthesis-related enzymes. The chromosome-level genomes of Physalis floridana Rydb., Physalis grisea (Waterf.) M. Martínez, and Physalis pruinosa L. have been recently published providing a valuable resource for genome editing in Physalis crops. Our review summarizes the recent progress in genetic diversity, molecular identification, phylogenetics, functional genes, and the application of omics in the genus Physalis and accelerates efficient utilization of this traditional herb.
Physalis L. is one of the largest genera within the Solanaceae family consisting of approximately 75–90 species, which are mostly distributed in tropical and temperate regions worldwide (Whitson and Manos, 2005; Pretz and Deanna, 2020). The most notable characteristic of the species in this genus is the calyx, which surrounds the fruit and increases in size as the fruit grows larger. The interest in Physalis species is mostly motivated by the economic importance of a subset of species that have been used in traditional medicine (Zhang and Tong, 2016). Most Physalis species have potential medicinal properties, including antibacterial, antileukemic, antipyretic, anti-inflammatory, immunomodulatory, and anticancer actions, and often have been used to treat various illnesses such as dermatitis, malaria, asthma, hepatitis, and liver disorders. Moreover, some Physalis species are extensively cultivated for their edible fruit or ornamental value in various countries. Recently, there has been a growing focus on the genus Physalis in molecular research related to taxonomy, systematics and evolution, genetic diversity, and omics. In this review, we aim to provide a comprehensive analysis of the genetic diversity, molecular identification, phylogenetics, functional genes, and the application of omics in the genus Physalis. The relevant references for this review were obtained from the PubMed database of NCBI and the Web of Science, which are widely recognized as leading databases for published articles and citations. The searches were conducted within a single day in May 2024. The term “Physalis” was utilized to search for instances in the title, abstract, and keywords. Articles focusing on authentication, genetic diversity, phylogenetics, functional genes, and omics were chosen for inclusion.
The accurate identification of germplasm resources is a crucial foundation for the systematic classification, population genetics, omics research, and molecular genetic breeding of Physalis plants. In the past, morphological methods were the primary means of identifying Physalis plants (Sinha, 1951; Axelius, 1996; Gonzalez et al., 2008). However, the morphological characteristics of Physalis plants are very similar, and these morphological traits are extremely susceptible to the restrictions of growth period and growth environment, which bring great difficulties to morphological identification methods (Whitson and Manos, 2005; Feng et al., 2016). With the development of biotechnology, various molecular markers have emerged and been widely used in the identification of plant species, varieties, and genotypes. Compared with morphological methods, DNA molecular markers are not easily affected by the external environment and growth period, and their identification has good stability and high accuracy (Schindel and Miller, 2005; Sarwat et al., 2012; Li et al., 2015).
Several chloroplast DNA regions (such as rbcL, atpF-atpH, ycf1, matK, rpoB, and psbA-trnH) and some nuclear ribosomal DNA (nrDNA) regions [such as internal transcribed spacer, ITS, and internal transcribed spacer 2 (ITS2)] have been advocated by some experts and the Consortium for the Barcode of Life (CBOL) as potential standard DNA barcodes for plant species identification (Schindel and Miller, 2005; Cbol Plant Working Group, 2009; Chen et al., 2010; China Plant et al., 2011; Chen et al., 2024). Feng et al. (2016) demonstrated the efficacy of nrDNA ITS2 regions for molecular identification in 45 Physalis species, as detailed in Table 1. The findings revealed a high rate of species authentication using the ITS2 sequence suggesting its potential as an efficient barcode for identifying Physalis species. Furthermore, the variability in secondary structures of ITS2 among most Physalis species, including differences in loop number, size, position, and degree of angles from the center of the spiral arm, presents a novel approach for identifying challenging-to-distinguish species based on their ITS2 sequence. Terrones et al. (2021) successfully used nrDNA ITS sequences as a DNA barcode to authenticate the two species of the genus Physalis [Physalis acutifolia (Miers) Sandwith and Physalis angulata L.] in the Iberian Peninsula of Spain (Terrones et al., 2021). Chloroplast psbA-trnH region, as one of the highly recommended candidate DNA barcodes, had also been successfully applied to the molecular identification of the species of the genus Physalis (Feng et al., 2018a).
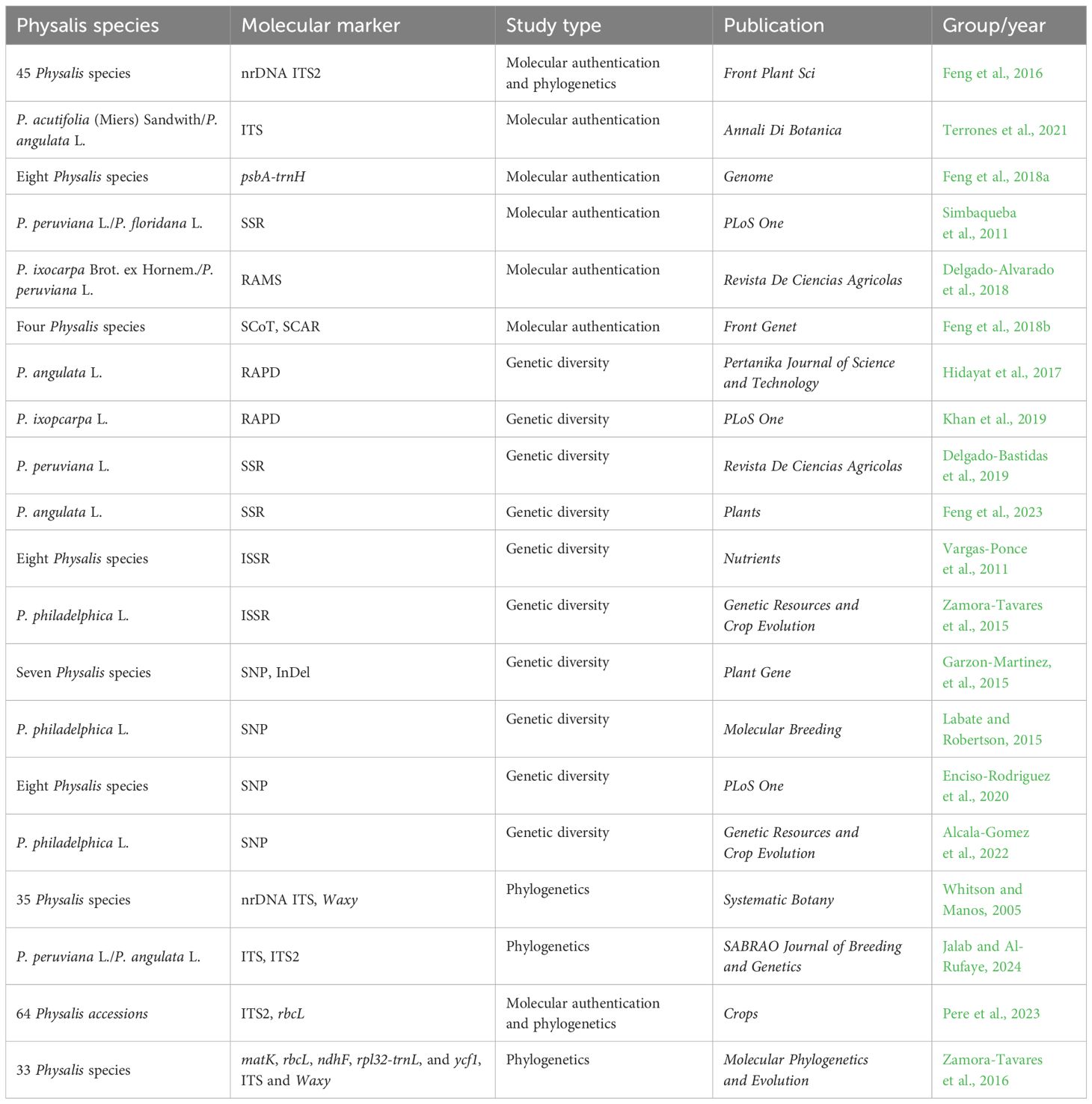
Table 1 Molecular techniques applied to Physalis authentication, genetic diversity, and phylogenetics.
Some DNA-based markers, such as simple sequence repeat (SSR), random amplified microsatellites (RAMS), start codon targeted (SCoT), and sequence-characterized amplified region (SCAR), have also shown excellent performance in plant molecular identification. Simbaqueba et al. (2011) identified 1,520 SSRs in the assembled leaf transcriptome of Physalis peruviana and developed 138 SSR primer pairs that successfully amplified in P. peruviana L. and Physalis floridana Rydb. genotypes, with a polymorphism rate of 22%. Delgado-Alvarado et al. (2018) applied RAMS markers to authenticate the varieties and landraces of Physalis ixocarpa Brot. ex Hornem. The results indicated that RAMS could be used as good specific markers not only to distinguish P. ixocarpa from its close relatives but also to provide specific fingerprints for the authentication of different varieties of P. ixocarpa. Feng et al. (2018b) developed four specific SCAR markers for P. angulata, Physalis minima L., Physalis pubescens L., and Physalis alkekengi var. franchetii (Mast.) Makino based on polymorphism analysis of SCoT molecular markers providing a new method for rapid and accurate molecular identification of the four Physalis species.
Research on genetic diversity is crucial for species management planning, as the preservation of diversity plays a vital role in conservation and the breeding of superior individuals. In recent years, there has been a focus on studying the genetic diversity of Physalis plants, with several related studies being reported. Various types of DNA molecular markers, including inter-simple sequence repeats (ISSR), random amplified polymorphic DNA (RAPD), SSR, insertion and deletion (InDel), and single nucleotide polymorphism (SNP) markers, have been utilized in numerous studies to assess the genetic diversity and population dynamics of Physalis plants. A comprehensive summary of these studies on the genetic diversity of Physalis plants can be found in Table 1.
RAPD markers, a relatively early molecular marker technology, have been widely utilized in studying genetic diversity in various plants due to their simple operation, high versatility, and cost effectiveness. Hidayat et al. (2017) used RAPD markers to assess the genetic diversity of 23 P. angulata plants from different regions of Bandung. Similarly, Khan et al. (2019) employed eight RAPD markers to determine the genetic diversity of 17 accessions of P. ixocarpa, with the results aligning with the ecological distribution of accessions and highlighting two accessions (P1512005 and PI360740) from Mexico and Ecuador as exhibiting the highest genetic diversity among P. ixocarpa accessions.
Microsatellites, also known as simple sequence repeats (SSRs), are designed based on conserved nucleotide sequences found on both sides of simple repeat sequences, widely distributed in plant genomes (Tautz, 1989). SSRs are co-dominant, multi-allelic, highly polymorphic, and have been widely used in various fields, including genetic diversity, phylogenetic studies, molecular identification, and genetic mapping (Poczai et al., 2013). Several SSR markers have been developed and extensively utilized in the investigation of genetic diversity within Physalis species (Simbaqueba et al., 2011; Wei et al., 2012; Delgado-Bastidas et al., 2019; Feng et al., 2023). In a study by Delgado-Bastidas et al. (2019), six SSR markers were employed to evaluate the genetic diversity of 40 genotypes of P. peruviana revealing that these genotypes were categorized into three populations. However, it was observed that the level of genetic diversity among the genotypes was notably low, with no discernible population structure. In a more recent study, Feng et al. (2023) developed a set of SSR markers based on chloroplast genome and applied them to assess the genetic diversity and population structure of P. angulata. The SSR analysis revealed that 16 populations of P. angulata formed four clusters displaying significant geography-related population structure as well as extensive admixture.
ISSR markers are molecular markers that utilize microsatellite oligonucleotides as primers, with two to four randomly selected nucleotides added to the 5′ or 3′ end of the SSR to facilitate annealing at specific sites. The result in PCR amplification of DNA fragments located between relatively spaced repeats that are complementary to the anchor primers (Zietkiewicz et al., 1994). The ISSR, which integrates the advantages of RAPD and SSR, not only exhibits excellent stability and polymorphism but also offers simplicity, rapidity, and efficiency. It has been successfully employed in assessing genetic diversity, genetic relationship, and molecular identification in plants (Wang et al., 2009; Kumar et al., 2018; Tyagi et al., 2020). The ISSR marker has been proven to be valuable in the analysis of genetic diversity and genetic relationships within Physalis plants (Vargas-Ponce et al., 2011; Zamora-Tavares et al., 2015). Vargas-Ponce et al. (2011) showed that 12 samples from eight Physalis species could be grouped into two clusters with an interspecific genetic similarity ranging from 0.48 to 0.58 based on ISSR analysis. Meanwhile, Zamora-Tavares et al. (2015) utilized 88 ISSR markers to study the genetic diversity and structure of nine Physalis philadelphica Lam. populations in western Mexico, revealing high genetic diversity among the samples and grouping the populations into two clusters based on structure analysis.
Single nucleotide polymorphism (SNP) is a widely utilized DNA marker technology that has been developed in recent years. It represents a common genetic variation caused by the alteration of a single nucleotide (A, T, C, and G) in the DNA sequence (Uppu et al., 2018). SNP markers are prevalent in genomes and hold significant value for applications such as plant genetic diversity analysis, genotype identification, high-density genetic map construction, and molecular marker-assisted breeding (Lu et al., 2018; Arca et al., 2020; Guo et al., 2021; Park et al., 2022). Additionally, SNP is one of the most popular molecular marker techniques used to study genetic diversity in Physalis plants, as demonstrated by several studies (Cely et al., 2015; Labate and Robertson, 2015; Enciso-Rodriguez et al., 2020; Alcala-Gomez et al., 2022). For example, Enciso-Rodriguez et al. (2020) identified 7,425 SNPs based on Genotyping-By-Sequencing (GBS) and utilized them to assess the diversity of P. peruviana and related taxa. Their findings revealed significant gene flow (FST: 0.01–0.05) in different subpopulations of P. peruviana. Similarly, Alcala-Gomez et al. (2022) investigated the genetic diversity of P. philadelphica using 270 SNP markers based on their study of 40 samples.
The taxonomy of Physalis is considered to be a highly complex issue within the Solanaceae due to the significant intraspecific morphological variation and substantial interspecific similarity (Axelius, 1996; Sullivan, 2004; Whitson and Manos, 2005; Olmstead et al., 2008; Pretz and Deanna, 2020). In recent years, molecular analyses have yielded new insights into this problem (Whitson and Manos, 2005; Olmstead et al., 2008; Feng et al., 2016; Zamora-Tavares et al., 2016; Feng et al., 2020; Pere et al., 2023). ITS regions of nrDNA are widely used for studying phylogenic relationships among angiosperms, including the genus Physalis, at the interspecific and infrageneric level. This is due to their biparental inheritance, simplicity, universality, intra-genome consistency, inter-genome variability, and high copy number (Whitson and Manos, 2005; Xiang et al., 2013; Feng et al., 2016; Zamora-Tavares et al., 2016; Pere et al., 2023; Jalab and Al-Rufaye, 2024). Whitson and Manos (2005) conducted a study on the phylogenetic relationships among 35 species of Physalis and the relationships among the genera of the subtribe Physalinae utilizing the sequence analysis of the nrDNA ITS region and the nuclear gene waxy. The findings revealed that the morphologically typical Physalis species formed a strongly supported clade. However, the morphologically atypical species, such as P. alkekengi L., Physalis carpenter Riddell, and Physalis microphysa A.Gray were found to be distantly related to any other Physalis species resulting in paraphyly within the genus. Zamora-Tavares et al. (2016) utilized five plastids (matK, rbcL, ndhF, rpl32-trnL, and ycf1) and two nuclear regions (ITS and waxy) to examine the phylogenetic relationships of 50 species within the Physalinae, which included 33 Physalis species. The study assessed the phylogenetic relationships among recognized genera in Physalinae, with a focus on identifying monophyletic groups and resolving the physaloid grade. Additionally, the study analyzed potential causes for recent divergence within Physalinae. All the aforementioned studies utilized single or a few gene sequence fragments from the plastid genome or nuclear genome to investigate the phylogeny of genus Physalis. Due to the limited length of these DNA sequences and their restricted genetic information, there are significant limitations in studying phylogenetic evolution using these methods. We are confident that the ongoing advancements in chloroplast genome and mitochondrial genome-sequencing technology will lead to a more refined and precise reconstruction of the phylogenetic tree of genus Physalis.
The inflated calyx syndrome, also known as the Chinese lantern, is a post-floral morphological novelty in Physalis plants (Hu and Saedler, 2007). During the fruit ripening process, the green calyx expands, inflates, and completely envelops the fruit (de Souza et al., 2022). Despite extensive research on this morphological feature of Physalis plants, only a limited number of functional genes involved in its development have been investigated and cloned from Physalis plants (Wilf et al., 2017). The ortholog of Solanum tuberosum MADS16 in P. pubescens, MPF2 is a floral tissue-specific expressed gene that is essential for the development of inflated calyx syndrome (He and Saedler, 2005). Furthermore, an MPF2-binding protein, MAGO NASHI, was identified using the yeast two-hybrid system. Two MAGO-encoding genes, PFMAGPO1 and PFMAGPO2, were discovered in P. floridana. These genes play a role in male fertility and the evolution of calyx development in Physalis (He et al., 2007). Promoter analysis of an MPF2-like gene revealed degenerative mutations in its core CArG-box indicating an interaction between floral development and hormone pathways during the development process of calyx inflation syndrome (Khan et al., 2012). MPF3, a core eudicot APETALA1-like MADS-domain protein, has been reported to act as a repressor of MPF2 during the development of floral calyx identity and inflated calyx syndrome in Physalis (Zhao et al., 2013).
Recently, CRISPR-Cas9-targeted mutagenesis technology was utilized for a forward genetics screen to identify the purported essential regulators of inflated calyx syndrome. For instance, the mutation of an AP2-like gene has been found to result in a lack of inflated calyx syndrome (He et al., 2023). This technological breakthrough positions Physalis as a new model crop for studying fruit development and ripening (Lopez-Gomollon, 2023).
Physalis fruits are increasingly gaining popularity due to their outstanding sensory and functional characteristics as a functional food (Avendaño et al., 2022). It is important to note that Physalis fruit serves as a significant supplementary source of bioactive compounds with high antioxidant activity (Vaillant et al., 2021). Fruit size is a critical quality characteristic of Physalis fruit, and there is significant variation in berry sizes among Physalis plants. Consequently, the Physalis genus is utilized as a model plant for identifying the regulators that may contribute to their variation in berry size (Wang et al., 2012). In P. philadelphica, the expression level of Physalis Organ Size 1 (POS1) gene is positively associated with variations in fruit size (Wang et al., 2014). POS1 plays a crucial role in regulating fruit size by controlling cell wall expansion in Physaleae (Wang et al., 2022). In P. floridana, the Cell Number Regulator 1 (CNR1) gene encodes a cell membrane-anchored modulator that negatively regulates fruit size through its interaction with an AGAMOUS-like ovary identity protein (PfAG2) (Li and He, 2015). Two C-class MADS-domain AGAMOUS-like genes, PfAG1 and PfAG2, in P. floridana play essential roles in regulating fruit size and the development process of Chinese lantern (Zhao et al., 2021). Recently, CRISPR-Cas9 technology has been used to target the CLV1 gene revealing its essential role in enhancing fruit size by increasing the number of locules in P. pruinosa L (Lemmon et al., 2018).
In addition to regulating the inflated calyx syndrome and fruit development, P. floridana MPF1 also influences plant architecture, seed development, and flowering time by regulating the expression of PFLFY, PFSOC1, and PFFT genes (He et al., 2010). The CRABS CLAW gene in P. floridana alters carpel meristem determinacy and carpel closure by mediating the neofunctionalization of GLOBOSA genes belonging to the floral B-function MADS-box family (Gong et al., 2021). Additionally, four core exon junction complex core genes in P. floridana—namely, PFMAGO, PFY14, PFeIF4AIII, and PFBTZ—have been found to play diverse developmental roles in carpel functionality and environmental stress responses. Furthermore, an intron retention in the transcript of DYT1 was detected in the mutated flowers of P. floridana indicating its significance in floral development (Gong et al., 2018). These works provide us with candidate genes for studying the growth and development of Physalis plants.
Physalis plants produce edible fruits containing numerous antioxidants and bioactive metabolites, including steroidal lactones, withanolides, and physalins (Popova et al., 2022). However, the biosynthesis pathways for these bioactive compounds remain largely unclear. Utilizing Physalis transcriptomes, several research groups have identified a variety of genes associated with terpenoid backbone and steroid biosynthesis pathways. In P. alkekengi, candidate genes for the oxidation at the C-15/18 positions of steroid backbone required in physalin biosynthesis include a CYP450 chloroplastic-like gene (unigene-ID: c13295_g2_i2) and an oxidoreductase-like gene (unigene-ID: c16207_g5_i1). Additionally, a gene encoding sterol reductase (c27112_g1_i1) has been identified to be involved in the biosynthesis of specialized metabolites in P. peruviana (Fukushima et al., 2016). Pa24ISO catalyzes the isomerization of 24-methylenecholesterol to 24-methyldesmosterol in the physalin biosynthesis process (Yang et al., 2022). DHCR7 from P. angulata was newly identified through heterologous expression. Heterologous expression of P. angulata DHCR7 in Saccharomyces cerevisiae confirmed its role in producing 24-methylene-cholesterol, a key substrate in the physalin and withanolide biosynthesis pathway (Yang et al., 2021). In P. angulata, DUF538 was predicted as a positive regulator and TAS14 as a negative regulator in the regulation of physalin biosynthesis (Zhan et al., 2020). These functional genes have potential implications for accelerating the breeding of high-yielding Physalis varieties rich in bioactive compounds (Figure 1).
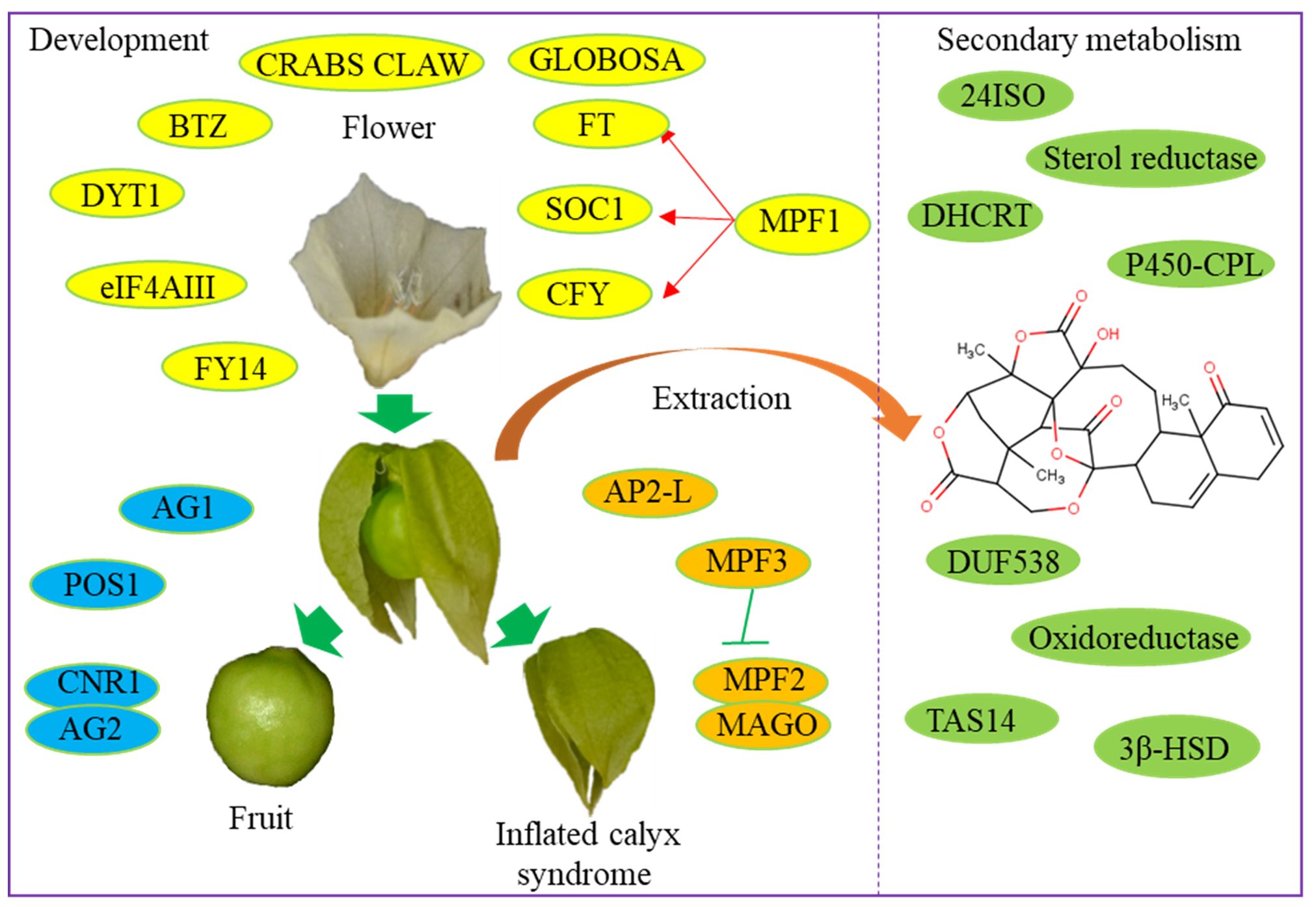
Figure 1 Functional genes involved in the development and secondary metabolism of Physalis varieties.
Several omic datasets of Physalis plants have been made available online offering extensive genetic information for the screening of functional genes and the identification of active compounds. In the present review, all the omic datasets of Physalis plants are summarized in Tables 2, 3.
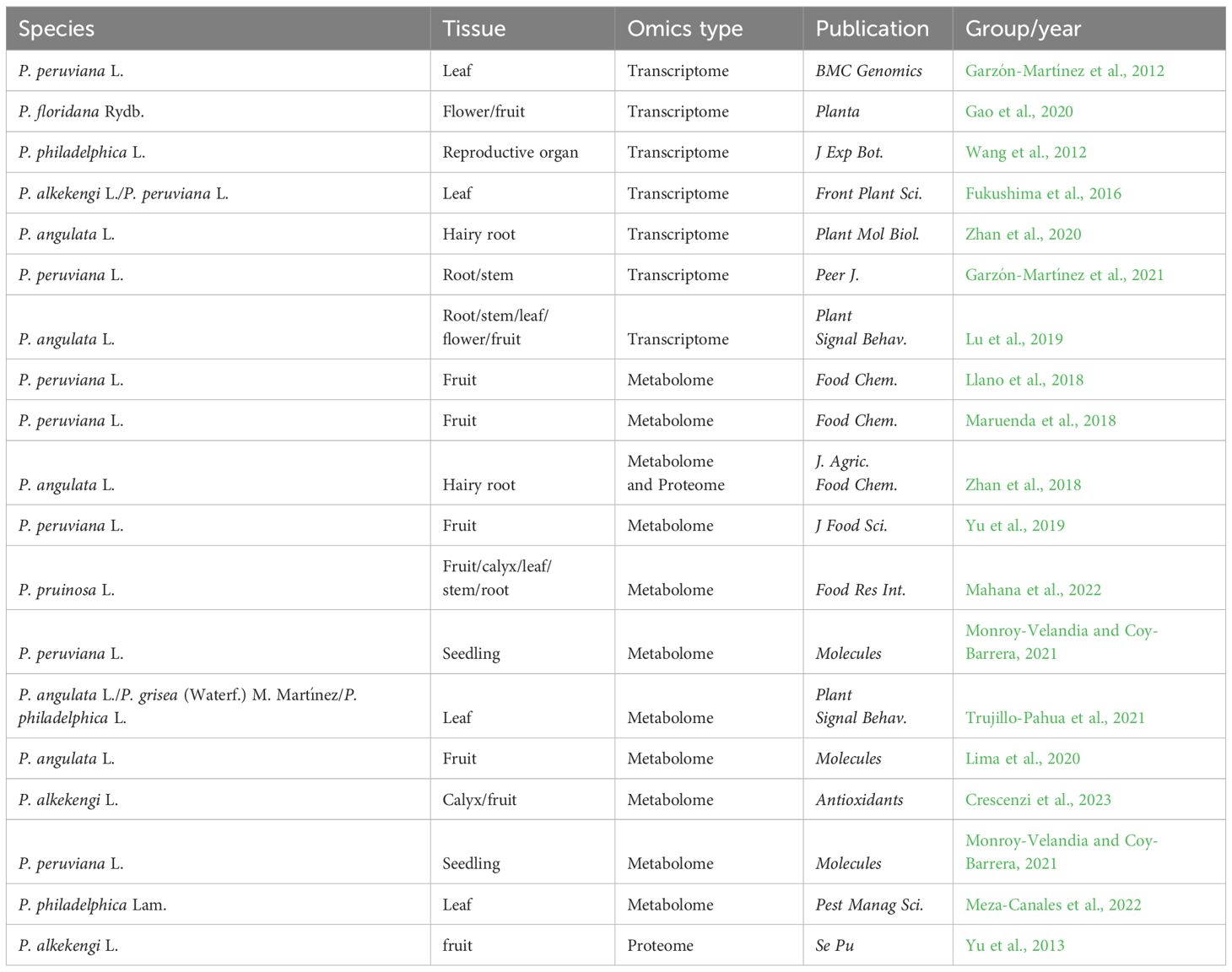
Table 2 The detailed information of all the transcriptomes, metabolomes. and proteomes of Physalis plants.
In recent years, advancements in the next-generation sequencing technology have enabled the identification of functional genes in non-model plants even without genomic data (Su et al., 2011). The first Physalis transcriptome was published in 2012 using fresh leaf tissue from the Colombian ecotype of Cape gooseberry (P. peruviana), as plant material, generating a number of assembled sequences and candidate markers (Garzón-Martínez et al., 2012). To study the Physalis–Fusarium oxysporum pathosystem, the transcriptome of P. peruviana was further utilized to identify genes related to immunity, including 74 resistance genes, 17 receptor-like kinase genes, 8 PAMP-triggered immunity genes, and 9 effector-triggered immunity genes (Enciso-Rodriguez et al., 2013). Comparative transcriptomic analysis of P. philadelphica with different sizes of reproductive organs resulted in the identification of 263 differentially expressed transcripts (Wang et al., 2012). Using the RNA-seq method, 75,221 genes of P. alkekengi and 54,513 genes of P. peruviana were identified. The authors discovered numerous genes potentially involved in each step of the terpenoid backbone and steroid biosynthesis pathway providing new insights into the intricate chemical and structural diversity of Physalis plants (Fukushima et al., 2016). In P. angulata, a well-known traditional Chinese medicine with various active compounds, transcriptomic approaches were used to screen genes involved in the biosynthesis of bioactive compounds (Zhan et al., 2020). A transcriptomic analysis revealed 468 unigenes involved in the flower–fruit transition process in P. floridana uncovering some potential genetic variations that contribute to the early stage of fruit development in Physalis (Gao et al., 2020). These studies in P. angulata can help spur our understanding of the biosynthetic pathways underlying key metabolites important to medicine and plant development.
Untargeted metabolomics is a recently developed method that offers a streamlined approach to systematically analyze and compare the differences in primary and secondary metabolites among different groups (Souard et al., 2018). Using untargeted metabolomics, Medina’s group identified several specifically accumulated withanolides and fatty acyl glycosides as molecular markers to differentiate between organic and conventional P. peruviana fruits (Llano et al., 2018). NMR-based metabolomic analysis revealed significant phytochemical variations in P. peruviana fruits (Maruenda et al., 2018). In P. angulata hairy roots, a comparative metabolomic analysis revealed variations in the contents of physalins D and H under MeJA treatment suggesting a possible regulatory mechanism underlying the MeJA-induced biosynthesis of active compounds (Zhan et al., 2018, 2020). LC-MS/MS-based metabolomic analysis revealed variations in carotenoid content during different growth stages of P. peruviana fruit (Yu et al., 2019). Metabolite profiling using UPLC-MS identified a total of 293 metabolites, including 61 terpenoids, 58 phenolic acids, and 53 flavonoids, in aqueous and ethanolic extracts of Amazonian fruits (including P. angulata) (Lima et al., 2020). Metabolomics, in combination with chemometrics, has identified several potential α-glucosidase and α-amylase inhibitory metabolites in P. pruinosa. Physangulide B, physaperuvin G, and neophysalin A were found to be positively correlated with α-glucosidase inhibition activity, while guaiacyl-primeveroside, phyperunolide C, and perulactone were found to be positively correlated with α-amylase inhibitory activity (Mahana et al., 2022). Using LC-ESI/LTQOrbitrap/MS followed by LC-ESI/LTQOrbitrap/MS/MS technique, 58 phytocompounds were identified in the calyx ad fruit of yellow P. alkekengi (Crescenzi et al., 2023). Metabolomic analysis has been utilized to investigate the responses of Physalis species to environmental stimuli. In P. peruviana, the upregulation of a free flavonol during different growth stages indicates a response to salt stress (Monroy-Velandia and Coy-Barrera, 2021). Metabolomic analysis of three different Physalis species revealed several species-specific metabolites following larval herbivory. In P. angulata, the response to herbivory is highlighted by the upregulating of various compounds, such as withanolide, α-trehalose, and cimiracemoside D. Pheophorbide A and azamacrocycle are common metabolites of P. grisea (Waterf.) M. Martínez and P. philadelphica that are responsive to herbivory (Trujillo-Pahua et al., 2021). Husk tomato (P. philadelphica) seedlings are susceptible to infestation by the whitefly Trialeurodes vaporariorum. A newly published metabolome study showed that P. philadelphica impairs whitefly development by inducing significant changes in metabolic profiles (Meza-Canales et al., 2022). Metabolomics researches provide us an opportunity to understand the differences in types and contents of active ingredients in Physalis plants. Metabolomic analysis is also an effective way to screen novel varieties with high medicinal ingredients.
MS/MS-based peptide sequencing techniques have been utilized for the large-scale identification and screening of differentially produced proteins (Yates et al., 1993). In 2013, protein extracted from P. alkekengi fruit was analyzed by nano-RPLC-MS/MS system with shotgun proteomics method providing the foundation for further investigation into the functional proteins in Physalis species (Yu et al., 2013). MeJA is commonly employed as a chemical elicitor to enhance the accumulation levels of various bioactive metabolites in plants (Liu et al., 2016). Proteomic analysis revealed that several terpenoid and steroid biosynthesis-related enzymes, such as CYP monooxygenases and 3β-hydroxysterioid dehydrogenase, might be the targets of the MeJA-induced active ingredient biosynthesis (Zhan et al., 2018). Enzyme engineering is currently a hot topic in biotechnology. Proteomic analysis helps us to identify key enzymes involved in the biosynthesis of active ingredients and improve their activities by enzyme engineering modifications.
The chloroplast plays crucial roles in various cellular functions, such as photosynthesis, signal transduction, and stress response (Martin Avila et al., 2016). The examination of the complete chloroplast genomes of the Physalis genus will be useful for in-deep genetic research. Currently, the complete chloroplast genomes of 12 Physalis species, including Physalis chenopodiifolia Lam., P. angulata, P. angulata var. villosa Bonati, P. alkekengi, P. minima, P. pubescens, P. peruviana, P. pruinosa, Physalis cordata Houst. ex Mill., P. philadelphica, Physalis macrophysa Rydb., and P. ixocarpa were available (Sandoval-Padilla et al., 2019; Zamora-Tavares et al., 2019; Feng et al., 2020; Zhan et al., 2022; Sandoval-Padilla et al., 2022a, b; Zhang et al., 2023) (Table 3). The complete chloroplast genomes mentioned above ranged in size from 156,578 to 157,007 bp, with the number of protein coding genes ranging from 79 to 80 and the number of tRNA genes ranging from 30 to 31 (Sandoval-Padilla et al., 2019; Zamora-Tavares et al., 2019; Feng et al., 2020; Sandoval-Padilla et al., 2022a, b). These publicly available chloroplast genomes enable effective phylogeography and phylogenetic studies of Physalis. Furthermore, a significant number of SSR loci have been identified providing precise molecular markers for investigating the intraspecific diversity of Physalis.
Genome-editing technologies have been developed to enhance the quality and yield of crops, improve adaptation to diverse environments, manipulate plant architecture and fruit size, and broaden the range of staple crops that can be cultivated (Scheben et al., 2017). Although most of the Physalis species have a similar chromosome number and structure to Solanaceae, genomic knowledge is essential for genome editing in Physalis crops. The genome of the orphan crop P. pruinosa was first sequenced and published in 2018 producing 66.3 Gb of raw data (Lemmon et al., 2018). To gain a deeper better understanding of the genetic variations that contribute to the origin and diversity of these distinctive traits, high-quality genomes of classic Physalis genus plants were published (Lu et al., 2021). P. floridana possesses an assembled genome size of 1,389 Mb, which serves as a valuable resource for breeding Physalis crops (Lu et al., 2021). Recently, a chromosome-scale references for P. grisea and its close relative P. pruinosa were published providing high-quality genome assemblies for genome editing in Physalis species (He et al., 2023). To fully understand the genomic variation of Physalis plants, a high-quality haplotype-resolved genome will greatly promote the understanding of the complex traits of Physalis plants.
Researchers have performed many studies on the screening of germplasm collections and identifying SNPs with continuously updating genetic techniques. Although several molecular markers have been developed, outdated platforms provide a limited ability to estimate the extent of Physalis genetic variability. In the future, a number of convenient and phenotype-based molecular markers will definitely be the direction for efficient genetic diversity analysis and germplasm resource identification. Genetic identification is a prerequisite for species conservation and resource utilization. Furthermore, the advancement of high-throughput sequencing technology will lead to an increasing number of published chloroplast genomes and mitochondrial genomes, thus contributing to the further development of phylogenetic tree reconstruction for genus Physalis.
Single-cell RNA sequencing (scRNA-seq) is a novel technology used to investigate cell heterogeneity at a high resolution (Bawa et al., 2022). Mass spectrometry (MS) imaging is a recently developed MS-based metabolomics approach to reveal the distribution of metabolites at the spatial level (Xiang et al., 2022; Zhan et al., 2024). However, scRNA-seq and MS imaging have not yet been applied to the study of Physalis plants. ScRNA-seq will provide novel insights into the inflated calyx syndrome and fruit development of Physalis plants. High-resolution MS imaging can be utilized to identify and visualize metabolic heterogeneity, including inflated calyx syndrome. Future researches on Physalis plants will move toward higher resolutions at both the temporal and spatial levels.
The recently sequenced genome of P. floridana acts as a starting point for genome-enabled research (Lu et al., 2021). As more species’ genomes are fully sequenced, new knowledge regarding evolutionary relationships, processes, and patterns, as well as the ability to map biosynthetic pathways through comparative means will emerge.
YJ: Writing – original draft, Data curation, Investigation. YYJ: Writing – original draft, Investigation, Methodology. YS: Writing – original draft, Investigation, Methodology. QZ: Writing – original draft, Investigation, Data curation. HW: Writing – original draft, Funding acquisition, Resources. CS: Methodology, Writing – review & editing. SF: Writing – original draft, Writing – review & editing, Project administration, Funding acquisition.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was funded by the National Natural Science Foundation of China (31970346, 32271905, and 32270382), Zhejiang Provincial Natural Science Foundation of China (LY20H280012 and LY23C160001), and the Hangzhou Scientific and Technological Program of China (20191203B02 and 20150932H04).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alcala-Gomez, G., Perez-Alquicira, J., Cabrera-Toledo, D., Cortes-Cruz, M., Zamora-Tavares, M. D., Vargas-Ponce, O. (2022). Genetic diversity and structure in husk tomato based (Physalis philadelphica Lam.) on SNPs: a case of diffuse domestication. Genet. Resour. Crop Evol. 69, 443–459. doi: 10.1007/s10722–021-01278–2
Arca, M., Mary-Huard, T., Gouesnard, B., Berard, A., Bauland, C., Combes, V., et al. (2020). Deciphering the genetic diversity of Landraces with high-throughput SNP genotyping of DNA bulks: Methodology and application to the maize 50k array. Front. Plant Sci. 11. doi: 10.1101/2020.05.19.103655
Avendaño, W. A., Muñoz, H. F., Leal, L. J., Deaquiz, Y. A., Castellanos, D. A. (2022). Physicochemical characterization of cape gooseberry (Physalis Peruviana L.) fruits ecotype Colombia during preharvest development and growth. J. Food Sci. 87, 4429–4439. doi: 10.1111/1750–3841.16318
Axelius, B. (1996). The phylogenetic relationships of the Physaloid genera (Solanaceae) based on morphological data. Amer J. Bot. 83, 118–124. doi: 10.1002/j.1537-2197.1996.tb13884.x
Bawa, G., Liu, Z., Yu, X., Qin, A., Sun, X. (2022). Single-cell RNA sequencing for plant research: Insights and possible benefits. Int. J. Mol. Sci. 23, 4497. doi: 10.3390/ijms23094497
Cbol Plant Working Group. (2009). A DNA barcode for land plants. Proc. Natl. Acad. Sci. U.S.A. 106, 12794–12797. doi: 10.1073/pnas.0905845106
Cely, J. A. B., Rodriguez, F. E., Almario, C. G., Meneses, L. S. B. (2015). Genetic variability of parentals and inter and interspecific F1 populations of Physalis Peruviana L. and P. floridana Rydb. Rev. Bras. Fruticultura 37, 179–192. doi: 10.1590/0100-2945-002/14
Chen, S., Yao, H., Han, J., Liu, C., Song, J., Shi, L., et al. (2010). Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS One 5, e8613. doi: 10.1371/journal.pone.0008613
Chen, Z. M., Gao, L., Wang, H. Z., Feng, S. G. (2024). Molecular identification and phylogenetic analysis of Cymbidium species (Orchidaceae) based on the potential DNA barcodes matK, rbcL, psbA-trnH, and internal transcribed spacer. Agronomy 14, 933. doi: 10.3390/agronomy14050933
China Plant, B. O. L. G., Li, D. Z., Gao, L. M., Li, H. T., Wang, H., Ge, X. J., et al. (2011). Comparative analysis of a large dataset indicates that internal transcribed spacer (ITS) should be incorporated into the core barcode for seed plants. Proc. Natl. Acad. Sci. U.S.A. 108, 19641–19646. doi: 10.1073/pnas.1104551108
Crescenzi, M. A., Serreli, G., Deiana, M., Tuberoso, C. I. G., Montoro, P., Piacente, S. (2023). Metabolite profiling, through LC-ESI/LTQOrbitrap/MS analysis, of antioxidant extracts from Physalis alkekengi L. Antioxidants (Basel) 12, 2101. doi: 10.3390/antiox12122101
Delgado-Alvarado, E. A., Almaraz-Abarca, N., Escamirosa-Tinoco, C., Uribe-Soto, J. N., Avila-Reyes, J. A., Torres-Ricario, R., et al. (2018). Potential of random amplified microsatellites (RAMS) to typify and discriminate varieties of Physalis ixocarpa Brot. ex Hornem. Emirates J. Food Agric. 30, 396–403. doi: 10.9755/ejfa.2018.v30.i5.1684
Delgado-Bastidas, N., Lagos-Santander, L. K., Lagos-Burbano, T. C. (2019). Genetic diversity of 40 genotypes of golden berry Physalis Peruviana L. using microsatellite markers. Rev. Cienc. Agricolas 36, 95–107. doi: 10.22267/rcia.1936E.110
de Souza, A. X., Riederer, M., Leide, J. (2022). Multifunctional contribution of the inflated fruiting calyx: Implication for cuticular barrier profiles of the Solanaceous genera Physalis, Alkekengi, and Nicandra. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.888930
Enciso-Rodriguez, F. E., Gonzalez, C., Rodriguez, E. A., Lopez, C. E., Landsman, D., Barrero, L. S., et al. (2013). Identification of immunity related genes to study the Physalis Peruviana–Fusarium oxysporum pathosystem. PloS One 8, e68500. doi: 10.1371/journal.pone.0068500
Enciso-Rodriguez, F. E., Osorio-Guarin, J. A., Garzon-Martinez, G. A., Delgadillo-Duran, P., Barrero, L. S. (2020). Optimization of the genotyping-by-sequencing SNP calling for diversity analysis in cape gooseberry (Physalis Peruviana L.) and related taxa. PLoS One 15, e0238383. doi: 10.1371/journal.pone.0238383
Feng, S. G., Jiang, M. Y., Shi, Y. J., Jiao, K. L., Shen, C. J., Lu, J. J., et al. (2016). Application of the ribosomal DNA ITS2 region of Physalis (Solanaceae): DNA barcoding and phylogenetic study. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01047
Feng, S. G., Jiao, K. L., Zhang, Z. H., Yang, S., Gao, Y. D., Jin, Y. Y., et al. (2023). Development of chloroplast microsatellite markers and evaluation of genetic diversity and population structure of cutleaf groundcherry (Physalis angulata L.) in China. Plants 12, 1755. doi: 10.3390/plants12091755
Feng, S. G., Jiao, K. L., Zhu, Y. J., Wang, H. F., Jiang, M. Y., Wang, H. Z. (2018a). Molecular identification of species of Physalis (Solanaceae) using a candidate DNA barcode: the chloroplast psbA-trnH intergenic region. Genome 61, 15–20. doi: 10.1139/gen-2017-0115
Feng, S. G., Zheng, K. X., Jiao, K. L., Cai, Y. C., Chen, C. L., Mao, Y. Y., et al. (2020). Complete chloroplast genomes of four Physalis species (Solanaceae): lights into genome structure, comparative analysis, and phylogenetic relationships. BMC Plant Biol. 20, 242. doi: 10.1186/s12870-020-02429-w
Feng, S. G., Zhu, Y. J., Yu, C. L., Jiao, K. L., Jiang, M. Y., Lu, J. J., et al. (2018b). Development of species-specific SCAR markers, based on a SCoT analysis, to authenticate Physalis (Solanaceae) species. Front. Genet. 9. doi: 10.3389/fgene.2018.00192
Fukushima, A., Nakamura, M., Suzuki, H., Yamazaki, M., Knoch, E., Mori, T., et al. (2016). Comparative characterization of the leaf tissue of Physalis alkekengi and Physalis Peruviana using RNA-seq and metabolite profiling. Front. Plant Sci. 7. doi: 10.3389/fpls.2016.01883
Gao, H. H., Li, J., Wang, L., Zhang, J. S., He, C. Y. (2020). Transcriptomic variation of the flower-fruit transition in Physalis and Solanum. Planta 252, 28. doi: 10.1007/s00425-020-03434-x
Garzon-Martinez, G. A., Garcia-Arias, F. L., Enciso-Rodriguez, F. E., Soto-Suarez, M., Gonzalez, C., Bombarely, A., et al. (2021). Combining transcriptome analysis and GWAS for identification and validation of marker genes in the Physalis peruviana-Fusarium oxysporum pathosystem. PeerJ. 9, e11135. doi: 10.7717/peerj.11135
Garzon-Martinez, G. A., Osorio-Guarin, J. A., Delgadillo-Duran, P., Mayorga, F., Enciso-Rodriguez, F. E., Landsman, D., et al. (2015). Genetic diversity and population structure in Physalis peruviana and related taxa based on InDels and SNPs derived from COSII and IRG markers. Plant Gene. 4, 29–37. doi: 10.1016/j.plgene.2015.09.003
Garzón-Martínez, G. A., Zhu, Z. I., Landsman, D., Barrero, L. S., Marino-Ramirez, L. (2012). The Physalis Peruviana leaf transcriptome: assembly, annotation and gene model prediction. BMC Genomics 13, 151. doi: 10.1186/1471-2164-13-151
Gong, P., Li, J., He, C. (2018). Exon junction complex (EJC) core genes play multiple developmental roles in Physalis floridana. Plant Mol. Biol. 98, 545–563. doi: 10.1007/s11103-018-0795-9
Gong, P., Song, C., Liu, H., Li, P., Zhang, M., Zhang, J., et al. (2021). Physalis floridana CRABS CLAW mediates neofunctionalization of GLOBOSA genes in carpel development. J. Exp. Bot. 72, 6882–6903. doi: 10.1093/jxb/erab309
Gonzalez, O. T., Torres, J. M. C., Cano, C. I. M., Arias, M. L., Arboleda, A. A. N. (2008). Morphologic characterization of forty six accessions of cape gooseberry (Physalis Peruviana L.), in Antioquia (Colombia). Rev. Bras. Fruticultura 30, 708–715. doi: 10.1590/S0100–29452008000300025
Guo, Z. F., Yang, Q. N., Huang, F. F., Zheng, H. J., Sang, Z. Q., Xu, Y. F., et al. (2021). Development of high-resolution multiple-SNP arrays for genetic analyses and molecular breeding through genotyping by target sequencing and liquid chip. Plant Commun. 2, 100230. doi: 10.1016/j.xplc.2021.100230
He, J., Alonge, M., Ramakrishnan, S., Benoit, M., Soyk, S., Reem, N. T., et al. (2023). Establishing Physalis as a Solanaceae model system enables genetic reevaluation of the inflated calyx syndrome. Plant Cell 35, 351–368. doi: 10.1093/plcell/koac305
He, C. Y., Saedler, H. (2005). Heterotopic expression of MPF2 is the key to the evolution of the Chinese lantern of Physalis, a morphological novelty in Solanaceae. Proc. Natl. Acad. Sci. U.S.A. 102, 5779–5784. doi: 10.1073/pnas.0501877102
He, C. Y., Sommer, H., Grosardt, B., Huijser, P., Saedler, H. (2007). PFMAGO, a MAGO NASHI-like factor, interacts with the MADS-domain protein MPF2 from Physalis floridana. Mol. Biol. Evol. 24, 1229–1241. doi: 10.1093/molbev/msm041
He, C. Y., Tian, Y., Saedler, R., Efremova, N., Riss, S., Khan, M. R., et al. (2010). The MADS-domain protein MPF1 of Physalis floridana controls plant architecture, seed development and flowering time. Planta 231, 767–777. doi: 10.1007/s00425-009-1087-z
Hidayat, T., Yosnata, L. J., Wulandari, L. T., Priyandoko, D., Aisyah, S. (2017). Random Amplified Polymorphism DNA Method to Authenticate Indonesian medicinal plant ciplukan (Physalis angulata; Solanaceae). Pertanika J. Sci. Technol. 25, 13–22.
Hu, J. Y., Saedler, H. (2007). Evolution of the inflated calyx syndrome in Solanaceae. Mol. Biol. Evol. 24, 2443–2453. doi: 10.1093/molbev/msm177
Jalab, A. Z., Al-Rufaye, Z. T. A. (2024). Phylogenetic analysis of ground cherry (Physalis) species. SABRAO J. Breed. Genet. 56, (1) 112–118. doi: 10.54910/sabrao2024.56.1.10
Khan, W., Bakht, J., Khan, B. M., Naeem, I., Uddin, M. N., Padder, B. A. (2019). Morphometric and genetic characterization of medicinally important accessions of Physalis ixocarpa Brot. Bangladesh J. Bot. 48, 105–111. doi: 10.3329/bjb.v48i1.47421
Khan, M. R., Hu, J., Ali, G. M. (2012). Reciprocal loss of CArG-boxes and auxin response elements drives expression divergence of MPF2-Like MADS-box genes controlling calyx inflation. PLoS One 7, e42781. doi: 10.1371/journal.pone.0042781
Kumar, A., Rodrigues, V., Mishra, P., Baskaran, K., Shukla, A. K., Shasany, A. K., et al. (2018). ISSR-derived species-specific SCAR marker for rapid and accurate authentication of Ocimum tenuiflorum L. Planta Med. 84, 117–122. doi: 10.1055/s-0043-116853
Labate, J., Robertson, L. (2015). Nucleotide diversity estimates of tomatillo (Physalis philadelphica) accessions including nine new inbred lines. Mol. Breed. 35, 1–10. doi: 10.1007/s11032-015-0302-9
Lemmon, Z. H., Reem, N. T., Dalrymple, J., Soyk, S., Swartwood, K. E., Rodriguez-Leal, D., et al. (2018). Rapid improvement of domestication traits in an orphan crop by genome editing. Nat. Plants 4, 766–770. doi: 10.1038/s41477-018-0259-x
Li, Z., He, C. Y. (2015). Physalis floridana cell number regulator1 encodes a cell membrane-anchored modulator of cell cycle and negatively controls fruit size. J. Exp. Bot. 66, 257–270. doi: 10.1093/jxb/eru415
Li, X., Yang, Y., Henry, R. J., Rossetto, M., Wang, Y., Chen, S. (2015). Plant DNA barcoding: from gene to genome. Biol. Rev. Camb Philos. Soc. 90, 157–166. doi: 10.1111/brv.12104
Lima, L. G. B., Montenegro, J., Abreu, J. P., Santos, M. C. B., Nascimento, T. P. D., Santos, M. D. S., et al. (2020). Metabolite profiling by UPLC-MS(E), NMR, and antioxidant properties of Amazonian fruits: Mamey apple (Mammea americana), camapu (Physalis angulata), and uxi (Endopleura uchi). Molecules 25, 342. doi: 10.3390/molecules25020342
Liu, J., Liu, Y., Wang, Y., Zhang, Z. H., Zu, Y. G., Efferth, T., et al. (2016). The combined effects of ethylene and MeJA onmetabolic profiling of phenolic compounds in Catharanthus roseus revealed by metabolomics analysis. Front. Physiol. 7. doi: 10.3389/fphys.2016.00217
Llano, S. M., Munoz-Jimenez, A. M., Jimenez-Cartagena, C., Londono-Londono, J., Medina, S. (2018). Untargeted metabolomics reveals specific withanolides and fatty acyl glycoside as tentative metabolites to differentiate organic and conventional Physalis Peruviana fruits. Food Chem. 244, 120–127. doi: 10.1016/j.foodchem.2017.10.026
Lopez-Gomollon, S. (2023). Physalis: A new model crop to understand plant diversity. Plant Cell 35, 338–339. doi: 10.1093/plcell/koac315
Lu, J., Liu, Y., Xu, J., Mei, Z., Shi, Y., Liu, P., et al. (2018). High-density genetic map construction and stem total polysaccharide content-related QTL exploration for Chinese endemic dendrobium (Orchidaceae). Front. Plant Sci. 9. doi: 10.3389/fpls.2018.00398
Lu, J. J., Luo, M. F., Wang, L., Li, K. P., Yu, Y. Y., Yang, W. F., et al. (2021). The Physalis floridana genome provides insights into the biochemical and morphological evolution of Physalis fruits. Hortic. Res. 8, 244. doi: 10.1038/s41438-021-00705-w
Lu, J., Xu, M., Cai, J., Yu, D., Meng, Y., Wang, H. (2019). Transcriptome-wide identification of microRNAs and functional insights inferred from microRNA-target pairs in Physalis angulata L. Plant Signal Behav. 14, 1629267. doi: 10.1080/15592324.2019.1629267
Mahana, A., Hammoda, H. M., Harraz, F. M., Shawky, E. (2022). Metabolomics combined to chemometrics reveals the putative alpha-glucosidase and alpha-amylase inhibitory metabolites of ground cherry (Physalis pruinosa L.). Food Res. Int. 161, 111903. doi: 10.1016/j.foodres.2022.111903
Martin Avila, E., Gisby, M. F., Day, A. (2016). Seamless editing of the chloroplast genome in plants. BMC Plant Biol. 16, 168. doi: 10.1186/s12870–016-0857–6
Maruenda, H., Cabrera, R., Canari-Chumpitaz, C., Lopez, J. M., Toubiana, D. (2018). NMR-based metabolic study of fruits of Physalis Peruviana L. grown in eight different Peruvian ecosystems. Food Chem. 262, 94–101. doi: 10.1016/j.foodchem.2018.04.032
Meza-Canales, I. D., Trujillo-Pahua, V., Vargas-Ponce, O., Ramirez-Romero, R., Montero-Vargas, J. M., Ordaz-Ortiz, J. J., et al. (2022). Systemic whitefly-induced metabolic responses in newly developed distal leaves of husk tomato plants (Physalis philadelphica) impairs whiteflies development. Pest Manag. Sci. 79, 368–380. doi: 10.1002/ps.7206
Monroy-Velandia, D., Coy-Barrera, E. (2021). Effect of salt stress on growth and metabolite profiles of cape gooseberry (Physalis Peruviana L.) along three growth stages. Molecules 26, 2756. doi: 10.3390/molecules26092756
Olmstead, R. G., Bohs, L., Migid, H. A., Santiago-Valentin, E., Garcia, V. F., Collier, S. M. (2008). A molecular phylogeny of the Solanaceae. Taxon 57, 1159–1181. doi: 10.1002/tax.574010
Park, J. S., Kang, M. Y., Shim, E. J., Oh, J., Seo, K. I., Kim, K. S., et al. (2022). Genome-wide core sets of SNP markers and Fluidigm assays for rapid and effective genotypic identification of Korean cultivars of lettuce (Lactuca sativa L.). Hortic. Res. 9, uhac119. doi: 10.1093/hr/uhac119
Pere, K., Mburu, K., Muge, E. K., Wagacha, J. M., Nyaboga, E. N. (2023). Molecular discrimination and phylogenetic relationships of Physalis species based on ITS2 and rbcL DNA barcode sequence. Crops 3, 302–319. doi: 10.3390/crops3040027
Poczai, P., Varga, I., Laos, M., Cseh, A., Bell, N., Valkonen, J. P., et al. (2013). Advances in plant gene-targeted and functional markers: a review. Plant Methods 9, 6. doi: 10.1186/1746-4811-9-6
Popova, V., Petkova, Z., Mazova, N., Ivanova, T., Petkova, N., Stoyanova, M., et al. (2022). Chemical composition assessment of structural parts (seeds, peel, pulp) of Physalis alkekengi L. fruits. Molecules 27, 5787. doi: 10.3390/molecules27185787
Pretz, C., Deanna, R. (2020). Typifications and nomenclatural notes in Physalis (Solanaceae) from the United States. Taxon 69, 170–192. doi: 10.1002/tax.12159
Sandoval-Padilla, I., Perez-Alquicira, J., Rodriguez, A., Zamora-Tavares, M. D., Vargas-Ponce, O. (2022a). The plastome of the husk tomato (Physalis philadelphica Lam., Solanaceae): a comparative analysis between wild and cultivated pools. Genet. Resour. Crop Evol. 69, 1391–1405. doi: 10.1007/s10722-021-01334-x
Sandoval-Padilla, I., Perez-Alquicira, J., Zamora-Tavares, M. D. P., Rodriguez, A., Cortes-Cruz, M., Alcala-Gomez, G., et al. (2019). Complete sequence of wild Physalis philadelphica chloroplast genome. Mitochondrial DNA B Resour 4, 3295–3297. doi: 10.1080/23802359.2019.1673231
Sandoval-Padilla, I., Zamora-Tavares, M. D. P., Ruiz-Sanchez, E., Pérez-Alquicira, J., Vargas Ponce, O. (2022b). Characterization of the plastome of Physalis cordata and comparative analysis of eight species of Physalis sensu stricto. PhytoKeys 210, 109–134. doi: 10.3897/phytokeys.210.85668
Sarwat, M., Nabi, G., Das, S., Srivastava, P. S. (2012). Molecular markers in medicinal plant biotechnology: past and present. Crit. Rev. Biotechnol. 32, 74–92. doi: 10.3109/07388551.2011.551872
Scheben, A., Wolter, F., Batley, J., Puchta, H., Edwards, D. (2017). Towards CRISPR/Cas crops - bringing together genomics and genome editing. New Phytol. 216, 682–698. doi: 10.1111/nph.14702
Schindel, D., Miller, S. E. (2005). DNA barcoding a useful tool for taxonomists. Nature 435, 17. doi: 10.1038/435017b
Simbaqueba, J., Sanchez, P., Sanchez, E., Nunez Zarantes, V. M., Chacon, M. I., Barrero, L. S., et al. (2011). Development and characterization of microsatellite markers for the Cape gooseberry Physalis Peruviana. PLoS One 6, e26719. doi: 10.1371/journal.pone.0026719
Souard, F., Delporte, C., Stoffelen, P., Thevenot, E. A., Noret, N., Dauvergne, B., et al. (2018). Metabolomics fingerprint of coffee species determined by untargeted-profiling study using LC-HRMS. Food Chem. 245, 603–612. doi: 10.1016/j.foodchem.2017.10.022
Su, Z., Ning, B., Fang, H., Hong, H., Perkins, R., Tong, W., et al. (2011). Next-generation sequencing and its applications in molecular diagnostics. Expert Rev. Mol. Diagn. 11, 333–343. doi: 10.1586/erm.11.3
Sullivan, J. R. (2004). The genus Physalis (Solanaceae) in the Southeastern United States. Rhodora 106, 305–326.
Tautz, D. (1989). Hypervariability of simple sequences as a general source for polymorphic DNA markers. Nucleic Acids Res. 17, 6463–6471. doi: 10.1093/nar/17.16.6463
Terrones, A., Moreno, J., Juan, A. (2021). DNA barcoding supports an easier identification of Alien plants: The case of the genus Physalis (Solanaceae) in the Iberian Peninsula (Spain). Annali Di Botanica 11, 105–119. doi: 10.13133/2239–3129/16618
Trujillo-Pahua, V., Vargas-Ponce, O., Rodriguez-Zaragoza, F. A., Ordaz-Ortiz, J. J., Delano-Frier, J. P., Winkler, R., et al. (2021). Metabolic response to larval herbivory in three Physalis species. Plant Signal Behav. 16, 1962050. doi: 10.1080/15592324.2021.1962050
Tyagi, R., Sharma, V., Sureja, A. K., Das Munshi, A., Arya, L., Saha, D., et al. (2020). Genetic diversity and population structure detection in sponge gourd (Luffa cylindrica) using ISSR, SCoT and morphological markers. Physiol. Mol. Biol. Plants 26, 119–131. doi: 10.1007/s12298-019-00723-y
Uppu, S., Krishna, A., Gopalan, R. P. (2018). A Review on methods for detecting SNP interactions in high-dimensional genomic data. IEEE/ACM Trans. Comput. Biol. Bioinform. 15, 599–612. doi: 10.1109/TCBB.2016.2635125
Vaillant, F., Corrales-Agudelo, V., Moreno-Castellanos, N., Ángel-Martín, A., Henao-Rojas, J. C., Muñoz-Durango, K., et al. (2021). Plasma metabolome profiling by high-performance chemical isotope-labelling LC-MS after acute and medium-term intervention with golden berry fruit (Physalis Peruviana L.), confirming its impact on insulin-associated signaling pathways. Nutrients 13, 3125. doi: 10.3390/nu13093125
Vargas-Ponce, O., Perez-Alvarez, L. F., Zamora-Tavares, P., Rodriguez, A. (2011). Assessing genetic diversity in Mexican Husk tomato species. Plant Mol. Biol. Rep. 29, 733–738. doi: 10.1007/s11105-010-0258-1
Wang, H. Z., Feng, S. G., Lu, J. J., Shi, N. N., Liu, J. J. (2009). Phylogenetic study and molecular identification of 31 Dendrobium species using inter-simple sequence repeat (ISSR) markers. Scientia Hortic. 122, 440–447. doi: 10.1016/j.scienta.2009.06.005
Wang, L., He, L., Li, J., Zhao, J., Li, Z., He, C. (2014). Regulatory change at Physalis organ size 1 correlates to natural variation in tomatillo reproductive organ size. Nat. Commun. 5, 4271. doi: 10.1038/ncomms5271
Wang, L., Li, Z., He, C. (2012). Transcriptome-wide mining of the differentially expressed transcripts for natural variation of floral organ size in Physalis philadelphica. J. Exp. Bot. 63, 6457–6465. doi: 10.1093/jxb/ers299
Wang, L., Liu, X., Li, Q., Xu, N., He, C. (2022). A lineage-specific arginine in POS1 is required for fruit size control in Physaleae (Solanaceae) via gene co-option. Plant J. 111, 183–204. doi: 10.1111/tpj.15786
Wei, J., Hu, X., Yang, J., Yang, W. (2012). Identification of single-copy orthologous genes between Physalis and Solanum lycopersicum and analysis of genetic diversity in Physalis using molecular markers. PLoS One 7, e50164. doi: 10.1371/journal.pone.0050164
Whitson, M., Manos, P. S. (2005). Untangling Physalis (Solanaceae) from the Physaloids: A two-gene phylogeny of the Physalinae. Systematic Bot. 30, 216–230. doi: 10.1600/0363644053661841
Wilf, P., Carvalho, M. R., Gandolfo, M. A., Cúneo, N. R. (2017). Eocene lantern fruits from Gondwanan Patagonia and the early origins of Solanaceae. Science 355, 71–75. doi: 10.1126/science.aag2737
Xiang, X. G., Schuiteman, A., Li, D. Z., Huang, W. C., Chung, S. W., Li, J. W., et al. (2013). Molecular systematics of Dendrobium (Orchidaceae, Dendrobieae) from mainland Asia based on plastid and nuclear sequences. Mol. Phylogenet. Evol. 69, 950–960. doi: 10.1016/j.ympev.2013.06.009
Xiang, L., Wang, F., Bian, Y., Harindintwali, J. D., Wang, Z., Wang, Y., et al. (2022). Visualizing the distribution of phthalate esters and plant metabolites in carrot by matrix-assisted laser desorption/ionization imaging mass spectrometry. J. Agric. Food Chem. 70, 15311–15320. doi: 10.1021/acs.jafc.2c06995
Yang, J., Li, C., Zhang, Y. (2021). Engineering of saccharomyces cerevisiae for 24-methylene-cholesterol production. Biomolecules 11, 1710. doi: 10.3390/biom11111710
Yang, J., Tian, J., Yang, Y., Zhu, Y., Li, C., Zhang, Y. (2022). RNAi of sterol delta24-isomerase implicated its involvement in Physalin biosynthesis in Physalis angulata L. Front. Plant Sci. 13. doi: 10.3389/fpls.2022.850711
Yates, J. R., 3rd, Speicher, S., Griffin, P. R., Hunkapiller, T. (1993). Peptide mass maps: a highly informative approach to protein identification. Anal. Biochem. 214, 397–408. doi: 10.1006/abio.1993.1514
Yu, Y., Chen, X., Zheng, Q. (2019). Metabolomic profiling of carotenoid constituents in Physalis Peruviana during different growth stages by LC-MS/MS technology. J. Food Sci. 84, 3608–3613. doi: 10.1111/1750-3841.14916
Yu, H., Yan, J., Guo, M., Jin, Y. (2013). Analysis of proteins in the extracts of Physalis alkekengi L. var. franchetii (Mast.) Makino using nanoflow reversed-phase liquid chromatography-tandem mass spectrometry. Se Pu 31, 362–366. doi: 10.3724/sp.j.1123.2012.11023
Zamora-Tavares, M. D., Martinez, M., Magallon, S., Guzman-Davalos, L., Vargas-Ponce, O. (2016). Physalis and physaloids: A recent and complex evolutionary history. Mol. Phylogenet. Evol. 100, 41–50. doi: 10.1016/j.ympev.2016.03.032
Zamora-Tavares, M. D. P., Sandoval-Padilla, I., Chavez Zendejas, A., Perez-Alquicira, J., Vargas-Ponce, O. (2019). Complete chloroplast genome of Physalis chenopodifolia Lam. (Solanaceae). Mitochondrial DNA B Resour 5, 162–163. doi: 10.1080/23802359.2019.1698364
Zamora-Tavares, P., Vargas-Ponce, O., Sanchez-Martinez, J., Cabrera-Toledo, D. (2015). Diversity and genetic structure of the husk tomato (Physalis philadelphica Lam.) in Western Mexico. Genet. Resour Crop Evol. 62, 141–153. doi: 10.1007/s10722-014-0163-9
Zhan, X. R., Liao, X. Y., Luo, X. J., Zhu, Y. J., Feng, S. G., Yu, C. N., et al. (2018). Comparative metabolomic and proteomic analyses reveal the regulation mechanism underlying MeJA-induced bioactive compound accumulation in cutleaf groundcherry ( Physalis angulata L.) Hairy Roots. J. Agric. Food Chem. 66, 6336–6347. doi: 10.1021/acs.jafc.8b02502
Zhan, X. R., Luo, X. J., He, J. Y., Zhang, C. C., Liao, X. Y., Xu, X. Y., et al. (2020). Bioactive compounds induced in Physalis angulata L. by methyl-jasmonate: an investigation of compound accumulation patterns and biosynthesis-related candidate genes. Plant Mol. Biol. 103, 341–354. doi: 10.1007/s11103-020-00996-y
Zhan, X. R., Zhang, Z. H., Zhang, Y., Gao, Y. D., Jin, Y. Y., Shen, C. J., et al. (2022). Complete plastome of Physalis angulata var. villosa, Gene organization, comparative genomics and phylogenetic relationships among Solanaceae. Genes (Basel) 13, 2291. doi: 10.3390/genes13122291
Zhan, X., Zang, Y., Ma, R., Lin, W., Li, X. L., Pei, Y., et al. (2024). Mass spectrometry-imaging analysis of active ingredients in the leaves of Taxus cuspidata. ACS omega 9 (16), 18634–18642. doi: 10.1021/acsomega.4c01440
Zhang, Z. H., Jin, Y. Y., Gao, Y. D., Zhang, Y., Ying, Q. C., Shen, C. J., et al. (2023). The complete chloroplast genomes of two Physalis species, Physalis macrophysa and P. ixocarpa: Comparative genomics, evolutionary dynamics and phylogenetic relationships. Agronomy 13, 135. doi: 10.3390/agronomy13010135
Zhang, W. N., Tong, W. Y. (2016). Chemical constituents and biological activities of plants from the genus Physalis. Chem. Biodiversity 13, 48–65. doi: 10.1002/cbdv.201400435
Zhao, J., Gong, P. C., Liu, H. Y., Zhang, M. S., He, C. Y. (2021). Multiple and integrated functions of floral C-class MADS-box genes in flower and fruit development of Physalis floridana. Plant Mol. Biol. 107, 101–116. doi: 10.1007/s11103-021-01182-4
Zhao, J., Tian, Y., Zhang, J. S., Zhao, M., Gong, P., Riss, S., et al. (2013). The euAP1 protein MPF3 represses MPF2 to specify floral calyx identity and displays crucial roles in Chinese lantern development in Physalis. Plant Cell 25, 2002–2021. doi: 10.1105/tpc.113.111757
Keywords: genetic diversity, molecular marker, inflated calyx syndrome, omics, Physalis
Citation: Jiang Y, Jin Y, Shan Y, Zhong Q, Wang H, Shen C and Feng S (2024) Advances in Physalis molecular research: applications in authentication, genetic diversity, phylogenetics, functional genes, and omics. Front. Plant Sci. 15:1407625. doi: 10.3389/fpls.2024.1407625
Received: 27 March 2024; Accepted: 07 June 2024;
Published: 27 June 2024.
Edited by:
Vladimir Orbovic, University of Florida, United StatesReviewed by:
Ashley N. Egan, Utah Valley University, United StatesCopyright © 2024 Jiang, Jin, Shan, Zhong, Wang, Shen and Feng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shangguo Feng, ZmVuZ3NnQGh6bnUuZWR1LmNu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.